Abstract
The photosynthesis machinery in chloroplast thylakoid membranes is comprised of multiple protein complexes and supercomplexes1,2. Here, we show a novel supramolecular organization of photosystem I (PSI) in the moss Physcomitrella patens by single-particle cryo-electron microscopy. The moss-specific light-harvesting complex (LHC) protein Lhcb9 is involved in this PSI supercomplex, which has been shown to have a molecular density similar to that of the green alga Chlamydomonas reinhardtii3. Our results show that the structural organization is unexpectedly different—two rows of the LHCI belt exist as in C. reinhardtii4, but the outer one is shifted toward the PsaK side. Furthermore, one trimeric LHC protein and one monomeric LHC protein position alongside PsaL/K, filling the gap between these subunits and the outer LHCI belt. We provide evidence showing that Lhcb9 is a key factor, acting as a linkage between the PSI core and the outer LHCI belt to form the unique supramolecular organization of the PSI supercomplex in P. patens.
The initial step of photosynthesis is absorption of light energy, which mainly occurs in LHC proteins in chloroplast thylakoid membranes of algae and plants. LHC proteins bind and coordinate the light-harvesting pigments around the photosynthetic reaction centres. They are generally categorized into two groups, LHCI and LHCII, depending on their different molecular affinities for the two photosystems, PSI and PSII, respectively5,6. Both photosystem core structures are highly conserved throughout the evolution of photosynthetic organisms7. However, the supramolecular organization comprised of each photosystem and its bound LHC proteins appears to be variable depending on light conditions as well as among different photosynthetic taxa8. It is therefore conceivable that light-harvesting antenna systems might be evolutionarily optimized in organization to achieve efficient excitation energy transfer to photosystems under different light environments9.
In the plant PSI supramolecular organization, four LHCI proteins are aligned laterally into a crescent shape (the so-called LHCI belt) and positioned along the PsaG/F/J/K interface of the PSI core, forming the PSI–LHCI supercomplex10–12. The analysis of LHCI-deficient lines of Arabidopsis thaliana has shown that the LHCI belt is made of two pairs of an LHCI heterodimer—Lhca1-Lhca4 and Lhca2-Lhca3, with the exception that Lhca5 can substitute partially for Lhca413. Compared to the plant PSI–LHCI supercomplex, the one in the green alga Chlamydomonas reinhardtii has been reported to harbour a larger antenna system14,15—up to nine LHCI proteins (Lhca1–9), forming a double-crescent-shaped antenna complex on the PsaG/F/J/K side of the PSI core4. The double-crescent-shaped LHCI complex is composed of two rows of the LHCI belt, in which the inner belt is seemingly similar to the one in the plant PSI–LHCI supercomplex, while the outer belt may contain up to five additional LHCI proteins positioned outside the inner LHCI belt relative to the PSI core.
The LHC genes of the moss Physcomitrella patens are more diverse than those found in A. thaliana and C. reinhardtii and of interest to study the evolutionary transition of photosystem antenna organization16–18. Interestingly, the P. patens genome has revealed a newly identified LHCII protein, Lhcb9. There are two lines of evidence showing that Lhcb9 can be associated with PSI3 and PSII19. It has been shown that the association of Lhcb9 with PSI is key to the formation of two differently sized PSI–LHCI supercomplexes in P. patens—a smaller and a larger one, with sizes similar to PSI in A. thaliana and C. reinhardtii, respectively3. Although the molecular composition of PSI in P. patens has been previously reported20, the detailed structural organization of its PSI–LHCI supercomplexes has not yet been elucidated. To determine the detailed supramolecular organization by comparative analysis, we performed single-particle electron microscopy of endogenous PSI–LHCI supercomplexes from A. thaliana, C. reinhardtii and P. patens.
We first performed a density gradient analysis using thylakoid membranes isolated similarly from wild type (WT) A. thaliana, C. reinhardtii and P. patens. The results confirmed the previous report3 that A. thaliana WT PSI–LHCI supercomplex (hereafter, At-PSI) and C. reinhardtii WT PSI–LHCI supercomplex (hereafter, Cr-PSI) each have a single PSI–LHCI, of relative lower and higher molecular density, respectively (Fig. 1a). On the other hand, the P. patens WT thylakoids contain two different molecular sizes of PSI–LHCI supercomplexes—a smaller and a larger one (hereafter, Pp-PSI-S and Pp-PSI-L, respectively), with densities similar to that of At-PSI and Cr-PSI, respectively (Fig. 1a). We visualized each PSI supercomplex by single-particle electron microscopy, using both negatively stained and frozen-hydrated samples.
