Abstract
Independent Component Analysis-based Automatic Removal of Motion Artifacts (ICA-AROMA; Pruim et al., 2015) is a robust approach to remove brain activity related to head motion within functional magnetic resonance imaging (fMRI) datasets. However, ICA-AROMA requires command line implementation and customized code to batch process large datasets. We developed a cross-platform, open-source graphical user Interface for Batch processing fMRI datasets using ICA-AROMA (INFOBAR). INFOBAR allows a user to search directories, identify appropriate datasets, and batch execute ICA-AROMA. INFOBAR also has additional data processing options and visualization features to support all researchers interested in mitigating head motion artifact in post-processing using ICA-AROMA.
Keywords: ICA-AROMA, GUI, Batch process, Python
Code metadata
| Current Code version | 2 |
| Permanent link to code / repository used of this code version | https://github.com/ElsevierSoftwareX/SOFTX_2020_214 |
| Legal Code License | GPLv3 |
| Code Versioning system used | git |
| Software Code Language used | python |
| Compilation requirements, Operating environments & dependencies | Python 3.5+, ICA-AROMA(https://github.com/maartenmennes/ICA-AROMA), FSL(https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL), python modules: future, Tkinter, threading, statistics, json, bs4, webbrowser |
| If available Link to developer documentation / manual | https://github.com/m-anand/INFOBAR/blob/master/Manual.txt |
| Support email for questions | manand.wk@gmail.com |
Software metadata
| Current software version | 2 |
| Permanent link to executables of this version | https://github.com/ElsevierSoftwareX/SOFTX_2020_214 |
| Legal Software License | GPLv3 |
| Computing platform / Operating System | Linux, macOS, Microsoft Windows (Windows Subsystem for Linux) |
| Installation requirements & dependencies | Python 3.5+, ICA-AROMA(https://github.com/maartenmennes/ICA-AROMA), FSL(https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL), python modules: future, Tkinter, threading, statistics, json, bs4, webbrowser |
| If available Link to user manual - if formally published include a reference to the publication in the reference list | https://github.com/m-anand/INFOBAR/blob/master/Manual.txt |
| Support email for questions | manand.wk@gmail.com |
1. Motivation and significance
Participant head motion is a common problem for resting-state and task-based functional magnetic resonance imaging (fMRI) paradigms. Task-based fMRI paradigms often introduce slight movements of participants’ head as they interact with equipment and complete tasks (e.g., memorizing and verbally repeating words, operating button boxes with their hands). Resultant task-correlated head motion can render the majority of fMRI data unusable as traditional head motion correction techniques (e.g. Nuisance regression [1,2]) may regress out variability that is statistically independent from the motion artifacts (i.e., the blood oxygen level dependent [BOLD] signal) or exclude high motion associated volumes entirely (e.g. spike regression [3] or scrubbing [4]). Further, even if there is no overt task to be completed, such as ‘resting-state’ fMRI, head motion can still be problematic as participants may not remain completely still for the duration of the scan sequence [2,5–7] especially in younger [8] or clinical populations where familiarization training is not as effective [9]. Specifically, head motion during resting-state fMRI compromises the validity of the data by inducing spurious temporal correlations between brain regions [4]. Recent advances in fMRI data processing techniques have resulted in the application of independent component analysis (ICA)-based methods for the identification of motion artifacts. An important blind source separation method, ICA maximizes the statistical independence of its estimated components, sometimes called factors, in order to separate a set of source signals from mixed signals. Utilizing ICA-based methods, a seminal paper validated the automated removal of motion artifacts (ICA-AROMA) as an effective technique to classify source variability originating from motion that preserves BOLD signal source variability, is generalizable across datasets, and when utilized appropriately is not subject to human error [10].
