Abstract
SIRT1 (Sir2) is an NAD+-dependent deacetylase that plays critical roles in a broad range of biological events, including metabolism, immune response, and aging1–5. While there is strong interest in stimulating SIRT1 catalytic activity, the homeostasis of SIRT1 at the protein level is poorly understood. Here, we report that macroautophagy (hereafter referred as autophagy), a catabolic membrane trafficking pathway that degrades cellular components through autophagosomes and lysosomes, mediates downregulation of mammalian SIRT1 protein during senescence and in vivo aging. In senescence, nuclear SIRT1 is recognized as an autophagy substrate and is subjected to cytoplasmic autophagosome-lysosome degradation, via the autophagy protein LC3. Importantly, the autophagy-lysosome pathway contributes to loss of SIRT1 during aging of several tissues related to the immune and hematopoietic system in mice, including spleen, thymus, and hematopoietic stem and progenitor cells, and in CD8+CD28- T cells from aged human donors. Our study reveals a mechanism in regulating the protein homeostasis of SIRT1, and suggests a potential strategy to stabilize SIRT1 to promote productive aging.
Sirtuins are an evolutionarily conserved family of NAD+-dependent deacetylases and ADP-ribosyltransferases that play important roles in a broad range of biological activities1, 2. Among mammalian sirtuins, SIRT1 is well-characterized to regulate several cellular and organismal processes, including metabolism and aging3–5. SIRT1 functions through deacetylation of its substrates, including histone substrates, such as acetylated histone H4K16 and H3K56, and non-histone targets, such as p532.
We have previously reported that the level of the SIRT1 orthologue in yeast, Sir2, declines upon replicative aging, which is a cause of yeast aging6. However, the regulatory mechanisms of SIRT1 protein homeostasis during mammalian aging remain unclear. Here, we investigated SIRT1 homeostasis in mammalian senescence and aging.
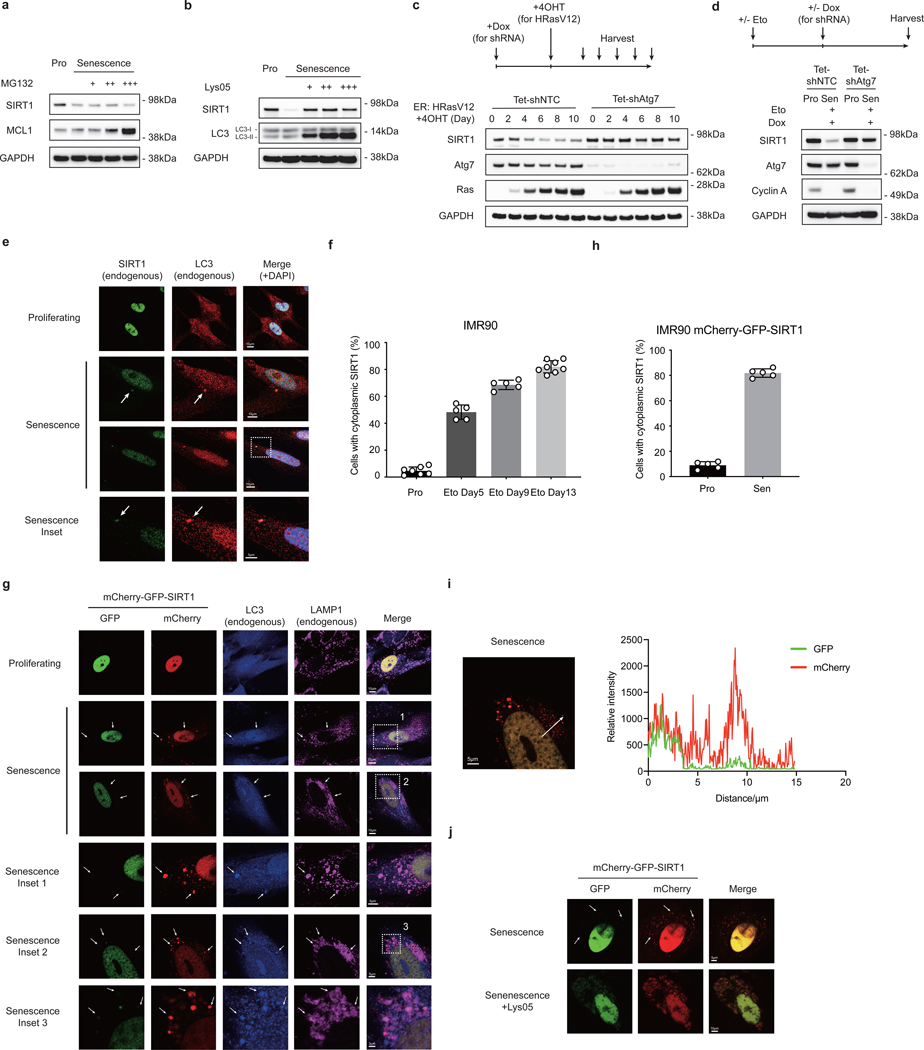
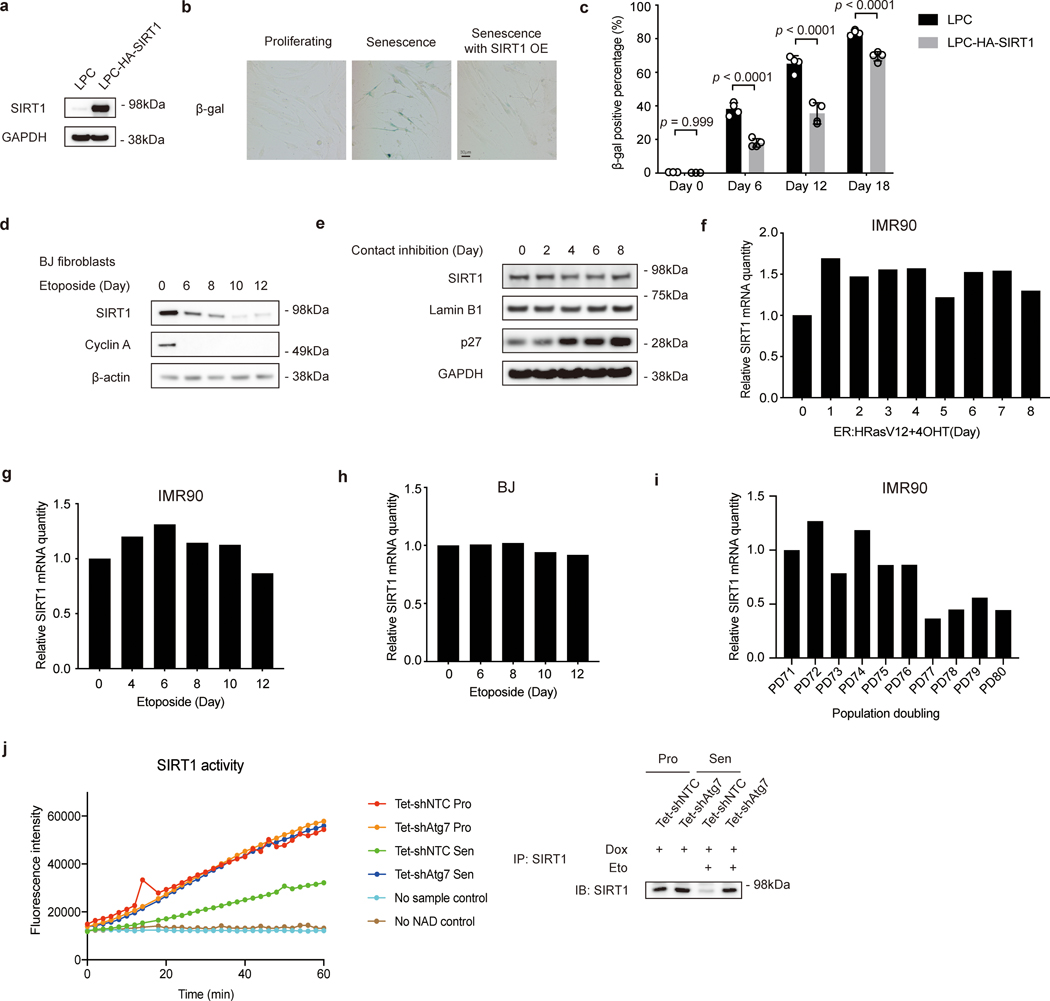
Cellular senescence is a stable form of cell cycle arrest induced by telomere shortening or by cellular stress7. Aged tissues are typically characterized by accumulation of senescent cells8, 9. Clearance of senescent cells delays age-related pathologies10, suggesting a critical association between cellular senescence and organismal aging. Overexpression of SIRT1 in primary human lung fibroblasts IMR90 (Extended Data Fig. 1a) delayed senescence, as shown by reduced senescence-associated beta-galactosidase (SA-β-gal) staining (Extended Data Fig. 1b-c), consistent with previous observations11, 12. We observed that SIRT1 protein levels gradually decreased in IMR90 cells upon replicative senescence (RS) (Fig. 1a), in stress-induced senescence triggered by activated oncogene HRasV12 expression (oncogene induced senescence, OIS) (Fig. 1b) and by the DNA damaging agent etoposide (Fig. 1c), and in senescent primary BJ neonatal foreskin fibroblasts (Extended Data Fig. 1d). In contrast, SIRT1 protein levels remained unchanged in quiescence induced by contact inhibition (Extended Data Fig. 1e), suggesting that SIRT1 protein is specifically lost in cellular senescence.
Figure 1. SIRT1 protein is reduced during cellular senescence.
a, Western blot showing SIRT1 expression in primary IMR90 fibroblasts with indicated population doublings; n = 2 independent experiments. PD: population doubling. b, Western blot showing SIRT expression in IMR90 cells stably expressing ER:HRasV12. Days of 4-hydroxytamoxifen (4OHT) induction are indicated; n = 2 independent experiments. Asterisk indicates SIRT1 band. c, Western blot showing SIRT1 expression in DNA damage-induced senescent cells. Cells were treated 100 μM etoposide for 48 hrs, harvested at indicated days after treatment; n = 2 independent experiments. d,e, Gene expression level of SIRT1 and CDKN2A in proliferating (Pro) and in oncogene-induced senescence (OIS) condition (d) and replicative senescence (RS) condition (e)13. FPKM: fragments per kilobase million. In d,e, mRNA levels are normalized against ACTB; unpaired two-tailed Students’ t-test; data are mean ± s.d.; n=3 biologically independent samples. Statistical information and unprocessed blots are provided as source data.
