Abstract
Circulating tumor DNA (ctDNA) has become an attractive biomarker in human oncology, and its use may be informative in canine cancer. Thus, we used droplet digital PCR or PCR for antigen receptor rearrangement, to explore tumor-specific point mutations, copy number alterations, and chromosomal rearrangements in the plasma of cancer-affected dogs. We detected ctDNA in 21/23 (91.3%) of histiocytic sarcoma (HS), 2/8 (25%) of oral melanoma, and 12/13 (92.3%) of lymphoma cases. The utility of ctDNA in diagnosing HS was explored in 133 dogs, including 49 with HS, and the screening of recurrent PTPN11 mutations in plasma had a specificity of 98.8% and a sensitivity between 42.8 and 77% according to the clinical presentation of HS. Sensitivity was greater in visceral forms and especially related to pulmonary location. Follow-up of four dogs by targeting lymphoma-specific antigen receptor rearrangement in plasma showed that minimal residual disease detection was concordant with clinical evaluation and treatment response. Thus, our study shows that ctDNA is detectable in the plasma of cancer-affected dogs and is a promising biomarker for diagnosis and clinical follow-up. ctDNA detection appears to be useful in comparative oncology research due to growing interest in the study of natural canine tumors and exploration of new therapies.
Subject terms: Cancer genetics, Cancer models, Haematological cancer, Sarcoma, Tumour biomarkers
Introduction
Cell-free DNA (cfDNA) circulates in the plasma and other body fluids, such as urine, cerebrospinal fluid, pleural fluid, and saliva, as short double-stranded DNA fragments of approximately 160 base pairs. In healthy individuals, cfDNA is released during cell death that occurs via apoptosis or necrosis or by active secretion. It is mainly derived from the hematopoietic lineage with minimal contribution from other tissues1–4. Since its first discovery in humans in 19485, the presence of cfDNA in the blood has been a known indicator of various pathological processes, such as inflammatory disease, sepsis, trauma, stroke, and cancer, and its concentration is correlated with the severity of disease and prognosis2,6–8. In patients with cancer, cfDNA is partially derived from tumor cells and is referred to as circulating tumor DNA (ctDNA). ctDNA carries tumor-related genetic and epigenetic alterations that are relevant to the development, progression, and resistance to therapy of cancer8.
Next-generation sequencing (NGS) has been used in human medicine to identify genetic alterations in ctDNA, which may include mutations in tumor suppressor genes such as TP53 in patients with head and neck cancer9, mutations in oncogenes such as EGFR or KRAS in patients with non-small-cell lung cancer10,11, and copy number alterations (CNA) and chromosomal rearrangements as observed in the variable-diversity-joining (VDJ) receptor gene sequences in lymphoma12. In the past few decades, strategies including the study of loss of heterozygosity (LOH), DNA methylation, DNA integrity, NGS, or digital PCR have been successfully developed to detect somatic alterations in the plasma of patients, making ctDNA a new powerful biomarker for cancer8,13. In addition to being a minimally invasive and robust approach, the analysis of ctDNA has several clinical applications, such as the early detection of cancer, prognosis, real-time monitoring of treatment response, and the identification of appropriate therapeutic targets and resistance mechanisms2,8,10,11,14.
In veterinary medicine, the use of cfDNA as a biomarker has recently gained attention, particularly in dogs. As in humans, it has been found that cfDNA has a short half-life in canine plasma (approximately 5 h)15, and various diseases cause an increase in its concentration, including immune-mediated hemolytic anemia16, sepsis, severe trauma, and inflammation17–19. Moreover, the concentration of cfDNA is correlated with the severity of various diseases and prognosis in dogs18 and is of potential interest in canine cancers. Studies have shown that dogs with lymphoid neoplasia and mammary carcinoma have higher plasma cfDNA concentrations than do controls19,20. In the last decade, the presence of recurrent somatic alterations has been identified in several canine cancers, such as multicentric lymphoma21,22, histiocytic sarcoma (HS)23–27, and oral malignant melanoma (OMM)28–32. The detection of cancer-specific recurrent somatic alterations in plasma may allow the development of novel minimally invasive biomarkers for the diagnosis, prognosis, and assessment of responses to treatment in veterinary medicine. Additionally, tests based on ctDNA detection may be useful in the field of comparative oncology research. Indeed, naturally occurring canine cancers have become relevant models for the study of rare human cancers, and have been used for the discovery of mutations for the development and screening of targeted therapies27.
In this proof-of-concept study, our objective was to examine the presence of ctDNA and whether several types of recurrent somatic alterations are detectable in the plasma of dogs with three types of malignancies that may serve as models for their human counterparts, i.e., histiocytic sarcoma27, oral malignant melanoma33, and multicentric lymphoma34. Further, we aimed to evaluate whether ctDNA can be used in veterinary medicine for cancer diagnosis and to evaluate responses to treatment with chemotherapy by monitoring minimal residual disease (MRD) in dogs with lymphoma.
Results
Characteristics of the cohort
Characteristics of the dogs
Plasma samples were collected from 49 dogs with histiocytic sarcoma (17 disseminated forms, 30 localized forms, and 2 unknown), 16 dogs with oral melanoma (OMM), and 25 dogs with multicentric lymphoma (including 18 high-grade B-cell, 2 high-grade T-cell, and 3 low-grade lymphomas) (Supplementary Table 1). Matched tumor samples were available from 45 dogs with HS, 10 with OMM, and 14 with multicentric high-grade B-cell lymphomas. Plasma samples from 19 healthy dogs (mean age at sampling was 7.4 years), 14 dogs with non-cancerous diseases (mean age at sampling was 7.3 years), and 10 dogs with various cancers (mean age at sampling was 6.9 years) were collected as controls. Among the healthy dogs and dogs with cancer, the most represented breeds were the Bernese mountain dog (9/19 and 45/100, respectively) and the flat-coated retriever (n = 8/19 and 7/100, respectively).
