Existing DNA methylation clocks are highly accurate in blood but are less precise when used in older samples or on brain tissue. Shireby et al. develop a novel epigenetic clock that performs optimally in human cortical tissue and has the potential to identify phenotypes associated with biological ageing in the brain.
Keywords: DNA methylation, age, cortex, brain, clock
Abstract
Human DNA methylation data have been used to develop biomarkers of ageing, referred to as ‘epigenetic clocks’, which have been widely used to identify differences between chronological age and biological age in health and disease including neurodegeneration, dementia and other brain phenotypes. Existing DNA methylation clocks have been shown to be highly accurate in blood but are less precise when used in older samples or in tissue types not included in training the model, including brain. We aimed to develop a novel epigenetic clock that performs optimally in human cortex tissue and has the potential to identify phenotypes associated with biological ageing in the brain. We generated an extensive dataset of human cortex DNA methylation data spanning the life course (n = 1397, ages = 1 to 108 years). This dataset was split into ‘training’ and ‘testing’ samples (training: n = 1047; testing: n = 350). DNA methylation age estimators were derived using a transformed version of chronological age on DNA methylation at specific sites using elastic net regression, a supervised machine learning method. The cortical clock was subsequently validated in a novel independent human cortex dataset (n = 1221, ages = 41 to 104 years) and tested for specificity in a large whole blood dataset (n = 1175, ages = 28 to 98 years). We identified a set of 347 DNA methylation sites that, in combination, optimally predict age in the human cortex. The sum of DNA methylation levels at these sites weighted by their regression coefficients provide the cortical DNA methylation clock age estimate. The novel clock dramatically outperformed previously reported clocks in additional cortical datasets. Our findings suggest that previous associations between predicted DNA methylation age and neurodegenerative phenotypes might represent false positives resulting from clocks not robustly calibrated to the tissue being tested and for phenotypes that become manifest in older ages. The age distribution and tissue type of samples included in training datasets need to be considered when building and applying epigenetic clock algorithms to human epidemiological or disease cohorts.
Introduction
Advancing age is associated with declining physical and cognitive function, and is a major risk factor for many human brain disorders including dementia and other neurodegenerative diseases (Harper, 2014; Sierra, 2019). Understanding the biological mechanisms involved in ageing will be a critical step towards preventing, slowing or reversing age-associated phenotypes. Because of substantial inter-individual variation in age-associated phenotypes, there is considerable interest in identifying robust biomarkers of ‘biological’ age, a quantitative phenotype that is thought to better capture an individual’s risk of age-related outcomes than actual chronological age (Jylhävä et al., 2019). Several data modalities have been used to generate estimates of biological age; these include measures of physical fitness (e.g. muscle strength) (Sosnoff and Newell, 2006), cellular phenotypes (e.g. cellular senescence) (Baker et al., 2011), genomic changes (e.g. telomere length) (Sanders and Newman, 2013; Jylhävä et al., 2017) and epigenetic mechanisms (e.g. DNA methylation) (Horvath, 2013).
Epigenetic mechanisms act to regulate gene expression developmentally via chemical modifications to DNA and histone proteins (Bernstein et al., 2007), conferring cell-type-specific patterns of gene expression and differing markedly between tissues and cell types (Mendizabal and Yi, 2016). There has been recent interest in the dynamic changes in epigenetic processes over the life course, and a number of ‘epigenetic clocks’ based on one specific epigenetic modification, DNA methylation (DNAm), have been developed that appear to be highly predictive of chronological age (Hannum et al., 2013; Horvath, 2013). The landmark DNAm clock was developed by Horvath (2013), who applied elastic net regression to Illumina DNAm array data from a large number of samples derived from a range of tissues (n = ∼8000 across 51 tissue and cell types), and generated a predictor based on DNAm at 353 CpG sites that is highly predictive of chronological age (Horvath, 2013). Given that changes in DNAm are known to index exposure to certain environmental risk factors (for example, tobacco smoking) (Elliott et al., 2014; Sugden et al., 2019) that are associated with diseases of old age, and variable DNAm is robustly associated with a number of age-associated disorders (Smith et al., 2016; Chuang et al., 2017; Chouliaras et al., 2018), there has been interest in the hypothesis that DNAm clocks might robustly quantify variation in biological age. Horvath’s DNAm age clock, for example, has been widely applied to identify accelerated epigenetic ageing, where DNAm age predictions deviate from chronological age such that individuals appear older than they really are, in the context of numerous health and disease outcomes (Horvath and Ritz, 2015; Levine et al., 2015; Marioni et al., 2015; McCartney et al., 2018). Although the original DNAm clocks were primarily developed to predict chronological age and are not robustly predictive of clinical health measures (e.g. blood pressure) (Quach et al., 2017), more recent DNAm clocks such as Levine’s ‘pheno age’ clock (Levine et al., 2018) incorporate surrogate measures of biological age and are more directly aimed at predicting mortality and healthspan. Since age is a major risk factor for dementia and other neurodegenerative brain disorders, there is particular interest in the application of epigenetic clock algorithms to these phenotypes, especially as differential DNAm in the cortex has been robustly associated with diseases including Alzheimer’s disease and Parkinson’s disease (Lunnon et al., 2014; Yu et al., 2015; Smith et al., 2016). Recent studies have reported an association between accelerated DNAm age and specific markers of Alzheimer’s disease neuropathology in the cortex (e.g. neuritic plaques, diffuse plaques and amyloid-β load) (Levine et al., 2015, 2018). Furthermore, among individuals with Alzheimer’s disease, DNAm age acceleration is associated with declining global cognitive functioning and deficits in episodic and working memory (Levine et al., 2015, 2018).
A strength of several existing epigenetic clocks is that they work relatively well across different types of sample; the Horvath multi-tissue clock, for example, can accurately predict age in multiple tissues across the life course. Importantly, as with any predictor, the composition of the training data used to develop the clock influences the generality of the model. To date, there has been limited research comparing the prediction accuracy and potential bias of existing clock algorithms across different tissues and ages. Recent analyses have highlighted potential biases when using Horvath’s clock in older samples (>∼60 years) and in samples derived from certain tissues, especially the CNS (El Khoury et al., 2019). This is important for the interpretation of studies of possible relationships between accelerated epigenetic age and age-related diseases affecting the human brain (e.g. neurodegenerative phenotypes); reported associations between accelerated DNAm age and disease may actually be a consequence of fitting a suboptimal predictor to available datasets. Potential confounders include differential changes in DNAm with age across tissues, and the age distribution of the samples used to train existing classifiers. Resolution of these biases requires the construction of specific DNAm clocks developed using data generated on the relevant tissue-type and including broad representation of the age spectrum they will be used to interrogate. Recently, a number of tissue-specific DNA methylation clocks have been described, including clocks designed for whole blood (Hannum et al., 2013; Zhang et al., 2019), muscle (Voisin et al., 2020), bone (Gopalan et al., 2019) and paediatric buccal cells (McEwen et al., 2019). Importantly, although these DNAm age estimators have increased predictive accuracy within the specific tissues in which they were built, they lose this precision when applied to other tissues (Zhang et al., 2019).
