Summary
CD103+CD8+ tumor-resident memory T cells (TRM) are important components of anti-tumor immunity. However, their role in response to cancer immunotherapy is not fully understood. The protocol describes how to isolate CD8+ T cells and autologous tumor cells from human lung tumors to study the functional activities of CD8+ T cells. Tumors are heterogeneous in terms of the quantity and quality of immune cell types, so the yield of TRM cells depends on the features of the tumor.
For complete details on the use and execution of this protocol, please refer to Corgnac et al. (2020).
Subject areas: Cell isolation, Flow cytometry/mass cytometry, Cancer, Immunology
Graphical Abstract
Highlights
-
•
Isolation of human CD8+ tumor-resident memory T cells
-
•
Isolation of tumor cells from fresh human lung tumor samples
-
•
Adaptable for isolation of diverse populations of tumor-infiltrating T cells
-
•
Suitable for in vitro expansion, functional tests, RNA-seq
CD103+CD8+ tumor-resident memory T cells (TRM) are important components of anti-tumor immunity. However, their role in response to cancer immunotherapy is not fully understood. The protocol describes how to isolate CD8+ T cells and autologous tumor cells from human lung tumors to study the functional activities of CD8+ T cells. Tumors are heterogeneous in terms of the quantity and quality of immune cell types, so the yield of TRM cells depends on the features of the tumor.
Before you begin
Background
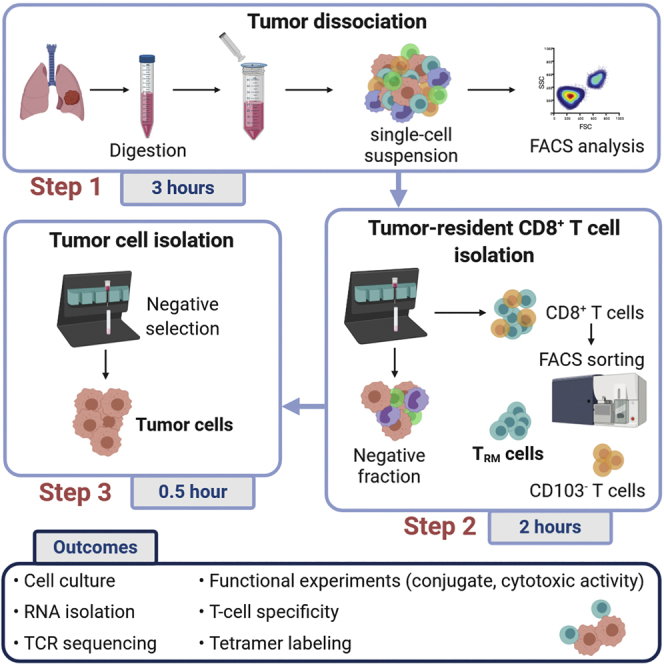
This protocol describes how to isolate tumor-resident memory CD8+ T cells (TRM) and autologous tumor cells from fresh human lung tumor samples. The procedure is divided in 3 main steps, which are realized in sequential manner; the tumor sample dissociation, the magnetic isolation of CD8+ tumor-infiltrating lymphocytes (TILs) and fluorescence-activated cell sorting (FACS) of TRM cells, then the isolation of autologous tumor cells. The second and the third steps could be skipped if one of the cell population is not needed. The procedure for CD8+ T cell isolation could be adapted for CD4+ T cells. This protocol is also compatible with renal tumor samples and other solid tumors of human and mouse origins.
Others methods of TIL isolation have been described in literature, such as a procedure to isolate bulk T cells or specific T cells from human tumors (Cohen et al., 2015) and B cells from breast human samples (Lu et al., 2020). These techniques use FACS to directly isolate cells of interest, without pre-separation with magnetic beads. A detailed method that describes a long-term expansion of tumor cells (Liu et al., 2017) could also be used. To isolate CD45+ TILs from mouse tumors, users can refer to the publication Perret et al., which also uses the magnetic sorting strategy (Perret et al., 2014).
Culture medium and buffers preparation
Timing: 30 min
-
1.
See Materials and equipment for preparation of needed materials
-
2.
Pre-warm culture media to 37°C and pre-heat an incubator to 37°C in which samples can be rotated in during incubation
Enzyme stock preparation
Timing: 20 min
-
3.Enzymes from Human Tumor Dissociation Kit (Miltenyi Biotec) are reconstituted from lyophilized powders according to manufacturer’s instructions and conserved for maximum 8 months at −20°C.
-
a.Enzyme H is reconstituted with 3 mL of Roswell Park Memorial Institute (RPMI) media. Prepare 400 μL aliquots to avoid repeated freeze-thaw cycles and store at −20°C.