Quantification of cfDNA in the plasma
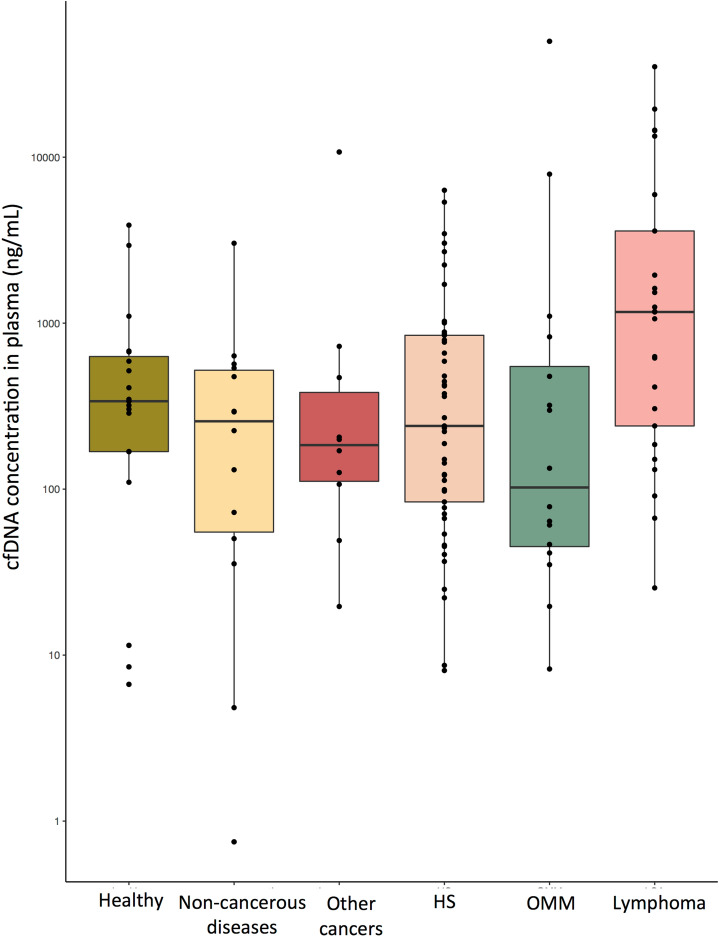
The cfDNA in the plasma of dogs was accurately quantified by the Droplet Digital PCR (ddPCR) method using the wild-type (wt) PTPN11 gene as a reference. The median cfDNA concentrations in healthy, HS, multicentric lymphoma, OMM, other cancers, and non-cancerous dogs were 339 ng/mL (mean 678 ng/mL; range 6.7–3895 ng/mL), 240 ng/mL (mean 827 ng/mL; range 8.1–6320 ng/mL), 1168 ng/mL (mean 4764 ng/mL; range 25.4–35,079 ng/mL), 106 ng/mL (mean 3833 ng/mL; range 8.2–49,910 ng/mL), 185 ng/mL (mean 1282 ng/mL; range 19.6–10,746 ng/mL), and 259 ng/mL (mean 454 ng/mL; range 0.7–3030 ng/mL), respectively (Fig. 1). Although dogs with multicentric lymphoma had higher concentrations of cfDNA in the plasma than the other groups, this was not significant (Kruskal–Wallis test, p = 0.53). Moreover, advanced clinical stage was not associated with higher quantities of cfDNA in our cohort of dogs with lymphomas (stage V vs. III–IV; Wilcoxon test, p = 0.45) or oral melanomas (stage IV vs. II–III; Wilcoxon test, p = 0.76). Similarly, the clinical presentation of HS was not linked to the quantity of cfDNA in the plasma (disseminated vs. localized forms, Wilcoxon test, p = 0.26; external vs. visceral forms, Wilcoxon test, p = 0.39).
Figure 1.
Quantification of cfDNA (ng/mL) in the plasma of dogs in different conditions: healthy, non-cancerous disease, various cancers, HS, OMM, and multicentric lymphoma (drawing produced by R studio software Version 1.1.463; Vienna, Austria60). The amount of DNA was determined via the Poisson distribution.
Taken together, these findings indicate that cfDNA is not a sensitive marker for tumor diagnosis and that the detection of tumor-specific genetic alterations is required to confirm the existence of a malignancy.
Detection of ctDNA in the plasma of dogs with cancer
Detection of ctDNA in canine histiocytic sarcoma by targeting tumor-specific mutations
The knowledge of tumor-specific and recurrent mutation hotspots is required to detect ctDNA by the screening of point mutations. PTPN11 is an oncogene involved in the MAPKinase pathway and is mutated in 23–56.7% of HS cases, mainly in two hotspots (E76K or G503V), depending on clinical presentation25–27.
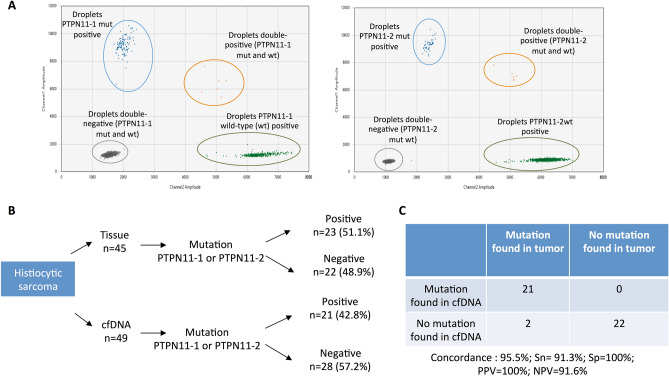
Our results showed that PTPN11 mutations were found in 23/45 (51.1%) of HS tumor samples. Among these 23 dogs, the same mutation was found in the plasma of 21/23 (91.3%). Further, for the 21 plasma samples with a detectable PTPN11 mutation, the variant allelic fraction ranged from 0.056–36% (Supplementary Table 1). No mutations were detected in the plasma of dogs with wt PTPN11 tumors (Fig. 2).
Figure 2.
(A) An example of data output from the ddPCR multiplex assay for absolute quantification of PTPN11 mutations (PTPN11-1 for E76K variant and PTPN11-2 for the G503V variant) in canine HS. Exemplary two-dimensional cluster plots from QuantaSoft software (version 1.7.4, Bio-Rad, freely available after registration: https://www.bio-rad.com/fr-fr/sku/1864011-quantasoft-software-regulatory-edition) in which PTPN11-1mut (left) or PTPN11-2mut (right) are plotted against the PTPN11-1 wild-type (wt) and PTPN11-2wt respectively. The droplets form clusters that should arrange orthogonally to each other and represent the PTPN11mut/PTPN11wt negative (double-negative droplets, grey), PTPN11mut positive (blue), PTPN11wt positive (green), and PTPN11mut/PTPN11wt positive samples (double-positive droplets, orange). (B) Flowchart summarizing the results of the ddPCR assay targeting the PTPN11 mutations (E76K and G503V) in the tumor and plasma samples from 49 dogs with HS. cfDNA: cell-free DNA. (C) Parameters of the test for the detection of the PTPN11 mutation in the plasma of dogs with HS. PPV positive predictive value, NPV negative predictive value.
Detection of ctDNA in canines with OMM by targeting copy number alterations (CNAs)
To detect ctDNA in OMM patients, we targeted the MDM2 and TRPM7 amplifications that are present in up to 50% of canines with OMM28,30–32. The copy number was determined in tumor and plasma samples via ddPCR. Of the ten OMM cases for which tumor samples were available, MDM2 amplification was found in 3/10 tumors and was detected in the plasma of 1/3 of the corresponding dogs (Table 1). Similarly, TRPM7 amplification was found in 8/10 tumors, but was detected in the plasma of only one dog (Table 1). These results indicate that specific tumor CNAs were detectable in the plasma of 2/8 (25%) dogs with OMM.
Table 1.
Results of the ddPCR assay to identify the CNA of the TRPM7 and MDM2 genes in tumors and plasma of dogs with OMM.
| Dog Id | Tumor | cfDNA |
|---|---|---|
| Gain detected: copy number (95% confidence interval) | Gain detected: copy number (95% confidence interval) | |
| 19006 | TRPM7: 37.90 (28.41–47.39) | TRPM7: 2.80 (2.58–3.03) |
| 18394 | TRPM7: 7.98 (7.71–8.25) | No |
| 18657 | TRPM7: 3.01 (2.89–3.14) | No |
| 18718 | MDM2: 21.26 (19.37–23.15) | MDM2: 3.02 (2.19–3.85) |
| TRPM7: 7.46 (7.17–7.75) | No | |
| 18395 | MDM2: 52.96 (45.74–60.17) | No |
| TRPM7: 8.78 (8.37–9.19) | No | |
| 18303 | TRPM7: 8.88 (8.55–9.20) | No |
| 18333 | MDM2: 4.30 (3.91–4.68) | No |
| TRPM7: 27.24 (25.94–28.54) | No | |
| 18848 | TRPM7: 9.12 (8.78–9.46) | No |
Eight dogs had at least one gene amplified in their tumors, and two dogs had a copy number imbalance in the plasma.
