Abstract
Inherited retinal diseases (IRDs) were first classified clinically by history, ophthalmoscopic appearance, type of visual field defects, and electroretinography (ERG). ERGs isolating the two major photoreceptor types (rods and cones) showed some IRDs with greater cone than rod retinal dysfunction; others were the opposite. Within the cone-rod diseases, there can be phenotypic variability, which can be attributed to genetic heterogeneity and the variety of visual function mechanisms that are disrupted. Most cause symptoms from childhood or adolescence, although others can manifest later in life. Among the causative genes for cone-rod dystrophy (CORD) are those encoding molecules in phototransduction cascade activation and recovery processes, photoreceptor outer segment structure, the visual cycle and photoreceptor development. We review 11 genes known to cause cone-rod disease in the context of their roles in normal visual function and retinal structure. Knowledge of the pathobiology of these genetic diseases is beginning to pave paths to therapy.
Keywords: Genotype, Phenotype, Photoreceptor, Retina, Vision
1. Introduction
1.1. Human photoreceptor types and topography
The process of vision in the human eye begins with the absorption of light by visual pigments in the photoreceptor outer segments, extensions from the rod and cone cells. Rods are the most numerous with 100 million in each eye while cones are far less numerous at about 6 million [1]. The micro-anatomy and physiology of these different photoreceptor types have been studied extensively as have their topography in the human adult retina (Fig. 1a,b) [1]. Researching these cells and their connectivity within the retina and into the brain has occupied scientists for more than a century and the pursuit has more recently led to discoveries of the genes underlying human vision [2], [3], [4], [5]. This progress has now advanced to the point where basic science discovery has provided the opportunity to identify the causes of human inherited retinal diseases (IRDs) [6]. Although there is interdependence of rods and cones and their pathways, this review focuses on the genetic diseases that begin in early life with more cone than rod involvement.
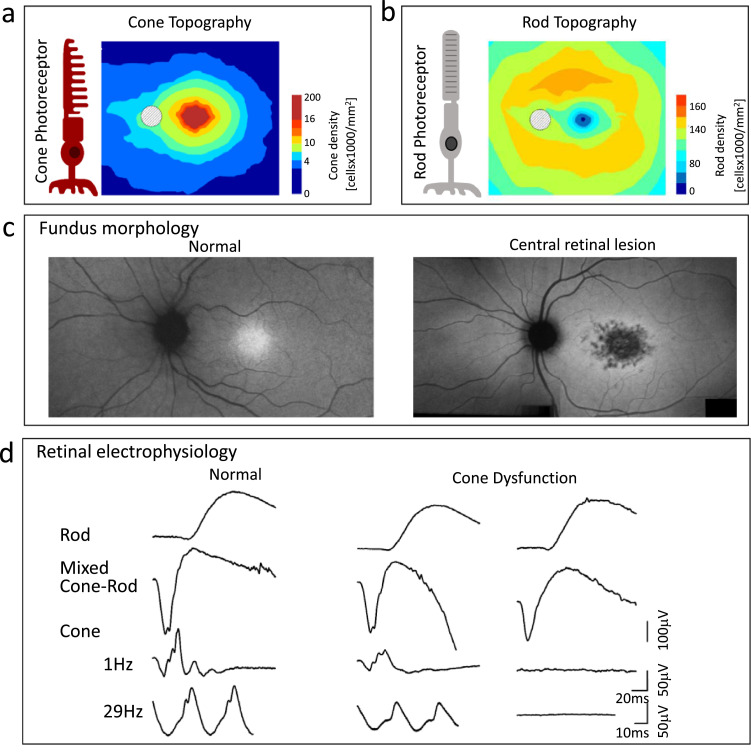
Fig. 1.
Photoreceptor topography and CORD clinical diagnostics. (a) Cone photoreceptor diagram and cone ONL thickness topography based on photoreceptor density map [1]. (b) Rod diagram and rod ONL thickness topography based on photoreceptor density map [1]. (c) En face near-infrared autofluorescence image showing normal fundus appearance (left), and central retinal lesion in a patient with ABCA4-retinopathy (right). (d) Retinal electrophysiology, specifically standard rod and cone ERGs, in a normal subject (left) and two patients with normal rod but reduced (middle) or non-detectable (right) cone signals. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
1.2. Cone-rod photoreceptor diseases – making the diagnosis
Phenotype by fundus morphology and vision: A report of a ‘peculiar form of retinitis pigmentosa’ (RP) 150 years ago has been considered an early description of a cone dystrophy (COD) [7]. The 35-year-old female patient had symmetrical pigmentary lesions in the macula, reduced vision to counting fingers, central scotomas, eccentric fixation, color vision complaints, no night vision symptoms and no constriction of the visual field which was ‘peculiar’ because these symptoms differed from other reports of RP [8,9]. In fact, the ‘peculiar form of RP’ was not RP (a misnomer for IRDs that tend to affect rod more than cone photoreceptors and show pigmentary disturbances in the fundus) but a retinal disease that negatively affected the macula with its high concentration of cone photoreceptors leading to cell loss or dysfunction and decreased visual acuity and color vision, the province of cones and cone pathways (Fig. 1c).
Phenotype by retinal electrophysiology: Although the funduscopic examination (made possible by the invention of the ophthalmoscope [10]) and a detailed history of visual symptoms are first steps toward diagnosis of a retinal disease, ancillary tests of vision and retinal function later served to refine or extend the diagnosis before molecular diagnostics of IRDs became possible. Specifically, the full-field electroretinogram (ERG), when recorded under conditions to isolate rod- and cone-mediated function, shows a normal or subnormal rod signal and a more reduced cone signal (Fig. 1d) [11,12]. Within the spectrum of diseases that mainly show cone dysfunction or degeneration (COD), there are those that progress to negatively affect rod function and structure as well. The nomenclature then becomes cone-rod dystrophy or degeneration (CORD). The phenotypic diversity by retinal function testing in CORD led to the hypothesis that the different disease patterns likely represented different molecular genetic causes and mechanisms and this was confirmed as we entered the era of gene discovery [11].
1.3. Cone-rod diseases – Mendelian genetic types for these phenotypes
All inheritance patterns are represented among the cone and cone-rod diseases. Currently, there are at least 30 genes associated with the clinical diagnosis of COD and CORD. X-linked inheritance is rare. Autosomal recessive inheritance accounts for 2/3 of these genes and close to 1/3 are autosomal dominantly inherited [13]. The association of COD and CORD phenotypes with a number of different genes provides an opportunity to try to understand not only the genetic heterogeneity but also the underlying disease mechanisms. Of the many genes known to be associated with COD and CORD, [13], we selected a subgroup of 11 genes to present in this review. These fit within four proposed disease pathways or categories. Structural and functional features of the disease expression in patients are illustrated for the 11 genetic diseases. Optical coherence tomography (OCT) cross-sections of the central retina through the fovea showed laminar abnormalities associated with various stages of the diseases; the histological laminar boundaries in relation to OCT have been well studied [14]. Rod and cone perimetry maps were used to illustrate the loss of visual sensitivity relative to normal mean data at each of 72 loci across the visual field.
