Abstract
Facioscapulohumeral muscular dystrophy (FSHD) arises from epigenetic changes that de-repress the DUX4 gene in muscle. The full-length DUX4 protein causes cell death and muscle toxicity, and therefore we hypothesize that FSHD therapies should center on inhibiting full-length DUX4 expression. In this study, we developed a strategy to accomplish DUX4 inhibition using U7-small nuclear RNA (snRNA) antisense expression cassettes (called U7-asDUX4). These non-coding RNAs were designed to inhibit production or maturation of the full-length DUX4 pre-mRNA by masking the DUX4 start codon, splice sites, or polyadenylation signal. In so doing, U7-asDUX4 snRNAs operate similarly to antisense oligonucleotides. However, in contrast to oligonucleotides, which are limited by poor uptake in muscle and a requirement for lifelong repeated dosing, U7-asDUX4 snRNAs can be packaged within myotropic gene therapy vectors and may require only a single administration when delivered to post-mitotic cells in vivo. We tested several U7-asDUX4s that reduced DUX4 expression in vitro and improved DUX4-associated outcomes. Inhibition of DUX4 expression via U7-snRNAs could be a new prospective gene therapy approach for FSHD or be used in combination with other strategies, like RNAi therapy, to maximize DUX4 silencing in individuals with FSHD.
Keywords: DUX4, FSHD, U7, snRNA, gene therapy, RNA silencing
Graphical Abstract

Treatments for facioscapulohumeral muscular dystrophy (FSHD) are designed to suppress the toxic DUX4 gene that underlies the disorder. This study describes a novel approach to inhibit DUX4 production at the mRNA level by using designed U7-antisense RNA expression cassettes to sterically hinder maturation and translation of DUX4 mRNA.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD) is among the most common muscular dystrophies, affecting up to 870,000 people worldwide.1 Disease symptoms typically arise in the second decade of life but can manifest at any age, with early-onset forms often showing increased severity compared with those arising later.2,3 Classical descriptions of FSHD include weakness in muscles of the face, shoulder girdle, and upper arms, but presentation is not uniform within the FSHD population. For example, roughly half of individuals have lower limb weakness, and although some people may maintain lifelong ambulation, others become severely debilitated and wheelchair dependent. FSHD is progressive, and symptoms often worsen with age, and although some individuals may die from FSHD-related respiratory complications, as a population, people affected by FSHD have a normal lifespan. Thus, FSHD can reduce quality of life while increasing health-related costs over a span of decades.4 Currently, there are no treatments available that alter the course of the disease or improve outcomes, and as a result, therapy development remains a critical need in the field.
FSHD is caused by aberrant de-repression of the DUX4 gene in muscle.5 DUX4 encodes a transcription factor that normally operates in early human development but is epigenetically silenced in adult muscle.6,7 When expressed in muscles, the DUX4 protein is toxic and activates genes associated with cell death, oxidative stress, impaired muscle differentiation, double-stranded RNA activation, immune responses, and atrophy.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 Potentially hundreds of DUX4 copies exist in the human genome, embedded within tandemly arrayed repetitive elements called D4Z4 repeats.21,22 Although each DUX4 copy could be transcribed, only one—located within the terminal D4Z4 repeat on the chromosome 4q35 subtelomere—is translated into toxic DUX4 protein in FSHD muscle.5 This FSHD-associated D4Z4 copy is unique because it contains a poly(A) signal the other copies lack. Structurally, all D4Z4 copies contain the entire DUX4 coding region in exon 1, followed by a small intron and a second untranslated exon.5,9 The terminal 4q35 D4Z4 copy in FSHD-permissive muscle has an additional intron followed by a third untranslated exon harboring the poly(A) signal.5 In addition to toxic full-length DUX4 protein, this locus has been reported to produce a second potential isoform called DUX4-short (DUX4 s), which lacks a portion of the C terminus containing a transactivation domain and is non-toxic.14
The most straightforward methods to treat FSHD should inhibit DUX4 gene expression in adult muscles. Several strategies to accomplish this are currently under development, including increasing the heterochromatin status of the DUX4 DNA locus to prevent its expression and inhibiting the DUX4 mRNA before it can be translated into protein.18,23, 24, 25, 26, 27, 28, 29 Indeed, our lab previously used gene therapy to deliver artificial microRNAs (miRNAs) engineered to knock down the DUX4 mRNA via RNA interference (RNAi) in vitro and in vivo.10,30,31 Other groups demonstrated that antisense oligonucleotides (ASOs), designed to mask splice sites and/or the poly(A) signal, could interfere with DUX4 mRNA production and suppress full-length DUX4 expression in FSHD cells.18,23, 24, 25,29 The system we describe here has some similarity to these ASO studies because we utilize antisense sequences in our designs. However, our approach is distinct because it uses a recombinant U7 small nuclear RNA (U7-snRNA) expression system to produce antisense sequences, overcoming some major limitations of ASOs, such as inefficient delivery to muscle and a requirement for lifelong administration.32
The U7-snRNA is a component of the small nuclear ribonucleoprotein complex (U7-snRNP), involved in 3′ end processing of histone pre-mRNAs in the nucleus.33 The wild-type U7 snRNA contains an antisense RNA sequence at its 5′ end, which normally base pairs with a histone pre-mRNA, as well as binding sites to seed the formation of 7 core Sm proteins. At the 3′ end is a hairpin structure that provides stability to the RNA. Importantly, the antisense sequence of U7-snRNA can be changed to retarget the RNP to other mRNAs, such as DUX4.34 When embedded into an snRNP complex, the U7-snRNA is protected from cellular enzymatic digestion, allowing it to remain in the nucleus for a longer time compared with ASOs and extending the effects of masking RNA sequences like splice sites, start codons, or poly(A) signals.35 Furthermore, U7-snRNA expression cassettes can be packaged into adeno-associated virus (AAV) particles to achieve widespread delivery of therapeutic antisense sequences at high efficiency into animal and human target tissues, including muscle.35 Thus, although the ASO approach for DUX4 suppression would require chronic, lifelong administration of chemically synthesized sequences, AAV-delivered U7-snRNAs are produced in target cells using endogenous transcriptional machinery, offering a chance to provide long-term DUX4 inhibition with one administration. In this study, we designed several U7-snRNAs containing antisense regions with sequence complementarity to the DUX4 start codon, splice sites and splice enhancers, or regulatory elements (3′ UTR and poly(A)) and investigated their efficacy for DUX4 silencing in HEK293 cells overexpressing DUX4 and in myotubes from individuals with FSHD expressing endogenous levels of DUX4. We show that the U7-snRNA system can reduce DUX4 and DUX4-associated outcomes in these human cell models, supporting further translation of the U7-snRNA system for gene therapy of FSHD.
