Abstract
Non-small-cell lung cancer (NSCLC) is one of the most devastating diseases worldwide. The study is aimed at identifying reliable prognostic biomarkers and to improve understanding of cancer initiation and progression mechanisms. RNA-Seq data were downloaded from The Cancer Genome Atlas (TCGA) database. Subsequently, comprehensive bioinformatics analysis incorporating gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and the protein-protein interaction (PPI) network was conducted to identify differentially expressed genes (DEGs) closely associated with NSCLC. Eight hub genes were screened out using Molecular Complex Detection (MCODE) and cytoHubba. The prognostic and diagnostic values of the hub genes were further confirmed by survival analysis and receiver operating characteristic (ROC) curve analysis. Hub genes were validated by other datasets, such as the Oncomine, Human Protein Atlas, and cBioPortal databases. Ultimately, logistic regression analysis was conducted to evaluate the diagnostic potential of the two identified biomarkers. Screening removed 1,411 DEGs, including 1,362 upregulated and 49 downregulated genes. Pathway enrichment analysis of the DEGs examined the Ras signaling pathway, alcoholism, and other factors. Ultimately, eight prioritized genes (GNGT1, GNG4, NMU, GCG, TAC1, GAST, GCGR1, and NPSR1) were identified as hub genes. High hub gene expression was significantly associated with worse overall survival in patients with NSCLC. The ROC curves showed that these hub genes had diagnostic value. The mRNA expressions of GNGT1 and NMU were low in the Oncomine database. Their protein expressions and genetic alterations were also revealed. Finally, logistic regression analysis indicated that combining the two biomarkers substantially improved the ability to discriminate NSCLC. GNGT1 and NMU identified in the current study may empower further discovery of the molecular mechanisms underlying NSCLC's initiation and progression.
1. Introduction
As one of the most devastating diseases worldwide, lung cancer causes nearly 1.6 million mortalities each year [1–3]. Approximately 85% of lung cancers are characterized as non-small-cell lung cancer (NSCLC) [4–6], which is typically classified into two subtypes, squamous cell carcinoma (SCC) and adenocarcinoma (AD), using standard pathology methods [7–10]. Tobacco smoking is the most common risk factor for lung cancer. Smoking is also associated with multiple risks, including worse tolerance of treatment, higher risk of failure and second primary tumors, and poorer quality of life. Indeed, it has become clear that the significant reduction in tobacco consumption would result in the prevention of a large fraction of lung cancer cases and other smoking-related diseases [11–13].
In addition, other factors such as air pollution, poor diet, occupational exposure, and hereditary factors have been reported in association with NSCLC in nonsmokers [14–16]. Over the past few years, newly developed cytotoxic agents, including paclitaxel, gemcitabine, and vinorelbine, have emerged to offer multiple therapeutic choices for patients with LUAD [17–20]. However, chemotherapy for advanced NSCLC is often considered ineffective or excessively toxic [21–23].
In an attempt to improve treatments for NSCLC, new therapeutic strategies, such as the development of noncytotoxic targeted agents, have emerged [24–27]. Moreover, the targeted therapies have significantly improved clinical outcomes in a subset of lung cancer patients whose tumors harbor EGFR [28], ALK [29, 30], and HER2 alterations [31–33].
Despite recent advances in cancer treatment, unfortunately, the current five-year survival rate of NSCLC remains unsatisfactory [34–37]. Thus, it is imperative to identify potential biomarkers and explore NSCLC's underlying biological mechanisms.
In recent years, bioinformatics analysis has been utilized as a powerful tool to explore novel prognostic and therapeutic biomarkers and to unveil the potential mechanisms of NSCLC [38–41]. For instance, a novel model including seven genes was reported to indicate a promising prognostic biomarker for lung SCC patients using integrated bioinformatics methods [41–43]. In addition, studies used comprehensive bioinformatics analysis to show that the cell cycle pathway may play a significant role in NSCLC in nonsmokers [44–47].
In the present study, RNA-Seq data were downloaded from The Cancer Genome Atlas (TCGA) database. Then, the EdgeR package was applied to uncover differentially expressed genes (DEGs) between NSCLC tissues and normal tissues. Using the resulting data, this study is aimed at unveiling the underlying molecular mechanism of NSCLC onset and progression through gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and the protein-protein interaction (PPI) network. Subsequently, cytoHubba, a novel Cytoscape plugin, was used to reveal the hub genes from 12 topological analysis methods. Furthermore, the prognostic and diagnostic values of the hub genes were further confirmed by survival analysis and receiver operating characteristic (ROC) curve analysis.
The screening revealed two key genes, GNGT1 and NMU, and the protein expressions of these genes were validated by the Human Protein Atlas online database at the system level. Their genetic alteration and coexpression were also revealed. Finally, a logistic regression model was built to evaluate the combined diagnostic capability of GNGT1 and NMU.
2. Materials and Methods
2.1. Downloading of TCGA Datasets and DEG Screening
The mRNA expression data of NSCLC patients were downloaded from the TCGA database (https://cancergenome.nih.gov/) [48]. The criteria used were as follows: primary site (lung), data category (Transcriptome Profiling), project ID (TCGA-LUAD and TCGA-LUSC), experimental strategy (RNA-Seq), and workflow type (HTSeq-counts). The other filters were kept as default. Practical Extraction and Reporting Language (Perl) was utilized to extract the sample information, generate the mRNA expression matrix, and annotate gene symbols. Finally, data from a cohort containing 1,145 samples were obtained from TCGA. Of these 1,145 samples, there were 108 normal tissue and 1,037 NSCLC samples, respectively. The EdgeR package from Bioconductor was used to screen the DEGs between normal tissue and NSCLC [49–51]. The adjusted P < 0.001, and fold change (FC) > 4 were set as the cutoff criteria.
2.2. DEG Functional Enrichment Analysis
Gene ontology (GO) analysis provides a standardized description of gene products in terms of molecular function (MF), biological process (BP), and cellular component (CC) [52]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database offering gene functional meanings and expressed proteins [53]. GO and KEGG enrichment analyses were conducted using the powerful online tool DAVID (DAVID, https://david.ncifcrf.gov/) and visualized by the R package “ggplot2” [54]. In addition, P < 0.05 was considered to indicate statistical significance.
2.3. Constructing the Protein-Protein Interaction Network
The Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) database, a database that integrates all functional interactions between proteins, was used to build the PPI network [55]. An interaction score of ≥0.4 was considered statistically significant.
2.4. Hub Gene Selection and Analysis
A Cytoscape plugin, Molecular Complex Detection (MCODE), was utilized to screen modules of PPI networks with a node score cutoff of 0.2, degree cutoff of 2, k-core of 2, and max depth of 100. A P value of <0.05 was considered statistically significant. Next, the DEGs were ranked by cytoHubba [56], which contains 12 algorithms: Maximal Clique Centrality, Edge Percolated Component, Betweenness, Density of Maximum Neighborhood Component, Degree, Bottleneck, Eccentricity, Closeness, Radiability, Maximum Neighborhood Component, Stress, and Clustering Coefficient. The MCODE and cytoHubba results were combined to identify the hub genes.
