Abstract
Purpose
The deformable nature of the liver can make focal treatment challenging and is not adequately addressed with simple rigid registration techniques. More advanced registration techniques can take deformations into account (eg, biomechanical modeling) but require segmentations of the whole liver for each scan, which is a time-intensive process. We hypothesize that fully convolutional networks can be used to rapidly and accurately autosegment the liver, removing the temporal bottleneck for biomechanical modeling.
Methods and Materials
Manual liver segmentations on computed tomography scans from 183 patients treated at our institution and 30 scans from the Medical Image Computing & Computer Assisted Intervention challenges were collected for this study. Three architectures were investigated for rapid automated segmentation of the liver (VGG-16, DeepLabv3 +, and a 3-dimensional UNet). Fifty-six cases were set aside as a final test set for quantitative model evaluation. Accuracy of the autosegmentations was assessed using Dice similarity coefficient and mean surface distance. Qualitative evaluation was also performed by 3 radiation oncologists on 50 independent cases with previously clinically treated liver contours.
Results
The mean (minimum-maximum) mean surface distance for the test groups with the final model, DeepLabv3 +, were as follows: μContrast(N = 17): 0.99 mm (0.47-2.2), μNon_Contrast(N = 19)l: 1.12 mm (0.41-2.87), and μMiccai(N = 30)t: 1.48 mm (0.82-3.96). The qualitative evaluation showed that 30 of 50 autosegmentations (60%) were preferred to manual contours (majority voting) in a blinded comparison, and 48 of 50 autosegmentations (96%) were deemed clinically acceptable by at least 1 reviewing physician.
Conclusions
The autosegmentations were preferred compared with manually defined contours in the majority of cases. The ability to rapidly segment the liver with high accuracy achieved in this investigation has the potential to enable the efficient integration of biomechanical model-based registration into a clinical workflow.
Introduction
The treatment of primary and secondary liver cancers normally involves surgery, chemoembolization, ablation, or external beam radiation therapy (RT), and requires 3-dimensional (3D) imaging, either computed tomography (CT) or magnetic resonance imaging. Specifically for RT, segmentation of the liver on CT imaging is required for treatment planning. More complex RT treatment regimens may include additional CT imaging for dose escalation and plan adaptation. Owing to the deformable nature of the liver, simple rigid registration techniques are suboptimal for 3D dose tracking. Biomechanical model-based deformable image registration have been shown to accurately model the deformation of the liver,1,2 but these methods require segmentations of the whole liver as the controlling region of interest. Thus, segmenting the liver in a consistent manner is particularly important. Manual contouring of the liver can take up to 30 minutes,3,4 and in the case of biomechanical models, inter/intraoperator contour variations may adversely affect the deformable image registration.
Fully convolutional networks (FCNs) have shown great promise in the ability to accurately segment 2- and 3D images with multiple classes in very short amounts of time.5, 6, 7, 8, 9 FCNs have been shown to accurately contour the liver in contrast-enhanced CT images,10 but less work on noncontrast CT images has been reported. When assessing the feasibility of these FCN models, Dice similarity coefficient (DSC) scores or volume differences are often reported as a sole metric. Unfortunately, such metrics can be relatively insensitive to erratic edge segmentations that would not be acceptable for clinical use, yet return a high score. For example, for mean surface distance (MSD), the sheer number of images present on a CT scan can result in a low value while hiding potentially clinically impactful errors. Therefore, the work described herein includes both quantitative and separate qualitative (blinded physician comparison) analyses of the results to determine actual clinical feasibility. We hypothesize that FCNs can be used to rapidly and accurately contour the liver on both contrast- and noncontrast-enhanced CT with minimal disruption of the treatment workflow.
Methods and Materials
Data
For this retrospective work, 155 consecutively acquired patients from our institution (The University of Texas MD Anderson Cencer Center) were collected under an institutional review board–approved protocol and 30 image sets were obtained from the Medical Image Computing & Computer Assisted Intervention (MICCAI) multiatlas challenge (data: https://www.synapse.org/#!Synapse:syn3193805/wiki/89480).11 Research data from our insitution is not available at this time.
