Abstract
SARS-CoV-2, the virus that causes COVID-19 consists of several enzymes with essential functions within its proteome. Here, we focused on repurposing approved and investigational drugs/compounds. We targeted seven proteins with enzymatic activities known to be essential at different stages of the viral cycle including PLpro, 3CLpro, RdRP, Helicase, ExoN, NendoU, and 2′-O-MT. For virtual screening, energy minimization of a crystal structure of the modeled protein was carried out using the Protein Preparation Wizard (Schrodinger LLC 2020-1). Following active site selection based on data mining and COACH predictions, we performed a high-throughput virtual screen of drugs and investigational molecules (n = 5903). The screening was performed against viral targets using three sequential docking modes (i.e., HTVS, SP, and XP). Virtual screening identified ∼290 potential inhibitors based on the criteria of energy, docking parameters, ligand, and binding site strain and score. Drugs specific to each target protein were further analyzed for binding free energy perturbation by molecular mechanics (prime MM-GBSA) and pruning the hits to the top 32 candidates. The top lead from each target pool was further subjected to molecular dynamics simulation using the Desmond module. The resulting top eight hits were tested for their SARS-CoV-2 anti-viral activity in-vitro. Among these, a known inhibitor of protein kinase C isoforms, Bisindolylmaleimide IX (BIM IX), was found to be a potent inhibitor of SARS-CoV-2. Further, target validation through enzymatic assays confirmed 3CLpro to be the target. This is the first study that has showcased BIM IX as a COVID-19 inhibitor thereby validating our pipeline.
Abbreviations: CoronaVirus Disease-2019 (COVID-19), SARS Corona Virus2 (SARS-CoV-2); CAD, computer-aided drug design; ExoN, exoribonuclease; COACH, Meta-server approach to protein-ligand binding site prediction; HTVS, high throughput virtual screening; MERS, Middle Eastern Respiratory Syndrome; PLpro, Papain-like proteinase; 3CLpro, 3-Chymotrypsin-Like Protease; 2′-O-MT, 2′-O-Methyltransferase; MOA, mechanism of action; RdRP, RNA-directed RNA polymerase; NendoU, Nonstructural Uridylate-specific endoribonuclease; SARS, Severe Acute Respiratory Syndrome; BIM IX, bisindolylmaleimide IX
Keywords: SARS-CoV-2; COVID-19; MM-GBSA; MD simulations; Virtual screening; Drug repurposing; PLpro, 3CLpro, RdRP, Helicase, ExoN, NendoU, and 2′-O-MT; Bisindolylmaleimide (BIM) IX; Anti-Viral assay; 3CLpro enzymatic assay
1. Introduction
A novel coronavirus was first reported in Wuhan, the capital city of the Hubei province in China in December of 2019 [1]. This pathogen has been named SARS-CoV-2 [2] and the disease caused by it is named, COVID-19 (Coronavirus Disease-2019). So far there have been 58 million COVID-19 cases including 1.3 million deaths worldwide [3].
The genome of coronaviruses consists of a single-stranded, positive-sense RNA, causing respiratory and enteric disease in mammals including humans. This family of viruses consists of a large genome, ranging from 28 to 32 kilobases [4]. Coronavirus family members are organized into three subsets based on antigenic and genetic facets: α-CoVs, β-CoVs, and γ-CoVs [5], [6]. MERS-CoV, SARS-CoV, and SARS-CoV-2 are all β-coronaviruses, where both SARS-CoV and CoV-2 are derived from the lineage B and MERS-CoV is derived from the lineage C β-coronavirus [7]. Interestingly, these viruses have similar genomic structures with functional proteins encoded at the 5′ end, and structural proteins encoded at the 3′ end of the genome [8]. β-Coronaviruses have accessory proteins dispersed throughout their structural genes with both SARS-CoV and SARS-CoV-2 having seven different accessory proteins, while MERS-CoV has five different accessory proteins [4], [8].
The discovery and development of novel compounds that specifically target SARS-CoV-2 will require an extended period of preclinical testing before they can enter clinical trials. Due to the lack of specific anti-SARS-CoV-2 therapeutics, there is an urgent need for faster treatment options. One approach is through screening already approved or experimental drugs that could be repurposed for SARS-CoV-2. In this study, we have used a multi-pronged drug discovery approach through rapid screening that is in-line with the World Health Organization’s (WHO) guidance to repurpose approved drugs having already demonstrated acceptable safety profiles.
2. Rational study design
Enzymes generally have binding sites that recognize small molecules and thus, are comparatively more druggable than non-enzymatic proteins. We have selected seven essential coronavirus enzymes as potential targets, namely 3CLpro, PLpro, RdRP, Helicase, NendoU, ExoN, and 2O-MT, and subjected them to virtual screenings (Fig. 2). Details of these targets are described in the following sections below. The 3D structures were accessed from among the available PDB X-ray crystal structures as well as from predicted structures available from the I-Tasser server [9]. The energy minimizations and accompanying relaxations of 3D structure models (crystal structures/modeled proteins) were carried out using the Protein Preparation Wizard followed by a short 20 ns MD simulation [10]. Following active site selection based on data mining and COACH predictions, we performed high-throughput virtual screening (HTVS) of compounds (n = 5903) approved by worldwide regulatory bodies including the FDA secured from the Zinc database or were advanced investigational molecules [11]. The screening was performed using the Virtual Screening Wizard [10] consisting of three sequential docking modes (HTVS, SP, and XP). Preliminary in-silico virtual screening identified 290 potential inhibitory compounds based on criteria including energy, docking parameters, ligand and binding site strain energies, and fit score. Compounds specific to each target protein were further analyzed for binding free energy perturbation by the molecular mechanics’ method using Prime MM-GBSA [10], followed by refining the hits to the best 32 lead candidates. A top-scoring lead from each target group was further subjected to a molecular dynamic simulation (MDS) using the Desmond module [10] to validate the screening pipeline (Overall Study Design; Fig. 1 ).
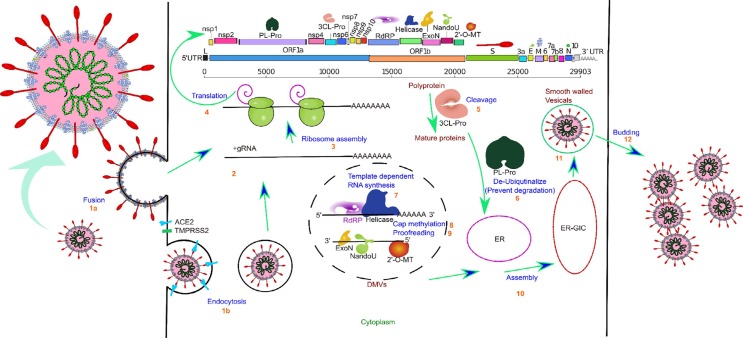
Fig. 2.
Graphical illustration of the SARS-CoV-2 life cycle along with target proteins used in HTVS. The infection cycle starts when the SARS-CoV-2 Spike protein binds to the Human ACE2 receptor. An S1-induced post-stable S2 conformation allows either viral-host cell fusion (1a) or endocytosis (1b). Fusion directly allows the viral RNA to enter the host cell (2), but endocytosis requires lysosomal degradation of coat and envelop for the release of viral nucleocapsid in the cytoplasm. The large viral script is known to encode 29 viral proteins (3), including the 7 essential nonstructural proteins that are selected as targets in our paper. A replicase is used to translate most of the viral genomic RNA to synthesize two replicase polyproteins, pp1a and pp1ab, and many small ORFs(4). The two major polyproteins are processed by two proteases, PLpro and 3CLpro(5), generating 16 nonstructural proteins. ExoN possesses a viral exoribonuclease activity that acts on both ssRNA and dsRNA in a 3′ to 5′ direction(9). Viral Helicase plays a critical role in viral replication by expediting appropriate folding (7). The enzyme 2′-O-MT methylates the viral 2′ end which is important for the virion to avoid host recognition of their RNA (8). RdRP is involved in viral-host cell replication through catalyzing template synthesis of polynucleotides in the 5′ to 3′ direction (7). NendoU is an Mn2+ dependent hexamer (dimer of trimer) enzyme with sparse functional information. The most prominent theory regarding NendoU is that the activity of this protein is responsible for protein interference with the innate immune system. For viral assembly of S, E, and M proteins in the endoplasmic reticulum, along with the N protein are combined with the (+) gRNA to become a helical nucleoprotein complex. They assemble to form a virus particle in the endoplasmic reticulum-Golgi apparatus compartment, this particle is then excreted from the cell through budding mediated by fusion of smooth-walled vesicles to the plasma membrane (11–12).
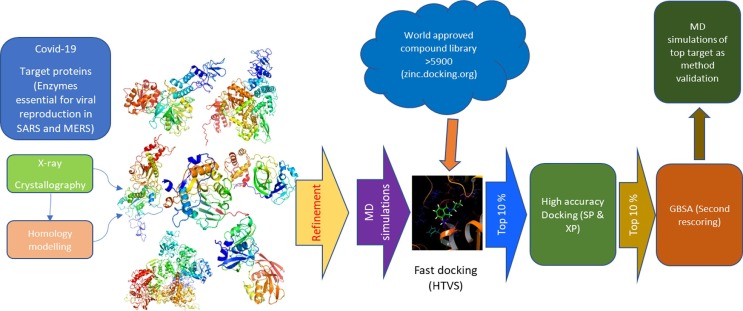
Fig 1.
Schematic road map of the overall study design. The protein models from various protein bank and other sources were optimized and relaxed by MD simulations. The relaxed structures were then mapped for active site and used to generate GLIDE Grid for HT-virtual screen with world approved drug libraries. The top 10% of these compounds were subjected to high accuracy docking (SP/XP) which were then further refined to the top 10%. This was followed by a secondary rescoring (GBSA). Top leads were subjected to MD simulations of the top compounds for each viral target tested as a methodological validation.
2.1. Drug repurposing for COVID-19
While drugs have initially been produced for use against a specific target and disease, drug repurposing offers a new and faster approach to initiate research-based methodologies. The utility of protein modeling and molecular docking has shown that approved drugs specified for certain indications can have a significant impact on other diseases [12]. For example, loperamide is an approved drug for controlling acute and chronic diarrhea that has exhibited inhibition of the MERS-CoV replication cycle [13]. Several studies are currently in progress exploring the use of antiviral drugs that were approved for influenza, hepatitis C virus, and human immunodeficiency virus (HIV) 1 against COVID-19, though with limited efficacy as well as at least in some cases added morbidity due to serious side effects. When a drug is repurposed with efficacy and safety demonstrated for other diseases, the timeline to availability to the patient population is reduced, the cost of production is lower, and the distribution channels are already in place.
2.2. Identifying SARS-CoV-2 proteins as essential targets for repurposable drugs
Out of many proteins (∼29) known to be produced by the virus, there are several critical non-structural proteins in SARS-CoV-2 that are valuable targets for antiviral drugs as illustrated below.
In the coronaviridae family, a replicase is used to translate most of the viral genomic RNA to synthesize two replicase polyproteins, pp1a and pp1ab. These two polyproteins are processed by two proteases, (1) Papain-Like Protease (PLpro) and (2), coronavirus 3-Chymotrypsin-Like Protease (3CLpro), generating 16 nonstructural proteins [14], [15], [16]. This proteolytic processing is essential for generating functional replication complexes [17]. As such, both PLpro and 3CLpro are promising antiviral targets and have already shown promise against COVID-19 in the drug combination therapy using lopinavir-ritonavir. PLpro has a core catalytic domain containing 316 amino acids. This protease cleaves the N-terminal region of the polyprotein to generate three different nonstructural proteins (1/2/3). PLpro is also suggested to have de-ubiquitinating activity, due to its structural similarities with cellular de-ubiquitinating enzymes [18] (Fig. 2). The enzyme 3CLpro contains a cysteine-histidine dimer within its active site that directs proteolytic activity. This protease can cleave 11 different sites of the replicase polyprotein to produce a mature protein that anchors replication/transcription complexes and releases mature NSPs. Structural analyses and computational screening with 3CLpro as a target have shown promising results for drug candidates against SARS-CoV [19], [20].
