Using CSF proteomics, Tijms et al. identify three Alzheimer’s disease subtypes that show: 1) hyperplasticity and increased BACE1 levels; 2) innate immune activation; and 3) blood-brain barrier dysfunction with low BACE1 levels. Future therapeutics may need tailoring to individual disease subtypes.
Keywords: Alzheimer’s disease, cerebrospinal fluid, proteomics, subtypes
Abstract
Alzheimer’s disease is biologically heterogeneous, and detailed understanding of the processes involved in patients is critical for development of treatments. CSF contains hundreds of proteins, with concentrations reflecting ongoing (patho)physiological processes. This provides the opportunity to study many biological processes at the same time in patients. We studied whether Alzheimer’s disease biological subtypes can be detected in CSF proteomics using the dual clustering technique non-negative matrix factorization. In two independent cohorts (EMIF-AD MBD and ADNI) we found that 705 (77% of 911 tested) proteins differed between Alzheimer’s disease (defined as having abnormal amyloid, n = 425) and controls (defined as having normal CSF amyloid and tau and normal cognition, n = 127). Using these proteins for data-driven clustering, we identified three robust pathophysiological Alzheimer’s disease subtypes within each cohort showing (i) hyperplasticity and increased BACE1 levels; (ii) innate immune activation; and (iii) blood–brain barrier dysfunction with low BACE1 levels. In both cohorts, the majority of individuals were labelled as having subtype 1 (80, 36% in EMIF-AD MBD; 117, 59% in ADNI), 71 (32%) in EMIF-AD MBD and 41 (21%) in ADNI were labelled as subtype 2, and 72 (32%) in EMIF-AD MBD and 39 (20%) individuals in ADNI were labelled as subtype 3. Genetic analyses showed that all subtypes had an excess of genetic risk for Alzheimer’s disease (all P > 0.01). Additional pathological comparisons that were available for a subset in ADNI suggested that subtypes showed similar severity of Alzheimer’s disease pathology, and did not differ in the frequencies of co-pathologies, providing further support that found subtypes truly reflect Alzheimer’s disease heterogeneity. Compared to controls, all non-demented Alzheimer’s disease individuals had increased risk of showing clinical progression (all P < 0.01). Compared to subtype 1, subtype 2 showed faster clinical progression after correcting for age, sex, level of education and tau levels (hazard ratio = 2.5; 95% confidence interval = 1.2, 5.1; P = 0.01), and subtype 3 at trend level (hazard ratio = 2.1; 95% confidence interval = 1.0, 4.4; P = 0.06). Together, these results demonstrate the value of CSF proteomics in studying the biological heterogeneity in Alzheimer’s disease patients, and suggest that subtypes may require tailored therapy.
Introduction
Alzheimer’s disease is a neurodegenerative disorder and the most common cause of dementia. The pathological hallmarks are amyloid plaques and tau neurofibrillary tangles in the brain. Biomarkers for amyloid and tau pathology are therefore part of the biological definition of Alzheimer’s disease (Dubois et al., 2007, 2014; Albert et al., 2011; Jack et al., 2011, 2018; Sperling et al., 2011). The current definition implies that Alzheimer’s disease is a single disease entity. However, individuals with Alzheimer’s disease show considerable variability in terms of clinical symptoms, age of onset, disease progression, cortical atrophy patterns, CSF levels of tau, and other pathological markers (Blennow and Wallin, 1992; Iqbal et al., 2005; van der Vlies et al., 2009; Wallin et al., 2010; Whitwell et al., 2012; Lam et al., 2013; Möller et al., 2013; Ossenkoppele et al., 2015; Smits et al., 2015; Hondius et al., 2016). Part of the heterogeneity in Alzheimer’s disease is explained by genetic variance (Ridge et al., 2016) indicating that multiple biological pathways are involved in Alzheimer’s disease, and these include processes related to amyloid and tau metabolism, the innate immune system, lipid processing, and synaptic functioning (Lambert et al., 2013; Kunkle et al., 2019; ,Jansen et al., 2020). It is likely that patients will require personalized medicine depending on the molecular processes involved, but at this point there are no tools to identify biological subtypes in Alzheimer’s disease in vivo. CSF contains many proteins that reflect (patho)physiological processes in the brain, and could provide insight into biological processes involved in Alzheimer’s disease.
Previous studies examining heterogeneity in Alzheimer’s disease based on CSF levels of targeted proteins amyloid, tau and p-tau and/or ubiquitin, suggest that at least three subtypes exist, mainly characterized by having low, intermediate or high tau levels (Iqbal et al., 2005; van der Vlies et al., 2009; Wallin et al., 2010). Unbiased or large-scale targeted proteomic CSF analyses have potential to further refine which biological processes become disrupted in Alzheimer’s disease. So far, Alzheimer’s disease proteomic studies mostly focussed on finding novel biomarkers by comparing patients with Alzheimer’s disease with control subjects (Maarouf et al., 2009; Meyer et al., 2018; for reviews see Pedrero-Prieto et al., 2020; Wesenhagen et al., 2020), and so it remains unclear whether pathophysiological subtypes within Alzheimer’s disease can be discovering with CSF proteomics. Furthermore, if genetic variance in Alzheimer’s disease risk genes contributes to interindividual variability in underlying disease mechanisms, it can be hypothesized that these should already be detectable in presymptomatic stages of Alzheimer’s disease.
In this study we used a data-driven dual clustering technique to identify biological subtypes of Alzheimer’s disease in CSF proteomics in two large independent Alzheimer’s disease cohorts [i.e. the European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery (EMIF-AD MBD) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI)] across the clinical spectrum. We defined Alzheimer’s disease by the presence of amyloid pathology as indicated by abnormal levels of CSF amyloid-β1–42, because abnormal amyloid-β1–42 CSF shows high concordance with the presence of amyloid and tau pathology upon neuropathological examination (Shaw et al., 2009). In contrast, CSF tau levels show more variability amongst patients, with up to 30% of individuals with pathologically confirmed Alzheimer’s disease showing normal levels of CSF tau (Shaw et al., 2009). Therefore, we used CSF total (t)-tau and phosphorylated (p)-tau as independent outcome markers. We further excluded patients that had evidence of known neurodegenerative disorders associated with amyloid aggregation other than Alzheimer’s disease. We first identified which proteins were associated with Alzheimer’s disease. Next, we used unsupervised clustering on these proteins to identify biological subtypes of Alzheimer’s disease. We interpreted Alzheimer’s disease subtype protein profiles in terms of biological processes through enrichment analyses, and performed post hoc analyses to characterize Alzheimer’s disease subtypes in terms of: clinical and biological characteristics known to be associated with Alzheimer’s disease i.e. established CSF markers (neurogranin, BACE1 activity, neurofilament light, VILIP, YKL-40, sTREM2), APOE genotype, Alzheimer’s disease polygenic risk scores, MRI markers for cortical atrophy, cognitive functioning and decline. Furthermore, we compared subtypes on vascular comorbidity using MRI markers for vascular damage. Finally, we compared subtypes on neuropathological measures that were available for a subset of individuals (ADNI only), and we assessed stability of proteomic subtypes over time for a subset of individuals who had longitudinal proteomics available (ADNI only).
Materials and methods
Participants
We selected individuals with CSF amyloid-β1–42, tau, and proteomics data from two independent multicentre Alzheimer’s disease studies, the EMIF-AD MBD (Bos et al., 2018) and the ADNI (adni.loni.usc.edu). Both cohorts included individuals with normal cognition, mild cognitive impairment (MCI) or Alzheimer’s disease-type dementia, as determined according to international consensus criteria (McKhann et al., 1984, 2011; Petersen et al., 1999; Winblad et al., 2004). Control was defined by normal cognition and normal CSF amyloid-β1–42 and tau biomarkers (see below), and Alzheimer’s disease pathological change was defined by abnormal CSF amyloid-β1–42 (Jack et al., 2018). Both studies excluded patients with any neurological disease other than suspected Alzheimer’s disease, such as Parkinson’s disease, dementia with Lewy bodies, frontotemporal dementia, progressive supranuclear palsy, corticobasal syndrome, normal pressure hydrocephalus, or vascular dementia. ADNI began in 2003 as a public-private collaboration under the supervision of principle investigator Michael W. Weiner, MD. The primary goal of ADNI is to study whether serial MRI, PET, other biological markers, and clinical and neuropsychological measures can be combined to measure the progression of MCI and early Alzheimer’s disease. See www.adni-info.org for the latest information. The institutional review boards of all participating institutions approved the procedures for this study. Written informed consent was obtained from all participants or surrogates.
