Abstract
Sex-specific differences in prevalence are well documented for many common, complex diseases, especially for immune-mediated diseases, yet the precise mechanisms through which factors associated with biological sex exert their effects throughout life are not well understood. We interrogated sex-specific transcriptional responses of peripheral blood leukocytes (PBLs) to innate immune stimulation by lipopolysaccharide (LPS) in 46 male and 66 female members of the Hutterite community, who practice a communal lifestyle. We identified 1217 autosomal and 54 X-linked genes with sex-specific responses to LPS, as well as 71 autosomal and one X-linked sex-specific expression quantitative trait loci (eQTLs). Despite a similar proportion of the 15 HLA genes responding to LPS compared to all expressed autosomal genes, there was a significant over-representation of genes with sex by treatment interactions among HLA genes. We also observed an enrichment of sex-specific differentially expressed genes in response to LPS for X-linked genes compared to autosomal genes, suggesting that HLA and X-linked genes may disproportionately contribute to sex disparities in risk for immune-mediated diseases.
Subject terms: Gene expression, Gene regulation
Introduction
Differences in both individual risk for and population prevalence of many common, complex diseases are due to genetic and environmental effects, and interactions between them. Sex can influence disease risk and prevalence by modifying both genetic and environmental effects. Thus, understanding the impact of sex as a modifier of responses to important environmental exposures will contribute to our understanding of sex disparities in disease risk, and potentially inform clinical management and therapeutic strategies (see1). The most striking examples of sex disparities are for immune-mediated diseases. For example, women represent approximately 80% of all autoimmune disease cases diagnosed in the United States each year2, and asthma is considerably more frequent among boys compared to girls in early life, but women have a higher prevalence of asthma compared to men starting post-puberty and continuing throughout life 3. Although hormonal differences between males and females may underlie some of these differences4,5, how sex modifies individual responses to environmental exposures that influence disease risk is not well understood.
Broadly speaking, age, environment, and genotype can all contribute to sex differences on disease in general, and on immune development and function in particular6. Age differences may reflect the effects of lifelong exposures but may also be a proxy for differences in hormone exposures between males and females across different life stages. While male hormones (androgens) generally suppress pro-inflammatory activity through actions such as decreased synthesis of TNF and TLR4 in macrophages in mice7,8, exposure to female hormones (estrogens) increases neutrophil numbers in the blood9 and augments Th2-type responses across species7. Environmental factors, such as diet and nutrition, may also have sex-specific effects. Studies of maternal nutrition have shown that vitamin or micronutrient supplementation provide a greater benefit to females compared to males10–12. Genetic factors can contribute due to differences in sex chromosome number or the effects of genotypes at both sex chromosome and autosomal loci that influence immune responses. In fact, autosomal variants, particularly those in interferon-related or immune pathways, have been implicated in sex-specific risk for asthma. In a genome-wide interaction study of sex-specific effects, a variant downstream of interferon regulatory factor 1 (IRF1) was associated with increased risk of childhood asthma in European American males13. In another study, genotypes for a variant in the interferon gamma (IFNG) gene were differentially associated with asthma in males compared to females14. This genotype by sex interaction was only observed in children who wheezed with a moderate to severe viral illness in the first three years of life, which is itself an independent risk factor for asthma15. Finally, a variant in the interleukin 10 (IL10) gene was associated with increased tetanus and diphtheria toxoid response in girls but not boys16.
Because genotypes at autosomal loci should not differ between males and females, such sex-specific effects must be in large part mediated by differences in the regulation of genes in sex-specific developmental trajectories and/or of responses to exposures. Therefore, a simpler approach to understanding potential mediators of sex-differences on disease risk and prevalence is to study the more proximal phenotype of gene expression than disease risk itself. To that end, a study of sex-dependent gene expression effects in 1000 sex- and age-balanced adults reported that 509 of 560 autosomal immune-related genes transcriptionally responded to at least one of six viral, bacterial, or fungal stimuli in whole blood cells differently in men and women17. Although genes on the X chromosome are often excluded from genome-wide studies, an analysis of the impact of the X chromosome and sex on regulatory variation identified an overall depletion of X-linked eQTLs compared to autosomal eQTLs, but an enrichment of sex-specific eQTLs among X-linked genes compared to the autosomal eQTLs18. These studies highlight the potentially important role for sex-specific gene regulation in immune cells, which could represent the biological underpinnings of sex-specific differences in immune responses and potentially for risk of diseases with underlying immune etiologies.
In this study, we interrogated sex-specific transcriptional responses of peripheral blood leukocytes (PBLs) to innate immune stimulation by lipopolysaccharide (LPS) in 112 members of the Hutterite community, a population of central European ancestry who live on large communal farms in the central plains of North America. Because of their communal lifestyle, Hutterites are exposed to a relatively uniform environment, particularly in early life during critical windows of immune development. This lifestyle, therefore, reduces the effects of environmental variation on immune development and disease risk, and potentially enhances sex-specific and genetic effects19. We report here the identification of hundreds of autosomal and X-linked genes with sex-specific responses to LPS, a potent stimulator of innate immune cells20, as well as almost 600 autosomal and 9 X-linked treatment-specific eQTLs and 71 autosomal and one X-linked sex-specific eQTLs. We also report an enrichment of genes with sex by treatment interactions among HLA genes compared to all autosomal genes and of sex-specific differentially expressed genes in response to LPS for X chromosome genes compared to autosomal genes. These results suggest that X-linked and HLA genes may disproportionately contribute to sex disparities in risk for immune-mediated diseases.
Results
Autosomal genes show sex by LPS treatment interaction effects on gene expression
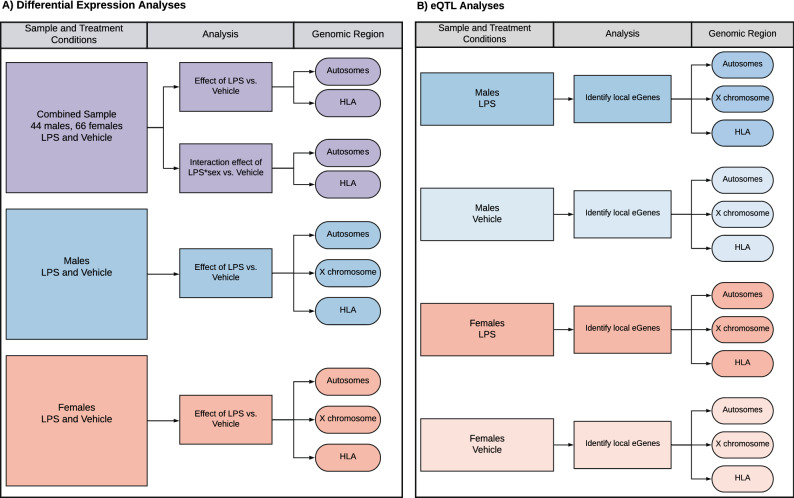
LPS is a component of the cell wall in gram negative bacteria and robustly induces activation of innate immune pathways through binding to the TLR4 receptor20. To assess sex-specific transcriptional differences in response to innate immune stimulation, we evaluated RNA sequences in PBLs after 30 h of treatment with media plus 0.1 µg/ml LPS or media alone in each of 112 individuals (66 females and 46 males; mean ages 33.1 and 32.5 years, respectively (Supplementary Fig. S1). Sex differences in gene expression responses were assessed using a mixed linear model that included a kinship matrix as a random effect to account for relatedness among individuals21. We performed two sets of analyses for autosomal genes. First, we tested for transcriptional responses in the combined sample, specifying treatment as a main effect and age and sex as covariates (main effects model); second, we included an interaction term to test for treatment by sex interactions as a main effect on response, using treatment, sex, and age as covariates (interaction model) (Fig. 1A, lavender box). In both analyses, transcriptional response to LPS was estimated as the gene expression difference between LPS and vehicle treatments. Because of the important role that HLA genes play in immune responses, and the complexity of the HLA region, we present all results for HLA genes separately at the end of the “Results” section.
Figure 1.
Study overview. Where shown, HLA and X-linked genes were analyzed together with the remaining autosomal genes but are discussed separately in the manuscript and are therefore shown here separate from the autosomes to follow the manuscript flow. (A) Flowchart of differential expression analysis. Analyses of autosomal genes in the combined sample (main effect and sex interaction) is shown in lavender, analyses of X-linked genes in males and females separately (autosomal genes included for comparison) are shown in blue and orange, respectively. (B) Flowchart of eQTL mapping in each of the four groups.