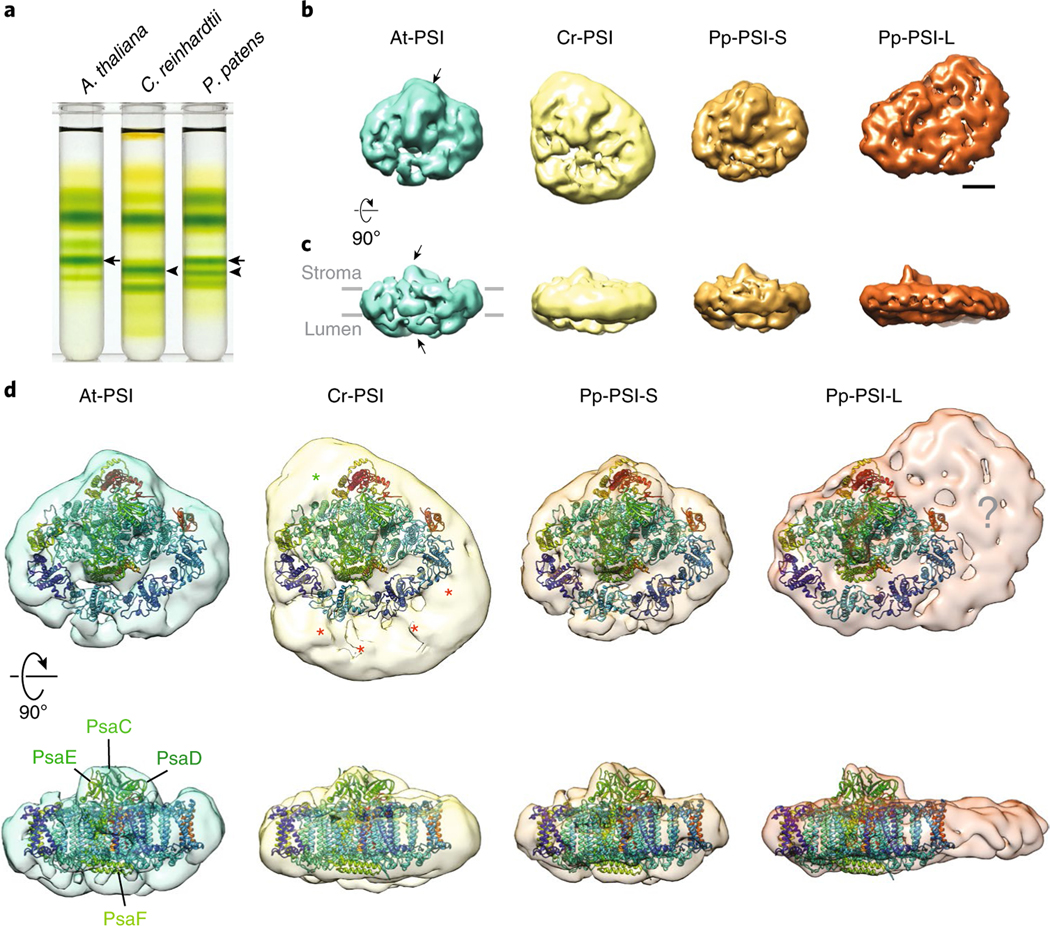
Fig. 1 |. single-particle negative-stained electron microscopy analysis of PSI-LHCI supercomplexes from A. thaliana, C. reinhardtii and P. patens.
a, Isolation of At-PSI, Cr-PSI, Pp-PSI-S and Pp-PSI-L from thylakoid membranes by maltose density gradient ultracentrifugation. Arrows and arrowheads indicate the locations of the bands corresponding to the smaller and larger PSI–LHCI supercomplexes, respectively. A representative result was shown from five technical replicates. b,c, The 3D reconstruction of each negatively stained PSI supercomplex in the same ‘top’ (b) and ‘side’ (c) views with respect to the membrane direction. Arrows point out the positions of the conserved iron-sulfur cluster comprised of the PsaC, D and E subunits recognizable in each structure. d, Comparison of the four PSI–LHCI supercomplexes based on the docking of plant PSI–LHCI (PDB 5L8R) and the relative positions of their additional LHCI densities relative to the core complex. Despite their similarity in size, there are notable differences in the organization of the enlarged antenna in Cr-PSI (stars) and Pp-PSI-L (question mark). Scale bar for b and c, 50 Å.
With between 19,000 and 56,000 single-particle images of each negatively stained PSI supercomplex, we generated two-dimensional (2D) class averages as well as three-dimensional (3D) class volumes using RELION (see Methods, Fig. 1b–d, Supplementary Fig. 1). Initial inspection of the At-PSI 2D electron microscopy data revealed class averages in good agreement with the known plant PSI–LHCI structure. This similarity was further confirmed by the 3D reconstruction showing the characteristic ‘open fan’ shape of the complex, with notable protrusions to accommodate PsaC, D and E on the stromal side and PsaF on the lumenal side (Fig. 1b–d). We subsequently used the 3D reconstruction of At-PSI as a reference structure for alignment and comparison with the other PSI–LHCI complexes. The 2D class averages of the Cr-PSI showed substantial additional density relative to that of At-PSI, which is in agreement with a previous study4 (Supplementary Fig. 1b). The 3D reconstruction of Cr-PSI showed that the extra density corresponds to a larger antenna system that is compatible with the double-crescent structure previously observed (Fig. 1d, red stars). Some additional density was also evident near the PsaH/I subunits (Fig. 1d, green star), which has been suggested to be a monomeric LHC protein, CP2921, but could potentially accommodate other subunits.