(3) viral non-structural protein (nsp) 14: Nsp14. Nsp14 has been implicated in SARS-CoV-2 as possessing two different activities: an exoribonuclease (ExoN) activity acting on both ssRNA and dsRNA in a 3′ to 5′ direction, and an N7-guanine methyltransferase activity (N7-MTase) [21], [22]. The activity of N7-MTase adds the N7-methyl guanosine cap during mRNA cap synthesis that is necessary for nsp16, activated by nsp10, to facilitate 2′-O-ribose methylation of the viral mRNA cap [23], [24]. 2′-O-Methylation is critical for CoV RNA to avoid host recognition [25], [26]. 2′-O-Methylation is further discussed below. ExoN activity has been suggested to be important for CoV replication and transcription, as well as for RNA proofreading during its replication. Using human CoV 229E and CoV murine hepatitis virus (MHV) respectively, Minskaia et al. (2006) and Eckerle et al. (2007) demonstrated that ExoN active-site mutants possess defects in viral RNA synthesis [22], [27]. Eckerle et al. (2007) additionally demonstrated that ExoN active-site mutants exhibit reduced replication fidelity [27]. Although there are no reports as of yet revealing drugs that target guanine-N7 methyltransferase (ExoN) of SARS-CoV or MERS-CoV, given that ExoN is important in coronaviruses for viral RNA synthesis and replication fidelity as well as for avoiding recognition of CoV RNA by the host, we assert it is to be a promising drug target (Fig. 2).
(4) nonstructural uridylate-specific endoribonuclease (NendoU) Nsp-15. NendoU activity is found in the N-terminal domain. This active site has been shown in MERS-CoV and SARS-CoV and is suggested to be a genetic marker common to coronaviruses. MERS-CoV Nsp16 appears to display unique features compared to its homologs. Nsp7/Nsp8 displays a higher binding affinity for Nsp15, also affecting enzymatic activity. Nsp15 from SARS-CoV appears to be an inhibitor of mitochondrial antiviral signaling adaptor, inducing apoptosis. Nsp15 activity is stimulated by manganese ions (Mn2+) and the enzymes generate 2′-3′ cyclic phosphate ends [28]. Nsp15 functions as a homohexamer, although the enzyme has some activity as a monomer [29]. The structures of MERS-Nsp15 and SARS-Nsp15 have been superimposed showing high homology [30]. Previous studies established that Nsp15 from both SARS-CoV and MHV can be stimulated by Mn2+ [31]. The spatial arrangements revealed that residues S290 and Y339 in MERS-Nsp15 correspond to residues S293 and Y342 in SARS-Nsp15, which are postulated to interact with the substrate and confer uridylate specificity [31], suggesting that there is conserved recognition for uridylate [30], [32], [33].
(5) 2′-O-methyltransferase (2′-O-MT). Following the addition of the N7-methyl guanosine cap, nsp16, activated by nsp10, mediates mRNA cap 2′-O-ribose methylation to the 5′-cap structure of viral mRNAs. This has been shown in SARS-CoV and MERS-CoV and shown to be universal to coronaviruses [34], [35]. 2′-O-Methylation is important for the host immune system to discern self RNA from non-self RNA. Thus, through 2′-O-methylation of viral RNA, coronaviruses can subvert host innate immune responses by avoiding host recognition of their RNA [35]. Menachery et al. (2014) have demonstrated the requirement of 2′-O-methyltransferase (2′-O-MT) activity for SARS-CoV pathogenesis by characterizing deficient strain to have reduced viral titers and viral replication in-vitro and in-vivo (less weight loss and reduced breathing dysfunction in mice) [35]. Similarly, the importance of nsp16 and thus the activity of 2′-O-methyltransferase (2′-O-MT), has also been shown for MERS-CoV pathogenesis. Menachery et al. (2017) introduced mutations in the MERS-CoV NSP16 conserved KDKE motif, which resulted in significant attenuation of the viral load relative to the controls both in-vitro and in-vivo [34]. Despite there being no publication to date detailing drugs that target 2′-O-Methyltransferase to treat SARS-CoV or MERS-CoV, we are interested in 2′-O-MT as a potential drug target because of its role in avoiding recognition of CoV RNA by the host and its importance for SARS-CoV and MERS-CoV pathogenesis (Fig. 2).
(6) viral helicase. Helicase is essential to viral genome replication and is therefore a potential target for antiviral drug development. Virus-encoded RNA helicases have important roles during viral life cycles for folding and replication of viral RNA [36]. As such, Nsp13 possesses NTPase and RNA helicases to facilitate hydrolysis of NTPs and unwind RNA (Fig. 2). In one study, it was demonstrated that myricetin and scutellarein are strong inhibitors of SARS-CoV helicase protein by affecting its ATPase activity [37]. Helicase is a multi-functional protein with a zinc-binding domain in the N-terminus displaying RNA and DNA duplex-unwinding activities with 5′ to 3′ polarity. The activity of helicase is dependent on magnesium (Mg2+). As such, bismuth salts have been shown to inhibit NTPase and RNA helicase activities of SARS-CoV-2 nsp13 [9], [38]. Sequence annotation by Ivanov et al have shown that SARS-CoV nsp13 is divided into three domains: (A) an N-terminal Zn(II) binding domain and (B) a hinge domain, and (C) a helicase domain [28], [36]. Little is known of the viral nsp-12 activity, but activity occurs on two domains, on the N-terminal subunit NiRAN, and motif BN on the C-terminus. These sites have been shown in SARS-CoV and are suggested to apply to coronaviruses in general [39], [40].
(7) RNA-directed RNA polymerase (RdRP). RDRP plays a critical RNA replication in RNA viruses due to its involvement in catalyzing template synthesis of polynucleotides in the 5′-3′ direction. Further, RdRP is essential for the initiation of RNA replication in the host cell, a key step in the RNA viruses infection cycle [41], [42]. Chien et al. (2020) have demonstrated the importance of RdRP activity for SARS-CoV pathogenesis [43]. They showed that without RdRP, there is a complete disruption of SARS-CoV -RNA replication and viral growth halted (Fig. 2). Additionally, these authors also suggested that the hepatitis C drug EPCLUSA (Sofosbuvir/Velpatasvir) would target the active site of RdRP to inhibit coronaviruses [44]. RdRP is an established drug target for the treatment of SARS-CoV due to its role in viral RNA replication and its importance for SARS-CoV pathogenesis RdRP is of particular interest as a drug target.
3. Results and discussion
Following in-silico HTVS, molecular modeling, MDS, and utilizing resources from the literature, we identified a series of potential lead molecules from the repurposable drug libraries against proteins that are critical for the ability of SARS-CoV-2 to infect and reproduce within the host cell (Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7 ). Additionally, all of the simulated hit-target complexes were found to be strongly interacting and exhibited highly stable binding indicating the potential of being potent inhibitors of key SARS-CoV-2 enzymes and therefore promising candidates or leads for further development. Outputs from these analyses are presented in tables (Table 1, Table 2, Table 3, Table 4, Table 5, Table 6, Table 7) and graphically (Top three interaction maps Supplementary Fig. 1,3,5,7,9,11&13), and are discussed in the following sections.
Table 1.
2′-O-methyltransferase (2′-O-MT) hits.
| Dock ID | Compound name | Indicated target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Maltotetraose | Substances that inhibit the growth or reproduction of BACTERIA | −58.44 | −11.31 | −1.54 | −6.96 | −2 | 0.09 | 0.1 |
| B | Natural Crocin and/or Crocetin | for treating neurodegenerative disorders of the central nervous system, e.g. nootropic agents, cognition enhancers, drugs for treating Alzheimer's disease or other forms of dementia | −53.89 | −10.36 | −1.68 | −6.05 | −1.84 | 0.14 | 0.07 |
| C | Paclitaxel | inhibits the disassembly of microtubules | −70.51 | −8.7 | −1.24 | −5.03 | −2 | 0.38 | 0.17 |
| D | NADH dianion | Dianion of NADH arising from deprotonation of the two diphosphate OH groups; major species at pH 7.3. | −61.72 | −8.55 | −1.56 | −5.69 | −2 | 0.74 | 0.14 |
| E | Iohexol | MRI contrasting agent/ histone acetyltransferase KAT2A (human) | −67.38 | −6.04 | −2.87 | −2.84 | −1.37 | 0.8 | 0.19 |
| F | Heparin | Anticoagulant | −86.41 | −12 | −4.02 | −12- | 4.02 | 1.1 | 0.07 |
Table 2.
RNA-dependent RNA polymerase hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Lactulose | Laxative & portal-systemic encephalopathy (PSE) | −67.99 | −15.22 | −2.9 | −11.26 | −2 | 0.11 | 0.19 |
| B | Framycetin/Paromomycin | 16S ribosomal RNA | −66.49 | −14.87 | −1.56 | −6.87 | −2 | 0.22 | 0.11 |
| C | Amikacin/Arbekacin | 16S ribosomal RNA | −73.4 | −14.06 | −1.87 | −10.24 | −2 | 0.19 | 0.11 |
| D | Bekanamycin | Not Available | −63.88 | −14.69 | −2.31 | −6.6 | −2 | 0.2 | 0.06 |
| E | Lividomycin A | Not Available | −64.68 | −14.6 | −2.22 | −6.56 | −2 | 0.18 | 0.11 |
| F | Lapatinib Ditosylate | Blocks phosphorylation of the epidermal growth factor receptor (EGFR), ErbB2, and the Erk-1 and-2 and AKT kinases; it also inhibits cyclin D protein levels in human tumor cell lines and xenografts. | −51.39 | −12.03 | −1.31 | −5.48 | −2 | 0.01 | 0.11 |
Table 3.
Uridylate-specific endoribonuclease (NendoU) Hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Daidzin | Aldehyde dehydrogenase, mitochondrial (human) | −68.65 | −14.56 | −1.9 | −6.32 | −2 | 0.05 | 0.11 |
| Ba | SCHEMBL24383, ZINC8143723 | Active principle of Strychnos potatorum Linn. Seed extracts. Known antiallergic, antianaphylactic and mast cell stabilization activity | −72.09 | −14.52 | −1.87 | −6.28 | −2 | 0.03 | 0.11 |
| Bb | Metrizamide | Resorbable, non-ionic contrast medium. | −71.41 | −14.26 | −1.78 | −6.32 | −2 | 0.23 | 0.11 |
| C | Haloperidol Glucuronide | Anti-depressant | −71.81 | −13.48 | −2.61 | −9.12 | −2 | 0.19 | 0.06 |
| D | 4-Hydroxyphenytoin glucuronide | Phenytoin metabolite by liver UDP-glucuronosyltransferase. | −74.49 | −13.5 | −1.87 | −7 | −2 | 0.18 | 0.19 |
| E | acetaminophen O-β-D-glucosiduronate | A β-D-glucosiduronate that is the conjugate base of acetaminophen O-β-D-glucosiduronic acid | −70.28 | −14.07 | −1.51 | −6.27 | −2 | 0.1 | 0.11 |
| F | p-Aminophenyl-alpha-D-Galactopyranoside | Heat-labile enterotoxin B chain | −71.44 | −14.06 | −1.87 | −5.82 | −2 | 0.02 | 0.11 |
Table 4.
Guanine-N7 methyltransferase (ExoN) Hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Diosmin | Diosmin, a flavone that can be found in the plant Teucrium gnaphalodes. Is an agonist at the human aryl hydrocarbon receptor | −68.66 | −13.26 | −1.66 | −6.01 | −2 | 0.11 | 0.11 |
| B | Hidrosmin | Capillary stabilizing agent | −71.34 | −13.24 | −2.01 | −5.57 | −2 | 0.05 | 0.11 |
| C | N-Desmethyl-4-hydroxy tamoxifen beta-D-glucuronide (E/Z Mixture) | N/A | −72.69 | −13.23 | −1.77 | −6.22 | −2 | 0.48 | 0.11 |
| D | Octane-1,3,5,7- tetracarboxylic acid | Carboxypeptidase A1 | −68.77 | −13.17 | −1.65 | −6.08 | −2 | 0.28 | 0.11 |
| E | Bempedoic acid | Metabolic activation affords ETC-1002-CoA, the pharmacologically active metabolite. ATP lyase (a.k.a. ATP synthase) plays an important part of cholesterol synthesis | −71.39 | −13.15 | −2.14 | −5.48 | −2 | 0.22 | 0.08 |
| F | Bisindolylmaleimide IX | PKC Inhibitor, Potential anti-cancer activity | −50.6 | −7.8 | −4.5 | −2.7 | −0.7 | 0.0 | 0.3 |
Table 5.
Helicase (HEL) Hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Inosine | Neurorestorative, anti-inflammatory, immunomodulatory, and cardioprotective effects. | −72.87 | −13.05 | −1.76 | −6.23 | −2 | 0.66 | 0.11 |
| B | 5-F-UMP | Bacterial Thymidylate synthase | −57.22 | −12.94 | −1.79 | −6.68 | −2 | 0.13 | 0.08 |
| C | Fenoterol | Agonist to Beta-2 adrenergic receptor | −57.22 | −12.94 | −1.79 | −6.68 | −2 | 0.13 | 0.08 |
| D | Didanosine | Anti-HIV | −72.87 | −12.94 | −1.76 | −6.12 | −2 | 0.66 | 0.11 |
| E | Doxifluridine | Not Available/Anti-cancer | −68.72 | −12.9 | −1.88 | −5.81 | −2 | 0.09 | 0.13 |
Table 6.