In the first analysis of treatment main effects, the vast majority of genes (10,368 of 12,475) were responsive to LPS compared to treatment with vehicle alone (false discovery rate [FDR] < 0.05) (Supplementary Fig. S2; Supplementary Table S1). In the second analysis of treatment by sex interaction effects, 1,055 genes showed significant interactions (FDR < 0.05) (Supplementary Table S2), 90% of which (949 of 1,055) were also among those responsive to LPS treatment in the first analysis, reflecting different effect sizes of response in males and females (Fig. 2). In fact, most genes with significant transcriptional responses to LPS in the first analysis showed the same direction of response in males and females, but the magnitude of response to treatment was generally larger in males compared to females, with 68% of genes showing stronger responses in males than females (Wilcoxon signed rank test P < 2.2 × 10–16) (Fig. 3). Males also had a larger absolute fold change in response to LPS compared to females for 912 of the 1055 (86%) genes showing interaction effects in the second analysis (Wilcoxon signed rank test P < 2.2 × 10–16) (Fig. 4). In large part, the stronger responses to LPS treatment in males result from overall lower gene expression in vehicle-treated cells. Among genes exhibiting significant responses to LPS treatment in the combined sample, gene expression levels in vehicle-treated cells were lower but not significantly different in males compared to females (Wilcoxon signed rank test P = 0.113), but among genes exhibiting sex by treatment interactions, expression levels in vehicle-treated cells were significantly lower in males (Wilcoxon signed rank test P < 2.2 × 10–16).
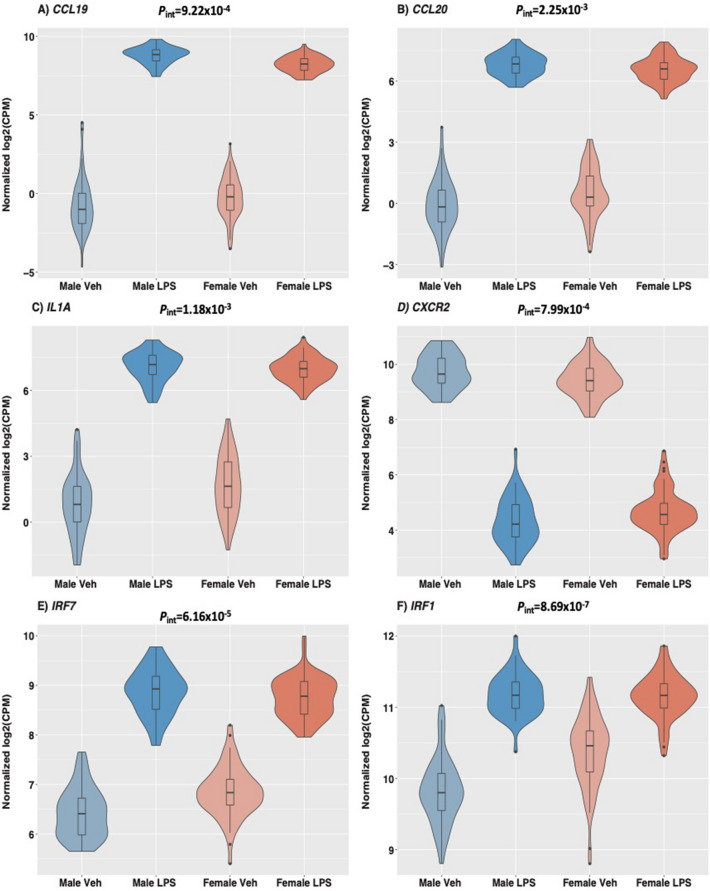
Figure 2.
Violin plots of exemplar genes with significant (FDR < 0.05) LPS treatment by sex interaction effects on gene expression indicating different responses to LPS treatment in males and females. Interaction p-values are shown. For each gene, expression levels for each treatment and sex are shown on the x-axis (male untreated, male LPS-treated, female untreated, female LPS-treated). Each boxplot inside the violin shows the 1st-3rd interquartile range; the horizontal line shows the median expression value. (A) CCL19, Chemokine ligand 19; (B) CCL20, chemokine ligand 20/macrophage inflammatory protein-3; (C) IL1A, Interleukin 1 alpha; (D) CXCR2, Interleukin 8 receptor, beta; (E) IRF7, Interferon regulatory factor 7; (F) IRF1, Interferon regulatory factor 1. Interaction P-values are shown. Note the different y-axis scales.
Figure 3.
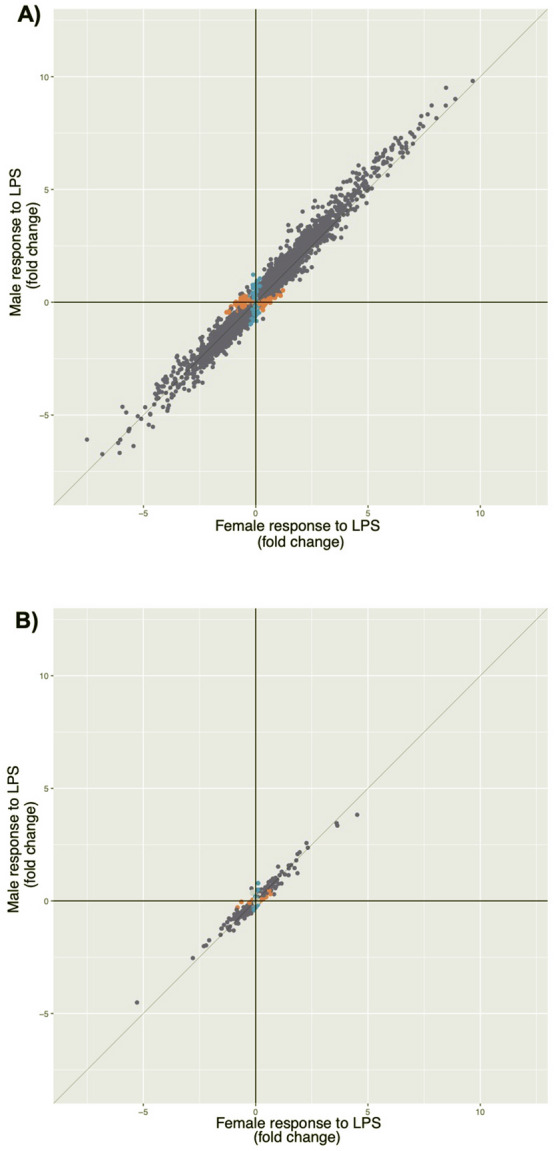
Scatterplot of fold change responses to LPS of genes located on (A) the autosomes (N = 12,475) and (B) the X chromosome (N = 393). Only genes with significant responses are shown. At FDR < 0.05, genes colored in blue were observed only in males, genes colored in orange were observed only females, and genes colored in purple were observed in both sexes.
Figure 4.
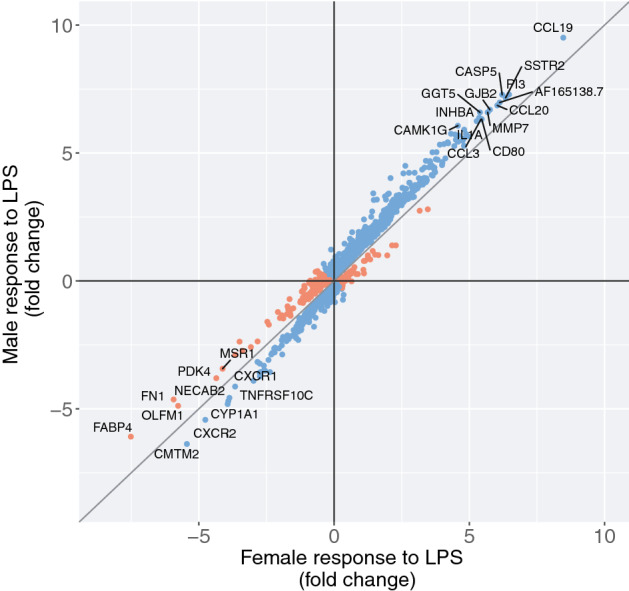
Scatterplot of fold change for genes with significant (FDR < 0.05) sex by LPS treatment interaction effects (n = 1055). Genes colored in blue have a larger absolute magnitude of response to LPS in males compared to females; genes colored orange have a larger absolute magnitude of response in females compared to males. Labeled genes have a fold change of > 6 or < − 4.
