Abstract
The mega-diversity of herbivorous insects is attributed to their co-evolutionary associations with plants. Despite abundant studies on insect-plant interactions, we do not know whether host-plant shifts have impacted both genomic adaptation and species diversification over geological times. We show that the antagonistic insect-plant interaction between swallowtail butterflies and the highly toxic birthworts began 55 million years ago in Beringia, followed by several major ancient host-plant shifts. This evolutionary framework provides a valuable opportunity for repeated tests of genomic signatures of macroevolutionary changes and estimation of diversification rates across their phylogeny. We find that host-plant shifts in butterflies are associated with both genome-wide adaptive molecular evolution (more genes under positive selection) and repeated bursts of speciation rates, contributing to an increase in global diversification through time. Our study links ecological changes, genome-wide adaptations and macroevolutionary consequences, lending support to the importance of ecological interactions as evolutionary drivers over long time periods.
Subject terms: Biodiversity, Biogeography, Evolutionary ecology, Molecular evolution, Phylogenetics
Arms races between herbivores and plants have likely affected their evolutionary histories, which could have led to their high diversity. Allio et al. find that butterflies shifting to new host plants have more adaptive molecular signatures across their genomes and show repeated bursts of speciation rates.
Introduction
Plants and phytophagous insects account for the majority of the documented species of terrestrial organisms1,2. To explain the high diversity of insects, a long held hypothesis states that their diversification is directly related to that of plants3,4. More than half a century ago, Ehrlich and Raven5 proposed a model in which a continual arms race of attacks by herbivorous insects and new defences by their host plants is linked to species diversification via the creation of new adaptive zones, later termed the ‘escape-and-radiate’ model6. According to Ehrlich and Raven5, these developments mainly correspond to toxic secondary compounds in plants, and the associated detoxification mechanisms in insects. This model would apply to all plants and plant-eating insects and could explain why these groups represent an important part of global biodiversity7,8.
Study of insect–plant interactions has progressed tremendously since then through a focus on host chemistry9, phylogenetics10,11 and genomics12–15. Divergence of key gene families13–16 and high speciation rates17–19 have been identified after host–plant shifts, with one example linking duplication of key genes to the ability to feed on new plants and increase diversification13. The emerging consensus from most phylogenetic studies indicates (1) strong phylogenetic conservatism of host–plant associations (related insect species tend to feed on plants that are also related), suggesting ancient and specialized biotic interactions20, and (2) enhanced diversification rates for clades shifting to new host–plant groups compared to those remaining on ancestral plants. Despite high levels of conservatism and specialization, bursts of insect diversification appear to mainly be a consequence of host shifts21, and this somewhat paradoxical conclusion can be understood by considering ecological as well as genetic mechanisms behind host shifts12,15. There are several ways—both direct and indirect—that interactions can influence speciation22, with or without host–plant-based divergent selection on reproductive barriers. One current debate is on the relative importance of radiations following shifts to new adaptive zones and elevated rates of speciation in groups with plastic and diverse host use23–25. Increasingly sophisticated use of time-calibrated phylogenies is being made to investigate the actual timing and rate of diversification and to link such events more conclusively to other factors that may have been important, whether biotic or abiotic18,19.
Genomic aspects of adaptation by herbivorous insects to their host plants have received significant attention26, but few studies have put their genomic data into phylogenetic perspectives. A seminal study by Edger et al.13 on the evolutionary arms race between Pierinae butterflies and their Brassicales host plants showed that shifts in diversification within the plants and their butterflies are associated with gradual changes in plant chemical defences and insect molecular counter adaptations. They identified the genomic mechanisms (gene and genome duplications) explaining the evolution of biosynthetic pathways associated with this arms race. More clues for host-encoded digestive and detoxification mechanisms come from a cross-taxonomic comparison of the gut microbiome of caterpillars with other insects and vertebrates27. The microbes in caterpillar guts are unusually at low densities, and reflect the abundance and composition of leaf-associated microbes in the caterpillar faeces, with high pH, simple gut structure, and fast transit times potentially preventing microbial colonization.
These recent results have illustrated the need for a multidisciplinary approach to studying the evolution of insect–plant interactions within a macroevolutionary and genomic framework. However, a major knowledge gap lies in our understanding of the evolutionary links and drivers of host–plant shifts, genome-wide signatures of adaptations and processes of species diversification28. As noted by Hembry and Weber29, this implies that the questions of if, when and how coevolution has an impact on macroevolutionary dynamics remain open challenges. Here we address this gap with an emblematic group that was instrumental in Ehrlich and Raven’s model—the swallowtail butterflies (Lepidoptera: Papilionidae). Swallowtail caterpillars feed on a range of different flowering families30, but a third of all species, including the tribes Zerynthiini (Parnassiinae), Luehdorfiini (Parnassiinae) and Troidini (Papilioninae), feeds exclusively on the birthwort family (Aristolochiaceae), which is one of the most toxic plant groups31. The Aristolochiaceae notoriously contain toxic aristolochic acids, which are known to be carcinogenic to many organisms, and Papilionidae are among the few that can feed on these plants32,33. By eating these toxic plants, the caterpillars sequester aristolochic acids that render both the caterpillars and the adults unpalatable for predators31. Interestingly, previous phylogenetic estimations of ancestral states indicated either that Aristolochiaceae was the ancestral host plant of Papilionidae34 or that Aristolochiaceae was colonized twice35, suggesting that the host–plant shifts have ancient origins and seem to be highly constrained as shown by the high level of host conservatism. Moreover, the arms race between Papilionidae and their host plants has been demonstrated at the molecular level with the evolution of a cytochrome P450 gene that plays a role in the detoxification of secondary plant compounds36. Some mutations can bypass the toxic defences of certain plants, providing survival and diversification on certain plants (and not others). Further studies have shown how changes in the use of host plants are associated with changes in the sequence, structure and function of P450. Results provide evidence that new P450 copies can appear for herbivores that colonize new hosts, supporting the hypothesis that interaction between herbivores and their host plants contributed to P450–gene diversification37. These studies provide convincing examples of host–plant shifts that may result in increased net diversification rate18,34 and specific changes in key genes that confer new abilities to feed on toxic plants36–38.
Here, we study the insect–plant interactions at macroevolutionary scale using genomic and diversification approaches within a phylogenetic context. Given the complexity of shifting to a new host plant, we can expect more widespread effects across the entire genome15,39,40, but this has remained difficult to demonstrate. Indeed, both comprehensive species-level phylogeny and genomic data are necessary to disentangle the origin of the arms race and to understand the underlying mechanisms of insect–plant interaction as a major driver of diversification. The swallowtail model offers a relevant opportunity to better understand the role played by ecological interactions over the long timescales shaping the astonishing diversity of herbivores41.
Results and discussion
Co-phylogenetic history of an insect–plant antagonistic interaction
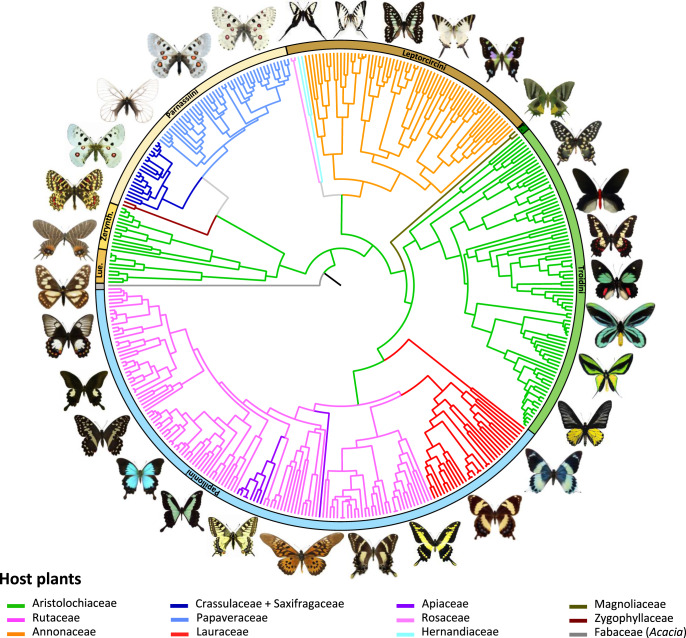
First, we created an extensive phylogenetic dataset including seven genetic markers for 71% of swallowtail species diversity (408 of ~570 described species, see ‘Methods’). This dataset leads to the assembly of the most complete and well-resolved dated phylogeny of swallowtail butterflies (79% of nodes with strong bootstrap support defined as ≥95%; Supplementary Figs. 1–3). Both tribe- and genus-level relationships are mostly consistent with previous results using multilocus datasets18,34,35,42–45. However, our species tree benefits from a phylogenomic backbone that we recently inferred at the genus level for the Papilionidae using genome-scale data46. Second, we compiled host–plant preferences for each swallowtail species in the dataset, and we performed ancestral-state estimations (see ‘Methods’). Phylogenetic estimates of ancestral host–plant preferences indicate that Aristolochiaceae were either the food plant of ancestral Papilionidae34 or were colonized twice35, suggesting an ancient and highly conserved association with Aristolochiaceae throughout swallowtail butterflies evolution. Using this robust time-calibrated phylogeny (Supplementary Figs. 1–3), we have traced the evolutionary history of food-plant use and infer that the family Aristolochiaceae was the ancestral host for Papilionidae (Fig. 1; relative probabilities = 0.915, 0.789 and 0.787 with three models; Supplementary Figs. 4 and 5). We further show that the genus Aristolochia was the ancestral host plant, as almost all Aristolochiaceae-associated swallowtails feed on Aristolochia (Supplementary Fig. 6). Across the swallowtail phylogeny, we recover only 14 host–plant shifts at the plant family level (14 nodes out of 407; Supplementary Figs. 4 and 5), suggesting strong evolutionary host–plant conservatism.
Fig. 1. Evolution of host–plant association through time shows strong host–plant conservatism across swallowtail butterflies.
Phylogenetic relationships of swallowtail butterflies, with coloured branches mapping the evolution of host–plant association, as inferred by a maximum-likelihood model (Supplementary Figs. 4 and 6). Additional analyses with two other maximum-likelihood and Bayesian models inferred the same host–plant associations across the phylogeny (Supplementary Fig. 5). Lue. Luehdorfiini, Zerynth. Zerynthiini, T. Teinopalpini. Pictures of butterflies made by Fabien Condamine.
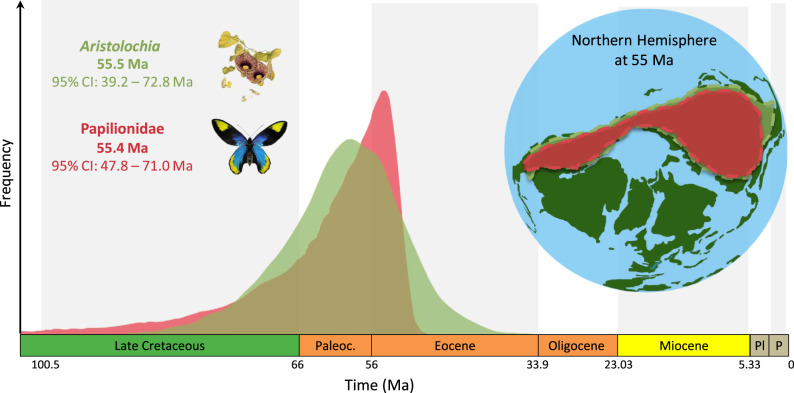
With the ancestor of swallowtails feeding on birthworts, evidence for synchronous temporal and geographical origins further links the genus Aristolochia and the family Papilionidae and supports the escape-and-radiate model. Reconstructions of co-phylogenetic history for other insect–plant antagonistic interactions have shown either synchronous diversification11 or herbivore diversification lagging behind that of their host plants10,47. We assembled a molecular dataset for ~49% of the species diversity of Aristolochiaceae (247 of ~502 described species; see ‘Methods’) and reconstructed their phylogeny (Supplementary Fig. 7), which is in agreement with previous works48–52. Divergence time estimates strongly suggest synchronous radiations of Papilionidae (55.4 million years ago [Ma], 95% credibility intervals (CIs): 47.8–71.0 Ma) and Aristolochia (55.5 Ma, 95% CIs: 39.2–72.8 Ma) since the early Eocene (Fig. 2 and Supplementary Figs. 3, 8 and 9). This result is robust to known biases in inferring divergence times, with slightly older ages inferred for both groups when using more conservative priors on clade ages (Supplementary Fig. 9). Such temporal congruence between Aristolochia and Papilionidae raises the question of whether both clades had similar geographical origins and dispersal routes. To characterize the macroevolutionary patterns of the Aristolochia/Papilionidae arms race in space, we assembled two datasets of current geographic distributions for all species included in the phylogenies of both Aristolochiaceae and Papilionidae. We reconstructed the historical biogeography of both groups, taking into account palaeogeographical events throughout the Cenozoic (see ‘Methods’). Along with the known fossil record of both groups53–57, these results suggest that both Papilionidae and Aristolochia were ancestrally co-distributed throughout a region, including West Nearctic (WN), East Palaearctic (EP), and Central America (CA) in the early Eocene, when Asia and North America were connected by the Bering land bridge (Fig. 2 and Supplementary Figs. 10 and 11). This combination of close temporal and spatial congruence provides strong evidence that Papilionidae and Aristolochia diversified concurrently through time and space until several swallowtail lineages shifted to the new host–plant families in the middle Eocene.