To address the mechanism of SIRT1 loss in senescence, we first examined whether SIRT1 is downregulated at the mRNA level by RNA-sequencing13 and RT-qPCR. SIRT1 mRNA levels did not decrease in OIS or DNA damage-induced senescence (Fig. 1d, Extended Data Fig. 1f-h). SIRT1 mRNA levels in RS cells were reduced, but to a lesser extent than the protein reduction (Fig. 1e, Extended Data Fig. 1i). These results suggest that mechanisms other than regulation of mRNA synthesis or stability are primarily involved in modulating SIRT1 protein homeostasis during cellular senescence.
We then investigated the possible mechanisms of SIRT1 protein downregulation. We first tested if the proteasome-mediated degradation is involved. Treatment with the proteasome inhibitor (MG132) failed to restore SIRT1 protein in senescent cells (Fig. 2a). We then tested whether lysosome-mediated degradation contributes to SIRT1 protein downregulation. Treatment with Lys05 (a dimeric form of chloroquine14) rescued the loss of SIRT1 protein in senescent cells (Fig. 2b), indicating that SIRT1 is degraded through lysosomes during senescence.
Figure 2. SIRT1 is subjected to autophagosome-lysosome degradation during cellular senescence.
a,b, Western blot showing SIRT expression in proliferating (Pro) and DNA-damage induced senescent IMR90 cells (day 6 after etoposide treatment) treated with MG132 (a) and Lys05 (b). MG132 were added at 0.125, 0.25 and 0.5 μM for 48 hrs. The proteasomal substrate MCL1 serves as control showing the effects of MG132. Lys05 were added at 2, 3 and 5 μM for 48 hrs. c,d, Western blot showing changes of SIRT1 expression in IMR90 expressing inducible hairpins of non-targeting control (shNTC) and shAtg7, in response to OIS induced by 4-hydroxytamoxifen (4OHT) for indicated days (c) and after the establishment of senescence induced by etoposide (d). Doxycycline (Dox) was added on day 4 after treatment of etoposide (d). In c,d, hairpin expression was induced by Dox treatment for 5 days; Sen: senescence. In a-d, each experiment has been repeated for two times. e,f, Confocal microscopy analysis of cytoplasmic SIRT1 in proliferating and senescent IMR90 at day 9 after etoposide treatment (e) and quantification of the percentage of cells with cytoplasmic SIRT1 puncta (f). Arrows indicate co-localization of cytoplasmic SIRT1 puncta and LC3. Eto: etoposide. g,h, Confocal microscopy analysis of IMR90 stably expressing mCherry-GFP-SIRT1 under proliferating and at day 14 after etoposide treatment (g) and quantification of the percentage of cells with cytoplasmic mCherry signals (h). Cells were co-stained with LC3 and LAMP1 antibodies. In f,h, data are mean ± s.d.; more than 500 cells; each data point (n) represents cells in 10 random fields, n = 7, 5, 5, 8 for respective conditions (f) and n = 5 for all conditions (h). i. Relative intensities of mCherry and GFP signals of a typical senescent cell as in (g) were quantified by LAS X Core software. In g-i, each experiment has been repeated for two times. j, Confocal microscopy analysis of senescent cells (etoposide) stably expressing mCherry-GFP-SIRT1 with and without Lys05 treatment. On day 6 after etoposide treatment, cells were treated with 5 μM Lys05 for 48 hrs; n=2 independent experiments. In g, j, arrows indicate cytoplasmic SIRT1 puncta with strong mCherry signals and fading GFP signals. Source Data are provided.
A central lysosomal-mediated process is autophagy, which is activated upon cellular senescence15–17. We recently showed that autophagy has a role in degrading nuclear components, such as Lamin B115. Therefore, we hypothesized that autophagy contributes to SIRT1 downregulation upon senescence.
To test this hypothesis, we first genetically knocked down an upstream autophagy protein, Atg7, to block autophagy activity, and then induced senescence to examine SIRT1 protein levels. Compared to the non-targeting control (NTC), Atg7 knockdown impaired SIRT1 downregulation upon senescence (Fig. 2c). Furthermore, we examined whether Atg7 knockdown in already-established senescent cells can restore SIRT1 protein levels. After induction of senescence by etoposide, Atg7 was then inactivated by shRNA. Atg7 knockdown did not reverse senescence-associated cell cycle arrest as indicated by Cyclin A loss, but rescued SIRT1 protein levels (Fig. 2d). Atg7 inactivation also rescued the deacetylation activity of SIRT1, which was correlated with the increased SIRT1 protein levels (Extended Data Fig. 1j). Taken together, these findings suggest that autophagy is required for SIRT1 degradation during cellular senescence.
To test whether SIRT1 is an autophagy substrate in senescence, we performed immunofluorescence staining of endogenous SIRT1. While in proliferating cells, SIRT1 was primarily localized in the nucleus, cytoplasmic SIRT1 puncta were observed in senescent cells (Fig. 2e-f). Importantly, the cytoplasmic SIRT1 puncta were co-localized with LC3, an autophagy protein required for recognition and delivery of substrates to autophagosomes (Fig. 2e)15. We then used an mCherry-GFP-SIRT1 tandem tag construct18 to investigate the autophagic trafficking of SIRT1. Due to the sensitivity of GFP to low pH, mCherry-only signals of the tandem tagged protein represent localization within acidic autolysosomes and lysosomes. Confocal imaging of mCherry-GFP-SIRT1 expressed in proliferating IMR90 showed predominantly nuclear localization with merged yellow color (Fig. 2g upper panels). In contrast, senescent cells showed cytoplasmic red-only SIRT1 puncta (Fig. 2g-i) that co-localized with LC3 or LAMP1 (Fig. 2g). Neutralizing lysosome pH with Lys05 prevented GFP quenching from mCherry-GFP-SIRT1 protein in senescent cells, and led to retention of its merged yellow signals in the cytoplasm (Fig. 2j), which further indicates that cytoplasmic SIRT1 is targeted by lysosomal degradation. Taken together, these results indicate that nuclear SIRT1 is subjected to cytoplasmic autophagy-lysosome degradation during cellular senescence.
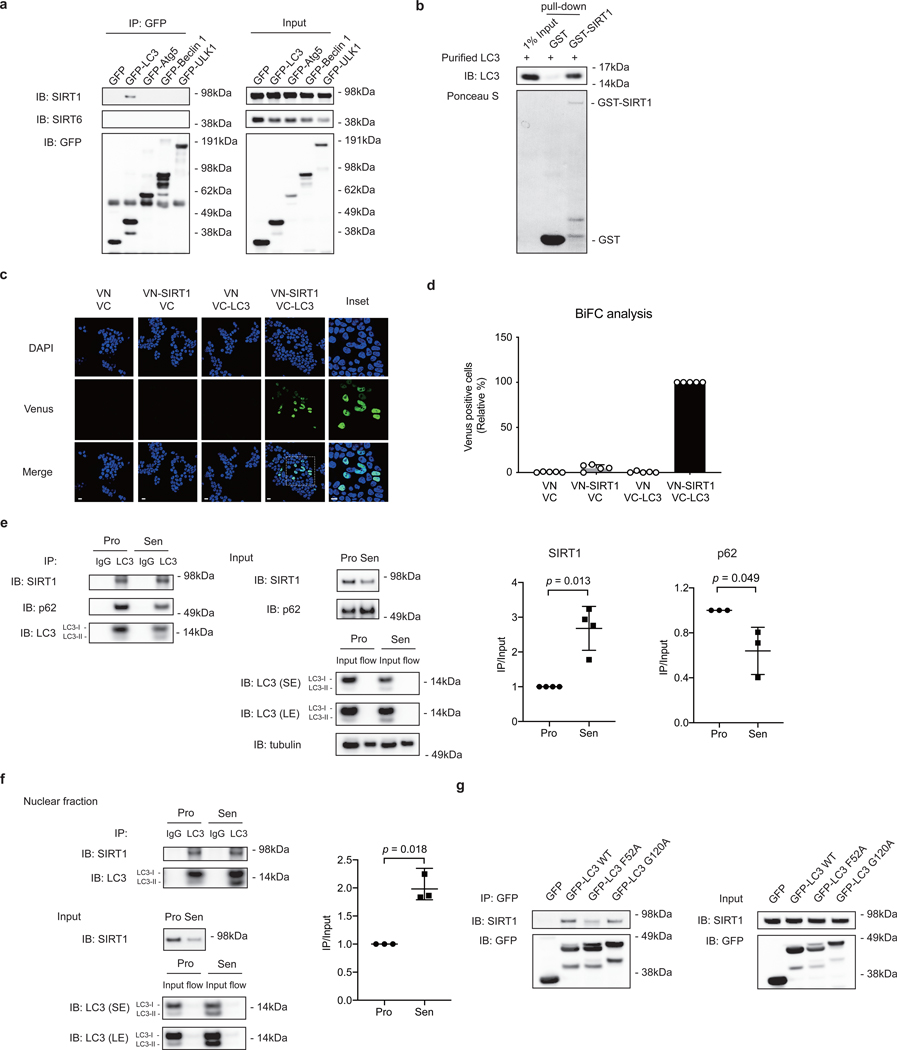
As binding to autophagy proteins is essential for degradation of autophagy substrates15, we investigated potential interactions between SIRT1 and several autophagy proteins by immunoprecipitation (IP). The results showed that SIRT1 associates with LC3, but not with other proteins in the autophagy cascade, including Atg5, Beclin 1, and ULK1 (Fig. 3a). Furthermore, GST pull-down using bacteria-expressed and purified GST-SIRT1 and LC3 proteins showed that SIRT1 directly binds to LC3 in vitro (Fig. 3b).
