Abstract
Background & Aims
Tristetraprolin (TTP) is a key post-transcriptional regulator of inflammatory and oncogenic transcripts. Accordingly, TTP was reported to act as a tumor suppressor in specific cancers. Herein, we investigated how TTP contributes to the development of liver inflammation and fibrosis, which are key drivers of hepatocarcinogenesis, as well as to the onset and progression of hepatocellular carcinoma (HCC).
Methods
TTP expression was investigated in mouse/human models of hepatic metabolic diseases and cancer. The role of TTP in nonalcoholic steatohepatitis and HCC development was further examined through in vivo/vitro approaches using liver-specific TTP knockout mice and a panel of hepatic cancer cells.
Results
Our data demonstrate that TTP loss in vivo strongly restrains development of hepatic steatosis and inflammation/fibrosis in mice fed a methionine/choline-deficient diet, as well as HCC development induced by the carcinogen diethylnitrosamine. In contrast, low TTP expression fostered migration and invasion capacities of in vitro transformed hepatic cancer cells likely by unleashing expression of key oncogenes previously associated with these cancerous features. Consistent with these data, TTP was significantly down-regulated in high-grade human HCC, a feature further correlating with poor clinical prognosis. Finally, we uncover hepatocyte nuclear factor 4 alpha and early growth response 1, two key transcription factors lost with hepatocyte dedifferentiation, as key regulators of TTP expression.
Conclusions
Although TTP importantly contributes to hepatic inflammation and cancer initiation, its loss with hepatocyte dedifferentiation fosters cancer cells migration and invasion. Loss of TTP may represent a clinically relevant biomarker of high-grade HCC associated with poor prognosis.
Keywords: AUBPs, NASH, HNF4A, Liver Cancer, Oncogenes
Abbreviations used in this paper: 5-AZA, 5-aza-2'deoxycytidine; ARE, adenylate-uridylate-rich element; AUBP, adenylate-uridylate-rich element binding protein; DEN, diethylnitrosamine; DMEM, Dulbecco modified Eagle medium; DMSO, dimethyl sulfoxide; EGR1, early growth response 1; EMT, epithelial-mesenchymal transition; EV, empty vector; FCS, fetal calf serum; FLX, floxed allele; GEO, gene expression omnibus; GEPIA, Gene Expression Profiling Interactive Analysis; GSEA, gene set enrichment analysis; HCC, hepatocellular carcinoma; HNF4α, hepatocyte nuclear factor 4 alpha; ICC, intrahepatic cholangiocarcinoma; IL6, interleukin 6; LTTPKO, liver-specific tristetraprolin knockout mice; MCD, methionine and choline-deficient; MPH, mouse primary hepatocyte; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; SD, standard deviation; siRNA, small interfering RNA; TCGA, the cancer genome atlas; TGFβ, transforming growth factor beta; TMA, tissue microarray; TSA, trichostatin A; TTP, tristetraprolin; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; UTR, untranslated region; VLDL, very low density lipoprotein
Graphical abstract
Summary.
The post-transcriptional regulator tristetraprolin harbors a dual role in liver cancer. Indeed, while tristetraprolin promotes hepatic inflammation and cancer initiation, it restrains cancer cells migration and invasion. Loss of TTP may represent a clinically relevant biomarker of high-grade hepatocellular carcinoma.
Trans-acting factors controlling the fate of messenger RNAs are key regulators of gene expression and contribute importantly to various pathophysiological processes. Among these factors are adenylate-uridylate-rich element (ARE) binding proteins (ARE-BPs or AUBPs), which are RNA-binding proteins targeting AU-rich sequences (eg, “AUUUA” motif) within the 3'-untranslated regions (UTRs) of mRNAs, thereby influencing their stabilities and/or their access to the translation machinery.1 Deregulated expression or activity of AUBPs is associated with inflammatory disorders and cancers. They control the expression of numerous oncogenes, tumor suppressors, and inflammatory mediators, therefore altering their cellular function with similar outcomes as activating or inhibitory genetic mutations. Deregulated activity of specific AUBPs, eg, HuR,2 TIA-1,3 CUGBP2,4 was associated with hepatocarcinogenesis; however, the molecular mechanisms behind their pathophysiological functions remain poorly understood currently.
The AUBP tristetraprolin (TTP, ZFP36) was previously suggested to have tumor suppressive activity in several cancers.5,6 TTP belongs to the TIS11 (12-O-tetradecanoylphorbol-13-acetate-induced sequence) family,7 with a sequence encompassing 2 cysteine-histidine zinc-finger motifs, which enable RNA binding, and 4 proline motifs allowing interactions with co-factor proteins.8 The ZFP36 gene fulfills criteria for an intermediate-early response gene because its expression is rapidly stimulated by various factors including proinflammatory cytokines and growth factors.9,10 In physiological conditions, TTP localizes in the cytoplasm, where it binds to transcripts and recruits them to small cytoplasmic granules called processing bodies (P-bodies), where mRNA decay occurs.11,12 Many TTP target mRNAs have been experimentally validated including inflammatory cytokines (eg, IL2, IL6, IL8, IL10, TNFA), factors involved in cell cycle (CDK1, NEK2, KIF11, CCND1), apoptosis (eg, HIF1A, SOX9, PDK1, BCL2), or metabolism (eg, HK2, PDK1, GPD2, DLAT).13,14 In the liver specifically, few and inconsistent information are available about the pathophysiological functions of TTP. Hepatic TTP down-regulation was reported to promote glucose tolerance and insulin sensitivity,15 while contributing to fibrosis development.16 Methylation of TTP promoter was further reported in cultured liver cancer cells and hepatocellular carcinoma (HCC) patients and suggested to confer resistance to transforming growth factor alpha (TGFα) antiproliferative action.17 However, deletion of TTP in mouse liver was also associated with reduced tumorigenesis.18 On the basis of our current knowledge, the role of TTP in the development of hepatic disorders preceding and promoting carcinogenesis, ie, inflammation and fibrosis, remains unclear and needs further investigations.
In this study, we have identified several AUBPs whose expression is significantly deregulated in HCC. Among them, TTP was found significantly repressed in poorly differentiated tumors and to exert a tight post-transcriptional control on genes involved in tumor progression to malignancy. Our data are consistent with a dual role for TTP in liver inflammation and carcinogenesis. Indeed, hepatic TTP promotes in vivo hepatic steatosis, inflammation, fibrosis, and cancer initiation, thus clearly indicating cancer-driving functions for TTP within an in vivo orthotopic environment. In contrast, TTP appears to restrain migration and invasion of transformed cancer cells, thus indicating a tumor suppressive function of TTP in late stages of hepatic carcinogenesis.
Results
TTP Down-Regulation Is Associated With High-Grade HCC and Poor Prognosis
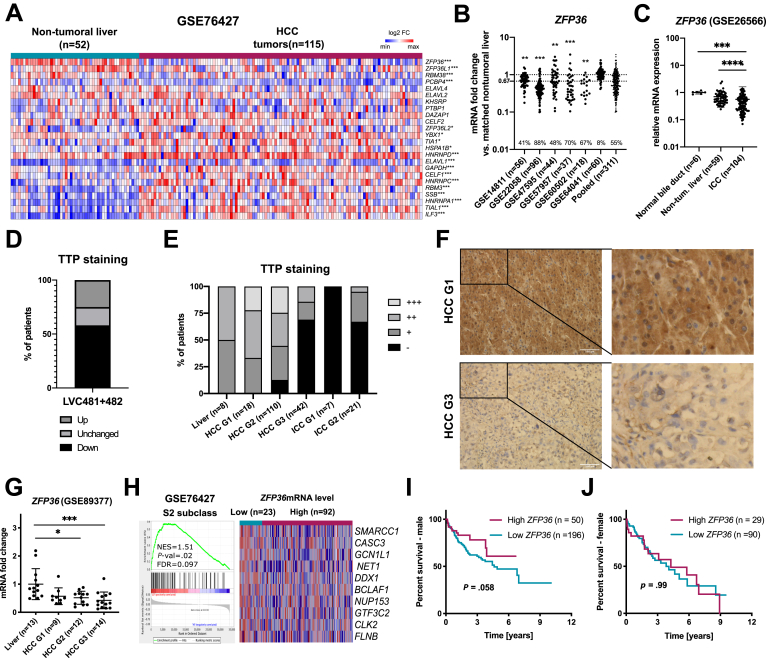
To uncover potential alterations of AUBP expression occurring in HCC, AUBPs mRNA expression levels were analyzed in a human transcriptomic dataset including paired HCC tumors and matched non-tumoral tissues (GSE76427; Figure 1A). Among 24 well-characterized AUBPs,13 several of them were up-regulated (eg, ILF3, TIAL1, HNRNPA1), whereas others were significantly down-regulated (eg, ZFP36, ZFP36L1, RBM38, PCBP4). Among the latter, ZFP36 (coding for TTP protein) was the most down-regulated in HCC, a feature further confirmed by additional human transcriptomic datasets of paired HCC versus matched non-tumoral tissues (Figure 1B). Similarly, ZFP36 down-regulation was also observed in human intrahepatic cholangiocarcinoma (ICC) (Figure 1C). On the basis of these bioinformatic analyses of mRNA expression in human HCC, we then investigated TTP protein expression in human HCC and ICC tissue microarrays (TMAs) (Figure 1D–F). Our data revealed a homogenous staining for TTP in hepatocytes and a significant down-regulation of TTP protein expression in 58% of HCC, as compared with matched non-tumoral tissues (TMA no: LVC481 and LVC482; Figure 1D). Further analyses of a third TMA including HCC and ICC of different grades indicated that TTP protein expression was predominantly lost in high-grade HCC (G 2/3) and both grade 1 (G1) and grade 2 (G2) ICC (TMA no: LV2161; Figure 1E and F), as further supported by analyses of publicly available human transcriptomic datasets (Figure 1G) and by gene set enrichment analysis (GSEA) of genes associated with poorly differentiated HCC having bad prognostic (S2 subclass, GSE76427), which are up-regulated in low ZFP36 expressing tumors (Figure 1H). Finally, TTP loss was associated with reduced overall survival in males but not in females (Figure 1I and J), suggesting a potential gender-dependent prognostic value for TTP loss.
Figure 1.
AUBPs are highly dysregulated in human HCC. (A) Heatmap showing log2 fold change expression levels of known AUBPs in non-tumoral vs tumoral liver tissue (GSE76427). (B) ZFP36 mRNA levels in human HCC GEO datasets. Data represented as mRNA fold change between tumoral and non-tumoral tissues. Percentages of patients bearing more than 67% reduction in ZFP36 levels are indicated. (C) ZFP36 mRNA expression in GEO Dataset of human cholangiocarcinoma (GSE26566). Data represented as fold change vs control. Significance level was determined with one-way analysis of variance with Tukey test for multiple comparisons. (D) Graph representation of TTP staining loss/gain in HCC tumors (TMA LVC481 and LVC482) reported as % of all patients per group. Intensity of TTP staining was assessed by 2 independent investigators using a staining scale (- no staining, + weak, ++ moderate, +++ strong staining). (E) Graph representation of TTP staining intensity in HCC and ICC tumors with regard to grade (LV2161). Intensity of TTP staining was assessed by 2 independent investigators using a staining scale (- no staining, + weak, ++ moderate, +++ strong staining). (F) Representative images of TTP staining intensity in HCC tumors of grade G1 and G3 in TMAs (TMA LV2161). (G) ZFP36 mRNA expression levels in human HCC with different grades (human GEO dataset GSE89377). Expression represented as fold change vs liver. P value based on one-way analysis of variance with Tukey test for multiple comparisons. (H) GSEA analysis of human HCC transcriptome data (GSE76427) for genes from HCC subclass S2 characterized by poor survival and low differentiation. Tumor samples were segregated on the basis of ZFP36 expression levels (low vs high ZFP36 expression, based on 20th percentile). Top 10 genes are represented for each gene set (expression normalized per row). Normalized enrichment score (NES), false discovery rate (FDR), and P value are displayed. A gene set was considered enriched at FDR <0.25. (I) Survival analysis in male HCC patients, based on ZFP36 mRNA expression levels (“Best separation method”, TCGA LIHC cohort, Human Protein Atlas). P value was calculated using a log-rank test (data retrieved from TCGA and Human Protein Atlas). (J) Survival analysis in female HCC patients, based on ZFP36 mRNA expression levels (“Best separation method”, TCGA LIHC cohort, Human Protein Atlas). P value was calculated using log-rank test (data retrieved from TCGA and Human Protein Atlas). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Transcriptional Activity of Hepatocyte Nuclear Factor 4 Alpha–Early Growth Response 1 Is Required for TTP Expression in Hepatocytes