Fig. 2. Synchronous temporal and geographic origin for swallowtails and birthworts.
Bayesian molecular divergence times with exponential priors estimate an early Eocene origin (~55 Ma) for both swallowtails and Aristolochia (alternatively, analyses with a uniform prior estimated an origin around 67 Ma for swallowtails and 64 Ma for Aristolochia; Supplementary Figs. 3, 8 and 9). Biogeographical maximum-likelihood models infer an ancestral area of origin comprising West Nearctic, East Palaearctic and Central America for both swallowtails and birthworts (Supplementary Figs. 10 and 11). Paleoc Paleocene, Pl Pliocene, P Pleistocene, Ma million years ago. Pictures of the plant and butterfly made by Fabien Condamine, and the world map made by Rémi Allio.
Host–plant shifts confer higher rates of diversification
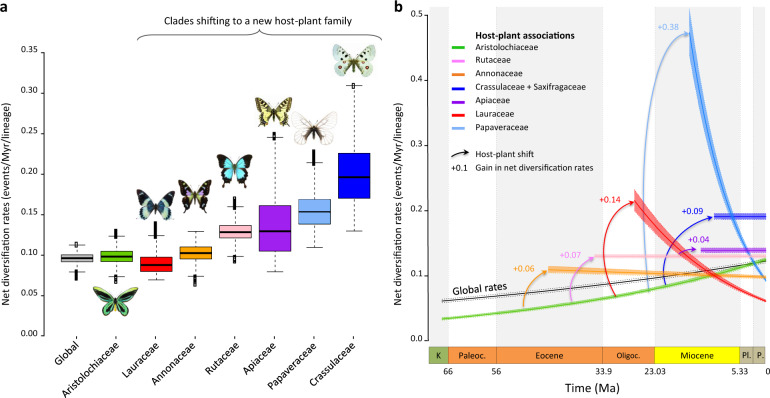
Our ancestral-state estimates and biogeographic analyses are consistent with a sustained arms race between Aristolochia and Papilionidae in the past 55 million years. According to the escape-and-radiate model, a host–plant shift should confer higher rates of species diversification for herbivores through the acquisition of novel resources to radiate into5,6 and/or the lack of competitors (Aristolochiaceae-feeder swallowtails have almost no competitors31). We tested the hypothesis that increases in diversification rates occurred in swallowtail lineages that shifted to new host plants. Given the uncertainty surrounding the inferences of macroevolutionary rates from phylogenies of extant species, we applied a suite of birth–death models to cross-validate the estimated rates of diversification LASER (Likelihood Analysis of Speciation and Extinction Rates), MuSSE (Multiple State Speciation Extinction), RPANDA (R: Phylogenetic ANalyses of DiversificAtion), BAMM (Bayesian analysis of macroevolutionary mixtures), CoMET (CPP on Mass-Extinction Times) and RevBayes; see ‘Methods’). We find evidence for (1) increases of diversification at host–plant shifts with trait-dependent birth–death models (as inferred with: MuSSE, Fig. 3a and Supplementary Fig. 12; RPANDA, Supplementary Fig. 13; and LASER, Supplementary Table 1) and (2) host–plant shifts contributing to a global increase through time with clade- and time-dependent birth–death models (as inferred with: RPANDA, Fig. 3b and Supplementary Fig. 13; BAMM, Supplementary Fig. 14; RevBayes, Supplementary Fig. 15; and CoMET, Supplementary Fig. 16). Although we should be cautious about the estimations of macroevolutionary rates58–63, all models concur that diversification rates increase through time either globally or due to recurrent host–plant shifts. Interestingly, these results contrast with the slowdown of diversification that is classically recovered in most phylogenies, often attributed to ecological limits and niche filling processes63. This sustained and increasing diversification during the Cenozoic may be explained by ecological opportunities not decreasing, due to a steady increase in host breadth for Papilionidae with new host–plant families colonized through time (Supplementary Fig. 17). Opening up new niches, which can also expand due to diversification increases of the host–plant families through time64–66, would allow a continuous increase in diversification rates through time in a dynamic biotic environment, lending support to the primary role of ecological interactions in clade diversification over long timescales—a long-contentious issue29. Nonetheless, when taking into account the possibility that rates may have been heterogeneous across the phylogeny, we find that the diversification of three lineages (those feeding on Annonaceae, Lauraceae and Papaveraceae) had early rates of speciation that are higher than the ancestral rates, but slowed down through time.
Fig. 3. Host–plant shifts lead to repeated bursts in diversification rates and a sustained overall increase in diversification through time.
a Diversification tends to be higher for clades shifting to new host plants, as estimated by trait-dependent diversification models. Boxplots represent Bayesian estimates of net diversification rates for clades feeding on particular host plants (see also Supplementary Fig. 12). b A global increase in diversification is recovered with birth–death models estimating time-dependent diversification (see also Supplementary Figs. 14 and 15). Taking into account rate heterogeneity by estimating host–plant and clade-specific diversification indicates positive gains of net diversification after shifting to new host plants (see also Supplementary Fig. 13). K Cretaceous, Paleoc. Paleocene, Oligoc. Oligocene, Pl Pliocene, P Pleistocene, Ma million years ago. Pictures of butterflies made by Fabien Condamine.
Interestingly, not all host–plant shifts led to evolutionary success in terms of extant species diversity. Given our rate estimations, we found significantly lower diversification rates than the rates on the ancestral host–plant Aristolochiaceae for three host–plant shifts (to Fabaceae, Magnoliaceae and to Zygophyllaceae; Supplementary Fig. 12). Altogether, these three host switches correspond to a very low proportion (~1%) of the total swallowtail diversity today. Indeed, a single species (Baronia brevicornis) feeds on the Fabaceae, the genus Hypermnestra (two species) feeds on Zygophyllaceae and the genus Teinopalpus (two species) feeds on Magnoliaceae. Hence, these are unsuccessful host–plant shifts from an evolutionary perspective (i.e. evolutionary dead-ends).
Genome-wide adaptations to host–plant shifts
Key innovations are often considered to underlie ecological opportunities and/or evolutionary success67, particularly in the case of chemically mediated interactions between butterflies and their host plants13. Studies on Papilionidae have provided strong examples of specific changes in key genes that confer new abilities to feed on toxic plants and allow host–plant shifts36,37. Adaptations of swallowtails to their hosts have particularly been assessed through the study of cytochrome P450 monooxygenases (P450s), which have a major role in detoxifying secondary plant compounds. New P450s appear to arise in swallowtails that colonize new hosts to bypass toxic defences, providing survival and diversification on some but not all plants15,36,37. This supports the hypothesis that insect–plant interactions contributed to P450-gene family diversification, with P450s being key innovations that explain the evolutionary and ecological success of phytophagous insects14,15,36,38,68,69. However, host–plant shifts not only alter single genes but may also influence unlinked genes40. Moreover, host–plant shifts can accompany changes of the abiotic environment, which may, in turn, require further biotic adaptation (new predators and/or competitors). But the macroevolutionary and genomic consequences of the evolutionary dynamics of host–plant shifts have not yet been demonstrated.
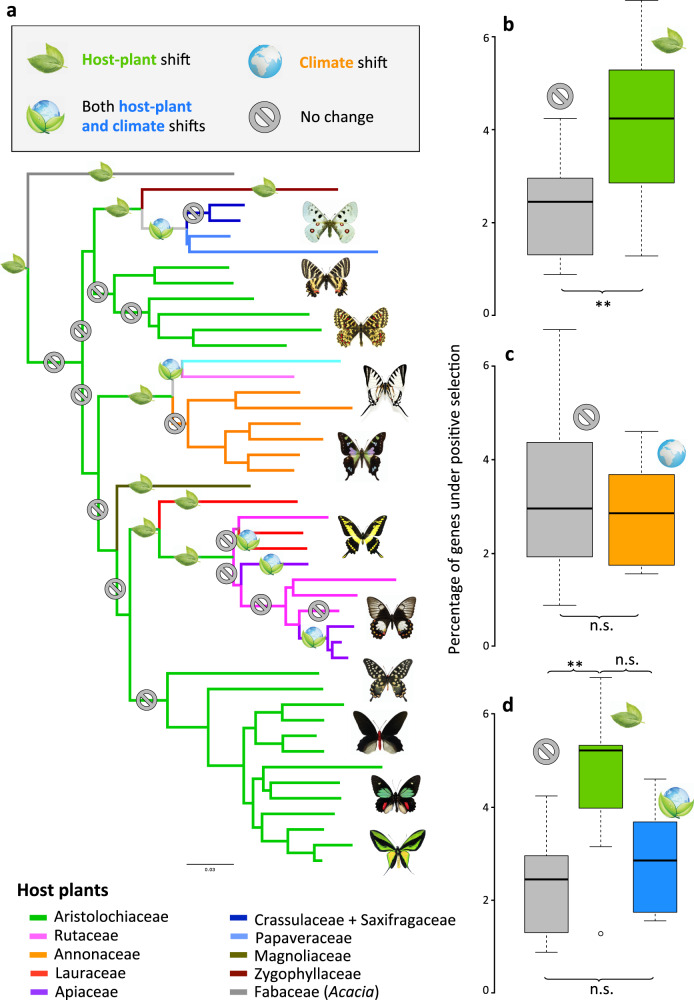
Relying on a genomic dataset comprising 45 genomes covering all swallowtail genera46,70–72, we constructed two specific datasets (Dataset 1: 520 genes and Dataset 2: 1533 genes; mean gene coverage = 26.7×; see ‘Methods’ and Supplementary Data 1). To test whether there are any genomic signatures of positive selection caused by host–plant shifts within swallowtails, we performed a comparative genomic survey of molecular adaptation between swallowtail lineages that shifted to new host plants compared to non-shifting lineages (see ‘Methods’). We selected 14 phylogenetic branches representing a host–plant shift and 14 phylogenetic branches with no change as negative controls73,74 (Fig. 4a). For a fair molecular comparison, each branch selected as a negative control was chosen to be as close as possible to a test branch representing a host–plant shift (i.e. sister groups; Supplementary Fig. 18). Among branches with host–plant shifts, five branches also had a shift in climate preference (represented by distributional changes from tropical to temperate conditions). Using a maximum-likelihood (ML) method, we estimated the ratio of non-synonymous substitutions (dN) other synonymous substitutions (dS) in all branches where a host–plant shift was identified relative to branches with no host–plant shift75,76 (see ‘Methods’). The dN/dS analyses on branches with host–plant shifts (combined or not with environmental shifts) showed more genes with a subset of codons evolving under positive selection (dN/dS > 1) in lineages shifting to a new plant family, although the difference was marginally non-significant for the smallest dataset and highly significant for the second dataset containing more genes (Fig. 4b, Supplementary Fig. S19 and Supplementary Table 2, P = 0.0501/0.0079 for the two datasets, respectively, Wilcoxon rank-sum test, see ‘Methods’ for the definition of the datasets). However, dN/dS analyses on branches with environmental shifts indicated a balanced number of genes under positive selection (Fig. 4c, Supplementary Fig. S19 and Supplementary Table 2, P = 0.336/0.8162 for the two datasets, respectively, Wilcoxon rank-sum test), suggesting a lower impact of environmental shifts than host–plant shifts. We then performed dN/dS analyses for branches with host–plant shifts only (not followed by environmental shifts) and found that swallowtail lineages shifting to a new host–plant family had significantly more genes under positive selection (4.41%/3.98% of genes under positive selection for the two datasets, respectively; Supplementary Table 2) than non-shifting lineages (3.02%/2.43% of genes under positive selection for the two datasets, respectively, Fig. 4d, Supplementary Fig. S19 and Supplementary Table 2, P = 0.0071/0.00156 for the two datasets, respectively, Wilcoxon rank-sum test). Surprisingly, the dual changes in environment and host–plant preferences did not spur molecular adaptation across swallowtail lineages compared to control branches (P = 1/0.4439 for the two datasets, respectively, Wilcoxon rank-sum test; Fig. 4d, Supplementary Fig. S19 and Supplementary Table 2). Comparing the proportion of genes under positive selection between the branches with dual changes and branches with host–plant shifts only shows a marginally significant difference with Dataset 1 and no difference with Dataset 2 (P = 0.0327/0.1471 for the two datasets, respectively, Wilcoxon rank-sum test; Fig. 4d and Supplementary Fig. S19). However, this result might be an artefact due to the use of a few branches to perform the statistical comparison. Although we did not control for the effect of multi-nucleotide mutations77, which should affect dN/dS analyses equally for control and host–plant shift branches, we checked individually the gene alignments and performed sensitivity analyses, which showed that our results are not driven either by an excess of misaligned regions or missing data and GC-content variations among species (see ‘Methods’ and Supplementary Figs. 20–26). Finally, given that fixing the topology for CodeML (see ‘Methods’) can spuriously inflate substitution rates on some branches78, we computed the proportion of genes under positive selection by selecting the gene trees from the largest dataset (Dataset 2) for which the focal branches were recovered (in agreement with the species tree). These analyses confirmed the previous results suggesting more genes under positive selection during host–plant shifts (P = 0.0444, Wilcoxon rank test; Supplementary Table 2).