In this study, we describe the development of a novel DNAm clock that is specifically designed for application in DNA samples isolated from the human cortex and is accurate across the lifespan including in tissue from older donors (aged over 60 years). We demonstrate that our clock outperforms existing DNAm-based predictors developed for other tissues, minimizing the potential for spurious associations with ageing phenotypes relevant to the brain.
Materials and methods
Datasets used to develop the novel cortical DNAm age clock
To develop and characterize our cortical DNAm age clock (‘DNAmClockCortical’) we collated an extensive collection of DNAm data from human cortex samples (Supplementary Tables 1 and 2), complementing datasets generated by our group (http://www.epigenomicslab.com) with publicly available datasets downloaded from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (Jaffe et al., 2016; De Jager et al., 2014; Lunnon et al., 2014; Pidsley et al., 2014; Smith et al., 2018, 2019; Wong et al., 2019) (Supplementary Tables 1 and 2). In each of these datasets DNAm was quantified across the genome using the Illumina 450K DNA methylation array, which covers >450 000 DNA methylation sites as previously described (Pidsley et al., 2013). To optimize the performance of the DNAmClockCortical and to avoid reporting over-fitted statistics, the samples were split into a ‘training’ dataset (used to determine the DNAm sites included in the model and their weighted coefficients) and a ‘testing’ dataset (used to profile the performance of the proposed model). To reduce the effects of experimental batch in our model, we maximized the number of different datasets included in the training data by combining the 10 cohorts and randomly assigning individuals within them to either the training or testing dataset in a 3:1 ratio (Table 1). In total, our training dataset (age range = 1–108 years, median = 57 years; Supplementary Fig. 1) comprised DNAm data from 1047 cortex samples (derived from 832 donors) and our testing dataset (age range = 1–108 years, median = 56 years; Supplementary Fig. 1) comprised DNAm data from 350 cortex samples (derived from 323 donors). Individuals with a diagnosis of Alzheimer’s disease and other major neurological phenotypes were excluded from our analysis given the previous associations between them and deviations in DNAm age (Levine et al., 2015, 2018).
Table 1.
Sample characteristics of the training (cortex), testing (cortex), independent test (cortex) and whole blood datasets used in the development and evaluation of DNAmClockCortical
| Dataset | Age, years |
Sex, n |
Illumina methylation array | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean | 1st Quartile | Median | 3rd Quartile | Range | Female | Male | |||
| Training | 1047 | 56.53 | 38.56 | 57 | 78 | 1–108 | 362 | 685 | 450K | De Jager et al., 2014; Lunnon et al., 2014; Pidsley et al., 2014; Jaffe et al., 2016; Smith et al., 2018, 2019; Wong et al., 2019 |
| Testing | 350 | 55.87 | 39.05 | 56 | 78 | 1–108 | 144 | 206 | 450K | De Jager et al., 2014; Lunnon et al., 2014; Pidsley et al., 2014; Jaffe et al., 2016; Smith et al., 2018, 2019; Wong et al., 2019 |
| Independent test | 1221 | 83.49 | 78 | 84 | 90 | 41–104 | 577 | 644 | EPIC | – |
| Blood | 1175 | 57.96 | 46 | 59 | 69 | 28–98 | 686 | 489 | EPIC | Hannon et al., 2018 |
Cortex independent test dataset
An ‘independent test’ cortical dataset was generated using post-mortem occipital and prefrontal cortex samples from the Brains for Dementia Research (BDR) cohort. BDR was established in 2008 and is a UK-based longitudinal cohort study with a focus on dementia research (Francis et al., 2018) coordinated by a network of six dementia research centres located around the UK. Post-mortem brains underwent full neuropathological dissection, sampling and characterization using a standardized protocol (Bell et al., 2008; Samarasekera et al., 2013). DNA was isolated from cortical tissue samples using the Qiagen AllPrep DNA/RNA 96 Kit (Qiagen, cat no. 80311) following tissue disruption using BeadBug 1.5 mm Zirconium beads (Sigma Aldrich, cat no. Z763799) in a 96-well deep well plate (Fisher Scientific, cat no. 12194162) shaking at 2500 rpm for 5 min. Genome-wide DNA methylation was profiled using the Illumina EPIC DNA methylation array (Illumina Inc), which interrogates >850 000 DNA methylation sites across the genome (Moran et al., 2016). After stringent data quality control (see below) the final independent test dataset consisted of DNAm estimates for 800 916 DNAm sites profiled in 1221 samples (632 donors; 610 prefrontal cortex; 611 occipital cortex; see Table 1 for more details). This dataset consists of predominantly older samples (age range = 41–104 years, median = 84 years; Supplementary Fig. 1).
Whole blood dataset
We recently generated DNAm data from whole blood obtained from 1175 individuals (age range = 28–98 years; median age = 59 years; Table 1) included in the UK Household Longitudinal Study (UKHLS) (https://www.understandingsociety.ac.uk/) (Hannon et al., 2018). The UKHLS was established in 2009 and is a longitudinal panel survey of 40 000 UK households from England, Scotland, Wales and Northern Ireland (Buck and McFall, 2011). For each participant, non-fasting blood samples were collected through venepuncture; these were subsequently centrifuged to separate plasma and serum, and samples were aliquoted and frozen at −80°C. DNAm data were generated using the Illumina EPIC DNA methylation array as described previously (Hannon et al., 2018). After stringent quality control (see below) the whole blood dataset consisted of data for 857 071 DNAm sites profiled in 1175 samples (Hannon et al., 2018).
DNA methylation data preprocessing
Unless otherwise reported, all statistical analysis was conducted in the R statistical environment (version 3.5.2; https://www.r-project.org/). Raw data for all datasets were used, prior to any quality control or normalization, and processed using either the wateRmelon (Pidsley et al., 2013) or bigmelon (Gorrie-Stone et al., 2019) packages. Our stringent quality control pipeline included the following steps: (i) checking methylated and unmethylated signal intensities and excluding poorly performing samples; (ii) assessing the chemistry of the experiment by calculating a bisulphite conversion statistic for each sample, excluding samples with a conversion rate <80%; (iii) identifying the fully methylated control sample was in the correct location (where applicable); (iv) multidimensional scaling of sites on the X and Y chromosomes separately to confirm reported sex; (v) using the 65 single nucleotide polymorphism (SNP) probes present on the Illumina 450K array and 59 on the Illumina EPIC array to confirm that matched samples from the same individual (but different brain regions) were genetically identical and to check for sample duplications and mismatches; (vi) use of the pfilter() function in wateRmelon to exclude samples with >1% of probes with a detection P-value > 0.05 and probes with >1% of samples with detection P-value > 0.05; (vii) using principal component analysis on data from each tissue to exclude outliers based on any of the first three principal components; and (viii) removal of cross-hybridizing and SNP probes (Chen et al., 2013). The subsequent normalization of the DNA methylation data was performed using the dasen() function in either wateRmelon or bigmelon (Pidsley et al., 2013; Gorrie-Stone et al., 2019).