To evaluate whether differences in cell composition between males and females and/or in response to treatment contribute to the observed differences, we estimated the relative proportions of 22 different mature hematopoietic populations and activation states using CIBERSORT22. We observed similar cell proportions between males and females in the LPS-treated cells among the seven most common cells types (> 5% on average in LPS and vehicle-treated samples) (Supplementary Table S3). Although not significant after multiple testing correction, estimated monocyte proportions were lower in males compared to females in vehicle-treated samples (proportions = 0.079 in females and 0.074 in males; P = 0.0074). To investigate whether this observation could account for the overall lower gene expression levels in the vehicle-treated cells from males (examples shown in Fig. 2), we examined the responses of 77 autosomal and 2 X-linked monocyte-associated genes22 and asked whether those genes were enriched among the sex by treatment interaction genes with lower expression among vehicle-treated cells from males. The proportion of monocyte genes was 0.013 (9/650) among genes with lower expression in males, similar to 0.012 (13/1,055) among all sex by treatment interaction genes (Fisher Exact Test P = 0.83). Overall, these results suggest that the lower baseline expression of most genes in males is not due to differences in cell composition between males and females.
LPS-responsive genes on the X chromosome are shared between males and females
Due to the different number of sex chromosomes in males and females and the abundance of immune-related genes on the X chromosome in humans23, it is possible that differential responses of X-linked genes to LPS could underlie some of the sexually-dimorphic risks for disease. To examine sex-specific responses of X-linked genes to LPS in PBLs and account for the different number of X chromosomes in males and females, we analyzed transcriptional response differences between cells treated with vehicle and with LPS in each sex separately. We tested both X-linked and autosomal genes in this manner in order to draw comparisons between them (Fig. 1A, blue and yellow boxes). Of the 393 X-linked genes detected as expressed in PBLs, 279 (71%) were responsive to LPS in females and 273 (69%) were responsive to LPS in males at an FDR < 0.05 (Table 1). Most of the LPS-responsive genes at FDR < 0.05 were shared by both sexes (237; 60%), but 23 genes (6%) were responsive only in males and 31 (8%) were responsive only in females at an FDR < 0.05 threshold in males or females, respectively, and at an FDR > 0.2 threshold in the opposite sex (Table 1 and Supplementary Table S4). Of the 237 LPS-responsive genes in both males and females, only one gene responded in opposite directions: ERCC excision repair 6 like (ERCC6L), a DNA helicase for spindle assembly, which increased expression in males (3.59-fold change) and decreased expression in females (0.65-fold change) in response to LPS. These results suggest that most of the changes in X-linked gene expression in response to LPS are shared between males and females, and unsurprisingly involve immune pathways.
Table 1.
Number of differentially expressed (DE) genes (FDR < 0.05) in response to LPS on the autosomes and the X chromosome.
| Category of genes | Autosomal | X-linked |
|---|---|---|
| Male-specific DE | 632 | 23 |
| Female-specific DE | 585 | 31 |
| Sex-specific | 1217 | 54 |
| DE in both sexes (FDR < 0.05) | 8549 | 237 |
| DE in both sexes (0.05 > FDR > 0.20) | 1036 | 30 |
| Neither DE (FDR > 0.20) | 1673 | 72 |
| Not specific | 11,258 | 339 |
| Sex-specific | 1217 | 54 |
| Not specific | 11,258 | 339 |
| Total | 12,475 | 393 |
DE genes considered sex-specific were significant at FDR < 0.05 in the primary sex and not significant at FDR < 0.20 in the opposite sex. There was an enrichment of sex-specific responses to LPS among X-linked genes (54/393, 13.7%) compared to autosomal genes (1217/12,475, 9.8%) (Fisher’s Exact Test P = 0.01).
A previous study in whole blood cells reported that in the absence of stimulation, more X-linked genes than autosomal genes showed sex-specific gene expression (54.8% vs. 48.4%)18. Our data showed similar findings in PBLs, with sex-specific gene expression for 22.4% (88/393) of X-linked genes and 5.3% (656/12,475) of autosomal genes in the absence of stimulation (vehicle-only) at FDR < 0.05 (Fisher’s Exact Test P < 2.2 × 10–16). We also observed a similar enrichment for sex-specific responses to LPS among X-linked genes (13.7 of X-linked vs. 9.8% of autosomal genes; Fisher’s Exact Test P = 0.013). Finally, of the 237 LPS-response genes on the X chromosome that were shared between males and females, 51.1% (121/237) had larger absolute fold changes in response to LPS in males compared to females, significantly less than the 72% among autosomal genes (Fisher’s Exact Test P = 4.56 × 10–11). Consistent with this finding and in contrast to the autosomal genes, there was no trend towards stronger responses to LPS among X-linked genes in males compared to females (not shown).
Although dosage compensation of X-linked genes in females should minimize sex-specific differences in gene expression, some X-linked genes escape inactivation24,25. Therefore, we examined sex differences in response to LPS separately for 36 genes that escape X inactivation in whole blood cells (Supplementary Table S5)26. Of the 36 genes, 26 were expressed in PBLs in our study. Of those, 19 (73%) escape genes were LPS-responsive in either males, females, or both (FDR < 0.05), and six of the 19 were sex-specific (1 in males and 5 in females) (see Supplementary Fig. S3 for examples). Overall, the proportion of LPS-responsive escape genes on the X chromosome is similar to the proportion of LPS-responsive X chromosome genes overall (73% of escape genes compared to 76% of all X chromosome genes expressed in PBLs; Fisher Exact Test P = 0.68). The proportion of escape genes with sex-specific effects (23%) is also similar to that of all X chromosome genes (14%) (Fisher Exact Test P = 0.24). Together, these results indicate that expression of genes that have been reported to escape X-inactivation in females is not contributing to the observed differences in response between males and females.
Expression quantitative trait loci (eQTLs) are unique to treatment condition and/or sex
To identify genetic variation that contributes to treatment or sex effects on transcriptional response to LPS, we mapped autosomal eQTLs in each of four conditions (see “Methods”): vehicle-treated cells in males, vehicle-treated cells in females, LPS-treated cells in males, and LPS-treated cells in females (Fig. 1B). We tested 3.2 million SNPs that had minor allele frequencies > 5% in both males and females and were ± 1 Mb of the transcription start site of each expressed gene. The sharing of eQTLs across conditions was assessed using multivariate adaptive shrinkage (mash)27. Mash estimates effect sizes jointly across datasets to identify both condition-specific and shared effects, using the betas and standard errors as input to identify patterns of sparsity, sharing, and correlations among effects. Significant eQTLs within each condition were defined by a < 0.05 local false sign rate (lfsr). This method avoids overestimating eQTLs due to linkage disequilibrium by only calculating lfsr values for the most significant eQTL per gene. Therefore, we focus on eGenes (and not eQTLs) from this point forward.
Across all conditions 2108 eGenes were identified: 895 in male vehicle-treated cells, 1123 in female vehicle-treated cells, 795 in LPS-treated male cells, and 1144 in LPS-treated female cells (Fig. 5). 496 autosomal and X-linked eGenes (24%) were shared across all four conditions (e.g., Fig. 6A). An additional 104 eGenes were shared across two conditions: 72 were sex-specific (56 in females and 16 in males, in both conditions, e.g., Fig. 6B) and 32 were treatment specific (12 in vehicle-treated and 20 in LPS-treated cells, regardless of sex, e.g., Fig. 6C,D). In contrast, 1,375 eGenes were significant in only one condition and in one sex (e.g., Fig. 6E,F). Additionally, while we observed fewer eGenes in males among autosomal genes, there was a significant enrichment of eGenes specific to males among X-linked genes (1,112 of 2,741 [40.5%] autosomal eGenes compared to 19 of 28 [67.8%] X-linked eGenes in males; Fisher’s Exact Test P = 0.006) (Fig. 5B).
Figure 5.
