Abstract
The mucosal barrier in combination with innate immune system are the first line of defense against luminal bacteria at the intestinal mucosa. Dysfunction of the mucus layer and bacterial infiltration are linked to tissue inflammation and disease. To study host-bacterial interactions at the mucosal interface, we created an experimental model that contains luminal space, a mucus layer, an epithelial layer, and suspended immune cells. Reconstituted PSIM formed an 880 ± 230 μm thick gel layer and had a porous structure. In presence of mucus, 7-fold less probiotic and non-motile VSL#3 bacteria transmigrated across the epithelial barrier compared to no mucus. The higher bacterial transmigration caused immune cell differentiation and increased the concentration of IL-8 and TNF-α (P < 0.01). Surprisingly, the mucus layer increased transmigration of pathogenic Salmonella and increased secretion of TNF-α and IL-8 (P < 0.05). Non-motile, flagella knockout Salmonella had lower transmigration and caused lower IL-8 and TNF-α secretion (P < 0.05). These results demonstrate that motility enables pathogenic bacteria to cross the mucus and epithelial layers, which could lead to infection. Using an in vitro co-culture platform to understand the interactions of bacteria with the intestinal mucosa has the potential to improve treatment of intestinal diseases.
Keywords: Mucus barrier, Motility, Flagella, Inflammation, In vitro mucosa model
1. INTRODUCTION
In the intestines, three major components mediate the host-response to pathogens and the gut microbiota: the mucosal barrier, the epithelial layer and tissue-resident immune cells (Turner, 2009). The interactions of these components in pathophysiological conditions are poorly understood. Two major disease states result from disruption of these protective mechanisms, acute enteropathogenic infections, and chronic inflammatory bowel diseases (IBD) (König et al., 2016; McGuckin et al., 2009). The mucus layer is the first line of defense, which plays a critical role in the prevention of enteropathogenic infections. In the healthy colon, the secreted mucus layer that covers the entire epithelium minimizes interactions of commensal bacteria with host cells (M. E. Johansson et al., 2008; M. E. V. Johansson et al., 2011). Dysfunction of the mucus layer and bacterial infiltration are linked to tissue inflammation and disease (Alipour et al., 2016; M. E. Johansson et al., 2010; Sovran et al., 2016; Swidsinski et al., 2005). During enteropathogenic infections, bacteria can penetrate intestinal regions of disrupted mucus (Croxen et al., 2013; Hering et al., 2015; Marteyn et al., 2012), but there is recent evidence that bacteria can also penetrate regions with intact mucus (Furter et al., 2019). A better understanding of the interaction between microbes and the protective host mechanisms could lead to improved treatment for intestinal diseases.
The responses of epithelial and immune cells to bacterial infiltration are difficult to decouple from in vivo measurements and are poorly understood. The epithelial barrier and cells of the innate immune system collectively function as the subsequent layers of protection after the mucus layer (Sansonetti, 2004). In the colon, tight junctions between epithelial cells prevents transmigration of pathogens into underlying tissue (Lee, 2015). Colonic epithelial cells produce chemokines, such as interleukin-8 (IL-8), that attract neutrophils and other effector cells of the innate and adaptive immune system (Kucharzik et al., 2005). Monocytes, which are innate immune cells, detect pathogens directly as well as respond to epithelial derived signals. One response to these stimulants is differentiation into macrophages and dendritic cells (Bain et al., 2014; Iliev et al., 2009). LPS and flagellin are common pathogen associated molecular patterns (PAMPs) that bind to toll-like receptors (TLR4 and TLR5) and activate the downstream pathways that cause pro-inflammatory cytokine secretion (Hayashi et al., 2001; Hoshino et al., 1999; Leon et al., 2008). Immune-cell-derived tumor necrosis factor (TNF-α) is a cause of systemic inflammation. Both TNF-α and IL-8 levels are significantly upregulated in the intestines of patients with IBD (Mazzucchelli et al., 1994; Neurath, 2014). Blocking IL-8 has the potential to prevent neutrophil infiltration, which is a major source of inflammation in the intestines, and is therefore an important therapeutic target (Natsui et al., 1997; Zhou & Liu, 2017).
A common enteropathogenic infection is Salmonellosis (Crum-Cianflone, 2008; Eng et al., 2015). Salmonella enterica subsp. enterica serovar Typhimurium is a foodborne pathogen that causes diarrhea and can often be fatal (Eng et al., 2015). In humans, infection with Salmonella or other enteropathogenic bacteria is linked to higher incidence of IBD (Garcia Rodriguez et al., 2006; Gradel et al., 2009; Porter et al., 2008). Studies have shown that attachment of Salmonella to mucus is a critical step for infection (Hallstrom & McCormick, 2011; McCormick et al., 1988; Nevola et al., 1987; Vimal et al., 2000). In mouse models, Furter et al. showed that Salmonella infects the colonic epithelium even when covered with a thick layer of mucus (Furter et al., 2019). This recent observation contradicts the common notion that the mucus barrier serves as a sticky net to trap the invading luminal microbes (Linden et al., 2008). How Salmonella penetrate the mucus barrier and the effects of this penetration on epithelial and immune cells is not fully understood.
Murine models are the most common method to study the gastrointestinal biology. However, studying the complex interactions between luminal microbes, tissue immune cells and the mucosal barrier in living animals is challenging. Alternately, in vitro platforms are well suited to studying these complex interactions. For example, three-dimensional biomaterial platforms have been developed to mimic the morphology of the epithelial layer (Chen et al., 2015; Wang et al., 2017). Epithelial and immune cell interactions are commonly measured in transwell-based platforms (Leonard et al., 2010; Noel et al., 2017). Flow based microfluidic platforms have enabled long-term co-culture of gut-representative microbial communities (Kim et al., 2012; Marzorati et al., 2014; Shah et al., 2016). Incorporation of mechanical forces within microfluidic platforms induced three-dimensional villus differentiation and allowed quantification of communication between bacteria-epithelial-immune cell compartments (Kim et al., 2012; Kim & Ingber, 2013; Kim et al., 2016).
The inclusion of a mucus layer is critical because of its role in mediating bacteria-epithelial cell interactions. Some cultured epithelial cells secrete mucus and form mucus layers that enable the quantification of drug and molecular interactions with mucus (Behrens et al., 2001; Mahler et al., 2009). Two recent studies demonstrated thick mucus layer secretion from primary human colonic epithelial cells using air-liquid interface in transwell culture or colon-on-a-chip microfluidic culture (Sontheimer-Phelps et al., 2019; Wang et al., 2019). Most cultured cell lines, however, do not generate a layer matching physiological thickness of colonic mucus. Due to insufficient mucus secretion from cultured human epithelial cells, porcine gastric mucin (PGM) has been used as human mucin substitute (Boegh et al., 2014; Boegh et al., 2015). PGM has been used to recreate the intestinal mucus layer by mixing with agar and forming a solid interface with luminal solution that supported bacterial attachment and growth (Marzorati et al., 2014; Shah et al., 2016). However, stomach mucus has different physicochemical properties than intestinal mucus and commercially available PGM does not undergo sol-gel transitions (Celli et al., 2005). An alternative to PGM is purified intestinal mucus. It has been shown that MUC2, which is purified from intestinal mucus, has antiviral activity and attenuates the virulence of pathogenic bacteria (Lieleg et al., 2012; Wheeler et al., 2019). We have developed a method to extract porcine small intestinal mucus (PSIM). Reconstituted PSIM undergoes sol-gel transitions with changes in pH and ion concentrations, similar to native intestinal mucus (Sharma et al., 2020). Low pH caused mucus aggregation and reduced bacterial transport, and moderate calcium concentrations formed microscopic aggregates that impeded molecular diffusion.
Here, we demonstrate a model of intestinal mucosa to quantify the role of mucus and epithelial barrier in regulating the host innate immune responses to probiotic and pathogenic bacteria. We hypothesize that mucus prevents the penetration of bacteria through the intestinal lining and that the motility of pathogenic bacteria affects their penetration and immune response. To decouple the host-microbe interactions, mucus and epithelial layers were developed in a physiological form on transwell membranes. The porous transwell membrane enabled molecular communication between human epithelial (HT-29) cells and human monocytic (THP-1) cells. The hypotheses were tested with commensal VSL#3 bacteria, wild-type SL1344 Salmonella, and a non-motile flhDC knockout strain of Salmonella. Bacterial penetration, immune cell differentiation, and cytokine release were quantified using fluorescent immunostaining, microscopic imaging, and ELISA. The effects of a mucus layer on bacterial transmigration and molecular diffusion were quantified using fluorescent spectroscopy. This intestinal mucosa model enabled the quantification of the differences in the integrity of mucosal barrier compared to epithelial barrier alone when challenged with live bacteria. This mucosal barrier model enabled the measurement of bacterial penetration, immune cell differentiation and cytokine release. Understanding the mechanisms of action of probiotics or pathogens when in contact with the mucosal barrier will lead to development of therapies for bacterial infections and immunological diseases of the intestine.