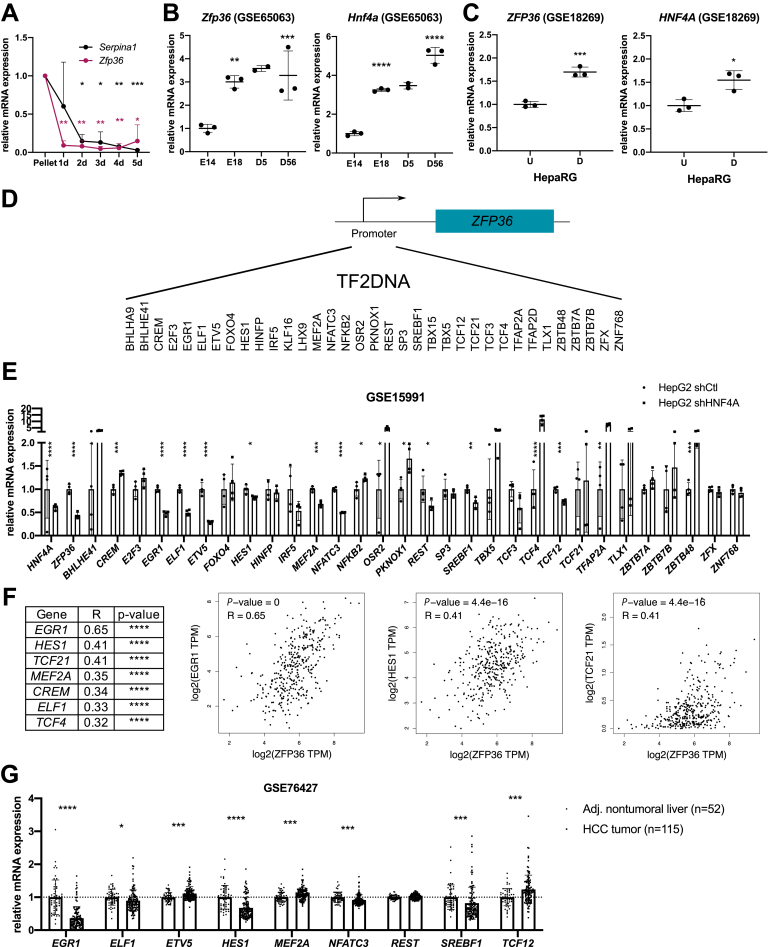
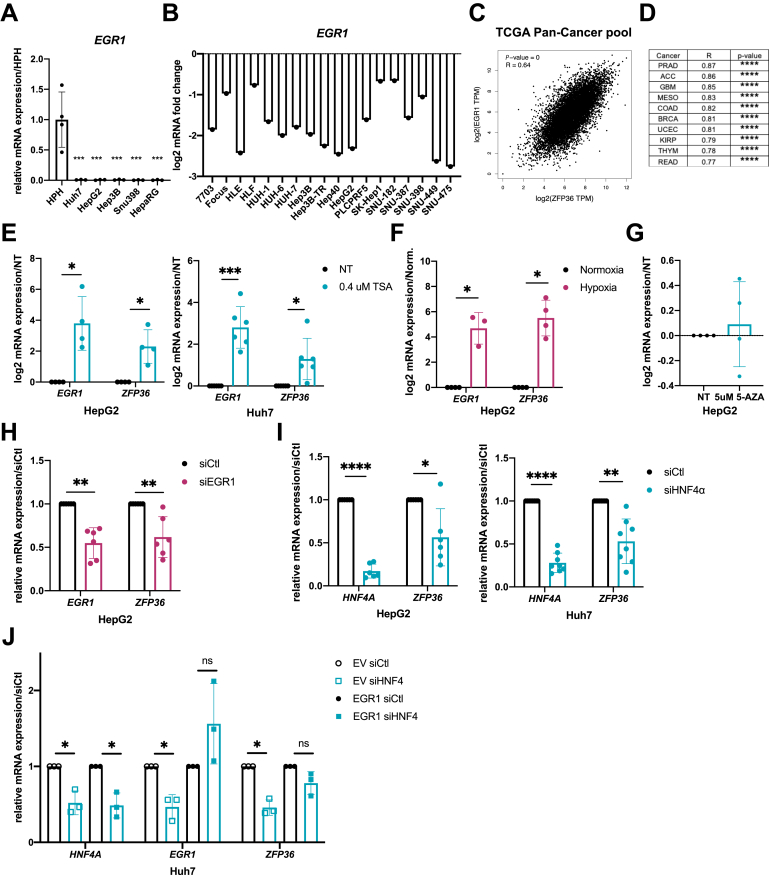
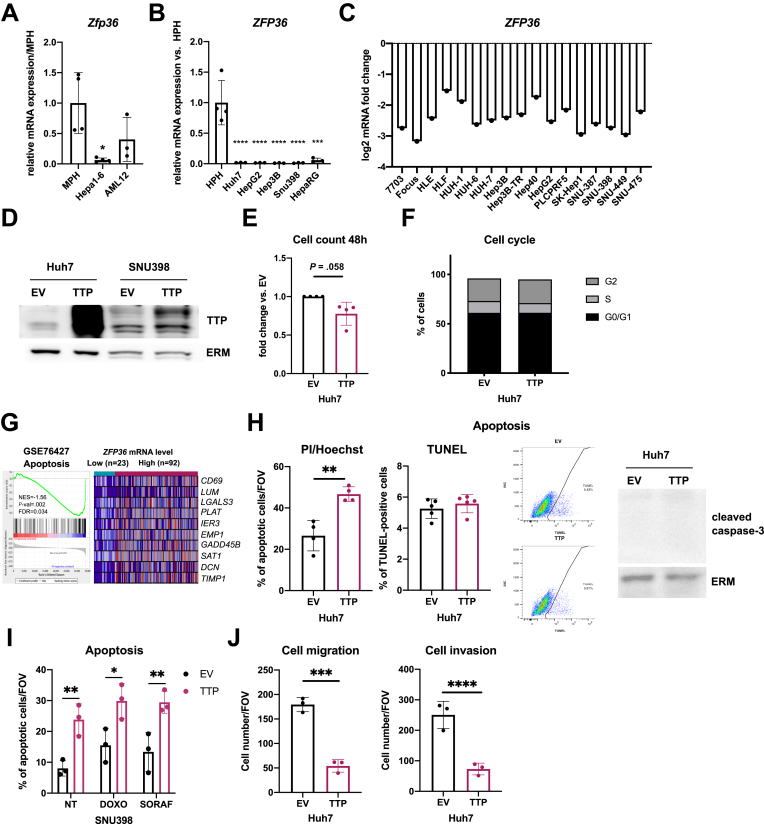
The reduction of TTP expression in high-grade HCC suggests a tight link between hepatocyte differentiation and TTP expression. In support of this hypothesis, a decreased TTP expression was observed during dedifferentiation of cultured mouse primary hepatocytes (MPH) in vitro (Figure 2A). Furthermore, in silico analyses of various gene expression omnibus (GEO) datasets indicated that TTP expression was increased with hepatocyte differentiation in mice during development (Figure 2B) and in vitro in HepaRG cells undergoing differentiation (Figure 2C). Several transcription factors are well-known to regulate hepatocytes differentiation (eg, FOXA2, the C/EBP family, HNF4α).19 Among them, hepatocyte nuclear factor 4 alpha (HNF4α) is of particular interest, although canonical HNF4α binding motifs are not present within TTP’s promoter (Figure 2D). Indeed, HNF4α regulates the expression of several other transcription factors predicted to control ZFP36 (TTP) transcription (eg, EGR1, ELF1, ETV5, NFATC3) as shown by the analysis of available transcriptomic data from HepG2 cells having HNF4A knockdown by short hairpin RNA (GSE15991) (Figure 2E). Consistent with these data, expression of several of these transcription factors under the control of HNF4α (eg, EGR1, ELF1, HES1) significantly correlates with ZFP36 expression in human liver cancer (The Cancer Genome Atlas [TCGA]-Liver Hepatocellular Carcinoma [LIHC] cohort, Figure 2F). Among them, EGR1 was considered for further analyses because it displayed the highest correlative factor with ZFP36 expression. Supporting the relevance of early growth response 1 (EGR1) as a HNF4α downstream transcription factor regulating ZFP36 expression, EGR1 was previously reported to (1) be under the transcriptional control of HNF4α20 and (2) regulate ZFP36 transcription.21 Further analyses of an HCC patients cohort (GSE76427) indicate that EGR1 is strongly repressed in human tumoral tissues as compared with adjacent non-tumoral tissues similarly to ZFP36 (Figure 2G), and its expression is also significantly down-regulated in human hepatic cancer cell lines (Figure 3A and B). Correlations between EGR1 and ZFP36 expression were found not only in HCC (Figure 2F) but also in other type of cancers (Figure 3C and D; Gene Expression Profiling Interactive Analysis [GEPIA] analyses of whole TCGA cohort of cancer patients, top 10 correlations are shown in Figure 3D). Finally, we observed that both ZFP36 and EGR1 were strongly induced in HepG2 and Huh7 cells subjected to histone deacetylase inhibition using trichostatin A (TSA) or in hypoxic HepG2 cells (Figure 3E and F). Of note, promoter methylation seems not to control ZFP36 expression in hepatic cancer cells, because incubation of HepG2 cells with demethylating agent 5-aza-2'-deoxycytidine (5-AZA) did not affect ZFP36 expression (Figure 3G).
Figure 2.
TTP levels correlate with differentiation status of hepatocytes and expression of HNF4α/EGR1 in HCC. (A) Serpina1 (AAT) and Zfp36 (TTP) mRNA expression in isolated murine primary hepatocytes before (Pellet) and after plating during 5 days. Data represented as relative expression vs pellet and normalized by 18S gene. (B) Zfp36 and Hnf4a mRNA expression fold change during liver development (E, embryonic; D, days after birth) (GSE65063). (C) ZFP36 and HNF4A mRNA expression fold change in undifferentiated (U) vs differentiated (D) human HepaRG cell line (GSE18269). (D) Potential transcription factors binding to promoter of ZFP36 retrieved from TF2DNA. (E) Potential transcription factors mRNA expression in control (shCtl) and HNF4A knockdown by shRNA in HepG2 cells (shHNF4α) (GSE15991 transcriptomic data). Data represent mean ± standard deviation (SD). The t test for comparison with Benjamini, Krieger and Yekutieli correction for multiple comparisons (α = 5%) was used. (F) Correlation analysis between mRNA expression of ZFP36 and the best candidates predicted to act as transcription factors for ZFP36 in HCC (Pearson coefficient, LIHC TCGA cohort, GEPIA software). (G) mRNA fold change of potential transcription factors for TTP in non-tumoral liver (n = 52) and HCC tumors (n = 115) (GSE76427). Data represented as mean ± SD. The t test for comparison of 2 groups was used. P value was corrected for multiple testing using the Benjamini, Krieger and Yekutieli procedure (α = 5%). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Figure 3.
TTP expression is regulated by HNF4α/EGR1 signaling axis. (A) EGR1 mRNA expression levels in HCC cell lines vs human primary hepatocytes (HPH) (n = 4 for HPH, n = 3 for other cell lines). Data represent mean ± SD. P value based on one-way analysis of variance with Tukey test for multiple comparisons. (B) EGR1 log2 mRNA expression in liver cell lines, retrieved from CellMiner. (C) Correlation analysis between mRNA expression of ZFP36 and EGR1 in TCGA cohort of cancer patients (Pearson coefficient, TCGA cohort, GEPIA software). (D) Correlation analysis between mRNA expression of ZFP36 and EGR1 in top 10 cancers (Pearson coefficient, TCGA cohort, GEPIA software). (E) EGR1 and ZFP36 log2 mRNA expression in HepG2 and Huh7 cells stimulated for 24 hours with 0.4 μmol/L TSA. (F) ZFP36 and EGR1 log2 mRNA expression in HepG2 cell line incubated for 24 hours in normoxic and hypoxic conditions. (G) ZFP36 mRNA expression fold change in HepG2 cells stimulated for 24 hours with 5 μmol/L 5-AZA. For (E–G), data represent mean ± SD of at least 3 independent experiments. Significance level was determined using unpaired t test. (H) EGR1 and ZFP36 mRNA expression in control and EGR1-silenced (by siRNAs) HepG2 cells 72 hours after transfection. (I) HNF4A and ZFP36 mRNA expression in control and HNF4A-silenced (by siRNAs) HepG2 and Huh7 cells 48 hours after transfection. (J) HNF4A, EGR1, and ZFP36 mRNA expression in control and HNF4A-silenced (by siRNAs) Huh7 cells transiently overexpressing (EGR1), or not (EV), the transcription factor EGR1. For (H–J), data represent mean ± SD of at least 3 independent experiments. Significance level was determined using unpaired t test. ∗∗∗P < .001, ∗∗P < .01, ∗P < .05. NT, non-treated cells.
To confirm the relevance of the control of ZFP36 expression by the HNF4α-EGR1 signaling axis, we further proceeded with silencing/overexpression approaches of these 2 factors in hepatic cells. Our data demonstrated that small interfering RNA (siRNA)-mediated silencing of EGR1 or HNF4A down-regulates ZFP36 expression in HepG2 and/or Huh7 cells, suggesting a strong link between these 2 factors (Figure 3H and I). Finally, down-regulation of ZFP36 after HNF4A silencing in Huh7 cells was prevented by the simultaneous overexpression of EGR1 in the same cells, thereby supporting EGR1 as a downstream effector of HNF4α (as also previously reported by others20, 21, 22) in regulating ZFP36 expression (Figure 3J).
Altogether, these results indicate that TTP loss in high-grade HCC is tightly associated with down-regulation of HNF4α and EGR1 and that the HNF4α-EGR1 signaling axis is a master regulator of TTP transcription.
TTP Fosters Hepatic Inflammation and Fibrosis in Mice
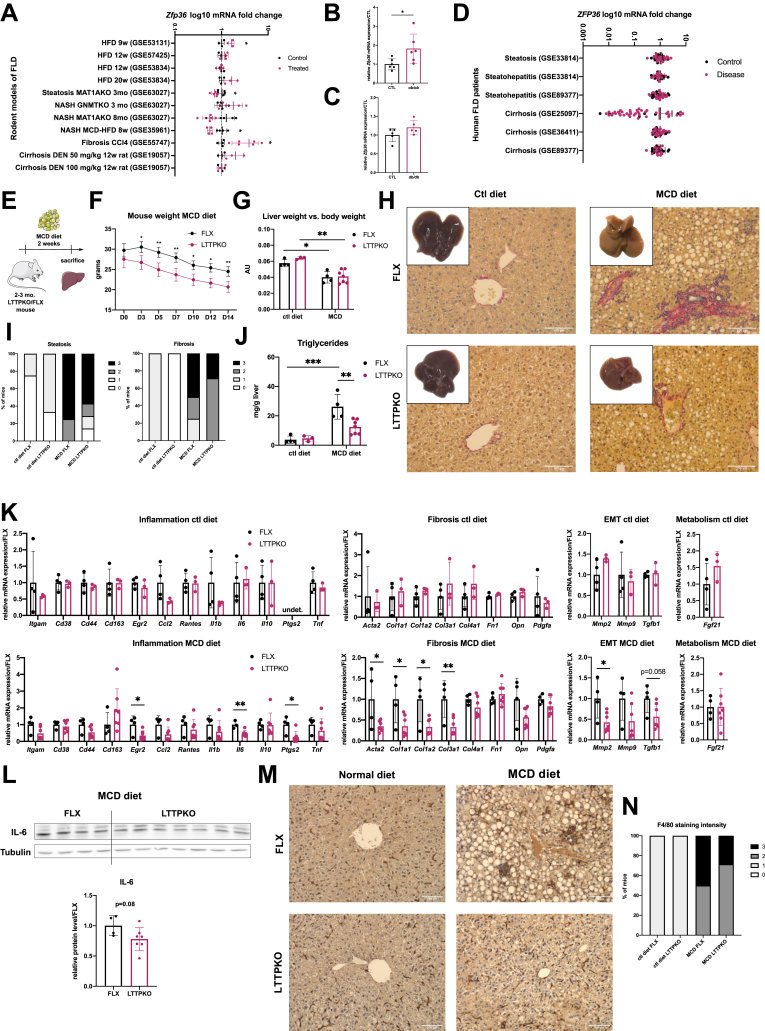
On the basis of previously described targets of TTP,13,14 it is likely that TTP is functionally relevant in the development of preneoplastic stages of the liver, setting a favorable landscape for HCC development. Of particular interest are hepatic inflammation and fibrosis, which are key drivers of HCC development, in the presence or absence of cirrhosis.23 In this regard, we first investigated whether hepatic ZFP36 expression was significantly altered with the development of steatosis, inflammation, fibrosis, and cirrhosis. As shown, both experimentally and bioinformatically through the analyses of publicly available transcriptomic GEO Datasets, ZFP36 expression slightly tends to either increase or decrease in transgenic or diet-induced mouse models of steatosis, inflammation, and fibrosis but with inconsistent trends in all studies considered (Figure 4A–C). Similarly, analyses of human transcriptomic GEO Datasets from human patients with hepatic steatosis, steatohepatitis, or cirrhosis were inconclusive (Figure 4D). Although ZFP36 expression may remain unchanged in nontransformed hepatocytes, alterations of its activity can significantly affect the development of hepatic inflammation and fibrosis. Therefore, to elucidate the functional relevance of TTP in hepatic steatosis, inflammation, and fibrosis, we submitted control (FLX) and hepatocytes-specific TTP knockout (LTTPKO) mice to a methionine/choline-deficient (MCD) diet. Administration of an MCD diet for 2 weeks in mice leads to the development of severe steatosis, inflammation, and fibrosis as illustrated in Figure 4. In mice fed an MCD diet we observed significant loss of body and liver weights (Figure 4E–G). Hepatic TTP deficiency significantly prevents in vivo MCD diet-induced histopathologic features of steatosis and fibrosis, as shown by histologic assessment of Picro Sirius red staining and lipid droplets content of the liver parenchyma (Figure 4H and I). In agreement with this phenotype, hepatic triglycerides levels were reduced in LTTPKO mice as compared with FLX control mice (Figure 4J), and the expression of key genes promoting inflammation (Il6, Ptgs2), macrophage infiltration (Egr2), fibrosis (Col1a1, Col1a1, Col3a1, Acta2), and epithelial-mesenchymal transition (EMT) (Mmp2, Tgfb1) were significantly down-regulated (Figure 4K). Of particular interest was the effect of TTP deletion on the expression of proinflammatory cytokine Il6, further confirmed at the protein level (Figure 4L), which was previously reported to promote hepatocarcinogenesis in a gender-dependent manner.24 Finally, we observed in TTP-deficient mice a slight decrease in macrophage infiltration, as shown by F4/80 histologic staining of liver sections, which corroborates with the reduction of Egr2 expression and a tendency for a down-regulation of Cd44 expression at the mRNA level (Figure 4K, M, N).