Contrast-enhanced computed tomography scans
Of the 155 consecutive patients from our institution, 62 patients received intravenous contrast CT using a quadriphasic protocol, enabling the visualization of the tumor and vasculature within the liver, with pixel spacing ranging from 0.5625 mm to 1.27 mm and slice thickness from 1.5 cm to 5 cm. For this cohort of patients, manual segmentations of the liver were defined on both scans by a graduate student (BMA) under the guidance, evaluation, and approval of a board-certified interventional radiologist (BCO) with expertise in treating cancers in the liver. Forty-two patients had multiple CT images and when this occurred, the patient images were kept within the training group. In total, there were 108 contrast-enhanced CT examinations.
Noncontrast computed tomography scans
The remaining 93 patients’ CT scans were without contrast injection and are unique patients from the original contrast-enhanced CT cohort of patients. These noncontrast CT scans were included in the training set to create a more robust model that could identify the liver in both contrast and noncontrast images. We found that creating a model using contrast images alone provided similarly high DSC scores on the test set (μ = 0.96; σ = 0.02), but inspection of the noncontrast-enhanced images often showed oversegmentation, including the heart, and undersegmentation when near a disease site. Therefore, we deemed including contrast and noncontrast image sets necessary. Three patients had multiple examinations, and when this occurred, the patient images were kept in the training cohort. In total, there were 97 noncontrast-enhanced CT examinations. Fifty-three of these image sets had previously defined manual contours, which were visually evaluated and edited when deemed necessary. The contours for the remaining 44 image sets are explained in the data preparation section.
Training/validation data
Seventy-two institutional contrast-enhanced CT image sets and 63 institutional noncontrast CT images were randomly selected from the data cohort as training sets, resulting in a total of 135 images. We ensured that all 42 patients with multiple examinations appeared solely within the training set (ie, no overlap between training/testing/validation sets). Nineteen contrast-enhanced CT images and 15 noncontrast CT image sets were randomly selected as a validation set to optimize the model parameters.
Test data
A test set of 66 images was created, composed of 30 MICCAI abdomen challenge image sets and 19 noncontrast-enhanced and 17 contrast-enhanced institutional image sets. The patients were never seen in either the training or validation phase of the model.
A breakdown of the data and distribution of images across training, testing, and validation is shown in Table 1. These images were acquired on a variety of scanners with varying imaging protocols. Aa detailed description of the imaging parameters of all data is shown in Table E1.
Table 1.
Data distribution
| Data set | Npatients | NImages | Distribution (images) |
||
|---|---|---|---|---|---|
| Train | Validation | Test | |||
| Contrast | 62 | 108 | 72 | 19 | 17 |
| Noncontrast | 93 | 97 | 63 | 15 | 19 |
| MICCAI | 30 | 30 | 0 | 0 | 30 |
Abbreviation: MICCAI = Medical Image Computing & Computer Assisted Intervention.
All patients with multiple examinations were kept in the training set.
Architecture
We investigated 3 architectures, 2 of which are 2-dimensional (2D) and built upon the ideas of transfer learning (eg, ability to take a previously trained network and maintain some of the abstract concepts for a new identification task), and 1 3D U-Net style architecture. The pretrained networks were learned on nonmedical images and then applied to our segmentation task. First, we investigated a 2D U-Net style architecture built on top of the Visual Geometry Group (VGG)–16 net,12 where 16 refers to the number of convolutional and fully connected layers. The VGG network was originally created to classify images from >1000 classes (eg, dog, cat, car). These images are nonmedical, but the features learned in the early layers are often abstract and can be useful to other tasks.
Studies have shown that algorithms pretrained on nonmedical images improve segmentation accuracies on medical images.13 Long et al adapted the VGG-16 architecture for pixel-wise segmentation by including transpose convolutions and skip connections.5 For our architecture, we used bilinear upsampling in lieu of transpose convolutions to help mitigate the issue of checkerboard artifact (https://distill.pub/2016/deconv-checkerboard/) and added concatenation layers. The VGG-16 architecture was investigated because the reduced number of parameters increases training and prediction efficiency, and Long et al. had similarly found negligible differences between the VGG-16 and -19 architectures.