CSF data
CSF samples were obtained as previously described (Shaw, et al., 2009; Toledo et al., 2013; Bos et al., 2018). CSF amyloid-β1–42, t-tau and p-tau levels were measured with INNOTEST ELISAsor INNOBIA AlzBio3 kit (Fujirebio) in EMIF-AD MBD (Bos et al., 2018), and in ADNI with the multiplex xMAP luminex platform (Luminex Corp) at the ADNI Biomarker Core laboratory at the University of Pennsylvania Medical Center. For the ADNI cohort, biomarker abnormality was defined by amyloid-β1–42 levels <192 pg/ml, and t-tau levels >93 pg/ml (Shaw et al., 2009). In the EMIF-AD MBD cohort, cut-offs for p-tau and t-tau were study-specific, as previously reported (Bos et al., 2018). For amyloid-β1–42 cut-offs, the studies in EMIF-AD MBD differed in methodologies used to determine cut-offs, which may lead to bias (Bertens et al., 2017). To minimize such bias across studies, we determined centre-specific cut-offs using unbiased Gaussian mixture modelling (Supplementary Table 1) (De Meyer, 2010; Bertens et al., 2017; Tijms et al., 2018). Cluster analyses were performed on proteomic data using the tandem mass tag (TMT) technique with 10 + 1 plexing in EMIF-AD MBD using high pH reverse phase HPLC for peptide prefractionation (Batth et al., 2014; Magdalinou et al., 2017) (Supplementary material). For EMIF-AD MBD, the median (interquartile range, IQR) analytical coefficient of variation across included proteins was 5.6 (3.8, 8.0) (see Supplementary Table 2 for protein-specific coefficients of variation). The EMIF-AD MBD mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2018) partner repository with the dataset identifiers PXD019910 and 10.6019/PXD019910. Normalized abundances with associated clinical data can be requested from the EMIF-AD MBD consortium (Bos et al., 2018). In the ADNI cohort, four proteins included were determined with ELISAs, 311 protein fragments determined with Multi Reaction Monitoring (MRM) targeted mass spectroscopy, and 83 proteins measured with the Rules Based Medicine (RBM) multiplex. Information on protein assessment and quality control is described at http://adni.loni.usc.edu/data-samples/biospecimen-data/. For ADNI MRM we used the quality-controlled finalized ‘normalized intensity’ data (Spellman et al., 2015) (for detailed explanation of the normalization procedure see the ‘Biomarkers Consortium CSF Proteomics MRM dataset’ in the ‘Data Primer’ document at adni.loni.ucla.edu). All protein (EMIF-AD MBD and ADNI) and protein fragment (ADNI) values were first normalized according to mean and standard deviation (SD) values of the control group. Then, for ADNI, protein fragments from MRM measurements were combined into a protein score when these correlated with r > 0.5, and fragments that did not correlate were omitted from the present analyses. Eleven proteins were measured by different platforms in ADNI, for which values were averaged if they correlated with r > 0.5, or we selected the protein as measured by MRM [mean r = 0.74; min r = −0.50, max r = 0.92; for one protein RBM was excluded, another protein (CST3) showed a strong anticorrelation between RBM and MRM of r = −0.85, and was excluded]. Only proteins that were observed in 100% of the sample were considered for subsequent analyses, resulting in a total 707 proteins in EMIF-AD MBD and 204 proteins in ADNI. A subset of individuals had additional protein measurements available, which we excluded from clustering to use as independent outcomes for subtype interpretation. In ADNI these were amyloid-β1–40 and amyloid-β1–38, measured with 2D-UPLC tandem mass spectrometry, BACE1 activity, and ELISA measures of neurogranin, neurofilament light, VILIP, YKL40, SNAP25 and sTREM2. In EMIF-AD MBD measurements were available for amyloid-β1–40, amyloid-β1–38, neurogranin, neurofilament light, and YKL-40 (Bos et al., 2018). A subset of 70 (29%) ADNI individuals had repeated MRM for 62 proteins [median five repeated measures; over median (IQR) of 6 (4.3, 6.7) years for cognitively normal subjects and median (IQR) of 4 (3.9, 6.0) years for Alzheimer’s disease patients], which we used to study the stability of proteomic subtypes. For these analyses, we first standardized the proteins levels according to the baseline mean and SD levels of the control group, and then we constructed proteomic profile scores by averaging levels of proteins specific for a subtype.
Genetic analyses
ADNI samples were genotyped using either the Illumina 2.5-M array (a byproduct of the ADNI whole-genome sequencing sample) or the Illumina OmniQuad array (Saykin et al., 2010). APOE genotype was assessed with two single nucleotide polymorphisms (SNPs) (rs429358, rs7412) that define the ε2, 3, and 4 alleles, using DNA extracted by Cogenics from a 3 ml aliquot of EDTA blood. EMIF-AD MBD samples were genotoyped at the USKH site using the Global Screening Array (Illumina, Inc) (see Hong et al., 2019 for more details on imputation preprocessing). In the ADNI cohort, SNPs were imputed using the 1000 Genomes reference panel, with the use of the Michigan imputation server. Genotype data were quality checked for gender mismatch, relatedness and ancestry. SNPs were excluded prior to data analyses if they had a minor allele frequency <2%, deviated significantly from Hardy-Weinberg equilibrium (P < 1 × 10−6) in the total sample of founder individuals, or had a call rate of <98%. We only used SNPs with <5% genotype missingness and removed samples with excess heterozygosity rate (>5 SD). After filtering, the genotype data in ADNI included 1 496 949 SNPs and 6 706 731 SNPs in EMIF-AD MBD. To control for population stratification, five principal components were computed on a subset of relatively uncorrelated (r2 < 0.2) SNPs (PC1–PC5). Polygenic risk scores for Alzheimer’s disease were calculated by adding the sum of each allele weighted by the strength of its association with Alzheimer’s disease risk using PRSice (Euesden et al., 2014). The strength of these associations was calculated previously by the International Genomics of Alzheimer’s project (IGAP) genome-wide association study (GWAS) (Lambert et al., 2013). Clumping was performed prior to calculating polygenic risk score to remove SNPs that are in linkage disequilibrium (r2 <0.1) within a slicing 1 Mbp window. After clumping we computed 14 PGRSs with varying SNP inclusion threshold (P < 10−30 to P < 0.5). Finally, we constructed specialized polygenic risk scores including only SNPs that corresponded to genes part of the GO pathways ‘innate immune response’ and ‘complement activation’ for SNP inclusion thresholds (P < 10−30 to P < 1). All polygenic risk scores were regressed on PC1–PC3.
Cluster analyses with non-negative matrix factorization
First, in each cohort we selected proteins for clustering that differed between the control and Alzheimer’s disease groups at uncorrected P < 0.10 using Kruskal-Wallis tests. As protein levels can change non-linearly with levels of neuronal injury and/or disease severity (De Leon et al., 2018; Duits et al., 2018), we repeated analyses stratifying Alzheimer’s disease individuals on disease stage (i.e. normal cognition, MCI and dementia), and on the presence of abnormal CSF levels of the neuronal injury marker t-tau. Next, we clustered these proteins with non-negative matrix factorization (NMF). NMF is a dual clustering approach that is based on decomposition of the data by parts, which reduces the dimensionality of data protein expression levels into fewer components that we consider protein profiles (Lee and Seung, 1999), and at the same time this algorithm groups subjects together into subtypes based on how well their protein expression levels match the protein profiles. A strength of NMF compared to correlation-based approaches is that it is able capture non-linear patterns associated with a certain subtype. To aid interpretation of the results, we labelled proteins according to which subtype showed the highest average levels. We used the R package NMF for clustering, with the ‘nonsmooth’ option that ensures sparse cluster solutions with enhanced separability (Gaujoux and Seoighe, 2010). The NMF algorithm is stochastic and therefore subject classification to a subtype can vary from run to run, based on the random initial conditions. We assessed stability of subtype classification over 50 different runs of NMF with the co-phonetic coefficient, with values ranging from 0 (i.e. unstable solution) to 1 (i.e. subjects are always classified the same). We tested up to five clusters, and the optimal number of clusters was determined as the number of clusters for which: (i) the co-phonetic correlation was high; (ii) fit compared to a lower cluster number solution was improved at least 2-fold over a random solution; and (iii) silhouette width of the cluster solution was > 0.5. Clustering analyses were performed separately for each cohort. We performed pathway enrichment analysis for proteins that were characteristic for each subtype using the online Panther application (Mi et al., 2013). We used the ENCODE and ChEA consensus transcription factor database in the Enrichr webserver to identify potential upstream drivers of subtype-specific protein alterations (Chen et al., 2013; Kuleshov et al., 2016). We selected pathways that were most consistently associated with the subtypes for visualization, and report all observed pathways in the Supplementary material. To determine cell type production we used the BRAIN RNASeq database (http://www.brainrnaseq.org) (Zhang et al., 2014). Proteins were labelled as being specifically produced by a certain cell type when levels were >50% of the total produced across cell types, as non-specific when none of the cell types was >50%, or as not detected when levels were all <0.2.
Statistical procedures for post hoc subtype comparisons
We performed the following post hoc comparisons of subtypes: CSF levels of t-tau, p-tau and other established Alzheimer’s disease CSF markers that were not included in the cluster analyses to provide further independent interpretation of the cluster solutions, age, gender, disease stage, APOE ε4 genotype, Alzheimer’s disease PGRS, pathological measures, cortical thickness measures from 34 cortical areas as defined by the Desikan-Killiany atlas (averaged over the left and right hemispheres) (see Bos et al., 2018 for EMIF-AD MBD and http://adni.loni.cule.edu/ for detailed documentation on variable specific methods in ADNI), vascular damage (visual ratings in EMIF-AD MBD, and white matter hyperintensity volumes in ADNI), Mini-Mental State Examination (MMSE) scores, level of education, neuropsychological test scores covering the memory (memory immediate and delayed recall scores on the logical memory subscale II of the Wechsler Memory Scale), language (Boston Naming Test, and animal fluency), visuospatial processing (Clock drawing) and attention/executive domains [digit span, Trail Making Test (TMT) A and TMT B]. All continuous variables (except for age, MMSE, and years of education) were standardized according to the mean and SD of the control group. Subtype comparisons were performed with general linear models in case of continuous variables with two-sided testing, and with Chi-square tests for discrete variables. Comparisons for continuous variables were performed without and with adjustment for age and sex, and cognitive measures were additionally adjusted for level of education. We used the R package ‘emmeans’ to obtain estimated marginalized means. Finally, we performed an exploratory random forest analysis on the subset of 92 proteins that were associated with Alzheimer’s disease in both EMIF-AD MBD and ADNI to identify the best subset of proteins that can separate the subtypes in the pooled sample and for each cohort separately. For this, we used a bootstrap approach that fitted the model in two-thirds of the dataset and tested classification performance in the left out one-third, repeated 10 000 times in order to estimate 95% confidence intervals (CI) for classification performances. ADNI data were downloaded on 30 March 2018. All analyses were performed in R v3.5.1 ‘Feather Spray’.
