Abstract
Background & Aims
The fragile X mental retardation protein (FMRP) affects multiple steps of the mRNA metabolism during brain development and in different neoplastic processes. However, the contribution of FMRP in colon carcinogenesis has not been investigated.
Methods
FMR1 mRNA transcript and FMRP protein expression were analyzed in human colon samples derived from patients with sporadic colorectal cancer (CRC) and healthy subjects. We used a well-established mouse model of sporadic CRC induced by azoxymethane to determine the possible role of FMRP in CRC. To address whether FMRP controls cancer cell survival, we analyzed cell death pathway in CRC human epithelial cell lines and in patient-derived colon cancer organoids in presence or absence of a specific FMR1 antisense oligonucleotide or siRNA.
Results
We document a significant increase of FMRP in human CRC relative to non-tumor tissues. Next, using an inducible mouse model of CRC, we observed a reduction of colonic tumor incidence and size in the Fmr1 knockout mice. The abrogation of FMRP induced spontaneous cell death in human CRC cell lines activating the necroptotic pathway. Indeed, specific immunoprecipitation experiments on human cell lines and CRC samples indicated that FMRP binds receptor-interacting protein kinase 1 (RIPK1) mRNA, suggesting that FMRP acts as a regulator of necroptosis pathway through the surveillance of RIPK1 mRNA metabolism. Treatment of human CRC cell lines and patient-derived colon cancer organoids with the FMR1 antisense resulted in up-regulation of RIPK1.
Conclusions
Altogether, these data support a role for FMRP in controlling RIPK1 expression and necroptotic activation in CRC.
Keywords: Colorectal Cancer, FMRP, Necroptosis, RIPK
Abbreviations used in this paper: AnnV, annexin V; AOM, azoxymethane; CRC, colorectal cancer; CREB, cyclic adenosine monophosphate responsive element-binding protein; DAPI, 4´,6-diamidino-2-phenylindole; FMRP, fragile X mental retardation protein; FXS, fragile X syndrome; HRP, horseradish peroxidase; KO, knockout; MLKL, mixed lineage kinase domain-like; NT, colonic samples derived from healthy mucosa of patients without cancer; PARP-1, poly (ADP-ribose) polymerase; PI, propidium iodide; RBP, RNA-binding protein; RIPK1, receptor-interacting protein kinase 1; RT-qPCR, real-time quantitative polymerase chain reaction; SD, standard deviation; SEM, standard error of the mean; T, tumor areas of human CRC samples; TUNEL, deoxyuride-5´-triphosphate biotin nick end labeling
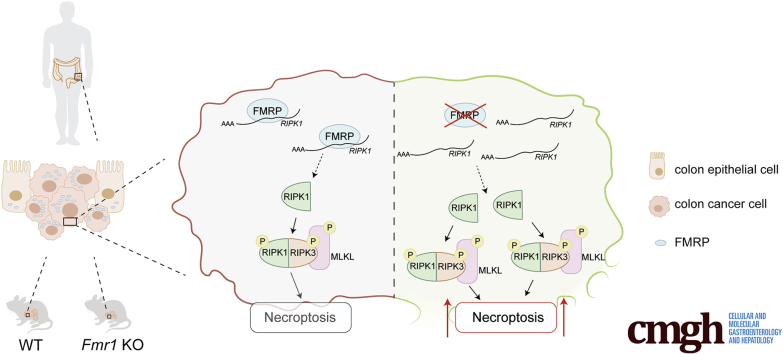
Graphical abstract
Summary.
The identification of a specific target as FMRP that could control directly the necroptosis pathway represents a novel attractive strategy to overcoming programmed cell death resistance in CRC.
Colorectal cancer (CRC) is one of the most common cancers worldwide, causing half-million deaths every year.1 CRC develops in a stepwise manner from normal mucosa to adenomatous polyps to carcinoma, a multistage process characterized by accumulation of genetic mutations, which confer a selective advantage to tumor cell growth.1 These changes, ultimately result in uncontrolled cell growth, resistance to cell death, and clonal tumor development.2 These mechanisms are dictated by alterations of oncogenic and/or tumor-suppressive signaling pathways responsible for the progression from normal mucosa to adenomatous polyp and then to carcinoma.2 During these sequential events driving the neoplastic phenotype, genetic and epigenetic changes that disrupt the balance between cell proliferation and cell death are crucial.2
In addition to a regulation at the level of transcription, the oncogenic and/or tumor-suppressive signaling are tightly regulated at posttranscriptional levels (eg, splicing, transport of the mRNA to the cytoplasm, turnover, storage, and translation) mainly by RNA-binding proteins (RBPs).3 RBPs are interesting in the context of cancer, because many cancer-related proteins are encoded by mRNAs whose expression levels are regulated by RBPs modulating both mRNA translation and turnover.3 Recent studies demonstrated the key contribution of several RBPs in the control of intestinal epithelial cell homeostasis and in response to injury.4 Among the different pathways involved in CRC, a series of evidence have highlighted that colon tumor cells highjack posttranscriptional mechanisms that enable a fast and robust adjustment of protein expression in response to intrinsic and extracellular signals, leading to cell adaptation to the local microenvironment.5 Dysregulated RBPs influence the expression and function of pro-tumorigenic and tumor-suppressor proteins, among others.5 Several studies have provided evidence that RBPs are abnormally expressed in cancer relative to adjacent normal tissues, and their expression correlates with patients’ prognosis.6,7 The fragile X mental retardation protein (FMRP), a RBP involved in multiple steps of mRNA metabolism, is gaining a pivotal importance in controlling the development and growth of different types of human cancer.8, 9, 10 Mutations or absence of FMRP cause fragile X syndrome (FXS), the most frequent form of inherited intellectual disability in humans.11 In brain, the absence of FMRP causes impaired structural and functional synaptic plasticity partially due to defects in locally synthesized proteins, cytoskeletal organization, and receptor mobility.11 FMRP can act as a negative regulator of translation and, in addition, modulates the stability, transport, or editing of the mRNAs depending on the identity of the target mRNA and the cellular context.11,12 Of note, several of the brain-identified FMRP-regulated mRNAs are involved in mechanisms controlling cancer progression and metastasis formation.13
In cancer tissues, FMRP is highly expressed in triple negative breast cancers and in aggressive skin cancer such as melanoma.14,15 In addition, a decreased incidence of different cancer types has been reported in two cohorts, Danish and British respectively, of patients with FXS, and a case report showed an unusual low growth of glioblastoma in a boy with FXS.16,17 Finally, FMRP promotes astrocytoma proliferation via the MEK/ERK signaling pathway.10 Overall, these data suggest that specific FMRP-regulated mechanisms might affect malignant transformation.
In this study, we assessed the role of FMRP in human sporadic CRC by using human cells and mouse models. We observed that the absence of FMRP is protective toward cancer progression and identified the underlying molecular mechanism based on the control of the receptor-interacting serine/threonine-protein kinase 1 (RIPK1), a key mediator of the necroptosis pathway.
Results
FMRP Is Up-regulated in Human CRC Tissues and Cell Lines
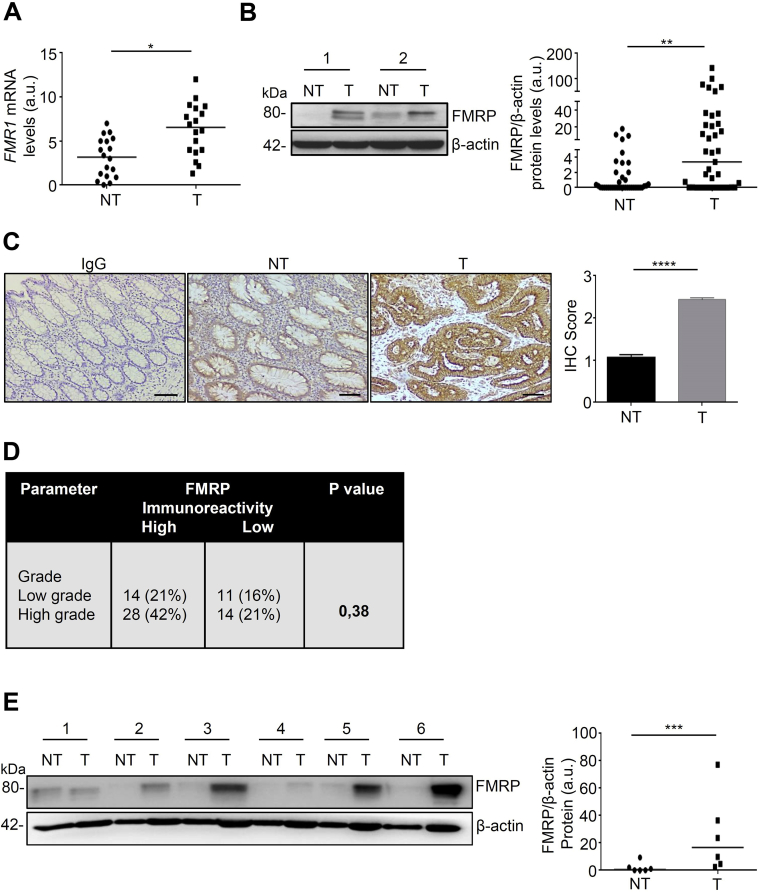
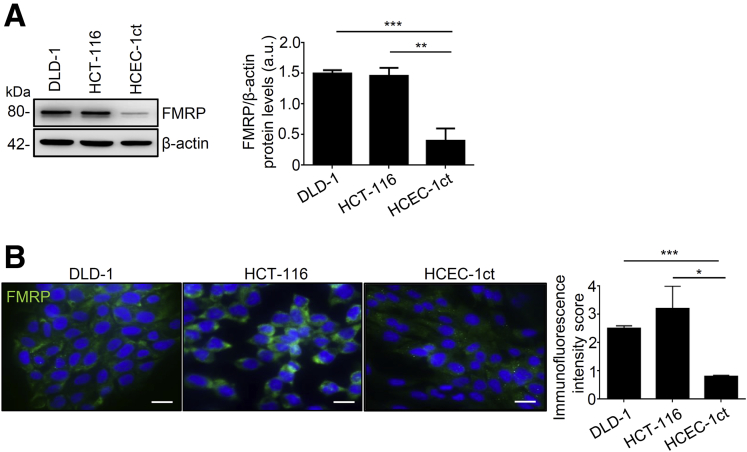
To address the question whether FMRP is involved in survival of patients with CRC, different publicly available datasets were screened for genetic alterations or aberrant protein expression of FMRP. FMR1 mRNA and FMRP protein were found highly expressed in different tissues and in cancer cell types (http://www.cbioportal.org/; https://www.proteinatlas.org/). The Kaplan-Meier analysis from the human protein atlas (https://www.proteinatlas.org/), consisting of 597 patients with CRC, showed a reduced disease-free survival in CRC patients with low expression of FMRP (5-year survival in high expression group, 69%; 5-year survival in low expression group, 59%). However, the analysis of the CRC available dataset on CBioPortal (http://www.cbioportal.org/; 3667 patients) reveals that patients with a nonfunctional or truncated FMRP proteins (49 patients) have a favorable outcome (5-year survival in the group with the mutated FMR1 gene, 70%; 57% in the group with no mutations in the FMR1 gene). Moreover, CRC available dataset from Cancer Genome Atlas (https://portal.gdc.cancer.gov/; 606 patients) reveals that patients with a mutation in the FMR1 gene (48 patients) have a favorable outcome (5-year survival for the group with the mutated FMR1 gene, 73%; 62% in the group with no mutations in the FMR1 gene). Although there are some discrepancies between FMRP expression, FMR1 gene mutations and survival of patients with CRC, the absence of a functional FMRP seems to be protective in cancer progression. We examined FMR1 mRNA and FMRP protein expression in tumor and normal samples. FMR1 mRNA expression was analyzed by real-time quantitative polymerase chain reaction (RT-qPCR) in tumor areas of human CRC samples (T) and in colonic samples derived from healthy mucosa of patients without cancer (NT). FMR1 mRNA expression was higher in cancer samples compared with NT, which showed a lower level of FMR1 mRNA (Figure 1A). We also analyzed FMRP protein levels by Western blotting and immunohistochemistry. Western blotting analysis showed that FMRP was highly expressed in approximately 60% of colon cancer samples (T) compared to healthy subjects (NT) (Figure 1B). These findings were confirmed by immunohistochemistry, revealing an increase of FMRP levels in human CRC samples compared with NT (Figure 1C). Moreover, the majority of patients with high-grade tumors revealed an overexpression of FMRP in CRC samples; however, because of the low number of human patients available, the analysis did not reach statistical significance (Figure 1D). FMRP expression was also analyzed by Western blotting in protein extracts from 6 matched pairs of human CRC and adjacent tissues and found significantly increased in CRC samples, compared with non-tumor mucosa (Figure 1E). In addition, FMRP was detected at high levels in 2 colon cancer cell lines DLD-1 and HCT-116 compared to non-cancer colonic epithelial cell line HCEC-1ct (Figure 2A and B). These data show that FMR1 mRNA and FMRP protein are overexpressed in colon cancer.