Second, we investigated the Deeplabv3+ network with an implementation in tensorflow, facilitated by the work presented here (https://github.com/bonlime/keras-deeplab-v3-plus). This network benefits from the robustness of spatial pyramid pooling and the sharp lines achieved from the encoder-decoder setup. Contrary to most encoder-decoder architectures, this saves memory by only implementing a single skip layer. Our code varies slightly from the original implementation in that all relu activations were converted to elu activations, and the dropout was removed, helping to remove some of the model instability seen in training.
Lastly, we investigated a 3D U-Net style architecture with and without atrous convolutions and residual and skip connections. We investigated varying numbers of layers from 2 to 5, the number of atrous convolutions, and the number of initial and maximum filters.
Data evaluation
Quantitative
The accuracy of the liver autosegmentation algorithm, compared with manual segmentation, was determined based on 2 metrics: DSC (Eq. 1) and MSD. A paired student t test was performed between the 3 models on the test data to evaluate the best final model.
| (1) |
Qualitative
A second, completely independent set of 50 patients (25 hepatocellular carcinoma [HCC] and 25 colorectal liver metastases [CLM]) who had received RT at The University of Texas MD Anderson Cancer Center, with clinically defined and approved manually delineated contours of the liver, was also obtained. A group of 3 radiation oncologists experienced in treating liver cancer (EK, GS, PD) were asked to blindly rate the generated contours versus the previously manually defined and approved clinical contours. The clinically defined and peer-reviewed contours had been created by the gastrointestinal radiation oncology group at our institution, which included the 3 radiation oncologists performing the comparisons. One of the radiation oncologists reviewed all 50 patients twice, with a 4-month gap between reviews to reduce repeat bias.
The first blinded review was performed alone and the second in the group. Both the previous manual and automatically generated contours were randomized by name as either A or B, and assigned a random color for comparison. The images and both contours were displayed without identifying the contour source in a RT treatment planning system (Raystation v6, Raysearch Laboratories, Stockholm, Sweden). The contours were judged based on 2 criteria: whether or not there was a preference for 1 contour over the other, and whether or not the contours were acceptable for immediate clinical use without any edit, needing minor edits, or needing major edits. The physicians specified that minor edits were contours where 2 to 4 slices would need editing and major edits where ≥5 slices needed editing or a clinically impactful part of the liver was not included. For the comparison, we first investigated if any of the 3 radiation oncologists believed that the contours were immediately clinically usable for RT planning. Then, we performed majority voting for each patient.
Training
Data preprocessing
All images were normalized using the mean (ie, 80) and standard deviation (SD; ie, 42) of the liver as found across the images in the training data set. Each image was normalized by subtracting the mean and dividing by the SD.
Initial optimizing
Training and validation was performed using Tensorflow 1.15.214 within Python. The Texas Advanced Computing Center, which incorporates a 16 GB Tesla K40 GPU, was used to facilitate the process of creating a model.15 The final training was performed on an in-house system using a 16 GB NVIDIA Quadro P5000 GPU with 24 CPU cores (3 GHz).
Data preparation
Owing to the smaller number of noncontrast CT images compared with contrast-enhanced CT images, an initial model was trained on the available images and used to generate new contours on the remaining 44 noncontrast image sets. These generated contours were manually edited to ensure an accurate final contour of the liver. With the new images, the model was retrained from the ground up. These patients were not included in the test set.
Parameter search
Learning rates for models can have a substantial impact on model performance. Learning rates that are too high lead to overfitting on the training set, and rates that are too low prevent the model from reaching a stable solution. We identified the minimum and maximum learning rates using our own adaptation of the learning rate finder (https://www.pyimagesearch.com/2019/08/05/keras-learning-rate-finder/). This was done for each version of the 3D model and the VGG16 and v3Plus models. The Adam optimizer with categorical cross entropy loss function was used across all 3 architectures.
