Abstract
The alga Euglena gracilis (E. gracilis) has recently gained attention as a health food, but its effects on human gut microbiota remain unknown. This study aimed to determine the effect of E. gracilis on gut microbiota and defecation due to modulation of microbiota composition in vitro and in vivo. The in vitro model simulating human colonic microbiota revealed that E. gracilis addition stimulated the growth of commensal Faecalibacterium. Further, E. gracilis addition enhanced butyrate production by Faecalibacterium prausnitzii. Paramylon, an insoluble dietary fibre that accumulates in E. gracilis and is the main component of E. gracilis, did not stimulate Faecalibacterium growth in vitro. Daily ingestion of 2 g of E. gracilis for 30 days increased bowel movement frequency as well as stool volume in 28 human participants. Collectively, these findings indicate that E. gracilis components other than paramylon, stimulate the growth of Faecalibacterium to improve digestive health as well as promote defecation by increasing butyrate production.
Subject terms: Biomarkers, Health care
Introduction
Photosynthetic microalgae are present in both marine and freshwater environments and have attracted attention owing to their potential applications in nutraceuticals1. Microalgae produce various compounds that can be consumed as food products. These compounds are not only used to supplement the diet but are also used in the prevention or treatment of diseases and/or disorders1,2. Euglena gracilis (E. gracilis) is a candidate fast-proliferating microalga, besides Chlorella and Spirulina, and research into its commercial cultivation is ongoing3. E. gracilis is a rich source of vitamins, minerals, unsaturated fatty acids, and a crystalline β-1,3-glucan polysaccharide called paramylon, which is considered a functional dietary fibre4,5. E. gracilis is often used as a dietary supplement. In recent years, the immunomodulatory effects of E. gracilis have been reported in both humans and animal models6. E. gracilis supplementation improved hyperglycemia in a type 2 diabetes mellitus rat model7 and provided protection against influenza virus infection in mice8. Furthermore, oral administration of paramylon relieved the symptoms of atopic dermatitis in mice9, exerted hepatoprotective effects via antioxidative action in rats10, and exerted preventive effects against colon cancer in mice11.
The gastrointestinal tract is colonised by a complex bacterial community, the intestinal microbiota, which interacts with the host and exerts a strong impact on host homeostasis and immunostasis. Therefore, intestinal microbiota is essential for maintaining host health12–14. Despite an explosion of interest in optimising the composition of intestinal microbiota by dietary means using probiotics15 or functional food products16, the effect of E. gracilis ingestion on human gut microbiota remains unknown.
The gut microbiota-modulating effects of diet are often investigated via in vivo studies in humans17. Information on the composition of colonic microbiota originates mainly from the analysis of faecal samples in human dietary intervention studies. However, this method is limited by the fact that the production of metabolites, such as short-chain fatty acids (SCFAs), cannot be measured at the actual site (the intestinal tract). An in vitro study method using a model culture system was previously developed in our laboratory to closely reproduce the microbial components in human faecal inoculum18. This in vitro human colonic microbiota model detected the decreased production of butyrate in ulcerative colitis patients18. Thus, combining in vitro and in vivo studies can help interpret the changes in human gut microbiota.
This study aimed to evaluate the effect of E. gracilis consumption on human colonic microbiota in healthy human subjects and validate the findings by analysing effects of adding E. gracilis or paramylon to an in vitro human colonic microbiota model.