Figure 1.
FMRP is overexpressed in human CRC tissues. (A) FMR1 mRNA levels detected by RT-qPCR in colonic samples from healthy subjects (NT) and tumor areas (T) from CRC patients (T); values were normalized to β-actin mRNA. Each point in the graph represents the value of FMR1 mRNA in a single patient (NT versus T, ∗P < .05, n = 18). (B) Left, FMRP levels in representative images of Western blotting from colonic samples derived from 39 healthy subjects (NT) and 39 patients with CRC (T). β-actin was used as a loading control. Right, quantitative analysis of FMRP/β-actin protein ratio as measured by densitometry scanning of Western blots. Each point in the graph indicates the value of FMRP/β-actin in each patient (values are expressed in arbitrary units (a.u.); NT versus T, ∗∗P < .01). (C) Left, representative images of immunohistochemistry (IHC) and (right) quantification of FMRP levels in colon sections derived from healthy subjects (NT) and tumor areas of CRC patients (T) (n = 10 each condition). Antibodies towards Immunoglobulin G (IgG) were used as a negative control (∗∗∗∗P < .0001). Scale bars, 100 μm. (D) Correlation between FMRP immunoreactivity and low or high risk of cancer progression in a cohort of individuals with CRC (n = 67). Number of cases with low-moderate FMRP or high FMRP expression and the percentage of FMRP-positive cases (%) are reported in each patient subgroup (CRC Low grade corresponds to stage I/II, High grade to stage III/IV). (E) Left, representative Western blotting of FMRP levels in tumoral (T) and non-tumoral (NT) areas of the same patients with sporadic CRC (n = 6). β-actin was used as loading control. Right, quantitative analysis of FMRP/β-actin protein as measured by densitometry scanning of Western blots. Values are expressed in arbitrary units (a.u.) (∗∗∗P < .001). Statistical analysis of the data was performed using Student t test, the Mann-Whitney test, and χ2 test.
Figure 2.
FMRP is highly expressed in human CRC cell lines. (A) Left, FMRP levels in representative images of Western blotting from DLD-1, HCT-116 CRC cell lines and the healthy colon epithelial cell line HCEC-1ct. β-actin was used as loading control. Right, quantitative analysis of FMRP/β-actin protein ratio as measured by densitometry scanning of Western blots (values are expressed in arbitrary units (a.u.), mean ± SD, of all experiments; DLD-1 versus HCEC-1ct, ∗∗∗P < .001; HCT-116 versus HCEC-1ct, ∗∗P < .01, n = 4). (B) Left, representative immunofluorescence of FMRP expression, right, quantification in DLD-1, HCT-116 and HCEC-1ct cells. DAPI (blue) and FMRP (green). (Mean ± SD of all experiments; DLD-1 versus HCEC-1ct, ∗∗∗P < .001; HCT-116 versus HCEC-1ct ∗P < .05, n = 5). Scale bars, 50 μm. Statistical analysis of the data was performed using Student t test and the Mann-Whitney test.
CREB Controls FMRP Expression in Human CRC
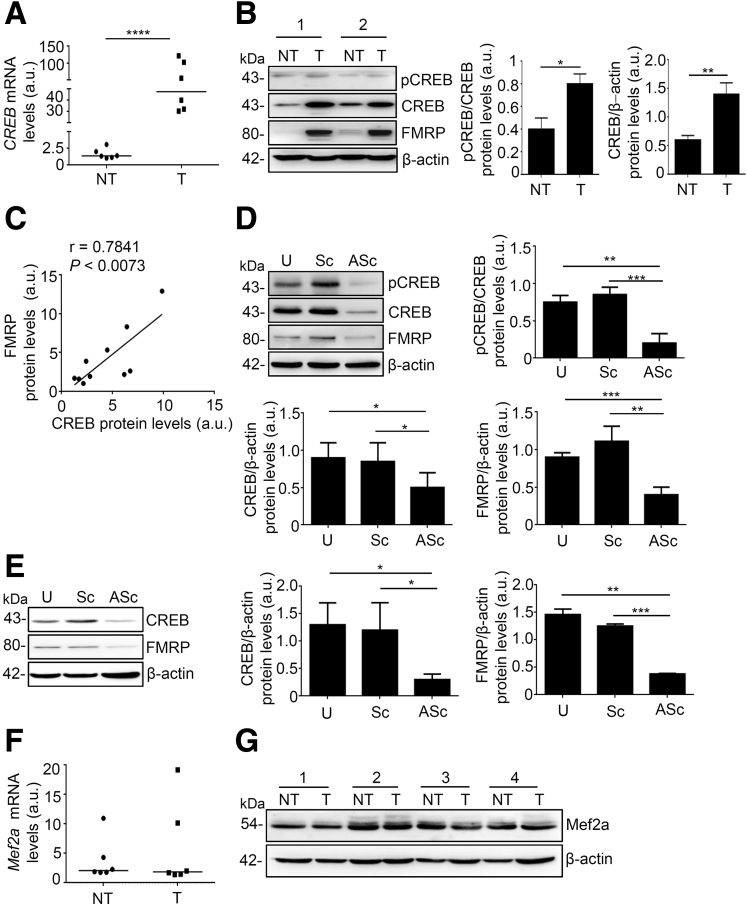
FMRP expression is modulated by the transcription factor cyclic adenosine monophosphate responsive element-binding protein (CREB),18 a protein closely associated with development and progression of human colon cancer.19 CREB mRNA and protein were significantly increased in the areas of the colon tumor, similarly CREB activity (Figure 3A and B). There is a clear direct relationship in individual samples between the expression of CREB and the amount of FMRP in CRC samples (Figure 3C). To investigate whether CREB levels were directly linked to FMRP levels in CRC, CREB expression was inhibited with a specific antisense oligonucleotide (ASc). In DLD-1 and HCT-116 cells CREB antisense oligonucleotide reduced CREB levels and showed a significant decrease of FMRP levels, whereas no effect was observed with the control oligonucleotide (Sc) (Figure 3D and E). The expression of the transcription factor Mef2, which is also involved in cancer and FMRP modulation,20 did not show any change (Figure 3F and G). These findings suggest that in human CRC cells CREB might positively control FMRP expression, consistent with previous studies showing that FMR1 is a CREB-regulated gene.21,22
Figure 3.
CREB controls FMRP expression in CRC. (A) CREB mRNA levels were evaluated by RT-qPCR in colonic samples from paired healthy subjects (NT) and patients with sporadic CRC (T); β-actin mRNA was used as loading control (NT versus T, ∗∗∗∗P < .0001, n = 6 each group). (B) pCREB, CREB, and FMRP expression was evaluated by Western blotting in paired colonic (mucosal) samples from healthy subjects and patients with sporadic CRC (n = 10 each group). Technical duplicates were performed for each individual analyzed. pCREB/CREB and CREB/β-actin protein ratio was measured by densitometry scanning of Western blots (values are expressed in arbitrary units (a.u.), mean ± SD of all experiments; NT versus T, ∗P < .05, ∗∗P < .01). (C) Correlation between FMRP and CREB expression levels in mucosal samples from CRC patients as in (B) (n = 10). Expression of FMRP positively correlates with CREB expression (r = 0.7841; P < .0073). (D) Left, upper panel, representative Western blotting of untreated DLD-1 cells (U) or transfected with CREB sense (Sc) or CREB antisense (ASc) oligonucleotide for 48 hours. Quantification of pCREB/CREB (top right panel), CREB/β-actin (lower left panel), and FMRP/β-actin (lower rightpanel) protein ratio, as measured by densitometry scanning of Western blots (values are expressed in arbitrary units (a.u.), mean ± SD; pCREB/CREB: U-cells and Sc-transfected cells versus ASc-transfected cells, ∗∗P < .01, ∗∗∗P < .001; CREB/β-actin: U-cells and Sc-transfected cells versus ASc-transfected cells, ∗P < .05; FMRP/β-actin: U-cells and Sc-transfected cells versus CREB ASc-transfected cells, ∗∗P < .01, ∗∗∗P < .001, n = 3 independent experiments/treatments). (E) Left, representative Western blotting of CREB levels in untreated HCT-116 cells (U) or transfected with CREB sense (Sc) or CREB antisense (ASc) oligonucleotide. (right) Quantification of CREB/β-actin (middle panel) and FMRP/β-actin (right panel) protein ratio (values are expressed in arbitrary units (a.u.), mean ± SD, n = 3; CREB/β-actin: U-cells and Sc-transfected cells versus ASc-transfected cells, ∗P < .05; FMRP/β-actin: U-cells and Sc-transfected cells versus CREB AS-transfected cells, ∗∗P < .01, ∗∗∗P < .001). (F) Mef2 mRNA detected by RT-qPCR and normalized to β-actin mRNA in colonic samples from heathy subjects (NT) and tumoral areas (T) of paired colonic samples as in (A) (n = 6). (G) Representative Western blotting of Mef2 protein expression in paired colonic samples from healthy subjects (NT) and patients with sporadic CRC (T) (n = 10). The Western blot shows 4 (of the 10 analyzed) paired colonic samples. Statistical analysis of the data was performed using Student t test and the Mann-Whitney test.
