Abstract
Sandalwood (Santalum album L.) is famous for its unique fragrance derived from the essential oil of heartwood, whose major components are santalols. To understand the mechanism underlying the biosynthesis of santalols, in this study, we cloned two related genes involved in the mevalonate pathway in S. album coding for acetyl-CoA C-acetyl transferase (AACT) and 3-hydroxy-3-methyglutary-CoA synthase (HMGS). These genes were characterized and functionally analyzed, and their expression profiles were also assessed. An AACT gene designated as SaAACT (GenBank accession No. MH018694) and a HMGS gene designated as SaHMGS (GenBank accession No. MH018695) were successfully cloned from S. album. The deduced SaAACT and SaHMGS proteins contain 415 and 470 amino acids, and the corresponding size of their open-reading frames is 1538 bp and 1807 bp, respectively. Phylogenetic trees showed that the SaAACT protein had the closest relationship with AACT from Hevea brasiliensis and the SaHMGS proteins had the highest homology with HMGS from Siraitia grosvenorii. Functional complementation of SaAACT and SaHMGS in a mutant yeast strain deficient in these proteins confirmed that SaAACT and SaHMGS cDNA encodes functional SaAACT and SaHMGS that mediate mevalonate biosynthesis in yeast. Tissue-specific expression analysis revealed that both genes were constitutively expressed in all examined tissues (roots, sapwood, heartwood, young leaves, mature leaves and shoots) of S. album, both genes showing highest expression in roots. After S. album seedlings were treated with 100 μM methyl jasmonate, the expression levels of SaAACT and SaHMGS genes increased, suggesting that these genes were responsive to this elicitor. These studies provide insight that would allow further analysis of the role of genes related to the sandalwood mevalonate pathway in the regulation of biosynthesis of sandalwood terpenoids and a deeper understanding of the molecular mechanism of santalol biosynthesis.
Subject terms: Biotechnology, Molecular biology, Physiology
Indian sandalwood (Santalum album L.) is a well-studied small hemi-parasitic evergreen tree that is widely distributed in tropical and subtropical regions of India, Indonesia, Australia and the Pacific Islands1,2. The scented heartwood of sandalwood and essential oil extracted from it have considerable economic value. Sandalwood is widely used in medicine3, wooden handicraft4 and cosmetics5. The value of sandalwood mainly depends on the proportion of heartwood and on the concentration and quality of essential oil extracted from the heartwood6,7. Indian sandalwood is considered to be one of the most valuable sandalwood species, usually yielding 3–8% essential oil8. Over 85% of S. album essential oil consists of sesquiterpenoids, including sesquiterpene alcohols (α-santalol, β-santalol, trans-α-bergamotol and epi-β-santalol) and their corresponding sesquiterpenes (α-santalene, β-santalene, trans-α-bergamotene and epi-β-santalene)8–12.
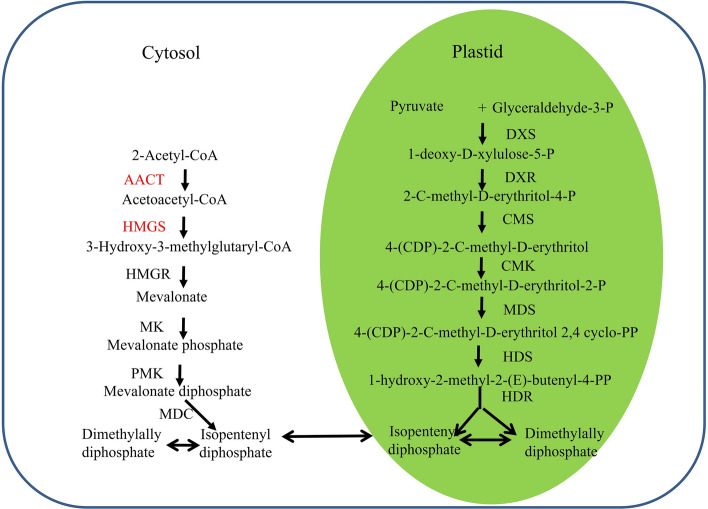
Like all other terpenoids, sesquiterpenoids are derived from two building blocks, isopentenyl diphosphate (IPP) and its allylic isomer, dimethylallyl diphosphate (DMAPP). IPP and DMAPP are formed via two pathways. One is the mevalonate (MVA) pathway, which is located in the cytoplasm, and the other is the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, which is located in plastids (Fig. 1)13–16. The MEP pathway is mainly involved in the biosynthesis of monoterpenoids, diterpenoids and other terpenoids such as hormones, plant pigments, and ubiquitins. The MVA pathway chiefly synthesizes sesquiterpenoids and triterpenoids, for example sterol, as well as other polyterpenoids17–19. Since the main components in sandalwood essential oils are sesquiterpenoids, the separation and identification of genes coding for MVA pathway-related enzymes and their functions are essential for further studies of the molecular mechanism of heartwood formation.
Figure 1.
Pathway of isoprenoids biosynthesis in plants (DXS, 1-deoxy-Dxylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; CMS, 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase). The Figure was drawn by ourselves after referring to previous studies (Laule et al. 2003; Miziorko et al. 2011; Tholl et al. 2015) and tagged the genes marked red were studied.
The MVA pathway generates IPP and DMAPP in six steps, as shown in Fig. 120–22. Acetyl coenzyme A (CoA) acetyltransferase (AACT) catalyzes the condensation of two molecules of acetyl-CoA to form acetoacetyl-CoA through Claisen condensation, and AACT is a regulatory enzyme involved in the biosynthesis of terpenoids under salt stress, promoting the production of squalene23. 3-Hydroxy-3-methyglutaryl-CoA synthase (HMGS) transforms acetyl-CoA and acetoacetyl-CoA to 3-hydroxy-3-methylglutaryl-CoA24, which is further converted into MVA by 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR). The analysis of HMGS in S. album is important to investigate the biosynthesis of α-, β-, epi-β-santalols and bergamotol. In view of their importance in the biosynthesis of terpenoids, the genes related to the MVA pathway have been successfully cloned from a number of plants, such as Arabidopsis thaliana20,25–28,30, Salvia miltiorrhiza31, Aquilaria sinensis32, Hevea brasiliensis33–35 and Ginkgo biloba36–38. In contrast, research related to sandalwood terpenoids has mainly focused on terpene synthase, which cyclizes farnesyl pyrophosphate into various cyclic sesquiterpenes such as endo-bergamotene, α-santalene, epi-β-santalene and β-santalene39–42, and cytochrome P450 oxygenase, which oxidizes α-, β-, epi-β-santalene and bergamotene to produce α-, β-, epi-β-santalols and bergamotol, respectively43,44. To identify the spatial patterns of sesquiterpenoid biosynthesis and to clone genes that encode the enzymes involved in sesquiterpene biosynthesis, transcriptomic analyses of S. album have been performed42. Srivastava et al. described the cloning and functional characterization of five genes encoding two sesquisabinene synthases (SaSQS1, SaSQS2), bisabolene synthase (SaBS), santalene synthase (SaSS), and farnesyl diphosphate synthase (SaFDS). There are no reports on the upstream genes of the MVA pathway in the biosynthesis of sesquterpenoids in the genus Santalum, including S. album.