2. Genes and disease mechanisms
2.1. Phototransduction pathway abnormalities
Mutations in genes that affect function of photoreceptors, such as those involved in the phototransduction cascade, are among the causes of COD and CORD. A generalised overview of the phototransduction cascade is given (Fig. 2a). Although rods and cones differ slightly in this process, including having rod- or cone-specific proteins or different concentrations of some of these proteins, there is general similarity [15,16].
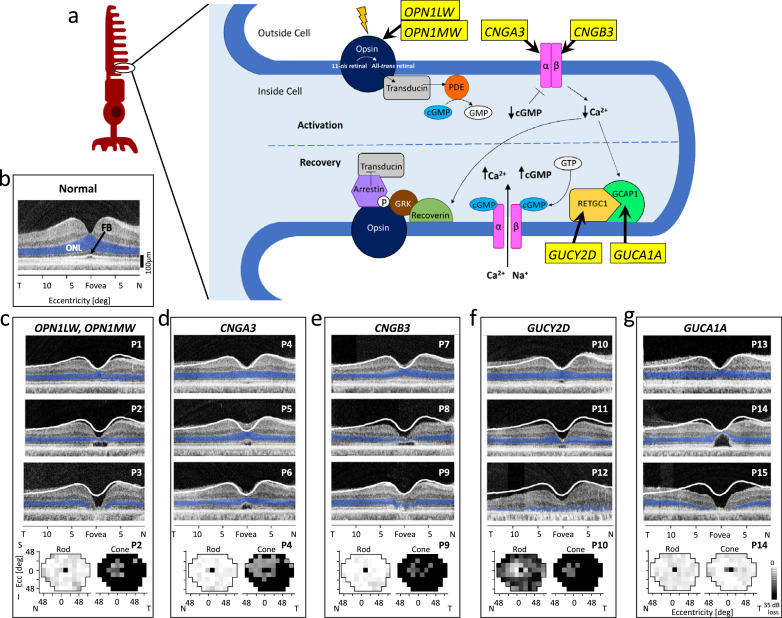
Fig. 2.
Phototransduction activation and recovery. (a) Overview of the normal phototransduction pathway occurring in photoreceptor OS disks. Key proteins are depicted, and associated genes that are discussed in this review are given in yellow boxes. A photon of light hits the opsin molecule and 11-cis-retinal is converted to all-trans retinal. Transducin is activated, which in turn activates a phosphodiesterase (PDE) which hydrolyzes cGMP. Reduction in intracellular cGMP results in closure of cGMP-gated ion channels, and subsequent reduction of intracellular Ca2+ culminates in the hyperpolarization of the photoreceptor. Low intracellular Ca2+ promotes recovery in two ways. Reduced Ca2+levels cause GCAP1 to activate RETGC1 to synthesize cGMP. Increased levels of cGMP cause cGMP-gated ion channels to reopen and the cell to become depolarized. Low Ca2+ levels also facilitate activation of recoverin, which allows G-protein coupled receptor kinases (GRK) to phosphorylate (P) the activated opsin. Arrestin then binds to the phosphorylated opsin, blocking its ability to further activate transducin, turning off the transduction. (b) OCT across the horizontal meridian through the fovea with highlighted (blue) ONL in a normal subject. FB, foveal bulge. (c-g) OCTs of patients representing each of the 5 genotypes causing abnormalities in phototransduction activation and recovery. White lines represent the lower limit of normal retinal thickness (−2 standard deviations from mean); ONL is highlighted in blue. At the bottom of each panel are rod (left) and cone (right) perimetry maps for patients with the genotype. N, nasal; T, temporal. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.1.1. Cone opsin mutations
OPN1LW and OPN1MW encode red- and green-cone opsins, respectively. The phototransduction cascade is initiated upon photoexcitation of the opsin by conversion of 11-cis retinal to all-trans retinal. OPN1LW and OPN1MW are located on the X chromosome and are configured in a head-to-tail tandem array consisting of a single OPN1LW gene at the 5′ end, followed by at least one OPN1MW gene [17]. A locus control region (LCR) upstream of OPN1LW governs opsin gene expression and the identity of the red-sensitive or green-sensitive cone types [18,19]. Mutations in OPN1LW and OPN1MW cause X-linked vision disorders such as Blue Cone Monochromacy (BCM), which affects approximately 1 in 100,000 individuals from birth [20,21].
Clinical examples of BCM (Fig. 2c): A normal OCT with highlighted outer nuclear layer (ONL; blue) and arrow pointing to foveal bulge (FB), representing foveal cone outer segments (COS), is shown for comparison to patient scans (Fig. 2b). BCM patients have congenitally impaired visual acuity, abnormal color vision, photosensitivity, nystagmus, and often myopia [21], [22], [23] OCTs from three BCM patients, P1 (age 12), P2 (age 51) and P3 (age 50), show different degrees of central retinal abnormalities (top to bottom) from near normal foveal outer nuclear layer (ONL) thickness but reduced COS, to thinned foveal ONL with cavitation at inner/outer segment (IS/OS) level, to loss of ONL at the fovea with atrophy. Rod and cone sensitivity maps for P2 show rod function within normal limits but severely reduced cone function.
2.1.2. Cone cyclic nucleotide-gated (CNG) channelopathies
The cone-specific cyclic guanosine monophosphate (cGMP)-gated cation channels are localised to the plasma membrane of the COS. They are tetramers composed of three α- and one β-subunit, with each subunit containing six transmembrane segments and a cytoplasmic cyclic nucleotide-binding domain [15,24]. The complex forms a pore in the plasma membrane through which cations, including Ca2+, can traverse (Fig. 2a). They are open when bound to cGMP in the dark state, and close upon initiation of the phototransduction cascade when cGMP levels decrease. During recovery, channels reopen when cGMP is replenished by activation of retinal membrane guanylyl cyclase (RETGC1). Mutations that affect cone CNG channels can result in dysregulation of cGMP and Ca2+ in cells, and accumulation of these molecules can cause cell death [25].
CNGA3: CNGA3 encodes the α-subunit of the cone CNG cation channel. Disease-causing mutations are autosomal recessive and can cause COD, CORD or achromatopsia (ACHM) [16,26,27]. Many are missense mutations that may cause altered function, loss of function or interfere with trafficking of the α-subunit [15,20,21,27–29].
CNGB3: CNGB3 encodes the β-subunit of the cone CNG cation channel. Mutations in CNGB3 cause >50% of ACHM in western populations, and together, CNGB3 and CNGA3 account for >75% of ACHM [15,21,27]. Most CNGB3 mutations are nonsense, insertions, deletions, or splice site mutations that produce truncated subunits or which can affect channel formation and function and may result in cell death [15,20,21].