-
b.Enzyme R is reconstituted with 2.7 mL RPMI media. Prepare 200 μL aliquots and store at −20°C.
-
c.Enzyme A is reconstituted with 1 mL of buffer A (supplied by the manufacturer). Prepare 100 μL aliquots and store at −20°C.
-
a.
Tumor sample collection
Timing: 1 h
-
4.The tumor needs to be fresh for assays therefore lab’s members have to be ready to process the sample immediately after receipt or at last 12 h after receipt.
-
a.The tumor sample is processed by the pathologist after surgery and placed into a sterile tube filled with serum free RPMI media and conserved at 4°C. The sample has to be entirely immersed in the medium.
-
a.
Note: Usually we perform the tumor dissociation the same day after surgery or maximum 1 day after. In the last case, the tumor sample has to be conserved in the medium at 4°C until processing.
CRITICAL: To ensure an optimal recovery of T cells and tumor cells, the resected tumor sample has to be handle and place in the medium at 4°C as fast as possible after the surgery.
CRITICAL: Appropriate safety protection should be followed according to the laboratory directive when working with live unfixed human samples. Human samples are considered to be potentially infectious and should be handled in Biosafety Level II cabinets using standard aseptic precautions.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD8a-Pacific Blue (RPA-T8) | BioLegend | Cat# 301023; RRID:AB_493110 |
| Anti-human CD103-FITC (Ber-ACT8) | BioLegend | Cat# 350204; RRID:AB_10639865 |
| Anti-human KLRG1-PE (13F12F2) | ebioscience | Cat# 12-9488-42; RRID:AB_2572716 |
| Anti-human CD3-Alexa700 (UCHT1) | BioLegend | Cat# 300424; RRID:AB_493741 |
| Anti-human CD69-APC-Cy7 (FN50) | BioLegend | Cat# 310914; RRID:AB_314849 |
| Anti-human CD103-BV711 (Ber-ACT8) | BD Biosciences | Cat# 563162; RRID:AB_2738039 |
| Anti-human CD49-PerCPefluor710 (TS2/7) | Thermo Fisher Scientific | Cat# 46-9490-41; RRID:AB_2573890 |
| Anti-human PD-1-PeCyanine7 (eBioJ105) | Thermo Fisher Scientific | Cat# 25-2799-42; RRID:AB_10853804 |
| FcR blocking reagent, human | Miltenyi Biotec | Cat#130-059-901 RRID: |
| Biological samples | ||
| Resected lung tumors and adjacent healthy lung tissue samples | Institut Mutualiste Montsouris and the Hôpital Marie Lannelongue | n/a |
| Chemicals, peptides, and recombinant proteins | ||
| Human serum | Jacques Boy | Cat#201021337 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat#A3840402 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| Sodium pyruvate | Thermo Fisher Scientific | Cat#11360039 |
| Trypan blue 0.4% | Thermo Fisher Scientific | Cat#15250061 |
| PBS | Thermo Fisher Scientific | Cat#14190169 |
| Recombinant IL-2 | Sanofi recherche | Cat#1A1 |
| TO-PRO3 iodide (642/661) | Thermo Fisher Scientific | Cat#T3605 |
| Critical commercial assays | ||
| CD8 microbeads, human | Miltenyi Biotec | Cat#130-045-201 |
| Tumor cells isolation kit, human | Miltenyi Biotec | Cat#130-108-339 |
| Human tumor dissociation kit | Miltenyi Biotec | Cat#130-095-929 |
| LS columns | Miltenyi Biotec | Cat#130-042-401 |
| Red blood cell lysis | Miltenyi Biotec | Cat#130-094-183 |
| Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat#00-5523-00 |
| Single-cell RNA purification kit | Norgen | Cat#51800 |
| LIVE/DEAD fixable blue dead cell stain kit | Thermo Fisher Scientific | Cat#L34962 |
| Software and algorithms | ||
| FlowJo V10 | Tree Star Inc | RRID: SCR_008520 |
| Other | ||
| 100 μm cell strainers | Falcon | Cat#352360 |
| 50 μm pre-separation strainers | Sysmex | Cat#04-004-2327 |
| Magnet Stand | Miltenyi Biotec | Cat#130-090-976 |
| BD FACSAria | BD Biosciences | N/A |
Materials and equipment
Culture medium
| Final concentration | Amount | |
|---|---|---|
| RPMI 1640 | n/a | 450 mL |
| Penicillin streptomycin | 40 U/mL | 2 mL |
| 10× sodium pyruvate | 1× | 5 mL |
| Human serum | 10% | 50 mL |
| Total | n/a | 500 mL |
Store at 4°C for up to 1 month.