Results
Euglena gracilis addition changed microbiota composition and enhanced butyrate production by Faecalibacterium in in vitro model culture system
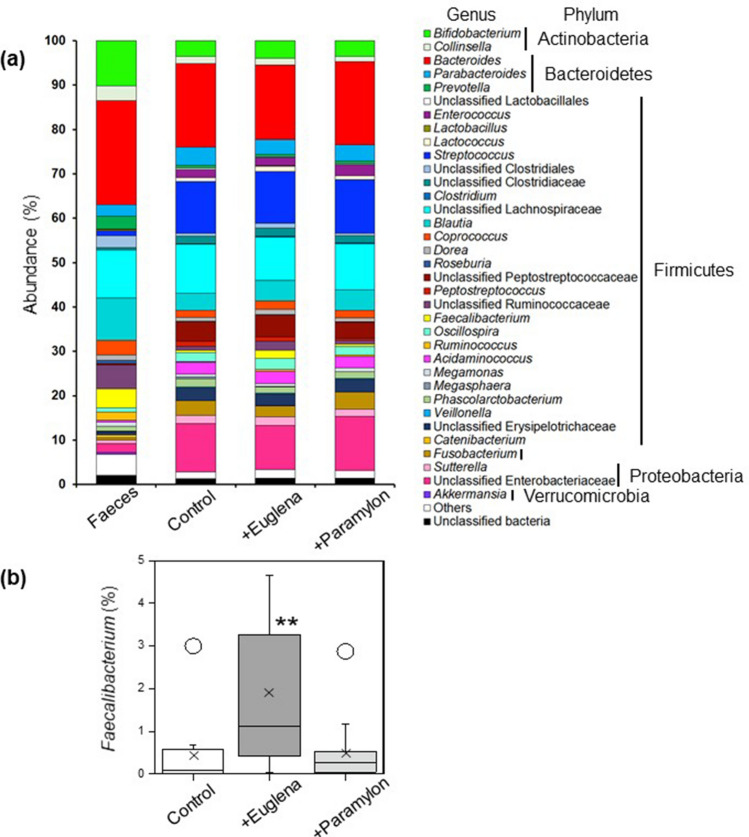
Faecal samples were obtained from 11 healthy subjects. Each sample was used as an inoculum and cultivated for 48 h to construct an in vitro human colonic microbiota model. Three microbiota models were constructed for each subject (without addition [Control], with 9 g/L E. gracilis [+Euglena], and with 3 g/L paramylon [+Paramylon]). Total bacterial DNA was extracted from the original faecal samples and the three corresponding groups (Control, +Euglena, and +Paramylon). Sequencing of bacterial 16S rRNA genes provided 8,699,439 reads (Supplementary Table S1). The corresponding in vitro models maintained the bacterial species richness of the original faeces; the Chao1 index showed no difference between faecal inoculums and corresponding in vitro models (P = 0.69, Mann–Whitney U test). Based on Shannon index data, bacterial diversity was slightly lower in the corresponding in vitro models than in the original faecal samples (P = 0.022, Mann–Whitney U test). Inverse Simpson scores showed a similar trend between faecal inoculums and corresponding in vitro models (P = 0.076, Mann–Whitney U test). Genus level bacterial composition was compared between corresponding in vitro models (Fig. 1a). The microbial composition of in vitro models with added E. gracilis or paramylon (+Euglena, or +Paramylon) was similar to that of the control model. However, in the in vitro model, E. gracilis addition (+Euglena) resulted in a significant increase (average 1.92%) in the relative abundance of Faecalibacterium bacteria than in the control conditions (average 0.47%) (P = 0.0038, paired Wilcoxon signed rank test; Fig. 1b). In contrast, compared with the control, paramylon addition (+Paramylon) did not change the relative abundance (average 0.54%) of Faecalibacterium in the in vitro model (P = 0.54, paired Wilcoxon signed rank test; Fig. 1b).
Figure 1.
Euglena gracilis addition changes microbiota composition. (a) Genus-level compositional view of bacteria in the original faecal samples (Faeces), in vitro human colonic microbiota models (Control), models with E. gracilis (+ Euglena), and models with paramylon (+ Paramylon) after 48 h of fermentation. The means of 11 samples from healthy human subjects are shown. Genera of lower abundance (< 1.0%) and lower similarity (< 97%) were included in Others and Unclassified Bacteria, respectively. (b) Box-and-whisker plots representing relative abundances of bacteria related to Faecalibacterium. **P < 0.01, n = 11, Wilcoxon signed rank test. Circles outside the whiskers represent outlier samples.