Results
DUX4-targeting U7-snRNAs reduce apoptosis and increase the viability of co-transfected HEK293 cells
Recombinant U7-snRNAs were developed previously to induce skipping of mutated exons as potential treatment for Duchenne muscular dystrophy36 and β-thalassaemia.37 In these studies, U7-snRNAs were used to restore the expression of frameshifted genes by skipping entire exons. In contrast, our goal here was to develop a novel gene silencing strategy by using U7-snRNAs to interfere with DUX4 pre-mRNA maturation or inhibit translational initiation.38,39 To do this, we developed recombinant U7-snRNAs targeting splice donor (SD), splice acceptor (SA), and splice enhancer (SE) sequences or the polyadenylation signal (PAS) in DUX4 exon 3 (Figures 1A and 1B). In addition, we generated two constructs (9 and 10) designed to sit atop the full-length DUX4 start codon and potentially interfere with translation. The structure of our DUX4-targeting U7-snRNAs (called U7-antisense [as]DUX4) is shown in Figure 1A, where the key feature for specificity is an antisense sequence modified to base-pair with various regions of the DUX4 pre-mRNA (Figure 1A). To choose effective sequences for interfering with correct splicing, we used the Human Splicing Finder tool (Figure S1) to predict potential SD, SA, and SE sites for all three DUX4 exons and within introns 1 and 2. We then designed U7-asDUX4s to target the highest-scoring sites (Figure 1B). For those U7-asDUX4s targeting the poly(A) signal or start codon, we ensured that the antisense sequences provided complete coverage of the cognate sites on the DUX4 mRNA. All U7-asDUX4 sequences and their important features are summarized in Table S1.
Figure 1.
U7-asDUX4 snRNAs protect HEK293 cells from DUX4-mediated death
(A) U7-snRNA structure consisting of a stabilizing hairpin loop, Sm binding region, and an antisense sequence complementary to a target site on the DUX4 pre-mRNA (see Table S1 for sequences). (B) Schematic drawing of 18 U7-asDUX4 constructs targeting different parts of DUX4 mRNA. ATG indicates the start codon. Exon 1 (Ex1) contains the entire DUX4 open reading frame with a small 3′ untranslated region (3′ UTR), whereas Ex2 and Ex3 contain only 3′ UTR sequences. (C) Caspase-3/7 assay for apoptosis. All DUX4-targeting U7-asDUX4 snRNA constructs significantly reduced caspase-3/7 activity except in cells treated with sequence 6. Fourteen of 18 constructs reduced caspase-3/7 activity more than 50% (exceptions were 1, 2, 6, and 18). Constructs 3, 4, 7, 8, 11, and 16 provided the best protection from DUX4-mediated cell death (relative caspase-3/7 activity from U7-asDUX4-3 (34 ± 12), U7-asDUX4-4 (26 ± 7), U7-asDUX4-7 (36 ± 8), U7-asDUX4-11 (30 ± 3), and U7-asDUX4-16 (33 ± 6). (D) Cell viability increased significantly in all U7-asDUX4-treated cells compared with those transfected with DUX4 alone, with U7-asDUX4-7 (95% ± 5%), U7-asDUX4-8 (85% ± 4%), and U7-asDUX4-4 (79% ± 8%), respectively, showing the most percentage of viable cells. For caspase-3/7 and cell viability, n = 3 independent experiments performed in triplicate (p < 0.01, ANOVA; n = 3 independent experiments performed in triplicate). ∗p ≤ 0.05, ∗∗p ≤ 0.01, ANOVA.
HEK293 cells do not normally express DUX4 protein or polyadenylated DUX4 mRNA but are susceptible to DUX4-induced cell death following transfection with a cytomegalovirus (CMV).DUX4 expression plasmid. We therefore initially assessed the efficacy of U7-asDUX4 expression plasmids by measuring apoptotic cell death using caspase-3/7 and cell viability assays as outcome measures in co-transfected HEK293 cells. We designed and tested 18 U7-asDUX4 sequences and found that 13 significantly reduced cell death (>50%) and increased viability (>50%) of co-transfected HEK293 cells (Figures 1C and 1D). Combining the two parameters, the most effective U7-asDUX4s were constructs 4, 7, and 8, targeting the exon 1-intron 1 junction or the PAS. Cells transfected with these constructs showed reduced relative caspase-3/7 activity (U7-asDUX4-4, 75% ± 7% reduction; U7-asDUX4-7, 60% ± 9% reduction; U7-asDUX4-8, 50% ± 8% reduction) and significantly increased viability (U7-asDUX4-4, 79% ± 8%; U7-asDUX4-7, 95% ± 5%; U7-asDUX4-8, 85% ± 4%) compared with only DUX4 (19% ± 2%) or DUX4 with nontargeting U7-snRNA (23% ± 0.5%) (Figures 1C and 1D). Because of their superior protective properties, we selected U7-asDUX4 constructs 4, 7, and 8 as our lead candidate sequences.
U7-asDUX4s significantly decrease DUX4 expression in transfected HEK293 cells
The reduction in DUX4-related cell death outcomes in samples treated with U7-asDUX4 plasmids suggested that these sequences operated to inhibit full-length DUX4 gene expression. To investigate the specificity of our lead U7-asDUX4 sequences to target and reduce overexpressed DUX4 mRNA in HEK293 cells, we first used the RNAscope in situ hybridization assay to detect DUX4 mRNA in co-transfected cells. Fixed cells were incubated with probes targeting DUX4, control transcripts, or negative control reagents and then treated with a diaminobenzidine (DAB) reagent that stains hybridized target mRNAs brown. As we reported previously, cells transfected with DUX4 expression plasmid alone showed abundant, spider-like brown signals when incubated with DUX4 probes as well as relatively low cell density consistent with death (Figure 2A).30 In contrast, the DUX4 probe signal was reduced significantly in cells co-transfected with DUX4 and our three lead U7-asDUX4 (Figures 2B–2D). Specifically, in U7-asDUX4-treated wells, there were significantly fewer DUX4-positive cells and/or reduced intensity of DUX4 staining in cells that still showed DUX4 signal (Figures 2B–2D). Positive and negative controls behaved as expected; we found little to no DUX4 signal in untransfected HEK293 cells (Figure 2E), whereas abundant signal was evident in cells stained with probes to the peptidylprolyl isomerase B (PPIB) gene (Figure 2F), a positive control for the RNAscope assay. Consistent with DUX4 knockdown, which provided some protection from cell death (Figure 1D), wells transfected with U7-asDUX4 plasmids had greater cell density compared with “DUX4-only”-transfected samples.
Figure 2.
U7-asDUX4 snRNAs significantly reduced overexpressed DUX4 mRNA in transfected HEK293 cells.
RNAscope assay: DUX4 mRNA signals appeared as brown, punctate dots in transfected cells. (A) Abundant DUX4 signal was detected in HEK293 cells co-transfected with CMV.DUX4 and a non-targeting U7 control plasmid. (B–D) There was a reduction in brown DUX4 signal after co-transfection of HEK293 cells with CMV.DUX4 and (B) U7-asDUX4-4, (C) U7-asDUX4-7, and (D) U7-asDUX4-8 plasmids. (E) Background signal with DUX4 probe in untransfected HEK293 cell line. (F) The housekeeping gene PPIB was detected in all HEK293 cells and served as a positive control for the assay. (G) The bacterial gene dapB probe was used a negative control for the RNAscope assay. (H) RNAscope quantification showed a significantly reduced DUX4-positive signal in DUX4-transfected cells co-expressing U7-asDUX4 snRNAs 4, 7, and 8. 40× objective. Scale bar, 50 μm. Quantification was performed as described previously.30 Two representative microscopic fields were counted from 3 independent experiments; each point represents quantification of one field. ∗∗p < 0.01, ANOVA.
