Abstract
Francisella tularensis is one of the most virulent pathogenic bacteria causing the acute human respiratory disease tularemia. While the mechanisms underlying F. tularensis pathogenesis are largely unknown, previous studies have shown that a F. novicida transposon mutant with insertions in a gene coding for a putative lysine decarboxylase was attenuated in mouse spleen, suggesting a possible role of its protein product as a virulence factor. Therefore, we set out to structurally and functionally characterize the F. novicida lysine decarboxylase, which we termed LdcF. Here, we investigate the genetic environment of ldcF as well as its evolutionary relationships with other basic AAT-fold amino acid decarboxylase superfamily members, known as key actors in bacterial adaptative stress response and polyamine biosynthesis. We determine the crystal structure of LdcF and compare it with the most thoroughly studied lysine decarboxylase, E. coli LdcI. We analyze the influence of ldcF deletion on bacterial growth under different stress conditions in dedicated growth media, as well as in infected macrophages, and demonstrate its involvement in oxidative stress resistance. Finally, our mass spectrometry-based quantitative proteomic analysis enables identification of 80 proteins with expression levels significantly affected by ldcF deletion, including several DNA repair proteins potentially involved in the diminished capacity of the F. novicida mutant to deal with oxidative stress. Taken together, we uncover an important role of LdcF in F. novicida survival in host cells through participation in oxidative stress response, thereby singling out this previously uncharacterized protein as a potential drug target.
Subject terms: Phylogenetics, Biological techniques, Mass spectrometry, Microbiology techniques, Proteomic analysis, Structure determination
Introduction
The Gram-negative bacterium Francisella tularensis is the etiological agent of tularemia1. This zoonotic disease can be contracted by humans through insect bites, contact with infected animal products, ingestion of polluted food or water and inhalation of contaminated aerosols. Respiratory tularemia resulting from aerosol uptake causes typical pneumonia symptoms with 30 to 60% fatality rate of untreated infections. Due to the ease of culture and the extremely high infectivity by airborne route, F. tularensis is considered as a dangerous bioweapon classified as a category A bioterrorism agent by the Centers for Disease Control and Prevention (CDC)2. Indeed, this bacterium, capable of surviving for weeks at low temperature in water, soil, grass or animal carcasses is one of the most infectious pathogens known because inhalation of as few as a dozen of organisms can suffice to cause illness and death. Yet, no licensed vaccine against tularemia is currently available, and the mechanisms underlying pathogenesis of F. tularensis are still largely unknown. The virulent strains are classified as F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, whereas the closely related F. novicida is considered avirulent for humans and is therefore a suitable working model3.
The pathogenicity of F. tularensis mainly relies on the Francisella Pathogenicity Island (FPI)4, a gene cluster encoding 17 proteins comprising a type VI-like secretion system machinery (T6SS)5,6. The regulation of Francisella virulence genes is complex and poorly understood but requires the expression of the macrophage growth locus protein A (MglA)7,8. This major transcriptional regulator associates with the stringent starvation protein A (SspA) to form a heterodimer able to interact with RNA polymerase and whose stability is tightly linked to inorganic polyphosphate9. Another FPI transcriptional regulator essential for intracellular bacterial growth and virulence is the Francisella effector of virulence regulation (FevR also known as PigR) which physically interacts with the MglA/SspA complex10. MglA is also involved in the regulation of genes outside FPI and more specifically in the Francisella oxidative stress response11.
Francisella is a facultative intracellular pathogen whose replication inside macrophages is mostly admitted to be at the heart of the bacterial pathogenesis. However, this bacterium is also capable of invading many other cell types such as dendritic cells and neutrophils1,12,13. Murine models of intranasal infection with different Francisella species demonstrated that alveolar macrophages were predominantly infected at 4–24 h post-infection (hpi) and that neutrophils serve as a replicative niche accounting for at least 50% of F. tularensis-infected cells from day 3 post-infection. The intracellular life cycle exposes Francisella to oxidative stress upon bacterial uptake, temporary residence in the phagosomes, and escape into the host cytoplasm for replication. Indeed, as a defense mechanism for the clearance of phagocytosed microorganisms, both macrophages and neutrophils produce reactive oxygen species (ROS), such as superoxide anions (O2·−), hydrogen peroxide (H2O2) and hydroxyl radicals (OH·), which in turn trigger bacterial killing by causing damage to macromolecules including DNA, proteins and membrane lipids14,15. Several factors including for example catalase (KatG), superoxide dismutase (SodB, SodC) and peroxiredoxin (AhpC)16–20, have been identified as allowing Francisella to cope with such oxidative stress and thus contributing to intracellular bacteria survival. Another factor that makes bacteria more resistant to ROS killing is a lower iron content21,22 .
A previous report showed that a F. novicida transposon mutant with insertions in cadA (FTT_0406), which encodes a putative aspartate aminotransferase fold (AAT-fold) pyridoxal 5′-phosphate (PLP)-dependent lysine decarboxylase (hereafter referred to as LdcF), was attenuated in mouse spleen23. Its role as virulence factor was further hypothesized from the comparative bioinformatic analysis of Francisella strains exhibiting different levels of pathogenicity24. Considering the current knowledge on the other members of the superfamily of AAT-fold PLP-dependent basic amino acid decarboxylases and their recognized involvement in bacterial physiology, stress responses and virulence, we set out to investigate the structure and function of LdcF, using the F. novicida model as a practicable surrogate of F. tularensis for experimental studies3.
Bacterial AAT-fold PLP-dependent basic amino acid decarboxylases are grouped into a superfamily termed LAOdc because these enzymes decarboxylate lysine (LdcI, LdcC and LdcA), arginine (AdcI) and ornithine (OdcI and OdcC) into corresponding polyamines (cadaverine, agmantine and putrescine) while consuming protons and producing CO225–27. The main role of LdcI, AdcI and OdcI (with I standing for acid stress-inducible LAOdcs) is to buffer the bacterial cytosol in acid stress response, while the primary function of LdcC, LdcA and OdcC is polyamine biosynthesis. As polycations, polyamines bind negatively charged macromolecules such as DNA, proteins and phospholipids28,29, thereby contributing to a remarkable diversity of processes such as DNA replication, gene expression, protein synthesis, stress and antibiotic resistance, siderophore synthesis, biofilm formation and virulence30. In this work, we investigate the genetic environment of Francisella ldcF and the evolutionary relationships of LdcF with the other superfamily members in the light of a recent exhaustive phylogenetic analysis of proteobacterial LAOdcs26. We determine its crystal structure and identify specific structural elements which distinguish LdcF from E. coli LdcI, the most thoroughly-studied Ldc. We consider functional implications of these structural differences, in particular in terms of nutrient stress response, and analyze the influence of ldcF inactivation on bacterial growth under a variety of stress conditions. These experiments demonstrate the involvement of LdcF in oxidative stress resistance in dedicated growth media, as well as in infected macrophages. Finally, a comparative mass spectrometry (MS)-based quantitative analysis of the proteome of the wild-type F. novicida versus the ΔldcF mutant provides elements for the explanation of LdcF involvement in defense against oxidative stress, virulence and survival in macrophages. Taken together, this study provides a structural and functional characterization of Francisella LdcF. It uncovers the important role of this previously uncharacterized protein in survival in the host cells through participation in oxidative stress response, thereby identifying LdcF as a potential drug target. Finally, the differential proteomic analysis opens up further avenues for mechanistic investigations of the LdcF mode of action.
Results
Bioinformatic analysis of LdcF and the ldc genetic environment
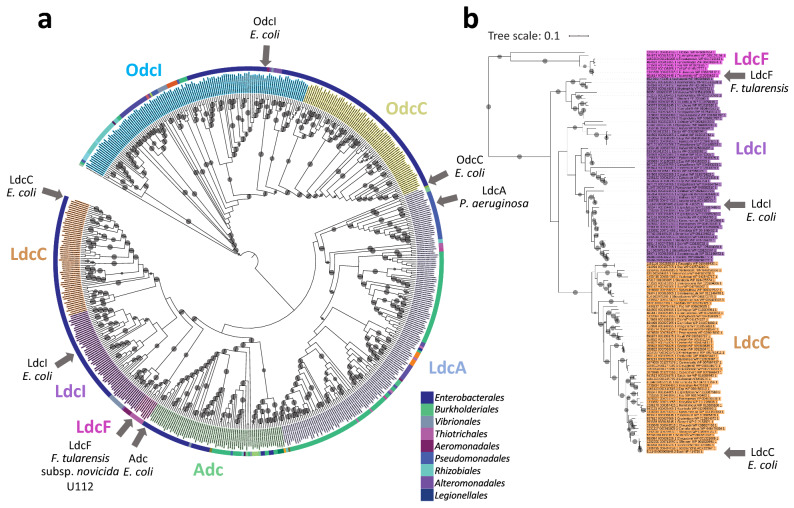
A genomic survey of 4,467 prokaryote complete proteomes identified a single LAOdc sequence in Francisellaceae, which we termed LdcF. LdcF sequences found in Francisellaceae strains display a high level of sequence identity (83%), and contain four functional regions, corresponding to a wing domain (Pfam ID: PF03709), a PLP-binding domain and a AAT-like domain (both corresponding to Pfam ID: PF01276), and a C-terminal domain (Pfam ID: PF03711) (Supplementary Fig. S1). This corresponds to the canonical organization of the wing-containing LAOdc superfamily26. Based on sequence comparison, LdcF proteins appear more similar to Escherichia coli LdcI (52.65% identity) and LdcC (48.18%) than to Pseudomonas aeruginosa LdcA (36.44%), E. coli AdcI (33.03%), E. coli OdcI (30.34%) and E. coli OdcC (27.28%). The phylogenetic analysis of 553 wing-containing LAOdcs present in 1,904 representative proteomes confirmed the specific relationship of Francisellaceae LdcF with the LdcI/C family (Fig. 1a and Supplementary Fig. S2). More precisely, Francisellaceae LdcF grouped robustly with a sequence from Legionella fallonii at the base of the clade corresponding to LdcI/C (ultrafast bootstrap = 100%, Fig. 1b). However, the long stem of the LdcI/C cluster reflects the large evolutionary distance between LdcF and LdcI/C sequences and thus their relative high divergence (Fig. 1b).
Figure 1.
Phylogenetic position of LdcF sequences within the wing-domain containing LAOdc family. (a) Tree showing the relationships of 553 WING-containing AAT-fold decarboxylase sequences. The tree is a cladogram, meaning that the length of the branches has no evolutionary significance. The cladogram is rooted according to Carriel et al.26. The colour of leaves corresponds to the LdcI, LdcC, AdcI, OdcC, OdcI, and LdcA subfamilies26. The group corresponding to Francisellaceae sequences (referred as to LdcF) is indicated in pink. F. tularensis, E. coli, and P. aeruginosa sequences are indicated by grey arrows. Grey circles at branches correspond to ultrafast bootstrap values > 95%. The taxonomy of species (Class) is represented by a coloured strip. (b) Phylogram corresponding to the LdcI, LdcC, and LdcF subtree (122 sequences). The scale bar corresponds to the average number of substitutions per site. The length of branches is proportional to genetic divergence.