Figure 4.
TTP loss in vivo restrains steatosis development, inflammation, and fibrosis induced by MCD diet. (A) ZFP36 levels of human NAFLD and NASH patients of GEO Datasets. Data are represented as means ± SD of fold change vs control. The t test for comparison of 2 groups was used. Adjusted P value based on t test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (Q = 5%). (B) Zfp36 expression in livers of ob/ob (n = 6 per group) mice vs control (CTL). (C) Zfp36 expression in livers of db/db (n = 5 per group) mice vs control CTL. (D) Zfp36 levels in GEO Datasets of mice models of NAFLD and NASH. Data are represented as means ± SD of fold change vs control. (E) Scheme of experimental protocol used to fed mice fed with MCD diet. Two- to 3-month-old LTTPKO and FLX littermates were fed with control or MCD diet for 2 weeks. (FLX CTL, n = 4; LTTPKO CTL, n = 3; FLX MCD, n = 4; LTTPKO MCD, n = 7). (F) Mouse body weight (g) during 2 weeks of MCD diet feeding (D, day). (G) Liver weight vs body weight of FLX and LTTPKO mice after 2 weeks of MCD diet feeding. (H) Representative livers anatomies and histologic liver sections stained with Picro Sirius red of FLX and LTTPKO mice fed with normal or MCD diet for 2 weeks. (I) Quantifications of liver parenchymal lipid droplets content (steatosis) and fibrosis (Picro Sirius red staining) in mice fed with normal or MCD diet for 2 weeks. Lipid droplets density and intensity of Picro Sirius red staining was assessed by 2 independent investigators (- no steatosis/fibrosis staining, + weak, ++ moderate, +++ strong steatosis/fibrosis) and reported as percentage of animal per each group. (J) Triglyceride measurement in livers of FLX and LTTPKO mice fed with normal or MCD diet for 2 weeks. (K) mRNA expression of inflammatory markers, fibrotic markers, EMT markers, and Fgf-21 in livers of FLX (n = 4) and LTTPKO (n = 7) mice fed with normal or MCD diet for 2 weeks. Data represent mean ± SD. The t test for comparison of 2 groups was used. (L) Representative Western blot and quantification of IL6 protein levels in FLX and LTTPKO mice fed with MCD diet for 2 weeks. Tubulin was used as loading control. (M) Representative F4/80 staining of FLX and LTTPKO mice fed with normal or MCD diet. (N) Quantification of F4/80 staining as % of all mice per group. Intensity was assessed by 2 independent investigators using a staining scale (- no staining, + weak, ++ moderate, +++ strong staining). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Together, these data demonstrate that in vivo in mice, hepatic TTP fosters the development of steatosis, inflammation, and fibrosis, which are all key drivers of hepatocarcinogenesis.
TTP Loss Reduces Tumor Burden in Vivo
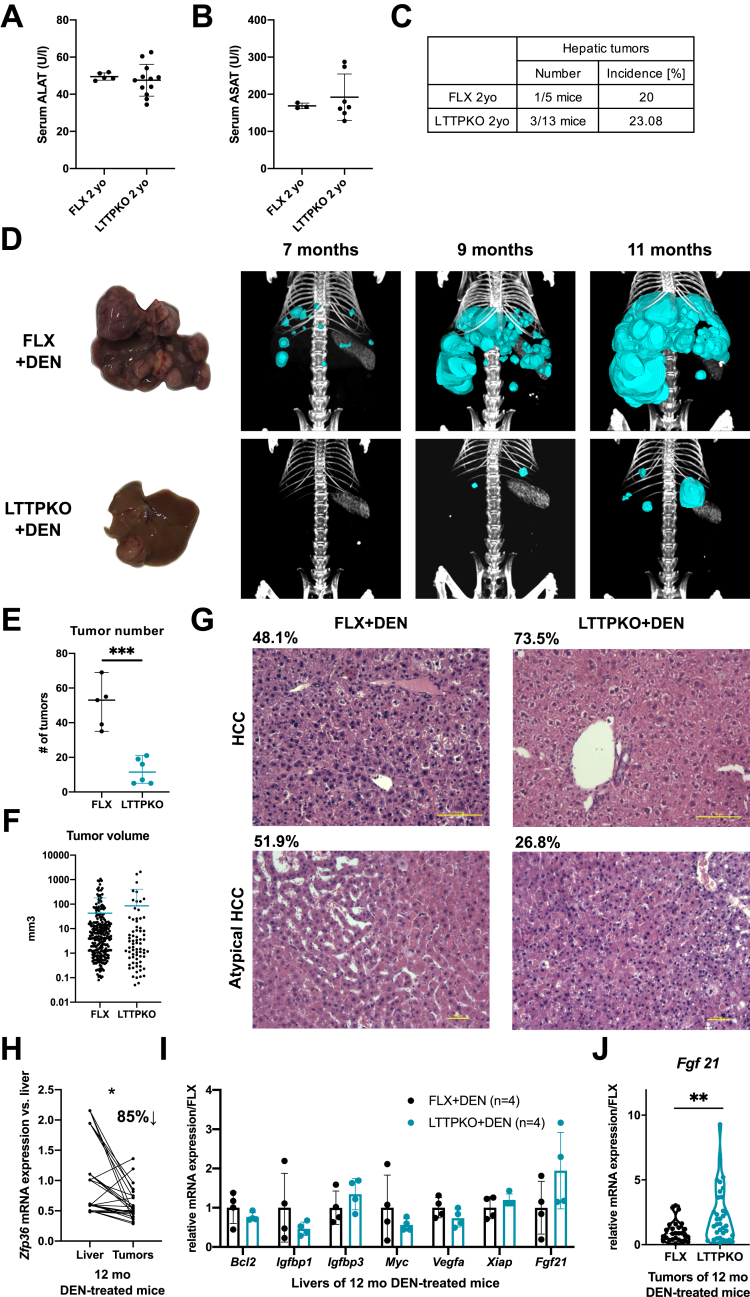
Tumor initiation and HCC development can be induced in mice over 1 year by injection of the carcinogen diethylnitrosamine (DEN). Deletion of TTP in the liver is asymptomatic and did not lead per se to spontaneous tumor development or hepatic damages (as assessed by serum alanine aminotransferase/aspartate aminotransferase measurements) with ageing in mice (over 2 years; Figure 5A–C). However, treatment of mice with DEN (single injection of 25 mg DEN/kg body weight at 15 days of age) induces multiple tumoral foci progressing to HCC by 1 year of age (Figure 5D). As expected from the outcomes of TTP loss in hepatic inflammation and fibrosis development, we observed a strong reduction of the number of tumors developing in LTTPKO mice as compared with control mice. Indeed, computed tomography scan imaging using ExiTron contrasting agent between 7 and 11 months of age before death indicated that the number of tumors was reduced by ≈5-fold in the absence of TTP (Figure 5D and E). However, the volume of the tumors was unchanged as compared with those developing in FLX mice, suggesting that TTP deficiency in hepatocytes was not affecting tumor growth in these conditions but likely initiation (Figure 5F). At the histologic level, 51.9% of tumoral nodules present in control mice were atypical hepatocellular tumors (tumors that do not fulfill the criteria for HCC in humans), which often contain intracytoplasmic hyaline bodies. The remaining 48.1% of tumors were diagnosed as classic HCC characterized by pseudoglandular and/or trabecular architecture, steatosis, and in few cases by steatohepatitis-like features. In LTTPKO mice, 26.5% and 73.5% of tumors were diagnosed as atypical hepatocellular tumor and HCC, respectively (typical histopathology in Figure 5G). Together, these data support an oncogenic function of TTP, which promotes hepatic tumor initiation in vivo in mice. However, the total absence of TTP in hepatocytes (LTTPKO mice), as compared with the partial down-regulation observed in around 85% of tumoral nodules of CTRL mice (Figure 5H), appears to foster tumor progression toward HCC, thus suggesting a dual role of TTP depending on the stage of carcinogenesis (initiation versus progression to malignancy). Because of the high number of cancer-related transcripts targeted by TTP, identifying the likely multiple factors relevant for carcinogenesis that might be affected in LTTPKO mice is challenging. However, a recent report highlighted some potential candidates, ie, BCL2, IGFBP1, IGFBP3, MYC, VEGFA, and XIAP, which are up-regulated in vitro in the absence of TTP and which may potentially affect carcinogenesis.18 We therefore analyzed expression of these factors in vivo in hepatic tumoral tissues from FLX and LTTPKO mice. Our data indicate that the expression of these factors is not up-regulated in vivo by TTP deficiency, some of them being even down-regulated, suggesting that TTP affects cancer development by regulating other cancer-related factors (Figure 5I). Interestingly, another well-established target of TTP, ie, FGF21,15 which has anti-inflammatory and tumor suppressive activities,25,26 was on the contrary found significantly up-regulated in tumoral tissues of LTTPKO mice (Figure 5J), but not in the non-tumoral livers (Figure 5I). Analyses of HCC patient cohorts (GSE76427, Figure 1A) further indicated that 33% of patients having low ZFP36 expression in tumors vs non-tumoral livers also have high FGF21 expression (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76427).
Figure 5.
TTP loss in vivo prevents DEN-induced HCC. (A) Detectable serum alanine aminotransferase levels in 2-year-old FLX (n = 5) and LTTPKO (n = 13) mice. (B) Detectable serum aspartate aminotransferase levels in 2-year-old FLX and LTTPKO mice. (C) Tumor incidence in 2-year-old FLX and LTTPKO mice. (D) Representative liver anatomies and computed tomography scan 3-dimensional reconstructions of FLX and LTTPKO livers illustrating tumoral nodules in 12-month-old mice exposed to DEN. (E) Mean tumor number in FLX (n = 5) and LTTPKO (n = 6) mice treated exposed to DEN based on analysis of computed tomography scans at 11 months. (F) Mean tumor volume of FLX (n = 5) and LTTPKO (n = 6) mice treated with DEN based on analysis of computed tomography scans at 11 months. (G) Representative hematoxylin/eosin staining of healthy and tumoral tissues sections from 12-month-old FLX and TTPKO mice exposed to DEN. Percentages of atypical or classic HCC in each group of mice are indicated. (H) Zfp36 expression levels in tumors of wild-type 12-month-old mice exposed to DEN (n = 9) vs their corresponding non-tumoral livers. Data represented as mean ± SD. The t test for comparison of 2 groups was used. (I) mRNA expression of potential TTP targets in non-tumoral livers of 12-month-old FLX (n = 4) and LTTPKO (n = 4) mice exposed to DEN. Data represented as mean ± SD. Significance level was determined using unpaired t test. (J) Fgf21 mRNA expression in isolated tumors of 12-month-old FLX (n = 9) and LTTPKO (n = 12) mice exposed to DEN. ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
These data indicate that TTP has an oncogenic function promoting tumor initiation but likely not tumor growth in vivo and that TTP-mediated inhibition of FGF21 expression might contribute to this oncogenic activity.
TTP Restrains Migration and Invasion Capacities of Transformed Cancer Cells in Vitro
Grade 3-like HCC and hepatic metastases in HCC mouse models are very rarely observed,27,28 but migration and invasive capacities of transformed human hepatic cancer cells can be investigated in vitro. Because ZFP36 expression is strongly reduced in mouse and human hepatic cancer cell lines, as well as in undifferentiated immortalized HepaRG cells (Figure 6A–C), we therefore overexpressed TTP in Huh7 or SNU398 (Figure 6D) to investigate TTP function in already transformed cancerous hepatic cells. Our data showed that TTP overexpression tends to slightly restrain proliferation of Huh7 cells (Figure 6E), but we could not detect major significant differences in fluorescence-activated cell sorter–based analyses of the cell cycle rate (Figure 6F). On the basis of GSEA enrichment analyses, which show an up-regulation of genes associated with apoptosis in high TTP expressing human tumors (Figure 6H), we further investigated whether TTP could promote apoptosis in human hepatic cancer cells. Analyses of the nuclear morphology of Huh7 cells overexpressing TTP or not by PI/Hoechst staining revealed a weak increase of apoptosis in cells overexpressing TTP, but this effect could not be confirmed by alternative approaches such as terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay or Western blot analyses of caspase-3 cleavage (Figure 6H). A similar effect on apoptosis was observed in SNU398 TTP-overexpressing cells by assessing nuclear morphology, but neither sorafenib or doxorubicin anti-cancer drugs could further sensitize SNU398 cells for apoptosis (Figure 6I). Consistent with the observed TTP loss in high-grade human HCC and metastatic signatures in poorly differentiated tumors of HCC patients (Figure 1E–H), migratory and invasive capacities of Huh-7 cells were in contrast significantly reduced by TTP overexpression, independently of the effect on proliferation (which was not significant during the migration/invasion assays) (Figure 6J).
Figure 6.
