Abstract
Background
Advanced gastric signet-ring cell carcinoma (SRCC) is a specific type of malignant gastric cancer (GC) with distinct poorer survival. Claudin18.2 (CLDN18.2) is a promising neo-biomarker for the treatment of GC. Clinical trials of CLDN18.2-targeted antibody and T cell-based immunotherapy providing promising prospects for the treatment of GC. The effect of antibody therapy depended on the expression rate of CLDN18.2 has been found in clinical trials. This study aimed to determine the prevalence and the therapeutic value of CLDN18.2 in advanced gastric SRCC.
Methods
Expression of CLDN18.2 in 105 formalin-fixed, paraffin-embedded (FFPE) tumor tissues was detected by immunohistochemistry (IHC) and evaluated according to FAST criteria. Next-generation sequencing (NGS) using 416 pan-cancer genes panel was performed to characterize the genomic landscape in 61 advanced gastric SRCC patients. Fisher’s exact test was used to determine gene differences in different CLDN18.2 expression levels.
Results
A total number of 105 advanced gastric SRCC samples were analyzed, of which 95.2% (100/105) were positive stained. Moderate-to-strong CLDN18.2 expression was observed in 64.8% (68/105) of all samples. In particularly, 21.0% (22/105) samples had positive staining in more than 90% tumor cells. No significance was found between CLDN18.2 expression and overall survival (OS). NGS results showed that single nucleotide variations (SNVs) could be frequently found in TP53 (26.2%), CDH1 (19.7%), MED12 (18.0%), PKHD1 (18.0%) and ARID1A (11.5%), besides, copy number variations (CNVs) were rich in NOTCH1 (18.0%) and FLT4 (9.8%) in SRCC samples. Moreover, SNVs in GRIN2A was found in 20% of the patients who had CLDN18.2 staining in <40% of tumor cells (P=0.043), indicating CLDN18.2 expression might be related to the aberration of GRIN2A in advanced gastric SRCC.
Conclusions
The highly expressed CLDN18.2 among advanced gastric SRCC patients that we found certified the value of CLDN18.2-targeted therapy in this specific type of GC. In addition, Analyses between CLDN18.2 expression and genetic abnormalities provided novel therapeutic options for advanced gastric SRCC.
Keywords: Gastric signet-ring cell carcinoma (gastric SRCC), CLDN18.2, next-generation sequencing (NGS)
Introduction
Gastric signet-ring cell carcinoma (SRCC) is an exceptional subtype of gastric adenocarcinoma which has abundant intracellular mucin accumulation and the crescent-shaped nucleus are displaced toward one side of the tumor cells (1). According to World Health Organization (WHO) classification: gastric cancer (GC) pathologic specimen with at least 50% of signet-ring cell is defined as SRCC. In recent decades, the overall morbidity of GC decreased worldwide, however, the decline is mainly caused by the decrease of intestinal type, while for diffuse type, especially gastric SRCC, is on the rise (2-4). Usually, SRCC is more prone to invasion and metastasis and have unique chemosensitivities, which led to a worse prognosis (3,5,6). However, SRCC treatment regimen are still controversial as SRCC is not specifically identified in most studies. For diffuse type GC, targeted therapies including anti-HER2 (7) and anti-VEGFR2 (8) provide few benefit in overall survival (OS). Immunotherapy may be a promising treatment as PD-L1 is overexpressed in about 23% of gastric SRCC (5).
CLDN18.2 is a splice variant of the membrane epithelial tight junctions protein Claudin18 (CLDN18) and has been identified as a promising biomarker for targeted therapy (9). CLDN18.2 has a restricted expression profile in normal tissues, it physiologically expresses only in the tight junction supramolecular complex of gastric mucosa. While upon malignant transformation, the changes in cell polarity lead to the exposure of CLDN18.2 epitopes (10), which are suitable for targeted therapy. Prior evidence demonstrates that except for GC, CLDN18.2 is aberrantly expressed in various primary tumors and metastases, including pancreatic, biliary, ovarian, and lung adenocarcinomas (11-14), which makes CLDN18.2 a pan-cancer target. At present, antibody [zolbetuximab (10,15-18), formerly called IMAB362] and chimeric antigen receptor engineered T cells (CAR-T) (19,20) targeting CLDN18.2 has been applied in clinical trials with promising results achieved. Correlation between higher CLDN18.2 expression and better therapeutic benefits has been found through antibody-based clinical trials. Previous studies have reported a relatively high expression rate of CLDN18.2 in diffuse type GC (9,21), however, no study on advanced gastric SRCC has been conducted. Therefore, this study aimed to establish the prevalence of CLDN18.2 expression in advanced gastric SRCC, and to determine the therapeutic value of CLDN18.2 in this specific type of GC.