Deriving a novel cortical DNAm age classifier
To build the DNAmClockCortical we implemented an elastic net regression model, using the methodology described by Horvath (2013). The elastic net model is designed for high dimensional datasets with more features than samples and where the features are potentially highly correlated (Zou and Hastie, 2005). As part of the methodology, the model selects the subset of features (i.e. DNAm sites) that cumulatively produce the best predictor of a provided outcome. Elastic net was implemented in the R package GLMnet (Friedman et al., 2010). It uses a combination of Ridge and LASSO (least absolute shrinkage and selection operator) regression. Ridge regression penalizes the sum of squared coefficients and has an (alpha) parameter of 0. LASSO regression penalises the sum of the absolute values of the coefficients and has an α parameter of 1. Elastic net is a convex combination of ridge and LASSO and, therefore, the elastic net α parameter was set to 0.5. The lambda value (the shrinkage parameter) was derived using 10-fold cross-validation on the training dataset (lambda = 0.0178). DNAm probes included in the analysis were limited to sites which were present on both the Illumina EPIC and Illumina 450K arrays, with no missing values across the training datasets (n probes = 383 547). Previous analyses have shown that the relationship between DNAm age (predicted age from epigenetic age estimators) and chronological age is logarithmic between 0 and 20 years and linear from 20 years plus (Horvath, 2013). Our data revealed a similar pattern and therefore chronological age was transformed (Supplementary Fig. 2). A transformed version of chronological age was regressed on DNAm levels at all included DNAm sites.
Implementing DNAm Age prediction
We applied the DNAmClockCortical (comprising 347 DNAm sites) to the testing, independent test and whole blood DNAm datasets. We then compared its performance to a number of existing DNAm clocks which have previously shown biases when applied to brain tissue and older samples (Horvath and Raj, 2018; El Khoury et al., 2019; Zhang et al., 2019): Horvath’s original multi-tissue clock (‘DNAmClockMulti’; 353 DNAm sites) (Horvath, 2013), Zhang’s elastic net blood and saliva-based DNAm clock (‘DNAmClockBlood’: 514 DNAm sites) (Zhang et al., 2019) and Levine’s second generation ‘pheno age’ DNAm Clock (‘DNAmClockPheno’; 513 DNAm sites) (Levine et al., 2018). Briefly, to predict DNAm age using the DNAmClockMulti we applied the agep() function in wateRmelon (Pidsley et al., 2013). Although this function does not contain the custom normalization method applied at the DNAm age calculator website (https://DNAmClock.genetics.ucla.edu/), both methods work similarly in brain and blood studies, providing the data have been preprocessed adequately (El Khoury et al., 2019). To predict age using the DNAmClockPheno (Levine et al., 2018), we also applied the agep() function, inputting a vector of the coefficients and the intercept using the data provided in the supplementary material of Levine et al’s paper. To predict DNAm age with the DNAmClockBlood, we used the authors’ published code (available on GitHub https://github.com/qzhang314/DNAm-based-age-predictor) (Zhang et al., 2019).
Determining the predictive accuracy of different DNAm clocks
DNAm age was estimated in the testing dataset (n = 350), independent test dataset (n = 1221) and whole blood dataset (n = 1175) using DNAmClockCortical, DNAmClockMulti, DNAmClockBlood and DNAmClockPheno. To compare and evaluate the predictive accuracy of these DNAm age predictors, estimates were assessed using two measures: Pearson’s correlation coefficient [r; a measure indicating the strength of the linear relationship between the actual (chronological age) and predicted (DNAm age) variables] and the root mean squared error (RMSE; square root of the mean differences between the actual and predicted variables), which quantifies the precision of the estimator.
Analysis against age
To test associations between DNAm age and chronological age, we fitted regression models to each dataset. As a subset of donors included in the testing and independent test datasets contributed data from multiple cortical regions, we used mixed effects linear regression models, implemented with the lme4 and lmerTest packages, where DNAm age was regressed against chronological age as a fixed effect and individual was included as a random effect. In the blood cohort, as there was only one sample per individual, we applied standard linear regression models. A second regression model was also fitted which additionally tested for associations with an age-squared term. In the whole blood dataset, we ran these analyses again in the subset of samples over 55 years old to make the results more comparable to those generated using the independent test dataset.
Analysis against biological and technical factors
To test associations between DNAm age and sex, post-mortem interval, experimental batch and neuronal cell proportion estimates [derived using the CETS algorithm (Guintivano et al., 2013)] we fitted regression models to the independent test dataset (n = 1221 cortical samples). We used mixed effects regression models implemented as described above. DNAm age was regressed against each variable in turn with age, age squared and derived cell proportion estimates (excluding the model looking for associations with cell proportions) as fixed effects and individual as random effect. In the analysis with post-mortem interval we included brain bank as a fixed effect.
Data availability
The datasets used for the training and testing samples are available for download from GEO (https://www.ncbi.nlm.nih.gov/geo/) using the following accession numbers: GSE74193; GSE59685; GSE80970; GPL13534 and GSE43414. The independent test data are available from the authors upon request or via the Dementias Platform UK (DPUK) data portal (https://portal.dementiasplatform.uk/). The whole blood DNA methylation data are available upon application through the European Genome-Phenome Archive under accession code EGAS00001001232. Analysis scripts used in this manuscript and code to run the clock are available on GitHub (https://github.com/gemmashireby/CorticalClock).
Results
Existing human cortex DNAm clock algorithms systematically underestimate age in older individuals
The performance of DNAm clocks is influenced by the characteristics (e.g. specific tissue type and age range) of the training data used to build the prediction algorithm. Applying predictors to datasets that differ in terms of these characteristics may lead to biases when estimating DNAm age, and confound phenotypic analyses using these variables (El Khoury et al., 2019). We found that existing DNAm clocks [i.e. the DNAmClockMulti (Horvath, 2013) the DNAmClockBlood (Zhang et al., 2019) and the DNAmClockPheno (Levine et al., 2018)] do not perform optimally in human cortex tissue (Supplementary Fig. 3), with notable differences between derived DNAm age and actual chronological age (i.e. the derived values do not lie along the y = x line; Fig. 1). In our testing dataset (n = 350 cortex samples; age range = 1–108 years; median age = 57 years), the DNAmClockMulti systematically underestimated DNAm age in individuals over ∼60 years old, and systematically overestimated it in individuals below ∼60 years old [Figs 1A(ii) and 2A(ii)]. In the older group (aged over 60 years), around 80% of samples had lower predicted DNAm ages than their actual chronological age. These deviations were also observed when looking at the mean differences between actual age and predicted DNAm age [referred to as Δ (delta) age), such that Δ age was positive for younger ages and vice versa for the older group (Supplementary Fig. 4A). Use of the DNAmClockBlood produced even more pronounced systematic underestimation of DNAm age in adults, beginning around 30 years [Figs 1A(iii) and 2A(iii)], and this trend was mirrored for Δ age (Supplementary Fig. 4A). Finally, the DNAmClockPheno severely under predicted age in the cortex, with 100% of samples being assigned a lower DNAm age than the actual chronological age [Figs 1A(iv), 2A(iv) and Supplementary Fig. 4A(iv)]. Similar biases in age prediction were seen in our independent test dataset (n = 1221 cortex samples; age range = 41 years to 104 years; mean age = 83.49 years), confirming the systematic underestimation of DNAm age in older donors (Figs 1B and 2B). As with the other clocks, Δ age captured these biases, with particularly poor performance evident when applying the DNAmClockPheno and the DNAmClockBlood to this dataset, in which Δ age was consistently below zero (where zero would represent perfect prediction; see Supplementary Fig. 4B).