Figure 3. SIRT1 associates with autophagy protein LC3.
a, IP of extracts from HEK293T cells expressing GFP-tagged constructs; n = 2 independent experiments. b, GST-SIRT1 pull-down of bacteria-expressed and purified LC3 protein; n = 2 independent experiments. c,d, BiFC analysis of SIRT1-LC3 interaction. HEK293T cells were transfected with indicated combinations of constructs. c, Cells were imaged by confocal microscopy; This experiment has been repeated for 3 times. Scale bar: 15 μm. d, Relative percentage of Venus-positive cells were quantified and normalized to VN-SIRT1+VC-LC3 condition (as 100%). Mean ± s.d.; more than 500 cells and n = 5 random fields. e, IP of extracts from proliferating and senescent IMR90 cells. Senescent cells were harvested for IP at day 8 after etoposide treatment. Excessive beads and antibodies were used in the IP to capture nearly 100% of LC3 protein in the lysates. Flow: flow-through. Western blot quantification: SIRT1 or p62 IP bands were normalized to LC3 IP and SIRT1 or p62 input bands. Mean ± s.d.; n = 3 independent experiments; paired two-tailed Students’ t-test. f, IP of nuclear extracts from proliferating and senescent IMR90 cells. Senescent cells were harvested for IP at day 8 after etoposide treatment. Excessive beads and antibodies were used in the IP to capture nearly 100% of LC3 protein in the lysates. Flow: flow-through. Western blot quantification: SIRT1 IP bands were normalized to LC3 IP and SIRT1 input bands. Mean ± s.d.; n = 3 independent experiments; paired two-tailed Students’ t-test. g, IP of HEK293T cells expressing GFP-tagged LC3 wild type (WT) or mutant constructs; n = 2 independent experiments. Statistical information and unprocessed blots are provided as source data.
To visualize the intracellular location of SIRT1-LC3 interaction, we performed a Bimolecular Fluorescence Complementation assay (BiFC)19. SIRT1 and LC3 were fused to the N-terminal (VN) or C-terminal (VC) domains of Venus to generate VN-SIRT1 and VC-LC3 fusion proteins. While the two fusion proteins do not fluoresce when co-transfected with VN or VC, co-expression of VN-SIRT1 and VC-LC3 exhibited fluorescence of Venus in the nucleus, showing that SIRT1 and LC3 interact in the nucleus (Fig. 3c-d).
Since our findings indicated that SIRT1 and LC3 interact at the basal state, we next investigated their interaction upon cellular senescence by performing co-IP in the two states. While the SIRT1 protein level in the input of senescent cells was reduced compared to proliferating cells, LC3 brought down similar levels of SIRT1 in senescent cells, suggesting that the LC3-SIRT1 interaction was enhanced during senescence (Fig. 3e, Extended Data Fig. 2a). We confirmed comparable and complete IP of LC3 from lysates of both proliferating and senescent cells (Fig. 3e). In contrast, LC3 interaction with another autophagy substrate, p62, was not enhanced in senescence compared to proliferating control (Fig. 3e). Moreover, contact inhibition did not enhance the LC3-SIRT1 interaction (Extended Data Fig. 2b-c). LC3-SIRT1 interaction, but not LC3-p62 interaction, was also increased in the nuclear fraction of senescent cells, compared to proliferating control (Fig. 3f, Extended Data Fig. 2d-f). These results suggest that SIRT1-LC3 association is specifically enhanced in cellular senescence, which potentially promotes SIRT1 degradation in senescent cells.
Next, we examined the potential mechanisms for the enhanced SIRT1-LC3 interaction during senescence. At least in some cases, LC3 interactions with autophagy substrates are known to be modulated by substrate phosphorylation20. We therefore performed an endogenous LC3 IP in proliferating cells with Lambda protein phosphatase treatment to reduce the overall protein phosphorylation. We observed that LC3-SIRT1 binding was drastically increased upon the phosphatase treatment, when overall phosphorylation levels were lowered (Extended Data Fig. 2g), indicating that dephosphorylation enhances SIRT1-LC3 binding. We subsequently evaluated the phosphorylation status of endogenous SIRT1 in senescent cells by mass spectrometry. The amounts of peptides injected for mass spectrometry were comparable between proliferating and senescence samples (Extended Data Fig. 2h). 5 out of 6 peptides of SIRT1 showed decreased phosphorylation levels in senescent samples (Extended Data Fig. 2i). The results suggest that SIRT1 overall phosphorylation levels are reduced in senescence, consistent with our observation that LC3-SIRT1 interaction is enhanced upon phosphatase treatment (Extended Data Fig. 2g).
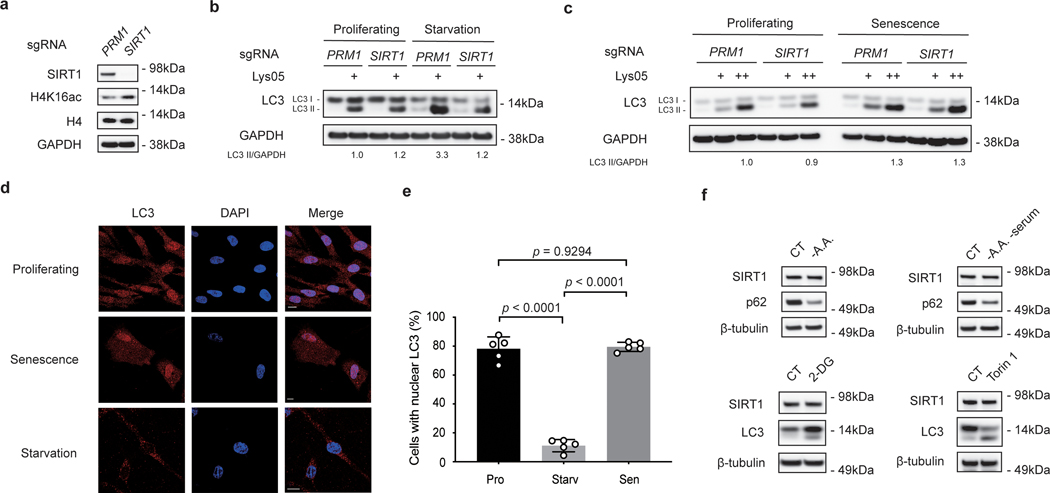
Previously, SIRT1 was shown to deacetylate LC3, which led to the exportation of LC3 from the nucleus to execute its roles in the cytoplasmic autophagy cascade during starvation21. Paradoxically, in the context of cellular senescence, we propose that LC3 may instead facilitate the degradation of SIRT1. To dissect the differential responses of LC3 and SIRT1 in starvation versus senescence, we disrupted SIRT1 and evaluated the consequences in starvation and senescence. CRISPR/Cas9-mediated gene inactivation of SIRT1 resulted in loss of SIRT1 protein, and corresponding elevation of histone H4K16 acetylation (Extended Data Fig. 3a). Upon starvation, LC3-II levels were induced in control cells (the induction was further revealed by Lys05 addition), while SIRT1 depletion suppressed induction of LC3-II levels (Extended Data Fig. 3b), which is consistent with previous literature21. In contrast, during senescence, depletion of SIRT1 did not suppress LC3-II induction (Extended Data Fig. 3c), suggesting that during senescence, a different pathway than SIRT1-mediated deacetylation of LC3 may be employed to induce LC3-II. Furthermore, immunofluorescence staining of endogenous LC3 in starved cells showed that LC3 was predominantly transported to the cytoplasm (Extended Data Fig. 3d-e). However, in senescent cells, while showing increased cytoplasmic localization, LC3 was not completely translocated out of the nucleus (Extended Data Fig. 3d-e). Lastly, in contrast to SIRT1 protein reduction upon cellular senescence, cells cultured in various nutrient-deprived conditions did not lose SIRT1 (Extended Data Fig. 3f). Taken together, these data demonstrate that nuclear SIRT1-LC3 interaction has distinct dynamics and functions in senescence compared to starvation. Hence, we went on to investigate the potential role of LC3 in reducing SIRT1 protein levels during cellular senescence in this study.
We subsequently identified the amino acid residues responsible for LC3-SIRT1 interaction. We first disrupted two essential residues on LC3, which are involved in substrate binding (F52A substitution)22 or in the lipidation process that facilitates association of LC3 with autophagic membranes (G120A substitution)23. While G120A did not affect SIRT1 binding, suggesting that LC3 lipidation is not required for SIRT1 interaction, the F52A substitution showed impaired interaction with SIRT1 (Fig. 3g).