Detection of ctDNA in canine multicentric lymphoma by targeting chromosomal rearrangements
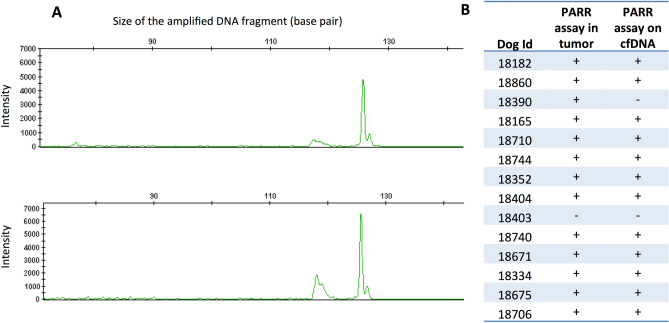
ctDNA can be identified by tumor-specific chromosomal rearrangements, as is performed for humans with multicentric lymphomas12. VDJ recombination of the genes for immunoglobulins and T-cell receptors occurs in the early stages of lymphocyte maturation and is specific to the lymphocyte clone. Thus, in lymphomas, the VDJ sequence is unique and constitutes a specific marker of the tumor clone12. To identify lymphoma-specific VDJ rearrangements, we performed PCRs for antigen receptor rearrangement (PARR) in 14 dogs with high-grade B-cell multicentric lymphoma (11 cases were classified as diffuse large B-cell lymphomas, and 3 were not classified). PARR was positive and detected a clonal rearrangement in the tumors from 13/14 dogs. To detect ctDNA, PARR was performed on cfDNA extracted from plasma at the time of diagnosis. All samples except one were positive and had the same clone in the tumor and plasma samples (Fig. 3). These results indicate that tumor-specific chromosomal rearrangements are detectable in the plasma of dogs with multicentric lymphomas and that a majority of canine lymphoma patients (92.3% in our cohort) had detectable ctDNA at the time of diagnosis.
Figure 3.
(A) Results of a PARR genotyping on the DNA extracted from a tumor sample (above) and plasma (below) from a dog with high-grade multicentric lymphoma (Id 18175) (drawing produced by GeneMapper Software 4.1: https://www.thermofisher.com/ order/catalog/product/4366925). The same clonal antigen receptor rearrangement (IgH major) was found with a high intensity in the tumor and the plasma. (B) Results of the PARR assay on canine lymphoma. The results on DNA extracted from the tumor and DNA extracted from plasma are shown. PARR was positive (clonal band) in the tumor and cfDNA for 12/13 dogs.
The potential clinical applications of the ctDNA biomarker in veterinary medicine and oncology research
ctDNA has become an attractive biomarker for monitoring human cancer patients, with clinical applications in early cancer detection, prognosis, real-time monitoring of treatment response, and identification of appropriate therapeutic targets and resistance mechanisms, among other things. At the same time, dogs are recognized as underutilized models for developing new therapies in high-grade B-cell lymphoma34,35 or rare human cancers such as HS27. Here, we illustrate two potential applications for ctDNA detection in veterinary medicine and comparative oncology for the diagnosis and monitoring of MRD.
The use of ctDNA in diagnostics: PTPN11 mutation in canine HS
The knowledge of a recurrent tumor-specific mutation may be useful in the development of non-invasive molecular diagnostic tools36,37. To assess the sensitivity and specificity of the PTPN11 mutation in the screening of plasma to diagnose HS, we tested the plasma of 49 dogs with HS, 19 healthy dogs, 14 dogs with non-cancerous diseases, and 51 dogs with other cancers (multicentric lymphoma, OMM, soft tissue sarcoma, Langerhans cell sarcoma, nephroblastoma, hemangiosarcoma, and ocular melanoma) (Supplementary Table 1). The PTPN11 mutation was detected in the plasma of 21/49 dogs with HS and 1 dog with a non-cancerous disease. The mutation was absent from the plasma of healthy dogs and those with other cancers. The screening for PTPN11 mutations in the plasma had a specificity of 98.8% and a sensitivity of 42.8% for the diagnosis of canine HS. However, the sensitivity varied according to the characteristics of HS and its clinical presentation. It was higher among the Bernese mountain dogs (53.8%; Fisher exact test p = 0.0026), visceral forms of HS (59.4%; Fisher exact test, p = 0.0042), and in the thoracic location (77%; Fisher exact test p = 0.01) (Supplementary Table 2). This was due to the higher frequency of PTPN11 mutations in Bernese mountain dogs (Fisher exact test, p = 0.0015), the visceral forms of HS (Fisher exact test, p = 0.008), and the presence of pulmonary involvement (Fisher exact test, p = 0.002) in this cohort.
The use of ctDNA for the monitoring of MRD: example of multicentric diffuse large B-cell lymphoma
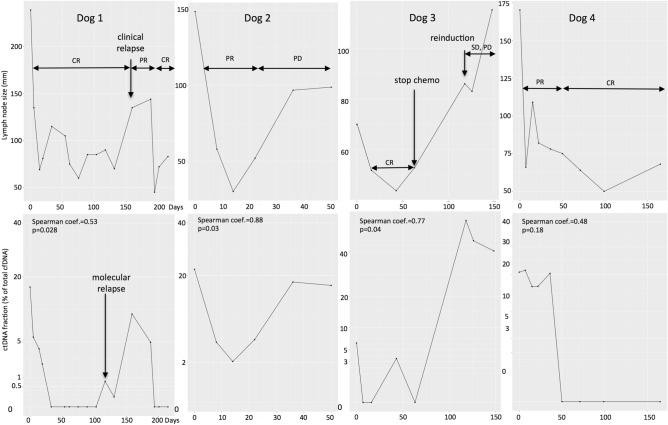
In lymphoma, the unique VDJ rearrangement of the tumor clone constitutes a “barcode” that is used for the follow-up of human lymphoma patients in clinical trials12. In this study, four dogs with multicentric diffuse large B-cell lymphoma were included prospectively, and their lymphoma-specific VDJ rearrangements were sequenced to perform ctDNA analysis during follow-up. Three dogs received cyclophosphamide, doxorubicin, vincristine, and prednisolone (CHOP)-based chemotherapy38–40, and one dog (dog 3) received L-asparaginase at induction of chemotherapy, followed by vincristine injections alone. In the absence of response or relapse, L-asparaginase and lomustine were used as rescue treatments. Before the initiation of chemotherapy, the ratio of ctDNA (% of cfDNA) was determined in all dogs (mean proportion: 15.5% of total cfDNA before the initiation of chemotherapy; min = 6.25%, max = 22%). The monitoring details of each case are summarized in Supplementary File 1 and represented in Fig. 4.
Figure 4.