In this study, we identified, for the first time, that the expression rate of CLDN18.2 in advanced gastric SRCC patients was relatively high. The survival analyses based on CLDN18.2 expression were performed. We also conducted next-generation sequencing (NGS) in 61 advanced gastric SRCC samples, genetic abnormalities in advanced gastric SRCC were profiled and the relation between CLDN18.2 expression and GRIN2A mutation was discovered.
We present the following article in accordance with the MDAR reporting checklist. (available at: http://dx.doi.org/10.21037/jgo-20-344).
Methods
Tissue specimens and ethical statement
A total of 105 formalin-fixed, paraffin-embedded (FFPE) tissue specimens with histology of advanced gastric SRCC for the testing of CLDN18.2 expression were collected at Nanjing Drum Tower Hospital. Patients at stage III were all administrated first-line 5-FU-based adjuvant chemotherapy after D2 gastrectomy, while patients at stage IV were treated by first-line 5-FU-based palliative chemotherapy. None of the patients had radiotherapy, chemotherapy or other medical intervention before specimen collection. Samples from stage III patients were curative surgical specimens, while samples from stage IV patients were palliative surgical specimens or gastroscope specimens. OS data was available in 86 cases; 61 of the tissues were selected for NGS.
Immunohistochemistry (IHC) and histologic assessment
All tissue samples were stained using rabbit monoclonal anti-CLDN18.2 antibody (Abcam, 222512) at 1/800 dilution. This antibody was designed to recognize human CLDN18.2 aa 1–100. After diluted antibody was added to the whole tissue surface, the slides were incubated at 4 °C overnight. Slides were then rewarmed at room temperature for 10 minutes and sufficiently washed with PBS for three times. Goat Anti-Rabbit IgG H&L (HRP) (Abcam, 205718) were used as secondary antibody, when the slides were dry, secondary antibody were dropped to cover the whole tissue and incubate in a 37 °C incubator for 30 minutes. Nuclear were stained with hematoxylin; 3% hydrogen peroxide were used to block endogenous peroxidases. Each patient’s slides were tested twice.
To determine CLDN18.2 expression status, tissue samples were analyzed according to the intensity of staining and the percentage of stained tumor cells. The intensity was classified into 4 grades: no membrane or cytoplasmic reactivity as 0, weak membrane or cytoplasmic reactivity as 1+, moderate membrane or cytoplasmic reactivity as 2+, and strong membrane or cytoplasmic reactivity as 3+. Samples showing any specific staining with ≥1+ intensity were defined as CLDN18.2 positive. According to FAST criterion, more than 40% of tumor tissues specific staining with ≥2+ intensity were defined as moderate-to-strong expression (21). Percentage of overall CLDN18.2 positive cells was considered by the estimated number of CLDN18.2 positive cells divided by the estimated overall number of tumor cells in each sample.
The pathological diagnosis of SRCC in our study were confirmed by two independent pathologists. The interpretation of IHC results were performed according to the FAST criterion.
NGS and data processing
61 out of 105 samples which had enough tumor tissue were selected for NGS. 15 had CLDN18.2 positive staining in <40% of tumor cells, 46 had CLDN18.2 positive staining in ≥40% of tumor cells. Genomic DNA from FFPE tissue specimens were extracted using QIAamp DNA FFPE Tissue Kit (Qiagen, Catalog no.56404) according to the manufacturer’s protocols. Hybridization-based enrichment was carried out with GeneseeqOneTM pan-cancer gene panel (416 cancer-relevant genes). Captured libraries by Dynabeads M-270 (Life Technologies, MA, USA) were amplified in KAPA HiFi HotStart ReadyMix (KAPA Biosystems, MA, USA) and quantified by qPCR using the KAPA Library Quantification kit (KAPA Biosystems, MA, USA) for sequencing. The libraries were paired-end sequenced on Illumina HiSeq4000 NGS platforms (Illumina, CA, USA) according to the manufacturer’s instructions. VarScan2 was employed for the detection of single nucleotide variations (SNVs). Copy number variations (CNVs) were detected by ADTEx.