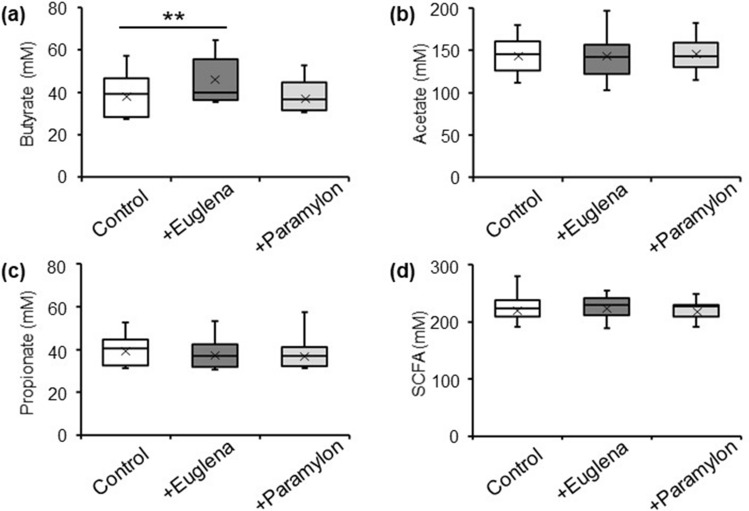
The SCFA production in the in vitro microbiota models (Control, +Euglena, and +Paramylon) was measured after 48 h of fermentation. The average butyrate production in E. gracilis addition (+Euglena) was 45.13 mM compared with 38.65 mM in the control. E. gracilis addition (9 g/L) significantly increased the butyrate production (P = 0.0098, paired Wilcoxon signed rank test), although no significant changes in acetate or propionate production were detected between in vitro models without or with E. gracilis (Control or +Euglena; P = 0.97 and 0.10, respectively, paired Wilcoxon signed rank test; Fig. 2). Paramylon addition (3 g/L) did not change the production of acetate, propionate, or butyrate (P = 0.52, 0.17, and 0.64, respectively, paired Wilcoxon signed rank test).
Figure 2.
Euglena gracilis addition enhanced butyrate concentrations. Production of (a) butyrate, (b) acetate, (c) propionate, and (d) short-chain fatty acids (SCFAs) (sum of lactate, succinate, acetate, propionate, and butyrate) in in vitro human colonic microbiota models (Control), models with E. gracilis (+ Euglena), and models with paramylon (+ Paramylon) after 48 h fermentation. **P < 0.01, n = 11, Wilcoxon signed rank test.
Euglena gracilis addition increased the growth of Faecalibacterium prausnitzii and enhanced butyrate production by Faecalibacterium prausnitzii in vitro
We investigated the effect of E. gracilis addition (9 g/L) on a single species, Faecalibacterium prausnitzii (F. prausnitzii). Real time PCR assay showed that E. gracilis addition increased the growth of F. prausnitzii (average 11.61 ± 10.34 copies/mL) at 72 h of culture compared with an average of 10.61 ± 10.22 copies/mL in samples without E. gracilis (P < 0.0001, two-tailed Student’s t-test; Fig. 3a). At the same time, E. gracilis addition increased the butyrate production by F. prausnitzii to an average of 5.67 ± 1.30 mM compared with 2.55 ± 0.23 mM in samples without E. gracilis (Control) (P = 0.015, two-tailed Student’s t-test; Fig. 3b).
Figure 3.
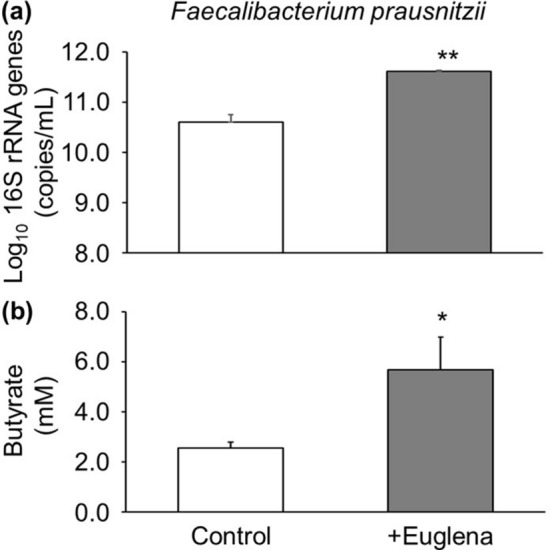
(a) Cells detected by real-time PCR and (b) concentration of butyrate produced by Faecalibacterium prausnitzii without Euglena gracilis (Control) and with E. gracilis addition (+ Euglena) after 72 h of cultivation. *P < 0.05, **P < 0.01, n = 3, two-tailed Student’s t-test.