Miltenyi buffer
| Final concentration | Amount | |
|---|---|---|
| PBS | 1× | 490 mL |
| Human serum | 2% | 10 mL |
| Total | n/a | 500 mL |
Store at 4°C for up to 1 month.
FACS buffer
| Final concentration | Amount | |
|---|---|---|
| PBS | 1× | 98 mL |
| FCS | 2% | 2 mL |
| Total | n/a | 100 mL |
Store at 4°C for up to 1 month.
Red blood cell lysis solution
| Final concentration | Amount | |
|---|---|---|
| RBC lysis 10× | 1× | 0.5 mL |
| Distilled water | n/a | 4.5 mL |
| Total | n/a | 5 mL |
Always prepare fresh red blood cell lysis solution. Do not store 1× red blood cell lysis solution.
Freezing solution
| Final concentration | Amount | |
|---|---|---|
| FCS | n/a | 900 μL |
| DMSO | 10% | 100 μL |
| Total | n/a | 1 mL |
Always prepare fresh freezing solution. Do not store.
LC medium
| Final concentration | Amount | |
|---|---|---|
| RPMI 1640 | n/a | 450 mL |
| Penicillin streptomycin | 40 U/mL | 2 mL |
| 10× sodium pyruvate | 1× | 5 mL |
| FCS | 10% | 50 mL |
| Total | n/a | 500 mL |
Store at 4°C for up to 1 month.
Culture medium IL-2
| Final concentration | Amount | |
|---|---|---|
| RPMI 1640 | n/a | 450 mL |
| Penicillin streptomycin | 40 U/mL | 2 mL |
| 10× sodium pyruvate | 1× | 5 mL |
| Human serum | 10% | 50 mL |
| Recombinant IL-2 | 50 U/mL to 100 U/mL | n/a |
| Total | n/a | 500 mL |
Store at 4°C for up to 1 month.
CRITICAL: Medium and buffers that contain human serum should be filtered through 0.2 μm membrane before use. Do not add Ethylenediaminetetraacetic acid (EDTA) in the Miltenyi buffer as it will cause clogging in the LS column if sample contains a lot of dead cells.
Alternatives: Alternatively, the human serum in the Miltenyi buffer could be replaced by bovine serum albumin (BSA) at 0.5% in phosphate buffered saline (PBS).
Step-by-step method details
Tumor dissociation
Timing: 3 h
This section describes how to process solid tumor samples into single-cell suspension in order to isolate each cell population. We recommend to process the sample few hours after the surgical collection, but the tumor can be stored into the medium tube at 4°C up to 24 h.
-
1.
Decontaminate working spaces under the hood and equipment (scissor and tweezers) with 70% ethyl alcohol before beginning.
-
2.Prepare enzyme digestion mix in a 15 mL tube:
-
a.Add 4.7 mL of RPMI medium without serum and any other supplements.
-
b.Add 200 μL of enzyme H, 100 μL of enzyme R and 25 μL of enzyme A into the tube.
-
c.Mix well by vortexing.
-
a.
-
3.
Cut the tumor into small pieces of approximately 2 mm3 and place it directly into the enzyme mix.
-
4.
Place the sample into a 37°C incubator under continuous rotation during 40 min.
Note: Usually, we obtained 4 to 12 cm3 tumor sample. The protocol is for around 6 cm3 tumor sample volume and we recommend splitting the tumor into two mix enzyme tubes if it is larger than 6 cm3.
-
5.
Place a 100 μm cell strainer on 50 mL tube and rinse the cell strainer with 1 mL of culture medium.
-
6.
Transfer digested tumor pieces on the cell strainer with the enzyme digestion medium.
-
7.Smash the tumor gently with the flat end of a 1 mL syringe plunger (remove the plunger from the syringe).
-
a.Regularly wash the cell strainer with culture medium during smashing of the tumor.
-
b.Continue to smash until the maximum of tumor tissue is dissociated.
-
c.Wash the cell strainer until reaching 48 mL of cell suspension volume in the 50 mL tube.
-
a.
CRITICAL: At this step, if the tumor is over smashed or smashed with too much force, there is a risk to decrease cell viability, especially for tumor cells, which are often sensitive. Depending on the tumor features, some tissue fragments with calcifications are not well digested and cannot be dissociated in single-cell suspension. These fragments are tough to dissect and have to be wasted after the washing step of the cell strainer.
-
8.
Centrifuge the cell suspension for 10 min at 400–500 × g.
-
9.
During the centrifugation, prepare 5 mL of Red Blood Cell (RBC) lysis solution 1×.