Clinical Examples of Channelopathies (Fig. 2d,e): ACHM is characterized by congenital loss of cone function resulting in reduced visual acuity, color blindness, photosensitivity, and nystagmus. Of interest, mutations in GNAT2, PDE6H, PDE6C and ATF6 also can cause ACHM [21,27,30]. OCTs from three CNGA3 patients P4 (age 18), P5 (age 28) and P6 (age 32) show a spectrum of foveal structure abnormalities: near normal ONL thickness, albeit with reduced COS (P4); reduced central ONL with IS/OS cavitation lesion (P5); and a more extensive cavitation lesion (P6) (Fig. 2d). P4 has normal rod sensitivity but reduced cone sensitivities across the field (right map); central-nasal field loci with moderate versus severe cone sensitivity loss suggest that this patient may exemplify an incomplete form of ACHM [28,30]. Three CNGB3 patients, P7 (age 13), P8 (age 65), and P9 (age 18) also show a range of structural changes: normal ONL thickness but reduced COS (P7); loss of foveal ONL thickness (and central retinal thickness) with a cavitation lesion (P8); and an atrophic lesion with barely detectable foveal ONL (P9) (Fig. 2e). P9 has a normal rod visual field but severely reduced cone function.
2.1.3. Phototransduction recovery kinetics
GUCY2D: GUCY2D encodes RETGC1, which is responsible for synthesizing cGMP (Fig. 2a). It is expressed primarily in the OS of both rods and cones, and at a higher concentration in cones, which may in part explain the effect of mutations in this gene on cones relative to rods [31]. GUCY2D mutations cause autosomal dominant COD or CORD and autosomal recessive Leber Congenital Amaurosis (LCA). The majority are missense mutations. Missense mutations in the dimerization domain of RETGC1, particularly the arginine at position 838, are a major cause of dominant GUCY2D-CORD. Such mutations result in a shift in the calcium sensitivity of the cGMP synthesis activity. Activation of RETGC1 at higher than normal levels of intracellular Ca2+ causes overproduction of cGMP and results in photoreceptor death [31]. This differs from mutations that cause LCA, which inhibit the ability of RETGC1 to synthesize cGMP [31].
GUCA1A: GUCA1A encodes guanylyl cyclase-activating protein (GCAP1), the intracellular calcium sensor that regulates the activity of RETGC1. In the dark, Ca2+is bound to the three active-metal binding sites in GCAP1, and Mg2+ replaces Ca2+ during phototransduction when levels of intracellular Ca2+ decline. This induces a conformational change in GCAP1 that activates RETGC1 to produce cGMP, facilitating photoreceptor recovery (Fig. 2a) [31,32]. Point mutations in GUCA1A can cause dominant COD, CORD and macular dystrophy [31]. Mutations such as the p.Gly68Arg variant lead to a shift in sensitivity to Ca2+such that RETGC1 is active at higher Ca2+ levels, causing overproduction of cGMP which can lead to cell death [31,33,34].
Clinical Presentation of autosomal dominant GUCY2D and GUCA1A (Fig. 2f,g): P10, A 26-yr-old GUCY2D patient shows normal ONL thickness across the central retina but with a small IS/OS defect at the fovea, while P11 (49 years) has thinned ONL and a slightly greater extent of IS/OS cavitation. Severe disease is illustrated by P12 (27 years) who has detectable but very limited ONL and no IS/OS lamination. P10’s rod map has a central visual area of relatively preserved function (correlating with the OCT result) while the cone map, with its limited central function has no measurable sensitivity in the periphery. GUCA1A disease shows some similarities and differences to the GUCY2D examples. P13 (16 years) retains central retinal ONL but definitely reduced thickness of IS/OS lamination. Thinned foveal ONL and a large cavitation of IS/OS abnormality are evident in P14 (35 years), but extracentral ONL has only moderate thinning. Foveal ONL loss is prominent in P15 (62 years). In contrast to the major dysfunction of the GUCY2D patient, P14’s rod and cone maps are within normal limits except for the reduced foveal cone function (correlating with foveal OCT results).
2.2. Photoreceptor outer segment structure – formation and maintenance
Genes integral to OS formation and maintenance are also implicated in COD and CORD: PROM1, CDHR1 and PRPH2.
PROM1: PROM1 encodes Prominin-1, a transmembrane glycoprotein composed of five transmembrane domains and two highly glycosylated extracellular loops. It is expressed in photoreceptors predominantly at the base of the outer segments in the protrusions of disk membranes (Fig. 3a) [35], [36], [37]. Prominin-1 interacts with cadherin related family member 1 (CDHR1) and actin to promote normal photoreceptor disk morphology [38]. Murine studies indicate that Prom1 mutations cause disruption of photoreceptor disk morphogenesis and mislocalisation of visual pigments that interfere with phototransduction and result in photoreceptor degeneration [38,39]. Zebrafish studies suggest that cones may be more severely affected than rods since mutations in Prom1 affected peripherin 2 (Prph2) localization and oligomerization, and Prph2 is expressed at higher levels in cones [40]. The majority of reported mutations in PROM1 cause autosomal recessive disease, but specific variants (p.Arg373Cys, p.Asp829Asn and p.Leu245Pro) cause dominant disease. Mutations result in a variety of phenotypes including CORD and bull's-eye maculopathy [43]. Autosomal recessive truncating, splice site and missense variants were postulated to result in total loss of function and were associated with a more severe phenotype with an earlier age of onset and a retina-wide effect whereas patients with an autosomal dominant variant showed a less severe later onset phenotype with primarily macular involvement [41].
Fig. 3.
Photoreceptor outer segment structure. (a) Structural elements of the photoreceptor OS are depicted with associated genes given in yellow boxes. (b-d) OCTs of 2 patients representing each of the 3 genotypes causing abnormalities in photoreceptor OS structure. ONL is highlighted in blue; lower limit of normal retinal thickness is demarcated by white lines. At the bottom of each panel are rod (left) and cone (right) perimetry maps for a patient with the genotype. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Clinical Examples of PROM1-related Disease (Fig. 3b): P16, a 5-yr-old with dominant PROM1 has near normal ONL thickness across the central retina but abnormal IS/OS lamination, consistent with his reduced visual acuity. An autosomal recessive PROM1-CORD patient (P17, age 33) shows little or no detectable ONL across the central retina and features of inner retinal thickening, the result of retinal remodeling secondary to photoreceptor loss [42]. Visual field maps of P17 (at the earlier age of 17) showed abnormal paracentral and central rod and cone function and no detectable peripheral sensitivities.
CDHR1: CDHR1 protein is a photoreceptor-specific protocadherin responsible for OS morphogenesis [36]. CDHR1 is localised to the base of rod and cone OS (Fig. 3a) [36,43]. In mice, the absence of CDHR1 expression resulted in disorganized OS and progressive photoreceptor death [43]. Cleavage of the six extracellular domains from the transmembrane and intracellular domains is an important process in OS assembly, and direct interaction with PROM1 ensures proper morphogenesis of OS disks [38,44]. Mutations in CDHR1 are associated with recessive CORD or COD [36,45]. There is genotypic and phenotypic heterogeneity, but macular involvement is common in CDHR1 mutations [45].