Fig. 4. Host–plant shifts promote higher molecular adaptations.
a Genus-level phylogenomic tree displaying branches with and without host–plant shifts, on which genome-wide analyses of molecular evolution are performed. b Number of genes under positive selection (dN/dS > 1) for swallowtail lineages shifting to new host–plant families (n = 14, green) or not (n = 14, grey). c Number of genes under positive selection for swallowtail lineages undergoing climate shifts (n = 5, orange) or not (n = 23, grey). d Number of genes under positive selection for swallowtail lineages shifting to new host plants (n = 9, green), shifting both host–plant and climate (n = 5, blue) or not (n = 14, grey). The proportion of genes was estimated with Dataset 2 (1533 genes, see Supplementary Fig. 19 for the results with Dataset 1 and 520 genes). This demonstrates genome-wide signatures of adaptations in swallowtail lineages shifting to new host–plant families. Genes under positive selection did not contain over- or under-represented functional GO categories (Supplementary Data 2). Wilcoxon rank-sum test: n.s. = not significant (P > 0.05), *P ≤ 0.05, **P ≤ 0.01. Pictures and icons made by Fabien Condamine.
We further studied the functional categories of positively selected genes by using gene ontology (GO) analyses (PANTHER and EggNOG; see ‘Methods’). Applied to the high-quality genomes of Papilio xuthus71 and Heliconius melpomene79, we found that ~70% of the genes could be associated with a gene function and ~30% lacked annotation, which suggests a gap of knowledge in the current insect database of gene function. Among the annotated genes, we found that genes under positive selection along branches with host shifts did not contain over- or under-represented functional GO categories: 252 out of 1213 GO categories represented by genes under positive selection (P > 0.05, Fisher’s exact test after false discovery rate correction; Supplementary Data 2). These results support the hypothesis that genome-wide signatures of adaptations are associated with host–plant shifts, and encourage enlarging the hypothesis that changes in only one or a few candidate gene families could be enough to act as key innovations for adaptation to new resources13,17. Despite a weak signal, it is striking that host–plant shifts left stronger genome-wide signatures than were associated with changing climate preferences. This result further suggests that the success of phytophagous insects involved widespread adaptations to biotic interactions than for shifts in the abiotic environment.
To conclude, establishing evolutionary links between ecological adaptations, genomic changes and species diversification over geological timescales remains a tremendous challenge28,80,81 with, for instance, important limitations due to the lack of knowledge in functional gene annotations in insects. However, the successful development of powerful analytical tools in conjunction with the increasing availability of insect genomes and improvements in genomic analyses82 have allowed the detection of more genes than those already known to be involved in detoxification pathways playing a role in long-term relationships between plants and insects. Our genome-wide analyses have also generated a list of candidate genes potentially involved in plant–insect interactions. This opens new research avenues for finding the functionality of genes potentially linked with the adaptation and diversification of phytophagous insects. We hope that our study will help move in that direction, and that it will provide perspectives for future investigations of other model groups.
Over a half-century ago, Ehrlich and Raven5 proposed that insect–plant interactions driven by diffuse coevolution over long evolutionary periods can be a major source of terrestrial biodiversity. Applied to a widely appreciated case in the insect–plant interactions theory, our study has been able to investigate genome-wide adaptive processes and corresponding macroevolutionary consequences in a comprehensive framework, suggesting that more genes could be involved in host–plant shifts than previously studied in the diversification of herbivorous insects. This result confirms the general belief in the insect–plant community that host–plant shifts are complex and would thus require a number of adaptations, which likely affect various genes beyond those directly linked to detoxification of the plant compounds36,39,40. By expanding the possible genes and gene families and identifying more adaptations than those gene families in detoxification pathways that were detected through antagonist interactions39, we show genomically wide-ranging co-evolutionary consequences40,83 for close relationships between insects and their larval host plants. Hence, genome-wide macroevolutionary consequences of key adaptations in new insect–plant interactions may be a general feature of the co-evolutionary interactions that have generated Earth’s diversity.
Methods
Time-calibrated phylogeny of Papilionidae
We assembled a supermatrix dataset with available data extracted from GenBank as of May 2017 (most of which has been generated by our research group), using five mitochondrial genes (COI, COII, ND1, ND5 and rRNA 16S) and two nuclear markers (EF-1a and Wg) for 408 Papilionidae species (~71% of the total species diversity) and 20 outgroup species. We aligned the DNA sequences for each gene using MAFFT 7.11084 with default settings (E-INS-i algorithm), and the alignments were checked for codon stops and eventually refined by eye with Mesquite 3.1 (available at: www.mesquiteproject.org). The best-fit partitioning schemes and substitution models for phylogenetic analyses were determined with PartitionFinder 2.1.185 using the greedy search algorithm and the Bayesian Information Criterion. All gene alignments were concatenated in a supermatrix, which is available in Figshare (see Data availability).
Phylogenetic relationships were estimated with both ML and Bayesian inference. ML analyses were carried out with IQ-TREE 1.6.886. We set the best-fit partitioning scheme (-ssp option) and used ModelFinder to determine the best-fit substitution model for each partition87 and then estimated model parameters separately for every partition88 such that all partitions shared the same set of branch lengths, but we allowed each partition to have its own evolution rate (-m TESTNEW option). For tree search parameters, we relied on a more thorough and slower nearest-neighbour interchange search to consider all possible nearest-neighbour interchanges instead of only those in the vicinity previously applied (-allnni option). Following the recommendation of IQ-TREE developers, we also set smaller perturbation strength (-pers 0.2) and a larger number of stop iterations (-nstop 500) to avoid local optima. We performed 2000 ultrafast bootstrap replicates to investigate nodal support across the topology, considering values ≥95 as strongly supported nodes89.
Estimating phylogenetic relationships for such a dataset is computationally intensive with Bayesian inference. The ML tree inferred with IQ-TREE was used as a starting tree for Bayesian inference as implemented in MrBayes 3.2.690. Rather than using a single substitution model per molecular partition, we sampled across the entire substitution-model space91 using reversible-jump Markov Chain Monte Carlo (rj-MCMC). Two independent analyses with one cold chain and seven heated chains, each run for 50 million generations, sampled every 5000 generations. Convergence and performance of Bayesian runs were evaluated using Tracer 1.7.192, the average deviation of split frequencies (ADSFs) between runs, the effective sample size (ESS) and the potential scale reduction factor (PSRF) values for each parameter. The runs had to have values of ADSF approaching zero, PSRF close to 1.0 and ESS >200 to be considered convergent. A 50% majority-rule consensus tree was built after conservatively discarding 25% of sampled trees as burn-in. Node support was evaluated with posterior probability considering values ≥0.95 as strong support93. All analyses were performed on the CIPRES Science Gateway computer cluster94, using BEAGLE95.
Dating inferences were performed using Bayesian relaxed-clock methods accounting for rate variation across lineages96. MCMC analyses implemented in BEAST 1.8.497 were employed to approximate the posterior distribution of rates and divergences times and infer their CIs. Estimation of divergence times relied on constraining clade ages through fossil calibrations. Swallowtail fossils are scarce, but five can unambiguously be attributed to the family. The oldest fossil occurrences of Papilionidae are the fossils †Praepapilio colorado and †Praepapilio gracilis53, both from the Green River Formation (Colorado, USA). The Green River Formation encompasses a 5 million years period between ~48.5 and 53.5 Ma, which falls within the Ypresian (47.8–56 Ma) in the early Eocene98. These fossils can be phylogenetically placed at the crown of the family as they share synapomorphies with all extant subfamilies55,99, and have proven to be reliable calibration points for the crown group18,34,46. Two other fossils belong to Parnassiinae, whose systematic position was assessed using phylogenetic analyses based on both morphological and molecular data in a total-evidence approach18. The first is †Thaites ruminiana100, a compression fossil from limestone in the Niveau du gypse d’Aix Formation of France (Bouches-du-Rhône, Aix-en-Provence, France) within the Chattian (23.03–28.1 Ma) of the late Oligocene54,101. †Thaites is sister to Parnassiini, and occasionally sister to Luehdorfiini + Zerynthiini18. Thus, we constrained the crown age of Parnassiinae with a uniform distribution bounded by a minimum age of 23.03 Ma. The second is †Doritites bosniaskii102, an exoskeleton and compression fossil from Italy (Tuscany) from the Messinian (5.33–7.25 Ma, late Miocene)54. †Doritites is sister to Archon (Luehdorfiini18), in agreement with Carpenter103. The crown of Luehdorfiini was thus constrained for divergence time estimation using a uniform distribution bounded with 5.33 Ma. Absolute ages of geological formations were taken from the latest update of the geological time scale.
We used a conservative approach to apply calibration priors with the selected fossil constraints by setting uniform priors bounded with a minimum age equal to the youngest age of the geological formation where each fossil was found. All uniform calibration priors were set with an upper bound equal to the estimated age of angiosperms (150 Ma104), which is more than three times older than the oldest Papilionidae fossil. This upper age is intentionally set as ancient to allow exploration of potentially old ages for the clade. Since the fossil record of butterflies is incomplete and biased105, caution is needed in using these fossil calibrations (effect shown in burying beetles106).
After enforcing the fossil calibrations, we set the following settings and priors: a partitioned dataset (after the best-fitting PartitionFinder scheme) was analysed using the uncorrelated log-normal distribution clock model, with the mean set to a uniform prior between 0 and 1, and an exponential prior (lambda = 0.333) for the standard deviation. The branching process prior was set to a birth–death107 process, using the following uniform priors: the birth–death mean growth rate ranged between 0 and 10 with a starting value at 0.1, and the birth–death relative death rate ranged between 0 and 1 (starting value = 0.5). We performed four independent BEAST analyses for 100 million generations, sampled every 10,000th, resulting in 10,000 samples in the posterior distribution, of which the first 2500 samples were discarded as burn-in. All analyses were performed on the CIPRES Science Gateway computer cluster94, using BEAGLE95. Convergence and performance of each MCMC run were evaluated using Tracer 1.7.192 and the ESS for each parameter (ESS > 200). We combined the four runs using LogCombiner 1.8.497. A maximum-clade credibility (MCC) tree was reconstructed, with median ages and 95% CIs. The BEAST files generated for this study are available in Figshare (see Data availability).
Estimating ancestral host–plant association
We inferred the temporal evolution of host–plant association up to the ancestral host plant(s) at the root of Papilionidae using three approaches: the ML implementation of the Markov k-state (Mk) model108, the ML Dispersal-Extinction-Cladogenesis (DEC) model109, and the Bayesian approach in BayesTraits110. These approaches require a time-calibrated tree and a matrix of character states (current host–plant preference) for each species in the tree. An extensive bibliographic survey was conducted to obtain primary larval host plants at the family level5,30,111–113. The host associations of species were categorized using the following 12 character states: (1) Annonaceae, (2) Apiaceae, (3) Aristolochiaceae, (4) Crassulaceae or Saxifragaceae (core Saxifragales), (5) Fabaceae, (6) Hernandiaceae, (7) Lauraceae; (8) Magnoliaceae, (9) Papaveraceae, (10) Rosaceae, (11) Rutaceae, and (12) Zygophyllaceae. The host–plant matrix of Papilionidae is available in Figshare (see Data availability).
Ancestral states for host–plant association were first reconstructed using the Mk model (one rate for all transitions between states) allowing any host shift to be equally probable. The Mk model does not allow multiple states for a species. The few species that use multiple host families were thus scored with the most frequent host association. The Mk model was performed with Mesquite 3.1 (available at: www.mesquiteproject.org). To estimate the support of any one character state over another, the most likely state was selected according to a decision threshold, such that if the log likelihoods between two states differ by two log-likelihood units, the one with lower likelihood is rejected108.
The DEC model was also used to reconstruct ancestral host–plant states109,114. As with the Mk model, we assumed that host–plant shifts occurred at equivalent probabilities between plant families and through time, which may not be true given that the host–plant families of Papilionidae did not originate at the same time (e.g. Aristolochiaceae originated ~108.07 Ma [95% CIs: 81.01–132.66 Ma]115, and Annonaceae originated ~98.94 Ma [95% CIs: 84.78–113.70 Ma]115). We used the estimated molecular ages of the different host–plant groups to constrain our inferences of ancestral host plants a posteriori. We preferred such an approach compared to a more constrained one in which the DEC model is informed with a matrix of host–plant appearances based on their estimated ages by implementing matrices of presence/absence of the character states through time (equivalent to the time-stratified palaeogeographic model, see below for inference of biogeographical history).