2.5. Survival Analysis of Hub Genes
Whether the expression level of hub genes was associated with overall survival was investigated using the Kaplan–Meier plotter (http://www.kmplot.com/). An online database is capable of assessing the effect of 54,675 genes on survival using 10,461 cancer samples, including samples from 2,437 lung cancer, 1,065 gastric cancer, 1,816 ovarian cancer, and 5,143 breast cancer patients. P < 0.05 (Cox) was considered statistically significant.
2.6. ROC Curve
The ROC curve analysis was applied to evaluate the specificity and sensitivity of the hub genes. The area under the curve (AUC) and P value were calculated. P < 0.05 was considered to denote statistical significance.
2.7. Validation of Hub Genes
The expression level of hub genes in LUAD was validated by Oncomine (https://www.oncomine.org/resource/login.html) [57]. The threshold was set as the following: P < 1E − 4, fold change > 2, and gene ranking in the top 10%.
2.8. Human Protein Atlas
The Human Protein Atlas (https://www.proteinatlas.org) is an online website that includes immunohistochemical data of nearly 20 types of tumors [58]. In our study, immunohistochemical images were used to directly compare the expression of biomarkers in normal and NSCLC tissues. The intensity of antibody staining indicated the protein expression of hub genes.
2.9. Genetic Alteration of Hub Genes
The cBio Cancer Genomics Portal (http://www.cbioportal.org/) is an open platform that provides visualization, analysis, and downloads of large-scale cancer genomic datasets for various cancer types [59]. Complex cancer genomic profiles can be easily obtained using the portal's query interface, enabling researchers to explore and compare genetic alterations across samples. cBioPortal was used to explore genetic alterations, coexpression, and overall survival of two hub genes, GNGT1 and NMU.
2.10. Statistical Analysis
SPSS version 23.0 (SPSS Inc., Chicago, IL, USA) was used to perform logistic regression analysis. ROC curves were generated to evaluate the diagnostic accuracy of GNGT1 and NMU, and AUC was used to evaluate sensitivity and specificity.
3. Results
3.1. Identification of DEGs in NSCLC
The workflow is shown in Figure 1(a). DEGs were identified using the criteria of P < 0.001 and FC > 4. A total of 1,411 DEGs were screened out between NSCLC and normal samples, including 1,362 upregulated genes and 49 downregulated genes (Figures 1(b) and 1(c)).
Figure 1.

Identification of DEGs in NSCLC. (a) Workflow for the identification of key pathways and genes between non-small-cell lung cancer and normal samples. (b) DEGs between LUAD tissue and normal tissue. The volcano plot showed 1,411 DEGs. The red dots represented the upregulated genes, while the green dots represented downregulated genes. DEGs: differentially expressed genes. (c) Heatmap of the 20 upregulated and downregulated DEGs. The red color represents high expression, and the blue color represents low expression.
3.2. Functional and Pathway Analysis of DEGs
To further investigate the specific function of these genes, all DEGs were uploaded to the online tool DAVID. GO analysis revealed that in terms of BP, the DEGs were associated with nucleosome assembly, transcription from RNA polymerase II promoter, telomere organization, flavonoid glucuronidation, and DNA replication-dependent nucleosome assembly.
When examined in terms of MF, DEGs were enriched in protein heterodimerization activity, retinoic acid-binding, hormone activity, glucuronosyltransferase activity, and extracellular ligand-gated ion channel activity. Regarding CC, the DEGs were mainly enriched in the extracellular region, cornified envelope, nucleosome, extracellular space, and intermediate filament. KEGG analysis found that the DEGs were predominantly involved in the Ras signaling pathway, nicotine addiction, steroid hormone biosynthesis, alcoholism, and systemic lupus erythematosus (Figure 2(a)).
Figure 2.

Functional and pathway analysis of DEGs. (a) GO and KEGG analysis of DEGs. P value is displayed on the x-axis, and GO function enrichment and KEGG pathway are shown on the y-axis. GO: gene ontology. KEGG: Kyoto Encyclopedia of Genes and Genomes. (b) The overlapping genes of different algorithms selected by cytoHubba. (c) The most significant modules obtained from the PPI network. PPI: protein-protein interaction.
3.3. PPI Network Construction, Module Analysis, and Hub Gene Selection
The PPI network was constructed using the STRING database and visualized in Cytoscape. The PPI network consisted of 787 nodes and 2,104 edges, including 1,362 upregulated genes and 49 downregulated genes. The overlapping genes of different algorithms selected by cytoHubba were GNGT1, GNG4, NMU, GCG, TAC1, GAST, NPSR1, and GCGR (Figure 2(b)). The top modules were then extracted from the PPI network (Figure 2(c)).
3.4. Survival Analysis
The Kaplan–Meier plotter was used to predict the prognostic value of the six identified hub genes. The results demonstrated that high expressions of GNGT1 (HR = 1.17 (1.03–1.33), logrank P = 0.017), GNG4 (HR = 1.42 (1.2–1.67), logrank P = 4.4e − 05), NMU (HR = 1.48 (1.3–1.68), logrank P = 2.5e − 09), GCG (HR = 1.15 (1.01–1.31), logrank P = 0.031), TAC1 (HR = 1.23 (1.08–1.39), logrank P = 0.0017), GAST (HR = 1.27 (1.12–1.44), logrank P = 0.00025), GCGR (HR = 0.79 (0.69–0.89), logrank P = 0.00022), and NPSR1 (HR = 1.21 (1.02–1.42), logrank P = 0.024) were associated with worse overall survival for NSCLC patients (Figure 3).
Figure 3.

The prognostic value of hub genes in NSCLC patients. Kaplan–Meier curve analysis between hub gene expression and prognosis in NSCLC patients from the KM plotter database.
3.5. ROC Curve
According to ROC curve analysis, the AUCs of GNGT1, GNG4, NMU, GCG, TAC1, GAST, GCGR1, and NPSR1 were 0.9027 (P < 0.0001), 0.8729 (P < 0.0001), 0.9323 (P < 0.0001), 0.559 (P < 0.0432), 0.6822 (P < 0.0001), 0.7426 (P < 0.0001), 0.816 (P < 0.0001), and NPSR1 0.8949 (P < 0.0001), respectively (Figure 4(a)).
Figure 4.