To identify the best of the various iterations of architectures in the 3D model, each architecture was trained for 40 epochs, the point at which performance appeared to plateau. R was then used to plot the validation loss, looking for trends that might indicate if more or less layers or filters would result in improved performance and ensure we are searching within a useful range (ie, if loss was decreasing with increasing layers, we would want to investigate adding more layers until increase; Fig 1). For final training, each model was trained for 100 epochs, or until performance on the validation set plateaued.
Figure 1.
Hyper-parameter searching for ideal UNet style architecture. Parameters varied were number of layers in depth (2-5), number of convolution layers (0-2) versus atrous layers, and maximum number of filters (16-32). For ease of viewing, convolution layer 2 is not shown.
Confidence threshold
The probability cutoff for the model to determine the liver from the background on a voxel-by-voxel basis was investigated from 0.05 to 0.95 in 0.05 increments for each model. The cutoff was decided for each model independently based on the validation set by finding the maximum peak of Dice. The output Dice appeared to trend similarly between contrast and noncontrast, and so a single cutoff was chosen to allow for the model to predict on any liver without having to worry about whether the threshold should change based on the presence/absence of contrast (Fig E1). Any pixel with a probability of being the liver greater than the cutoff was added to the binary output mask. Because the liver is a single continuous organ, an automatic step was implemented to remove all but the largest continuous binary structure.
Qualitative continuity
The qualitative evaluation of 50 patients, which required a significant amount of expert users’ time, was performed using the prediction model that had been trained on data including the MICCAI challenge data. To determine whether the newly trained model was equivalent and the qualitative results would hold for the new model, we performed a MSD comparison between the contours generated from the original and final DeepLabV3+ predicted liver models.
Results
The prediction cutoff based on the validation data for VGG16, v3Plus, and 3D models were found to be 0.4, 0.3, and 0.3, respectively. With this cutoff, the 3 models were used to predict on the 63 test CT image sets (33 institutional, 30 MICCAI). The volumetric comparison (DSC and MSD) between the predicted and ground-truth volumes for these patients are summarized in Table 1. The MSD mean (minimum-maximum) were contrast (n = 17): VGG16: 1.25 mm (0.60-2.95), V3Plus: 0.99 mm (0.47-2.2), 3DUNet: 4.66 mm (2.35-13.88); Noncontrast (n = 19): VGG16: 1.37 mm (0.69-2.93); V3Plus: 1.12 mm (0.41-2.87), 3DUNet: 5.20 mm (1.94-17.92); and Miccai (n = 30): VGG16: 1.80 mm (0.65-7.02), V3Plus: 1.48 mm (0.82-3.96), and 3DUNet: 5.15 mm (3.08-9.07; Table 2).
Table 2.
Test results by group for each model
| Test data |
Mean (minimum, maximum) |
||||||
|---|---|---|---|---|---|---|---|
| Dice similarity coefficient |
Mean surface distance (mm) |
||||||
| Model name |
Model name |
||||||
| Data set | N_Images | 3D Unet | VGG_16 | V3_Plus | 3D Unet | VGG_16 | V3_Plus |
| Contrast | 17 | 0.87 (0.72, 0.92) | 0.96 (0.93,0.97) | 0.96 (0.95, 0.98) | 4.66 (2.35, 13.88) | 1.25 (0.60, 2.95) | 1.02 (0.46, 1.89) |
| Noncontrast | 19 | 0.86 (0.74, 0.93) | 0.95 (0.91, 0.97) | 0.96 (0.91, 0.98) | 5.20 (1.94, 17.92) | 1.37 (0.69, 2.93) | 1.18 (0.41, 3.21) |
| MICCAI | 30 | 0.85 (0.74, 0.91) | 0.95 (0.90, 0.97) | 0.95 (0.90, 0.97) | 5.15 (3.08, 9.07) | 1.80 (0.65, 7.02) | 1.54 (0.90, 3.68) |
Abbreviation: MICCAI = Medical Image Computing & Computer Assisted Intervention.