Results
We included 127 control subjects with normal cognition and normal CSF amyloid-β1–42 and tau, and 425 individuals with Alzheimer’s disease across the clinical spectrum [89 (21%) normal cognition, 195 (46%) MCI, and 141 (33%) Alzheimer’s disease-type dementia]. Compared to controls, individuals with Alzheimer’s disease more often carried an APOE ε4 allele, had lower MMSE scores, and more often abnormal CSF p-tau and t-tau in both cohorts (Table 1). Other characteristics were similar between groups in both cohorts, except that individuals with Alzheimer’s disease were older than controls in the EMIF-AD MBD. Relative to controls, individuals with Alzheimer’s disease showed differential CSF levels for 556 of 707 proteins (79%) measured in EMIF-AD MBD and 149 of 204 (73%) proteins measured in ADNI (Supplementary Table 2). These Alzheimer’s disease-specific proteins were considered for cluster analyses with NMF within in each cohort.
Table 1.
Participant descriptions
| Descriptive | EMIF-AD MBD |
ADNI |
||
|---|---|---|---|---|
| Controls (n = 82) | Alzheimer’s disease (n = 228) | Controls (n = 45) | Alzheimer’s disease (n = 197) | |
| Cognitive status, n (%) | ||||
| Intact cognition | 82 (100) | 57 (25) | 45 (100) | 32 (16) |
| MCI | 0 (0) | 92 (40)* | 0 (0) | 103 (52) |
| Dementia | 0 (0) | 79 (35) | 0 (0) | 62 (31) |
| Age in years, mean (SD) | 61.1 (7)** | 68.1 (8)*,*** | 75.8 (6) | 74.9 (7) |
| Female, n (%) | 47 (57) | 126 (55)* | 23 (51) | 81 (41) |
| Years of education, mean (SD) | 11.9 (3.5)** | 11.2 (3.5)* | 15.6 (3) | 15.6 (3) |
| MMSE, mean (SD)a | 28.6 (1.3)** | 25.6 (3.9)*** | 29.2 (0.6) | 26.1 (2.6)*** |
| APOE e4, at least one allele (%)b | 14 (22)** | 140 (52) | 4 (8) | 129 (65)*** |
| Hippocampal volume, mean (SD)c,e | 0 (1) | −1.4 (1.5)*,*** | 0 (1) | −1.7 (1.4)*** |
| Amyloid-β1–42 pg/ml, mean (SD)f | 0 (1) | −2.8 (1.5)*,*** | 247.5 (29.2) | 139.1 (23.1)*** |
| t-tau pg/ml, mean (SD)f | 0 (1) | 4.4 (4.7)*** | 57.1 (13.1) | 114.3 (54.9)*** |
| Abnormal t-tau, n (%)g | 0 (0) | 151 (66)*** | 0 (0) | 115 (58)*** |
| p-tau pg/ml, mean (SD)d,f | 0 (1) | 2.1 (2.5)*** | 20.3 (9.4) | 39.1 (17.5)*** |
| Abnormal p-tau, n (%)g | 7 (8.5) | 149 (65)*,*** | 9 (20) | 168 (85)*** |
MMSE = Mini-Mental State Examination.
aData missing for one individual;
b16 individuals;
c159 individuals; and
dfive individuals.
eScaled according to the mean and SD values in controls.
fBased on Luminex in ADNI and scaled for EMIF according to cohort-specific controls as previously described (Bos et al., 2018).
gCut-off points to define abnormal levels for ADNI: t-tau > 93 pg/ml, p-tau > 21 pg/ml (Shaw et al., 2009), and cohort specific for EMIF-AD MBD as previously described (Bos et al., 2018).
Groups were compared with χ2 tests or t-test where appropriate.
P < 0.05 for across cohort comparisons between Alzheimer’s disease groups.
P < 0.05 for across cohort comparisons between controls.
P < 0.01 × 10−30 for controls versus Alzheimer’s disease within cohort.
Cohort differences in continuous amyloid-β1-42, t-tau and p-tau levels are based on cohort scaled values according to control group levels.
Three biological Alzheimer’s disease subtypes detected in CSF proteomic data
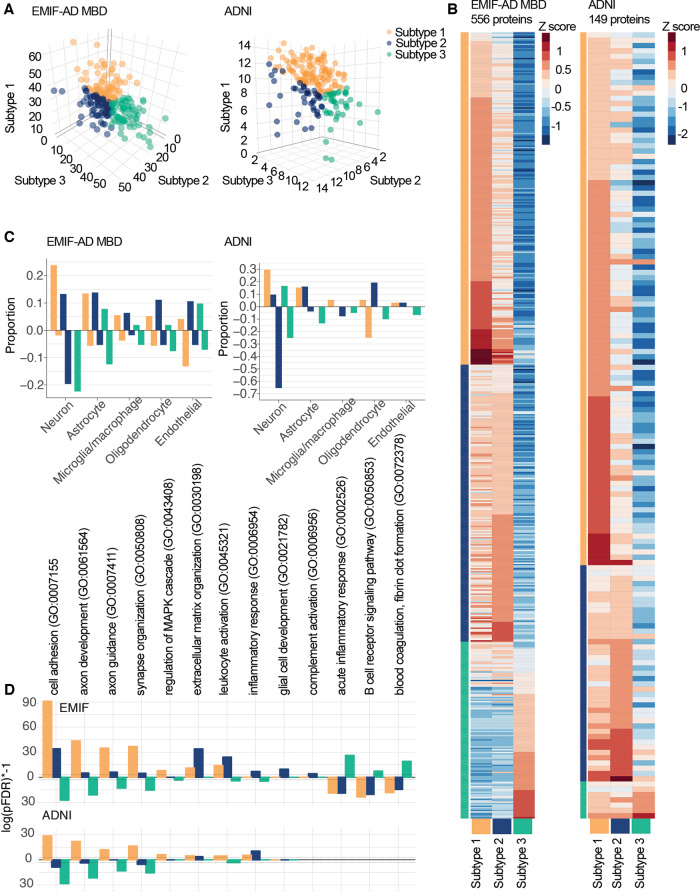
According to our fit criteria, three clusters best described the CSF proteomic data in both cohorts (Supplementary Table 3). Repeating clustering of proteins using a Louvain modularity algorithm on a weighted protein co-expression network also resulted in three protein clusters, which showed good correspondence with the NMF protein clusters of 80% in EMIF-AD MBD and 86% in ADNI (Supplementary Tables 5A and B). A 3D plot of subject loadings on clusters revealed a subset of five individuals with extreme loadings in EMIF-AD MBD (Supplementary Fig. 1). These individuals did not show differences with other Alzheimer’s disease individuals in terms of sample characteristics (Supplementary Table 4). To avoid potential overfitting, we repeated cluster analyses excluding these individuals, and a three-cluster solution remained most optimal. We next labelled individuals according to the subtype they scored highest on (Fig. 1A). In both cohorts, the majority of individuals were labelled as having subtype 1 [80 (36%) in EMIF-AD MBD, 117 (59%) in ADNI]; 71 (32%) in EMIF-AD MBD and 41 (21%) in ADNI were labelled as subtype 2; and 72 (32%) in EMIF-AD MBD and 39 (20%) individuals in ADNI were labelled as subtype 3. A subset of 92 proteins was measured in both EMIF-AD MBD and ADNI, which showed consistent subtype differences in levels for 84–98% of proteins across the cohorts (Supplementary Fig. 2 and Supplementary Table 5A). An exploratory random forest analysis on this subset of 92 overlapping proteins identified a subset of five proteins (SLITRK1, NEO1, EXTL2, TNFRSF21, IGFBP2) that best separate the subtypes, with an overall accuracy of 80% (95% CI: 74–86%) in the pooled sample, and similar performance within cohorts (EMIF: 80%, 95% CI: 71%–88%; ADNI: 80%, 95% CI: 71%–89%), further supporting that subtype definitions are robust. Individuals with subtype 1, compared to controls, had significantly higher levels for the majority of proteins in both cohorts [EMIF-AD MBD: 309 (56%); ADNI: 92 (65%); P-values ranging between 0.02 × 10−21 and 0.049] (Fig. 1B and Supplementary Table 5A and B). The predominant cell types producing these proteins were neurons and astrocytes in both cohorts (Fig. 1C and Supplementary Table 6). GO pathway analyses for proteins increased in subtype 1 showed in both EMIF-AD MBD and ADNI enrichment for processes related to the MAPK/ERK cascade, synaptic structure and function, axonal development, and glucose metabolism, suggesting that subtype 1 shows neuronal hyperplasticity (Fig. 1D and Supplementary Table 7) (77% processes enriched in ADNI overlapped with those enriched in EMIF-AD MBD). Subtype 2 also showed mostly higher protein levels than controls [EMIF-AD MBD: 202 (36%); ADNI: 31 (21%); P-values ranging between 0.01 × 10−16 and 0.049]. The predominant cell types producing these proteins were oligodendrocytes, neurons and astrocytes. GO pathway analyses for proteins specifically