Clinical Examples of CDHR1 CORD (Fig. 3c): Siblings with CDHR1 mutations were clinically diagnosed as CORD in their 3rd decade of life. By ages 45 (P18) and 38 (P19), visual loss was severe and there was no detectable ONL by OCT and inner retinal thickening indicating retinal remodeling. There was no measurable rod and cone function by visual field maps in P19 at age 39.
PRPH2: PRPH2 (previously known as RDS) encodes peripherin 2 (PRPH2), a photoreceptor-specific tetraspanin membrane protein vital to OS disk morphogenesis, stability and maintenance in rods and cones [36,37]. The protein is composed of four transmembrane domains, two intradiscal loops, the D1 and D2 loops, and cytoplasmic N and C termini. Peripherin 2 is localised to the rims of photoreceptor disks (Fig. 3a) [36]. PRPH2 disease has genetic and phenotypic heterogeneity [36,37,46]. In CORD patients, mutations are most often associated with autosomal dominant inheritance [36,46]. Although genotype-phenotype correlations have been elusive, the majority of PRPH2-CORD patients have mutations in exon 1, and the D2 loop of the protein, important for mediating protein-protein interactions, is a mutational hotspot in exon 1. Additionally, p.Arg172Trp, a common mutation in PRPH2 that can cause a disease with early cone involvement, is generally associated with earlier age of onset compared to other PRPH2 variants [46].
Clinical Examples of PRPH2 Maculopathy (Fig. 3d): P20, a 34-yr-old with pattern dystrophy had central visual distortion but otherwise normal vision; OCT showed a subfoveal deposit but no other abnormalities. The patient's mother had a similar history and fundus appearance. A 39-yr-old with complaints of distortion (P21) had an initial clinical diagnosis of macular degeneration; a PRPH2 mutation was later identified. Other family members had been diagnosed as having macular degeneration. At age 51, P21 showed subfoveal deposits as well as reduced ONL thickness in regions of the macula but not at the fovea. P22 (55 years) had progressive reduction in visual acuity over previous decades but no clinically apparent macular degenerative disease (no OCT shown); rod and cone visual field maps were within normal limits.
2.3. Visual cycle
Mutations in ABCA4, critical to the visual cycle, cause cone-rod disease. ABCA4 is a 50-exon gene that encodes a 2273 amino acid ATP-binding cassette transporter protein primarily expressed in rod and cone OS's. ABCA4 is localised to the rim regions of the OS disk membranes. The main function of ABCA4 is the transport of N-retinylidene-PEs (N-ret-PEs) from the intradiscal side of the disk membrane to the cytoplasmic side to facilitate normal metabolism of all-trans retinal and 11-cis retinal. ABCA4’s role in the visual cycle is in transporting all-trans retinal bound to PE to the cytoplasmic side where it is acted upon by retinol dehydrogenase (RDH) to enter the photopigment regeneration cycle (Fig. 4a) [47].
Fig. 4.
Visual cycle and ABCA4 retinopathy. (a) Schematic of a photoreceptor OS disk and location of ABCA4 molecules. Normal function of ABCA4 in the visual cycle is shown. 11-cis retinal is isomerized to all-trans retinal which dissociates from opsin. A portion of the all-trans retinal can diffuse from the inner disk membrane to the cytoplasmic side of the membrane, and the remainder binds to phosphatidylethanolamine (PE) to form N-trans-retinylidene-PE (N-t-R-PE) in the inner leaflet of the disk membrane. ABCA4 flips the N-t-R-PE from the intradiscal side of the membrane to the cytoplasmic side where PE then dissociates from all-trans retinal, and all-trans retinal can enter the visual cycle pathway. (b) Ultra-wide-angle en face near-infrared (NIR) autofluorescence image of right eye of a 21-yr-old ABCA4-retinopathy patient (P23); inset (defined on larger image by black box): short-wavelength (SW) autofluorescence image showing lipofuscin accumulation. (c) Upper 3 panels: OCTs of 3 patients with ABCA4 mutations illustrating (top to bottom) different degrees of central photoreceptor loss. ONL is highlighted in blue, white lines represent lower limit of normal retinal thickness. Lower: Rod (left) and cone (right) perimetry maps for 2 ABCA4 patients. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
ABCA4 mutations cause mislocalised, impaired or nonfunctional protein which impedes or inhibits clearance of excess N-ret-PEs from photoreceptor disks [47]. N-Ret-PEs form cytotoxic bisretinoids that accumulate in the disks and are subsequently converted to N-retinylidene-N-retinylethanolamine (A2E), a component of lipofuscin, in the RPE upon phagocytosis of OS tips. This process results in aggregation of lipofuscin in the RPE and causes the atrophy of the RPE and secondary photoreceptor cell death [47], [48], [49]. Cone photoreceptors may be uniquely vulnerable to primary cytotoxicity from buildup of bisretinoids in OS's when ABCA4 is nonfunctional [50]. Mutations in ABCA4, well-known as the cause of the early-onset recessive maculopathy Stargardt disease, is also estimated to cause ~60% of autosomal recessive CORD [16,51,52].
Clinical Examples of ABCA4 Retinopathy (Fig. 4b,c): An ultra-wide near-infrared autofluorescence image of P23, a 21-yr-old with ABCA4 mutations (causing Stargardt disease) shows that the RPE disease distribution extends from the central retina into surrounding extramacular retina [53]. The inset, from the macula, uses short-wavelength autofluoresence to capture the lipofuscin accumulation characteristic of ABCA4-retinopathy. This patient had reduced acuities, a central scotoma and rod and cone sensitivities within normal limits in the peripheral field. ABCA4-CORD examples are also shown (Fig. 4c). P24, a 15-yr-old with photosensitivity and reduced acuity, had borderline normal rod and cone ERGs but macular pigmentary changes and slightly attenuated retinal vessels. OCT indicated a normal photoreceptor nuclear layer but central retinal IS/OS lamination abnormalities. P25 at 9 years of age had a history of failed school vision tests due to reduced acuities, photosensitivity, a normal rod but severely reduced cone ERG, and a bull's eye appearance in the macula. OCT at age 14 had reduced retinal thickness, no detectable foveal ONL and only limited ONL in the extracentral retina scanned. At age 17, vision maps in this patient showed about 1 log unit of rod sensitivity loss with greater losses more centrally; central and peripheral cone losses were pronounced, but there was a residual midperipheral island of impaired cone function. P26, a 17-yr-old, had longstanding reduced acuity, normal rod signals but slightly reduced cone ERGs, and OCT with total retinal thickness loss and barely detectable central retinal ONL. Rod and cone vision maps revealed a large central scotoma surrounded by reduced sensitivities into the periphery.