Euglena gracilis ingestion increased the relative abundance of Faecalibacterium in human gut flora and promoted defecation
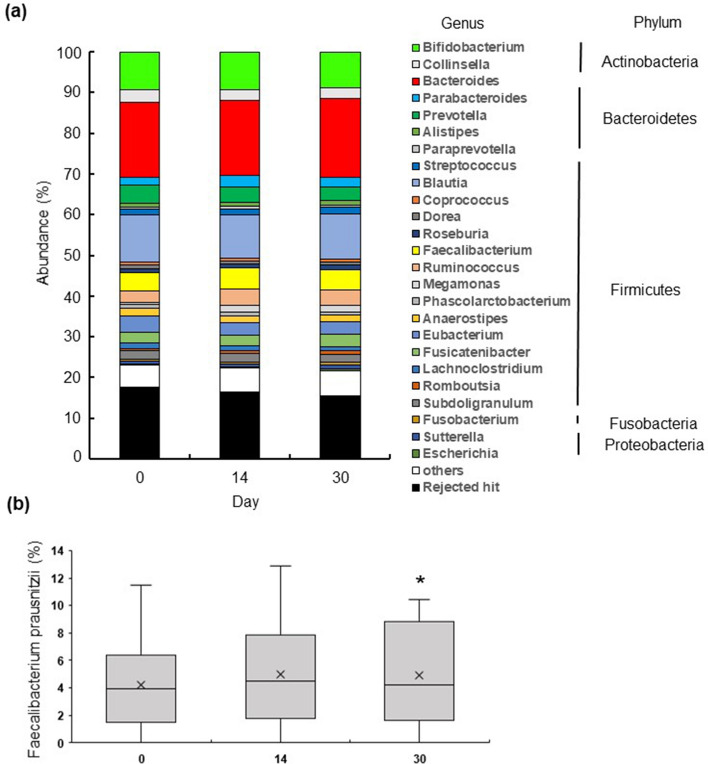
We further examined the changes in the intestinal microflora before and after continuous E. gracilis ingestion in 28 human participants (Fig. 4a). Sequencing of bacterial 16S rRNA provided 3,047,422 reads (Supplementary Table S2). Importantly, 30 days after the beginning of the ingestion of E. gracilis (2 g/day) the bacterial species richness was maintained (vs. the baseline); the Chao1 and Shannon indexes and the Inverse Simpson score were not significantly different between days 0 and 30 (P = 0.76, 0.84 and 0.45, respectively; Kruskal–Wallis test). Comparison of faecal samples collected before (0 days) and at 14 and 30 days after ingesting E. gracilis (2 g/day) showed that the relative abundance of bacteria related to the Faecalibacterium genus was unchanged after 14 days (P = 0.18, paired Wilcoxon signed rank test). However, the relative abundance of Faecalibacterium upon E. gracilis ingestion (average 5.10%) significantly increased after 30 days compared with that before E. gracilis ingestion (0 days; average 4.35%) (P = 0.04, paired Wilcoxon signed rank test; Fig. 4b).
Figure 4.
Euglena gracilis ingestion increases the proportion of the Faecalibacterium genus in the gut flora. (a) Genus-level compositional view of bacteria in human faecal samples after 0, 14, and 30 days of E. gracilis ingestion. (b) The relative abundances of the Faecalibacterium genus. *P < 0.05, n = 28, Wilcoxon signed rank test.
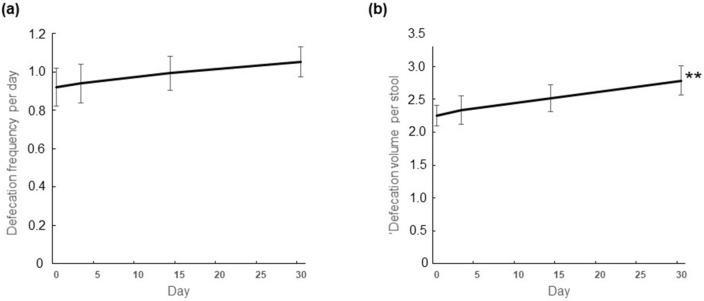
We simultaneously examined the changes in daily defecation quantity before and after daily ingestion of E. gracilis (2 g/day) in 28 human participants using the Bristol Stool Form Scale19. All outcomes were compared before and after E. gracilis ingestion. The number of bowel movements per day increased from an average of 0.92 ± 0.10 to an average of 1.05 ± 0.08 (P = 0.069, paired Wilcoxon signed rank test, Fig. 5a) and the amount of stool per bowel movement increased significantly from an average of 2.25 ± 0.16 to an average of 2.79 ± 0.23 (P = 0.009, paired Wilcoxon signed rank test, Fig. 5b), respectively.