Statistical analysis
SPSS was used for all statistical analysis. GraphPad Prism 7.0 (GraphPad Software) was used for presenting the statistical result graphs. Survival analysis were obtained using the Kaplan-Meier method and compared with the log-rank test. Fisher’s exact test were used to determine NGS results. A P value of less than 0.05 was considered significant.
Ethical statement
The study was approved by the Ethics Committee of Nanjing Drum Tower Hospital (No. 2016-196-01). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Because of the retrospective nature of the study, the requirement for informed consent was waived.
Results
CLDN18.2 highly expressed in advanced gastric SRCC
Tissue samples from 105 advanced gastric SRCC patients were collected and stained with anti-CLDN18.2 antibody. Positive staining was detected in 95.2% (100/105) of all SRCC tumor tissues, which is much higher than general gastric adenocarcinoma (77%) (9). In accordance with the FAST clinical trial (NCT01630083), ≥2+ membrane staining intensity in ≥40% tumor cells was defined as moderate-to-strong CLDN18.2 expression. In our study, moderate-to-strong CLDN18.2 expression was observed in 64.8% (68/105) of all samples (Table 1). In particularly, 21.0% (22/105) samples had CLDN18.2 staining in ≥90% tumor cells (Table 1). There is no connection of CLDN18.2 expression with gender or stage (Table 1 and Table S1). However, staining intensity ≥2+ in ≥90% of tumor cells often occurred in younger patients (<60 years old) (P=0.042).
Table 1. CLDN18.2 expression in advanced gastric SRCC.
| Factors | Total cases (%) | Staining intensity ≥2+ in ≥40% of cells | Staining intensity ≥2+ in ≥90% of cells | |||
|---|---|---|---|---|---|---|
| N (%) | P value | N (%) | P value | |||
| All samples | 105 (100.0) | 68 (64.8) | – | 22 (21.0) | – | |
| Gender | ||||||
| Male | 71 (67.6) | 50 (70.4) | 0.079 | 17 (23.9) | 0.276 | |
| Female | 34 (32.4) | 18 (52.9) | 5 (14.7) | |||
| Age | ||||||
| ≥60 | 38 (36.2) | 22 (57.9) | 0.267 | 4 (10.5) | 0.042* | |
| <60 | 67 (63.8) | 46 (68.7) | 18 (26.9) | |||
| Stage | ||||||
| III | 92 (87.6) | 59 (64.1) | 0.719 | 19 (20.7) | 0.841 | |
| IV | 13 (12.4) | 9 (69.2) | 3 (23.1) | |||
*, represent for P<0.05. SRCC, signet-ring cell carcinoma.