U7-asDUX4 snRNAs reduce full-length DUX4 protein in transfected HEK293 cells
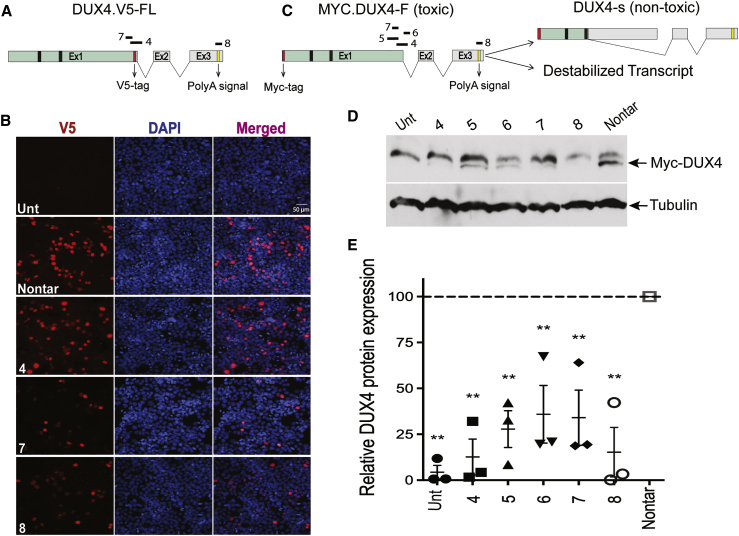
To confirm full-length DUX4 mRNA knockdown, we next determined the efficiency of U7-asDUX4 to suppress DUX4 protein expression. To do this, we first co-transfected cells with our three lead U7-asDUX4 constructs or a non-targeting control along with a CMV.DUX4 expression plasmid. To facilitate protein detection, we used a full-length DUX4 construct containing an in-frame COOH-terminal fusion of the V5 epitope tag (CMV.DUX4.V5-full-length [FL]) (Figure 3A). To detect DUX4 protein, we then stained cells with fluorescence-labeled antibodies to the V5 tag. We found reduced DUX4.V5 signal in cells co-transfected with U7-asDUX4 sequence 7 or 8 compared with cells that received a non-targeting control U7-snRNA (Figure 3B). Interestingly, U7-asDUX4-4 did not affect DUX4.V5 protein levels because its binding site was disrupted by the V5 epitope tag, serving as an inadvertent control for specificity.
Figure 3.
U7-asDUX4 snRNAs reduce DUX4 protein production in transfected HEK293 cells
(A) Schematic of the FL DUX4 expression construct containing a C-terminal V5 epitope tag. The 42-bp DNA sequence encoding the 14-amino-acid V5 tag disrupted the U7-asDUX4-4 target site. Black bars in Ex1 indicate relative locations of DNA binding homeodomains 1 and 2 (HOX1 and HOX2) but are not to scale. Introns 1 and 2 are indicated as ∨ symbols. (B) Anti-V5 immunofluorescence staining of HEK293 cells co-transfected with CMV.DUX4.V5-FL, where the DUX4.V5 signal appears as red fluorescence. The blue DAPI stain (4′,6-diamidino-2-phenylindole) shows HEK293 nuclei. The U7-asDUX4-7 and U7-asDUX4-8 constructs reduced DUX4.V5 protein staining compared with cells treated with non-targeting U7-snRNAs. Although the U7-asDUX4-4 sequence was functional in other assays, it did not reduce DUX4.V5 protein expression because of disruption of its binding site by the V5 tag. 40× objective. Scale bar, 50 μm. (C) The Myc-DUX4-FL construct used for the western blot assay and possible mechanisms of DUX4 inhibition by lead U7-asDUX4 targeting of DUX4 (discussed in the text). DUX4 s is a non-toxic potential isoform of DUX4 that lacks the C-terminal transactivation domain. (D) Western blot results demonstrated reduced DUX4 protein in U7-asDUX4-treated cells compared with those transfected with non-targeting U7-snRNA. Two bands, 52 kDa and 60 kDa, appeared in western blots after using the anti-Myc antibody on CMV.myc-DUX4-transfected HEK293 cell extracts. The 60-kDa protein band was detected in untransfected cells and migrates at approximately the size of endogenous Myc protein. Consistent with our prior immunofluorescence, cell death, and RNAscope results, U7-asDUX4 snRNAs reduced transfected DUX4 expression compared with non-targeting controls. Western blots were performed three times using protein extracts from three independent experiments (raw blots shown in Figure S3). Tubulin was used as a normalizer. (E) Quantification of the western blot. DUX4 protein signal intensity was reduced significantly in cells treated with U7-asDUX4-4 (87% ± 10%) and U7-asDUX4-8 (85% ± 14%) compared with the nontargeting controls. U7-asDUX4-5 and -7 target similar splice junction sites, and U7-asDUX4- 6 targets an intron1 SD site. We tested these three constructs here to determine whether any could induce DUX4 s production by altering correct splicing of FL DUX4 mRNA. No DUX4 short protein band (22 kDa) was detected using this western blot assay. ∗∗p ≤ 0.01, ANOVA; n = 3 independent experiments, where each experiment was normalized to its respective non-targeting control.
Next, to additionally confirm DUX4 protein knockdown by our lead U7-snRNAs, we performed similar co-transfection experiments in HEK293 cells but used western blots as an outcome measure. In addition, because the C-terminal V5 tag disrupted the U7-asDUX4-4 binding site, we utilized a different DUX4 expression construct in this set of experiments. Specifically, we generated a CMV.DUX4 expression plasmid containing an amino-terminal, in-frame Myc epitope tag (Figure 3C). We also reasoned that this construct would allow us to determine whether the U7-asDUX4 sequences designed to mask the FL DUX4 mRNA SD/SA near the 3′ end of exon 1 would bias splicing to produce a truncated and non-toxic DUX4-s protein isoform (Figure 3C).14 We performed western blotting using protein extracts from HEK293 cells co-transfected with plasmids expressing Myc.DUX4 and five different U7-asDUX4 constructs, including our 3 leads and sequences 5 and 6, which were designed to base-pair near the exon 1-intron 1 junction. Using a Myc epitope antibody, we detected the FL Myc-DUX4 protein band (52 kDa) in all transfected cells, and all samples contained a larger non-DUX4 protein that migrated at the size of endogenous c-Myc protein (∼60 kDa). Consistent with our previous experiments, our lead constructs, U7-asDUX4 sequences 4, 7, and 8, significantly reduced DUX4 protein by 87% ± 10%, 66% ± 15%, and 85% ± 14% (n = 3 independent experiments; Figures 3D and 3E; Figure S2). We did not detect evidence of DUX4 s production by western blot (predicted size, 22 kDa), suggesting that the reduction of FL DUX4 gene expression by U7-asDUX4 sequences designed to mask DUX4 splice sites (4, 5, 6, and 7) did not operate by shifting splicing patterns to favor the DUX4 s isoform. Similarly, we found no evidence of a shorter DUX4 s transcript using 3′ rapid amplification of cDNA ends (RACE) RT-PCR (Figure S3). We also noted that the non-specific upper band on these western blots showed variable expression. Because DUX4 has been shown to activate Myc, it is possible that changing DUX4 levels could affect the abundance of that upper band if it is Myc.15 However, we did not note a strong correlation between residual DUX4 and Myc abundance in these experiments.