We next analysed both Pp-PSI-S and Pp-PSI-L. The 2D class averages for the smaller complex, Pp-PSI-S, showed that the overall structure was very similar to that of At-PSI, and correspondingly the 3D reconstruction also showed the characteristic plant PSI architecture, with four LHCI proteins bound to the PsaG/F/J/K side of the PSI core (Fig. 1b–d). Although P. patens lacks an orthologue of A. thaliana Lhca417, which is suggested to stabilize the At-PSI structure13, it is interesting that the Pp-PSI-S structure is still similar to that of At-PSI at the resolution of this study. On the other hand, despite having a molecular density similar to that of Cr-PSI (Fig. 1a, arrowheads), the larger complex, Pp-PSI-L, showed a very different PSI supercomplex organization (Fig. 1b–d). We could reliably dock the known crystal structure of the plant PSI–LHCI supercomplex into the 3D reconstruction of Pp-PSI-L (Fig. 1d). The resulting model shows one layer of LHCI belt assigned at the PsaG/F/J/K side of the PSI core, just as in At-PSI, but not the double-crescent LHCI structure seen in Cr-PSI. Interestingly, however, it appeared that a large, additional density was present at the PsaL/A/K side of the PSI core (Fig. 1d, question mark). An upper part of this additional density appears to be an LHCII trimer, similar to what has previously been seen for the PSI–LHCI supercomplex in A. thaliana in State 2 conditions22, where the excitation energy is balanced between the two photosystems under preferential excitation of PSII by increasing the PSI antenna size. The other additional extra density next to PsaK and Lhca3 would fit four or five LHC protein monomers.
Previously, it has been shown that the lack of Lhcb9 causes the loss of Pp-PSI-L, and the hexahistidine-tagged Lhcb9 copurified with the PSI–LHCI supercomplex and with LHCII proteins such as Lhcb4 and Lhcbm3. To further investigate the protein composition of the additional density of Pp-PSI-L, we analysed the purified Pp-PSI-S and Pp-PSI-L by mass spectrometry. The results indicated that both Pp-PSI-S and Pp-PSI-L contained the same set of PSI subunits as expected, but Pp-PSI-L additionally contained Lhcb9.1, Lhcb4 and Lhcbm proteins, which are missing in Pp-PSI-S (Supplementary Fig. 2). Thus, these LHCII proteins are likely to correspond to the additional density present in Pp-PSI-L.
As a way to probe this potential assignment, we used the P. patens knockout line of Lhcb9.1/9.2 (PpΔLhcb9) to isolate the PSI–LHCI supercomplex. In this mutant, the Pp-PSI-L was missing, reproducing the results of a previous report3 (Supplementary Fig. 3a). To examine whether the lack of Lhcb9 affects the supramolecular organization of Pp-PSI-S, we performed single-particle electron microscopy analysis of the PSI–LHCI supercomplex from PpΔLhcb9 (PpΔb9-PSI). The additional density found in Pp-PSI-L was completely lost, as expected, and the 3D reconstruction of PpΔb9-PSI was very similar to that of Pp-PSI-S, suggesting that the lack of Lhcb9 does not affect the structural organization of Pp-PSI-S (Supplementary Fig. 3b). Therefore, Lhcb9 is most probably located within the additional density present in Pp-PSI-L and directly interacting with the PSI core, acting as the linker to connect the additional LHC proteins.