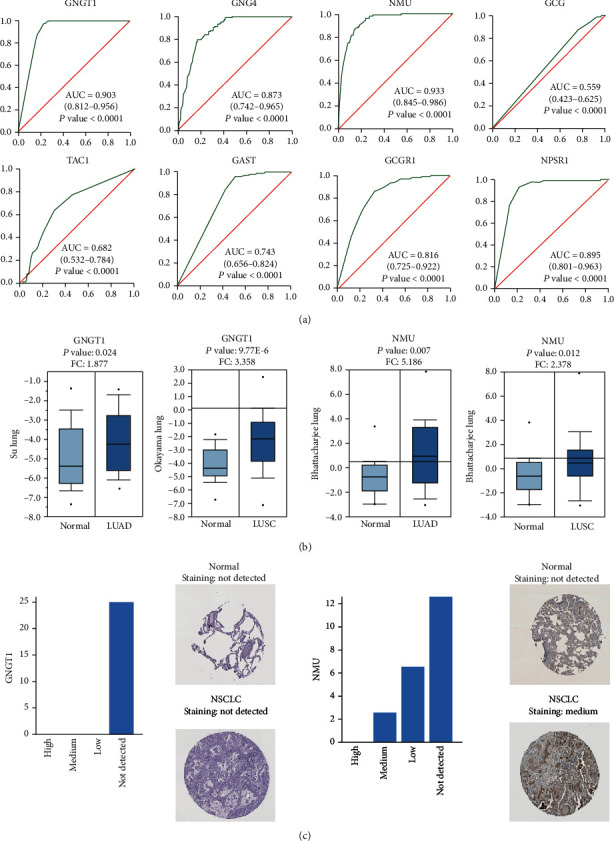
The expression and prognostic value of four hub genes in NSCLC patients. (a) The ROC curves of hub genes. AUC and P values of each hub gene are displayed in the plot. ROC: receiver operating characteristic. AUC: area under the curve. (b) Expression levels of significant genes compared between different types of NSCLC and normal tissues from the Oncomine platform. Fold changes and P values of each hub gene are displayed in the plot. (c) Immunohistochemical analysis of GNGT1 and NMU in normal tissues and NSCLC tissues from the Human Protein Atlas.
3.6. Validating Hub Gene Expression
The Oncomine database was used to validate the expression of hub genes. The results demonstrated that GNGT1 had high expression in LUAD (P: 0.024, FC: 1.877) and LUSC (P: 9.77E-6, FC: 3.358). In Bhattacharjee's study, NMU showed high expression in LUAD (P: 0.007, FC: 5.186) and LUSC (P: 0.012, FC: 2.378) (Figure 4(b)).
3.7. Human Protein Atlas
After studying the mRNA expression of hub genes in NSCLC, we tried to explore the protein expression of hub genes using the Human Protein Atlas. The results revealed that NMU protein was not expressed in normal lung tissues, whereas medium expression of NMU protein was observed in the NSCLC tissues. However, GNGT1 was not detected in either normal lung tissues or NSCLC tissues (Figure 4(c)).
3.8. Genetic Alteration of Hub Genes
The two hub genes altered in 22 (4%) of the 584 patients, and the frequency of alteration of each hub gene, are shown in Figure 5(a). GNGT1 and NMU were altered most often (2.7% and 1.7%, respectively), with mutation, amplification, and mRNA upregulation as the main types of alterations observed (Figure 5(b)). The expression of GNGTA was correlated with NMU (Spearman: 0.13, P = 2.415e − 3; Pearson = 0.13, P = 4.821e − 3) (Figure 5(c)). Patients with CYP1A2 and GSTA3 alteration had worse overall survival than patients without CYP1A2 and GSTA3 alteration (P = 0.465) (Figure 5(d)).
Figure 5.

The expression and prognostic value of GNGT1 and NMU in NSCLC patients. (a) Genetic alteration of GNGT1 and NMU genes in NSCLC patients. (b) Illustration of the alteration frequency of GNGT1 and NMU genes in NSCLC patients. (c) Coexpression between GNGT1 and NMU. (d) Overall survival analysis for GNGT1 and NMU genes in NSCLC patients. (e) Combined diagnosis of GNGT1 and NMU genes in NSCLC patients.
Notably, according to the ROC curve analysis, the AUC of GNGT1 was 0.903 (P < 0.0001). For NMU, the AUC was 0.932 (P < 0.0001). The AUC was largest when GNGT1 was combined with NMU (AUC = 0.969, P < 0.0001) (Figure 5(e)).
4. Discussion
Elucidating the molecular mechanisms of the initiation and development of NSCLC would benefit the early diagnosis and targeted therapy efforts [60–63]. In this study, we identified 1,362 upregulated genes and 49 downregulated genes and selected GNGT1, GNG4, NMU, GCG, TAC1, GAST NPSR1, and GCGR as hub genes using Molecular Complex Detection (MCODE) and cytoHubba. These genes were primarily enriched in terms of the Ras signaling pathway, steroid hormone biosynthesis, nicotine addiction, alcoholism, steroid hormone biosynthesis, and systemic lupus erythematosus.
The Ras signaling pathway is closely related to the occurrence and progression of most human tumors [64–67]. The activation of RAS-RAF-MEK-MAPK in gene transcription regulation can promote proliferation, migration, and angiogenesis of cancer cells [68–70]. RAS-PI3K interaction is an important signaling node and potential therapeutic target in EGFR-mutant lung cancer [71–73]. In addition, steroid hormones were not previously considered to be involved with lung function [74–76]. However, numerous studies have reported that steroid hormones are important in normal lung development and function [77], as well as in the pathogenesis of pulmonary diseases, including lung cancer [78–81].
Cigarette smoking is a well-known risk factor for the occurrence and progression of malignant diseases [82–85]. Nicotine, the major constituent in cigarette smoke, plays key roles in cancer progression [86–89]. Nicotine likely promotes lung cancer cell proliferation by upregulating HIF-1α and SOCC components [90–93]. It was demonstrated that nicotine increased NSCLC cell proliferation through nicotinic acetylcholine receptor-mediated signals [94–97]. Nicotine can also induce the expression of embryonic stem cell factor Sox2, which is indispensable for self-renewal and the maintenance of stem cell properties in NSCLC cells [98–100].
Several studies have been conducted to investigate the association between alcohol and lung cancer. Some studies have reported that alcohol is linked to a number of human diseases, including cancers [101–103]. Interestingly, another report shows that alcohol has nothing to do with lung cancer [104]. Thus, conducting further experiments is necessary to confirm whether lung cancer is attributable to alcohol abuse. All in all, the findings of these studies are consistent with our results.
In the current study, the expressions of GNGT1 and NMU were low both in the Oncomine and TCGA databases, indicating that GNGT1 and NMU may play a role as oncogenes. The transducin γ-subunit gene (GNGT1) has been localized to human chromosome 7 [104] and is associated with various forms of cancer [105–108]. GNGT1 exerts effects in different tissues regulating cell proliferation, migration, adhesion, and apoptosis [109–111]. One study showed that GNGT1 could serve as a marker of medulloblastoma [112]. GNGT1 can be utilized to differentiate gastrointestinal stromal tumor and leiomyosarcoma, two cancers that have very similar histopathology, but require very different treatments [113–115]. In the current study, GNGT1 was significantly upregulated and high mRNA expression of GNGT1 was associated with poor overall survival in NSCLC patients. Furthermore, KEGG analysis showed that GNGT1 was involved in the Ras signaling pathway. Therefore, it is reasonable to regard GNGT1 as a hub gene of NSCLC. Further studies are needed to better understand GNGT1's association with NSCLC.