2. RESULTS
2.1. PSIM formed a gel layer on top of epithelial cells without affecting cell viability.
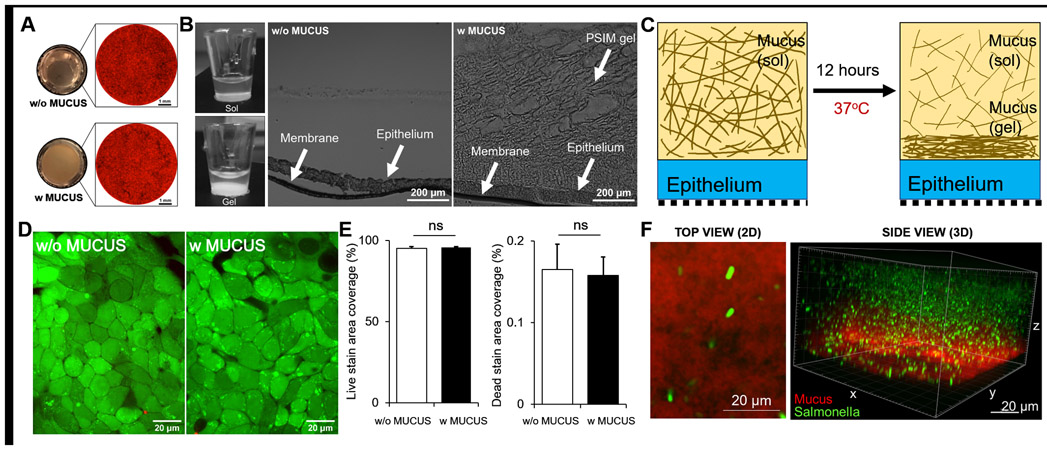
We have developed a method to mechanistically study the host-microbe interactions at the mucosal interface of the intestines. A mucus gel layer was reconstituted on top of epithelial cells growing on transwell membranes (Fig. 1). Human colonic epithelial cells (HT-29) genetically labeled to express red fluorescent protein (RFP) covered the entire surface of the transwell membrane and coverage was not affected by the presence of PSIM (Fig. 1A). PSIM (20 mg/ml) was added to the epithelial cells and after 12 hours it formed an opaque gel layer in situ that was structurally porous (Fig. 1B). This 12-hour incubation at 37 °C reduced the thickness of the PSIM layer (Fig. 1C). Initially, the total height of solubilized PSIM (100 μl/well) layered on top of epithelial cells was approximately 2.7 mm. Final thickness of the gel layer formed was 880 ± 230 μm (Fig. 1B). The PSIM gel layer formation was robust and the structure was maintained after cryo-freezing and histological sectioning. PSIM formed a tightly bound gel layer and did not affect the viability of HT-29 cells (Fig. 1D). No significant difference was observed in the live (green) and dead (red) cell area coverage between the presence and absence of a PSIM layer (Fig. 1E). DAPI (4′,6-diamidino-2-phenylindole) staining of PSIM (red) showed that the PSIM gel layer has a fibrous structure (Fig. 1F). When added, fluorescent wild type Salmonella (green) were suspended in the three-dimensional mucus matrix (Fig. 1F).
Figure 1. Biocompatibility of epithelial and mucus gel layers formed on a transwell membrane.
(A) Macroscopic, bottom-view images of the translucent membrane without mucus (top left) and the opaque mucus layer after gel formation (bottom left). Tiled microscopic image of the transwell membrane with RFP-expressing HT-29 cells covering the entire membrane without (top right) and with a mucus layer (bottom right). (B) Side view of transwell inserts demonstrating sol (left, top) to gel (left, bottom) transition of PSIM at 0 and 12 hours. Microscopic image of a 20 μm thick histological cross-section of the transwell membrane covered with HT-29 cell layer (middle) and PSIM gel layer formed on top (right). (C) A mucus gel layer formed 12 hours after addition of solubilized PSIM (20 mg/ml) and incubation at 37 °C. (D) Live (Green) and Dead (Red) cell staining of the epithelial cell layer on a transwell membrane. (E) Quantified area covered by GFP positive and RFP positive signals showed no significant difference with or without mucus (n = 3). (F) Confocal microscope image of a DAPI-stained PSIM gel layer (red) with embedded GFP-expressing Salmonella (green).
2.2. PSIM is not immunogenic to epithelial or immune cells.
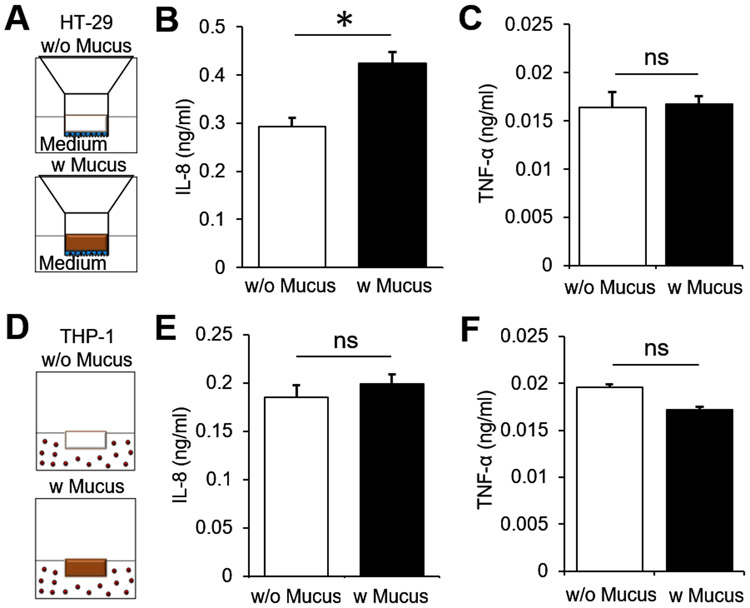
To test its immunogenicity with human-derived cell lines, reconstituted PSIM was directly added to the monocultured HT-29 and THP-1 cells. PSIM mostly did not induce a pro-inflammatory cytokine and chemokine response from HT-29 and THP-1 cells. HT-29 cells were cultured on the apical side of 24-well transwell inserts (Fig. 2A) and IL-8 and TNF-α were quantified in the basolateral medium. HT-29 cells secreted a small but significantly higher amount of IL-8 in response to PSIM when compared to HT-29 cells alone (P < 0.05, Fig. 2B). There was no difference in the amount of TNF-α secreted from HT-29 with and without PSIM application (Fig. 2C). THP-1 cells were cultured in suspension in 24 well plates and IL-8 and TNF-α quantified in the culture medium (Fig. 2D). PSIM did not induce IL-8 or TNF-α secretion from THP-1 cells compared to cells without PSIM (Fig. 2E, F).
Figure 2. Immunological response of epithelial and immune cells to PSIM.
(A) Schematic depiction of the transwell insert placed in 24 multi well plates. HT-29 cells (blue) were cultured on the apical side of the transwell membrane and cell culture medium was added to the apical (100 μl) and basolateral (600 μl) side of the cells (w/o mucus). Reconstituted mucus at 20 mg/ml concentration (brown, 100 μl) layered on top of the epithelial cells (w mucus). (B) THP-1 cells cultured in 24 multi well plates and cell culture media (w/o mucus, 100 μl) or reconstituted mucus (w/ mucus, 100 μl) added directly to the culture medium (600 μl). (C) Mucus increased chemokine (IL-8) secretion from HT-29 cells (*, P < 0.05; n = 3) and (D) no change in cytokine (TNF-α) secretion. Mucus did not cause a significant change in (E) IL-8 or (F) TNF-α concentration in THP-1 culture medium (n = 3).
2.3. Immunological responses from epithelial and immune cells are stimulant dependent.
When cultured independently, HT-29 and THP-1 cells respond differentially after direct contact with bacterial antigen LPS, a set of 8 live probiotic bacteria (VSL#3), and pathogenic bacteria Salmonella. Intestinal epithelial cells have regular incidence of bacterial contact, but immune cells only come in direct contact with live bacteria during bacterial infection events. The amounts of pro-inflammatory chemokine (IL-8) and cytokine (TNF-α) secreted from epithelial and immune cells were stimulant dependent. Salmonella induced a robust chemokine response in HT-29 cells, causing approximately 24-fold higher IL-8 secretion compared to unstimulated cells (P < 0.01, Fig. 3A). Direct contact of HT-29 cells with LPS or VSL#3 did not increase IL-8 secretion (Fig. 3A). LPS-stimulated THP-1 cells secreted 61-fold more IL-8 compared to unstimulated cells (P < 0.01, Fig. 3B). Direct contact with VSL#3 and Salmonella each caused an approximately 30-fold increase in IL-8 secretion from THP-1 cells compared to bacteria-free controls (P < 0.01, Fig. 3B).
Figure 3. Stimulation of HT-29 and THP-1 monocultures by LPS and live bacteria.
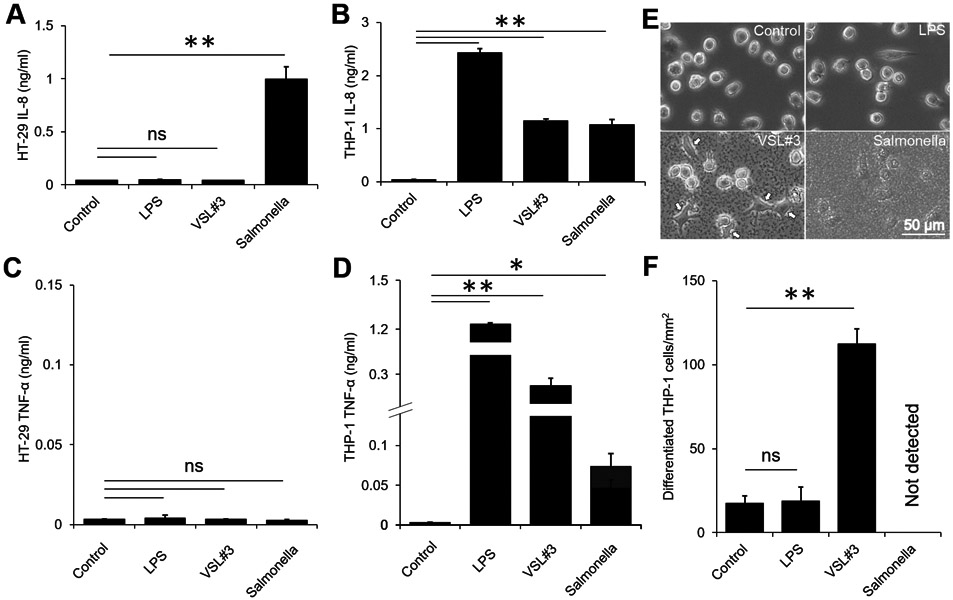
(A) IL-8 secretion from HT-29 cells was significantly increased by direct contact with Salmonella (**, P < 0.01; n = 3). LPS (10 μg/ml) and VSL#3 caused no significant change in IL-8 secretion. (B) LPS and direct contact with VSL#3 and Salmonella caused IL-8 secretion from THP-1 cells (**, P < 0.01; n = 3). (C) LPS, VSL#3 or Salmonella did not cause any TNF-α secretion from HT-29 cells. (D) LPS and direct contact with VSL#3 and Salmonella caused TNF-α secretion from THP-1 cells (*, P < 0.05; **, P < 0.01; n = 3). (E) Direct contact with VSL#3 caused a change in THP-1 cell morphology (arrows). LPS and Salmonella did not change THP-1 morphology. Images were taken 12 hours after adding a stimulant. (F) The number of differentiated THP-1 cells per squared millimeter was significantly higher with VSl#3 compared to controls (**, P < 0.01; n = 3).