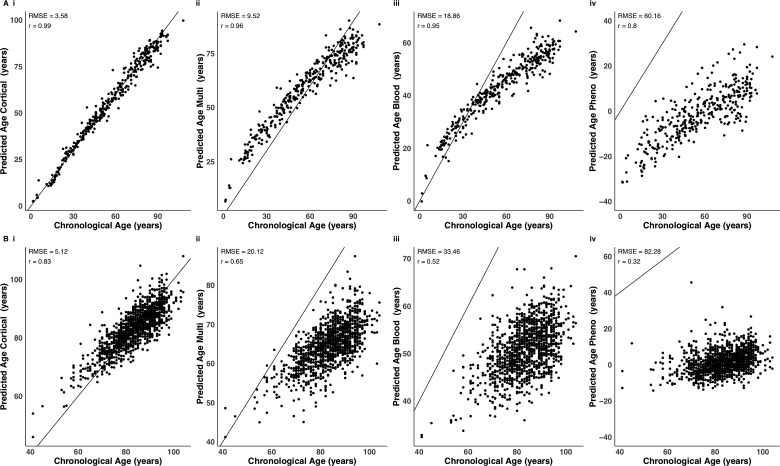
Figure 1.
Comparison of chronological age with DNA methylation age derived using four DNA methylation age clocks. Shown are comparisons of chronological age with predicted age in (A) the testing dataset (n = 350 cortical samples) and (B) the independent test dataset (n = 1221 cortical samples). DNAm age was predicted using four DNA methylation age clocks: (i) our novel DNAmClockCortical; (ii) Horvath’s DNAmClockMulti; (iii) Zhang’s DNAmClockBlood and (iv) Levine’s DNAmClockPheno. The x-axis represents chronological age (years) and the y-axis represents predicted age (years). Each point on the plot represents an individual sample. Our cortical clock out-performed the three alternative DNAm clocks across all accuracy statistics. DNA methylation age estimates derived using the DNAmClockMulti [A(ii) testing and B(ii) independent test] and the DNAmClockBlood [A(iii) testing and B(iii) independent test] appear to have a non-linear relationship with chronological age.
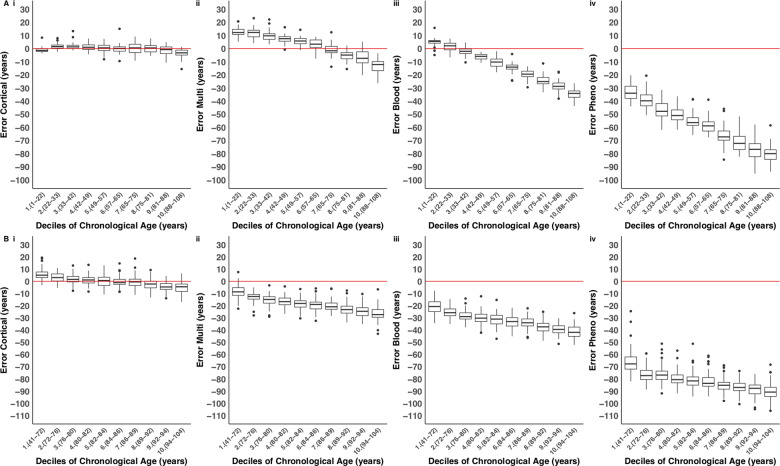
Figure 2.
The cortical DNA methylation age clock has elevated accuracy in human cortex samples across the lifespan. Shown is the distribution of the error (DNA methylation age − chronological age) for each age decile in (A) the testing dataset (n = 350 cortical samples), and (B) the independent test dataset (n = 1221 cortical samples) for each of the four DNA methylation age clocks: (i) our novel DNAmClockCortical; (ii) Horvath’s DNAmClockMulti; (iii) Zhang’s DNAmClockBlood and (iv) Levine’s DNAmClockPheno. Deciles were calculated by assigning chronological age into 10 bins and are represented along the x-axis by the numbers 1 to 10, followed by parentheses, which display the age range included in each decile. The ends of the boxes are the upper and lower quartiles of the errors, the horizontal line inside the box represents the median deviation and the two lines outside the boxes extend to the highest and lowest observations. Outliers are represented by points beyond these lines. The red horizontal line represents perfect prediction (zero error). Our novel DNAmClockCortical [A(i) testing and B(i) independent test] consistently had the smallest error across the age groups, shown by the tightness of the box plot distributions along the zero-error line. The DNAmClockMulti over-predicted younger ages [deciles 1–5 in A(ii)], shown by box plot distributions that are above the zero-error line, and under predicted older ages [deciles 8–10 in A(ii) and deciles 1–10 in B(ii)], shown by box plot distributions below the zero-error line. The DNAmClockBlood [A(iii) testing and B(iii) independent test] and the DNAmClockPheno [A(iv) testing and B(iv) independent test] consistently under predicted age, with under prediction of DNA methylation age increasing with chronological age.
Developing a novel DNAm clock for the human cortex based on DNA methylation sites
To alleviate the biases observed when applying existing DNAm clocks to data generated on older human cortex samples, we focused on building a DNAm clock using relevant tissue samples from donors that spanned a broad range of ages and included a large number of samples from older donors (Supplementary Fig. 1). We developed our novel cortical DNAm clock (DNAmClockCortical) using an elastic net regression, regressing chronological age against DNAm levels across 383 547 sites quantified in 1047 cortex samples (see ‘Materials and methods’ section). This approach identified a set of 347 DNAm sites, which in combination, optimally predict age in the human cortex. The sum of DNAm levels at these sites weighted by their regression coefficients provides the DNAmClockCortical age estimate (Supplementary Table 3). Of note, the majority of sites selected for our cortex clock were novel and not present in existing DNAm clock algorithms; only five of the sites overlap with the DNAmClockMulti (composed of 353 DNAm sites), 15 with the DNAmClockBlood (comprising 514 DNAm sites), and five with the DNAmClockPheno (comprising 513 DNAm sites) (Supplementary Table 4).