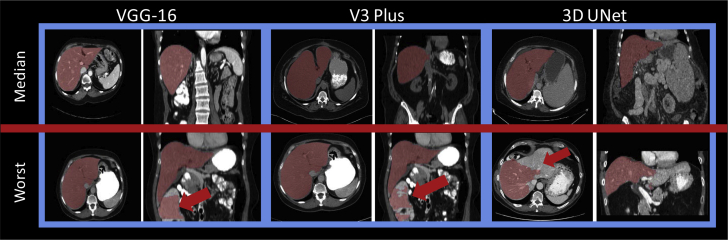
A paired t test between the VGG16 and V3Plus model found the V3Plus model to be significantly better (P = 1e-6), and a paired t test between the V3Plus model and the 3DUnet found the V3Plus to be significantly better (P = 1e-27). We suspect that the predictions on the contrast-enhanced images are better than the noncontrast scans for the 2D models because the major failings of the 2D models are where the segmentation goes too far inferior into the bowel. With contrast, differentiation from the liver and bowel is easier. In the 3D model, contrast-enhanced scans actually appeared to do more poorly where the model appeared to arbitrarily not segment part of the liver. Predictions overlayed onto CT scans are shown for the median and worst case for each architecture in Figure 2. Box plots showing the results (DSC, MSD, and Hausdorff distance) of each model for each group are in the Figures E1-E9.
Figure 2.
Predictions (red) overlayed on top of computed tomography scans for median and worst cases for each architecture. Red arrows indicate regions of failure. (A color version of this figure is available at https://doi.org/10.1016/j.adro.2020.04.023.)
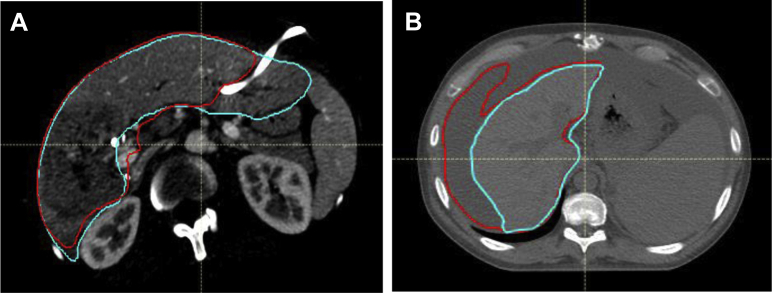
A summary of the qualitative results of the V3Plus model prediction is shown in Table 3. In 41 of 50 cases (82%), at least 1 physician preferred the automatically generated contours to the clinically drawn contours. In 48 of 50 cases (96%), at least 1 radiation oncologist deemed the automatically generated contours immediately clinically usable. The 2 cases deemed not clinically usable are shown in Figure 3. Compared with the manual segmentations, the automatically generated segmentations were preferred in 32 of 50 cases (64%) upon visual inspection. The V3Plus predictions were created in a median time of <0.1 seconds per slice or approximately 9 seconds for a 90-second image scan on both contrast and noncontrast livers on a 16GB GPU computer.
Table 3.
Consensus model results for the 3 reviewing radiation oncologists
| Reviewers | Majority or one? | Preference |
Clinically usable |
Minor edits |
Major edits |
||||
|---|---|---|---|---|---|---|---|---|---|
| Auto | Manual | Auto | Manual | Auto | Manual | Auto | Manual | ||
| 1a, 1b, 2, 3 | Majority voting | 60% (30/50) | 40% (20/50) | 81%∗ (40.5/50) | 89%∗ (44.5/50) | 33%∗ (16.5/50) | 45%∗ (22.5) | 19%∗ (9.5/50) | 11%∗ (5.5/50) |
| At least 1 vote | 82% (41/50) | 64% (32/50) | 96% (48/50) | 100% (50/50) | 86% (43/50) | 96% (48/50) | 58% (29/50) | 52% (26/50) | |
| 1b, 2, 3 | Majority Voting | 62% (31/50) | 38% (19/50) | 76% (38/50) | 82% (41/50) | 42% (21/50) | 50% (25/50) | 24% (12/50) | 18% (19/50) |
| At least 1 vote | 76% (38/50) | 64% (32/50) | 88% (44/50) | 96% (48/50) | 76% (38/50) | 88% (44/50) | 58% (29/50) | 52% (26/50) | |
When specifying reviewers, 1a is Reviewer 1’s initial review and 1b is their review with a 4-month time gap to reduce bias. Majority voting implies at least half of the reviewers agreed on a case-by-case basis, and ties were split. At least 1 implies that at least 1 reviewer voted in the manner listed.