ICA-AROMA involves three consecutive steps that include a probabilistic Independent Component Analysis (ICA), component classification, followed by identification and removal of motion related components through regression-based denoising. Briefly, probabilistic ICA is achieved using Multivariate Exploratory Linear Decomposition into Independent Component’s (MELODIC) within FSL to automatically estimate the number of independent components [11], a single, robust classifier based on four theoretically-motivated temporal and spatial features is employed to identify motion-related components, and linear regression is subsequently is used to remove resultant spurious/motion-related factors within the data. When compared to alternative techniques, including scrubbing, extensive nuisance regression, spike regression, and ICA-based X-noiseifier denoising (ICA-FIX [12]), ICA-AROMA demonstrated superiority as evidenced by high activation sensitivity and consistent resting-state network reproducibility, while preserving the data’s temporal degrees of freedom for increased statistical power within higher-level analyses [13]. ICA-AROMA also provides the advantage of not requiring classifier (re) training and preserves the temporal autocorrelation of the data overcoming key limitations of prior methods.
Currently, ICA-AROMA is implemented as a command line tool whereby users must call a python script to perform motion correction using this technique [14]. However, this command line tool is currently designed to process one imaging dataset at a time which can scale up in effort with large datasets. Without expertise to develop project specific command line solutions for batch processing large datasets through ICA-AROMA, neuroimaging researchers may not be able to manage large datasets due to limitations in the needed programming background. For instance, researchers from various disciplines have begun utilizing gross motor movement paradigms during fMRI to discover neural mechanisms important for lower extremity motor control [15–27]. Due to the gross nature of the movements being studied, these paradigms are often accompanied by head motion artifacts that necessitate correction techniques, such as ICA-AROMA. Even without task implementation, as is the case with standard resting-state fMRI, head motion is common in children and clinical populations., Head motion in this case can lead to spurious findings, and needs to be managed appropriately [4]. Clinical researchers may not be equipped with the programming skills to batch process large datasets, resulting in significant manual and inefficient effort being invested in this one step to manage movement artifacts. Further, some researchers prefer graphical user interfaces (GUIs) over command line implementation, especially considering that users commonly perform the necessary pre-and post-ICA-AROMA processing steps using a GUI within the fMRI Software Library (FSL: The Oxford Centre for Functional MRI of the Brain, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom) [28]. Specifically, the intermediate step to perform motion artifact removal using ICA-AROMA requires the use of command line, which disrupts workflow, and can be difficult for investigators that do not have the needed coding expertise. Currently, no such open-source cross-platform GUI for ICA-AROMA exists. The Interface for batch processing ICA-AROMA (INFOBAR) provides a standardized open source, cross-platform, user-friendly interface for identifying and batch-processing large datasets, along with quality checking the data and aggregating motion stats in a useful and comprehensive format.
2. Software description
INFOBAR was developed in Python (www.python.org) to visualize and batch process datasets with ICA-AROMA. The GUI uses Tkinter, the native TCL/TK toolkit in python, to ensure pre-built cross platform compatibility across Linux, Windows sub-system for Linux (WSL), and macOS. It should be noted that since ICA-AROMA (underlying program of INFOBAR) relies on FSL which runs natively in Linux/macOS, these are the recommended platforms for INFOBAR.
The software incorporates search and filter functions to identify datasets and individual dataset status tracking to let users identify the processing status of their database. It also displays individual and group’s cumulative motion stats.
2.1. Software architecture
INFOBAR is divided into two windows, a main window for search and execution tasks and a viewer for visualization of outputs from pre- and post-processed datasets (generated by FSL). Specifically, after pre-processing (i.e., prior to ICA-AROMA), Infobar shows the output from MCFLIRT, which includes the rotation, translation and displacement of the brain during fMRI. Following post-processing (i.e., after ICA-AROMA and first-level analysis), Infobar shows the model fit to the peak voxel activity and the primary zstat lightbox image for further quality control evaluation. The main window as shown in Fig. 1 has a control panel and a results table and a status-bar which provides the users with useful status updates. A settings menu entry allows the users to set options for processing through ICA-AROMA as shown in Fig. 2.
