Abstract
Chalcone is a polyphenolic compound found abundantly in natural plant components. They have been acclaimed as potential antitumor compounds in multiple tumor cells. However, not much attention has been paid to elucidate its antitumor mechanism of action. Here, chalcone was demonstrated to trigger endoplasmic reticulum (ER) stress-induced apoptosis through sulfonation of IRE1α by ER-localized NADPH oxidase 4 (NOX4). IRE1α-sulfonation at a cysteine residue was shown to induce “regulated IRE1α-dependent decay” (RIDD) of mRNA rather than specific splicing of XBP1. The IRE1α sulfonation-induced RIDD degraded miR-23b, enhancing the expression of NOX4. The expression of NOX4 was also upregulated in breast, and prostate cancer tissue. In chalcone-administered mice in vivo, tumor growth was regressed by the consistent mechanisms “NOX4-IRE1α sulfonation-RIDD”. Similarly, NOX4 activation and IRE1α sulfonation were also highly increased under severe ER stress conditions. Together, these findings suggest chalcone as a lead anticancer compound where it acts through NOX4-IRE1α-RIDD-miR-23b axis providing a promising vision of chalcone derivatives’ anticancer mechanism.
Keywords: Apoptosis, Chalcone, IRE1α sulfonation, NOX4, RIDD, miR-23b
1. Introduction
Chalcones (1,3-diphenyl-2-propene-1-one) are a group of plant-derived polyphenolic compounds constituting classes flavonoids known for their biological and antitumor activities [1,2]. Hence, chalcones received a great deal of interest in cancer research. The growing interest in chalcones guided the research, and several reports are available on chalcone derivatives with notable growth inhibitory activities in tumor cell lines [[3], [4], [5], [6]]. Developing chalcone derivatives as a prodrug or lead compound to overcome cancer or cancer resistance needs an in-depth study.
ER is a main subcellular organelle with a major role in the folding process for secretory and membrane proteins, lipid biosynthesis, and calcium and redox homeostasis [7,8]. However, the accumulation of unfolded or misfolded proteins in the lumen of the ER induces adaptive and/prolonged or pathological ER stress response. In response to ER stress, inositol-requiring kinase/endonuclease-1 alpha (IRE1α), PERK (protein kinase RNA‐like ER kinase), and ATF6 (activating transcription factor‐6) as three branches of unfolded protein response (UPR) pathways activated by the release of the immunoglobulin heavy chain‐binding protein (BiP/GRP78). IRE1α is activated by oligomerization and trans-autophosphorylation, resulting in splicing of the mRNA encoding X‐box binding protein‐1 (XBP1) by ribonuclease activity. The spliced isoform of XBP1 (XBP1s) is translocated to nucleus and induces essential genes related to ER folding capacity and ER associated protein degradation (ERAD) for ER homeostasis. Under the pathological ER stress, the IRE1α leads to cell death by activating c‐Jun N‐terminal kinases (JNK) pathway or degrading a subset of endogenous mRNA, a process termed “regulated IRE1α-dependent decay (RIDD) [9,10]. Another branch, activated PERK phosphorylates the eukaryotic translation initiation factor 2 alpha (eIF2α), resulting in global translational attenuation, reducing ER protein folding task. However, the PERK pathway also induces a proapoptotic mediator CHOP (CCAAT-enhancer-binding protein homologous protein; also known as GADD153 or DDIT3).
Prolonged or pathological ER stress is related to various diseases including type 2 diabetes, neurodegeneration, atherosclerosis, liver disease, endothelial impairment, and cancer [[11], [12], [13]]. Apart from the adaptive ER stress response, prolonged or pathological ER stress is related to ROS accumulation or redox imbalance, leading to post-translational modifications (PTMs) of proteins and its subsequent alteration of protein function [[14], [15], [16]]. Among the various PTMs of proteins, sulfenylation and sulfonation as oxidations of specific cysteine residues by endogenous or exogenous ROS have been reported with the evidence of protein dysfunction [17]. Interestingly, 442 S-sulfenylation sites in 392 proteins in mouse liver and over 1000 S-sulfenylation sites in more than 700 proteins in human cells were identified [18,19]. Redox PTMs of thiols at cysteine are classified as reversible which are oxidized S-sulfenylation, or irreversible which are oxidized S-sulfonation, largely depending on the degree of oxidation [20].
ER-derived H2O2 is originated from NOX4, leading to ER stress [21] and more specifically, IRE1α sulfenylation has been reported to be activated by NOX-ER and mitochondrial ROS axis linked to cell death [22]. Thus, considering the cancer characteristics where it requires high protein synthesis for cell proliferation, protein redox imbalance-based protein dysfunction and the ultimate cell death could be potential anti-cancer therapeutic mechanisms. Hence, we made an effort to elucidate the molecular mechanism behind the antitumor activity of chalcone and its derivatives through NOX4-IRE1α sulfonation.
2. Materials and methods
2.1. Reagents
trans-Chalcone, N-acetyl cysteine (NAC), tunicamycin (Tu), thapsigargin (Tg), rotenone, thallium trifluoroacetate, allopurinol, mefenamic acid, apocynin, n-ethylmalemide, iodacetamide were purchased from Sigma Aldrich (St Louis, MO, USA). Z-VAD-fmk was purchased from Enzo-life Technologies (Farmingdale, NY, USA). GKT137831 was obtained from Cayman Chemical (Michigan, USA). Hoechst 33342 and ER-Tracker™ Green were purchased from Invitrogen (Invitrogen, CA, USA).
2.2. Cell culture and transfection
Human fibrosarcoma cells (HT1080), MDA-MB-231, and PC-3 were obtained from the Korean Cell Line Bank (Seoul, Republic of Korea). Cells were maintained in Dulbecco's modified Eagle's medium and RPMI 1640 medium supplemented with 10% fetal bovine serum and 100 units/mL penicillin/streptomycin. MCF10A, and RWPE-1 cells were obtained from the ATCC (American Type Culture Collection, VA, USA). MCF10A was maintained in Mammary Epithelial Cell Growth Medium BulletKitTM (Lonza, Basel, Switzerland) with 100 ng/mL cholera toxin. RWPE-1 cell was maintained in Keratinocyte SFM (Gibco). All cells were cultured at 37 °C in 5% CO2 incubator. Cell lines were routinely tested for the absence of Mycoplasma by PCR using e-Myco™ plus Mycoplasma PCR Detection Kit (Intron Biotechnology, Korea). Cell transfection was performed using Lipofectamine 3000 (Invitrogen) and Viromer® RED (Lipocalyx, Halle, Germany) according to the manufacturer's instructions. For pC1-Hyper-Reder HT1080 stable cell line, cells were transfected and selected with 500 μg/mL G418 (Geneticin, Gibco) for 2 weeks and maintained with 100 μg/mL G418.
2.3. Plasmid
IRE1α plasmid was kindly donated by Yong Liu (University of the Chinese Academy of Sciences, China). Mutant constructs of IRE1α were generated using QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) or Muta-Direct™ Site Directed Mutagenesis Kit (iNtRON biotechnology, Korea) according to the manufacturer's protocols. All constructs were used after confirming by DNA sequencing. mCherry-ER-3 (#55041) and Hyper-Red (#48249) plasmid were purchased from Addgene. For Hyper-RedER plasmid constructs, we cloned pC1 vector with NheⅠ and EcoRⅠ containing calreticulin signal sequence at N-terminal and KDEL sequence at the C-terminal of Hyper-Red. Plasmid expressing miR-23b was constructed by inserting synthesized human miR-23b sequence into pBIC-A vector (Bioneer, Republic of Korea).
2.4. Cell viability assay
Cell viability assay was performed with the trypan blue staining as described previously [23]. Cell viability was calculated the percentage of viable cells by dividing the number of viable cells by the number of total cells.
2.5. Animal studies
Six to eight weeks BklNbt:BALB/c/nu/nu old nude mice (Damul, Daejeon, Republic of Korea) were housed (n = 5 per cage) in a fully climate-controlled room at constant temperature and humidity on a 12 h light: dark cycle with food and water ad libitum. Animal welfare and experimental procedures were performed strictly in accordance with the care and use of laboratory animals (Jeonbuk National University IACUC, Jeonju, Republic of Korea; CBNU 2016-56, CBNU 2019-047). For tumor xenografts, HT1080 and PC-3 cells were injected subcutaneously into the flank of each mouse at 5 × 106 cells mixed with matrigel (1:1) in 0.1 mL of saline. MDA-MB-231 cells were injected at 1 × 107 cells mixed with matrigel (1:1) in 0.1 mL of saline. When the tumor reached approximately 100 mm3 (7–10 days after inoculation), mice were assigned randomly. Chalcone was dissolved in DMSO as stock. Five mg/kg chalcone diluted in saline was administered intraperitoneally for 5 days per week for 3 weeks. Tumor volume in mm3 was calculated using the formula: (shortest diameter)2 × longest diameter/2.
2.6. Small interfering RNA (siRNA)
The 21-nucleotide siRNA duplexes for NOX4 (50507-2, 50507-3), IRE1α (1171246-1), CHOP (1649-1, 1648-2), NOX1 (27035-1, 27035-2) and control siRNA were purchased from Bioneer. The transfection of siRNA oligonucleotides was performed with RNAiMax (Invitrogen) in Opti-MEM I Reduced Serum Medium (Invitrogen) according to the manufacturer's recommendations. After 12 h incubation, the medium was replaced with fresh medium and was used for assays at 48 h.