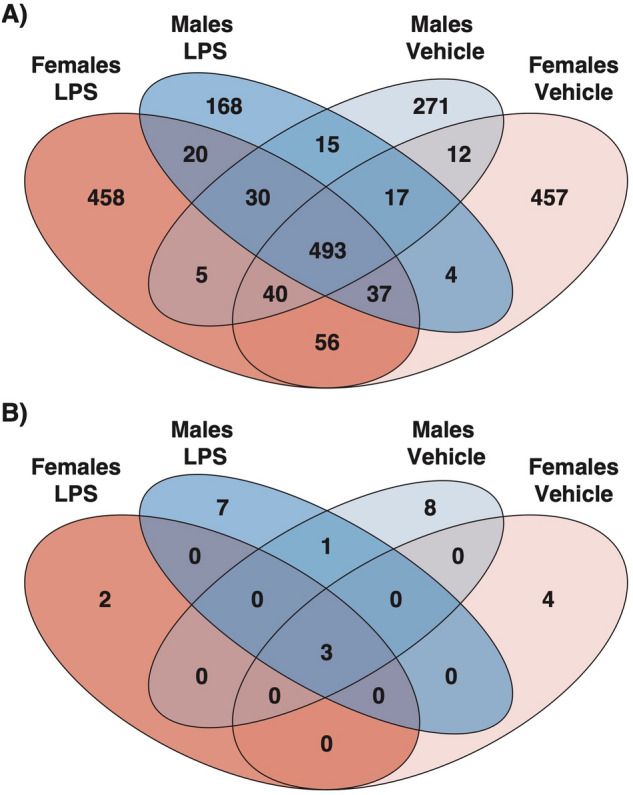
Venn Diagrams with (A) Number of significant eGenes by condition (lfsr < 0.05) on the autosomes, and (B) number of significant eGenes by condition (lfsr < 0.05) on the X chromosome.
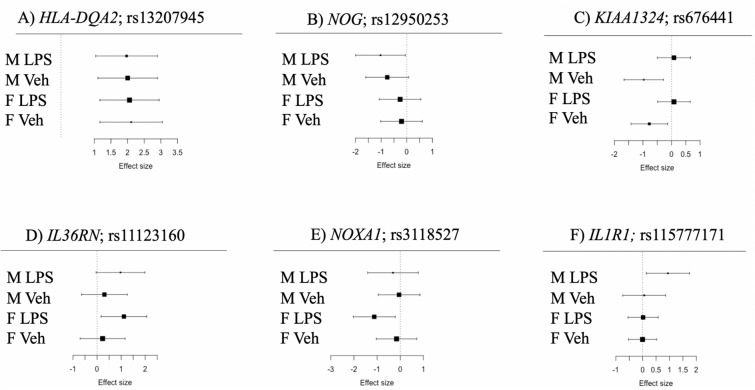
Figure 6.
Meta-analysis forest plots showing the effect size (x-axis) of exemplar gene-SNP pairs for each treatment condition (y-axis; M LPS: expression data in males in the LPS-treated samples; M Veh: expression data in males in the untreated controls; F LPS: expression data in females in the LPS-treated samples; F Veh: expression data in females in the untreated controls). Black boxes show the mean effect size; and the size of the box is proportional to the inverse of the standard error (larger boxes indicate a smaller standard error). The gray lines show the 95% confidence intervals. All eQTLs shown are significant in at least one condition at an lfsr < 0.05. (A) Example of an eQTL shared across all conditions. HLA-DQA2, Human leukocyte antigen DQ alpha 2. (B) Example of a sex-specific eQTL (males), NOG, Noggin. (C) Example of a treatment-specific eQTL (vehicle only), KIAA1324, estrogen-induced gene 121 protein. (D) Example of a treatment-specific eQTL (LPS only). IL36RN, interleukin 36 receptor antagonist. (E) Example of a treatment specific and sex specific eQTL (Female LPS only), NOXA1, NADPH oxidase activator 1. (F) Example of a treatment specific and sex specific eQTL (Male LPS only), IL1R1, interleukin 1 receptor type 1.
Of the 2108 eGenes, 171 (8%) were genes that also showed sex by treatment interaction effects on transcriptional responses to LPS. There was a significant depletion of eGenes with opposite directions of effect between males and females eGenes exhibiting sex by treatment interactions (4.1%) relative to all eGenes (9.8%) (Fisher Exact Test P = 0.013). This indicates that eQTLs for genes with sex by treatment interaction effects on response primarily modify the strength of the response rather than the directionality.
HLA Genes are enriched for sex interactions and sex-interacting eQTLs
Genetic variation at HLA loci show striking associations with autoimmune and allergic diseases28,29, immune-mediated diseases that are also sexually dimorphic with respect to prevalence, severity, and/or age of onset3. Previous studies have reported sex-specific associations of HLA alleles with immune-related diseases and traits30–32, but sex-specific gene expression studies have not focused on HLA genes. Due to the highly polymorphic nature of HLA genes, standard mRNA mapping pipelines that align reads to a reference genome can yield biased estimates of HLA transcript abundance. To address this potential bias, we mapped reads of HLA genes to personalized reference sequences matched to each individual’s HLA alleles, as described in Aguiar et al.33 (see “Methods”).
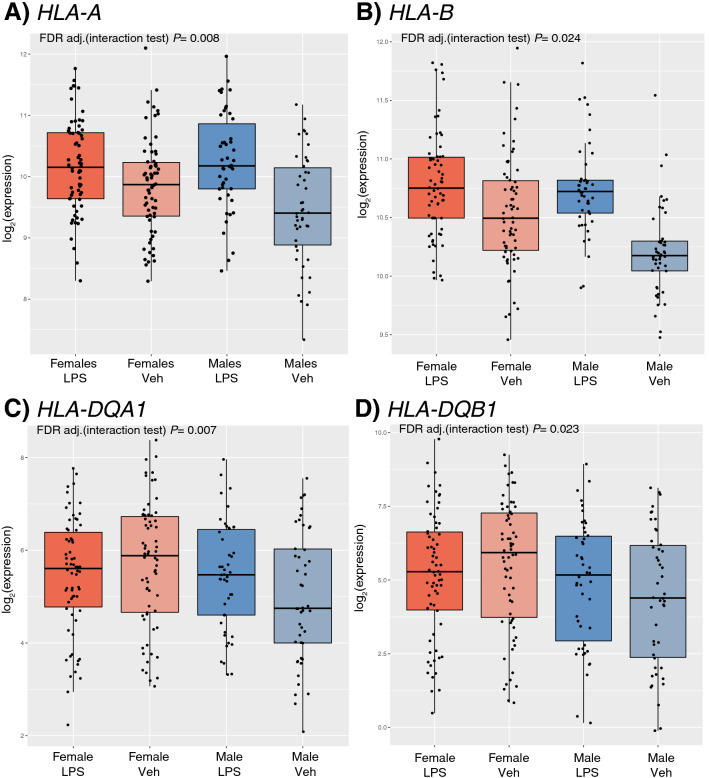
In total, 15 HLA genes in the class I and class II regions were expressed in PBLs, of which 10 responded to LPS treatment in the combined sample at FDR < 0.05 (Table 2), a lower but similar proportion compared to all expressed autosomal genes (67% vs. 83%; Fisher’s Exact Test P = 0.16). For the 10 LPS-responsive HLA genes, responses were similar in males and females (defined by an FDR < 0.05 threshold in one sex and an FDR > 0.2 threshold in the opposite sex). However, seven HLA genes showed sex by treatment interactions, significantly more than that observed for all autosomal genes (7/15, 47% vs. 1055/12,475, 8%, respectively; Fisher’s Exact Test P = 1.1 × 10–4) (see Fig. 7 for examples). Only two genes with sex by treatment interactions, HLA-DQA1 and HLA-DQB1, showed opposite directions of response (females: − 0.237 and − 0.322 log fold change, respectively; males: 0.429 and 0.463 log fold change, respectively; interaction FDR = 0.007 and 0.023 respectively) (Fig. 7C,D).
Table 2.