Papain-like proteinase (PLpro) Hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | Troxerutin | Not known | −73.57 | −13.5 | −1.62 | −2 | −7.17 | 0.19 | 0.09 |
| B | 4-O-(4,6-Dideoxy-4-{[4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-Beta-D-lyxo-hexopyranosyl)-alpha-D-erythro-hexopyranose | Pancreatic alpha-amylase (DB03495) | −61.8 | −13.79 | −1.53 | −2 | −5.92 | 0.05 | 0.11 |
| C | 4,6-Dideoxy-4-{[4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-lyxo-hexopyranosyl-(1->4)-alpha-D-threo-hexopyranosyl-(1->6)-alpha-L-threo-hexopyranose | Pancreatic alpha-amylase (DB02889) | −75.87 | −13.3 | −1.72 | −2 | −5.36 | 0.25 | 0.11 |
| D | Hyaluronic acid | Supplement | −64.36 | −13.24 | −2.04 | −2 | −5.21 | 0.4 | 0.11 |
| E | Monoxerutin | Not known | −64.68 | −14.6 | −13.92 | −2 | −6.56 | 0.02 | 0.11 |
| F | Anastrozole | Non-steroidal aromatase inhibitor (AI) | −61.81 | −14.48 | −12.65 | −2 | −2 | 0.19 | 0.06 |
Table 7.
Main proteinase (3CLpro)Hits.
| Dock ID | Compound name | Normal target | Glide Energy | DockScore | LipophilicEvdW | HBond | Electro | ExposPenal | RotPenal |
|---|---|---|---|---|---|---|---|---|---|
| A | XAV-939 | Not known; Anti-cancer | −71.17 | −12.66 | −5.74 | −5.98 | −1.46 | 0.37 | 0.15 |
| B | Crocin | Arthritis | −63.96 | −11.56 | −3.24 | −6.68 | −2 | 0.23 | 0.14 |
| C | Iopromide | non-ionic X-ray contrast agent | −77.95 | −11.07 | −5.34 | −4.64 | −1.63 | 0.39 | 0.15 |
| D | Troxerutin | Chronic Venous Insufficiency | −64.53 | −10.69 | −4.69 | −4.29 | −1.42 | 0 | 0.11 |
| E | Isoquercetin | Anti-cancer | −73.56 | −10.31 | −4.89 | −4.42 | −1.71 | 0.54 | 0.17 |
| F | Danoprevir | Inhibitor of the HCV NS3/4A protease | −66.83 | −10.03 | −4.73 | −4.01 | −1.44 | 0.04 | 0.11 |
| G | Cefoperazone | Cephalosporin; antibacterial | −79.52 | −7.8 | −3.14 | −4.22 | −1.67 | 0.7 | 0.19 |
| H | Nevirapine | Non-nucleoside reverse transcriptase inhibitor (NNRTI) | −63.17 | −7.79 | −3.31 | −3.25 | −1.09 | 0.11 | 0.14 |
| I | Pentostatin | Adenosine deaminase; anti-cancer | −81.01 | −7.12 | −3.78 | −3.74 | −2 | 2.09 | 0.19 |
| J | Cladribine | Synthetic purine nucleoside that acts as an antineoplastic agent | −78.93 | −6.86 | −5.19 | −1.48 | −1.09 | 0.85 | 0.19 |
| K | Bisindolylmaleimide IX | PKC Inhibitor, Potential anti-cancer activity | −56.2 | −5.9 | −5.1 | −0.6 | −0.6 | 0.1 | 0.3 |
2′-O-methyltransferase (2′-O-MT) hits (Table 1). Maltotetraose is an oligosaccharide of four units of alpha-D-glucopyranose linked by alpha-(1–4) bonds. This is a sugar variant of a substance that is found to inhibit the growth and reproduction of bacteria [45], [46]. Ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 1.’ The 20 ns long MD simulation revealed the interaction to be a very strong one validating the pipeline (Supplementary Fig. 2). Crocin/Crocetin are the major bioactive ingredients of saffron. There have been several studies that have shown the various efficacies of these drugs, including as neurotropic and chemotherapeutic agents [47], [48]. Crocin/Crocetin are popular agents to be tested in clinical settings due to their anti-oxidative properties [49]. Paclitaxel is a diterpene alkaloid natural product and belongs to a family of drugs that target tubulin leading to an abnormality of the mitotic spindle assembly, chromosome segregation, and consequently defects of cell division [50]. Paclitaxel is one of the most widely used anticancer drugs for the treatment of various cancers [51]. Other studies have shown that low doses of paclitaxel show promise in treating some non-cancer diseases including renal and hepatic fibrosis and artery restenosis [52], [53], [54]. NADH dianion is a species of NADH that arises from the deprotonation of the two diphosphates OH groups. Iohexol is a compound most well-known as a nonionic, water-soluble radiographic contrast medium used especially for renal disease determination. This compound is absorbed from the cerebrospinal fluid into the bloodstream and is eliminated by renal excretion. Heparin is a very interesting hit as it has been reported in multiple studies to increase the likelihood of survival of terminal COVID-19 patients [55], [56] by a mechanism that is not well understood but likely involves heparin’s activity of reducing hypoxia as well via inhibiting the cytokine storm [57]. There is a minor report suggesting that hepcidin hormone mimics the spike protein of SARS-CoV2, [58], and heparin is known to interfere with hepcidin [59]. This suggests that heparin’s anti-COVID activity may present a multifaceted therapy option. MMGBSA re-ranking brought this molecule to the bottom despite a high glide energy score due to penalties exacted due to extra-active site exposure of the bulky compound.
RNA-directed RNA polymerase (RdRP) hits (Table 2). The SARS-CoV-2 RdRP is predicted to bind tightly to aminoglycosides. Ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 3.’ Drug screening reveals many hits from this class of antibiotics including Framycetin, Paromomycin, Amikacin, Arbekacin, Bekanamycin, & Lividomycin A. While aminoglycosides have a binding affinity for RNA and have been reported to inhibit binding of RdRP to the decoding loop in case of influenza [60] the complementary RNA binding site of RdRP has also been reported to be susceptible to aminoglycosides such as neomycin B which targets HCV RdRP [61]. Interestingly, the highest-scoring among the aminoglycoside hits is Paromomycin which is an antiparasitic used to treat amoebiasis, visceral leishmaniasis, and cryptosporidiosis in immunocompromised patients. With the potential to treat pulmonary tuberculosis as well, the range of indications of this drug is indeed very wide and if found effective against COVID-19, it could prove to be a valuable addition to the arsenal of combination therapies. The Paromomycin RdRP complex was simulated for 20 ns and the interaction was highly stable throughout the simulation validating the pipeline for RdRP (Supplementary Fig. 4).
Lactulose is a synthetic disaccharide of galactose and fructose which can be produced by the isomerization of lactose. This compound has been used for treating bacterial infections, constipation, and cancer. An important note on lactulose is that it is not hydrolyzed by mammalian enzymes, therefore, ingested lactulose passes through the stomach and small intestine without degradation [62]. Framycetin is an aminoglycoside antibiotic isolated from Streptomyces lavendula that shows broad-spectrum antibacterial activity. This drug has been used as a therapeutic against a variety of cancers [63], [64]. Amikacin is an aminoglycoside antibiotic that is on the WHO list of essential medicines. It is a prokaryotic translation inhibitor that binds to the 16S ribosomal subunit. Bekanamycin is another aminoglycoside that inhibits prokaryotic translation by binding to the highly conserved A site of 16S rRNA in the 30S ribosomal subunit. This compound has the lowest antibacterial activity of the aminoglycosides in clinical use and manifests a moderate level of toxicity, therefore, it is no longer used as a first-line antibiotic[65]. Lividomycin A is an aminoglycoside that shows antibiotic activity against several of the Gram+/− bacteria by inhibiting protein synthesis [66]. Lapatinib ditosylate is the salt of Lapatinib, a synthetic quinazoline that blocks the phosphorylation of various epidermal growth factor receptors and inhibits cyclin D protein levels in human tumor xenografts and cell lines [67], [68].
Uridylate-specific endoribonuclease (NendoU) Hits (Table 3). Ligand interaction maps of the top three hits are shown in ‘Supplementary Fig. 5.’ Daidzin is an isoflavone natural product found in several Legumimosae such as the Japanese Kudzu root and is the 7-O-glucoside of the isoflavone daidzein. It is shown to have anticancer and antiallergenic activities [69]. Daidzein and Haloperidol have been identified in multiple virtual screenings [70], [71]. MD Simulation of the Daidzin-NendoU complex for 20 ns revealed a stable and energetically favorable interaction and validated the pipeline for the NendoU virtual screening (Supplementary Fig. 6). SCHEMBL24383 (Active principle of Strychnos potatorum Linn. seed extracts) has been extensively used to combat respiratory diseases including asthma, chronic obstructive pulmonary disease (COPD), and bronchitis. Interestingly, given its predicted anti-COVID-19 activity, it may result in a dual therapeutic potential drug. Metrizamide is a non-ionic iodine-based radiocontrast agent that is widely used in lumbar myelography [72]. Haloperidol glucuronide is a metabolite of the commonly prescribed antipsychotic drug on the WHO’s list of Essential Medicines. Haloperidol is used in the treatment of schizophrenia, mania in bipolar, delirium, and other neurological diseases [73], [74] and is on the WHO’s List of Essential Medicines. 4-Hydroxy phenytoin glucuronide is a metabolite of the widely used antiepileptic phenytoin. The adverse effects of phenytoin can range from moderate diseases including gingival hyperplasia to severe more effects including toxic epidermal necrolysis and teratogenic effects [75]. Similarly, acetaminophen O-β-D-glucose iduronate is a metabolite of acetaminophen generated in the liver by UDP-glucuronyltransferase. It is highly water-soluble and is excreted through the kidneys. Acetaminophen was recommended by WHO in the case of COVID-19 due to a concern over ibuprofen being an ACE2 inhibitor that might increase viral entry [76]. The advisory was subsequently revised to state that both medications are appropriate [77]. A metabolite of acetaminophen could prove to be an added advantage of already being used as a drug. p-Aminophenyl-alpha-D-galactopyranoside is an experimental phenolic glycoside that competitively binds to heat-labile enterotoxin B pentamers by mimicking host cell receptors (intracellular adenylyl cyclase) [78].
Guanine-N7 methyltransferase (ExoN) Hits (Table 4). Ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 7.’ Diosmin is a flavone glycoside that is mostly found in citrus fruits. It is a non-prescribed dietary supplement that is primarily used for venous diseases, although there is limited clinical data to support its efficacy. Common adverse effects include mild gastrointestinal disturbances and cardiac arrhythmias [79]. Interestingly other studies have reported diosmin to be a coronavirus main protease inhibitor. MD Simulations for 20 ns with Diosmin-ExoN complex revealed a very strong binding establishing the target specificity of Diosmin and validated the screening pipeline with ExoN (Supplementary Fig. 8). Hidrosmin is a synthetic bioflavonoid that is used to treat chronic venous insufficiency of the lower limbs [80]. A crude preparation of Diosmin and Hidrosmin has been determined to be effective against COVID-19 [81]. N-Desmethyl-4-hydroxy tamoxifen beta-D-glucuronide (E/Z Mixture) has also been identified as a strong binding ligand. Tamoxifen has also been proposed as an anti-COVID-19 drug as it can induce autophagy associated with the unfolded protein response to kill infected cells and thus contain the virus [82]. Octane-1,3,5,7-tetracarboxylic acid B belongs to the class of zinc ion binding compounds that target Carboxypeptidase A1.