2.7. Immunoblotting
Cells were lysed with RIPA buffer containing protease and phosphatase inhibitor cocktail (Sigma). The supernatant was collected after centrifugation at 12,000×g for 30 min at 4 °C. Samples containing 20–40 μg were separated on a polyacrylamide gel and transferred onto PVDF membrane (Bio-Rad, Hercules, CA, USA), which was blocked with 5% non-fat dry milk in 0.05% Tween-Tris-buffered saline (T-TBS) for 60 min at room temperature. The blot was incubated with the primary antibody overnight at 4 °C. The following primary antibodies were used as follows the antibodies against p-IRE1α (Abcam, MA, USA, ab124945), cysteine-sulfonate (Abcam, ab176487), IRE1α (Cell signaling, MA, USA, 3294), p-eIF2α (Cell signaling, 3597), eIF2α (Cell signaling, 9722), ATF4 (Cell signaling, 11815), CHOP (Cell signaling, 2895), cleaved caspase-7 (Cell signaling, 9492), PARP (Cell signaling, 9532), DYKDDDDK tag (Flag, Cell signaling, 8146), p-PERK (Santa Cruz, sc-32577), PERK (Santa Cruz, sc-13073), GRP78 (Santa Cruz, 13539), β-actin (Santa Cruz, sc-47778), Ki-67 (Santa Cruz, sc-15402), NOX4 (Invitrogen, PA5-72816), NOX1 (Invitrogen, PA5-38031), NOX2 (Abcam, ab129068), NOX3 (Abcam, ab81864), and ATP5F1B (Complex V, Invitrogen, A21351). Blots were washed with TBS-T buffer and probed again with species-specific horseradish peroxidase-conjugated secondary antibodies. Protein signal was visualized with enhanced chemiluminescence reagents.
2.8. Immunoprecipitation
Five hundred μg of cell lysates prepared in lysis buffer (Cell signaling) containing complete protease and phosphatase inhibitor cocktail (Sigma) were incubated with the antibody overnight at 4 °C. Protein A/G Sepharose beads (Sigma) were added and incubated for an additional 1 h. Immunoprecipitates were washed five times with PBS-T buffer or PBS before being resolved by SDS-PAGE and were immunoblotted with the indicated antibodies.
2.9. Subcellular fractionation
ER microsome and mitochondrial isolations were performed as described previously [24]. Briefly, cells were re-suspended with iso-osmotic buffer (0.32 M sucrose, 1 mM MgCl2, 10 mM Tris-HCl [pH 7.4]) and lysed with 20 passes using a Dounce homogenizer. The homogenate was centrifuged at 1000×g for 10 min at 4 °C, and then supernatant was removed to a new tube. The supernatant was centrifuged at 10,000×g for 30 min at 4 °C, and then supernatant was removed to another new tube. Pellet was added with PBS (mitochondria). The supernatant was centrifuged at 100,000×g for 1 h at 4 °C using an SW32.1 rotor in an L8-80 M ultracentrifuge (Beckman-Coulter) and then, supernatant was discarded. The pellets (ER fractions) were stored at -80 °C until use.
2.10. NADPH-dependent oxidoreductase (NOX) activity assay
NOX activity was measured by the lucigenin-enhanced chemiluminescence method [25]. ER micorsomes were added to phosphate buffer containing 5 μM lucigenin (Sigma), and 100 μM NADPH (Sigma). Luminescence indicative of NOX-derived ROS production was expressed relative to total protein content.
2.11. Sulfenylation and sulfonation
Sulfenylation was performed as described previously [22]. Briefly, cells were lysed in lysis buffer (50 mM HEPES, 50 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100) supplemented with 1 mM DCP-BIO1 (Kerafast, Boston, MA, USA), 0.1 mM N-Ethyl maleimide, 0.1 mM iodoacetamide and protease inhibitors (Sigma). Samples were sonicated, and then kept on ice for 30 min. After centrifugation at 12,000×g for 20 min at 4 °C, the supernatant was transferred to a new tube. The samples were rotated for 1 h at room temperature to allow for the labeling of sulfenic acids. And then, the protein was precipitated by acetone and centrifuged at 12,000×g for 5 min. The pellet was washed by 70% acetone and suspended in a non-supplemented lysis buffer. One mg of total protein was added to 50 μL slurry of streptavidin beads, and then were rotated over-night at 4 °C. The beads were centrifuged at 1000×g for 3 min, and beads were washed three times with 1 mL of lysis buffer. And then the beads were eluted in 30 μL of reducing 2 x LDS buffer (Life technologies). For detection of sulfonation of IRE1α, immunoprecipitation using anti-cysteine-sulfonate (Abcam, ab176487) from lysates was performed, and then applied of anti-IRE1α antibody (cell signaling, 3294).
2.12. ROS measurement
For ROS detection, DCFDA Cellular ROS Assay Kit (Abcam) was used and measured according to the manufacturer's instructions.
2.13. Apoptosis detection
Apoptosis Assay Kit (Abcam) was used and detected according to the manufacturer's protocol.
2.14. Oxyblot assay
For immunoblot detection of carbonyl proteins by the oxidative stress, OxyBlot™ Protein Oxidation Detection Kit (Millipore) was used and performed according to the manufacturer's recommendations.
2.15. Lipid peroxidation
ER lipid peroxidation was determined to measure the formation of malondialdehyde (MDA) using a Lipid Peroxidation Colorimetric/Fluorometric Assay Kit (BioVision) according to the manufacturer's protocol.
2.16. Real-time PCR analysis
Total RNA was isolated from cells using Trizol reagent (Invitrogen). First-strand cDNA was synthesized with a SuperScript III first-strand synthesis system (Invitrogen) using 3 μg of RNA following the manufacturer's instructions. The primer pairs for target and reference used in this study are listed in Table S1. The real-time PCR mixture (10 μL) contained 1 μL of cDNA, 5 pmol/μL primers, and 5 μL of Power SYBR green PCR master mix (Applied Biosystems). The reaction conditions using a PE Biosystems ABI PRISM 7700 Sequence Detection System were as follows: initial denaturation at 95 °C for 5 min, followed by 40 cycles at 94 °C for 10 s, 51–55 °C for 10 s, and 72 °C for 30 s. The data were normalized to GAPDH to determine relative expression levels. For Taqman assays, the real-time PCR mixture (10 μL) contained 1 μL of cDNA, 5 pmol/μL primers, 250 nM of probe, and 5 μL of TaqMan Fast advanced master mix (Thermo Scientific). The reaction conditions using a PE Biosystems ABI PRISM 7700 Sequence Detection System were as follows: initial denaturation at 50 °C for 20 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. For miRNA assays, total RNA was reverse transcribed using TaqMan™ MicroRNA Reverse Transcription Kit (4366596, Thermo), and TaqMan™ Advanced miRNA cDNA Synthesis Kit (A28007, Thermo) following the manufacturer's instructions. The primers for miR-23b-3p (483150, Thermo), miR-23b-5p (002126, Thermo), miR-25 (000403, Thermo) and miR-146a (002163, Thermo) and U6 snRNA (001973, Thermo) as reference used in this study.
2.17. In vitro IRE1α-mediated miR-23b and XBP-1 cleavage assay
In vitro cleavage of miR-23b was performed as described previously with modifications [26]. In brief, pBIC-miR23b plasmids were linearized with BamHⅠ containing T7 promoter region. To recover ultrapure DNA from the restriction enzyme cutting was used AccuPrep® PCR/Gel DNA Purification Kit (Bioneer). Transcription of large-scale RNAs of miR-23b was performed using the T7 RiboMax Express RNA Production System (Promega). In vitro-transcribed RNA (5 μg) was incubated with or without 0.5 μg recombinant human IRE1α protein (IRE1c, SignalChem) or ATP (2 mmol/L final concentration) in cleavage assay buffer (20 mM HEPES, pH 7.0, 70 mM NaCl, 2 mM ADP, 2 mM MgCl2, 5 mM DTT, 5% glycerol). The reaction products were resolved on 1.2% denaturing agarose gels after incubation at 37 °C for 30 min.
In vitro XBP-1 cleavage assay was performed as described previously [27]. 5′-Carboxyfluorescein (FAM)- and 3′-Black Hole Quencher (BHQ)-labeled XBP1 single stem-loop minisubstrate (5′FAM GAACAAGAUAUCCGCAGCAUAUACAGUUC3′BHQ) was purchased from Bioneer. IRE1c was incubated with H2O2 for 1 h in cleavage assay buffer at room temperature in the dark, followed by incubation with 50 μM RNA XBP1 substrate for 20 min at 30 °C. The cleavage products were resolved by 19% urea PAGE after quenched by adding urea to a final concentration of 4 M.
2.18. Transwell cell migration and invasion assay
Cell migration was measured using Transwell inserts (BD Biosciences, Franklin Lakes, NJ, USA). Cells (2 × 104) were added to the top Transwell chamber with chalcone and were allowed to migrate for 48 h. Migrated cells were fixed with 4% formaldehyde at room temperature for 5 min and then stained with crystal violet for 5 min. For invasion assays, BD BioCoat Matrigel Invasion Chambers in 24-well cell culture inserts (BD Biosciences) were used with 5% FBS as the chemoattractant in the lower chamber. Cells were allowed to invade for 48 h.
2.19. Immunohistochemistry
Immunohistochemistry was performed as described previously, in formalin-fixed, paraffin-embedded tissue samples [28]. The 5 μm sections were de-paraffinized, rehydrated, and subjected to heat-induced epitope retrieval. Slides were incubated with various primary antibodies at 4°C overnight after blocking with 1% goat serum and then washed with TBS-T buffer. Slides underwent color development with AEC and hematoxylin counterstaining. Positive Ki-67 staining was calculated as the percentage of positive cells per field and normalized by the total cancer cell number in each field.
2.20. Membrane fluidity
ER membrane fluidity was determined to measure the formation of pyrenedecanoic acid (PDA) excimers using a Membrane Fluidity Kit (Axxora, Enzo Life Sciences) as described previously [29]. ER microsomes were resuspended in PBS including 10 μM PDA and 0.08% pluronic F127. After incubation for 20 min at 25 °C for PDA incorporation into the membranes, the microsomes were washed three times with PBS and resuspended in fresh PBS. Endpoint fluorescence was measured using a SpectraMax M5 microplate reader (Molecular Devices) at an excitation wavelength of 360 nm. PDA monomer and excimer emissions were detected at 400 and 470 nm, respectively. Since increased membrane fluidity favors PDA excimer formation, excimer emission intensity was normalized to monomer emission intensity in order to quantify relative membrane fluidity.
2.21. Immunofluorescence
Cells were seeded onto confocal dish (SPL) and rinsed with PBS, fixed in ice-cold methanol for 20 min, and then washed twice in PBS. The cells were blocked with blocking buffer (Thermo Scientific) for 30 min and then incubated overnight at 4 °C with the primary antibody. After washing twice in PBS, cells were incubated with fluorescein isothiocyanate- or tetramethylrhodamine-conjugated secondary antibody for 1 h at room temperature. The specimens were mounted with ProLong Gold anti-fade reagent and stained with 4-,6-diamidino-2-phenylindole (Invitrogen). Images were obtained by confocal laser scanning microscope LSM 880 (Carl Zeiss) installed in the Center for University-Wide Research Facilities (CURF) at Jeonbuk National University.