U7-asDUX4s significantly decrease endogenous DUX4 expression in myotubes from individuals with FSHD
Our results in HEK293 cells suggested that several U7-asDUX4 snRNAs could reduce FL DUX4 expression and offer protection from cell death in an overexpression model. Next we assessed the ability of lead U7-asDUX4 snRNAs (4, 7, and 8) to decrease endogenous DUX4 mRNA in myotubes from individuals with FSHD using RNAscope in situ hybridization. We previously used RNAscope to detect DUX4 in FSHD myotubes.30 Consistent with prior reports, we found that DUX4 staining was only present in a small percentage of cells at any given time, but importantly, we were also able to quantify DUX4 knockdown following delivery of an artificial DUX4-targeted miRNA (mi405).30,40 We therefore used RNAscope to determine whether U7-asDUX4 snRNAs could reduce endogenous DUX4 signal in myotubes derived from individuals with FSHD, supporting the potential translatability of this approach. To do this, we used electroporation to transfect FSHD muscle cells, which typically yields ∼50%–70% transfection efficiency (Figure S4). Consistent with our previous results, untransfected 15A FSHD myotubes showed brown DUX4 signals, whereas those transfected with U7-asDUX4 sequences 4, 7, and 8 had significantly reduced RNAscope signals (Figures 4A–4H). This supported that these three U7-asDUX4s lead to destabilization and degradation of endogenous DUX4 mRNA in FSHD muscle cells.
Figure 4.
U7-asDUX4 constructs reduce endogenous DUX4 and DUX4-associated biomarkers in myotubes derived from individuals with FSHD
(A) FSHD 15A myotubes demonstrated higher amounts of DUX4 mRNA signal compared with cells treated with U7-asDUX4s. Arrows indicate an example of the DUX4-positive brown signal. (B–D) DUX4 expression in FSHD 15A myotubes was reduced or absent in 15A cells transfected with (B) U7-asDUX4-4, (C) U7-asDUX4-7, and (D) U7-asDUX4-8. (E) A very weak or absent signal was present in unaffected 15V myotubes, which served as a negative control for RNAscope staining using the DUX4 probe. (F) 15A myotubes stained with the housekeeping gene PPIB as a positive control for the RNAscope assay. (G) 15A myotubes stained with the bacterial dapB gene probe, which served as a negative control for the assay. 100× objective. Scale bar, 20 μm. (H) Quantification of the DUX4 RNAscope signal was performed as described previously.30 3–4 representative microscopic fields were counted from 3 independent experiments; each point represents quantification of one field. ∗∗p < 0.01, ANOVA. The DUX4 signal was absent or very low in unaffected 15V cells as well as affected 15A cells transfected with lead U7-asDUX4 snRNA plasmids compared with untreated, affected 15A samples. ∗∗p ≤ 0.01, ANOVA. (I) Knockdown of DUX4-activated biomarkers by U7-asDUX4 sequences. The plots show significant reductions in ZSCAN4, PRAMEF12, MBD3L2, and TRIM43 in U7-asDUX4-treated FSHD 15A myotubes compared with controls transfected with non-targeting snRNA. n = 4 independent experiments performed in triplicate or, in some cases, duplicate. ∗∗p ≤ 0.01, ANOVA.
U7-asDUX4 snRNAs decrease DUX4-activated biomarker expression in FSHD myotubes
With the emergence of prospective FSHD therapies came a need in the FSHD field to develop clinical outcome measures and biomarkers that could be used to establish therapeutic efficacy.41, 42, 43, 44 Although DUX4 expression is the most direct measure of target engagement by a prospective drug or gene therapy, it is difficult to detect and relatively scarce in FSHD muscle biopsies. Thus, DUX4 expression in human muscle biopsies is currently not a reliable outcome measure for FSHD clinical trials, and several groups have now turned to examining DUX4-activated biomarkers as an indirect measure of DUX4 expression. At least 67 different genes contain regulatory regions with DUX4 binding sites and are consistently activated upon DUX4 expression. However, recent studies suggest that only a few biomarkers are needed to represent the entire set.17,19,20,45, 46, 47, 48 We selected four biomarkers in this study (ZSCAN4, PRAMEF12, TRIM43, and MBD3L2) because they are established DUX4 target genes and FSHD disease biomarkers and consistently show differential expression between FSHD and healthy control cells in our experiments.19,20,24,26,30 We therefore tested the ability of our lead U7-asDUX4 snRNAs to suppress these DUX4-activated biomarkers in cells from individuals with FSHD. To do this, we transfected 15A myoblasts from individuals with FSHD with U7-asDUX4 snRNA-4, -7, and -8 as well as a non-targeting control. We then differentiated cells into myotubes for 7 days and performed quantitative RT-PCR to measure expression of the DUX4-activated human biomarkers TRIM43, MBD3L2, PRAMEF12, and ZSCAN4. All four biomarkers were present in untreated 15A myotubes and reduced significantly in U7-asDUX4-treated 15A cells (Figure 4I).
Discussion
The FSHD field has made great strides in the last decade or so by identifying DUX4 as a primary target for therapeutic intervention, generating numerous cell and animal models, producing several prospective therapeutic strategies, and working to define outcome measures for clinical trials. Despite this progress, there are still no approved treatments for FSHD, and therapeutic development remains a critical need in the field. We propose that FSHD therapies should focus on inhibiting DUX4, and several strategies that target the gene, mRNA, and/or protein could be utilized.24, 25, 26, 27, 28, 29,31,49 Our lab has been primarily focused on attacking the DUX4 mRNA, and we previously demonstrated the safety and efficacy of DUX4 silencing using RNAi-based gene therapy delivered by AAV vectors in pre-clinical studies.31 This strategy is now being translated. However, because even very small amounts of DUX4 protein may be toxic in muscle cells, we believe that it is beneficial to develop additional DUX4 silencing strategies employing alternative mechanisms, which could be used alone or in combinatorial therapies, to help maximize DUX4 silencing in muscles of individuals with FSHD. Here we describe a new method to accomplish DUX4 silencing using designed U7-asDUX4 snRNAs.