TTP inhibits migration and invasive capacities of human transformed HCC cells. (A) Zfp36 mRNA expression in MPH, cancer (Hepa1-6), and immortalized (AML-12) mouse cells (n = 4 for MPH and Hepa1-6, n = 3 for AML12). (B) ZFP36 mRNA expression levels in HCC and immortalize (HepaRG) hepatic cell vs human primary hepatocytes (HPH) (n = 4 for HPH, n = 3 for other cell lines). (C) ZFP36 log2 mRNA expression in liver cell lines, retrieved from CellMiner. (D) Representative Western blot showing TTP protein levels in control (transfection with EV) and TTP-overexpressing (TTP) Huh7 and SNU398 cells. ERM was detected as loading control. (E) Percentage of viable TTP-overexpressing Huh7 cells compared with cells transfected with EV, 48 hours after transfection, n = 4. (C) Cell cycle analysis of Huh7 cells transfected with TTP-overexpressing plasmid vs EV. (G) GSEA analysis of human HCC transcriptome data (GSE76427) for genes associated with apoptosis. Tumor samples were segregated on the basis of TTP expression levels (low vs high ZFP36 expression, based on 20th percentile). Top 10 genes are represented for each gene set (expression normalized per row). Normalized enrichment score (NES), false discovery rate (FDR), and P value are displayed. A gene set was considered enriched at FDR <0.25. (H) Percentage of apoptotic control (EV) and TTP-overexpressing (TTP) Huh7 cells 48 hours after transfection evaluated by nucleus morphology after staining with Hoechst-33342 and propidium iodide (n = 4, left panel), by TUNEL assay (n = 5, central panels including representative dot plots of fluorescence intensity [TUNEL] and side scatter [SSC-A]), or by caspase-3 protein cleavage (representative Western of right panel, Ezrin/Radixin/Moesin (ERM) was used as loading control). (K) Percentage of apoptotic SNU398 cells evaluated by nucleus morphology after staining with Hoechst-33342 and propidium iodide, 48 hours after transfection with TTP-overexpressing plasmids followed or not by 24-hour incubation with doxorubicin (DOXO)/sorafenib (SORAF). (L) Number of Huh7 cells/field of view that migrated through the Boyden chamber membrane coated (invasion, right panel) or not coated (migration, left panel) with Matrigel 48 hours after transfection with TTP-overexpressing plasmid. All experiments were performed with n = 3 replicates unless otherwise specified. Data represent mean ± SD. Significance level was determined with one-way analysis of variance with Tukey test for multiple comparisons. ∗∗∗P < .001, ∗∗P < .01, ∗P < .05. ERM,▪▪▪ .
Together these data suggest that loss of TTP in late stages/high grades HCC significantly impacts the metastatic potential of tumors by promoting cell migration and invasion.
Cancer-Related TTP Targets Altered by TTP Loss in High-Grade HCC
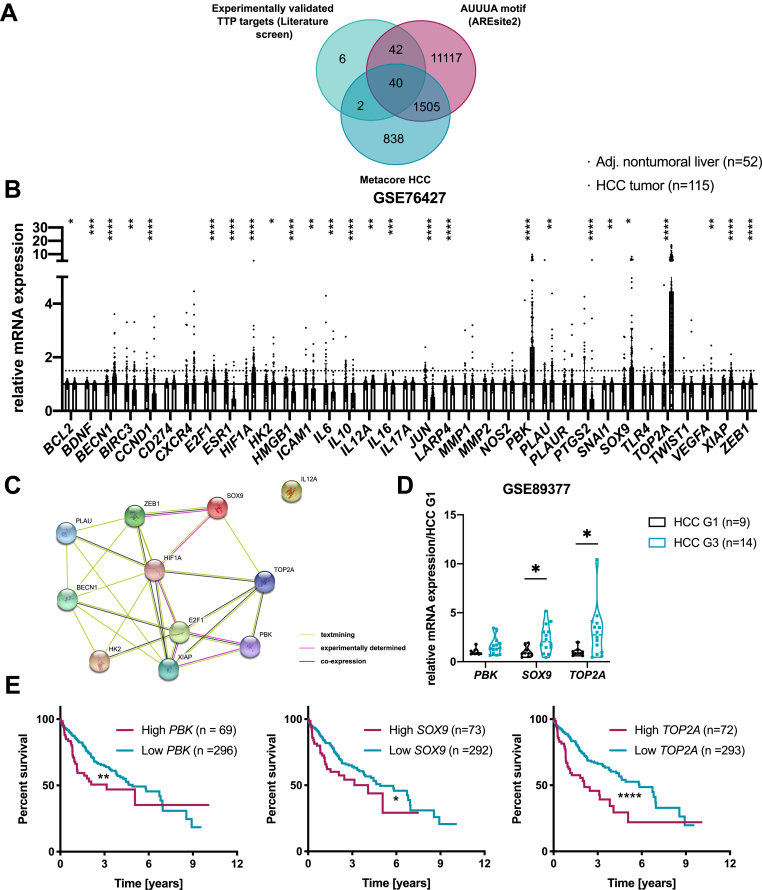
TTP promotes mRNA decay of numerous cancer-related transcripts.13 To identify potential TTP targets promoting cancer cells migration/invasion in its absence, we performed a literature-based screening of experimentally validated TTP targets involved in cancer and compared it with HCC-associated genes and proteins (MetaCore), as well as transcripts bearing the canonical TTP binding site (AUUUA pentamer, retrieved from AREsite2). As shown in Figure 7A, 40 potential cancer-related TTP targets were identified, and their expression levels were then assessed in HCC versus non-tumoral tissues of a cohort of patients (GSE76427, Figure 7B). STRING analysis of significantly induced candidates in this cohort revealed an interconnected network of interactions and/or coexpression patterns between them (Figure 7C) and tight connections with cancer-related processes and pathways (KEGG pathway analysis, Table 1). We further analyzed the expression of candidates increased by more than 1.5-fold (PBK, SOX9, and TOP2A) in a second cohort of HCC patients segregated by grades, which revealed either a trend (PBK) or a significant up-regulation (SOX9 and TOP2A) of these TTP targets in HCC grade 3 as compared with grade 1 (GSE89377, Figure 7D). Consistent again with specific up-regulations of PBK, SOX9, and TOP2A in low TTP expressing high-grade HCC, these factors were previously reported to contribute to invasion and metastasis of HCC and other cancers,29,30 and their increased expression is associated with poor clinical outcomes (Figure 7E). Together, these data indicate that in addition to FGF21, TTP loss in high-grade HCC fosters the expression of a whole set of key cancer-related factors, including in particular PBK, SOX9, and TOP2A, which likely contribute to tumor malignancy and metastasis formation.
Figure 7.
Bioinformatic predictions of cancer-promoting factors potentially targeted by TTP in HCC. (A) Venn diagram showing TTP targets identified by crossing experimentally validated TTP targets with genes containing an AUUUA motif (based on AREsite2) and HCC-related factors (MetaCore analyses). (B) mRNA fold change of 40 potential targets in HCC tumors (n = 115) versus non-tumoral liver (n = 52) (GSE76427 transcriptomic dataset). (C) String analysis of interactions between 11 potential TTP targets (STRING database). (D) mRNA fold change of candidates up-regulated >1.5-fold in GSE76427 dataset from (B) (PBK, SOX9, and TOP2A) in HCC G1 (n = 9) and HCC G3 tumors (n = 14) of second human transcriptomic dataset (GSE89377). Data represent mean ± SD. (E) Survival analysis of HCC patients, patients stratified based on PBK, SOX9, and TOP2A mRNA expression levels (80th percentile). P value was calculated using a log-rank test (data retrieved from TCGA and Human Protein Atlas). For transcriptomics analyses of GEO datasets the P value was corrected for multiple testing using the Benjamini, Krieger and Yekutieli procedure (α = 5%). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
Table 1.
KEGG Pathways Enriched in the 40 Potential Targets of TTP
| Term ID | Term description | FDR | Proteins involved |
|---|---|---|---|
| hsa04137 | Mitophagy - animal | 0.00044 | BECN1, E2F1, HIF1A |
| hsa05215 | Prostate cancer | 0.00077 | E2F1, PLAU, ZEB1 |
| hsa05206 | MicroRNAs in cancer | 0.0018 | E2F1, PLAU, ZEB1 |
| hsa04215 | Apoptosis - multiple species | 0.0024 | BECN1, XIAP |
| hsa05167 | Kaposi's sarcoma-associated herpesvirus infection | 0.0024 | BECN1, E2F1, HIF1A |
| hsa05200 | Pathways in cancer | 0.0024 | E2F1, HIF1A, IL12A, XIAP |
| hsa01524 | Platinum drug resistance | 0.0066 | TOP2A, XIAP |
| hsa05230 | Central carbon metabolism in cancer | 0.0066 | HIF1A, HK2 |
| hsa04064 | NF-kappa B signaling pathway | 0.01 | PLAU, XIAP |
| hsa04066 | HIF-1 signaling pathway | 0.01 | HIF1A, HK2 |
| hsa05222 | Small cell lung cancer | 0.01 | E2F1, XIAP |
| hsa05145 | Toxoplasmosis | 0.0105 | IL12A, XIAP |
| hsa04140 | Autophagy - animal | 0.0126 | BECN1, HIF1A |
| hsa05202 | Transcriptional misregulation in cancer | 0.0209 | PLAU, ZEB1 |
| hsa05205 | Proteoglycans in cancer | 0.0258 | HIF1A, PLAU |
| hsa05166 | HTLV-I infection | 0.0389 | E2F1, XIAP |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Conclusions
Alterations of post-transcriptional regulators of gene expression have been associated with a wide variety of inflammatory diseases and cancers.31, 32, 33 Indeed, through their ability to control mRNA stability or translation, trans-acting factors such as TTP can alter the expression of whole networks of transcripts governing liver physiology and/or contributing to the development of liver inflammation and cancer. Our study highlights a dual role for TTP as a key hepatic post-transcriptional regulator that promotes inflammation/fibrosis and tumor initiation in the liver and restrains migration and invasiveness of HCC cells. Our study further demonstrates that TTP expression is controlled by an HNF4α-EGR1 signaling axis and that down-regulation of TTP is strongly associated with hepatocyte dedifferentiation, therefore representing a reliable biomarker for high-grade HCC.
Although the genetic landscape of HCC has been well-characterized, main mutations featuring HCC are currently not therapeutically targetable (eg, TP53, AXIN1). Therefore, deepening our knowledge of non-genomic alterations strongly contributing to initiation and progression of hepatic carcinogenesis is crucial to identify new therapeutic targets and/or biomarkers. Similar to microRNAs, AUBPs represent an important class of post-transcriptional regulators that determine the fate of hundreds of mRNAs by controlling either mRNA stability or mRNA access to the translation machinery.34, 35, 36 Approximately 5%–8% of protein-coding transcripts contain an ARE within their 3'UTR,37 thus highlighting the importance of such regulation but also rendering very challenging the identification of single or multiple AUBP targets relevant for the pathophysiological process under study. Our in silico analyses of available databases, as well as our experimental data, did not allow us to firmly conclude that TTP is consistently down-regulated in all mouse and human models of nonalcoholic fatty liver disease (NAFLD)/nonalcoholic steatohepatitis (NASH) (Fig. 4A–D). These observations further support our conclusions that TTP loss is mostly a late event occurring in high-grade HCC. However, the loss of TTP in vivo strongly restrains hepatic inflammation/fibrosis development, eg, in mice fed an MCD diet, thereby pointing to TTP as a key factor in these pathologic processes. In this regard, although the MCD diet model is not perfectly recapitulating all features of the heterogenous human NAFLD/NASH disease, this diet induces similar pathologic features in mice as those observed in patients with severe NASH, including (1) perisinusoidal/perivenular fibrosis, (2) lobular/periportal inflammation, (3) the ductular response intensity, (4) decreased autophagy, (5) activation of the Hedgehog signaling, and (6) hepatic oxidative and endoplasmic reticulum stress.38, 39, 40 In addition, mice fed an MCD diet share a significantly high number of deregulated transcripts with human NASH and display down-regulation of genes for fatty acid esterification and very low density lipoprotein (VLDL) secretion.38 Therefore, mice fed an MCD diet represent a pertinent animal model (among those currently available) mimicking severe NASH usually progressing toward HCC in humans41 to investigate the role and function of TTP in this pathologic process. On the basis of our data showing a strong inhibition of steatosis, inflammation, and fibrosis in TTPKO mice fed an MCD diet, it is likely that TTP activity rather than its expression is modulated in early stages of liver diseases. Consistent with this hypothesis, AUBPs can undergo post-translational modifications, which control their subcellular localization and thus their ability to bind to specific mRNA targets.35,42 For instance, TTP phosphorylation by the p38 mitogen-activated protein kinase-activated kinase (MK2) impairs its ability to recruit deadenylases.43 Accordingly, treatment of HCC cells with an MK2 inhibitor and 5-AZA was shown to impair proliferation of HCC cells in a TTP-dependent manner.44 Because of the multiple cellular targets of TTP, which may also change depending on the pathophysiological context, assessing alterations in its activity remains currently unfeasible with available methodologies. However, it remains that our in vivo experimental data with LTTPKO mice uncover an important role of TTP expression or activity in hepatic inflammation and fibrosis, which are key drivers for the onset of HCC.45
In agreement with the concept that the role and regulation of TTP are highly dependent on the cell type and pathophysiological context, down-regulation of TTP expression was observed in liver tissues from distinct cohorts of diabetic humans and mice.15 TTP down-regulation in these pathophysiological conditions might be induced as a protective mechanism developed to fight insulin resistance and glucose intolerance. Indeed, TTP deficiency in the liver was further demonstrated to protect mice against glucose intolerance and insulin resistance associated with diet-induced obesity, and up-regulation of Fgf21, a validated target of TTP, was shown to importantly contribute to this protective effect of TTP deficiency.15 Glucose intolerance and insulin resistance associated with hepatic steatosis usually precede more severe stages of liver disorders such as inflammation and fibrosis. These data are thus in line with the outcomes of hepatic TTP deficiency regarding the development of inflammation/fibrosis that we observed in our study. However, abrogation of TTP expression in other cell types was reported to lead to totally opposite inflammatory phenotypes. Indeed, constitutive knockout of TTP in the whole mouse leads to the development of severe inflammatory syndrome,46 as opposed to the protective effect of TTP deletion in hepatocytes specifically. The most straightforward explanation for these discrepant phenotypes likely resides on different sets of mRNAs controlled by TTP in different cell types, eg, immune cells, and pathophysiological conditions, but further studies are now required to confirm this hypothesis. Finally, an additional layer of complexity in the interpretation of the hepatic pathologic outcomes associated with alterations in TTP expression/activity appears to result from gender differences. Our in silico analyses indeed revealed that a reduced overall survival was associated with TTP down-regulation in male but not in female HCC patients. We further observed that liver-specific TTP deletion prevents the expression of the proinflammatory cytokine interleukin 6 (IL6), which was previously reported to promote hepatocarcinogenesis in a gender-dependent manner.24 The tumorigenic effect of IL6 in the liver was shown to be dependent on the protective effect of ERα signaling,24 but whether TTP regulates IL6 expression by modulating ERα signaling in hepatocytes, as suggested in breast cancer cells,47 remains to be investigated.