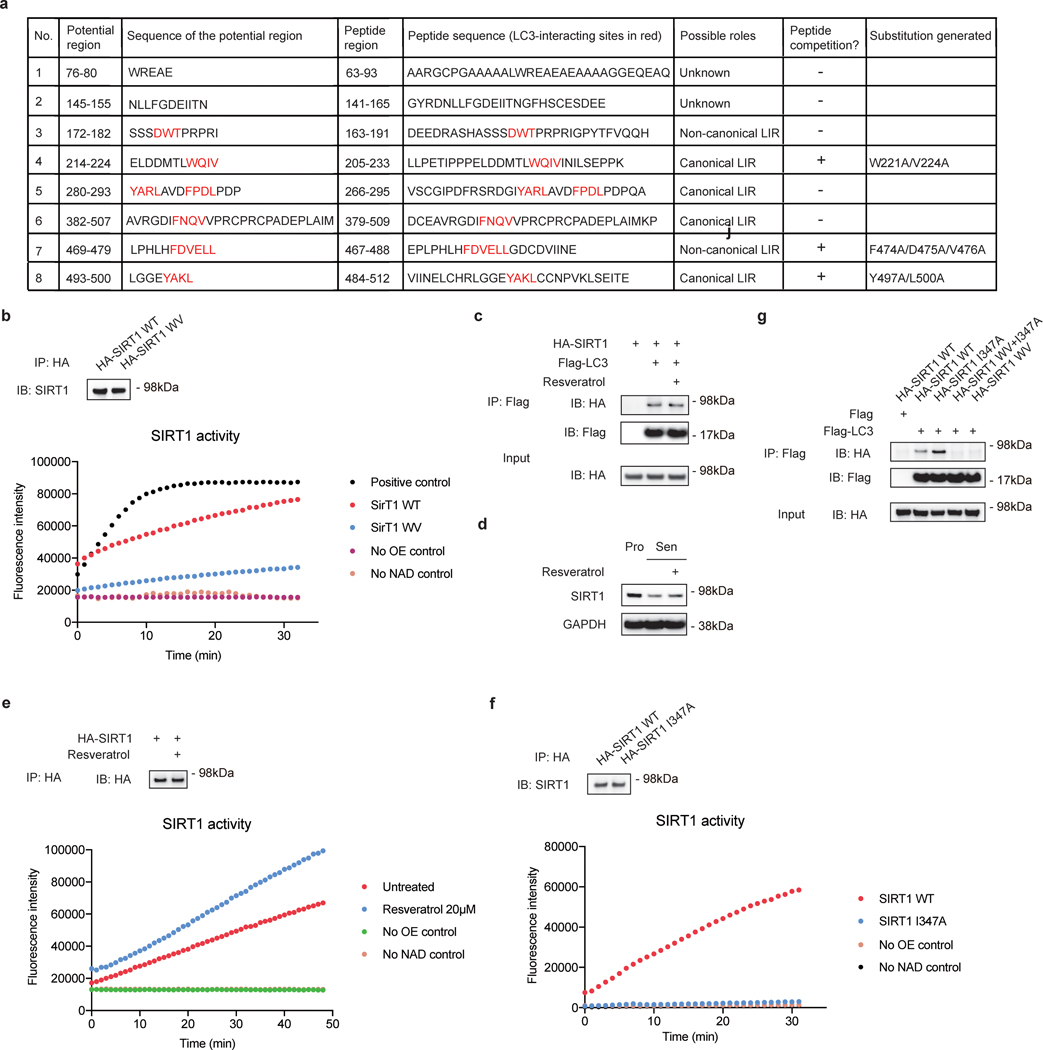
We next investigated the region within SIRT1 that interacts with LC3. Many autophagy substrates bind to LC3 through an LC3-interacting region (LIR) motif. LIR motifs typically have a core consensus sequence of [W/F/Y]XX[L/I/V], and acidic-charged residues (E or D) at the N-terminus of the LIR motif further facilitate the binding20. The aromatic residue [W/F/Y] binds to one hydrophobic pocket in LC3, while the hydrophobic residue [L/I/V] binds to another hydrophobic pocket containing the crucial F52 residue20. Therefore, reduced binding between SIRT1 and LC3 F52A implies that SIRT1 may bind to LC3 through an LIR or LIR-like motif. We identified LC3-binding regions on SIRT1 employing a peptide array approach that has been used to characterize LC3 interaction24–26. Using the array, we identified eight regions in SIRT1 that potentially associate with LC3 (Fig. 4a, peptide locations shown in Fig. 4e, peptide sequences shown in Extended Data Fig. 4a). To narrow down the essential regions, we performed peptide competition IP by adding ~30 amino-acid synthetic peptides that correspond to the individual regions (Extended Data Fig. 4a) into SIRT1-LC3 IP assays, to examine their abilities to compete with the full-length SIRT1-LC3 interaction. Using this assay, we identified three target LIR/LIR-like motifs, where the addition of synthetic peptides reduced full-length SIRT1-LC3 binding (Fig. 4b). To further confirm the role of these motifs in LC3 binding, we individually substituted the key residues of the three LIR/LIR-like motifs within full-length SIRT1 (Extended Data Fig. 4a), and performed IP between LC3 and full-length SIRT1 substitutions. The IPs showed that the W221A/V224A double substitution (hereafter referred as WV mutant), which disrupts the aromatic and hydrophobic positions of an LIR motif at amino acid position 221–224 of SIRT1, had impaired interaction with LC3, while substitutions of the other two motifs, F474A/D475A/V476A triple substitution and Y497A/L500A double substitution, did not affect LC3 binding (Fig. 4c). The crucial LIR motif (221–224) is located at the N-terminal region of SIRT1, with acidic E214, D216 and D217 residues present at the N-terminus of this motif (Fig. 4e); substitution of these residues also impaired LC3 binding (Fig. 4d). Moreover, introducing WV substitution on a synthetic peptide spanning 205–233 showed impaired ability to compete with full-length SIRT1-LC3 interaction (Fig. 4f). Overall, these results show that the region 221–224 on SIRT1 is essential for SIRT1-LC3 interaction.
Figure 4. SIRT1 interacts with LC3 through a LIR motif.
a, SIRT1 peptide array showing potential interaction regions with LC3. 20-mer peptides covering full-length SIRT1 with a moving window of 3 residues were synthesized and incubated on a cellulose membrane. The array was probed with GST-LC3B. Eight potential regions for LC3 binding were boxed and numbered. b, IP of HEK293T cells expressing HA-tagged SIRT1 and Flag-tagged LC3, with addition of DMSO or 500 μM synthetic SIRT1 peptides as indicated; n = 3 independent experiments. c, IP of HEK293T cells expressing HA-tagged SIRT1 WT, W221A/V224A (WV), F474A/D475A/V476A or Y497A/L500A substitutions; n = 2 independent experiments. d, IP of HEK293T cells expressing Flag-tagged LC3 and HA-tagged SIRT1 WT, WV or E214A/D216A/D217A (EDD) mutants; n = 2 independent experiments. e, Scheme of SIRT1 showing the location of the 205–233 peptide. Potential amino acid residues involved in LC3 binding are labeled in red, including the core LIR motif WQIV. Location of 205–233 peptide is labeled in blue; locations of other peptides tested are labeled in grey. f, IP of HEK293T cells expressing HA-tagged SIRT1 and Flag-tagged LC3, with addition of DMSO or 500 μM synthetic SIRT1 peptides as indicated; n = 2 independent experiments. g, IMR90 cells stably expressing HA-tagged SIRT1 I347A or WV+I347A mutants were infected with HRasV12 retrovirus, selected by antibiotics and harvested at indicated days for western blotting; n = 2 independent experiments. h-i, Senescent IMR90 cells expressing mCherry-GFP-SIRT1 I347A or WV+I347A mutants were analyzed. Senescence was initiated by etoposide treatment for indicated days. h, Cells at day 7 after etoposide treatment were imaged by confocal microscopy. Scale bar: 5 μm. i, Percentages of cells with cytoplasmic SIRT1 puncta were quantified. Data are mean ± s.d.; more than 500 cells were counted; each data point (n) represents cells in 10 random fields, n = 5 for all conditions; two-way ANOVA with Sidak’s multiple comparisons test. Statistical information and unprocessed blots are provided as source data.
We next asked whether this LIR motif is required for degradation of SIRT1 in senescent cells. The 221–224 LIR motif resides adjacent to the catalytic domain of SIRT1, and a broader region covering this motif (183–229) has been reported to bind to SIRT1 activating compounds27. Therefore, we assayed the deacetylation activity of the SIRT1 WV mutant, and found that the mutant maintained approximately 36% deacetylation activity compared to wild-type SIRT1 (Extended Data Fig. 4b). Treatment of a SIRT1-activating compound, resveratrol, did not disrupt SIRT1-LC3 interaction or significantly rescue SIRT1 protein levels during senescence (Extended Data Fig. 4c-e). Since SIRT1 catalytic activity suppresses senescence11, 12 (Extended Data Fig. 1a-c), we introduced a previously characterized catalytic dead mutation, I347A28, to eliminate potential differences in inducing senescence when stably expressing the various SIRT1 mutants in IMR90. The SIRT1 I347A mutant showed no deacetylation activity compared to wild-type protein (Extended Data Fig. 4f), while still maintaining its interaction with LC3 (Extended Data Fig. 4g).
To assess the degradation of the I347A mutant and the W221A/V224A/I347A (WV+I347A) mutant, we stably expressed HA-tagged SIRT1 I347A and WV+I347A mutants in IMR90, and examined their levels upon senescence. While p16 induction was at similar levels in cells expressing either mutants, indicating similar induction of senescence, the WV+I347A mutant showed impaired downregulation compared to I347A mutant (Fig. 4g). Confocal imaging of mCherry-GFP-tagged SIRT1 I347A and WV+I347A mutants in IMR90 showed that at the same timepoints for senescence induction, cells with the WV+I347A mutant had stronger fluorescence intensities of nuclear SIRT1, and there were fewer cells exhibiting cytoplasmic SIRT1 puncta compared to cells with the I347A mutant (Fig. 4h-i). Taken together, these data show that the SIRT1-LC3 interaction is required for nuclear SIRT1 degradation in senescence.