There was no increase in the level of TNF-α secreted by HT-29 cells after direct contact with LPS or live bacteria compared to unstimulated cells (Fig. 3C). TNF-α secretion from THP-1 cells was significantly increased when cultured with LPS or live bacteria (Fig. 3D). LPS induced the highest TNF-α secretion and the level of TNF-α was 342-fold higher compared to unstimulated THP-1 cells (P < 0.01, Fig. 3D). Direct contact of THP-1 cells with VSL#3 (P < 0.01) and Salmonella (P < 0.05) caused 77 and 20-fold increases in TNF-α, respectively (Fig. 3D).
The morphology of THP-1 cells changed significantly when cultured in direct contact with VSL#3 (Fig. 3E). Change in morphology indicates the differentiation into macrophage and dendritic-like cells. At 12 hours post infection with Salmonella, THP-1 cell differentiation or morphology change was not detectable, and the cells were mostly round (Fig. 3E). This response was similar to the stimulation with LPS that also did not induce a change in morphology (Fig. 3E). THP-1 cells stimulated with VSL#3 induced approximately 6-fold increase in the number of differentiated cells per millimeter squared area (P < 0.01, Fig. 3F). Collectively, the results show that epithelial and immune cell activation and immunological responses are stimulant dependent.
2.4. A PSIM gel layer prevents LPS induced immunogenic responses.
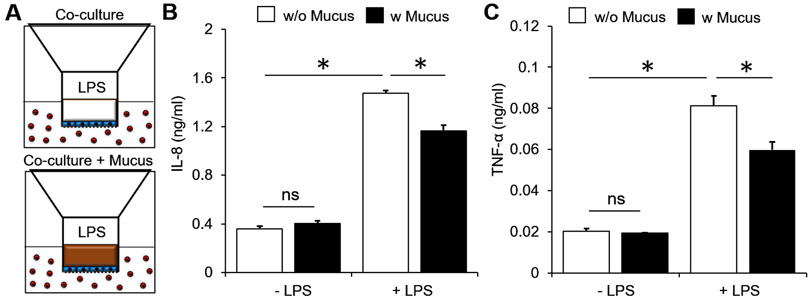
In the co-culture platform, we hypothesized that the mucus layer would act as a physical barrier for molecular diffusion. Mucus layers prevented pro-inflammatory chemokine and cytokine secretion by human epithelial and immune cell co-culture in response to bacterial products. To test this hypothesis, epithelial-immune cell co-culture was used with and without mucus (Fig. 4A). Co-culture of the HT-29 and THP-1 cells did not affect the basolateral levels of IL-8 and TNF-α when compared to monocultures (compare without mucus and without LPS in Fig. 4B, C to Fig. 2B, C, E, F). The addition of PSIM on the apical side of the epithelial layer caused no change in the levels of IL-8 and TNF-α (Fig. 4B, C). LPS increased the concentrations of both IL-8 (P < 0.05, Fig. 4B) and TNF-α (P < 0.05, Fig. 4C) 4-fold in the absence of a PSIM layer. The presence of a mucus layer significantly reduced the levels of IL-8 (P < 0.05, Fig. 4B) and TNF-α (P < 0.05, Fig. 4C) after LPS stimulation. The total amounts of IL-8 and TNF-α from the co-cultures were significantly lower compared to direct stimulation of THP-1 cells by LPS (P < 0.05, compare Fig. 4B, C to Fig. 3B, D). Collectively, the results show that the epithelial barrier attenuates immune cell stimulation by preventing direct contact of immune cells with bacteria derived molecules, and the mucus layer further increases this barrier function.
Figure 4. Barrier function of PSIM layer to molecular diffusion.
(A) HT-29 cells (blue) on the apical side of the membrane co-cultured with THP-1 cells (red) in 24 multi-well plates in the absence (top) and presence (bottom) of a PSIM layer (brown). (B,C) The concentrations of IL-8 (B) and TNF-α (C) from the co-cultures did not change by adding a PSIM layer, significantly increased by adding LPS to the apical side without mucus, and was significantly attenuated by adding PSIM layer prior to LPS stimulation (*, P < 0.05; n = 3).
2.5. A PSIM gel layer is a barrier to migration of live probiotic bacteria.
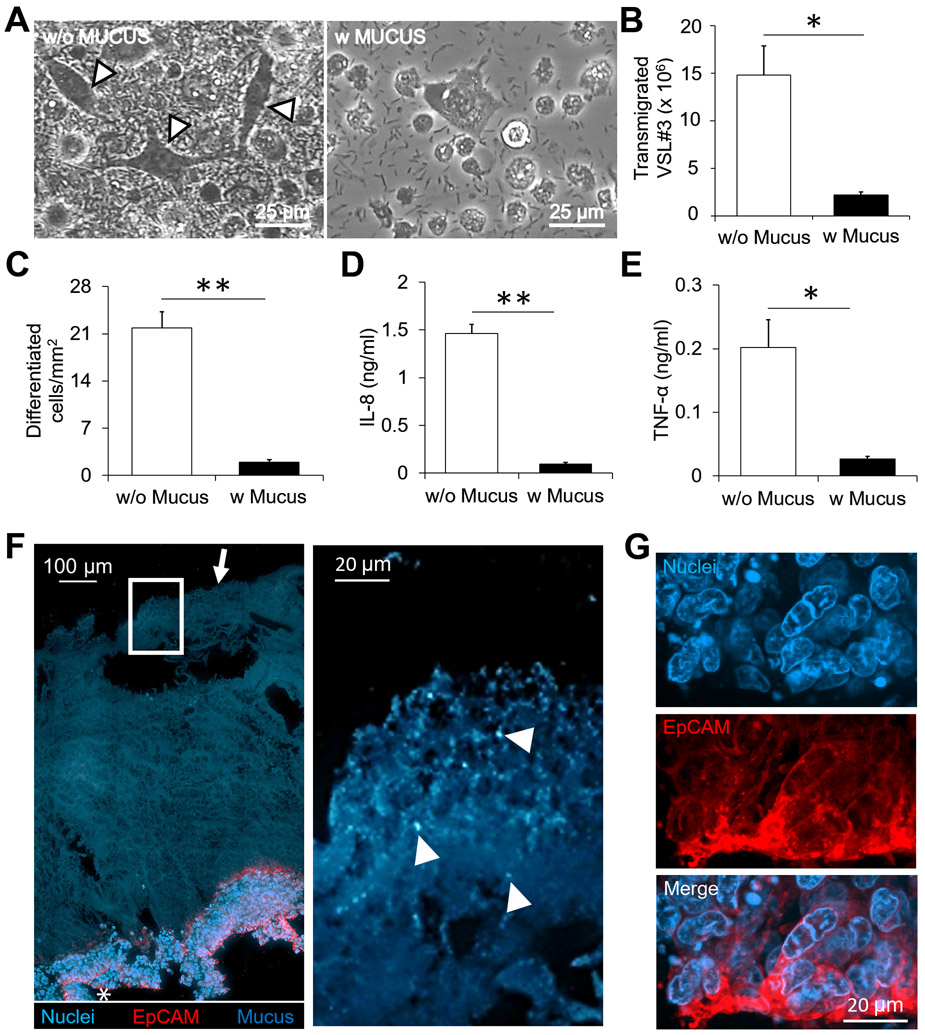
To investigate the barrier function of the mucus layer in presence of commensal bacteria, we added a commercial probiotic mix VSL#3 to the HT-29 and THP-1 co-culture and measured the bacterial transmigration across the epithelial layer and the corresponding cytokine and chemokine responses. Without mucus, bacteria transmigrate across the epithelial layer to the bottom well (Fig. 5A). The mucus layer reduced bacterial transmigration by 7-fold (P < 0.05, Fig. 5B). Bacterial transmigration in the absence of a mucus layer induced THP-1 cell differentiation characterized by changes in morphology from round to elongated (Fig. 5A). A 11-fold higher number of differentiated cells per millimeter squared surface area were present without the mucus layer when compared to the conditions containing a mucus layer (P < 0.01, Fig. 5C). Due to increased bacterial loads, significantly higher concentrations of both IL-8 and TNF-α were measured in the bottom well (P < 0.01, Fig. 5D and P < 0.05, Fig. 5E). The mucus layer reduced the IL-8 concentration by 16-fold (Fig. 5D) and TNF-α concentration by 7-fold (Fig. 5E). Immunohistostaining showed the spatial arrangement of the epithelial cell layer expressing epithelial cell adhesion molecule (EpCAM) at the base and the mucus layered on top of the epithelial surface. VSL#3 bacteria that stained positive for nuclear stain DAPI were mostly seen on the outer surface of the mucus layer and few or no bacteria were present deeper in the mucus layer (Fig. 5F). In addition, no bacteria were present in the epithelial layer (Fig. 5G). Collectively, the results show that a reconstituted mucus layer reduced bacterial transmigration, thereby preventing direct contact between commensal bacteria and epithelial cells. The barrier effect leads to reduced epithelial damage by bacteria and reduced proinflammatory cytokine and chemokine responses from the underlying immune cells.