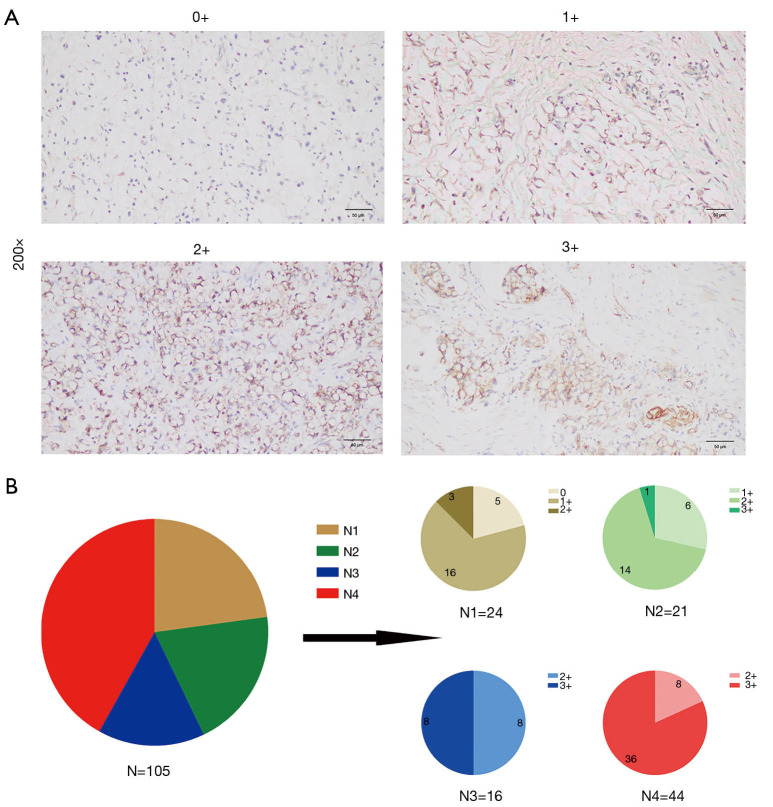
Micrographs of representative stained tissues were shown in Figure 1A according to the 0–3+ staining intensity classification, of all samples, 5 (4.8%) were 0+, 22 (21.0%) were 1+, 33 (31.4%) were 2+ and 45 (42.9%) were 3+. The percentage distributions of overall CLDN18.2 positive cells in the 105 samples were as follows: 24 (22.9%) had 0–25% CLDN18.2 positive tumor cells; 21 (20.0%) had 26–50%; 16 (15.2%) had 51–75% and 44 (41.9%) had 76–100% (Figure 1B). The increased proportion of 3+ samples were observed as the percentage level went up (Figure 1B), which indicated that there might be a positive correlation between staining intensity and the percentage of positive cells.
Figure 1.
Expression of CLDN18.2 in advanced gastric SRCC. (A) Micrographs of representative stained tissues: 0+, 1+, 2+ and 3+ staining intensity. The magnification was 200×. (B) The graph depicted the distribution of CLDN18.2 staining percentages and intensities in tumor cells from advanced gastric SRCC patient samples. N, total number of cases; N1, number of cases that had 0–25% CLDN18.2 positive tumor cells; N2, number of cases that had 26–50% CLDN18.2 positive tumor cells; N3, number of cases that had 51–75% CLDN18.2 positive tumor cells; N4, number of cases that had 76–100% CLDN18.2 positive tumor cells. SRCC, signet-ring cell carcinoma.
Relationship of OS and CLDN18.2 expression in advanced gastric SRCC patients
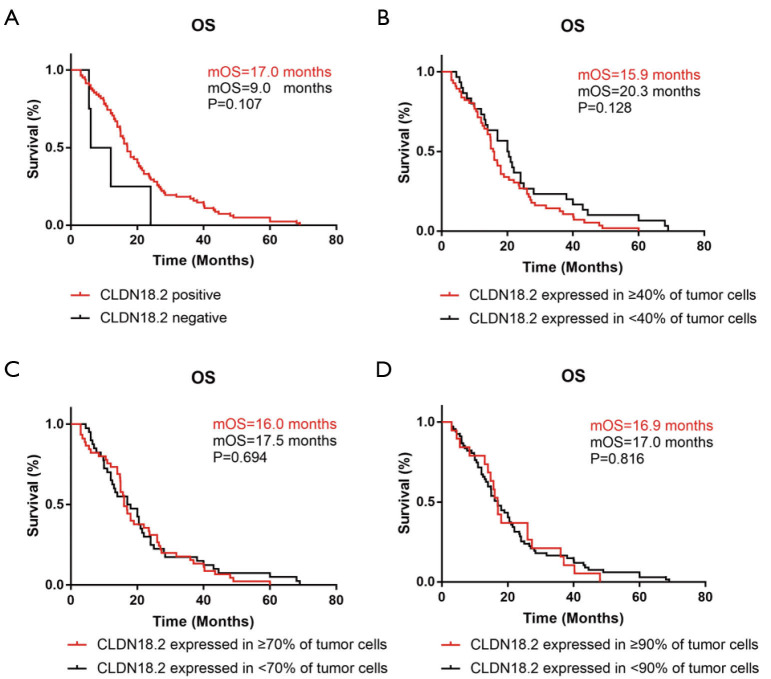
OS was analyzed by the Kaplan-Meier method in 86 cases whose survival time were available. Different grouping patterns were set up according to previous studies (9,21) by different percentage of overall CLDN18.2 positive cells. The correlation between OS and CLDN18.2 expression were shown in Figure 2. There were no significant differences of OS by different CLDN18.2 expression levels in advanced gastric SRCC (log-rank test). This result indicated that CLDN18.2 expression was not a prognostic risk factor in advanced gastric SRCC patients.