The U7-asDUX4 constructs we developed have some parallels to ASOs, which have been tested previously tested in FSHD models.18,23, 24, 25,29 A significant amount of knowledge about ASOs for treatment of muscle disease comes from their use in preclinical and clinical studies for treating Duchenne muscular dystrophy (DMD) and myotonic dystrophy (DM1).50, 51, 52, 53, 54 In the case of DMD, ASO uptake is facilitated by porous cell membranes associated with loss of the dystrophin glycoprotein complex.55, 56, 57 In contrast, intact muscle membranes in DM1 patients serve as a barrier to efficient ASO delivery, suggesting that improved delivery mechanisms are needed when treating muscle diseases without membrane defects.58 Similar to DM1, membrane damage is not a feature of muscles of individuals with FSHD; thus, delivering ASOs to FSHD muscles could be challenging. In addition, DUX4 is rare and not present in most FSHD myonuclei at any given time, but when it turns on at sufficiently toxic levels, it could rapidly kill cells expressing it. Thus, it is likely that therapies designed to inhibit DUX4 will need to be present within target muscles prior to DUX4 activation. Accomplishing this with ASOs will require repetitive, lifelong systemic dosing to produce optimal therapeutic efficacy.25 In contrast to ASOs, U7-snRNAs can be packaged as DNA expression cassettes within AAV vectors, many of which have natural myotropism. Thus, AAV-delivered U7-asDUX4 snRNAs should express the therapeutic antisense RNAs as long as the vector is present within a target muscle cell and the promoter is active. In stable, post-mitotic muscle, this could theoretically provide lifelong protection following a single administration, although the persistence of AAV transduction in human muscle still requires more study. In addition, U7-snRNAs are produced in the host nucleus, facilitating access to DUX4 pre-mRNA, and their association with the U7-snRNPs increases their resistance to degradation.59 These properties increase their effectiveness and reduce the necessity for repeated administration, especially for diseases that require lifelong treatment, such as FSHD. In the next step of our study, it will be important to assess the long-term safety and efficacy of our lead U7-asDUX4 sequences, or combinations thereof, using AAV vectors in our FSHD mouse models.11 Part of this will include investigation of sequence-specific off-target effects. Nucleotide BLAST results using U7-asDUX4-4 and -7 against the human transcriptome yielded no significant hits, whereas sequence 8 has partial sequence complementarity to some human transcripts, including ZNF91 (80%), NAA16 (60%), CYB5A (53%), CDH19 (53%), and STEAP2 (50%) and 46% sequence complementarity with TCAIM LIPK, FAM177A1, MAN1A1, XCR1, and the FREM2 and ZDHHC3 non-coding RNAs. These predicted changes could be confirmed experimentally using RNA sequencing (RNA-seq) and/or western blotting to detect protein changes.
U7 snRNAs are transcribed in the nucleus and then exported to the cytoplasm, where they assemble with Sm and Lsm proteins. The assembled U7-snRNP can remain in the cytoplasm or be imported back into the nucleus. In the nucleus, they are associated with splicing machinery, whereas in the cytoplasm, they associate with P bodies, which normally function in mRNA turnover.60 Similarly, mRNAs can be detected in the nucleus and the cytoplasm because they are transcribed and matured in the nucleus and then transported to the cytoplasm for translation. The sequences we identified as our lead constructs target the exon 1-intron1 junction (U7-asDUX4-4 and -7) or the DUX4 poly(A) signal (U7-asDUX4-8). Targeting the splice junction is a new approach, but using antisense sequences to bind the DUX4 PAS has been done previously using chemically synthesized ASOs, which have been shown to reduce DUX4 and DUX4-activated biomarkers in vitro and in vivo.18,23, 24, 25 Polyadenylation is an important process required for stabilizing nascent mRNAs and coordinating mRNA transit through nuclear pores to the cytoplasm for translation. Chemically synthesized DNA-based ASOs may operate by forming DNA:RNA hybrids and activating RNase H against the target transcript, but it is also possible that published ASO sequences designed to base-pair with the DUX4 poly(A) signal could operate by masking the signal and preventing polyadenylation, leading to DUX4 mRNA destabilization. Because the antisense portion of our PAS-targeting U7-asDUX4-8 was composed of RNA, it should not be able to reduce DUX4 expression via RNase H, which requires base-pairing of DNA:RNA molecules. It is therefore more likely that U7-asDUX4-8 operates by recruiting snRNP proteins to the DUX4 transcript and sterically hinders poly(A) machinery, destabilizing DUX4 mRNA. Thus, sequence 8 might work in the nucleus, whereas sequences 4 and 7, which are located near the exon 1 splice junction, could operate in the nucleus or the cytoplasm.
In conclusion, we identified three lead U7-asDUX4 constructs that significantly reduced DUX4 and DUX4-associated outcomes in co-transfected cells and myotubes derived from individuals with FSHD. Our findings provide a proof of concept for DUX4 silencing using recombinant U7-asDUX4 as a treatment for FSHD. Translating this approach will require evaluating efficacy and safety in FSHD mouse models following AAV-mediated delivery, which is now ongoing in our laboratory.
Materials and methods
Designing DUX4 targeting U7-snRNAs
The Human Splicing Finder version 3.1 program from Marseille University (http://umd.be/Redirect.html) was used to predict potential SA, SD, and SE sites at the end of DUX4 exon 1 (coding sequence) and within the untranslated exons 1 and 2. For designing U7-snRNAs against DUX4 (called U7-asDUX4s), we selected 18 high-scoring target sites with the fewest number of CpGs and one non-targeting region (Table S1). Predicted off-target matches were determined by BLAST, using each sequence against the human genome database (http://blast.ncbi.nlm.nih.gov//blast.ncbi.nlm.nih.gov/Blast.cgi). The expression cassettes of all U7-asDUX4s, containing a mouse U7 promoter, were synthesized and cloned into the pUCIDT plasmid (Integrated DNA Technologies). Sequences were also designed to bind the DUX4 start codon and poly(A) signal via reverse complementary base-pairing (Table S1). The non-targeting control snRNA antisense sequence is 5′-GTCATGTCGCGTGCCCCGGTGGTCGACACGTCGG-3′.
Cell culture
HEK293 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin at 37°C in 5% CO2. Affected and unaffected immortalized human myoblasts derived from an individual with FSHD and an unaffected relative (15Abic and 15Vbic)40,61 were expanded in DMEM supplemented with 16% medium 199, 20% fetal bovine serum, 1% penicillin/streptomycin, 30 ng/mL zinc sulfate, 1.4 mg/mL vitamin B12, 55 ng/mL dexamethasone, 2.5 ng/mL human growth factor, 10 ng/mL fibroblast growth factor, and 20 mM HEPES. Cells were maintained as myoblasts and differentiated prior to measuring DUX4 mRNA and DUX4-activited biomarkers by qRT-PCR and RNAscope. To differentiate myoblasts into myotubes, transfected myoblasts were switched to differentiation medium composed of a 4:1 ratio of DMEM:medium 199 supplemented with 15% KnockOut Serum (Thermo Fisher Scientific), 2 mM L-glutamine, and 1% antibiotics/antimycobiotics for up to 7 days before harvesting.
Viability assay
HEK293 cells (250,000 cells/well) were co-transfected (Lipofectamine 2000, Invitrogen) with an expression plasmid from which FL DUX4 pre-mRNA (DUX4-FL) was transcribed from the CMV promoter (CMV.DUX4-FL) along with plasmids expressing U7-asDUX4 snRNAs or the non-targeting U7-snRNA at a 1:6 ratio using the protocol. The cells were trypsinized 48 h after transfection and collected in 1 mL of growth medium. Automated cell counting was performed using Countess cell counting chamber slides. The results were confirmed with traditional cell counting using a hemocytometer and trypan blue staining. Three independent experiments were performed, and data were reported as a mean of total cell number ± SEM per group.