Figure 5. Barrier function and immune modulation by a PSIM gel layer in response to VSL#3.
(A) VSL#3 transmigrated across the epithelial layer and caused THP-1 cell differentiation identified by elongated morphologies (arrows). Images were taken 24 hours after addition of bacteria. (B) The mucus layer significantly reduced the number of transmigrated VSL#3 bacteria (*, P < 0.05; n = 3). (C-E) The mucus layer significantly reduced the number of differentiated cells per mm2 (C; **, P < 0.01; n = 3), the concentration of IL-8 (D; (**, P < 0.01; n = 3), and the concentration of TNF- α (E; *, P < 0.05; n = 3) in the co-culture. (F) A 20 μm thick histological section (Left) stained with EpCAM (Red) and DAPI (Blue). The PSIM gel layer is auto fluorescent (Blue, arrow). Expanded image (Right) shows DAPI-stained VSL#3 bacteria embedded at the outer surface of the PSIM gel layer (Arrows). (G) Expanded images of the epithelial layer (white star) showing nuclei and EpCAM staining.
2.6. A PSIM gel layer enhances the transmigration of Salmonella.
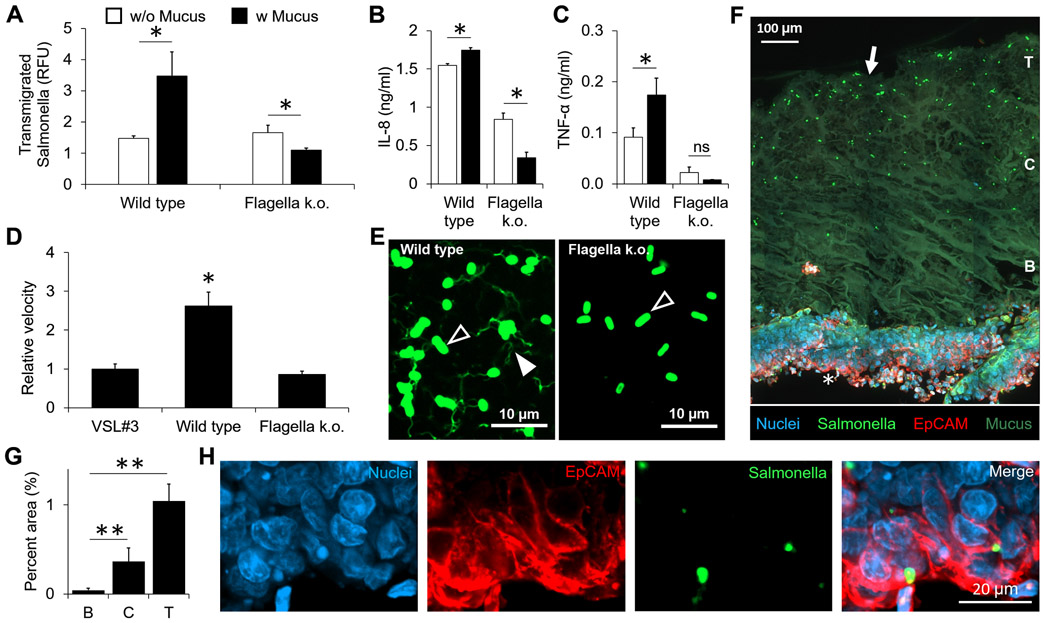
To test if active motility enables bacteria to transmigrate across the mucus layer, we used motile Salmonella. Surprisingly, the presence of mucus significantly increased the transmigration of wild type Salmonella and 3-fold more Salmonella transmigrated across the epithelial cell layers in presence of mucus (P < 0.05, Fig. 6A). In the HT-29 and THP-1 co-culture, wild type Salmonella significantly increased the amounts of IL-8 and TNF-α in the presence of mucus and an epithelial layer compared to an epithelial layer alone (P < 0.05, Fig. 6B, C). To test if these effects are controlled by flagella, a flagellar knockout (ΔflhDC) strain of Salmonella was created. The knockout caused a loss in Salmonella motility and the mean velocity was similar to VSL#3. Mean velocity of wild type Salmonella was significantly higher compared to the knockout strain and VSL#3 (P < 0.05, Fig. 6D). The presence of a mucus layer reduced the transmigration of knockout Salmonella across the epithelial cell layer (P < 0.05, Fig. 6A). The amount of IL-8 was significantly lower in presence of a mucus layer compared to epithelial cells alone for knockout Salmonella (Fig. 6B). The concentration of TNF-α in the medium with knockout Salmonella with or without a mucus layer (Fig. 6C) was equivalent to basal levels from an unstimulated co-culture of cells (Fig. 4C). Immunostaining showed the flagella of wild type Salmonella and the lack of flagella of knockout Salmonella (Fig. 6E). Histological sections of the mucosal layers on transwell membranes showed wild type Salmonella embedded deeper into the mucus layer (Fig. 6F) than VSL#3 (Fig. 5F). Within the gel layer, a significantly higher number of bacteria were found at the top and center of the mucus layer compared to the area proximal to the epithelial surface (Fig. 6G). Although less, Salmonella were also clearly present in the epithelial layer (Fig. 6H). Collectively, these results show that motile Salmonella, in presence of mucus, utilize flagellar motility for higher transmigration across the mucus and epithelial layers. Higher transmigration caused significantly higher immune responses from the epithelial-immune cell co-culture.
Figure 6. Mucus gel layer is not protective against motile Salmonella.
(A) In the presence of a mucus layer, the number of transmigrated wild type Salmonella increased (*, P < 0.05; n = 3). Mucus decreased the number of transmigrated flagella knockout (k.o.) Salmonella (*, P < 0.05). (B) The mean velocity of wild type Salmonella was greater than VSL#3 and knockout (ΔflhDC) Salmonella (*, P < 0.05; n = 5). (C, D) The concentrations of IL-8 (C) and TNF-α (D) in the co-culture in response to wild type and knockout Salmonella with and without mucus (*, P < 0.05; n = 3). (E) Fluorescent microscope images showing the cell bodies (black arrows) and flagella (white arrow) of the wild type (left) and the knockout (right). Only cell bodies are present for the knockout. (F) A histological section stained for EPCAM-1 (Red), nuclei (Blue), Salmonella (Green), and the auto fluorescent PSIM gel layer (Green, arrow). Depth into the mucus layer is indicated as T: top surface, C: center, B: bottom. (G) Bacterial density in the mucus layer as a function of depth quantified as the area covered by positive signal from Salmonella stain (**, P < 0.01; n = 3). (H) Expanded images of the epithelial layer (white star) showing nuclei, Salmonella and EpCAM staining.
3. DISCUSSION
To gain a deeper understanding of how commensal and pathogenic bacteria interact with the mucosal lining of the intestine, we created an in vitro model of this barrier which consisted of a mucus layer, an epithelial cell layer and cells of immune system. These components represent essential complexities of the mucosal barrier. With this model we showed that the mucosal layer decreased the penetration of LPS and non-motile bacteria (Figs. 4&5). All bacteria tested in this study induced the production of cytokines and chemokines by THP-1 monocytes (Fig. 3B, D). Additionally, Salmonella induced the production of IL-8 by HT-29 epithelial cells. In contrast, VSL#3 did not induce epithelial IL-8 production (Fig. 3A, C). Higher production of IL-8 in epithelial and monocytes indicates activation of pro-inflammatory signaling. These results indicate that epithelial cells tolerate contact with commensal bacteria but respond to pathogens, whereas innate immune cells respond to all bacterial contact.
This intestinal mucosa model was built upon the commonly used transwell platform, which enabled creation of epithelial and mucus gel layers, and co-culture with bacteria and immune cells (Fig. 1). The mucus was not cytotoxic (Fig. 1) and was biocompatible to human epithelial and immune cells (Fig. 2). The thickness of the mucus gel layer can be adjusted to match the thickness of mucus in different compartments of the gastro-intestinal tract (Fig. S1). The mucus layer caused spatial segregation of live commensal bacteria away from the epithelial layer and enabled co-culture studies up to 24 hours. The mucosa model demonstrated key mucus-layer-associated processes that have been reported in animal models but are difficult to reproduce in vitro.
In this model, both mucus and epithelial cells formed a physical barrier to molecular diffusion and bacterial penetration. The amounts of IL-8 and TNF-α produced in the co-culture system without mucus (Fig. 4B, C) were lower than THP-1 cells directly stimulated with same amount of LPS (Fig. 3B, D). When mucus was present, these amounts were further reduced (Fig. 4). In the absence of the mucus layer, more bacteria crossed the epithelial cell layer (Fig. 5D) and more monocytes differentiated (Fig. 5E). This dependence indicates that the mucus layer helps maintain the integrity of epithelial barrier in presence of commensal bacteria. The lack of production of TNF-α by epithelial cells indicates that inflammation is only induced after a breach in epithelial barrier and direct contact of live bacteria with immune cells.
As a barrier, the mucus layer was more effective at preventing penetration of commensal bacteria (Fig. 5) than immunogenic molecules (Fig. 4). The transmigration of non-motile commensal bacteria (VSL#3) across the epithelial layer was significantly lower in presence of mucus (Fig. 5). With mucus, the amounts of IL-8 and TNF-α induced by VSL#3 were very low (Fig. 5), similar to the amounts produced by unstimulated epithelial and immune cells (-LPS in Fig. 4). The difference in penetration between bacteria and molecules was most likely because of the structure of intestinal mucus. The mucus gel layer was porous as shown by histological sectioning and fluorescent confocal microscopy (Fig. 1B, F). After gelation, intestinal mucus forms islands of aggregated protein that permit easier transmigration of small molecules than microbes and particulates (Sharma et al., 2020).