Finally, the Bayesian approach implemented in BayesTraits 3.0.1110 was performed to provide a cross-validation of ML analyses. This approach automatically detects shifts in rates of evolution for multistate data using rj-MCMC. The number of parameters and priors was set by default. We ran the rj-MCMC for ten million generations and sampled states and parameters every 1000 generations (burn-in of 10,000 generations). We specifically estimated ancestral states at 21 nodes as well as at the root of Papilionidae. For this analysis, we used a set of 100 trees randomly taken from the dating analysis to probe the robustness of our ancestral-state estimation across topological uncertainty.
The results of these inferences determined the host–plant family(ies) that was (were) the most likely ancestral host(s) at the origin of Papilionidae, indicating (1) which plant phylogeny to reconstruct for studying the macroevolution of the arms race, and (2) the evolution of ancestral host–plant association along the phylogeny to identify the tree branches where shifts occurred and test for genome-wide changes.
The Mk and BayesTraits models always inferred with high support (relative probability = 0.915 and 0.789, respectively) that Aristolochiaceae is the ancestral host plant at the crown of Papilionidae. With the unconstrained DEC model, we found that the ancestral host–plant preference for Papilionidae was always composed of Aristolochiaceae, but also included another family (either Fabaceae, Hernandiaceae or Zygophyllaceae, which are only fed upon by Baronia, Lamproptera and Hypermnestra, respectively). As the sister lineage to all other Papilionidae, Baronia is the only species that feeds on Fabaceae. More precisely, only one species of Fabaceae is consumed: Vachellia cochliacantha (formerly Acacia cochliacantha; recent changes in Acacia taxonomy116). However, Vachellia diverged from its sister clade in the Eocene, ~50 Ma, and diversified in the Miocene between 13 and 17 Ma117, which substantially postdate the origin of Papilionidae. Therefore, this result suggests that the family Aristolochiaceae represents the most likely candidate as the ancestral host plant of Papilionidae. Hernandiaceae are consumed by Lamproptera (occasionally by Papilio homerus, Graphium codrus, G. doson and G. empedovana113). More precisely, the host plants of Lamproptera belong to the genus Illigera. This plant genus diverged from its sister genus 48 Ma115 and started diversifying 27 Ma118. The derived phylogenetic position of Lamproptera and the age of its use as a host plant make it very unlikely that Hernandiaceae could constitute the ancestral host plant for Papilionidae. Similarly, the family Zygophyllaceae is consumed by Hypermnestra, most specifically it feeds on the genus Zygophyllum in Central Asia. The genus Zygophyllum is not monophyletic, but Asian Zygophyllum appeared 19.6 Ma119. Applying the same rationale, we are able to discard Zygophyllaceae as a candidate ancestral host plant for Papilionidae. To further refine our ancestral host–plant estimates, we built a presence–absence matrix of plant families based on clade origins estimated in molecular dating studies. Thereby, the age of the different plants can be used to constrain the inference of ancestral host plants. Under such a constrained model, Aristolochiaceae is always recovered as the most likely ancestral host plant for Papilionidae. It is also interesting that almost all Aristolochiaceae feeders have Aristolochia as host plants, and tests to determine which genus of Aristolochiaceae was originally consumed by Papilionidae showed that it was Aristolochia.
Time-calibrated phylogeny of the ancestral host: the Aristolochiaceae
Estimation of ancestral host–plant relationships indicated that the family Aristolochiaceae was the ancestral host for Papilionidae. We refer to Aristolochiaceae in its traditional circumscription including the genera Asarum, Saruma, Thottea and Aristolochia. The Angiosperm Phylogeny Group120 proposes that Aristolochiaceae also includes the holoparasitic genera Hydnora and Prosopanche (Hydnoraceae), as well as the monotypic family Lactoridaceae from the Juan Fernandez Islands of Chile (Lactoris fernandeziana). The conclusion of Angiosperm Phylogeny Group (APG)120 is based on an online survey121 rather than on primary data and this is why we disagree with their argumentation as well as the resulting conclusion of APG given available resilient primary molecular phylogenomic data. However, arguments based on morphology and anatomy122–125, genetics49,50,126–129, molecular divergence time115,129, and conservation considerations (Tod Stuessy, personal communication with S.W., July 2019) favour splitting them into four families: Aristolochiaceae (Aristolochia and Thottea), Asaraceae (Asarum and Saruma), Hydnoraceae (Hydnora and Prosopanche), and Lactoridaceae (Lactoris), collectively called the perianth-bearing Piperales. Therefore, we extracted and assembled a supermatrix dataset with available data from GenBank for the perianth-bearing Piperales and its sister lineage, the perianth-less Piperales including Saururaceae and Piperaceae (as of May 2017, most of which has been generated by our research group). We obtained four chloroplast genes (matK, rbcl, trnL and trnL-trnF) and one nuclear marker (ITS) for 247 species of perianth-bearing Piperales (~49% of the total species diversity130) and six outgroups from perianth-less Piperales. We could not include the two genera Hydnora and Prosopanche (Hydnoraceae) because available genetic data do not overlap those of perianth-bearing Piperales126,128,131,132. We applied the same analytical procedure that we did for Papilionidae. DNA sequences for each gene were aligned using MAFFT 7.11084 with default settings (E-INS-i algorithm and Q-INS-I to take into account secondary structure). Resulting alignments were checked for codon stops and eventually refined by eye with Mesquite 3.1 (available at: www.mesquiteproject.org). The best-fit partitioning schemes and substitution models for phylogenetic analyses were determined with PartitionFinder 2.1.185. All gene alignments were concatenated into a supermatrix; the final dataset is available in Figshare (see Data availability).
Phylogenetic relationships were estimated with Bayesian inference as implemented in MrBayes 3.2.690. Rather than using a single substitution model per molecular partition, we sampled across the entire substitution-model space91 using rj-MCMC. Two independent analyses with one cold chain and seven heated chains, each was run for 50 million generations, sampled every 5000 generations. Convergence and performance of Bayesian runs were evaluated using Tracer 1.7.192 and the ESS, ADSF and PSRF criteria. Once convergence was achieved, a 50% majority-rule consensus tree was built after discarding 25% of the sampled trees as burn-in.
Bayesian relaxed-clock methods were used that accounted for rate variation across lineages96. MCMC analyses implemented in BEAST 1.8.497 were employed to approximate the posterior distribution of rates and divergences times and infer their CIs. Estimation of divergence times relied on constraining clade ages through fossil calibrations. Three unambiguous fossils from perianth-bearing Piperales (Aristolochiaceae sensu lato), and one corresponding to the family Saururaceae were used. First, we relied on the fossil record of the monotypic family Lactoridaceae (L. fernandeziana)126,129, a shrub endemic to the cloud forest of the Juan Fernández Islands archipelago of Chile. The oldest pollen fossil for the group is †Lactoripollenites africanus133,134 from the Turonian/Campanian (72.1–89.8 Ma) of the Orange Basin in South Africa. This fossil confers a minimum age of 72.1 Ma for the stem node of L. fernandeziana. Second, the oldest and only pollen record of the Aristolochiaceae was recently described from Late Cretaceous sediments of Siberia: †Aristolochiacidites viluiensis56 from the Timerdyakh Formation of the latest Campanian to earliest Maastrichtian (66–72.1 Ma) in the Vilui Basin (Russia). Because inaperturate pollen grains in combination with this unique exine configuration and fitting size can be observed in extant members of Aristolochiaceae, this fossil provides a minimum age of 66 Ma for the family. The third fossil belongs to the genus Aristolochia and described as †Aristolochia austriaca57 from the Pannonian (late Miocene) in the Hollabrunn–Mistelbach Formation (Austria). Based on a thorough morphological leaf comparison, this fossil is assigned to a species group including Aristolochia baetica and Aristolochia rotunda, which then confers a minimum age of 7.25 Ma for the clade. Finally, we used the fossil †Saururus tuckerae135 from the Princeton Chert of Princeton in British Columbia (Canada), which is part of the Princeton Group, Allenby Formation dated with stable isotopes to the middle Eocene136. This fossil has been phylogenetically placed as sister to extant Saururus species136, hence providing a minimum age of 44.3 Ma for the stem node of Saururus. Absolute ages of geological formations were taken from the latest update of the geological time scale.
We set the following settings and priors: a partitioned dataset (after the best-fitting PartitionFinder scheme) was analysed using the uncorrelated log-normal distribution clock model, with the mean set to a uniform prior between 0 and 1, and an exponential prior (lambda = 0.333) for the standard deviation. The branching process prior was set to a birth–death107 process, using the following uniform priors: the birth–death mean growth rate ranged between 0 and 10 with a starting value at 0.1, and the birth–death relative death rate ranged between 0 and 1 (starting value = 0.5). We performed four independent BEAST analyses for 100 million generations, sampled every 10,000th, resulting in 10,000 samples in the posterior distribution of which the first 2500 samples were discarded as burn-in. All analyses were performed on the CIPRES Science Gateway computer cluster94, using BEAGLE95. Convergence and performance of each MCMC run were evaluated using Tracer 1.7.192 and the ESS for each parameter. We combined the four runs using LogCombiner 1.8.497. The MCC tree was reconstructed with median age and 95% CI. The BEAST files generated for this study are available in Figshare (see Data availability).
Dual biogeographic history of Papilionidae and Aristolochiaceae
We estimated the ancestral area of origin and geographic range evolution for both clades using the ML approach of DEC model109 as implemented in the C++ version137,138 that is available at: https://github.com/champost/DECX. To infer the biogeographic history of a clade, DEC requires a time-calibrated tree, the current distribution of each species for a set of geographic areas, and a time-stratified geographic model that is represented by connectivity matrices for specified time intervals spanning the entire evolutionary history of the group.
The geographic distribution for each species in Papilionidae30,112,113 and Aristolochiaceae was categorized as present or absent in each of the following areas: (1) WN, (2) East Nearctic, (3) CA, (4) South America, (5) West Palaearctic, (6) EP, (7) Madagascar, (8) Indonesia and Wallacea, (9) India, (10) Africa and (11) Australasia. The resulting matrices of species distribution for the two groups are available in Figshare (see Data availability).
A time-stratified geographic model was built using connectivity matrices that take into account palaeogeographic changes through time, with time slices indicating the possibility or not for a species to access a new area138. Based on palaeogeographical reconstructions139–141, we created a connectivity matrix for each geological epoch that represented a period bounded by major changes in tectonic and climatic conditions thought to have affected the distribution of organisms. The following geological epochs were selected: (1) 0–5.33 Ma (Pliocene to present), (2) 5.33–23.03 Ma (Miocene), (3) 23.03–33.9 Ma (Oligocene), (4) 33.9–56 Ma (Eocene) and (5) 56 Ma to the origin of the clade (Palaeocene to Late Cretaceous). For each of these five time intervals, we specified constraints on area connectivity by coding 0 if any two areas are not connected or 1 if they are connected in a given time interval. We assumed a conservative dispersal matrix with equal dispersal rates between areas through time142.
Impact of host–plant shifts on swallowtail diversification
We tested the effect of host–plant association on diversification by estimating speciation and extinction rates with five methods to cross-test hypotheses and corroborate results. Analyses were performed on 100 dated trees randomly sampled from the Bayesian dating analyses to take into account the uncertainty in age estimates. We used the following approaches: (1) ML-based trait-dependent diversification143,144; (2) ML-based time-dependent diversification145; (3) Bayesian analysis of macroevolutionary mixture146; (4) Bayesian branch-specific diversification rates147; and (5) Bayesian episodic birth–death model148. It is worth mentioning that each method differs at several points in their estimation of speciation and extinction rates. For instance, trait-dependent birth–death models estimate constant speciation and extinction rates144, whereas time-dependent birth–death models estimate clade-specific speciation and extinction rates and their variation through time145,147. Therefore, we expect some differences in the values of estimated diversification rates that are inherent to each approach. Our diversification analyses should be seen as complementary to the inferred diversification trend rather than corroborating the values and magnitude of speciation and extinction rates.
First, we computed the probability of obtaining a clade as large as size n, given the crown age of origin, the overall net diversification rate of the family, and an extinction rate as a fraction of speciation rate following the approach in Condamine et al.34 relying on the method of moments149. We used the R-package LASER 2.3150 to estimate the net diversification rates of Papilionidae and six clades shifting to new host plants with the bd.ms function (providing crown age and total species diversity). Then, we used the crown.limits function to estimate the mean expected clade size for each clade shifting to new host plants given clades’ crown age and overall net diversification rates, and we finally computed the probability to observe such clade size using the crown.p function. All rate estimates were calculated with three ε values (ε = 0/0.5/0.9), knowing that the extinction rate in swallowtails is usually low34 (supported by the results of this study).