Reduction of FMRP Results in a Better Outcome of CRC
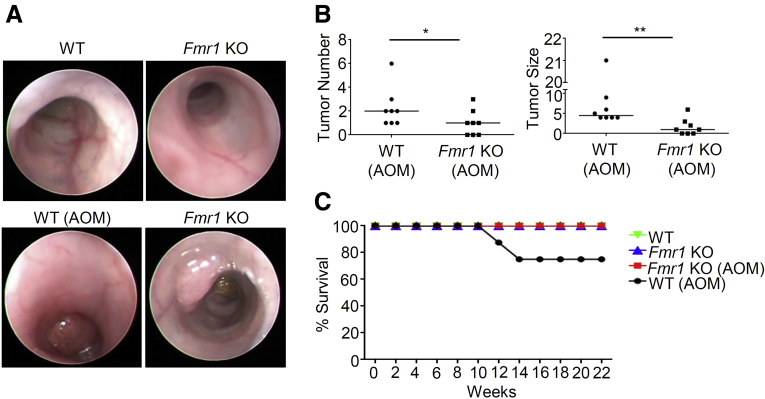
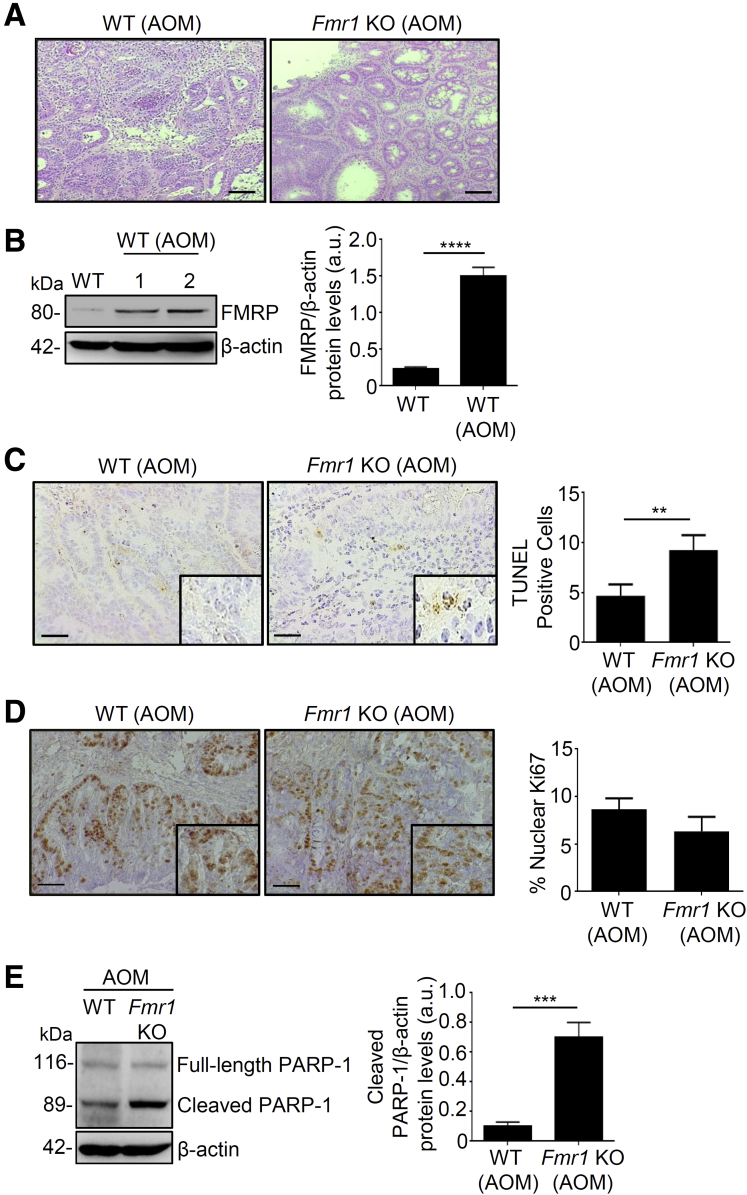
To determine the possible role of FMRP in CRC, we used a well-established mouse model of sporadic CRC induced by azoxymethane (AOM).23,24 Wild-type (WT) and Fmr1 knockout (Fmr1 KO) mice were injected intraperitoneally with AOM and monitored for tumor formation. Endoscopy performed on week 21 after the end of the AOM treatment showed that WT mice developed large tumors, as previously reported.24 In contrast, the number and size of the tumors generated in the Fmr1 KO mice were significantly reduced (Figure 4A and B). These results were confirmed by direct assessment of tumors in mice sacrificed on week 22. This difference in tumor size and number was accompanied by a decrease of viability by 20% in AOM WT mice compared to all the other groups, as shown in the survival curve (Figure 4C). Histologic evaluation revealed that in the absence of AOM treatment, the cellular morphology and organization of the colon from the Fmr1 KO was comparable to WT mice (data not shown). However, in AOM-treated animals, tumors excised at 22 weeks showed that WT mice developed well-differentiated tumors, whereas Fmr1 KO animals had a preserved normal tissue architecture nearby dysplastic areas (Figure 5A). Western blotting analysis showed an increase of FMRP expression in the tumor areas of WT AOM-treated compared to non-treated mice (Figure 5B), consistent with the observations in human CRC samples. To investigate whether the decreased tumorigenesis observed in the Fmr1 KO animals on AOM treatment was due to either an increase in cell death or a decrease in tumor cell proliferation, we performed a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay and evaluated the number of Ki67-positive cells. In addition, we assessed the expression of the active (cleaved form) poly (ADP-ribose) polymerase (PARP-1), a nuclear enzyme involved in cell death programs.25 The Fmr1 KO AOM mice showed an increased number of TUNEL-positive cells (Figure 5C), whereas no differences were observed in the number of Ki67-positive cells (Figure 5D). Furthermore, Fmr1 KO AOM mice displayed an increased expression of cleaved PARP-1 compared to WT AOM mice (Figure 5E). These observations indicate that FMRP could amplify the tumor resistance to cell death, raising the possibility that FMRP can play an important role in colon carcinogenesis.
Figure 4.
Colorectal cancer formation is reduced in Fmr1 KO mice. (A) Representative images of endoscopic examination performed in WT and Fmr1 KO mice at week 21 after injection with AOM. (B) Graphs show the number and size of colon tumors in WT and Fmr1 KO AOM mice (WT AOM versus Fmr1 KO AOM, ∗P < .05, ∗∗P < .01, n = 8 for each group). (C) Kaplan-Meier survival analysis of WT and Fmr1 KO mice AOM treated or not. Statistical analysis of the data was performed using the Student t test and the Mann-Whitney test.
Figure 5.
Fmr1 KO mice show decreased colorectal tumorigenesis compared to WT mice. (A) Representative staining with hematoxylin-eosin of tumor area from WT and Fmr1 KO mice AOM-treated. Scale bars, 100 μm. (B) Left, representative Western blotting of FMRP expression in colon tissue from WT and WT AOM-treated mice. β-actin was used as a loading control. Right, quantification of FMRP/β-actin protein ratio (values are expressed in arbitrary units (a.u.), mean ± SD of all experiments; WT versus WT AOM, ∗∗∗∗P < .0001, n = 8). (C) Left, representative images of TUNEL staining of colonic sections from WT and Fmr1 KO AOM-treated mice. Right, quantification of the number of TUNEL+ cells (WT AOM versus Fmr1 KO AOM, mean ± SD, ∗∗P < .01, n = 8 each group). Scale bars, 50 μm. (D) Left, representative Ki67 staining of colonic sections from WT and Fmr1 KO AOM-treated mice. Right, quantification of the % of Ki67+ cells (mean ± SD, n = 8 KO mice and n = 6 WT mice). (E)Left, representative Western blotting of uncleaved (full-length) and cleaved PARP-1 in colonic sections derived from WT and Fmr1 KO AOM-treated mice. Three independent experiments were performed (n = 8 for each group). β-actin was used as loading control. Right, quantification of the cleaved PARP-1/β-actin protein ratio (values are expressed in arbitrary units (a.u.), mean ± SD; WT AOM versus Fmr1 KO AOM, ∗∗∗P < .001). Statistical analysis of the data was performed using Student t test and the Mann-Whitney test.
FMRP Affects Survival in Human CRC Cells
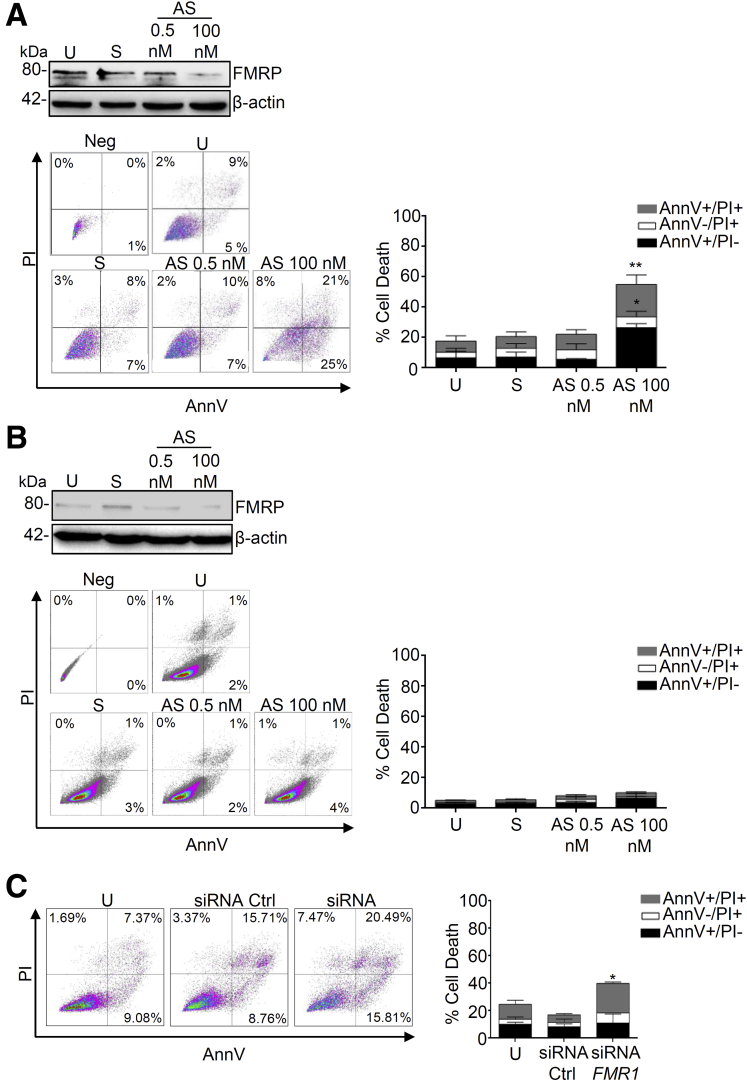
To address whether FMRP controls cancer cell survival, we analyzed cell death in CRC human epithelial cell lines in presence or absence of FMRP. Treatment of DLD-1 and HCEC-1ct cells with a specific FMR1 RNA antisense oligonucleotide (AS) significantly reduced FMRP expression, the use of sense oligonucleotide (S) did not affect FMRP level (Figure 6A and B). DLD-1 cells treated with the FMR1 AS showed an induced susceptibility to spontaneous cell death; in particular the majority of the DLD-1 cells appear AnnexinV (AnnV)+ or AnnV+ propidium iodide (PI)+, the typical flow cytometry label of programmed cell death (Figure 6A). This effect was cancer cell type specific and did not occur in normal human epithelial colon cells HCEC-1ct (Figure 6B). Similar results were obtained silencing FMR1 in DLD-1 cells using a different approach, namely a specific FMR1 small interfering RNA (Figure 6C).
Figure 6.
Knockdown of FMRP triggers cell death in CRC cell lines. (A) Left upper panel, representative Western blot showing FMRP expression in (A) DLD-1 or (B) HCEC-1ct cells untreated (U) or transfected with FMR1 sense oligonucleotide (S) or FMR1 antisense oligonucleotide (AS). Left lower panel, representative dot plot of Annexin V (AnnV) and propidium iodide (PI)-positive (A) DLD-1 or (B) HCEC-1ct cells treated as indicated above. Right, quantification of the percentage of AnnV and/or PI-positive (A) DLD-1 or (B) HCEC-1ct cells (mean ± SEM, n = 5; U-cells and S-transfected cells versus AS-transfected cells, ∗P < .05, ∗∗P < .01, n = 5). (C) Left, representative dot plot of AnnV and PI-positive DLD-1 cells untreated (U) or transfected with siCTRL or siFMR1. Right, quantification of the percentage of AnnV and/or PI-positive DLD-1 cells in untreated (U) or transfected with siCTRL or siFMR1 (U-cells and siCTRL-transfected cells versus siFMR1-transfected cells, mean ± SEM, ∗P < .05, n = 5). Statistical analysis of the data was performed using the Mann-Whitney test.