Fig. 1.
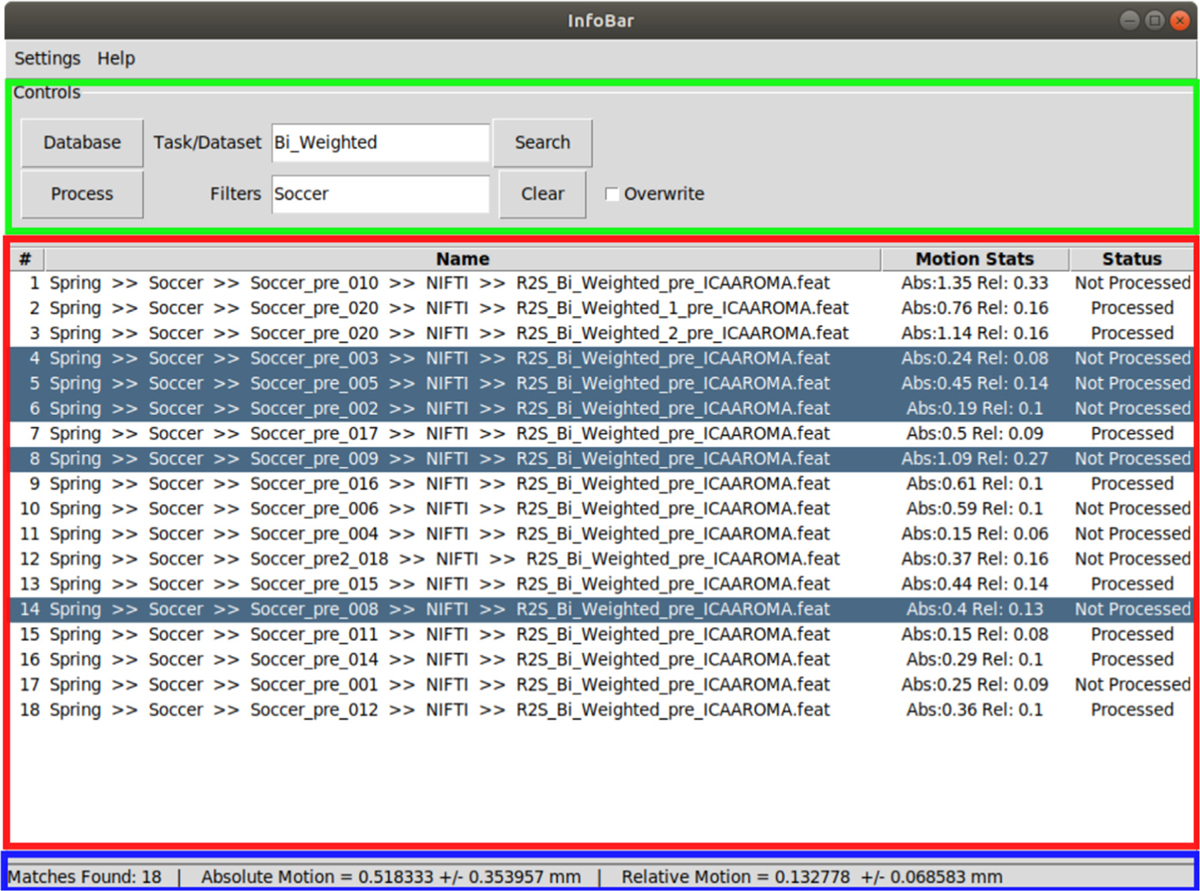
The main window of the GUI showing the control panel on the top (green), results table in the middle (red) and status bar at the bottom (blue).
Fig. 2.