Increased prediction accuracy of the novel cortex clock in cortical tissue compared to existing DNAm clocks
We used DNAmClockCortical to estimate DNAm age in both the testing (n = 350 cortex samples) and independent test (n = 1221 cortex samples) datasets, and compared the estimates to those derived using DNAmClockMulti, DNAmClockBlood and DNAmClockPheno. The DNAmClockCortical predicted age accurately in the testing dataset and there was a strong correlation between DNAm age and age [r = 0.99; Table 2 and Fig. 1A(i)]. In the independent test dataset, which consisted predominantly of older samples, our clock also performed well and was highly correlated with age (r = 0.83), outperforming DNAmClockMulti (r = 0.65), DNAmClockBlood (r = 0.52), and DNAmClockPheno (r = 0.32) [Table 2 and Fig. 1B(i)]. The most striking differences were in the accuracy of the DNAmClockCortical in comparison to previously developed DNAm clocks; it outperformed the three other clocks we tested across all accuracy statistics in both cortical datasets (Table 2). The biggest differences in accuracy can be seen in the independent test dataset (Figs 1B, 2B and Supplementary Fig. 4B), in which the RMSE was 15 years more accurate when using the DNAmClockCortical (RMSE: 5 years) than the DNAmClockMulti (RMSE: 20 years), 28 years more accurate than the DNAmClockBlood (RMSE: 33 years) and 77 years more accurate than the DNAmClockPheno (RMSE: 82 years). This is further supported by analysing how much of the variation in DNAm age is explained by age, where the DNAmClockCortical was the best fitting model in both cortical datasets (testing dataset R2 = 0.98 independent test sample R2 = 0.65) in comparison to the three other clocks, with age explaining the least variance in DNAm age estimated using the DNAmClockPheno (testing dataset R2 = 0.65; independent test sample R2 = 0.10) (Table 2). The DNAmClockPheno was consistently the most inaccurate at estimating age in the cortical datasets (RMSE: testing 60 years; independent test 82 years), followed by DNAmClockBlood (RMSE: testing = 19 years; independent test = 33 years) and the DNAmClockMulti (RMSE: testing = 10 years; independent test = 20 years) (Table 2).
Table 2.
Our novel cortex clock outperforms existing DNAm age algorithms in human cortex samples
|
Testing dataset
(n = 350) |
Independent test dataset
(n = 1221) |
|||||||
|---|---|---|---|---|---|---|---|---|
| DNAmClockCortical | DNAmClockMulti | DNAmClockBlood | DNAmClockPheno | DNAmClockCortical | DNAmClockMulti | DNAmClockBlood | DNAmClockPheno | |
| Correlation (r) | 0.99 | 0.96 | 0.95 | 0.8 | 0.83 | 0.65 | 0.52 | 0.32 |
| RMSE, years | 3.58 | 9.52 | 18.86 | 60.16 | 5.12 | 20.12 | 33.46 | 82.28 |
Accuracy statistics between DNAm age estimates and chronological age using our DNAmClockCortical, Horvath’s multi-tissue clock (DNAmClockMulti) (Horvath, 2013), Zhang’s elastic net blood clock (DNAmClockBlood) (Zhang et al., 2019) and Levine’s Pheno Age clock (DNAmClockPheno) (Levine et al., 2018) in both the testing (n = 350 cortical samples) and the independent test (n = 1221 cortical samples) datasets. RMSE = root mean squared error.
The relationship between age and DNAmClock plateaus in old age
By definition, DNAm age is correlated with chronological age, meaning age is a potential confounder for analyses of Δ age; not adequately controlling for age increases the likelihood that false positive associations will be identified (El Khoury et al., 2019). To assess associations between DNAm age and chronological age we used a mixed effects regression model (see ‘Materials and methods’ section) and found that estimates from all four DNAm age clocks were significantly associated with age in the testing dataset (Bonferroni P < 0.005; Table 3). Many studies of Δ age in health and disease control for age by using a linear model to regress out its effect (Marioni et al., 2015; McKinney et al., 2018) although one of the assumptions of this approach is that the prediction accuracy of the DNAm clock is consistent across the life course. If the accuracy varies non-linearly with chronological age, then simply including age as a linear covariate in association analyses will not sufficiently negate the confounding effect of age. We therefore sought to formally test the extent to which the prediction accuracy of the four clocks correlates with age by including an age squared term in the regression model. In the testing dataset all four clocks had a significant age squared term (Table 3), indicating that their predictive accuracy varies as a function of age. Specifically, all clocks were associated with a plateau where the difference between DNAm age and chronological age becomes larger as actual age increases (Fig. 2). Importantly, however, of the three first generation clocks the coefficient for the age squared term was smallest for the DNAmClockCortical (beta = −1.64 × 10−3, P = 1.94 × 10−7), again highlighting that bespoke clocks can be used to minimize bias in subsequent analyses.
Table 3.
The relationship between DNAm age and age (age and age2) using different DNAm clock algorithms
| Testing dataset |
Independent test dataset |
Blood dataset |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | R2 | Beta | SE | P | R2 | Beta | SE | P | R2 | |
| DNAmClockCortical | ||||||||||||
| DNAm age versus age | 1.14 | 3.39 × 10−2 | 2.86 × 10−108 | 0.98 | 1.03 | 0.17 | 5.31 × 10−9 | 0.65 | 0.58 | 0.06 | 5.37 × 10−20 | 0.78 |
| DNAm age versus age2 | −1.64 × 10−3 | 3.08 × 10−4 | 1.94 × 10−7 | 1.57 × 10−3 | −2.39 × 10−3 | 1.08 × 10−3 | 2.80 × 10−2 | 1.47 × 10−3 | −2.05 × 10−4 | 5.34 × 10−4 | 0.70 | 1.61 × 10−4 |
| DNAmClockMulti | ||||||||||||
| DNAm age versus age | 1.08 | 4.14 × 10−2 | 3.17 × 10−83 | 0.93 | 0.68 | 0.16 | 3.51 × 10−5 | 0.42 | 0.75 | 0.06 | 6.01 × 10−30 | 0.80 |
| DNAm age versus age2 | −3.81 × 10−3 | 3.75 × 10−4 | 2.45 × 10−21 | 0.02 | −1.76 × 10−3 | 1.02 × 10−3 | 8.50 × 10−2 | 1.39 × 10−3 | −1.15 × 10−3 | 5.49 × 10−4 | 0.04 | 5.66 × 10−4 |
| DNAmClockBlood | ||||||||||||
| DNAm age versus age | 0.82 | 3.41 × 10−2 | 1.30 × 10−74 | 0.90 | 0.64 | 0.18 | 3.00 × 10−4 | 0.26 | 1.14 | 0.05 | 9.50 × 10−111 | 0.94 |
| DNAm age versus age2 | −3.16 × 10−3 | 3.09 × 10−4 | 1.81 × 10−21 | 0.02 | −2.08 × 10−3 | 1.09 × 10−3 | 5.70 × 10−2 | 2.30 × 10−3 | −2.26 × 10−3 | 3.90 × 10−4 | 8.47 × 10−9 | 1.61 × 10−3 |
| DNAmClockPheno | ||||||||||||
| DNAm age versus age | 0.57 | 6.89 × 10−2 | 3.19 × 10−15 | 0.65 | −0.35 | 0.23 | 1.27 × 10−1 | 0.10 | 0.63 | 0.08 | 1.86 × 10−13 | 0.75 |
| DNAm age versus age2 | −1.79 × 10−3 | 6.25 × 10−4 | 4.47 × 10−3 | 0.01 | 3.53 × 10−3 | 1.42 × 10−3 | 1.40 × 10−2 | 0.01 | 6.22 × 10−4 | 7.21 × 10−4 | 0.39 | 5.41 × 10−5 |
DNAm age was estimated using our novel cortical clock (DNAmClockCortical), Horvath’s multi-tissue clock (DNAmClockMulti) (Horvath, 2013), Zhang’s elastic net blood clock (DNAmClockBlood) (Zhang et al., 2019) and Levine’s Pheno Age clock (DNAmClockPheno) (Levine et al., 2018) in the ‘testing’ dataset (n = 350 cortical samples), the ‘independent test’ dataset (n = 1221 cortical samples) and the blood dataset (n = 1175 whole blood samples).