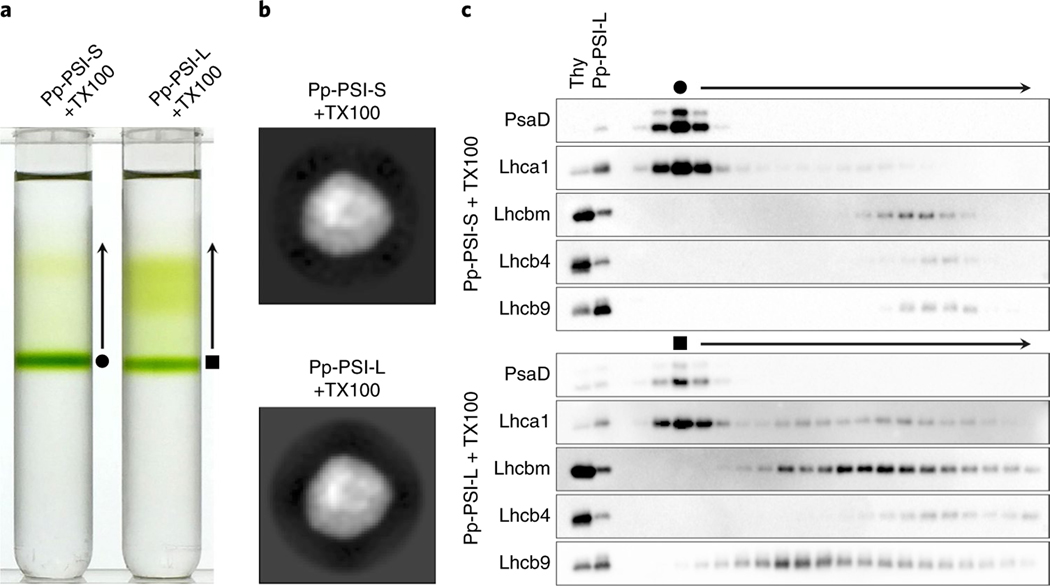
The previous study also suggested that Pp-PSI-L contains more LHCI proteins per PSI core than does Pp-PSI-S3. To verify the involvement of LHCI proteins in the additional density of Pp-PSI-L, we attempted to separate the additional LHC proteins from Pp-PSI-L without removing the regular LHCI belt from the PSI core. The fractions of Pp-PSI-S and Pp-PSI-L were collected from the first ultracentrifugation and subjected to a second ultracentrifugation with maltose gradients containing 0.03% (w/v) Triton X-100 (TX100) to substitute for n-dodecyl-α -D-maltoside (α -DM) (see Methods for detail). A distinct green band appeared at a similar position in the gradients for both the TX100-treated Pp-PSI-S and Pp-PSI-L (Fig. 2a). Single-particle electron microscopy analysis confirmed that the distinct band in both corresponded to the intact PSI–LHCI supercomplex (that is, one PSI core and one LHCI belt; Fig. 2b). Thus, substituting detergents resulted in separation of the additional density from Pp-PSI-L but kept the standard unit of the PSI–LHCI supercomplex intact. Mass spectrometry data supported these results (Supplementary Fig. 4). Apparently, the TX100-treated Pp-PSI-L generated the additional green, lighter fraction (Fig. 2a). The mass spectrometry analysis indicated that LHCI, Lhcb9, Lhcb4 and Lhcbm proteins were the main components of this lighter fraction (Supplementary Table 1). Moreover, the immunoblot analysis revealed that LHCI and Lhcb9 were located within the same fraction, indicating that LHCI and Lhcb9 interact with each other (Fig. 2c), as previously suggested3. We conclude that the additional density in Pp-PSI-L contains the Lhcb9-LHCI belt complex and an LHCII trimer, forming a larger supercomplex than Pp-PSI-S.
Fig. 2 |. The protein composition of the additional antenna complex of Pp-PSI-L.
a, The separation of the additional antenna complex from Pp-PSI-L. The fractions of Pp-PSI-S and Pp-PSI-L were collected from the first density gradient ultracentrifugation and re-subjected to a second ultracentrifugation using maltose gradients containing 0.03% (w/v) TX100, which separated the additional antenna complex from the PSI but kept the standard unit of PSI–LHCI supercomplex intact. A representative result was shown from three technical replicates. b, The single-particle negative-stained electron microscopy 2D analysis of the protein in the distinct green bands produced from the TX100-treated Pp-PSI-S and Pp-PSI-L (filled circle and filled square, respectively). Both indicate the structure equivalent to Pp-PSI-S, suggesting that only the additional antenna complex is removed after TX100 treatment. Representative images were shown from 60 class averages. The dataset sizes for each structure were summarized in Supplementary Table 2. c, Immunoblot analysis showing the location of PSI, LHCI, Lhcb9, Lhcb4 and Lhcbm. The filled circle and filled square indicate the fraction of the distinct green band that appeared as a result of the second ultracentrifugation shown in a. Arrows indicate fractions lighter than the distinct green bands shown in a. Thy, 0.5 μg Chl thylakoids as a control. Next to thylakoids, the same amount of Pp-PSI-L (0.5 μg Chl) was also loaded as a control. Representative results were shown from 2–3 technical replicates.