-
10.
Discard the supernatant and keep a small volume of medium into the tube (around 500 μL).
-
11.
Dissociate gently the pellet by tapping the tube.
-
12.
Resuspend slowly the pellet with 5 mL of RBC lysis solution using a 5 mL pipette and make 5× up and down.
-
13.
Incubate 3 min at 20°C–25°C.
-
14.
Centrifuge the cell suspension for 5 min at 400–500 × g.
-
15.
Discard supernatant and resuspend the pellet in 20 mL culture medium.
-
16.
Count the number of cells and the viability with trypan blue solution.
-
17.Centrifuge the cell suspension for 5 min at 400–500 × g, discard the supernatant, and resuspend the pellet in 10 mL of culture medium.
-
a.Set apart around 0.5–1 million of total cells for flow cytometry staining in a 5 mL tube.
-
a.
-
18.
Centrifuge the cell suspension for 5 min at 400–500 × g, discard the supernatant and proceed to column cell isolation (see Isolation of tumor-resident memory T cells).
Pause point: It is possible to keep cells in culture for 24 h before the isolation of CD8+ T cells; however, the viability could be reduced. For cell culture, plate cells in a 6-well cell culture plate at 2 million/mL in rIL2 culture medium, and place cells in 37°C incubator. Another possibility is to freeze cells in freezing media at 10–20 million cells/mL. However, this process will also give rise to cell mortality. The tumor cell population is the most sensitive cell subset.
-
19.For flow cytometry staining, centrifuge cell suspension (0.5–1 million of total cells) for 5 min at 400–500 × g, discard the supernatant.
-
a.Resuspend the cell pellet in 200 μL of PBS.
-
b.Centrifuge the cell suspension for 5 min at 400–500 × g, discard the supernatant.Note: During the following steps, cells have to be protected from light with aluminum foil.
-
c.During the spin, prepare LIVE/DEAD Fixable blue viability staining by diluting 0.1 μL of concentrated stock with 100 μL of PBS.
-
d.Resuspend the cell pellet with 100 μL of diluted LIVE/DEAD Fixable blue and incubate 25 min at 20°C–25°C .
-
e.During the incubation, prepare the antibody mix (see mix 1).
-
f.Add 100 μL of FACS buffer and centrifuge 5 min at 400–500 × g, discard supernatant.
-
g.Resuspend the cell pellet with 25 μL of Fc block solution diluted 1/10 with FACS buffer. Incubate 10 min at 20°C–25°C.
-
h.Add directly on cell suspension the 25 μL of antibody mix 1 (concentrated 2×). Incubate 25 min on ice.
-
i.Wash by adding 300 μL of FACS buffer in the tube. Centrifuge 5 min at 400–500 × g, discard supernatant.
-
j.Resuspend in 200 μL FACS buffer and filter cell suspension on a 50 μm cell strainer before analysis with Fortessa 488/640/405/561/355 (BD Biosciences ) flow cytometer.
-
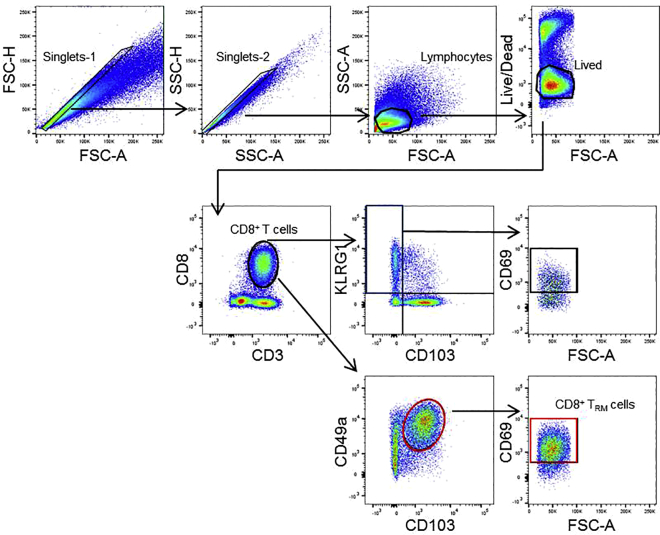
k.See Figure 1 for the gating strategy of TRM cell analysis with Flowjo software.
-
a.
