Abstract
Background
Previous studies showed that suppression of pyruvate carboxylase (PC) expression in highly invasive breast cancer cell line, MDA-MB-231 inhibits cell growth as a consequence of the impaired cellular biosynthesis. However, the precise cellular mechanism underlying this growth restriction is unknown.
Methods
We generated the PC knockdown (PCKD) MDA-MB-231 cells and assessed their phenotypic changes by fluorescence microscopy, proliferation, apoptotic, cell cycle assays and proteomics.
Results
PC knockdown MDA-MB-231 cells had a low percentage of cell viability in association with accumulation of abnormal cells with large or multi-nuclei. Flow cytometric analysis of annexin V-7-AAD positive cells showed that depletion of PC expression triggers apoptosis with the highest rate at day 4. The increased rate of apoptosis is consistent with increased cleavage of procaspase 3 and poly (ADP-Ribose) polymerase. Cell cycle analysis showed that the apoptotic cell death was associated with G2/M arrest, in parallel with marked reduction of cyclin B levels. Proteomic analysis of PCKD cells identified 9 proteins whose expression changes were correlated with the degree of apoptosis and G2/M cell cycle arrest in the PCKD cells. STITCH analysis indicated 3 of 9 candidate proteins, CCT3, CABIN1 and HECTD3, that form interactions with apoptotic and cell cycle signaling networks linking to PC via MgATP.
Conclusions
Suppression of PC in MDA-MB-231 cells induces G2/M arrest, leading to apoptosis. Proteomic analysis supports the potential involvement of PC expression in the aberrant cell cycle and apoptosis, and identifies candidate proteins responsible for the PC-mediated cell cycle arrest and apoptosis in breast cancer cells.
General significance
Our results highlight the possibility of the use of PC as an anti-cancer drug target.
Keywords: Pyruvate carboxylase, Breast cancer, Metabolism, Apoptosis
Highlights
-
•
Suppression of PC in MDA-MB-231 decreases cell growth and increases apoptosis.
-
•
Impaired G2/M cell cycle arrest is associated with increased apoptosis.
-
•
Proteomic analysis identified 9 proteins whose expression levels were associated with cell cycle and apoptosis.
-
•
STITCH analysis showed 3 of 9 proteins directly links with apoptotic and cell cycle network.
1. Introduction
The tricarboxylic acid (TCA) cycle functions as both an oxidative hub to oxidize acetyl-CoA and a biosynthetic hub for the synthesis of nucleotides, amino acids and fatty acids [1]. Glutaminloysis and pyruvate carboxylation are important anaplerotic reactions which maintain TCA cycle integrity by replenishing its intermediates upon the removal for the biosynthesis of biomolecules. While glutaminolysis produces α-ketoglutarate from glutamine [2], pyruvate carboxylation produces oxaloacetate from pyruvate [3]. Recent studies showed that these two anaplerotic reactions are essential to support growth of many cancers [4,5]. Depending on their genetic background and stress conditions, most cancers switch these two pathways to support their survival. Non-small cell lung cancer (NSCLC), breast cancer and low grade glioblastoma bearing loss-of-function mutation of IDH1/2 gene use pyruvate carboxylation to support their growth [[6], [7], [8], [9]]. In NSCLC, suppression of pyruvate carboxylase (PC) expression inhibits tumor growth both in vitro and in xenograft model [7,10]. In murine breast cancer model, suppression of PC in 4T1 breast cancer inhibits its ability to metastasize to the lung, underscoring the importance of PC in establishment of growth at the secondary tumor sites [11].
Previous studies by our group showed that PC expression is overexpressed in breast cancer tissue patients [8], and suppression of PC expression in highly metastatic breast cell line, MDA-MB-231, impairs biosynthesis of amino acids, nucleotides and fatty acids from TCA cycle activity, resulting in growth inhibition [12]. Although these studies show the importance of PC in supporting various biosynthetic pathways, it is unknown how depletion of these anabolic supplies restricts cell growth. Here we show that suppression of PC expression in MDA-MB-231 cell line induces G2/M cell cycle arrest, accompanied by increased apoptosis. Proteomics analysis of PC knockdown cells identified several proteins which interact with key proteins in cell cycle and apoptosis.
2. Materials and methods
2.1. Generation of PC knockdown cell lines
Stable PC knockdown (PCKD) MDA-MB-231 cell lines were generated by transfecting MDA-MB-231 cell line (ATCC: HTB26) cells with a shRNA construct targeted to human PC as described previously [12]. Two stable PC KD, 4B#3, and 4B#4 and the scrambled control (SC) cell lines were isolated and used for the subsequent analysis. PC KD cell lines were cultured in DMEM (Gibco) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) and penicillin/streptomycin (Gibco), at 37 °C with a 5% (v/v) CO2.