Second, we relied on the state-dependent speciation and extinction (SSE) model, in which speciation and extinction rates are associated with phenotypic evolution of a trait along a phylogeny143. In particular, we used the MuSSE144 implemented in the R-package diversitree 0.9-10151, which allows multiple character states to be studied. Larval host–plant data were taken from previous works5,18,30,34,112,113,152. The following ten host–plant character states and corresponding ratios of sampled species in the tree of all known species for each character (sampling fractions) were used: 1 = Aristolochiaceae (110/152), 2 = Annonaceae (69/138), 3 = Lauraceae (33/39), 4 = Apiaceae (9/10), 5 = Rutaceae (119/163), 6 = Crassulaceae (19/19), 7 = Papaveraceae (44/44), 8 = Fabaceae (1/1), 9 = Zygophyllaceae (2/2), and 10 = Magnoliaceae (2/2). Data at a lower taxonomic level than plant family were not used because of the large number of multiple associations exhibited by genera that could alter the phylogenetic signal. We assigned a single state to each species by selecting the food plant with the maximum number of collections for each species. We did not employ multiple states per species, which represents a lesser problem because (1) few swallowtail species feed on multiple plant families, (2) current shared-state models can only model two states, and (3) the addition of multi-plant states to the MuSSE analysis would have greatly increased the number of parameters. We performed both ML and Bayesian MCMC analyses (10,000 steps) performed using an exponential (1/(2 × net diversification rate)) prior with starting parameter values obtained from the best-fitting ML model and resulting speciation, extinction and transition rates. After a burn-in of 500 steps, we estimated posterior density distribution for speciation, extinction and transition rates. There have been concerns about the power of SSE models to infer diversification dynamics from a distribution of species traits153–155, hence other birth–death models were used to corroborate the results obtained with SSE models.
Third, to provide an independent assessment of the relationship between diversification rates and host specificity, we used the ML approach of Morlon et al.145 implemented in the R-package RPANDA 1.3156. This is a birth–death method in which speciation and/or extinction rates may change continuously through time. This method has the advantage of not assuming a constant extinction rate over time (unlike BAMM146), and allows clades to have declining diversity since extinction can exceed speciation, meaning that diversification rates can be negative145. For each clade that shifted to a new host family, we designed and fitted six diversification models: (1) a Yule model, where speciation is constant and extinction is null; (2) a constant birth–death model, where speciation and extinction rates are constant; (3) a variable speciation rate model without extinction; (4) a variable speciation rate model with constant extinction; (5) a rate-constant speciation and variable extinction rate model; and (6) a model in which both speciation and extinction rates vary. Models were compared by computing the ML estimate of each model and the resulting Akaike information criterion corrected by sample size. We then plotted rates through time with the best-fit model for each clade, and the rates for the family as a whole for comparison purpose.
Fourth, we performed models that allow diversification rates to vary among clades across the whole phylogeny. BAMM 2.5146,157 was used to explore for differential diversification dynamic regimes among clades differing in their host–plant feeding. BAMM can automatically detect rate shifts and sample distinct evolutionary dynamics that explain the diversification dynamics of a clade without a priori hypotheses on how many and where these shifts might occur. Evolutionary dynamics can involve time-variable diversification rates; in BAMM, speciation is allowed to vary exponentially through time while extinction is maintained constant: subclades in a tree may diversify faster (or slower) than others. This Bayesian approach can be useful in detecting shifts of diversification potentially associated with key innovations157. BAMM analyses were run with four MCMC for 20 million generations, sampling every 20,000th and with three different values (1, 5 and 10; Supplementary Table 3) of the compound Poisson prior (CPP) to ensure the posterior is independent of the prior158. We accounted for non-random incomplete taxon sampling using the implemented analytical correction; we set a sampling fraction per genus based on the known species diversity of each genus. Mixing and convergence among runs (ESS > 200 after 15% burn-in) were assessed with the R-package BAMMtools 2.1159 to estimate (1) the mean global rates of diversification through time, (2) the estimated number of rate shifts evaluating alternative diversification models comparing priors and posterior probabilities and (3) the clade-specific rates through time when a distinct macroevolutionary regime is identified.
Fifth, BAMM has been criticized for incorrectly modelling rate shifts on extinct lineages, that is, unobserved (extinct or non-sampled) lineages inherit the ancestral diversification process and cannot experience subsequent diversification-rate shifts158,160. To solve this, we used a Bayesian approach implemented in RevBayes 1.0.10161 that models rate shifts consistently on extinct lineages by using the SSE framework147,158. Although there is no information of rate shifts for unobserved/extinct lineages in a phylogeny including extant species only, these types of events must be accounted for in computing the likelihood. The number of rate categories is fixed in the analysis but RevBayes allows any number to be specified, thus allowing direct comparison of different macroevolutionary regimes.
Finally, we evaluated the impact of abrupt changes in diversification using the Bayesian episodic birth–death model of CoMET148 implemented in the R-package TESS 2.1162. These models allow detection of discrete changes in speciation and extinction rates concurrently affecting all lineages in a tree, and estimate changes in diversification rates at discrete points in time, but can also infer mass extinction events (sampling events in which the extant diversity is reduced by a fraction163). Speciation and extinction rates can change at those points, but remain constant within time intervals. In addition, TESS uses independent CPPs to simultaneously detect mass extinction events and discrete changes in speciation and extinction rates, while TreePar estimates the magnitude and timing of speciation and extinction changes independently to the occurrence of mass extinctions (i.e. the three parameters cannot be estimated simultaneously due to parameter identifiability issues163). We performed two independent analyses allowing and disallowing mass extinction events. Bayes factor comparisons were used to assess the model fit between models with varying number and time of changes in speciation/extinction rates and mass extinctions.
Detecting genome-wide adaptations during host–plant shifts
We analysed genomic sequence data in swallowtail butterflies that have independently shifted to new ecological (biological) traits. Similar approaches have been conducted on mammals164,165 and birds166, but have been rarely implemented on arthropod groups over such a long geological time scale. Here, we estimated swallowtail molecular evolution with whole-genome data and compared selection regimes on protein-coding genes along independent branches with or without host–plant shift and/or environmental shift.
For these analyses, we studied 45 whole genomes46 covering all 32 genera of the family Papilionidae: 41 of which were previously generated by our research group added to four genomes already available70–72. In summary, raw reads (Sequence Read Archive: SRR8954507-SRR8954549) were cleaned using Trimmomatic 0.33167, and assembled into contigs and scaffolds with SOAPdenovo-63mer 2.04168 to obtain whole-genome assemblies (30× average read depth46). All coding DNA sequences (CDS) were retrieved from the high-quality annotated genome of P. xuthus71. To annotate the sequences of all our genomes, a BLAST search using all available CDS of P. xuthus was performed at the amino-acid level (using tblastn). For each species, the recovered genes were aligned one by one with P. xuthus using TranslatorX169. This method performs alignment at the amino-acid level and preserves the open reading frame. All sites showing intraspecific variation were set to N, to conservatively avoid false informative sites. Any contamination was removed using CroCo 0.1170 and orthologous proteins were identified with OrthoFinder 2.2.0171. Finally, CDS alignments were strongly cleaned from misaligned sequences (gene by gene) using HMMCleaner 1.8172. A last cleaning step was performed using trimAl 1.2rev59173, which is designed to trim alignments for large-scale phylogenomic analyses. The resulting dataset comprised 6621 genes in at least four sampled species (median of 32% of missing data), which was used to reconstruct a robust phylogenomic tree of Papilionidae46 (Supplementary Fig. 18).
We used this genomic dataset of 45 species representing all genera in which the resulting genus-level swallowtail phylogenomic tree46 accurately represents the evolutionary associations with host plants as estimated using the ancestral-state analyses applied to the species-level phylogeny34 (Fig. 1 and Supplementary Figs. 4 and 5). We thus transferred the inference of ancestral host–plant shifts on the phylogenomic tree and selected the branches representing a host–plant shift and/or a shift of climate preference (in general from tropical to temperate conditions; Supplementary Fig. 10). We also selected branches with no change as negative controls73. As a result, 14 branches are selected to measure the impact of a host–plant shift and 14 branches are selected as controls (Supplementary Fig. 18). Within these 14 branches with an ecological change, nine branches represent host–plant shifts only, and five branches correspond to shifts in both host plant and environment (from tropical to temperate conditions). To test the impact of these different changes on the genomes, two datasets were created, Dataset 1 and 2. Given the low quality of the genomes of Allancastria cerisyi and Parnassius imperator, these two genomes were discarded for the downstream analyses. We first selected the genes from the 6621-gene dataset for each focal branch using three criteria: (1) the dataset is composed only of orthologous protein-coding genes (OrthoFinder 2.2171), (2) the species needed to accurately define the branch were available (i.e. crown node of the clade) and (3) for each branch, one species per tribe was available, and therefore include a different number of genes per branch. Thus, for Dataset 1, only the genes containing sequences for the species needed to generate all focal branches were selected. This stringent selection leads to Dataset 1, comprising only 520 genes but the same genes for all branches (no missing genes). For Dataset 2, the genes were selected for each branch independently (i.e. for a given branch, a gene was selected if the sequence needed to generate that branch was present). This second selection leads to 1439 genes per branch on average among a total of 1533 genes, which were selected at least once for one branch. The genomic dataset is available in Figshare (see Data availability).
We studied the ratio (ω) of dN/dS to find genes under positive selection76,174. The dN/dS ratio is traditionally used to estimate selective pressure from protein-coding sequences. If host–plant shifts have no effect on the selection of a given gene, we expect a dN/dS = 1 and the selective regime is considered neutral. However, if host–plant shifts result in positive selection on coding genes, the ratio increases such that dN/dS > 1. Finally, it is possible that host–plant shifts lead to purifying selection, thus reducing the number of dN and resulting in dN/dS < 1. Here we focused on the adaptation of Papilionidae to host–plant shifts, that is, outgroups are not studied. We tested if branches representing inferred host–plant shifts along the phylogeny of swallowtails have more genes with dN/dS > 1 than lineages that did not have an inferred shift. The branch-site models allow ω to vary both among sites in the protein and across branches on the tree and aim to detect positive selection affecting a few sites along particular lineages. The approach described by Zhang et al.175 was chosen to determine genome-wide selection regimes as performed with two ML models: (1) a null model assuming two site classes, one with dN/dS < 1 and one with dN/dS = 1 (model = 2, NSsites = 2, fix_omega = 1, omega = 1) and (2) an alternative model adding a third site class with dN/dS > 1 (model = 2, NSsites = 2, fix_omega = 0, omega = 1.5). The fit for including positive selection is tested using a likelihood ratio test comparing the null model with the alternative model with one degree of freedom76,176. If the alternative model is better suited to host-shift branches, it is more likely the gene was under positive selection during the host–plant shifts. For each gene and for each branch, both the null and alternative models using CodeML were implemented in PAML 4177 with a fixed topology (as inferred with the phylogenomic dataset46) and the nucleotide alignment of each gene. To test the robustness of the estimations, we used a false discovery rate test to control false positives178. Finally, for each branch, we reported the number of genes under positive selection (i.e. for which the alternative model including the site class with dN/dS > 1 have a better likelihood) on the total gene number. The proportion of genes under positive selection was compared with associated control branches for branches representing host–plant shifts, environmental shifts or both plant and environmental shifts using the non-parametric, Wilcoxon rank-sum test179.
Sensitivity analyses
We performed several control analyses to ensure that the signal of more genes under positive selection in host–plant shifts branches is not artefactual.
First, it has been shown that the choice of the tree is an important factor for the branch-site analysis of positive selection180. Indeed, constraining the topology for a given gene may lead to overestimating the number of substitution events for the constrained branches78 and so could lead to overestimating the dN/dS ratio. Estimating dN/dS over thousands of gene trees would make the branch comparison not equal between control and test branches. Indeed, in a given gene tree it is likely and expected that the species topology is not always recovered, which results in a different number of branches compared to the species tree. For instance, the host–plant shift to Annonaceae might disappear in certain proportions of genes. We thus decided to estimate dN/dS on a fixed species tree topology for all genes to be sure to be able to measure this ratio for each gene that must be present in the topology for the focal branches. However, given that this issue can lead to a bias in our analysis, we decided to compute the number of gene trees that did not recover the branches of interest. We then checked whether the branches leading to a host–plant shift were more often unrecovered than the control branches without shift. Overall the control branches were less often recovered than host–plant shift branches (P = 0.030, Wilcoxon rank-sum test; data presented in Supplementary Table 4), which suggests that if gene tree/species tree discordance leads to an overestimation of positive selection, then this overestimation is higher for control branches than for host–plant shift branches. Finally, we filtered out the gene trees for which the focal branches were recovered in agreement with the species tree and used these genes to re-estimate the proportion of genes under positive selection among this new set of genes. We found that the P value remains significant (P = 0.0444, Wilcoxon rank-sum test; more genes during host–plant shifts than along control branches, Supplementary Table 2). Then, we specifically focused on missing data and GC-content variation among genes known to bias dN/dS estimations. Missing data are prone to introducing misaligned regions that could create false positives in branch-site likelihood method for detecting positive selection181–183. Variations in GC content are known to impact the estimation of dN/dS mainly through the process of GC-biased gene conversion (gBGC184–186).