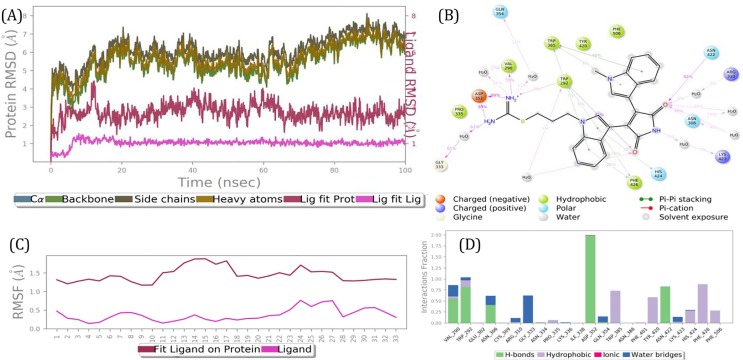
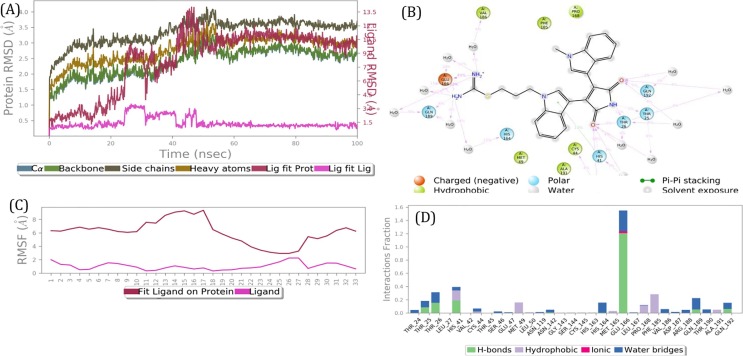
Bempedoic acid is a prodrug that is converted to its active form in the liver. It is an FDA approved treatment for hypercholesterolemia and has few adverse effects. This compound inhibits adenosine triphosphate citrate lyase, an enzyme within the cholesterol biosynthesis pathway [83]. Bisindolylmaleimide IX (BIM IX) a was strongly bound to ExoN active site for a 100 ns simulation (Figs. 4 & 5(B)) and the stable complex had a tight binding with the whole molecule involved in interactions with 3CLpro active site side-chain amino acid residues (Figs. 4(B) & 5(B)).
Fig. 4.
Results of a 100 ns MD simulation. (A) Root mean square deviations difference between guanine-N7 methyltransferase (ExoN) and bound ligand BIM IX (<4 Å). Graph obtained for RMSF value of ligand (purple line) from the protein back bone (green line). The ligand was tightly bound to the active site throughout the simulation. The complex progressed towards a more stable state during the simulation. This suggests two binding conformers of same ligand within the binding site. (B) Schematic 2D representation of bound ligand interactions of BIM IX throughout the simulation. (C) Root mean square fluctuation between the binding site of target protein and interacting ligand. (D) Critical protein ligand contacts of amino acid side chain residues with the interaction properties. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
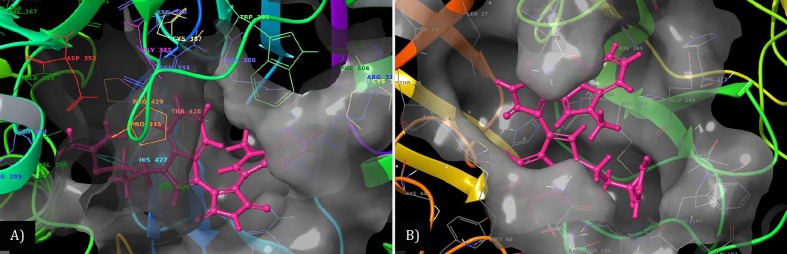
Fig. 5.
Post simulation 3D interaction representation for BIM IX (Pink licorish). A) 3CLpro active site and B) ExoN active site). The active site is depicted as translucent surface model covering binding side chain aminoacid recidues as thiner line labeled and colored individually. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Helicase (HEL) Hits (Table 5). Ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 9.’ The FDA approved drug ivermectin recently showed significant anti- SARS-CoV-2 antiviral activity, suggesting its therapeutic potential [84]. While the target of ivermectin in SARS-CoV-2 is yet to be confirmed, it has been reported that its antiviral activity in the case of flaviviruses, particularly the Australian Kunjin virus (West Nile virus variant) [85] is based on targeting viral helicases. While our screening did not show ivermectin as a top helicase inhibitor, its binding activity established viral helicase as an important target in SARS-CoV-2. We found several small-molecule compounds including inosine, 5-F-UMP, fenoterol, didanosine, and doxifluridine as potential helicase inhibitors. While Inosine, 5-F-UMP, and fenoterol have a high affinity towards the ATP-Nucleic acid interface, our top target against Helicase is inosine. Inosine is a nucleoside commonly found in tRNAs and is essential for proper translation of the genetic code in wobble base pairs. It has been tried in various clinical settings, most notably in multiple sclerosis [86]. MD Simulations for 20 ns with the Helicase-Inosine docked complex revealed a very strong binding establishing the target specificity of Inosine and validated the screening pipeline with HEL (Supplementary Fig. 10). 5-F-UMP is a pyrimidine ribonucleoside 5′-monophosphate having 5-fluorouracil as the pyrimidine component. It shows antineoplastic activity by being inorporated in RNA and it inhibits RNA processing, thereby inhibiting cell growth. Fenoterol is a β adrenoreceptor agonist that is used as an inhaled bronchodilator asthma medication [87]. Didanosine is particularly interesting as it is a reverse transcriptase (HIV) inhibitor [88]. Also, Didanosine has been profiled as a beneficial drug in the case of the COVID-19 type of lung fibrosis by matching single-cell RNA sequencing data [89]. Inhibition of inosine-5′-monophosphate dehydrogenase (IMPDH) has been shown to control SARS-CoV-2 replication [90], which could be due to inosine accumulation. Didanosine is also a medication used to slow the progress of HIV/AIDS and is a nucleoside analog of adenosine. There are several common adverse effects associated with this medication including diarrhea, vomiting, and peripheral neuropathy [91]. Fenoterol has already been recommended to reduce severe pulmonary symptoms of COVID-19 patients [92]. Doxifluridine has been suggested as a possible SARS-CoV-2 inhibitor based on similarities with active antivirals already tested by artificial intelligence [93]. Doxifluridine is a nucleoside analog prodrug that interferes with RNA transcription by competing with uridine triphosphate for incorporation into the RNA strand. It is used as a cytostatic agent in chemotherapy in several countries in Asia [94].
Papain-like proteinase (PLpro) Hits (Table 6). The ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 11.’ Troxerutin is a naturally occurring flavonoid that has been reported to show promise as a vasoprotective agent by improving hepatic homeostasis [95]. There have been many in silico drug screening efforts with the coronavirus Main Protease that have predicted troxerutin as a potential inhibitor of 3Clpro [96]. Also, the famous ‘Chai-Ling decoction’ a Chinese medicine herbal formulation with reported protection from SARS, MERS, and now COVID-19 has troxerutin as a major component [97], [98]. MD Simulations for 20 ns with troxerutin-PLpro docked complex revealed a very strong binding establishing the target specificity of troxerutin and validated the screening pipeline with PLpro (Supplementary Fig. 12). The identified hit 4-O-(4,6-dideoxy-4-{[4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-beta-D-lyxo-hexopyranosyl)-alpha-D-erythro-hexopyranose is an aminocyclitol glycoside, as well as 4,6-dideoxy-4-{[4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-aapha-D-lyxo-hexopyranosyl-(1->4)-alpha-D-threo-hexopyranosyl-(1->6)-alpha-L-threo-hexopyranose have been shown to target pancreatic alpha-amylase [99]. Both of these compounds belong to the class of aminocyclitol glycosides similar to streptomycin. This class of compounds has been used previously to target the RNA metabolism of viruses [100]. Hyaluronic acid is a non-sulfated glycosaminoglycan (GAG) and is composed of repeating polymeric disaccharides of D-glucuronic acid and N-acetyl-D-glucosamine linked by a glucuonidic β (1 → 3) bond [101]. It is a key molecule involved in skin moisture that can retain water. Monoxerutin is another flavonoid which has been patented to be used against Hepatitis C [102]. Anastrozole is an aromatase inhibitor approved by the FDA to treat postmenopausal women diagnosed with hormone receptor-positive breast cancer at advanced-stage or early-stage after surgery [103], [104].
Main proteinase (3CLpro) Hits (Table 7). Ligand interaction maps of the top 3 hits are shown in ‘Supplementary Fig. 13.’ XAV-939 is a beta-catenin signaling inhibitor that has been shown to have promise in treating prostate cancer [105]. XAV-939 is also an effective inhibitor of PARP and Wnt pathway. PARPs enhance IFNγ production and can halt viral infections [106], [107]. Thus, a dual-mode of action is expected of XAV-939 if it is found to inhibit the main protease as well. MD Simulation of XAV-939-3CLpro docked complex for 100 ns revealed a stable and energetically favorable interaction and validated the pipeline for the 3CLpro virtual screening (Supplementary Fig. 14). Crocin is a carotenoid diester that is responsible for the color of saffron and is popularly used in India as a treatment for arthritis and psychological disorders [108]. Loperamide is used to treat gastrointestinal symptoms including diarrhea with few side effects and is on the WHO's List of Essential Medicines. Isoquercetin is a flavonoid natural product that can be isolated from various plant species and has shown promise as an anti-cancer agent [109]. Danoprevir is a 15-membered ring macrocyclic peptidomimetic inhibitor of the hepatitis C protease NS3/4A [110]. Other virtual drug screenings have reported danoprevir as a possible coronavirus main protease inhibitor [111]. In clinical trials, danoprevir had a positive effect on recovery and faster discharge of COVID-19 patients [112]. Cefoperazone is a cephalosporin antibiotic. It is one of the few cephalosporin antibiotics effective in treating Pseudomonas bacterial infections [113]. In China, most of the treatment regimens reported using cefoperazone to prevent secondary infections in COVID-19 patients [114], [115]. Nevirapine is a non-nucleoside reverse-transcriptase inhibitor [116]. This compound is FDA approved for use in adult patients infected with HIV-1. Nevirapine has also been revealed in various in-silico drug screening with the main protease [117], [118]. Pentostatin is a purine analog that is widely used as a treatment for hairy cell leukemia [119]. Similar to pentostatin, cladribine is also a therapy for hairy cell leukemia. And also like nevirapine, cladribine has been predicted to block main protease 3CLpro in many reports [118], [120]. Bisindolylmaleimide IX a PKC inhibitor, strongly bound to 3CLpro for a 100 ns simulation (Figs. 3 & 5(A)), and the stable complex exhibited a tight binding with the whole molecule involved in interactions with 3CLpro active site side-chain amino acid residues (Figs. 3(B) & 5(A)).
Fig. 3.
Results of a 100 nano seconds (ns) MD simulation. (A) Root mean square deviations difference between the Main protease (3CLPro) and bound ligand BIM IX (<4 Å). Graph obtained for RMSF value of ligand (purple line) from the protein back bone (green line). It revealed that there was a major conformational change of the ligand at around 50 ns without loss of the ligand. This suggests two binding conformers of same ligand within the binding site. (B) Schematic 2D representation of bound ligand interactions of BIM IX throughout the simulation. (C) Root mean square fluctuation between the binding site of target protein and interacting ligand. (D) Critical protein ligand contacts of amino acid side chain residues with the interaction properties. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
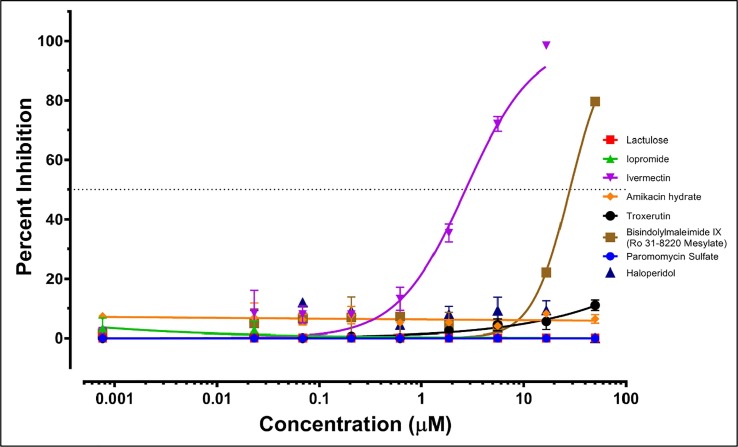
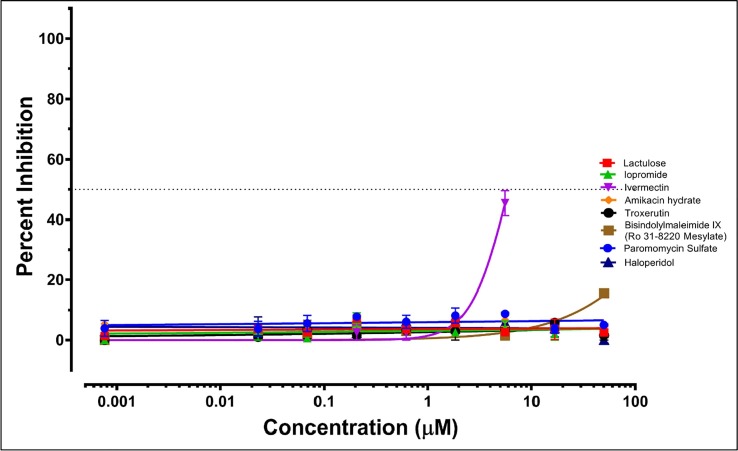
In-vitro anti-SARS-CoV-2 testing of select drug compounds. All compounds unless stated otherwise were procured from Selleck Chemicals LLC (Houston, TX). As per screening results, Troxerutin (3CLpro & PLpro) and BIM IX (3CLpro & Exon) had possible dual targets, while other targets were shortlisted based on novelty and scores. Mild anti-SARS-CoV-2 activity at higher concentrations; at 25 µM Amikacin (7.2%), Troxerutin (2.8%), Paromomycin (2.3%%) & Haloperidol (9.8%%) had non-translatable activity (Fig 6, Fig 7 ). Lactulose & Iopromide had no observed inhibitory activity, which could be due to low intracellular accumulation as Lactulose is anti-diarrhea and Iopromide is an MRI contrast agent. This supports the predicted interaction and MOA and can be used to improve or screen more members from the same chemotypes and similar pharmacophore. Lactulose (Anti-Diarrhea) and Iopromide (MRI contrast) both showed no activity at the concentration range tested. This could be due to a lack of cell permeability as both are not used for intracellular functions. An inhibitory concentration (IC50) was estimated by curve fitting over linear regression of Log10 drug concentrations vs. normalized data (percentage, Table 8 ). The IC50 was plotted on the variable slope of the drug responses (Fig 6, Fig 7).