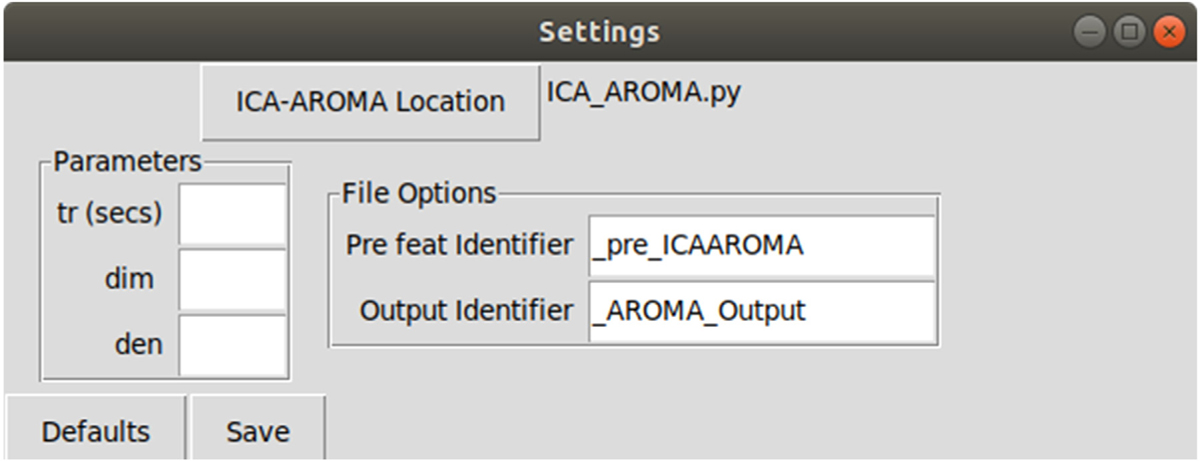
Settings menu allows the user to set path to ICA-AROMA, and configure various options provided by the program.
2.2. Preprocessing
Datasets need to be preprocessed through FSL before removing motion artifacts which involve the following steps, as reported in [10]:
Head movement correction by volume-realignment to the middle volume using MCFLIRT.
Global 4D mean intensity normalization.
Spatial smoothing (e.g., 6 mm full width at half maximum [FWHM]).
Importantly, no temporal filtering should be applied at this stage of processing.
2.3. Software functionality
Operation of INFOBAR is divided into two steps. The first is generating the process queue. The user starts off by selecting the database location (by clicking “Database” button). The database location would contain the .feat folders generated by FSL. The search is agnostic of the hierarchy of dataset locations within a given database. The user can choose to find all datasets in the database (by clicking “Search”) and display them in the results table. If the .feat folders associated with datasets have characteristic identifiers or names, those can be entered (into the text-field “Task/Dataset”). Datasets can be further filtered at directory levels by entering filters separated by a semicolon (;) (in the text-field “Filters”). Datasets can be deleted from the queue by selecting them and pressing ‘d’. These features are designed to help reduce large datasets into manageable units as needed.
The results table shows the location of each dataset in the database, the absolute and relative motion stats (from FSL’s pre-processed files) [29] and a status indicating the processing status of the dataset. The software labels datasets that have already been processed through ICA-AROMA as “Processed” and others as “Not Processed”. The mean and standard deviation of the absolute and relative motion for all datasets displayed in the table are presented in the status bar. The datasets in the table form the process queue.
The second step is batch processing the queue with INFOBAR. Clicking ‘Process’ runs the datasets through ICA-AROMA and places the outputs in the dataset’s directory. The user can also select specific datasets to be processed. If no datasets are selected, then all datasets in the data table will be processed. If previously processed datasets are present, the program will skip them. If the user wishes to process them again, then the “Overwrite” option can be selected. Processing is multithreaded, thereby substantially saving processing time.
ICA-AROMA provides options to change the default TR, dimensionality reductions and de-noising techniques. The functionality is available through a settings panel where the following user defined settings can be set:
ICA-AROMA location: Select the location of the ICA_AROMA.py file from the original program. Default location is installation directory.