2.22. PLA
PLA assay was performed using the Duolink® In Situ Detection Reagents (Sigma) according to the manufacturer's instructions. Immunofluorescent images were analyzed using confocal laser scanning microscope LSM 880 (Carl Zeiss).
2.23. RNA-seq analysis
RNA-seq analyses of HT1080 cells were performed by Ebiogen (Seoul, Korea). Total RNA was isolated using Trizol reagent (Invitrogen). RNA quality was assessed by Agilent 2100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies, Amstelveen, The Netherlands), and RNA quantification was performed using ND-2000 Spectrophotometer (Thermo Inc., DE, USA). For control and test RNAs, the construction of library was performed using QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen, Inc., Austria) according to the manufacturer's instructions. In brief, each 500 ng total RNA were prepared and an oligo-dT primer containing an Illumina-compatible sequence at its 5′ end was hybridized to the RNA and reverse transcription was performed. After the degradation of the RNA template, second strand synthesis was initiated by a random primer containing an Illumina-compatible linker sequence at its 5′ end. The double-stranded library was purified by using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The finished library is purified from PCR components. High-throughput sequencing was performed as single-end 75 sequencing using NextSeq 500 (Illumina, Inc., USA). Later, QuantSeq 3′ mRNA-Seq reads were aligned using Bowtie2 (Langmead and Salzberg, 2012). Bowtie2 indices were either generated from the genome assembly sequence or the representative transcript sequences for aligning to the genome and transcriptome. The alignment file was used for assembling transcripts, estimating their abundances, and detecting differential expression of genes. Differentially expressed genes were determined based on counts from unique and multiple alignments using coverage in Bedtools (Quinlan AR, 2010). The RC (Read Count) data were processed based on quantile normalization method using EdgeR within R (R development Core Team, 2016) using Bioconductor (Gentleman et al., 2004). Gene classification was based on searches done by DAVID (http://david.abcc.ncifcrf.gov/), Medline databases (http://www.ncbi.nlm.nih.gov/), Enrichr (https://maayanlab.cloud/Enrichr/) [30,31], and Gene Set Enrichment analysis (https://www.gsea-msigdb.org/gsea/).
2.24. S-sulfenylation/sulfonation of synthetic peptides
Two peptides (Peptide 1 sequence: KAMISDFGLCKKLAVGRHSF, Peptide 2 sequence: KENPTYTVDIFSAGCVFYYVISEGSHPFGK) predicted from the kinase domain of human IRE1α were synthesized and purchased from GL Biochem (Shanghai) Ltd (China). MALDI MS measurements were performed on a NanoLC/HR Mass (Waters, SYNAPT G2-Si HDMS) equipped with a pulsed nitrogen laser (337 nm) in reflector mode. MALDI results were obtained from installed in the Center for University-Wide Research Facilities (CURF) at Jeonbuk National University.
2.25. LC‐MS/MS analysis
The LC-MS/MS results were obtained from installed in the Center for University-Wide Research Facilities (CURF) at Jeonbuk National University. The LC-MS/MS system comprised of Waters XEVO-TQS#WAA250 (Wilmslow, UK) binary pump, vacuum degasser and auto-sampler system connected to a tandem quadrupole MS equipped with electrospray ionization (ESI) source with Waters technology. The MS/MS system was performed under positive ESI and multiple reactions monitoring (MRM) mode to identify the compounds of interest. The MS operational parameters were as follows: argon as a collision gas, capillary voltage at 15 kV, gas flow 0.16 mL/min, source temperature at 150 °C, and collision energies of 4 and 20 Ev. Detections of the analytes were performed using MRM mode to monitor the precursor-to-product ion transitions of 209 > 130.95 m/z for chalcone. Mass data were collected and processed using the Waters Acquity SM Postrun Report software (Version 1.50.2736, waters, Inc., USA).
2.26. Statistical analysis
Publicly available Gene Expression Omnibus (GEO) datasets for breast cancer (GSE31448), and prostate cancer (GSE3325) were used for bioinformatics analysis. GEO2R provided from NCBI was used for NOX4 expression analysis. All statistical analyses were performed using GraphPad 8.0 (GraphPad Software, Inc., San Diego, CA) software. The results are presented as mean ± SD, and Student's t-tests, one-way ANOVA with Tukey post hoc test, and Two-Way ANOVA followed by Bonferroni post hoc test were applied to test and control conditions. Significance was set at *, p < 0.05; **, p < 0.01; ***, p < 0.001. In each case, the statistical test used is indicated, and the number of experiments is stated in the legend of each figure.
3. Results
3.1. Chalcone shows anticancer effect through cell death
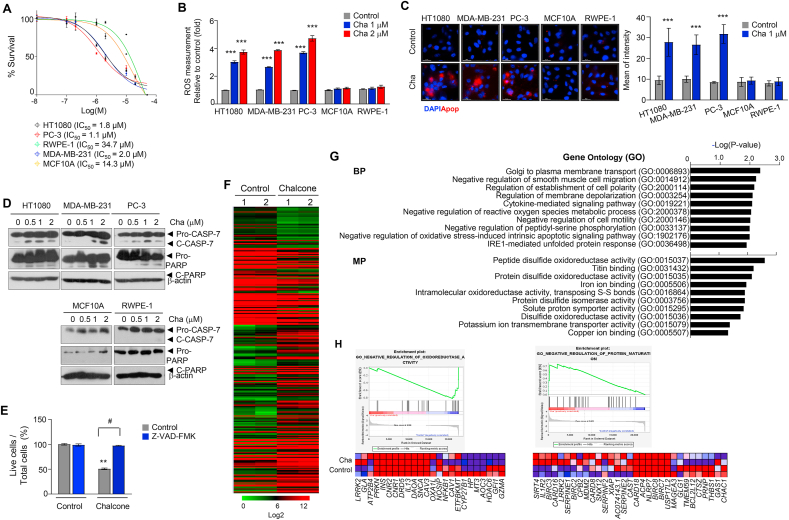
The structure of the chalcone is presented in Fig. S1A. First, cancer cell death analysis was carried out to assess the antitumor effects of chalcone on three cancer cell lines (HT1080, a fibrosarcoma cell line; MDA-MB-231, a breast triple-negative cell line that does not express the genes for estrogen receptor, progesterone receptor, and HER2/neu; and PC-3 cells, a prostate cell line that does not express the genes for androgen receptor and prostate-specific antigen [32]), and two non-diseased cell lines (MCF10A, human breast normal epithelial cells; and RWPE-1, human prostate normal epithelial cells). Chalcone treatment was found to selectively inhibit cell growth in cancer cell lines with an observed IC50 of ≤2 μM and a selectivity index of at least seven to thirty folds compared with control cell types (Fig. 1A). Next, a dose-dependent increase in ROS levels was observed in the chalcone-treated HT1080, MDA-MB-231 and PC-3 cells, non-diseased cell lines (Fig. 1B). The fluorescence of apopxin green, an apoptosis indicator, was increased in the presence of chalcone (Fig. 1C). Cleaved caspase-7 and PARP expressions were also increased in the chalcone-treated cancer cells (Fig. 1D). Correspondingly, the treatment of pan-caspase inhibitor Z-VAD-fmk significantly reduced chalcone-induced cell death (Fig. 1E). Moreover, the migration and invasion capacity of HT1080, MDA-MB-231 and PC-3 cells were found to be lesser in the chalcone-treated condition compared with control condition (Figs. S1B–C). Collectively, these results indicated that chalcone induces cell death via apoptosis.
Fig. 1.
Chalcone inhibits cell growth. (A) Cell survival curves for three cancers and two non-diseased cell lines treated with chalcone for 72 h (n = 3 independent experiments). (B) Cells were incubated with different concentrations of chalcone and then, ROS was measured using detection reagent (n = 3 independent experiments). (C) Apoptotic cells were measured using green fluorescence dye (Apopxin) in five cell lines. Scale bar, 25 μm. (D) Immunoblotting using antibodies against indicated proteins was performed using cell lysates from cells treated with different concentrations of chalcone. (E) Cell viability in HT1080 cells treated with 1 μM of chalcone or co-cultured with 20 μM of Z-VAD-fmk. Z-VAD-fmk was added every 6 h. (F) Hierarchical clustering of genes expressed in control versus chalcone treatment HT1080 cell. (G) Enrichr enrichment results of upregulated differentially expressed genes. (H) GSEA enrichment plots of gene clusters that are enriched in chalcone treated-cells. Heat map of positively enriched genes in chalcone treated-cells compared to control. Data represent mean ± SD. Statistical differences were detected with two-Way ANOVA followed by Bonferroni's test. Asterisks indicate significant differences from the control. Hashtag indicates significant differences from the chalcone. Cha; chalcone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To investigate the effect of chalcone on various biological function gene sets, we analyzed expression profiles using RNA-sequence of hierarchical clustering differentiated control and chalcone-treatments samples (Fig. 1F). Gene ontology (GO) pathway analysis with Enrichr [30,31] or Gene Set Enrichment analysis (GSEA) were carried out for upregulated differentially expressed genes (DEGs). Biological process enrichment suggests upregulated DEGs were significantly enriched in golgi to plasma membrane transport, negative regulation of smooth muscle cell migration, cytokine-mediated signaling pathway, negative regulation of reactive oxygen species metabolic process, and negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway, IRE1-mediated unfolded protein response (Fig. 1G). Molecular process enrichment suggests that upregulated DEGs were significantly enriched in peptide/protein disulfide oxidoreductase activity, intramolecular oxidoreductase activity, transposing S-S bonds, protein disulfide isomerase activity, and potassium ion transmembrane transporter activity (Fig. 1G). In GESA, genes involved in negative regulation of oxidoreductase and protein maturation were significantly enriched in chalcone-treated cells (Fig. 1H). Thus, chalcone-treated cells exhibit expression of genes that promote ROS, oxidative stress, unfolded protein response, and ER stress.