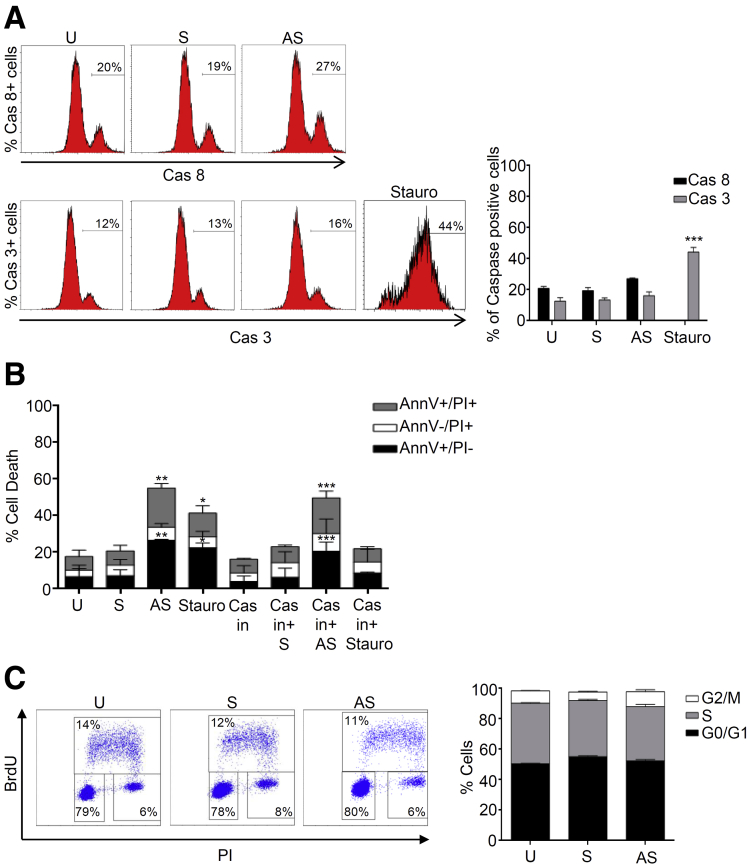
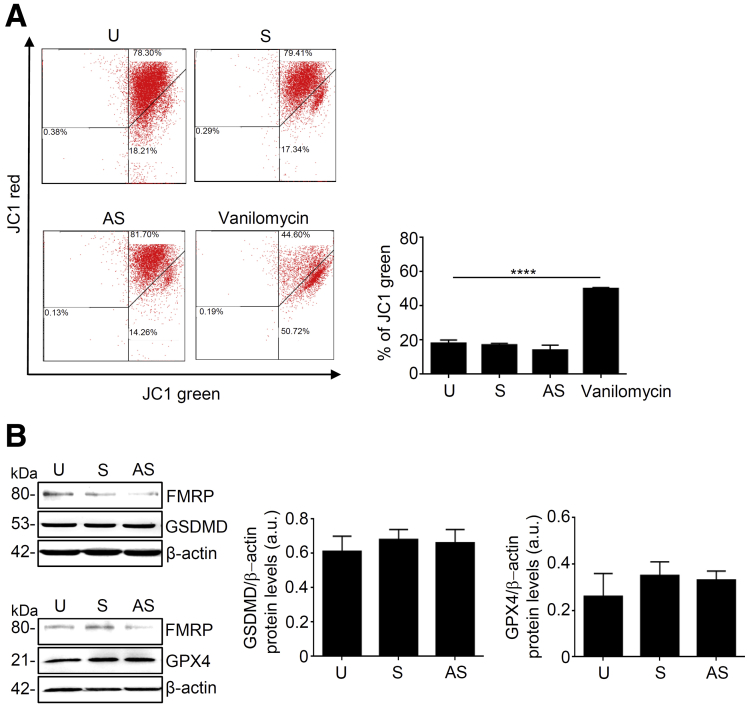
To dissect the molecular mechanism observed in FMR1 AS oligonucleotide-induced cell death, we analyzed the activation of caspase 8 and caspase 3, which play a role in the initiation and execution of cell apoptosis.26 Treatment of DLD-1 cells with FMR1 AS oligonucleotide did not alter the percentage of activated caspase 3 or caspase 8 positive cells (Figure 7A). Staurosporin, a well-known inducer of apoptosis, significantly increased the percentage of activated caspase 3 positive cells (Figure 7A). Furthermore, a pretreatment of cells with a pan-caspase inhibitor did not alter FMR1 AS oligonucleotide-induced cell death (Figure 7B). To verify whether FMR1 AS oligonucleotide-induced cell death was secondary to cell growth arrest, we analyzed the cell cycle in DLD-1 cell lines. The FMR1 AS oligonucleotide treatment did not affect cell cycle progression (Figure 7C). In addition, the treatment of DLD-1 cells with FMR1 AS oligonucleotide did not alter mitochondrial membrane potential, the expression of gasdermin D and glutathione peroxidase 4, two key molecules that regulate the pyroptosis or ferroptosis pathways, respectively (Figure 8A and B).
Figure 7.
FMRP-triggered CRC cell death is caspase activation and cell cycle-independent. (A) Left, representative dot plot showing percentage of activated caspase 8 (Cas8) or activated caspase 3 (Cas3) positive DLD-1 cells untreated (U) or transfected with FMR1 sense oligonucleotide (S) or antisense oligonucleotide (AS) for 36 hours. Staurosporin (Stauro) was used as positive control. Right, percentage of activated caspase3+ cells (grey bar) or activated caspase 8+ cells (black bar) measured by flow cytometry (U-cells, S-transfected cells, and AS-transfected cells versus Stauro-treated cells, mean ± SD, n = 3; ∗∗∗P < .001). (B) Percentage of AnnV and/or PI-positive DLD-1 cells preincubated with pan-caspase inhibitor (Cas in) Z-VAD-FMK untreated (U) or transfected with FMR1 sense oligonucleotide (S) or antisense oligonucleotide (AS) (mean ± SEM, n = 4; U-cells and S-transfected cells versus AS-transfected cells, ∗∗P < .01; U-cells and S-transfected cells versus Stauro, ∗P < .05; Cas in-cells, Cas in+S-cells and Cas in+Stauro-cells versus Cas in+AS-cells, ∗∗∗P < .001). (C) Left, representative flow cytometric analysis of cell cycle progression in DLD-1 cells treated with FMR1 sense (S) or antisense (AS) oligonucleotide. Right, percentages of cells in the different phases of cell cycle (mean ± SD, n = 3). Statistical analysis of the data was performed using the Mann-Whitney test. BrdU, bromodeoxyuridine.
Figure 8.
FMRP knockdown does not affect mitochondrial membrane potential, pyroptosis or ferroptosis pathways. (A) DLD-1 cells untreated (U) or transfected with FMR1 sense (S) or FMR1 antisense (AS) oligonucleotide or positive control vanilomycin, stained with JC-1, and analyzed by flow cytometry. Mitochondrial membrane potential loss was observed as a decrease in JC-1 red fluorescence and an increase in JC-1 green fluorescence. Representative dot plot (U cells, S cells and AS cells versus vanilomycin cells, mean ± SEM, n = 3, ∗∗∗∗P < .0001). (B) Left, representative Western blotting of Gasdermin D (GSDMD, upper panel) and Glutathione peroxidase 4 (GPX4, lower panel) in DLD-1 cells untreated (U) or transfected with FMR1 sense (S) or FMR1 antisense (AS) oligonucleotide. β-actin was used as loading control. Right, quantification of GSDMD/β-actin (middlepanel) and GPX4/β-actin (rightpanel) protein ratio (values are expressed in arbitrary units (a.u.), mean ± SD). Statistical analysis of the data was performed using the Mann-Whitney test. GPX4, glutathione peroxidase 4; GSDMD, gasdermin D.
Altogether, these findings suggest that FMRP influences CRC cell death without affecting the apoptotic pathway or cell cycle.
FMRP Regulates the Necroptotic Pathway
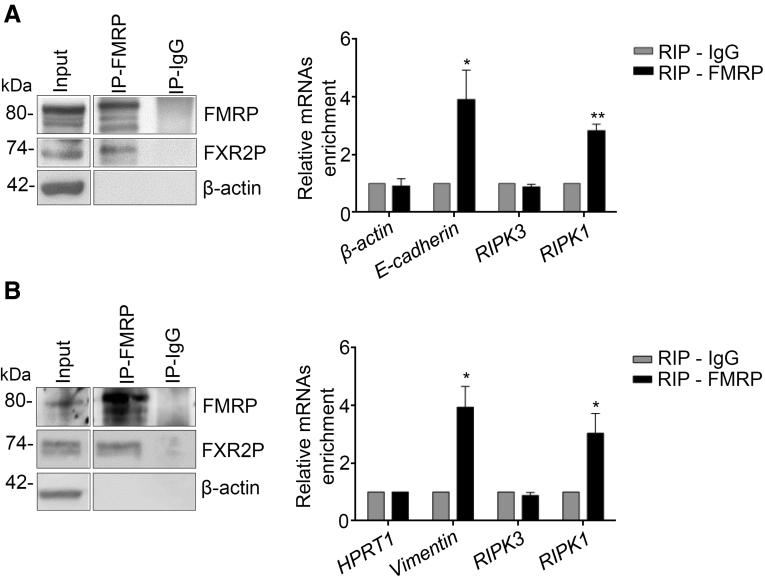
Necroptosis is a regulated necrotic cell death modality in a caspase-independent manner and is mainly mediated by Receptor-Interacting Protein Kinase 1 (RIPK1), RIPK3, and mixed lineage kinase domain-like protein (MLKL).27, 28, 29 This core complex, called the necrosome, mediates downstream executing molecules and events such as reactive oxygen species burst, plasma membrane permeabilization, and cytosolic adenosine triphosphate reduction that drives to the irreversible necroptosis-executing mechanisms.30,31 Next, we explored the possibility that the induced cell death in the absence of FMRP could be due to the activation of necroptosis pathway and evaluated whether FMRP binds the mRNAs encoding the core components of the necroptosis complex, RIPK1 and RIPK3. FMRP was immunoprecipitated from human CRC samples and DLD-1 cell lines, and the association of candidate mRNAs tested by RT-qPCR (Figure 9A and B). A significant enrichment in the FMRP complex from human CRC tissues and DLD-1 cell lines, was detected for RIPK1 mRNA but not for RIPK3 mRNA (Figure 9A and B); β-actin, hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNAs were used as negative controls, while Vimentin and E-cadherin mRNAs as positive controls.15 These data suggest that FMRP binds RIPK1 mRNA and thus possibly affects RIPK1 synthesis, an initial core element of the necroptosis pathway. Although FMRP could act at the level of mRNA stability and/or mRNA translation, the stability of RIPK1 mRNA seems not to be affected (data not shown).
Figure 9.
RIPK1 mRNA is part of the FMRP complex. (A) Left, representative Western blotting of FMRP immunoprecipitation from colonic samples derived from 3 CRC patients. FXR2P, a known FMRP interactor, is detected as part of the FMRP complex; β-actin was used as a negative control. Input (1/20) of the total extract, FMRP immunoprecipitation (IP-FMRP), and mock immunoprecipitation (IP-IgG). Right, quantification of β-actin, E-cadherin, RIPK3, and RIPK1 mRNA enrichment by RT-qPCR. β-actin and E-cadherin mRNAs were used as negative and positive controls, respectively (mean ± SEM, n = 3; E-cadherin: RIP-IgG versus RIP-FMRP; ∗P < .05; RIPK1: RIP-IgG versus RIP-FMRP, ∗∗P < .01). (B) Left, representative Western blotting of FMRP immunoprecipitation from DLD-1 cells. FXR2P, a known FMRP interactor, is detected as part of the FMRP complex; β-actin is used as a negative control. Input (1/20) of the total extract, FMRP immunoprecipitation (IP-FMRP), and mock immunoprecipitation (IP-IgG). Right, quantification of HPRT1, Vimentin, RIPK3, and RIPK1 mRNA enrichment, by RT-qPCR, in FMRP immunoprecipitation/total protein extracted from DLD-1 cells. HPRT1 and Vimentin mRNAs were used as negative and positive controls, respectively (mean ± SEM, n = 3; Vimentin: RIP-IgG versus RIP-FMRP, ∗P < .05; RIPK1: RIP-IgG versus RIP-FMRP, ∗P < .05). Statistical analysis of the data was performed using Student t test.