In the present study, we cloned two related genes, named as SaAACT and SaHMGS, which are involved in the MVA pathway, by RACE technology. This, the first such study for Indian sandalwood, provides more detailed insight into the molecular biology of the santalol biosynthetic pathway. The structure and function of SaAACT and SaHMGS were analyzed by bioinformatics analyses, functional complementation was performed in yeast, and their expression level in tissues was analyzed by fluorescence quantitative PCR. These studies provide guidance for further analyses of the roles of genes related to the sandalwood MVA pathway in the regulation of biosynthesis of sandalwood terpenoids and a deeper understanding of the molecular mechanism of santalol biosynthesis.
Results
Molecular cloning and characterization of the cDNA of SaAACT and SaHMGS
After sequencing PCR products, it was shown that full-length SaAACT is 1538 bp long and contains a 1248 bp open reading frame (ORF) that encodes 415 deduced amino acid residues while full-length SaHMGS is 1807 bp long and contains a 1413 bp ORF that encodes 470 deduced amino acid residues. A BLASTn search of SaAACT and SaHMGS with other plant species showed that both genes are highly homologous to AACT and HMGS genes from other plant species. SaAACT nucleotide sequences showed 80%, 80%, 79%, 79%, and 79% identity with Camellia oleifera, Catharanthus roseus, Populus trichocarpa, Euphorbia helioscopia and H. brasiliensis, respectively (Table 1). SaHMGS nucleotide sequences showed 82% identity with C. sinensis, Panax ginseng, H. brasiliensis and Siraitia grosvenorii and 81% similarity with Platycodon grandiflorus (Table 1). These results reveal that SaAACT and SaHMGS belong to the AACT and HMGS gene families, respectively. Therefore, these genes were designated as SaAACT (GenBank accession No. MH018694) and SaHMGS (GenBank accession No. MH018695).
Table 1.
Physicochemical properties of deduced proteins in Santalum album.
| Proteins | Molecular weight (kD) | Theoretical isoelectric point | Number of acidic amino acids | Number of basic amino acids | Instability index | Aliphatic index | Total average hydropathicity |
|---|---|---|---|---|---|---|---|
| SaAACT | 43.1326 | 8.71 | 37 | 42 | 32.06 | 95.01 | 0.139 |
| SaHMGS | 51.9982 | 6.29 | 54 | 50 | 37.79 | 75.13 | -0.242 |
Bioinformatics analysis of the deduced SaAACT and SaHMGS proteins
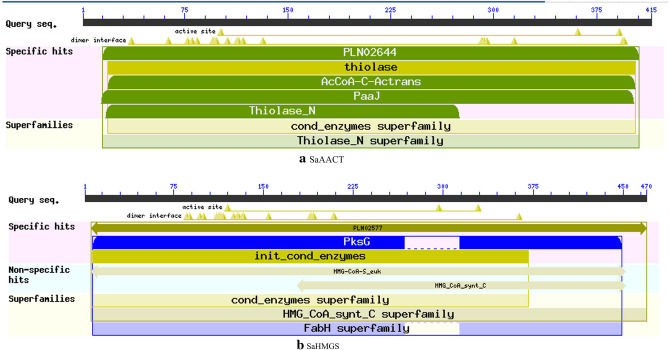
The relative molecular weight, theoretical isoelectric point, instability index, aliphatic index and grand average of hydropathicity of the deduced SaAACT and SaHMGS proteins, which were predicted by ExPASy, are shown in Table 2. The relative molecular weight of SaAACT is 43 kDa, similar to previously reported AACT38, which had a molecular weight of 41.5 kDa. The relative molecular weight of SaHMGS is 52 kDa, similar to previously reported HMGS45, with a molecular weight of 53 kDa. The theoretical isoelectric point of SaAACT and SaHMGS are 8.71 and 6.29, respectively. SaAACT is a stable hydrophobic protein and SaHMGS is a stable hydrophilic protein. Well, SaAACT and SaHMGS have no transmembrane domain or signal peptide. SaAACT has a thiolase (like) domain from aa 19-403 and acetyl-CoA acyltransferase activity from aa 15-406 (Fig. 2a). Residues of two cystines (Cys101, Cys391) and one histidine (His361) are present in SaAACT46, and these are highly conserved in AACT among the thiolases from different sources, and are important for its catalytic activity (Fig. 3A, marked with an asterisk). One highly conserved domain (NVHGGAVSIGHPIGCSG) (aa 351-367) at the C-terminal end (Fig. 3a, marked with a red box), which is also present in SaAACT, is a characteristic sequence of thiolase. SaHMGS has significant HMGS activity from aa 4-407 (Fig. 2b). Residues of one cystine (Cys119), one tyrosine (Tyr296) and one aspartic acid (Asn329) are present in SaHMGS47,48. They are highly conserved in HMGS from different sources, and are important for its catalytic activity (Fig. 3b, marked with an asterisk). One conserved motif (NxD/NE/VEGI/VDx (2) NACF/YxG) (aa 108-123), which was present in SaHMGS, is a characteristic sequence of HMGS37,49. These findings confirm that SaAACT and SaHMGS have similar catalytic functions to the corresponding AACT and HMGS from other plants.
Table 2.
Nucleotide sequences of target genes and similarity to genes from other plant species.
| a: SaAACT | ||
|---|---|---|
| Species | Accession number | Identity (%) |
| Camellia oleifera | GU594059.1 | 80 |
| Catharanthus roseus | JF739870.1 | 80 |
| Populus trichocarpa | XM002308719.2 | 79 |
| Euphorbia helioscopia | KP995935.1 | 79 |
| Hevea brasiliensis | JN036531.1 | 78 |
| b: SaHMGS | ||
|---|---|---|
| Species | Accession number | Identity (%) |
| Camellia sinensis | JQ390224.1 | 82 |
| Panax ginseng | GU565098.1 | 82 |
| Hevea brasiliensis | JN036533.1 | 82 |
| Siraitia grosvenorii | HQ128555.1 | 82 |
| Platycodon grandiflorus | KC439366.1 | 81 |
Figure 2.
Conserved domain of SaAACT (a) and SaHMGS (b) in S. album. Which was predicted by the Conserved Domains database in NCBI using my own gene sequences.
Figure 3.
Multiple alignments of SaAACT (a) and SaHMGS (b) deduced amino acid sequences with other corresponding homologous proteins. The figures were a comparison diagram and cluster analysis of my own gene sequences with related sequences of other species by CLUSTALX 2.0 and MEGA.