3.2. Chalcone stimulates the unfolded protein response by ROS
In the above results, ROS levels were increased in chalcone-treated cells (Fig. 1B). Furthermore, the level of oxidatively modified proteins showing accumulated carbonyl groups introduced into protein side chains by the oxidative reaction were elevated in the chalcone-treated group as compared with control (Fig. S2A). In our data, negative regulation of reactive oxygen species metabolic process, protein disulfide oxidoreductase activity, and IRE1-mediated unfolded protein response related genes were upregulated in chalcone-treated cells (Fig. 1F and G). Thus, it can be presumed that ROS participates in inducing ER stress.
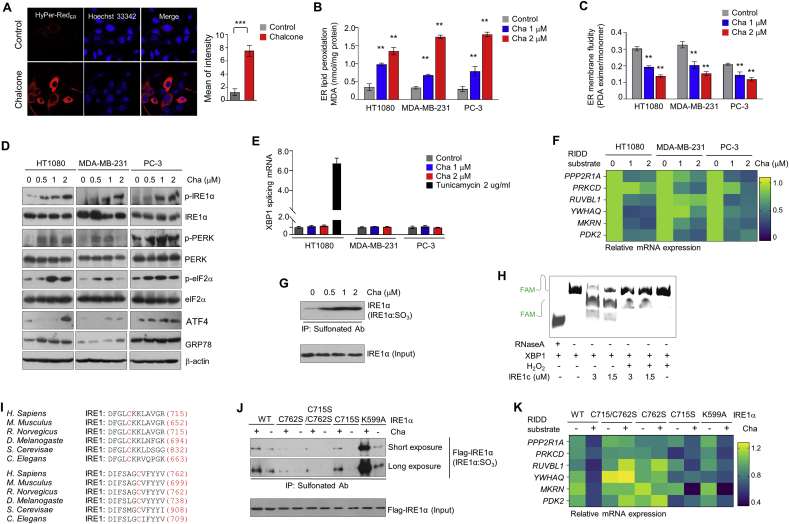
To determine whether chalcone generates ROS in the ER, HyPer-RedER is constructed, a genetically encoded fluorescent hydrogen peroxide sensor that can be targeted to the ER compartments. Observations confirmed that HyPer-RedER was fully localized in the ER (Fig. S2B). When HyPer-RedER expressed cells were treated with chalcone, the cells displayed increased HyPer-RedER fluorescence than untreated cells (Fig. 2A). ER-generating ROS by chalcone increased lipid peroxidation, which refers to the oxidative degradation of lipids, in which ER membrane acquired damage (Fig. 2B). In addition, ER membrane fluidity was also significantly reduced in chalcone-treated cells (Fig. 2C).
Fig. 2.
Chalcone mediates ER stress-induced apoptosis by ROS. (A) Images showing cell transfected with ER-targeted HyPer-Red (HyPer-RedER). Right: quantification of a mean of intensity from HyPer-RedER (n = 5 independent experiments). (B) ER lipid peroxidation was measured in cells treated or untreated with chalcone (n = 3 independent experiments). (C) ER membrane fluidity was measured on cells (n = 3 independent experiments). (D) Immunoblotting using antibodies against indicated proteins was performed using cell lysates from three cells treated with different concentrations of chalcone. (E) The mRNA expression of splicing XBP1 were measured by qRT-PCR (n = 3 independent experiments). (F) Heatmap of the mRNA expression of the indicated genes as RIDD substrates represented (n = 3 independent experiments). (G) Immunoprecipitation using anti-sulfonate Ab was performed in cells with the treatment of chalcone. (H) Urea polyacrylamide gel electrophoresis of sXBP1 substrate with FAM cleaved by different doses of IRE1c by H2O2 donor. RNase A was used as a control. (I) Conservation of C715 and C762 residues within the IRE1α kinase domain in various organisms. (J) Immunoprecipitation using anti-sulfonate Ab was analyzed in cells with transiently transfected IRE1α wild type (WT) and IRE1α mutant plasmid tagging to flag (C715S/C762S, C762S, C715S, and K599A). (K) Heatmap of the mRNA expression of RIDD substrates of the indicated genes represented (n = 3 independent experiments). Data represent mean ± SD. Statistical differences were detected with two-tailed unpaired Student's t-tests (A), and two-way ANOVA followed by Bonferroni's test (B, C, E). Asterisks indicate significant differences from the control. Cha; chalcone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To check whether ER ROS by chalcone induced the ER stress, relative levels of several ER stress-inducible UPR proteins were assessed, including phospho-IRE1α, phospho-PERK, phospho-eIF-2α, GRP78, and ATF4. As illustrated in Fig. 2D, the levels of UPR-associated proteins were markedly increased in cells treated with chalcone as compared to control cells. When small interfering RNA (siRNA) of IRE1α were applied to inhibit ER stress, chalcone-induced cell death was significantly recovered in IRE1α depleted cells (Fig. S2C), indicating that chalcone might be regulating the cell death through IRE1α.
Further, active form of XBP-1 (spliced XBP1, sXBP1) is inspected which was generated by removing a 26-bp intron within unspliced XBP-1 (XBP-1u) mRNA as an endoribonuclease function of activated IRE1α protein. For accuracy detection of sXBP1 mRNA, real-time PCR was performed using sXBP1 Taqman probes as previously reported [33]. Tunicamycin (Tu), a potent ER stress inducer that blocks N-linked glycosylation of proteins, was used as a positive control. While phosphorylation of IRE1α was increased, sXBP1 was not increased in the chalcone-treated cells compared with Tu-treated cells (Fig. 2E). Upon activation, IRE1α has also been reported to degrade a subset of endogenous mRNA, a process termed “regulated IRE1α-dependent decay (RIDD), which leads to cell death [9,10]. To identify the involvement of RIDD, expression levels of the known IRE1α-RIDD target genes PPP2R1A, PRKCD, RUVBL1, YWHAQ, MKRN, and PDK2 following the treatment with chalcone were assessed. Surprisingly, the expression of IRE1α-RIDD targeted mRNAs were decreased in a chalcone concentration-dependent manner (Fig. 2F). Collectively, these results suggest that ER stress is involved in chalcone-induced cancer cell death.
3.3. Chalcone mediates cysteine sulfonation of IRE1α
The modification of IRE1α cysteine, such as sulfenylation or sulfonation due to ROS generation was considered to discover the reason for above results. Protein cysteine thiols, which are oxidized by H2O2 to form the cysteine sulfenic acids (-SOH), generally referred to sulfenylation [22]. These can be further oxidized under high ROS conditions and form sulfinic or sulfonic acids. The latter is referred to as sulfonation (Fig. S2D). Therefore, to investigate whether IRE1α is sulfenylated by ROS upon treatment of chalcone, biotin pull-down assay was conducted using a biotin-linked Dimedone derivative (DCP-BIO1), which can specifically detect the modification of cysteine. The assay showed that the IRE1α is sulfenylated (IRE1α:SOH) by chalcone treatment (Fig. S2E). In addition, immunoprecipitation was accomplished with anti-sulfonate antibody to detect further oxidized form. IRE1α was highly sulfonated (IRE1α:SO3) in a chalcone concentration-dependent manner (Fig. 2G).
To assess whether sulfonation of IRE1α affects splicing of XBP1, we performed in vitro cleavage assay using a fluorescence resonance energy transfer (FRET)–quenched XBP1 RNA minisubstrate and a purified cytosolic portion of the human IRE1α (amino acids 465–977, IRE1c) after treatment of H2O2. IRE1c was sulfonated by applying to H2O2 as chemical donor in in vitro assay (Fig. S2F), leading to impairment of IRE1c-mediated XBP1 cleavage (Fig. 2H). Next, we investigated the relevant sulfenylated cysteine residues in IRE1α by alignment amino acid sequences of several species including H. sapiens, M. musculus, R. norvegicus, D. melanogaste, S. cerevisae, and C. elegans. In C. elegans, C663 is sulfenlyated in a previous report [22], and this site is located within the IRE1α kinase domain. While alignment of IRE1α amino acid sequences, we also found C709 another conserved site at the IRE1α kinase domain. These sites matched to C715 and C762 in human IRE1α (Fig. 2I). To confirm whether two cysteine sites of IRE1α is modified by ROS, we were performed the MALDI MS analysis. In the presence of a H2O2 as chemical donor, mass spectrometry (MS) analysis revealed sulfenylation of both (C715) and (C762) on human IRE1α peptides (Fig. S2G). Thus, we constructed cysteine residues of IRE1α to serine that mutation of both sites (C715S/C762S); one site (C715S or C762S), and a kinase site (K599A). Cells transfected with IRE1α WT revealed increased sulfonation by chalcone, but not in the mutant IRE1α C715S/C762S (Fig. 2J), suggesting that IRE1α is sulfonated at C716 and C762. Cells transfected with IRE1α K599A showed increased sulfonation as compared to IRE1α WT under treatment of chalcone, indicating that sulfonation of IRE1α by chalcone is independent of phosphorylation of IRE1α.
To identify whether this mutation affects IRE1α enzyme activity, the status of XBP-1s and RIDD by real-time PCR were confirmed. Splicing of XBP1 was clearly increased in cells transfected with IRE1α C715S/C762S under the chalcone treatment compared with IRE1α WT cells. (Fig. S2H). In RIDD assay, the RNA decay of the representative genes, PPP2R1A, PRKCD, RUVBL1, YWHAQ, MKRN, and PDK2, not occurred in IRE1α C715S/C762S and IRE1α C762S-overexpressed cells compared with IRE1α-overexpressed cells under chalcone treatment conditions (Fig. 2K). These results suggest that chalcone induces sulfonation of IRE1α, thereby the activated RIDD results in cell death promotion.