increased in subtype 2 showed enrichment for innate immune response, extracellular matrix organization and oligodendrocyte development, hence these individuals may be characterized as having innate immune activation (54% of the processes enriched in ADNI overlapped with those enriched in EMIF-AD MBD). Compared to controls, subtype 3 individuals showed mostly decreased proteins [415 (75%) in EMIF-AD MBD; 120 (81%) in ADNI; P-values ranging between 0.02 × 10−22 and 0.049] that mirrored the increases observed in subtype 1, which suggests that type 3 has neuronal hypoplasticity. Another group of proteins was specifically increased in subtype 3 compared to control subjects [76 (14%) in EMIF-AD MBD; 6 (4%) in ADNI], including albumin and immunoglobulin proteins, of which higher CSF levels have been reported with blood–brain barrier dysfunction (Dayon et al., 2019). GO pathway analyses for proteins specifically increased in subtype 3 were performed for EMIF-AD MBD only, as the number of subtype 3-specific increased proteins in ADNI was too low (note that for proteins showing subtype 3-specific decreases 99% of the pathways enriched in ADNI overlapped with EMIF-AD MBD, and these pathways involved 66% of those enriched for subtype 1 increased proteins). Subtype 3 increased proteins were enrichmed for acute inflammation, B-cell activation, blood coagulation-related processes, lipid processing, and lipoprotein clearance, which together suggest that this subtype may be characterized as having blood–brain barrier dysfunction. Subtype 3 also showed enrichment for complement activation, but for a different group of proteins than observed in subtype 2: C6, C8A, C8B and C9, which are part of the terminal pathway of the complement system (Veerhuis et al., 2011; Orsini et al., 2014) (Supplementary Fig. 3). We searched the ENCODE and ChEA consensus transcriptional factor database using the Enrichr server to identify potential upstream drivers for proteins that were highest in each subtype and significantly different from controls. The strongest transcriptional factors associated with increased proteins specifically in subtype 1 in both EMIF-AD MBD and ADNI were REST and SUZ12, which are transcriptional repressors that are important for neuronal development and synaptic plasticity (Rodenas-Ruano et al., 2012) (Supplementary Table 8). The top transcription factors associated with subtype 2 did not show consistent patterns between EMIF-AD MBD and ADNI, and no significant transcription factors were observed for subtype 3-specific increases (tested in EMIF-AD MBD only). Longitudinal proteomics was available for a subset in ADNI [n = 70 (29%), including 23 controls and 47 Alzheimer’s disease: 31 with subtype 1; nine with subtype 2; seven with subtype 3], including only proteins associated with subtypes 1 and 2. Proteomic profile scores that summarized levels of proteins that were associated with either subtype 1 (52 proteins) or 2 (12 proteins) in the discovery dataset remained stable over time in all subtypes, as none of the slopes differed from 0 (all subtypes P > 0.10, Supplementary Table 9). This suggests that subtype definitions remained stable over time.
Figure 1.
Cluster results. (A) Subject loadings on subtype scores (orange: subtype 1, hyperplasticity; blue: subtype 2, innate immune activation; green: subtype 3, blood–brain barrier dysfunction) for EMIF-AD MBD (left) and ADNI (right). Each dot shows how an individual matches all three proteomic subtypes at the same time, e.g. the right-most green dot is a subject who shows very high loading on the subtype 3 axis, and very low loadings on subtypes 1 and 2 axes. (B) Heat map of subtype average Z-scores (according to the mean and SD of controls). Labels not shown, see Supplementary Table 5 for list of proteins. (C) Proportion of cell type production for protein levels higher (positive proportions) or lower than control subjects (negative proportions). (D) Selected subset of GO pathways that show subtype-specific enrichment with log(pFDR) positive values for proteins with higher levels than controls, and negative values for proteins with lower levels than controls (see Supplementary Table 7 for complete list of enriched pathways).
Genetic comparisons of subtypes
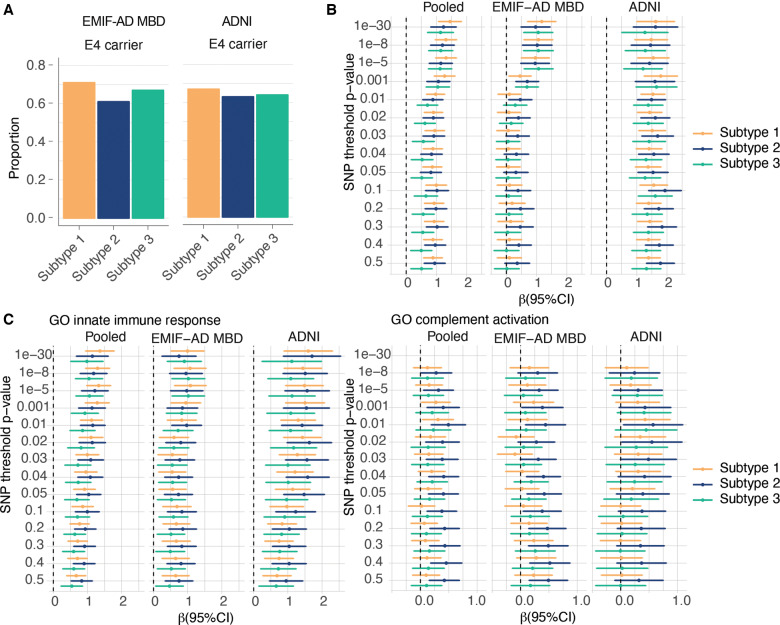
Subtypes showed similar proportions of APOE ε4 carriers in both cohorts (Fig. 2A and Supplementary Table 10; all P's > 0.05). Relative to controls, all subtypes had an excess of Alzheimer’s disease genetic risk (Fig. 2B and Supplementary Table 11; P-values ranging between 0.02 × 10−13 and 0.004). For SNP inclusion thresholds 0.1 to 0.5, subtype 2 individuals showed higher Alzheimer’s disease polygenic risk score than subtype 1 and 3, but these associations lost significance after adjusting for age and sex. Because subtype 2 individuals were associated with innate immune response, which has been previously associated with top Alzheimer’s disease risk SNPs, we compared subtypes on polygenic risk score for innate immune response and complement activation, and found for the majority of SNP inclusion thresholds the highest scores for subtype 2 (Fig. 2C and Supplementary Table 11; P-values compared to controls ranging between 0.02 × 10−7 and 0.045; P-values compared to the other subtypes ranging between 0.004 and 0.045). These effects remained largely unchanged after adjusting for age and sex.
Figure 2.
Genetic factor comparisons between subtypes. (A) Proportion of APOE e4 carriers according to subtype. (B) Effect sizes (95% CI) of Alzheimer’s disease polygenic risk scores for increasing SNP inclusion P-value thresholds, comparing Alzheimer’s disease subtypes to the control group (normal cognition, normal CSF amyloid and tau levels). (C) Comparisons of Alzheimer’s disease subtypes in Alzheimer’s disease polygenic risk specific for GO innate immune response (left) and GO complement activation (right), for increasing SNP inclusion P-value thresholds. (B and C) The pooled sample, and the cohorts separately. The dotted vertical lines in B and C indicate mean scores for the control group. See Supplementary Tables 10 and 11 for test statistics of all comparisons.