Phylogenetic analysis of the deduced SaAACT and SaHMGS proteins
To better understand the evolutionary relationships between deduced SaAACT and SaHMGS proteins with other AACTs and HMGSs from angiosperms, gymnosperms, fungi, and bacteria, MEGA 7 was used to construct two phylogenetic trees with the neighbor-joining (NJ) method. The first phylogenetic tree revealed that angiosperm AACTs were clustered in one group where SaAACT exhibited the highest homology with AACT from H. brasiliensis, while AACT of G. biloba formed a distinct group of gymnosperms, and the AACT gene from fungi and bacteria were clustered as a different group (Fig. 4a). The second phylogenetic tree indicated that angiosperm HMGSs were clustered in one group where SaHMGS exhibited the closest homology with HMGS from S. grosvenorii, while HMGS of Pinus sylvestris and Taxus × media formed a distinct group of gymnosperms and the AACT gene from fungi and bacteria were clustered as a separate group (Fig. 4b). These results suggest that SaAACT and SaHMGS share a common evolutionary base with other plant AACT and HMGS proteins based on their conserved structure and sequence characteristics.
Figure 4.
Phylogenetic analysis of target S. album proteins (a: SaAACT; b: SaHMGS).
Sub-cellular localization of SaAACT and SaHMGS proteins
Predicted sub-cellular localization of SaAACT and SaHMGS proteins by PSORT showed that SaAACT has the highest probability scores for peroxisomes (0.78), followed by the cytosol (0.11) and mitochondria (0.11) whereas SaHMGS exists mainly in the cytosol (0.61) but is also distributed in the nucleus (0.22) and mitochondria (0.17). To further verify the sub-cellular localization of SaAACT and SaHMGS protein, sub-cellular localization of SaAACT-YFP (yellow fluorescent protein) and SaHMGS-YFP were studied using a modified polyethylene glycol method to transform Arabidopsis thaliana protoplasts with SaAACT-YFP and SaHMGS-YFP constructs. In A. thaliana, AACT2, which is involved in the MVA pathway, is localized in the cytosol and nucleus whereas AACT1, which may be involved in fatty acid degradation, is located in peroxisomes50,51. We found that SaAACT proteins were located in the nucleus (Fig. 5). Sub-cellular localization of SaHMGS-YFP showed that SaHMGS proteins were located in the cytosol (Fig. 5), like BjHMGS1 of Brassica juncea52. Our results suggest that SaAACT and SaHMGS cloned in this study may be involved in MVA pathway in S. album.
Figure 5.
Subcellular localization of SaAACT and SaHMGS proteins in A. thaliana mesophyll protoplasts. Yellow fluorescence indicates SaAACT-YFP and SaHMGS-YFP fusion protein signal. Blue signal indicates chlorophyll (Chl) autofluorescence and red signal indicates m-Cherry fluorescence. The merged images represent a digital combination of Chl autofluorescence, YFP fluorescent and m-Cherry protein fluorescence images. Fluorescence was excited for YFP at 514 nm, for Chl at 543 nm and for m-Cherry at 587 nm. Scale bar of SaHMGS-YFP and YFP = 5 μm and scale bar of SaAACT-YFP = 8 μm.
Functional complementation of SaAACT and SaHMGS in yeast
In yeast, the MVA pathway is a biosynthetic pathway that is essential for survival, and disrupting MVA pathway genes in yeast strains can be fatal53,54. The disrupted strains with empty pYES2 could not grow on either YPG expression medium or YPD non-expression medium (Fig. 6a). YPL028W harbored pYES2-SaAACT and YML126C harbored pYES2-SaHMGS, which grew well on YPG medium. However, neither YPL028W, which harbored pYES2-SaAACT, nor YML126C, which harbored pYES2-SaHMGS, could grow on YPD medium (Fig. 6b). These results indicate that SaAACT and SaHMGS have AACT and HMGS activity, respectively.
Figure 6.
Functional complementation of two Santalum album genes (a: SaAACT; b: SaHMGS) in yeast (Saccharomyces cerevisiae). WT, wild type.
Tissue-specific and MeJA treatment expression analysis of SaAACT and SaHMGS
To explore the tissue-specific expression pattern of SaAACT and SaHMGS genes in S. album, total RNA was isolated from different tissues, including roots, heartwood, sapwood, young leaves, mature leaves and shoots. quantitative real-time PCR (qRT-PCR) was also performed. The results of qRT-PCR are shown in Fig. 7. SaAACT and SaHMGS were constitutively expressed in all tissues. The SaAACT gene was differentially expressed in various tissues and exhibited the highest level of expression in roots, followed by mature leaves and heartwood, approximately 13.44-, 3.66- and 3.55-fold higher than young leaves, respectively. A similar expression pattern of the AACT gene was found in Bacopa monnieri and G. biloba, in which the BmAACT and GbAACT genes were highly expressed in roots, followed by stems and leaves38,55. As shown in Fig. 7, SaHMGS was highly expressed in roots, approximately 4.22-fold more than in young leaves. The expression level of SaHMGS in mature leaves, sapwood, heartwood, shoots and young leaves showed few differences. In contrast, in Taxus × media, TmHMGS was expressed in needles and stems at a similar level, but no expression was detected in roots45.
Figure 7.
Gene expression levels of SaAACT (a) and SaHMGS (b) in different organs of Santalum album using qRT-PCR. The gene expression level of SaAACT and SaHMGS in the young leaves was set to 1. Data from qRT-PCR are means ± SD (standard deviation) from triplicate experiments (n = 3). Different letters indicate significant differences (p < 0.05) following one-way ANOVA, using Duncan’s multiple range test.
Methyl jasmonate (MeJA) is a small signaling molecule that can regulate secondary metabolism in plants when applied exogenously56. MeJA is closely related to terpene metabolism57. MeJA treatment can effectively induce the expression of AACT and HMGS genes and terpenoid biosynthesis in S. miltiorrhiza58, G. biloba38 and Tripterygium wilfordii59. In this study, we measured the transcript level of SaAACT and SaHMGS in S. album roots, shoots and leaves after treatment with 100 μM MeJA (Fig. 8). SaAACT and SaHMGS expression were significantly induced by MeJA. The trend of the change in transcript level of SaAACT in S. album roots, shoots and leaves after MeJA treatment was consistent, all increasing gradually and peaking at 24 h compared with control seedlings in which SaAACT transcript level decreased slowly after MeJA treatment. With regard to the SaHMGS gene, the trends in the change of transcript level of SaAACT in S. album roots, shoots and leaves after MeJA treatment were different. In roots, expression level increased rapidly after seedlings were treated with MeJA for 6 h, then gradually decreased. In shoots, expression increased slowly then peaked at 48 h after MeJA treatment. In leaves, expression level gradually increased and peaked at 12 h after MeJA treatment, then gradually decreased.
Figure 8.
Level of SaAACT (a,b,c) and SaHMGS (d,e,f) transcripts in Santalum album roots (a,d), shoots (b,e) and leaves (c,f) after induction by 100 mM MeJA. The expression level of SaAACT and SaHMGS genes in the untreated control (CK) was set to 1. Data from qRT-PCR are means ± SD (standard deviation) from triplicate experiments (n = 3).