3.4. Chalcone induces ROS through NADPH oxidase 4
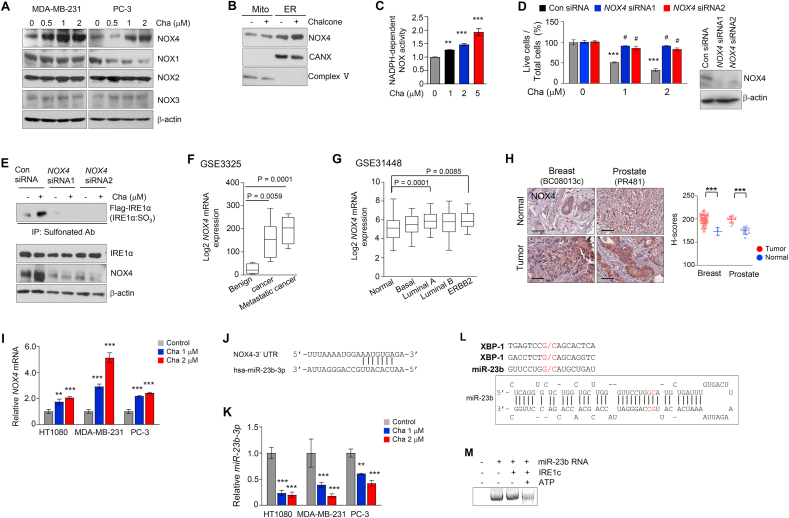
We sought to identify the enzyme implication in ER-generated ROS. ROS induced by chalcone was dramatically reduced through the ROS scavenger NAC and apocynin, an inhibitor of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase, but not through rotenone plus thallium trifluoroacetate a mitochondria-dependent ROS inhibitor, allopurinol; a xanthine oxidase-dependent ROS inhibitor or mefenamic acid; a cyclooxygenase-dependent ROS inhibitor (Fig. S3A). Since sulfonation of proteins is formulated through oxidizing of cysteine thiols by H2O2, predicting the predominant role of H2O2-producing NADPH-dependent oxidoreductase (NOX) might contribute to the oxidation of cysteine thiols and the subsequent protein sulfonation [34]. NOX4 produces approximately 90% of H2O2 and forms 10% superoxide, whereas the other NOX enzymes including NOX1 and NOX3 predominantly release superoxide [34]. Among these NOX family proteins, NOX4 expression was highly increased in chalcone-treated cells (Fig. 3A). ROS induced by chalcone was dramatically reduced in cells with GKT137831, an inhibitor of NOX1/4 (Fig. S3B), indicating that NOX4 is required for ROS production during chalcone treatment.
Fig. 3.
Degradation of miR-23b by IRE1α-RIDD increased NOX4 expression in ER. (A) Immunoblotting of NOX family was performed using cell lysates treated with different concentrations of chalcone in MDA-MB-231 and PC-3. (B) Immunoblotting of NOX4 was performed in subcellular fractionation of PC-3 cells treated with chalcone. (C) NOX activity was measured by chemiluminescence (n = 3 independent experiments). (D) Live cells was confirmed in PC-3 cells with NOX4 siRNA transfection compared with cells transfected with scrambled siRNA oligonucleotides (n = 3 independent experiments). Right; immunoblotting was performed to validate knockdown of NOX4. (E) Immunoprecipitation using anti-sulfonate antibody was performed on MDA-MB-231 cells with NOX4 siRNA, scrambled siRNA upon the treatment of chalcone. (F-G) NOX4 expression was analyzed using the Gene Expression Omnibus database from NCBI. Prostate (GSE3325, F), and Breast (GSE31448, G) datasets are presented. (H) Representative immunohistochemical staining of NOX4 on tissue microarrays. Right; quantification data of NOX4 expression. Scale bar, 100 μm. (brown: positive antibody staining, blue: hematoxylin for nuclear staining). (I) The mRNA expression of NOX4 was measured by qRT-PCR (n = 3 independent experiments). (J) Sequence motif of miR-23b-3p binding sites to NOX4. (K) miR-23b-3p expression was detected after treatment of chalcone (n = 3 independent experiments). (L) Sequence motif for the IRE1α cleavage sites (upper) and predicted secondary structures of miR-23b with their potential IRE1α cleavage sites (G/C sites marked with red arrows). (M) In vitro IRE1α mediated miR-23b cleavage assay. In vitro–transcribed miR-23b RNA was incubated with or without recombinant IRE1α protein in the presence or absence of ATP in the reaction buffer. The cleavage reaction products were resolved on an agarose gel. Data represent mean ± SD. Statistical differences were detected with one-way ANOVA followed by Tukey post hoc test (C, F, G, H), and two-way ANOVA followed by Bonferroni's test (D, I, K). Asterisks indicate significant differences from the control. Hashtag indicates significant differences from the chalcone. Cha; chalcone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Since NOX4 is localized in mitochondria and the ER [35], localization of NOX4 to the ER in HT1080 cells was confirmed (Fig. S3C). To determine in which subcellular compartment NOX4 protein-expression increases during the treatment of chalcone, immunoblot assays were carried with subcellular fractionation. Chalcone-induced upregulation of NOX4 protein occurred in microsomes, but not in the mitochondria (Fig. 3B). Moreover, NADPH-dependent NOX enzyme activity was also significantly increased (Fig. 3C), showing that ER-localized NOX4 is upregulated in the chalcone-treated cells.
To demonstrate the involvement of NOX1 and 4 in chalcone-induced cell death, NOX4 available RNA interference (RNAi) was applied to the current study. The cells with knockdown of NOX4, showed higher cell viability compared to control cells by reducing ROS generation (Fig. 3D, Figs. S3D–E). Sulfonation of IRE1α was abrogated in cells transfected with siRNA of NOX4 compared to control (Fig. 3E). Thus, these results suggest that chalcone induced-cell death is associated with ROS through NOX4.
We analyzed NOX4 mRNA expression profiling datasets in cancer samples related above results from the NCBI Gene Expression Omnibus database (NCBI/GEO). NOX4 was significantly overexpressed in breast (GSE31448), and prostate (GSE3325) cancers compared with its normal level (Fig. 3F and G). Next, we confirmed the expression levels of NOX4 using tissue microarrays, and obtained the same results that NOX4 expression was increased in cancer tissues (Fig. 3H), indicating high expression of NOX4 may be target marker for chalcone or derivative of chalcone.
3.5. Degradation of miR-23b by IRE1α RNase activity induces NOX4 expression
To identify which factor increase of NOX4 expression in chalcone-treatment conditions, we focused microRNAs (miRNAs), short non-coding RNAs, which inhibited protein translation by binding to a complementary target mRNA. We first confirmed increased NOX4 mRNA expression in chalcone treatment conditions (Fig. 3I). The expression of NOX4 is down-regulated by miRNAs as miR-23b, miR-25, and miR-146a [36,37]. Thus, we examined the expression levels of homo sapiens (hsa)-mir-23b-3p, hsa-mir-25b (miR-25), and hsa-mir-146a (miR-146a) (Fig. 3J). MiR-23b-3p was decreased under chalcone treatment of three cell lines as compared with control cells (Fig. 3K). MiR-25 was only slightly decreased in PC-3 cells and miR-146a was not detected in all cells (data not shown).
IRE1α RNase was activated under ER stress-induced cleavage of miRNAs [38]. To demonstrate whether miR-23b is degraded by IRE1α RNase activity, we performed in vitro IRE1α mediated miRNA cleavage assay. In the miR-23b sequence, G/C splicing sites was predicted to be within the stem-loop of the secondary structures (Fig. 3L). miR-23b incubated with the recombinant IRE1c protein was degraded in the presence of ATP (Fig. 3M), indicating that IRE1α suppressed the production of miR-23b through the RIDD pathway. Moreover, miR-23b was significantly recovered in IRE1α depleted chalcone-treated cells compared with control siRNA-chalcone treated cells (Fig. S3F). Since sulfonation of IRE1α by ROS through NOX4 induced IRE1α RIDD activity in the above results, the status of miR-23b was confirmed in transiently transfected IRE1α mutant cells. Under chalcone-treated conditions, miR-23b was clearly decreased in cells transfected with IRE1α WT, whereas miR-23b was not changed in cells transfected with C715S/C762S and IRE1α C762S (Fig. S3G). These results suggest that miR-23b is degraded by increasing RIDD activity through sulfonation of IRE1α under chalcone treatment conditions, resulting in an increase of NOX4 expression.
3.6. Chalcone suppresses the tumor growth in vivo by inducing apoptosis
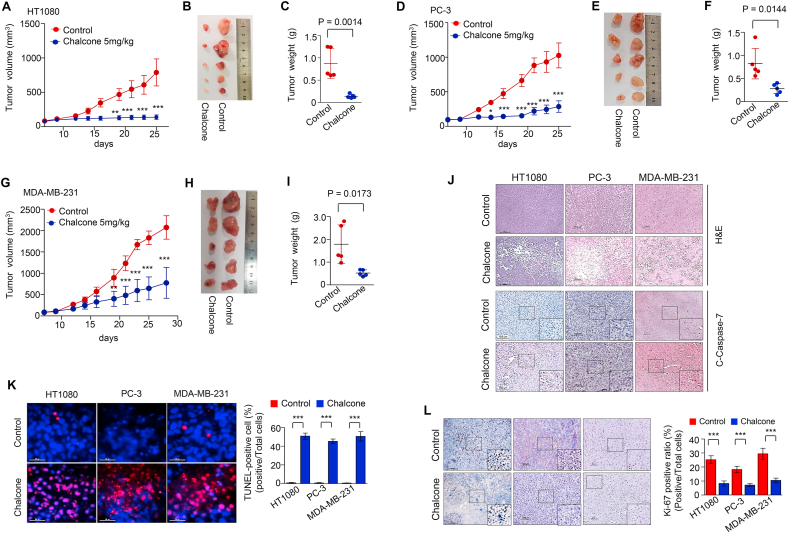
To elucidate the potential of chalcone as a therapeutic agent, in vivo tests were conducted on xenograft mouse model. The cells of the exponential phase were injected subcutaneously into flank of BALB/c nude mice. One week later, when the tumors began to enlarge, about 100 mm3, the mice were divided randomly into two groups: control and chalcone (5 mg/kg) group. Tumor-bearing mice were intraperitoneally injected with chalcone once daily for 5 days per week for indicated days. During the study, tumor growth in chalcone-treated mice was reduced as compared with control mice (Fig. 4A–I). By day 25, tumor weight in control and chalcone-treated groups of HT1080 were 874 ± 339 and 140 ± 43 mg, respectively (Fig. 4C). Tumor weight in control and chalcone-treated groups of PC-3 were 821 ± 331 and 279 ± 107 mg, respectively (Fig. 4F). Tumor weight in control and chalcone-treated groups of MDA-MB-231 were 1790 ± 844 and 522 ± 137 mg, respectively (Fig. 4I). Tumor cell morphological study revealed a large number of intact cells in control, whereas higher cell death was observed in the chalcone-treated condition (Fig. 4J). The expression of cleaved caspase-7 and TUNEL positive cells were increased in the chalcone-treated mice as compared with the control counterparts (Fig. 4J and K). In addition, the proliferation index in tumor sections was demonstrated by Ki-67 immunoreactivity, a marker of cell proliferation, which was decreased in chalcone-treated mice than those of control tissues (Fig. 4L). These results suggest that chalcone inhibits tumor growth in vivo through induction of apoptosis. To test the chalcone's bioavailability in the in vivo model, we performed a pharmacokinetic study of this compound following intraperitoneal administration of the dose (5 mg/kg). The pharmacokinetic parameters of chalcone are listed in Table S2. The Cmax values and Tmax were 6.167 ng/mL and 0.583 h. Taken together, chalcone or derivatives with improvements in pharmacokinetic properties may provide a new therapeutic approach for patients with prostate or breast.