Other biological and clinical subtype characterization
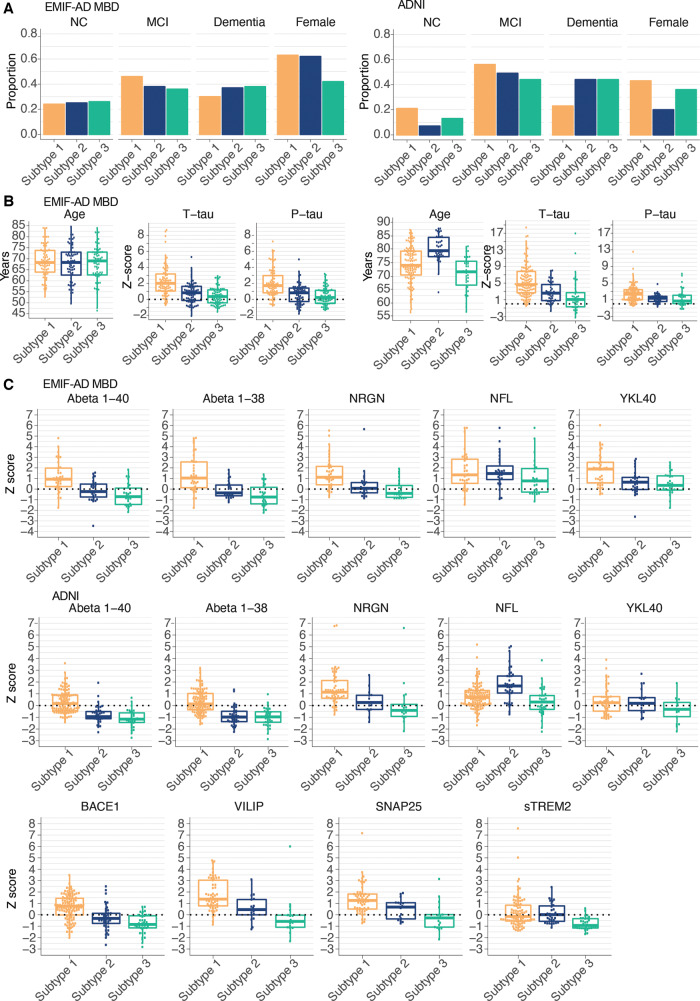
We next compared subtypes on clinical characteristics and established Alzheimer’s disease CSF markers. In EMIF-AD MBD, subtypes had comparable proportions of disease stages and sex, and comparable age (Fig. 3A). In ADNI, individuals with subtype 1 (hyperplasticity) less often had dementia (compared to subtype 2, P = 0.02; compared to subtype 3, P = 0.02), and individuals with subtype 2 (innate immune activation) were older and more often male. In both cohorts, t-tau and p-tau CSF levels were highest and most often abnormal in the subtype 1 (hyperplasticity; Fig. 3B and Supplementary Table 10), intermediate for subtype 2 (innate immune activation), and the lowest and most often normal in subtype 3 (blood–brain barrier dysfunction). Other neuronal injury markers such as neurogranin (both cohorts), VILIP and SNAP25 (ADNI only) were consistently highest in subtype 1 (hyperplasticity), and lowest in subtype 3 (blood–brain barrier dysfunction) (Fig. 3C and D). NEFL levels were comparable across subtypes in EMIF-AD MBD, but were increased in subtype 2 (innate immune activation) in ADNI, which remained after additional correction for age and sex. Subtype 1 (hyperplasticity) further showed higher levels of proteins associated with amyloid precursor protein (APP) processing (i.e. higher levels of amyloid-β1–40 and amyloid-β1–38 in both cohorts, and higher levels of BACE1 activity in ADNI). Subtype 3 (blood–brain barrier dysfunction) showed the lowest concentrations for those markers. Both subtypes 1 (hyperplasticity) and 2 (innate immune activation) showed higher levels of inflammation markers YKL40 and sTREM2 (ADNI only) than subtype 3 (blood–brain barrier dysfunction). Since some of these markers can increase with disease severity, we repeated subtype comparisons stratified for disease stage (normal cognition, MCI and dementia). Results showed largely similar subtype profiles (all Pinteraction > 0.05; Supplementary Figs 4 and 5).
Figure 3.
Clinical stage, age, sex and other CSF markers comparisons between subtypes. (A) Proportions of disease stage (NC = normal cognition), and females according to Alzheimer’s disease subtypes and cohort. (B) Distributions of age, t-tau and p-tau levels according to Alzheimer’s disease subtypes and cohort. (C) Distributions of CSF markers not included in clustering according to subtype and cohort, including additional CSF biomarkers that were available in ADNI only for a subset of individuals. See Supplementary Table 10 for test statistics of all comparisons.
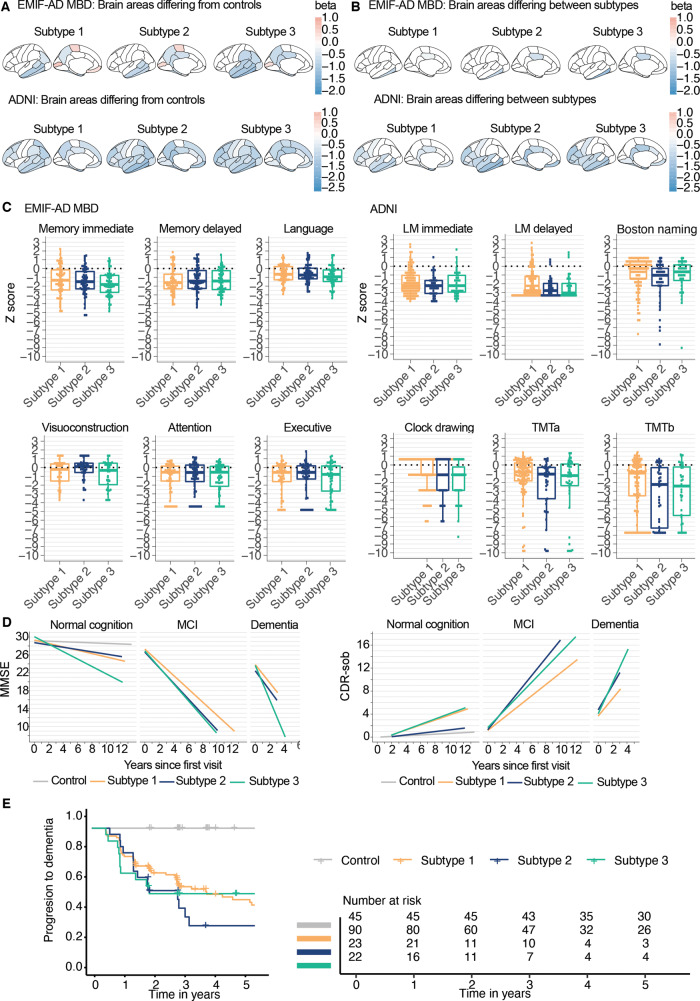
Atrophy, vascular damage, cognitive profiles and pathological comparisons
Atrophy relative to controls was most pronounced in the hippocampus, medial and lateral temporal cortex and the precuneus for all subtypes (Fig. 4A, Supplementary Table 12 and Supplementary Fig. 6). Compared to subtype 1 (hyperplasticity), individuals with subtypes 2 (innate immune activation) and 3 (blood–brain barrier dysfunction) showed more atrophy in the posterior cingulate in both cohorts (Fig. 4B). In ADNI, subtype 2 (innate immune activation) showed more atrophy than subtype 1 (hyperplasticity) in the inferior temporal gyrus, insula, isthmus cingulate, rostral middle frontal and temporal pole. Visual ratings for vascular damage on MRI in EMIF-AD MBD showed that subtype 3 (blood–brain barrier dysfunction) more often had a lacunar infarct (n = 10, 22%) than subtype 2 (n = 1, 2%; P = 0.003) and subtype 1 (n = 4, 7.5%; P = 0.04). No differences between subtypes were observed in white matter intensity load (Fazekas score of 3), or the presence of more than one microbleed (Supplementary Table 10). In ADNI, white matter hyperintensity volumes were larger in subtype 3 (blood–brain barrier dysfunction; 1.2 ± 2.7 cm3) and subtype 2 (innate immune activation; 1.3 ± 1.4 cm3) compared to subtype 1 (hyperplasticity; 0.85 ± 3.0 cm3; subtype 1 versus subtype 2, P = 0.0004; subtype 1 versus subtype 3, P = 0.01; subtype 2 versus subtype 3, P = 0.44). Subtypes showed largely similar scores on cognitive tests (Fig. 4C and Supplementary Table 13). Repeating analyses stratified on disease stage, showed that individuals with subtype 3 in the dementia stage scored worse on the TMT (A) than the other two subtypes (Pinteraction = 0.004; both subtype 1 versus subtype 3, and subtype 2 versus subtype 3, P < 0.001) (Supplementary Table 13 and Supplementary Fig. 7). Worsening over time on Clinical Dementia Rating Scale Sum of Boxes (CDRsob) was steeper for subtype 2 compared to subtype 1 in MCI (P = 0.01; Fig. 4E, F and Supplementary Table 14), and for subtype 3 compared to subtype 1 in dementia (P = 0.02). Compared to subtype 1, individuals without dementia and subtype 2 showed increased risk of progression to dementia, also after correcting for age, sex, level of education and tau levels [hazard ratio (HR) (95% CI) subtype 2 versus subtype 1 = 2.5 (1.2, 5.1), P = 0.01], and subtype 3 at trend level [HR = 2.1 (1.0, 4.4), P = 0.06] (Fig. 4G and Supplementary Table 15). For a subset of 20 (10%) ADNI individuals with neuropathological information, we found similar pathological scores for amyloid and tau for subtypes, and they showed similar frequencies of occurring co-pathologies, such as Lewy body pathology, TDP-43 and hippocampal sclerosis (Supplementary Table 16).
Figure 4.
Cortical thickness and cognition comparisons between subtypes. (A) All brain areas where differences compared to controls were observed. (B) Brain areas with a significant main effect for subtype; all beta values reflect volumetric differences of subtypes compared to controls. (C) Comparisons of cognitive profiles between subtypes. (D) Changes over time on MMSE (left), and CDRsob (right) in ADNI only. (E) Cumulative progression to dementia curves for subtypes, in ADNI only. All cortical thickness and neuropsychological test values are standardized according to the mean and SD values of the control subjects. See Supplementary Tables 10 and 12–15 for test statistics of all comparisons.
Discussion
Understanding biological heterogeneity in patients with Alzheimer’s disease is critical for treatment development. We proteomically defined three Alzheimer’s disease pathophysiological subtypes that were associated with distinct biological processes, i.e. hyperplasticity, innate immune activation and blood–brain barrier dysfunction, and these subtypes were robustly observed in two large independent cohorts. These biological subtypes of Alzheimer’s disease showed pronounced differences in levels of proteins associated with processes known to be deregulated in Alzheimer’s disease, including APP processing, neuronal injury, and inflammation. All subtype-specific alterations in CSF protein levels could both be increased or decreased for subsets of proteins, indicating that differences cannot be explained by trivial aspecific changes in CSF composition. All subtypes had an excess genetic risk for Alzheimer’s disease, and pathological measures did not show a difference in the presence of comorbidities, providing further support that these differences reflect heterogeneity within Alzheimer’s disease. A particularly novel finding is the observation of the blood–brain barrier dysfunction subtype that showed mostly abnormally low concentrations of proteins associated with APP processing, as well as t-tau and p-tau levels. Together, these results demonstrate the potential for CSF proteomics to identify which biological processes are disrupted in individual patients with Alzheimer’s disease, and suggest that individuals might require specific treatments depending on their subtype.