Figure 2.
OS was irrelated with CLDN18.2 expression. (A) OS in CLDN18.2 positive patients and negative patients, median OS (mOS) was 17.0 months compared to 9.0 months, P=0.107. (B) mOS was 15.9 months in patients who had CLDN18.2 expression in ≥40% of tumor cells and 20.3 months in patients who had CLDN18.2 expression in <40% of tumor cells, P=0.128. (C) mOS was 16.0 months in patients who had CLDN18.2 expression in ≥70% of tumor cells and 17.5 months in patients who had CLDN18.2 expression in <70% of tumor cells, P=0.694. (D) mOS was 16.9 months in patients who had CLDN18.2 expression in ≥90% of tumor cells and 17.0 months in patients who had CLDN18.2 expression in <90% of tumor cells, P=0.816. OS, overall survival.
GRIN2A mutation was related to CLDN18.2 expression
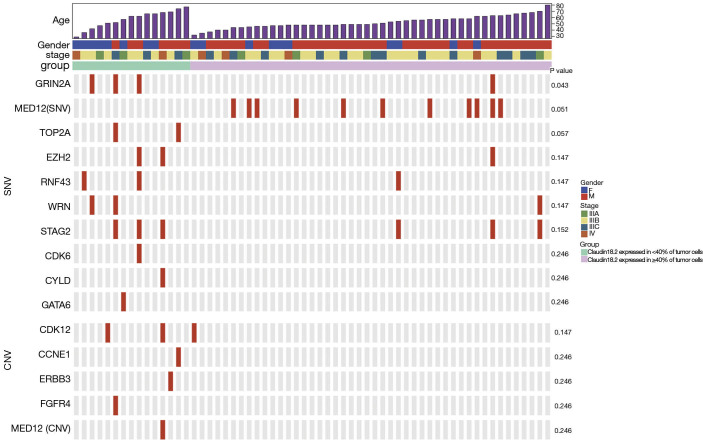
To explore the relation between CLDN18.2 expression and genetic abnormalities, 61 specimens were examined using a gene panel that covers entire exons in 416 cancer-relevant genes (Table S2). Genetic aberrations identified in all 61 samples revealed several common SNVs and CNVs in advanced gastric SRCC (Table S3 and Table S4). Briefly, the top 5 genes with the highest SNVs rate were TP53 (26.2%), CDH1 (19.7%), MED12 (18.0%), PKHD1 (18.0%) and ARID1A (11.5%). The top 2 genes with the highest CNVs rate were NOTCH1 (18.0%) and FLT4 (9.8%). Furthermore, specimens for NGS were divided into two groups based on CLDN18.2 expression. Of them, 15 were CLDN18.2 expressed in <40% of tumor cells, 46 were CLDN18.2 expressed in ≥40% of tumor cells. After the analyses of genetic aberrations in these two groups, one gene named GRIN2A were found significantly different between the two groups (Figure 3, Table S5 and Table S6). Three (20.0%) of the 15 samples that had CLDN18.2 expression in <40% of tumor cells harboring GRIN2A mutation, while only 1 (2.2%) of the 46 moderate-to-strong CLDN18.2 expression samples had GRIN2A mutation (P=0.043), indicating GRIN2A variation was more likely to occur in patients who had lower CLDN18.2 staining. Since anti-CLDN18.2 antibody was not effective enough in CLDN18.2 low expression patients, mutant GRIN2A might be a potential therapeutic target in those patients. Other detected SNVs (MED12, TOP2A, EZH2, RNF43, WRN, STAG2, CDK6, CYLD and GATA6) or CNVs (CDK12, CCNE1, ERBB3, FGFR4 and MED12) were not statistically significant (Figure 3, Table S5 and Table S6).