Fig. 4.
Chalcone suppresses tumor growth in vivo. (A-C) Tumor volume (A), representative tumor photographs (B), and tumor weight (C). HT1080 cells (cells mixed with Matrigel, 1:1) were subcutaneously implanted into the mice. Mice were injected intraperitoneally with 5 mg/kg chalcone and control (0.01% DMSO with saline), and tumor volume was measured for 25 days (n = 5 mice per group). (D-I) Tumor volume (D, G), tumor photo (E, H), and tumor weight (F, I). PC-3 cells (D-F) and MDA-MB-231 cells (G-I) were mixed with Matrigel (1:1) were subcutaneously implanted into the mice. Mice were injected intraperitoneally with 5 mg/kg chalcone and control and tumor volume was measured for 28 days (n = 5 mice per group). (J) H&E staining, and immunohistochemistry of cleaved caspase-7 in xenograft tumors with or without chalcone treatment. Scale bar, 100 μm. (K) TUNEL staining indicating cell apoptosis in tumor tissues (n = 5 mice per group). Scale bar, 25 μm. (L) Immunohistochemistry of Ki-67 was performed in xenograft tumors with or without chalcone treatment (n = 5 mice per group). Scale bar, 100 μm. Data represent mean ± SD. Statistical differences were detected with two-tailed unpaired Student's t-tests (C, F, I), and two-way ANOVA followed by Bonferroni's test (A, D, G, K, L). Asterisks indicate significant differences from the control. Cha; chalcone.
3.7. Chalcone consistently activated NOX4-IRE1α sulfonation-RIDD-miR-23b axis in vivo
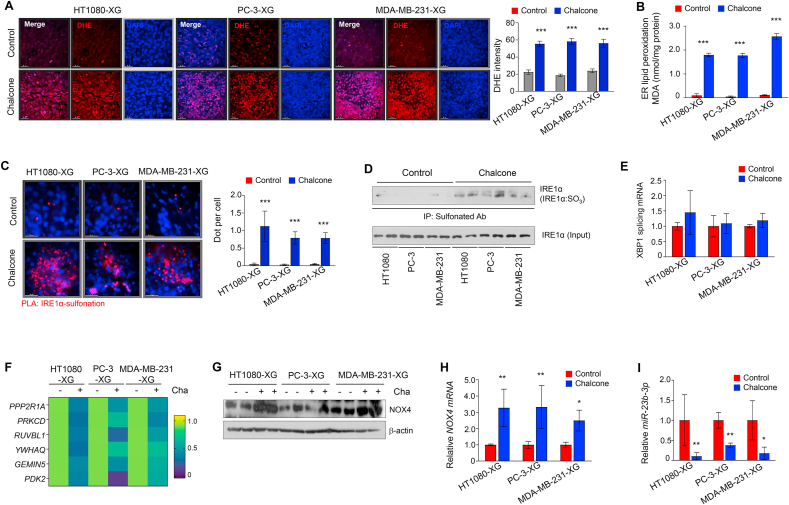
To identify that tumor growth in vivo is regressed by a similar mechanism in vitro, we performed several investigations in tumor samples from generating xenograft mouse model. First, ROS levels and lipid peroxidation were increased in all chalcone treated xenograft tumor (Fig. 5A and B). Next, to assess the status of IRE1α sulfonation, we conducted proximity ligation assay (PLA) using antibody of IRE1α and cysteine-sulfonate. As shown in Fig. 5C, red dot as the interaction between IRE1α and sulfonation protein was increased in chalcone-treated xenograft tumor. Additionally, IRE1α was highly sulfonated (IRE1α:SO3) in xenograft tumor by immunoprecipitation with anti-sulfonate antibody (Fig. 5D). Consistently in vitro results, RIDD, not splicing of XBP1, occurred in chalcone-treated xenograft tumor (Fig. 5E and F). Further, the protein and mRNA expression of NOX4 were increased by reduction of miR-23b-3p in xenograft tumor (Fig. 5G–I). These results suggest that the IRE1α sulfonation-RIDD by chalcone contributes to the inhibition of tumor growth in vitro and in vivo.
Fig. 5.
Chalcone activated NOX4-IRE1α sulfonation-RIDD axis in vivo. (A) DHE staining in tumor samples from generating xenograft (XG) mouse model with or without chalcone treatment. Right; quantification data of DHE intensity (n = 5 mice per group). Scale bar, 25 μm. (B) ER lipid peroxidation was measured in xenograft tumors (n = 5 mice per group). (C) PLA assay between IRE1α and sulfonation (red dot) in xenograft tumors (n = 5 mice per group). Right; quantification data of red dots (n = 5 mice per group). Scale bar, 25 μm. (D) Immunoprecipitation using anti-sulfonate antibody was performed in xenograft tumors. (E) The mRNA expression of splicing XBP1 were measured by qRT-PCR (n = 5 mice per group). (F) Heatmap of the mRNA expression of the indicated genes as RIDD substrates represented (n = 5 mice per group). (G) Immunoblotting of NOX4 was performed using cell lysates from xenograft tumors. (H-I) The mRNA expression of NOX4 (H) and miR-23b-3p (I) were measured by qRT-PCR (n = 5 mice per group). Data represent mean ± SD. Statistical differences was detected with two-way ANOVA followed by Bonferroni's test. Asterisks indicate significant differences from the control. Cha; chalcone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.8. Severe ER stress induces cysteine sulfonation of IRE1α, thereby increasing RIDD
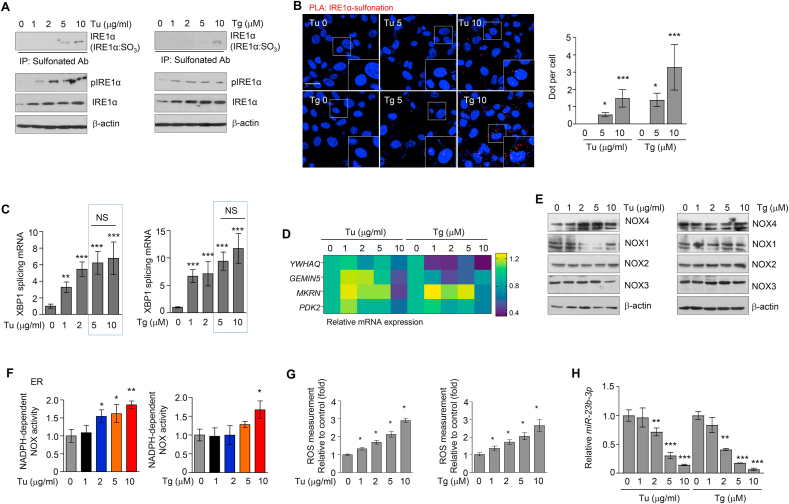
To further understand the relationship between sulfonation of IRE1α and RIDD, we explored the status of IRE1α sulfonation upon degree of ER stress strength under Tu and thapsigargin (Tg), a Ca2+-ATPase inhibitor treatment. As shown in Fig. 6A, sulfonation of IRE1α was only detected in a high concentration of Tu and Tg, -treated cells, while phosphorylation of IRE1α was represented in all the concentrations. In PLA results, red dot as the interaction between IRE1α and sulfonation protein was increased in high ER stress-induced cells compared with control cells (Fig. 6B). Splicing of XBP1 was highly increased in cells treated with 1–5 μg/mL of Tu and 1–5 μM of Tg, but no longer increased in cells treated with 10 μg/mL Tu and 10 μM Tg (Fig. 6C). RIDD, instead of sXBP1, has occurred in the high concentration of Tu- and Tg-treated conditions (Fig. 6D), indicating the chalcone-induced IRE1α sulfonation and RIDD is similar to the high ER stress-inducing condition.
Fig. 6.
ER stress-induced IRE1α sulfonation. (A) Immunoprecipitation using anti-sulfonate antibody was performed in HT1080 cells with the treatment depends on the concentration of tunicamycin (Tu, left) and thapsigargin (Tg, right). (B) PLA assay between IRE1α and sulfonation (red dot) in HT1080 cells (n = 3 independent experiments). Scale bar, 20 μm. (C) The mRNA expression of splicing XBP1 was measured by qRT-PCR (n = 3 independent experiments). (D) Heatmap of the mRNA expression of the indicated genes as RIDD substrates represented (n = 3 independent experiments). (E) Immunoblotting of NOX family was performed using cell lysates treated with different concentrations of Tu and Tg for 24 h. (F) NOX activity was measured in ER fraction of cells treated with Tu and Tg. (G) ROS in cells were incubated with different concentrations of Tu and Tg for 12 h (n = 3 independent experiments). (H) miR-23b-3p expression was detected after treatment of Tu and Tg (n = 3 independent experiments). Data represent mean ± SD. Statistical differences were detected with one-way ANOVA followed by Tukey post hoc test. Asterisks indicate significant differences from the control. NS; non-significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Further, NOX4 activity and expressions were examined to confirm whether NOX4 is implicated in sulfonation of IRE1α under severe ER stress. In Fig. 6E, the protein expression of NOX4, not NOX1-3, was highly increased in the high concentrations of Tu- and Tg-treated conditions. In addition, NOX enzyme activity was also increased under the high concentration of Tu- and Tg-treated cells (Fig. 6F), indicating that NOX4 is upregulated in response to severe ER stress. ROS levels were also increased by Tu and Tg treatment in a concentration-dependent manner (Fig. 6G). Consistently, miR-23b-3p was decreased in Tu- and Tg-treated cells compared with control cells (Fig. 6H). Collectively, these results suggest that high ROS caused by severe stress induced IRE1α sulfonation, leading to RIDD for cell death.