Previous studies that clustered targeted proteins amyloid, t-tau, p-tau and/or ubiquitin CSF levels suggested three to five Alzheimer’s disease subtypes that were characterized by having low, intermediate and high tau values (Iqbal et al., 2005; van der Vlies et al., 2009; Wallin et al., 2010), and our proteomically defined subtypes show a similar distinction in tau levels. We further show, with our large-scale proteomic analyses, which biological processes may underlie interindividual differences in tau levels. Individuals with Alzheimer’s disease with the hyperplasticity subtype showed high levels for the majority of proteins, and pathways enriched in both cohorts showed involvement of regulation of the MAPK/ERK cascade, glucose metabolism, synaptic structure and function, and axonal development, all processes important for synaptic plasticity. This hyperplasticity subtype also showed higher levels of markers presumed to reflect neuronal injury, i.e. t-tau, p-tau, and neurogranin in both cohorts (as well as VILIP, and SNAP25, which was only measured in ADNI), which could reflect more severe neuronal damage (Fagan and Perrin, 2012; Brinkmalm et al., 2014). However, this is unlikely because these proteins were already increased in individuals with Alzheimer’s disease and normal cognition, when atrophy was less severe compared to the other subtypes. An alternative explanation could be increased synaptic activity as this can lead to increased tau (Pooler et al., 2013; Yamada et al., 2014) and amyloid secretion (Cirrito et al., 2005; Bero et al., 2011). Hyperactive neurons have been reported in Alzheimer’s disease with concurrent increased tau and amyloid levels (Roberson et al., 2011; Palop and Mucke, 2016). Aberrant increases in neuronal activation can be caused by amyloid oligomers, which disrupt the balance of excitation and inhibition of neuronal circuits (Palop and Mucke, 2016). Furthermore, subtype 1-specific proteomic increases in both EMIF and ADNI converged on the transcription factors REST and SUZ12, which are transcription suppressors that are known to interact with each other (Tsai et al., 2010) and play an important role in regulating neuronal development and synaptic plasticity (Ballas and Mandel, 2005; Rodenas-Ruano et al., 2012; Yang et al., 2012). These factors have previously been identified as key regulators in a sporadic Alzheimer’s disease iPSC model (Meyer et al., 2019), in which neurons also showed increase excitability, and increased tau phosphorylation and amyloid-β1–40 concentrations, which is in line with our observations and interpretation of subtype 1. Finally, proteins increased in subtype 1 also showed enrichment for glucose metabolism, which has been reported in two recent proteomic studies in tissue and in CSF that also observed clusters of proteins associated with glucose metabolism to be involved in Alzheimer’s disease, as well as clusters of synaptic proteins (Higginbotham et al., 2019; Johnson et al., 2020). In those studies, and our results, glucose metabolism pathways mostly included proteins that were specific for neurons and/or astrocytes. Neurons can increase neuronal glycolysis during neuronal stimulation (Díaz-García et al., 2017). Although neurons use most of the brain’s energy, glucose metabolism is predominantly driven by astrocytes, which themselves consume only up to 15% of energy in the brain, and can regulate glucose metabolism upon neuronal activation (Magistretti and Allaman, 2015). These findings indicate that non-neuronal cell types may be dysregulated and/or have detrimental effects on neurons in subtype 1.
The second subtype had a proteomic profile that also indicated upregulation of plasticity-related processes like subtype 1, but less pronounced. Subtype 2 further showed higher levels of proteins that—in both cohorts—pointed to involvement of the innate immune system, extracellular matrix organization and oligodendrocyte development. Oligodendrocytes are important for axonal myelination. In ADNI, subtype 2 showed increased white matter hyperintensity volume, and was increased in both EMIF-AD MBD and ADNI NEFL, suggesting axonal damage in this subtype. Furthermore, individuals with subtype 2 showed high levels of complement proteins C1q B chain and C4a in both cohorts, as well as C1q A chain, C1q C chain, C1s, and C1r in EMIF-AD MBD (not measured in ADNI), which are early components of the classical complement pathway (Veerhuis et al., 2011; Orsini et al., 2014). Alzheimer’s disease PGRSs restricted to genes involved in innate immune response and complement activation were mostly higher in subtype 2 in both cohorts, suggesting that these CSF proteomic alterations reflect genetic effects. Higher concentrations of C1q and C4 in Alzheimer’s disease brains have been reported in pathological studies (Veerhuis et al., 2011; Dejanovic et al., 2018), and therefore higher concentrations of C4a may indicate complement activation in this subtype. Amyloid-β fibrils are known to activate the complement pathway by binding to the C1q complex (Rogers et al., 1992; Webster et al., 2002). Complement activation may also play a role in neuronal injury in Alzheimer’s disease, because complement proteins can accumulate at synapses and tag these for phagocytosis by activated microglia (Pooler et al., 2013; Orsini et al., 2014; Yamada et al., 2014). Knocking out C1q in APP transgenic mice or blocking C1q in Tau-P301S transgenic mice attenuates both complement activation and neuronal injury (Pooler et al., 2013; Orsini et al., 2014; Yamada et al., 2014), possibly by preventing inappropriate microglia activation. Together, the biological processes specific for subtype 2 seem to be associated with activated microglia, which may contribute to neuronal dysfunction. Alternatively, these processes may be associated with activation or dysregulation of astrocytes, because microglia secreting C1q can induce so-called ‘A1 reactive’ astrocytes that lose the ability to facilitate plasticity processes that promote cell survival and accelerate death of neurons and oligodendrocytes (Liddelow et al., 2017).
In both cohorts, subtype 3 had low and more often normal t-tau and p-tau CSF levels compared to the other subtypes, together with abnormally low levels for the majority of other proteins. The generally low levels of t- and p-tau raise the question as to whether these individuals have Alzheimer’s disease (Jack et al., 2018). Our results provide strong support that in fact these individuals do have Alzheimer’s disease, because: (i) these individuals had abnormal amyloid levels; (ii) the most severe atrophy included typical Alzheimer’s disease regions, such as the medial temporal lobe; (iii) non-demented individuals with subtype 3 showed increased risk for clinical progression; and (iv) they have an excess of genetic risk for Alzheimer’s disease. The percentage of individuals classified as having subtype 3 is in line with previous studies reporting pathologically confirmed Alzheimer’s disease, which show that up to 30% of individuals can have normal CSF tau levels (Shaw et al., 2009). Thus, CSF tau levels may reflect other processes in addition to neurofibrillary tau tangles, and conversely, normal levels do not exclude underlying tau pathology. Given the relationship of tau levels and neuronal activity discussed above, low tau levels in this subtype may reflect hypoplasticity. Alternatively, low levels of tau suggest less neuronal injury. However, this explanation seems implausible, because subtype 3 had—in both cohorts—widespread atrophy and significantly higher levels of the axonal damage marker NEFL. Furthermore, in EMIF-AD MBD only (because that cohort used an untargeted proteomics technique), a large group of proteins that were increased in subtype 3 have previously been reported to correlate with the CSF/plasma albumin ratio, which is a marker for blood–brain barrier integrity (Dayon et al., 2019), and this suggests that subtype 3 may have blood–brain barrier dysfunction (Sagare et al., 2012; Yamazaki and Kanekiyo, 2017; Sweeney et al., 2018). Blood–brain barrier dysfunction disrupts glucose metabolism, which can impair neuronal activity and plasticity processes (Yamazaki and Kanekiyo, 2017; Sweeney et al., 2018). This would explain why proteins involved in synaptic structure and function were decreased in subtype 3, consistently observed in both cohorts, and suggest that these individuals have hypoplasticity. Another subset of proteins specifically increased in subtype 3 (in EMIF-AD MBD only) was enriched for lipid processing, clearance and regulation, and these included apoC1. ApoC1 is produced by astrocytes (Petit-Turcotte et al., 2001; Abildayeva et al., 2008), can inhibit receptor-mediated clearance of lipoproteins containing APOE (Sehayek and Eisenberg, 1991; Shachter, 2001), and has been observed in amyloid plaques (Abildayeva et al., 2008). This suggests that vascular factors might play a role in amyloid pathogenesis, possibly contributing to reduced clearance of aggregated amyloid. Alternatively, amyloid might aggregate in the vasculature, which could lead to blood–brain barrier dysfunction.
The proteomic subtypes we discovered could have implications for treatment: subtype 1 showed the highest levels of BACE1 activity (measured in ADNI only) and products of amyloid metabolism (amyloid-β1–40 and amyloid-β1–38; measured in both cohorts) and so it can be hypothesized that particularly this subtype will benefit from treatments that target APP processing, such as BACE1 inhibitors, whereas this type of treatment may be harmful for individuals with subtype 3 that showed decreased levels of BACE1 activation. Individuals with subtype 2 may potentially benefit from therapeutic strategies that target microglia and astrocyte activation. Subtype 3 may benefit from therapies that protect the vasculature. Future research should further study treatment effects on CSF proteomic profiles, and whether effects are subtype-dependent.