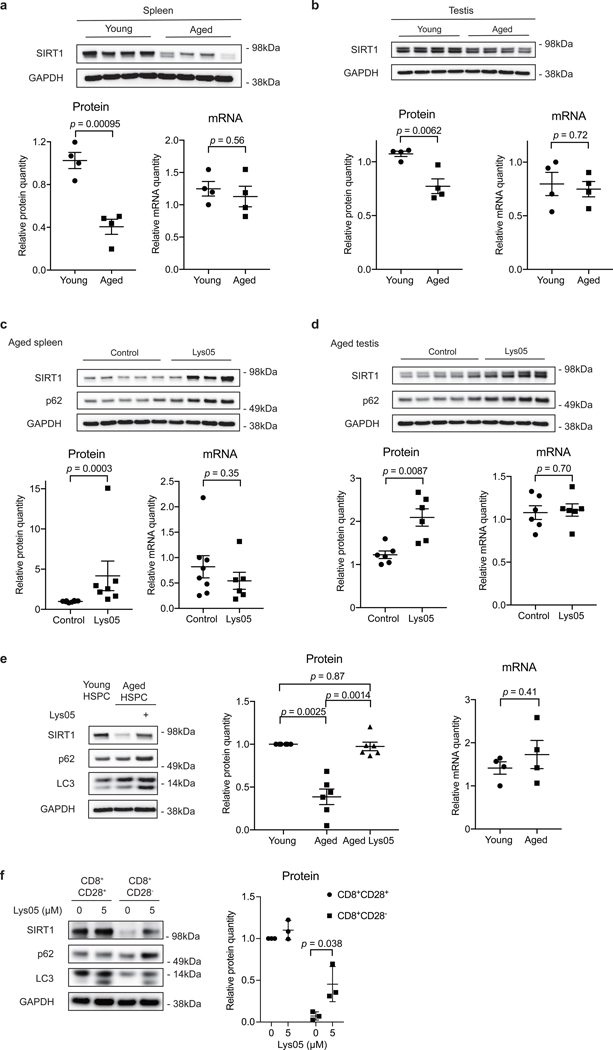
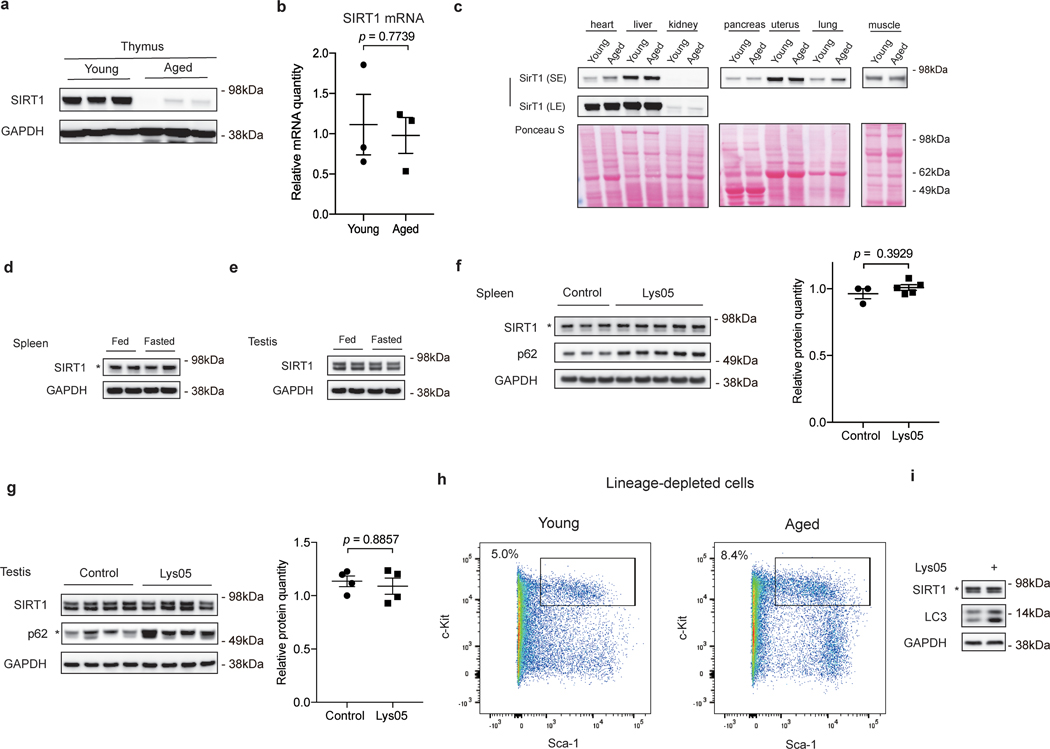
In addition to senescence of primary fibroblasts, we also investigated SIRT1 protein levels upon natural aging in mice. We dissected several organs and tissues from young (2–4 months old) and naturally aged (19–26 months old) C57BL/6 mice, and analyzed SIRT1 protein and mRNA quantities. Aged spleens, testes and thymus showed reduced SIRT1 protein levels, compared to young samples (Fig. 5a-b, Extended Data Fig. 5a). The decline in SIRT1 protein was not seen in other tissues, including heart, liver, kidney, pancreas, uterus, lung and muscle (Extended Data Fig. 5c). In contrast, SIRT1 protein levels were not reduced in spleens and testes of mice subjected to 24 h fasting (Extended Data Fig. 5d-e), which is consistent with previous reports29, indicating that SIRT1 protein downregulation is specific to aging of some tissues. SIRT1 mRNA levels in aged spleens, testes and thymus were not significantly different from young samples (Fig. 5a-b, Extended Data Fig. 5b). Next, to examine the role of autophagy/lysosome in SIRT1 downregulation, we injected aged mice intraperitoneally with Lys05 or PBS, and then harvested corresponding tissues for analysis. Autophagy inhibition was confirmed by accumulation of p62 from treated mice in spleens and testes (but not thymus, possibly due to issues in drug uptake in aged thymus) (Fig. 5c-d). Spleens and testes from Lys05-injected mice showed elevated SIRT1 protein levels, compared to samples from PBS-injected group; the trends of SIRT1 protein were consistent with changes in p62 levels (Fig. 5c-d). SIRT1 mRNA levels remained unaltered upon Lys05 treatments (Fig. 5c-d). Notably, Lys05 treatments in young mice did not affect SIRT1 protein levels (Extended Data Fig. 5f-g), suggesting that the lysosomal degradation of SIRT1 is specific in aged tissues. Taken together, these results indicate that SIRT1 protein is downregulated through lysosomal-mediated degradation during aging in several mouse tissues.
Figure 5. SIRT1 undergoes lysosomal degradation during aging in mouse and human.
a,b, Spleens (a) and testes (b) from young (2–4 months) and aged (19–26 months) C57BL/6 mice were analyzed by western blotting and RT-qPCR. Data are mean ± s.e.m.; unpaired two-tailed Students’ t-test; n = 4 animals. c,d, Spleens (c) and testes (d) were analyzed by western blotting and RT-qPCR for SIRT1 expression, from aged (19–24 months) mice subjected to daily i.p. injection of 10 mg/kg Lys05 in PBS or PBS control in 100 μL volume for two weeks. Data are mean ± s.e.m.; two-tailed Mann-Whitney test. For spleen protein, control group n = 8 animals, Lys05 group n = 7 animals; RNA, control group n = 8 animals, Lys05 group n = 6 animals. For testis protein and RNA, control group n=6 animals, Lys05 group n=6 animals. e.HSPC populations were isolated from young (2–4 months) and aged (20–26 months) C57BL/6 mice, cultured with or without 2 μM Lys05 for 24 hours and analyzed by western blotting and RT-qPCR. For protein, data are mean ± s.e.m.; one-way ANOVA coupled with Tukey’s test; n=6 independent experiments. For RNA, data are mean ± s.e.m.; unpaired two-tailed Students’ t-test; n=4 independent experiments. f. Freshly sorted CD8+CD28+ (control) and CD8+CD28- T cells were treated with Lys05 at doses of 0 and 5 μM for 14 hours, and then were harvested and analyzed by western blotting. Donor age: 53, 54 and 66. Data are mean ± s.d.; unpaired two-tailed Students’ t-test; n = 3 human donors. In a-f, western blot quantification: SIRT1 bands were normalized to GAPDH bands, for testes both bands of SIRT1 were considered. In a-e, RT-qPCR quantification: data were normalized to 18S. Statistical information and unprocessed blots are provided as source data.
Furthermore, we examined SIRT1 in hematopoietic stem and progenitor cells (HSPCs). It has been reported that SIRT1 deletion in young HSPCs results in aging-like phenotypes30. To determine whether SIRT1 levels change during HSPC aging, we isolated Lin-Sca-1+c-Kit+ (LSK) cells from young and aged mice to represent HSPC populations (Extended Data Fig. 5h), and examined SIRT1 protein and mRNA levels. Aged LSK cells displayed significantly decreased SIRT1 protein levels compared to young cells (Fig. 5e), while young and aged mRNA levels were not significantly distinct (Fig. 5e). Importantly, short-term Lys05 treatment of aged LSK cells led to elevation of SIRT1 protein levels compared to control cells from the same pool of aged mice (Fig. 5e). Lys05 treatment of young LSK cells did not change SIRT1 protein levels (Extended Data Fig. 5i). These results indicate that SIRT1 downregulation during HSPC aging is mediated by a lysosome-mediated pathway.
The above studies implicate loss of SIRT1 in the aging of the immune and hematopoietic system. To explore this notion further, we investigated T cells from aged human donors. The accumulation of terminally differentiated CD8+CD28- memory T cells is one of the hallmarks of immune aging31. These cells are highly cytotoxic but have reduced proliferative response to antigen-specific activation. SIRT1 protein levels are reported to be downregulated in CD8+CD28- T cells, while the mRNA levels remain unaltered32. Therefore, we asked whether autophagy-lysosome pathway contributes to loss of SIRT1 in CD8+CD28- T cells from aged human donors. Consistent with previous studies, SIRT1 protein levels were markedly reduced in CD8+CD28- T cells, compared to CD8+CD28+ T cells (Fig. 5f). Importantly, transient treatment of these cells with low-dose Lys05 increased SIRT1 protein levels (Fig. 5f). Because MG132 treatment failed to increase SIRT1 in these cells32, our results indicate that SIRT1 is degraded at least in part through the autophagy-lysosome pathway during T cell aging in humans.
In conclusion, we report that autophagy contributes to downregulation of SIRT1 protein during cellular senescence, during aging of several mouse tissues and cells, and in human CD8+CD28- T cells. While loss of SIRT1 protein has also been previously observed in several cellular models of senescence and aging11, 12, 33, 34, our work demonstrates that SIRT1 is a nuclear substrate of autophagy, which involves SIRT1 recognition by LC3 and transport to cytoplasmic autophagosomes for degradation upon cellular senescence. In addition to Lamin B1, our discovery of a second major nuclear substrate of LC3-mediated selective autophagy indicates that the nuclear autophagy pathway may have a general role in cell physiology.
While we showed that SIRT1 dephosphorylation in senescence could be the trigger of its degradation (Extended Data Fig. 2h-i), we note that the SIRT1 region 199–236, which includes the WQIV motif responsible for LC3 interaction, was not detected in mass spectrometry samples due to the high charge sequence at the region. Moreover, the specific dephosphorylation site on SIRT1 remains to be identified. Future studies are needed to characterize the post-translational modification(s) and signaling pathway that trigger SIRT1 degradation through autophagy during senescence.
Autophagy, as a quality control pathway, is generally considered to delay functional decline associated with aging; elevation of basal autophagy activity promotes healthspan and longevity in mice35. Our study suggests that selective degradation of certain autophagy substrates in certain tissues, like SIRT1, may paradoxically be detrimental during aging. One plausible explanation for the discrepancy could be the differences between bulk autophagy and selective autophagy. While stimulation of the overall autophagy flux is beneficial for cell and tissue fitness, selective degradation of nuclear constituents, such as Lamin B1 and SIRT1, leads to arrest of cell cycles and induction of inflammation. Further studies are needed to dissect the mechanisms underlying cytoplasmic versus nuclear autophagy and their roles in senescence and aging.