The log fold change responses of HLA genes expressed in PBLs to LPS treatment in the combined (n = 112), female (n = 66), and male (n = 46) samples.
| Gene | Log Fold Change (Combined Sample) |
Log Fold Change (Females) |
Log Fold Change (Males) |
FDR adj. P-value (response test in combined sample) | FDR adj. P-value (response test in females only) | FDR adj. P-value (response test in males only) | FDR adj. P-value (sex by treatment interaction test) |
|---|---|---|---|---|---|---|---|
| HLA-F | 0.551 | 0.450 | 0.695 | 1.81 × 10–16 | 6.25 × 10–8 | 3.72 × 10–9 | 0.093 |
| HLA-A | 0.504 | 0.314 | 0.778 | 1.94 × 10–11 | 0.003 | 9.25 × 10–4 | 0.008 |
| HLA-E | 0.051 | − 0.005 | 0.132 | 0.083 | 0.935 | 0.265 | 0.105 |
| HLA-C | 0.052 | − 0.030 | 0.171 | 0.203 | 0.579 | 0.234 | 0.091 |
| HLA-B | 0.360 | 0.239 | 0.534 | 8.23 × 10–11 | 0.017 | 0.004 | 0.024 |
| HLA-DRA | 0.458 | 0.335 | 0.634 | 2.25 × 10–9 | 3.21 × 10–4 | 2.91 × 10–6 | 0.135 |
| HLA-DRB1 | − 0.519 | − 0.768 | − 0.160 | 1.61 × 10–7 | 0.001 | 0.062 | 0.014 |
| HLA-DQA1 | 0.036 | − 0.237 | 0.429 | 0.706 | 0.639 | 0.611 | 0.007 |
| HLA-DQB1 | − 0.001 | − 0.322 | 0.463 | 0.995 | 0.353 | 0.994 | 0.023 |
| HLA-DQA2 | − 0.261 | − 0.397 | − 0.065 | 0.105 | 0.062 | 0.808 | 0.528 |
| HLA-DOB | 0.767 | 0.700 | 0.863 | 7.66 × 10–12 | 4.84 × 10–7 | 6.43 × 10–6 | 0.608 |
| HLA-DMB | − 0.839 | − 1.006 | − 0.599 | 7.55 × 10–18 | 1.63 × 10–12 | 4.48 × 10–6 | 0.042 |
| HLA-DMA | − 0.806 | − 0.976 | − 0.563 | 2.29 × 10–20 | 7.32 × 10–14 | 3.31 × 10–7 | 0.009 |
| HLA-DOA | − 0.254 | − 0.164 | − 0.384 | 0.001 | 0.051 | 0.014 | 0.371 |
| HLA-DPB1 | − 0.686 | − 0.765 | − 0.573 | 4.54 × 10–17 | 0.107 | 0.859 | 0.304 |
Also presented are FDR-adjusted p-values for four tests: (1) test of significant response in the combined sample, (2) test of significant response in females, (3) test of significant response in males, and (4) test for sex by treatment interactions in the combined sample. Genes are presented in order from the telomeric to the centromeric end of the short arm of chromosome 6. Significant FDR-adjusted p-values < 0.05 are denoted with italics text.
Figure 7.
Boxplots illustrating four HLA genes with significant sex by treatment interactions. (A) HLA-A and (B) HLA-B are examples of genes where the magnitude of response differs between males and females. (C) HLA-DQA1 and (D) HLA-DQB1 show opposite directions of response to LPS treatment in males and females.
Seven of the 15 HLA genes were eGenes (Supplementary Table S6), which is greater than the proportion of eGenes among autosomal genes (47% vs 18%; Fisher’s Exact Test P = 0.010). The eQTLs for the seven HLA eGenes showed the same direction of effect across all four conditions, and six of the seven were eGenes in all four conditions. Only the eQTL for HLA-DRA was specific to the Female/LPS condition. The proportion of HLA eGenes with eQTLs that shared directionality across all four conditions was significantly greater than that for all autosomal eGenes (86% for HLA eGenes vs 22% for all autosomal genes; Fisher’s Exact Test P = 7.51 × 10–4).
Discussion
Gene expression is an important intermediate phenotype for elucidating mechanisms contributing to sex-biased phenotypic differences because both sex and genotype significantly impact gene expression levels34–36. In previous studies, sex-specific eQTLs for autosomal genes were in strong linkage disequilibrium with variants associated with sexually dimorphic complex traits35,36, suggesting a molecular mechanism for sex differences in disease prevalence, severity, and/or onset. Genetic differences at sex chromosome loci may also contribute, although only two previous studies examined the effect of sex on X chromosome eQTLs in untreated whole blood cells18,35. The sample sizes for both studies were considerably larger than ours, resulting in greater power to detect eQTLs with smaller effects, while our study in treated cells allowed us to interrogate sex-specific responses of X-linked genes to immune stimulation. Moreover, relatively few genome-wide association studies of immune-related traits have included X chromosome variation and, of those, few significant associations were reported37–41. Here, we report for the first time that the vast majority of X-linked eGenes (22/25; 88%) are sex or condition specific. Our results suggest that genetic differences at genes on the X chromosome or their responses to immune stimulation may indeed contribute to observed differences between males and females in immune responses and potentially to risk for immune-mediated diseases.
To assess sex-specific immune responses to a potent innate stimulator, we focused our studies on members of the Hutterite community. We hypothesized that the unique, communal Hutterite lifestyle might enhance the effects of sex on transcriptional responses that are due to genetic differences or to developmental trajectories that differ between males and females, and minimize effects due to differences in environmental exposures during early childhood42. We observed 1,055 significant LPS-treatment by sex interaction effects (FDR < 0.05) on gene expression responses of PBLs from 112 Hutterites, indicating that gene expression response to LPS stimulation is indeed shaped by sex. A direct comparison of these findings to similar studies is not possible owing to differences in study design. For example, previous studies of LPS exposure in humans were not population-based43–46, did not look at sex-specific differences in response44,46, and/or did not include the X chromosome44. One additional study assessed sex-specific differences in gene expression in the presence of immune stimulators but did not include LPS and focused on only a panel of immune-response genes17, and one study included X-linked genes in sex-specific eQTL studies but did not examine stimulated cells18.
Moreover, ours is first study of sex specific responses that used personalized HLA read alignments to generate unbiased gene expression data at these highly polymorphic loci. Previous studies of sex differences in HLA gene expression35,47–49 did not account for this potential bias, and gene expression studies that did use personalized alignments of HLA genes did not examine sex differences50,51. Using unbiased estimates of HLA gene expression levels and genome-wide approaches to identify sex-specific patterns, we made the novel observation of an enrichment of LPS responses exhibiting sex by treatment interactions among class I and II HLA genes compared to all autosomal genes: 47% of HLA genes showed sex by treatment interactions whereas these interactions were observed in 8% of all autosomal genes (P = 1.1 × 10–4). This difference in interactions was observed despite HLA genes being no more likely to respond to LPS treatment than autosomal genes overall. Together these results suggest that HLA genes play a disproportionate role in sex-specific immune responses, potentially mediating risk for sexually dimorphic immune-mediated diseases.
Among the seven HLA genes showing sex by treatment interaction effects on response, HLA-DQA1 and HLA-DQB1 were the only genes that responded to LPS treatment in opposite directions in females and males (Fig. 7). The similarity in HLA-DQA1 and HLA-DQB1 response sizes and directions within each sex, as well as their physical proximity to each other in the class II region, suggest the presence of a cis, sex-specific co-regulatory mechanism of response to LPS. In fact, the HLA-DQ2 molecule, encoded by the HLA-DQA1*05 and HLA-DQB1*02 alleles, have shown sex-specific associations with both type 1 diabetes52 and celiac disease53, two autoimmune diseases. The opposite response of these two genes to a potent immune stimulator in our study could provide insight into these, and potentially other, HLA associations with immune-mediated diseases.
Overall, we observed that most genes with significant treatment by sex interactions on the autosomes had a greater average fold change in males, compared to females. This was largely driven by lower gene expression levels in the vehicle-treated cells and larger responses to LPS treatment in males, resulting in similar gene expression levels between males and females in LPS-treated cells. This is consistent with previous studies reporting stronger cytokine responses to LPS in males compared to females46,54,55. For example, a study of sex differences in cytokine secretion in response to LPS reported significantly greater TNF, IL-1, IL-8, and IFN γ production in males compared to females (n = 72 and 82, respectively, both adjusted and unadjusted for cell counts and independent of age effects56). The authors concluded that the greater cytokine responses in males were largely due to differences in the slope of the response curve, which was greater (steeper) in males after 30 h of exposure to LPS, in line with our results, which were also after a 30-h exposure. It is possible that hormones or other intrinsic differences between males and females act in combination with duration of exposure. For example, a recent study of gene expression responses to inactivated influenza virus showed that females have larger immune gene responses compared to males after 24 h of exposure, but that most of these genes showed opposite expression levels (either up or down) between males and females after 36 h of exposure57. That is, differences in immune response between males and females may change both in magnitude and in direction with different exposure lengths. Because of the logistical constraints of field work, we only measured responses to one exposure (LPS) and at one time point (30 h). Therefore, our conclusions cannot be generalized to other exposure times or to other stimuli.