A potential limitation of this study is that proteins specifically increased in subtype 3 were mostly observed in the EMIF-AD MBD cohort, because that study used an untargeted approach, whereas those proteins were not measured in ADNI, because that study selected a limited number of proteins with brain enriched expression patterns. However, subtype 3 in ADNI showed a hypoplasticity response, similar to that of subtype 3 in EMIF-AD MBD, suggesting that they share common pathophysiological processes. Furthermore, the random forest analysis showed that in principle, it is possible to use these proteins as robust subtype markers, independent of cohort or measuring technique. Still, further analyses focussing on single biomarkers are necessary since in ADNI, proteins were preselected, while in EMIF, an untargeted technique was used, and so not all top markers in EMIF for specific subtypes were available in ADNI. Also, new studies are necessary for further validation of these subtypes, using independent sets of proteins for subtyping and for characterization to avoid potential inflation of classification accuracy due to circularity. Furthermore, although many of the proteins specifically increased in subtype 3 were previously reported to be correlated to blood–brain barrier function (Dayon et al., 2019), future analyses should further verify this by measuring both CSF and plasma albumin. Furthermore, the EMIF-AD MBD and ADNI cohorts are a clinical multicentre and a research multicentre study, respectively, and differed in their baseline characteristics, most notably ADNI participants being on average 10 years older. However, it is unlikely cohort-specific and age effects explain the subtypes, as the subtypes showed highly similar proteomic profiles across the cohorts. Furthermore, MRI scans in EMIF-AD MBD were acquired in clinical routine, in contrast to the harmonized scanning protocol in ADNI. This may have made it more difficult to detect differences in atrophy patterns amongst subtypes in combination with relatively small sample sizes (Zhang et al., 2016; Scheltens et al., 2017; Ten Kate et al., 2018). Another point of consideration is that co-pathology often occurs with older age, which may influence proteomic subtype definitions (Beach et al., 2012). Our analyses in a subset of the ADNI cohort showed that subtypes did not differ in the occurrence of co-pathology, suggesting that it is unlikely that this has driven the subtypes. Still, those analyses need further replication in larger samples with combined tissue and CSF proteomic data. Our subtypes were defined by their proteomic profiles, and at this point single markers from our analyses should not be used in practice until thoroughly validated, which we aim to pursue in future studies. A strength of our study is that we were able to replicate the biological subtypes that we detected with CSF proteomics in two independent cohorts, and even though different methods were used to measure proteins, we observed similar processes to be involved in Alzheimer’s disease, supporting the robustness of our findings.
In conclusion, we have identified a hyperplasticity, innate immune activation and a blood–brain barrier dysfunction subtype in Alzheimer’s disease using CSF proteomics. The most important implication of our results is that currently existing—and even failed—treatments may be beneficial for specific subtypes, and that CSF proteomics may serve as a stratification tool to investigate this further.
Funding
This work has been supported by ZonMW Memorabel grant programme #73305056 (B.M.T.) and #733050824 (B.M.T. and P.J.V.), the Swedish Research Council (#2018-02532, H.Z.), the European Research Council (#681712, H.Z.) and Swedish State Support for Clinical Research (#ALFGBG-720931, H.Z.), the Alzheimerfonden (#AF-930934, J.G.) and Dtiftelsen Gamla tjänarirror (J.G.), and the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement #115372 (P.J.V., H.Z.). Statistical analyses were performed at the VUmc Alzheimer Center that is part of the neurodegeneration research program of the Neuroscience Campus Amsterdam. EMIF-AD MBD proteomic analyses were performed at the Department of Psychiatry and Neurochemistry, the Sahlgrenska Academy at the University of Gothenburg, Sweden. The VUmc Alzheimer Center is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. H.Z. is a Wallenberg Academy Fellow. F.B. is supported by the NIHR biomedical research centre at UCLH. The Leuven cohort was funded by Stichting Alzheimer Onderzoek (#11020, #15005, #13007) and the Vlaamse Impulsfinanciering voor Netwerken voor Dementie-onderzoek (IWT #135043).
Data were used for this project of which collection and sharing was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Competing interests
B.M.T., P.J.V., J.G., H.Z. and K.B. are the inventors of the CSF proteomic profile definition of Alzheimer’s disease subtypes and have a patent pending (#19165795.6; Applicant: Stichting VUmc). H.Z. has served at scientific advisory boards for Roche Diagnostics, Wave, Samumed and CogRx, has given lectures in symposia sponsored by Biogen and Alzecure, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg (all unrelated to the submitted work). S.L. is currently an employee at Janssen R&D and has in the past five years provided consultancy to Eisai, SomaLogic, Merck and Optum Labs. F.B. is a consultant for Biogen, Bayer, Merck, Roche, Novartis, Lundbeck, and IXICO; has received sponsoring from European Commission–Horizon 2020, National Institute for Health Research–University College London Hospitals Biomedical Research Centre, Biogen, TEVA and Novartis. The other authors declare no competing interests with the content of this article.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- EMIF-AD MBD =
European Medical Information Framework for Alzheimer’s Disease Multimodal Biomarker Discovery
- NMF =
non-negative matrix factorization
- MCI =
mild cognitive impairment
- SNP =
single nucleotide polymorphism
Using CSF proteomics, Tijms et al. identify three Alzheimer’s disease subtypes that show: 1) hyperplasticity and increased BACE1 levels; 2) innate immune activation; and 3) blood-brain barrier dysfunction with low BACE1 levels. Future therapeutics may need tailoring to individual disease subtypes.
References
- Abildayeva K, Berbée JFP, Blokland A, Jansen PJ, Hoek FJ, Meijer O, et al. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J Lipid Res 2008; 49: 856–69. [DOI] [PubMed] [Google Scholar]
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Mandel G.. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol 2005; 15: 500–6. [DOI] [PubMed] [Google Scholar]
- Batth TS, Francavilla C, Olsen JV.. Off-line high-pH reversed-phase fractionation for in-depth phosphoproteomics. J Proteome Res 2014; 13: 6176–86. [DOI] [PubMed] [Google Scholar]
- Beach TG, Monsell SE, Phillips LE, Kukull W.. Accuracy of the clinical diagnosis of Alzheimer Disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J Neuropathol Exp Neurol 2012; 71: 266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bero AW, Yan P, Roh JH, Cirrito JR, Stewart FR, Raichle ME, et al. Neuronal activity regulates the regional vulnerability to amyloid-β deposition. Nat Neurosci 2011; 14: 750–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertens D, Tijms BM, Scheltens P, Teunissen CE, Visser PJ.. Unbiased estimates of cerebrospinal fluid β-amyloid 1–42 cutoffs in a large memory clinic population. Alzheimers Res Ther 2017; 9: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blennow K, Wallin A.. Clinical heterogeneity of probable Alzheimer's disease. J GeriatrPsychiatry Neurol 1992; 5: 106–13. [DOI] [PubMed] [Google Scholar]
- Bos I, Vos S, Vandenberghe R, Scheltens P, Engelborghs S, Frisoni G, et al. The EMIF-AD Multimodal Biomarker Discovery study: design, methods and cohort characteristics. Alzheimers Res Therapy 2018; 10: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmalm A, Brinkmalm G, Honer WG, Frölich L, Hausner L, Minthon L, et al. SNAP-25 is a promising novel cerebrospinal fluid biomarker for synapse degeneration in Alzheimer’s disease. Mol Neurodegener 2014; 9: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, Yamada KA, Finn MB, Sloviter RS, Bales KR, May PC, et al. Synaptic activity regulates interstitial fluid amyloid-β levels in vivo. Neuron 2005; 48: 913–22. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013; 14: 128–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayon L, Cominetti O, Wojcik J, Galindo AN, Oikonomidi A, Henry H, et al. Proteomes of paired human cerebrospinal fluid and plasma: relation to blood–brain barrier permeability in older adults. J Proteome Res 2019; 18: 1162–74. [DOI] [PubMed] [Google Scholar]
- De Leon MJ, Pirraglia E, Osorio RS, Glodzik L, Saint-Louis L, Kim HJ, et al. The nonlinear relationship between cerebrospinal fluid Aβ42 and tau in preclinical Alzheimer’s disease. PLoS One 2018; 13: e0191240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer G. Diagnosis-independent Alzheimer disease Biomarker signature in cognitively normal elderly people. Arch Neurol 2010; 67: 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejanovic B, Huntley MA, De Mazière A, Meilandt WJ, Wu T, Srinivasan K, et al. Changes in the synaptic proteome in tauopathy and rescue of tau-induced synapse loss by C1q antibodies. Neuron 2018; 100: 1322–36.e7. [DOI] [PubMed] [Google Scholar]
- Díaz-García CM, Mongeon R, Lahmann C, Koveal D, Zucker H, Yellen G.. Neuronal stimulation triggers neuronal glycolysis and not lactate uptake. Cell Metab 2017; 26: 361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, DeKosky ST, Barberger-Gateau P, Cummings J, et al. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS–ADRDA criteria. Lancet Neurol 2007; 6: 734–46. [DOI] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol 2014; 13: 614–29. [DOI] [PubMed] [Google Scholar]