Figure 5.
Euglena gracilis ingestion promotes defecation. (a) The frequency of defecation per day and (b) the volume of defecation per stool. One defecation volume is equivalent to one medium-sized chicken egg (about 60 g). Mean ± SEM, **P < 0.01, n = 28, Wilcoxon signed rank test.
Discussion
This study investigated the effect of E. gracilis consumption on human gut microbiota and defecation characteristics in human subjects. Both in vitro and in vivo experiments revealed that E. gracilis consumption significantly increased the relative abundance of bacteria belonging to the Faecalibacterium genus. However, this effect did not appear to be due to paramylon consumption. F. prausnitzii, the only known species in the genus Faecalibacterium, is one of the most abundant butyrate-producing bacteria in the gastrointestinal tract20. Compared with fermentation without E. gracilis, butyrate production increased after fermenting with E. gracilis due to the increase in Faecalibacterium. At this time, there was no difference in other butyric acid producing bacteria, such as Roseburia, both in vitro and in vivo. Thus, the changes in the flora caused by E. gracilis may be specific to Faecalibacterium. Previous studies reported that butyrate improves insulin resistance21 and that increasing the abundance of butyrate-producing bacteria may be a feasible strategy to counteract type 2 diabetes22. Our results corroborate with those of a study, which showed that E. gracilis supplementation decreased blood glucose concentration in a type 2 diabetes mellitus rat model7. It was shown that components in E. gracilis other than paramylon enhanced Faecalibacterium. E. gracilis is also a source of vitamins, minerals, and unsaturated fatty acids4,5; these components would promote the growth of Faecalibacterium. Another possibility is that E. gracilis indirectly stimulates Faecalibacterium. Our previous study showed that E. gracilis consumption increases Bifidobacterium in the gut microbiota of mice23. Here, the relative abundance of Bifidobacterium did not significantly increase in vitro or in vivo by E. gracilis consumption (P = 0.46 and 0.65, respectively, paired Wilcoxon signed rank test; Figs. 1 and 4). Thus, E. gracilis consumption may stimulate acetate production by Bifidobacterium, which in turn, could be consumed by Faecalibacterium to enhance its growth and/or butyrate formation. A cross-feeding process between Bifidobacterium adolescentis and F. prausnitzii was previously reported using fructooligosaccharide as a carbon source24.
Further, stool volume increased in participants who ingested E. gracilis compared with those who consumed non-supplemented food. Our previous results have also shown increased stool volumes in individuals who consumed E. gracilis25. These findings were also consistent with those of Kawano et al., who found that cholesterol residence time in the gut was shorter in rats fed a diet containing cholesterol and E. gracilis compared with that in rats fed a diet containing cholesterol alone26.
Dietary fibre intake can increase stool frequency in patients with constipation27. Paramylon in E. gracilis is insoluble and not digested or absorbed by the body; thus, it is believed to exert effects similar to those of dietary fibre. In addition, increased butyrogenesis via F. prausnitzii enhancement caused by E. gracilis consumption corresponded with that in a previous study, which showed butyrate supplementation potentially reduces difficulties in bowel movement28. Thus, E. gracilis consumption could have a beneficial effect on constipation, such as in reducing pain during defecation. Further, butyrate produced in the gut increases the number of specific CD8+ cells that eliminate the influenza virus and control infection29. These findings are consistent with studies, which have shown that E. gracilis supplementation regulates immunity6,8, and that the contribution of E. gracilis to immunity may not be because of paramylon alone.
In summary, our work shows that consuming E. gracilis stimulates Faecalibacterium growth and butyrate production. This could be a mechanism to improve stool volume. One of the limitations of this study is the small sample size in the in vivo analysis. Therefore, it is necessary to increase the sample size in future studies to validate the findings of the present study. Further experiments are also needed to identify components other than paramylon in E. gracilis. Nevertheless, this study demonstrates the potential use of E. gracilis as a novel prebiotic.