2.2. Proliferation assay
2 × 105 of PC KD or SC MDA-MB-231 cells were plated and grown in 6-well plates containing complete DMEM medium for 24 h before the medium was changed to MEM (5 mM glucose) without non-essential amino acids. All cell lines were maintained at 37 °C for 5 days. Viable cell count was assessed by staining with 0.2% trypan blue.
2.3. Immunofluorescent staining
1.0 × 104 cells were plated and grown into 24-well plate containing complete MEM supplemented with serum and antibiotics for 3 days. SC and PCKD cell lines were washed with PBS and fixed with 0.2% (w/v) paraformaldehyde for 15 min and stained with 1:500 dilution of Alexa Flour™ 546 phalloidin (Invitrogen) in PBST [PBS with 0.1% (v/v) Tween-20] for 30 min. The cells were washed and counter stained with 0.5 μg/ml Hoechst 33342 (Cell signaling) for 2 min and observed under a fluorescent microscope (Olympus IX83 inverted microscope).
2.4. Cell cycle and apoptosis assays
5 × 105 cells were plated into 6-well plate containing MEM supplemented with serum and antibiotics and grown for 4 days. At each time point, cells were trypsinized and subjected to cell cycle analysis using Muse Cell Cycle Assay Kit (Merck) following the manufacturer protocols. Apoptotic cell death was performed using Muse Annexin V and Dead Cell Assay Kit (Merck).
2.5. SDS-PAGE and Western blotting
30 μg of whole cell lysate was separated on 7.5–12% SDS-PAGE and Western blotting as previously described [12]. Cyclin-B was detected using anti-cyclin B (Sigma). Apoptotic markers including, caspase-3 and poly (ADP-ribose) polymerase (PARP) were detected using anti-caspase-3 (Cell Signaling) and anti-PARP (Cell Signaling) antibodies, respectively. Anti-β-actin antibody (Sigma), was used as the loading control.
2.6. Protein extraction, in-gel digestion, LC-MS/MS and, protein quantitation and identification
SC and PC KD clones 4B#3 and 4B#4, were cultured in MEM medium for 4 days before cells were scraped in 2 ml of ice-cold PBS, and centrifuged at 3,000 rpm for 5 min. Protein extraction, gel electrophoresis and liquid chromatography-tandem mass spectrometry (GeLC MS/MS) were performed as described previously [13].
Protein quantitation was performed with DeCyder MS 2.0 Differential Analysis software (DeCyderMS, GE Healthcare). The acquired LC-MS data were converted, and the PepDetect module was used for automate peptide detection, charge state assignments and quantitation. The MS/MS data were analyzed and searched against the NCBI human database using the Mascot software version 2.2 (Matrix Science, London, UK). The subcellular distribution and biological processes were assigned to protein identification according to the Uniprot (http://www.uniprot.org). A Venn diagram was generated using jvenn web application [14]. Hierarchical clustering heat map was analyzed by Multiexperiment Viewer (MeV) program, version 4.8, with the Pearson correlation distance metric [15]. Protein-protein interaction network was analyzed using STITCH 5.0 database (http://stitch.embl.de/).
2.7. Statistical analysis
Statistical analysis was performed using two-way ANOVA and paired student's t-test using Graphpad Prism Software.
3. Results
3.1. Suppression of pyruvate carboxylase in MDA-MB-231 induces apoptotic cell death
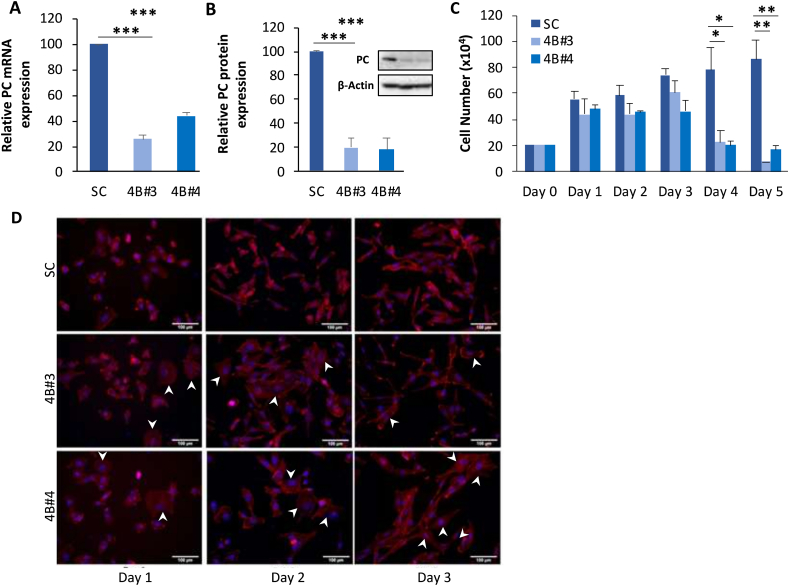
Both PC KD 4B#3 and 4B#4 clone retained PC mRNA approximately 25% and 40% and PC protein approximately 20% of the SC cells, respectively (Fig. 1A and 1B). Proliferation assay showed that both PC KD clones grew at the similar rate during the first two days, however they started to grow more slowly at day 3. At day 4, growth rate of both PC KD cell lines was only 25% of the SC cells. At day 5, PC KD clone 4B#3 showed a further reduction of cell numbers while clone 4B#4 remained unchanged (Fig. 1C). Examination of the morphology of both PC KD cell lines showed that the spindle shape of the cells becomes less prominent. Their sizes were larger from day 1 and became more obvious at day 2. Hoechst staining showed that the nuclei of PC KD cells were slightly larger than those of SC cells (Fig. 1D). Both PC KD clones were mostly detached at day 4 (Fig. 1D).