Cell death assay
HEK293 cells (42,000 cells/well) were plated on a 96-well plate 16 h prior to transfection. The next morning, cells were co-transfected (Lipofectamine 2000, Invitrogen) with CMV.DUX4-FL and U7-asDUX4 snRNAs or a non-targeting U7-snRNA expression plasmid at a 1:6 molar ratio. Cell death was measured using the Apo-ONE homogeneous caspase-3/7 assay (Promega, Madison, WI) 48 h after transfection using a fluorescent plate reader (Spectra Max M2, Molecular Devices, Sunnyvale, CA). Three individual assays were performed in triplicate, and data were averaged per experiment and reported as mean caspase activity ± SEM relative to our control assay, which was transfected with CMV.DUX4-FL only.
Western blot assay
For this experiment, DUX4 expression plasmids were used with and without epitope tags (CMV.Myc-DUX4-FL, which contained a Myc epitope tag fused to the DUX4 N terminus, or CMV-DUX4-FL). HEK293 cells were co-transfected at a 1:6 ratio of DUX4:U7asDUX4 expression plasmids. Twenty hours after transfection, cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) supplemented with a cocktail containing protease inhibitors. Protein concentration was determined using the Lowry protein assay kit (Bio-Rad). 25 μg of each protein sample was run on a 12% SDS-polyacrylamide gel. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes via a semi-dry transfer method, blocked in 5% non-fat milk, and incubated with primary monoclonal mouse anti-DUX4 (1:500; P4H2, Novus Biologicals), mouse anti-Myc (R95125, Invitrogen), or rabbit polyclonal anti-α-tubulin antibodies (1:1,000; ab15246, Abcam, Cambridge, MA) overnight at 4°C. The next day, following multiple washes, blots were probed with horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (1:100,000; Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. Protein bands were developed on X-ray films after short incubation in Immobilon chemiluminescent HRP substrate (Millipore, Billerica, MA). Protein quantification was assessed by ImageJ software (National Institutes of Health, Bethesda, Maryland, USA; https://imagej.nih.gov/ij/).
Immunofluorescence staining
V5 epitope-tagged DUX4 protein in treated and untreated cells was visualized using V5 immunofluorescence staining.10 HEK293 cells were transfected with a plasmid carrying a FL DUX4 sequence consisting of the coding and 3′ UTR sequences but engineered to express DUX4 protein with an in-frame carboxy-terminal V5 epitope fusion. Twenty hours after transfection, cells were fixed in 4% paraformaldehyde (PFA) for 20 min, and nonspecific antigens were blocked with 5% BSA in PBS supplemented with 0.2% Triton X-100. The cells were incubated at 4°C, overnight in rabbit polyclonal anti-V5 primary antibody (1:2,500, Abcam, ab9116). The following day, cells were washed with PBS, incubated with goat anti-rabbit Alexa 594 secondary antibodies (1:2,500, Invitrogen), and mounted with Vectashield mounting medium containing DAPI (Vector Laboratories, Burlingame, CA).
RNAscope assay and quantification
Detecting overexpressed DUX4 in HEK293 cells
We used RNAscope in situ hybridization assay to measure DUX4 mRNA levels following co-transfection of CMV.DUX4-FL and U7.asDUX4 expression plasmids in HEK293 cells (1:6 ratio). Specifically, HEK293 cells were seeded in triplicate on glass coverslips in 24-well plates at a density of 120,000 cells per well 16 h prior to transfection. The next morning, upon reaching 70% confluency, cells were co-transfected with 250 ng of CMV.DUX4-FL expression plasmid (Lipofectamine 2000, Thermo Fisher Scientific), according to the manufacturer’s instructions. Sixteen hours after transfection, cells were fixed with 4% PFA, and RNAscope staining was performed following the manufacturer’s instructions (ACDBio; detailed below).
Detecting endogenous DUX4 in human FSHD myotubes
To determine the specificity of U7-asDUX4 snRNAs for targeting endogenous DUX4 mRNA, 15Abic FSHD myoblasts (15A, 500,000 cells/reaction) were transfected with U7-asDUX4 expression plasmids via electroporation (Lonza, VVPD-1001) and then differentiated into myotubes for up to 7 days. RNAscope staining was performed as described previously.30 The cells were fixed in 4% PFA and dehydrated/rehydrated with ethanol gradients. Endogenous peroxidase activity was blocked by hydrogen peroxide treatment. Protease III was added to increase the permeability of fixed cells for RNAscope probes. The cells were treated with a DUX4-specific RNAscope probe (ACDBio, catalog number 498541) or probes to detect the positive control housekeeping gene peptidylprolyl isomerase B (PPIB) and negative control bacterial gene dihydrodipicolinate reductase (dapB). Following probe incubation, cells were treated with several signal amplification steps using RNAscope 2.5 HD Assay Brown according to the manufacturer’s protocol (ACDBio). The cells were counterstained with 50% Gill’s hematoxylin I (catalog number HXGHE1LT, American Master Tech Scientific) for 2 min at room temperature, followed by several washes. After mounting, images were captured using an Olympus DP71 microscope. DUX4 RNAscope signals were quantified using ImageJ-Fiji software as described previously.30
Quantitative real-time PCR analysis of DUX4 biomarkers
15A FSHD myoblasts were transfected as described in RNAscope assay and quantification above and differentiated into myotubes. Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol, and yield was measured by Nanodrop. Isolated RNA was then DNase treated (DNA-Free, Ambion, TX), and cDNA was generated with a high-capacity cDNA reverse transcription kit (Applied Biosystems) using random hexamer primers. Subsequent cDNA samples were then used as a template for the TaqMan Assay using pre-designed TRIM43, MBD3L2, PRAMEF12, and ZSCAN4 (biomarkers of DUX4 activity) and human RPL13A control primer/probe sets (Applied Biosystems). All data were normalized to the non-targeting U7-snRNA-transfected cells. Data were generated from two independent experiments performed in triplicate for each biomarker.
Statistical analysis
All statistical analyses (caspase-3/7 assay, cell viability assay, RNAscope, western blot, and qRT-PCR) were performed in GraphPad Prism 5 (GraphPad, La Jolla, CA) using the indicated statistical tests.
Acknowledgments
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Center of Research Translation in Muscular Dystrophy Therapeutic Development grant 1P50AR070604. During the course of this study, A.R.’s salary was supported by a post-doctoral fellowship from the Friends of FSH Research Foundation. During the course of this study, G.A.-C.’s salary was supported by a post-doctoral grant from the FSHD Society (grant 5888746560). The authors thank members of the Harper laboratory for assistance and support and Dr. Charles Emerson, Dr. Kathryn Wagner, and the University of Massachusetts Wellstone Muscular Dystrophy Program for providing the human immortalized myoblasts used in our studies.
Author contributions
A.R. designed U7-asDUX4s, performed or directed all experiments, created figures and legends, and wrote the paper. G.A.-C. performed the RNAscope assay and quantified the corresponding results. N.K.T cloned the Myc-DUX4 plasmid, contributed to western blots, and assisted with manuscript writing and editing. N.W. contributed to construct design. S.Q.H. conceptualized and obtained funding for the project, designed U7-asDUX4s, and edited the manuscript.
Declaration of interests
The sequences and methods described here were included in a provisional patent application filed on December 1, 2020 (USPTO serial no. 63/120,190). S.Q.H., A.R., and N.W. are listed as inventors.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.004.