Methods
Euglena gracilis preparation
E. gracilis was prepared at euglena Co., Ltd. (Tokyo, Japan). Components of E. gracilis were measured as described previously7. E. gracilis is composed of 52.2% carbohydrates, 28.6% protein, and 13.0% fat. Approximately 70–80% of the carbohydrates in E. gracilis was paramylon. Paramylon was isolated as described previously30. E. gracilis and paramylon used in this study were dry powders.
Faecal collection and model culture system operation
Fresh faecal samples were obtained from 11 healthy human volunteers who had no history of antibiotic treatment for more than six months and provided written informed consent prior to sample collection. Immediately following collection, each faecal sample was stored using an anaerobic culture swab system (212550 BD BBL Culture Swab, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and used within 24 h. All experimental protocols were approved by the Institutional Ethics Review Board of Kobe University and in accordance with the guidelines approved by the Medical Ethics Committee at Kobe University.
The model culture system used a multichannel fermenter (Bio Jr. 8; ABLE, Tokyo, Japan) as previously described18. Inoculum was prepared by suspending each faecal sample in phosphate buffer (2 mL, 0.1 M, pH 6.5, consisting of a 68.5:31.5 mixture of 0.1 M NaH2PO4 and 0.1 M Na2HPO4) supplemented with 1% l-ascorbic acid (Wako Pure Chemical Industries, Osaka, Japan). Fermentation was initiated by the inoculation of autoclaved Gifu anaerobic medium (Nissui Pharmaceutical Co., Tokyo, Japan) with the faecal suspension (100 μL). We prepared three types of media for each human faecal inoculum, medium without addition (Control), medium containing E. gracilis (9 g/L) (+Euglena), and medium containing paramylon (3 g/L) (+Paramylon). Aliquots of fermentation cultures were sampled through the side projection of the vessel. Faecal samples and fermentation cultures were stored at − 20 °C until further use. The concentrations of SCFAs (acetate, propionate, and butyrate) were measured using a high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan) equipped with an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA, USA) and a RID-10A refractive index detector (Shimadzu).
Faecalibacterium strain culture
Faecalibacterium prausnitzii JCM31915 was obtained from the Japan Collection of Microorganisms. F. prausnitzii was cultured in autoclaved Gifu anaerobic medium at 37 °C under anaerobic conditions (10% H2, 10% CO2, and 80% N2) for three days. For comparison, F. prausnitzii was similarly cultured by further adding autoclaved E. gracilis (9 g) in deionised distilled water (1 L).
Real time PCR analysis
Real time PCR was performed to quantify total bacteria, using the LightCycler 96 system (Roche, Basal, Switzerland) with a universal primer set (5′-ACTCCTACGGGAGGCAGCAGT-3′ and 5′-GTATTACCGCGGCTGCTGGCAC-3′)31. PCR amplification was performed as per the methods described previously18.
Study design for Euglena gracilis ingestion
This study was conducted from September 2017 to December 2017. Selected subjects were healthy (i.e., not undergoing any treatment for ailments) individuals between the ages of 40 and 60 years. The purpose and content of the study were explained thoroughly via written and oral explanation, and 28 individuals who agreed to participate and provided written informed consent were enrolled as the target population for analysis. The study protocol was approved by the Japan Conference of Clinical Research (Approval Date: 16 October 2016, Approval No. 183).
The test was designed and performed as an open study. Each faecal sample collected on day 0 was used as the baseline, and thereafter, faecal samples were collected, and the frequency of defecation and volume of faecal samples was evaluated with reference to the Bristol Stool Form Scale19. After day 0, each participant ingested E. gracilis (2 g/day) in capsule form and were requested to continuously ingest the same amount each day for 30 days. Faecal samples were collected on 14 and 30 days of ingestion.
DNA was extracted from faecal samples as previously described32. The 16S rRNA sequencing using the MiSeq system (Illumina, San Diego, CA, USA) was performed as previously described32. The V3–V4 hypervariable regions of 16S rRNA were amplified from microbial genomic DNA by PCR using bacterial universal primers (341f33/806r34) and the dual-index method35. Barcoded amplicons were sequenced using the paired-end 2 × 284-bp cycle of the MiSeq system using the MiSeq Reagent Kit v. 3 (600 Cycle) chemistry.