HCC induced by DEN in mice displays histologic and genetic signatures similar to human HCC with poor prognosis,48 is furthermore dependent on inflammation,49 and presents gender disparity in terms of HCC incidence (100% in male mice versus 10%–30% in female mice50) as seen in humans.51 Consistent with the reduced inflammation/fibrosis and IL6 expression in liver-specific TTP knockout mice fed an MCD diet, induction of HCC was also strongly reduced in TTP-deficient mice treated with DEN. Our data indicate that TTP deficiency mostly restrains tumor initiation but does not significantly impact further tumoral proliferation, which was again in accordance with the reported effect of IL6.52
During the preparation of our manuscript, a study reporting a similar inhibitory effect of TTP deficiency on HCC development was published.18 However, in this study, Krohler et al18 used a different protocol of DEN induction of liver cancers and analyzed their data at a different stage of tumorigenesis. They indeed treated mice with lower doses of DEN and analyzed tumor development only after 6 months. In these conditions, tumoral foci are just starting to grow, and later stages of the disease are not present.18,53 Because we show herein that TTP is lost mostly at advanced stages of HCC, we therefore choose to investigate cancer development at later time points, ie, 11 months after DEN induction, with higher doses than in the study by Krohler et al, which allow us to examine the outcomes of TTP deficiency in advanced stages of the disease. In the same study potential targets of TTP (ie, BCL2, IGFBP1, IGFBP3, MYC, VEGFA, XIAP) were identified in in vitro cultured cells and suggested to contribute to the inhibitory effects of TTP deficiency in hepatic cancer occurrence.18 However, we could not confirm up-regulation of these candidates in vivo in hepatic tissues of TTP-deficient mice submitted to our protocol of DEN-induced HCC, suggesting that in vivo TTP targets might be different, again supporting the concept that TTP activity, as those of other AUBPs, is highly cell-specific and context-dependent. In this regard, we observed an increased expression of a previously identified TTP target, Fgf21, in tumoral tissue.15 FGF21 was previously reported to have a significant anti-inflammatory and tumor suppressive activity in the liver.25,26 FGF21 induction may therefore contribute to the reduced tumor burden observed in LTTPKO mice, thereby providing a first mechanism supporting the oncogenic activity of TTP in cancer initiation. Further studies should now assess experimentally the relevance of FGF21 in restraining cancer initiation in the absence of TTP. However, we expect that because of the pleiotropic action of TTP on numerous transcripts, the outcomes of TTP expression and activity on tumor initiation likely result from a complex interplay between multiple factors regulated by TTP.
Although TTP deficiency clearly restrains tumor initiation in vivo, our data also indicate that TTP down-regulation occurs in human high-grade HCC and is associated with poor prognosis and metastatic invasion. Consistent with human data, the histologic characterization of tumors developing in our DEN-treated mice further indicates that incidence of tumors with well-established human-like HCC features is higher in liver-specific TTP knockout mice as compared with control mice, although the number of tumoral nodules is lower. However, because grade 3-like HCC and hepatic metastasis in HCC mouse models are very rarely observed,27,28 confirming in vivo such a tumor suppressive role of TTP in late stages of hepatocarcinogenesis, as it has been suggested in other types of cancers,54 is challenging. In this study, we provided several lines of experimental evidence supporting a tumor suppressive activity of TTP in late stages of hepatic carcinogenesis. First, our analyses of migration/invasion properties of transformed human cancer cells that express low TTP levels showed that migration/invasion capacities of these cells are significantly reduced when TTP is overexpressed, as previously shown with colorectal cancer cells,13 and consistent with the reduction of TTP expression during EMT in various cancers.55 Then, we uncovered specific oncogenes, eg, PBK, SOX9, and TOP2A, potentially repressed by TTP, which are (1) overexpressed in hepatic transformed cells with low TTP expression, (2) up-regulated in high-grade HCC tumors from patients and associated with poor clinical outcomes, and (3) previously reported to contribute to invasion and metastasis of HCC and other cancers.29,30,56
One important finding in our study is that TTP (ZFP36) expression is regulated by a HNF4α/EGR1 signaling axis. The relevance of ZFP36 regulation by HNF4α in NAFLD/NASH is supported by several studies, which are consistent with our observations that TTP deficiency prevents steatosis in mice fed an MCD diet (potentially by restoring VLDL secretion). Indeed, HNF4α is known to regulate gluconeogenesis and lipid metabolism in hepatocytes, ie, VLDL, cholesterol uptake, as well as regulation enzymes responsible for fatty acid metabolism.57,58 Liver-specific HNF4AKO mice fed a high-fat diet were reported to have higher susceptibility for the development of HCC by increasing hepatic lipid accumulation and increasing IL6 expression, a major factor contributing to the development of HCC.59 HNF4α was also described as a key factor in NASH, in particular by regulating HNF1α, another transcription factor controlling lipid metabolism.60 Although no direct mechanistic and functional links have been currently experimentally demonstrated between HNF4α and TTP in NAFLD/NASH development, it is possible that the impact of HNF4α on NAFLD/NASH is in part related to TTP. Indeed, among the multiple potential targets of TTP are found genes involved in lipid and cholesterol metabolism. Along this line, bone marrow deletion of TTP was reported to lower triglyceride, total cholesterol, and VLDL/LDL serum levels, as well as to cause hepatic steatosis and to regulate many genes involved in lipid metabolism and inflammation (eg, SREBP1, SAA1, CCR2).61 Finally, because TTP and HuR share the same targets62 and HuR is involved in the development of hepatic steatosis,63 TTP down-regulation may also indirectly promote steatosis by not competing anymore with HuR for common targets. Our data indicate also that HNF4α is an indirect regulator of ZFP36 expression by promoting the expression of other transcription factors inducing ZFP36 expression, in particular EGR1. Regulation of EGR1 by HNF4α was previously reported in hepatic cancer cells,20 and EGR1 is known to target ZFP36 in other organs.21,22 Loss of HNF4α and down-regulation of EGR1 in transformed and undifferentiated hepatocytes could therefore contribute to ZFP36 down-regulation and the resulting cascade of events. However, we cannot exclude that other mechanisms contribute to the loss of TTP in high-grade HCC. Methylation of a single CpG island within ZFP36 promoter was previously reported to inhibit TTP expression.17 However, in our hands, treatment of HCC cell lines with the DNA methyltransferase inhibitor 5-AZA did not induce TTP expression (Fig. 3G). Other mechanisms, such as microRNA-dependent down-regulation of TTP expression (eg, miR-29a in melanoma)64 or constitutive degradation of TTP protein by the proteasome,65 were not investigated here but will need consideration in future studies.
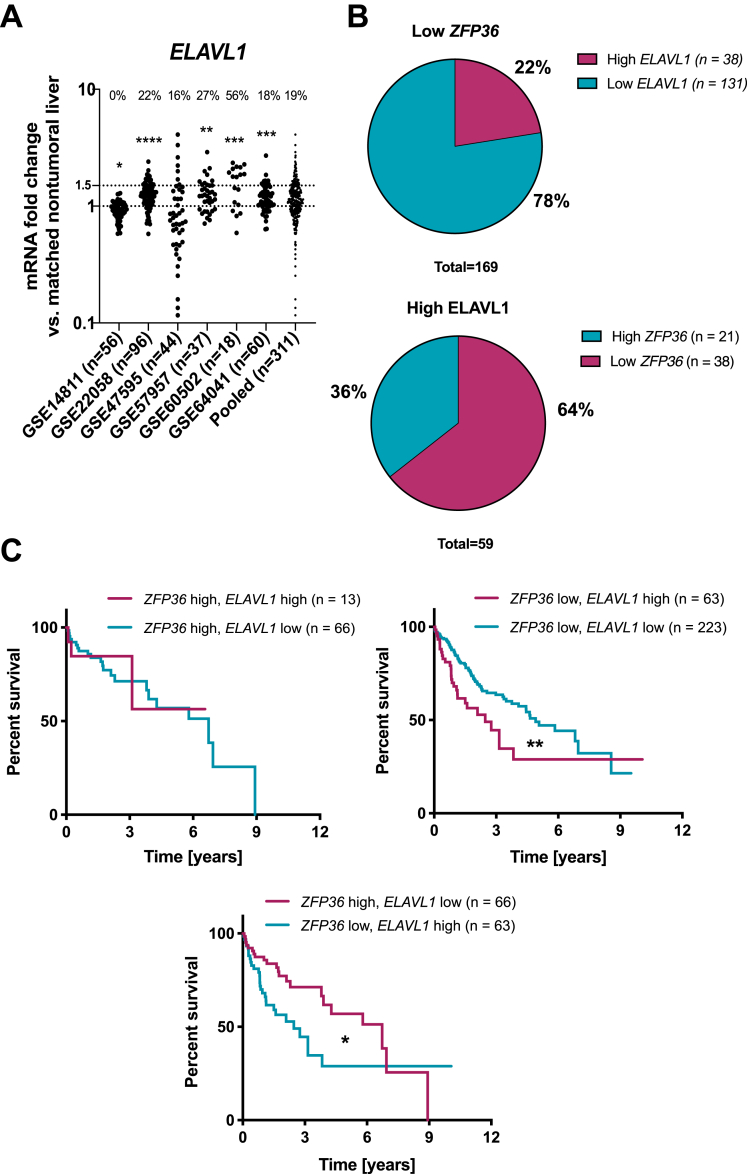
There is still a long way to precisely decipher the pleiotropic molecular mechanisms by which post-transcriptional regulators such as TTP control pathophysiological processes, eg, development and progression of fatty liver disease and hepatic cancers. TTP may indeed also affect gene expression through other non-canonical mechanisms, irrespective of its binding to AU-rich sequence in 3'UTR of mRNAs. In this regard, evidence indicates that TTP has a nuclear function still poorly characterized but likely deeply impacting inflammatory process and carcinogenesis.66,67 Non-coding RNAs may also be under the direct, or indirect, control of TTP as illustrated by the case of the TTP-dependent regulation of the RNA-binding protein Lin28, which fosters colorectal cancer development by down-regulating let-7 miRNA.68 Finally, the importance of other post-transcriptional regulators, which may compete with TTP, eg, for the same binding sites on target mRNAs, should not be underestimated. This is the case of the stabilizing AUBP HuR, which can prevent the binding of TTP to its mRNA targets as evidenced for COX-2 in colorectal cancer.32 Interestingly, HuR (ELAVL1) is frequently overexpressed in cancer cells concomitantly with TTP loss,32 which in HCC patients is associated with a worsening of the prognosis (Figure 8).
Figure 8.
Relative expressions of TTP (ZFP36) and HuR (ELAVL1) in HCC patients. (A) ELAVL1 mRNA levels in human HCC GEO datasets. Data are represented as mRNA fold change between tumoral and non-tumoral tissues. Percentages of patients having more than 1.5-fold increase in ELAVL1 expression in tumors are indicated. (B) Percentage of HCC patients having low ZFP36 and high ELAVL1 levels based on GEO datasets from (A). (C) Survival analyses of HCC patients stratified based on ZFP36 and ELAVL1 mRNA expression levels (80th percentile). P value was calculated using a log-rank test (data retrieved from TCGA and Human Protein Atlas). ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
TTP may represent a novel, relevant, and attractive therapeutic target for HCC, as previously suggested for HuR69 in other cancers. However, because of the dual role of this post-transcriptional regulator in cancer initiation and progression to malignancy, an in-depth analysis of TTP activity and networks of targets, which might be different at distinct steps of carcinogenesis, is required before considering this regulator for therapeutic purposes. However, TTP is also interesting in a clinical setting as a novel and reliable biomarker of poorly differentiated tumors (grade 2/3 HCC) with poor prognosis. Biomarkers to efficiently diagnose/characterize HCC tumor grading and prognosis from biopsies are tremendously needed, because current markers are insufficient to predict patient outcome. In this context, assessing routinely TTP expression in HCC samples might be useful to refine HCC diagnosis and gender-dependent prognosis.
Methods
Animals
Animal housing
Mice were housed at 23°C in a 12-hour day and night cycle in cages containing enrichment (disposable house and cotton cocoons) with ad libitum access to food (SAFE-150 diet; SAFE, Augy, France) and water. All experiments were performed on male mice, and mice were anesthetized with isoflurane (Rothacher-Medical, Heitenried, Switzerland; cat.#ISO250) before death by decapitation. The db/db and control mice were obtained from Charles River Laboratories (C57BLKS/J). Liver samples from ob/ob and control mice (B6. V-Lepob/JRj) were obtained from Prof Françoise Rohner-Jeanrenaud (University of Geneva, Switzerland). LTTPKO mice (C57BL/6J, AlbCre/Zfp36flox/flox) were provided by Prof Perry Blackshear. All experiments were carried out with LTTPKO mice and Zfp36flox/flox littermates (FLX mice) as a control group. Experiments were performed in accordance with standards of the Animal Research Reporting of In Vivo Experiments (ARRIVE, https://www.nc3rs.org.uk/arrive-guidelines). All experiments were ethically approved by the Geneva Health head office and were conducted in agreement with the Swiss guidelines for animal experimentation.
DEN treatment
DEN-induced HCC was performed by injecting 25 mg/kg DEN (Sigma-Aldrich, St Louis, MO; cat. #N0258-1G) intraperitoneally to 15-day-old LTTPKO and their FLX control littermates. Animals were killed 11 months after injection (at ∼12 months of age). Blood and tissue samples were collected.
MCD
Eight- to 12-week-old LTTPKO and their FLX control littermates were fed an MCD (ssniff Spezialdiäten GmbH, Soest, Germany; cat. #E15653-94) (22 kJ% fat, 14 kJ% protein, 64 kJ% carbohydrates) or a standard diet for 2 weeks. Animals were then killed, and blood and tissue samples were collected.
Computed tomography
Tumor number, size, and volume were measured by using Quantum GX microCT (PerkinElmer, Waltham, MA). Mice were imaged at 7, 9, and 11 months after DEN injection. Before the first scan, a single injection of 100 μL ExiTron nano 12000 (Viscover, Berlin, Germany; cat. #130-095-698) diluted 1:1 in 0.9% NaCl was performed retro-orbitally. Analysis of images was performed by using OsiriX MD v.10.0.1.
Cell Cultures, Transfections, and Primary Hepatocytes Isolations
Huh7 cells were provided by S. Kirkland (Imperial College, London, UK) and R. D. Beauchamp (Vanderbilt University Medical Center, Nashville, TN). HepG2 human hepatoma cell line was purchased from ATCC (Manassas, VA). SNU398 and Hep3B were obtained from the lab of Prof Caroline Gest (Inserm U1053, University of Bordeaux, France). HepaRG cells were previously generated by Christiane Guillouzo, Philippe Gripon, and Christian Trepo70,71 and provided by Biopredic International. Hepa-1-6 and AML12 cells were provided by Prof Manlio Vinciguerra (Institute for Liver and Digestive Health, University College, London, UK).
Huh7, HepG2, Hep3B, and Hepa1-6 cells were cultured in Dulbecco modified Eagle medium (DMEM) (glucose 1 g/L; Gibco, Waltham, MA; cat. #21885025) supplemented with 10% fetal calf serum (FCS) (Gibco; cat. #10270106) and 1% penicillin–streptomycin solution (Gibco; cat. #15140122). SNU398 were cultured in RPMI GlutaMAX medium (Gibco; cat. #61870010) supplemented with 10% FCS and 1% penicillin–streptomycin solution. AML12 cells were cultured in DMEM F12 medium (Gibco; cat. #31331028) supplemented with 5 μg/mL insulin (Eli Lilly, Vernier, Switzerland), 40 mg/mL dexamethasone (Sigma-Aldrich; cat. #D8893-1MG), and 10% FCS. HepaRG cells were cultured in William’s medium (Gibco; cat. #32551020) supplemented with 10% FCS, 10–9 mol/L insulin, and 10-6 mol/L hydrocortisone (Sigma-Aldrich; cat. #H2270-100MG). Differentiation of HepaRG was performed as follows: cells were maintained at full confluence for 14 days, after which their growth medium was supplemented with 2% dimethyl sulfoxide (DMSO) for additional 14 days.