Neuromedin U (NMU) has been reported to exhibit early alterations associated with cancer, including lung cancer, pancreatic cancer, breast cancer, renal cancer, and endometrioid endometrial carcinoma, through promoting migration, invasion, glycolysis, a mesenchymal phenotype, a stem cell phenotype of cancer cells, and resistance to the antitumor immune response [116–118]. It is overexpressed in pancreatic cancer and increases the cancer invasiveness through the hepatocyte growth factor c-Met pathway [119–121]. A role has also been implicated for NMU in human breast cancer and endometrial cancer [122–124]. The protein encoded by NMU can amplify ILC2 to drive allergic lung inflammation [125]. NMU is regulated by RhoGDI2, a metastasis inhibitor, which can be used as a target for lung metastasis. The expression of NMU is negatively correlated with prognosis in most types of cancer [126–128]. In the present study, the higher mRNA and protein expression of NMU were negatively correlated with overall survival. Therefore, our results are in line with these previous studies, which indicated that NMU may be directly or indirectly important in NSCLC development.
Moreover, to explore the predictive ability of GNGT1 and NMU, logistic regression analysis was performed. The logistic regression analysis showed a probabilistic nonlinear regression, which has functions in discrimination and prediction. Notably, according to logistic regression analysis, the AUC of the ROC curve of GNGT1 was 0.903 (P < 0.0001), and the AUC of NMU was 0.932 (P < 0.0001). Combining the two biomarkers enabled a relatively high capacity for discrimination between NSCLC and normal patients, with an AUC of 0.969, indicating that the combined test of GNGT1 combined with NMU was superior to testing for either gene individually, with better clinical accuracy and higher diagnostic value. Therefore, it is of high scientific value to use a logistic regression model as a diagnostic model for NSCLC.
In conclusion, our results identified two hub genes, GNGT1 and NMU, as prognostic target genes, and highlighted their probable role in NSCLC. Nevertheless, a few limitations to this study should be acknowledged. Because all the data analyzed in the current study were retrieved from the online databases, further independent experiments are required to validate our findings and to explore the molecular mechanism of the hub genes in NSCLC development and progression.
Acknowledgments
This study was supported partly by grants from the National Natural Science Foundation of China (81972214, 81772932, 82071956, 81472202, and 81302065), Shanghai Talents Development Foundation (2017103), Shanghai Science and Technology Commission (18441903500, 14DZ1940606), Shanghai Academic/Technology Research Leader (18XD1403000), and National Key R&D Program of China (2016YFC0104303).
Contributor Information
Geng-Xi Jiang, Email: jiang9909@hotmail.com.
Da Fu, Email: fu800da900@126.com.
Fei Yu, Email: yufei021@sina.com.
Data Availability
All data generated or analyzed during this study are included in this article.
Additional Points
Impact Statement. GNGT1 and NMU identified in the current study may empower further discovery of the molecular mechanisms underlying NSCLC's initiation and progression.
Conflicts of Interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors' Contributions
Y.-S.M., D.F., and F.Y. designed the study. All authors collected data, performed the statistical analyses, and interpreted the data. J.-J.Z., D.F., and F.Y. wrote the manuscript. J.-J.Z., J.H., and Y.-S.M. contributed equally to this work. All authors contributed to the final version of the manuscript and approved the final manuscript. Jia-Jia Zhang, Jiang Hong, and Yu-Shui Ma contributed equally to this work.
References
- 1.Yu F., Liu J. B., Wu Z. J., et al. Tumor suppressive microRNA-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 14. Cancer Letters. 2018;427:74–84. doi: 10.1016/j.canlet.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Guo L., Zhang Y., Wei R., Wang C., Feng M. Lipopolysaccharide-anchored macrophages hijack tumor microtube networks for selective drug transport and augmentation of antitumor effects in orthotopic lung cancer. Theranostics. 2019;9:6936–6948. doi: 10.7150/thno.37380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J., Colombet M., Soerjomataram I., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. European Journal of Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Fu S., Zhao J., et al. Transbronchoscopic patient biopsy-derived xenografts as a preclinical model to explore chemorefractory-associated pathways and biomarkers for small-cell lung cancer. Cancer letters. 2019;440:180–188. doi: 10.1016/j.canlet.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Han Y., Guo W., Ren T., et al. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer letters. 2019;440:116–125. doi: 10.1016/j.canlet.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Hong S. K., Lee H., Kwon O. S., et al. Large-scale pharmacogenomics based drug discovery for ITGB3 dependent chemoresistance in mesenchymal lung cancer. Molecular Cancer. 2018;17(1):p. 175. doi: 10.1186/s12943-018-0924-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouclier C., Simon M., Laconde G., et al. Stapled peptide targeting the CDK4/Cyclin D interface combined with Abemaciclib inhibits KRAS mutant lung cancer growth. Theranostics. 2020;10:2008–2028. doi: 10.7150/thno.40971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbuzenko O. B., Kuzmov A., Taratula O., Pine S. R., Minko T. Strategy to enhance lung cancer treatment by five essential elements: inhalation delivery, nanotechnology, tumor-receptor targeting, chemo- and gene therapy. Theranostics. 2019;9(26):8362–8376. doi: 10.7150/thno.39816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen M., Zhao X., Zhao L., et al. Met is involved in TIGAR-regulated metastasis of non-small-cell lung cancer. Molecular Cancer. 2018;17:p. 88. doi: 10.1186/s12943-018-0839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin A., Wei T., Meng H., Luo P., Zhang J. Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Molecular Cancer. 2019;18:p. 139. doi: 10.1186/s12943-019-1062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Huang W. C., Huang B., et al. Clinical characteristics and prognosis of COVID-19 patients with initial presentation of lung lesions confined to a single pulmonary lobe. American Journal of Translational Research. 2020;12(11):7501–7509. [PMC free article] [PubMed] [Google Scholar]