Fig. 1.
Suppression of PC expression reduces cell growth. (A) qRT-PCR of PC mRNA and (B) Western blot of PC protein in PC KD clones (4B#3 and 4B#4) and SC cells. (C) Growth of PC KD and SC cells over 4 days (*p < 0.05, **p < 0.01, ***p < 0.005). (D) Cell morphology of PCKD and SC cells were assessed by staining with phalloidin and Hoescht. White arrows indicate the large PC KD cells.
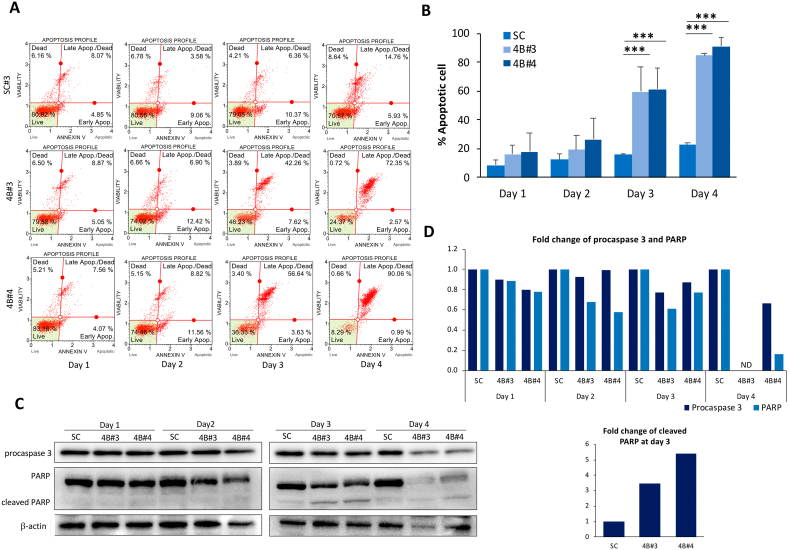
To examine whether the reduced cell growth of PC KD cells was associated with a programmed cell death, an apoptosis assay was performed. As shown in Fig. 2A and B, both PC KD cells had similar proportion of live and dead cells at day 1. However, both PC KD clones had an increased number of apoptotic cells at day 2, and more apparent at day 3. At day 4, both PC KD clones had the proportion of apoptotic and dead cells approximately 4-fold higher than that of the SC cells (Fig. 2B). Although cleaved caspase 3 was not observed, the levels of procaspase 3 were reduced by 20–30% at day 3 and more apparent at day 4 (Fig. 2C-D). The failure to detect cleaved caspase 3 may be attributed to its rapid turn-over between days 3 and 4. Beside caspase 3 activation, we also detected the cleavage of poly(ADP-ribose) polymerase (PARP), another marker of apoptosis. Similar to procaspase 3, the level of full length PARP of PC KD clones was reduced by 40–50% at day 2 and day 3, consistent with 3-5-fold increase of cleaved PARP at day 3. However, both full length and cleaved PARP were barely detectable at day 4. (Fig. 2C-D). The rapid increase of both PARP and procaspase 3 cleavages at days 3 and 4 is in agreement with the marked reduction of cell growth at day 4 (Fig. 1C) and the highest number of apoptotic cells (Fig. 2B).
Fig. 2.
Apoptotic cell death of PC KD MDA-MB-231 cells. (A) distribution of cell population in live, early apoptosis, late apoptosis and dead cell subpopulations, respectively. (B) Percentages of apoptotic cells are shown as the means ± SD from three independent experiments. ***p < 0.005. (C) Representative of Western immunoblot of procaspase 3, PARP and leaved PARP in PC KD clones 4B#3 and 4B#4, and SC cell lines at various time points. (D) Relative abundance of procaspase 3, PARP and cleaved PARP in PC KD clones. Intensity of these protein bands was normalized with that of β-actin as relative expression. The relative value of procaspase 3, PARP and cleaved PARP was relative to that of the scrambled control cell line, which was arbitrarily set to 1 at each time point. ND = Non-detectable.