3.9. New synthetic chalcone derivatives mediates cysteine sulfonation of IRE1α
To determine whether sulfonation of IRE1α also occurs in different chalcone derivatives, newly synthesized derivatives with high efficacy were selected through the cell viability test. IRE1α sulfonation was detected in chalcone derivatives by immunoprecipitation with an anti-sulfonate antibody (Fig. S3H). These results suggest that IRE1α sulfonation is the core anticancer mechanism of chalcone structure-based compounds.
4. Discussion
In this study, chalcone, a key structure of isoflavonoids, was applied to the cancer model where ER stress and ROS accumulation were highly increased through NOX4, leading to cell death. Chalcone-induced ROS production, especially H2O2 through NOX4, upregulated the classical ER stress response. Notably, IRE1α sulfonation along with the RIDD phenomenon were highly increased, whereas the sXBP1 was not activated, a suggested key mechanism in the IRE1α-linked anticancer therapy. Also, miR-23b as a substrate for IRE1α RNase activity was identified, resulting in the degradation of miR-23b in chalcone-treated condition. Reducing miR-23b expression induced NOX4 expression, thereby caused ROS accumulation (Fig. S4). The IRE1α sulfonation and NOX4-associated redox disturbance are suggested to be a novel mechanism to explain chalcone and the structure analogue-induced anticancer effect.
Here, chalcone showed anticancer effect through IRE1α sulfonation-RIDD-cell death in breast and prostate cancer cells. This newly identified mechanism of chalcone might be extended into other chalcone derivatives, including Bis-Chalcone and 2′-Hydroxy-2,3,5′-trimethoxychalcone (DK143) [39,40]. As one subclass of flavonoids [41], chalcones (1,3-diphenyl-2-propene-1-one) are open-chain flavonoids composed of two phenyl rings linked by a three-carbon enone moiety [1,2]. Since it has been reported that chalcone possesses anticancer effect [5], new derivatives by changing the chalcone structure have been continuously developed to enhance the anticancer effect. Its derivatives such as 2′-hydroxy-2,4,6′-trimethoxychalcone and 2-hydroxy-4-methoxy-2′,3′-benzochalcone have been documented as potential anticancer agents [3,4]. Also, S17 and DK143 have been reported to induce cancer cell death with the ER stress and DR5-involved mechanism, respectively [6,39]. Likewise, newly synthesized compounds induce cell death where IRE1α sulfonation is involved (Fig. S3H).
IRE1α phosphorylation and Xbp-1 splicing axis have been explained as a protective/adaptive response in the ER stress condition. IRE1α phosphorylation-Xbp-1 signaling is a main theme in the ER stress-adaptive signaling, whereas Xbp-1-non-linked IRE1α phosphorylation has been suggested to be related to RNA decay, selectively recognizing RNA containing consensus sequence ‘CUGCAG’ and degraded mRNAs encoding proteins, and cell death. In this chalcone-induced cancer cell death study, Xbp-1-non-linked IRE1α phosphorylation and its sulfenylation – sulfonation as post-translational modifications (PTMs) are demonstrated (Fig. 2D, G). The intermediate step “sulfenylation” has been suggested to be a counterbalance to the IRE1α phosphorylation through p38 MAPK and antioxidant processes [22]. Even the sulfenylated IRE1α population has ever been reported to be unphosphorylated [22], and chalcone response seems to influence more IRE1α sulfenylation and sulfonation rather than its phosphorylation. IRE1α sulfonation state seems to be more related to the RIDD-linked specific IRE1α function. Moreover, the IRE1α mutant cells “K599A” rather increased the sulfonation of IRE1α, indicating that at least the phosphorylation is not required for sulfonation (Fig. 2J and K). These distinctly disclose core point “no involvement of Xbp-1 splicing at ER stress-induced cancer cell death”.
Correspondingly, under mild ER stress and ROS, the initial step in IRE1α cysteine oxidation is the conversion of the thiol group (SH) to sulfenic acid (SOH) [22,42]. However, the conversion of the thiol group (SH) to sulfonic acid (SO3H) was highly induced in severe ER stress and ROS accumulation condition. IRE1α sulfonation induced RIDD rather than Xbp-1 splicing similarly to chalcone treatment conditions, suggesting that abnormal modification of proteins such as sulfonation has damaging effect on cells. Likewise, when parkin is sulfonated, this protein becomes insoluble, probably linked to neuronal dysfunction [43]. The other protein, Dj-1's sulfonation has been also reported to lose cytoprotective function [44]. Under severe stress, the Xbp-1 splicing might be overwhelmed/overcome by the parallel RNA decay process [45]. Therefore, IRE1α sulfonation-RIDD axis may have played an important role in ER stress-mediated cell death.
ER-localized NOX4 and the resultant ROS accumulation stimulated IRE1α sulfonation, ultimately leading to cell death in chalcone-treated cells and ER stressed cells. Considering that IRE1α sulfenylation and the subsequent sulfonation are related to ROS and ER stress, ER-localized NOX4 is suggestive to be related to ER stress-induced cell death [42,46]. In this study, NOX4 is suggested to contribute to the chalcone-induced ROS, IRE1α sulfonation, and cell death. NOX is a membrane-bound enzyme complex and consists of 7 distinct isoforms in humans, including NOX1, 2, 3, and 5, and Duox1, and 2 [47]. Under the ER folding, task is overloaded, intra-ER electron uncoupling and electron leak are hyper-activated, altering the protein folding process and delaying the ER oxidative folding process, called ER stress [48]. In this study, the silencing of NOX4 by applying siRNA or function inhibition by GKT137831, an inhibitor of NADPH oxidase, significantly inhibited ROS and cell death in the chalcone-treated cells (Fig. 3D, Fig. S3B, Fig. S3E). Furthermore, NOX4 expression was upregulated in breast, prostate cancer (Fig. 3F–H), suggesting NOX4 to be a cancer target in chalcone-anticancer therapy. Consistently, IRE1α sulfonation is also significantly controlled in the NOX4 knock-down condition, uncovering the NOX4-ROS-IRE1α sulfonation-RIDD axis in the chalcone-applied anticancer mechanism-based study.
Recently, the expression of miRNAs and its regulation of target genes has been paid attention as an important factor in the ER stress response [49,50]. For example, GRP78 was directly suppressed by miR-181a-5p, miR-199a-5p, and miR-30d, and XBP1 was negatively regulated by inducing of miR-30c-2-3p through PERK activation [51,52]. IRE1α activation reduced levels of select miR-17-5p, miR-34a-5p, miR-96-5p, and miR-125b-5p, resulting in increased expression of Caspase-2 mRNA leading to cell death [53]. Besides, IRE1α RNase cleaves miR-17 to repress the expression of TXNIP mRNA, leading to activation of the NLRP3 inflammasome and IL-1β expression [54]. In this study, IRE1α RNase activation by chalcone caused decay of miR-23b, reducing the translation of NOX4-mRNA (Fig. 3I-L), and in this manner, increased ROS generation in the ER. Reducing of NOX4 expression by miR-23b protected GABAergic neurons from cell death, showing the amelioration of neuropathic pain [37]. Therefore, it is suggested that miR-23b inhibitor or IRE1α RNase activation may have therapeutic value in treating cancer.
In summary, this study reports that NOX4 as an important source for the production of ROS, thereby sulfonation of IRE1α mediates ER stress-induced apoptosis. These findings provide the hope of supporting a preclinical rationale and molecular basis for applying chalcone compound or its derivatives as an effective chemotherapeutic agent.
Author contributions
HKK, TAR and KRB performed the experiments. HYL, KRB, and MC performed pharmacokinetics of chemical. JHA and JEJ screened the new chalcone derivatives. HJC and HRK conceived and supervised the study. HKK and HJC designed, analyzed the data, prepared the figures, and co-wrote the manuscript.
Funding
This research was supported by the Korean National Research Foundation (NRF), Republic of Korea (NRF-2017R1E1A1A01073796, 2017M3A9G7072719, 2017M3A9E4047243 and 2020R1I1A1A01069205).
Declaration of competing interest
The authors declare that they have no conflict of interest with this article.