Figure 3.
A representative genomic landscape in different CLDN18.2 expression levels. Each row represented one gene, and each column represented one sample. A P value of less than 0.05 was considered significant. SNV, single nucleotide variation; CNV, copy number variation.
Discussion
In this study, we first tested CLDN18.2 expression in 105 FFPE tumor tissues from advanced SRCC patients by IHC method. The results demonstrated that CLDN18.2 was highly expressed in gastric SRCC, which made a promising prospect for the clinical use of zolbetuximab in advanced gastric SRCC. Survival analyses based on different grouping patterns showed no significance between CLDN18.2 expression and OS. In addition, NGS found that GRIN2A mutation was related to CLDN18.2 expression level.
CLDN18.2 had been identified as a highly selective cell lineage marker since its expression in normal tissues was strictly confined to differentiated epithelial of the gastric mucosa. Besides, the retained expression of CLDN18.2 had been found in a significant proportion of primary GCs (any positive 77%, ≥2+ in ≥60% of cells 56%) and its metastases (lymph node 66%, ovarian 96%), which made CLDN18.2 become one of the most notable targets in GC studies (9). Zolbetuximab (IMAB362) was a first-in-class monoclonal antibody specific targeted to CLDN18.2, recently, a multicentre phase II clinical study of zolbetuximab found that all responders had ≥70% CLDN18.2 expression in tumor cells, which suggested the correlation between higher CLDN18.2 expression and better therapeutic benefit (17). Besides, another phase II trial (FAST; NCT01630083) had shown that patients with ≥2+ membrane staining intensity in ≥40% tumor cells could get benefits from zolbetuximab therapy (15). In this study, high expression rate of CLDN18.2 in advanced gastric SRCC patients had been found for the first time. Significant difference of CLDN18.2 expression between primary GCs and advanced gastric SRCC (77% vs. 95.24%) hinted that CLDN18.2-based targeted therapy had great potential in the treatment of advanced gastric SRCC. Moreover, since PD-L1 overexpressed in gastric SRCC (5), antibodies targeting PD-1/PD-L1 combined with CLDN18.2 antibody zolbetuximab might be an effective treatment for advanced gastric SRCC.
In the survival analyses, 4 grouping patterns were set up according to the percentage of overall CLDN18.2 positive cells. However, no relationship had been found between OS and CLDN18.2 expression, which exactly indicated that the high expression of CLDN18.2 was irrelevant to a poor prognosis in advanced gastric SRCC. No survival analysis of CLDN18.2 expression in advanced gastric SRCC has been reported before, however, some previous studies on general GC suggested that as disease progressing, the expression of CLDN18.2 decreasing, which contributed to the increased invasive potential of the tumor cells (22,23). However, according to FAST clinical trial and other studies (9,21), no significant correlation between the expression of CLDN18.2 and the progression or prognosis of GC had been found, which consisted with our findings. According to TCGA database (Figure S1), no statistical difference in OS was discovered between CLDN18.2 high and low expression groups as well (P=0.14), however, disease free survival (DFS) was statistically significant in those two groups (P=0.0062), which indicating CLDN18.2 high expression might be related with the progression of the disease. Further studies were needed for clarifying the role of CLDN18.2 in the prognosis of GC.
NGS was performed to explore the genomic landscape for advanced gastric SRCC and to discover the correlated genes in different CLDN18.2 expression groups. Our results confirmed several common gene abnormalities in advanced gastric SRCC, such as TP53, CDH1, MED12, PKHD1, ARID1A and NOTCH1. We also found that CLDN18.2 expression might have some relation with GRIN2A mutation. GRIN2A was the encoding gene of GluN2A, which was a subunit of the N-methyl-D-aspartate receptor (NMDAR) (24), and participated in the transport of calcium ions. GRIN2A was one of the key genes in epilepsy researches, however, only a few researches had been done on GRIN2A in cancer field. The existing studies had shown that GRIN2A mutation was frequently found in melanoma (25) and induced the loss of tumor suppressor function (26,27). Besides, NMDAR signal had been found to be related with brain metastasis of breast cancer (28). Here, in our study, we found for the first time that GRIN2A mutation had some potential relation with CLDN18.2 expression in advanced gastric SRCC. GRIN2A mutation was enriched in patients with lower CLDN18.2 expression, indicating NMDAR inhibitors were probably useful in those patients. However, supporting evidence seems relatively weak due to the small sample size of GRIN2A mutation in our study, further and more comprehensive researches were needed to clarify the relevant mechanism.