MPH were isolated as previously described.72 Briefly, mice were anesthetized with ketamine/xylazine, and their liver was perfused through the portal vein with a collagenase-containing solution (Sigma-Aldrich; cat. #C5138). Cells were then purified by density gradient centrifugation using Percoll (GE Healthcare, Danderyd, Sweden; cat. #17-0891-01). MPH were counted and plated on collagen-coated dishes in William’s medium supplemented with 10% FCS, 50 μmol/L hydrocortisone, and 5 μg/mL insulin.
Plasmid transfection
Cells were seeded at a density of 40,000 cells/cm2. Transfection using Lipofectamine 3000 (Invitrogen, Carlsbad, CA; cat. # L3000-008) was performed 24 hours later according to the manufacturer’s protocol with 500 ng plasmid per well of 6-well plate. Cells were lysed and processed for RNA and protein isolation 48 hours after transfection.
The TTP-overexpressing plasmid (pcDNA3 Myc2- Hs TTP WT) was a gift from Julian Downward (Addgene plasmid # 107008; http://n2t.net/addgene:107008; RRID: Addgene_107008) and used together with a corresponding control plasmid.73 The pcDNA3.1 EGR1-overexpressing plasmid was provided by Prof Dan A. Dixon, University of Kansas Cancer Center, University of Kansas.
siRNA transfection
Cells were seeded at a density of 20,000 cells/cm2 and transfected 24 hours later with 10–20 nmol/L siRNAs for EGR1, HNFA, or AllStars negative control siRNA (Qiagen, Hombrechtikon, Switzerland) using INTERFERIN (Polyplus Transfection, Illkirch, France; cat. #409-10) according to manufacturer’s protocol. Cells were isolated for RNA and protein 48 hours after transfection. For experiments in which cells were transfected with siRNAs for HNF4α and EGR1 overexpressing constructs, cells were first treated with siRNAs against HNF4A for 48 hours and then transfected with the EGR1-overexpressing plasmids for an additional 24 hours before analysis. SiRNAs used in this study are described in Table 2.
Table 2.
SiRNA Used in Experiments Included in the Article
| siRNAs | Provider | Catalogue number |
|---|---|---|
| Human siEGR1-1 | Qiagen, Switzerland | SI00030688 |
| Human siHNF4A-6 | Qiagen, Switzerland | SI03053785 |
| Human siHNF4A-7 | Qiagen, Switzerland | SI03083773 |
| All Stars Negative Control siRNA | Qiagen, Switzerland | 1027280 |
Drug treatments
For anti-cancer drugs treatment, cells were seeded at a density of 20,000 cells/cm2 and treated with 5 μmol/L sorafenib or 0.5 μmol/L doxorubicin (Sigma-Aldrich; cat. # D1515-10mg) for 24 hours. In case of cells overexpressing TTP, cells were seeded and transfected as described in “Plasmid transfection” and treated with the compounds 24 hours later. For TSA (Sigma-Aldrich; cat. # T1952-200UL) and 5-AZA (Sigma-Aldrich; cat. #A3656-5MG) treatment, cells were seeded at a density of 20,000 cells/cm2 and treated with 400 nmol/L TSA or 5 μmol/L 5-AZA or equivalent amount of DMSO (BioChemica; cat. #A1584,0100) for the designated period.
Liver Triglycerides Measurement
Liver tissues were dissolved in 1 mL hexane/isopropanol (3:2) and lysed in a TissueLyser II (Qiagen) at 20 Hz. Lysates were then centrifugated at 13,000g at 4°C for 15 minutes, and supernatants were dried in an Eppendorf Concentrator Plus (Eppendorf, Hamburg, Germany) for 30 minutes at 60°C. The dried triglycerides were finally dissolved in 300 μL PBS with 2% Triton-X100 (AppliChem, Darmstadt, Germany; cat. #A4975.0500). Triglycerides quantification was performed using a Triglycerides GPO-PAP kit (Roche/Hitachi; cat. #11730711 216) according to the protocol provided by the manufacturer.
Cell Cycle Analysis, Proliferation, Migration/Invasion, and Apoptosis Assays
Cell proliferation assay
The 40,000 cells/cm2 were seeded and transfected on the following day using a pcDNA3 (empty vector [EV]) or TTP-overexpressing (TTP) plasmid. Twenty-four or 48 hours later the cells were detached using Accutase (Chemie Brunschwig, Basel, Switzerland; cat. # AT-104-100ml) and counted using a Neubauer counter.
Cell cycle analysis
The 40,000 cells/cm2 were seeded and transfected on the following day using a pcDNA3 (EV) or TTP-overexpressing (TTP) plasmid. Twenty-four hours later the cells were detached using Accutase, counted, and fixed with 75% ethanol. After overnight incubation at 4°C, the cells were washed and stained with propidium iodide/RNase buffer (BD Pharmingen, Franklin Lakes, NJ; cat. #550825) according to the manufacturer’s instructions. Acquisition of 10,000 cells/sample was performed on Accuri C6 (BD Biosciences, San Jose, CA) flow cytometer using FL2A and FL2H channels. Data were analyzed using FlowJo v10 software (BD Biosciences).
Analysis of cell death by PI/Hoechst staining
The 40,000 cells/cm2 were seeded and transfected on the following day using an EV or TTP plasmid. Forty-eight hours later cells were stained for 15 minutes at 37°C with Hoechst 33342 (1 μg/mL, Sigma-Aldrich; cat. #14533) and propidium iodide (1 μg/mL, Sigma-Aldrich; cat. #P4864) and analyzed using a fluorescent microscope (Evos FL Cell Imaging System; Life Technologies). At least 100 cells in 3 separate fields of view were counted for each condition. For cells treated with anti-cancer drugs, cells were transfected with plasmids, 24 hours later were treated with drugs, and additional 24 hours later were analyzed.
TUNEL analysis
TUNEL assay (TUNEL Assay Kit – FITC, Abcam, Cambridge, UK; cat. #ab66108) was performed according to the manufacturer’s protocol. In brief, 40,000 cells/cm2 were seeded, and 48 hours after transfection cells were trypsinized, washed, and fixed with 1% paraformaldehyde. Cells were then transferred to 70% ethanol, washed, and stained, as described in the manufacturer’s protocol. Cells were analyzed with BD LSRFortessa analyzer (BD Biosciences). For paraffin-embedded samples, the samples were deparaffinized, treated with 20 μg/mL proteinase K (Chemie Brunschwig; cat. #GEXPRK01-I5), and then stained, as described in the protocol. Slides were then imaged using a fluorescent microscope (Evos FL Cell Imaging System; Life Technologies).
Migration and invasion assays
The 40,000 cells/cm2 were seeded and transfected on the following day using an EV or TTP plasmid. Twenty-four hours later the cells were detached using Accutase, counted, and reseeded in 48-well Micro Chemotaxis Chambers (Neuro Probe Inc, Gaithersburg, MD). For invasion assay membranes were coated with growth factors reduced Matrigel (Corning, Corning, NY; cat. #354230). For both types of assays the lower chambers were filled with DMEM (4.5 g/L glucose) supplemented with 10 ng/mL TGFβ (Peprotech, Rocky Hill, NJ; cat. # 100-21), whereas the upper chambers were filled with serum-free DMEM (1 g/L glucose) and 50,000 cells/chamber. After 24-hour incubation at 37°C, membranes were fixed in 70% ethanol and stained with hematoxylin for 10 minutes. Cells that crossed the membrane were counted in at least 3 fields of view using ImageJ software and the Cell Counter plug in.
Real-Time Polymerase Chain Reaction
RNA was isolated using Trizol reagent (Life Technologies; cat. #15596-018). Reverse transcription was performed on 0.5–1 μg RNA using High-Capacity RNA-to-cDNA Kit (Life Technologies; cat. #4387406) following the manufacturer’s guidelines. Real-time polymerase chain reaction analysis was performed using PowerUp SYBR Green Master Mix (Life Technologies; cat. #A25778) on StepOnePlus and QuantStudio systems (Life Technologies) using the software provided by the manufacturer. Primer sequences are described in Table 3. Results were normalized with housekeeping genes and expressed as ΔΔCt compared with control condition.
Table 3.
Sequences of Primers Used for Quantitative Polymerase Chain Reaction Analysis
| Gene | Forward | Reverse |
|---|---|---|
| Mouse primers | ||
| 18s | 5′-ACATCCAAGGAAGGCAGCAG-3′ | 5′-TTTTCGTCACTACCTCCCCG-3′ |
| Acta2 | 5′-AAAAAAAACCACGAGTAACAAATCAA-3′ | 5′-TCAGCGCCTCCAGTTCCT-3′ |
| Bcl2 | 5′-TCGCAGAGATGTCCAGTCAG-3′ | 5′-ATCTCCCTGTTGACGCTCTC-3′ |
| Ccl2 | 5′-GTCCCTGTCATGCTTCTG-3′ | 5′-TTAACTGCATCTGGCTGAG-3′ |
| Cd163 | 5′-ATGGGTGGACACAGAATGGTT-3′ | 5′-CAGGAGCGTTAGTGACAGCAG-3′ |
| Cd38 | 5′-TCTCTAGGAAAGCCCAGATCG-3′ | 5′-GTCCACACCAGGAGTGAGC-3′ |
| Cd44 | 5′-ACTTTGCCTCTTGCAGTTGAG-3′ | 5′-TTTCTCCACATGGAATACACCTG-3′ |
| Col1a1 | 5′-GCT CCT CTT AGG GGC CAC T-3′ | 5′-CCA CGT CTC ACC ATT GGG G-3′ |
| Col1a2 | 5′- CACCCCAGCGAAGAACTCAT-3′ | 5′-TCTCCTCATCCAGGTACGCA-3′ |
| Col3a1 | 5′- CCTGGCTCAAATGGCTCAC-3′ | 5′-GACCTCGTGTTCCGGGTAT-3′ |
| Col4a1 | 5′-TCCGGGAGAGATTGGTTTCC-3′ | 5′-CTGGCCTATAAGCCCTGGT-3′ |
| Egr2 | 5′-GCCAAGGCCGTAGACAAAATC-3′ | 5′-CCACTCCGTTCATCTGGTCA-3′ |
| Fgf21 | 5′-CAGTCCAGAAAGTCTCCTG-3′ | 5′-GATCAAAGTGAGGCGATCC-3′ |
| Fn1 | 5′-ATCTCGGAGCCATTTGTTCCT-3′ | 5′-CCAGGTCTACGGCAGTTGTCA-3′ |
| Gak | 5′-CTGCCCACCAGGCATTTG-3′ | 5′-CCATGTCACATACATATTCAATGTACCT-3′ |
| Igfbp1 | 5′-CTGGACAGCTTCCACCTGAT-3′ | 5′-GTTGGGCTGCAGCTAATCTC-3′ |
| Igfbp3 | 5′-GACAGAATACGGTCCCTG-3′ | 5′-TTTCTGCCTTTGGAAGGG-3′ |
| Il1b | 5′-GACAACTGCACTACAGGC-3′ | 5′-CATGGAGAATATCACTTGTTGG-3′ |
| Il6 | 5′-AGTTGCCTTCTTGGGACTGAT-3′ | 5′-TCCACGATTTCCCAGAGAAC-3′ |
| Il10 | 5′-CTTTCAAACAAAGGACCAGC-3′ | 5′-CCAAGTAACCCTTAAAGTCCT-3′ |
| Itgam (CD11b) | 5′-ATGGACGCTGATGGCAATACC-3′ | 5′-TCCCCATTCACGTCTCCCA-3′ |
| Mmp2 | 5′-GACATACATCTTTGCAGGAGACAAG-3′ | 5′-TCTGCGATGAGCTTAGGGAAA-3′ |
| Mmp9 | 5′-CCTGGAACTCACACGACATCTTC-3′ | 5′-TGGAAACTCACACGCCAGAA-3′ |
| Myc | 5′-CACTCACCAGCACAACTACG-3′ | 5′-GTTCCTCCTCTGACGTTCCA-3′ |
| Opn | 5′-CTCAGAGGAGAAGCTTTACAG-3′ | 5′-GGACACAGCATTCTGTGG-3′ |
| Pdgfa | 5′-GAGGAAGCCGAGATACCC C-3′ | 5′-TGCTGTGGATCTGACTTCGAG-3′ |
| Ppia | 5′-CAAATGCTGGACCAAACACAA-3′ | 5′-GCCATCCAGCCATTCAGTCT-3′ |
| Ptgs2 | 5′-CATCCCCTTCCTGCGAAGTT-3′ | 5′-CATGGGAGTTGGGCAGTCAT-3′ |
| Rantes (Ccl5) | 5′-CTCACCATATGGCTCGGA-3′ | 5′-TTCCTTCGAGTGACAAACAC-3′ |
| Serpina1 | 5′-CCCGGATCTTCAACAATGG-3′ | 5′-TTATGCACAGCCTTGCTG-3′ |
| Tgfb | 5′- GCCTGAGTGGCTGTCTTTTGA-3′ | 5′- GCTGAATCGAAAGCCCTGTATT-3′ |
| Tnfa | 5′-AGGCTGCCCCGACTACGT-3′ | 5′-GACTTTCTCCTGGTATGAGATAGCAAA-3′ |
| Vegf | 5′-GATCATGCGGATCAAACCT-3′ | 5′-CTTTCTTTGGTCTGCATTCAC-3′ |
| Xiap | 5′-CGGCGCTTAGTTAGGACTGGA-3′ | 5′-TGCTGAAACAGGACTACTACTTGG-3′ |
| Zfp36 | 5′-TGAGCTGTCACCCTCACCTA-3′ | 5′-ACTTGTGGCAGAGTTCCGTT-3′ |
| Human primers | ||
| EGR1 | 5′-GCCTGCGACATCTGTGGAA-3′ | 5′-GCCGCAAGTGGATCTTGGTA-3′ |
| HNF4A | 5′-CTCCTGCAGATTTAGCCG-3′ | 5′-CTGTCCTCATAGCTTGACC-3′ |
| PPIA | 5′-ATGGTCAACCCCACCGTGT-3′ | 5′-TCTGCTGTCTTTGGGACCTTGTC-3′ |
| ZFP36 | 5′-TCCACAACCCTAGCGAAGAC-3′ | 5′-GAGAAGGCAGAGGGTGACAG-3′ |
Western Blot
Cells/tissues were lysed in cold RIPA buffer (50 mmol/L Tris-HCl, pH 6.8, 100 mmol/L dithiothreitol, 2% sodium dodecyl sulfate, 0.1% bromophenol blue, 10% glycerol). Protein lysates were then centrifuged at 12,000g for 10 minutes, and the supernatant was collected. Protein content was determined using a bicinchoninic acid protein assay kit (Pierce Biotechnology, Waltham, MA; cat. #23225). Ten μg of lysates per sample was separated on 5%–20% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and blotted onto nitrocellulose membranes (Amersham, Little Chalfont, UK; cat. #RPN303D). Membranes were blocked for 1 minute at room temperature in polyvinyl alcohol (Sigma-Aldrich; P8136-250G) and further incubated with primary antibodies overnight at 4°C. Membranes were washed with PBS-Tween 20 (AppliChem; cat. #A4974,0500) and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour at room temperature. Detection and quantitation of blots were performed using the ECL Advance reagent (Amersham; cat. #RPN2135) and PXI/PXI Touch from Syngene (Synoptics Group, Cambridge, UK) and quantified with ImageJ software. For detailed description of antibodies used, see Table 4.