Course of disease for four dogs with lymphoma that were treated using chemotherapy (dog 1: 18182, dog 2: 18710, dog 3: 18680, dog 4: 18744) (drawing produced by R studio software Version 1.1.463; Vienna, Austria60). For each dog, the figures above represent the tumor burden with lymph node size (sum of longest diameter of at least 2 lymph nodes; in mm). The figures below represent the ctDNA measurement (% of total cfDNA in plasma) during the disease. For each case, a Spearman correlation analysis was performed to assess the correlation between the tumor burden and ctDNA quantity (coefficient and p value). CR complete response, PR partial response, SD stable disease, PD progressive disease.
In all dogs examined, the quantity of ctDNA decreased dramatically with the initiation of chemotherapy and followed the clinical response, reflecting the chemosensitivity of canine high-grade lymphoma. The complete response observed in three dogs (dogs 1, 3, and 4) matched molecular remission. Similarly, the progressive disease observed in three dogs (dogs 1, 2, and 3) matched increases in ctDNA quantity. Clinical relapse was identified in one dog (dog 1), and the molecular relapse was detected 42 days earlier. Finally, there was a significant correlation between lymph node size and ctDNA quantity in three dogs (dogs 1, 2, and 3). Altogether, these results suggest that ctDNA kinetics during chemotherapy treatment in dogs with multicentric lymphoma correlate with lymph node size and reflect the response to treatment.
Discussion
cfDNA is an attractive biomarker for use in human medicine, particularly in oncology, and it has been evaluated in different canine diseases16,17,19,20,41–44. The cfDNA concentration in dogs is correlated with disease severity and prognosis16,17,20,44. In our study, the small sample size may be a limiting factor in identifying a link between clinical stage and cfDNA quantity in dogs with multicentric lymphoma and OMM, and larger cohorts are needed to further explore this hypothesis. The cfDNA concentrations we identified were in agreement with those from previous studies, although previous studies have reported highly variable cfDNA concentrations ranging from a median of 4–583 ng/mL in healthy dogs. This inter-study variability in the results for healthy dogs may arise due to the use of different extraction kits and quantification methods16,20,42,44 and limits the use of cfDNA as a global biomarker for cancer diagnosis.
In human oncology, current studies are focused on the analysis of DNA released from tumor cells in the plasma (ctDNA), which is much more cancer-specific. Currently, a ctDNA assay for the detection of EGFR mutations in patients with non-small-cell lung cancer (NSCLC) has been approved by the Food and Drug Administration, and ctDNA assays for EGFR in NSCLC and for KRAS in colorectal cancer are available for commercial use in Europe3,4,45. As in humans, ctDNA has been found in some canine cancers, such as lymphoma20, mammary carcinoma41, and more recently, pulmonary adenocarcinoma43. Further, a previous study demonstrated that detection of the BRAF mutation in urine is effective in the diagnosis of canine urothelial and prostatic carcinomas46. In our study, we focused on identifying ctDNA in plasma as a liquid biopsy. Our results showed that ctDNA was detected in canine HS, OMM, and high-grade multicentric lymphoma. Additionally, the high frequency of ctDNA detection among dogs with HS and lymphoma indicates that ctDNA may serve as a promising biomarker for these hematopoietic malignancies.
We identified ctDNAs thanks to several tumor-associated alterations and showed that point mutations, chromosomal rearrangements, and CNAs were detectable in the plasma of dogs with cancer. We used different methods to identify the targeted alterations, and these have certain advantages and drawbacks. It is important to highlight that the performance of ctDNA detection largely depends on the level of cfDNA in the patient’s plasma, the fraction of ctDNA, and the sensitivity of the method used. As a large amount of DNA is released into the plasma of dogs with lymphoma (median cfDNA of 1168 ng/mL), we found that ctDNA was detected in 92.3% (12/13) of dogs at the time of diagnosis. These results are similar to those of a previous study that found ctDNA in 78% (7/9) of dogs with multicentric lymphoma20 using the same method. In dogs with HS, cfDNA concentrations were much lower (median 240 ng/mL); however, the use of the highly sensitive ddPCR method enabled us to detect the PTPN11 mutation in the plasma of 91.3% (21/23) dogs with tumors that had mutant PTPN11. The assay had positive and negative predictive values of 100% and 91.6%, respectively, for the detection of PTPN11 mutations. Finally, low cfDNA levels may be an issue in the study of ctDNA in OMM. The most recurrent genetic alterations in cancer are CNAs targeting oncogenes such as MDM2 and TRPM728,30–32, and we identified these tumor-associated amplifications in cfDNA via ddPCR in 1/3 and 1/8 cases, respectively. The detection of CNAs in cfDNA is challenging and depends on the level of amplification of the gene in the tumor, which in turn may involve sequencing at high depths41,47 and is not yet cost-effective for veterinary medicine. In these conditions, the analysis of ctDNA for the targeting of tumor-specific CNA is inappropriate in canine melanoma.
Further, we evaluated the potential clinical application of ctDNA analysis in veterinary oncology. Two aspects were investigated: the value of ctDNA analysis as a tool for HS diagnosis and MRD follow-up in dogs with lymphoma. The diagnosis of canine HS may be challenging because of hidden tumor locations (e.g., lungs, mediastinum, and central nervous system) and the potential difficulty in differentiating it from reactive inflammatory disease or other malignancies48. Thus, we assessed the value of a ctDNA test to diagnose HS using a noninvasive method. In our study population of 133 dogs, we showed that the screening of PTPN11 in the plasma for the diagnosis of HS was specific (98.8%) and sensitive (42.8%), with positive and negative predictive values of 95.5%, and 74.8%, respectively. We found that the PTPN11 mutation was highly specific to HS and was not detected in the plasma of dogs with other cancers. A single diagnosis was classified as a false positive, that of a 12-year-old Bernese mountain dog sampled at the time of euthanasia in old age, and which was classified in the “non-cancerous diseases” group. However, the old dog may have been suffering from undiagnosed or early-stage HS, though a necropsy was not performed to confirm this hypothesis. Another explanation may be the accumulation of somatic variants in the healthy dog due to “clonal hematopoiesis” as is observed in humans4. In humans, this increases with age and is correlated with the risk of further development of hematological cancer. Such accumulation has not been investigated in dogs, and further studies are needed in predisposed breeds to look for PTPN11 mutations in the plasma of apparently healthy dogs to determine whether clonal hematopoiesis precedes HS development. The relatively low sensitivity of this test (42.8%) is because PTPN11 is not mutated in all HS cases (e.g., only 50% of cases in this study). Those mutations are especially found in Bernese mountain dogs25 and the visceral/disseminated forms27. In our study, all cases with PTPN11 mutations were found in Bernese mountain dogs. Moreover, we targeted only the two most frequent PTPN11 mutations (E76K and G503V), and some HS cases may have other PTPN11 mutations27,49. Recently, mutations in KRAS and BRAF were found in 3–7% and less than 1%, respectively, of canine HS cases and were found to be exclusive to the PTPN11 mutation25,27. Thus, the sensitivity of this diagnostic test can be improved by integrating the other mutations for PTPN11, KRAS, and BRAF in the ddPCR assay. The best sensitivity was obtained in the subgroup of dogs with visceral forms and pulmonary involvement (10/13, 77%). Therefore, the screening of PTPN11 mutations in the plasma may be useful for the diagnosis of HS, especially in cases of pulmonary involvement and in Bernese mountain dogs. Further studies are needed to define the value of this test in a wider population by including more breeds and to explore the potential of this biomarker in early-stage diagnosis and follow-up treatment of dogs with HS. In the future, this test may be considered to guide treatment procedures toward targeted therapy in veterinary oncology practice. Indeed, we and others have shown that PTPN11 mutations are associated with the activation of the MAPK pathway, and in vitro tests have shown that canine HS cell lines are sensitive to MEK inhibitors27,49,50. Our ctDNA assay had a sensitivity of 91.3% and a specificity of 100% for the detection of PTPN11 mutations in the plasma of dogs carrying PTPN11-mutated HS, and it could efficiently select dogs that may benefit from this targeted therapy. Finally, as the same mutations for PTPN11 are found in disseminated human HS27, the identification and kinetic quantification of such driver mutations in the plasma are key to proposing and evaluating new targeted therapies in canine models for rare and aggressive human cancer, with benefits for both veterinary and human medicine.