The downregulation of SIRT1 in senescent cells may be associated with biological functions of cellular senescence. SIRT1 was reported to negatively regulate the expression of several senescence-associated secretory phenotype (SASP) factors, including IL-6, IL-8, and IL-1β36. Given the critical functions of SASP in immuno-surveillance of pre-malignant cells, we speculate that the downregulation of SIRT1 is a programmed event of senescence to alarm the immune system, to restrain tumorigenesis. This notion is consistent with the distinct dynamics of SIRT1-LC3 interaction in starvation versus senescence (Extended Data Fig. 3a-f); that is, since the pro-inflammatory response is not induced during starvation, SIRT1 is not degraded. While acute induction of senescence restrains tumorigenesis, the accumulation of senescent cells during aging promotes chronic inflammation and age-associated diseases. Hence, although the loss of SIRT1 in senescence serves as a potential tumor-suppressive mechanism, SIRT1 loss in aged tissues may promote aging and age-related pathologies, consistent with the “antagonistic pleiotropy hypothesis”.
Immunosenescence is an important aspect of aging9. The decline of the immune system, such as dysfunction of HSCs and T cells, contributes to age-related diseases37. SIRT1 is a critical regulator of hematopoietic lineage generation from HSPC, and the skewed myeloid and lymphoid differentiation upon aging is associated with defective immune response30. Besides, SIRT1-FOXO1 axis has been shown to contribute to the immune dysfunction in human CD8+CD28- T cells, accumulation of which is a common event in aging and age-related diseases32. In addition, reduction of SIRT1 protein in critical immune organs, including spleen and thymus (Fig. 5a, Extended Data Fig. 5a), further suggest the deterioration of the adaptive immune system during aging. Thus, restoring SIRT1 protein level may provide a critical means to reprogram the aged immune cells, shedding light on potential therapeutic intervention to delay the process of immune aging/senescence. Stabilizing SIRT1 protein level, such as via interrupting SIRT1-LC3 interaction, could be a new direction for the design of anti-aging compounds.
Methods
Cell lines, culture and treatments
IMR90, BJ and HEK293T cells were described previously15, 38, 39. IMR90 and BJ within population doubling of 40 were used, except for replicative senescence experiments. Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin (Invitrogen). IMR90 and BJ cells were cultured under physiological oxygen (3%). Stable cell lines were made by retrovirus or lentivirus infection as previously described15, 39.
For replicative senescence, cells were subjected to continuous passaging to induce senescence. For oncogene-induced senescence, cells expressing ER:HRasV12 were treated with 4OHT (4-hydroxytamoxifen) to induce HRasV12 expression. For DNA damage-triggered senescence, cells were treated with 100 μM etoposide for 48 hours.
For contact inhibition, cells were continuously cultured at 100% confluency to induce growth arrest and quiescence. Fresh media was replenished every two days to maintain cellular metabolic activities.
For amino acid starvation, cells were cultured in Hank’s buffer (with calcium and glucose) supplemented with 10% dialyzed FBS and 1% HEPES (Invitrogen). For amino acid and serum starvation, cells were cultured in Hank’s buffer with 1% HEPES. For 2-DG treatment, cells were incubated in complete media supplemented with 10 mM 2-DG. For Torin 1 treatment, cells were incubated in complete media supplemented with 250 nM Torin 1.
For MG132 treatment, doses of 0.125–0.5 μM were used for 48-hour treatment. For Lys05 treatment, doses of 2–5 μM were used for 48-hour treatment.
Reagents and antibodies
The following reagents were used: 4-hydroxytamoxifen (Sigma-Aldrich), etoposide (Sigma-Aldrich), 2-DG (Sigma-Aldrich), Torin 1 (Selleckchem), Resveratrol (Sigma-Aldrich), MG132 (Calbiochem), Lys05 (a gift from Dr. Ravi Amaravadi and Dr. Jeffery D. Winkler, and was also purchased from MedKoo Biosciences, Inc).
The antibodies used in this study were described in Supplementary Table 1.
Plasmids
GFP, GFP-LC3 wild type and mutants, Flag-LC3, GFP-Atg5, GFP-Beclin 1, GFP-ULK1, Tet-pLKO-puro-shAtg7 and constructs used for BiFC assay were described previously15. SIRT1 sequence was cloned into LPC-HA, LPC-Flag, pETDuet-GST and previously described pBabe-mCherry-GFP vectors15. All SIRT1 mutants were cloned from SIRT1 sequence and were verified by DNA sequencing.
The following CRISPR guide sequences were cloned into lentiCRISPR v2 vector. PRM1: CACCGGACAAAGAAGTCGCAGACGA, AAACTCGTCTGCGACTTCTTTGTCC; SIRT1: CACCGCTCCCCGGCGGGGGACGACG, AAACCGTCGTCCCCCGCCGGGGAGC. The constructs were incorporated into IMR90 cells through lentivirus infection for sgRNA knockout.
Western blotting
Western blotting was performed as previously described15 with modification. In brief, cells were lysed in buffer containing 20 mM Tris pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, supplemented with 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) and benzonase (Novagen) at 12.5 U/mL. For lysis, cells were rotated at 4 °C for 30 min, supplemented with 1% SDS for boiling, and then the supernatants were subjected to electrophoresis and transfer to 0.2 μm nitrocellulose membrane. Membrane was incubated with primary antibodies in TBS supplemented with 0.1% Tween 20 at 4 °C for overnight, probed with HRP-conjugated secondary antibodies, and then was developed with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher) and imaged by Amersham Imager 600.
RNA-sequencing and RT-qPCR
The results of RNA-sequencing were based upon data from Rai et al.(referenced GEO Accession: GSE52848, GSE53356)13. For proliferating cells, IMR90 cells at PD32 were used. For replicative senescence, IMR90 cells at PD88 were used. For oncogene-induced senescent cells, IMR90 cells expressing ER:HRasV12 construct were induced by 4OHT. For RNA-sequencing data analysis, quantification of fragments per kilobase million were used and were normalized to ACTB.
For RT-qPCR, RNA was extracted using Qiagen RNeasy Mini Kit (#74104). Reverse transcription was performed using Applied Biosystems High-Capacity RNA-to-cDNA kit (#4387406). cDNA was quantified using standard procedures on a 7900HT Fast-Real-Time PCR (ABI).
The following primers were used for RT-qPCR analysis. SIRT1 (human): TGACTTCAGGTCAAGGGATGG, GGGAAGTCTACAGCAAGGCG; GAPDH (human): CAGCCTCAAGATCATCAGCA, TGTGGTCATGAGTCCTTCCA; SIRT1(mouse) pair 1: GCCGCGGATAGGTCCATATA, GTGCCAATCATGAGATGTTGCT; pair 2: AGCGATCGGCTACCGAGA, TTAGTGAGGAGTCCATCGGTCA; 18S (mouse): GTAACCCGTTGAACCCCATT, CCATCCAATCGGTAGTAGCG.
Immunoprecipitation
Immunoprecipitation (IP) was performed as previously described15 with slight modification. In brief, cells were lysed in buffer containing 20 mM Tris pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, 10% glycerol, supplemented with 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) at 4 °C for 1 hour. The supernatant was incubated with antibody-conjugated Dynabeads (Life Technologies) at 4 °C for overnight, and was then washed and boiled with NuPAGE loading dye for western blotting analysis in the next day.
For Lambda-treated samples, cell lysates were incubated with Lambda at 30°C for 10min; for control samples, cell lysates were incubated with lysis buffer at 30°C for 10min. The lysates were then subjected to IP overnight at 4°C.
Bacteria expression and GST pull-down
GST-tagged constructs were transformed into BL21-CodonPlus E. coli for protein expression, and purified with glutathione beads (Life Technologies). LC3 were cloned into GST construct with a TEV protease recognition site between GST and the LC3 sequence. After glutathione bead purification, the protein was further digested with His-tagged TEV protease and purified with Ni-NTA beads (Qiagen).
GST pull-down was performed as previously described15. In brief, bacteria lysates containing GST-tagged proteins were incubated with glutathione beads at 4 °C for 2 hours for pre-binding. Purified proteins were diluted in binding buffer (20 mM Tris, pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, supplemented with 1:100 Halt Protease inhibitor cocktail), pre-cleared with glutathione beads at 4 °C for 1 hour, and then incubated with pre-bounded glutathione beads at 4 °C for overnight. The next day, beads were washed and boiled with NuPAGE loading dye for western blotting analysis.
Immunofluorescence
Immunofluorescence was performed as previously described15 with slight modification. In brief, cells on the coverslips were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in PBS. Cells were incubated with primary antibodies in 5% BSA in PBS supplemented with 0.1% Tween 20 first at room temperature for 30 min, and then at 4 °C for overnight. The next day, cells were probed with Alexa Fluor-conjugated secondary antibody (Life Technologies), washed and mounted with ProLong Gold (Life Technologies). The slides were imaged with Leica TCS SP8 fluorescent confocal microscope. If cells were subjected to DAPI staining, cells were incubated with 1 μg/ml DAPI in PBS for 5 min, and washed with PBS before mounting.
Bimolecular Fluorescence Complementation assay
HEK293T cells were plated on coverslips and transfected with 50–100 μg BiFC constructs for 2 days. The coverslips were fixed with 4% paraformaldehyde in PBS, stained with DAPI, mounted with ProLong Gold (Life Technologies) and then were subjected to microscopy imaging.
Cell nuclear fractionation
Cell fractionation was performed using a previously described protocol with modification40. 1 × 106 cells were trypsinized and washed with PBS for two times. Cell were resuspended in 1.2 mL HEPES-Sucrose-Ficoll-Digitonin solution (HSFD, 20 mM HEPES-KOH, 6.25% Ficoll, 0.27 M sucrose, 3 mM CaCl2, 2 mM MgCl2. pH7.4) supplemented with 300 μg/mL digitonin and 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo Scientific), rotated at 4°C for 10 min, and then centrifuged at 1000 × g for 3 min. The supernatant (cytoplasmic fraction) was removed, and the pellet was washed twice with PBS. The pellet was then resuspended in 1.2 mL buffer containing 20 mM Tris pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, 10% glycerol, supplemented with 1:1000 benzonase (Millipore) and 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) and rotated at 4 °C for 1 hour. The solution was centrifuged at 15,000 × g for 5 min and the supernatant was collected as nuclear fraction.