Therefore, LdcF sequences could represent a new Ldc family. In line with this hypothesis, the genomic context of ldcF is very different of those of ldcI and ldcC (Fig. 2). In particular, in many genomes, ldcI and ldcC are present in vicinity of lpxD, fabZ, lpxA, lpxB, rnhB, dnaE and accA genes involved in lipid synthesis and DNA replication. Furthermore, most ldcI are clustered with cadB and cadC, encoding the lysine-cadaverine antiporter and the transcriptional regulator of the cadBA operon respectively. In contrast, Francisellaceae ldcF are surrounded by lolC and lolD on the one hand, and gcvT, gcvH, and gcvP on the other hand, involved in lipid transport and glycine cleavage system, respectively. Altogether, these data underlie differences between LdcF and other LAOdcs. Accordingly, LdcF may constitute a new family of LAOdcs phylogenetically related to LdcI/C but presenting a different genomic context.
Figure 2.
Genomic context of LdcI, LdcC, and LdcF coding genes (black arrows) in a subsample of representative species. Other conserved neighbour genes are highlighted with colour. The taxonomy of species (Class) is indicated in brackets.
Structural characterization of Francisella novicida LdcF
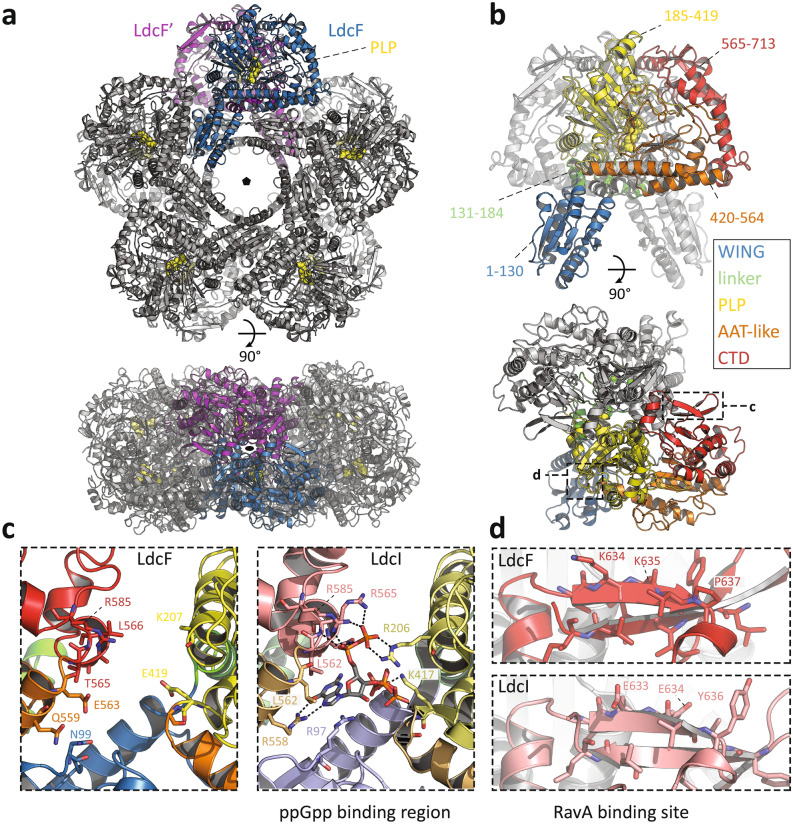
Having shown that the LdcF family is distinct from LdcI/C and LdcA, and considering that the structures of E. coli LdcI, E. coli LdcC and P. aeruginosa LdcA solved by either X-ray crystallography or cryo-EM are available, we decided to gain structural insights into F. novicida LdcF and to compare its structure with those of the related families. LdcF was purified to homogeneity, and its lysine decarboxylase activity was assessed at pH 6.5 and 37 °C using a 2,4,6-trinitrobenzensulfonic acid colorimetric assay31(see “Methods”). The initial activity rate in nanomoles cadaverine produced per minute and per microgram of enzyme was measured to be ~ 5 nmoles cadaverine min−1 µg−1 LdcF (Supplementary Fig. S3). The observation that this activity rate is 30 times smaller than that of E. coli LdcI at the same conditions31,32 may indicate that, similarly to LdcI and related enzymes33,34, optimal LdcF activity is pH, salt, and temperature-dependent. We were able to determine the structure of F. novicida LdcF from X-ray diffraction data collected to a resolution of 3.4 Å (Supplementary Table S1 and Supplementary Fig. S4). The structure was solved by molecular replacement (MR), using the crystal structure of the decameric E. coli LdcI (PDB ID: 3N75)32 as a starting model (see “Methods”). All Ldcs are pentamers of dimers arranged around a central pore, thereby forming a D5-symmetric decamer32,35,36. The LdcF crystal structure contains one LdcF pentamer in the crystallographic asymmetric unit, while an LdcF decamer is generated by a two-fold crystallographic symmetry axis perpendicular to the pentamer pore (Fig. 3a).
Figure 3.
Crystal structure of the F. novicida lysine decarboxylase LdcF. (a) Front (upper panel) and side view (lower panel) of decameric LdcF, with one highlighted dimer coloured blue and purple, while other dimers are coloured light and dark grey. The covalently bound pyridoxal phosphate (PLP) cofactor is shown as yellow spheres. (b) Front (upper panel) and side view (lower panel) of an LdcF dimer extracted from the decamer shown in (a). In one monomer, different domains are coloured according to a rainbow scheme (WING domain: blue, linker: green, PLP-binding domain: yellow, AAT-like domain: orange, C-terminal domain: red), with accompanying annotated amino acid residue ranges. (c) Comparison between the AAT-like domains (termed ppGpp binding domain in E. coli LdcI) of F. novicida LdcF (left) and E. coli LdcI (right). Residues of E. coli LdcI involved in ppGpp binding, and the corresponding residues in the AAT-like domain of F. novicida LdcF are annotated and shown as sticks. Domains are coloured as in (b), but using lighter tints for E. coli LdcI. (d) Comparison between the RavA-binding site in E. coli LdcI, and the corresponding region in F. novicida LdcF. Residues of E. coli LdcI involved in RavA binding, and the corresponding residues of F. novicida LdcF are annotated and shown as sticks.
In agreement with the relationship between LdcF and LdcI families disclosed by the phylogenetic analysis and the relatively high level of sequence conservation (see above and Fig. 4), structural alignment between F. novicida LdcF and E. coli LdcI dimers extracted from their respective decameric crystal structures demonstrates a high overall similarity, with a root-mean-square-deviation (RMSD) of 1.043 Å over 1,223 aligned atoms. Like LdcI and other LAOdcs, the LdcF monomer is organized in three different structural domains (Fig. 3b): A N-terminal wing domain involved in stabilization of the ring assembly though inter-dimer contacts (residues 1–130), a central core domain which contains a covalently bound PLP cofactor (residues 131–564) and a C-terminal domain (residues 565–713), which partially constitutes an entry channel into the active site. The core domain (Fig. 3b) encompasses a linker region (residues 113–184), a PLP-binding domain consisting of a seven-stranded β-sheet surrounded by eight α-helices (residues 185–419), and an AAT-like domain which harbors an antiparallel four-stranded β-sheet and three α-helices near the dimerization interface (residues 420–564).
Figure 4.
Alignment of F. novicida LdcF, E. coli LdcI and LdcC, and P. aeruginosa LdcA using Clustal Omega. Partially and fully conserved residues are annotated with ‘:’ and ‘*’ respectively. Domains are coloured according to a rainbow scheme (WING domain: blue, linker: green, PLP-binding domain: yellow, AAT-like domain: orange, C-terminal domain: red), and secondary structure elements are annotated. ppGpp and RavA-interaction sites are highlighted using red and blue transparent boxes respectively.
While the overall structures of LdcF and LdcI are very similar, some notable differences were found in both the AAT-like and the C-terminal domains. In E. coli Ldcs, the AAT-like domain is referred to as the ppGpp-binding domain due to its interaction with the stringent response alarmone ppGpp, which causes a strong inhibition of the lysine decarboxylase activity. The ppGpp binding site was actually discovered serendipitously upon building of the E. coli LdcI atomic model into the X-ray crystallography map, because under conditions of LdcI overexpression and purification used, the strongly-bound ppGpp was co-purified with LdcI32. Later, the enzymatic activity of both E. coli LdcI and LdcC was shown to be strongly inhibited by ppGpp37. In the case of LdcF, no additional density was present in the corresponding site. Moreover, a comparison of the ppGpp binding pocket in LdcI with the equivalent region in LdcF (Fig. 3c) revealed that, despite the overall high sequence conservation, only two out of 10 ppGpp-interacting residues are conserved between the two proteins. More importantly, 7 of the amino acid substitutions in LdcF result either in a change in charge or polarity, or in a change from hydrophobic to polar or vice versa, revealing that, contrary to LdcI and LdcC but similarly to P. aeruginosa LdcA35, LdcF is most likely not inhibited by ppGpp.
The C-terminal domain of the E. coli LdcI but not LdcC is known to interact with the MoxR AAA + ATPase RavA. The molecular determinant of the LdcI-RavA interaction resides in the C-terminal two-standed β-sheet of LdcI36,38 which was shown to be specifically evolved for RavA binding, contrary to OdcIC, AdcI, LdcA and even the closer related LdcC35,36. This LdcI-specific interaction leads to the formation of a huge cage-like LdcI-RavA complex38–40 proposed to enable enterobacteria, such as E. coli, Salmonella and Vibrio, to withstand acid stress even under conditions of nutrient deprivation eliciting stringent response41. Indeed, interaction with RavA was shown to maintain LdcI enzymatic activity upon starvation by preventing ppGpp binding to LdcI particles engaged in the LdcI-RavA complex41. Based on a medium-resolution cryo-EM structure of LdcI cross-linked with the LdcI-binding domain of RavA36, residues Glutamate 634 (E634), Tyrosine 636 (Y636) and Tyrosine 697 (Y697) are likely to be key players in the LdcI-RavA interaction. These residues are substituted in LdcF by Lysine (K635), Proline (P637) and Glutamate (E698) residues respectively, resulting in an impairment of a putative RavA interaction (Fig. 3d). This result is consistent with the absence of an orthologue of RavA in the Francicellae genome and further highlights the specific evolutionary tailoring of LdcI for RavA binding.