The number of missing data (‘N’ and ‘-’) sites and GC content at the third codon position (GC3) were computed using a home-made C++ program created with BIO++ library187. Mean GC content and missing data were calculated per gene and for each branch. For a given branch, mean GC3 and missing data were computed for the species of a clade for which the branch is the root. All statistics and graphical representations were performed using the R-packages tidyverse188 and cowplot189. We found that genes under positive selection (PSgenes, nDataset 1 = 142, nDataset 2 = 407) have significantly more missing data and GC3 than genes not under positive selection (NSgenes, nDataset 1 = 378, nDataset 2 = 1126; P = 0.001/0.02 for the two datasets, respectively, Mann–Whitney test; Supplementary Fig. 20). This result confirms that branch-site likelihood methods for detecting positive selection are sensitive to missing data, probably because of misaligned sites181,182, and that GC content that may be influenced by gBGC184,185.
Missing data were, however, heterogeneously distributed among species, ranging from <1% in P. xuthus to 45% in Hypermnestra helios (Supplementary Fig. 21). The difference in missing data between branches with (n = 14, mean missing Dataset 1 = 13.4%, mean missingDataset 2 = 14.1%) or without host–plant shifts (n = 14, mean missingDataset 1 = 12.8%, mean missingDataset 2 = 12.7%) is not significant (P = 0.83/1.00 for the two datasets, respectively, Mann–Whitney test; Supplementary Fig. 22). In addition, there is no correlation between the number of genes under positive selection and the amount of missing data (P = 0.33/0.20 for the two datasets, respectively, Spearman’s correlation test; Supplementary Fig. 23). For GC3, we also found variation between species ranging from 37% in Parnassius smintheus to 44% in Papilio antimachus (Supplementary Fig. 24). Similarly to missing data, we found no significant difference between plant-shift and no plant-shift branches (P = 0.63/0.63 for the two datasets, Mann–Whitney test; Supplementary Fig. 25) and there is no correlation between the number of genes under positive selection and GC3 (P = 0.20/0.1362 for the two datasets, respectively, Spearman’s correlation test; Supplementary Fig. 26).
Despite the known fact that false positives can increase with the amount of missing data, our control analyses indicate that variations in missing data and GC content do not drive the signal that more genes are under positive selection in branches that have undergone a host–plant shift. Additionally to these controls, we checked by eyes all the gene alignments at the amino-acid level for genes under positive selection in branches with and without host–plant shifts using SeaView 4190. Misaligned regions, which could lead to biased dN/dS ratios191, were not significantly more detected for genes under positive selection in branches with host–plant shifts. In some cases, we found ourselves in complicated situations to discriminate between false- and true-positive selected genes.
Overall, given our alignment checks and sensitivity analyses, we do not see any reason for biased dN/dS ratios in genes along branches with or without host–plant shifts. False-positive and false-negative genes can be present in the two categories of branches, but, in any cases, the general pattern observed is likely to remain conserved.
Gene ontology
To annotate proteins of our alignment, we used the two different approaches implemented in PANTHER 14192 (available at: http://pantherdb.org/) and EggNOG 5.0193,194 (available at: http://eggnog5.embl.de/#/app/home). We used the HMM Scoring tool to assign PANTHER family (library version 14.1192) to the protein of P. xuthus (assembly Pxut_1.0); similar results were obtained using another high-quality annotated genome (from H. melpomene) as reference (assembly ASM31383v2). We performed the statistical overrepresentation test implemented on the PANTHER online website, relying on the GO categories in the PANTHER GO-Slim annotation dataset including Molecular function, Biological process and Cellular component. First, we tested if positively selected genes have over- or under-represented functional GO categories as compared to the whole set of genes (option “PANTHER Generic Mapping”). Second, we tested if positively selected genes involving a host–plant shift along the 14 branches have over- or under-represented functional categories. These statistical comparisons were performed with the Fisher’s exact test using the false discovery rate correction to control for false positives. Independently, we used the eggNOG-mapper v2193 (https://github.com/eggnogdb/eggnog-mapper) and the associated Lepidoptera database (LepNOG, including the genomes of Bombyx mori, Danaus plexippus and Heliconius melpomene194) to annotate the proteins of our dataset. EggNOG uses precomputed orthologous groups and phylogenies from the database to transfer functional information from fine-grained orthologs only. We used the diamond method as recommended193. Finally, we reported the GO families inferred for the proteins of the Dataset 2.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This project has received funding from the Marie Curie International Outgoing Fellow under the European Union’s Seventh Framework Programme (project BIOMME, agreement no. 627684), a PICS grant from the CNRS (project PASTA), an ‘Investissement d’Avenir’ grant from the Agence Nationale de la Recherche (project CASMA, CEBA, ref. ANR-10-LABX-25-01) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (project GAIA, agreement no. 851188) to F.L.C.; a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2018-04920) to F.A.H.S.; and a German Research Foundation grant (WA 2461/9-1) to S.W. We are grateful to Sophie Dang, Troy Locke and Corey Davis at the Molecular Biology Service Unit of the University of Alberta for their help, assistance, and advice on next-generation sequencing. The analyses benefited from the Montpellier Bioinformatics Biodiversity (MBB) platform services. Finally, we are grateful to Seth Bybee, Frédéric Delsuc, Claude dePamphilis, Krushnamegh Kunte, Conrad Labandeira, Harald Letsch, Sören Nylin, Timothy O’Hara, Susanne Renner and Chris Wheat for helpful comments and discussions on earlier drafts of the study.
Source data
Author contributions
F.L.C. and F.A.H.S. designed and conceived the research. R.A. and F.L.C. assembled the phylogenetic data for swallowtail butterflies. S.W., O.A.P.-E., G.C., F.L.C. and R.A. assembled the phylogenetic data for birthworts. R.A. and F.L.C. analysed the phylogenetic data. R.A. and F.L.C. performed the estimations of the ancestral states. F.L.C. performed the diversification analyses. A.-L.C. and F.L.C. generated the genomic data. R.A. and B.N. assembled and analysed the genomic data. All authors contributed to the interpretation and discussion of results. R.A. and F.L.C. drafted the paper with substantial input from all authors.
Data availability
Supermatrix datasets (for phylogenetic analyses), phylogenetic trees, host–plant preferences, species geographic distributions, and gene alignments (for dN/dS analyses) that are necessary for repeating the analyses described here have been made available through the Figshare digital data repository (10.6084/m9.figshare.12278402). Source data are provided with this paper.
Code availability
Bioinformatic scripts used to perform the analyses described here are available through the Figshare digital data repository (10.6084/m9.figshare.12278402).
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Niklas Wahlberg, and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rémi Allio, Email: rem.allio@yahoo.fr.
Fabien L. Condamine, Email: fabien.condamine@gmail.com
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-20507-3.
References
- 1.Becerra JX. On the factors that promote the diversity of herbivorous insects and plants in tropical forests. Proc. Natl Acad. Sci. USA. 2015;112:6098–6103. doi: 10.1073/pnas.1418643112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stork NE. How many species of insects and other terrestrial arthropods are there on earth? Annu. Rev. Entomol. 2018;63:31–45. doi: 10.1146/annurev-ento-020117-043348. [DOI] [PubMed] [Google Scholar]
- 3.Grimaldi, D. A. & Engel, M. S. Evolution of the Insects (Cambridge University Press, 2005).
- 4.Strong, D. R., Lawton, J. H. & Southwood, R. Insects on Plants: Community Patterns and Mechanisms (Harvard University Press, 1984).
- 5.Ehrlich PR, Raven PH. Butterflies and plants: a study in coevolution. Evolution. 1964;18:586–608. doi: 10.1111/j.1558-5646.1964.tb01674.x. [DOI] [Google Scholar]
- 6.Thompson JN. Concepts of coevolution. Trends Ecol. Evol. 1989;4:179–183. doi: 10.1016/0169-5347(89)90125-0. [DOI] [PubMed] [Google Scholar]
- 7.Mitter C, Farrell B, Wiegmann B. The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am. Nat. 1988;132:107–128. doi: 10.1086/284840. [DOI] [Google Scholar]
- 8.Farrell BD. ‘Inordinate fondness’ explained: why are there so many beetles? Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 9.Berenbaum, M. & Specialization, P. F. Chemical Mediation of Host-plant Specialization: The Papilionid Paradigm. Specialization, Speciation, and Radiation: The Evolutionary Biology of Herbivorous Insects (University of California Press, 2008).
- 10.Winter S, Friedman ALL, Astrin JJ, Gottsberger B, Letsch H. Timing and host plant associations in the evolution of the weevil tribe Apionini (Apioninae, Brentidae, Curculionoidea, Coleoptera) indicate an ancient co-diversification pattern of beetles and flowering plants. Mol. Phylogenet. Evol. 2017;107:179–190. doi: 10.1016/j.ympev.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Kergoat GJ, et al. Opposite macroevolutionary responses to environmental changes in grasses and insects during the Neogene grassland expansion. Nat. Commun. 2018;9:5089. doi: 10.1038/s41467-018-07537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheat CW, et al. The genetic basis of a plant–insect coevolutionary key innovation. Proc. Natl Acad. Sci. USA. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edger PP, et al. The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl Acad. Sci. USA. 2015;112:8362–8366. doi: 10.1073/pnas.1503926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calla B, et al. Cytochrome P450 diversification and hostplant utilization patterns in specialist and generalist moths: Birth, death and adaptation. Mol. Ecol. 2017;26:6021–6035. doi: 10.1111/mec.14348. [DOI] [PubMed] [Google Scholar]
- 15.Nallu S, et al. The molecular genetic basis of herbivory between butterflies and their host plants. Nat. Ecol. Evol. 2018;2:1418–1427. doi: 10.1038/s41559-018-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karageorgi M, et al. Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature. 2019;574:409–412. doi: 10.1038/s41586-019-1610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo RK, Warren AD, Collins SC, Kodandaramaiah U. Hostplant change and paleoclimatic events explain diversification shifts in skipper butterflies (Family: Hesperiidae) BMC Evol. Biol. 2017;17:174. doi: 10.1186/s12862-017-1016-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Condamine FL, Rolland J, Höhna S, Sperling FAH, Sanmartín I. Testing the role of the red queen and court jester as drivers of the macroevolution of apollo butterflies. Syst. Biol. 2018;67:940–964. doi: 10.1093/sysbio/syy009. [DOI] [PubMed] [Google Scholar]
- 19.Letsch H, et al. Climate and host-plant associations shaped the evolution of ceutorhynch weevils throughout the Cenozoic. Evolution. 2018;72:1815–1828. doi: 10.1111/evo.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forister ML, et al. The global distribution of diet breadth in insect herbivores. Proc. Natl Acad. Sci. USA. 2015;112:442–447. doi: 10.1073/pnas.1423042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler IS, Mitter C, Scheffer SJ. Repeated climate-linked host shifts have promoted diversification in a temperate clade of leaf-mining flies. Proc. Natl Acad. Sci. USA. 2009;106:18103–18108. doi: 10.1073/pnas.0904852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomicki G, Weber M, Antonelli A, Bascompte J, Kiers ET. The impact of mutualisms on species richness. Trends Ecol. Evol. 2019;34:698–711. doi: 10.1016/j.tree.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Janz N. Ehrlich and Raven revisited: mechanisms underlying codiversification of plants and enemies. Annu. Rev. Ecol. Evol. Syst. 2011;42:71–89. doi: 10.1146/annurev-ecolsys-102710-145024. [DOI] [Google Scholar]
- 24.Suchan T, Alvarez N. Fifty years after Ehrlich and Raven, is there support for plant–insect coevolution as a major driver of species diversification? Entomol. Exp. Appl. 2015;157:98–112. doi: 10.1111/eea.12348. [DOI] [Google Scholar]
- 25.Endara M-J, et al. Coevolutionary arms race versus host defense chase in a tropical herbivore-plant system. Proc. Natl Acad. Sci. USA. 2017;114:E7499–E7505. doi: 10.1073/pnas.1707727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon J-C, et al. Genomics of adaptation to host-plants in herbivorous insects. Brief. Funct. Genomics. 2015;14:413–423. doi: 10.1093/bfgp/elv015. [DOI] [PubMed] [Google Scholar]
- 27.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. Caterpillars lack a resident gut microbiome. Proc. Natl Acad. Sci. USA. 2017;114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hua X, Bromham L. Darwinism for the genomic age: connecting mutation to diversification. Front. Genet. 2017;8:12. doi: 10.3389/fgene.2017.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hembry, D. H. & Weber, M. G. Ecological interactions and macroevolution: a new field with old roots. Annu. Rev. Ecol. Evol. Syst. 51, (2020).
- 30.Scriber, J. M., Tsubaki, Y. & Lederhouse, R. C. Swallowtail Butterflies: Their Ecology and Evolutionary Biology (Scientific Publishers, 1995).