Peptide array and peptide competition
For peptide array, the experiment was performed following previously described procedure26. 20-amino acid peptides were made throughout full-length SIRT1 with a 3-amino acid shift. Peptides were synthesized and immobilized on a cellular membrane using INTAVIS MultiPep automated peptide synthesizer (INTAVIS Bioanalytical Instruments AG, Germany). GST-LC3B was used as an overlay probe for the binding, and interaction signals were detected using specific GST antibody, developed by chemiluminescence and imaged by imager.
For peptide competition IP, ~30-amino acid peptides were synthesized by GenScript and were dissolved with DMSO. 500 μM peptides or same volume of DMSO were added in the IP system at the overnight incubation step. The other steps of the IP were performed the same as the regular procedure.
Mouse experiments
Mice in C57BL/6 background were used in this study. Aged mice were obtained from NIA aged rodent colonies. Mice were housed under a 12-h light and 12-h dark cycle, with lights on at 7 a.m. and lights off at 7 p.m. Water and standard chow were provided ad libitum following regulations and guidelines of the University of Pennsylvania. Tumor-free mice with both sexes were included in the study.
For Lys05 injection experiment, mice were injected intraperitoneally with 10 mg/kg Lys05 in PBS or PBS alone in 100 μL volume daily for two weeks, and then were subjected to euthanasia and tissue harvesting. All procedures were approved and performed following regulations and guidelines of the University of Pennsylvania.
For LSK cell isolation and culture, mice were euthanized and dissected to excise the tibia, femur and hip bones. Bone marrow cells were harvested and subjected to lineage depletion using Lineage Cell Depletion Kit (Miltenyi Biotec, 130–090-858) following the manufacturer’s instructions. Briefly, harvested cells were incubated with biotinylated monoclonal antibodies cocktail against lineage specific cells (CD5, CD11b, B220, Gr1and Ter119). Cells were then washed and incubated with anti-biotin Magnetic beads followed by magnetic separation on autoMACS Pro Separator (Miltenyi Biotec, 130–092-545). Eluted Lineage depleted cells were stained with surface markers to define c-Kit and Sca-1 population. LSK cells were sorted using BD FACS Aria flow cytometer with DiVa software (Becton Dickinson). Forward and side scatter parameters were used to exclude the doublets. Purity was maintained at > 95–97%. Sorted LSK cells were cultured in StemSpan SFEM II (STEMCELL Technologies, 09600) supplemented with 10% fetal bovine serum (Sigma-Aldrich, 12103C), penicillin/streptomycin, L-glutamine (Gibco, 15140–122), 2-mercaptoethanol, 100 ng/mL SCF (PeproTech, 250–03), 20 ng/mL mTPO (PeproTech, 315–14), 20 ng/mL IL3 (PeproTech, 213–13) and 20 ng/mL IL6 (PeproTech, 216–16), at the cell concentration of 1 million cells/mL. For Lys05 treatment, at 24 hours post-isolation, cells were collected, resuspended in media with 2 μM Lys05 at the cell concentration of 1 million cells/mL, and incubated for 24 hours. Cells were then harvested for analysis. The gating strategy is illustrated in Supplementary Fig. 1a.
Senescence-associated beta-galactosidase (β-gal) assay
Assay were performed using Cellular Senescence Assay Kit (Millipore, KAA002), and were imaged and quantified under regular light microscope.
SIRT1 deacetylation activity assay
Cells were lysed in buffer containing 20 mM Tris pH 7.5, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, 10% glycerol, supplemented with 1:100 Halt protease and phosphatase inhibitor cocktail (Thermo Scientific) at 4 °C for 1 hour. The supernatant was incubated with antibody-conjugated agarose beads (Thermo Fisher) at 4 °C for overnight. The next day, beads were washed 10 min for 4 times at 4 °C, added with 5 μL buffer, and were subjected to SIRT1 activity detection using SIRT1 Activity Assay Kit (Abcam, ab156065). The deacetylation activity of the samples was determined by the average slopes of fluorescence intensity curves, which were calculated through lineage regression analyses.
Mass Spectrometry
Elution samples of IP were separated by 4–12% Bis-Tris NuPAGE and stained using G-250. The SIRT1 bands (confirmed by Western blotting) were analyzed in standard protocol of in-gel digestion, C18 stage tip and label-free quantitative LC-MS/MS. Tryptic peptides were run on a Thermo-Fisher Orbitrap Fusion equipped with a Dionex UHPLC. Samples were separated on a home-packed capillary column (75 μm * 20 cm) containing C18-AQ resin (3 μm) at a flow rate of 400 nl/min. Mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in 80% acetonitrile. A gradient of 70 min was preceded by a 2-min loading period (2% B). The mobile phase was programmed to 5% B in 3 min, then 5% to 30% B in 40 min, followed by an increase to 55% buffer B by 60 min. Column washing was set at 99% B for 6 min before returning to the initial conditions. Full scan mass range of m/z 300–1800 was analyzed in the Orbitrap at 60,000 FWHM (200 m/z) resolution and 4.0e5 AGC target value with maximum injection time to 100 ms. Determined charge states between 2 and 6 were required, and 30 s dynamic exclusion window was used with isotopes excluded. MS/MS was performed in the ion trap in the Rapid mode with the TopSpeed mode (3 s) using data-dependent acquisition. HCD collision energy was set to 30%, AGC target to 1.0e4 and maximum injection time to 150 ms.
Raw files were analyzed by MaxQuant 1.6.0.16 against SIRT2 and common contaminant database. The search included fixed modification of carbamidomethyl cysteine and variable modifications of methionine oxidation, N-terminal acetylation and serine/threonine/tyrosine phosphorylation. All other values used default settings. The msms.txt generated by MaxQuant was sent to Skyline software for label-free quantification. All peptides from SIRT1 were manually checked before extracting the peak area for computing relative abundance of unmodified and phosphorylated status.
Human T cell population sorting and culture
Blood samples were collected at the Vitalant in San Francisco, CA. All donors were age 50 or older, provided written informed consent, and were de-identified. Total human CD8+ T cells were enriched using the RosetteSep Human CD8+ T Cell Enrichment Cocktail (Stemcell Technologies, 15063). Then, the following antibodies were used to stain CD8+CD28+ and CD8+CD28- T cells: fixable viability dye eFlour506 (Invitrogen, 65–0866-14), CD3-PECy5 (BD Biosciences, 555334), CD8-V450 (BD Biosciences, 560347), and CD28-PE (Invitrogen, 12–0289-42). Sorting was performed on an ARIA II (BD Biosciences). The gating strategy is illustrated in Supplementary Fig. 1b.
T cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, and penicillin–streptomycin. Lys05 was used at 5 μM for 14 h.
Statistics and Reproducibility
Unpaired two-tailed Student’s t-test and Mann-Whitney test was used for comparison between two groups. One-way ANOVA coupled with Tukey’s multiple comparison test or two-way ANOVA coupled with Sidak’s multiple comparisons tests were used for comparisons over two groups. All bar graphs show mean values with error bars (s.d. or s.e.m., as indicated in figure legends). 95% confidence intervals were used, and significance was considered when p value was less than 0.05. The number of times the experiment was repeated has been indicated in the figure legends.
Data Availability
RNA-sequencing data were referenced to GEO under accession number GSE52848 and GSE53356. Mass spec data have been deposited in ProteomeXchange with the primary accession code PXD020081. Source data are provided in the Source Data files. The authors declare that the data that support the findings of this study are available within the manuscript. No restriction on data availability applies. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Extended Data
Extended Data Fig. 1. Characterization of SIRT1 role and mRNA levels in cellular senescence.
a, Western blot showing SIRT1 expression in IMR90 cells stably expressing LPC vector or LPC-HA-SIRT1 construct; n = 3 independent experiments. b, β-gal-stained cells at day 9 post etoposide treatment were imaged by microscopy; n = 2 independent experiments. OE: overexpression. c, Percentages of β-gal staining-positive cells were quantified at indicated days after etoposide treatment. Data are mean ± s.d.; more than 500 cells and 5 fields were counted; two-way ANOVA with Sidak’s multiple comparisons test (p values). d, Western blot showing SIRT1 expression in primary BJ fibroblasts treated with 100 μM etoposide for 48 hrs in time course as indicated; n = 2 independent experiments. e, Western blot showing SIRT1 expression in IMR90 cells cultured at 100% confluency for indicated days; n = 2 independent experiments. f-i, RT-qPCR time course analysis of proliferating and senescent IMR90 cells induced by overexpressing ER:HRasV12 treated with 4OHT (f), by etoposide for 48 hrs (g) and BJ cells treated with etoposide for 48 hrs (h), and in IMR90 replicative senescent cells (i). Data were normalized to GAPDH; the bar indicates the average of three technical replicates. PD, population doubling. j, SIRT1 deacetylation activity assay of endogenous SIRT1 protein immunoprecipitated from extracts of proliferating or senescent IMR90 cells expressing inducible hairpins of shNTC and shAtg7. For senescent cells, cells were induced by Dox for 5 days, and then were subjected to etoposide treatment for 48hrs; Cells at Day 8 after etoposide treatment were harvested for analysis. For proliferating cells, cells were induced by Dox for 5 days and then were harvested for analysis. Proteins loaded were analyzed by western blotting. This experiment has been repeated for two times. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 2. Characterization of SIRT1-LC3 interaction.
a, IP of extracts from proliferating and senescent IMR90 cells. Quantification: SIRT1 IP bands were normalized to LC3 IP and SIRT1 input bands. Mean ± s.d.; n = 6 independent experiments; paired two-tailed Students’ t-test. b, IP of extracts from proliferating and contact-inhibited IMR90 cells (at 100% confluency for 8 days). This experiment has been repeated once. Excessive beads and antibodies were used in the IP to capture nearly 100% of LC3 protein in the lysates. Flow: flow-through. c, IP of extracts from proliferating and contact-inhibited cells; n = 3 independent experiments. d, IP of nuclear extracts from proliferating and senescent cells. Excessive beads and antibodies were used to capture nearly 100% of LC3 protein in the lysates. This experiment has been repeated once. Flow: flow-through. e, IP of nuclear extracts from proliferating and senescent cells; n = 2 independent experiments. In a,d,e, senescent cells were harvested at day 8 after etoposide treatment. f, Western blotting of nuclear (Nuclear) and cytoplasmic (Cyto) extracts from proliferating and senescent cells; n = 2 independent experiments. g, Endogenous LC3 IP of IMR90 cell extracts with or without protein phosphatase Lambda treatment. Quantification: SIRT1 IP bands were normalized to LC3 IP and SIRT1 input bands. Mean ± s.d.; paired one-tailed Students’ t-test; n = 5 independent experiments. h-i, Mass spectrometry analysis of SIRT1 immunoprecipitated from proliferating and senescent IMR90 cells. h, Boxplot showing the peptide intensity distribution of SIRT1. N = 54 peptides; p value = 0.25; unpaired two-tailed Student’s t-test. The median of the data was indicated as the line in the box, and edges stand for the 25th/75th percentile. i, Phosphorylated peptides identified by mass spec and their phosphorylation levels in proliferating and senescence states. AA: amino acid. Source data are provided.