Antibody mix 1
| Antibody | Initial concentration | Final concentration | Amount |
|---|---|---|---|
| FACS buffer | n/a | n/a | 25 μL |
| CD8-Pacific Blue | 0.5 mg/mL | 1/100 | 0.5 μL |
| CD3-Alexa700 | 0.5 mg/mL | 1/100 | 0.5 μL |
| CD103-FITC or BV711 | 0.05 mg/mL | 1/100 | 0.5 μL |
| CD69-APC-Cy7 | 0.2 mg/mL | 1/100 | 0.5 μL |
| CD49a-PerCPefluor710 | 0.025 mg/mL | 1/100 | 0.5 μL |
| KLRG1-PE | 0.05 mg/mL | 1/200 | 0.25 μL |
| PD-1-PECy7 | 0.1 mg/mL | 1/100 | 0.5 μL |
Pause point: The stained cell could be keep 12–20 h at 4°C before flow cytometer analyses. To keep the cells intact, we recommend fixing the cells with 100 μL of diluted FoxP3 fixation/permeabilization solution according to the manufacturer protocol. Wash cell suspension with 100 μL of FACS buffer, centrifuge, and resuspend in 200 μL of FACS buffer. Keep at 4°C up to 24 h until flow cytometer analysis.
Figure 1.
Gating strategy for analyses of tumor-infiltrating CD8+CD103+ TRM cells by flow cytometry
Isolation of tumor-resident memory T cells
Timing: 2 h
This section describes the protocol leading to the isolation of the CD8+CD103+ TRM cell population. First, it consists in a positive selection of total CD8+ T cell population with magnetic bead Miltenyi system and it is followed by the FACS isolation of CD8+CD103+ T cells.
CRITICAL: This section has to be processed under biosafety hood and in sterile conditions.
-
20.
Resuspend total cells in cold Miltenyi buffer at the concentration of 10 × 106 total cells for 80 μL.
-
21.
Add 20 μL of human CD8 microbeads for 10 × 106 total cells and mix well with up and down pipetting. Incubate at 4°C during 15 min.
Note: Always wait until the column reservoir is empty before proceeding to the next step.
-
22.Attach the magnet to the magnetic stand and place the LS column in the magnet field of a suitable Miltenyi magnet.
-
a.Add the 50 μm pre-separation filter in the LS column.
-
b.Equilibrate the column by pipetting 3 mL cold Miltenyi Buffer into the 50 μm pre-separation filter. Wait until the column reservoir is empty.
-
c.Place a 15 mL tube under the column to collect unlabeled cells. The unlabeled CD8-negative cells will be in the flow through from the column.
-
a.
-
23.
After incubation with microbeads, wash cells by adding 5 mL cold Miltenyi buffer. Centrifuge the cell suspension for 5 min at 400–500 × g and aspirate the supernatant completely.
-
24.
Resuspend cells in 1 mL cold Miltenyi buffer (or 500 μL if less than 10 million cells).
-
25.
Apply cell suspension through the cell strainer in the LS column. If the cell suspension is dense or contains a lot of dead cells, split the volume to pass cell suspension through two cell strainers. Troubleshooting 1.
-
26.
When cell suspension is passed through the column, wash by adding 3 mL of cold Miltenyi buffer. Keep collecting the flow through into the 15 mL tube. Troubleshooting 2.
Optional: The negative fraction contains tumor cells for further cell separation (see the section Isolation of tumor cells).
-
27.
When the column reservoir is empty, perform washing steps by adding 3 mL of Miltenyi buffer. Repeat step 26 three times.
-
28.
When the column reservoir is empty, remove the LS column from the magnet and place it in a new collection 15 mL tube.
-
29.
To recover the positive CD8+ fraction, add 5 mL of Miltenyi buffer in the LS column and immediately flush out the cells by pushing the plunger into the column.
-
30.
Centrifuge the cell suspension for 5 min at 400–500 × g, aspirate the supernatant completely, and resuspend the cell pellet in 3 mL culture medium.
-
31.
Count the number of cells in the positive fraction and the viability with trypan blue solution.
-
32.
Centrifuge the CD8+ cell suspension for 5 min at 400–500 × g.
Optional: It is possible to keep CD8+ cells in culture to amplify them in culture medium supplemented with 50 U/mL of recombinant (r)IL-2. Plate cells in a 96-V-well culture plate at 10,000 cells/200 μL/well and place it in 37°C incubator. Another possibility is to freeze cells; resuspend total cells in 1 mL in freezing media and place the tube at −80°C.
Note: All cells and reagents should be kept on ice during this section of the protocol to prevent cell death.
-
33.
While the cells spin, prepare the following antibody mix 2 (for up to 20 million cells, scale up accordingly)
Antibody mix 2
| Antibody | Initial concentration | Final concentration | Amount |
|---|---|---|---|
| Culture medium | n/a | n/a | 100 μL |
| CD8-Pacific Blue | 0.5 mg/mL | 1/100 | 1 μL |
| CD103-FITC | 0.05 mg/mL | 1/100 | 1 μL |
| KLRG1-PE | 0.05 mg/mL | 1/200 | 0.5 μL |
CRITICAL: We recommend avoiding sorting the cells with anti-CD3 antibody as it could result in non-specific activation of T cells and consequently biased results.