Values of 0.5 were split ties/.
Figure 3.
(A) Presence of high-contrast biliary stent causing autosegmentation to underestimate liver, requiring edits, and (B) ascites misidentified as liver. Teal: ground truth; red: auto segmentation. (A color version of this figure is available at https://doi.org/10.1016/j.adro.2020.04.023.)
The difference between the V3Plus model used for qualitative evaluation and the final optimized V3Plus model, excluding patients who needed major edits (eg, liver ascites and stent, which were also not usable in the new model), was a median MSD of 1.02 mm (SD = 0.41) with a maximum MSD of 2.2 mm.
Discussion
This work presents a comparison of 3 state-of-the-art automatic segmentation methods for the liver with the addition of more informative, qualitative metrics of segmentation efficacy. A potential limitation is that the 2-dimensional algorithm could undersegment the liver when large disease is present on the boundary of the liver. Incorrect segmentation was often due to the 2D model’s difficulty in identifying the most inferior aspects of the liver, where a small liver size could cause the model to undersegment or continue onward into the bowel. The main failure of the 3D model was undersegmenting the liver in certain patients. A better view of each model’s failures can be seen in the worst case for each architecture (Fig 2). A comparison of our results to the recent literature is shown in Table 4.
Table 4.
Comparison of proposed method versus recent liver segmentation methods
| Method | Source | Test size | Dice |
|---|---|---|---|
| Proposed | Contrast | 17 | 0.96 |
| Noncontrast | 19 | 0.96 | |
| MICCAI | 30 | 0.95 | |
| 20167 | Contrast | 20 | 0.94 |
| 20178 | Contrast | 127 | 0.96 |
| Noncontrast | 13 | 0.96 | |
| 201716 | Contrast | 150 | 0.95 |
| 201717 | Contrast | 7 | 0.94 |
| 201818 | Contrast | 129 | 0.95 |
| Contrast | 20 | 0.94 |
Abbreviation: MICCAI = Medical Image Computing & Computer Assisted Intervention.
A variety of automatic liver segmentation architectures have been researched with promising results7, 8, 9,15, 16, 17 and a variety of methods (cascaded 3D, pretrained 2D networks, residual networks), but there is often no inclusion of human elements to validate the efficacy of the model segmentations. A comparison of other present techniques is shown in Table 3, but only our method includes the qualitative assessment of the created liver segmentation compared with previous clinical segmentations. This is particularly important in the liver where the size of the organ makes volume metrics, such as DSC, insensitive to potentially clinically impactful mis-segmentations. The 2 cases deemed not clinically usable (Fig 3) had DSC scores of 0.94 and 0.72.
The combination of cascaded UNet and 3D conditional random fields7 showed positive results (0.94 DSC) in segmenting the liver on contrast-enhanced images, but was limited to a 20-patient test set, of which 15 patients (75%) had HCC. A unique system of a 3D convolutional neural network for an initial probability map, followed by probability density function refinement,8 was presented with similar DSC scores for both contrast and noncontrast image scans; however, the results are from a 5-fold cross validation of abdominal CT scans and not a withheld test set. A multiorgan abdominal segmentation 3D FCN15 presented results from a training on 281 contrast-enhanced CT scans, with an external test set of 150 contrast-enhanced CT scans from another hospital, with a mean Dice score of 0.954. The created model is able to segment the liver, spleen, and pancreas, but does not include noncontrast segmentation capabilities. A combination of axial, sagnital, and corronal 2D views combined with 2D FCNs and voxel-based voting, showed benefits of 3D information with 2D efficiency17; however, the withheld data set was limited to 7 contrast-enhanced CT patients. The method by Roth et al18 used a unique multiscale pyramid 3D FCN with 2 image resolution sizes and Dice coefficient loss. This architecture was able to segment multiple organs and segmentations similar to those presented by our own algorithm, but is limited to contrast-enhanced CT scans.