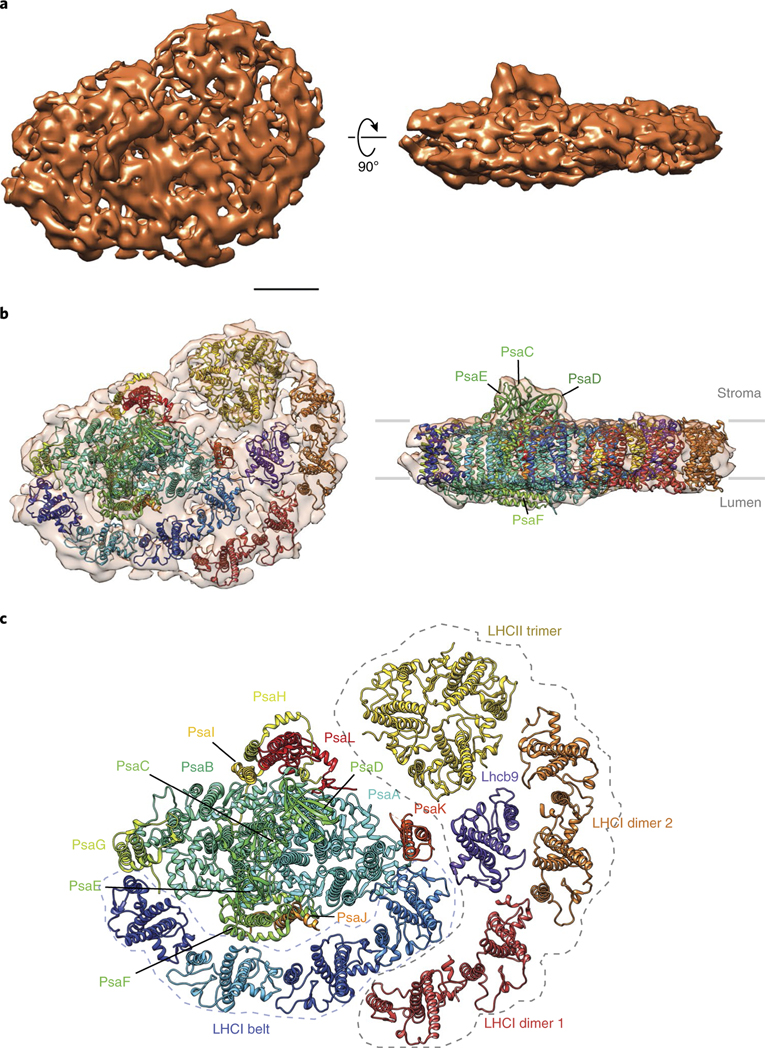
Our observations suggest that: (1) a trimeric LHCII can bind to the PsaL/A/K side of PSI (Fig. 1d), (2) Lhcb9 is associated with the PSI–LHCI supercomplex3, (3) the lack of Lhcb9 causes the loss of Pp-PSI-L (Supplementary Fig. 3 and see ref. 3) and (4) Lhcb9 and the LHCI belt form a protein subcomplex (Fig. 2). To determine the localization of each LHC protein in the additional density of Pp-PSI-L, we performed single-particle cryo-electron microscopy analysis (see Methods and Supplementary Fig. 5 for details). The results validated the overall structure observed by negative-stained single-particle electron microscopy. Known crystal structures of plant PSI and several LHC proteins (that is, trimeric, dimeric and monomeric LHC proteins) were fitted into the 11.6-Å resolution 3D density map of Pp-PSI-L (Fig. 3, see Methods for details). The docking matched the protein composition from previous crystal structures of plant PSI 11,12 and mass spectrometry data (Supplementary Figs. 2 and 4). In particular, the additional density contained one clear LHCII-like trimer contacting an unassigned density adjacent to PsaA that could putatively be attributed to the N terminus of the nearby PsaL and/or PsaH, which were not resolved in the crystal structure. More interestingly, another set of LHCI belt-like organization, a dimer of LHCI-like dimers, was also found, but positioned toward the PsaK side of the PSI core, forming an arc between Lhca3 of PSI on one end and the LHCII trimer at the other while contacting both. An eighth monomeric LHC protein density was found at the core of the LHCI antenna, between the LHCII trimer and the additional LHCI belt-like structure, contacting PSI mostly through PsaK and Lhca3. Although it is not definitive, we propose that this central location probably corresponds to Lhcb9 on the basis of the observations described above. The association of a trimeric LHCII with PSI is unlikely to be the primary factor for the new antenna organization we are describing, because such association is also observed in both C. reinhardtii21 and A. thaliana22, yet the organization we see for Pp-PSI-L has not been observed for those species. We propose that it is Lhcb9 that plays a key role in forming the unique PSI antenna organization in Pp-PSI-L due to its strategic interactions with both the PsaK side and the LHCI belt (Figs. 2 and 3).
Fig. 3 |. The supramolecular organization of Pp-PSI-L observed by single-particle cryo-electron microscopy analysis.
a,b, 3D reconstruction and docking of Pp-PSI-L in the top (left) and side views (right panels). Scale bar for a and b, 50 Å. c, Fitted crystal structures showing the detailed organization of the extended LHCI antenna with respect to the PSI complex (dotted line contour). Lhca1–4 from the plant PSI crystal structure (5L8R) were split into two distinct rigid body dimers (Lhca1–4 as dimer 1 in red and Lhca2–3 as dimer 2 in orange) to be fitted into the additional LHCI belt density of Pp-PSI-L. It should be noted, however, that the orthologue of A. thaliana Lhca4 is missing in P. patens. Thus, the location for Lhca4 is replaced by another yet unidentified LHCI protein. In addition to the second belt, an LHCII-like trimer contacts PsaA, K and L, and the putative Lhcb9 central to the antenna. See Supplementary Fig. 8 for putative components of the unassigned density in Pp-PSI-L.