- 31.Nishida R. Sequestration of defensive substances from plants by Lepidoptera. Annu. Rev. Entomol. 2002;47:57–92. doi: 10.1146/annurev.ento.47.091201.145121. [DOI] [PubMed] [Google Scholar]
- 32.Schmeiser HH, Stiborovà M, Arlt VM. Chemical and molecular basis of the carcinogenicity of Aristolochia plants. Curr. Opin. Drug Discov. Dev. 2009;12:141–148. [PubMed] [Google Scholar]
- 33.Poon SL, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med. 2013;5:197ra101. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- 34.Condamine FL, Sperling FAH, Wahlberg N, Rasplus J-Y, Kergoat GJ. What causes latitudinal gradients in species diversity? Evolutionary processes and ecological constraints on swallowtail biodiversity. Ecol. Lett. 2012;15:267–277. doi: 10.1111/j.1461-0248.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 35.Simonsen TJ, et al. Phylogenetics and divergence times of Papilioninae (Lepidoptera) with special reference to the enigmatic genera Teinopalpus and Meandrusa. Cladistics. 2011;27:113–137. doi: 10.1111/j.1096-0031.2010.00326.x. [DOI] [PubMed] [Google Scholar]
- 36.Berenbaum MR, Favret C, Schuler MA. On defining ‘Key Innovations’ in an adaptive radiation: cytochrome P450s and Papilionidae. Am. Nat. 1996;148:S139–S155. doi: 10.1086/285907. [DOI] [Google Scholar]
- 37.Cohen MB, Schuler MA, Berenbaum MR. A host-inducible cytochrome P-450 from a host-specific caterpillar: molecular cloning and evolution. Proc. Natl Acad. Sci. USA. 1992;89:10920–10924. doi: 10.1073/pnas.89.22.10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Schuler MA, Berenbaum MR. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: specificity and substrate encounter rate. Proc. Natl Acad. Sci. USA. 2003;100(Suppl.):14593–14598. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JN. Variation in preference and specificity in monophagous and oligophagous swallowtail butterflies. Evolution. 1988;42:118–128. doi: 10.1111/j.1558-5646.1988.tb04112.x. [DOI] [PubMed] [Google Scholar]
- 40.Thompson JN, Wehling W, Podolsky R. Evolutionary genetics of host use in swallowtail butterflies. Nature. 1990;344:148–150. doi: 10.1038/344148a0. [DOI] [Google Scholar]
- 41.Berenbaum, M. R. & Feeny, P. P. in Specialization, Speciation, and Radiation: The Evolutionary Biology of Herbivorous Insects (ed. Tilmon, K.) 2–19 (University of California Press, 2008).
- 42.Zakharov EV, Caterino MS, Sperling FAH. Molecular phylogeny, historical biogeography, and divergence time estimates for swallowtail butterflies of the genus Papilio (Lepidoptera: Papilionidae) Syst. Biol. 2004;53:193–215. doi: 10.1080/10635150490423403. [DOI] [PubMed] [Google Scholar]
- 43.Braby M, Trueman J, Eastwood R. When and where did troidine butterflies (Lepidoptera: Papilionidae) evolve? Phylogenetic and biogeographic evidence suggests an origin in remnant Gondwana in the Late Cretaceous. Invertebr. Syst. 2005;19:113–143. doi: 10.1071/IS04020. [DOI] [Google Scholar]
- 44.Condamine FL, Silva-Brandão KL, Kergoat GJ, Sperling FA. Biogeographic and diversification patterns of Neotropical Troidini butterflies (Papilionidae) support a museum model of diversity dynamics for Amazonia. BMC Evol. Biol. 2012;12:82. doi: 10.1186/1471-2148-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Condamine FL, et al. Deciphering the evolution of birdwing butterflies 150 years after Alfred Russel Wallace. Sci. Rep. 2015;5:11860. doi: 10.1038/srep11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allio R, et al. Whole genome shotgun phylogenomics resolves the pattern and timing of swallowtail butterfly evolution. Syst. Biol. 2020;69:38–60. doi: 10.1093/sysbio/syz030. [DOI] [PubMed] [Google Scholar]
- 47.McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl Acad. Sci. USA. 2009;106:7083–7088. doi: 10.1073/pnas.0810618106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi D, Setoguchi H. Molecular phylogeny and taxonomic implications of Asarum (Aristolochiaceae) based on ITS and matK sequences. Plant Species Biol. 2018;33:28–41. doi: 10.1111/1442-1984.12189. [DOI] [Google Scholar]
- 49.Wanke S, et al. Evolution of Piperales—matK gene and trnK intron sequence data reveal lineage specific resolution contrast. Mol. Phylogenet. Evol. 2007;42:477–497. doi: 10.1016/j.ympev.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Neinhuis C, Wanke S, Hilu KW, Müller K, Borsch T. Phylogeny of Aristolochiaceae based on parsimony, likelihood, and Bayesian analyses of trnL-trnF sequences. Plant Syst. Evol. 2005;250:7–26. doi: 10.1007/s00606-004-0217-0. [DOI] [Google Scholar]
- 51.Wanke S, González F, Neinhuis C. Systematics of pipevines: combining morphological and fast‐evolving molecular characters to investigate the relationships within subfamily Aristolochioideae. Int. J. Plant Sci. 2006;167:1215–1227. doi: 10.1086/508024. [DOI] [Google Scholar]
- 52.González F, et al. Present trans-Pacific disjunct distribution of Aristolochia subgenus Isotrema (Aristolochiaceae) was shaped by dispersal, vicariance and extinction. J. Biogeogr. 2014;41:380–391. doi: 10.1111/jbi.12198. [DOI] [Google Scholar]
- 53.Durden, C. J. & Rose, H. Butterflies from the Middle Eocene: The Earliest Occurrence of Fossil Papilionoidea (Lepidoptera) (Prarce-Sellards Ser. Tax. Mem. Mus., 1978).
- 54.Sohn J, Labandeira C, Davis D, Mitter C. An annotated catalog of fossil and subfossil Lepidoptera (Insecta: Holometabola) of the world. Zootaxa. 2012;3286:1–132. doi: 10.11646/zootaxa.3286.1.1. [DOI] [Google Scholar]
- 55.de Jong R. Estimating time and space in the evolution of the Lepidoptera. Tijdschr. voor Entomol. 2007;150:319–346. doi: 10.1163/22119434-900000233. [DOI] [Google Scholar]
- 56.Hofmann C-C, Zetter R. Upper Cretaceous sulcate pollen from the Timerdyakh formation, Vilui Basin (Siberia) Grana. 2010;49:170–193. doi: 10.1080/00173134.2010.512364. [DOI] [Google Scholar]
- 57.Meller B. The first fossil Aristolochia (Aristolochiaceae, Piperales) leaves from Austria. Palaeontol. Electron. 2014;17:1–17. [Google Scholar]
- 58.Nee S, May RM, Harvey PH. The reconstructed evolutionary process. Philos. Trans. R. Soc. Lond. Ser. B. 1994;344:305–311. doi: 10.1098/rstb.1994.0068. [DOI] [PubMed] [Google Scholar]
- 59.Nee S. Birth-death models in macroevolution. Annu. Rev. Ecol. Evol. Syst. 2006;37:1–17. doi: 10.1146/annurev.ecolsys.37.091305.110035. [DOI] [Google Scholar]
- 60.Rabosky DL, Lovette IJ. Explosive evolutionary radiations: Decreasing speciation or increasing extinction through time? Evolution. 2008;62:1866–1875. doi: 10.1111/j.1558-5646.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 61.Crisp MD, Cook LG. Explosive radiation or cryptic mass extinction? Interpreting signatures in molecular phylogenies. Evolution. 2009;63:2257–2265. doi: 10.1111/j.1558-5646.2009.00728.x. [DOI] [PubMed] [Google Scholar]
- 62.Quental TB, Marshall CR. Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol. Evol. 2010;25:434–441. doi: 10.1016/j.tree.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 63.Morlon H. Phylogenetic approaches for studying diversification. Ecol. Lett. 2014;17:508–525. doi: 10.1111/ele.12251. [DOI] [PubMed] [Google Scholar]
- 64.Xue B, et al. Accelerated diversification correlated with functional traits shapes extant diversity of the early divergent angiosperm family Annonaceae. Mol. Phylogenet. Evol. 2020;142:106659. doi: 10.1016/j.ympev.2019.106659. [DOI] [PubMed] [Google Scholar]
- 65.Folk RA, et al. Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proc. Natl Acad. Sci. USA. 2019;116:10874–10882. doi: 10.1073/pnas.1817999116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun M, et al. Recent accelerated diversification in rosids occurred outside the tropics. Nat. Commun. 2020;11:3333. doi: 10.1038/s41467-020-17116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Losos JB. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 2010;175:623–639. doi: 10.1086/652433. [DOI] [PubMed] [Google Scholar]
- 68.Cheng T, et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 2017;1:1747–1756. doi: 10.1038/s41559-017-0314-4. [DOI] [PubMed] [Google Scholar]
- 69.Rane RV, et al. Detoxifying enzyme complements and host use phenotypes in 160 insect species. Curr. Opin. Insect Sci. 2019;31:131–138. doi: 10.1016/j.cois.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 70.Cong Q, Borek D, Otwinowski Z, Grishin NV. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 2015;10:910–919. doi: 10.1016/j.celrep.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X, et al. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 2015;6:8212. doi: 10.1038/ncomms9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishikawa H, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat. Genet. 2015;47:405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- 73.Thomas GWC, Hahn MW. Determining the null model for detecting adaptive convergence from genomic data: a case study using echolocating mammals. Mol. Biol. Evol. 2015;32:1232–1236. doi: 10.1093/molbev/msv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou Z, Zhang J. No genome-wide protein sequence convergence for echolocation. Mol. Biol. Evol. 2015;32:1237–1241. doi: 10.1093/molbev/msv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge University Press, 1983).
- 76.Yang, Z. Computational Molecular Evolution (Oxford University Press, 2006).
- 77.Venkat A, Hahn MW, Thornton JW. Multinucleotide mutations cause false inferences of lineage-specific positive selection. Nat. Ecol. Evol. 2018;2:1280–1288. doi: 10.1038/s41559-018-0584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mendes FK, Hahn MW. Gene tree discordance causes apparent substitution rate variation. Syst. Biol. 2016;65:711–721. doi: 10.1093/sysbio/syw018. [DOI] [PubMed] [Google Scholar]
- 79.Dasmahapatra KK, et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walden N, et al. Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae. Nat. Commun. 2020;11:3795. doi: 10.1038/s41467-020-17605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGee MD, et al. The ecological and genomic basis of explosive adaptive radiation. Nature. 2020;586:75–79. doi: 10.1038/s41586-020-2652-7. [DOI] [PubMed] [Google Scholar]
- 82.Thomas GWC, et al. Gene content evolution in the arthropods. Genome Biol. 2020;21:15. doi: 10.1186/s13059-019-1925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Medeiros, B. A. S. & Farrell, B. D. Evaluating species interactions as a driver of phytophagous insect divergence. bioRxiv10.1101/842153 (2019).
- 84.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 86.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ronquist F, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huelsenbeck JP, Larget B, Alfaro ME. Bayesian phylogenetic model selection using reversible jump Markov Chain Monte Carlo. Mol. Biol. Evol. 2004;21:1123–1133. doi: 10.1093/molbev/msh123. [DOI] [PubMed] [Google Scholar]
- 92.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Douady CJ, Delsuc F, Boucher Y, Doolittle WF, Douzery EJP. Comparison of bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 2003;20:248–254. doi: 10.1093/molbev/msg042. [DOI] [PubMed] [Google Scholar]
- 94.Miller MA, et al. A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evol. Bioinforma. 2015;11:EBO.S21501. doi: 10.4137/EBO.S21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ayres DL, et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 2012;61:170–173. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith ME, Singer B, Carroll A. 40Ar/39Ar geochronology of the Eocene Green River Formation, Wyoming. Geol. Soc. Am. Bull. 2003;115:549–565. doi: 10.1130/0016-7606(2003)115<0549:AGOTEG>2.0.CO;2. [DOI] [Google Scholar]
- 99.de Jong R. Fossil butterflies, calibration points and the molecular clock (Lepidoptera: Papilionoidea) Zootaxa. 2017;4270:1–63. doi: 10.11646/zootaxa.4270.1.1. [DOI] [PubMed] [Google Scholar]
- 100.Scudder SH. Fossil butterflies. Mem. Am. Assoc. Adv. Sci. 1875;1:1–99. [Google Scholar]
- 101.Rasnitsyn, A. P. & Zherikhin, V. V. in History of Insects 437–446 (Kluwer Academic Publishers, 2002).
- 102.Rebel H. Doritites bosniaskii. Sitzungsberichte der akademie der wissenschaften. Mathematischen-Naturwissenschaftliche classe. Abt. 1 Mineral. Biol. Erdkd. 1898;1:734–741. [Google Scholar]
- 103.Carpenter, F. Treatise on Invertebrate Paleontology: Arthropoda 4. Superclass Hexapoda (Geological Society of America, 1992).