All reviewing radiation oncologists agreed that 2 of 25 patients with HCC whole liver contours required major edits due to the algorithm’s difficulty identifying the liver around the disease site. An investigation into the 2 cases requiring major edits by qualitative assessment (Fig 3) showed that in case A, the segmentation was cut off on some slices due to the biliary stent, and in case B, the algorithm had difficulty distinguishing between the liver and ascites. There were no cases with stents or ascites in the training set. We hypothesize that including more patients with these occurrences in the training set could improve the model’s ability to accurately autosegment the livers for these types of patients; however, this needs to be further investigated. Between the 2 patient cohorts (25 HCC and 25 CLM) of the qualitative data, 25 of 25 patients with CLM (100%) automatic contours were deemed clinically usable without edit by at least 1 reviewing physician.
Conclusions
The current work represents a clinically applicable method to implement rapid automated liver segmentation with minimal temporal impact on the clinical workflow. The V3Plus model with minor tweaks has demonstrated accuracy in the generation of liver segmentation for both contrast-enhanced and noncontrast CT scans, as presented by our quantitative and qualitative assessments on completely withheld data in <1 minute per patient. The development of an accurate, efficient, and robust autosegmentation algorithm for both contrast and noncontrast CT images can enable the use of deformable registration algorithms that rely on segmentation of the liver into near real-time image guidance processes. This model has further application wherever whole liver segmentations are required (eg, patient-specific liver mass estimation in radiopharmaceutical therapy liver dosimetry.
Acknowledgments
The authors thank the Gastrointestinal Radiation Oncology group, especially Grace L. Smith, MD, PhD, MPH, and Prajnan Das, MD, MS, MPH, at MD Anderson Cancer Center for their time and input in the creation of this model.
Footnotes
Sources of support: Brian Anderson is supported as a fellow through funding from the Society of Interventional Radiology Allied Scientist Grant. Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under award numbers 1R01CA221971, R01CA235564. The authors would like to acknowledge funding and support from the Helen Black Image Guided Fund and the Image Guided Cancer Therapy Research Program at The University of Texas MD Anderson Cancer Center. Dr Eugene Koay was supported by institutional funds from the MD Anderson Cancer Moonshots program, the NIH (U54CA143837 and U01CA196403), and the Andrew Sabin Family Fellowship. The authors would also like to recognize the Medical Image Computing and Computer Assisted Intervention (MICCAI), and the Texas Advanced Computing Center (TACC, http://www.tacc.utexas.edu) at The University of Texas at Austin for providing computing resources that contributed to the research results reported with this paper.
Disclosures: During the manuscript work, Mr Dustin Gress’ employment changed from UT MD Anderson Cancer Center to the American College of Radiology (ACR). The ACR has a Data Science Institute (DSI, acrdsi.org), but none of Mr Gress’ interactions with ACR DSI as part of his job duties were related in any way to the work of this manuscript. Furthermore, there was no influence from ACR or DSI. Mr William Erwin reports grants from Oncosil Medical Ltd., IPSEN Pharmaceuticals S.A.S., Advanced Accelerator Applications International SA, and Y-mAbs Therapeutics A/S outside of the submitted work. Dr Bruno Odisio received grants from Siemens Healthineers and other incentives from Koo Foundation outside of the submitted work. Dr Eugene J. Koay reports grants from the National Cancer Institute, Stand Up to Cancer, Project Purple, Pancreatic Cancer Action Network, and Philips Health Care during the conduct of the study, as well as other incentives from Taylor and Francis, LLC, outside of the submitted work. Dr Kristy K. Brock reports research funding and a Licensing Agreement with RaySearch Laboratories. None of the other authors have any conflicts of interest to report.