- Parameters for ICA-AROMA [14]:
- tr (secs): TR in seconds. If this is not specified, the TR will be extracted from the header of the fMRI file using ‘fslinfo’. In that case, make sure the TR in the header is correct!
- dim: Dimensionality reduction into a defined number of dimensions when running MELODIC (default is 0; automatic estimation)
- den: Type of denoising strategy (default is nonaggr). Options include:
- no: only classification, no denoising
- nonaggr: non-aggressive denoising, i.e. partial component regression (default)
- aggr: aggressive denoising, i.e. full component regression
- both: both aggressive and non-aggressive de-noising (two outputs)
- File Options:
- Pre feat Identifier: If the preprocessed files are stored with a unique keyword identifier for record keeping, this keyword can be specified here. This keyword will be replaced by the Output Identifier keyword. If the keyword is not found, the folder name will simply be augmented with the Output Identifier. Default value is ‘_pre_ICAAROMA’
- Output Identifier: Keyword to replace Pre feat identifier. Default value is ‘_AROMA_Output’.
Settings tab also allows the user to select the location of the ICA-AROMA program file for function call. The settings are saved in a JSON file. The settings panel is shown in Fig. 2.
INFOBAR incorporates a viewer that allows the user to see motion graphs generated by FSL in one single location. Left clicking a preprocessed dataset in the table displays the MCFLIRT translation, rotation, and mean displacement graphs in the Viewer. Right/middle-clicking or pressing ‘w’ on a selected dataset after processing through INFOBAR allows the user to scroll through and view special maps of the independent components associated with motion identified by the algorithm. Further, when the first-level analysis has been completed in FSL on the processed data, the new. feat directory will be labeled in INFOBAR as post-processed and right/middle-clicking/ pressing ‘w’ the dataset displays the z-stat and model fit. Sample views are demonstrated in Fig. 3.
Fig. 3.
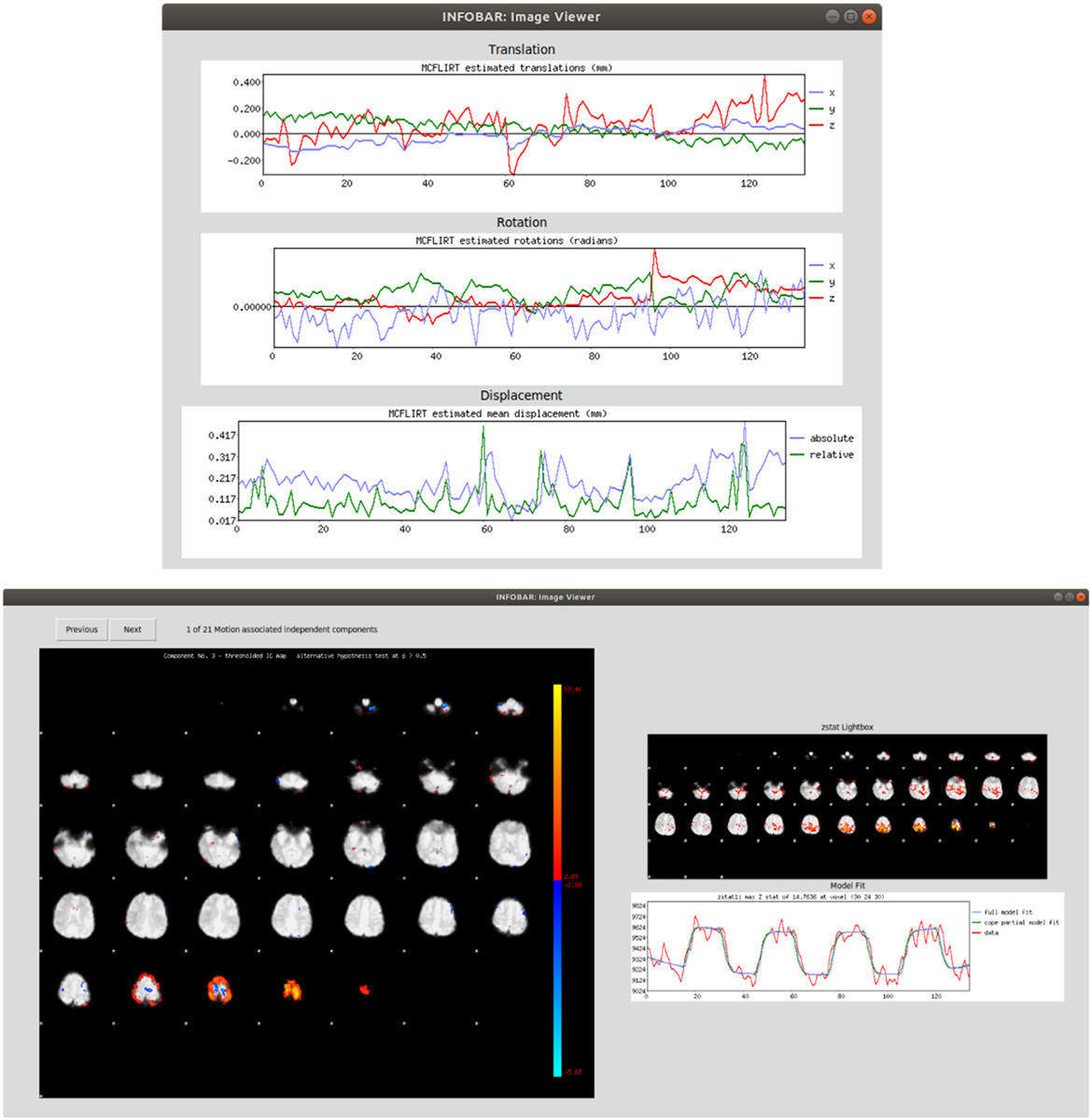