FMRP Regulates Cell Death Modulating the RIPK/MLKL Pathway
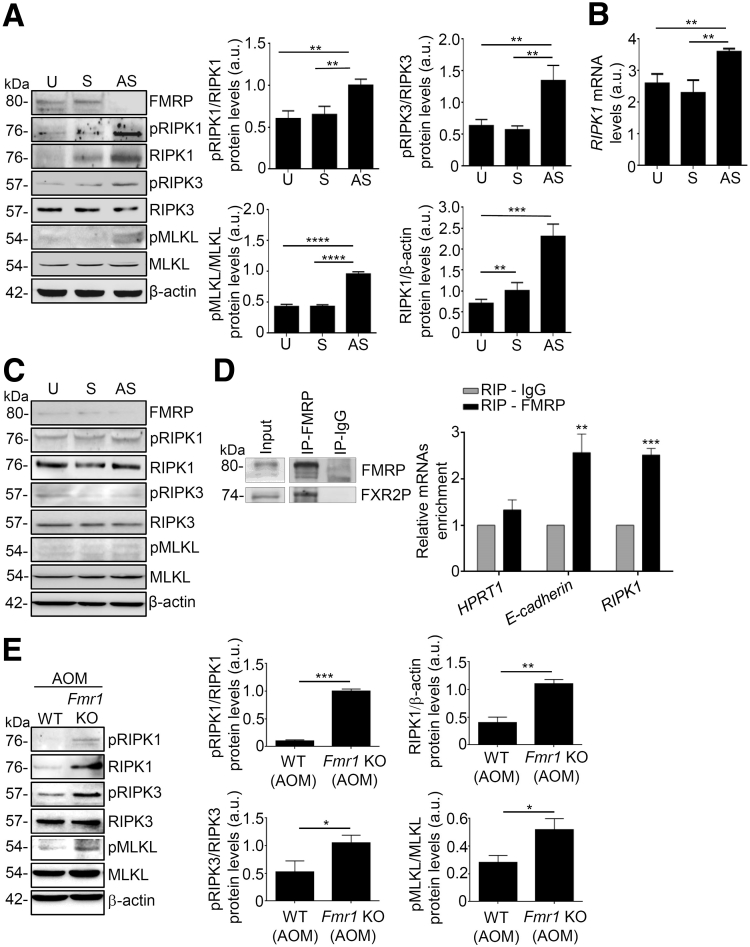
To examine whether FMRP is responsible for inhibiting RIPK1 signaling in CRC, we explored RIPK1 expression in DLD-1 cell lines treated with FMR1 AS oligonucleotide. Treatment of DLD-1 with the FMR1 AS but not with the sense oligonucleotide inhibited FMRP expression (Figure 10A). Reduction of FMRP levels was associated with an increase in phosphorylation of RIPK1, RIPK3, and MLKL proteins (Figure 10A). Of note, FMR1 AS treatment led to a significant increase of RIPK1 protein and mRNA (Figure 10A and B). In the healthy colon cell line HCEC-1ct FMRP is expressed at low levels and no changes in the expression/phosphorylation of RIPK1, RIPK3, and MLKL were observed upon FMR1 mRNA silencing (Figure 10C). Moreover, we evaluated the presence of RIPK1 mRNA in the FMRP complex in the murine colon tissue. As shown in Figure 10D, a significant enrichment of RIPK1 mRNA was detected after FMRP immunoprecipitation, suggesting that FMRP may regulate necroptosis in vivo during colon tumorigenesis. The analysis of the colon tumor area from the Fmr1 KO AOM mice revealed an increased level of phosphorylated RIPK1, RIPK3, and MLKL proteins (Figure 10E). Furthermore, we exploited the effect of FMR1 mRNA silencing for the ability to support growth of patient-derived human colon cancer organoids. Treatment of tumor organoids with FMR1 AS significantly reduced FMRP expression, which led to a higher expression of RIPK1 (Figure 11A).
Figure 10.
FMRP regulates the necroptotic pathway in human CRC and AOM-treated mice. (A) Left, representative Western blotting showing FMRP, pRIPK1, RIPK1, pRIPK3, RIPK3, pMLKL, MLKL, and β-actin in DLD-1 cells untreated (U) or treated with FMR1 sense (S) or antisense (AS) oligonucleotide. Right, quantification of pRIPK1, pRIPK3, pMLKL, and RIPK1 proteins in DLD-1 cells as measured by densitometry of Western blotting (values are expressed in arbitrary units (a.u.), mean ± SD, n = 4; pRIPK1/RIPK1: U-cells and S-transfected cells versus AS-transfected cells, ∗∗P < .01; pRIPK3/RIPK3: U-cells and S-transfected cells versus AS-transfected cells, ∗∗P < .01; pMLKL/MLKL: U-cells and S-transfected cells versus AS-transfected cells, ∗∗∗∗P < .0001; RIPK1/β-actin: U-cells and S-transfected cells versus AS-transfected cells, ∗∗P < .01; ∗∗∗P < .001). (B) RIPK1 mRNA levels in DLD-1 cells untreated (U) or treated with FMR1 sense (S) or antisense (AS) oligonucleotide, normalized to β-actin mRNA (mean ± SD, n = 4; U-cells and S-transfected cells versus AS-transfected cells, ∗∗P < .01). (C) Representative Western blotting showing FMRP, pRIPK1, RIPK1, pRIPK3, RIPK3, pMLKL, MLKL, and β-actin in HCEC-1ct cells untreated (U) or transfected with FMR1 sense (S) or antisense (AS). (D) Left, representative Western blotting of FMRP immunoprecipitation from colon samples of WT mice. FXR2P, a known FMRP interactor, is detected as part of the FMRP complex. Input (30 μg) of the total extract, FMRP immunoprecipitation (IP-FMRP), and mock immunoprecipitation (IP-IgG). Right, quantification of Hprt1, E-cadherin, and RIPK1 mRNAs enrichment by RT-qPCR. Hprt1 and E-cadherin mRNAs are negative and positive controls, respectively (mean ± SEM, n = 3 mice; E-cadherin: RIP-IgG versus RIP-FMRP, ∗∗P < .01; RIPK1: RIP-IgG versus RIP-FMRP, ∗∗∗P < .001). (E) Left, representative Western blotting showing pRIPK1, RIPK1, pRIPK3, RIPK3, pMLKL, MLKL, and β-actin in colon tissue of WT and Fmr1 KO AOM-treated mice. Right, quantification of pRIPK1, pRIPK3, pMLKL, and RIPK1 proteins in colon tissue of WT and Fmr1 KO AOM-treated mice as measured by densitometry of Western blotting (values are expressed in arbitrary units (a.u.), mean ± SD, n = 8 mice per group; ∗P < .05, ∗∗P < .01, ∗∗∗P < .001). Statistical analysis of the data was performed using Student t test and the Mann-Whitney test.
Figure 11.
FMRP-induced cell death is dependent from RIPK/MLKL complex activation. (A) Representative immunostaining images of FMRP (upper panels) and RIPK1 (lower panels) in human CRC organoids untreated (U) or transfected with sense (S) or FMR1 antisense (AS) oligonucleotide, n = 2. (B) Percentage of AnnV and/or PI-positive DLD-1 cells pretreated with a specific RIPK1 inhibitor. Left, DLD-1 treated with RIPK1-specific inhibitor (NEC1). Right, DLD-1 cells treated with MLKL-specific inhibitor (NSA) and then transfected with FMR1 sense (S) or antisense (AS) oligonucleotide (mean ± SD, n = 4; NEC1 plus S-transfected cells and NEC1 plus AS-transfected cells versus AS-transfected cells, ∗∗∗P < .001; NSA plus S-transfected cells and NSA plus AS-transfected cells versus AS-transfected cells, ∗∗∗P < .001, ∗∗∗∗P < .0001). Statistical analysis of the data was performed using Student t test and the Mann-Whitney test.
Finally, to further evaluate whether FMRP-induced cell death was dependent from RIPK/MLKL complex activation, human CRC cell lines were incubated with RIPK1-specific inhibitor (NEC1) or MLKL-specific inhibitor (NSA). No difference in cell death was observed in DLD-1 cells incubated with FMR1 AS oligonucleotide in presence of NEC1 or NSA inhibitors (Figure 11B). These data indicate that FMR1 AS oligonucleotide-induced cell death in CRC cells is due to the RIPK/MLKL intracellular signaling cascade.
Discussion
The intestinal epithelium features a rapid turnover of cells.32 The lifespan is based on a dynamic equilibrium that is regulated by several factors and that allows proliferation, migration, differentiation, and senescence of the cells.32 This equilibrium can be disturbed during inflammation or injury that results from cellular stress mediated by infectious organisms, radiation, inflammatory disease, or harmful events.2 These events trigger a rapid protective and regenerative response that is regulated by several intracellular and extracellular factors.2 Prolonged injury together with genetic alterations can result in malignant transformation.2 A similar process occurs in the development of CRC, which results from a combination of genetic, epigenetic and environmental factors.33 Compelling evidence indicates that CRC cells manifest enhanced activation of various intracellular signals that ultimately promote the expression of molecules involved in programmed resistance to cell death or in cell growth.27,34
Among the factors that ensure the correct development of intestinal cells are the family of RNA binding proteins (RBPs) able to act in a rapid and efficient manner to alter gene expression, especially but not only during changes in the microenvironment.35 Increasing evidence indicates that the response and adaptation of intestinal epithelium to various types of injuries and to malignant transformation are mediated by RBPs.35 A single RBP can bind to hundreds, if not thousands, of targets, and a combination of several RBPs interactions contribute to cellular identity in healthy, but also pathological conditions as cancer and neurodevelopment. In cancer, RBPs regulate a number of mRNAs that encode for proteins involved in tumorigenesis.5,36 In the specific case of CRC, several RBPs have been shown to be dysregulated and also associated with survival rate of cancer patients.35,37
For example, IGF2 mRNA-binding protein 1 (IMP1), CUGBP Elav-Like Family Member 1 (CELF1), and Human antigen R (HUR) constitute a new set of regulatory RBPs, playing a role in intestinal homeostasis, adaptation to injury, and participation in malignant transformation.35
Here we show that FMRP expression is significantly increased in human CRC. Although the correlation of FMRP expression with colon cancer patient outcome requires a larger sample size, previous findings indicate that FMRP levels are predictive of poor survival in multiple solid tumors.14,15 The analysis of different cancer atlases (http://www.cbioportal.org/; https://www.proteinatlas.org/) revealed a high FMRP expression level in CRC tissues, further confirming and extending our observation. In addition, patients with a mutation in the FMR1 gene, encoding a nonfunctional or trunkated FMRP protein, have a favorable outcome (http://www.cbioportal.org/). Therefore, absence of a functional FMRP seems to be protective in cancer. An independent public dataset of human protein atlas (https://www.proteinatlas.org/) showed a reduced disease-free survival in CRC patients with low expression of FMRP. In conclusion, the analysis of different datasets revealed some discrepancies about protection vs risk, suggesting that further analyses on a larger cohort of patients with CRC are required to evaluate whether FMRP levels correlate with prognostic indicators of aggressive CRC, metastases probability, and response to cancer therapies.
The expression of the FMR1 gene is regulated at multiple levels. The FMR1 gene contains binding sites for different transcription factors,21,38 including CREB binding site.21 CREB has been shown to affect colonic tumorigenesis, and neoplastic progression. Suppression of CREB activity in cancer cells may also have a therapeutic effect.19 Our data indicate that CREB is overexpressed in human CRC, and positively regulates FMRP expression in human colon cancer cells.
Using a well-established mouse model of CRC, we show that FMRP controls colon cancer progression. In AOM-treated Fmr1 KO mice, colonic tumor incidence and size were significantly reduced compared with WT AOM-treated mice. However, because FMRP is relatively ubiquitously expressed, we cannot exclude the possibility that the anti-cancer effect detected in the Fmr1 KO mice is also partly due to the function of other mucosal cell types (eg, immune cells). In addition, different analyses on the colon tissue of the AOM-treated Fmr1 KO mice revealed an increased presence of tumor cell death suggesting a pro-tumorigenic function of FMRP leading to epithelial cancer cells survival.