Fig 6.
SARS CoV2 anti-viral entry assay. Synchronized infections were conducted for viral entry as described in Materials and Methods. Each curve shows a dose-response to the indicated 8 drug compounds (Color-coded; key inset). The results are presented as the PFU formed in the presence of drug as a percentage of the PFU formed and each plotted value is the mean with ± standard deviations of an experiment performed in triplicate.
Fig 7.
SARS-CoV2 Anti-viral spread assay. Synchronized infections were conducted for a viral spread as described in Materials and Methods. Each curve shows a dose-response to the indicated 8 drug compounds (Color-coded; key inset). The results are presented as the PFU formed in the presence of drug as a percentage of the PFU formed and each plotted value is the mean with ± standard deviations of an experiment performed in triplicate.
Table 8.
Results of anti-SARS-CoV-2 testing. Note: ‘Very wide’ means some activity was observed at high concentrations too staggering to plot.
| Test compound | IC50_Viral Entry blocking assay (µM) | Percent inhibition at 50 µM (%) | IC-50_Viral Spread assay (µM) | Percent inhibition at 50 µM (%) | Cytotoxicity (µM) | Predicted Target(s) |
|---|---|---|---|---|---|---|
| *Bisindolylmaleimide IX | 28.18 | 80 | 381.5 | 15 | Nontoxic within tested conc. range | 3CLpro and ExoN |
| Ivermectin | 2.702 | 99 | 6.015 | 46 | Toxic above 50 | Probably Helicase |
| *Haloperidol | (Very wide) | 10 | (Very wide) | 03 | Non-toxic within tested conc. range | NendoU and 3CLpro |
| Troxerutin | (Very wide) | 11 | (Very wide) | 03 | Nontoxic within tested conc. range | 3CLpro and PLpro |
| Amikacin hydrate | (Very wide) | 08 | (Very wide) | 03 | Nontoxic within tested conc. range | RdRP |
| Paromomycin | ∼99.86 | 05 | (Very wide) | 02 | Nontoxic within tested conc. range | RdRP |
| Lactulose | nil | 01 | nil | 03 | Nontoxic within tested conc. range | RdRP |
| Iopromide | nil | 02 | nil | 02 | Nontoxic within tested conc. range | 3CLpro |
NT, Not tested
* Targets validated.
* Compounds with validated MOA through in-vitro assay.
Bisindolylmaleimide derivatives are widely used as inhibitors of protein kinase C (PKC) isoforms. Bisindolylmaleimide IX (BIM IX), also known as Ro 31-8220, is an advanced investigational molecule and one of the most commonly used PKC inhibitor standards [121]. BIM IX is an imidothio carbamic ester, as well as a bis-indole maleimide. Several reported pharmacological effects of BIM IX; stimulation of c-Jun-N-terminal kinase (JNK) expression, an activator of p38 MAPK through phosphorylation; activation of glycogen synthase activity in adipocytes, among other effects [122], [123], [124]. BIM IX shows potent inhibition of PKC α, PKC βI, PKC βII, PKC γ, and PKC ε [125]. BIM IX also inhibits MSK1, MAPKAPK1, RSK, GSK3β, and S6K1 with a potency similar to that for PKC [125]. BIM IX significantly decreases apoE secretion from primary human macrophages by inhibiting vesicular transport of apoE to the plasma membrane without significantly affecting apoE mRNA or apoE protein levels [125]. Bim IX has been extensively studied in several animals and human tissue types showing a safe profile [126], [127], [128]. In the context of SARS CoV2, the anti-inflammatory and anti-bronchitis profile of this molecule may prove to be an additional therapeutic advantage [127]. While the effect of PKC inhibition on this particular virus is not known, but the SARS-CoV pathogenesis profile is known to be greatly dependent on PKC [129], [130].
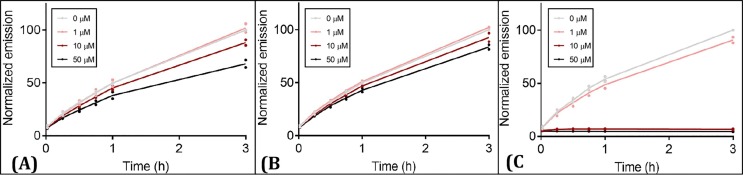
Validation of 3CLpro target through enzyme inhibition assay. To characterize the mechanistic aspects of BIM IX activity, biochemical studies of the 3CLpro enzyme-inhibitor complex were performed. We used a peptide substrate with a C-terminal 7-amino-4-methyl-coumarin (AMC) with a fluorogenic reporter group to perform kinetic assays of 3CLpro inhibition. BIM IX and Haloperidol were tested at four concentrations and the rate of substrate cleavage was recorded at different time points (Fig. 8 ). Haloperidol had a very mild activity, while BIM IX had significant activity. Finally, BIM IX concentrations were varied relative to a fixed concentration of the peptide substrate and monitored over time. Inhibition was observed with an IC50 value of 113.7 ± 5.2 µM (Fig. 9 ).
Fig 8.
Preliminary compound screening for 3CLpro inhibitors. Fluorescence-based biochemical activity assay for (A) BIM IX (B) Haloperidol and (C) Disulfiram (+ve control) using the TVLQ-AMC probe substrate. Each point is the mean of compounds screened in duplicate (1–50 µM) or a DMSO control (0 µM) for n = 2 replicate screens. Emission is normalized to 0 µM at 3 h to identify compounds with inhibitory activity toward 3CLpro.
Fig 9.
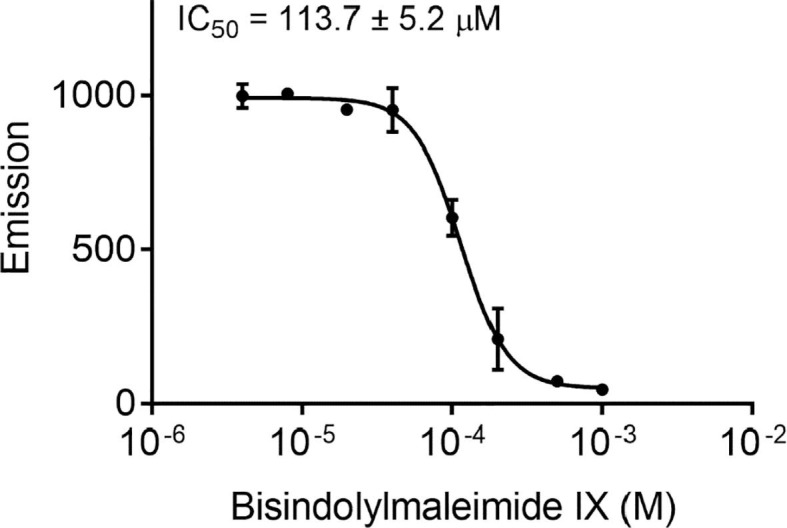
3CLpro inhibition assay. Fluorescence-based biochemical activity assay for BIM IX using the TVLQ-AMC probe substrate. All values are mean ± standard deviation with n = 3 replicates.
4. Conclusion
The rapid spread of SARS-CoV-2 continues to create havoc in health systems and economies, affected every nation around the world. As this is a novel coronavirus, vaccines are being developed at an unprecedented pace and we must also resort to the rapid development of therapeutics for COVID-19. There has been an international focus on the potential efficacy of repurposable drug candidates including remdesivir and hydroxychloroquine, among others. With each controlled clinical study that comes out, it becomes more evident that nations were too quick to push certain compounds, consequently reducing the supply to patients who needed them to treat already indicated diseases [131]. There is a possibility that these compounds may still have a place in the clinical realm as a treatment for COVID-19, but not as stand-alone therapies as currently utilized. Until a SARS-CoV-2 specific compound is developed and clinically approved, the best way is to find treatment through a multifaceted approach of repurposing as well as developing new drugs. This philosophy is at the forefront of our work, first by screening approved compounds for repurposing potential, and by identifying the best possible combinations providing a possible treatment for SARS-CoV-2.
In this context, we commenced a CADD through the HTVS approach using a large pool of world approved drug libraries to identify potential drugs for immediate deployment (Fig. 1). Our in-silico studies have shortlisted a series of repurposable drugs that can be utilized in clinical trials. From our eight selected approved compounds that underwent in-vitro studies against SARS-CoV-2, we found two compounds with high inhibitory activity. One of these was ivermectin, which showed significant viral inhibition, confirming the results of a previous study [84]. Like previous reports, our in-vitro studies found that ivermectin has nonselective toxicity to the ATCC E6 Vero cells at ≤ 50 μM and 16.67 μM based on the number of nuclei counted. This shows that high doses (>16.67 μM) of ivermectin have high cytotoxicity as reported by other groups [132], [133], and this must be considered when preparing to use this drug in COVID-19 clinical trials as its currently approved dosage is very low (25 mg) [84]. The second compound with significant inhibitory activity against in-vitro SARS-CoV-2 was bisindolylmaleimide IX. Further, BIM IX specifically blocked 3CLpro in-vitro enzymatic assay confirming the robustness of our in-silico prediction pipeline. Higher activity in entry assays of 3CLpro inhibitors points to its high activity as once it is expressed in sufficient amounts partial inhibition has a diminished effect or viral replication. This is a critique for high throughput screening assays (spread assay format) as they might miss inhibitors for this target. The target-specific high throughput screenings (both enzymetic and anti-viral) are needed to be designed with parallel toxicity screens for better drug repurposing screens. Overall, this is the first study that showcases the possibility of using bisindolylmaleimide IX specific derivatives and an urgent need to further evaluate their suitability to advance to clinical use to treat COVID-19.
5. Methods
Protein (Receptor) structures. The crystal structures of NSP3 (PDB ID- 6 W02), NSP9(PDB ID- 6W4B), NSP15 (PDB ID- 6VWW) from SARS CoV-2 were obtained from Protein Data Bank [134]. The crystal structures of COVID-19 main protease (NSP5) in complex with Z44592329 (ID:5r83) were obtained from the Protein Structure Database of Europe [135]. QHD43415 [9]: the I-TASSER models were obtained for NSP3(QHD43415_3), NSP5 (QHD43415_5), NSP11(QHD43415_11), NSP12 (QHD43415_12), NSP13(QHD43415_13), and NSP14 (QHD43415_14) [9].
Target Proteins (Receptors) preparation. Proteinpreparationwas performed usingthe ProteinPreparation Wizard in Maestro [10]. All co-crystallized atoms including calcium and chlorine were deleted. Co-crystalized ligands within the active site were not deleted because they were used for grid generation. All water with less than 3 hydrogen bonds (Sample water orientation) with ‘non-waters’ i.e. receptor protein or ligand were deleted. Each of the seven models developed was subjected to preparation by the Wizard in Maestro [10]. Each of the complexes was optimized with minimized hydrogens of altered species and then minimized using the OPLS3e force field [10], [136]. Further, the models were subjected to MD simulation (MDS) after solvation in water and 0.15 M NaCl (physiological solution). A simulation box covering the entire enzyme system was introduced with a 10 Å buffer space. The simulation was run for 100 ns at 300 K and standard pressure (1.01325 bar). The target complex trajectory analysis for interacting residues and was conducted using the Desmond software [10].