3.2. Suppression of pyruvate carboxylase in MDA-MB-231 induces G2/M arrest
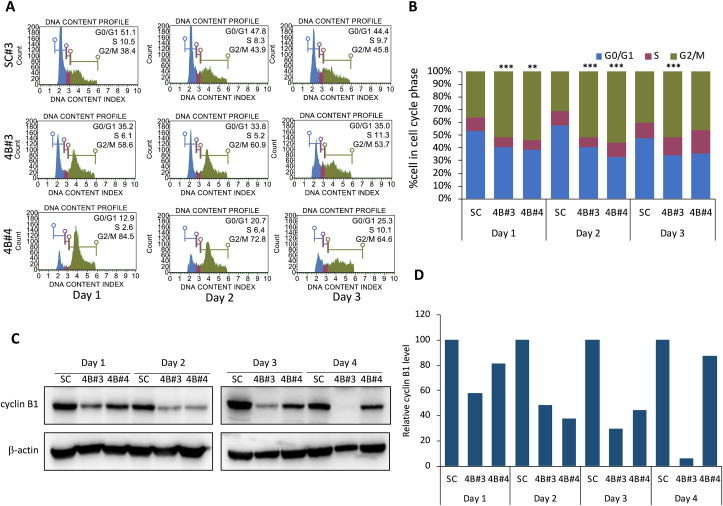
As apoptosis and cell proliferation are connected, some cell cycle regulators can influence both cell division and programmed cell death [16,17]. We assessed whether suppression of PC in MDA-MB-231 cells perturbed cell cycle progression. For the SC cell line, the proportion of cells in G0/G1, S, G2/M phases were relatively unchanged throughout 3 days as shown in Fig. 3A and B. However, both PC KD cell lines had the proportion of cells in G2/M phase almost double that of the SC cell line (70–79% vs 40%) while the proportion of cells in G0/G1 phase was decreased by half of the SC cell line (15–21% vs 46%) since day 1. At days 2 and 3, the proportion of PC KD cells was mostly in G2/M albeit slightly lower than on day 1, but this was still higher than the SC cell line (50–60% vs 40%). Because cyclin B is an important regulator which allows cells to progress from G2 to M phase, lack of cyclin B would result in G2/M arrest [18]. Western blot analysis showed that cyclin B level was markedly reduced in both PC KD clones since day 1 and became more apparent at days 2 and 3 (Fig. 2C), consistent with the highest proportion of cells distributing in G2/M phase. This result indicates that suppression of PC expression induces G2/M arrest.
Fig. 3.
Impaired G2/M2 cell cycle arrest of PC KD MDA-MB-231 cells. (A) The DNA content profiles were plotted versus percentage of cell count from days 1–3. (B) Percentage of cells distributed in G0/G1, S and G2/M phases. The data shown is a representative of three independent experiments. (***p < 0.005). (C) Representative of Western immunoblot of cyclin B in PC KD clones 4B#3 and 4B#4, and SC cell lines at various time points. (D) Intensity of cyclin B band of each PC KD clone was normalized with that of β-actin of each day as relative expression. The relative value of cyclin B of PC KD was relative to that of the scrambled control cell line, which was arbitrarily set to 1 at each time point. ND = Non-detectable.
3.3. Protein expression profiles of PCKD cells associated with cell cycle arrest and apoptosis
Because suppression of PC expression induces G2/M arrest, leading to programmed cell death, we used proteomics to identify cellular pathways that may be associated with apoptosis and cell cycle arrest. A two-way hierarchical clustering analysis showed that 5,286 proteins were differentially expressed between both PC KD and SC cell lines cultured at days 1–4, respectively (Fig. 4A). The number of differentially expressed proteins in these three clones is shown in a Venn diagram. As shown in Fig. 4B, there were 452 proteins expressed in both PCKD cell lines but not expressed in the SC cells. Among these 452 proteins, there were 5 proteins whose expression changes were correlated with the degree of apoptosis, and 4 proteins whose expression changes were correlated with G2/M cell cycle arrest during 4 days of culture, as demonstrated in Fig. 2, Fig. 3, respectively. The function and subcellular distribution of these proteins are shown in Table 1. STITCH analysis showed that there were 6 proteins including calcineurin binding protein 1 (CABIN1), HECT domain containing E3 ubiquitin protein ligase (HECTD3), chaperonin containing TCP1 subunit-3 (gamma) (CCT3), PR domain containing-15 (PRDM15), chromodomain helicase DNA binding protein 2 (CHD2) and lactotransferrin (LTF), found in the database. The interaction of these proteins with classical signaling molecules in the cell cycle and apoptosis, including caspases, BCL-2-associated X protein (Bax), B-cell lymphoma 2 (BCl-2), cyclin, cyclin-dependent kinases (CDKs) and tumor suppressor p53 (TP53) is shown in Fig. 4C. Three proteins, PRDM15, CHD2, LTF were not associated with the cell cycle and apoptotic network, whereas the three proteins, CABIN1, HECTD3, CCT3, together with PC and MgATP were associated with the signaling network of cell cycle and apoptosis. This interaction model supports the involvement of candidate proteins in apoptosis and cell cycle arrest observed in PC KD MDA-MB-231 cells
Fig. 4.