Resistance to cell death is a crucial hallmark acquired during cancer progression. The identification of deregulated pathways affecting cell death has led to the development of therapeutic strategies that have been used in CRC patients.34 In healthy tissues, programmed cell death plays a pivotal role in the development and maintenance of tissue homeostasis.39 During the last 2 decades, several functional studies established that cell death serves as a natural barrier to cancer development.40 CRC cells evolved a variety of strategies to limit or circumvent programmed cell death. Tumor cells may block the apoptosis process by increasing expression of antiapoptotic regulators such as Bcl-2 and Bcl-xL, by down-regulating proapoptotic factors (Bax, Bim, Puma), or by short-circuiting the extrinsic ligand-induced death pathway.2 RBPs can modulate the expression of genes implicated in cell survival,35,36 and this prompted us to suppose that the pro-survival effect of FMRP could be controlled by inhibition of caspase/apoptotic mechanisms. However, the inhibition of caspase or cell cycle did not have an effect on cell death after a decrease of FMRP. In addition, inhibition of 2 key pathways of programmed cell death such as ferroptosis or pyroptosis was not influenced by the levels of FMRP. These findings suggest that anti-survival effect of FMR1 AS oligonucleotide is not due to the apoptotic, ferroptotic, pyroptotic mechanisms or secondary to cell cycle arrest. Necroptosis, a caspase-independent cell death mechanism, could be an alternative way to eradicate resistant cell death in cancer cells.27 Here we demonstrated that CRC cells incubated with FMR1 AS oligonucleotide regain a normal activity of the necroptosis machinery that drives to programmed cell death. Specific immunoprecipitation experiments show that FMRP binds RIPK1 mRNA, suggesting that FMRP acts as a regulator of the necroptosis pathway ultimately affecting RIPK1 synthesis. Of note, the application of high-throughput approaches allowed the identification of hundreds of putative FMRP mRNA targets in brain (>1000) and in non-neuronal HEK293 cells (>6000).41, 42, 43, 44 FMRP has 4 RNA-binding domains: 2 Tudor domains in the N-terminus region, 2 K homology domains in the central region, and an RGG box in the C-terminal region of FMRP, this last crucial for the interaction with some mRNAs containing a G-quartet/G-rich structure.45,46 So far, FMRP can bind mRNAs directly or indirectly via different types of sequences/structures.47 In this study we detect, for the first time, the presence of RIPK1 mRNA in the FMRP complex, indicating that FMRP could regulate its metabolism. Interestingly, using a webserver for prediction of RNA containing G-quadruplex, namely G4Catchall, we found putative G-quadruplex structures in RIPK1 mRNA that represent a possible FMRP binding site. Although this is a predictive approach, the indication is promising, and future studies should further investigate whether FMRP could directly bind RIPK1 mRNA.48
While our study was ongoing, Zhuang et al49 showed that FMRP plays a central role in the inhibition of tumor necrosis factor (TNF)–mediated necroptosis during infection and liver disease. The authors demonstrated that FMRP is important for regulating key molecules in TNF receptor 1–dependent necroptosis including CYLD, c-FLIPS, and JNK, which contribute to prolonged RIPK1 expression and necrosome activation. Therefore, our findings together with the above-mentioned previous observations strengthen the hypothesis of targeting FMRP as an anti-cancer strategy affecting both RIPK1 expression and the TNF-mediated necropoptosis.
Cancer cells are able to eradicate necroptosis machinery by down-regulating the necroptotic core pathway and activating downstream executing molecules and events.27,31 The identification of a regulator such as FMRP could explain the molecular mechanism that allows to down-regulate RIPK1 expression in colon cancer. Moreover, our results are consistent with the observation that overexpression of RIPK1 suppresses proliferation, migration, and invasion of human CRC cell lines.50,51
One of the most largely used chemotherapic approaches occurs through the proapoptotic therapy (eg, using cisplatin, carboplatin, paclitaxel, 5-fluorouracil, and gemcitabine), but the efficacy of this therapy is limited by drug resistance and toxic effects. The discovery of necroptosis as an inducible, alternative form of programmed cell death has opened up novel and exciting perspectives to kill resistant cancer cells. Therefore, the control of necroptosis by defined signal transduction pathways offers the opportunity to target this cellular process for anti-cancer therapy.
Several strategies exist to trigger necroptosis in other human cancer types. The natural compound shikonin has been shown to bypass deficiencies in apoptosis pathways,52 whereas SMAC mimetics and the alkaloid staurosporine induce necroptosis in acute myeloid leukemia and different carcinoma cell lines.53 Moreover, traditional chemotherapeutic or molecular targeted drugs approved for marketing or in clinical trials have been recently identified as cancer necroptosis inducers such as TRAIL, obatoclax, or 3-bromopyruvate plus chloroquine.27,54,55 These drugs have been proven to be safe for use in human, and induction of necroptosis in cancer cells does not have toxic effect in healthy cells or lead to severe side effects in vivo. Therefore, the identification of a specific target such as FMRP that could control the necroptosis pathway may further enhance the specificity and selectivity of pro-necroptosis strategies. In conclusion, our data indicate that down-regulation of FMRP drives colon cancer cells to switch to necroptosis and represents a novel attractive strategy to overcoming programmed cell death resistance in CRC.
Methods
Patients and Human Samples
Samples of human CRC areas were derived from 67 patients who had undergone colonic resection for sporadic CRC, whereas healthy (normal) mucosa samples include colonic mucosal biopsy from 67 patients with irritable bowel syndrome from Tor Vergata University Hospital (Rome, Italy). FMRP and FMR1 mRNA expression was evaluated by immunohistochemistry, Western blotting, and RT-qPCR. In addition, paired tissue samples were derived from the tumoral area and the macroscopically unaffected, adjacent colonic mucosa of 6 patients who underwent colon resection for sporadic CRC at the Tor Vergata University Hospital (Rome, Italy) and used for FMRP expression by Western blotting. Patients with sporadic CRC received neither radiotherapy nor chemotherapy before surgery. Written informed consent was obtained from all patients. The study protocol was approved by the Tor Vergata University Hospital Review Board (protocol number 129/17).
Experimental Model of CRC
Mice were housed in a ventilated, temperature-controlled (23°C) room. A 12-h light/dark cycle was used, and food and water were available ad libitum. Starting at 6 weeks of age, male FVB.129P2 WT and Fmr1 KO mice were injected with the alkylating agent AOM (10 mg/kg; Sigma-Aldrich, Milan, Italy) intraperitoneally once a week for 5 weeks to induce tumor formation.23 Mice were monitored for tumor formation and were endoscopically screened 1 week before being euthanized using a high-resolution endoscopic system. At week 22, after last AOM injection, mice were sacrificed by cervical dislocation, and colonic tissues were collected for the different analyses. All experiments using animals were performed according to Italian and European legislation on animal experimentation (protocol number: 1138/2016-PR, 494/2017-PR).
Mouse Endoscopy
Colonoscopy was performed blinded to the genotype by using the COLOVIEW (Karl Storz, Tuttlingen, Germany) high-resolution mouse endoscopic system.56 The number of tumors was counted during endoscopic examination and performed at week 21 after the last AOM injection. All tumors were evaluated on the basis of their size and scored as previously described.56 Tumors were graded as follows: grade 1 (very small but detectable tumor), grade 2 (tumor covering up to one-eighth of the colonic circumference), grade 3 (tumor covering up to one-fourth of the colonic circumference), grade 4 (tumor covering up to half of the colonic circumference), and grade 5 (tumor covering more than half of the colonic circumference).
Immunohistochemistry
All reagents were purchased from Sigma-Aldrich (Milan, Italy) unless specified. Immunohistochemistry was performed on formalin-fixed, paraffin-embedded sections of normal tissues and tumoral samples from CRC patients. Sections were deparaffinized and dehydrated through xylene and ethanol, and the antigen retrieval was performed in Tris-EDTA citrate buffer (pH 7.8) for 30 minutes in a thermostatic bath at 98°C (Dako Agilent Technologies, Santa Clara, CA). Immunohistochemical staining was performed using a monoclonal antibody directed against human FMRP (final dilution 1:5000; LifeSpan BioSciences, Seattle, WA) incubated at room temperature for 1 hour, followed by a biotin-free horseradish peroxidase (HRP) polymer detection technology with 3,3′diaminobenzidine as a chromogen MACH 4 Universal HRP-Polymer Kit (Biocare Medical, Pacheco, CA). Immunohistochemistry was performed on colonic cryosections of WT and Fmr1 KO mice. The slices were incubated with a mouse monoclonal antibody directed against mouse Ki67 clone MIB-5 (final dilution 1:100, Dako, Agilent Technologies) at room temperature for 30 minutes, followed by biotin-free HRP polymer detection Ultravision Detection System (Thermo Scientific, Waltham, MA) with 3,3´diaminobenzidine as a chromogen (Dako, Agilent Technologies). Histopathologic analysis was performed on mouse colonic cryosections derived from WT and Fmr1 KO mice in tumor and peritumor areas after hematoxylin and eosin (H&E) staining.
TUNEL Assay
In colonic cryosections derived from WT and Fmr1 KO mice, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was performed to detect apoptotic cells using the in situ Cell Death Detection kit (Roche Applied Science, Penzberg, Germany) according to the manufacturer’s instructions.
RNA Extraction, RT-qPCR
RNA was extracted using PureLink mRNA mini kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. RNA (1 μg per sample) was reverse transcribed into complementary DNA (cDNA), and this was amplified using the following conditions: denaturation for 1 minute at 95°C; annealing for 30 seconds at 59°C for human FMR1; 60°C for human CREB and human/mouse β-actin; 61°C for human RIPK1, 30 seconds of extension at 72°C. Gene expression was calculated using the ΔΔCt formula.
Sequences of the primer used were the following: human FMR1 (forward 5´-GTTGAGCGGCCGAGTTTGTCAG-3´, reverse 5´-CCCACTGGGAGAGGATTATTTGGG-3´); human CREB (forward 5´-CCACTGATGGACAGCAGATC-3´, reverse 5´-CGGACCTCTCTCTTTCGTG-3´); human RIPK1 (forward 5´-CACAAGGACCTGAAGCCTGAA-3´, reverse 5´-TGCTTGTTTTGAGCTGTAGCC-3´); human and mouse β-actin (forward 5´-AAGATGACCCAGATCATGTTTGAGACC-3´, reverse 5´-AGCCAGTCCAGACGCAGGAT-3´).
Protein Extraction and Western Blotting
Total proteins were extracted in lysis buffer with 10 mmol/L HEPES (pH 7.9), 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.2 mmol/L ethylene glycol-bis (β-aminoethyl ether)-N,N,N´,N´-tetraacetic acid, and 0.5% Nonidet P40 supplemented with 1 mmol/L dithiothreitol, 10 mg/mL aprotinin, 10 mg/mL leupeptin, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L Na3VO4, and 1 mmol/L NaF. Lysates were clarified by centrifugation and separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Membranes were incubated with antibodies against anti-human FMRP, CREB, MEF2a (Cell Signaling, Danvers, MA); pRIPK3, RIPK3, pMLKL, MLKL, RIPK1 (Abcam, Cambridge, UK); pRIPK1 (SAB, Maryland, WA) (final dilution 1:1000); anti-mouse FMRP, pRIPK3, RIPK3, pMLKL, MLKL, RIPK1 (Abcam); pRIPK1 (SAB), and PARP-1 (Cell Signaling) (final dilution 1:1000), followed by a secondary antibody conjugated to HRP (Dako, Agilent Technologies). A mouse anti-β-actin antibody was used to detect β-actin and for normalization. A computer-assisted scanning densitometry was used to analyze the intensity of the immunoreactive bands.
Immunofluorescence
CRC cell lines and HCEC-1ct were fixed by 3.7% formaldehyde for 10 minutes at 4°C and permeabilized with 0.1% Triton for 10 minutes at room temperature, and nonspecific labeling was blocked (bovine serum albumin 1%, Tween 0.1%, glycine 2%) for 1 hour at room temperature. Anti-FMRP monoclonal antibody (1:500; Cell Signaling) was incubated overnight at 4°C. After washing with phosphate-buffered saline (PBS) 1 time, the secondary antibody goat anti-rabbit Alexa 488 (1:2000; Invitrogen, Carlsbad, CA) was incubated for 1 hour at room temperature. Cells were washed with PBS1X, mounted using Prolong gold antifade reagent with DAPI (Invitrogen), and analyzed by a Leica DMI4000 B (Wetzlar, Germany) microscope with Leica application suite software (V4.6.2).