Another promising application of ctDNA analysis in veterinary medicine is the follow-up of MRD during chemotherapy to assess treatment efficacy. In our pilot study, we found that ctDNA levels were correlated with the clinical evaluation of canine multicentric diffuse large B-cell lymphomas, and in the one dog that experienced a relapse under treatment, ctDNA was detected 42 days before clinical relapse. These results are concordant with the studies of MRD targeting circulating tumor cells by real-time PCR, which showed that clinical relapse could be anticipated by molecular evaluation until 86 days51. Further studies are needed to explore the potential of ctDNA in monitoring MRD in canine lymphomas and to demonstrate its utility in the treatment course in dogs. Nevertheless, these findings as well as the detection of ctDNA 1 year after the surgical excision of a mammary carcinoma due to lung metastasis41 suggest that ctDNA analysis in veterinary oncology may serve as an objective parameter for treatment response and earlier relapse detection.
In conclusion, this study shows that several somatic alterations can be found in the plasma of dogs with HS, OMM, and multicentric lymphoma, indicating the presence of ctDNA in canine cancers, especially in hematopoietic malignancies. We explored two potential applications of this minimally invasive assay. First, the HS signature of PTPN11 mutations and their detection in plasma may be used as a ctDNA diagnostic test to confirm the occurrence of HS, especially in visceral forms. Second, the search for lymphoma-specific rearrangements in the plasma of affected dogs appears to be a promising personalized method to assess treatment efficacy and follow-up. ctDNA has numerous applications in human medicine and appears to be a promising noninvasive tool in veterinary medicine with potential for future comparative oncology studies.
Methods
Sample collection
Blood and tissue biopsy samples from dogs were collected by veterinarians through the Cani-DNA BRC (http://dog-genetics.genouest.org). For each dog, 2–5 mL of blood was taken during a routine examination or the standard care protocol for the dog, with written consent from the owners. Blood was collected in ethylenediaminetetraacetic acid (EDTA) or Streck tubes (Streck Cell-Free DNA BCT Genomax). Plasma was separated from the blood cells and cell debris by centrifugation at 1500 × g for 10 min followed by a spin of 16,000×g for 10 min and was stored at − 20 °C before DNA purification. The work with canine samples was approved by the CNRS ethical board, France (35-238-13), and the methods were performed per the relevant regulations. The diagnosis of cancer was confirmed via histopathology or cytology by a board-certified pathologist (JA, LD).
Case selection for monitoring of MRD
The study on dogs with a clinical follow-up to study MRD was performed from October 2017 to October 2018. Dogs were prospectively enrolled in the study if they were diagnosed by histopathology with diffuse large B-cell lymphoma (DLBCL) and if they had started a chemotherapy protocol. Clinical staging was performed based on clinical examination, CT scan, and fine-needle aspiration for the detection of liver, spleen, and thorax involvement and bone marrow aspiration cytology for evaluating bone marrow infiltration. Evaluation of the response to treatment and relapse was performed at each visit according to the criteria described for multicentric lymphoma by VCOG (Veterinary Cooperative Oncology Group)52 based on clinical examination, with lymph node measurement and lymph node aspiration cytology performed at the discretion of the clinician. Plasma was sampled before the administration of chemotherapy every week during the first month, and subsequently every 2–4 weeks until relapse or death occurred.
DNA extraction
Tumor DNA was extracted from RNAlater- or FFPE-preserved tumor samples using the DNA Nucleospin Tissue or FFPE Tissue DNA kits (MACHEREY NAGEL, Düren, Germany) according to the manufacturer’s protocol. cfDNA was extracted from 2 mL of plasma with NucleoSnap DNA Plasma (MACHEREY NAGEL, Düren, Germany), and eluted in 50 µL of elution buffer according to the manufacturer’s protocol. When less than 2 mL of plasma was available, the volume was made equivalent using PBS (Thermo Fisher Scientific, Waltham, MA, USA).
PARR analysis
For dogs diagnosed with high-grade multicentric lymphoma, paired samples (tumor DNA and plasma DNA) were analyzed by PCR for antigen receptor rearrangement (PARR). M13-tailed primers were used to target the immunoglobulin heavy chain genes in B lymphocytes and the T-cell receptor γ in T lymphocytes (Table 2)18,53,54. Amplification was performed using the Type-it Multiplex PCR Master Mix (QIAGEN, Hilden, Germany), as described previously55 using the following conditions: denaturation at 92 °C for 5 min, 35 cycles of 95 °C for 8 s, 49 °C for 10 s, and 72 °C for 15 s (for IgH major, IgH minor, and TCR Burnett), or using 30 cycles of 95 °C for 45 s, 48 °C for 30 s, and 72 °C for 15 s (for IgH Tamura, T Yagihara VA, and Vb)55. The PCR amplicons were sequenced using the 3130 ABI sequencer (Applied Biosystems), and clonal profiles were determined using the Genemapper Software.
Table 2.
Primers used in the study.