Target library preparation. A virtual library of n = 5903 drug compounds was downloaded from the latest Zinc database [11] under the ‘world’ subset (approved drugs in major jurisdictions, including the FDA, i.e DrugBank approved). The database was then checked for redundancy, and duplicates were removed. Ligand preparation was performed using LigPrep, which generated variations of the ligands, eliminated reactive species, and optimized the ligands. Optimization was performed under the OPLS3e force field. EPIK minimization was performed on possible states at pH 7.0 ± 2.0. Tautomers were generated for each ligand retaining specific chirality combinations with a maximum of 32 structures per ligand [10].
Receptor grid generation. COACH analysis was performed to determine the location and size of the active site [137]. The shape and properties of the receptor are represented on a grid to ensure that possible active compounds are not missed. The centroid of the COACH predicted binding pocket residues and was used to generate grids using default values of protein atom scaling (1.0 Å) within a cubic box and ligand docking set to a length of 20 Å. The force field employed for grid generation was OPLS3e [136]. Rotatable groups were automatically determined by the Receptor Grid Generation wizard of the Glide module [10]. COACH predicted binding sites for each target were as follows; Exoribonuclease (ExoN); Based on the active site similarity with PDB ID: 5C8T, S-Adenosyl-L-Methionine (SAM) binding site was selected 292, 333, 335, 352, 353, 366, 367, 368, 385, 386, 387 & 426 PLpro (Papain-like proteinase); Based on PDB similarity with SARS-CoV Plpro model PDB ID: 3mj5B and catalytic triad of Cys–His–Asp the amino acid grid site selected was 209, 223, 230, 233, 234, 235, 236, 237, 238, 239, 293, 318, 321, 324, 325, 327, 349, 350, 352, 353, 354, 356 & 763. 3-Chymotrypsin-Like Protease (3CLpro); Based on PDB id: 2q6gB the substrate-binding site was selected comprising of amino acid numbers 25, 26, 27, 41, 49, 140, 141, 142, 143, 144, 145, 163, 164, 165, 166, 168, 172, 187, 189, 190 & 192. 2′-O-methyltransferase (2′-O-MT); Based on similarity with PDB id. 4N48 the SAM binding site was predicted to be 46, 74, 133, 134, 170 & 203 and was used for grid selection RNA-directed RNA polymerase (RdRP); Based on PDB hit; 3ol9I with similar active site the nucleic acid binding site was selected comprising of amino acid side chain residues 500, 501, 507, 512, 543, 545, 557, 559, 560, 569, 580, 589, 590, 591, 592, 682, 683, 684, 685, 686, 758, 857, 860, 864 & 914 Nonstructural Uridylate-specific endoribonuclease (NendoU): The COACH hit was PDB id:2C1W and the active site selected was 234, 249, 289, 291, 292, 293, 332, 334, 339, 340, 341 & 342 & Helicase (Hel): based on the similarity of active site with hit PDB id: 4N0O nucleic acid binding site was selected comprising of amino acid side chain residues 177, 178, 179, 180, 181, 197, 214, 309, 310, 311, 334, 335, 336, 337, 338, 339, 408, 409, 410, 411, 412, 413, 414, 485, 486, 516, 534, 554 & 560
High-Throughput Virtual Screening (HTVS). The screening was performed with default parameters including ionization states, Epik state penalties within the Glide (Grid-based ligand docking from energetics) module of Schrödinger suite [138]. The scaling factor was maintained at a default of 0.8 and a partial charge cut-off was limited to 0.15. The OPLS3e force field was used during the docking process. The HTVS ligand docking was the first to be performed, followed by SP and XP docking on the top 10% of scoring hits from each previous step. The XP docking aids in removing false positives and the scoring function is much stricter than the HTVS. The greater the XP Glide score, the better the calculated affinity of the hit in binding to the protein target. Further, the estimation of free binding energies for the best hit-docked complexes using MM force fields and implicit solvation was performed using the molecular mechanics/generalized Born surface area (MM-GBSA) method within the virtual screening workflow of Schrödinger suite [10]. The binding energy was calculated based on the following equation.
ΔG = E_complex(minimized) − (E_ligand(minimized) + E_receptor(minimized))
The leads were ranked based on the binding free energy calculation for their respective protein-ligand complexes.
Molecular Dynamics Simulation (MDS). To determine the stability of the ligand-receptor complex especially during flexibility of the binding site and to determine the energetic strain on the docked ligand that could result in immediate fly off (<10 ns). The top hit for each target was subjected to a 20 ns MD simulation (100 ns for XAV-939 + 3CLpro, BIM IX + 3CLpro, & BIM IX + ExoN) as described above in the Target/receptor Preparation section to validate the interaction and thereby the HTVS pipeline and MM-GBSA performance. If the top hit flew off immediately due to major conformation change in the receptor site then the receptor was stimulated alone for 20 ns to reach a stable state and screening was repeated with the resulting minimized structure.
In-vitro microneutralization assay. BIM IX was shortlisted for its predicted affinity for both 3CLpro (Main protease) and ExoN (Exonuclease) active site. Ivermectin was included as a positive control. Other shortlisted molecules included RdRP hits (paromomycin, amikacin, and lactulose), 3CLpro hits (iopromide & troxerutin), and one NendoU hit (haloperidol).
In vitro studies on SARS-CoV-2 were performed at the United States Army Medical Research Institute for Infectious Diseases (USAMRIID). Both entry and spread assays were performed on ATCC Vero E6 cells infected with the SARS-CoV-2 virus. Entry Assay: Vero E6 cells, in 96 well plates, were pre-treated with 14 different test compounds starting at a concentration of 50 μM with 3 fold dilutions down to 0.76 nM for approximately 1hr at 37 °C. SARS-CoV-2/MT020880 was then added to the test compound treated cells for 1 h. at 37 °C at an MOI of 0.4. After 1 h., the cells were washed with PBS before adding additional compounds back in fresh culture media to the cells for 24 h. at 37 °C. Cell media was removed and washed with PBS and plates were submerged in the formal fixing solution for 24 h., then permeabilized with 0.2% Triton-X for 10 min at RT and treated with blocking solution (3% BSA/PBS). Infected cells were detected using a primary detection antibody recognizing the SARS-CoV-2 nucleocapsid protein (Sino Biological, 40143-R001) and Alexa Fluor 488 conjugated antibody (goat α rabbit), then counterstained with DAPI for visualization of the cell nuclei. Infected cells were enumerated using the Operetta high content imaging instrument and data analysis was performed using the Harmony software (Perkin Elmer). Spread Assay: A similar protocol was utilized to the entry assay described above with the following modifications. The virus was used at an MOI of 0.02 and the assay was incubated for 48 hrs. rather than 24 hr. after infection of the cells.
Compound screening to identify 3CLpro inhibitors. Assays were performed in duplicate at room temperature in 96-well black plates at 25 °C. Reactions containing varying concentrations of inhibitor (1–50 µM) and 3CLpro enzyme (0.4 µM) in Tris-HCl pH 7.3, 1 mM EDTA were incubated for approximately five minutes. Reactions were then initiated with TVLQ-AMC probe substrate (40 µM), shaken linearly for 5 s, and then measured for fluorescence emission intensity (excitation λ: 364 nm; emission λ: 440 nm) over time (1 min-3 h) on a Synergy Neo2 Hybrid. Each assay contained 6–8 positive control wells (DMSO replacing inhibitor) and 2 negative control wells (assay components without 3CLpro).
3CLpro enzyme inhibition assay. Fluorescence-based biochemical assay for inhibitors of 3CLpro. Inhibition assays were performed in 96-well black plate in triplicate at 25 °C. Reactions containing varying concentrations of inhibitor (0–1000 µM) and 3CLpro enzyme (0.4 µM) in Tris-HCl pH 7.3, 1 mM EDTA were incubated for approximately five minutes. Reactions were then initiated with TVLQ-AMC probe substrate (40 µM), shaken linearly for 5 s, and then measured continuously for fluorescence emission intensity (excitation λ: 364 nm; emission λ: 440 nm). Data were fit using nonlinear regression (dose-response inhibition, variable slope) analysis in GraphPad Prism 7.0.
Acknowledgments
Acknowledgments
The authors sincerely thank the Department of Medicine, Loyola University Medical Center, and Stritch School of Medicine for supporting the Drug Discovery Program and the High Computing Platform. This project was also supported by the Research Funding Committee (RFC), Loyola University Chicago (PK, LU#213627). Opinions, discussions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense.
Declaration of Competing Interest
The authors declare no conflict of interest.
Author contributions
YG and PK designed the study and performed in-silico analysis. DM, RM, CMP, DJI, and HH assisted in data mining, virtual screening, related in-silico analysis, and table generations for drug characteristics. DM, CMP, and YG covered the epidemiology, virus, genome, and target protein background. PK, AB, and RD contributed to the clinical aspects of the study. DPB, YG, and BR contributed to the chemical and molecular aspects of leads. ASH, SEZ, and JMD performed in-vitro testing against the SARS-CoV-2 virus and toxicity data. BD, KAJ, RSK, and SA synthesized fluorescent probe for 3CLpro and performed biochemical assays and assisted with the mechanistic aspects, and validated the viral target and lead interactions. All authors contributed to writing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2021.01.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.W.H. Organization, others, Coronavirus disease (COVID-19): situation report, Weekly Epidemiology Update 2020, (2020).
- 4.Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019 doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pradesh U, Upadhayay PDD, Vigyan PC, Coronavirus infection in equines: A review, Asian J Anim Vet Adv. 9 (2014) 164–76.
- 6.Ahmad T., Rodriguez-Morales A.J. Emergence of COVID-19 (formerly 2019-novel Coronavirus): a new threat from China. Rev. Panam. Enfermedades Infecc. 2020:37–38. [Google Scholar]
- 7.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. Zhang, W. Zheng, X. Huang, E.W. Bell, X. Zhou, Y. Zhang, Protein structure and sequence re-analysis of 2019-nCoV genome does not indicate snakes as its intermediate host or the unique similarity between its spike protein insertions and HIV-1, ArXiv Prepr. ArXiv200203173. (2020). [DOI] [PMC free article] [PubMed]
- 10.L. Schrodinger, Small-Molecule Drug Discovery Suite 2020-1. 2020, 2020.
- 11.Irwin J.J., Shoichet B.K. ZINC- a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov. Today. 2018;23:661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillaiyar T., Manickam M., Jung S. Middle East respiratory syndrome-coronavirus (MERS-CoV): an updated overview and pharmacotherapeutics. Med. Chem. 2015;5:361–372. [Google Scholar]
- 14.C.S.G. of the International, others, The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol. (2020) 1. [DOI] [PMC free article] [PubMed]
- 15.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020 doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 using direct RNA sequencing and tandem mass spectrometry reveals evidence for a cell passage induced in-frame deletion in the spike glycoprotein that removes the furin-like cleavage site. BioRxiv. 2020 doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Astuti I., et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020 doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar V., Shin J.S., Shie J.-J., Ku K.B., Kim C., Go Y.Y., Huang K.-F., Kim M., Liang P.-H. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y., Cai H., Xiang N., Tien P., Ahola T., Guo D. others, Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Ahola T. others, Coronavirus nsp10/nsp16 methyltransferase can be targeted by nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H. others, 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. others, Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.I. Imbert, R. Ulferts, J. Ziebuhr, B. Canard, SARS coronavirus replicative enzymes: structures and mechanisms, in: Mol. Biol. SARS-Coronavirus, Springer, 2010: pp. 99–114.
- 30.Zhang L., Li L., Yan L., Ming Z., Jia Z., Lou Z., Rao Z. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by Middle East respiratory syndrome coronavirus. J. Virol. 2018;92:e00893–18. doi: 10.1128/JVI.00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K.M. Baig, SITE DIRECTED MUTAGENESIS OF SEVERE ACUTE RESPIRATORY SYNDROME (SARS) CORONAVIRUS NSP 15 ENDORIBONUCLEASE, PhD Thesis, 2006.