Supplemental information
References
- 1.Deenen J.C., Arnts H., van der Maarel S.M., Padberg G.W., Verschuuren J.J., Bakker E., Weinreich S.S., Verbeek A.L., van Engelen B.G. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology. 2014;83:1056–1059. doi: 10.1212/WNL.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastorello E., Cao M., Trevisan C.P. Atypical onset in a series of 122 cases with FacioScapuloHumeral Muscular Dystrophy. Clin. Neurol. Neurosurg. 2012;114:230–234. doi: 10.1016/j.clineuro.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klinge L., Eagle M., Haggerty I.D., Roberts C.E., Straub V., Bushby K.M. Severe phenotype in infantile facioscapulohumeral muscular dystrophy. Neuromuscul. Disord. 2006;16:553–558. doi: 10.1016/j.nmd.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Statland J.M., Tawil R. Facioscapulohumeral Muscular Dystrophy. Continuum (Minneap. Minn.) 2016;22:1916–1931. doi: 10.1212/CON.0000000000000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camaño P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017;49:935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppée F. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A.M., Leo O. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace L.M., Garwick S.E., Mei W., Belayew A., Coppee F., Ladner K.J., Guttridge D., Yang J., Harper S.Q. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giesige C.R., Wallace L.M., Heller K.N., Eidahl J.O., Saad N.Y., Fowler A.M., Pyne N.K., Al-Kharsan M., Rashnonejad A., Chermahini G.A. AAV-mediated follistatin gene therapy improves functional outcomes in the TIC-DUX4 mouse model of FSHD. JCI Insight. 2018;3:e123538. doi: 10.1172/jci.insight.123538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowaljow V., Marcowycz A., Ansseau E., Conde C.B., Sauvage S., Mattéotti C., Arias C., Corona E.D., Nuñez N.G., Leo O. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Snider L., Asawachaicharn A., Tyler A.E., Geng L.N., Petek L.M., Maves L., Miller D.G., Lemmers R.J., Winokur S.T., Tawil R. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., Miller D.G. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shadle S.C., Zhong J.W., Campbell A.E., Conerly M.L., Jagannathan S., Wong C.J., Morello T.D., van der Maarel S.M., Tapscott S.J. DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet. 2017;13:e1006658. doi: 10.1371/journal.pgen.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dmitriev P., Bou Saada Y., Dib C., Ansseau E., Barat A., Hamade A., Dessen P., Robert T., Lazar V., Louzada R.A.N. DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med. 2016;99:244–258. doi: 10.1016/j.freeradbiomed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Rickard A.M., Petek L.M., Miller D.G. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 2015;24:5901–5914. doi: 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S.D., Coppée F., Belayew A. The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLoS ONE. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Z., Snider L., Balog J., Lemmers R.J., Van Der Maarel S.M., Tawil R., Tapscott S.J. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 2014;23:5342–5352. doi: 10.1093/hmg/ddu251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eidahl J.O., Giesige C.R., Domire J.S., Wallace L.M., Fowler A.M., Guckes S.M., Garwick-Coppens S.E., Labhart P., Harper S.Q. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum. Mol. Genet. 2016;25:4577–4589. doi: 10.1093/hmg/ddw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyle R., Wright T.J., Clark L.N., Hewitt J.E. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 22.Gabriëls J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 23.Marsollier A.C., Ciszewski L., Mariot V., Popplewell L., Voit T., Dickson G., Dumonceaux J. Antisense targeting of 3′ end elements involved in DUX4 mRNA processing is an efficient therapeutic strategy for facioscapulohumeral dystrophy: a new gene-silencing approach. Hum. Mol. Genet. 2016;25:1468–1478. doi: 10.1093/hmg/ddw015. [DOI] [PubMed] [Google Scholar]
- 24.Chen J.C., King O.D., Zhang Y., Clayton N.P., Spencer C., Wentworth B.M., Emerson C.P., Jr., Wagner K.R. Morpholino-mediated Knockdown of DUX4 Toward Facioscapulohumeral Muscular Dystrophy Therapeutics. Mol. Ther. 2016;24:1405–1411. doi: 10.1038/mt.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ansseau E., Vanderplanck C., Wauters A., Harper S.Q., Coppée F., Belayew A. Antisense Oligonucleotides Used to Target the DUX4 mRNA as Therapeutic Approaches in FaciosScapuloHumeral Muscular Dystrophy (FSHD) Genes (Basel) 2017;8:93. doi: 10.3390/genes8030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek A., Zhang Y., Woodman K.G., Huang S., DeSimone A.M., Cohen J., Ho V., Conner J., Mead L., Kodani A. Applying genome-wide CRISPR-Cas9 screens for therapeutic discovery in facioscapulohumeral muscular dystrophy. Sci. Transl. Med. 2020;12:eaay0271. doi: 10.1126/scitranslmed.aay0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himeda C.L., Jones T.I., Jones P.L. CRISPR/dCas9-mediated Transcriptional Inhibition Ameliorates the Epigenetic Dysregulation at D4Z4 and Represses DUX4-fl in FSH Muscular Dystrophy. Mol. Ther. 2016;24:527–535. doi: 10.1038/mt.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSimone A.M., Leszyk J., Wagner K., Emerson C.P., Jr. Identification of the hyaluronic acid pathway as a therapeutic target for facioscapulohumeral muscular dystrophy. Sci. Adv. 2019;5:eaaw7099. doi: 10.1126/sciadv.aaw7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim K.R.Q., Maruyama R., Echigoya Y., Nguyen Q., Zhang A., Khawaja H., Sen Chandra S., Jones T., Jones P., Chen Y.W., Yokota T. Inhibition of DUX4 expression with antisense LNA gapmers as a therapy for facioscapulohumeral muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2020;117:16509–16515. doi: 10.1073/pnas.1909649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amini Chermahini G., Rashnonejad A., Harper S.Q. RNAscope in situ hybridization-based method for detecting DUX4 RNA expression in vitro. RNA. 2019;25:1211–1217. doi: 10.1261/rna.070177.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace L.M., Saad N.Y., Pyne N.K., Fowler A.M., Eidahl J.O., Domire J.S., Griffin D.A., Herman A.C., Sahenk Z., Rodino-Klapac L.R., Harper S.Q. Pre-clinical Safety and Off-Target Studies to Support Translation of AAV-Mediated RNAi Therapy for FSHD. Mol. Ther. Methods Clin. Dev. 2017;8:121–130. doi: 10.1016/j.omtm.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma A. Recent Advances in Antisense Oligonucleotide Therapy in Genetic Neuromuscular Diseases. Ann. Indian Acad. Neurol. 2018;21:3–8. doi: 10.4103/aian.AIAN_298_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ideue T., Adachi S., Naganuma T., Tanigawa A., Natsume T., Hirose T. U7 small nuclear ribonucleoprotein represses histone gene transcription in cell cycle-arrested cells. Proc. Natl. Acad. Sci. USA. 2012;109:5693–5698. doi: 10.1073/pnas.1200523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schümperli D., Pillai R.S. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goyenvalle A., Babbs A., van Ommen G.J., Garcia L., Davies K.E. Enhanced exon-skipping induced by U7 snRNA carrying a splicing silencer sequence: Promising tool for DMD therapy. Mol. Ther. 2009;17:1234–1240. doi: 10.1038/mt.