In summary, the objective of this study was to assess the expression status of CLDN18.2 in advanced gastric SRCC patients, and to determine the therapeutic value of CLDN18.2 in this particular GC subtype. The high expression rate of CLDN18.2 in advanced gastric SRCC that we found provided the basic information for the CLDN18.2-based targeted therapy in advanced gastric SRCC patients. NGS results revealed the relation between GRIN2A mutation and CLDN18.2 expression, which provided possible innovative targeted therapy direction in advanced gastric SRCC.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper.
Funding: This work was funded by grants from the National Major Projects for Major New Drugs Innovation and Development (2019ZX09301-150) and Fund for Distinguished Young Scholars of Jiangsu Province (BK20190001).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experimental methods were carried out in accordance with the approved guidelines. This study was conducted with the approval of the Ethics Committee of Nanjing Drum Tower Hospital (No. 2016-196-01). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Because of the retrospective nature of the study, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-344
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-344
Peer Review File: Available at http://dx.doi.org/10.21037/jgo-20-344
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at: http://dx.doi.org/10.21037/jgo-20-344). The authors have no conflicts of interest to declare.
References
- 1.Shu Y, Zhang W, Hou Q, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun 2018;9:2447. 10.1038/s41467-018-04907-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu M, Yang Z, Feng Q, et al. The characteristics and prognostic value of signet ring cell histology in gastric cancer: A retrospective cohort study of 2199 consecutive patients. Medicine (Baltimore) 2016;95:e4052. 10.1097/MD.0000000000004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Cai H, Sheng W, et al. Clinicopathological Characteristics and Survival Outcomes of Primary Signet Ring Cell Carcinoma in the Stomach: Retrospective Analysis of Single Center Database. PLoS One 2015;10:e0144420. 10.1371/journal.pone.0144420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamboat ZM, Tang LH, Vinuela E, et al. Stage-stratified prognosis of signet ring cell histology in patients undergoing curative resection for gastric adenocarcinoma. Ann Surg Oncol 2014;21:1678-85. 10.1245/s10434-013-3466-8 [DOI] [PubMed] [Google Scholar]
- 5.Pernot S, Voron T, Perkins G, et al. Signet-ring cell carcinoma of the stomach: Impact on prognosis and specific therapeutic challenge. World J Gastroenterol 2015;21:11428-38. 10.3748/wjg.v21.i40.11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taghavi S, Jayarajan SN, Davey A, et al. Prognostic significance of signet ring gastric cancer. J Clin Oncol 2012;30:3493-8. 10.1200/JCO.2012.42.6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. 10.1016/S0140-6736(10)61121-X [DOI] [PubMed] [Google Scholar]
- 8.Wilke H, Muro K, Van Cutsem E, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol 2014;15:1224-35. 10.1016/S1470-2045(14)70420-6 [DOI] [PubMed] [Google Scholar]
- 9.Sahin U, Koslowski M, Dhaene K, et al. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res 2008;14:7624-34. 10.1158/1078-0432.CCR-08-1547 [DOI] [PubMed] [Google Scholar]
- 10.Sahin U, Schuler M, Richly H, et al. A phase I dose-escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro-oesophageal junction cancer. Eur J Cancer 2018;100:17-26. 10.1016/j.ejca.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 11.Micke P, Mattsson JS, Edlund K, et al. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int J Cancer 2014;135:2206-14. 10.1002/ijc.28857 [DOI] [PubMed] [Google Scholar]