Table 4.
Antibodies Used for Western Blot, Immunohistochemistry, and Other Cellular Assays
| Protein targeted | Host | Provider | Catalogue number |
|---|---|---|---|
| Primary antibodies | |||
| Cleaved caspase-3 | Rabbit | Cell Signaling, US | #9661S |
| COX-2 | Goat | Santa Cruz Biotechnology, US | sc-1745 |
| EGR2 | Rabbit | Abcam, UK | ab245228 |
| ERM | Rabbit | Cell Signaling, US | 3726 |
| InVivoMab F4/80 | Rat | Bio X Cell, US | BE0206 |
| IL6 | Mouse | Santa Cruz Biotechnology, US | sc-57315 |
| TTP | Rabbit | Aviva Systems Biology, US | ARP38303_P050 |
| Normal rabbit immunoglobulin G | Goat | Cell Signaling, US | 2729 |
| AffiniPure Fab fragment goat anti-mouse immunoglobulin G (H+L) | Goat | Jackson ImmunoResearch, US | 115-007-003 |
| Secondary antibodies | |||
| HRP-conjugated anti-rabbit | Goat | Biorad, Switzerland | 170-6515 |
| Mouse TrueBlot ULTRA: anti-mouse immunoglobulin HRP | Rat | Rockland, US | 18-8817-30 |
| Anti-goat | Rabbit | Sigma-Aldrich, US | A-5420 |
| ImPRESS HRP goat anti-rat | Mouse | Vector Labs, US | MP-7444 |
HRP, horseradish peroxidase.
Histology and Human Tissue Microarrays
Histologic sections preparation and staining
Liver tissues were fixed in 4% paraformaldehyde overnight and washed with phosphate-buffered saline. Then, the specimens were dehydrated and embedded in paraffin. Five-μm tissue sections were stained with hematoxylin (Merck; cat. #1.0402.0025) and eosin (Sigma-Aldrich; cat. #E4382) (for morphologic analysis) or Picro Sirius Red (Sigma-Aldrich; cat. #365548) and hematoxylin (for analysis of fibrosis) and mounted with coverslips. H&E slides were further analyzed by a pathologist (Dr Claudio De Vito) blinded to mouse genotype.
Human tissue microarray staining
Human tissue microarrays (US Biomax, Derwood, MD) were stained against TTP according to the Abcam IHC-Paraffin protocol and visualized with DAB Substrate Kit (Abcam; cat. # ab64238). In brief, slides were deparaffinized, rehydrated, and heated in citrate buffer for antigen retrieval. Then, they were incubated in 0.3% Triton X-100 in TBS for 15 minutes, blocked with 10% goat serum in bovine serum albumin (AppliChem; cat. #A1391,0100) for 2 hours at room temperature, and incubated overnight at 4°C with anti-TTP rabbit antibody diluted 1:100. On the following day, slides were incubated in 0.3% H2O2 solution (Acros Organics/ThermoFisher; cat. #202465000) for 15 minutes to block endogenous peroxidase and then incubated with anti-rabbit antibody conjugated with horseradish peroxidase (dilution 1:500) at room temperature for 1 hour. Each step was followed by rinsing in 0.025% Triton X-100 in TBS. Finally, the slides were visualized by 3-minute incubation with the DAB Substrate kit and counterstained with hematoxylin for 5 minutes. The resulting colorations were characterized by 2 researchers who were blinded to clinical data of the specimen, using a “–“ to “+++” qualitative scale, with “–“ signifying no staining and “+++” intense staining.
F4/80 staining
Paraffin-embedded sections were deparaffinized and rehydrated and treated with 20 μg/mL proteinase K solution. Sections were permeabilized with 0.3% Triton X-100 solution for 10 minutes and blocked sequentially with AffiniPure Fab fragment goat anti-mouse immunoglobulin G (H+L) (Jackson ImmunoResearch, West Grove, PA) for 30 minutes, 2.5% ImPRESS goat serum (Vector Labs, Burlingame, CA) for 30 minutes and 0.3% H2O2 for 15 minutes. Incubation with F4/80 antibody (Bio X Cell, Lebanon, NH) was performed overnight, followed by ImmPRESS HRP anti-rat immunoglobulin G antibody (Vector Labs) for 30 minutes and staining with DAB Substrate kit. Semiquantitative quantifications were performed as described in the “Human tissue microarray staining” section. Antobodies are described in Table 4.
Bioinformatics Analysis
Analysis of GEO datasets
Microarray data obtained from GEO (https://www.ncbi.nlm.nih.gov/gds) were used to compare mRNA levels in various models of liver diseases in mice and humans. The data were analyzed either by GEO2R web tool (https://www.ncbi.nlm.nih.gov/geo/geo2r) or ShinyGEO (https://gdancik.shinyapps.io/shinyGEO/). P values were calculated on the basis of log2 expression and corrected for multiple testing using the Benjamini, Krieger and Yekutieli procedure (alpha = 5%). For multiple available probes, only the one rendering most significant results was considered. All GEO datasets are represented in Tables 5 and 6.
Table 5.
Transcriptomic Datasets Used for Rodent and Human NAFLD/NASH
| GEO ID | Description | Stage | Method | PMID |
|---|---|---|---|---|
| Rodent NAFLD/NASH | ||||
| GSE53131 | 3 WT mice fed a chow or HFD (60 Kcal% fat) for 9 weeks | Steatosis | Microarray | 24618914 |
| GSE57425 | 3 WT mice fed a chow or HFD (60 Kcal% fat) for 12 weeks | Steatosis | Microarray | — |
| GSE53834 | 3 WT mice fed a chow or HFD for 12 weeks | Steatosis | Microarray | 25284781 |
| GSE53834 | 3 WT mice fed a chow or HFD for 20 weeks | Steatosis | Microarray | 25284781 |
| GSE63027 | 5 WT vs 5 MAT1A KO mice (3 months old) | Steatosis | Microarray | 25993042 |
| GSE63027 | 5 WT vs 5 GNMT KO mice (3 months old) | NASH | Microarray | 25993042 |
| GSE63027 | 5 WT vs 5 MAT1A KO mice (8 months old) | NASH | Microarray | 25993042 |
| GSE35961 | 4 WT mice fed a chow or HFD + MCD for 8 weeks | NASH | Microarray | 23028442 |
| GSE55747 | 4 healthy WT mice vs 4 CCl4-treated mice | Fibrosis | Microarray | — |
| GSE27641 | 3 healthy Wistar rats vs 6 DEN-treated rats (50 mg/kg) | Cirrhosis | Microarray | 24677197 |
| GSE27641 | 3 healthy Wistar rats vs 6 DEN-treated rats (100 mg/kg) | Cirrhosis | Microarray | 24677197 |
| Human NAFLD/NASH | ||||
| GSE33814 | 13 healthy livers vs 19 steatotic livers | Steatosis | Microarray | 23071592 |
| GSE33814 | 13 healthy livers vs 12 steatotic livers | NASH | Microarray | 23071592 |
| GSE25097 | 6 healthy livers vs 40 cirrhotic livers | Cirrhosis | Microarray | 22634754 |
| GSE36411 | 21 healthy livers vs 21 cirrhotic livers | Cirrhosis | Microarray | — |
| GSE89377 | 13 healthy livers vs 12 cirrhotic livers | Cirrhosis | Microarray | — |
HFD, high fat diet; WT, wild-type.
Table 6.
Transcriptomic Datasets Used for Rodent and Human HCC/ICC
| GEO ID | Description | Method | PMID |
|---|---|---|---|
| Rodent HCC | |||
| GSE63574 | 5 HCCs from 8-month-old DEN-treated mice vs 4 normal livers | Microarray | 26161998 |
| GSE51188 | 3 HCCs from 9-month-old DEN-treated mice vs 6 non-tumoral tissues | Microarray | 24367269 |
| GSE34760 | 7 HCCs from 1-year-old DEN-treated mice vs 4 normal livers | Microarray | 22342966 |
| GSE73498 | 3 HCCs from DEN-treated rats vs 3 normal livers from untreated rats | Microarray | 22194203 |
| GSE66717 | 4 tumors from 10-month-old LPTENKO mice vs 3 normal livers | Microarray | 26627606 |
| GSE70681 | 3 tumors from 15-month-old LPTENKO mice vs 5 normal livers | Microarray | — |
| GSE2127 | 3 hepatic tumors vs non-tumoral tissues from 22-month-old Txnipmut mice | Microarray | 16607285 |
| GSE19004 | 4 tumors from Rb, p130, and p107 triple KO mice vs 4 normal livers from wild-type mice | Microarray | — |
| GSE25457 | 5 HCCs vs nontumoral tissues from 18-month-old MAT1AKO mice | Microarray | 21562899 |
| GSE26538 | 6 spontaneous HCCs vs non-tumoral tissues | Microarray | 21571946 |
| GSE29813 | 6 spontaneous HCCs vs non-tumoral tissues | Microarray | 23262642 |
| GSE31431 | 4 HCCs from 48-week-old PDGF overexpressing mice vs 4 normal livers | Microarray | 22651928 |
| GSE54054 | 3 tumors from 15-month-old aflatoxin-treated mice vs 3 normal livers | Microarray | 26035378 |
| GSE67316 | 6 HCCs from BDCA-treated mice vs 6 normal livers | Microarray | 26289556 |
| GSE77503 | 4 tumors from Akt/βCat-overexpressing mice vs 3 non-tumoral tissues | Microarray | 26844528 |
| GSE83596 | 5 tumors vs non-tumoral tissues from 20-week-old STAMTM mice | Microarray | — |
| GSE63027 | 4 tumors from 8-month-old GNMTKO mice vs 5 normal livers | Microarray | 25993042 |
| Human HCC | |||
| GSE14811 | 56 HCCs vs matched non-tumoral tissue | Microarray | 15607117 |
| GSE22058 | 96 HCCs vs matched non-tumoral tissue | Microarray | 20739924, 22403344 |
| GSE47595 | 44 HCCs vs matched non-tumoral tissue | Microarray | 24498002 |
| GSE57957 | 37 HCCs vs matched non-tumoral tissue | Microarray | 25093504 |
| GSE60502 | 18 HCCs vs matched non-tumoral tissue | Microarray | 25376302 |
| GSE64041 | 60 HCCs vs matched non-tumoral tissue | Microarray | 27499918 |
| GSE76427 | 115 HCCs vs 52 non-tumoral tissue | Microarray | 29117471 |
| GSE89377 | 13 normal livers vs 9 HCCs grade 1, 12 HCCs grade 2, and 14 HCCs grade 3 | Microarray | — |
| Human ICC | |||
| GSE26566 | 104 ICCs vs 59 non-tumoral livers and 6 normal bile ducts | Microarray | 22178589 |
GSEA
GSEA was performed on human RNA microarray datasets obtained from GEO Database. Patients were segregated into 2 cohorts (TTPHI and TTPLO, 80th percentile) that were based on TTP expression levels. Enrichment score was calculated on the basis of 1000 permutations of type “phenotype,” and genes were ranked on the basis of signal-to-noise ratio. A P value of .05 and false discovery rate <0.25 were used as determinant of significant gene set enrichment.
Survival analyses
Survival and RNA-seq data of HCC patients were obtained from Human Protein Atlas database (https://www.proteinatlas.org/) and TCGA (https://portal.gdc.cancer.gov/) and further processed in GraphPad Prism 8 (San Diego, CA) to obtain Kaplan-Meier survival curves based on TTP expression levels. The patients were segregated into TTPHI and TTPLO groups on the basis of 80th percentile of expression values.
Targets analysis
For identification of potential tumor suppressors targeted by TTP, a literature screen of established TTP targets was performed in PubMed. The obtained list was further crossed a list of HCC-related genes and proteins downloaded from MetaCore (https://portal.genego.com/) and genes containing an ARE motif (AUUUA) retrieved from AREsite2 (http://rna.tbi.univie.ac.at/AREsite2/welcome).
Interactome analysis
STRING software (http://string-db.org) was used to determine interactions between TTP targets. Minimum required interaction score was set for low confidence interactions (0.15).
Correlation analysis
Correlation analyses between mRNA levels of TTP and other proteins in human HCC (LIHC cohort) were determined with GEPIA software (http://gepia.cancer-pku.cn/detail.php?clicktag=correlation###) using Pearson coefficient.
Transcription factor binding sites
Potential transcription factors binding TTP were predicted by TF2DNA software (http://www.fiserlab.org/tf2dna_db/).
Additional tools
Venn diagrams were constructed using an online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Heatmaps were constructed using Morpheus software (https://software.broadinstitute.org/morpheus/).
For all in silico analyses performed on human samples, no ethical approval is required as they are based on publicly available, anonymized, and previously approved studies.
Statistical Analysis
Data were reported as mean ± standard deviation. To assess the significance of the difference in means between 2 groups of unpaired data, two-tailed Student t test was used. In case of fold change comparisons, one-sample t and Wilcoxon tests were used. Analysis of variance test was used for comparison of 3 or more groups of samples. Log-rank test was used for survival analyses. The χ2 test was used to evaluate the independence of different variables. P values were represented on graphs as follows: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. All statistical analyses were performed in GraphPad Prism 8 unless otherwise specified.
Illustrations
Graphical illustrations for the figures were made using Servier Medical Art (Suresnes, France).
Acknowledgments
The authors thank Prof Caroline Guest (Inserm, Bordeaux University) for providing us with Hep3B and SNU-398 cells, Prof Francoise Jeanrenaud-Rohner (Geneva University) for providing us with liver tissue samples from control and ob/ob mice, and Prof Dan A. Dixon (Department of Molecular Biosciences, University of Kansas, University of Kansas Cancer Center) for providing the EGR1-overexpressing plasmid.
CRediT Authorship Contributions
Dobrochna Dolicka (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal; Writing – original draft: Equal)
Cyril Sobolewski, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Supervision: Equal; Visualization: Equal; Writing – original draft: Equal),
Monika Gjorgjieva, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Marta Correia de Sousa (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Flavien Berthou, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Claudio De Vito, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Didier J. Colin, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Olivia Bejuy (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Margot Fournier (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Christine Maeder (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Perry J. Blackshear, MD, DPhil (Resources: Supporting)
Laura Rubbia-Brandt, Prof (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting)
Michelangelo Foti (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was supported by the Swiss National Science Foundation (grant no. 310030-172862 and CRSII3-160717 to M. Foti) and the Geneva Cancer League (grant no. 1514 to M. Foti and 1711 to C. Sobolewski) in Switzerland. P.J. Blackshear was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, United States.