Our study had other limitations. Although the reduced variation in environmental exposures due to the Hutterite lifestyle likely improved our power to detect treatment by sex interaction effects on transcriptional responses, the sample was still small for identifying treatment-specific eQTLs. As a result, we may have missed treatment-specific eQTLs with more modest effects, or genes whose interaction effects differ between children and adults. In addition, the individuals in our study were generally healthy (no autoimmune diseases and only 11.6% with asthma) so we cannot evaluate whether these responses would have differed among individuals with immune-mediated diseases. Finally, we did not measure sex hormones and therefore could not directly assess their effects on LPS responses or attribute sex-specific responses to hormonal differences between males and females. However, by adjusting for age in all analyses, and including children and adults, we should have reduced the effects of age-related differences, which would include changes in hormone levels throughout the lifecycle.
Despite these limitations, our study demonstrated that transcriptional response to innate immune cell stimulation by LPS is markedly shaped by sex. We show that genetic regulation of both autosomal and X-linked transcriptional responses to innate immune stimuli differ between males and females, and suggest that these genes may contribute to sex disparities in disease risk. Moreover, most eQTLs (70%) were specific to treatment and/or sex, suggesting genetic variation plays an important role in sex-specific responses to immune stimuli. Altogether, our study adds to the steadily increasing number of investigations of the impact of sex on immune response, and provides novel insights into potential molecular mechanisms underlying sex differences in the prevalence and course of many immune-mediated diseases.
Methods
Study population
This study was conducted in 143 Hutterites (7–76 years old) with RNA-seq data who are a subset of the > 1400 Hutterites who have participated in our population-based studies of complex phenotypes58–62. The Hutterites are of central European ancestry, and the participants in our studies live on communal farms in South Dakota and are related to each other through multiple lines of descent in a single 3671-member pedigree with 64 founders. Informed consent for these studies was obtained from the adult participants and parents of children under 18; written assent was obtained from all children. This study was approved by the University of Chicago Institutional Review Board. All methods were carried out in accordance with relevant guidelines and regulations.
Collection of whole blood samples
One milliliter of whole blood was drawn into each of two TruCulture (Myriad RBM; Austin, Texas) tubes containing either proprietary TruCulture media alone or media + 0.1 µg/ml LPS during field trips to South Dakota. Samples were processed in a make-shift lab on the Hutterite colonies as follows. TruCulture tubes were incubated upright in a dry heat block at 37 °C for 30 h. Following incubation, supernatants were separated from the cell pellets, and then aliquoted and flash-frozen on dry ice. The remaining cells were washed twice with Buffer EL (Qiagen; Hilden, Germany) and resuspended in 350 µl RLT Buffer (Qiagen) and frozen on dry ice. Frozen samples were shipped on dry ice overnight to our lab in Chicago and stored at − 80 °C. DNA and RNA were extracted from thawed cell pellets using AllPrep DNA/RNA Mini Kits (Qiagen).
RNA sequencing
RNA-seq libraries were made from all RNA samples using the TruSeq Library kit (Illumina; San Diego, California); quality and concentration of libraries were assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies; Santa Clara, California) and quantitative PCR using the Kapa library quantification kit (Kapa Biosystems; Wilmington, Massachusetts). Samples were sequenced in pools of 16–18 samples across three flow cells of an Illumina HiSeq 2500; 81 samples with low read count were re-sequenced on three flow cells on the same machine. VerifyBamID63 detected one sample swap; these two samples were relabeled and retained in downstream analyses. Read quality was assessed with multiQC64, and mapped to hg19 and genes were counted using STAR v2.5.265. Read counts for the same sample sequenced multiple times were summed together, and samples with > 7 million uniquely mapped reads were retained. Genes were included either if they were expressed or had counts per million (CPM) values > 1 in 20 or more subjects. Trimmed Means of M-values (TMM) normalization and voom transformation were used to correct for differences in library sizes and the mean–variance trend, respectively, resulting in log2 CPM values66. TMM normalization and voom transformation were performed in males and females separately for X chromosome genes. Individuals missing vehicle control or LPS-treated match were removed. In the remaining 224 samples (matched LPS-treated and vehicle control in 112 individuals) confounding technical effects were assessed in the normalized expression data using principal components analysis (PCA), and sequencing pool and RNA extraction batch were adjusted using the function RemoveBatchEffect() from the R package limma67.
HLA typing and read mapping
Of the 112 individuals with normalized expression data, 95 previously had their HLA haplotypes determined for nine polymorphic HLA loci (HLA-A, HLA-B, HLA-C, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRB1, HLA-E, and HLA-G) using methods described in68. For the remaining 17 individuals, the Type 1 Diabetes Genetics Consortium (T1DGC) HLA reference panel69 was used to impute HLA haplotypes from their genotype data using SNP2HLA v1.0.370. HLA-DPA1 had not been haplotyped and was dropped from further consideration. HLA-DRB5 was also excluded from the analyses due to copy number variation.
Reads were then mapped to a personalized index determined by each individual’s HLA alleles using STAR v2.6.1a and quantified using salmon v0.8.271 as described in Aguiar et al.33. Two individuals (1 male and 1 female) had poor quality expression data for the haplotyped loci, so all analyses for these loci are based on a sample of 110 individuals. Additionally, HLA-G had low expression across the entire sample and was not included in any analysis. Raw read counts of each individual’s two alleles at the eight remaining polymorphic HLA loci were then summarized to yield a total read count for each individual at every locus. The sums were then TMM normalized and voom transformed as described above.
Analysis of gene expression
Differential gene expression on the autosomes and eQTL studies (below) were examined using Genome-wide Efficient Mixed-Model Association (GEMMA)21, which is an exact method for testing associations with mixed models. Importantly, GEMMA allows inclusion of a genetic relatedness matrix (GRM) based on identity by descent (IBD) to account for the inbreeding and relatedness (population structure) in the sample (n = 112 paired samples, 46 males, 66 females). Treatment by sex interaction was tested in GEMMA using an interaction term as a main effect, and treatment, sex, and age were included as covariates. Differences in gene expression by treatment across sex were assessed in GEMMA with treatment as a main effect, and sex and age included as covariates. Treatment by sex interaction was also tested using treatment as a main effect and sex included in the “-gxe” flag and yielded similar results to the treatment by sex interaction term analysis method presented in the results. Because immune responses differ by age, we questioned whether some of the transcriptional differences between males and females could be due in part to differences in the age distributions between males and females in our sample (Supplementary Fig. S1); however, the age distributions of males and females were not significantly different (Kolmogorov–Smirnov P = 0.108).
As with the autosomes, differential expression on the X chromosome was examined using GEMMA for the 393 genes expressed in PBLs. Males and females were analyzed separately using linear mixed models. LPS treatment was included as a fixed effect, age was included as a covariate. The analysis was conducted twice: once using an autosomal GRM and a second time using a GRM solely based on X-linked variants, as described72. Since the results were highly correlated (Supplementary Fig. S4), we chose to move ahead with the autosomal GRM since the X chromosome GRM included fewer samples (25 males and 61 females, compared to 46 males and 66 females using the autosomal GRM).
Significance was assessed in GEMMA using the score test, and multiple testing correction was adjusted using the methods of Benjamini and Hochberg73. Genes differentially expressed at a false discovery rate (FDR) of 5% were analyzed using Advaita Bio’s iPathway Guide (https://www.advaitabio.com/ipathwayguide). This software analysis tool implements the “Impact Analysis” approach that takes into consideration the direction and type of all signals on a pathway, the position, role and type of every gene, etc. as described in74–77. The list of all genes detected as expressed in PBLs was used as the reference gene panel for analyses.
Genotype data and Expression quantitative trait loci (eQTL) analysis
Genotypes were imputed using PRIMAL, a pedigree-based imputation method with high accuracy in the Hutterites78. For these studies, we selected SNPs within 1 Mb of the transcription start site in all genes defined as expressed in the RNAseq sample (12,475 genes), had a minor allele frequency (MAF) < 5% in both males and females, and a call rate > 95%. This resulted in 3.7 million SNPs available for eQTL studies.
To correct for relatedness between Hutterites in our study, local eQTLs were identified using GEMMA21, which included a kinship matrix based on identity by descent (IBD). eQTLs were identified across four conditions: (1) vehicle-treated male PBLs, (2) LPS-treated male PBLs, (3) vehicle-treated female PBLs, (4) LPS-treated females. Significance and sharing of eQTLs was calculated using multivariate adaptive shrinkage (mash)27. Briefly, mash estimates effects sizes jointly, across multiple conditions, allowing for both condition specific and shared effects using effect sizes and standard errors as input. Using the most significant eQTL per gene, mash learns the typical patterns of sparsity, sharing, and correlations within the data. These putative covariance matrices are then applied to a larger data set to produce improved effect estimates. As recommended by the authors of mash and mashr, the R package that executes the mash analysis, we taught the model the correlation structure of the results using a “random” subset of the data consisting of the most significant eQTL per gene, in addition to a random selection of 1 out of every 50 tested SNPs for each gene for a total of 655,822 gene-SNP pairs. We then trained the model to calculate the data-driven covariances using a “strong” subset of the data that consisted only of the most significant eQTL per gene. The mash model was then fit to the “random” subset of data, and the fitted model was used to calculate posterior summaries for the “strong” set of results.