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goyenvalle A., Wright J., Babbs A., Wilkins V., Garcia L., Davies K.E. Engineering multiple U7snRNA constructs to induce single and multiexon-skipping for Duchenne muscular dystrophy. Mol. Ther. 2012;20:1212–1221. doi: 10.1038/mt.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nualkaew T., Jearawiriyapaisarn N., Hongeng S., Fucharoen S., Kole R., Svasti S. Restoration of correct βIVS2-654-globin mRNA splicing and HbA production by engineered U7 snRNA in β-thalassaemia/HbE erythroid cells. Sci. Rep. 2019;9:7672. doi: 10.1038/s41598-019-43964-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biferi M.G., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Bos C., Voit T., Ferry A., Barkats M. A New AAV10-U7-Mediated Gene Therapy Prolongs Survival and Restores Function in an ALS Mouse Model. Mol. Ther. 2017;25:2038–2052. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wein N., Vulin A., Falzarano M.S., Szigyarto C.A., Maiti B., Findlay A., Heller K.N., Uhlén M., Bakthavachalu B., Messina S. Translation from a DMD exon 5 IRES results in a functional dystrophin isoform that attenuates dystrophinopathy in humans and mice. Nat. Med. 2014;20:992–1000. doi: 10.1038/nm.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P., Jr. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LoRusso S., Johnson N.E., McDermott M.P., Eichinger K., Butterfield R.J., Carraro E., Higgs K., Lewis L., Mul K., Sacconi S., ReSolve Investigators and the FSHD CTRN18 Clinical trial readiness to solve barriers to drug development in FSHD (ReSolve): protocol of a large, international, multi-center prospective study. BMC Neurol. 2019;19:224. doi: 10.1186/s12883-019-1452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mul K., Vincenten S.C.C., Voermans N.C., Lemmers R.J.L.F., van der Vliet P.J., van der Maarel S.M., Padberg G.W., Horlings C.G.C., van Engelen B.G.M. Adding quantitative muscle MRI to the FSHD clinical trial toolbox. Neurology. 2017;89:2057–2065. doi: 10.1212/WNL.0000000000004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L.H., Friedman S.D., Shaw D., Snider L., Wong C.J., Budech C.B., Poliachik S.L., Gove N.E., Lewis L.M., Campbell A.E. MRI-informed muscle biopsies correlate MRI with pathology and DUX4 target gene expression in FSHD. Hum. Mol. Genet. 2019;28:476–486. doi: 10.1093/hmg/ddy364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong C.J., Wang L.H., Friedman S.D., Shaw D., Campbell A.E., Budech C.B., Lewis L.M., Lemmers R.J.F.L., Statland J.M., van der Maarel S.M. Longitudinal measures of RNA expression and disease activity in FSHD muscle biopsies. Hum. Mol. Genet. 2020;29:1030–1043. doi: 10.1093/hmg/ddaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., Tapscott S.J. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagannathan S., Shadle S.C., Resnick R., Snider L., Tawil R.N., van der Maarel S.M., Bradley R.K., Tapscott S.J. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet. 2016;25:4419–4431. doi: 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Heuvel A., Mahfouz A., Kloet S.L., Balog J., van Engelen B.G.M., Tawil R., Tapscott S.J., van der Maarel S.M. Single-cell RNA sequencing in facioscapulohumeral muscular dystrophy disease etiology and development. Hum. Mol. Genet. 2019;28:1064–1075. doi: 10.1093/hmg/ddy400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma V., Harafuji N., Belayew A., Chen Y.W. DUX4 differentially regulates transcriptomes of human rhabdomyosarcoma and mouse C2C12 cells. PLoS ONE. 2013;8:e64691. doi: 10.1371/journal.pone.0064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas L.A., Valentine E., Accorsi A., Maglio J., Shen N., Robertson A., Kazmirski S., Rahl P., Tawil R., Cadavid D. p38α Regulates Expression of DUX4 in a Model of Facioscapulohumeral Muscular Dystrophy. J. Pharmacol. Exp. Ther. 2020;374:489–498. doi: 10.1124/jpet.119.264689. [DOI] [PubMed] [Google Scholar]
- 50.Wein N., Vulin A., Findlay A.R., Gumienny F., Huang N., Wilton S.D., Flanigan K.M. Efficient Skipping of Single Exon Duplications in DMD Patient-Derived Cell Lines Using an Antisense Oligonucleotide Approach. J. Neuromuscul. Dis. 2017;4:199–207. doi: 10.3233/JND-170233. [DOI] [PubMed] [Google Scholar]
- 51.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., De Kimpe S.J., Eagle M., Guglieri M., Hood S. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 52.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendell J.R., Goemans N., Lowes L.P., Alfano L.N., Berry K., Shao J., Kaye E.M., Mercuri E., Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal effect of eteplirsen versus historical control on ambulation in Duchenne muscular dystrophy. Ann. Neurol. 2016;79:257–271. doi: 10.1002/ana.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrell S.T., Carrell E.M., Auerbach D., Pandey S.K., Bennett C.F., Dirksen R.T., Thornton C.A. Dmpk gene deletion or antisense knockdown does not compromise cardiac or skeletal muscle function in mice. Hum. Mol. Genet. 2016;25:4328–4338. doi: 10.1093/hmg/ddw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aoki Y., Nagata T., Yokota T., Nakamura A., Wood M.J., Partridge T., Takeda S. Highly efficient in vivo delivery of PMO into regenerating myotubes and rescue in laminin-α2 chain-null congenital muscular dystrophy mice. Hum. Mol. Genet. 2013;22:4914–4928. doi: 10.1093/hmg/ddt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heemskerk H., de Winter C., van Kuik P., Heuvelmans N., Sabatelli P., Rimessi P., Braghetta P., van Ommen G.J., de Kimpe S., Ferlini A. Preclinical PK and PD studies on 2′-O-methyl-phosphorothioate RNA antisense oligonucleotides in the mdx mouse model. Mol. Ther. 2010;18:1210–1217. doi: 10.1038/mt.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu-Motohashi Y., Komaki H., Motohashi N., Takeda S., Yokota T., Aoki Y. Restoring Dystrophin Expression in Duchenne Muscular Dystrophy: Current Status of Therapeutic Approaches. J. Pers. Med. 2019;9:1. doi: 10.3390/jpm9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-Barriga A., Kranzen J., Croes H.J., Bijl S., van den Broek W.J., van Kessel I.D., van Engelen B.G., van Deutekom J.C., Wieringa B., Mulders S.A., Wansink D.G. Cell membrane integrity in myotonic dystrophy type 1: implications for therapy. PLoS ONE. 2015;10:e0121556. doi: 10.1371/journal.pone.0121556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brun C., Suter D., Pauli C., Dunant P., Lochmüller H., Burgunder J.M., Schümperli D., Weis J. U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping. Cell. Mol. Life Sci. 2003;60:557–566. doi: 10.1007/s000180300047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J.L., Gall J.G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA. 2007;104:11655–11659. doi: 10.1073/pnas.0704977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadler G., Rahimov F., King O.D., Chen J.C., Robin J.D., Wagner K.R., Shay J.W., Emerson C.P., Jr., Wright W.E. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat. Struct. Mol. Biol. 2013;20:671–678. doi: 10.1038/nsmb.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.