Higher cortical DNAm age is associated with decreased neuronal cell proportions
Many sample-related and technical factors can influence analyses of DNAm in post-mortem cortex tissue including sex, neuronal cell proportions, post-mortem interval and experimental batch effects. To assess associations between DNAm age and these variables we used a mixed effects regression model (see ‘Materials and methods’ section) and after correcting for multiple comparisons (P < 0.005) found no association between DNAm age and sex (P = 0.03), post-mortem interval (P = 0.54) or batch (P = 0.38) (Supplementary Table 5). In contrast there was a significant association between neuronal cell proportion estimates derived from the DNAm data (beta = −8.72, P = 9.57 × 10−36; Supplementary Table 5) and DNAm age, indicating that individuals who are predicted as older using the DNAmClockCortical have lower neuronal cell proportions. This correlation is not surprising as other clocks have been widely reported to associate with differences in cell-type proportions (Horvath and Ritz, 2015; Levine et al., 2018) and it is known that the proportion of neuronal cells in the cortex changes with age. This result highlights the importance of, where possible, including cellular proportion variables as a covariate in any downstream analyses performed using DNAm clocks on tissue from the human brain.
The cortical clock loses accuracy when applied to non-cortical tissues
To assess the specificity of the novel cortex clock we next applied each of the DNAm age clocks to a large whole blood DNAm dataset (n = 1175; age range = 28–98 years; mean age = 57.96 years). Although the DNAmClockCortical performed remarkably well on whole blood (r = 0.88), with a similar predictive ability to the DNAmClockMulti (r = 0.90) (Fig. 3 and Supplementary Fig. 5), there was a non-linear relationship between DNAm age and age estimated using this clock and a systematic under prediction of DNAm age in samples from individuals aged over 60 years [Fig. 3A(i) and B(i)]. The DNAmClockBlood performed best on the blood dataset (r = 0.97), with age explaining the highest proportion of variation in DNAm age (R2 = 0.94), outperforming the three other clocks (Table 4, Fig. 3 and Supplementary Figs 5 and 6), and providing further support for the notion that epigenetic clocks work optimally for the tissue-type on which they are calibrated. Of note, when limiting the age range of samples included in the blood cohort to be more comparable to the independent test dataset (age range limited to >55 years), the relationship between estimated and actual age is considerably lower for the three non-blood-specific clocks (r ∼ 0.7) and the DNAmClockBlood (r = 0.88), reflecting the lower variability of age across samples in the dataset (Supplementary Fig. 7).
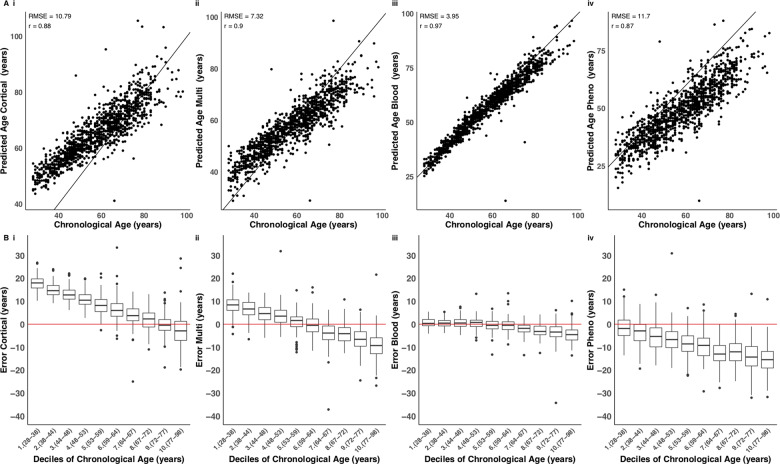
Figure 3.
The blood-based DNA methylation clock performs best in data derived from whole blood samples.(A) Shown is a comparison of DNA methylation age estimates against chronological age in a large whole blood dataset (n = 1175), where DNAm age derived using four DNA methylation age clocks: (i) our novel DNAmClockCortical; (ii) Horvath’s DNAmClockMulti; (iii) Zhang’s DNAmClockBlood and (iv) Levine’s DNAmClockPheno. The x-axis represents chronological age (years), the y-axis represents predicted age (years). Each point on the plot represents an individual in the whole blood dataset. Our novel clock does not predict as well in blood compared to the cortex, although it has a similar predictive ability to Horvath’s clock. The distribution of the error (DNA methylation age − chronological age) is presented in B for each decile for each of the four DNA methylation clocks. Deciles were calculated by assigning chronological age into 10 bins and are represented along the x-axis by the numbers 1 to 10, followed by parentheses, which display the age range included in each decile. The ends of the boxes are the upper and lower quartiles of the errors, the horizontal line inside the box represents the median deviation and the two lines outside the boxes extend to the highest and lowest observations. Outliers are represented by points beyond these lines. The red horizontal line represents perfect prediction (zero error).
Table 4.
The cortex clock is less accurate at estimating DNA methylation age algorithms in blood compared to cortex tissue samples
| DNAmClockCortical | DNAmClockMulti | DNAmClockBlood | DNAmClockPheno | |
|---|---|---|---|---|
| Correlation (r) | 0.88 | 0.90 | 0.97 | 0.87 |
| RMSE, years | 10.79 | 7.32 | 3.95 | 11.70 |
Accuracy statistics between DNAm age estimates and chronological age using using our DNAmClockCortical, Horvath’s multi-tissue clock (DNAmClockMulti) (Horvath, 2013), Zhang’s elastic net blood clock (DNAmClockBlood) (Zhang et al., 2019) and Levine’s Pheno Age clock (DNAmClockPheno) (Levine et al., 2018) in our blood dataset (n = 1175 whole blood samples). RMSE = root mean squared error.
Discussion
Existing DNAm age clocks have been widely used for predicting age and exploring accelerated ageing in disease, although there is evidence of systematic underestimation of DNAm age in older samples, particularly in the brain (El Khoury et al., 2019). We developed a novel epigenetic age model specifically for human cortex, DNAmClockCortical, built using an extensive collection of DNAm data from >1000 human cortex samples. Our model dramatically outperforms existing DNAm-based biomarkers for age prediction in data derived from the human cortex.