In vitro phenotypic analysis of the ΔldcF mutant
The physiological significance of LdcF was investigated through the construction of a F. novicida FTN_0504 deletion mutant (ΔldcF). Before proceeding with a comprehensive phenotypic analysis, we checked whether ldcF deletion affected bacterial fitness. When grown on PolyViteX-enriched chocolate agar (PVX-CHA) plates, F. novicida wild-type (WT) and ΔldcF displayed similar colony morphology (Supplementary Fig. S5a). Accordingly, bacterial division and metabolism of both strains were found unchanged whether the protein was expressed or not (Supplementary Figs. S5b–d, S6). We then investigated a putative role of LdcF in bacterial tolerance to acidic pH exposure for 1 h but observed no difference between the WT and the deletion mutant (Supplementary Fig. S7). Bacterial growth was then examined in liquid Modified Mueller–Hinton (MMH) medium previously adjusted at different pH values ranging from 2.5 to 10 (Fig. 5a). Under all conditions tested, the replication curves for the WT and the deletion mutant were strictly similar. No growth was observed for the extreme acidic or alkaline pH values tested, while in the range of pH values from 4 to 8, bacteria grew and reproduced best at pH 6.6. At the 24 h time point the survival of bacteria was further evaluated by plating serial dilutions of each bacterial suspensions on PVX-CHA plates. For each pH tested, comparable numbers of colony forming units (cfu) were found for both strains, thus confirming that bacterial viability was not altered upon ldcF deletion (Supplementary Table S2). The replication rate of both strains was also found identical at 25 °C or at 37 °C, which correspond to the temperatures in tick and mammal hosts, respectively (Supplementary Fig. S8).
Figure 5.
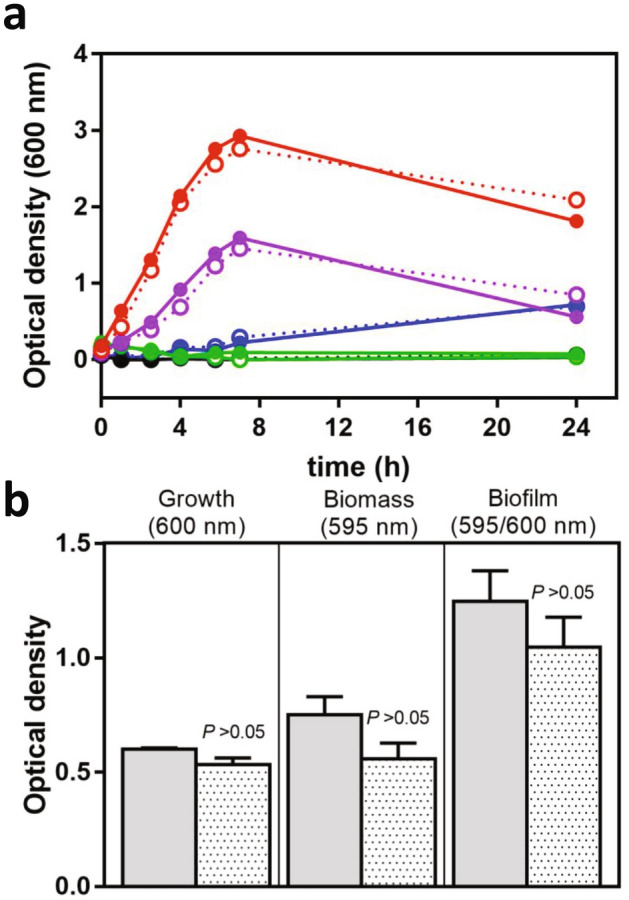
Growth and biofilm formation of F. novicida. (a) F. novicida WT (solid lines) and ΔldcF (dotted lines) were grown under shaking at 37 °C in MMH adjusted at pH 2.5 (green), pH 4 (blue), pH 6.6 (red), pH 8 (purple) or pH 10 (black) and the bacterial growth was monitored by OD600nm measurement. Results are representative of three independent trials. (b) F. novicida WT (grey columns) and ΔldcF (dotted columns) were grown for 24 h under static conditions at 37 °C in a 96-wells plates. The bacterial growth was evaluated by measurement of OD600nm and the biofilm biomass was further determined by OD595nm after Crystal violet staining. This graph corresponds to mean ± s.e.m. of three independent experiments, with at least 4 technical replicates each.
Besides growth fitness and acid stress response, another physiological process in which polyamine products of LAOdcs are likely to be involved is biofilm formation27,30,42–44. Yet, as assessed by crystal violet staining, no significant difference between the amount of biofilm produced by F. novicida WT and ΔldcF strains could be documented (Fig. 5b). We also investigated whether LdcF activity is promoting antibiotic resistance by determining the minimum inhibitory concentrations (MICs) that were found unchanged for either ciprofloxacin (0.064 µg/mL ; n = 3) or gentamicin (1 µg/mL ; n = 3). In addition, we examined the rate at which these antibiotics kill bacteria – the minimum duration for killing (MDK) metric—as a quantitative indicator of antibiotic tolerance45,46. Again, the MDK99 values corresponding to the time required to kill 99% of the bacterial populations including WT, ΔldcF and ΔldcF-complemented (ΔldcF::ldcF) strains exposed either to ciprofloxacin (Supplementary Fig. S9a) or to gentamicin (Supplementary Fig. S9b) were very similar, thus discarding the involvement of LdcF in antibiotic tolerance. This result was confirmed by the Minimal Bactericidal Concentration (MBC)/MIC ratios found to be below 32, i.e. the value defined by the Clinical Laboratory Standards Institute (CLSI) guidelines as the tolerance threshold47.
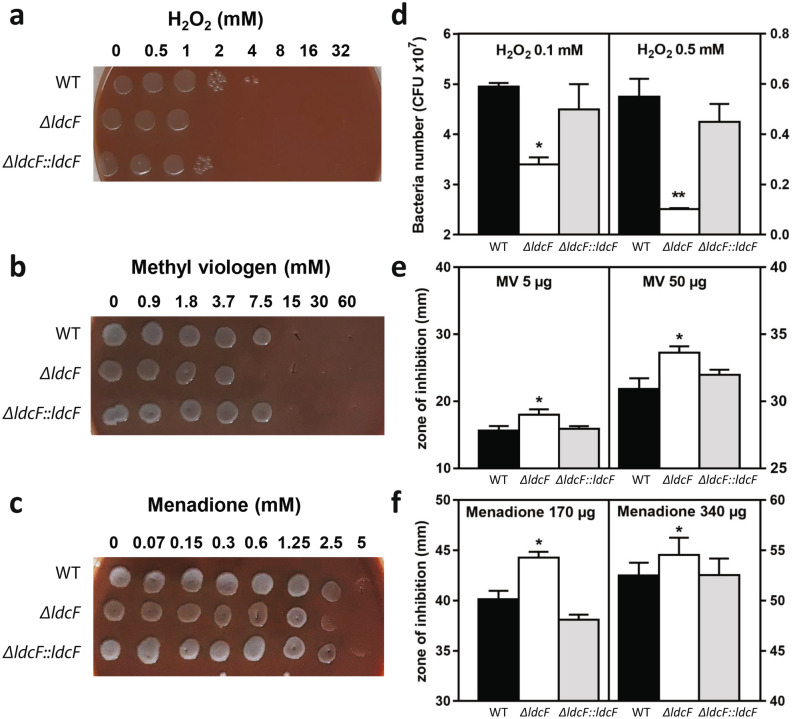
The same set of strains was then tested for susceptibility to oxidative stress. Interestingly, results obtained from spot plating assays indicated that LdcF significantly contributed to survival of bacteria exposed to hydrogen peroxide or to the redox-cycling drugs methyl viologen (MV) and menadione (MD). The ∆ldcF mutant was indeed found systematically less resistant to ROS exposure than the other strains, whereas complementation restored the WT phenotype (left panels in Fig. 6). Under such experimental conditions, and while the incubation of ΔldcF with MD was accompanied with a moderate but reproducible inhibition of growth (Fig. 6c), MV (Fig. 6b) was found even more efficient than H2O2 (Fig. 6a). The enhanced sensitivity of the LdcF-deleted strain to H2O2 was accurately confirmed by a lower number of cfu when ΔldcF was exposed to this reagent as compared to the value obtained with the WT (Fig. 6d). Because cfu counting is a time-consuming approach not fully appropriate to evaluate the effect of compounds on short incubation periods, the extent of ∆ldcF susceptibility to MV and MD was further validated through the disk diffusion assays. Thus, the diameter of inhibition zone, which is related to the susceptibility of the isolate, was significantly higher for ∆ldcF than for the WT strain when the disks were impregnated either with both compounds (Fig. 6e,f).
Figure 6.
Sensitivity of F. novicida to oxidative stress. Exponential growth phase bacteria diluted in MMH were exposed to increasing concentration of (a) H2O2 (b) methyl viologen or (c) menadione for 1 h at 37 °C and 3 µl of the cell suspensions were spotted on PVX-CHA plates. These pictures are representative of at least 3 distinct experiments performed in duplicate each. The antibacterial activity of oxidative compounds was also quantified by (d) cfu counting from a cell suspension containing 108 bacteria incubated for 1 h under shaking in presence of H2O2 or by disk diffusion assays with (e) methyl viologen or menadione (f) as detailled in materials and methods section. Histograms correspond to the mean ± s.e.m. of at least 3 distinct experiments performed in duplicate (*P < 0.05, **P < 0.01).
In vivo phenotypic analysis of the ΔldcF mutant
We next evaluated the consequences of ldcF deletion on bacterial replication in macrophages. The uptake of F. novicida WT, ΔldcF and ΔldcF::ldcF strains into J774 cells, estimated upon macrophage infection with a MOI of 100, was found identical for the three strains which displayed equivalent intracellular growth profiles over the first 24 h (Fig. 7a). However, at 48 hpi and beyond, the number of viable intracellular ∆ldcF cells was found significantly lower than observed for the WT strain (∆ldcF: 2.63 × 108 ± 0.47 × 108, n = 8 vs WT: 4.08 × 109 ± 0.64 × 109, n = 8 ; P < 0.0005), and the effect was reversed with the ΔldcF::ldcF (1.27 × 1010 ± 0.35 × 1010, n = 4). As assessed by measuring lactate dehydrogenase activity, this reduced level of recovered viable bacteria was not related with an increased host cell lysis that would result in the release of bacteria into the extracellular medium (WT: 30.6% ± 4.92% vs ∆ldcF: 25.95% ± 3.29% and ΔldcF::ldcF: 30.79% ± 7.9%; P > 0.05; n = 3, measured at 72 hpi). Together, these data suggest that the deletion mutant displays a reduced capacity to escape macrophage killing mechanisms. Their failure to survive the antibacterial activities of host macrophages most probably relies on an intricate overlapping network of signals combining pro-inflammatory and immune responses as well as metabolic response of the infected cell. However, considering the role of macrophage oxidative burst in pathogen clearance, we then evaluated the ROS level in infected J774 cells. Our results demonstrate that macrophages infected with the ∆ldcF strain, which is less resistant to oxidative stress, display a higher ROS activity than cells infected either with the WT or the complemented strain (Fig. 7b), an effect that could result either from an impaired degradation or from an increased production of ROS.
Figure 7.