References
- 1.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez-Chantada M., Fernandez-Ramos D., Embade N., Martinez-Lopez N., Varela-Rey M., Woodhoo A., Luka Z., Wagner C., Anglim P.P., Finnell R.H., Caballeria J., Laird-Offringa I.A., Gorospe M., Lu S.C., Mato J.M., Martinez-Chantar M.L. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramaniam K., Ooi L.L., Hui K.M. Transcriptional down-regulation of IGFBP-3 in human hepatocellular carcinoma cells is mediated by the binding of TIA-1 to its AT-rich element in the 3'-untranslated region. Cancer Lett. 2010;297:259–268. doi: 10.1016/j.canlet.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Xiao Z., Ching Chow S., Han Li C., Chun Tang S., Tsui S.K., Lin Z., Chen Y. Role of microRNA-95 in the anticancer activity of Brucein D in hepatocellular carcinoma. Eur J Pharmacol. 2014;728:141–150. doi: 10.1016/j.ejphar.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Brennan S.E., Kuwano Y., Alkharouf N., Blackshear P.J., Gorospe M., Wilson G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fallahi M., Amelio A.L., Cleveland J.L., Rounbehler R.J. CREB targets define the gene expression signature of malignancies having reduced levels of the tumor suppressor tristetraprolin. PloS One. 2014;9:e115517. doi: 10.1371/journal.pone.0115517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varnum B.C., Lim R.W., Sukhatme V.P., Herschman H.R. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:119–120. [PubMed] [Google Scholar]
- 8.Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016;22:373–382. doi: 10.1261/rna.054833.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varnum B.C., Ma Q.F., Chi T.H., Fletcher B., Herschman H.R. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco F.F., Sanduja S., Deane N.G., Blackshear P.J., Dixon D.A. Transforming growth factor beta regulates P-body formation through induction of the mRNA decay factor tristetraprolin. Mol Cell Biol. 2014;34:180–195. doi: 10.1128/MCB.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson B.A., Stehn J.R., Yaffe M.B., Blackwell T.K. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J Biol Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 12.Sandler H., Kreth J., Timmers H.T., Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legrand N., Dixon D.A., Sobolewski C. AU-rich element-binding proteins in colorectal cancer. World Journal of Gastrointestinal Oncology. 2019;11:71–90. doi: 10.4251/wjgo.v11.i2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanduja S., Blanco F.F., Dixon D.A. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdisciplinary Reviews RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawicki K.T., Chang H.C., Shapiro J.S., Bayeva M., De Jesus A., Finck B.N., Wertheim J.A., Blackshear P.J., Ardehali H. Hepatic tristetraprolin promotes insulin resistance through RNA destabilization of FGF21. JCI Insight. 2018;3 doi: 10.1172/jci.insight.95948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J.C., Luo S.Z., Liu T., Lu L.G., Xu M.Y. linc-SCRG1 accelerates liver fibrosis by decreasing RNA-binding protein tristetraprolin. FASEB J. 2019;33:2105–2115. doi: 10.1096/fj.201800098RR. [DOI] [PubMed] [Google Scholar]
- 17.Sohn B.H., Park I.Y., Lee J.J., Yang S.J., Jang Y.J., Park K.C., Kim D.J., Lee D.C., Sohn H.A., Kim T.W., Yoo H.S., Choi J.Y., Bae Y.S., Yeom Y.I. Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology. 2010;138:1898–1908. doi: 10.1053/j.gastro.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 18.Krohler T., Kessler S.M., Hosseini K., List M., Barghash A., Patial S., Laggai S., Gemperlein K., Haybaeck J., Muller R., Helms V., Schulz M.H., Hoppstadter J., Blackshear P.J., Kiemer A.K. The mRNA-binding protein TTP/ZFP36 in hepatocarcinogenesis and hepatocellular carcinoma. Cancers. 2019;11 doi: 10.3390/cancers11111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Bonzo J.A., Gonzalez F.J., Wang L. Diurnal regulation of the early growth response 1 (Egr-1) protein expression by hepatocyte nuclear factor 4alpha (HNF4alpha) and small heterodimer partner (SHP) cross-talk in liver fibrosis. J Biol Chem. 2011;286:29635–29643. doi: 10.1074/jbc.M111.253039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobolewski C., Sanduja S., Blanco F.F., Hu L., Dixon D.A. Histone deacetylase inhibitors activate tristetraprolin expression through induction of early growth response protein 1 (EGR1) in colorectal cancer cells. Biomolecules. 2015;5:2035–2055. doi: 10.3390/biom5032035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florkowska M., Tymoszuk P., Balwierz A., Skucha A., Kochan J., Wawro M., Stalinska K., Kasza A. EGF activates TTP expression by activation of ELK-1 and EGR-1 transcription factors. BMC Mol Biol. 2012;13:8. doi: 10.1186/1471-2199-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starley B.Q., Calcagno C.J., Harrison S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 24.Naugler W.E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A.M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 25.Bao L., Yin J., Gao W., Wang Q., Yao W., Gao X. A long-acting FGF21 alleviates hepatic steatosis and inflammation in a mouse model of non-alcoholic steatohepatitis partly through an FGF21-adiponectin-IL17A pathway. Br J Pharmacol. 2018;175:3379–3393. doi: 10.1111/bph.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singhal G., Kumar G., Chan S., Fisher F.M., Ma Y., Vardeh H.G., Nasser I.A., Flier J.S., Maratos-Flier E. Deficiency of fibroblast growth factor 21 (FGF21) promotes hepatocellular carcinoma (HCC) in mice on a long term obesogenic diet. Mol Metab. 2018;13:56–66. doi: 10.1016/j.molmet.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newell P., Villanueva A., Friedman S.L., Koike K., Llovet J.M. Experimental models of hepatocellular carcinoma. J Hepatol. 2008;48:858–879. doi: 10.1016/j.jhep.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heindryckx F., Colle I., Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J Exp Pathol. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Q.X., Zhong S., He L., Jia X.J., Tang H., Cheng S.T., Ren J.H., Yu H.B., Zhou L., Zhou H.Z., Ren F., Hu Z.W., Gong R., Huang A.L., Chen J. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90–102. doi: 10.1016/j.canlet.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C., Qu Y., Xiao H., Xiao W., Liu J., Gao Y., Li M., Liu J. LncRNA SNHG3 promotes clear cell renal cell carcinoma proliferation and migration by upregulating TOP2A. Exp Cell Res. 2019;384:111595. doi: 10.1016/j.yexcr.2019.111595. [DOI] [PubMed] [Google Scholar]
- 31.Guo J., Qu H., Chen Y., Xia J. The role of RNA-binding protein tristetraprolin in cancer and immunity. Med Oncol. 2017;34:196. doi: 10.1007/s12032-017-1055-6. [DOI] [PubMed] [Google Scholar]
- 32.Young L.E., Sanduja S., Bemis-Standoli K., Pena E.A., Price R.L., Dixon D.A. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trojanowicz B., Brodauf L., Sekulla C., Lorenz K., Finke R., Dralle H., Hoang-Vu C. The role of AUF1 in thyroid carcinoma progression. Endocr Relat Cancer. 2009;16:857–871. doi: 10.1677/ERC-08-0234. [DOI] [PubMed] [Google Scholar]
- 34.Lykke-Andersen J., Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doller A., Schlepckow K., Schwalbe H., Pfeilschifter J., Eberhardt W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 2010;30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J.Y., Bergman N., Sadri N., Schneider R.J. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakheet T., Williams B.R., Khabar K.S. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34(Database issue):D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machado M.V., Michelotti G.A., Xie G., Almeida Pereira T., Boursier J., Bohnic B., Guy C.D., Diehl A.M. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PloS One. 2015;10 doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.Y., Garcia-Carbonell R., Yamachika S., Zhao P., Dhar D., Loomba R., Kaufman R.J., Saltiel A.R., Karin M. ER stress drives lipogenesis and steatohepatitis via caspase-2 activation of S1P. Cell. 2018;175:133–145 e15. doi: 10.1016/j.cell.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiers J.L., Malhi H. Endoplasmic reticulum stress in metabolic liver diseases and hepatic fibrosis. Semin Liver Dis. 2019;39:235–248. doi: 10.1055/s-0039-1681032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendez-Sanchez N., Cruz-Ramon V.C., Ramirez-Perez O.L., Hwang J.P., Barranco-Fragoso B., Cordova-Gallardo J. New aspects of lipotoxicity in nonalcoholic steatohepatitis. Int J Mol Sci. 2018;19:2034. doi: 10.3390/ijms19072034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark A.R., Dean J.L. The control of inflammation via the phosphorylation and dephosphorylation of tristetraprolin: a tale of two phosphatases. Biochemical Society Transactions. 2016;44:1321–1337. doi: 10.1042/BST20160166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clement S.L., Scheckel C., Stoecklin G., Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran D.D.H., Koch A., Allister A., Saran S., Ewald F., Koch M., Nashan B., Tamura T. Treatment with MAPKAP2 (MK2) inhibitor and DNA methylation inhibitor, 5-aza dC, synergistically triggers apoptosis in hepatocellular carcinoma (HCC) via tristetraprolin (TTP) Cellular Signalling. 2016;28:1872–1880. doi: 10.1016/j.cellsig.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Alison M.R., Nicholson L.J., Lin W.R. Chronic inflammation and hepatocellular carcinoma: recent results in cancer research. Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2011;185:135–148. doi: 10.1007/978-3-642-03503-6_8. [DOI] [PubMed] [Google Scholar]
- 46.Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., Schenkman D.I., Gilkeson G.S., Broxmeyer H.E., Haynes B.F., Blackshear P.J. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 47.Barrios-Garcia T., Gomez-Romero V., Tecalco-Cruz A., Valadez-Graham V., Leon-Del-Rio A. Nuclear tristetraprolin acts as a corepressor of multiple steroid nuclear receptors in breast cancer cells. Mol Genet Metab Rep. 2016;7:20–26. doi: 10.1016/j.ymgmr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J.S., Chu I.S., Mikaelyan A., Calvisi D.F., Heo J., Reddy J.K., Thorgeirsson S.S. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 49.Maeda S., Kamata H., Luo J.L., Leffert H., Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Nakatani T., Roy G., Fujimoto N., Asahara T., Ito A. Sex hormone dependency of diethylnitrosamine-induced liver tumors in mice and chemoprevention by leuprorelin. Jpn J Cancer Res. 2001;92:249–256. doi: 10.1111/j.1349-7006.2001.tb01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bosch F.X., Ribes J., Diaz M., Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 52.Park E.J., Lee J.H., Yu G.Y., He G., Ali S.R., Holzer R.G., Osterreicher C.H., Takahashi H., Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolba R., Kraus T., Liedtke C., Schwarz M., Weiskirchen R. Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab Anim. 2015;49(Suppl):59–69. doi: 10.1177/0023677215570086. [DOI] [PubMed] [Google Scholar]
- 54.Sanduja S., Blanco F.F., Young L.E., Kaza V., Dixon D.A. The role of tristetraprolin in cancer and inflammation. Front Biosci. 2012;17:174–188. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon N.A., Jo H.G., Lee U.H., Park J.H., Yoon J.E., Ryu J., Kang S.S., Min Y.J., Ju S.A., Seo E.H., Huh I.Y., Lee B.J., Park J.W., Cho W.J. Tristetraprolin suppresses the EMT through the down-regulation of Twist1 and Snail1 in cancer cells. Oncotarget. 2016;7:8931–8943. doi: 10.18632/oncotarget.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W., Wu Y., Hou B., Wang Y., Deng D., Fu Z., Xu Z. A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives tumor growth and metastasis in hepatocellular carcinoma through the Wnt/beta-catenin pathway. Molecular Oncology. 2019;13:2194–2210. doi: 10.1002/1878-0261.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayhurst G.P., Lee Y.H., Lambert G., Ward J.M., Gonzalez F.J. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinez-Jimenez C.P., Kyrmizi I., Cardot P., Gonzalez F.J., Talianidis I. Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol. 2010;30:565–577. doi: 10.1128/MCB.00927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fekry B., Ribas-Latre A., Baumgartner C., Mohamed A.M.T., Kolonin M.G., Sladek F.M., Younes M., Eckel-Mahan K.L. HNF4alpha-deficient fatty liver provides a permissive environment for sex-independent hepatocellular carcinoma. Cancer Res. 2019;79:5860–5873. doi: 10.1158/0008-5472.CAN-19-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baciu C., Pasini E., Angeli M., Schwenger K., Afrin J., Humar A., Fischer S., Patel K., Allard J., Bhat M. Systematic integrative analysis of gene expression identifies HNF4A as the central gene in pathogenesis of non-alcoholic steatohepatitis. PloS One. 2017;12 doi: 10.1371/journal.pone.0189223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saaoud F., Wang J., Iwanowycz S., Wang Y., Altomare D., Shao Y., Liu J., Blackshear P.J., Lessner S.M., Murphy E.A., Wang H., Yang X., Fan D. Bone marrow deficiency of mRNA decaying protein tristetraprolin increases inflammation and mitochondrial ROS but reduces hepatic lipoprotein production in LDLR knockout mice. Redox Biol. 2020:101609. doi: 10.1016/j.redox.2020.101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherjee N., Jacobs N.C., Hafner M., Kennington E.A., Nusbaum J.D., Tuschl T., Blackshear P.J., Ohler U. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014;15:R12. doi: 10.1186/gb-2014-15-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z., Zong C., Jiang M., Hu H., Cheng X., Ni J., Yi X., Jiang B., Tian F., Chang M.W., Su W., Zhu L., Li J., Xiang X., Miao C., Gorospe M., de Cabo R., Dou Y., Ju Z., Yang J., Jiang C., Yang Z., Wang W. Hepatic HuR modulates lipid homeostasis in response to high-fat diet. Nat Commun. 2020;11:3067. doi: 10.1038/s41467-020-16918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gebeshuber C.A., Zatloukal K., Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Reports. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang L., Yu Z., Zhang Z., Ma W., Song S., Huang G. Interaction with pyruvate kinase M2 destabilizes tristetraprolin by proteasome degradation and regulates cell proliferation in breast cancer. Scientific Reports. 2016;6:22449. doi: 10.1038/srep22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schichl Y.M., Resch U., Hofer-Warbinek R., de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor G.A., Thompson M.J., Lai W.S., Blackshear P.J. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol Endocrinol. 1996;10:140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- 68.Kim C.W., Vo M.T., Kim H.K., Lee H.H., Yoon N.A., Lee B.J., Min Y.J., Joo W.D., Cha H.J., Park J.W., Cho W.J. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012;40:3856–3869. doi: 10.1093/nar/gkr1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Hjelmeland A.B., Nabors L.B., King P.H. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther. 2019;20:979–988. doi: 10.1080/15384047.2019.1591673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parent R., Marion M.J., Furio L., Trepo C., Petit M.A. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology. 2004;126:1147–1156. doi: 10.1053/j.gastro.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 71.Gripon P., Rumin S., Urban S., Le Seyec J., Glaise D., Cannie I., Guyomard C., Lucas J., Trepo C., Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peyrou M., Bourgoin L., Poher A.L., Altirriba J., Maeder C., Caillon A., Fournier M., Montet X., Rohner-Jeanrenaud F., Foti M. Hepatic PTEN deficiency improves muscle insulin sensitivity and decreases adiposity in mice. J Hepatol. 2015;62:421–429. doi: 10.1016/j.jhep.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 73.Coelho M.A., de Carne Trecesson S., Rana S., Zecchin D., Moore C., Molina-Arcas M., East P., Spencer-Dene B., Nye E., Barnouin K., Snijders A.P., Lai W.S., Blackshear P.J., Downward J. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity. 2017;47:1083–1099 e6. doi: 10.1016/j.immuni.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]