- 12.Reda M., Ngamcherdtrakul W., Gu S., et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Letters. 2019;467:9–18. doi: 10.1016/j.canlet.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasparello J., Lomazzi M., Papi C., et al. Efficient delivery of microRNA and antimiRNA molecules using an argininocalix [4] arene macrocycle. Molecular Therapy-Nucleic Acids. 2019;18:748–763. doi: 10.1016/j.omtn.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nong J., Gong Y., Guan Y., et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nature Communications. 2018;9(1):p. 3114. doi: 10.1038/s41467-018-05327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowling C. M., Zhang H., Chonghaile T. N., Wong K.-K. Shining a light on metabolic vulnerabilities in non-small cell lung cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2021;1875(1) doi: 10.1016/j.bbcan.2020.188462.188462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z., Zheng X., Jiao D., et al. LunX-CAR T cells as targeted therapy for non-small cell lung cancer. Molecular Therapy-Oncolytics. 2020;17:361–370. doi: 10.1016/j.omto.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Yu J., Wang Q., et al. Tartrate-resistant acid phosphatase 5/ACP5 interacts with p53 to control the expression of SMAD3 in lung adenocarcinoma. Molecular Therapy-Oncolytics. 2020;16:272–288. doi: 10.1016/j.omto.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giulietti M. V., Vespa A., Ottaviani M., et al. Personality (at intrapsychic and interpersonal level) associated with quality of life in patients with cancer (lung and colon) Cancer Control. 2019;26 doi: 10.1177/1073274819880560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Yin Z., Fan J., Zhang S., Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal transduction and targeted therapy. 2019;4:p. 47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taschauer A., Polzer W., Alioglu F., et al. Peptide-targeted polyplexes for aerosol-mediated gene delivery to CD49f-overexpressing tumor lesions in lung. Molecular Therapy-Nucleic Acids. 2019;18:774–786. doi: 10.1016/j.omtn.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christian W. J., Vanderford N. L., McDowell J., et al. Spatiotemporal analysis of lung cancer histological types in Kentucky, 1995-2014. Cancer Control. 2019;26, article 1073274819845873 doi: 10.1177/1073274819845873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matés J. M., Campos-Sandoval J. A., Santos-Jiménez J. L., Márquez J. Dysregulation of glutaminase and glutamine synthetase in cancer. Cancer Letters. 2019;467:29–39. doi: 10.1016/j.canlet.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Dutta R. K., Chinnapaiyan S., Unwalla H. Aberrant microRNAomics in pulmonary complications:implications in lung health and diseases. Molecular Therapy-Nucleic Acids. 2019;18:413–431. doi: 10.1016/j.omtn.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi H. R., Song I. A., Oh T. K. Association of opioid use in the week before death among patients with advanced lung cancer having sepsis. Cancer Control. 2019;26, article 1073274819871326 doi: 10.1177/1073274819871326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago C. P., Keuthan C. J., Boye S. L., Boye S. E., Imam A. A., Ash J. D. A drug-tunable gene therapy for broad-spectrum protection against retinal degeneration. Molecular Therapy. 2018;26:2407–2417. doi: 10.1016/j.ymthe.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchini F., Portioli E., Ferlenghi F., et al. Cell-targeted c(AmpRGD)-sunitinib molecular conjugates impair tumor growth of melanoma. Cancer Letters. 2019;446:25–37. doi: 10.1016/j.canlet.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Caine M., Chung T., Kilpatrick H., et al. Evaluation of novel formulations for transarterial chemoembolization: combining elements of Lipiodol emulsions with drug-eluting beads. Theranostics. 2019;9(19):5626–5641. doi: 10.7150/thno.34778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C., Yi C., Yi Y., et al. lncRNA PVT1 promotes tumorigenesis of colorectal cancer by stabilizing miR-16-5p and interacting with the VEGFA/VEGFR1/AKT axis. Molecular Therapy-Nucleic Acids. 2020;20:438–450. doi: 10.1016/j.omtn.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N., Nan A., Chen L., et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Molecular Cancer. 2020;19(1):p. 101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M., Xu Y., Zhao J., et al. Concurrent driver gene mutations as negative predictive factors in epidermal growth factor receptor-positive non-small cell lung Cancer. EBioMedicine 2019;42:304-310. Nicot C. RNA-Seq reveal the circular RNAs landscape of lung cancer. Molecular Cancer. 2019;18:p. 183. doi: 10.1016/j.ebiom.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vellanki S. H., Cruz R. G. B., Jahns H., et al. Natural compound Tetrocarcin-A downregulates Junctional Adhesion Molecule-A inconjunction with HER2 and inhibitor of apoptosis proteins and inhibits tumorcell growth. Cancer Letters. 2019;440:23–34. doi: 10.1016/j.canlet.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 32.Yu S., Zhang J., Yan Y., et al. A novel asymmetrical anti-HER2/CD3 bispecific antibody exhibits potent cytotoxicity for HER2-positive tumor cells. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 355. doi: 10.1186/s13046-019-1354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan B., Zhao J., Zhou C., et al. Co-occurring alterations of ERBB2 exon 20 insertion in non-small cell lung cancer (NSCLC) and the potential indicator of response to afatinib. Frontiers in Oncology. 2020;10:p. 729. doi: 10.3389/fonc.2020.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Chen D., Yi Y., et al. Histone deacetylase inhibitor sensitizes ERCC1-high non-small-cell lung cancer cells to cisplatin via regulating miR-149. Molecular Therapy-Oncolytics. 2020;17:448–459. doi: 10.1016/j.omto.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W., Yang J., Saren G., et al. HDAC6-specific inhibitor suppresses Th17 cell function via the HIF-1α pathway in acute lung allograft rejection in mice. Theranostics. 2020;10(15):6790–6805. doi: 10.7150/thno.44961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen P., Wu Q., Feng J., et al. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduction and Targeted Therapy. 2020;5:p. 51. doi: 10.1038/s41392-020-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T., Liu Y., Zhang W., et al. A rapid liquid biopsy of lung cancer by separation and detection of exfoliated tumor cells from bronchoalveolar lavage fluid with a dual-layer "PERFECT" filter system. Theranostics. 2020;10:6517–6529. doi: 10.7150/thno.44274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Deng Q., Lv Z., et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Molecular Cancer. 2019;18:p. 181. doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Hu Z., Zheng X., Jiao D., et al. LunX-CAR T Cells as a targeted therapy for non-small cell lung cancer. Molecular Therapy-Oncolytics. 2020;17:361–370. doi: 10.1016/j.omto.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Wang Z., Tong F., Dong X., Wu G., Zhang R. lncRNA UCA1 promotes gefitinib resistance as a ceRNA to target FOSL2 by sponging miR-143 in non-small cell lung cancer. Molecular Therapy-Nucleic Acids. 2020;19:643–653. doi: 10.1016/j.omtn.2019.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Zhang M., Zhang L., Li Y., et al. Exome sequencing identifies somatic mutations in novel driver genes in non-small cell lung cancer. Aging (Albany NY) 2020;12(13):13701–13715. doi: 10.18632/aging.103500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X. H., Yan X., Zhang Q. H., et al. Direct targeting of HSP90 with daurisoline destabilizes β-catenin to suppress lung cancer tumorigenesis. Cancer Letters. 2020;489:66–78. doi: 10.1016/j.canlet.2020.05.024. [DOI] [PubMed] [Google Scholar]