-
34.
Aspirate supernatant, resuspend the cell pellet with the antibody mix and incubate for 20 min at 4°C (protect the tube from light).
-
35.
Add 3 mL of culture medium and centrifuge the CD8+ cell suspension for 5 min at 400–500 × g.
-
36.
Resuspend cells in culture medium at a concentration of approximately 10 million cells/mL.
-
37.
Filter the cell suspension in a 50 μm cell strainer.
-
38.
Add to the cell suspension TO-PRO3 iodide at a final concentration of 0.1 μM just prior to cell sorting.
Note: We use an aliquot of stained cells for area scaling and setting sort gates.
Note: The TO-PRO3 iodide is added just prior to FACS in order to prevent overstaining that can occur with prolonged exposure (not more than 1 h) of cells to TO-PRO3 iodide.
-
39.
Prepare two 5 mL tubes containing 1 mL of culture medium to collect the sorted cell populations.
-
40.
Sort cells on a BD FACS Aria fusion 488/561/640/405/355 (Becton Dickinson).
Note: A 85 μm nozzle is used with 45 psi pressure and frequency fixed at 60 kH. Adjust the sorting rate at 4,000 events/s.
-
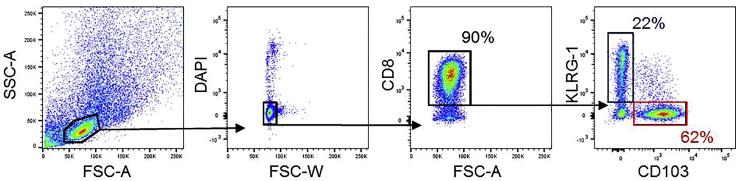
41.
Sort desired CD8+CD103+ and CD8+KLRG1+ (or CD8+CD103−) populations, according to the gating strategy (Figure 2), into culture medium in sterile, capped tubes and transfer onto ice.
Pause point: For culture of T cell populations, centrifuge cell suspension in conic 15 mL tube or 1.5 mL Eppendorf tube and resuspend in culture medium with rIL-2 (final concentration at 50 U/mL) at 0.2 to 0.5 million/mL and transfer in culture plate.
Note: T cells can be cultured and amplify for several days in culture medium with rIL-2. The culture medium needs to be replaced every 4 or 5 days.
Pause point: For RNA sequencing analyses, add 2 mL PBS in the tube containing the sorted cells and centrifuge cell suspension in conic 15 mL tube or 1 mL Eppendorf tube for 10 min at 500 × g. Remove completely and carefully the supernatant with a 200 μL pipette without touching the cell pellet. Resuspend cell pellet directly in 100 μL buffer RL (Kit Norgen) supplemented with 10% β-mercaptoethanol, and mix by pipetting up and down a few times. Proceed with the RNA purification (Kit Norgen) according to the manufacturer protocol, or keep the cell lysate at −80°C until RNA extraction. (https://norgenbiotek.com/sites/default/files/resources/Single-Cell-RNA-Purification-Kit-Insert-PI51800-3-M14.pdf)
Figure 2.
Gating strategy for sorting CD8+CD103+ and CD8+KLRG1+ cells by FACS
Isolation of tumor cells
Timing: 30 min
This section describes the isolation of tumor cells from the CD8-negative cell fraction obtained previously. The isolation of tumor cells is based on Tumor Cell Isolation Kit from Miltenyi, designed for the enrichment of untouched tumor cells and depletion of non-tumor cells (see manufacturer protocol at https://www.miltenyibiotec.com/FR-en/products/tumor-cell-isolation-kit-human.html#130-108-339). The isolation of CD8+ T cells and autologous tumor cells enable to perform in vitro functional experiments to assess the tumor specificity of CD8+ T cell populations, as such cytotoxicity with chromium51 release assay or conjugate formation.
-
42.
Determine the number of cells in the CD8-negative fraction.
-
43.
Centrifuge the cell suspension for 5 min at 400–500 × g. Aspirate the supernatant completely.
-
44.
Resuspend up to 10 million total cells in 60 μL of cold Miltenyi buffer (scale up according to cell number).
-
45.
Add 20 μL of Non-tumor cell depletion Cocktail A and 20 μL of Non-tumor cell depletion Cocktail B for up to 10 million cells (scale up according to the cell number), mix well with up and down pipetting and incubate for 15 min at 4°C.