The overlapping paired-end sequencing reads were merged using the fastq-join programme with default settings36. Reads were processed using quality and chimera filtering as follows. Reads with quality value score of 20 for more than 99% of the sequence were extracted; chimeric sequences were removed using USEARCH v. 6.137. Nonchimeric reads were submitted for 16S rDNA-based taxonomic analysis using the Ribosomal Database Project v. 2.11 and the TechnoSuruga Lab Microbial Identification database DB-BA10.0 (TechnoSuruga Laboratory, Shizuoka, Japan)38,39.
Statistical analysis
Data were compared between groups using the Wilcoxon signed rank test, Mann–Whitney U test, two-tailed Student’s t-test, or Kruskal–Wallis test using JMP v. 12 (SAS Institute, Cary, NC, USA) or R v. 3.4.1 (The R Foundation, Vienna, Austria). Results with P < 0.05 were considered statistically significant.
Supplementary Information
Acknowledgements
The authors thank Ayami Fujino, Yasunobu Takeshima, Yasuko Koura, and Kimiko Enda of Kobe University and TechnoSuruga Laboratory Co., Ltd., for providing analytical support.
Author contributions
A.N., K.S., and D.S. designed the study. A.N., K.S., D.S., and K.Y. performed the experiments and data analysis. K.S. and A.K. contributed to sample collection and supervised the work. A.N., K.S., D.S., and A.K. drafted and revised the manuscript. All authors read and approved the final manuscript.
Data availability
The raw sequencing data generated in this in vivo study were deposited into the DDBJ, EMBL, and Genbank databases (http://getentry.ddbj.nig.ac.jp/) under the PSUB ID:PSUB013826; the data referring to the in vitro study were deposited into the MG-RAST server40 (http://metagenomics.anl.gov) in a file named ‘Model Culture System of Human Colonic Microbiota_Euglena’ under the accession numbers mgm4908075.3–mgm4908118.3.
Competing interests
AN, KY, and KS are employees of euglena Co., Ltd. All other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ayaka Nakashima, Email: nakashima@euglena.jp.
Kengo Sasaki, Email: sikengo@people.kobe-u.ac.jp.
Supplementary Information
The online version contains supplementary material availlable at 10.1038/s41598-020-80306-0.
References
- 1.García JL, de Vicente M, Galán B. Microalgae, old sustainable food and fashion nutraceuticals. Microb. Biotechnol. 2017;10:1017. doi: 10.1111/1751-7915.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Zorita S, et al. Anti-obesity effects of microalgae. Int. J. Mol. Sci. 2019;21:41. doi: 10.3390/ijms21010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K. Large-scale cultivation of Euglena. Adv. Exp. Med. Biol. 2017;979:285–293. doi: 10.1007/978-3-319-54910-1_14. [DOI] [PubMed] [Google Scholar]
- 4.Gissibl A, Sun A, Care A, Nevalainen H, Sunna A. Bioproducts from Euglena gracilis: synthesis and applications. Front. Bioeng. Biotechnol. 2019;7:108. doi: 10.3389/fbioe.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugimoto R, et al. Euglena extract suppresses adipocyte-differentiation in human adipose-derived stem cells. PLoS ONE. 2018;13:e0192404. doi: 10.1371/journal.pone.0192404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishibashi K, et al. Effects of Euglena gracilis EOD-1 ingestion on salivary IgA reactivity and health-related quality of life in humans. Nutrients. 2019;11:1144. doi: 10.3390/nu11051144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimada R, et al. Oral administration of green algae, Euglena gracilis, inhibits hyperglycemia in OLETF rats, a model of spontaneous type 2 diabetes. Food. Funct. 2016;7:4655–4659. doi: 10.1039/C6FO00606J. [DOI] [PubMed] [Google Scholar]
- 8.Nakashima A, et al. Oral administration of Euglena gracilis Z and its carbohydrate storage substance provides survival protection against influenza virus infection in mice. Biochem. Biophys. Res. Commun. 2017;494:379–383. doi: 10.1016/j.bbrc.2017.09.167. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama A, et al. Oral administration of paramylon, a beta-1,3-D-glucan isolated from Euglena gracilis Z inhibits development of atopic dermatitis-like skin lesions in NC/Nga mice. J. Vet. Med. Sci. 2010;72:755–763. doi: 10.1292/jvms.09-0526. [DOI] [PubMed] [Google Scholar]