| Primer targeted region | Primer sequence | Purpose | |
|---|---|---|---|
| IgH major | cacgacgttgtaaaacgacCAGCCTGAGAGCCGAGGACAC | PARR | |
| TGAGGAGACGGTGACCAGGGT | |||
| IgH minor | cacgacgttgtaaaacgacCAGCCTGAGAGCCGAGGACAC | PARR | |
| TGAGGACACAAAGAGTGAGG | |||
| TCR | cacgacgttgtaaaacgacCGTGTACTACTGCGCTGCCTGG | PARR | |
| TGTGCCAGGACCAAGCACTTTGTT |
| Primer targeted region | MIQE context | Purpose | Elongation temperature (°C) |
|---|---|---|---|
| PTPN11 hot spot 1 (E76K) | GATTCAGAACACTGGTGATTACTATGACTTGTATGGAGGGGAAAAGTTTGCCACGTTGGCT[G/A]AGTTGGTCCAGTATTATATGGAACATCACGGACAATTAAAAGAGAAGAATGGAGATGTTAT | ddPCR: mutation detection of PTPN11 E76K | 58 |
| PTPN11 hot spot 2 (G503V) | GGTGTTGACTGCGACATTGATGTTCCCAAAACCATTCAGATGGTGCGGTCTCAGAGGTCAG[G/T]GATGGTCCAGACAGAAGCACAGTACCGATTCATTTATATGGCTGTCCAGCATTACATTGAA | ddPCR: mutation detection of PTPN11 G503V | 58 |
| TRPM7 | CATTACCTGTGTTTACTCCTCCAGTTAAAATCCAGGCTCCGGTTGTAACTGCAGCTTTAATAAGACCCTTTCCAAGCAGCTGCTTGATTCGTGGGTGAAGCTCAAATTTCTGCATGCCTCCAT | ddPCR: Copy Number determination | 58 |
| MdM2 | TGATCTTCTAGGAGATTTGTTTGGAGTGCCAAGCTTCTCTGTGAAAGAGCACAGGTAAATTCTTCAGTTTAGTCCGTTGTAAAAAGCCAGCTGGGCAAACATTTCAGTTTACCTCCTCCTTTT | ddPCR: Copy Number determination | 58 |
| chr9 control | GCAGTCACCCCATCATCGTCCCGGCAAGGAAGCTTGTGCACAAACGCCAGCATCTGGTACTTGGCCCTGATGAATTCTGACTTGATGGGGCTGGGCAGGGCGCATAGGATGCGACGGTGAGCA | ddPCR: Copy Number determination | 58 |
| Specific probe for dog 1 | F: CGGCtgTcTATTACTGTGCg | ddPCR MRD follow-up | 60 |
| R: CTGCACCAGGACAAACAGAA | |||
| 56-FAM/TGTCACCCA/ZEN/ATTGGAGCTGATCCC | |||
| Specific probe for dog 2 | F: ACTACCTCCGTGCTTTTGGT | ddPCR MRD follow-up | 60 |
| R: CGTGTCCTCGGCTCTCAG | |||
| 56-FAM/CGGCCGTAT/ZEN/ATTATTGTGTGAAGGC | |||
| Specific probe for dog 3 | F: ACACGGCCGTCTATTACTGT | ddPCR MRD follow-up | 60 |
| R: GTTCCTGGCCCCAATAGTCA | |||
| 56-FAM/ACGGCCGTC/ZEN/TATTACTGTTTCGGG | |||
| Specific probe for dog 4 | F: CGACGGTAGCTACGGTAACAAT | ddPCR MRD follow-up | 60 |
| R: GGTCTCACCCGCACAGTAAT | |||
| 56-FAM/AGACCGACG/ZEN/GTAGCTACGGTAACA | |||
| chr26 control | F: TGAGACAACCCTCCC | ddPCR MRD follow-up | 60 |
| R: CTTCTCTCGCTGAGAAAG | |||
| 5HEX/CTGCCCGCT/ZEN/CTTTGATCCTGAGTC |
Design of primers for digital droplet PCR (ddPCR)
ddPCR assays were designed using the Bio-Rad Droplet Digital PCR Assays and tools available online for mutation (https://www.bio-rad.com/digital-assays/#/assays-create/mutation) or copy number determination (https://www.bio-rad.com/digital-assays/#/assays-create/cnd) to detect the PTPN11 mutations (E76K and G503V) or determine MDM2, TRPM7, and CFA 9 copy numbers, respectively. The region of CFA 9 was used as an internal control, as it was previously shown to have a high copy number stability in OMM28,56. The Bio-Rad MIQE Context locations of primers and probes are shown in Table 2.
For each dog with multicentric lymphoma examined for MRD, a PARR assay was performed on the tumor DNA as described above to isolate the clonal antigen receptor rearrangement. The PCR product was sequenced using a 3130 ABI sequencer with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The sequences overlapping the CDR3 (complementarity-determining region 3) were used to design clone-specific primers and FAM fluorescent probes for ddPCR analysis using the PrimerQuest Tool (Integrated DNA Technologies, Coralville, IA, USA). A region of CFA26 was used as the control gene, as it was previously found to be relatively stable in canine lymphomas57 (Table 2).
ddPCR protocol and analysis
The ddPCRs assay was performed as previously described by Lodrini et al.58 or Mochizuki et al.46. Briefly, a 20 μL reaction volume containing the droplet Supermix (Bio-Rad Laboratories, Hercules, CA, USA) with a final concentration of 500 nM of forward and reverse primers, 250 nM of VIC- and FAM-labeled probes, and 8 µL of eluted cfDNA as input (mean: 15.2 ng) or 80 ng of DNA from tumor tissue isolated from each patient sample. Next, the PCR reaction mixtures were partitioned into an emulsion of approximately 20,000 droplets using a QX200 ddPCR droplet generation system (Bio-Rad). PCR was conducted using the GeneAmp PCR system 9700 thermocycler (Applied Biosystems) using the following program: 95 °C for 10 min; 40 cycles of 94 °C for 30 s, and an elongation temperature of 60 s, and subsequently 98 °C for 10 min. After PCR, the droplets were analyzed on a QX200 Droplet Reader (Bio-Rad). The concentrations of the target sequences were calculated using the Poisson distribution in the software Version 1.7.4 (Bio-Rad). Wild-type control DNA and the non-template control reactions were included in each experiment. To ensure experiment quality, wells with total droplet counts less than 8000 were considered invalid, and the experiment was repeated to obtain a sufficient number of droplets.
The PTPN11 mutation frequency in cfDNA was determined by calculating the fraction of the mutant PTPN11 molecule concentration A (copies/μL) relative to the wt PTN11 reference molecule concentration B (copies/μL) (fraction = A/(A + B)). The estimation of the false-positive rate was determined by performing three experiments for each assay using the wt samples, in which the total numbers of detected mutated positive droplets were used to determine the thresholds above which the positive droplets in patient samples were considered as true positives. According to the input quantity, only reactions containing at least one, two, three, and four positive droplets for 0–7 ng, 7–15 ng, 15–100 ng and > 100 ng of input, respectively, were classified as positive in the ddPCR analysis.
In canine OMM, MDM2 and TRPM7, copy numbers were determined using the QuantaSoft analysis software (version 1.7.4, Bio-Rad), which calculated the ratio of the target molecule concentration A (copies/μL) to the reference molecule concentration B (copies/μL) multiplied by the number of reference copies in the canine genome (copy number = (A/B)*2). These genes were considered amplified when the copy number was ≥ 3 and the confidence interval was > 2. The same method was used to quantify MRD in the plasma of dogs with lymphoma (A: lymphoma-specific antigen receptor rearrangement, and B: CFA 26 region, control, and ctDNA% = (A/B) × 100).
Statistical analysis
Statistical analyses were performed using R studio software (Version 1.1.463; Vienna, Austria)59,60. The normal distribution of the data was evaluated using the Shapiro–Wilk test. When the data were not normally distributed, non-parametric tests were performed. The Wilcoxon test was used to compare the mean cfDNA concentrations and WHO stage in lymphoma and melanoma cases. A Kruskal–Wallis test was used to compare the cfDNA concentration in plasma between the different clinical groups. The Fisher exact or Chi-square tests were performed to evaluate associations between the presence of PTPN11 mutations and clinical parameters. Finally, Spearman correlation tests were used to evaluate the correlation between lymph node size and ctDNA fraction in dogs with multicentric lymphoma that were receiving chemotherapy.