Discussion
The commercial value of sandalwood lies mainly in its fragrant heartwood. The final purpose of planting sandalwood is to harvest high quality heartwood in great quantities. However, slow growth rates, susceptibility to diseases and variation in sandalwood oil yield hamper sandalwood production43. Researchers have attempted to synthesize santalol by chemical approaches60–62, but yield is very low and industrial production is not economic. In 1998, sandalwood was listed on the red list of threatened plants as a vulnerable plant by the International Union for Conservation Nature and Natural Resources63. Currently there are no reports for S. album on the genes upstream of the MVA pathway involved in sesquiterpenoid biosynthesis.
This study is the first to clone and characterize the SaAACT and SaHMGS genes encoding AACT and HMGS of the MVA pathway and analyze their tissue expression levels. A 1538 bp full-length cDNA of the SaAACT gene was isolated from S. album. The deduced SaAACT protein contained 415 amino acids and weighed 43 kDa. In addition, an 1807 bp full-length cDNA of the SaHMGS gene was isolated from S. album. The deduced SaHMGS protein contained 470 amino acids and weighed 52 kDa. Bioinformatics analysis showed that SaAACT and SaHMGS genes have high similarity with other corresponding genes in other plants. The deduced amino acids of SaAACT and SaHMGS have typical motifs and domains of AACT and HMGS proteins. Multiple alignments and phylogenetic analyses showed that SaAACT and H. brasiliensis AACT were located in the same clade, while SaHMGS and S. grosvenorii HMGS were clustered together in a separate clade, indicating a close genetic relationship between these protein pairs. These results suggest that SaAACT and SaHMGS are conserved across different plants based on their conserved structure and sequence characteristics. Functional complementation experiments in yeast showed that SaAACT and SaHMGS encode functional AACT and HMGS, respectively. The expression of HMGS genes is related to the accumulation of terpenoids64. Overexpression of AACT and HMGS genes can increase the level of sesquiterpenoids in many plants. Previous studies showed that the expression patterns of AACT and HMGS genes vary considerably in different plants50,60. The qRT-PCR results showed that SaAACT and SaHMGS genes were constitutively expressed in all the tested tissues but were differentially expressed in various tissues. The transcript level of SaAACT in roots was higher than in other tissues followed by mature leaves and heartwood, and the lowest expression level was in young leaves, which is consistent with a previous study55. The transcript level of SaHMGS in roots was higher than in other tissues, and the lowest expression level was in sapwood, which contrasts to SmHMGS reported previously45,58. Our results revealed that SaAACT and SaHMGS were expressed in all tissues, but at higher levels in roots. Many studies have shown that the yield of terpenoids is positively correlated with the expression of AACT and HMGS genes. Previous studies demonstrated that the accumulation of terpenoids increased in AACT overexpressing transgenic plants65,66. In Ganoderma lucidum, triterpene content increased in Gl-aact overexpressing transformants65. Overexpression of A. thaliana AACT in Taraxacum brevicorniculatum latex increased sterol levels by about five-fold66. An Escherichia coli strain that was transformed with a codon-optimized HMGS gene exhibited significantly more bisabolene production than control bacteria67. BjHMGS overexpressing transgenic plants significantly increased the total sterol content in leaves and seedlings68. Our results revealed that the expression levels of SaAACT and SaHMGS were highest in roots followed by heartwood. This trend is consistent with the chemical composition analysis in earlier reports69,70, which showed that roots and heartwood are used to extract essential oils. These results support the relationship between transcription levels of SaAACT and SaHMGS and the content of sesquiterpenoids, indicating that SaAACT and SaHMGS may play an important role in the production of sesquiterpenoids in S. album. Our results show that the transcript levels of SaAACT and SaHMGS in roots, shoots and leaves of S. album seedlings increased after MeJA treatment, implying that the putative SaAACT and SaHMGS were responsive to the elicitor, MeJA, and could be induced, at least at the transcriptional level. Studying the biosynthesis of terpenoids at the molecular level is an important way to understand the mechanism of heartwood formation. The expression profiles of AACT and HMGS genes, which code for key enzymes in the biosynthesis of sesquiterpenes in plants, suggest that MeJA treatment may be an effective way to induce a high yield of sesquiterpenes in S. album. However, the regulatory mechanism of AACT and HMGS genes in the biosynthesis of sesquiterpenes in S. album needs to be clarified through additional studies.
The findings of our study are not only helpful to further understand the biosynthesis of santalols, but also provide a theoretical basis for further studies on the prokaryotic expression of related proteins. This study provides a molecular resource for increasing the content of santalols by genetic engineering.
Materials and methods
Young leaves of five-year-old sandalwood trees (S. album) growing in South China Botanical Garden, Guangzhou, were used to isolate the SaAACT and SaHMGS genes. The young and mature leaves, shoots, heartwood, sapwood and roots were harvested to test the tissue-specific expression of SaAACT and SaHMGS genes by qRT-PCR. All collected samples were wrapped in tin foil, frozen immediately in liquid nitrogen, and then stored at −70 °C until further use.
Total RNA extraction from sandalwood leaves and first-strand cDNA synthesis
Total RNA of sandalwood leaves was extracted using Column Plant RNAOUT (Tiandz, Beijing, China) according to the manufacturer’s protocol. The quantity and quality of isolated RNA was measured with a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, NC, USA). The integrity of isolated RNA was detected on a 1% agarose gel. High quality RNA was stored in DEPC-treated water at – 70 °C for future use. First strand cDNA were obtained with the PrimeScript first-strand cDNA synthesis kit (Takara Bio Inc., Dalian, China). Synthesized cDNA was stored at − 20 °C and served as the template for downstream reactions. For qRT-PCR, total RNA was isolated from different tissues (i.e.: roots, heartwood, sapwood, young leaves, mature leaves, and shoots) and first strand cDNA was synthesized as described above.
Isolation and cloning of full-length SaAACT and SaHMGS cDNA by RACE
The full-length cDNA sequence of SaAACT and SaHMGS genes was isolated by 5′ and 3′ rapid amplification of cDNA ends (RACE)-PCR with the SMARTer RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer’s protocol. Gene-specific primers for 3′ RACE-PCR were designed on the basis of the initial data of AACT and HMGS unigenes in the S. album transcriptome71. Based on data from the partial SaAACT and SaHMGS sequence generated from 3′ RACE-PCR, gene-specific primers were designed for 5′ RACE-PCR to obtain the remaining sequences of SaAACT and SaHMGS. PCR products were purified by gel DNA purification kits (Tiangen, Beijing, China) and ligated into the pMD18-T vector (Takara Bio Inc., Dalian, China). Recombined plasmids were transformed into DH5α-competent E. coli cells (Takara Bio Inc., Dalian, China) and sequenced.