- 104.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. N. Phytol. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 105.Sohn J-C, Labandeira CC, Davis DR. The fossil record and taphonomy of butterflies and moths (Insecta, Lepidoptera): implications for evolutionary diversity and divergence-time estimates. BMC Evol. Biol. 2015;15:12. doi: 10.1186/s12862-015-0290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Toussaint EFA, Condamine FL. To what extent do new fossil discoveries change our understanding of clade evolution? A cautionary tale from burying beetles (Coleoptera: Nicrophorus). Biol. J. Linn. Soc. 2016;117:686–704. doi: 10.1111/bij.12710. [DOI] [Google Scholar]
- 107.Gernhard T. The conditioned reconstructed process. J. Theor. Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 108.Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- 109.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 110.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 111.Igarashi S. The classification of the Papilionidae mainly based on the morphology of their immature stages. Lepid. Sci. 1984;34:41–96. [Google Scholar]
- 112.Collins, N. M. & Morris, M. Threatened Swallowtail Butterflies of the World: the IUCN Red Data Book (IUCN, 1985).
- 113.Tyler, H. A., Brown, K. S. & Wilson, K. H. Swallowtail Butterflies of the Americas: A Study in Biological Dynamics, Ecological Diversity, Biosystematics, and Conservation (Scientific Publishers, 1994).
- 114.Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. doi: 10.1111/j.0014-3820.2005.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 115.Massoni J, Couvreur TL, Sauquet H. Five major shifts of diversification through the long evolutionary history of Magnoliidae (Angiosperms) BMC Evol. Biol. 2015;15:49. doi: 10.1186/s12862-015-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kyalangalilwa B, Boatwright JS, Daru BH, Maurin O, van der Bank M. Phylogenetic position and revised classification of Acacia s.l. (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Bot. J. Linn. Soc. 2013;172:500–523. doi: 10.1111/boj.12047. [DOI] [Google Scholar]
- 117.Miller JT, Murphy DJ, Ho SYW, Cantrill DJ, Seigler D. Comparative dating of Acacia: combining fossils and multiple phylogenies to infer ages of clades with poor fossil records. Aust. J. Bot. 2013;61:436–445. doi: 10.1071/BT13149. [DOI] [Google Scholar]
- 118.Michalak I, Zhang L-B, Renner SS. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. J. Biogeogr. 2010;37:1214–1226. doi: 10.1111/j.1365-2699.2010.02306.x. [DOI] [Google Scholar]
- 119.Wu S-D, et al. Evolution of asian interior arid-zone biota: Evidence from the diversification of asian Zygophyllum (Zygophyllaceae) PLoS ONE. 2015;10:e0138697. doi: 10.1371/journal.pone.0138697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chase MW, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- 121.Christenhusz MJM, Vorontsova MS, Fay MF, Chase MW. Results from an online survey of family delimitation in angiosperms and ferns: recommendations to the Angiosperm Phylogeny Group for thorny problems in plant classification. Bot. J. Linn. Soc. 2015;178:501–528. doi: 10.1111/boj.12285. [DOI] [Google Scholar]
- 122.Gonzáles F, Rudall PJ, Furness CA. Microsporogenesis and systematics of Aristolochiaceae. Bot. J. Linn. Soc. 2001;137:221–242. doi: 10.1111/j.1095-8339.2001.tb01119.x. [DOI] [Google Scholar]
- 123.González F, Rudall P. The questionable affinities of Lactoris: evidence from branching pattern, inflorescence morphology, and stipule development. Am. J. Bot. 2001;88:2143–2150. doi: 10.2307/3558375. [DOI] [PubMed] [Google Scholar]
- 124.Isnard S, et al. Growth form evolution in Piperales and its relevance for understanding angiosperm diversification: An integrative approach combining plant architecture, anatomy, and biomechanics. Int. J. Plant Sci. 2012;173:610–639. doi: 10.1086/665821. [DOI] [Google Scholar]
- 125.Wagner ST, et al. Major trends in stem anatomy and growth forms in the perianth-bearing Piperales, with special focus on Aristolochia. Ann. Bot. 2014;113:1139–1154. doi: 10.1093/aob/mcu044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nickrent DL, et al. Molecular data place Hydnoraceae with Aristolochiaceae. Am. J. Bot. 2002;89:1809–1817. doi: 10.3732/ajb.89.11.1809. [DOI] [PubMed] [Google Scholar]
- 127.Kelly LM, González F. Phylogenetic relationships in Aristolochiaceae. Syst. Bot. 2003;28:236–249. [Google Scholar]
- 128.Naumann J, et al. Single-copy nuclear genes place haustorial Hydnoraceae within piperales and reveal a cretaceous origin of multiple parasitic angiosperm lineages. PLoS ONE. 2013;8:e79204. doi: 10.1371/journal.pone.0079204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salomo K, et al. The emergence of earliest angiosperms may be earlier than fossil evidence indicates. Syst. Bot. 2017;42:607–619. doi: 10.1600/036364417X696438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Christenhusz MJM, Byng JW. The number of known plants species in the world and its annual increase. Phytotaxa. 2016;261:201–217. doi: 10.11646/phytotaxa.261.3.1. [DOI] [Google Scholar]
- 131.Naumann J, et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae) Genome Biol. Evol. 2016;8:345–363. doi: 10.1093/gbe/evv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jost M, Naumann J, Rocamundi N, Cocucci AA, Wanke S. The first plastid genome of the Holoparasitic genus Prosopanche (Hydnoraceae) Plants. 2020;9:306. doi: 10.3390/plants9030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zavada MS, Benson JM. First fossil evidence for the primitive angiosperm family Lactoricidae. Am. J. Bot. 1987;74:1590–1594. doi: 10.1002/j.1537-2197.1987.tb12150.x. [DOI] [Google Scholar]
- 134.Gamerro JC, Barreda V. New fossil record of Lactoridaceae in southern South America: a palaeobiogeographical approach. Bot. J. Linn. Soc. 2008;158:41–50. doi: 10.1111/j.1095-8339.2008.00860.x. [DOI] [Google Scholar]
- 135.Smith SY, Stockey RA. Establishing a fossil record for the perianthless Piperales: Saururus tuckerae sp. nov. (Saururaceae) from the Middle Eocene Princeton Chert. Am. J. Bot. 2007;94:1642–1657. doi: 10.3732/ajb.94.10.1642. [DOI] [PubMed] [Google Scholar]
- 136.Massoni J, Doyle J, Sauquet H. Fossil calibration of Magnoliidae, an ancient lineage of angiosperms. Palaeontol. Electron. 2015;18:1–25. [Google Scholar]
- 137.Smith SA. Taking into account phylogenetic and divergence-time uncertainty in a parametric biogeographical analysis of the Northern Hemisphere plant clade Caprifolieae. J. Biogeogr. 2009;36:2324–2337. doi: 10.1111/j.1365-2699.2009.02160.x. [DOI] [Google Scholar]
- 138.Beeravolu, C. R. & Condamine, F. L. An extended maximum likelihood inference of geographic range evolution by dispersal, local extinction and cladogenesis. bioRxiv10.1101/038695 (2016). [DOI] [PubMed]
- 139.Scotese CR. A continental drift flipbook. J. Geol. 2004;112:729–741. doi: 10.1086/424867. [DOI] [Google Scholar]
- 140.Blakey RC. Gondwana paleogeography from assembly to breakup—a 500 m.y. odyssey. Geol. Soc. Am. Spec. Pap. 2008;441:1–28. [Google Scholar]
- 141.Seton M, et al. Global continental and ocean basin reconstructions since 200 Ma. Earth Sci. Rev. 2012;113:212–270. doi: 10.1016/j.earscirev.2012.03.002. [DOI] [Google Scholar]
- 142.Chacón J, Renner SS. Assessing model sensitivity in ancestral area reconstruction using Lagrange: a case study using the Colchicaceae family. J. Biogeogr. 2014;41:1414–1427. doi: 10.1111/jbi.12301. [DOI] [Google Scholar]
- 143.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst. Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 144.FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- 145.Morlon H, Parsons TL, Plotkin JB. Reconciling molecular phylogenies with the fossil record. Proc. Natl Acad. Sci. USA. 2011;108:16327–16332. doi: 10.1073/pnas.1102543108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rabosky DL, et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- 147.Höhna, S. et al. A Bayesian approach for estimating branch-specific speciation and extinction rates. bioRxiv10.1101/555805 (2019).
- 148.May MR, Höhna S, Moore BR. A Bayesian approach for detecting the impact of mass-extinction events on molecular phylogenies when rates of lineage diversification may vary. Methods Ecol. Evol. 2016;7:947–959. doi: 10.1111/2041-210X.12563. [DOI] [Google Scholar]
- 149.Magallon S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 150.Rabosky DL. Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006;60:1152–1164. doi: 10.1111/j.0014-3820.2006.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 151.FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 2012;3:1084–1092. doi: 10.1111/j.2041-210X.2012.00234.x. [DOI] [Google Scholar]
- 152.Scriber, J. M. in Chemical Ecology of Insects (eds Bell, W. J. & Cardé, R. T.) 159–202 (Springer US, 1984).
- 153.Davis MP, Midford PE, Maddison W. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol. Biol. 2013;13:38. doi: 10.1186/1471-2148-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maddison WP, FitzJohn RG. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 2015;64:127–136. doi: 10.1093/sysbio/syu070. [DOI] [PubMed] [Google Scholar]
- 155.Rabosky DL, Goldberg EE. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst. Biol. 2015;64:340–355. doi: 10.1093/sysbio/syu131. [DOI] [PubMed] [Google Scholar]
- 156.Morlon H, et al. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol. 2016;7:589–597. doi: 10.1111/2041-210X.12526. [DOI] [Google Scholar]
- 157.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE. 2014;9:e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl Acad. Sci. USA. 2016;113:9569–9574. doi: 10.1073/pnas.1518659113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rabosky DL, et al. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 2014;5:701–707. doi: 10.1111/2041-210X.12199. [DOI] [Google Scholar]
- 160.Rabosky DL, Mitchell JS, Chang J. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst. Biol. 2017;66:477–498. doi: 10.1093/sysbio/syx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Höhna S, et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 2016;65:726–736. doi: 10.1093/sysbio/syw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Höhna S, May MR, Moore BR. TESS: an R package for efficiently simulating phylogenetic trees and performing Bayesian inference of lineage diversification rates. Bioinformatics. 2016;32:789–791. doi: 10.1093/bioinformatics/btv651. [DOI] [PubMed] [Google Scholar]
- 163.Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl Acad. Sci. USA. 2011;108:6187–6192. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Partha R, et al. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife. 2017;6:e25884. doi: 10.7554/eLife.25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu J, Yonezawa T, Kishino H. Rates of molecular evolution suggest natural history of life history traits and a Post-K-Pg nocturnal bottleneck of placentals. Curr. Biol. 2017;27:3025–3033. doi: 10.1016/j.cub.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 166.Zhang G, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Simion P, et al. A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 2018;16:28. doi: 10.1186/s12915-018-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Di Franco A, Poujol R, Baurain D, Philippe H. Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. BMC Evol. Biol. 2019;19:21. doi: 10.1186/s12862-019-1350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 175.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 176.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 177.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 178.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 179.Bauer DF. Constructing confidence sets using rank statistics. J. Am. Stat. Assoc. 1972;67:687–690. doi: 10.1080/01621459.1972.10481279. [DOI] [Google Scholar]
- 180.Diekmann Y, Pereira-Leal JB. Gene tree affects inference of sites under selection by the branch-site test of positive selection. Evol. Bioinforma. 2015;11:11–17. doi: 10.6026/97320630011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mallick S, Gnerre S, Muller P, Reich D. The difficulty of avoiding false positives in genome scans for natural selection. Genome Res. 2009;19:922–933. doi: 10.1101/gr.086512.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- 183.Jordan G, Goldman N. The effects of alignment error and alignment filtering on the sitewise detection of positive selection. Mol. Biol. Evol. 2012;29:1125–1139. doi: 10.1093/molbev/msr272. [DOI] [PubMed] [Google Scholar]
- 184.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 185.Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 186.Ratnakumar A, et al. Detecting positive selection within genomes: the problem of biased gene conversion. Philos. Trans. R. Soc. Ser. B. 2010;365:2571–2580. doi: 10.1098/rstb.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guéguen L, et al. Bio++: efficient extensible libraries and tools for computational molecular evolution. Mol. Biol. Evol. 2013;30:1745–1750. doi: 10.1093/molbev/mst097. [DOI] [PubMed] [Google Scholar]
- 188.Wickham, H. & Grolemund, G. R for Data Science: Import, Tidy, Transform, Visualize, and Model Data (O’Reilly Media, Inc., Canada, 2016).
- 189.Wilke, C. O. cowplot: streamlined plot theme and plot annotations for ‘ggplot2.’ CRAN Repos. 2, R2 (2016).
- 190.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 191.Redelings B. Erasing errors due to alignment ambiguity when estimating positive selection. Mol. Biol. Evol. 2014;31:1979–1993. doi: 10.1093/molbev/msu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huerta-Cepas J, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Supermatrix datasets (for phylogenetic analyses), phylogenetic trees, host–plant preferences, species geographic distributions, and gene alignments (for dN/dS analyses) that are necessary for repeating the analyses described here have been made available through the Figshare digital data repository (10.6084/m9.figshare.12278402). Source data are provided with this paper.
Bioinformatic scripts used to perform the analyses described here are available through the Figshare digital data repository (10.6084/m9.figshare.12278402).