- 43.de Man Y., Atsma F., Oosterveld-Vlug M. G., et al. The intensity of hospital care utilization by Dutch patients with lung or colorectal cancer in their final months of life. Cancer Control. 2019;26, article 1073274819846574 doi: 10.1177/1073274819846574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang G., Chen Q., Xiao J., Zhang H., Wang Z., Lin X. Identification of genes and analysis of prognostic values in nonsmoking females with non-small cell lung carcinoma by bioinformatics analyses. Cancer Management and Research. 2018;Volume 10:4287–4295. doi: 10.2147/CMAR.S174409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., Lu B., Ren S., et al. Long Noncoding RNA LINC01116 contributes to gefitinib resistance in non-small cell lung cancer through regulating IFI44. Molecular Therapy-Nucleic Acids. 2020;19:218–227. doi: 10.1016/j.omtn.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Z., Liu Z., Jiang X., et al. Depleting PTOV1 sensitizes non-small cell lung cancer cells to chemotherapy through attenuating cancer stem cell traits. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 341. doi: 10.1186/s13046-019-1349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong H. G., Li H., Xiao Y., et al. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 278. doi: 10.1186/s13046-019-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Z., Fu P., Yu Z., Zhen F. X., Yanhong Gu Y. H. Comprehensive analysis of lncRNA-miRNA- mRNA network ascertains prognostic factors in patients with colon cancer. Technology in Cancer Research & Treatment. 2019;18, article 1533033819853237 doi: 10.1177/1533033819853237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson M. D., McCarthy D. J., Smyth G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun X., Wu K., Cook D. PKgraph:an R package for graphically diagnosing population pharmacokinetic models. Computer Methods and Programs in Biomedicine. 2011;104(3):461–471. doi: 10.1016/j.cmpb.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Iratxeta C., Bork P., Andrade-Navarro M. A. Update of the G2D tool for prioritization of gene candidates to inherited diseases. Nucleic Acids Research. 2007;35:W212–W216. doi: 10.1093/nar/gkm223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chin C. H., Chen S. H., Wu H. H., Ho C. W., Ko M. T., Lin C. Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC systems biology. 2014;8:p. S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhodes D. R., Yu J., Shanker K., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie X. Q., Wang M. J., Li Y., et al. miR-124 intensified oxaliplatin-based chemotherapy by targeting CAPN2 in colorectal cancer. Molecular Therapy-Oncolytics. 2020;17:320–331. doi: 10.1016/j.omto.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoshikawa N., Sakai A., Takai S., Suzuki H. Targeting extracellular miR-21-TLR7 signaling provides long-lasting analgesia in osteoarthritis. Molecular Therapy-Nucleic Acids. 2020;19:199–207. doi: 10.1016/j.omtn.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao C., Sun X., Wu Z., et al. A novel benzofuran derivative Moracin N induces autophagy and apoptosis through ROS generation in lung cancer. Frontiers in Pharmacology. 2020;11:p. 391. doi: 10.3389/fphar.2020.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demircan N. C., Akın Telli T., Başoğlu Tüylü T., et al. QT interval prolongation related to afatinib treatment in a patient with metastatic non-small-cell lung cancer. Current Problems in Cancer. 2020;27:p. 100594. doi: 10.1016/j.currproblcancer.2020.100594. [DOI] [PubMed] [Google Scholar]
- 61.Malinina A., Dikeman D., Westbrook R., et al. IL10 deficiency promotes alveolar enlargement and lymphoid dysmorphogenesis in the aged murine lung. Aging Cell. 2020;19, article e13130 doi: 10.1111/acel.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong S., Hong Z., Huang L. S., et al. IL-1β suppression of VE-cadherin transcription underlies sepsis-induced inflammatory lung injury. The Journal of Clinical Investigation. 2020;8:p. 136908. doi: 10.1172/JCI136908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu K., Park D., Magis A. T., et al. Small molecule KRAS agonist for mutant KRAS cancer therapy. Molecular Cancer. 2019;18:p. 85. doi: 10.1186/s12943-019-1012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen P., Li X., Zhang R., et al. Combinative treatment of β-elemene and cetuximab is sensitive to KRAS mutant colorectal cancer cells by inducing ferroptosis and inhibiting epithelial-mesenchymal transformation. Theranostics. 2020;10(11):5107–5119. doi: 10.7150/thno.44705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shan G., Shao B., Liu Q., et al. circFMN2 sponges miR-1238 to promote the expression of LIM-homeobox gene 2 in prostate cancer cells. Molecular Therapy-Nucleic Acids. 2020;21:133–146. doi: 10.1016/j.omtn.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Jin H., Jang Y., Cheng N., et al. Restoration of mutant K-Ras repressed miR-199b inhibits K-Ras mutant non-small cell lung cancer progression. Journal of Experimental & Clinical Cancer Research. 2019;38(1):p. 165. doi: 10.1186/s13046-019-1170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin Y., Liu M., Sa R., Fu H., Cheng L., Chen L. Mouse models of thyroid cancer: bridging pathogenesis and novel therapeutics. Cancer Letters. 2020;469:35–53. doi: 10.1016/j.canlet.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 68.Wu C., Guo E., Ming J., et al. Radiation-induced DNMT3B promotes radioresistance in nasopharyngeal carcinoma through methylation of p53 and p21. Molecular Therapy-Oncolytics. 2020;17:306–319. doi: 10.1016/j.omto.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu R., Han Q., Zhang J. STAT3: a key signaling molecule for converting cold to hot tumors. Cancer Letters. 2020;489:29–40. doi: 10.1016/j.canlet.2020.05.035. [DOI] [PubMed] [Google Scholar]
- 70.Han C., Choi B. K., Kim S. H., et al. Polymorphic region-specific antibody for evaluation of affinity-associated profile of chimeric antigen receptor. Molecular Therapy-Oncolytics. 2020;17:293–305. doi: 10.1016/j.omto.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim Y. J., Baek D. S., Lee S., et al. Dual-targeting of EGFR and Neuropilin-1 attenuates resistance to EGFR-targeted antibody therapy in KRAS-mutant non-small cell lung cancer. Cancer Letters. 2019;466:23–34. doi: 10.1016/j.canlet.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Casal-Mouriño A., Ruano-Ravina A., Torres-Durán M., et al. Lung cancer survival in never-smokers and exposure to residential radon: results of the LCRINS study. Cancer Letters. 2020;487:21–26. doi: 10.1016/j.canlet.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 73.Xu F., Chen J. X., Yang X. B., et al. Analysis of lung adenocarcinoma subtypes based on immune signatures identifies clinical implications for cancer therapy. Molecular Therapy-Oncolytics. 2020;17:241–249. doi: 10.1016/j.omto.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin W., Xu Y., Chen X., et al. Radiation-induced small extracellular vesicles as "carriages" promote tumor antigen release and trigger antitumor immunity. Theranostics. 