Note: CD8-negative fraction cells could be kept at 4°C for 3 h before cell isolation; however, longer storage could result in reduction of cell viability.
Note: For next steps, always wait until the column reservoir is empty before proceeding to the next step.
-
46.During incubation, place the LS column in the magnet field of a Miltenyi magnet.
-
a.Add the 50 μm pre-separation filter in the LS column.
-
b.Equilibrate the LS column by pipetting 3 mL cold Miltenyi Buffer into the pre-separation filter. Wait until the column reservoir is empty.
-
c.Place a 15 mL tube under the LS column to collect unlabeled cells. The unlabeled tumor cells will be in the flow through from the column.
-
a.
-
47.Bring up the cell suspension to 500 μL of cold Miltenyi buffer for up to 10 million cells (scale up according to the cell number).
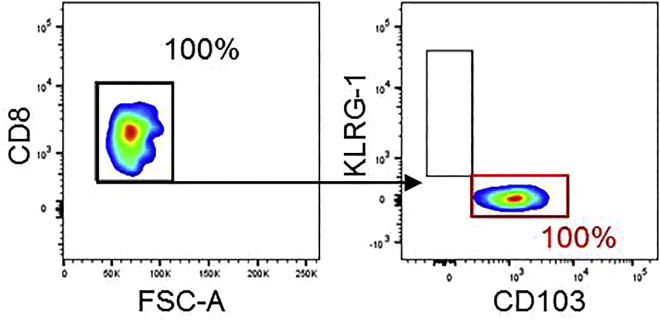
Figure 3.
Purity of sorted CD8+ TRM cells post-FACS -
48.
Apply cell suspension through the cell strainer in the LS column.
-
49.
When the column reservoir is empty, perform washing steps by adding 1 mL of Miltenyi buffer in the column. Keep collecting the flow through from the washing steps into the 15 mL tube containing the unlabeled cells.
-
50.
Repeat step 49 once.
Optional: The labeled fraction containing non-tumor cells can be recovered by removing the column from the magnet, placing it into a new 15 mL tube, adding 3 mL of Miltenyi buffer, and immediately flushing out the cells by pushing the plunger into the column.
-
51.
Determine the cell number and viability with trypan blue solution of the untouched tumor cells.
-
52.
Centrifuge the tumor cell suspension for 5 min at 400–500 × g and discard the supernatant completely.
-
53.
Resuspend in LC medium at 200,000 tumor cells/mL and plate them in 6/12 or 24-well culture plate depending of the number of cells.
-
54.
Place the cells in incubator 37°C, 5% CO2.
Note: The number of tumor cells recovered is extremely dependent of the tumor piece. A strong variability between patient tumors exists both for the cancer cell number and for immune cell infiltration.
Note: For the majority of tissue samples, isolated tumor cells usually attached to the plastic culture plate after a few hours of culture. However, depending of the tumor sample, it appends that tumor cells do not attach on the plastic, reflecting a weak viability. To detach tumor cells, we recommend to remove the culture medium, wash with 2–3 mL of PBS, remove the remaining medium and then apply PBS with EDTA diluted at 1/1000. Incubate 2–3 min at 37°C and monitor the detachment of tumor cells under microscope. Perform gently up and down pipetting with a 1 mL pipette to continue detaching adherent cells. Stop the EDTA by adding LC medium and transfer cell suspension in 15 mL tube to centrifuge and resuspend in the desired volume of culture medium.
Expected outcomes
Table 1 details the step-by-step expected cell recovery outcomes. The obtained number of cells for each population is extremely dependent of the tumor sample composition. Usually, for tumors that are highly infiltrated by immune cells, the yield of tumor cells obtained is weak.
Table 1.
Expected number of cells after each main step
| Outcome | Result |
|---|---|
| Total cells post-tumor dissociation | 1 to 100 million of cells depending of the tumor sample, 50%–80% viability |
| CD8+ cells post-MACS isolation | 0.2 to 30 million, purity 50%–95% |
| Tumor cells post-MACS isolation | 0.1 to 20 million, purity 80%–95% |
| CD8+CD103+ TRM cells post-FACS | 10,000 to 500,000 cells, purity 95%–100% (Figure 3) |
Regarding the RNA extraction from TRM cells, the expected RNA yield after purification is ranged from 15 to 140 ng for 70,000 to 250,000 cell numbers.