Protein expression profiles of PC KD and SC MDA-MB-231 cells, and their potential interaction with PC, apoptotic and cell cycle regulators. A) Protein expression profiles in SC (columns 1–4), PC KD 4B#3 (columns 5–8) and 4B#4 (columns 9–12) cells from days 1–4, respectively. Green, black and red colors represent low, moderate and high levels of expression, respectively. B) Numbers of proteins expressed in PC KD 4B#3 and 4B#4 and SC cells. C) STITCH 5.0 analysis indicates the association of candidate proteins whose expression levels are correlated with the degree of apoptosis and G2/M arrest. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
A list of 9 proteins whose expression are correlated with increased apoptosis and G2/M arrest in PC KD MDA-MB-231 cells cultured from day 1 to day 4.
| No. | IDs | Proteins associated with apoptosis | Q-Scores | Biological process | Subcellular distribution |
|---|---|---|---|---|---|
| 1 | Q9Y6J0 CABIN_HUMAN | Calcineurin-binding protein cabin-1 | 0.97750 | Signaling | Nucleus |
| 2 | Q5T447 HECD3_HUMAN | E3 ubiquitin-protein ligase HECTD3 | 0.97495 | Protein degradation | Perinuclear region of cytoplasm |
| 3 | P02788 TRFL_HUMAN | Lactotransferrin | 0.03279 | Regulatory protein | Secreted |
| 4 | P20963 CD3Z_HUMAN | T-cell surface glycoprotein CD3 zeta-chain | 0.97409 | Signaling | membrane |
| 5 | O14647 CHD2_HUMAN | Chromodomain-helicase-DNA-binding protein 2 | 0.97462 | DNA replication | Nucleus |
| Proteins associated with G2/M arrest | |||||
| 1 | P49368 TCPG_HUMAN | T-complex protein 1 subunit gamma | 0.97552 | Protein folding, | cytoplasm |
| (chaperonin containing TCP1, subunit 3 (gamma); CCT3) | DNA maintenance | ||||
| 2 | Q6UB99 ANR11_HUMAN | Ankyrin repeat domain-containing protein 11 | 0.97155 | Chromatin regulator | Nucleus |
| 3 | P05783 K1C18_HUMAN | Keratin, type I cytoskeletal 18 | 0.97325 | cell cycle, signaling | Nucleus |
| 4 | P57071 PRD15_HUMAN | PR domain zinc finger protein 15 | 0.97820 | Transcription | Nucleus |
4. Discussion
Metabolism provides biosynthetic precursors for lipids, nucleotides and energy during proliferation while cell cycle regulators also control expression of metabolic enzymes [19]. G1, G2 and M checkpoints are crucial steps which ensure cells can progress to each phase of the cell cycle without any defects [17]. Failure of those checkpoints can trigger apoptosis, eliminating the abnormal cells. On the contrary, disruption of key metabolic enzymes can also perturb cell cycle progression, indicating a tight connection between metabolism and the cell cycle. In G1 phase, glycolysis is an important metabolic pathway that generates ATP to support synthesis of many proteins and enzymes required for DNA replication [20]. A high level of c-Myc also enhances expression of key glycolytic enzymes, sustaining a high rate of glycolysis throughout G1 phase [21]. Once entering S-phase, a high activity of cyclin E/E2F increases expression of thymidine kinase and DNA polymerase, enhancing the rate of DNA synthesis [22]. High activities of pentose phosphate pathway and glutaminolysis during this period also provide ribose-5-phosphate and glutamate that support DNA synthesis. In G2 phase, the de novo lipogenesis is activated, allowing cells to produce membrane phospholipids. He et al. (2018) [23] showed that pharmacological inhibition of acetyl-CoA carboxylase in human renal carcinoma cell lines triggers G2/M arrest, inhibiting cell growth. Similarly, inhibition of fatty acid synthase, another rate-limiting enzyme of de novo lipogenesis inhibits growth of melanoma cell line as a result of G2/M arrest [24]. It is unclear why suppression of PC expression in MDA-MB-231 cells specifically induces G2/M-arrest but it is possible that the oxaloacetate produced by PC may be used to enable the transfer of mitochondrial acetyl-CoA to the cytosol in order to maintain the citrate pool which is a precursor of fatty acid synthesis [25]. This evidence is supported by recent studies which demonstrate that pharmacological inhibition of PC activity or silencing PC expression impairs de novo lipogenesis both in non-cancer and cancer cells [12,26]. Our finding that knocking down of PC expression in MDA-MB-231 cells triggers programmed-cell death is well-supported by the very recent study. Li et al. [27] showed that inhibition of PC activity with small molecule, ZY-444 in MDA-MB-231 cells inhibits their growth via caspase-3-dependent apoptosis.