2.4. Photoreceptor development
The gene regulatory networks that mediate cellular differentiation in the retina are complex. We offer a simplified diagram of the interactions of some of the key regulators of photoreceptor cell fate in the vertebrate retina (Fig. 5a); mutations in these developmental genes can cause inherited photoreceptor diseases. Two of these genes, CRX and NR2E3, are discussed as they relate to the very different human diseases of cones that are associated with them.
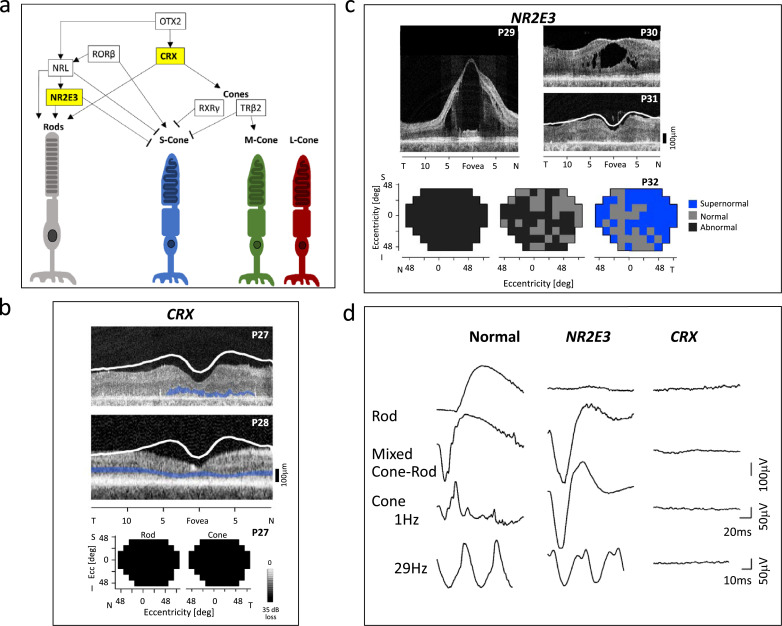
Fig. 5.
Photoreceptor development. (a) A simplified depiction of the regulatory network that governs photoreceptor differentiation. OTX2, CRX and RORβ act to promote both rod and cone differentiation. NRL and NR2E3 promote rod-specific gene expression and NR2E3 also suppresses S-cone-specific gene expression. TRβ2 promotes M-cone-specific gene expression and works with RXRγ to suppress S-cone-specific gene expression [54,55,58]. Yellow boxes indicate the 2 development genes discussed in this review. (b) OCTs of 2 patients with a CRX mutation causing LCA (P27) and dominant CORD (P28) and maps showing no measurable rod and cone function in the LCA patient. ONL is highlighted in blue; lower limit of normal retinal thickness is marked with the white line. (c) OCTs across the horizontal meridian through the fovea of 3 patients with NR2E3 mutations. Maps of the visual field of an NR2E3 patient illustrating abnormal rod function (left), abnormal and normal loci of L/M cone function (middle), and supernormal and normal loci of S-cone function. (d) Comparison of ERGs in a normal subject, an NR2E3 patient and a CRX patient. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
CRX: The cone-rod homeobox protein (CRX) plays an important role in promoting photoreceptor cell fate and regulates expression of both rod- and cone-specific genes [54,55]. Missense mutations primarily affect the homeobox domain responsible for binding DNA whereas frameshift and nonsense mutations are clustered in the last exon and are predicted to affect the domains involved in transcriptional regulation; mutations cause variable effects on DNA binding ability and CRX function [56]. Mutations in CRX can cause, for example, dominant CORD and dominant LCA [57].
Clinical Examples of CRX LCA and CORD (Fig. 5b,d): P27, a 7-yr-old patient with nystagmus, strabismus, and severely impaired vision noted from 2 months of age, was clinically diagnosed with LCA; an ERG at this age showed a small waveform to a bright flash of light but no other detectable signals (Fig. 5d, waveforms at right). A molecular diagnosis subsequently revealed a heterozygous CRX mutation. The OCT at age 30 showed reduced retinal thickness and very limited ONL and outer retinal lamination; increased inner retinal thickness indicated retinal remodeling. There was no measurable rod and cone function across the visual field. P28, a 28-yr-old patient with dominant CORD, had longstanding visual complaints (mainly central) but retained measurable acuities and peripheral visual fields. OCT at this age had reduced retinal thickness, and ONL was detectable across the central retina.
NR2E3: NR2E3 is an orphan nuclear receptor which has dual functions of promoting expression of rod-specific genes and suppressing S-cone-specific genes. CRX and NRL interact to induce its expression during development [54,55,59]. Mutations are commonly found in the DNA-binding domains or ligand-binding domains. The initial evidence leading to understanding of the role of NR2E3 in photoreceptor development came from the clinical observation that an autosomal recessive retinopathy was a unique disorder characterized by nightblindness early in life, reduced L/M cone function, cystic maculopathy, and a surprising excess of S-cone function and structure [59], [60], [61], [62], [63], [64], [65], [66], [67]. The ‘enhanced S-cone syndrome’ (ESCS), like other genetic cone diseases in this review, has reduced L/M cone cells and dysfunction. Unlike any other retinopathy, however, ESCS-NR2E3 has increased function and structure of the minority S-cone photoreceptor.
Clinical Examples of NR2E3 Retinopathy (Fig. 5c,d): The ERG of an NR2E3 patient (Fig. 5d, waveforms in middle column) differs from normal in that the dark-adapted blue flash stimulus that elicits a rod signal in a normal retina produced no signal. Also, in contrast to the normal mixed cone-rod signal from a bright white flash, there is a large negative waveform in NR2E3. Unlike the normal cone system ERG (mainly L/M receptors) elicited with a white flash, light-adapted, the NR2E3 large negative waveform in the light-adapted state is almost identical to the dark-adapted response to this stimulus. The flicker cone ERG, however, is abnormally reduced in amplitude versus normal. This pattern of ERGs was proven to derive from reduced rod and L/M cone system responses but hyperfunctioning S-cones [60,63].
OCTs of NR2E3 patients P29 (20 years) and P30 (23 years) illustrate different degrees of cystic maculopathy associated with this disease. P31, a 66-yr-old patient with very reduced vision has a thinned retina with indistinct outer lamination. Visual field maps from P32, a 33-yr-old NR2E3 patient (Fig. 5b), show rod sensitivities (left) abnormally reduced throughout; L/M cone sensitivities were either abnormal or within normal limits (middle); and S-cone sensitivities are supernormal at most loci or fall within normal limits in the remainder (right).