Supplementary Information
Acknowledgements
The authors acknowledge the technicians, pathologists, veterinarians, and nurses that contributed to the project (particularly Drs. A. Garand, F. Allais, and A-S Guillory, IGDR, Rennes), the Cani-DNA Biological Resource Center (Biosit, Rennes: http://dog-genetics.genouest.org) that provided the samples, Stéphane Dréano for the sequencing facilities, and Yann Le Cunff for help in statistics. The authors acknowledge Laurent Griscom (IGDR, Rennes) who assisted them in proofreading the manuscript.
Author contributions
Conception and design: A.P., and B.H. Development of methodology: A.P., J.A.D., E.C., A.L., and B.H. Acquisition of data: A.P., J.A.D., P.De.F., E.C., N.B., R.U., M.R., A.B., R.F., L.D., P.D., J.A., A.L., and B.H. Analysis and interpretation of data: A.P., J.A.D., T.D., C.K., P.De.F., and B.H. Writing, review, and/or revision of the manuscript: A.P., J.A.D., P.De.F., T.D., J.A., C.A., and B.H.
Funding
AP is funded by a PhD fellowship from the FHU CAMIn University-Hospital Federation Cancer, Microenvironment & Innovation (2018–2020) of the University of Rennes 1, France. This study was partly funded by the American Kennel Club Health Foundation CHF grant 2446. The Cani-DNA BRC (http://dog-genetics.genouest.org), is funded through PIA1 funding (2012–2022): the CRB-Anim infrastructure, ANR-11-INBS-0003.
Data availability
The data from the article is available in the supplementary material provided with the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material availlable at 10.1038/s41598-020-80332-y.
References
- 1.Stewart CM, Tsui DWY. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018;228–229:169–179. doi: 10.1016/j.cancergen.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler TM, Spellman PT, Gray J. Circulating-tumor DNA as an early detection and diagnostic tool. Curr. Opin. Genet. Dev. 2017;42:14–21. doi: 10.1016/j.gde.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 3.De Rubis G, Krishnan SR, Bebawy M. Circulating tumor DNA—Current state of play and future perspectives. Pharmacol. Res. 2018;136:35–44. doi: 10.1016/j.phrs.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Merker JD, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 5.Mandel P, Metais P. Les acides nucleiques du plasma sanguin chez 1 homme. C. R. Seances Soc. Biol. Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 6.Saukkonen K, et al. Cell-free plasma DNA as a predictor of outcome in severe sepsis and septic shock. Clin. Chem. 2008;54:1000–1007. doi: 10.1373/clinchem.2007.101030. [DOI] [PubMed] [Google Scholar]
- 7.Saukkonen K, et al. Association of cell-free plasma DNA with hospital mortality and organ dysfunction in intensive care unit patients. Intensive Care Med. 2007;33:1624–1627. doi: 10.1007/s00134-007-0686-z. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 9.van Ginkel JH, Huibers MMH, van Es RJJ, de Bree R, Willems SM. Droplet digital PCR for detection and quantification of circulating tumor DNA in plasma of head and neck cancer patients. BMC Cancer. 2017;17:428. doi: 10.1186/s12885-017-3424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeung KT, More S, Woodward B, Velculescu V, Husain H. Circulating tumor DNA for mutation detection and identification of mechanisms of resistance in non-small cell lung cancer. Mol. Diagn. Ther. 2017;21:375–384. doi: 10.1007/s40291-017-0260-5. [DOI] [PubMed] [Google Scholar]
- 11.Wu T-H, Hsiue EH-C, Yang JC-H. Opportunities of circulating tumor DNA in lung cancer. Cancer Treat. Rev. 2019;78:31–41. doi: 10.1016/j.ctrv.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Kwok M, Wu SP, Mo C, Summers T, Roschewski M. Circulating tumor DNA to monitor therapy for aggressive B-cell lymphomas. Curr. Treat. Options Oncol. 2016;17:47. doi: 10.1007/s11864-016-0425-1. [DOI] [PubMed] [Google Scholar]
- 13.Alcaide M, et al. Multiplex droplet digital pcr quantification of recurrent somatic mutations in diffuse large B-cell and follicular lymphoma. Clin. Chem. 2016;62:1238–1247. doi: 10.1373/clinchem.2016.255315. [DOI] [PubMed] [Google Scholar]
- 14.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 15.Wilson IJ, et al. Kinetics of plasma cell-free DNA and creatine kinase in a canine model of tissue injury. J. Vet. Int. Med. 2018;32:157–164. doi: 10.1111/jvim.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffery U, Ruterbories L, Hanel R, LeVine DN. Cell-free DNA and DNase activity in dogs with immune-mediated hemolytic anemia. J. Vet. Intern. Med. 2017;31:1441–1450. doi: 10.1111/jvim.14808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letendre J-A, Goggs R. Measurement of plasma cell-free DNA concentrations in dogs with sepsis, trauma, and neoplasia. J. Vet. Emerg. Crit. Care (San Antonio) 2017;27:307–314. doi: 10.1111/vec.12592. [DOI] [PubMed] [Google Scholar]
- 18.Burnett RC, et al. Diagnosis of canine lymphoid neoplasia using clonal rearrangements of antigen receptor genes. Vet. Pathol. 2003;40:32–41. doi: 10.1354/vp.40-1-32. [DOI] [PubMed] [Google Scholar]
- 19.Beffagna G, et al. Circulating cell-free DNA in dogs with mammary tumors: Short and long fragments and integrity index. PLoS ONE. 2017;12:e0169454. doi: 10.1371/journal.pone.0169454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer DMW, et al. Quantification of plasma DNA as a prognostic indicator in canine lymphoid neoplasia. Vet. Comp. Oncol. 2007;5:145–155. doi: 10.1111/j.1476-5829.2007.00122.x. [DOI] [PubMed] [Google Scholar]
- 21.Elvers I, et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 2015;25:1634–1645. doi: 10.1101/gr.194449.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas R, et al. Refining tumor-associated aneuploidy through ‘genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk. Lymphoma. 2011;52:1321–1335. doi: 10.3109/10428194.2011.559802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedan B, et al. Molecular cytogenetic characterization of canine histiocytic sarcoma: A spontaneous model for human histiocytic cancer identifies deletion of tumor suppressor genes and highlights influence of genetic background on tumor behavior. BMC Cancer. 2011;11:201. doi: 10.1186/1471-2407-11-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy K, et al. Genome-wide DNA copy number analysis and targeted transcriptional analysis of canine histiocytic malignancies identifies diagnostic signatures and highlights disruption of spindle assembly complex. Chromosome Res. 2019;27:179–202. doi: 10.1007/s10577-019-09606-0. [DOI] [PubMed] [Google Scholar]
- 25.Takada M, et al. Activating mutations in PTPN11 and KRAS in canine histiocytic sarcomas. Genes. 2019;10:505. doi: 10.3390/genes10070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaiwong T, Sirivisoot S, Takada M, Yuzbasiyan-Gurkan V, Kiupel M. Gain-of-function mutation in PTPN11 in histiocytic sarcomas of Bernese mountain dogs. Vet. Comp. Oncol. 2018;16:220–228. doi: 10.1111/vco.12357. [DOI] [PubMed] [Google Scholar]
- 27.Hédan B, et al. PTPN11 mutations in canine and human disseminated histiocytic sarcoma. Int. J. Cancer. 2020;147:1657–1665. doi: 10.1002/ijc.32991. [DOI] [PubMed] [Google Scholar]