(Top) Left clicking a dataset shows the translation, rotation and displacement graphs generated through FSL, (Bottom) Right/middle-clicking or pressing ‘w’ on the dataset shows independent components associated with motion, as well as post-processed primary zstat and model fit curves, if data has been post-processed.
3. Illustrative example
A team of investigators has collected fMRI data on a large cohort of subjects from several sports and across different semesters of a school year. The fMRI experiment also involved multiple tasks resulting in multiple datasets per subject. A trained neuroimaging analyst completes the appropriate preprocessing steps within FSL to prepare the data for ICA-AROMA. Using INFOBAR, any member on the team, irrespective of their familiarity with command line tools can further process this data through ICA-AROMA. The user can choose to load the data for only one semester or one task (e.g. Bi_Weighted) across semesters for just one sport (e.g. Soccer). Movement statistics shown in the status bar can be tracked across different sub-datasets, to compare how one task might differ from another, or to compare how one subgroup may differ from another subgroup. A quick quality assurance check of the data can be performed by clicking through the datasets to visualize the motion graphs in the viewer. Clicking ‘Process’ will pass the queue through the ICA-AROMA script. Following the completion of ICA-AROMA, the researcher would then complete post-processing in FSL (including Nuisance regression: WM, CSF & linear trend, and high-pass filtering). Using INFOBAR, the user can now view the model fit to the peak voxel activity and the primary zstat lightbox image for further quality control evaluation. For example, peak voxel activity in a region without theoretical or physiological support (e.g., near the skull) and/or if model fit was poor, could be indicative that the motion parameters utilized were inappropriate. In these cases, the user can choose to reprocess the data through ICA-AROMA using a different set of processing parameters (e.g. aggressive denoising) by changing the ‘denoise’ setting in the settings panel to ‘aggr’, and repeating the previous steps. The process flow has been visualized in Fig. 4.
Fig. 4.