- Duits FH, Brinkmalm G, Teunissen CE, Brinkmalm A, Scheltens P, van der Flier WM, et al. Synaptic proteins in CSF as potential novel biomarkers for prognosis in prodromal Alzheimer’s disease. Alzheimers Res Ther 2018; 10: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O'Reilly PF.. PRSice: polygenic Risk Score software. Bioinformatics 2014; 31: 1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Perrin RJ.. Upcoming candidate cerebrospinal fluid biomarkers of Alzheimer’s disease. Biomarkers Med 2012; 6: 455–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C.. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010; 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham L, Ping L, Dammer E, Duong D, Zhou M, Gearing M, et al.. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer's Disease. bioRxiv 2019; 806752. doi:10.1101/806752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondius DC, van Nierop P, Li KW, Hoozemans JJM, van der Schors RC, van Haastert ES, et al. Profiling the human hippocampal proteome at all pathologic stages of Alzheimer's disease. Alzheimers Dement 2016; 12: 654–68. [DOI] [PubMed] [Google Scholar]
- Hong S, Prokopenko D, Dobricic V, Kilpert F, Bos I, Vos SJB, et al. Genome-wide association study of Alzheimer’s disease CSF biomarkers in the EMIF-AD Multimodal Biomarker Discovery dataset. bioRxiv 2019; 1: 412–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Flory M, Khatoon S, Soininen H, Pirttila T, Lehtovirta M, et al. Subgroups of Alzheimer's disease based on cerebrospinal fluid molecular markers. Ann Neurol 2005; 58: 748–57. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr, Albert MS, Knopman DS, Mckhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer‘s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement 2018; 14: 535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet 2020; 51: 404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ECB, Dammer EB, Duong DM, et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med 2020; 26: 769–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44: W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet 2019; 51: 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B, Masellis M, Freedman M, Stuss DT, Black SE.. Clinical, imaging, and pathological heterogeneity of the Alzheimer's disease syndrome. Alzheimers Res Ther 2013; 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J-C Ibrahim-Verbaas CA Harold D Naj AC Sims R Bellenguez C, . et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013; 45: 1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS.. Learning the parts of objects by non-negative matrix factorization. Nature 1999; 401: 788–91. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017; 541: 481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarouf C, Andacht T, Kokjohn T, Castano E, Sue L, Beach T, et al. Proteomic analysis of Alzheimers disease cerebrospinal fluid from neuropathologically diagnosed subjects. Curr Alzheimer Res 2009; 6: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalinou NK, Noyce AJ, Pinto R, Lindstrom E, Holmén-Larsson J, Holtta M, et al. Identification of candidate cerebrospinal fluid biomarkers in parkinsonism using quantitative proteomics. Parkinsonism Relat Disord 2017; 37: 65–71. [DOI] [PubMed] [Google Scholar]
- Magistretti PJ, Allaman I.. A cellular perspective on brain energy metabolism and functional imaging. Neuron 2015; 86: 883–901. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM.. Clinical diagnosis of Alzheimer‘s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984; 34: 939. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Feldman HM, Lu T, et al. REST and neural gene network dysregulation in iPSC models of Alzheimer's disease. Cell Rep 2019; 26: 1112–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P-F, Savard M, Poirier J, Labonte A, Rosa-Neto P, Weitz TM, et al. Bi-directional association of cerebrospinal fluid immune markers with stage of Alzheimer's disease pathogenesis. J Alzheimers Dis 2018; 63: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD.. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller C, Vrenken H, Jiskoot L, Versteeg A, Barkhof F, Scheltens P, et al. Different patterns of gray matter atrophy in early- and late-onset Alzheimer’s disease. Neurobiol Aging 2013; 34: 2014–2022. [DOI] [PubMed] [Google Scholar]
- Orsini F, De Blasio D, Zangari R, Zanier ER, De Simoni M-G.. Versatility of the complement system in neuroinflammation, neurodegeneration and brain homeostasis. Front Cell Neurosci 2014; 8: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer's disease. Hum Brain Mapp 2015; 36: 4421–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Mucke L.. Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat Rev Neurosci 2016; 17: 777–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrero-Prieto CM, García-Carpintero S, Frontiñán-Rubio J, et al. A comprehensive systematic review of CSF proteins and peptides that define Alzheimer's disease. Clin Proteom 2020; 17: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2018; 47: D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E.. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol 1999; 56: 303. [DOI] [PubMed] [Google Scholar]
- Petit-Turcotte C, Stohl SM, Beffert U, Cohn JS, Aumont N, Tremblay M, et al. Apolipoprotein C-I expression in the brain in Alzheimer's disease. Neurobiol Disease 2001; 8: 953–963. [DOI] [PubMed] [Google Scholar]
- Pooler AM, Phillips EC, Lau DHW, Noble W, Hanger DP.. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep 2013; 14: 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge PG, Hoyt KB, Boehme K, Mukherjee S, Haines JL, Mayeux R, et al. Assessment of the genetic variance of late-onset Alzheimer's disease. Neurobiol Aging 2016; 41: 200.e13–200.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, et al. Amyloid-/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer's disease. J Neurosci 2011; 31: 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS.. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci 2012; 15: 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc Natl Acad Sci USA 1992; 89: 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagare AP, Bell RD, Zlokovic BV.. Neurovascular dysfunction and faulty amyloid-peptide clearance in Alzheimer disease. Cold Spring Harb Perspect Med 2012; 2: a011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer's disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimer's Dement 2010; 6: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens NME, Tijms BM, Koene T, Barkhof F, Teunissen CE, Wolfsgruber S, et al. Cognitive subtypes of probable Alzheimer's disease robustly identified in four cohorts. Alzheimers Dement 2017; 13: 1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehayek E, Eisenberg S.. Mechanisms of inhibition by apolipoprotein C of apolipoprotein E-dependent cellular metabolism of human triglyceride-rich lipoproteins through the low density lipoprotein receptor pathway. J Biol Chem 1991; 266: 18259–18267. [PubMed] [Google Scholar]
- Shachter NS. Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Curr Opin Lipidol 2001; 12: 297–304. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol 2009; 65: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits LL, Pijnenburg YAL, van der Vlies AE, Koedam ELGE, Bouwman FH, Reuling IEW, et al. Early onset APOE E4-negative Alzheimer’s disease patients show faster cognitive decline on non-memory domains. Eur Neuropsychopharmacol 2015; 25: 1010–1017. [DOI] [PubMed] [Google Scholar]
- Spellman DS, Wildsmith KR, Honigberg LA, Tuefferd M, Baker D, Raghavan N, et al. Development and evaluation of a multiplexed mass spectrometry based assay for measuring candidate peptide biomarkers in Alzheimer's Disease Neuroimaging Initiative (ADNI) CSF. Proteomics Clin Appl 2015; 9: 715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer‘s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Sagare AP, Zlokovic BV.. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 2018; 14: 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Kate M Dicks E Visser PJ Van Der Flier WM Teunissen CE Barkhof F, . et al. Atrophy subtypes in prodromal Alzheimer’s disease are associated with cognitive decline. Brain 2018; 141: 3443–56. doi: 10.1093/brain/awy264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms BM, Willemse EAJ, Zwan MD, Mulder SD, Visser PJ, van Berckel BNM, et al. Unbiased approach to counteract upward drift in cerebrospinal fluid amyloid-β 1–42 Analysis Results. Clin Chem 2018; 64: 576–585. [DOI] [PubMed] [Google Scholar]
- Toledo JB, Xie SX, Trojanowski JQ, Shaw LM.. Longitudinal change in CSF Tau and Aβ biomarkers for up to 48 months in ADNI. Acta Neuropathol 2013; 126: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-C, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlies AE, Verwey NA, Bouwman FH, Blankenstein MA, Klein M, Scheltens P, et al. CSF biomarkers in relationship to cognitive profiles in Alzheimer disease. Neurology 2009; 72: 1056–1061. [DOI] [PubMed] [Google Scholar]
- Veerhuis R, Nielsen HM, Tenner AJ.. Complement in the brain. Mol Immunol 2011; 48: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin AK, Blennow K, Zetterberg H, Londos E, Minthon L, Hansson O.. CSF biomarkers predict a more malignant outcome in Alzheimer disease. Neurology 2010; 74: 1531–1537. [DOI] [PubMed] [Google Scholar]
- Webster S, Bradt B, Rogers J, Cooper N.. Aggregation state-dependent activation of the classical complement pathway by the amyloid β peptide. J Neurochem 2002; 69: 388–398. [DOI] [PubMed] [Google Scholar]
- Wesenhagen KEJ, Teunissen CE, Visser PJ, Tijms BM.. Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer's disease: a literature review. Crit Rev Clin Lab Sci 2020; 57: 86–98. [DOI] [PubMed] [Google Scholar]
- Whitwell J, Dickson D, Murray M, Petersen R, Jack C, Josephs K.. MRI in pathologically-defined hippocampal sparing and limbic predominant atypical variants of Alzheimer's disease. Alzheimers Dement 2012; 8: P160–P161. [Google Scholar]
- Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund L-O, et al. Mild cognitive impairment–beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. J Intern Med ; 2004; 256: 240–246. [DOI] [PubMed] [Google Scholar]
- Yamada K, Holth JK, Liao F, Stewart FR, Mahan TE, Jiang H, et al. Neuronal activity regulates extracellular tau in vivo. J Exp Med 2014; 211: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Kanekiyo K.. Blood-brain barrier dysfunction and the pathogenesis of Alzheimer’s Disease. Int J Mol Sci 2017; 18: 1965–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YJ, Baltus AE, Mathew RS, et al. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 2012; 151: 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, Yeo BTT, et al. Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer’s disease. Proc Natl Acad Sci USA 2016; 113: E6535–E6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.