Because PC maintains TCA cycle activity by replenishing oxaloacetate, suppression of PC in several types of cancers can potentially disrupt the cellular ATP pool. Depletion of cellular ATP is well known to perturb cell cycle and trigger apoptosis [28,29]. Expression of CCT3, CABIN1 and HECTD3, possibly together with signaling molecules in the apoptotic and cell cycle network, in response to PC suppression may also contribute to the depletion of ATP. CCT3 is part of the chaperonin-containing T-complex, assisting protein folding [30]. This complex is involved in cell proliferation and tumorigenesis of many types of cancers [[31], [32], [33], [34]]. Suppression of CCT3 expression down-regulates CDC42 and cyclin D3, and upregulates cyclin-dependent kinase-2 and -6 that inhibit growth and promote apoptosis in gastric cancer [33]. CABIN1 is an inhibitor of calcineurin, which regulates Ca2+-triggered cell death [35]. CABIN1 has recently been identified as a negative regulator of p53 [36,37]. Suppression of CABIN1 promotes cell death in a p53-dependent manner [36]. CABIN1 has recently been shown as one of the proteins which mediates cisplatin-resistance in cervical, ovarian, NSCLC and nasopharyngeal cancer cell lines. Suppression of CABIN1 expression sensitizes these cells to cisplatin-mediated apoptosis [38]. HECTD3 is implicated in ubiquitination of proteins involved in apoptosis, drug resistance and immune response [39]. Specifically, HECTD3 inhibits activity of apoptosis-related proteins, i.e. MALT1, caspase-8, and caspase-9 [39]. HECTD3 is overexpressed in many breast cancer cell lines where it supports survival by inhibiting caspase 3 [40]. Similar to CABIN1, overexpression of HECTD3 is associated with cisplatin-resistance human breast cancer and cervical cell lines [41], and esophageal squamous carcinoma through inhibition of caspase 9 [42]. While many studies show the direct or indirect roles of CCT3, CABIN1 and HECTD3 in cell cycle, apoptosis, and tumorigenesis, the exact link between PC and these candidates warrants further investigations.
In summary, we showed that suppression of PC expression in MDA-MB-231 cells induces G2/M cell cycle arrest, leading to apoptosis. Proteomic analysis identifies candidate proteins responsible for the PC-mediated cell cycle arrest and apoptosis in breast cancer cells. Our results highlight the possibility of the use of PC as an anti-cancer drug target.
Declaration of competing interest
The authors declare no conflicts of interest.
CRediT authorship contribution statement
Khanti Rattanapornsompong: Conceptualization, Investigation, Data curation, Methodology, Writing - original draft, Writing - review & editing. Janya Khattiya: Data curation, Methodology. Phatchariya Phannasil: Methodology, Supervision, Writing - review & editing. Narumon Phaonakrop: Resources. Sittiruk Roytrakul: Resources, Supervision, Writing - review & editing. Sarawut Jitrapakdee: Funding acquisition, Conceptualization, Resources, Supervision, Writing - review & editing. Chareeporn Akekawatchai: Conceptualization, Investigation, Data curation, Methodology, Resources, Supervision, Writing - original draft, Writing - review & editing.
Acknowledgements
The authors thank Professor John Wallace, University of Adelaide, Australia for critical reading of the manuscript. This work was supported by the International Research Network grant IRN59W0003 from the Thailand Research Fund to S.J. K.R. was supported by the Science Achievement Scholarship of Thailand and IRN59W0003. The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Sarawut Jitrapakdee, Email: sarawut.jit@mahidol.ac.th.
Chareeporn Akekawatchai, Email: ejareepo@tu.ac.th.