- 10.Sugiyama A, et al. Hepatoprotective effects of paramylon, a beta-1,3-D-glucan isolated from Euglena gracilis Z, on acute liver injury induced by carbon tetrachloride in rats. J. Vet. Med. Sci. 2009;71:885–890. doi: 10.1292/jvms.71.885. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, et al. Antitumor activity of the β-glucan paramylon from Euglena against preneoplastic colonic aberrant crypt foci in mice. Food. Funct. 2013;4:1685–1690. doi: 10.1039/c3fo60256g. [DOI] [PubMed] [Google Scholar]
- 12.Lazar V, et al. Aspects of gut microbiota and immune system interactions in infectious disease, immunopathology, and cancer. Front. Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010;7:503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laparra JM, Sanz Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol. Res. 2010;61:219–225. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Graf D, et al. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Health. Dis. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki K, et al. Construction of a model culture system of human colonic microbiota to detect decreased Lachnospiraceae abundance and butyrogenesis in the feces of ulcerative colitis patients. Biotechnol. J. 2019;14:e1800555. doi: 10.1002/biot.201800555. [DOI] [PubMed] [Google Scholar]
- 19.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand. J. Gastroenterol. 1997;32:920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 20.Miquel S, et al. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Gao Z, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia L, et al. Anti-diabetic effects of Clostridium butyricum CGMCC0313.1 through promoting the growth of gut butyrate-producing bacteria in type 2 diabetic mice. Sci. Rep. 2017;7:7046. doi: 10.1038/s41598-017-07335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okouchi R, et al. Simultaneous intake of Euglena gracilis and vegetables exerts synergistic anti-obesity and anti-inflammatory effects by modulating the gut microbiota in diet-induced obese mice. Nutrients. 2019;11:204. doi: 10.3390/nu11010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG, Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS. Microbiol. Lett. 2015;362:fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 25.Asayama Y, Suzuki K, Nakashima A, Shioya N, Sugimura H. The study of the effects of food containing Euglena gracilis on improvement in constipation. Jpn. Pharmacol. Ther. 2017;45:1359–1364. [Google Scholar]
- 26.Kawano Y, et al. Effects of Euglena cells on the absorption and tissue distribution of dietary cholesterol in rats. J. Jpn. Soc. Nutr. Food. Sci. 1987;40:193–198. doi: 10.4327/jsnfs.40.193. [DOI] [Google Scholar]
- 27.Yang J, Wang HP, Zhou L, Xu CF. Effect of dietary fiber on constipation: a meta analysis. World J. Gastroenterol. 2012;18:7378–7383. doi: 10.3748/wjg.v18.i48.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pituch A, Walkowiak J, Banaszkiewicz A. Butyric acid in functional constipation. Prz. Gastroenterol. 2013;8:295–298. doi: 10.5114/pg.2013.38731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trompette A, et al. Dietary fiber confers protection against flu by shaping Ly6c-patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 30.Inui H, Miyatake K, Nakano Y, Kitaoka S. Wax ester fermentation in Euglena gracilis. FEBS Lett. 1982;150:89–93. doi: 10.1016/0014-5793(82)81310-0. [DOI] [Google Scholar]
- 31.Nordeste R, et al. Molecules produced by probiotics prevent enteric colibacillosis in pigs. BMC Vet. Res. 2017;13:335. doi: 10.1186/s12917-017-1246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/AEM.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hisada T, Endoh K, Kuriki K. Inter-and intra-individual variations in seasonal and daily stabilities of the human gut microbiota in Japanese. Arch. Microbiol. 2015;197:919–934. doi: 10.1007/s00203-015-1125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma. J. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 37.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasai C, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer F, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data generated in this in vivo study were deposited into the DDBJ, EMBL, and Genbank databases (http://getentry.ddbj.nig.ac.jp/) under the PSUB ID:PSUB013826; the data referring to the in vitro study were deposited into the MG-RAST server40 (http://metagenomics.anl.gov) in a file named ‘Model Culture System of Human Colonic Microbiota_Euglena’ under the accession numbers mgm4908075.3–mgm4908118.3.