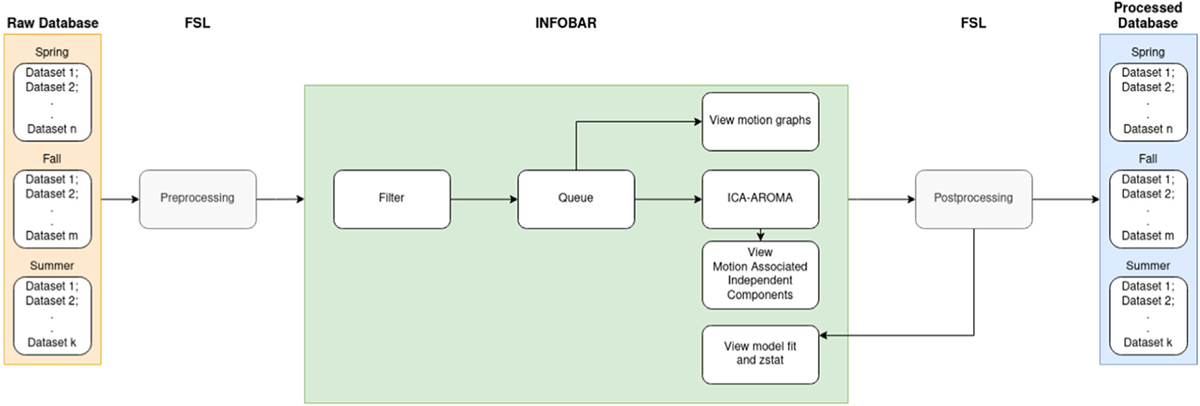
Process flow chart representing the example case.
4. Impact
Previously, a researcher would have to use the command line method to perform these tasks. This would require the user to manually enter the details per dataset and process them individually. Further, to find the motion stats and to visualize pre-processed motion graphs and/or post-processed zstat and model fit data, the user would have to access the reports generated by FSL individually by accessing the respective directories. INFOBAR provides a central location to perform all these tasks. It is useful for any researcher who wishes to integrate ICA-AROMA into their fMRI analysis pipelines to manage motion artifacts in neuroimaging data. Since ICA-AROMA is applied as an intermediate step in an analysis that would commonly be completed in FSL, it provides a user-friendly GUI to reduce workflow disruption. Its processing tracking will be useful for record keeping on the status of datasets. The ability to track motion statistics for large groups will also prevent significant manual effort and chances of error. Finally, the ability to quickly visualize the pre- and post-processed data’s images from a central location will further streamline the process.
5. Conclusion
INFOBAR provides a convenient implementation to remove head motion artifacts using ICA-AROMA. It supports processing large datasets through ICA-AROMA with a cross-platform, point and click operation while equipping the user with motion stats and data quality assurance capabilities. This GUI implementation of ICA-AROMA brings INFOBAR at parity in terms of user friendliness with FSL, one of the most utilized GUI tools for brain imaging data processing. The ability to quality check data from a single location can facilitate optimized methods with more efficient analyses without required knowledge of command line implementation for batch processing.