3. Current and future therapies
The diseases discussed in this review were categorised as ‘incurable’ in past decades, but therapeutic strategies are now slowly emerging given progress in understanding the causative genes and disease mechanisms. Gene augmentation therapy is one strategy that has been used to treat mainly autosomal or X-linked recessive IRDs. This is reasonable to consider for diseases where there are remaining photoreceptors to treat, such as CNGA3, CNGB3, ABCA4, and BCM patients. Clinical trials to treat ACHM caused by mutations in CNGA3 or CNGB3 are ongoing, and previously, there was a trial to treat ABCA4 Stargardt disease [70]. Whereas we now know how to diagnose most cone-rod IRDs, we have only limited experience in the design and conduct of treatment trials for these groups of patients. In addition to knowing the disease-causing gene and mutation there should also be careful consideration of the phenotype and stage of disease to treat. A key decision to make is whether the therapeutic goal is to improve visual function or try to preserve vision and slow progression of disease [68], [69], [70]. After improved vision was the consensus result of subretinal gene therapy trials in RPE65-LCA, there seems to have been an assumption that a similar strategy in other IRDs would also lead to significant efficacy within weeks. Subsequent early phase gene therapy trials, however, were relatively disappointing in terms of efficacy, although showing safety [70]. Future clinical trials should follow the lead of the two efficacious gene-based therapies to date (the two forms of Leber congenital amaurosis: RPE65-LCA and CEP290-LCA) and measure, not assume, the potential for efficacy in patient groups targeted for therapy. The potential for improving visual function in these two LCA subtypes was calculated using the relationship between OCT and visual thresholds [68], [69], [70]. If improvement of vision is determined to be an unrealistic goal, then planning of the trial takes a different direction. Greater understanding of the natural history of the disorder, with disease-specific outcomes and not only conventional time-honored parameters, becomes critical so there is a reasonable timeline to conduct the trial and determine if progression is slowed and treatment efficacy is achieved.
There are, of course, other complexities to confront. Dominant, gain of function mutations are more difficult to treat using traditional gene augmentation therapy because correction would also likely need to be accompanied by suppression of a toxic gene product. Recently, a single AAV vector was developed that inactivated mutant rhodopsin and replaced it with a functional gene in a canine model [71]. Using a CRISPR/Cas9 platform packaged in AAV vector as another option for dominant disease is being explored to treat GUCY2D-CORD [72]. The delivery method also needs to be considered; intravitreal gene delivery is being developed as an alternative to subretinal gene delivery for some IRDs and should be considered for diseases with already fragile foveas, as in BCM or ACHM [70], and there are advances in suprachoroidal gene transfer [73]. For diseases with severe degeneration where there may be very few or no photoreceptors left to treat, gene therapy would not be a viable strategy. For example, for CRX disease, regenerative medicine may be a more suitable treatment approach. In addition to regenerative medicine, other non-gene-based methods for treating cone-rod IRDs are being considered such as visual cycle and immune complement system modulators and factors that target the cell death pathways which may have implications for many disease types; some of these non-gene-based methods target pathways common to multiple diseases [74,75]. Research should continue on mechanisms of cone and rod cell death in these diseases so that the pathways of differential photoreceptor cell death can be better understood and potentially targeted for therapeutic intervention.
4. Search strategy and selection criteria
The starting point for many of the searching strategies for this review was the combined knowledge base of the authors of this work. From there, PubMed was the primary database used to identify further resources. Searches for general cone inherited retinal disease were made as were searches for individual genes reviewed in this manuscript.
Declaration of Competing Interest
The authors declare no competing interests or conflicts of interest.
Acknowledgements
No funding was received for this review.
Footnotes
Ethics committee approval: The Institutional Review Board of the University of Pennsylvania approved this research; informed consent was obtained and procedures were performed in accordance with the tenets of the Declaration of Helsinki.
References
- 1.Curcio C.A., Sloan K.R., Kalina R.E., Hendrickson A.E. Human photoreceptor topography. J Comp Neurol. 1990;292(4):497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- 2.Müller H. Über einige Verhältnisse der Netzhaut bei Menschen und Tieren. Verhandlungen der Physikalisch-Medizinischen Gesellschaft zu Würzburg. 1854;4:96–100. [Google Scholar]
- 3.Schultze M.J. Max Cohen; Bonn: 1866. Über den gelben fleck der retina, seinen einfluss auf normales Sehen und auf Farbenblindheit. [Google Scholar]
- 4.Cajal S.R. La rétine des vertébrés. Cellule. 1893;9:119–257. [Google Scholar]
- 5.Wässle H., Boycott B.B. Functional architecture of the mammalian retina. Physiol Rev. 1991;71(2):447–480. doi: 10.1152/physrev.1991.71.2.447. [DOI] [PubMed] [Google Scholar]
- 6.Yan W., Peng Y.R., van Zyl T. Cell atlas of the human fovea and peripheral retina. Sci Rep. 2020;10(1):9802. doi: 10.1038/s41598-020-66092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krill A.E. Vol. 2. Harper & Row Publishing Co.; Hagerstown: 1977. p. 421. (Cone degenerations. Krill's hereditary retinal and choroidal disease). [Google Scholar]
- 8.Knapp H. Peculiar form of retinitis pigmentosa. Trans Am Ophthalmol Soc. 1870;1(7):120–122. [PMC free article] [PubMed] [Google Scholar]
- 9.Donders F. Beitrage zur pathologischen Anatomie des Auges. 2. Pigmentbildung in der Netzhaut. Graefes Arch Ophthalmol. 1857;3:139–150. [Google Scholar]
- 10.Helmholtz H. Förstner'sche Verlagsbuchhandlung; Berlin: 1851. Beschreibung eines Augen-Spiegels. [Google Scholar]
- 11.Yagasaki K., Jacobson S.G. Cone-rod dystrophy. Phenotypic diversity by retinal function testing. Arch Ophthalmol. 1989;107(5):701–708. doi: 10.1001/archopht.1989.01070010719034. [DOI] [PubMed] [Google Scholar]
- 12.Szlyk J.P., Fishman G.A., Alexander K.R., Peachey N.S., Derlacki D.J. Clinical subtypes of cone-rod dystrophy. Arch Ophthalmol. 1993 Jun;111(6):781–788. doi: 10.1001/archopht.1993.01090060069025. [DOI] [PubMed] [Google Scholar]
- 13.Retnet, the Retinal Information Network [Internet]. University of Texas-Houston health science center [updated 2020 September 3; cited 2020 October 15]. Available from: https://sph.uth.edu/RetNet/