Limitations
NSCLC tumors are variable between patients and do not permit to obtain standardized results. The number of TRM cells recovered after this protocol is dependent of the tumor infiltration by immune cells. A too small tumor sample size or a defective conservation of the fresh tumor sample could lead to a small yield of CD8+ T cells and tumor cells. Highly necrotic tumor sample is not usable for this protocol. A limited number of TRM cells (>10,000 cells) could be enough for RNA extraction, but insufficient for further applications or functional experiments.
Troubleshooting
Problem 1
The pre-separation 50 μm cell strainer is clogged after adding the cell suspension.
Potential solution
The cell suspension could be very dense depending of the tumor composition or, if a high proportion is composed of dead cells. It is essential to remove the maximum of aggregate before adding cells to the separation column. Pass half of the cell suspension through cell strainer and use a new cell strainer to pass the rest of the cells. The same volume of cells could eventually pass through two consecutive cell strainers.
Problem 2
The cell suspension does not go through the LS column, meaning the column is clogged.
Potential solution
It is preferable to filter cell suspension before applying onto the LS column. If the cell strainer is also clogged, there is a strong probability that the column will be clogged as well. Make sure that the cell suspension is not viscous due to cell death and DNA release. If the cell suspension is too viscous it will most probably result in a very few cell recovery. If the cell suspension does not go through the LS column, recover the volume of cell suspension on the top of the column, filter on a 50 μm cell strainer and apply on new pre-washed LS column. Cells blocked on the first column could be recovered by flushing out with syringe, but the purity will not be achieved.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stéphanie Corgnac (stephanie.corgnac@gustaveroussy.fr).
Materials availability
This protocol does not generate unique reagents.
Data and code availability
This protocol does not generate data.
Acknowledgments
This work was supported by grants from the Association pour la Recherche sur le Cancer (ARC; grant numbers SIGN’IT20181007792, PJA20161204720, and PJA 20181208049), the Institut National du Cancer (INCa; PLBIO016-080 grant number 10557), Ligue contre le Cancer (Comité des Yvelines, grant number 9FI12414QLCZ and Comité du Val de Marne, 2019, grant number R19054LL), and Bristol-Myers Squibb (BMS, France; grant number CA209-942). S.C. was supported by a grant from the Groupement des Entreprises françaises dans la Lutte contre le Cancer (GEFLUC; grant number 2016-R16180LL) and Cancéropôle Ile de France and was a recipient of a fellowship from Fondation Recherche Medicale (FRM), Gustave Roussy (SIRIC-SOCRATE), and INCa (PLBIO016-080 grant number 10557). We are grateful to Pierre Validire from Institut Mutualiste de Montsouris and Vincent De Montpréville and Caroline Communaux from the CRB of Marie Lannelongue Hospital for their precious help with fresh human NSCLC tumors. We also thank Philippe Rameau and Cyril Catelain from the cytometry facility (Plateforme d’Imagerie-Cytométrie) of Gustave Roussy. Graphical abstract was created with Biorender.
Author contributions
Conception and design, collection and assembly of data (performance of experiments), data analysis and interpretation, manuscript writing, and funding acquisition, S.C.; FACS sorting, performance of experiments, and manuscript writing, Y.L.; data interpretation, manuscript writing, and funding acquisition, F.M.-C.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Stéphanie Corgnac, Email: stephanie.corgnac@gustaveroussy.fr.
Fathia Mami-chouaib, Email: fathia.mami-chouaib@gustaveroussy.fr.
References
- Cohen C.J., Gartner J.J., Horovitz-Fried M., Shamalov K., Trebska-Mcgowan K., Bliskovsky V.V., Parkhurst M.R., Ankri C., Prickett T.D., Crystal J.S. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corgnac S., Malenica I., Mezquita L., Auclin E., Voilin E., Kacher J., Halse H., Grynszpan L., Signolle N., Dayris T. CD103+CD8+ Trm cells accumulate in tumors of anti-PD-1-responder lung cancer patients and are tumor-reactive lymphocytes enriched with Tc17. Cell Rep. Med. 2020;1:100127. doi: 10.1016/j.xcrm.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Krawczyk E., Suprynowicz F.A., Palechor-Ceron N., Yuan H., Dakic A., Simic V., Zheng Y.L., Sripadhan P., Chen C. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017;12:439–451. doi: 10.1038/nprot.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Liao J.Y., Su S. Protocol for single-cell analysis of tumor-infiltrating B cells isolated from human breast cancer tissue before and after neo-adjuvant chemotherapy. STAR Protoc. 2020;1:100040. doi: 10.1016/j.xpro.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret R., Sierro S.R., Botelho N.K., Corgnac S., Donda A., Romero P. Analysis of tumor-infiltrating lymphocytes following CD45 enrichment. Bio Protoc. 2014;4:E1218. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol does not generate data.