Supplementary Information
Acknowledgements
The authors would like to thank Xiang Zhou, Catherine Igartua, Mark Abney, and Matthew Stephens for statistical advice and helpful comments. This work was supported by NIH Grant R01 HL085197 and a grant supplement on sex effects. M.M.S was supported by NIH training Grants T32 GM007197 and T32 HL007605. M.C. was supported by R01 HL129735.
S.M.C. and C.B. are supported by UG3 OD023282. K.M.M. is supported by F31 HL143891, and E.E.T is supported by R01 HL113395.
Author contributions
M.S. collected data, wrote the manuscript and performed data analysis. M.C. wrote the manuscript and performed data analysis. K.M.M. and S.M.C. performed data processing. C.B., R.N. and K.N. processed samples. C.O. originated the study, wrote the manuscript, and provided materials. E.E.T. wrote the manuscript and performed data analysis.
Data availability
The gene expression dataset generated during the current study will be available in the dbGaP repository.
Code availability
The analysis scripts for this project are available at https://github.com/mstein3/TreatmentResp_RNAseq.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Michelle M. Stein and Mitch Conery.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80145-z.
References
- 1.Khramtsova EA, Davis LK, Stranger BE. The role of sex in the genomics of human complex traits. Nat. Rev. Genet. 2019;20(3):173–190. doi: 10.1038/s41576-018-0083-1. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson DL, Gange SJ, Rose NR, Graham NM. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin. Immunol. Immunopathol. 1997;84(3):223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- 3.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 2008;9(12):911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah R, Newcomb DC. Sex bias in asthma prevalence and pathogenesis. Front. Immunol. 2018;9:2997. doi: 10.3389/fimmu.2018.02997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han YY, Forno E, Celedon JC. Sex steroid hormones and asthma in a nationwide study of U.S. adults. Am. J. Respir. Crit. Care Med. 2020;201(2):158–166. doi: 10.1164/rccm.201905-0996OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein SL, Flanagan KL. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16(10):626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 7.Straub RH. The complex role of estrogens in inflammation. Endocr. Rev. 2007;28(5):521–574. doi: 10.1210/er.2007-00018. [DOI] [PubMed] [Google Scholar]
- 8.Rettew JA, Huet-Hudson YM, Marriott I. Testosterone reduces macrophage expression in the mouse of toll-like receptor 4, a trigger for inflammation and innate immunity. Biol. Reprod. 2008;78(3):432–437. doi: 10.1095/biolreprod.107.063545. [DOI] [PubMed] [Google Scholar]
- 9.Jilma B, Eichler HG, Breiteneder H, Wolzt M, Aringer M, Graninger W, et al. Effects of 17 beta-estradiol on circulating adhesion molecules. J. Clin. Endocrinol. Metab. 1994;79(6):1619–1624. doi: 10.1210/jcem.79.6.7527406. [DOI] [PubMed] [Google Scholar]
- 10.Friis H, Gomo E, Nyazema N, Ndhlovu P, Krarup H, Kaestel P, et al. Effect of multimicronutrient supplementation on gestational length and birth size: A randomized, placebo-controlled, double-blind effectiveness trial in Zimbabwe. Am. J. Clin. Nutr. 2004;80(1):178–184. doi: 10.1093/ajcn/80.1.178. [DOI] [PubMed] [Google Scholar]
- 11.Thorne-Lyman AL, Fawzi WW. Vitamin A and carotenoids during pregnancy and maternal, neonatal and infant health outcomes: A systematic review and meta-analysis. Paediatr. Perinat. Epidemiol. 2012 doi: 10.1111/j.1365-3016.2012.01284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osrin D, Vaidya A, Shrestha Y, Baniya RB, Manandhar DS, Adhikari RK, et al. Effects of antenatal multiple micronutrient supplementation on birthweight and gestational duration in Nepal: Double-blind, randomised controlled trial. Lancet. 2005;365(9463):955–962. doi: 10.1016/s0140-6736(05)71084-9. [DOI] [PubMed] [Google Scholar]
- 13.Myers RA, Scott NM, Gauderman WJ, Qiu W, Mathias RA, Romieu I, et al. Genome-wide interaction studies reveal sex-specific asthma risk alleles. Hum. Mol. Genet. 2014;23(19):5251–5259. doi: 10.1093/hmg/ddu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loisel DA, Tan Z, Tisler CJ, Evans MD, Gangnon RE, Jackson DJ, et al. IFNG genotype and sex interact to influence risk of childhood asthma. J. Allergy Clin. Immunol. 2011;128(3):524–531. doi: 10.1016/j.jaci.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 2008;178(7):667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baynam G, Zhang G, Khoo SK, Sly P, Holt P, Goldblatt J, et al. Gender-specific effects of cytokine gene polymorphisms on childhood vaccine responses. Vaccine. 2008;26(29–30):3574–3579. doi: 10.1016/j.vaccine.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Piasecka B, Duffy D, Urrutia A, Quach H, Patin E, Posseme C, et al. Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proc. Natl. Acad. Sci. USA. 2018;115(3):E488–E497. doi: 10.1073/pnas.1714765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukurba KR, Parsana P, Balliu B, Smith KS, Zappala Z, Knowles DA, et al. Impact of the X Chromosome and sex on regulatory variation. Genome Res. 2016;26(6):768–777. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat. Genet. 2006;38(2):218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- 20.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 1999;274(16):10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: When a chromosome makes the difference. Nat. Rev. Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 24.Balaton BP, Brown CJ. Escape artists of the X chromosome. Trends Genet. 2016;32(6):348–359. doi: 10.1016/j.tig.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 26.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550(7675):244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urbut SM, Wang G, Carbonetto P, Stephens M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 2019;51(1):187–195. doi: 10.1038/s41588-018-0268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat. Rev. Immunol. 2018;18(5):325–339. doi: 10.1038/nri.2017.143. [DOI] [PubMed] [Google Scholar]
- 29.Schoettler N, Rodriguez E, Weidinger S, Ober C. Advances in asthma and allergic disease genetics: Is bigger always better? J. Allergy Clin. Immunol. 2019;144(6):1495–1506. doi: 10.1016/j.jaci.2019.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher SA, Hampe J, Macpherson AJ, Forbes A, Lennard-Jones JE, Schreiber S, et al. Sex stratification of an inflammatory bowel disease genome search shows male-specific linkage to the HLA region of chromosome 6. Eur. J. Hum. Genet. 2002;10(4):259–265. doi: 10.1038/sj.ejhg.5200792. [DOI] [PubMed] [Google Scholar]
- 31.Ivanova R, Henon N, Lepage V, Charron D, Vicaut E, Schachter F. HLA-DR alleles display sex-dependent effects on survival and discriminate between individual and familial longevity. Hum. Mol. Genet. 1998;7(2):187–194. doi: 10.1093/hmg/7.2.187. [DOI] [PubMed] [Google Scholar]
- 32.Morrison BA, Ucisik-Akkaya E, Flores H, Alaez C, Gorodezky C, Dorak MT. Multiple sclerosis risk markers in HLA-DRA, HLA-C, and IFNG genes are associated with sex-specific childhood leukemia risk. Autoimmunity. 2010;43(8):690–697. doi: 10.3109/08916930903567492. [DOI] [PubMed] [Google Scholar]