- 32.Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei Y., Moore C.B., Liesman R.M., O’Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.-P.-Y. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., Leist S.R., Yount B.L., Graham R.L., McAnarney E.T., Stratton K.G., Cockrell A.S. others, Middle east respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. MSphere. 2017;2:e00346–17. doi: 10.1128/mSphere.00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adedeji A.O., Singh K., Sarafianos S.G. Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Cell. Mol. Biol. Noisy–Gd. Fr. 2012;58:114. [PMC free article] [PubMed] [Google Scholar]
- 37.Yu M.-S., Lee J., Lee J.M., Kim Y., Chin Y.-W., Jee J.-G., Keum Y.-S., Jeong Y.-J. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Shu, M. Huang, D. Wu, Y. Ren, X. Zhang, Y. Han, J. Mu, R. Wang, Y. Qiu, D.-Y. Zhang, others, SARS-coronavirus-2 nsp13 possesses NTPase and RNA helicase activities, (n.d.). [DOI] [PMC free article] [PubMed]
- 39.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., et al. Structural and biochemical characterization of nsp12-nsp7-nsp8 core polymerase complex from COVID-19 virus. BioRxiv. 2020 doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap Y., Zhang X., Andonov A., He R. Structural analysis of inhibition mechanisms of aurintricarboxylic acid on SARS-CoV polymerase and other proteins. Comput. Biol. Chem. 2005;29:212–219. doi: 10.1016/j.compbiolchem.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posthuma C.C., te Velthuis A.J., Snijder E.J. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res. 2017;234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chien M., Anderson T.K., Jockusch S., Tao C., Li X., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju J., Li X., Kumar S., Jockusch S., Chien M., Tao C., Morozova I., Kalachikov S., Kirchdoerfer R., Russo J.J. Nucleotide analogues as inhibitors of SARS-CoV polymerase. BioRxiv. 2020 doi: 10.1002/prp2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiyer P.V. Amylases and their applications. Afr. J. Biotechnol. 2005;4 [Google Scholar]
- 46.Takasaki Y., Shinohara H., Tsuruhisa M., Hayashi S., Imada K. Maltotetraose-producing amylase from Bacillus sp. MG-4. Agric. Biol. Chem. 1991;55:1715–1720. [Google Scholar]
- 47.Abdullaev F.I. Cancer chemopreventive and tumoricidal properties of saffron (Crocus sativus L.) Exp. Biol. Med. 2002;227:20–25. doi: 10.1177/153537020222700104. [DOI] [PubMed] [Google Scholar]
- 48.Stern S.A., Zink B., Wang X., Mertz M. The effects of resuscitation with trans-sodium crocetinate in a model of combined hemorrhagic shock and traumatic brain injury. Acad. Emerg. Med. 2002;9:415. [Google Scholar]
- 49.Tseng T.-H., Chu C.-Y., Huang J.-M., Shiow S.-J., Wang C.-J. Crocetin protects against oxidative damage in rat primary hepatocytes. Cancer Lett. 1995;97:61–67. doi: 10.1016/0304-3835(95)03964-x. [DOI] [PubMed] [Google Scholar]
- 50.Brito D.A., Yang Z., Rieder C.L. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J. Cell Biol. 2008;182:623–629. doi: 10.1083/jcb.200805072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E.K. Rowinsky MD, The development and clinical utility of the taxane class of antimicrotubule chemotherapy agents, Annu. Rev. Med. 48 (1997) 353–374. [DOI] [PubMed]
- 52.Zhou J., Zhong D.-W., Wang Q.-W., Miao X.-Y., Xu X.-D. Paclitaxel ameliorates fibrosis in hepatic stellate cells via inhibition of TGF-β/Smad activity. World J. Gastroenterol. WJG. 2010;16:3330. doi: 10.3748/wjg.v16.i26.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun L., Zhang D., Liu F., Xiang X., Ling G., Xiao L., Liu Y., Zhu X., Zhan M., Yang Y. others, Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. J. Pathol. 2011;225:364–377. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tepe G., Zeller T., Albrecht T., Heller S., Schwarzwälder U., Beregi J.-P., Claussen M.C.D., Oldenburg A., Scheller B., Speck U. Local delivery of paclitaxel to inhibit restenosis during angioplasty of the leg. N. Engl. J. Med. 2008;358:689–699. doi: 10.1056/NEJMoa0706356. [DOI] [PubMed] [Google Scholar]
- 55.Tang N., Bai H., Chen X., Gong J., Li D., Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemost. 2020 doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi C., Wang C., Wang H., Yang C., Cai F., Zeng F., Cheng F., Liu Y., Zhou T., Deng B., et al. The potential of low molecular weight heparin to mitigate cytokine storm in severe COVID-19 patients: a retrospective clinical study. MedRxiv. 2020 doi: 10.1111/cts.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Negri E.M., Piloto B., Morinaga L.K., Jardim C.V.P., Lamy S.A.E.-D., Ferreira M.A., D’Amico E.A., Deheinzelin D. Heparin therapy improving hypoxia in COVID-19 patients-a case series. MedRxiv. 2020 doi: 10.3389/fphys.2020.573044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Ehsani, Distant sequence similarity between hepcidin and the novel coronavirus spike glycoprotein: a potential hint at the possibility of local iron dysregulation in COVID-19, ArXiv Prepr. ArXiv200312191. (2020). [DOI] [PMC free article] [PubMed]
- 59.Poli M., Girelli D., Campostrini N., Maccarinelli F., Finazzi D., Luscieti S., Nai A., Arosio P. Heparin: a potent inhibitor of hepcidin expression in vitro and in vivo. Blood J. Am. Soc. Hematol. 2011;117:997–1004. doi: 10.1182/blood-2010-06-289082. [DOI] [PubMed] [Google Scholar]
- 60.Kim H., Lee M.-K., Ko J., Park C.-J., Kim M., Jeong Y., Hong S., Varani G., Choi B.-S. Aminoglycoside antibiotics bind to the influenza A virus RNA promoter. Mol. Biosyst. 2012;8:2857–2859. doi: 10.1039/c2mb25333j. [DOI] [PubMed] [Google Scholar]
- 61.Manvar D., Pandey N., Pandey V.N. Anti-HCV drug development. Abstr. Res. 2011;90:A21–A78. [Google Scholar]
- 62.Ruttloff H., Täufel A., Krause W., Haenel H., Täufel K. Die intestinal-enzymatische Spaltung von Galakto-Oligosacchariden im Darm von Tier und Mensch mit besonderer Berücksichtigung von Lactobacillus bifidus 2. Mitt. Zum intestinalen Verhalten der Lactulose. Food/Nahrung. 1967;11:39–46. [Google Scholar]
- 63.Hu G. Neomycin inhibits the angiogenic activity of fibroblast and epidermal growth factors. Biochem. Biophys. Res. Commun. 2001;287:870–874. doi: 10.1006/bbrc.2001.5668. [DOI] [PubMed] [Google Scholar]
- 64.K. Kishimoto, S. Yoshida, S. Ibaragi, N. Yoshioka, G.-F. HU, A. Sasaki, Neamine inhibits oral cancer progression by suppressing angiogenin-mediated angiogenesis and cancer cell proliferation, Anticancer Res. 34 (2014) 2113–2121. [PMC free article] [PubMed]
- 65.Lounis N., Ji B., Truffot-Pernot C., Grosset J. Which aminoglycoside or fluoroquinolone is more active against Mycobacterium tuberculosis in mice? Antimicrob. Agents Chemother. 1997;41:607–610. doi: 10.1128/aac.41.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamaguchi M., Eda J., Kobayashi F., Mitsuhashi S. Mode of action of lividomycin on protein synthesis in Escherichia coli. Antimicrob. Agents Chemother. 1973;4:380–382. doi: 10.1128/aac.4.3.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robidoux A., Tang G., Rastogi P., Geyer C.E., Jr, Azar C.A., Atkins J.N., Fehrenbacher L., Bear H.D., Baez-Diaz L., Sarwar S. others, Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14:1183–1192. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 68.Alba E., Albanell J., De La Haba J., Barnadas A., Calvo L., Sanchez-Rovira P., Ramos M., Rojo F., Burgues O., Carrasco E. others, Trastuzumab or lapatinib with standard chemotherapy for HER2-positive breast cancer: results from the GEICAM/2006-14 trial. Br. J. Cancer. 2014;110:1139–1147. doi: 10.1038/bjc.2013.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato K., Takahashi S., Cui L., Toda T., Suzuki S., Futakuchi M., Sugiura S., Shirai T. Suppressive effects of dietary genistin and daidzin on rat prostate carcinogenesis. Jpn. J. Cancer Res. 2000;91:786–791. doi: 10.1111/j.1349-7006.2000.tb01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.B. Pendyala, A. Patras, In silico Screening of Food Bioactive Compounds to Predict Potential Inhibitors of COVID-19 Main protease (Mpro) and RNA-dependent RNA polymerase (RdRp), (2020).
- 71.K. Sharp, D. Dange, others, Computational Drug Simulation: A step to the possible cure of COVID-19, (2020).
- 72.Ekholm S., Foley M., Kido D., Morris T. Lumbar myelography with metrizamide in rabbits: an investigation of contrast media penetration and resorption. Acta Radiol. Diagn. (Stockh.) 1984;25:517–522. doi: 10.1177/028418518402500612. [DOI] [PubMed] [Google Scholar]
- 73.Kudo S., Ishizaki T. Pharmacokinetics of haloperidol. Clin. Pharmacokinet. 1999;37:435–456. doi: 10.2165/00003088-199937060-00001. [DOI] [PubMed] [Google Scholar]
- 74.Schuckit M.A. Recognition and management of withdrawal delirium (delirium tremens) N. Engl. J. Med. 2014;371:2109–2113. doi: 10.1056/NEJMra1407298. [DOI] [PubMed] [Google Scholar]
- 75.Thorn C.F., Aklillu E., McDonagh E.M., Klein T.E., Altman R.B. PharmGKB summary: caffeine pathway. Pharmacogenet. Genomics. 2012;22:389. doi: 10.1097/FPC.0b013e3283505d5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.M. Day, Covid-19: ibuprofen should not be used for managing symptoms, say doctors and scientists, British Medical Journal Publishing Group, 2020. [DOI] [PubMed]
- 77.FitzGerald G.A. Misguided drug advice for COVID-19. Science. 2020;367:1434–1436. doi: 10.1126/science.abb8034. [DOI] [PubMed] [Google Scholar]
- 78.Fan E., Merritt E.A., Zhang Z., Pickens J.C., Roach C., Ahn M., Hol W.G. Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives. Acta Crystallogr. D Biol. Crystallogr. 2001;57:201–212. doi: 10.1107/s0907444900016814. [DOI] [PubMed] [Google Scholar]
- 79.Thanapongsathorn W., Vajrabukka T. Clinical trial of oral diosmin (Daflon®) in the treatment of hemorrhoids. Dis. Colon Rectum. 1992;35:1085–1088. doi: 10.1007/BF02253000. [DOI] [PubMed] [Google Scholar]
- 80.Fermoso J., Legido A.G., Del Pino J., Valiente R. Therapeutic value of hidrosmin in the treatment of venous disorders of the lower limbs. Curr. Ther. Res. 1992;52:124–134. [Google Scholar]
- 81.F. Meneguzzo, R. Ciriminna, F. Zabini, M. Pagliaro, Accelerated production of hesperidin-rich citrus pectin from waste citrus peel for prevention and therapy of COVID-19, (2020).
- 82.S. Nabirotchkin, A.E. Peluffo, J. Bouaziz, D. Cohen, Focusing on the unfolded protein response and autophagy related pathways to reposition common approved drugs against COVID-19, (2020).
- 83.Pinkosky S.L., Newton R.S., Day E.A., Ford R.J., Lhotak S., Austin R.C., Birch C.M., Smith B.K., Filippov S., Groot P.H. others, Liver-specific ATP-citrate lyase inhibition by bempedoic acid decreases LDL-C and attenuates atherosclerosis. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;104787 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mastrangelo E., Pezzullo M., De Burghgraeve T., Kaptein S., Pastorino B., Dallmeier K., de Lamballerie X., Neyts J., Hanson A.M., Frick D.N. others, Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 2012;67:1884–1894. doi: 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spitsin S., Hooper D., Leist T., Streletz L., Mikheeva T., Koprowski H. Inactivation of peroxynitrite in multiple sclerosis patients after oral administration of inosine may suggest possible approaches to therapy of the disease. Mult. Scler. J. 2001;7:313–319. doi: 10.1177/135245850100700507. [DOI] [PubMed] [Google Scholar]
- 87.Svedmyr N. Fenoterol: A Beta2-adrenergic Agonist for Use in Asthma; pharmacology, pharmacokinetics, clinical efficacy and adverse effects. Pharmacother. J. Hum. Pharmacol. Drug Ther. 1985;5:109–126. doi: 10.1002/j.1875-9114.1985.tb03409.x. [DOI] [PubMed] [Google Scholar]
- 88.Kahn J.O., Lagakos S.W., Richman D.D., Cross A., Pettinelli C., Liou S., Brown M., Volberding P.A., Crumpacker C.S., Beall G. others, A controlled trial comparing continued zidovudine with didanosine in human immunodeficiency virus infection. N. Engl. J. Med. 1992;327:581–587. doi: 10.1056/NEJM199208273270901. [DOI] [PubMed] [Google Scholar]
- 89.Alakwaa F.M. Repurposing didanosine as a potential treatment for COVID-19 using single-cell RNA sequencing data. Msystems. 2020;5 doi: 10.1128/mSystems.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bukreyeva N., Mantlo E.K., Sattler R.A., Huang C., Paessler S., Zeldis J. The IMPDH inhibitor merimepodib suppresses SARS-CoV-2 replication in vitro. BioRxiv. 2020 [Google Scholar]
- 91.McLaren C., Datema R., Knupp C., Buroker R. didanosine. Antivir. Chem. Chemother. 1991;2:321–328. [Google Scholar]
- 92.Klimke A., Hefner G., Will B., Voss U. Hydroxychloroquine as an aerosol might markedly reduce and even prevent severe clinical symptoms after SARS-CoV-2 infection. Med. Hypotheses. 2020;109783 doi: 10.1016/j.mehy.2020.109783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.M. Moskal, W. Beker, R. Roszak, E.P. Gajewska, A. Wo\los, K. Molga, S. Szymkuć, B.A. Grzybowski, Suggestions for second-pass anti-COVID-19 drugs based on the Artificial Intelligence measures of molecular similarity, shape and pharmacophore distribution., (2020).