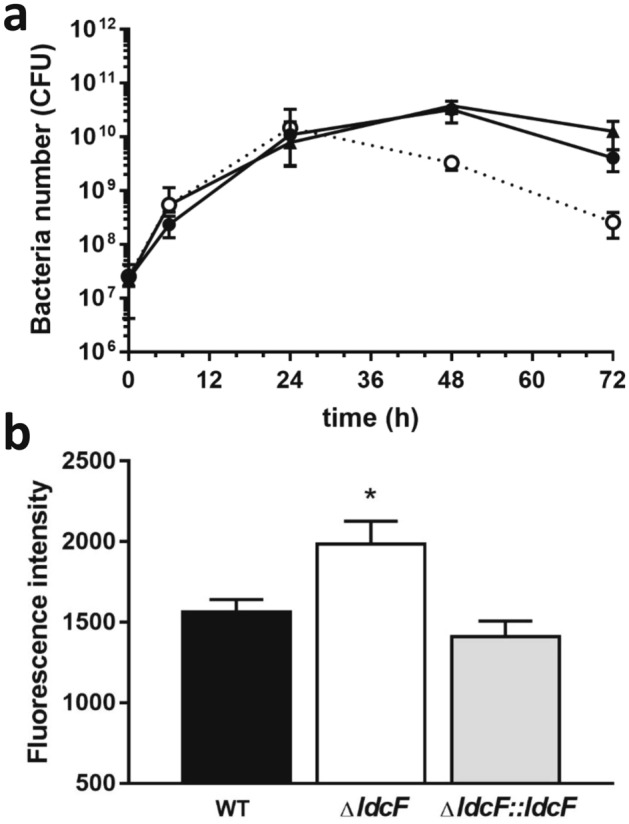
Replication of F. novicida strains within the J774 macrophage-like cell line. (a) F. novicida WT (black circles, solid line), ΔldcF (white circles, dotted line) and ΔldcF::ldcF (black triangles, solid line) were inoculated at a MOI of 100:1 and intracellular bacteria were enumerated by cfu counting at different times post infection (b) Production of ROS evaluated at 24 h after infection of macrophages with a MOI of 1,000:1 using the redox-sensitive dye DCFA detected by fluorescence spectroscopy. Data correspond to mean ± s.e.m. of 4 distinct experiments and after subtraction of background values obtained with uninfected macrophages. *P < 0.05.
Comparative proteomics reveals proteins impacted by ldcF deletion
A MS-based quantitative proteomic comparison was performed on whole-cell extracts of F. novicida WT and ΔldcF strains to identify proteins for which abundance was altered by ldcF deletion. A bioinformatics analysis reliably identified and quantified 1,263 different proteins from the 1,854 protein-coding ORFs annotated in the F. novicida genome (Supplementary Table S3; PXD016591). An ensuing statistical analysis revealed that expression levels of 80 proteins were significantly affected by ldcF deletion. Equal numbers of proteins were expressed at lower or higher abundance in ΔldcF compared to the WT (Table 1). Consistent with the lack of LdcF in the deletion mutant, the 2,3,4,5-tetrahydropyridine-2,6-carboxylate N-succinyltransferase (FTN_1727, DapD) involved in lysine biosynthesis was found to be downregulated. Surprisingly, although all proteins encoded by the FPI (FTN_1309 to FTN_1326) 5 were detected in the MS-based quantitative proteomic assay (Supplementary Table S3), none of them showed altered expression levels in the ΔldcF strain. In contrast, the amount of the major transcriptional regulator MglA (FTN_1290, MglA), which is described as a FPI gene regulator8,48, was significantly reduced in the ΔldcF mutant (Table 1).
Table 1.
List of proteins differentially expressed in ΔldcF.
| Gene name | Locus | Description | Proteomic data | |
|---|---|---|---|---|
| Log 2FC | P value | |||
| Downregulated proteins in ΔldcF | ||||
| cadA | FTN_0504 | Lysine decarboxylase | − 6.135487177 | 6.5901E−12 |
| udp | FTN_0652 | Uridine phosphorylase | − 2.442473795 | 0.000355044 |
| uvrB | FTN_1176 | Excinuclease ABC subunit B | − 2.359970258 | 1.5872E−07 |
| yhiP | FTN_0885 | Proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H + symporter | − 2.233739832 | 1.28816E−08 |
| – | FTN_1453 | Two-component regulator, sensor histidine kinase | − 2.225623368 | 0.000269144 |
| – | FTN_0705 | Abortive infection bacteriophage resistance protein | − 1.909345488 | 0.006797082 |
| – | FTN_0655 | N6-adenine-specific methylase | − 1.645819843 | 0.009836595 |
| – | FTN_1348 | Acetyltransferase | − 1.493386099 | 3.68028E−07 |
| – | FTN_0898 | Amino acid permease | − 1.424644622 | 3.5435E−06 |
| panD | FTN_1354 | Aspartate 1-decarboxylase | − 1.405501066 | 0.006567341 |
| – | FTN_0862 | Hypothetical protein | − 1.276557703 | 0.000969217 |
| ung | FTN_1486 | Uracil-DNA glycosylase | − 1.261354286 | 4.40904E−07 |
| – | FTN_0308 | Membrane protein of unknown function | − 1.255628185 | 0.000806353 |
| – | FTN_1272 | Proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H + symporter | − 1.243209089 | 1.20616E−05 |
| dapD | FTN_1727 | 2,3,4,5-tetrahydropyridine-2,6-carboxylate N-succinyltransferase | − 1.185622324 | 0.000977552 |
| – | FTN_1258 | Hypothetical protein | − 1.11437438 | 0.000177817 |
| hsdM | FTN_1152 | Type I restriction-modification system, subunit M (methyltransferase) | − 1.098834276 | 0.000436818 |
| – | FTN_1316 | Hypothetical protein | − 1.075883638 | 0.005337719 |
| – | FTN_1628 | LysR family transcriptional regulator | − 1.010217775 | 0.001268379 |
| hdsR | FTN_0710 | Type I restriction-modification system, subunit R (restriction) | − 0.974133834 | 0.00046316 |
| – | FTN_1212 | Glycosyl transferases group 1 family protein | − 0.967248679 | 7.07496E−06 |
| – | FTN_1397 | Hypothetical protein | − 0.962399859 | 6.53792E−05 |
| – | FTN_0976 | ThiF family protein | − 0.962086728 | 6.76869E−05 |
| waaG | FTN_1218 | Glycosyl transferase, group 1 | − 0.942030987 | 0.000643833 |
| pilE4 | FTN_0389 | Type IV pili, pilus assembly protein | − 0.94007106 | 2.01122E−05 |
| – | FTN_1440 | Hypothetical protein | − 0.935671185 | 0.007635418 |
| ubiC | FTN_0386 | Chorismate pyruvate lyase | − 0.934045712 | 6.57717E−05 |
| – | FTN_0137 | Hypothetical protein | − 0.928404833 | 3.73274E−06 |
| mglA | FTN_1290 | Macrophage growth locus, protein A | − 0.926959218 | 6.24086E−07 |
| – | FTN_1697 | Galactose mutarotase | − 0.901042767 | 0.008192095 |
| – | FTN_1148 | Glycoprotease family protein | − 0.816650922 | 0.005012835 |
| galP1 | FTN_0687 | Major facilitator superfamily galactose-proton symporter | − 0.746905316 | 5.94864E−05 |
| – | FTN_1459 | Short chain dehydrogenase | − 0.723695755 | 0.001285646 |
| – | FTN_1254 | Hypothetical protein | − 0.716291799 | 0.00237079 |
| – | FTN_1266 | ABC transporter membrane protein | − 0.66256701 | 0.001562237 |
| – | FTN_0923 | Hypothetical protein | − 0.654493997 | 0.002785307 |
| yhbG | FTN_0902 | ABC transporter, ATP-binding protein | − 0.652797305 | 3.32447E−05 |
| rpsF | FTN_0951 | 30S ribosomal protein S6 | − 0.641616628 | 5.78051E−05 |
| yrbI | FTN_0905 | 3-Deoxy-d-manno-octulosonate 8-phosphate phosphatase | − 0.627622272 | 0.001032099 |
| – | FTN_1547 | Hypothetical protein | − 0.608477024 | 0.000859667 |
| Upregulated proteins in ΔldcF | ||||
| rimM | FTN_1561 | Ribosome maturation factor rimM | 0.626513088 | 0.000555064 |
| apaH | FTN_0561 | Diadenosine tetraphosphatase | 0.629366731 | 0.000669224 |
| – | FTN_0118 | S49 family serine peptidase | 0.635972422 | 0.006578849 |
| – | FTN_1468 | Putative deoxyribonucleotide triphosphate pyrophosphatase | 0.640919668 | 0.000191944 |
| – | FTN_0089 | Allophanate hydrolase subunit 2 | 0.672742647 | 0.000997506 |
| rnc | FTN_1463 | Ribonuclease 3 | 0.681039745 | 0.001289497 |
| – | FTN_0789 | Putative rhodanese, sulfurtransferase | 0.68404703 | 0.000345296 |
| secF | FTN_1094 | Preprotein translocase subunit SecF | 0.695399198 | 3.15477E−05 |
| murD | FTN_0542 | UDP-N-acetylmuramoylalanine–d-glutamate ligase | 0.715198106 | 5.10756E−05 |
| xthA | FTN_0838 | Exodeoxyribonuclease III | 0.726407142 | 1.90678E−05 |
| sun | FTN_1347 | tRNA and rRNA cytosine-C5-methylases, sun protein | 0.73058117 | 0.00042405 |
| – | FTN_0872 | Small conductance mechanosensitive ion channel (MscS) family protein | 0.78091602 | 2.90828E−05 |
| – | FTN_1387 | Hypothetical protein | 0.812379067 | 0.00883337 |
| ispD | FTN_0623 | 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase | 0.824072741 | 5.66929E−06 |
| – | FTN_0041 | Hypothetical protein | 0.828287322 | 0.002571557 |
| mltA | FTN_1286 | Membrane-bound lytic murein transglycosylase | 0.843953091 | 0.000666109 |
| – | FTN_1080 | Phosphosugar binding protein | 0.857709173 | 0.000844453 |
| – | FTN_1015 | Isochorismatase family protein | 0.860701266 | 0.000470586 |
| pilW | FTN_0307 | Type IV pilus assembly protein | 0.903906623 | 0.004575204 |
| – | FTN_1061 | Acid phosphatase, HAD superfamily protein | 0.951666713 | 0.001807004 |
| pilV | FTN_0413 | Type IV pili, pilus assembly protein | 0.975215693 | 0.001866241 |
| murQ | FTN_1504 | N-acetylmuramic acid 6-phosphate etherase | 0.994710363 | 1.2722E−06 |
| tdh | FTN_0625 | l-Threonine 3-dehydrogenase | 1.02107175 | 2.99619E−05 |
| ruvA | FTN_1025 | Holliday junction ATP-dependent DNA helicase RuvA | 1.125358669 | 5.65196E−07 |
| – | FTN_1506 | Hypothetical protein | 1.20406212 | 0.000656496 |
| putP | FTN_0299 | Proline/Na + symporter | 1.206589507 | 0.001401361 |
| – | FTN_0452 | Hypothetical protein | 1.274923951 | 4.08167E−08 |
| – | FTN_0006 | Hypothetical protein | 1.345660313 | 1.02878E−06 |
| – | FTN_0004 | Aspartate/glutamate transporter | 1.352713527 | 0.003023321 |
| – | FTN_0829 | Hypothetical protein | 1.454738617 | 6.98601E−07 |
| – | FTN_1388 | Oxidoreductase | 1.515433578 | 0.000565317 |
| – | FTN_1267 | ABC transporter ATP-binding protein | 1.74578111 | 3.84443E−09 |
| lptC | FTN_0904 | Lipopolysaccharide export ABC transporter periplasmic protein | 1.999604273 | 1.63282E−05 |
| rnpA | FTN_0075 | Ribonuclease P protein component | 2.366923256 | 0.000708316 |
| – | FTN_0384 | Hypothetical protein | 2.702678582 | 0.002040018 |
| – | FTN_0987 | tRNA-dihydrouridine synthase | 2.830069785 | 5.08771E−06 |
| – | FTN_1386 | Hypothetical protein | 2.845246288 | 4.58158E−09 |
| – | FTN_0722 | l-lysine 2,3-aminomutase | 3.467288507 | 1.01829E−10 |
| – | FTN_1220 | Lipopolysaccharide synthesis sugar transferase | 3.613912551 | 0.000821341 |
| recB | FTN_1357 | ATP-dependent exoDNAse (exonuclease V) beta subunit | 3.825585172 | 1.16584E−11 |
The protein deleted in the mutant was labeled in bold.