Flow Cytometry Analysis
Cells were untreated or transfected with either FMR1 sense oligonucleotide (S) (final concentration 100 nmol/L) or FMR1 antisense oligonucleotide (AS) (final concentration 0.5 nmol/L and 100 nmol/L) and were incubated with necrosulfonamide (final concentration 1μmol/L; Calbiochem, Milan, Italy) or necrostatin1 (final concentration 10 μmol/L; Cayman Chemical, Ann Arbor, MI). After 24 hours cells were collected, washed 2 times in AnnV buffer, stained with FITC-AnnV (final dilution 1:100; Immunotools, Friesoyte, Germany) according to the manufacturer’s instructions, and incubated with 5 mg/mL PI for 30 minutes at 4°C. Cell death was quantified by flow cytometry; viable cells were considered as AnnV–/PI– cells.
Cells untreated or transfected with either FMR1S or FMR1AS oligonucleotide (final concentration 100 nmol/L) were incubated with Q-VD-OPh (final concentration 1μmol/L; R&D Systems, Minneapolis, MN). After 36 hours cells were collected, and caspase activation was quantified by flow cytometry using the specific antibody for cleaved caspase 3 and caspase 8 (final concentration 1:100; Biovision, Milpitas, CA). Staurosporin was used as apoptotic cell death positive control (final concentration 1 μmol/L; Sigma-Aldrich).
For cell cycle distribution, cells were untreated or transfected with either FMR1 S or FMR1 AS oligonucleotide (final concentration 100 nmol/L). After 48 hours, cells were pulsed with 10 mol/L bromodeoxyuridine for 60 minutes, fixed in 70% cold ethanol, and stored at 20°C for at least 3 hours. Next, cells were denatured in 2 mol/L HCl and stained with anti–bromodeoxyuridine monoclonal antibody (Immunotech, Marseille, France), followed by fluorescein isothiocyanate (FITC)–conjugated secondary anti-mouse immunoglobulin G (Molecular Probes, Milan, Italy). Cells were stained with 100 g/mL PI and analyzed by flow cytometry.
To assess mitochondrial membrane potential, we used 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimi-dazolylcarbocyanine iodide (JC-1) dye according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA).
Analysis was performed using the Kaluza software (Beckman Coulter Life Sciences, Pasadena, CA).
Cell Culture, FMR1 and CREB Silencing
All reagents were purchased from Sigma-Aldrich (Milan, Italy) unless otherwise specified. Human CRC cell line DLD-1 was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in RPMI 1640 and McCoy’s 5A medium, respectively. All media were supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin (both from Lonza, Verviers, Belgium). The healthy (normal) human colon epithelial cell line (HCEC-1ct) was obtained from EVERCYTE GmbH (Vienna, Austria) and cultured in ColoUp medium (EVERCYTE GmbH). Cells were maintained in a 37°C, 5% CO2, fully humidified incubator. Phosphorothioate single-stranded oligonucleotide of the human FMR1 complementary DNA sequence was synthesized in the antisense orientation (5´-TCCACCACCAGCTCCTCCAT-3´). CRC cell lines and HCEC-1ct were transfected with either FMR1 antisense (AS) (final concentration 0.5–100 nmol/L) or FMR1 sense (S) oligonucleotide (5´-AACACGTCTATACGC-3´, final concentration 100 nmol/L) for 24, 36, and 48 hours using Opti-MEM medium and lipofectamine 3000 reagent (both from Thermo Fisher Scientific) according to the manufacturer’s instructions. CRC cell lines were transfected with either CREB antisense (ASc) (5´-GCATCTCCACTCTGCTGGTT-3´) or CREB sense (Ss) (5´-AACACGTCTATACGC-3´ at final concentration 200 nmol/L) for 24/48 hours. siRNA-mediated silencing of FMRP was performed with FMR1-specific siRNAs (Thermo Fisher Scientific) (AM 16708, ID nos 10824, 10919 and 11010). As a non-specific control, a scrembled siRNA was used (Thermo Fisher Scientific, no. 4390843). DLD-1 cells were transfected using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer's instructions for 48 hours.
Intestinal Crypt Isolation, Organoid Formation, and Immunostaining
Surgically resected intestinal tissues were obtained from colon cancer patients who underwent colon resection for sporadic CRC (all with TNM stages II–III) at the Tor Vergata University Hospital (Rome, Italy). Intestinal tissues were washed in Hanks’ balanced salt solution (HBSS) (Lonza) containing 1% penicillin/streptomycin (Lonza) and chopped into approximately 5-mm pieces. Tissue fragments were placed in a tube, incubated in Advanced Dulbecco modified Eagle medium/F12 (DMEM/F12) medium (Gibco, Monza, Italy) with 15 mmol/L EDTA (Sigma-Aldrich), and rocked at 4°C for 30 minutes. Large chunks of tissue were then removed, and remaining crypts were centrifuged at 200g for 3 minutes, embedded in Matrigel (Becton Dickinson, Franklin Lakes, NJ), and seeded in warmed 48-well plates. Matrigel was allowed to solidify for 15 minutes at 37°C and overlaid with complete medium (advanced DMEM/F12 supplemented with 1% penicillin/streptomycin, 1% amphotericin B [Lonza], 0.1% gentamycin [Lonza], 1× B27 [Invitrogen], HEPES [15 mmol/L; Lonza], 1× GlutaMAX-I [Gibco], rh-EGF [100 ng/mL; R&D Systems], rh-Noggin [100 ng/mL; R&D Systems], rh-R-Spondin [50 ng/mL; R&D Systems], and nicotinamide [10 mmol/L; Sigma-Aldrich]). The whole medium was replaced every 3 days. Organoids were transfected with either FMR1 antisense (AS) (final concentration 200 nmol/L) or FMR1 sense (S) oligonucleotide (final concentration 200 nmol/L) for 48 hours using Opti-MEM medium and lipofectamine 3000 reagent (both from Thermo Fisher Scientific) according to the manufacturer’s instructions. Culture medium was removed, and organoids were washed with PBS and incubated with organoid harvesting solution (Trevigen, Winooski, VT) for 1 hour at 4°C with gentle shaking. Released organoids from depolymerized Matrigel were then collected and transferred into a Tissue-Tek Cryomold (Sakura Finetek Europe B.V., Alphen aan den Rijn, the Netherlands) containing optimal cutting temperature (OCT), frozen, and stored at –80°C. Immunohistochemical staining was performed using a monoclonal antibody directed against human RIPK1 (final dilution 1:250; Abcam), incubated at room temperature for 1 hour, followed by a biotin-free HRP-polymer detection technology with 3,3′diaminobenzidine as a chromogen MACH 4 Universal HRP-Polymer Kit (Biocare Medical). Sections were counterstained with hematoxylin, dehydrated, and mounted.
RNA Immunoprecipitation
All reagents were purchased from Sigma-Aldrich unless specified. Human tumor samples, DLD-1 cells and colon mouse tissues were lysed in ice-cold buffer (250 mmol/L NaCl, 20 mmol/L Tris/HCl pH 7.4, 10 mmol/L MgCl2, 1% Triton X-100, 10 μL/mL protease inhibitor cocktail [PIC; Roche Applied Science, Penzberg, Germany], 10 μL/mL phosphatase inhibitor cocktails II and III, and 40 U/mL RNaseOUT [Invitrogen]). Dynabeads (Invitrogen) were incubated with a specific anti-FMRP antibody57 or anti-rabbit immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, CA) in presence of 1% bovine serum albumin (BSA) for 1 hour at room temperature. The beads were washed in wash buffer (250 mmol/L NaCl, 10 mmol/L Tris-HCl pH 7.4, 10 mmol/L MgCl2, and 0.1% Triton X-100), and 800 μg of protein extract derived from 3 different human tumor samples, 800 μg of protein extract derived from DLD-1 cells, and 5 mg of protein extracts derived from colon tissues of 9 mice were added to the Dynabeads and incubated for 1–2 hours at 4°C. Proteins and RNA were eluted in Laemmli buffer and TRIzol, respectively. The co-immunoprecipitated RNA was extracted and analyzed by RT-qPCR using the StepOne Plus 7500 instrument (Life Technologies, Carlsbad, CA). Sequences of the primers used were the following: human HPRT1 (forward 5´-TGCTGAGGATTTGGAAAGGGT-3´, reverse 5´-TCGAGCAAGACGTTCAGTCC-3´); human β-actin (forward 5´-ACCGAGCGCGGCTACAG-3´, reverse 5´-CTTAATGTCACGCACGATTTCC-3´); human E-cadherin (forward 5´-CGAGAGCTACACGTTCACGG-3´, reverse 5´-CTTTGTCGACCGGTGCAATC-3´); human Vimentin (forward 5´-GCTTCAGAGAGAGGAAGCCG-3´, reverse 5´-AAGGTCAAGACGTGCCAGAG-3´); human RIPK3 (forward 5´-CAGTGTGCAACAGGCAGAAC-3´, reverse 5´-TCAGTCCTTCTAAGCCGGGA-3´); human RIPK1 (forward 5´-CACAAGGACCTGAAGCCTGAA-3´, reverse 5´-TGCTTGTTTTGAGCTGTAGCC-3´); mouse Hprt1 (forward 5´-CAGCCCCAAAATGGTTAAGGTTGC-3´, reverse 5´-TCCAACAAAGTCTGGCCTGTATCC-3´); mouse E-cadherin (forward 5´-GTGACGCTGAAGTCCATGG-3´, reverse 5´-TTCAGAGGCAGGGTCGCG-3´); mouse RIPK1 (forward 5´-GTCCACCGCCCGTCCT-3´, reverse 5´-GCTCAGAATCTCCAACACACC-3´).
Statistical Analysis
Values are expressed as mean ± standard deviation (SD) or ± standard error of the mean (SEM). Statistical analysis of the data was performed by using Student t test, Mann-Whitney test, or χ2 test. GraphPad Prism 6 (GraphPad Software, La Jolla, CA) was used for statistical and graphical data evaluations. P values <.05 were considered statistically significant.
Acknowledgments
The authors thank Vittoria Mariano for her help in making the graphical abstract using Adobe Illustrator and Laura Pacini and Maria Giulia Farace for advice and guidance. The authors are grateful to Giulia Cencelli and Nuria Dominguez-Iturza for critical reading of the manuscript.
CRediT Authorship Contributions
Antonio Di Grazia (Data curation: Lead; Investigation: Lead; Methodology: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Irene Marafini (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Writing – review & editing: Equal),
Giorgia Pedini (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Equal)
Davide Di Fusco (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Equal)
Federica Laudisi (Data curation: Equal; Formal analysis: Equal)
Vincenzo Dinallo (Data curation: Equal; Formal analysis: Equal)
Eleonora Rosina (Data curation: Equal; Formal analysis: Equal; Writing - review & editing: Equal)
Carmine Stolfi (Data curation: Equal; Formal analysis: Equal)
Eleonora Franzè (Data curation: Equal; Formal analysis: Equal)
Pierpaolo Sileri (Data curation: Supporting)
Giuseppe Sica (Data curation: Supporting)
Giovanni Monteleone (Funding acquisition: Supporting; Methodology: Supporting; Writing – review & editing: Supporting),
Claudia Bagni (Conceptualization: Equal; Formal analysis: Equal; Methodology: Equal; Funding acquisition: Equal; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead),
Ivan Monteleone (Conceptualization: Lead; Data curation: Equal; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the following funds: Associazione Italiana Sindrome X Fragile, PRIN201789LFKB, and Etat de Vaud Switzerland to CB and by Nogra Pharma Ltd (Dublin, Ireland) to IMo. The funders had no role in study design, data analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Claudia Bagni, Email: claudia.bagni@uniroma2.it.