Bioinformatics analysis of SaAACT and SaHMGS
The nucleotide and amino acid sequences of SaAACT and SaHMGS were analyzed by bioinformatics methods, and their physical and chemical characteristics, transmembrane domain, signal peptide, and subcellular localization were predicted by corresponding bioinformatics software. Sequence assembly was performed with DNAstar (http://www.dnastar.com). Nucleotide sequences, deduced amino acid sequences and ORFs were analyzed, and sequences were compared through a BLAST database search (http://www.ncbi.nlm.nih.gov). Protein molecular weight and theoretical isoelectric point, instability index, aliphatic index and grand average of hydropathicity were calculated by ExPASy (http://www.expasy.ch/tools/). Protein conserved domains and active sites were predicted by the Conserved Domains database in NCBI (http://www.ncbi.mlm.nih.gov/Structure/cdd/wrpsb.cgi) and by Prosit (http://prosite.expasy.org/) in ExPASy. Transmembrane domains were analyzed on the TMHMM Server (http://www.cbs.-dtu.dk/services/TMHMM/). Signal peptides were analyzed by SignalP (http://www.cbs.dtu.dk/services/SignalP/). Protein subcellular localization was predicted by PSORT II (http://psort.org/).
Molecular evolution analyses of SaAACT and SaHMGS proteins
A phylogenetic tree of SaAACT and SaHMGS proteins from S. album and other plants, including angiosperms, gymnosperms, fungi, and bacteria, was constructed by the NJ method72 with CLUSTALX 2.0 (Conway Institute, University College Dublin, Dublin, Ireland) and MEGA 773. The calculation of evolutionary distance was based on the Poisson correction method with 1000 bootstrap repeats.
Subcellular localization of SaAACT and SaHMGS
A vector pSAT6-EYFP containing the ORF of enhanced yellow fluorescent protein (EYFP) was used in this study. The cDNA encoding SaAACT and SaHMGS were amplified with two pairs of primers, YFP-AACT-F and YFP-AACT-R, and YFP-HMGS-F and YFP-HMGS-R, respectively (Table 3). The PCR products were digested by EcoR I and BamH I restriction enzymes. The digested fragment was ligated into EcoR I- and BamH I-digested pSAT6-EYFP vector to generate pSAT6-EYFP-SaAACT and pSAT6-EYFP-SaHMGS fusion constructs. The fusion expression vectors and the pSAT6-EYFP vector were transformed into A. thaliana mesophyll protoplasts followed a method described previously74. After 16–24 h of incubation at 22 °C, YFP fluorescence in transformed protoplasts of A. thaliana was observed using a confocal laser-scanning microscope (Leica TCS SP8 STED 3X, Wetzlar, Germany).
Table 3.
Specific primers used in this study.
| Primer purpose | Primer name | Primer sequence (5′ → 3′) |
|---|---|---|
| 5′ RACE | AACT-5(1)/(2) | CACAAACACCCATGCCAAAATCA/TGCTTTCCATGCCACCAGCTACA |
| HMGS-5(1)/(2) | AGTCTCGCTGCCTACTTCCATCC/TCCAGTGCTTCCTGTTGAACACA | |
| 3′ RACE | AACT-3(1)/(2) | GAGGAAGCTCCGACCAAGTTTTA/GGAGCTGTATCTCTGGGACATC |
| HMGS-3(1)/(2) | CTCAGTCAGCATGCCTAAACCT/CATTGTTGCCGGCTCTGTTC | |
| ORF | AACT-O(F)/(R) | ATGGCTCCATCCGGGACGAAAGC/TAGCTTTGAAGGTCCTACTCTCA |
| HMGS-O(F)/(R) | ATGGCTTCGCAGCAGAAGAAT/TTAATGGCCATTGGGAATTGAACCA | |
| qRT-PCR | q-AACT-F/R | CGGGACGAAAGCCTCTTATT/AGGGAACCAAGAAAGTCACC |
| q-HMGS-F/R | TCTCGCCATTGATGTCTACTTT/CCCAATGGTGTACTTCCCTTTA | |
| Functional complementation | pYES2-AACT-F | CAGTGTGCTGGAATTCATGGCTCCATCCGGGACG |
| pYES2-AACT-R | CATGCTCGAGCGGCCGCTAGCTTTGAAGGTCCTACTCTCAAG | |
| pYES2-HMGS-F | CAGTGTGCTGGAATTCATGGCTTCGCAGCAGAAGA | |
| pYES2-HMGS-R | CATGCTCGAGCGGCCGCATGGCCATTGGGAATTGAACCA | |
| Subcellular localization | YFP-AACT-F | CCGGAATTCATGGCTCCATCCGGGACGAAAGC |
| YFP-AACT-R | CGCGGATCCTAGCTTTGAAGGTCCTACTCTCA | |
| YFP-HMGS-F | CCGGAATTCATGGCTTCGCAGCAGAAGAA | |
| YFP-HMGS-R | CGCGGATCCATGGCCATTGGGAATTGAACCA |
Functional complementation of SaAACT and SaHMGS in yeast
To determine the function of SaAACT and SaHMGS, two ergosterol auxotrophic strains (Dharmacon, Chicago, IL, USA) of Saccharomyces cerevisiae that lacked the AACT or HMGS allele, named YPL028W (ΔERG10) and YML126C (ΔERG13), respectively, were used for the experiment. The pYES2 vectors, which contain a yeast galactose-dependent promoter, were used as carriers for target genes in this study. The coding regions of SaAACT and SaHMGS were amplified with two pairs of primers: pYES2-AACT-F and pYES2-AACT-R, and pYES2-HMGS-F and pYES2-HMGS-R, respectively (Table 3). The forward primers contained the EcoR I restriction site, and the reverse primers contained the Not I restriction site. The pYES2 vector was digested with EcoR I and Not I, and then PCR products and the digested fragment of the pYES2 vector were ligated into recombined vector pYES2-SaAACT and pYES2-SaHMGS by the In-Fusion HD Cloning Kit (Clontech, Palo Alto, CA, USA). The two constructed plasmids, pYES2-SaAACT and pYES2-SaHMGS, were extracted and transformed into YPL028W (ΔERG10) and YML126C (ΔERG13) with the Frozen-EZ Yeast Transformation II Kit (Zymo Research, Irvine, CA, USA). S. cerevisiae strain ΔERG10, which lacks the AACT allele, and the S. cerevisiae strain ΔERG13, which lacks the HMGS allele, are both haploid yeast strains59,75. The transformed heterozygous diploid cells were forced to sporulate on YPG medium (1% yeast extract, 2% bacto-peptone, and 2% galactose)38, thereby obtaining viable transformed haploid cells by dissecting tetrads. Haploid transformed cells bearing both the disrupted allele and the plasmid-borne MVA pathway genes were selected on minimal medium SC (-Ura) (6.7% yeast nitrogen base without amino acid, 2% galactose)37. The transformed diploid cells were induced to sporulate and subsequently formed haploid cells containing pYES2-SaAACT and pYES2-SaHMGS. To further observe their growth, transformed haploid strains YPL028W and YML126C were grown separately on YPD (1% yeast extract, 2% bacto-peptone, 2% glucose)76 and YPG.