Acknowledgements
We thank Yong Liu (University of the Chinese Academy of Sciences, China) for kindly providing IRE1α plasmid.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101853.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dimmock J.R., Elias D.W., Beazely M.A., Kandepu N.M. Bioactivities of chalcones. Curr. Med. Chem. 1999;6(12):1125–1149. [PubMed] [Google Scholar]
- 2.Echeverria C., Santibanez J.F., Donoso-Tauda O., Escobar C.A., Ramirez-Tagle R. Structural antitumoral activity relationships of synthetic chalcones. Int. J. Mol. Sci. 2009;10(1):221–231. doi: 10.3390/ijms10010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin S.Y., Lee M.S., Lee D.H., Lee D.Y., Koh D., Lee Y.H. The synthetic compound 2 '-hydroxy-2,4,6 '-trimethoxychalcone overcomes P-glycoprotein-mediated multi-drug resistance in drug-resistant uterine sarcoma MES-SA/DX5 cells. J. Kor. Soc. Appl. Biol. Chem. 2015;58(1):105–109. [Google Scholar]
- 4.Shin S.Y., Kim J.H., Yoon H., Choi Y.K., Koh D., Lim Y., Lee Y.H. Novel antimitotic activity of 2-hydroxy-4-methoxy-2',3'-benzochalcone (HymnPro) through the inhibition of tubulin polymerization. J. Agric. Food Chem. 2013;61(51):12588–12597. doi: 10.1021/jf4034688. [DOI] [PubMed] [Google Scholar]
- 5.Won S.J., Liu C.T., Tsao L.T., Weng J.R., Ko H.H., Wang J.P., Lin C.N. Synthetic chalcones as potential anti-inflammatory and cancer chemopreventive agents. Eur. J. Med. Chem. 2005;40(1):103–112. doi: 10.1016/j.ejmech.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S.Y., Li T.Y., Zhang L., Wang X.Y., Dong H.Q., Li L.L., Fu D.J., Li Y.C., Zi X.L., Liu H.M., Zhang Y.B., Xu H.D., Jin C.Y. A novel chalcone derivative S17 induces apoptosis through ROS dependent DR5 up-regulation in gastric cancer cells. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10400-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Wang M., Wey S., Zhang Y., Ye R., Lee A.S. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxidants Redox Signal. 2009;11(9):2307–2316. doi: 10.1089/ars.2009.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolz A., Wolf D.H. Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochim. Biophys. Acta. 2010;1803(6):694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39(5):245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Tam A.B., Koong A.C., Niwa M. Ire1 has distinct catalytic mechanisms for XBP1/HAC1 splicing and RIDD. Cell Rep. 2014;9(3):850–858. doi: 10.1016/j.celrep.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenna S., Han R., Trojanowska M. Endoplasmic reticulum stress and endothelial dysfunction. IUBMB Life. 2014;66(8):530–537. doi: 10.1002/iub.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozcan L., Tabas I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu. Rev. Med. 2012;63:317–328. doi: 10.1146/annurev-med-043010-144749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S., Kaufman R.J. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012;197(7):857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B. Antioxidants in human health and disease. Annu. Rev. Nutr. 1996;16:33–50. doi: 10.1146/annurev.nu.16.070196.000341. [DOI] [PubMed] [Google Scholar]
- 16.Cecarini V., Gee J., Fioretti E., Amici M., Angeletti M., Eleuteri A.M., Keller J.N. Protein oxidation and cellular homeostasis: emphasis on metabolism. Biochim. Biophys. Acta. 2007;1773(2):93–104. doi: 10.1016/j.bbamcr.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 17.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 18.Gould N.S., Evans P., Martinez-Acedo P., Marino S.M., Gladyshev V.N., Carroll K.S., Ischiropoulos H. Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 2015;22(7):965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Gupta V., Carroll K.S., Liebler D.C. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer P.A., Duan J., Qian W.J., Marcinek D.J. The measurement of reversible redox dependent post-translational modifications and their regulation of mitochondrial and skeletal muscle function. Front. Physiol. 2015;6:347. doi: 10.3389/fphys.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu R.F., Ma Z., Liu Z., Terada L.S. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol. Cell Biol. 2010;30(14):3553–3568. doi: 10.1128/MCB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hourihan J.M., Moronetti Mazzeo L.E., Fernandez-Cardenas L.P., Blackwell T.K. Cysteine sulfenylation directs IRE-1 to activate the SKN-1/Nrf2 antioxidant response. Mol. Cell. 2016;63(4):553–566. doi: 10.1016/j.molcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strober W. Trypan blue exclusion test of cell viability. Curr. Protoc. Im. 2015;111 doi: 10.1002/0471142735.ima03bs111. A3 B 1-A3 B 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H.R., Lee G.H., Ha K.C., Ahn T., Moon J.Y., Lee B.J., Cho S.G., Kim S., Seo Y.R., Shin Y.J., Chae S.W., Reed J.C., Chae H.J. Bax Inhibitor-1 Is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J. Biol. Chem. 2008;283(23):15946–15955. doi: 10.1074/jbc.M800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Freyhaus H., Huntgeburth M., Wingler K., Schnitker J., Baumer A.T., Vantler M., Bekhite M.M., Wartenberg M., Sauer H., Rosenkranz S. Novel Nox inhibitor VAS2870 attenuates PDGF-dependent smooth muscle cell chemotaxis, but not proliferation. Cardiovasc. Res. 2006;71(2):331–341. doi: 10.1016/j.cardiores.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Wang J.M., Qiu Y., Yang Z.Q., Li L., Zhang K. Inositol-requiring enzyme 1 facilitates diabetic wound healing through modulating MicroRNAs. Diabetes. 2017;66(1):177–192. doi: 10.2337/db16-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Calay E.S., Fan J., Arduini A., Kunz R.C., Gygi S.P., Yalcin A., Fu S., Hotamisligil G.S. METABOLISM. S-Nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science. 2015;349(6247):500–506. doi: 10.1126/science.aaa0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkhitko A., Myachina F., Morrison T.A., Hindi K.M., Auricchio N., Karbowniczek M., Wu J.J., Finkel T., Kwiatkowski D.J., Yu J.J., Henske E.P. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc. Natl. Acad. Sci. U. S. A. 2011;108(30):12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ersoy B.A., Maner-Smith K.M., Li Y., Alpertunga I., Cohen D.E. Thioesterase-mediated control of cellular calcium homeostasis enables hepatic ER stress. J. Clin. Invest. 2018;128(1):141–156. doi: 10.1172/JCI93123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A., Enrichr Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai S., Sun Y., Squires J.M., Zhang H., Oh W.K., Liang C.Z., Huang J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate. 2011;71(15):1668–1679. doi: 10.1002/pros.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiuolo J., Bulotta S., Verderio C., Benfante R., Borgese N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc. Natl. Acad. Sci. U. S. A. 2011;108(19):7832–7837. doi: 10.1073/pnas.1101379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nisimoto Y., Diebold B.A., Cosentino-Gomes D., Lambeth J.D. Nox4: a hydrogen peroxide-generating oxygen sensor. Biochemistry. 2014;53(31):5111–5120. doi: 10.1021/bi500331y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassegue B., San Martin A., Griendling K.K. Biochemistry, physiology, and pathophysiology of NADPH oxidases in the cardiovascular system. Circ. Res. 2012;110(10):1364–1390. doi: 10.1161/CIRCRESAHA.111.243972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fierro-Fernandez M., Miguel V., Lamas S. Role of redoximiRs in fibrogenesis. Redox Biol. 2016;7:58–67. doi: 10.1016/j.redox.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im Y.B., Jee M.K., Choi J.I., Cho H.T., Kwon O.H., Kang S.K. Molecular targeting of NOX4 for neuropathic pain after traumatic injury of the spinal cord. Cell Death Dis. 2012;3:e426. doi: 10.1038/cddis.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassler J., Cao S.S., Kaufman R.J. IRE1, a double-edged sword in pre-miRNA slicing and cell death. Dev. Cell. 2012;23(5):921–923. doi: 10.1016/j.devcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee D.H., Jung Jung Y., Koh D., Lim Y., Lee Y.H., Shin S.Y. A synthetic chalcone, 2'-hydroxy-2,3,5'-trimethoxychalcone triggers unfolded protein response-mediated apoptosis in breast cancer cells. Canc. Lett. 2016;372(1):1–9. doi: 10.1016/j.canlet.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Sansalone L., Veliz E.A., Myrthil N.G., Stathias V., Walters W., Torrens, Schurer S.C., Vanni S., Leblanc R.M., Graham R.M. Novel curcumin inspired bis-chalcone promotes endoplasmic reticulum stress and glioblastoma neurosphere cell death. Cancers. 2019;11(3) doi: 10.3390/cancers11030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlikova B., Tasdemir D., Golais F., Dicato M., Diederich M. Dietary chalcones with chemopreventive and chemotherapeutic potential. Genes Nutr. 2011;6(2):125–147. doi: 10.1007/s12263-011-0210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camargo L.L., Harvey A.P., Rios F.J., Tsiropoulou S., Da Silva R.N.O., Cao Z., Graham D., McMaster C., Burchmore R.J., Hartley R.C., Bulleid N., Montezano A.C., Touyz R.M. Vascular nox (NADPH oxidase) compartmentalization, protein hyperoxidation, and endoplasmic reticulum stress response in hypertension. Hypertension. 2018;72(1):235–246. doi: 10.1161/HYPERTENSIONAHA.118.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng F., Yao D., Shi Y., Kabakoff J., Wu W., Reicher J., Ma Y., Moosmann B., Masliah E., Lipton S.A., Gu Z. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahmed K., Boukhenouna S., Karim L., Andrews T., Lin J., Powers R., Wilson M.A., Lin C.R., Messier E., Reisdorph N., Powell R.L., Tang H.Y., Mason R.J., Criner G.J., Kosmider B. The effect of cysteine oxidation on DJ-1 cytoprotective function in human alveolar type II cells. Cell Death Dis. 2019;10(9):638. doi: 10.1038/s41419-019-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiramatsu N., Chiang W.C., Kurt T.D., Sigurdson C.J., Lin J.H. Multiple mechanisms of unfolded protein response-induced cell death. Am. J. Pathol. 2015;185(7):1800–1808. doi: 10.1016/j.ajpath.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carnesecchi S., Deffert C., Donati Y., Basset O., Hinz B., Preynat-Seauve O., Guichard C., Arbiser J.L., Banfi B., Pache J.C., Barazzone-Argiroffo C., Krause K.H. A key role for NOX4 in epithelial cell death during development of lung fibrosis. Antioxidants Redox Signal. 2011;15(3):607–619. doi: 10.1089/ars.2010.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahoo S., Meijles D.N., Pagano P.J. NADPH oxidases: key modulators in aging and age-related cardiovascular diseases? Clin. Sci. (Lond.) 2016;130(5):317–335. doi: 10.1042/CS20150087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grek C., Townsend D.M. Protein disulfide isomerase superfamily in disease and the regulation of apoptosis. Endoplasm. Reticulum Stress Dis. 2014;1(1):4–17. doi: 10.2478/ersc-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurel M., Chevet E. Endoplasmic reticulum stress signaling: the microRNA connection. Am. J. Physiol. Cell Physiol. 2013;304(12):C1117–C1126. doi: 10.1152/ajpcell.00061.2013. [DOI] [PubMed] [Google Scholar]
- 50.Bartoszewska S., Kochan K., Madanecki P., Piotrowski A., Ochocka R., Collawn J.F., Bartoszewski R. Regulation of the unfolded protein response by microRNAs. Cell. Mol. Biol. Lett. 2013;18(4):555–578. doi: 10.2478/s11658-013-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su S.F., Chang Y.W., Andreu-Vieyra C., Fang J.Y., Yang Z., Han B., Lee A.S., Liang G. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene. 2013;32(39):4694–4701. doi: 10.1038/onc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrd A.E., Aragon I.V., Brewer J.W. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. J. Cell Biol. 2012;196(6):689–698. doi: 10.1083/jcb.201201077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Upton J.P., Wang L., Han D., Wang E.S., Huskey N.E., Lim L., Truitt M., McManus M.T., Ruggero D., Goga A., Papa F.R., Oakes S.A. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338(6108):818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner A.G., Upton J.P., Praveen P.V., Ghosh R., Nakagawa Y., Igbaria A., Shen S., Nguyen V., Backes B.J., Heiman M., Heintz N., Greengard P., Hui S., Tang Q., Trusina A., Oakes S.A., Papa F.R. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metabol. 2012;16(2):250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.