References
- 1.Owen O.E., Kalhan S.C., Hanson R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 2.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Canc. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lao-On U., Attwood P.V., Jitrapakdee S. Roles of pyruvate carboxylase in human diseases: from diabetes to cancers and infection. J. Mol. Med. (Berl.) 2018;96:237–247. doi: 10.1007/s00109-018-1622-0. [DOI] [PubMed] [Google Scholar]
- 4.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2 doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izquierdo-Garcia J.L., Cai L.M., Chaumeil M.M., Eriksson P., Robinson A.E., Pieper R.O., Phillips J.J., Ronen S.M. Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase. PloS One. 2014;9 doi: 10.1371/journal.pone.0108289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sellers K., Fox M.P., Bousamra M., 2nd, Slone S.P., Higashi R.M., Miller D.M., Wang Y., Yan J., Yuneva M.O., Deshpande R., Lane A.N., Fan T.W. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phannasil P., Thuwajit C., Warnnissorn M., Wallace J.C., MacDonald M.J., Jitrapakdee S. Pyruvate carboxylase is up-regulated in breast cancer and essential to support growth and invasion of MDA-MB-231 cells. PloS One. 2015;10 doi: 10.1371/journal.pone.0129848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng T., Sudderth J., Yang C., Mullen A.R., Jin E.S., Mates J.M., DeBerardinis R.J. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson S.M., Papagiannakopoulos T., Olenchock B.A., Heyman J.E., Keibler M.A., Luengo A., Bauer M.R., Jha A.K., O'Brien J.P., Pierce K.A., Gui D.Y., Sullivan L.B., Wasylenko T.M., Subbaraj L., Chin C.R., Stephanopolous G., Mott B.T., Jacks T., Clish C.B., Vander Heiden M.G. Environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metabol. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinde A., Wilmanski T., Chen H., Teegarden D., Wendt M.K. Pyruvate carboxylase supports the pulmonary tropism of metastatic breast cancer. Breast Cancer Res. 2018;20:76. doi: 10.1186/s13058-018-1008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phannasil P., Ansari I.H., El Azzouny M., Longacre M.J., Rattanapornsompong K., Burant C.F., MacDonald M.J., Jitrapakdee S. Mass spectrometry analysis shows the biosynthetic pathways supported by pyruvate carboxylase in highly invasive breast cancer cells. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2017;1863:537–551. doi: 10.1016/j.bbadis.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akekawatchai C., Roytrakul S., Kittisenachai S., Isarankura-Na-Ayudhya P., Jitrapakdee S. Protein profiles associated with anoikis resistance of metastatic MDA-MB-231 breast cancer cells. Asian Pac. J. Cancer Prev. APJCP. 2016;17:581–590. doi: 10.7314/apjcp.2016.17.2.581. [DOI] [PubMed] [Google Scholar]
- 14.Bardou P., Mariette J., Escudie F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeed A.I., Bhagabati N.K., Braisted J.C., Liang W., Sharov V., Howe E.A., Li J., Thiagarajan M., White J.A., Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 16.Paulovich A.G., Toczyski D.P., Hartwell L.H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 17.Pucci B., Kasten M., Giordano A. Cell cycle and apoptosis. Neoplasia. 2000;2:291–299. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavet O., Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Icard P., Fournel L., Wu Z., Alifano M., Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem. Sci. 2019;44:490–501. doi: 10.1016/j.tibs.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Buchakjian M.R., Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat. Rev. Mol. Cell Biol. 2010;11:715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 21.Dang C.V., Le A., Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Canc. Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., Mc Henry K.T., Pinchback R.M., Ligon A.H., Cho Y.J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M.S., Weir B.A., Tanaka K.E., Chiang D.Y., Bass A.J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F.J., Sasaki H., Tepper J.E., Fletcher J.A., Tabernero J., Baselga J., Tsao M.S., Demichelis F., Rubin M.A., Janne P.A., Daly M.J., Nucera C., Levine R.L., Ebert B.L., Gabriel S., Rustgi A.K., Antonescu C.R., Ladanyi M., Letai A., Garraway L.A., Loda M., Beer D.G., True L.D., Okamoto A., Pomeroy S.L., Singer S., Golub T.R., Lander E.S., Getz G., Sellers W.R., Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He D., Sun X., Yang H., Li X., Yang D. TOFA induces cell cycle arrest and apoptosis in ACHN and 786-O cells through inhibiting PI3K/Akt/mTOR pathway. J. Canc. 2018;9:2734–2742. doi: 10.7150/jca.26374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho T.S., Ho Y.P., Wong W.Y., Chi-Ming Chiu L., Wong Y.S., Eng-Choon Ooi V. Fatty acid synthase inhibitors cerulenin and C75 retard growth and induce caspase-dependent apoptosis in human melanoma A-375 cells. Biomed. Pharmacother. 2007;61:578–587. doi: 10.1016/j.biopha.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Ballard F.J., Hanson R.W. The citrate cleavage pathway and lipogenesis in rat adipose tissue: replenishment of oxaloacetate. J. Lipid Res. 1967;8:73–79. [PubMed] [Google Scholar]