Expression profiling of SaAACT and SaHMGS in different tissues and their induction by MeJA
The expression levels of SaAACT and SaHMGS in different tissues (root, sapwood, heartwood, young leaves, mature leaves, and shoots) and expression profiles after MeJA (Aladdin, Shanghai, China) treatment was detected by qRT-PCR. Two-month-old young seedlings (6–8 leaves) of S. album were sprayed with 100 μM MeJA until the leaf surfaces were wet. Samples were then collected at 0 h, 2 h, 6 h, 12 h, 24 h, 48 h and 72 h after treatment. About 1.0 μg of total RNA was reverse transcribed into first-strand cDNA using the PrimeScript RT Reagent Kit (Takara Bio Inc., Dalian, China) according to the manufacturer’s protocols. The reaction was performed by ABI7500 fluorescence quantitative PCR (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) using the iTaq Universal SYBR Green Supermix as buffer (Applied Biosystems). Primer design, the reaction system and the reaction procedure were performed according to the manufacturer’s instructions. The housekeeping gene, β-actin, was selected as the internal control77. PCR amplification was performed under the following conditions: 95 °C for 30 s, followed by 35 cycles of 95 °C for 15 s and 60 °C for 60 s. Melting curve analyses were conducted. All experiments were performed in triplicate and the mean value was analyzed. Gene expression analysis using the 2−ΔΔCT method78 was used to normalize the relative gene expression of the transcripts in different tissues. Significant differences (p < 0.05) were assessed by one-way ANOVA, using Duncan’s multiple range test. The results were represented by different letters.
Author contributions
M.Y.N., Y.P.X., H.F.Y., Y.Y.Z., X.H.Z., Y.L., J.A.TdS. and G.H.M. designed the experiment and provided guidance for the study. MYN and YPX prepared samples for all analyses. H.F.Y. and Y.Y.Z. participated in statistical analyses. M.Y.N. and J.A.Td.S. co-wrote the manuscript. G.H.M. interpreted the data and experimental results. All authors read and approved the manuscript for publication.
Funding
This work was financially supported by the National Key Research Plan of China (Grant Number: 2016YFC050304) and the National Natural Science Foundation of China (Grant Numbers 31470685, 31270720 and 31100498) and a Guangdong key Science and Technology project (2015B020231008).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harbaugh DT, Baldwin BG. Phylogeny and biogeography of the sandalwoods (Santalum, Santalaceae): repeated dispersals throughout the Pacific. Am. J. Bot. 2007;94:1028–1040. doi: 10.3732/ajb.94.6.1028. [DOI] [PubMed] [Google Scholar]
- 2.Teixeira da Silva JA, Kher MM, Soner D, Page T, Zhang XH, Nataraj M, Ma GH. Sandalwood: basic biology, tissue culture, and genetic transformation. Planta. 2016;243:847–887. doi: 10.1007/s00425-015-2452-8. [DOI] [PubMed] [Google Scholar]
- 3.Benencia F, Courrèges MC. Antiviral activity of sandalwood oil against Herpes simplex viruses-1 and -2. Phytomed. 1999;6:119–123. doi: 10.1016/S0944-7113(99)80046-4. [DOI] [PubMed] [Google Scholar]
- 4.Solle HRL, Semiarti E. Micropropagation of sandalwood (Santalum album L.) endemic plant from East Nusa Tenggara. Indonesia. AIP Conf Proceed. 2016;1744:020026. doi: 10.1063/1.4953500. [DOI] [Google Scholar]
- 5.Birkbeck AA. The synthesis of fragrant natural products from Santalum album L.: (+)-(Z)-α-santalol and (-)-(Z)-β-santalol. Chimia. 2017;71:823–835. doi: 10.2533/chimia.2017.823. [DOI] [PubMed] [Google Scholar]
- 6.Brand JE, Fox JED, Pronk G, Cornwell C. Comparison of oil concentration and oil quality from Santalum spicatum and S. album plantations, 8–25 years old, with those from mature S. spicatum natural stands. Aust. For. 2007;70:235–241. doi: 10.1080/00049158.2007.10675025. [DOI] [Google Scholar]
- 7.Subasinghe SMCUP, Samarasekara SC, Millaniyage KP, Hettiarachchi DS. Heartwood assessment of natural Santalum album, populations for agroforestry development in Sri Lanka. Agrofor. Syst. 2017;91:1157–1164. doi: 10.1007/s10457-016-0001-5. [DOI] [Google Scholar]
- 8.Jones CG, Plummer JA, Barbour EL. Non-destructive sampling of Indian sandalwood (Santalum album L.) for oil content and composition. J. Essent. Oil Res. 2007;19:157–164. doi: 10.1080/10412905.2007.9699250. [DOI] [Google Scholar]
- 9.Howes MJ, Simmonds MSJ, Kite GC. Evaluation of the quality of sandalwood essential oils by gas chromatography-mass spectrometry. J. Chromatogr. A. 2004;1028:307–312. doi: 10.1016/j.chroma.2003.11.093. [DOI] [PubMed] [Google Scholar]
- 10.Demole E, Demole C, Enggist P. A chemical investigation of the volatile constituents of east Indian sandalwood oil (Santalum album L.) Helvet. Chim. Acta. 2010;59:737–747. doi: 10.1002/hlca.19760590304. [DOI] [Google Scholar]
- 11.Baldovini N, Delasalle C, Joulain D. Phytochemistry of the heartwood from fragrant Santalum species: a review. Flav. Frag. J. 2011;26:7–26. doi: 10.1002/ffj.2025. [DOI] [Google Scholar]
- 12.Hasegawa T, Toriyama T, Ohshima N, Tajima Y, Mimura I, Hirota K, Nagasaki Y, Yamada H. Isolation of new constituents with a formyl group from the heartwood of Santalum album L. Flav. Frag. J. 2015;26:98–100. doi: 10.1002/ffj.2020. [DOI] [Google Scholar]
- 13.Lichtenthaler HK. The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Ann. Rev. Plant Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 14.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 15.Lange BM, Rujan T, Martin W, Croteau R. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Nat. Acad. Sci. USA. 2000;97:13172–13177. doi: 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzuyama T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotech. Biochem. 2002;66:1619–1627. doi: 10.1271/bbb.66.1619. [DOI] [PubMed] [Google Scholar]
- 17.Estévez JM, Cantero A, Reindl A, Reichler S, León P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001;276:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- 18.Dubey VS, Bhalla R, Luthra R. An overview of the non-mevalonate pathway for terpenoid biosynthesis in plants. J. Biosci. 2003;28:637–646. doi: 10.1007/BF02703339. [DOI] [PubMed] [Google Scholar]
- 19.Gutensohn M, Orlova I, Nguyen TTH, Davidovich-Rikanati R, Ferruzzi MG, Sitrit Y, Lewinsohn E, Pichersky E, Dudareva N. Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J. 2013;75:351–363. doi: 10.1111/tpj.12212. [DOI] [PubMed] [Google Scholar]