- 12.Wöll S, Schlitter AM, Dhaene K, et al. Claudin 18.2 is a target for IMAB362 antibody in pancreatic neoplasms. Int J Cancer 2014;134:731-9. 10.1002/ijc.28400 [DOI] [PubMed] [Google Scholar]
- 13.Keira Y, Takasawa A, Murata M, et al. An immunohistochemical marker panel including claudin-18, maspin, and p53 improves diagnostic accuracy of bile duct neoplasms in surgical and presurgical biopsy specimens. Virchows Arch 2015;466:265-77. 10.1007/s00428-014-1705-4 [DOI] [PubMed] [Google Scholar]
- 14.Shinozaki A, Shibahara J, Noda N, et al. Claudin-18 in biliary neoplasms. Its significance in the classification of intrahepatic cholangiocarcinoma. Virchows Arch 2011;459:73-80. 10.1007/s00428-011-1092-z [DOI] [PubMed] [Google Scholar]
- 15.Dudov A, Pecheniy A, Rusyn A, et al. Final results of the FAST study, an international, multicenter, randomized, phase II trial of epirubicin, oxaliplatin, and capecitabine (EOX) with or without the anti-CLDN18.2 antibody IMAB362 as first-line therapy in patients with advanced CLDN18.2+ gastric and gastroesophageal junction (GEJ) adenocarcinoma. Ann Oncol 2016;27:vi207-42. [Google Scholar]
- 16.Singh P, Toom S, Huang Y. Anti-claudin 18.2 antibody as new targeted therapy for advanced gastric cancer. J Hematol Oncol 2017;10:105. 10.1186/s13045-017-0473-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Türeci O, Sahin U, Schulze-Bergkamen H, et al. A multicentre, phase IIa study of zolbetuximab as a single agent in patients with recurrent or refractory advanced adenocarcinoma of the stomach or lower oesophagus: the MONO study. Ann Oncol 2019;30:1487-95. 10.1093/annonc/mdz199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Türeci Ӧ, Mitnacht-Kraus R, Wöll S, et al. Characterization of zolbetuximab in pancreatic cancer models. Oncoimmunology 2018;8:e1523096. 10.1080/2162402X.2018.1523096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhan XB, Wang B, Li ZH, et al. Phase I trial of Claudin 18.2-specific chimeric antigen receptor T cells for advanced gastric and pancreatic adenocarcinoma. J Clin Oncol 2019;37:abstr 509.
- 20.Jiang H, Shi Z, Wang P, et al. Claudin18.2-Specific Chimeric Antigen Receptor Engineered T Cells for the Treatment of Gastric Cancer. J Natl Cancer Inst 2019;111:409-18. 10.1093/jnci/djy134 [DOI] [PubMed] [Google Scholar]
- 21.Rohde C, Yamaguchi R, Mukhina S, et al. Comparison of Claudin 18.2 expression in primary tumors and lymph node metastases in Japanese patients with gastric adenocarcinoma. Jpn J Clin Oncol 2019;49:870-6. 10.1093/jjco/hyz068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oshima T, Shan J, Okugawa T, et al. Down-regulation of claudin-18 is associated with the proliferative and invasive potential of gastric cancer at the invasive front. PLoS One 2013;8:e74757. 10.1371/journal.pone.0074757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun KH, Kim JH, Jung JH, et al. Expression of claudin-7 and loss of claudin-18 correlate with poor prognosis in gastric cancer. Int J Surg 2014;12:156-62. 10.1016/j.ijsu.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 24.Traynelis SF, Wollmuth LP, McBain CJ, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 2010;62:405-96. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 2011;43:442-6. 10.1038/ng.810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prickett TD, Zerlanko BJ, Hill VK, et al. Somatic mutation of GRIN2A in malignant melanoma results in loss of tumor suppressor activity via aberrant NMDAR complex formation. J Invest Dermatol 2014;134:2390-8. 10.1038/jid.2014.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’mello SA, Flanagan JU, Green TN, et al. Evidence That GRIN2A Mutations in Melanoma Correlate with Decreased Survival. Front Oncol 2014;3:333. 10.3389/fonc.2013.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng Q, Michael IP, Zhang P, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 2019;573:526-31. 10.1038/s41586-019-1576-6 [DOI] [PMC free article] [PubMed] [Google Scholar]