- 14.Curcio C., Messinger J.D., Sloan K.R. Human chorioretinal layer thicknesses measured in macula-wide, high resolution histologic sections. Invest Ophthalmol Vis Sci. 2011;52(7):3943–3954. doi: 10.1167/iovs.10-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michalakis S., Becirovic E., Biel M. Retinal cyclic nucleotide-gated channels: from pathophysiology to therapy. Int J Mol Sci. 2018;19(3):749. doi: 10.3390/ijms19030749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill J.S., Georgiou M., Kalitzeos A., Moore A.T., Michaelides M. Progressive cone and cone-rod dystrophies: clinical features, molecular genetics and prospects for therapy. Br J Ophthalmol. 2019;103(5):711–720. doi: 10.1136/bjophthalmol-2018-313278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathans J., Thomas D., Hogness D.S. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232(4747):193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Macke J.P., Merbs S.L. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9(3):429–440. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- 19.Smallwood P.M., Wang Y., Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc Natl Acad Sci U S A. 2002;99(2):1008–1011. doi: 10.1073/pnas.022629799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roosing S., Thiadens A.A., Hoyng C.B., Klaver C.C., den Hollander A.I., Cremers F.P. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Aboshiha J., Dubis A.M., Carroll J., Hardcastle A.J., Michaelides M. The cone dysfunction syndromes. Br J Ophthalmol. 2016;100(1):115–121. doi: 10.1136/bjophthalmol-2014-306505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cideciyan A.V., Hufnagel R.B., Carroll J. Human cone visual pigment deletions spare sufficient photoreceptors to warrant gene therapy. Hum Gene Ther. 2013;24(12):993–1006. doi: 10.1089/hum.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Silva S.R., Arno G., Robson A.G. The X-linked retinopathies: physiological insights, pathogenic mechanisms, phenotypic features and novel therapies [published online ahead of print, 2020 Aug 26] Prog Retin Eye Res. 2020 doi: 10.1016/j.preteyeres.2020.100898. [DOI] [PubMed] [Google Scholar]
- 24.Zhong H., Molday L.L., Molday R.S., Yau K.W. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature. 2002;420(6912):193–198. doi: 10.1038/nature01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J., Morris L., Thapa A. cGMP accumulation causes photoreceptor degeneration in CNG channel deficiency: evidence of cGMP cytotoxicity independently of enhanced CNG channel function. J Neurosci. 2013;33(37):14939–14948. doi: 10.1523/JNEUROSCI.0909-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohl S., Marx T., Giddings I. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet. 1998;19(3):257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- 27.Remmer M.H., Rastogi N., Ranka M.P., Ceisler E.J. Achromatopsia: a review. Curr Opin Ophthalmol. 2015;26(5):333–340. doi: 10.1097/ICU.0000000000000189. [DOI] [PubMed] [Google Scholar]
- 28.Zelinger L., Cideciyan A.V., Kohl S. Genetics and disease expression in the CNGA3 form of achromatopsia: steps on the path to gene therapy. Ophthalmology. 2015;122(5):997–1007. doi: 10.1016/j.ophtha.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho L.S., Vandenberghe L.H. Understanding cone photoreceptor cell death in achromatopsia. Adv Exp Med Biol. 2016;854:231–236. doi: 10.1007/978-3-319-17121-0_31. [DOI] [PubMed] [Google Scholar]
- 30.Hirji N., Aboshiha J., Georgiou M., Bainbridge J., Michaelides M. Achromatopsia: clinical features, molecular genetics, animal models and therapeutic options. Ophthalmic Genet. 2018;39(2):149–157. doi: 10.1080/13816810.2017.1418389. [DOI] [PubMed] [Google Scholar]
- 31.Sharon D., Wimberg H., Kinarty Y., Koch K.W. Genotype-functional-phenotype correlations in photoreceptor guanylate cyclase (GC-E) encoded by GUCY2D. Prog Retin Eye Res. 2018;63:69–91. doi: 10.1016/j.preteyeres.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Peshenko I.V., Dizhoor A.M. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains. J Biol Chem. 2007;282(30):21645–21652. doi: 10.1074/jbc.M702368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palczewski K., Sokal I., Baehr W. Guanylate cyclase-activating proteins: structure, function, and diversity. Biochem Biophys Res Commun. 2004;322(4):1123–1130. doi: 10.1016/j.bbrc.2004.07.122. [DOI] [PubMed] [Google Scholar]
- 34.Peshenko I.V., Cideciyan A.V., Sumaroka A. A G86R mutation in the calcium-sensor protein GCAP1 alters regulation of retinal guanylyl cyclase and causes dominant cone-rod degeneration. J Biol Chem. 2019;294(10):3476–3488. doi: 10.1074/jbc.RA118.006180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collison F.T., Fishman G.A., Nagasaki T. Characteristic ocular features in cases of autosomal recessive PROM1 cone-rod dystrophy. Invest Ophthalmol Vis Sci. 2019;60(6):2347–2356. doi: 10.1167/iovs.19-26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg A.F., Moritz O.L., Williams D.S. Molecular basis for photoreceptor outer segment architecture. Prog Retin Eye Res. 2016;55:52–81. doi: 10.1016/j.preteyeres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tebbe L., Kakakhel M., Makia M.S., Al-Ubaidi M.R., Naash M.I. The interplay between Peripherin 2 complex formation and degenerative retinal diseases. Cells. 2020;9(3):784. doi: 10.3390/cells9030784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Chen Y., Lillo C. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest. 2008;118(8):2908–2916. doi: 10.1172/JCI35891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zacchigna S., Oh H., Wilsch-Bräuninger M. Loss of the cholesterol-binding protein prominin-1/CD133 causes disk dysmorphogenesis and photoreceptor degeneration. J Neurosci. 2009;29(7):2297–2308. doi: 10.1523/JNEUROSCI.2034-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Z., Hu X., Reilly J. Deletion of the transmembrane protein Prom1b in zebrafish disrupts outer-segment morphogenesis and causes photoreceptor degeneration. J Biol Chem. 2019;294(38):13953–13963. doi: 10.1074/jbc.RA119.008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cehajic-Kapetanovic J., Birtel J., McClements M.E. Clinical and molecular characterization of PROM1-related retinal degeneration. JAMA Netw Open. 2019;2(6) doi: 10.1001/jamanetworkopen.2019.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobson S.G., Cideciyan A.V., Sumaroka A. Remodeling of the human retina in choroideremia: rab escort protein 1 (REP-1) mutations. Invest Ophthalmol Vis Sci. 2006;47(9):4113–4120. doi: 10.1167/iovs.06-0424. [DOI] [PubMed] [Google Scholar]
- 43.Rattner A., Smallwood P.M., Williams J. A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron. 2001;32(5):775–786. doi: 10.1016/s0896-6273(01)00531-1. [DOI] [PubMed] [Google Scholar]
- 44.Rattner A., Chen J., Nathans J. Proteolytic shedding of the extracellular domain of photoreceptor cadherin. J. Biol. Chem. 2004;279(40):42202–42210. doi: 10.1074/jbc.M407928200. [DOI] [PubMed] [Google Scholar]