- 33.Aguiar VRC, Cesar J, Delaneau O, Dermitzakis ET, Meyer D. Expression estimation and eQTL mapping for HLA genes with a personalized pipeline. PLoS Genet. 2019;15(4):e1008091. doi: 10.1371/journal.pgen.1008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimas AS, Nica AC, Montgomery SB, Stranger BE, Raj T, Buil A, et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012;22(12):2368–2375. doi: 10.1101/gr.134981.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao C, Joehanes R, Johnson AD, Huan T, Esko T, Ying S, et al. Sex- and age-interacting eQTLs in human complex diseases. Hum Mol Genet. 2014;23(7):1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen JJ, Wang YF, Yang W. Sex-Interacting mRNA- and miRNA-eQTLs and their implications in gene expression regulation and disease. Front. Genet. 2019;10:313. doi: 10.3389/fgene.2019.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang D, Gao F, Slavney A, Ma L, Waldman YY, Sams AJ, et al. Accounting for eXentricities: Analysis of the X chromosome in GWAS reveals X-linked genes implicated in autoimmune diseases. PLoS ONE. 2014;9(12):e113684. doi: 10.1371/journal.pone.0113684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li YR, Li J, Zhao SD, Bradfield JP, Mentch FD, Maggadottir SM, et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 2015;21(9):1018–1027. doi: 10.1038/nm.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47(12):1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Y, Andersen G, Yorgov D, Ferrara TM, Ben S, Brownson KM, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016;48(11):1418–1424. doi: 10.1038/ng.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JC, Biasci D, Roberts R, Gearry RB, Mansfield JC, Ahmad T, et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat. Genet. 2017;49(2):262–268. doi: 10.1038/ng.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss LA, Abney M, Cook EH, Jr, Ober C. Sex-specific genetic architecture of whole blood serotonin levels. Am. J. Hum. Genet. 2005;76(1):33–41. doi: 10.1086/426697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbasi A, de Paula VR, Bischof F, Walter M, Movassaghi M, Berchtold NC, et al. Sex-specific variation in signaling pathways and gene expression patterns in human leukocytes in response to endotoxin and exercise. J. Neuroinflamm. 2016;13(1):289. doi: 10.1186/s12974-016-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luan L, Patil NK, Guo Y, Hernandez A, Bohannon JK, Fensterheim BA, et al. Comparative transcriptome profiles of human blood in response to the toll-like receptor 4 ligands lipopolysaccharide and monophosphoryl lipid A. Sci. Rep. 2017;7:40050. doi: 10.1038/srep40050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gandoura S, Weiss E, Rautou PE, Fasseu M, Gustot T, Lemoine F, et al. Gene- and exon-expression profiling reveals an extensive LPS-induced response in immune cells in patients with cirrhosis. J. Hepatol. 2013;58(5):936–948. doi: 10.1016/j.jhep.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Khan HN, Perlee D, Schoenmaker L, van der Meer AJ, Franitza M, Toliat MR, et al. Leukocyte transcriptional signatures dependent on LPS dosage in human endotoxemia. J. Leukoc. Biol. 2019;106(5):1153–1160. doi: 10.1002/JLB.4A0219-050R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gautam Y, Afanador Y, Abebe T, Lopez JE, Mersha TB. Genome-wide analysis revealed sex-specific gene expression in asthmatics. Hum. Mol. Genet. 2019 doi: 10.1093/hmg/ddz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jansen R, Batista S, Brooks AI, Tischfield JA, Willemsen G, van Grootheest G, et al. Sex differences in the human peripheral blood transcriptome. BMC Genomics. 2014;15:33. doi: 10.1186/1471-2164-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syrett CM, Sierra I, Berry CL, Beiting D, Anguera MC. Sex-specific gene expression differences are evident in human embryonic stem cells and during in vitro differentiation of human placental progenitor cells. Stem Cells Dev. 2018;27(19):1360–1375. doi: 10.1089/scd.2018.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Filosi M, Kam-Thong T, Essioux L, Muglia P, Trabetti E, Spooren W, et al. Transcriptome signatures from discordant sibling pairs reveal changes in peripheral blood immune cell composition in Autism Spectrum Disorder. Transl. Psychiatry. 2020;10(1):106. doi: 10.1038/s41398-020-0778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez-Arcelus M, Baglaenko Y, Arora J, Hannes S, Luo Y, Amariuta T, et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet. 2020;52(3):247–253. doi: 10.1038/s41588-020-0579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen BH, Chiang CH, Lin SR, Chao MG, Tsai ST. The influence of age at onset and gender on the HLA-DQA1, DQB1 association in Chinese children with insulin dependent diabetes mellitus. Hum. Immunol. 1999;60(11):1131–1137. doi: 10.1016/s0198-8859(99)00106-8. [DOI] [PubMed] [Google Scholar]
- 53.Megiorni F, Mora B, Bonamico M, Barbato M, Montuori M, Viola F, et al. HLA-DQ and susceptibility to celiac disease: Evidence for gender differences and parent-of-origin effects. Am. J. Gastroenterol. 2008;103(4):997–1003. doi: 10.1111/j.1572-0241.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- 54.Aomatsu M, Kato T, Kasahara E, Kitagawa S. Gender difference in tumor necrosis factor-alpha production in human neutrophils stimulated by lipopolysaccharide and interferon-gamma. Biochem. Biophys. Res. Commun. 2013;441(1):220–225. doi: 10.1016/j.bbrc.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 55.Kim-Fine S, Regnault TR, Lee JS, Gimbel SA, Greenspoon JA, Fairbairn J, et al. Male gender promotes an increased inflammatory response to lipopolysaccharide in umbilical vein blood. J. Matern. Fetal. Neonatal. Med. 2012;25(11):2470–2474. doi: 10.3109/14767058.2012.684165. [DOI] [PubMed] [Google Scholar]
- 56.Aulock SV, Deininger S, Draing C, Gueinzius K, Dehus O, Hermann C. Gender difference in cytokine secretion on immune stimulation with LPS and LTA. J. Interferon Cytokine Res. 2006;26(12):887–892. doi: 10.1089/jir.2006.26.887. [DOI] [PubMed] [Google Scholar]
- 57.Wen F, Guo J, Li Z, Huang S. Sex-specific patterns of gene expression following influenza vaccination. Sci. Rep. 2018;8(1):13517. doi: 10.1038/s41598-018-31999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N. Engl. J. Med. 2008;358(16):1682–1691. doi: 10.1056/NEJMoa0708801.P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ober C, Nord AS, Thompson EE, Pan L, Tan Z, Cusanovich D, et al. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J. Lipid Res. 2009;50(5):798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao TC, Du G, Han L, Sun Y, Hu D, Yang JJ, et al. Genome-wide association study of lung function phenotypes in a founder population. J. Allergy Clin. Immunol. 2014;133(1):248–255. doi: 10.1016/j.jaci.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Igartua C, Mozaffari SV, Nicolae DL, Ober C. Rare non-coding variants are associated with plasma lipid traits in a founder population. Sci. Rep. 2017;7(1):16415. doi: 10.1038/s41598-017-16550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mozaffari SV, DeCara JM, Shah SJ, Sidore C, Fiorillo E, Cucca F, et al. Parent-of-origin effects on quantitative phenotypes in a large Hutterite pedigree. Commun. Biol. 2019;2:28. doi: 10.1038/s42003-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91(5):839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weitkamp LR, Ober C. Ancestral and recombinant 16-locus HLA haplotypes in the Hutterites. Immunogenetics. 1999;49(6):491–497. doi: 10.1007/s002510050525. [DOI] [PubMed] [Google Scholar]
- 69.Rich SS, Concannon P, Erlich H, Julier C, Morahan G, Nerup J, et al. The type 1 diabetes genetics consortium. Ann. N. Y. Acad. Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 70.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8(6):e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan L, Ober C, Abney M. Heritability estimation of sex-specific effects on human quantitative traits. Genet. Epidemiol. 2007;31(4):338–347. doi: 10.1002/gepi.20214. [DOI] [PubMed] [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57(1):11. [Google Scholar]
- 74.Ahsan S, Draghici S. Identifying significantly impacted pathways and putative mechanisms with iPathwayGuide. Curr. Protoc. Bioinform. 2017;57:7. doi: 10.1002/cpbi.24. [DOI] [PubMed] [Google Scholar]
- 75.Donato M, Xu Z, Tomoiaga A, Granneman JG, Mackenzie RG, Bao R, et al. Analysis and correction of crosstalk effects in pathway analysis. Genome Res. 2013;23(11):1885–1893. doi: 10.1101/gr.153551.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Livne OE, Han L, Alkorta-Aranburu G, Wentworth-Sheilds W, Abney M, Ober C, et al. PRIMAL: Fast and accurate pedigree-based imputation from sequence data in a founder population. PLoS Comput. Biol. 2015;11(3):e1004139. doi: 10.1371/journal.pcbi.1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression dataset generated during the current study will be available in the dbGaP repository.
The analysis scripts for this project are available at https://github.com/mstein3/TreatmentResp_RNAseq.