An unexpected result was that the presence of mucus increased the penetration of Salmonella through the intestinal barrier (Fig. 6). In comparison, mucus reduced the penetration of LPS (Fig. 3), and non-motile commensal bacteria (Fig. 5). Salmonella transmigration followed complete penetration of the mucus layer and the epithelial barrier (Fig. 6A). This effect was surprising because the penetration of Salmonella through mucus and an epithelial layer was greater than through an epithelial layer alone (Fig. 6A-E), which was considerably thinner (Fig. 1B). Flagella were required for this increase in transmigration through mucus (Fig. 6A-E), showing that this phenomenon is dependent on bacterial motility. Two explanations for this observation are that mucus increases the motility of Salmonella by (a) attracting flagellated Salmonella or (b) inducing the production of flagella.
The intestinal barrier responded in distinct ways to immunogenic molecules. The primary immunogenic molecules in Salmonella are LPS and flagellin, which are detected by toll-like receptors (TLR) 4 and 5, respectively (Chow et al., 1999; Hayashi et al., 2001). When LPS was applied to THP-1 monocytes, they did not differentiate (Fig. 3F). The LPS successfully crossed both the mucus and epithelial barriers as indicated by the secretion of IL-8 and TNF-α (Fig. 4). This result shows that monocyte differentiation after contact with bacteria (Fig. 3E) is controlled by a mechanism that is independent of LPS and TLR4. When knockout (ΔflhDC) Salmonella were applied to the intestinal barrier model, the THP-1 monocytes did not produce TNF-α (Fig. 6C). This result suggests that the production of TNF-α is mediated by TLR5, the primary toll-like receptor to detect the flagella of Salmonella. Similar to LPS, no THP-1 cell differentiation was detected in presence of Salmonella. Monocyte differentiation into macrophage and dendritic-like cell types is a critical step in their anti-bacterial response (Serbina et al., 2008). Our results showed that VSL#3 induced a robust differentiation response in THP-1 cells compared to Salmonella (Fig. 3F). Studies have shown that Salmonella has evolved to evade detection by certain cells of the immune system and is also known to have an intracellular lifestyle (Riquelme et al., 2011; Rydstrom & Wick, 2007). These factors could explain why the THP-1 cell differentiation to macrophage and dendritic-like cells was not detected with Salmonella.
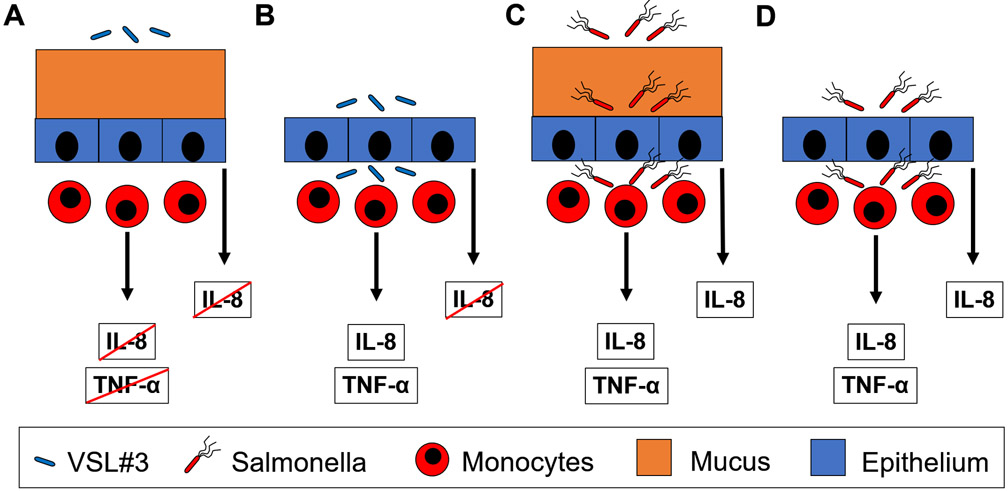
Combined, these results paint a picture of how the intestinal barrier could react to contact with bacteria (Fig. 7). In the healthy condition of an intact mucus layer and a lumen filled primarily with commensal bacteria (condition 1, Fig. 7A), no bacteria penetrate the barrier layer and no cytokines are produced. When there is physical damage to the mucus layer (condition 2, Fig. 7B), commensal bacteria penetrate the epithelial layer and trigger the production of cytokines and chemokines by monocytes (Fig. 5F, G). After this contact, the monocytes differentiate into macrophages and dendritic cells (Fig. 5C, E). Chemokines, e.g. IL-8, recruit neutrophils to contain the infection. When motile Salmonella are present in the lumen in significant amounts (condition 3), the mucus layer does not prevent penetration (Fig. 7C). In this condition, the epithelial layer produces chemokines (Fig. 3A) to recruit neutrophils before the pathogens cross the epithelial barrier. Once this barrier is breached, monocytes are activated, producing a similar immune response to an infection of commensal bacteria (Fig. 3B, D and 6C, D).
Figure 7. The mucus layer regulates the extent of host-bacterial interactions and the resultant immune responses.
(A) The mucus layer acts as a barrier to VSL#3 and prevents any immune response. (B) In the absence of a mucus barrier, VSL#3 breach the epithelial barrier and induce immune responses (TNF-α and IL-8). (C) Despite the presence of a mucus layer, Salmonella breach the epithelial barrier and induce chemokine (IL-8) responses from epithelial cells and cytokine (TNF-α) and chemokine (IL-8) responses from immune cells. (D) In the absence of a mucus layer, Salmonella breach the epithelial barrier and induce immune responses, similar to in the presence of mucus.
The intestinal mucosa model presented here provided insights into the role of mucus and epithelial barrier against bacteria and bacteria-derived molecular factors in presence of innate immune cells. We envision that this platform will be useful for screening host response against pathogenic, opportunistic, and probiotic strains of bacteria. Many of the obtained results could only have been obtained with an in vitro system that includes the three essential components of the mucosal barrier: mucus, epithelial cells and immune cells. For example, it was found that the epithelial barrier is a key component of the innate immune response to bacterial infection, especially in the detection of pathogens. A better understanding of the relation between microbial pathogens, the intestinal barrier and the immune system has the potential to improve treatment of infections and immunological diseases.
4. MATERIALS and METHODS
All chemicals and supplies were purchased from Fisher Scientific and Sigma Aldrich unless otherwise specified.
Human cell culture
HT-29 (ATCC, HTB-38) adherent cells were cultured in McCoy’s 5A medium (Sigma Aldrich) supplemented with 2.2 g/L sodium bicarbonate. THP-1 (ATCC, TIB-202) suspension cells were cultured in RPMI-1640 medium (containing 0.05 mM β-Mercaptoethanol). HEK293T/17 (ATCC, CRL-11268) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM). All media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and referred to as complete medium in the methods. Cells were maintained in a 37 °C humidified incubator with 5% CO2.
RFP-expressing HT-29 cells were made by lentivirus transfection. For lentivirus production, HEK293T/17 cells grown in a 10 cm dish were transfected with four plasmids acquired from addgene: (4 μg pLV-mcherry (Cat #36084), 2 μg pMDLg/pRRE (Cat #12251), 2 μg pRSV-Rev (Cat #12253) and 2 μg pMD2.G (Cat #12259), using the standard calcium-phosphate co-precipitation method. Virus was harvested 24 h post transfection and filtered through 0.45 μm filter. For transduction, HT-29 cells cultured in six well plates were cultured in equal volumes of the virus containing medium and complete McCoy’s 5A medium. Cells were monitored for RFP expression and sub-cultured after most cells expressed RFP when checked for fluorescence qualitatively.
PSIM extraction and purification
PSIM was extracted as described before (Sharma et al., 2020) with changes in sterilization procedure. Briefly, intact small intestine tubules were cut into 2-meter lengths and rinsed with water to eliminate food particles from the lumen. The tubules were drained gently, and the lumen was filled with 350 ml of 0.01 M NaOH. The ends of the tubules were tied and incubated for 24 hours at 4 °C. After incubation the solubilized material was drained by squeezing and collected. The extract was subjected to centrifugation at 20,000 x g (Thermo Scientific) at 4 °C for 2 hours to remove insoluble debris. The supernatant was collected without disturbing the precipitate. The clear supernatant was subjected to sol-gel transition by adjusting the pH to 4 using 2 M HCl. The aggregates formed at pH 4 were subjected to centrifugation at 200 x g for 15 minutes and resolubilized in sterile DI water by adjusting the pH value to 8. The solution was passed through a 40 μm strainer and subjected to dialysis using a membrane with molecular size cut-off equal to 14 kDa. The dialyzed mucus extract was sterilized by adding 1% (v/v) chloroform under constant stirring at 4 °C for 72 hours. The solution (30 ml aliquots) was transferred to 50 ml conical polypropylene tubes, frozen to −80 °C, and freeze dried (Labconco Freezone 2.5L freeze dry system). The freeze-dried mucus was reconstituted aseptically in serum- and antibiotic-free McCoy’s 5A medium. The solutions were sterilized in 15 ml polypropylene tubes by adding 10% chloroform and allowing to sit at 4 °C for 24 hours without shaking. The chloroform solution formed a distinct layer at the bottom and mucus solutions were collected without disturbing the interface under aseptic conditions. The solutions were aliquoted in 1.5 ml polypropylene tubes and stored at −80 °C until use.
Bacteria culture
Probiotic VSL#3 bacteria were acquired from a commercial source (VSL Pharmaceuticals, Inc.) and stored at 4 °C until use. For co-culture experiments, the freeze-dried bacteria were aseptically reconstituted in serum- and antibiotic-free McCoy’s 5A medium at a cell density of 1010 CFU/ml. Bacterial cells were incubated at 37 °C for 1 hour to recover. After 1 hour, the cell density was diluted to required cell density by serial dilution.