- 28.Wong K, et al. Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat. Commun. 2019;10:353. doi: 10.1038/s41467-018-08081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillard M, et al. Naturally occurring melanomas in dogs as models for non-UV pathways of human melanomas. Pigment Cell Melanoma Res. 2014;27:90–102. doi: 10.1111/pcmr.12170. [DOI] [PubMed] [Google Scholar]
- 30.Poorman K, et al. Comparative cytogenetic characterization of primary canine melanocytic lesions using array CGH and fluorescence in situ hybridization. Chromosome Res. 2015;23:171–186. doi: 10.1007/s10577-014-9444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendricks WPD, et al. Somatic inactivating PTPRJ mutations and dysregulated pathways identified in canine malignant melanoma by integrated comparative genomic analysis. PLoS Genet. 2018;14:e1007589. doi: 10.1371/journal.pgen.1007589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prouteau A, et al. Prognostic value of somatic focal amplifications on chromosome 30 in canine oral melanoma. Vet. Comp. Oncol. 2020;18:214–223. doi: 10.1111/vco.12536. [DOI] [PubMed] [Google Scholar]
- 33.Prouteau A, André C. Canine melanomas as models for human melanomas: Clinical, histological, and genetic comparison. Genes. 2019;10:501. doi: 10.3390/genes10070501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito D, Frantz AM, Modiano JF. Canine lymphoma as a comparative model for human non-Hodgkin lymphoma: Recent progress and applications. Vet. Immunol. Immunopathol. 2014;159:192–201. doi: 10.1016/j.vetimm.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeBlanc AK, et al. Perspectives from man’s best friend: National academy of medicine’s workshop on comparative oncology. Sci. Transl. Med. 2016;8:324ps5. doi: 10.1126/scitranslmed.aaf0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki H, Kennedy K, Shapiro SG, Breen M. BRAF mutations in canine cancers. PLoS ONE. 2015;10:e0129534. doi: 10.1371/journal.pone.0129534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiley C, Wise CF, Breen M. Novel noninvasive diagnostics. Vet. Clin. N. Am. Small Anim. Pract. 2019;49:781–791. doi: 10.1016/j.cvsm.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Garrett LD, Thamm DH, Chun R, Dudley R, Vail DM. Evaluation of a 6-month chemotherapy protocol with no maintenance therapy for dogs with lymphoma. J. Vet. Intern. Med. 2002;16:704–709. doi: 10.1111/j.1939-1676.2002.tb02411.x. [DOI] [PubMed] [Google Scholar]
- 39.Moore AS, et al. Evaluation of a discontinuous treatment protocol (VELCAP-S) for canine lymphoma. J. Vet. Intern. Med. 2001;15:348–354. doi: 10.1111/j.1939-1676.2001.tb02328.x. [DOI] [PubMed] [Google Scholar]
- 40.Sorenmo K, et al. Outcome and toxicity associated with a dose-intensified, maintenance-free CHOP-based chemotherapy protocol in canine lymphoma: 130 cases. Vet. Comp. Oncol. 2010;8:196–208. doi: 10.1111/j.1476-5829.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 41.Beck J, et al. Genome aberrations in canine mammary carcinomas and their detection in cell-free plasma DNA. PLoS ONE. 2013;8:e75485. doi: 10.1371/journal.pone.0075485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagawa M, Shimbo G, Inokuma H, Miyahara K. Quantification of plasma cell-free DNA levels in dogs with various tumors. J. Vet. Diagn. Investig. 2019;31:836–843. doi: 10.1177/1040638719880245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorch G, et al. Identification of recurrent activating HER2 mutations in primary canine pulmonary adenocarcinoma. Clin. Cancer Res. 2019;25:5866–5877. doi: 10.1158/1078-0432.CCR-19-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burnett DL, Cave NJ, Gedye KR, Bridges JP. Investigation of cell-free DNA in canine plasma and its relation to disease. Vet. Q. 2016;36:122–129. doi: 10.1080/01652176.2016.1182230. [DOI] [PubMed] [Google Scholar]
- 45.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mochizuki H, Shapiro SG, Breen M. Detection of BRAF mutation in urine DNA as a molecular diagnostic for canine urothelial and prostatic carcinoma. PLoS ONE. 2015;10:e0144170. doi: 10.1371/journal.pone.0144170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KCA, et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 48.Withrow S, Vail D, Page R. Withrow & MacEwen’s Small Animal Clinical Oncology. 5. Amsterdam: Elsevier; 2012. [Google Scholar]
- 49.Tani H, et al. Canine histiocytic sarcoma cell lines with SHP2 p.Glu76Gln or p.Glu76Ala mutations are sensitive to allosteric SHP2 inhibitor SHP099. Vet. Comp. Oncol. 2020;18:161–168. doi: 10.1111/vco.12524. [DOI] [PubMed] [Google Scholar]
- 50.Takada M, et al. Targeting MEK in a translational model of histiocytic sarcoma. Mol. Cancer Ther. 2018;17:2439–2450. doi: 10.1158/1535-7163.MCT-17-1273. [DOI] [PubMed] [Google Scholar]
- 51.Gentilini F, Turba ME, Forni M. Retrospective monitoring of minimal residual disease using hairpin-shaped clone specific primers in B-cell lymphoma affected dogs. Vet. Immunol. Immunopathol. 2013;153:279–288. doi: 10.1016/j.vetimm.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Vail DM, et al. Response evaluation criteria for peripheral nodal lymphoma in dogs (v10)—A veterinary cooperative oncology group (VCOG) consensus document. Vet. Comp. Oncol. 2010;8:28–37. doi: 10.1111/j.1476-5829.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, et al. Development of the polymerase chain reaction assay based on the canine genome database for detection of monoclonality in B cell lymphoma. Vet. Immunol. Immunopathol. 2006;110:163–167. doi: 10.1016/j.vetimm.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Yagihara H, et al. Genomic organization of the T-cell receptor gamma gene and PCR detection of its clonal rearrangement in canine T-cell lymphoma/leukemia. Vet. Immunol. Immunopathol. 2007;115:375–382. doi: 10.1016/j.vetimm.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Gautier, A. & Ibisch, C. Validation d’un test de clonalité pour le diagnostic des lymphomes canins par analyse moléculaire des réarrangements du récepteur à l’antigène (PARR) (2014).
- 56.Hitte C. & André C. Integrated genetic analysis of canine mucosal melanoma from three predisposed breeds. (2019).
- 57.Tsai P-C, Breen M. Array-based comparative genomic hybridization-guided identification of reference genes for normalization of real-time quantitative polymerase chain reaction assay data for lymphomas, histiocytic sarcomas, and osteosarcomas of dogs. Am. J. Vet. Res. 2012;73:1335–1343. doi: 10.2460/ajvr.73.9.1335. [DOI] [PubMed] [Google Scholar]
- 58.Lodrini M, et al. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget. 2017;8:85234–85251. doi: 10.18632/oncotarget.19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gómez-Rubio, V. ggplot2—Elegant graphics for data analysis (2nd Edition). J. Stat. Softw.77. 10.18637/jss.v077.b02 (2017).
- 60.Bunn, A. & Korpela, M. An Introduction to dplR. (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from the article is available in the supplementary material provided with the article.