The KEGG (Kyoto Encyclopedia of Genes and Genomes) database49 includes 99 pathways for F. novicida (https://www.genome.jp/kegg-bin/show_organism?org=ftn). Interestingly, following KEGG annotation, only 23 of the 80 proteins affected by the ldcF deletion were assigned to functional categories. These 23 proteins can be roughly grouped into a limited number of distinct functional pathways, including bacterial metabolism, DNA proofreading and repair, and pathways related to oxidative stress. One group of the KEGG-annotated differentially expressed proteins (5 out of 23) is associated with DNA proofreading and repair. This group is composed of two proteins involved in base excision repair pathways (FTN_1486, Ung and FTN_0838, XthA), one involved in nucleotide excision repair (FTN_1176, UvrB), and two involved in homologous recombination pathways (FTN_1025, RuvA and FTN_1357, RecB). Two of these proteins (UvrB and Ung), which displayed reduced expression levels in ΔldcF, are enzymes considered to be prokaryotic defense systems involved in virulence through their protection of bacterial DNA50,51. Other proteins that were downregulated in ΔldcF may also help bacteria to deal with DNA damage, although they lack functional KEGG assignment. These proteins included enzymes from type I restriction-modification systems (FTN_1152, HsdM; FTN_0710, HsdR)52 and the N6-adenine-specific methylase (FTN_0655). In contrast, some other DNA repair proteins were expressed at higher levels following ldcF deletion. An example is exodeoxyribonuclease III (XthA), a negative regulator of homologous recombination under log phase growth conditions, of which the overexpression can also result in unrepaired DNA damage.
The KEGG pathway annotation of proteins for which expression levels were significantly altered in ΔldcF also revealed several metabolic and transport pathways that could play a role in bacterial replication. Specifically, the deletion mutant’s reduced capacity to deal with the host immune system and survive within macrophages may be partly related to the observed decrease in uridine phosphorylase levels (FTN_0652, Udp), as previously suggested using Drosophila melanogaster as an experimental model53. Similarly, UbiC (FNT_0386) catalyzes the first step of ubiquinone (or coenzyme Q) biosynthesis involved in electron transport chains and is considered as a lipid-soluble antioxidant in prokaryotes; its expression was reduced, and could thus impact F. novicida’s oxidative defense54. These findings are in good agreement with a possible role of LdcF in the activation of the SOS-response, and are underscored by the increased expression levels measured for RuvA (FTN_1025, Holliday junction ATP-dependent DNA helicase) and RecB (FTN_1357, ATP-dependent exoDNAse) that could help F. novicida to cope with oxidative stress53.
Discussion
The phylogenetic analysis and amino acid sequence comparisons presented here indicate that the unique lysine decarboxylase identified within Francisella proteomes, i.e. LdcF (previously annotated as CadA) is more closely related to E. coli LdcI and LdcC than to P. aeruginosa LdcA or E. coli AdcI, OdcI and OdcC. However, similarly to most of the P. aeruginosa strains26 and unlike E. coli, Francisella genomes lack the presence of a RavA orthologue shown to alleviate inhibition of E. coli LdcI by the alarmone ppGpp32. Consistently, as in LdcA35, the C-terminal β-strands of LdcF display different amino acids at locations corresponding to the RavA binding site in E. coli LdcI. Furthermore, our structural analysis of F. novicida LdcF demonstrates that eight out of the 10 residues involved in LdcI interaction with ppGpp in E. coli show either a reverse in charge or change in hydrophobicity, which reveals that, again similarly to LdcA35, it is highly unlikely that LdcF would be inhibited by ppGpp. These observations underlying major differences between LdcF and LdcI are consistent with the absence of a RavA orthologue in Francisella genomes.
Examination of the ldcF genetic environment, which is highly conserved within different Francisella species, suggests that despite the high sequence identity and strong structural similarity with LdcI, LdcF expression is differently regulated. Notably, genes encoding both CadB, the putative cadaverine transport protein, and CadC, the pH sensor and membrane-bound transcriptional regulator of the cadBA operon55 are missing in the ldcF gene cluster. Upstream of ldcF are lolC and lolD which encode two components of the ABC transporter complex involved in lipoprotein transport and membrane biogenesis and are described as essential genes in F. tularensis56. The downstream genes belong to the glycine cleavage system (GCS) and were found significantly upregulated in the virulent F. tularensis type A Schu S4 strain inside macrophages57. Our observations are therefore in agreement with the data on the in vivo negative selection of F. novicida transposon mutants that pointed out the importance of GCS genes together with lolD and ldcF (FTT_0405 to FTT_0409) in the intracellular growth and/or virulence of F. novicida23.
The combined analysis of the genomic context of Francisella ldcF, the structure of F. novicida LdcF and the phylogenetic relationships between the new LdcF family and other proteobacterial LAOdcs raised questions about the LdcF regulation and functional activity. Our experiments showed that the growth rates of F. novicida WT and ΔldcF are very similar over a broad range of basic to acidic pHs, thus ruling out a strict role of LdcF in acid tolerance and buffering of the bacterial cytosol upon acid stress. Deletion of ldcF also failed to affect temperature-dependent bacterial growth. In contrast, in comparison with the WT, ΔldcF displayed a significantly lower resistance to oxidative stress. The capacity of cadaverine to scavenge oxygen radicals, thus providing bacteria with a higher tolerance towards oxidative stress, was previously reported for E. coli58 and Vibrio vulnificus59,60. Importantly, these results are corroborated by a greater survival of both the WT and ΔldcF::ldcF strains in infected macrophages which contain a lower amount of ROS than those infected with ΔldcF. Such a survival strategy could be shared with the bacterium that possesses the closest LdcF relative, i.e. L. fallonii, which replicates within the protozoan host Acanthamoeba in aquatic environments and must face oxidative and acidic stress conditions during its stationary phase of growth18,61.
To better understand the mechanism by which removal of ldcF and a subsequent defect in cadaverine synthesis altered the oxidative stress resistance, we performed an extensive quantitative comparison of the protein contents between the F. novicida WT and the ΔldcF strains. This analysis identified 80 proteins for which expression levels were altered following ldcF deletion. Among them, we were surprised not to observe any ROS scavenging enzymes. Indeed, similarly to several other bacterial species, to cope with oxidative stress, Francisella utilize enzymes such as SodB, SodC, KatG and the recently identified AhpC16,17,20,62,63 to convert harmful ROS into innocuous products19. Moreover, expression levels of other factors contributing to ROS defense mechanisms, such as the efflux pump EmrA1 (FTL_0687), involved in SodB and KatG secretion64, or the F. tularensis (FTL_1014) oxidative stress regulator OxyR16,18, also displayed similar expression levels in WT and ΔldcF strains. However, one of the low-abundance proteins in ΔldcF was an ABC transporter (FTN_0902; FTL_1065, YhbG). Interestingly, this transporter was recently reported to be down-regulated in a ΔoxyR mutant of F. tularensis LVS displaying an enhanced sensitivity to oxidative stress18. Furthermore, the reduced UbiC content compared to the WT strain could also contribute to the diminished capacity of the F. novicida ΔldcF strain to survive oxidative attack from ROS. Indeed, altered UbiC levels could indirectly promote ROS accumulation54. In addition to ROS-neutralizing enzymes, bacteria can also counteract ROS damage using their DNA damage-responsive genes. The products of these genes initiate DNA repair pathways to recognize and correct ROS-induced and other mismatches. An interesting hallmark of the F. novicida ΔldcF proteome is the significant changes in levels of proteins involved in DNA repair processes potentially reducing the bacteria’s capacity to deal with oxidative stress. Our results also indicate that MglA expression was significantly reduced in the mutant strain. Interestingly, in addition to ensuring the regulation of Francisella virulence factors – which were unchanged in ΔldcF compared to the WT strain – MglA has been reported to play a key role in the intracellular growth of F. tularensis and its adaptation to oxidative stress8,11.
While the relationships between the proteomic observations and Francisella ROS defense mechanisms are not straightforward and a thorough understanding of the link between ldcF inactivation and the changes in the protein expression pattern requires further investigations, our observations provide an important evidence of the LdcF involvement in F. novicida oxidative stress resistance. By suppressing lysine decarboxylation, the ldcF deletion promotes the accumulation of lysine and the decrease of cadaverine, which both should have a direct impact on the bacterial physiology. Lysine harvesting was indeed described as a powerful preventive metabolic antioxidant strategy displayed by microbial cells65, an effect most probably reverted when this amino acid accumulates. The contribution of polyamines in several bacterial infections has long been described66, and some studies have specifically emphasized their relevance in F. tularensis virulence. For example, an increased expression of ornithine decarboxylase was observed in F. tularensis infected mice67. The relevance of spermine within host cells infected by Francisella, and specifically the capacity of this polyamine to elicit transcriptional changes in F. tularensis, leading in turn to altered host cell activation, has also been reported68. While never investigated, it could be hypothesized that cadaverine could also exert transcriptional control on genes implicated in Francisella resistance against oxidative attack. Taken together, our work provides a biochemical and structural framework to further explore LdcF as a potential virulence factor and its involvement in Francisella oxidative stress resistance.
While LAOdcs are long-recognised as drug targets, and development of specific mechanism-based Ldc inhibitors is an active research field69–72, we envision that the LdcF structure and the functional findings presented in this work will empower further investigations aimed at design of new LdcF-based therapeutic approaches against tularemia.
Methods
Bioinformatic analyses
Sequences of AAT-fold decarboxylases were retrieved from NCBI: LdcI (NP_418555.1), LdcC (NP_414728.1), AdcI (NP_418541.1), OdcC (NP_417440.1), and OdcI (NP_415220.1) from Escherichia coli str. K-12 substr. MG1655 and LdcA (NP_250509.1) from P. aeruginosa PAO1. These sequences were used as seeds to query a local database containing 4,467 complete proteomes of prokaryotes (Supplementary Table S4) from the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov) with the BLASTP 2.2.6 software73 and with HMM-profile based approaches with the HMMER package v3.1b1 (default parameters)74. Finally, searches for unannotated sequences were performed with TBLASTN (default parameters) on the complete genome sequences corresponding to the 4,467 proteomes using default parameters. Sequences with an e-value lower than 10–4 were retrieved and aligned using MAFFT v.775. The resulting multiple alignment was visually inspected with AliView 1.2576. Doubtful sequences were systematically verified using reciprocal best reciprocal blast hit. This led to the identification of 4,091 AAT-fold decarboxylase sequences, 13 of which were unannotated or annotated as pseudogenes (Supplementary Table S5).