- 20.Laule O, Fürholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange BM. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv. Bioch. Engin. Biotech. 2015;148:63–106. doi: 10.1007/10_2014_295. [DOI] [PubMed] [Google Scholar]
- 23.Soto G, Stritzler M, Lisi C, Alleva K, Pagano ME, Ardila F, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J. Exp. Bot. 2011;62:5699–5711. doi: 10.1093/jxb/err287. [DOI] [PubMed] [Google Scholar]
- 24.Luskey KL, Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J. Biol. Chem. 1985;260:10271–10277. doi: 10.1016/S0021-9258(17)39242-6. [DOI] [PubMed] [Google Scholar]
- 25.Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A. Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme-A reductase. Plant Mol. Biol. 1990;13:627–638. doi: 10.1007/BF00016018. [DOI] [PubMed] [Google Scholar]
- 26.Riou C, Tourte Y, Lacroute F, Karsta F. Isolation and characterization of a cDNA encoding Arabidopsis thaliana, mevalonate kinase by genetic complementation in yeast. Gene. 1994;148:293–297. doi: 10.1016/0378-1119(94)90701-3. [DOI] [PubMed] [Google Scholar]
- 27.Montamat F, Guilloton M, Karst F, Delrot S. Isolation and characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl-coenzyme A synthase. Gene. 1995;167:197–201. doi: 10.1016/0378-1119(95)00642-7. [DOI] [PubMed] [Google Scholar]
- 28.Lluch MA, Masferrer A, Arró M, Boronat A, Ferrer A. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Mol. Biol. 2000;42:365–376. doi: 10.1023/A:1006325630792. [DOI] [PubMed] [Google Scholar]
- 29.Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J. Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol. Biol. 2007;63:97–108. doi: 10.1007/s11103-006-9075-1. [DOI] [PubMed] [Google Scholar]
- 30.Ahumada I, Cairó A, Hemmerlin A, González V, Pateraki I, Bach TJ, Rodríguez-Concepción M, Campos N, Boronat A. Characterisation of the gene family encoding acetoacetyl-CoA thiolase in Arabidopsis. Func. Plant Biol. 2008;35:1100–1111. doi: 10.1071/FP08012. [DOI] [PubMed] [Google Scholar]
- 31.Dai ZB, Cui GH, Zhou SF, Zhang XN, Huang LQ. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J. Plant Physiol. 2011;168:148–157. doi: 10.1016/j.jplph.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Xu YH, Zhang Z, Wang MX, Wei JH, Chen HJ, Gao ZH, Sui C, Luo HM, Zhang XL, Yang Y, Meng H, Li WL. Identification of genes related to agarwood formation: transcriptome analysis of healthy and wounded tissues of Aquilaria sinensis. BMC Genom. 2013;14:227. doi: 10.1186/1471-2164-14-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chye ML, Tan CT, Chua NH. Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensis: hmg1 and hmg3 are differentially expressed. Plant Mol. Biol. 1992;19:473–484. doi: 10.1007/BF00023395. [DOI] [PubMed] [Google Scholar]
- 34.Sirinupong N, Suwanmanee P, Doolittle RF, Suvachitanont W. Molecular cloning of a new cDNA and expression of 3-hydroxy-3-methylglutaryl-CoA synthase gene from Hevea brasiliensis. Planta. 2005;221:502–512. doi: 10.1007/s00425-004-1463-7. [DOI] [PubMed] [Google Scholar]
- 35.Sando T, Takaoka C, Mukai Y, Yamashita A, Hattori M, Ogasawara N, Fukusaki E, Kobayashi A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 2008;72:2049–2060. doi: 10.1271/bbb.80165. [DOI] [PubMed] [Google Scholar]
- 36.Shen GA, Pang YZ, Wu WS, Liao ZH, Zhao LX, Sun XF, Tang KX. Cloning and characterization of a root-specific expressing gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Ginkgo biloba. Mol. Biol. Rep. 2006;33:117–127. doi: 10.1007/s11033-006-0014-7. [DOI] [PubMed] [Google Scholar]
- 37.Tao TT, Chen QW, Meng XX, Yan JP, Xu F, Chang J. Molecular cloning, characterization, and functional analysis of a gene encoding 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Matricaria chamomilla. Genes Genom. 2016;38:1179–1187. doi: 10.1007/s13258-016-0463-x. [DOI] [Google Scholar]
- 38.Chen QW, Yan JP, Meng XX, Xu F, Zhang WW, Liao YL, Qu JW. Molecular cloning, characterization, and functional analysis of acetyl-CoA C-acetyltransferase and mevalonate kinase genes involved in terpene trilactone biosynthesis from Ginkgo biloba. Molecules. 2017;22:74. doi: 10.3390/molecules22010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones CG, Keeling CI, Ghisalberti EL, Barbour EL, Plummer JA, Bohlmann J. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch. Biochem. Biophys. 2008;477:121–130. doi: 10.1016/j.abb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J. Biol. Chem. 2011;286:17445–17454. doi: 10.1074/jbc.M111.231787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rani A, Ravikumar P, Reddy MD, Kush A. Molecular regulation of santalol biosynthesis in Santalum album L. Gene. 2013;527:642–648. doi: 10.1016/j.gene.2013.06.080. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava PL, Daramwar PP, Krithika R, Pandreka A, Shankar SS, Thulasiram HV. Functional characterization of novel sesquiterpene synthases from Indian sandalwood, Santalum album. Sci. Rep. 2015;5:10095. doi: 10.1038/srep10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz-Chavez ML, Moniodis J, Madilao LL, Jancsik S, Keeling CI, Barbour EL, Ghisalberti EL, Plummer JA, Jones CG, Bohlmann J. Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE. 2013;8:e75053. doi: 10.1371/journal.pone.0075053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Celedon JM, Chiang A, Yuen MMS, Diaz-Chavez ML, Madilao LL, Finnegan PM, Barbour EL, Bohlmann J. Heartwood-specific transcriptome and metabolite signatures of tropical identity sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 2016;86:289–299. doi: 10.1111/tpj.13162. [DOI] [PubMed] [Google Scholar]
- 45.Kai G, Miao Z, Zhang L, Zhao D, Liao Z, Sun X, Zhao L, Tan K. Molecular cloning and expression analyses of a new gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Taxus× media. Biol. Plant. 2006;50:359–366. doi: 10.1007/s10535-006-0050-0. [DOI] [Google Scholar]
- 46.Anderson VE, Bahnson BJ, Wlassics ID, Walsh CT. The reaction of acetyldithio-CoA, a readily enolized analog of acetyl-CoA with thiolase from Zoogloea ramigera. J. Biol. Chem. 1990;265:6255–6261. doi: 10.1016/S0021-9258(19)39318-4. [DOI] [PubMed] [Google Scholar]
- 47.Price AC, Choi KH, Heath RJ, Li ZM, White SW, Rock CO. Inhibition of β-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin structure and mechanism. J. Biol. Chem. 2001;276:6551–6559. doi: 10.1074/jbc.M007101200. [DOI] [PubMed] [Google Scholar]