Funding sources
The authors would like to acknowledge funding support from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases, USA grants U01 AR067997 and R01 AR076153.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Friston KJ, Williams S, Howard R, Frackowiak RSJ, Turner R. Magn Reson Med 1996;35:346–55. [DOI] [PubMed] [Google Scholar]
- [2].Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, et al. Neuroimage 2013;64:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lemieux L, Salek-Haddadi A, Lund TE, Laufs H, Carmichael D. Magn Reson Imaging 2007;25:894–901. [DOI] [PubMed] [Google Scholar]
- [4].Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Neuroimage 2012;59:2142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maknojia S, Churchill NW, Schweizer TA, Graham SJ. Front Neurosci 2019;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Power JD, Schlaggar BL, Petersen SE. Neuroimage 2015;105:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Parkes L, Fulcher B, Yücel M, Fornito A. Neuroimage 2018;171:415–36. [DOI] [PubMed] [Google Scholar]
- [8].Yuan W, Altaye M, Ret J, Schmithorst V, Byars AW, Plante E, et al. Hum Brain Mapp 2009;30:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li J, Li Q, Dai X, Li J, Zhang X. Medicine (Baltimore) 2019;98:e14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pruim RHR, Mennes M, van Rooij D, Llera A, Buitelaar JK, Beckmann CF. Neuroimage 2015;112:267–77. [DOI] [PubMed] [Google Scholar]
- [11].Beckmann CF, Smith SM. IEEE Trans Med Imaging 2004;23:137–52. [DOI] [PubMed] [Google Scholar]
- [12].Salimi-Khorshidi G, Douaud G, Beckmann CF, Glasser MF, Griffanti L, Smith SM. Neuroimage 2014;90:449–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pruim RHR, Mennes M, Buitelaar JK, Beckmann CF. Neuroimage 2015;112:278–87. [DOI] [PubMed] [Google Scholar]
- [14].Mennes M, ICA-AROMA, 2015, GitHub repository: https://github.com/maartenmennes/ICA-AROMA.
- [15].Grooms DR, Diekfuss JA, Ellis JD, Yuan W, Dudley J, Foss KDB, et al. J Neuroimaging 2019;29:580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fontes EB, Okano AH, De Guio F, Schabort EJ, Min LL, Basset FA, et al. Br J Sports Med 2015;49:556–60. [DOI] [PubMed] [Google Scholar]
- [17].Newton JM, Dong Y, Hidler J, Plummer-D’Amato P, Marehbian J, Albistegui-DuBois RM, et al. Neuroimage 2008;43:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hollnagel C, Brügger M, Vallery H, Wolf P, Dietz V, Kollias S, et al. J Neurosci Methods 2011;201:124–30. [DOI] [PubMed] [Google Scholar]
- [19].Raisbeck LD, Diekfuss JA, Grooms DR, Schmitz R. J Sport Rehabil 2020;29:441–7. [DOI] [PubMed] [Google Scholar]
- [20].Grooms DR, Kiefer AW, Riley MA, Ellis JD, Thomas S, Kitchen K, et al. J Sport Rehabil 2018;27:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grooms DR, Page SJ, Nichols-Larsen DS, Chaudhari AMW, White SE, Onate JA. J Orthop Sports Phys Ther 2017;47:180–9. [DOI] [PubMed] [Google Scholar]
- [22].Grooms DR, Page SJ, Onate JA. J Athl Train 2015;50:1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lepley AS, Grooms DR, Burland JP, Davi SM, Kinsella-Shaw JM, Lepley LK. Exp Brain Res 2019;237:1267–78. [DOI] [PubMed] [Google Scholar]
- [24].Ikeda T, Matsushita A, Saotome K, Hasegawa Y, Sankai Y. In: IEEE int. conf. intell. robot. syst.; 2012, p. 311–6. [Google Scholar]
- [25].Thijs Y, Vingerhoets G, Pattyn E, Rombaut L, Witvrouw E. Knee surgery. Sport Traumatol Arthrosc 2010;18:1145–9. [DOI] [PubMed] [Google Scholar]
- [26].Gilat M, Bell PT, Ehgoetz Martens KA, Georgiades MJ, Hall JM, Walton CC, et al. Neuroimage 2017;152:207–20. [DOI] [PubMed] [Google Scholar]
- [27].Xiao X, Lin Q, Lo WL, Mao YR, Shi XC, Cates RS, et al. Behav Neurol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, et al. Neuroimage 2004;23:S208–19. [DOI] [PubMed] [Google Scholar]
- [29].Sacca V, Sarica A, Novellino F, Barone S, Tallarico T, Filippelli E, et al. 2018;12:58–61. [Google Scholar]