2020;10(11):4871–4884. doi: 10.7150/thno.43539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie J., Zhang W., Liang X., et al. RPL32 promotes lung cancer progression by facilitating p53 degradation. Molecular Therapy-Nucleic Acids. 2020;21:75–85. doi: 10.1016/j.omtn.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keller M., Dubois F., Teulier S., et al. NDR2 kinase contributes to cell invasion and cytokinesis defects induced by the inactivation of RASSF1A tumor-suppressor gene in lung cancer cells. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 158. doi: 10.1186/s13046-019-1145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Waarde A., Rybczynska A. A., Ramakrishnan N. K., Ishiwata K., Elsinga P. H., Dierckx R. A. Potential applications for sigma receptor ligands in cancer diagnosis and therapy. Biochimica et Biophysica Acta. 2015;1848:2703–2714. doi: 10.1016/j.bbamem.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 78.Jiang L., Wang R., Fang L., et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics. 2019;9(9):2460–2474. doi: 10.7150/thno.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C., Liu W. R., Zhang B., et al. LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 andphosphorylation of AKT in EGFR-mutant lung cancers. Cancer Letters. 2020;486:58–70. doi: 10.1016/j.canlet.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 80.Fernandes E., Sores J., Cotton S., et al. Esophageal, gastric and colorectal cancers: looking beyond classical serological biomarkers towards glycoproteomics-assisted precision oncology. Theranostics. 2020;10:4903–4928. doi: 10.7150/thno.42480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen M., Xu Z., Xu W., et al. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 149. doi: 10.1186/s13046-019-1161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang J., Jeong S. H., Lee K., et al. Exacerbation of symptomatic arthritis by cigarette smoke in experimental arthritis. PLoS One. 2020;15(3, article e0230719) doi: 10.1371/journal.pone.0230719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galland S., Martin P., Fregni G., Letovanec I., Stamenkovic I. Attenuation of the pro-inflammatory signature of lung cancer-derived mesenchymal stromal cells by statins. Cancer Letters. 2020;484:50–64. doi: 10.1016/j.canlet.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 84.Liu H., Xue Y. C., Deng H., et al. MicroRNA modification of Coxsackievirus B3 decreases its toxicity, while retaining oncolytic potency against lung cancer. Molecular Therapy-Oncolytics. 2020;16:207–218. doi: 10.1016/j.omto.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiao Y., Wang Z., Tan F., et al. Enhancer reprogramming within pre-existing topologically associated domains promotes TGF-β-induced EMT and cancer metastasis. Molecular Therapy. 2020;28:2083–2095. doi: 10.1016/j.ymthe.2020.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Q., Wang Q., Li D., et al. Co-administration of 20(S)-protopanaxatriol (g-PPT) and EGFR-TKI overcomes EGFR- TKI resistance by decreasing SCD1 induced lipid accumulation in non-small cell lung cancer. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 129. doi: 10.1186/s13046-019-1120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fan M., Han Y., Gao S., et al. Ultrasmall gold nanoparticles in cancer diagnosis and therapy. Theranostics. 2020;10(11):4944–4957. doi: 10.7150/thno.42471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duan S., Li J., Tian J., et al. Crosstalk between let-7a-5p and BCL-xL in the initiation of toxic autophagy in lung cancer. Molecular Therapy-Oncolytics. 2019;15:69–78. doi: 10.1016/j.omto.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ben Q., An W., Sun Y., et al. A nicotine-induced positive feedback loop between HIF1A and YAP1 contributes to epithelial-to-mesenchymal transition in pancreatic ductal adenocarcinoma. Journal of Experimental & Clinical Cancer Research. 2020;39(1):p. 181. doi: 10.1186/s13046-020-01689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiong Y., He L., Shay C., et al. Nck-associated protein 1 associates with HSP90 to drive metastasis in human non-small-cell lung cancer. Journal of Experimental & Clinical Cancer Research. 2019;38(1):p. 122. doi: 10.1186/s13046-019-1124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen J., Liu A., Lin Z., et al. Downregulation of the circadian rhythm regulator HLF promotes multiple-organ distant metastases in non-small cell lung cancer through PPAR/NF-κb signaling. Cancer Letters. 2020;482:56–71. doi: 10.1016/j.canlet.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 92.Li Z., Zeng C., Nong Q., et al. Exosomal leucine-rich-Alpha2-glycoprotein 1 derived from non-small-cell lung cancer cells promotes angiogenesis via TGF-β signal pathway. Molecular Therapy-Oncolytics. 2019;4:313–322. doi: 10.1016/j.omto.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou L., Jiang Y., Luo Q., Li L., Jia L. Neddylation: a novel modulator of the tumor microenvironment. Molecular Cancer. 2019;18(1):p. 77. doi: 10.1186/s12943-019-0979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Z., Zhang N., Li A., et al. Insulin-like growth factor-1 receptor induces immunosuppression in lung cancer by upregulating B7-H4 expression through the MEK/ERK signaling pathway. Cancer Letters. 2020;485:14–26. doi: 10.1016/j.canlet.2020.04.013. [DOI] [PubMed] [Google Scholar]
- 95.Kim S., Maeng J. Y., Hyun S. J., et al. Extracellular vesicles from human umbilical cord blood plasma modulate interleukin-2 signaling of T cells to ameliorate experimental autoimmune encephalomyelitis. Theranostics. 2020;10:5011–5028. doi: 10.7150/thno.42742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen X., Jia Y., Zhang Y., Zhou D., Sun H., Ma X. α5-nAChR contributes to epithelial-mesenchymal transition and metastasis by regulating Jab1/Csn5 signalling in lung cancer. Journal of Cellular and Molecular Medicine. 2020;24(4):2497–2506. doi: 10.1111/jcmm.14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fang L., Ly D., Wang S. S., et al. Targeting late-stage non-small cell lung cancer with a combination of DNT cellular therapy and PD-1 checkpoint blockade. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 123. doi: 10.1186/s13046-019-1126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou W., Liu Y., Gao Y., et al. MICAL2 is a novel nucleocytoplasmic shuttling protein promoting cancer invasion and growth of lung adenocarcinoma. Cancer Letters. 2020;483:75–86. doi: 10.1016/j.canlet.2020.04.019. [DOI] [PubMed] [Google Scholar]