A phylogeny of WING-containing AAT-fold decarboxylase sequences was inferred using maximum likelihood. To limit taxonomic redundancy, the phylogenetic analysis was performed on a subset of 1,905 representative proteomes by selecting randomly one representative strain per species. The 553 WING-containing AAT-fold decarboxylase sequences contained in these representative proteomes were aligned with MAFFT using the L-INS-i option and trimmed with BMGE v1.1 with matrix substitution BLOSUM30 (589 amino acid positions kept after trimming)77. The maximum likelihood tree was inferred with IQ-TREE 1.6.1278. IQ-TREE identified the LG + R10 as the best suited evolutionary model according to the Bayesian information78. The robustness of the inferred tree was assessed using the ultrafast bootstrap (1,000 replicates implemented in IQ-TREE). The genomic context figure has been generated by GeneSpy 1.179 and phylogeny figures by iTOL80.
The percentage of identity between LdcF sequences and LdcI (NP_418555.1), LdcC (NP_414728.1), AdcI (NP_418541.1), OdcC (NP_417440.1), and OdcI (NP_415220.1) from Escherichia coli str. K-12 substr. MG1655 and LdcA (NP_250509.1) from P. aeruginosa PAO1 has been computed using the Needleman and Wunsch algorithm implemented at the NCBI (default parameters).
Bacterial strains and growth conditions
The strain F. novicida CIP56.12 (Centre de Ressources Biologiques de l'Institut Pasteur, Paris, France) and the ldc mutants were grown on PVX-CHA plates (bioMérieux, Marcy l'Étoile, France) incubated at 37 °C in a 5% CO2-enriched atmosphere. Liquid cultures were carried out at 25 °C or 37 °C under agitation at 180 rpm in MMH medium, as indicated. For the growth of the complemented strain ΔldcF::ldcF, liquid and solid media were supplemented with kanamycin (10 µg/mL).
Cloning, expression and purification of LdcF
The sequences of all primers used in this study are given in Supplementary Table S6. The gene encoding LdcF (FTN_0504) was amplified by PCR from genomic DNA using High-Fidelity PCR master mix (Phusion, Finnzymes) and gene-specific primers FTN_0504F/R encoding a Tobacco Etch Virus (TEV) site. The resulting product was cloned into the pDONR 201 vector and subsequently subcloned into pDEST-17 using the Gateway cloning system from Invitrogen following the manufacturer’s instructions. Integrity of the N-terminal 6xHis-tagged construct was confirmed by DNA sequencing (Eurofins, Ebersberg, Germany). Protein expression was started by picking 5–10 colonies of freshly transformed E. coli C41(DE3) strain in 2 mL LB medium supplemented with ampicillin (100 µg/mL). After 3 h at 37 °C under shaking the bacterial suspension was transferred in 50 mL medium for additional 3 h, then diluted in 400 mL up to an optical density OD600 nm of approximately 0.5 and expression was induced overnight at 16 °C by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After centrifugation (5,000×g, 20 min) the bacteria were resuspended in lysis buffer (50 mM Tris pH 7.9, 300 mM NaCl, 0.1 mM pyridoxal 5′-phosphate (PLP) (Sigma), 0.5% CHAPS (Sigma), 5% glycerol, 2 mM ß-mercaptoethanol, 1 mM PMSF, Complete Protease Inhibitor (Roche Diagnostics) and 10 mM imidazole), then disrupted by sonication. The bacterial lysate was then heated for 5 min at 70 °C, centrifuged (20,000 rpm, 30 min, 4 °C) and applied onto a Ni2+-NTA column (Qiagen) for affinity purification. After extensive washing (50 mM Tris pH 7.9, 300 mM NaCl, 0.1 mM PLP, 5% glycerol, 2 mM ß-mercaptoethanol, and 20 mM imidazole) the protein was eluted with the same buffer supplemented with 300 mM imidazole. The fractions containing LdcF were pooled and dialyzed overnight at 4 °C against 50 mM HEPES pH 7, 25 mM NaCl, 0.1 mM PLP, 1 mM DTT and further purified on a Superose 6 size exclusion chromatography column (GE Healthcare, UK) and using the NGC Chromatography System (Bio-Rad). The peak fractions were analyzed on 10% SDS-PAGE and Coomassie blue staining and concentrated for crystallization without His-tag removal (Supplementary Fig. S10). In addition to the N-terminally His-tagged LdcF, a C-terminally His-tagged construct was also evaluated. The lysine decarboxylase activity of purified LdcF was assessed at pH 6.5 and 37 °C using a 2,4,6-trinitrobenzensulfonic acid assay as described31,39, with 8 mM lysine and 500 nM LdcF in the initial solutions before the mixture. Purified recombinant protein (1 mg/mL) was also used for rabbit immunization and production of a polyclonal antibody (Biotem, France) which appeared to also recognize E. coli LdcI but not AdcI, illustrating the close relationships between LdcF and LdcI.
Crystallization, data collection and structure determination of LdcF
Prior to setting up crystallization trials, purified LdcF (50 mM HEPES pH 7, 25 mM NaCl, 0.1 mM PLP, 1 mM DTT) was concentrated to 3 mg/mL. Extensive crystallization trials that included 576 conditions tested in 96-well sitting drop vapor diffusion plates with drop volumes of 200 nL (100 nL protein solution + 100 nL reservoir solution, T: 20 °C) were performed at the high-throughput crystallization facility at the EMBL Grenoble outstation, France. LdcF initially crystallized in a condition containing 25% Ethylene Glycol (Crystal Screen HT, condition E4, Hampton Research). After manual optimization in 24-well hanging drop vapor diffusion plates (Molecular Dimensions, 1 µL protein solution + 1 µL reservoir solution, T: 20 °C), large brick shaped crystals appeared after several weeks of incubation, which displayed a bright yellow colour due to present PLP. LdcF crystals were scooped directly from the crystallization plates and were subsequently flash-cooled in liquid nitrogen.
Diffraction data was collected at beamline ID-29 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, and was processed using the XDS package81 in space group C2 2 21 (a = 165.25 Å, b = 318.21 Å, c = 183.98 Å, α = β = γ = 90). The structure of LdcF was solved by maximum-likelihood molecular replacement using Phaser82 in the Phenix software package83, starting from the crystal structure of E. coli LdcI (PDB ID: 3n75) as an initial model32 , with side-chains trimmed using the Schwarzenbacher method84 in Sculptor from the Phenix package. The molecular replacement solution was subsequently refined in Phenix, using reciprocal- and real-space refinement, with noncrystallographic symmetry (NCS) restraints, occupancy refinement, individual B-factor refinement with TLS (translation liberation screw), and optimized x-ray/stereochemistry and x-ray/ADP weights. Several rounds of alternating refinement in Phenix and manual building in Coot85 were performed, followed by a final refinement in Phenix.
Construction of the FTN_0504 knock-out strain
The F. novicida ldcF chromosomal deletion mutant was generated by allelic exchange as previously described86. Briefly, around 650 bp of both the 5′ and 3′ regions of FTN_0504 were amplified from genomic F. novicida DNA using Phusion DNA polymerase and fused with a kanamycin resistance cassette. The second round overlapping PCR was carried out with the primers FTN_0504ForUp and FTN_0504DownRev and using the mixture of the three previous PCR amplicons as template. Following purification from agarose gel (QIAquick Gel Extraction Kit, Qiagen), the resulting 2200 bp fragment of interest (1 µg) was used to transform chemically competent F. novicida U112 spread on PVX-CHA containing kanamycin (15 μg/mL). The antibiotic-resistance marker was further deleted through Flp-mediated excision and using the pKEK1112 temperature sensitive plasmid. The final mutant sentitive to both kanamycin and tetracycline was checked for loss of FTN_0504 by PCR product direct sequencing using appropriate primers (Eurofins).
Construction of the LdcF complementation plasmid
The gene encoding the F. novicida LdcF WT was amplified using a primer pair on which NotI and AgeI restriction sites have been engineered and cloned into the pFNLTP6 shuttle plasmid downstream of the gro promoter87 using a DNA ligation kit (TAKARA BIO INC.). Following restriction enzyme digestion and sequencing, the resulting LdcF complementation plasmid designed pFNLTP6-ldcF was introduced into chemically competent F. novicida. Transformed colonies selected on PVX-CHA containing kanamycin (15 μg/mL) appeared after 2 days of incubation. Complementation was confirmed by PCR on purified DNA and by western-blot with the anti-LdcF antibody on whole bacterial extracts.
Rezasurin assay
The metabolic activity of F. novicida wild-type and ΔldcF was estimated through their capacity to reduce resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide; λmax = 600 nm ; Sigma-Aldrich) into the pink fluorescent compound resorufin (λmax = 570 nm). This oxidation–reduction indicator of mitochondrial function was conveniently used to evaluate cell viability of several bacterial species88 including F. tularensis, F. novicida and F. philomiragia89,90. Exponential growth phase bacteria grown in MMH at 37 °C under shaking were transferred (200 µL) into a 96-well plate. After addition of 20 µL of resazurin (0.2 mg/mL) the microtiter plate was incubated for 1 h at 37 °C, and the cell viability was determined by OD570nm − OD600nm measurement (Tecan Plate reader). The experiment was repeated thrice with 6 replicates for each condition.
Biofilm assay
Assessment of the biofilm formation was carried out using crystal violet assay and following optimized protocol for F. novicida starting from a 200 µL bacterial inoculum (1.107 cells/mL) in flat-bottom 96-well plates. The biofilm biomass was quantified following 24 h incubation at 37 °C in a 5% CO2 incubator without shaking90.
Evaluation of antimicrobial susceptibility
Antibiotic susceptibility was assessed by MIC, MBC/MIC and time-dependent killing assays, as previously described89. Briefly, the MICs were determined through the broth microdilution method following the CLSI recommended guidelines47 but using MMH as culture medium and with final antibiotic concentrations ranging from 0.125 to 64 μg/mL for gentamicin and from 0.002 to 1 μg/mL for ciprofloxacin. Plate counting of serial dilutions of the wells where no bacterial growth was visually observed allowed to estimate the MBC and to calculate the MBC/MIC ratio as a tolerance criterion47. The antibiotic tolerance was also evaluated using the method based on Minimum Duration for Killing 99% of the population—MDK metric45 starting from bacterial cultures grown in MMH to an OD600nm of 0.5 prior addition of 25 times the MIC of gentamicin (25 µg/mL) or ciprofloxacin (1.6 µg/mL). Bacterial suspension were kept at 37 °C under shaking and the number of tolerant cells was assessed at different time points following antibiotic addition, through cfu counting. Each sample was processed in duplicate and results were expressed as a percent of viable bacteria relative to unexposed population.