- 48.Theisen MJ, Misra I, Saadat D, Campobasso N, Miziorko HM, Harrison DHT. 3-hydroxy- 3-methylglutaryl-CoA synthase intermediate complex observed in “real-time”. Proc. Nat. Acade. Sci. USA. 2004;101:16442–16447. doi: 10.1073/pnas.0405809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campobasso N, Pate M, Wilding IE, Kallender H, Rosenberg M, Gwynn MN. Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase. J. Biol. Chem. 2004;279:44883–44888. doi: 10.1074/jbc.M407882200. [DOI] [PubMed] [Google Scholar]
- 50.Ahumada I, Cairó A, Hemmerlin A, González V, Pateraki I, Bach TJ, Rodríguez-Concepción M, Campos N, Boronat A. Characterization of the gene family encoding acetoacetyl-CoA thiolase in Arabidopsis. Func. Plant Biol. 2008;35:1100–1111. doi: 10.1071/FP08012. [DOI] [PubMed] [Google Scholar]
- 51.Nagegowda DA. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010;584:2965–2973. doi: 10.1016/j.febslet.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 52.Nagegowda DA, Ramalingam S, Hemmerlin A, Bach TJ, Chye ML. Brassica juncea HMG-CoA synthase: localization of mRNA and protein. Planta. 2005;221:844–856. doi: 10.1007/s00425-005-1497-5. [DOI] [PubMed] [Google Scholar]
- 53.Trocha PJ, Sprinson DB. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch. Biochem. Bioph. 1976;174:45–51. doi: 10.1016/0003-9861(76)90322-2. [DOI] [PubMed] [Google Scholar]
- 54.Servouse M, Karst F. Regulation of early enzymes of ergosterol biosynthesis in Saccharomyces cerevisiae. Biochem. J. 1986;240:541–547. doi: 10.1042/bj2400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vishwakarma RK, Ruby SS, Sonawane PD, Srivastava S, Kumari U, Kumar RJS, Khan BM. Molecular cloning, biochemical characterization, and differential expression of an acetyl-CoA C-acetyltransferase gene (AACT) of brahmi (Bacopa monniera) Plant Mol. Biol. Rep. 2013;31:547–557. doi: 10.1007/s11105-012-0523-6. [DOI] [Google Scholar]
- 56.Pauwels L, Morreel K, Witte ED, Lammertyn F, Montagu MV, Boerjan W, Inzé D, Goossens A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Nati. Acad. Sci. USA. 2008;105:1380–1385. doi: 10.1073/pnas.0711203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Ann. Rev. Phytopath. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Yan XM, Wang J, Li SS, Liao P, Kai GY. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 2011;33:953–961. doi: 10.1007/s11738-010-0627-2. [DOI] [Google Scholar]
- 59.Liu YJ, Zhao YJ, Zhang M, Su P, Wang X, Zhang XJ, Gao W, Huang LQ. Cloning and characterisation of the gene encoding, 3-hydroxy-3-methylglutaryl-CoA synthase in Tripterygium wilfordii. Molecules. 2014;19:19696–19707. doi: 10.3390/molecules191219696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brocke C, Eh M, Finke A. Recent developments in the chemistry of sandalwood odorants. Chem. Biodiv. 2008;5:1000–1010. doi: 10.1002/cbdv.200890080. [DOI] [PubMed] [Google Scholar]
- 61.Muratore A, Clinet JC, Dunach E. Synthesis of new exo- and endo-3,8-dihydro-β-santalols and other norbornyl-derived alcohols. Chem. Biodiv. 2010;7:623–638. doi: 10.1002/cbdv.200900119. [DOI] [PubMed] [Google Scholar]
- 62.Zhang YY, Yan HF, Niu MY, Cheng QW, Zhang XH, Teixeira da Silva JA, Ma GH. Multiple strategies for increasing yields of essential oil and obtaining sandalwood terpenoids by biotechnological methods in sandalwood. Trees. 2018;32:21–29. [Google Scholar]
- 63.Oldfield, S., Lusty, C., MacKinven, A. The World List of Threatened Trees, WCMC, IUCN, 632 (1998).
- 64.Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 65.Fang X, Shi LA, Ren A, Jiang AL, Wu FL, Zhao MW. The cloning, characterization and functional analysis of a gene encoding an acetyl-CoA acetyltransferase involved in triterpene biosynthesis in Ganoderma lucidum. Mycoence. 2013;54:100–105. [Google Scholar]
- 66.Pütter KM, Deenen NV, Unland K, Prüfer D, Gronover CS. Isoprenoid biosynthesis in dandelion latex is enhanced by the overexpression of three key enzymes involved in the mevalonate pathway. BMC Plant Biol. 2017;17:88. doi: 10.1186/s12870-017-1036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS. Identification and microbial production of a terpene-based advanced biofuel. Nat. Comm. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, H., Nagegowda, A. D., Rawat, R., Bouvier-Navé, P., Guo, D. J., Bach, T. J. & Chye, M. L. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotech. J. 31–42 (2012). [DOI] [PubMed]
- 69.Burdock GA, Carabin IG. Safety assessment of sandalwood oil (Santalum album L.) Food Chem. Toxicol. 2008;46:421–432. doi: 10.1016/j.fct.2007.09.092. [DOI] [PubMed] [Google Scholar]
- 70.Zhang XH, Teixeira da Silva JA, Jia YX, Yan J, Ma GH. Essential oil composition from roots of Santalum album L. J Essent Oil Bear. Plants. 2012;15:1–6. doi: 10.1080/0972060X.2012.10644011. [DOI] [Google Scholar]
- 71.Zhang XH, Teixeira da Silva JA, Niu MY, Li MZ, He CM, Zhao JT, Zeng SJ, Duan J, Ma GH. Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Sci. Rep. 2017;7:42165. doi: 10.1038/srep42165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 73.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 75.Hiser L, Basson ME, Rine J. ERG10 from Saccharomyces cerevisiae encodes acetoacetyl-CoA thiolase. J. Biol. Chem. 1994;269:31383–31389. doi: 10.1016/S0021-9258(18)31705-8. [DOI] [PubMed] [Google Scholar]
- 76.Albers E, Larsson C. A comparison of stress tolerance in YPD and industrial lignocellulose-based medium among industrial and laboratory yeast strains. J. Indus. Microbiol. Biotech. 2009;36:1085–1091. doi: 10.1007/s10295-009-0592-1. [DOI] [PubMed] [Google Scholar]
- 77.Zhang XH, Berkowitz O, Teixeira da Silva JA, Zhang MH, Ma GH, Whelan J, Duan J. RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Front. Plant Sci. 2015;6:661. doi: 10.3389/fpls.2015.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 79.Zhang XH, Niu MY, Teixeira da Silva JA, Zhang YY, Yuan YF, Jia YX, Xiao YY, Li Y, Fang Y, Zeng SJ, Ma GH. Identification and functional characterization of three new terpene synthase genes involved in chemical defense and abiotic stresses in Santalum album. BMC Plant Biol. 2019;19:115. doi: 10.1186/s12870-019-1720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.