Wild type Salmonella strain SL1344 was used for live motile bacterial experiments. The ΔflhDC SL1344 strain was created using a modified lambda red recombination procedure (Mosberg et al., 2010). SL1344 was transformed with plasmid pkd46 by electroporation. Single colonies were inoculated into 50 ml of LB supplemented with 100 μg/ml of carbenicillin and grown at 30 °C. Once the absorbance at 600 nm (OD600) of the culture reached 0.1, arabinose was added to the culture to a final concentration of 20 mM and grown until the OD600 was equal to 0.8. Bacteria were centrifuged at 3000xg for 10 minutes and washed twice with nanopure water (Millipore). Bacterial pellet was resuspended in 400 μl of nanopure water. In a polypropylene tube, 1 μg of DNA (PCR amplification of pkd4 using the primers, 5’-FOFZCACGGGGTGCGGCTACGTCGCACAAAAATAAAGTTGGTTATTCTGGgtcttgagcgattgtgtaggc-3’ and 5’-ZZFFACAGCCTGTTCGATCTGTTCATCCAGCAGTTGTGGAATAATATCGGaattagccatggtccatatgaatatc-3’) was mixed with 50 μl of resuspended Salmonella. Bacteria were electroporated in a 1 mm cuvette (Fisher Scientific catalog # FB101) with the following settings on the electroporation system (BIORAD, GenePulser Xcell): Voltage = 1.8 kV, Capacitance = 25 μF, Resistance = 200 Ω, and 5 ms time constant. Primers were purchased from Invitrogen, where FOFZ are phosphorothioated nucleotides corresponding to ATAC and ZZFF correspond to TTAA. After electroporation, bacteria were recovered in 1 ml of LB for 2 hours at 37 °C. The recovery solution was plated on agar plates supplemented with kanamycin (50 μg/ml) and incubated at 37 °C overnight. Positive colonies were regrown on kanamycin plates overnight at 43 °C in order to eliminate pkd46 from the bacteria. Salmonella were transformed with a plasmid encoding for GFP under the control of constitutive Plac promoter using electroporation as discussed above. The cultures were stored at −80 °C in 25% glycerol solution.
For experiments, frozen stocks of the Salmonella strains were inoculated in LB medium and grown overnight at 37 °C. Small amount of overnight culture was inoculated in fresh LB medium and allowed to grow to a density of 4 x 108 CFU/ml and used for further experiments. To prepare bacteria for transwell co-culture experiments, bacteria culture was subjected to centrifugation at 2500 x g for 10 minutes. The bacterial cell pellet was washed three times with PBS by gentle pipetting multiple times and repeating the centrifugation step in between. Final pellet was reconstituted in serum- and antibiotic-free McCoy’s 5A medium and the required cell density was adjusted by serial dilution.
Epithelial cell seeding in transwell inserts
For transwell assays, HT-29 cells were detached from culture flasks using trypsin and seeded at a density of 105 cells per insert in 3 μm pore size polycarbonate membrane transwell inserts (CLS3415, Corning). Inserts were cultured for 7 days in 24 well plates in complete McCoy’s medium. The culture medium was changed 24 hours after cell seeding and every 48 hours after that. After day 7, the culture medium was aspirated from the transwell inserts. The inserts were washed three times with PBS on both the apical and basolateral sides. To check for incomplete cell coverage, inserts were filled with 250 μl of PBS and the change in liquid level was checked for 5 minutes. The inserts with no change in liquid level were used for further experiments. To check for coverage microscopically, inserts were seeded with RFP-expressing HT-29 cells. After 7 days of culture, the inserts were imaged using EVOS FL Auto imaging system (Life technologies). Images captured using a 10x objective were tiled using in-built EVOS software.
Mucus layer formation and cytotoxicity of PSIM on epithelial cells
To form mucus layers, HT-29 cells were cultured for 7 days in the transwell inserts and layered with 100 μl of sterile PSIM (20 mg/ml). Inserts were incubated in a 37 °C humidified incubator with 5% CO2 for 12 hours.
For testing cytotoxicity of PSIM, HT-29 cells were cultured for 7 days in the transwell inserts and washed three times with phosphate buffer saline (PBS). The inserts were transferred to a new 24 well plate and 600 μl of serum- and antibiotic-free McCoy’s 5A medium was added to the basolateral side. To form the mucus layers, 100 μl of thawed PSIM was added to the apical side. For controls, 100 μl of serum- and antibiotic-free McCoy’s 5A medium was added to the apical side. The plates were incubated for 24 hours at 37 °C. The viability of HT-29 cells after application of the PSIM layer was performed using a viability/cytotoxicity assay kit (Cat # L3224, Invitrogen). The transwell inserts were transferred to a fresh 24-well plate containing 600 μl of phosphate buffer saline (PBS) and rocked gently. The PBS was replaced three times to wash the bottom surface of the transwell membrane. Finally, the wash solution was replaced with PBS containing 2 μm Calcein AM and 4 μm Ethidium homodimer-1 and incubated for 30 minutes at room temperature. Fluorescent images were acquired using Zeiss Spinning Disk Axio Observer Z1 microscope (Carl Zeiss) with a C-Apochromat 63x oil immersion objective and Zen software. To quantify area coverage, fluorescent images of the stained cells were background subtracted and segmented to isolate green (Live) and red (Dead) signal. The percent area covered by positive signal from green and red channels was quantified relative to the total image area. Images from three independent samples were used for quantification.
To visualize bacteria embedded in the mucus gel, 103 CFU/ml wild type GFP Salmonella were mixed in the PSIM solution prior to adding it to transwell inserts covered with HT-29 cells (as above). After incubation at 37°C for 12 h, the cells and mucus in the inserts were fixed using 10% formalin solution in phosphate buffer saline for 20 minutes. The membranes were cut from the inserts and washed in PBS three times. The mucus gel layer was separated from the membrane and the cells, transferred onto a coverslip and stained with DAPI, which identified the mucus gel under UV light. GFP-expressing Salmonella embedded in PSIM gel were imaged using Zeiss Spinning Disk Axio Observer Z1 microscope (Carl Zeiss). Z-stack images were captured with a 63x oil immersion objective at 1 μm height intervals using Zen software. Three-dimensional rendering and image processing were performed using Imaris (Bitplane, Belfast, UK).
Immunological response of epithelial and immune cells to PSIM
For testing immunogenicity of PSIM, HT-29 cells cultured for 7 days in the transwell inserts were washed three times with phosphate buffer saline (PBS). The inserts were transferred to a new 24 well plate and 600 μl of serum- and antibiotic-free McCoy’s 5A medium was added to the basolateral side. Mucus layers were formed by adding 100 μl of thawed PSIM (20 mg/ml) to the apical side. For controls, 100 μl of serum- and antibiotic-free McCoy’s 5A medium was added to the apical side. The plates were incubated for 24 hours at 37 °C. The media from the basolateral side was collected from three independent samples. Manufacturers protocol was followed to quantify human IL-8 (Cat # DY 208) and human TNF-α (Cat # DY 210) concentrations using DuoSet ELISA assay kit (R&D Systems). Assays were calibrated to the standard curve created for serially diluted IL-8 and TNF-α standard solutions on each plate per assay.
Cultured THP-1 cells were washed three times with PBS. After centrifugation, 105 THP-1 cells were suspended in serum- and antibiotic-free medium (600 μl); seeded into 24 well plates; and 100 μl of thawed PSIM (20 mg/ml) was directly added to the medium. After 24 hours of incubation at 37 °C, the media were collected and the concentrations of human IL-8 and TNF-α were quantified using ELISA assays. Samples from three independent experiments were used for analysis.
Direct stimulation of HT-29 and THP-1 monocultures
Twenty-four well tissue culture treated plates were seeded with 105 HT-29 cells per well and cultured for 4 days with a media change every day. After 4 days, the cells were washed with PBS three times and replaced with 450 μl serum- and antibiotic-free McCoy’s medium. To each well, 50 μl of medium was added that contained bacterial endotoxin lipopolysaccharide (LPS, 10 μg/ml) from Escherichia coli O55:B5 (Cat # L6529), VSL#3 (2 x 109 CFU/ml), or Salmonella (2 x 105 CFU/ml). Media alone (50 μl) was added to control wells. After 12 hours of incubation at 37 °C, the media were collected from three independent experimental conditions. Human IL-8 and TNF-α concentrations were quantified using ELISA assays.
Using a similar procedure as above, 105 THP-1 cells were suspended in serum- and antibiotic-free medium (600 μl); seeded into 24 well plates; and stimulated with 50 μl LPS (10 μg/ml), VSL#3 (2 x 109 CFU/ml), and Salmonella (2 x 105 CFU/ml). After 12 hours of incubation at 37 °C, the medium was collected and the concentrations of human IL-8 and TNF-α were quantified using ELISA assays. Results from three independent experiments were used analysis.
Construction of in vitro mucosal lining model
Epithelial cells were grown on transwell inserts and checked for coverage and leakage using PBS as described above. Mucus layers was formed by adding 100 μl of sterile PSIM (20 mg/ml) and incubating at 37 °C for 12 hours. For controls without mucus, 100 μl serum- and antibiotic-free McCoy’s 5A medium was added to the apical side of the inserts. In cultures of THP-1 cells, the medium was removed by centrifugation and the cells were washed with PBS three times. The cell pellet was suspended in serum- and antibiotic-free McCoy’s 5A medium. The medium in the basolateral side of the transwell inserts was replaced with 600 μl of THP-1 cell suspension at a density of 105 cells/well.
LPS interaction with in vitro mucosal lining model
The in vitro mucosal lining model was challenged by adding 50 μl per well LPS (10 μg/ml) from Escherichia coli O55:B5 (Cat # L6529) to the apical side of the transwell inserts in the presence and absence of a pre-formed PSIM layer. After 24 hours of incubation with LPS, the medium from the bottom well was collected for quantification of human IL-8 and TNF-α using ELISA. Samples from three independent conditions were used for analysis.