Supplementary material for this article can be found at https://doi.org/10.1016/j.adro.2020.04.023.
Supplementary data
References
- 1.Brock K.K., Sharpe M.B., Dawson L.A., Kim S.M., Jaffray D.A. Accuracy of finite element model-based multiorgan deformable image registration. Med Phys. 2005;32:1647–1659. doi: 10.1118/1.1915012. [DOI] [PubMed] [Google Scholar]
- 2.Velec M., Moseley J.L., Svensson S., Hårdemark B., Jaffray D.A., Brock K.K. Validation of biomechanical deformable image registration in the abdomen, thorax, and pelvis in a commercial radiotherapy treatment planning system. Med Phys. 2017;44:3407–3417. doi: 10.1002/mp.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermoye L., Laamari-Azjal I., Cao Z. Liver segmentation in living liver transplant donors: Comparison of semiautomatic and manual methods. Radiology. 2005;234:171–178. doi: 10.1148/radiol.2341031801. [DOI] [PubMed] [Google Scholar]
- 4.Chartrand G., Cresson T., Chav R., Gotra A., Tang A., DeGuise J. SEMI-automated liver CT segmentation using Laplacian meshes. IEEE. 2014;1:641–644. [Google Scholar]
- 5.Long J., Shelhamer E., Darrell T. Fully convolutional networks for semantic segmentation. IEEE. 2015;1:3431–3440. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 6.Roth H.R., Oda Hirohisa, Zhou X. An application of cascaded 3D fully convolutional networks for medical image segmentation. Comput Med Imaging Graph. 2018;66:90–99. doi: 10.1016/j.compmedimag.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Christ P.F., Elshaer M.E.A., Ettlinger F. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. arXiv. 2016 1610.02177. [Google Scholar]
- 8.Hu P., Wu F., Peng J., Bao Y., Chen F., Kong D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J Comput Assist Radiol Surg. 2017;12:399–411. doi: 10.1007/s11548-016-1501-5. [DOI] [PubMed] [Google Scholar]
- 9.Bobo M.F., Bao S., Huo Y. Fully convolutional neural networks improve abdominal organ segmentation. Proc SPIE Int Soc Opt Eng. 2018;10574 doi: 10.1117/12.2293751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu P., Wu F., Peng J., Liang P., Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676–8698. doi: 10.1088/1361-6560/61/24/8676. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z., Lee C.P., Heinrich M.P. Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans Biomed Eng. 2016;63:1563–1572. doi: 10.1109/TBME.2016.2574816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2015 1409.1556. [Google Scholar]
- 13.van Opbroek A., Ikram M.A., Vernooij M.W., de Bruijne M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans Med Imaging. 2015;34:1018–1030. doi: 10.1109/TMI.2014.2366792. [DOI] [PubMed] [Google Scholar]
- 14.Abadi M., Barham P., Chen J. TensorFlow: Large-scale machine learning on heterogeneous distributed systems. arXiv. 2016 1605.08695. [Google Scholar]
- 15.Roth H.R., Oda H., Hayashi Y. Hierarchical 3D fully convolutional networks for multi-organ segmentation Hierarchical 3D fully convolutional networks. arXiv. 2017 1704.06382. [Google Scholar]
- 16.The University of Texas at Austin Texas Advanced Computing Center (TACC) http://www.tacc.utexas.edu Available at:
- 17.Zhou X., Takayama R., Wang S., Hara T., Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44:5221–5233. doi: 10.1002/mp.12480. [DOI] [PubMed] [Google Scholar]
- 18.Roth H.R., Shen C., Oda H. A multi-scale pyramid of 3D fully convolutional networks for abdominal multi-organ segmentation. arXiv. 2018 1806.02237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.