As the PSI supercomplex in C. reinhardtii is larger than that of A. thaliana, it is tempting to consider that the presence of two different sizes of PSI supercomplexes in P. patens might be related to an evolutionary advantage of this system18. However, the actual reason for the presence of two different sizes of PSI supercomplexes in P. patens remains to be determined. It has been shown that the additional antenna complex in Pp-PSI-L increases the total functional antenna size of PSI by about 40%, while maintaining efficient energy transfer3. It has also been shown that Pp-PSI-L becomes less abundant under high-light conditions3 (reconfirmed in Supplementary Fig. 6), suggesting that the presence of Pp-PSI-L may be beneficial for increasing light-harvesting capacity under low-light conditions. Previously, the binding of a trimeric LHCII with PSI was observed as a result of state transitions21,22, which are short-term responses to the redox change of the plastoquinone pool in thylakoid membranes. The primary cause for forming Pp-PSI-L is unlikely to be state transitions, because the PpΔLhcb9 line is still able to perform state transitions without forming Pp-PSI-L3. However, it could be related to a long-term acclimation to conditions in which increased PSI photochemistry is required.
Our preliminary results revealed an intriguing relationship between the formation of Pp-PSI-L and the external carbon source (Supplementary Fig. 7), which is known to affect the balance between chloronema and caulonema development in P. patens23,24—the chloronema cells are more photosynthetically active, whereas the caulonema cells contain fewer and smaller chloroplasts and serve to expand the colony25. Our observations suggest that formation of Pp-PSI-L occurs when caulonema development is promoted. Surprisingly, the gene expression level of Lhcb9.1 is also upregulated when caulonema growth is favoured26. Photosynthesis in caulonema cells is not well understood yet because of fewer and smaller chloroplasts, suggesting that a different mode of photosynthesis (for example, photoheterotrophy) might operate. Such a mode of photosynthesis might affect the redox state of the plastoquinone pool, which is also suggested to control the expression of Lhcb927. These observations may lead us to future studies that explore the physiological and functional advantages of Pp-PSI-L formation during substrate colonization.
Methods
Strains and growth conditions.
A. thaliana WT (ecotype Columbia-0) was grown on soil at 25 °C under ~80 μmol photons m−2 s−1 with a 10-h light and 14-h dark cycle for 5 weeks. C. reinhardtii WT (4A+) was grown under ~50 μmol photons m−2 s−1 in Tris acetate phosphate liquid medium28 at 25 °C until mid-log phase. P. patens WT (Gransden 2004) and ΔLhcb9 (ΔLhcb9.1/9.2) 3 lines were grown on a layer of cellophane overlaid on BCDAT agar medium supplemented with glucose (5 g l−1)29 at 25 °C under continuous light at ~ 50 μmol photons m−2 s−1.
Isolation of PSI–LHCI supercomplexes and protein analysis.
For each sample, thylakoid membranes were first purified by sucrose cushion centrifugation as described previously3,30. To isolate PSI–LHCI supercomplex, thylakoid membranes (100 μg chlorophyll (Chl) at 0.5 mg Chl per ml) were solubilized with 1% (w/v) α -DM (Anatrace) for 30 min on ice with gentle agitation, unsolubilized membranes were removed by centrifugation at 20,000g for 5 min at 4 °C and the solubilized membrane samples were subjected to maltose density gradient ultracentrifugation (0.1–1.3 M maltose with 25 mM MES-NaOH (pH 6.5) and 0.03% (w/v) α -DM) at 154,300g (SW 41 Ti rotor, Beckman Coulter) for 24 h at 4 °C. The separated bands of PSI–LHCI supercomplexes were collected dropwise from the bottom of the tube and immediately used for single-particle electron microscopy analysis. To increase the purity of the samples, 4–6 density gradient tubes were prepared as with the first ultracentrifugation and the collected fractions (for example, Pp-PSI-L) were diluted with 25 mM MES-NaOH (pH 6.5) and 0.03% (w/v) α -DM, concentrated using centrifugal filters (Amicon Ultra, 100 kDa MWCO) and re-subjected to a second maltose density gradient ultracentrifugation as described above. To separate the additional antenna complex from Pp-PSI-L, the concentrated PpPSI-L was subjected to a second ultracentrifugation as described above, except that 0.03% (w/v) TX100 was used instead of 0.03% (w/v) α -DM. Protein electrophoresis and immunoblot analysis were done as described previously3. Chl concentration was measured according to the previous method31.
Electron microscopy.