VSL#3 interaction with in vitro mucosal lining model
The in vitro mucosal lining model was challenged by adding 50 μl per well of VSL#3 culture (2 x 109 CFU/ml) to the apical side of the transwell inserts. After 24 hours of incubation at 37 °C, the transwell inserts were removed and brightfield images of the bottom well containing THP-1 cells were captured using EVOS FL Auto imaging system (Life technologies). The contrast of the images was uniformly enhanced. Post-imaging, the medium was collected from the bottom well. Half of the medium was used for quantification of human IL-8 and human TNF-α with ELISA. The other half was used for bacterial quantification by serial dilution, plating on solid agar plates and incubated at 37 °C for 24 hours. The colonies were manually counted and converted to CFU based on the dilution factor. The THP-1 cell morphology change was manually quantified using ImageJ (NIH). Differentiated immune cell were identified as cells having stretched or dendrite-like morphology. The number of cells was normalized by the total area of the images. Data was analyzed for three independent experiments.
Salmonella interaction with in vitro mucosal lining model
The in vitro mucosal lining model was challenged by adding 50 μl per well of GFP-labeled Salmonella (2 x 105 CFU/ml) to the apical side of the transwell inserts. After 12 hours of incubation at 37 °C, the transwell inserts were removed. Using a plate reader (Synergy H1, BioTek Instruments, Inc.), the GFP intensity was quantified in the bottom well plate to measure of bacterial cell density. The medium was collected and used for quantification of human IL-8 and human TNF-α with ELISA. Measurements and analysis were performed for three independently conducted experiments.
Bacterial motility analysis
Aqueous motility of wild type Salmonella, flagella knockout Salmonella and VSL#3 was quantified using fluorescent microscopy. Salmonella strains expressing GFP were grown in LB until OD600 was in the range 0.6 – 0.8. A 20 μl droplet containing 107 CFU/ml was added onto a glass slide and covered with a coverslip. Fluorescent microscope images were captured every 0.141 s for 60 s. Bacterial velocity was analyzed using an automated particle tracking program, Trackmate in ImageJ (NIH). The average velocity of all the tracks was calculated per sample. To measure the velocity of VSL#3, the bacteria were inoculated in serum- and antibiotic-free McCoy’s 5A medium and allowed to recover at 37°C for 1 hour. The cells were nuclei stained using Hoechst 33342 (NucBlue, Thermo Fisher, Cat # R37605). A 20 μl droplet containing 107 CFU/ml was added onto a glass slide and covered with a coverslip. Fluorescent microscope images were captured every 0.5 s for 60 s. The average velocity was calculated as described above. Measurements were recorded and results analyzed for five independent experiments.
Cryopreservation, histological sectioning, and immunofluorescence staining
To image and wild type and flagella knockout Salmonella, bacteria were grown to an OD600 of 0.8. Twenty μl droplets were applied to glass slides and allowed to dry in a biosafety cabinet for approximately six hours.
For histological sectioning, the transwell membranes with intact epithelial and mucus layers were cut from the inserts and fixed using 10% formalin solution in phosphate buffer saline for 20 minutes. Fixed membranes were embedded in optimum cutting temperature (OCT) medium and flash frozen in liquid nitrogen. Using a cryostat (CRYOSTAR NX70, Thermo Scientific), 30 μm thick cross-sections were cut and collected on ColorMark Plus glass slides for imaging. Brightfield images were acquired using an EVOS FL Auto imaging system (Life technologies) and the thickness of the mucus layers were measured manually using ImageJ (NIH). Images from three independent samples were used for measuring the thickness.
For immunofluorescent staining of slides with dried Salmonella, FITC-conjugated anti-Salmonella antibody (1:200, ab69253, Abcam) was used. Fluorescent images were acquired using Zeiss Spinning Disk Axio Observer Z1 microscope (Carl Zeiss) with a C-Apochromat 63x oil immersion objective and Zen software (Carl Zeiss). For immunofluorescent staining of histological cross-sections two antibodies were used: FITC-conjugated anti-Salmonella antibody and Alexa Fluor 594 conjugated anti-human CD326 (EpCAM) antibody (1:200, Cat # 324228, BioLegend). Both samples on glass slides were blocked in the blocking solution (1% BSA, 22.52 mg/ml in PBST (PBS + 0.1% Tween20)) for 1 hour at room temperature. Slides were incubated with antibodies diluted in 1% BSA in PBST for 1 hour at room temperature and washed three times with PBS. Histological slices were counter-stained with DAPI (1:10,000, D9542, Sigma Aldrich) to identify cell nuclei. Fluorescent images were acquired using Zeiss Spinning Disk Axio Observer Z1 microscope (Carl Zeiss) with a LD C-Apochromat 40x oil immersion objective and Zen software (Carl Zeiss).
The stained histological images were analyzed using ImageJ. To quantify Salmonella density in the mucus gel layer, images were tiled to capture from the top of the mucus to the bottom of the epithelial layer. Tiled images from three planes deep into the mucus layer on the glass slides were captured with 10 μm intervals. These three tiled images were projected on to a single plane. Projected images were uniformly segmented to isolate bacteria morphologically. The mucus layer height was divided into three regions of equal height (T: top; C: center; B: bottom). Images were collected from three independent samples. The amount of Salmonella in each region was measured as the percent area with FITC over the total area using ImageJ.
Statistical analysis
A two-tailed Student’s t-test assuming unequal variances was used for statistical analysis of pairwise comparison. One-way ANOVA with Bonferroni post hoc test was used for multiple comparisons. All data are presented as a mean ± standard deviation for three or more independent experimental conditions. Differences were considered significant at P < 0.05, with * denoting P < 0.05, and ** denoting P < 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Shelly Peyton for assistance with and access to confocal microscopy instruments and software.
Grant Support: This work was supported by the National Cancer Institute of the National Institutes of Health grant R00 CA163671 to JL and R01CA188382 to NSF.
Footnotes
Disclosures: No conflicts of interest exist.
REFERENCES
- Alipour M, Zaidi D, Valcheva R, Jovel J, Martinez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA, Carroll MW, Huynh HQ, & Wine E (2016). Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J Crohns Colitis, 10(4), 462–471. doi: 10.1093/ecco-jcc/jjv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain CC, Bravo-Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S, Malissen B, Osborne LC, Artis D, & Mowat AM (2014). Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nature Immunology, 15, 929. doi: 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens I, Stenberg P, Artursson P, & Kissel T (2001). Transport of Lipophilic Drug Molecules in a New Mucus-Secreting Cell Culture Model Based on HT29-MTX Cells. Pharmaceutical Research, 18(8), 1138–1145. doi: 10.1023/a:1010974909998 [DOI] [PubMed] [Google Scholar]
- Boegh M, Baldursdottir SG, Mullertz A, & Nielsen HM (2014). Property profiling of biosimilar mucus in a novel mucus-containing in vitro model for assessment of intestinal drug absorption. Eur J Pharm Biopharm, 87(2), 227–235. doi: 10.1016/j.ejpb.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Boegh M, Garcia-Diaz M, Mullertz A, & Nielsen HM (2015). Steric and interactive barrier properties of intestinal mucus elucidated by particle diffusion and peptide permeation. Eur J Pharm Biopharm, 95(Pt A), 136–143. doi: 10.1016/j.ejpb.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Celli J, Gregor B, Turner B, Afdhal NH, Bansil R, & Erramilli S (2005). Viscoelastic Properties and Dynamics of Porcine Gastric Mucin. Biomacromolecules, 6(3), 1329–1333. doi: 10.1021/bm0493990 [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, Li C, Isberg RR, Kumamoto CA, Mecsas J, & Kaplan DL (2015). Robust bioengineered 3D functional human intestinal epithelium. Sci Rep, 5, 13708. doi: 10.1038/srep13708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Young DW, Golenbock DT, Christ WJ, & Gusovsky F (1999). Toll-like Receptor-4 Mediates Lipopolysaccharide-induced Signal Transduction. Journal of Biological Chemistry, 274(16), 10689–10692. doi: 10.1074/jbc.274.16.10689 [DOI] [PubMed] [Google Scholar]
- Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, & Finlay BB (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clinical microbiology reviews, 26(4), 822–880. doi: 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum-Cianflone NF (2008). Salmonellosis and the gastrointestinal tract: more than just peanut butter. Current gastroenterology reports, 10(4), 424–431. doi: 10.1007/s11894-008-0079-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng S-K, Pusparajah P, Ab Mutalib N-S, Ser H-L, Chan K-G, & Lee L-H (2015). Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Frontiers in Life Science, 8(3), 284–293. doi: 10.1080/21553769.2015.1051243 [DOI] [Google Scholar]
- Furter M, Sellin ME, Hansson GC, & Hardt WD (2019). Mucus Architecture and Near-Surface Swimming Affect Distinct Salmonella Typhimurium Infection Patterns along the Murine Intestinal Tract. Cell Rep, 27(9), 2665–2678.e2663. doi: 10.1016/j.celrep.2019.04.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Rodriguez LA, Ruigomez A, & Panes J (2006). Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology, 130(6), 1588–1594. doi: 10.1053/j.gastro.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Gradel KO, Nielsen HL, Schonheyder HC, Ejlertsen T, Kristensen B, & Nielsen H (2009). Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology, 137(2), 495–501. doi: 10.1053/j.gastro.2009.04.001 [DOI] [PubMed] [Google Scholar]
- Hallstrom K, & McCormick B (2011). Salmonella Interaction with and Passage through the Intestinal Mucosa: Through the Lens of the Organism. Frontiers in Microbiology, 2(88). doi: 10.3389/fmicb.2011.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, & Aderem A (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature, 410(6832), 1099–1103. doi: 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- Hering NA, Fromm A, Kikhney J, Lee I-FM, Moter A, Schulzke JD, & Bücker R (2015). Yersinia enterocolitica Affects Intestinal Barrier Function in the Colon. The Journal of Infectious Diseases, 213(7), 1157–1162. doi: 10.1093/infdis/jiv571 [DOI] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, & Akira S (1999). Cutting Edge: Toll-Like Receptor 4 (TLR4)-Deficient Mice Are Hyporesponsive to Lipopolysaccharide: Evidence for TLR4 as the Lps Gene Product. The Journal of Immunology, 162(7), 3749–3752. [PubMed] [Google Scholar]