- 94.Schaaf L., Dobbs B., Edwards I., Perrier D. The pharmacokinetics of doxifluridine and 5-fluorouracil after single intravenous infusions of doxifluridine to patients with colorectal cancer. Eur. J. Clin. Pharmacol. 1988;34:439–443. doi: 10.1007/BF01046699. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Z.-F., Fan S.-H., Zheng Y.-L., Lu J., Wu D.-M., Shan Q., Hu B. Troxerutin improves hepatic lipid homeostasis by restoring NAD+-depletion-mediated dysfunction of lipin 1 signaling in high-fat diet-treated mice. Biochem. Pharmacol. 2014;91:74–86. doi: 10.1016/j.bcp.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 96.L. Mittal, A. Kumari, M. Srivastava, M. Singh, S. Asthana, Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach, (2020). [DOI] [PMC free article] [PubMed]
- 97.Liu X.-Y. Therapeutic effect of chai-ling-tang (sairei-to) on the steroid-dependent nephrotic syndrome in children. Am. J. Chin. Med. 1995;23:255–260. doi: 10.1142/S0192415X95000304. [DOI] [PubMed] [Google Scholar]
- 98.Yang L., Li Y.-T., Miao J., Wang L., Fu H., Li Q., Wen W.-B., Zhang Z.-Y., Song R.-W., Liu X.-G., et al. Network pharmacology studies on the effect of Chai-Ling decoction in coronavirus disease 2019. Med. Res. 2020;5:145–159. [Google Scholar]
- 99.Brayer G.D., Sidhu G., Maurus R., Rydberg E.H., Braun C., Wang Y., Nguyen N.T., Overall C.M., Withers S.G. Subsite mapping of the human pancreatic α-amylase active site through structural, kinetic, and mutagenesis techniques. Biochemistry. 2000;39:4778–4791. doi: 10.1021/bi9921182. [DOI] [PubMed] [Google Scholar]
- 100.Elson-Schwab L., Tor Y. Targeting HIV-1 RNA with Aminoglycoside antibiotics and their derivatives. Aminoglycoside Antibiot. 2007:267–287. [Google Scholar]
- 101.Weissmann B., Meyer K. The structure of hyalobiuronic acid and of hyaluronic acid from umbilical Cord 1, 2. J. Am. Chem. Soc. 1954;76:1753–1757. [Google Scholar]
- 102.Stauder G., Ransberger K. Treatment of hepatitis C virus infection using a protease and a flavonoid. Google Patents. 2001 [Google Scholar]
- 103.Sanford M., Plosker G.L. Anastrozole. Drugs. 2008;68:1319–1340. doi: 10.2165/00003495-200868090-00007. [DOI] [PubMed] [Google Scholar]
- 104.Lønning P.E. Clinical pharmacokinetics of aromatase inhibitors and inactivators. Clin. Pharmacokinet. 2003;42:619–631. doi: 10.2165/00003088-200342070-00002. [DOI] [PubMed] [Google Scholar]
- 105.Stakheev D., Taborska P., Strizova Z., Podrazil M., Bartunkova J., Smrz D. The WNT/β-catenin signaling inhibitor XAV939 enhances the elimination of LNCaP and PC-3 prostate cancer cells by prostate cancer patient lymphocytes in vitro. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-41182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grunewald M.E., Chen Y., Kuny C., Maejima T., Lease R., Ferraris D., Aikawa M., Sullivan C.S., Perlman S., Fehr A.R. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alhammad Y.M., Fehr A.R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12:384. doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khalatbari-mohseni A., Banafshe H.R., Mirhosseini N., Asemi Z., Ghaderi A., Omidi A. The effects of crocin on psychological parameters in patients under methadone maintenance treatment: a randomized clinical trial. Subst. Abuse Treat. Prev. Policy. 2019;14:9. doi: 10.1186/s13011-019-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.di Camillo Orfali G., Duarte A.C., Bonadio V., Martinez N.P., De Araújo M.E.M.B., Priviero F.B.M., Carvalho P.O., Priolli D.G. Review of anticancer mechanisms of isoquercitin, World. J. Clin. Oncol. 2016;7:189. doi: 10.5306/wjco.v7.i2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gane E.J., Rouzier R., Wiercinska-Drapalo A., Larrey D.G., Morcos P.N., Brennan B.J., Le Pogam S., Nájera I., Petric R., Tran J.Q. others, Efficacy and safety of danoprevir-ritonavir plus peginterferon Alfa-2a–ribavirin in hepatitis C virus genotype 1 prior null responders. Antimicrob. Agents Chemother. 2014;58:1136–1145. doi: 10.1128/AAC.01515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sekhar T. Virtual Screening based prediction of potential drugs for COVID-19. Preprints. 2020 [Google Scholar]
- 112.Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y., et al. First clinical study using HCV protease inhibitor danoprevir to treat naive and experienced COVID-19 patients. MedRxiv. 2020 doi: 10.1097/MD.0000000000023357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johnson C.A., Zimmerman S., Reitberg D., Whall T., Leggett J., Craig W. Pharmacokinetics and pharmacodynamics of cefoperazone-sulbactam in patients on continuous ambulatory peritoneal dialysis. Antimicrob. Agents Chemother. 1988;32:51–56. doi: 10.1128/aac.32.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang G., Hu C., Luo L., Fang F., Chen Y., Li J., Peng Z., Pan H. Clinical features and outcomes of 221 patients with COVID-19 in Wuhan, China. MedRxiv. 2020 doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Y. Li, R. Zhao, S. Zheng, X. Chen, J. Wang, X. Sheng, J. Zhou, H. Cai, Q. Fang, F. Yu, others, Early release-lack of vertical transmission of severe acute respiratory syndrome coronavirus 2, China, (2020). [DOI] [PMC free article] [PubMed]
- 116.Ena J., Amador C., Benito C., Pasquau F. Pharmacological and clinical evidence of nevirapine immediate-and extended-release formulations. HIVAIDS Auckl. NZ. 2012;4:169. doi: 10.2147/HIV.S35564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O. Kadioglu, M. Saeed, H. Johannes Greten, T. Efferth, Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning, (n.d.). [DOI] [PMC free article] [PubMed]
- 118.Lee V.S., Chong W.L., Sukumaran S.D., Nimmanpipug P., Letchumanan V., Goh B.H., Lee L.-H., Zain S.M., Abd Rahman N. Computational screening and identifying binding interaction of anti-viral and anti-malarial drugs: toward the potential cure for SARS-CoV-2. Prog. Drug Discov Biomed. Sci. 2020;3 [Google Scholar]
- 119.L.A. Andritsos, N. Dunavin, G. Lozanski, J.A. Jones, J.S. Blachly, D.M. Lucas, J.C. Byrd, E. Kraut, M.R. Grever, Reduced dose pentostatin for initial management of hairy cell leukemia patients who have active infection or risk of hemorrhage is safe and effective, Haematologica. (2014) haematol-2014. [DOI] [PMC free article] [PubMed]
- 120.L. Zhang, R. Zhou, Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase, (2020). [DOI] [PubMed]
- 121.Wilkinson S.E., Parker P., Nixon J. Isoenzyme specificity of bisindolylmaleimides, selective inhibitors of protein kinase C. Biochem. J. 1993;294:335–337. doi: 10.1042/bj2940335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Beltman J., McCormick F., Cook S.J. The selective protein kinase C inhibitor, Ro-31-8220, inhibits mitogen-activated protein kinase phosphatase-1 (MKP-1) expression, induces c-Jun expression, and activates Jun N-terminal kinase. J. Biol. Chem. 1996;271:27018–27024. doi: 10.1074/jbc.271.43.27018. [DOI] [PubMed] [Google Scholar]
- 123.Beltman J., Erickson J.R., Martin G.A., Lyons J.F., Cook S.J. C3 toxin activates the stress signaling pathways, JNK and p38, but antagonizes the activation of AP-1 in rat-1 cells. J. Biol. Chem. 1999;274:3772–3780. doi: 10.1074/jbc.274.6.3772. [DOI] [PubMed] [Google Scholar]
- 124.Standaert M., Bandyopadhyay G., Antwi E., Farese R. RO 31–8220 activates c-Jun N-terminal kinase and glycogen synthase in rat adipocytes and L6 myotubes. Comparison to actions of insulin. Endocrinology. 1999;140:2145–2151. doi: 10.1210/endo.140.5.6699. [DOI] [PubMed] [Google Scholar]
- 125.NCATS Inxight: Drugs — RO-31-8220, (n.d.). https://drugs.ncats.io/drug/W9A0B5E78O (accessed July 4, 2020).
- 126.Alessi D.R. The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett. 1997;402:121–123. doi: 10.1016/s0014-5793(96)01510-4. [DOI] [PubMed] [Google Scholar]
- 127.Chopra L., Twort C., Ward J. Differences in sensitivity to the specific protein kinase C inhibitor Ro31-8220 between small and large bronchioles of the rat. Br. J. Pharmacol. 1994;113:1237–1242. doi: 10.1111/j.1476-5381.1994.tb17130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haralabopoulos G., Grant D., Kleinman H., Maragoudakis M. Thrombin promotes endothelial cell alignment in Matrigel in vitro and angiogenesis in vivo. Am. J. Physiol.-Cell Physiol. 1997;273:C239–C245. doi: 10.1152/ajpcell.1997.273.1.C239. [DOI] [PubMed] [Google Scholar]
- 129.Ji H.-L., Song W., Gao Z., Su X.-F., Nie H.-G., Jiang Y., Peng J.-B., He Y.-X., Liao Y., Zhou Y.-J., et al. SARS-CoV proteins decrease levels and activity of human ENaC via activation of distinct PKC isoforms. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2009;296:L372–L383. doi: 10.1152/ajplung.90437.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peng J.-B., He Y.-X., Liao Y., Zhou Y.-J., Tousson A., Ji S.H.-L., Song W., Gao Z., Su X.-F., Nie H.-G. others, SARS-CoV proteins decrease levels and activity of human. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;296:L372–L383. doi: 10.1152/ajplung.90437.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pan H., Peto R., Karim Q.A., Alejandria M., Restrepo A.M.H., Garcia C.H., Kieny M.P., Malekzadeh R., Murthy S., Preziosi M.-P., et al. Repurposed antiviral drugs for COVID-19; interim WHO SOLIDARITY trial results. MedRxiv. 2020 [Google Scholar]
- 132.Momekov G., Momekova D. Ivermectin as a potential COVID-19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol. Biotechnol. Equip. 2020;34:469–474. [Google Scholar]
- 133.Schmith V.D., Zhou J., Lohmer L.R. The approved dose of ivermectin alone is not the ideal dose for the treatment of COVID-19. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z., et al. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2016:gkw1000. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.PDBe-KB: a community-driven resource for structural and functional annotations, Nucleic Acids Res. 48 (2020) D344–D353. [DOI] [PMC free article] [PubMed]
- 136.Roos K., Wu C., Damm W., Reboul M., Stevenson J.M., Lu C., Dahlgren M.K., Mondal S., Chen W., Wang L. others, OPLS3e: extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- 137.Wu Q., Peng Z., Zhang Y., Yang J. COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018;46:W438–W442. doi: 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein- ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.