Ivan Monteleone, Email: ivan.monteleone@uniroma2.it.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bisogno L.S., Keene J.D. RNA regulons in cancer and inflammation. Curr Opin Genet Dev. 2018;48:97–103. doi: 10.1016/j.gde.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parham L.R., Williams P.A., Chatterji P., Whelan K.A., Hamilton K.E. RNA regulons are essential in intestinal homeostasis. Am J Physiol Gastrointest Liver Physiol. 2019;316:G197–G204. doi: 10.1152/ajpgi.00403.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira B., Billaud M., Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends in Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Vo D.T., Subramaniam D., Remke M., Burton T.L., Uren P.J., Gelfond J.A., de Sousa Abreu R., Burns S.C., Qiao M., Suresh U., Korshunov A., Dubuc A.M., Northcott P.A., Smith A.D., Pfister S.M., Taylor M.D., Janga S.C., Anant S., Vogel C., Penalva L.O. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol. 2012;181:1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wurth L., Papasaikas P., Olmeda D., Bley N., Calvo G.T., Guerrero S., Cerezo-Wallis D., Martinez-Useros J., Garcia-Fernandez M., Huttelmaier S., Soengas M.S., Gebauer F. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 2016;30:694–707. doi: 10.1016/j.ccell.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Tang Y., Ye L., Liu B., Liu K., Chen J., Xue Q. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J Cancer Res Clin Oncol. 2003;129:43–51. doi: 10.1007/s00432-002-0396-4. [DOI] [PubMed] [Google Scholar]
- 9.Qie S., Majumder M., Mackiewicz K., Howley B.V., Peterson Y.K., Howe P.H., Palanisamy V., Diehl J.A. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma. Nature Communications. 2017;8:1534. doi: 10.1038/s41467-017-01199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing Z., Zeng M., Hu H., Zhang H., Hao Z., Long Y., Chen S., Su H., Yuan Z., Xu M., Chen J. Fragile X mental retardation protein promotes astrocytoma proliferation via the MEK/ERK signaling pathway. Oncotarget. 2016;7:75394–75406. doi: 10.18632/oncotarget.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagni C., Tassone F., Neri G., Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhogal B., Jepson J.E., Savva Y.A., Pepper A.S., Reenan R.A., Jongens T.A. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nature Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasciuto E., Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158:1446–1446.e1. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 14.Zalfa F., Panasiti V., Carotti S., Zingariello M., Perrone G., Sancillo L., Pacini L., Luciani F., Roberti V., D’Amico S., Coppola R., Abate S.O., Rana R.A., De Luca A., Fiers M., Melocchi V., Bianchi F., Farace M.G., Achsel T., Marine J.C., Morini S., Bagni C. The fragile X mental retardation protein regulates tumor invasiveness-related pathways in melanoma cells. Cell Death & Disease. 2017;8:e3169. doi: 10.1038/cddis.2017.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luca R., Averna M., Zalfa F., Vecchi M., Bianchi F., La Fata G., Del Nonno F., Nardacci R., Bianchi M., Nuciforo P., Munck S., Parrella P., Moura R., Signori E., Alston R., Kuchnio A., Farace M.G., Fazio V.M., Piacentini M., De Strooper B., Achsel T., Neri G., Neven P., Evans D.G., Carmeliet P., Mazzone M., Bagni C. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Molecular Medicine. 2013;5:1523–1536. doi: 10.1002/emmm.201302847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz-Pedersen S., Hasle H., Olsen J.H., Friedrich U. Evidence of decreased risk of cancer in individuals with fragile X. Am J Med Genet. 2001;103:226–230. [PubMed] [Google Scholar]
- 17.Kalkunte R., Macarthur D., Morton R. Glioblastoma in a boy with fragile X: an unusual case of neuroprotection. Arch Dis Child. 2007;92:795–796. doi: 10.1136/adc.2006.103382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H., Morishita Y., Miura D., Naranjo J.R., Kida S., Zhuo M. Roles of CREB in the regulation of FMRP by group I metabotropic glutamate receptors in cingulate cortex. Molecular Brain. 2012;5:27. doi: 10.1186/1756-6606-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordonaro M., Lazarova D.L. CREB-binding protein, p300, butyrate, and Wnt signaling in colorectal cancer. World J Gastroenterol. 2015;21:8238–8248. doi: 10.3748/wjg.v21.i27.8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Giorgio E., Hancock W.W., Brancolini C. MEF2 and the tumorigenic process, hic sunt leones. Biochimica et Biophysica Acta Reviews on Cancer. 2018;1870:261–273. doi: 10.1016/j.bbcan.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Smith K.T., Nicholls R.D., Reines D. The gene encoding the fragile X RNA-binding protein is controlled by nuclear respiratory factor 2 and the CREB family of transcription factors. Nucleic Acids Res. 2006;34:1205–1215. doi: 10.1093/nar/gkj521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwu W.L., Wang T.R., Lee Y.M. FMR1 enhancer is regulated by cAMP through a cAMP-responsive element. DNA Cell Biol. 1997;16:449–453. doi: 10.1089/dna.1997.16.449. [DOI] [PubMed] [Google Scholar]
- 23.Nambiar P.R., Girnun G., Lillo N.A., Guda K., Whiteley H.E., Rosenberg D.W. Preliminary analysis of azoxymethane induced colon tumors in inbred mice commonly used as transgenic/knockout progenitors. Int J Oncol. 2003;22:145–150. [PubMed] [Google Scholar]
- 24.Papanikolaou A., Wang Q.S., Papanikolaou D., Whiteley H.E., Rosenberg D.W. Sequential and morphological analyses of aberrant crypt foci formation in mice of differing susceptibility to azoxymethane-induced colon carcinogenesis. Carcinogenesis. 2000;21:1567–1572. [PubMed] [Google Scholar]
- 25.Degterev A., Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008;9:378–390. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- 26.Danial N.N., Korsmeyer S.J. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 27.Su Z., Yang Z., Xie L., DeWitt J.P., Chen Y. Cancer therapy in the necroptosis era. Cell Death Differ. 2016;23:748–756. doi: 10.1038/cdd.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He G.W., Gunther C., Thonn V., Yu Y.Q., Martini E., Buchen B., Neurath M.F., Sturzl M., Becker C. Regression of apoptosis-resistant colorectal tumors by induction of necroptosis in mice. J Exp Med. 2017;214:1655–1662. doi: 10.1084/jem.20160442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong Y., Fan Z., Luo G., Yang C., Huang Q., Fan K., Cheng H., Jin K., Ni Q., Yu X., Liu C. The role of necroptosis in cancer biology and therapy. Molecular Cancer. 2019;18:100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanden Berghe T., Linkermann A., Jouan-Lanhouet S., Walczak H., Vandenabeele P. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 31.Linkermann A., Green D.R. Necroptosis. N Engl J Med. 2014;370:455–465. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 33.Fearon E.R. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 34.Waldner M.J., Neurath M.F. The molecular therapy of colorectal cancer. Molecular Aspects of Medicine. 2010;31:171–178. doi: 10.1016/j.mam.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Chatterji P., Rustgi A.K. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med. 2018;24:490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurth L., Gebauer F. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta. 2015;1849:881–886. doi: 10.1016/j.bbagrm.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Legrand N., Dixon D.A., Sobolewski C. AU-rich element-binding proteins in colorectal cancer. World Journal of Gastrointestinal Oncology. 2019;11:71–90. doi: 10.4251/wjgo.v11.i2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwemmle S., de Graaff E., Deissler H., Glaser D., Wohrle D., Kennerknecht I., Just W., Oostra B.A., Doerfler W., Vogel W., Steinbach P. Characterization of FMR1 promoter elements by in vivo-footprinting analysis. Am J Hum Genet. 1997;60:1354–1362. doi: 10.1086/515456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evan G.I., Vousden K.H. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 40.Lowe S.W., Cepero E., Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 41.Ascano M., Jr., Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M., Williams Z., Ohler U., Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W., Licatalosi D.D., Richter J.D., Darnell R.B. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyashiro K.Y., Beckel-Mitchener A., Purk T.P., Becker K.G., Barret T., Liu L., Carbonetto S., Weiler I.J., Greenough W.T., Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 44.Sawicka K., Hale C.R., Park C.Y., Fak J.J., Gresack J.E., Van Driesche S.J., Kang J.J., Darnell J.C., Darnell R.B. FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. eLife. 2019;8 doi: 10.7554/eLife.46919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Marino D., Achsel T., Lacoux C., Falconi M., Bagni C. Molecular dynamics simulations show how the FMRP Ile304Asn mutation destabilizes the KH2 domain structure and affects its function. J Biomol Struct Dyn. 2014;32:337–350. doi: 10.1080/07391102.2013.768552. [DOI] [PubMed] [Google Scholar]
- 46.Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 47.Bagni C., Zukin R.S. A synaptic perspective of fragile X syndrome and autism spectrum disorders. Neuron. 2019;101:1070–1088. doi: 10.1016/j.neuron.2019.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doluca O. G4Catchall: a G-quadruplex prediction approach considering atypical features. J Theor Biol. 2019;463:92–98. doi: 10.1016/j.jtbi.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Zhuang Y., Xu H.C., Shinde P.V., Warfsmann J., Vasilevska J., Sundaram B., Behnke K., Huang J., Hoell J.I., Borkhardt A., Pfeffer K., Taha M.S., Herebian D., Mayatepek E., Brenner D., Ahmadian M.R., Keitel V., Wieczorek D., Haussinger D., Pandyra A.A., Lang K.S., Lang P.A. Fragile X mental retardation protein protects against tumour necrosis factor-mediated cell death and liver injury. Gut. 2020;69:133–145. doi: 10.1136/gutjnl-2019-318215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng X., Song Q., Yu A., Tang H., Peng Z., Wang X. Receptor-interacting protein kinase 3 is a predictor of survival and plays a tumor suppressive role in colorectal cancer. Neoplasma. 2015;62:592–601. doi: 10.4149/neo_2015_071. [DOI] [PubMed] [Google Scholar]
- 51.Moriwaki K., Bertin J., Gough P.J., Orlowski G.M., Chan F.K. Differential roles of RIPK1 and RIPK3 in TNF-induced necroptosis and chemotherapeutic agent-induced cell death. Cell Death & Disease. 2015;6:e1636. doi: 10.1038/cddis.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han W., Li L., Qiu S., Lu Q., Pan Q., Gu Y., Luo J., Hu X. Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol Cancer Ther. 2007;6:1641–1649. doi: 10.1158/1535-7163.MCT-06-0511. [DOI] [PubMed] [Google Scholar]
- 53.Steinhart L., Belz K., Fulda S. Smac mimetic and demethylating agents synergistically trigger cell death in acute myeloid leukemia cells and overcome apoptosis resistance by inducing necroptosis. Cell Death & Disease. 2013;4 doi: 10.1038/cddis.2013.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meurette O., Rebillard A., Huc L., Le Moigne G., Merino D., Micheau O., Lagadic-Gossmann D., Dimanche-Boitrel M.T. TRAIL induces receptor-interacting protein 1-dependent and caspase-dependent necrosis-like cell death under acidic extracellular conditions. Cancer Res. 2007;67:218–226. doi: 10.1158/0008-5472.CAN-06-1610. [DOI] [PubMed] [Google Scholar]
- 55.Basit F., Cristofanon S., Fulda S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013;20:1161–1173. doi: 10.1038/cdd.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stolfi C., Rizzo A., Franze E., Rotondi A., Fantini M.C., Sarra M., Caruso R., Monteleone I., Sileri P., Franceschilli L., Caprioli F., Ferrero S., MacDonald T.T., Pallone F., Monteleone G. Involvement of interleukin-21 in the regulation of colitis-associated colon cancer. J Exp Med. 2011;208:2279–2290. doi: 10.1084/jem.20111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferrari F., Mercaldo V., Piccoli G., Sala C., Cannata S., Achsel T., Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]