- Iliev ID, Spadoni I, Mileti E, Matteoli G, Sonzogni A, Sampietro GM, Foschi D, Caprioli F, Viale G, & Rescigno M (2009). Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut, 58(11), 1481–1489. doi: 10.1136/gut.2008.175166 [DOI] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjovall H, & Hansson GC (2010). Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS One, 5(8), e12238. doi: 10.1371/journal.pone.0012238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, & Hansson GC (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A, 105(39), 15064–15069. doi: 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Larsson JMH, & Hansson GC (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proceedings of the National Academy of Sciences of the United States of America, 108, 4659–4665. doi: 10.1073/pnas.1006451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Huh D, Hamilton G, & Ingber DE (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip, 12(12), 2165–2174. doi: 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- Kim HJ, & Ingber DE (2013). Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integrative Biology, 5(9), 1130–1140. doi: 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- Kim HJ, Li H, Collins JJ, & Ingber DE (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proceedings of the National Academy of Sciences of the United States of America, 113(1), E7–E15. doi: 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, & Brummer R-J (2016). Human Intestinal Barrier Function in Health and Disease. Clinical and translational gastroenterology, 7(10), e196–e196. doi: 10.1038/ctg.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharzik T, Hudson JT 3rd, Lügering A, Abbas JA, Bettini M, Lake JG, Evans ME, Ziegler TR, Merlin D, Madara JL, & Williams IR (2005). Acute induction of human IL-8 production by intestinal epithelium triggers neutrophil infiltration without mucosal injury. Gut, 54(11), 1565–1572. doi: 10.1136/gut.2004.061168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH (2015). Intestinal Permeability Regulation by Tight Junction: Implication on Inflammatory Bowel Diseases. Intestinal Research, 13(1), 11–18. doi: 10.5217/ir.2015.13.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon CG, Tory R, Jia J, Sivak O, & Wasan KM (2008). Discovery and Development of Toll-Like Receptor 4 (TLR4) Antagonists: A New Paradigm for Treating Sepsis and Other Diseases. Pharmaceutical Research, 25(8), 1751–1761. doi: 10.1007/s11095-008-9571-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard F, Collnot EM, & Lehr CM (2010). A three-dimensional coculture of enterocytes, monocytes and dendritic cells to model inflamed intestinal mucosa in vitro. Mol Pharm, 7(6), 2103–2119. doi: 10.1021/mp1000795 [DOI] [PubMed] [Google Scholar]
- Lieleg O, Lieleg C, Bloom J, Buck CB, & Ribbeck K (2012). Mucin biopolymers as broad-spectrum antiviral agents. Biomacromolecules, 13(6), 1724–1732. doi: 10.1021/bm3001292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden SK, Sutton P, Karlsson NG, Korolik V, & McGuckin MA (2008). Mucins in the mucosal barrier to infection. Mucosal Immunology, 1(3), 183–197. doi: 10.1038/mi.2008.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler GJ, Shuler ML, & Glahn RP (2009). Characterization of Caco-2 and HT29-MTX cocultures in an in vitro digestion/cell culture model used to predict iron bioavailability. The Journal of Nutritional Biochemistry, 20(7), 494–502. doi: 10.1016/j.jnutbio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Marteyn B, Gazi A, & Sansonetti P (2012). Shigella: a model of virulence regulation in vivo. Gut microbes, 3(2), 104–120. doi: 10.4161/gmic.19325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzorati M, Vanhoecke B, De Ryck T, Sadaghian Sadabad M, Pinheiro I, Possemiers S, Van den Abbeele P, Derycke L, Bracke M, Pieters J, Hennebel T, Harmsen HJ, Verstraete W, & Van de Wiele T (2014). The HMI module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol, 14, 133. doi: 10.1186/1471-2180-14-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucchelli L, Hauser C, Zgraggen K, Wagner H, Hess M, Laissue JA, & Mueller C (1994). Expression of interleukin-8 gene in inflammatory bowel disease is related to the histological grade of active inflammation. The American Journal of Pathology, 144(5), 997–1007. [PMC free article] [PubMed] [Google Scholar]
- McCormick BA, Stocker BA, Laux DC, & Cohen PS (1988). Roles of motility, chemotaxis, and penetration through and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestine of streptomycin-treated mice. Infect Immun, 56(9), 2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Eri R, Simms LA, Florin TH, & Radford-Smith G (2009). Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis, 15(1), 100–113. doi: 10.1002/ibd.20539 [DOI] [PubMed] [Google Scholar]
- Mosberg JA, Lajoie MJ, & Church GM (2010). Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics, 186(3), 791–799. doi: 10.1534/genetics.110.120782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsui M, Kawasaki K, Takizawa H, Hayashi SI, Matsuda Y, Sugimura K, Seki K, Narisawa R, Sendo F, & Asakura H (1997). Selective depletion of neutrophils by a monoclonal antibody, RP-3, suppresses dextran sulphate sodium-induced colitis in rats. J Gastroenterol Hepatol, 12(12), 801–808. doi: 10.1111/j.1440-1746.1997.tb00375.x [DOI] [PubMed] [Google Scholar]
- Neurath MF (2014). Cytokines in inflammatory bowel disease. Nature Reviews Immunology, 14(5), 329–342. doi: 10.1038/nri3661 [DOI] [PubMed] [Google Scholar]
- Nevola JJ, Laux DC, & Cohen PS (1987). In vivo colonization of the mouse large intestine and in vitro penetration of intestinal mucus by an avirulent smooth strain of Salmonella typhimurium and its lipopolysaccharide-deficient mutant. Infect Immun, 55(12), 2884–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel G, Baetz NW, Staab JF, Donowitz M, Kovbasnjuk O, Pasetti MF, & Zachos NC (2017). A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Scientific reports, 7, 45270–45270. doi: 10.1038/srep45270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CK, Tribble DR, Aliaga PA, Halvorson HA, & Riddle MS (2008). Infectious gastroenteritis and risk of developing inflammatory bowel disease. Gastroenterology, 135(3), 781–786. doi: 10.1053/j.gastro.2008.05.081 [DOI] [PubMed] [Google Scholar]
- Riquelme SA, Wozniak A, Kalergis AM, & Bueno SM (2011). Evasion of host immunity by virulent Salmonella: implications for vaccine design. Curr Med Chem, 18(36), 5666–5675. doi: 10.2174/092986711798347333 [DOI] [PubMed] [Google Scholar]
- Rydstrom A, & Wick MJ (2007). Monocyte recruitment, activation, and function in the gut-associated lymphoid tissue during oral Salmonella infection. J Immunol, 178(9), 5789–5801. doi: 10.4049/jimmunol.178.9.5789 [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ (2004). War and peace at mucosal surfaces. Nature Reviews Immunology, 4, 953. doi: 10.1038/nri1499 [DOI] [PubMed] [Google Scholar]
- Serbina NV, Jia T, Hohl TM, & Pamer EG (2008). Monocyte-mediated defense against microbial pathogens. Annual review of immunology, 26, 421–452. doi: 10.1146/annurev.immunol.26.021607.090326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, Zenhausern F, & Wilmes P (2016). A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nature Communications, 7, 11535. doi: 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Kwak J-G, Kolewe KW, Schiffman JD, Forbes NS, & Lee J (2020). In vitro reconstitution of an intestinal mucus layer shows that cations and pH control the pore structure that regulates its permeability and barrier function. ACS Applied Bio Materials. doi: 10.1021/acsabm.9b00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A, Chou DB, Tovaglieri A, Ferrante TC, Duckworth T, Fadel C, Frismantas V, Sutherland AD, Jalili-Firoozinezhad S, Kasendra M, Stas E, Weaver JC, Richmond CA, Levy O, Prantil-Baun R, Breault DT, & Ingber DE (2019). Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cellular and Molecular Gastroenterology and Hepatology. doi: 10.1016/j.jcmgh.2019.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovran B, Lu P, Loonen LM, Hugenholtz F, Belzer C, Stolte EH, Boekschoten MV, van Baarlen P, Smidt H, Kleerebezem M, de Vos P, Renes IB, Wells JM, & Dekker J (2016). Identification of Commensal Species Positively Correlated with Early Stress Responses to a Compromised Mucus Barrier. Inflamm Bowel Dis, 22(4), 826–840. doi: 10.1097/MIB.0000000000000688 [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, & Lochs H (2005). Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. Journal of Clinical Microbiology, 43(7), 3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR (2009). Intestinal mucosal barrier function in health and disease. Nature Reviews Immunology, 9(11), 799–809. doi: 10.1038/nri2653 [DOI] [PubMed] [Google Scholar]
- Vimal DB, Khullar M, Gupta S, & Ganguly NK (2000). Intestinal mucins: the binding sites for Salmonella typhimurium. Mol Cell Biochem, 204(1-2), 107–117. doi: 10.1023/a:1007015312036 [DOI] [PubMed] [Google Scholar]
- Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, Magness ST, & Allbritton NL (2017). A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials, 128, 44–55. doi: 10.1016/j.biomaterials.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim R, Sims CE, & Allbritton NL (2019). Building a Thick Mucus Hydrogel Layer to Improve the Physiological Relevance of In Vitro Primary Colonic Epithelial Models. Cellular and Molecular Gastroenterology and Hepatology, 8(4), 653–655.e655. doi: 10.1016/j.jcmgh.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler KM, Cárcamo-Oyarce G, Turner BS, Dellos-Nolan S, Co JY, Lehoux S, Cummings RD, Wozniak DJ, & Ribbeck K (2019). Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nature Microbiology. doi: 10.1038/s41564-019-0581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GX, & Liu ZJ (2017). Potential roles of neutrophils in regulating intestinal mucosal inflammation of inflammatory bowel disease. Journal of Digestive Diseases, 18(9), 495–503. doi: 10.1111/1751-2980.12540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.