PSI–LHCI supercomplexes from each strain were prepared for electron microscopy in negative stain on continuous carbon film-coated Cu grids (400 mesh, Electron Microscopy Sciences). Then, 4 μl of each sample, diluted to approximately 30 nM without maltose, was incubated for 30 s on grids pretreated in a Solarus plasma cleaner (Gatan, Inc.) and subsequently stained with four 40-μl drops of 1% uranyl formate (SPI) before blotting and drying. Cryo-electron microscopy samples were prepared on carbon film-coated C-Flats (Protochips). Samples were diluted to 30–100 nM, determined by trial and error for optimal particle distribution for imaging, with 25 mM MES-NaOH (pH 6.5), 0.03% (w/v) α -DM and 1% trehalose (w/v). Plasma-cleaned grids were mounted in a Vitrobot (FEI) equilibrated at 10 °C and 100% relative humidity with light turned off. Next, 4 μl of diluted sample was incubated for 1 min before blotting and immediately plunge-frozen in liquid ethane. Frozen grids were transferred to a DH626 cryoholder (Gatan, Inc.) and observed at liquid nitrogen temperature. Both negative stain and cryo-electron microscopy samples were loaded into a Tecnai 20F transmission electron microscope (FEI) and imaged at an acceleration voltage of 120 kV on an UltraScan4000 charged coupled device camera (Gatan, Inc.) with Leginon, at a calibrated magnification of 109,100 (1.38 Å per pixel at the specimen) and a total dose of 35 and 25 electrons per pixel (for negative stain and cryoelectron microscopy, respectively).
Electron microscopy data processing.
Contrast transfer function parameters were determined with CTFFIND332 for all dataset micrographs and particles were selected automatically using Gautomatch (version 0.53, http://www.mrc-lmb.cam.ac.uk/kzhang/) (Supplementary Table 2). All subsequent 3D and 2D classification and refinement steps were performed using RELION (version 1.4)33. Briefly, data were sorted using an initial step of 3D classification into 3–5 classes starting with an initial model filtered down to 60-Å resolution. The A. thaliana PSI data were classified starting with a low-pass filtered map simulated from the Pisum sativum PSI crystal structure (Protein Data Bank (PDB) 5L8R). After sorting, a final 3D electron microscopy density map was calculated for At-PSI. This map was used as initial reference for all subsequent negative stain data sets processed. The lowpass filtered Pp-PSI-L density map from negative stain electron microscopy was used as the initial model for the 3D classification of the cryo-electron microscopy data. Each 3D class was analysed by 2D classification and selected or rejected for further processing by visual inspection, rejecting impurities and low-quality classes (Supplementary Figs. 1 and 5). Culled and pooled same-complex classes were further refined, then postprocessed with a soft mask around the density. Final reconstructions were low-pass filtered at the final resolution determined by the Fourier shell correlation (FSC) at 0.143 cutoff in RELION with a B-factor of − 1,500. To further assess the quality of the 3D map, we used 3D FSC to estimate the effect of preferential orientations34, which yielded an anisotropy score of 0.88 and a global resolution of 11.6 Å from the half maps, identical to the RELION estimate at 0.143 cutoff. Comparison of 3D FSC-filtered maps to our original map filtered in RELION did not indicate any noticeable preferential orientation artefact. Rigid body docking of known crystal structures into our final density map was performed using UCSF Chimera35, as well as final volume rendering and map alignments. Density maps were aligned to each other for comparison based on best cross-correlation alignment of the maps in the common PSI area of the densities. Docking of the full Pp-PSI-L cryo-electron microscopy map was done in a stepwise manner, starting with the largest model and finishing with the smallest. We used Chimera fitmap highest scoring fit (correlation and overlap), subsequently subtracting the region from the map with vop subtract. Following this approach, we sequentially docked the P. sativum PSI–LHCI supercomplex (PDB 5L8R), an LHCII trimer (PDB 2BHW), an LHCI belt (PDB 5L8R) and an LHC monomer (Lhcb1 from PDB 2BHW) into our 3D density. The second LHCI tetramer was docked as two dimers to match more closely the different arc in the extended Pp-PSI-L antenna, and P. sativum Lhca3 and Lhcb1 monomers were used in place of Lhcb9 as its closest homologue with published crystal structure. Remaining unassigned densities after subtraction of the main subunits were compared to putative candidate sizes with Chimera molmap.
Mass spectrometry.
Samples of trypsin-digested proteins were analysed using a Synapt G2-Si high-definition ion mobility mass spectrometer equipped with an electrospray ionization source and connected in line with an Acquity M-class ultra-performance liquid chromatograph (Waters, Milford, MA). The ultraperformance liquid chromatograph was equipped with trapping (Symmetry C18: 180 μm × 20 mm, 5 μm, Waters) and analytical (HSS T3: 75 μm × 250 mm, 1.8 μm, Waters) columns. Solvent A was 99.9% water/0.1% formic acid, and solvent B was 99.9% acetonitrile/0.1% formic acid (v/v). The elution programme consisted of isocratic flow at 1% (v/v) B for 2 min, a linear gradient from 1% to 35% B over 30 min, a linear gradient from 35% to 85% B over 2 min, isocratic flow at 85% B for 4 min, a linear gradient from 85% to 1% B over 2 min and isocratic flow at 1% B for 20 min, at a flow rate of 300 nl min-1. Ion mobility-enabled high-definition mass spectrometry and tandem mass spectrometry data were acquired in the positive ion mode and in continuum format, operating the time-of-flight analyser in resolution mode, with a scan time of 0.5 s, over the range m/z = 50–2,000. An optimized wave velocity of 850 m s−1 was used for the travelling wave ion mobility cell. Collision-induced dissociation was performed in the ion transfer cell with a collision energy ramp from 30 to 75 V. Data acquisition was controlled using MassLynx software (version 4.1) and data analysis was performed using Protein Lynx Global Server software (version 3.0.2, Waters).