There are several potential causes of the systematic underestimation of DNAm age in the cortex, especially in samples from older donors (aged over 60 years), when using existing DNAm clocks such as Horvath’s DNAmClockMulti (Horvath, 2013), Zhang’s DNAmClockBlood (Zhang et al., 2019) and Levine’s DNAmClockPheno (Levine et al., 2018). First, it may be a consequence of the distribution of ages in the training data used in existing clocks; these clocks were derived using samples containing a relatively small proportion of samples from human brain and/or from older subjects. Second, as there is evidence for cell-type and tissue-specific patterns of DNAm (Mendizabal et al., 2019), the observed imprecision may reflect a consequence of underfitting the model across tissues. Third, the relationship between DNA methylation and age may not be linear across the lifespan, and a non-linear model is needed to capture attenuated effects in older samples. This would be comparable to the transformation required to accurately predict DNAm age for younger samples (0–20 years), where the association between age and with DNA methylation is of larger magnitude.
Our data suggest that both tissue-specificity and the age of samples included in the training dataset influence the precision of DNAm age estimators, as shown by the increase in accuracy when using our cortical clock relative to existing clocks in human cortex tissue samples. This notion is further supported by the accuracy we found using the blood-based estimators on a large blood dataset. Our observations suggest that tissue type has a major influence on the accuracy of DNAm age clocks, and to accurately predict age it is important to use a clock calibrated specifically for the tissue from which samples have been derived. Our data demonstrate that the performance of existing DNAm clocks varies considerably across ages and is diminished in samples from older donors. This is particularly important to consider when assessing DNAm age in the context of diseases and phenotypes that are associated with older age such as dementia and other types of neurodegenerative disease. Our results show that it is important to use a clock that has been trained using samples from the relevant age group; the training data used in the development of the DNAmClockCortical included a good representation of older samples, meaning it overcomes the systematic underestimation of DNAm age in the older that was observed with existing clocks. It is also important to consider the distribution of ages in the training dataset (e.g. minimum, maximum, median, first and third quartiles), as this can influence the predictor and lead to biases if not representative of the datasets it will be applied to.
The importance of developing tissue-specific estimators is supported by other recently developed tissue-specific clocks including DNAm age predictors for whole blood (Zhang et al., 2019), human skeletal muscle (Voisin et al., 2020) and human bone (Gopalan et al., 2019), which all out perform pan-tissue clocks in samples from the specific tissues in which they were trained. It is known that DNA methylation patterns are distinct between tissue and cell types (Mendizabal et al., 2019), and it is therefore not surprising that DNAm age estimation models would differ in accuracy across tissue types. As technologies for profiling DNAm in purified cell populations from bulk tissue become more accessible, future clocks should be developed for purified populations of individual cell-types to overcome issues of cellular heterogeneity in complex tissues such as the brain. Furthermore, our finding that the DNAmClockCortical, like other clocks, is associated with the proportion of specific cell-types in a given tissue sample highlights the importance of covarying for cellular heterogeneity in all subsequent analyses using values derived from epigenetic clocks.
Although a pan-tissue estimator such as Horvath’s DNAmClockMulti has clear general utility, the trade-off between accuracy and practicality needs to be taken into consideration depending on the hypothesized question being tested. Applying one model across multiple tissues may lead to a suboptimal fit (for example, when applying a linear model where there is non-linearity), and the performance of such a clock would need to be tested in individual tissue types. To assess the linearity of DNAm age predictors we investigated the association between DNAm age, and age squared. Of note, as age explains less of the variation in DNAm age in the second generation clocks (where the primary aim is to predict health outcomes) including the DNAmClockPheno, adding an age-squared term may be an unsuitable measure to address non-linearity where these predictors are applied. Adding the squared variable allowed us to more accurately model the effect of age in the three first generation clocks (where the primary aim is to predict age), which could have a non-linear relationship with DNAm age. The DNAmClockCortical was the most linear in terms of fitting DNAm age against actual age. Although age squared terms were significantly associated with DNAm age in the testing data using all estimators, the higher significance of the age squared term in the cortex-specific clock suggests that of all the clocks, our model is the least biased. However, as indicated by the relationship between DNAm and age squared, we need to consider the possibility that fitting a linear model might not be the best approach, and to account for this possibility we recommend that future age-acceleration analyses control for age squared terms. Because of the nature of DNAm clocks, Δ age estimated using existing clocks is highly correlated with chronological age (El Khoury et al., 2019). If age is not controlled for it could lead to spurious associations with health outcomes, which are driven by age and not the variable of interest. Furthermore, as the prediction is less precise in older individuals, even where DNAm is regressed on chronological age, the residual may still be associated with age, potentially leading to false positive associations. Recent studies have found associations between accelerated DNAm age in human brain and neurodegenerative phenotypes (Levine et al., 2015, 2018). Our findings suggest that previous associations with age-associated phenotypes may have been confounded by a lack of robust calibration to estimate DNAm age in human cortex from older donors; caution is warranted in interpreting reported results that have been generated using a non-tissue specific predictor. Future work will focus on applying our novel DNAmClockCortical to existing cohorts with DNAm data and detailed measures of neuropathology. While DNAm age is a useful indicator of age, it may not be the best indicator of health disparities between individuals with brain disorders.
In summary, we show that previous epigenetic clocks systematically underestimate age in older samples and do not perform as well in human cortex tissue. We developed a novel epigenetic age model specifically for human cortex. Our findings suggest that previous associations between predicted DNAm age and neurodegenerative phenotypes may represent false positives resulting from suboptimal calibration of DNAm clocks for the tissue being tested and for phenotypes that manifest at older ages. The age distribution and tissue type of samples included in training datasets need to be considered when building and applying epigenetic clock algorithms to human epidemiological or disease cohorts.
Supplementary Material
Acknowledgements
We would like to gratefully acknowledge all donors and their families for the tissue provided for this study. Human post-mortem tissue was obtained from the South West Dementia Brain Bank, London Neurodegenerative Diseases Brain Bank, Manchester Brain Bank, Newcastle Brain Tissue Resource and Oxford Brain Bank, members of the Brains for Dementia Research (BDR) Network. We wish to acknowledge the neuropathologists at each centre and BDR Brain Bank staff for the collection and classification of the samples.