Spot plating assay
To accurately compare the oxidative stress responses of the different strains, two-fold serial dilutions of the drugs were prepared in MMH using 2 mL Eppendorf tubes, then dispensed in a 96 well plate (100 µL/well). Exponential phase bacteria diluted to a OD600nm of 0.2 were then added (100 µL) in the wells containing drug dilutions. Posititive (without drug) and negative (without bacteria) controls were also included. Following 1 h incubation at 37 °C in a 5% CO2 incubator without shaking, 3 μL aliquots were spotted PVX-CHA plates further incubated at 37 °C for 48 h to 72 h before being photographed. All strains were processed in parallel in each experiment which included two independent replicates spotted twice.
Disk diffusion method
A protocol essentially identical to that recently described for F. tularensis LVS and SCHU S4 strains was applied20. PVX-CHA plates were inoculated onto the entire surface with early stationary phase bacteria by using a sterile cotton swabs. Sterile cellulose disks (diameter 6 mm, D. Dutscher, France) were placed with sterile forceps and slightly pressed onto the gelose surface (up to 3 discs per plate), then impregnated with 10 µL of methyl viologen dichloride hydrate (Sigma) or menadione (Sigma). After 48 h incubation at 37 °C the diameter of the zone of complete inhibition (in mm) around each disk was measured thrice from numerized agar plate pictures and using ImageJ software.
Western-blot analysis of LdcF expression
Whole bacterial extracts were resolved by 12% polyacrylamide SDS-PAGE before transfer on nitrocellulose membrane (Trans-Blot Turbo, Bio-Rad). Western-blot analysis was performed using a polyclonal antibody towards LdcF produced in this study (1:10,000) and a rabbit secondary antibody coupled to peroxidase (1:10,000; Jackson ImmunoResearch, Baltimore, PA, USA). The mouse anti-Francisella-IglC (1:10,000; Jackson ImmunoResearch, Baltimore, PA, USA) was used as positive control. Detection was carried out with enhanced chemiluminescence (Clarity Western ECL, Bio-Rad) using the Bio-Rad Chemidoc XRS + System.
Mass spectrometry-based quantitative proteomic analyses
Bacterial cultures were grown in MMH at 37 °C under shaking to late logarithmic phase. Five independent replicates were prepared for each sample type. Aliquots corresponding to 1 × 109 cells were centrifuged (7500×g; 10 min; 4 °C) and the resulting pellets were resuspended in 50 µL BugBuster Reagent to improve the release of soluble proteins before addition of 50 µL Laemmli buffer and storage at − 20 °C until further use. The protein concentrations estimated by the method of Bradford from the same amount of bacteria resuspended in distilled water and heated at 70 °C for 30 min was found homogenous ranging between 180 and 210 µg. Samples solubilized in Laemmli buffer were stacked in the top of a SDS-PAGE gel (4–12% NuPAGE, Life Technologies) and stained with Coomassie blue R-250 before in-gel digestion using modified trypsin (Promega, sequencing grade) as previously described91. Resulting peptides were analyzed by online nanoliquid chromatography coupled to tandem MS (UltiMate 3000 and Q Exactive Plus, Thermo Scientific). Peptides were sampled on a 300 µm × 5 mm PepMap C18 precolumn and separated on a 75 µm × 250 mm C18 column (ReproSil-Pur 120 C18-AQ 1.9 µm, Dr. Maisch GmbH) using a 120-min gradient. MS and MS/MS data were acquired using Xcalibur (Thermo Scientific).
Data were processed automatically using Mascot Distiller software (version 2.7.1.0, Matrix Science). Peptides and proteins were identified using Mascot (version 2.6) through concomitant searches against the F. novicida U112 database from MicroScope92, the classical contaminants database (homemade) and the corresponding reversed databases. The Proline software93 was used to filter the results: minimum peptide length of 7 amino acids, conservation of rank 1 peptides, peptide-spectrum match identification FDR < 1% as calculated on scores by employing the reverse database strategy, minimum peptide score of 25, and minimum of 1 specific peptide per identified protein group. Proline was then used to perform a compilation, grouping and MS1-based label-free quantification of the protein groups from the different samples.
Statistical analysis was performed using ProStaR94. Proteins identified in the reverse and contaminant databases, proteins identified with only 1 peptide, proteins identified by MS/MS in less than 3 replicates of one condition and proteins exhibiting less than 4 quantification values in one condition were discarded. After log2 transformation, abundance values were normalized using the variance stabilizing normalization procedure before missing value imputation (slsa method for POV and DetQuantile with quantile and factor set to 1 for MEC). Statistical testing was conducted using limma. Differentially expressed proteins were sorted out using a log2 (fold change) cut-off of ± 0.6 and P-values < 0.01 (FDR inferior to 2% according to the Benjamini–Hochberg estimator).
Macrophage culture, infection, and cytotoxicity assay
Murine macrophage J774 cells were grown in DMEM GlutaMAX supplemented with 10% fetal calf serum and 1% penicillin/streptomycin at 37 °C, 5% CO2. One day before infection with bacteria, confluent flask of cells was trypsinized and seeded into 96-well plates at a concentration of 1.5 × 104 cells/well and using antibiotic-free culture medium. The next day, host cells at 60–80% confluency were washed with 200 µL PBS and infected at a multiplicity of infection (MOI) of 100 with exponential growth phase bacteria. After centrifugation (350 × g; 5 min), the microtiter plate was incubated for 2 h at 37 °C, 5% CO2. The cell monolayer was then washed with PBS and remaining extracellular bacteria were killed by the addition of 10 μg/mL gentamicin (37 °C, 5% CO2) for 1 h. The antibiotic was removed by two washings with PBS and infected J774 incubated with the complete antibiotic-free culture medium. At a given time points after infection, cells were lysed by addition of 100 µL of Triton X-100 (0.5%) and the amount of viable bacteria was assessed though cfu counting from serial dilutions of lysed samples on PVX-CHA plates incubated at 37 °C for 24–48 h. Macrophage killing was measured using the CytTox 96 kit (Promega) following the manufacturer's instructions.
Measurement of intracellular ROS in infected macrophages
J774 cells seeded in a 96-well plate were infected as described above with the different F. novicida strains and using MOI 1,000:1 with 6 technical replicates per condition. Unifected macrophages were used as negative controls. After 24 h incubation at 37 °C the monolayers were washed twice with 100 µL PBS and incubated for 45 min with 20 µM DCFA reagent (Abcam) following the manufacturer’s instructions. The ROS were detected by fluorescence spectroscopy (Tecan Plate reader) using excitation and emission wavelengths of 485 and 535 nm, respectively.
Statistical analysis
All data correspond to at least 3 biological replicates. Otherwise indicated they were analyzed with Student’s t-tests and using the GraphPad PRISM software. The number of independent data points and P values are reported in figure legends.
Supplementary Information
Acknowledgements
Authors are very grateful to Thomas Henry for providing advices and plasmids for the construction of the F. novicida ΔldcF mutant and to Maighread Gallagher-Gambarelli for her advice on English language usage. This work has been supported by the FINOVI Foundation (Grant AO12-02) to PR and by the European Union’s Horizon 2020 research and innovation programme under grant agreement No 647784 to IG. This work used the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). Proteomic experiments were partly supported by ProFI (ANR-10-INBS-08-01 grant). J.F. was supported by a long-term EMBO fellowship (ALTF441-2017) and a Marie Curie fellowship (RespViRALI 789385). P.S.G. was supported by a PhD grant from ARC1 Santé Rhône-Alpes Auvergne. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
P.R., I.G., and Y.C. designed, conducted and supervised experiments. P.S.G. and C.B.A. performed the phylogenetic investigations and interpretation; J.F. and I.G. designed and performed structural investigations; J.N.D. and Y.C. were involved in proteomics; C.S., J.N. and P.R. were in charge of the microbiology aspects; A.F., K.H. and C.M. performed protein purifications and enzymatic assays; P.R. and I.G. analyzed and interpreted the whole of the data. P.R., I.G. and J.F. wrote the main manuscript text. P.S.G., C.B.A. and Y.C. performed manuscript editing. J.F., C.S. and P.S.G. created the figures. All authors commented on the manuscript and approved the final version.
Data availability
Crystallographic coordinates and structure factors for the crystal structure of F. novicida LdcF have been deposited in the wwPDB with accession code PDB: 6Y3X. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (PubMed ID: 30395289) partner repository with the dataset identifier PXD016669.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jan Felix and Claire Siebert.
Contributor Information
Irina Gutsche, Email: irina.gutsche@ibs.fr.
Patricia Renesto, Email: patricia.renesto@univ-grenoble-alpes.fr.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-79611-5.