- 99.Wei C., Dong X., Lu H., et al. LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 95. doi: 10.1186/s13046-019-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei C., Dong X., Lu H., et al. Novel lncRNA-IUR suppresses Bcr-Abl-induced tumorigenesis through regulation of STAT5-CD71 pathway. Molecular Cancer. 2019;18:p. 84. doi: 10.1186/s12943-019-1013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tian M., Wu X., Lu T., et al. Phytochemical analysis, antioxidant, antibacterial, cytotoxic, and enzyme inhibitory activities of Hedychium flavum rhizome. Frontiers in Pharmacology. 2020;11:p. 572659. doi: 10.3389/fphar.2020.572659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee H., Son Y. S., Lee M. O., et al. Low-dose interleukin-2 alleviates dextran sodium sulfate-induced colitis in mice by recovering intestinal integrity and inhibiting AKT-dependent pathways. Theranostics. 2020;10(11):5048–5063. doi: 10.7150/thno.41534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luo X., Li N., Zhao X., et al. DHRS2 mediates cell growth inhibition induced by Trichothecin in nasopharyngeal carcinoma. Journal of Experimental & Clinical Cancer Research. 2019;38:p. 300. doi: 10.1186/s13046-019-1301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scherer S. W., Feinstein D. S., Oliveira L., Tsui L. C., Pittler S. J. Gene structure and chromosome localization to 7q21.3 of the human rod photoreceptor transducin gamma-subunit gene (GNGT1) Genomics. 1996;35(1):241–243. doi: 10.1006/geno.1996.0346. [DOI] [PubMed] [Google Scholar]
- 105.Ong O. C., Hu K., Rong H., Lee R. H., Fung B. K. Gene structure and chromosome localization of the GγcSubunit of human cone G-protein (GNGT2) Genomics. 1997;44(1):101–109. doi: 10.1006/geno.1997.4814. [DOI] [PubMed] [Google Scholar]
- 106.Chen H., Leung T., Giger K. E., et al. Expression of the G protein gammaT1 subunit during zebrafish development. Gene Expression Patterns. 2007;7:574–583. doi: 10.1016/j.modgep.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lagman D., Callado-Pérez A., Franzén I. E., Larhammar D., Abalo X. M. Transducin duplicates in the zebrafish retina and pineal complex:differential specialisation after the teleost tetraploidisation. PLoS One. 2015;10, article e0121330 doi: 10.1371/journal.pone.0121330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sucheston-Campbell L. E., Clay-Gilmour A. I., Barlow W. E., et al. Genome-wide meta-analyses identifies novel taxane-induced peripheral neuropathy-associated loci. Pharmacogenetics and Genomics. 2018;28(2):49–55. doi: 10.1097/FPC.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mulligan T., Farber S. A. Central and C-terminal domains of heterotrimeric G protein gamma subunits differentially influence the signaling necessary for primordial germ cell migration. Cellular Signalling. 2011;23(10):1617–1624. doi: 10.1016/j.cellsig.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J. L., Liu M., Zhang C. N., Li E. C., Fan M. Z., Huang M. X. Transcriptomic analyses of tributyltin-induced sexual dimorphisms in rare minnow (Gobiocypris rarus) brains. Ecotoxicology and Environmental Safety. 2018;156:18–24. doi: 10.1016/j.ecoenv.2018.02.049. [DOI] [PubMed] [Google Scholar]
- 111.Mucaki E. J., Zhao J. Z. L., Lizotte D. J., Rogan P. K. Predicting responses to platin chemotherapy agents with biochemically-inspired machine learning. Signal Transduction and Targeted Therapy. 2019;4(1):p. 1. doi: 10.1038/s41392-018-0034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zou R., Zhang D., Lv L., et al. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. Journal of Translational Medicine. 2019;17(1):p. 45. doi: 10.1186/s12967-019-1790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu B. X., Huang G. J., Cheng H. B. Comprehensive analysis of core genes and potential mechanisms in rectal cancer. Journal of Computational Biology. 2019;26(11):1262–1277. doi: 10.1089/cmb.2019.0073. [DOI] [PubMed] [Google Scholar]
- 114.de-la-Torre A., Silva-Aldana C. T., Muñoz-Ortiz J., et al. Uveitis and multiple sclerosis: potential common causal mutations. Molecular Neurobiology. 2019;56:8008–8017. doi: 10.1007/s12035-019-1630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harten S. K., Esteban M. A., Shukla D., Ashcroft M., Maxwell P. H. Inactivation of the von Hippel-Lindau tumour suppressor gene induces Neuromedin U expression in renal cancer cells. Molecular Cancer. 2011;10:p. 89. doi: 10.1186/1476-4598-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fruzangohar M., Ebrahimie E., Adelson D. L. A novel hypothesis-unbiased method for Gene Ontology enrichment based on transcriptome data. PLoS One. 2017;12, article e0170486 doi: 10.1371/journal.pone.0170486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee J., Snyder E. R., Liu Y., et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nature Communications. 2017;8:p. 14686. doi: 10.1038/ncomms14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martinez V. G., Crown J., Porter R. K., O'Driscoll L. Neuromedin U alters bioenergetics and expands the cancer stem cell phenotype in HER2-positive breast cancer. International Journal of Cancer. 2017;140:2771–2784. doi: 10.1002/ijc.30705. [DOI] [PubMed] [Google Scholar]
- 119.Ensho T., Maruyama K., Mori K., et al. Neuromedin U precursor-related peptide (NURP) exerts neuromedin U-like sympathetic nerve action in the rat. Biochemical and Biophysical Research Communications. 2017;492(3):412–418. doi: 10.1016/j.bbrc.2017.08.084. [DOI] [PubMed] [Google Scholar]
- 120.Yang X., Wang C. C., Lee W. Y. W., Trovik J., Chung T. K. H., Kwong J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Letters. 2018;413:23–34. doi: 10.1016/j.canlet.2017.10.028. [DOI] [PubMed] [Google Scholar]
- 121.Chen X. G., Ma L., Xu J. X. Abnormal DNA methylation may contribute to the progression of osteosarcoma. Molecular Medicine Reports. 2018;17:193–199. doi: 10.3892/mmr.2017.7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watteyne J., Peymen K., Van der Auwera P., et al. Neuromedin U signaling regulates retrieval of learned salt avoidance in a C. elegans gustatory circuit. Nature Communications. 2020;11:p. 2076. doi: 10.1038/s41467-020-15964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaushik A. C., Mehmood A., Wei D. Q., Dai X. Systems biology integration and screening of reliable prognostic markers to create synergies in the control of lung cancer patients. Frontiers in Molecular Biosciences. 2020;7:p. 47. doi: 10.3389/fmolb.2020.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jowett G. M., Neves J. F. Commentary: neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Frontiers in Pharmacology. 2018;9:p. 230. doi: 10.3389/fphar.2018.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peng S., Lu Y., Li P., et al. The short interference RNA (siRNA) targeting NMUR2 relieves nociception in a bone cancer pain model of rat through PKC-ERK and PI3K-AKT pathways. Biochemical and Biophysical Research Communications. 2019;512:616–622. doi: 10.1016/j.bbrc.2019.03.067. [DOI] [PubMed] [Google Scholar]
- 126.Lu L., Wang R., Luo M. An optical brain-to-brain interface supports rapid information transmission for precise locomotion control. Science China. Life Sciences. 2020;63:875–885. doi: 10.1007/s11427-020-1675-x. [DOI] [PubMed] [Google Scholar]
- 127.Maruyama K., Kaiya H., Miyazato M., Murakami N., Nakahara K., Matsuda K. Purification and identification of native forms of goldfish neuromedin U from brain and gut. Biochemical and Biophysical Research Communications. 2019;517(3):433–438. doi: 10.1016/j.bbrc.2019.07.108. [DOI] [PubMed] [Google Scholar]
- 128.Lu L., Ren Y., Yu T., et al. Control of locomotor speed, arousal, and hippocampal theta rhythms by the nucleus incertus. Nature Communications. 2020;11(1):p. 262. doi: 10.1038/s41467-019-14116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.