- 45.Stingl K., Mayer A.K., Llavona P. CDHR1 mutations in retinal dystrophies. Sci Rep. 2017;7(1):6992. doi: 10.1038/s41598-017-07117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves M.J., Goetz K.E., Guan B. Genotype-phenotype associations in a large PRPH2-related retinopathy cohort. Hum Mutat. 2020;41(9):1528–1539. doi: 10.1002/humu.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molday R.S. Insights into the molecular properties of ABCA4 and its role in the visual cycle and Stargardt disease. Prog Mol Biol Transl Sci. 2015;134:415–431. doi: 10.1016/bs.pmbts.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Tsin A., Betts-Obregon B., Grigsby J. Visual cycle proteins: structure, function, and roles in human retinal disease. J Biol Chem. 2018;293(34):13016–13021. doi: 10.1074/jbc.AW118.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremers F.P.M., Lee W., Collin R.W.J., Allikmets R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations [published online ahead of print, 2020 Apr 9] Prog Retin Eye Res. 2020 doi: 10.1016/j.preteyeres.2020.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conley S.M., Cai X., Makkia R., Wu Y., Sparrow J.R., Naash M.I. Increased cone sensitivity to ABCA4 deficiency provides insight into macular vision loss in Stargardt's dystrophy. Biochim Biophys Acta. 2012;1822(7):1169–1179. doi: 10.1016/j.bbadis.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molday L.L., Rabin A.R., Molday R.S. ABCR expression in foveal cone photoreceptors and its role in stargardt macular dystrophy. Am J Ophthalmol. 2000;130(5):689. doi: 10.1016/s0002-9394(00)00756-x. [DOI] [PubMed] [Google Scholar]
- 52.Fishman G.A., Stone E.M., Eliason D.A., Taylor C.M., Lindeman M., Derlacki D.J. ABCA4 gene sequence variations in patients with autosomal recessive cone-rod dystrophy. Arch Ophthalmol. 2003;121(6):851–855. doi: 10.1001/archopht.121.6.851. [DOI] [PubMed] [Google Scholar]
- 53.Cideciyan A.V., Swider M., Schwartz S.B., Stone E.M., Jacobson S.G. Predicting progression of ABCA4-associated retinal degenerations based on longitudinal measurements of the leading disease front. Invest Ophthalmol Vis Sci. 2015;56(10):5946–5955. doi: 10.1167/iovs.15-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaroop A., Kim D., Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11(8):563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H.J., Ratnapriya R., Cogliati T., Kim J.W., Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Prog Retin Eye Res. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran N.M., Chen S. Mechanisms of blindness: animal models provide insight into distinct CRX-associated retinopathies. Dev Dyn. 2014;243(10):1153–1166. doi: 10.1002/dvdy.24151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hull S., Arno G., Plagnol V. The phenotypic variability of retinal dystrophies associated with mutations in CRX, with report of a novel macular dystrophy phenotype. Invest Ophthalmol Vis Sci. 2014;55(10):6934–6944. doi: 10.1167/iovs.14-14715. [DOI] [PubMed] [Google Scholar]
- 58.Brzezinski J.A., Reh T.A. Photoreceptor cell fate specification in vertebrates. Development. 2015;142(19):3263–3273. doi: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright A.F., Reddick A.C., Schwartz S.B. Mutation analysis of NR2E3 and NRL genes in Enhanced S Cone Syndrome. Hum Mutat. 2004;24(5):439. doi: 10.1002/humu.9285. [DOI] [PubMed] [Google Scholar]
- 60.Jacobson S.G., Marmor M.F., Kemp C.M., Knighton R.W. SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. Invest Ophthalmol Vis Sci. 1990;31(5):827–838. [PubMed] [Google Scholar]
- 61.Marmor M.F., Jacobson S.G., Foerster M.H., Kellner U., Weleber R.G. Diagnostic clinical findings of a new syndrome with night blindness, maculopathy, and enhanced S cone sensitivity. Am J Ophthalmol. 1990;110(2):124–134. doi: 10.1016/s0002-9394(14)76980-6. [DOI] [PubMed] [Google Scholar]
- 62.Jacobson S.G., Román A.J., Román M.I., Gass J.D., Parker J.A. Relatively enhanced S cone function in the Goldmann-Favre syndrome. Am J Ophthalmol. 1991;111(4):446–453. doi: 10.1016/s0002-9394(14)72379-7. [DOI] [PubMed] [Google Scholar]
- 63.Hood D.C., Cideciyan A.V., Roman A.J., Jacobson S.G. Enhanced S cone syndrome: evidence for an abnormally large number of S cones. Vis Res. 1995;35(10):1473–1481. doi: 10.1016/0042-6989(95)98727-q. [DOI] [PubMed] [Google Scholar]
- 64.Haider N.B., Jacobson S.G., Cideciyan A.V. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24(2):127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 65.Cepko C. Giving in to the blues. Nat Genet. 2000;24(2):99–100. doi: 10.1038/72887. [DOI] [PubMed] [Google Scholar]
- 66.Milam A.H., Rose L., Cideciyan A.V. The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc Natl Acad Sci U S A. 2002;99(1):473–478. doi: 10.1073/pnas.022533099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garafalo A.V., Calzetti G., Cideciyan A.V. Cone vision changes in the enhanced S-cone syndrome caused by NR2E3 gene mutations. Invest Ophthalmol Vis Sci. 2018;59(8):3209–3219. doi: 10.1167/iovs.18-24518. [DOI] [PubMed] [Google Scholar]
- 68.Jacobson S.G., Aleman T.S., Cideciyan A.V. Identifying photoreceptors in blind eyes caused by RPE65 mutations: prerequisite for human gene therapy success. Proc Natl Acad Sci U S A. 2005;102(17):6177–6182. doi: 10.1073/pnas.0500646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cideciyan A.V., Jacobson S.G. Leber Congenital Amaurosis (LCA): potential for improvement of vision. Invest Ophthalmol Vis Sci. 2019;60(5):1680–1695. doi: 10.1167/iovs.19-26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garafalo A.V., Cideciyan A.V., Héon E. Progress in treating inherited retinal diseases: early subretinal gene therapy clinical trials and candidates for future initiatives. Prog Retin Eye Res. 2020;77 doi: 10.1016/j.preteyeres.2019.100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cideciyan A.V., Sudharsan R., Dufour V.L. Mutation-independent rhodopsin gene therapy by knockdown and replacement with a single AAV vector. Proc Natl Acad Sci U S A. 2018;115(36):E8547–E8556. doi: 10.1073/pnas.1805055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCullough K.T., Boye S.L., Fajardo D. Somatic gene editing of GUCY2D by AAV-CRISPR/Cas9 alters retinal structure and function in mouse and macaque. Hum Gene Ther. 2019;30(5):571–589. doi: 10.1089/hum.2018.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen J., Kim J., Tzeng S.Y. Suprachoroidal gene transfer with nonviral particles. Sci Adv. 2020;6(27):1–10. doi: 10.1126/sciadv.aba1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rahman N., Georgiou M., Khan K.N., Michaelides M. Macular dystrophies: clinical and imaging features, molecular genetics and therapeutic options. Br J Ophthalmol. 2020;104(4):451–460. doi: 10.1136/bjophthalmol-2019-315086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vighi E., Trifunović D., Veiga-Crespo P. Combination of cGMP analogue and drug delivery system provides functional protection in hereditary retinal degeneration. Proc Natl Acad Sci U S A. 2018;115(13):E2997–E3006. doi: 10.1073/pnas.1718792115. [DOI] [PMC free article] [PubMed] [Google Scholar]