Funding
G.S. was supported by a PhD studentship from the Alzheimer’s Society. E.H., J.M., and L.C.S. were supported by Medical Research Council grant K013807. M.K. was supported by the University of Essex and ESRC (grant RES-596-28-0001). Data analysis was undertaken using high-performance computing supported by a Medical Research Council (MRC) Clinical Infrastructure award (M008924). DNA methylation data generated in the Brains for Dementia Research cohort was supported by the Alzheimer’s Society and Alzheimer’s Research UK (ARUK). Measurement of DNA methylation in The UK Household Longitudinal Study was funded through an enhancement to Economic and Social Research Council (ESRC) grant ES/N00812X/1. The BDR is jointly funded by Alzheimer's Research UK and the Alzheimer's Society in association with the Medical Research Council. The South West Dementia Brain Bank is part of the Brains for Dementia Research program, jointly funded by Alzheimer's Research UK and Alzheimer's Society, and is also supported by BRACE (Bristol Research into Alzheimer's and Care of the Elderly) and the Medical Research Council.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- DNAm =
DNA methylation;
- DNAmClockBlood/Cortical/Multi/Pheno =
blood/cortical/multi-tissue/pheno DNA methylation age clock
References
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011; 479: 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JE, Alafuzoff I, Al-Sarraj S, Arzberger T, Bogdanovic N, Budka H, et al. Management of a twenty-first century brain bank: experience in the BrainNet Europe consortium. Acta Neuropathol 2008; 115: 497–507. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES.. The mammalian epigenome. Cell 2007; 128: 669–81. [DOI] [PubMed] [Google Scholar]
- Buck N, McFall S. Understanding Society: design overview. Longitudinal and Life Course Studies 2011; 3: 5–17. [Google Scholar]
- Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8: 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Pishva E, Haapakoski R, Zsoldos E, Mahmood A, Filippini N, et al. Peripheral DNA methylation, cognitive decline and brain aging: pilot findings from the Whitehall II imaging study. Epigenomics 2018; 10: 585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y-H, Paul KC, Bronstein JM, Bordelon Y, Horvath S, Ritz B.. Parkinson’s disease is associated with DNA methylation levels in human blood and saliva. Genome Med 2017; 9: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury LY, Gorrie-Stone T, Smart M, Hughes A, Bao Y, Andrayas A, et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol 2019; 20: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics 2014; 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis PT, Costello H, Hayes GM.. Brains for dementia research: evolution in a longitudinal brain donation cohort to maximize current and future value. J Alzheimers Dis 2018; 66: 1635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R.. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gopalan S, Gaige J, Henn BM. DNA methylation-based forensic age estimation in human bone. bioRxiv 2019. doi: 10.1101/801647.
- Gorrie-Stone TJ, Smart MC, Saffari A, Malki K, Hannon E, Burrage J, et al. Bigmelon: tools for analysing large DNA methylation datasets. Bioinformatics 2019; 35: 981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics 2013; 8: 290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Gorrie-Stone TJ, Smart MC, Burrage J, Hughes A, Bao Y, et al. Leveraging DNA-methylation quantitative-trait loci to characterize the relationship between methylomic variation, gene expression, and complex traits. Am J Hum Genet 2018; 103: 654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49: 359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S. Economic and social implications of aging societies. Science 2014; 346: 587–91. [DOI] [PubMed] [Google Scholar]
- Horvath S, Raj K.. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018; 19: 371–84. [DOI] [PubMed] [Google Scholar]
- Horvath S, Ritz BR.. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany, NY) 2015; 7: 1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14: R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016; 19: 40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci 2014; 17: 1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jylhävä J, Jiang M, Foebel AD, Pedersen NL, Hägg S.. Can markers of biological age predict dependency in old age? Biogerontology 2019; 20: 321–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jylhävä J, Pedersen NL, Hägg S.. Biological age predictors. EBioMedicine 2017; 21: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Bennett DA, Horvath S.. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany, NY) 2015; 7: 1198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany, NY) 2018; 10: 573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci 2014; 17: 1164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 2015; 44: 1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney DL, Stevenson AJ, Walker RM, Gibson J, Morris SW, Campbell A, et al. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimers Dement 2018; 10: 429–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen LM, O’Donnell KJ, McGill MG, Edgar RD, Jones MJ, MacIsaac JL, et al. The PedBE clock accurately estimates DNA methylation age in pediatric buccal cells. Proc Natl Acad Sci USA 2019; 117: 23329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, Lin H, Ding Y, Lewis DA, Sweet RA.. DNA methylation age is not accelerated in brain or blood of subjects with schizophrenia. Schizophr. Res 2018; 196: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I, Berto S, Usui N, Toriumi K, Chatterjee P, Douglas C, et al. Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol 2019; 20: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I, Yi SV.. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation. Hum Mol Genet 2016; 25: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S, Arribas C, Esteller M.. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics 2016; 8: 389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Viana J, Hannon E, Spiers H, Troakes C, Al-Saraj S, et al. Methylomic profiling of human brain tissue' ' supports a neurodevelopmental origin for schizophrenia. Genome Biol 2014; 15: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Wong Cc Y, Volta M, Lunnon K, Mill J, Schalkwyk LC.. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 2013; 14: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany, NY) 2017; 9: 419–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasekera N, Al-Shahi Salman R, Huitinga I, Klioueva N, McLean CA, Kretzschmar H, et al. Brain banking for neurological disorders. Lancet Neurol 2013; 12: 1096–105. [DOI] [PubMed] [Google Scholar]
- Sanders JL, Newman AB.. Telomere length in epidemiology: a biomarker of aging, age-related disease, both, or neither? Epidemiol Rev 2013; 35: 112–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F. Geroscience and the challenges of aging societies. Aging Med 2019; 2: 132–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Smith RG, Condliffe D, Hannon E, Schalkwyk L, Mill J, et al. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol. Aging 2016; 47: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Smith RG, Pishva E, Hannon E, Roubroeks JAY, Burrage J, et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin Epigenetics 2019; 11: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RG, Hannon E, De Jager PL, Chibnik L, Lott SJ, Condliffe D, et al. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement 2018; 14: 1580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnoff JJ, Newell KM.. Are age-related increases in force variability due to decrements in strength? Exp Brain Res 2006; 174: 86–94. [DOI] [PubMed] [Google Scholar]
- Sugden K, Hannon EJ, Arseneault L, Belsky DW, Broadbent JM, Corcoran DL, et al. Establishing a generalized polyepigenetic biomarker for tobacco smoking. Transl Psychiatry 2019; 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S, Harvey NR, Haupt LM, Griffiths LR, Ashton KJ, Coffey VG, et al. An epigenetic clock for human skeletal muscle. J Cachexia Sarcopenia Muscle 2020; 11: 887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCY, Smith RG, Hannon E, Ramaswami G, Parikshak NN, Assary E, et al. Genome-wide DNA methylation profiling identifies convergent molecular signatures associated with idiopathic and syndromic autism in post-mortem human brain tissue. Hum Mo. Genet 2019; 28: 2201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, et al. Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 2015; 72: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Vallerga CL, Walker RM, Lin T, Henders AK, Montgomery GW, et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med 2019; 11: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T.. Regularization and variable selection via the elastic net. J R Stat Soc B 2005; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used for the training and testing samples are available for download from GEO (https://www.ncbi.nlm.nih.gov/geo/) using the following accession numbers: GSE74193; GSE59685; GSE80970; GPL13534 and GSE43414. The independent test data are available from the authors upon request or via the Dementias Platform UK (DPUK) data portal (https://portal.dementiasplatform.uk/). The whole blood DNA methylation data are available upon application through the European Genome-Phenome Archive under accession code EGAS00001001232. Analysis scripts used in this manuscript and code to run the clock are available on GitHub (https://github.com/gemmashireby/CorticalClock).