References
- 1.Sjostedt A. Tularemia: History, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 2.Oyston PC, Sjostedt A, Titball RW. Tularaemia: Bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 3.Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 2014;4:35. doi: 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nano FE, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broms JE, Sjostedt A, Lavander M. The Role of the Francisella Tularensis pathogenicity island in Type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 2010;1:136. doi: 10.3389/fmicb.2010.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauriano CM, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guina T, et al. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. J. Bacteriol. 2007;189:6580–6586. doi: 10.1128/JB.00809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrench AP, et al. MglA/SspA complex interactions are modulated by inorganic polyphosphate. PLoS ONE. 2013;8:e76428. doi: 10.1371/journal.pone.0076428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog. 2009;5:e1000641. doi: 10.1371/journal.ppat.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honn M, Lindgren H, Sjostedt A. The role of MglA for adaptation to oxidative stress of Francisella tularensis LVS. BMC Microbiol. 2012;12:14. doi: 10.1186/1471-2180-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Nasr A, et al. Critical role for serum opsonins and complement receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in phagocytosis of Francisella tularensis by human dendritic cells (DC): uptake of Francisella leads to activation of immature DC and intracellular survival of the bacteria. J. Leukoc. Biol. 2006;80:774–786. doi: 10.1189/jlb.1205755. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz JT, et al. Natural IgM mediates complement-dependent uptake of Francisella tularensis by human neutrophils via complement receptors 1 and 3 in nonimmune serum. J. Immunol. 2012;189:3064–3077. doi: 10.4049/jimmunol.1200816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinkead LC, Allen LA. Multifaceted effects of Francisella tularensis on human neutrophil function and lifespan. Immunol. Rev. 2016;273:266–281. doi: 10.1111/imr.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steiner DJ, Furuya Y, Jordan MB, Metzger DW. Protective role for macrophages in respiratory Francisella tularensis infection. Infect. Immunity. 2017;85:e0064. doi: 10.1128/IAI.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honn M, Lindgren H, Bharath GK, Sjostedt A. Lack of OxyR and KatG results in extreme susceptibility of Francisella tularensis LVS to oxidative stress and marked attenuation in vivo. Front. Cell. Infect. Microbiol. 2017;7:14. doi: 10.3389/fcimb.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindgren H, et al. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immunity. 2007;75:1303–1309. doi: 10.1128/IAI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Z, et al. Elucidation of a mechanism of oxidative stress regulation in Francisella tularensis live vaccine strain. Mol. Microbiol. 2016;101:856–878. doi: 10.1111/mmi.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabadi SM, et al. Antioxidant defenses of Francisella tularensis modulate macrophage function and production of proinflammatory cytokines. J. Biol. Chem. 2016;291:5009–5021. doi: 10.1074/jbc.M115.681478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alharbi A, et al. Role of peroxiredoxin of the AhpC/TSA family in antioxidant defense mechanisms of Francisella tularensis. PLoS ONE. 2019;14:e0213699. doi: 10.1371/journal.pone.0213699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lofgren S, Tarnvik A, Thore M, Carlsson J. A wild and an attenuated strain of Francisella tularensis differ in susceptibility to hypochlorous acid: a possible explanation of their different handling by polymorphonuclear leukocytes. Infect Immunity. 1984;43:730–734. doi: 10.1128/IAI.43.2.730-734.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren H, et al. Iron content differs between Francisella tularensis subspecies tularensis and subspecies holarctica strains and correlates to their susceptibility to H(2)O(2)-induced killing. Infect. Immunity. 2011;79:1218–1224. doi: 10.1128/IAI.01116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss DS, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Champion MD, et al. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 2013;67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 26.Carriel D, et al. A novel subfamily of bacterial AAT-fold basic amino acid decarboxylases and functional characterization of its first representative: Pseudomonas aeruginosa LdcA. Genome Biol. Evol. 2018;10:3058–3075. doi: 10.1093/gbe/evy228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael AJ. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016;473:2315–2329. doi: 10.1042/BCJ20160185. [DOI] [PubMed] [Google Scholar]
- 28.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/MR.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabor H, Tabor CW. Spermidine, spermine, and related amines. Pharmacol. Rev. 1964;16:245–300. [PubMed] [Google Scholar]
- 30.Michael AJ. Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 2016;291:14896–14903. doi: 10.1074/jbc.R116.734780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanjee U, Houry WA. An assay for measuring the activity of Escherichia coli inducible lysine decarboxylase. J. Vis. Exp. 2010;46:e2094. doi: 10.3791/2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanjee U, et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Efficient production of enantiopure d-lysine from l-lysine by a two-enzyme cascade system. Catalysts. 2016;6:168. doi: 10.3390/catal6110168. [DOI] [Google Scholar]
- 34.Deng J, et al. Identification and molecular characterization of a metagenome-derived L-lysine decarboxylase gene from subtropical soil microorganisms. PLoS ONE. 2017;12:e0185060. doi: 10.1371/journal.pone.0185060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandiah E, et al. Structure, function, and evolution of the Pseudomonas aeruginosa lysine decarboxylase LdcA. Structure. 2019;27:1842–1854. doi: 10.1016/j.str.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Kandiah E, et al. Structural insights into the Escherichia coli lysine decarboxylases and molecular determinants of interaction with the AAA+ ATPase RavA. Sci. Rep. 2016;6:24601. doi: 10.1038/srep24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanjee U, Gutsche I, Ramachandran S, Houry WA. The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry. 2011;50:9388–9398. doi: 10.1021/bi201161k. [DOI] [PubMed] [Google Scholar]
- 38.Malet H, et al. Assembly principles of a unique cage formed by hexameric and decameric E. coli proteins. Elife. 2014;3:e03653. doi: 10.7554/eLife.03653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snider J, et al. Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J. Biol. Chem. 2006;281:1532–1546. doi: 10.1074/jbc.M511172200. [DOI] [PubMed] [Google Scholar]
- 40.Jessop M, et al. Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex. Commun. Biol. 2020;3:46. doi: 10.1038/s42003-020-0772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Bakkouri M, et al. Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc. Natl. Acad. Sci. USA. 2010;107:22499–22504. doi: 10.1073/pnas.1009092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J. Biol. Chem. 2010;285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohinai Z, et al. Bacterial lysine decarboxylase influences human dental biofilm lysine content, biofilm accumulation, and subclinical gingival inflammation. J. Periodontol. 2012;83:1048–1056. doi: 10.1902/jop.2011.110474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karatan E, Michael AJ. A wider role for polyamines in biofilm formation. Biotechnol. Lett. 2013;35:1715–1717. doi: 10.1007/s10529-013-1286-3. [DOI] [PubMed] [Google Scholar]
- 45.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 46.Brauner A, Shoresh N, Fridman O, Balaban NQ. An experimental framework for quantifying bacterial tolerance. Biophys. J. 2017;112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barry AL, Craig WA, Nadler H, Reller LB, Sanders CC, Swenson JM. Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline. Wayne: Clinical and Laboratory Standards Institute; 1999. [Google Scholar]
- 48.Dai S, Mohapatra NP, Schlesinger LS, Gunn JS. Regulation of Francisella tularensis virulence. Front. Microbiol. 2010;1:144. doi: 10.3389/fmicb.2010.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morita R, et al. Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems. J. Nucleic Acids. 2010;2010:179594. doi: 10.4061/2010/179594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn WC, et al. Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision. Nat. Chem. Biol. 2019;15:607–614. doi: 10.1038/s41589-019-0289-3. [DOI] [PubMed] [Google Scholar]
- 52.Gallagher LA, McKevitt M, Ramage ER, Manoil C. Genetic dissection of the Francisella novicida restriction barrier. J. Bacteriol. 2008;190:7830–7837. doi: 10.1128/JB.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moule MG, Monack DM, Schneider DS. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 2010;6:e1001065. doi: 10.1371/journal.ppat.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soballe B, Poole RK. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology. 2000;146(Pt 4):787–796. doi: 10.1099/00221287-146-4-787. [DOI] [PubMed] [Google Scholar]
- 55.Brameyer S, et al. DNA-binding directs the localization of a membrane-integrated receptor of the ToxR family. Commun. Biol. 2019;2:4. doi: 10.1038/s42003-018-0248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS., Jr Revisiting the Gram-negative lipoprotein paradigm. J. Bacteriol. 2015;197:1705–1715. doi: 10.1128/JB.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wehrly TD, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 2009;11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chattopadhyay MK, Tabor CW, Tabor H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U S A. 2003;100:2261–2265. doi: 10.1073/pnas.2627990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JS, Choi SH, Lee JK. Lysine decarboxylase expression by Vibrio vulnificus is induced by SoxR in response to superoxide stress. J. Bacteriol. 2006;188:8586–8592. doi: 10.1128/JB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang IH, Kim JS, Kim EJ, Lee JK. Cadaverine protects Vibrio vulnificus from superoxide stress. J. Microbiol. Biotechnol. 2007;17:176–179. [PubMed] [Google Scholar]
- 61.Magnet A, et al. Vectorial role of Acanthamoeba in Legionella propagation in water for human use. Sci. Total Environ. 2015;505:889–895. doi: 10.1016/j.scitotenv.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 62.Bakshi CS, et al. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 2006;188:6443–6448. doi: 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melillo AA, et al. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J. Bacteriol. 2009;191:6447–6456. doi: 10.1128/JB.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma Z, et al. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol. Microbiol. 2014;91:976–995. doi: 10.1111/mmi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olin-Sandoval V, et al. Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature. 2019;572:249–253. doi: 10.1038/s41586-019-1442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008;68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x. [DOI] [PubMed] [Google Scholar]
- 67.Andersson H, et al. Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J. Med. Microbiol. 2006;55:263–271. doi: 10.1099/jmm.0.46313-0. [DOI] [PubMed] [Google Scholar]
- 68.Carlson PE, Jr, et al. Global transcriptional response to spermine, a component of the intramacrophage environment, reveals regulation of Francisella gene expression through insertion sequence elements. J. Bacteriol. 2009;191:6855–6864. doi: 10.1128/JB.00995-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poso H, McCann PP, Tanskanen R, Bey P, Sjoerdsma A. Inhibition of growth of Mycoplasma dispar by DL-alpha-difluoromethyllsine, a selective irreversible inhibitor of lysine decarboxylase, and reversal by cadaverine (1,5-diaminopentane) Biochem. Biophys. Res. Commun. 1984;125:205–210. doi: 10.1016/S0006-291X(84)80355-1. [DOI] [PubMed] [Google Scholar]
- 70.Berkowitz DB, Jahng WJ, Pedersen ML. Alpha-vinyllysine and alpha-vilylarginine are time-dependent inhibitors of their cognate decarboxylases. Bioorg. Med. Chem. Lett. 1996;6:2151–2156. doi: 10.1016/0960-894X(96)00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karukurichi KR, de la Salud-Bea R, Jahng WJ, Berkowitz DB. Examination of the new alpha-(2'Z-fluoro)vinyl trigger with lysine decarboxylase: The absolute stereochemistry dictates the reaction course. J. Am. Chem. Soc. 2007;129:258–259. doi: 10.1021/ja067240k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCune CD, et al. Synthesis and deployment of an elusive fluorovinyl cation equivalent: access to quaternary alpha-(1'-Fluoro)vinyl amino acids as potential PLP enzyme inactivators. J. Am. Chem. Soc. 2017;139:14077–14089. doi: 10.1021/jacs.7b04690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsson A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia PS, Jauffrit F, Grangeasse C, Brochier-Armanet C. GeneSpy, a user-friendly and flexible genomic context visualizer. Bioinformatics. 2019;35:329–331. doi: 10.1093/bioinformatics/bty459. [DOI] [PubMed] [Google Scholar]
- 80.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 81.Kabsch W. Xds. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D. 2004;60:1229–1236. doi: 10.1107/S0907444904010145. [DOI] [PubMed] [Google Scholar]
- 85.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lauriano CM, Barker JR, Nano FE, Arulanandam BP, Klose KE. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 2003;229:195–202. doi: 10.1016/S0378-1097(03)00820-6. [DOI] [PubMed] [Google Scholar]
- 87.Maier TM, et al. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pettit RK, et al. Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob. Agents Chemother. 2005;49:2612–2617. doi: 10.1128/AAC.49.7.2612-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siebert C, et al. Francisella tularensis: FupA mutation contributes to fluoroquinolone resistance by increasing vesicle secretion and biofilm formation. Emerg. Microbes Infect. 2019;8:808–822. doi: 10.1080/22221751.2019.1615848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siebert C, et al. Francisella novicida and F. philomiragia biofilm features conditionning fitness in spring water and in presence of antibiotics. PLoS ONE. 2020;15:e0228591. doi: 10.1371/journal.pone.0228591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Casabona MG, Vandenbrouck Y, Attree I, Coute Y. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics. 2013;13:2419–2423. doi: 10.1002/pmic.201200565. [DOI] [PubMed] [Google Scholar]
- 92.Vallenet D, et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2019;48:D579–D589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bouyssie D, et al. Proline: An efficient and user-friendly software suite for large-scale proteomics. Bioinformatics. 2020;36:3148–3155. doi: 10.1093/bioinformatics/btaa118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wieczorek S, et al. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics. 2017;33:135–136. doi: 10.1093/bioinformatics/btw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic coordinates and structure factors for the crystal structure of F. novicida LdcF have been deposited in the wwPDB with accession code PDB: 6Y3X. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (PubMed ID: 30395289) partner repository with the dataset identifier PXD016669.