Extended Data Fig. 3. Characterization of SIRT1 deacetylation role in starvation and senescence.
a-c, IMR90 cells that undergo CRISPR/Cas9-mediated gene inactivation of non-targeting control (PRM1, as PRM1 is involved in spermatogenesis and is not expressed in IMR90 cells) or SIRT1 were analyzed under starvation and senescence conditions. This experiment has been repeated for two times. a, Cells were analyzed by western blotting. b, Cells were subjected to 250 μM Torin 1 and 5 μM Lys05 treatment for 24 hrs, and analyzed by western blotting. Relative LC3-II intensities to GAPDH were quantified. c, Cells at day 6 after etoposide-treated senescence were subjected to 2 μM or 5 μM Lys05 treatments for 24 hrs, and analyzed by western blotting. Relative LC3-II intensities to GAPDH were quantified. d-e, IMR90 cells under proliferating, starvation (Torin 1 250 μM for 24 hrs) and senescence (induced by etoposide treatment, harvested at day 7) conditions were stained with LC3 antibody and analyzed. d, Cells were imaged by confocal microscopy. Scale bar: 10 μm. e, Percentages of cells with nuclear LC3 signals were quantified. Starv: starvation. Mean ± s.d.; more than 500 cells were counted; each data point (n) represents cells in 10 random fields, n = 5 for all conditions; one-way ANOVA coupled with Turkey’s multiple comparisons test. f, IMR90 were treated as indicated ways for 24 hrs and analyzed by western blotting; n = 2 independent experiments. CT: control. A.A.: amino acids. 2-DG: treatment of 10 mM 2-DG. Torin 1: treatment of 250 μM Torin 1. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 4. Characterization of SIRT1 mutants and peptides.
a, Information of potential SIRT1-LC3 interaction regions identified in the peptide array as in Fig. 4a, and the corresponding synthetic peptides and mutants. Key amino acid residues are labeled in red. Potential region: LC3-binding regions on SIRT1 identified in the peptide array as in Fig. 4a. Peptide region: synthetic peptides tested in the peptide competition IP as in Fig. 4b. Peptide competition: results of the peptide competition IP as in Fig. 4b. Substitution generated: SIRT1 mutants tested in the IP as in Fig. 4c. b, SIRT1 deacetylation activity assay of SIRT1 WT or WV mutant immunoprecipitated from extracts of HEK293T expressing corresponding HA-tagged constructs. Proteins loaded were analyzed by western blotting. This experiment has been repeated for two times. c, IP of HEK293T cells expressing HA-SIRT1 and Flag-LC3 constructs; n = 2 independent experiments. Cells were pre-treated with 20 μM resveratrol for 6 hrs. d, IMR90 cells at day 6 after etoposide-initiated senescence were subjected to treatment with 20 μM resveratrol for 48 hrs; n = 2 independent experiments. Cells were then harvested for western blotting. e, HEK293T cells expressing HA-SIRT1 and Flag-LC3 were treated with 20 μM resveratrol for 6 hrs, and were then harvested for SIRT1 activity assay. This experiment has been repeated for two times. f, SIRT1 deacetylation activity assay of SIRT1 WT or I347A mutant immunoprecipitated from extracts of HEK293T expressing corresponding HA-tagged constructs. Proteins loaded were analyzed by western blotting; n = 2 independent experiments. g, IP of HEK293T cell lysates expressing Flag-tagged LC3 and HA-tagged SIRT1 WT or I347A, WV+I347A, or WV mutants. This experiment has been repeated for two times. Statistical information and unprocessed blots are provided as source data.
Extended Data Fig. 5. Analysis of SIRT1 in mouse tissues and HSPCs.
a,b, Thymus from young (2–4 months) and aged (19–26 months) C57BL/6 mice were lysed and analyzed by western blotting (a) and RT-qPCR (b); n = 3 biologically independent animals in each group. RT-qPCR data were normalized to 18S; mean ± s.e.m.; unpaired two-tailed Students’ t-test. c. Indicated organs and tissues of young (3 months) and aged (19 months) mice were dissected and analyzed by western blotting; n = 2 independent experiments. SE: short exposure; LE: long exposure. d,e, Young (3 months) mice were fed or fasted for 24 hrs. Spleens (d) and Testes (e) were harvested for western blotting; n = 2 biologically independent animals in each group. f,g, Young (2–4 months) mice were subjected to daily i.p. injection of 10 mg/kg Lys05 in PBS or PBS control in 100 μL volume for two weeks. Spleens (f) and testes (g) were analyzed by western blotting. Western blot quantification: SIRT1 bands were normalized to GAPDH bands. For spleens, data are mean ± s.e.m.; control group n = 3 animals, Lys05 group n = 5 animals; two-tailed Mann-Whitney test. For testes, data are mean ± s.e.m.; n = 4 animals; two-tailed Mann-Whitney test. h, Representative flow cytometry plots of cell sorting of lineage-depleted bone marrow cells from young and aged mice to isolate Lin-Sca-1+c-Kit+ cells (HSPC populations). Boxes indicate cell populations isolated. i, HSPC populations were isolated from young (2–4 months) mice, cultured with or without 2 μM Lys05 for 24 hours and analyzed by western blotting. This experiment has been repeated once. Statistical information and unprocessed blots are provided as source data.
Supplementary Material
Acknowledgement
We acknowledge Dr. Jasmine Zhao for help with confocal microscopy and Dr. Lifeng Zhang for help with cell sorting by flow cytometry. We thank Dr. Peter Klein, Dr. E. John Wherry, Dr. Foteini Mourkioti, Dr. Maria Grazia Vizioli for help with mouse experiments, Dr. Benjamin Garcia and Dr. Ronen Marmorstein for help with SIRT1-LC3 interaction mapping experiments, and Nandhini Raman and the Gladstone Flow Cytometry Core for assistance with FACS. C.X. is supported by Glenn/AFAR Scholarship for Research in the Biology of Aging from American Federation of Aging Research. Z.D. is supported by NIH K99AG053406. S.L.B. and P.D.A. are supported by NIH P01AG031862–11.
Footnotes
Competing Interests Statement
The authors declare no competing financial and non-financial interests.
References
- 1.Hall JA, Dominy JE, Lee Y & Puigserver P. The sirtuin family’s role in aging and age-associated pathologies. J Clin Invest 123, 973–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haigis MC & Sinclair DA Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5, 253–295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin SJ, Defossez PA & Guarente L. Requirement of NAD and SIR2 for lifespan extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Tissenbaum HA & Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Rogina B & Helfand SL Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101, 15998–16003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang W et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campisi J & d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8, 729–740 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Chimenti C et al. Senescence and death of primitive cells and myocytes lead to premature cardiac aging and heart failure. Circ Res 93, 604–613 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Otin C, Blasco MA, Partridge L, Serrano M & Kroemer G. The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker DJ et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J et al. SIRT1 overexpression antagonizes cellular senescence with activated ERK/S6k1 signaling in human diploid fibroblasts. PLoS One 3, e1710 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki T, Maier B, Bartke A & Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell 5, 413–422 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Rai TS et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev 28, 2712–2725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaravadi RK & Winkler JD Lys05: a new lysosomal autophagy inhibitor. Autophagy 8, 1383–1384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou Z et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narita M et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerland LM et al. Association of increased autophagic inclusions labeled for beta-galactosidase with fibroblastic aging. Exp Gerontol 38, 887–895 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Pankiv S et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282, 24131–24145 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Kerppola TK Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 37, 465–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birgisdottir AB, Lamark T & Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 126, 3237–3247 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Huang R et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell 57, 456–466 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Noda NN et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Kraft LJ, Nguyen TA, Vogel SS & Kenworthy AK Size, stoichiometry, and organization of soluble LC3-associated complexes. Autophagy 10, 861–877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alemu EA et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J Biol Chem 287, 39275–39290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skytte Rasmussen M et al. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy 13, 834–853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansen T et al. Methods for Studying Interactions Between Atg8/LC3/GABARAP and LIR-Containing Proteins. Methods Enzymol 587, 143–169 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Dai H et al. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat Commun 6, 7645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davenport AM, Huber FM & Hoelz A. Structural and functional analysis of human SIRT1. J Mol Biol 426, 526–541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escande C et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest 120, 545–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rimmele P et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports 3, 44–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huff WX, Kwon JH, Henriquez M, Fetcko K & Dey M. The Evolving Role of CD8(+)CD28(−) Immunosenescent T Cells in Cancer Immunology. Int J Mol Sci 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeng MY et al. Metabolic reprogramming of human CD8(+) memory T cells through loss of SIRT1. J Exp Med 215, 51–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zu Y et al. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ Res 106, 1384–1393 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Chen H et al. SIRT1 ameliorates age-related senescence of mesenchymal stem cells via modulating telomere shelterin. Front Aging Neurosci 6, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez AF et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayakawa T et al. SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS One 10, e0116480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montecino-Rodriguez E, Berent-Maoz B & Dorshkind K. Causes, consequences, and reversal of immune system aging. J Clin Invest 123, 958–965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 38.Shah PP et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27, 1787–1799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dou Z et al. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 50, 29–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L & Fang J. Macromolecular crowding effect is critical for maintaining SIRT1’s nuclear localization in cancer cells. Cell Cycle 15, 2647–2655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data were referenced to GEO under accession number GSE52848 and GSE53356. Mass spec data have been deposited in ProteomeXchange with the primary accession code PXD020081. Source data are provided in the Source Data files. The authors declare that the data that support the findings of this study are available within the manuscript. No restriction on data availability applies. All other data supporting the findings of this study are available from the corresponding author on reasonable request.