- 26.Si Y., Shi H., Lee K. Impact of perturbed pyruvate metabolism on adipocyte triglyceride accumulation. Metab. Eng. 2009;11:382–390. doi: 10.1016/j.ymben.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Q., He Y., Wang X., Zhang Y., Hu M., Guo W., He Y., Zhang T., Lai L., Sun Z., Yi Z., Liu M., Chen Y. Targeting pyruvate carboxylase by a small molecule suppresses breast cancer progression. Adv. Sci. 2020;7:1903483. doi: 10.1002/advs.201903483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweet S., Singh G. Accumulation of human promyelocytic leukemic (HL-60) cells at two energetic cell cycle checkpoints. Cancer Res. 1995;55:5164–5167. [PubMed] [Google Scholar]
- 29.Martin D.S., Bertino J.R., Koutcher J.A. ATP depletion + pyrimidine depletion can markedly enhance cancer therapy: fresh insight for a new approach. Cancer Res. 2000;60:6776–6783. [PubMed] [Google Scholar]
- 30.Gruber R., Levitt M., Horovitz A. Sequential allosteric mechanism of ATP hydrolysis by the CCT/TRiC chaperone is revealed through Arrhenius analysis. Proc. Natl. Acad. Sci. U. S. A. 2017;114:5189–5194. doi: 10.1073/pnas.1617746114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boudiaf-Benmammar C., Cresteil T., Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PloS One. 2013;8 doi: 10.1371/journal.pone.0060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X., Cheng S., Wang W. Suppression of CCT3 inhibits malignant proliferation of human papillary thyroid carcinoma cell. Oncol Lett. 2018;15:9202–9208. doi: 10.3892/ol.2018.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L.J., Zhang L.S., Han Z.J., He Z.Y., Chen H., Li Y.M. Chaperonin containing TCP-1 subunit 3 is critical for gastric cancer growth. Oncotarget. 2017;8:111470–111481. doi: 10.18632/oncotarget.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Zhang X., Lin J., Chen Y., Qiao Y., Guo S., Yang Y., Zhu G., Pan Q., Wang J., Sun F. CCT3 acts upstream of YAP and TFCP2 as a potential target and tumour biomarker in liver cancer. Cell Death Dis. 2019;10:644. doi: 10.1038/s41419-019-1894-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M.J., Jo D.G., Hong G.S., Kim B.J., Lai M., Cho D.H., Kim K.W., Bandyopadhyay A., Hong Y.M., Kim D.H., Cho C., Liu J.O., Snyder S.H., Jung Y.K. Calpain-dependent cleavage of cain/cabin1 activates calcineurin to mediate calcium-triggered cell death. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9870–9875. doi: 10.1073/pnas.152336999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang H., Choi S.Y., Cho E.J., Youn H.D. Cabin1 restrains p53 activity on chromatin. Nat. Struct. Mol. Biol. 2009;16:910–915. doi: 10.1038/nsmb.1657. [DOI] [PubMed] [Google Scholar]
- 37.Choi S.Y., Jang H., Roe J.S., Kim S.T., Cho E.J., Youn H.D. Phosphorylation and ubiquitination-dependent degradation of CABIN1 releases p53 for transactivation upon genotoxic stress. Nucleic Acids Res. 2013;41:2180–2190. doi: 10.1093/nar/gks1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z.Z., Lu H.P., Chao C.C. Identification and functional analysis of genes which confer resistance to cisplatin in tumor cells. Biochem. Pharmacol. 2010;80:262–276. doi: 10.1016/j.bcp.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Q., Li F., Cheng Z., Kong Y., Chen C. The role of E3 ubiquitin ligase HECTD3 in cancer and beyond. Cell. Mol. Life Sci. 2020;77:1483–1495. doi: 10.1007/s00018-019-03339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Kong Y., Zhou Z., Chen H., Wang Z., Hsieh Y.C., Zhao D., Zhi X., Huang J., Zhang J., Li H., Chen C. The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death Dis. 2013;4:e935. doi: 10.1038/cddis.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., Chen X., Wang Z., Zhao D., Chen H., Chen W., Zhou Z., Zhang J., Zhang J., Li H., Chen C. The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia. 2013;15:39–48. doi: 10.1593/neo.121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Wu X., Li L., Liu Y., Xu C., Su D., Liu Z. The E3 ligase HECTD3 promotes esophageal squamous cell carcinoma (ESCC) growth and cell survival through targeting and inhibiting caspase-9 activation. Canc. Lett. 2017;404:44–52. doi: 10.1016/j.canlet.2017.07.004. [DOI] [PubMed] [Google Scholar]