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The 3D cryo-EM density map of Pp-PSI-L has been deposited in the Electron Microscopy Data Bank under accession code EMD-9107, and with the Protein Data Bank (PDB) under accession code 6MEM. The data are available from the corresponding authors (M.I. and K.K.N.) upon request.
Supplementary Material
Acknowledgements
The QB3/Chemistry Mass Spectrometry Facility at the University of California, Berkeley receives support from the National Institutes of Health (grant 1S10OD020062-01). This work was supported by the US Department of Energy, Office of Science, through the Photosynthetic Systems programme in the Office of Basic Energy Sciences. E.N. and K.K.N. are investigators of the Howard Hughes Medical Institute.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41477-018-0271-1.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dekker JP & Boekema EJ Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12–39 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Nelson N & Yocum CF Structure and function of photosystems I and II. Annu. Rev. Plant Biol 57, 521–565 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Iwai M. et al. Light-harvesting complex Lhcb9 confers a green alga-type photosystem I supercomplex to the moss Physcomitrella patens. Nat. Plants 1, 14008 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Drop B. et al. Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J. Biol. Chem 286, 44878–44887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Wobbe L, Bassi R & Kruse O. Multi-level light capture control in plants and green algae. Trends Plant Sci. 21, 55–68 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Nelson N & Junge W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem 84, 659–683 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Büchel C. Evolution and function of light harvesting proteins. J. Plant Physiol 172, 62–75 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Neilson JA & Durnford DG Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res 106, 57–71 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Jensen PE et al. Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 1767, 335–352 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Qin X, Suga M, Kuang T & Shen JR Structural basis for energy transfer pathways in the plant PSI–LHCI supercomplex. Science 348, 989–995 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Mazor Y, Borovikova A, Caspy I & Nelson N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 3, 17014 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Wientjes E, Oostergetel GT, Jansson S, Boekema EJ & Croce R. The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. J. Biol. Chem 284, 7803–7810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germano M. et al. Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525, 121–125 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Kargul J, Nield J & Barber J. Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii. Insights into light harvesting for PSI. J. Biol. Chem 278, 16135–16141 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Rensing SA et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Alboresi A, Caffarri S, Nogue F, Bassi R & Morosinotto T. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS ONE 3, e2033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwai M & Yokono M. Light-harvesting antenna complexes in the moss Physcomitrella patens: implications for the evolutionary transition from green algae to land plants. Curr. Opin. Plant Biol 37, 94–101 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Alboresi A, Gerotto C, Cazzaniga S, Bassi R & Morosinotto T. A red-shifted antenna protein associated with photosystem II in Physcomitrella patens. J. Biol. Chem 286, 28978–28987 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busch A. et al. Composition and structure of photosystem I in the moss Physcomitrella patens. J. Exp. Bot 64, 2689–2699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drop B, Yadav KNS, Boekema EJ & Croce R. Consequences of state transitions on the structural and functional organization of photosystem I in the green alga Chlamydomonas reinhardtii. Plant J. 78, 181–191 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Kouřil R. et al. Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44, 10935–10940 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Olsson T, Thelander M & Ronne H. A novel type of chloroplast stromal hexokinase is the major glucose-phosphorylating enzyme in the moss Physcomitrella patens. J. Biol. Chem 278, 44439–44447 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Thelander M, Olsson T & Ronne H. Effect of the energy supply on filamentous growth and development in Physcomitrella patens. J. Exp. Bot 56, 653–662 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Schumaker KS & Dietrich MA Programmed changes in form during moss development. Plant Cell 9, 1099–1107 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiyama T. et al. Involvement of PpDof1 transcriptional repressor in the nutrient condition-dependent growth control of protonemal filaments in Physcomitrella patens. J. Exp. Bot 63, 3185–3197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinnola A. et al. A LHCB9-dependent photosystem I megacomplex induced under low light in Physcomitrella patens. Nat Plants. 10.1038/s41477-018-0270-2 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Gorman DS & Levine RP Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 54, 1665–1669 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M & Hasebe M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 7, 9–17 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Iwai M, Takahashi Y & Minagawa J. Molecular remodeling of photosystem II during state transitions in Chlamydomonas reinhardtii. Plant Cell 20, 2177–2189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porra RJ, Thompson WA & Kriedemann PE Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989). [Google Scholar]
- 32.Mindell JA & Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan YZ et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D cryo-EM density map of Pp-PSI-L has been deposited in the Electron Microscopy Data Bank under accession code EMD-9107, and with the Protein Data Bank (PDB) under accession code 6MEM. The data are available from the corresponding authors (M.I. and K.K.N.) upon request.