Abstract
Background
Immune checkpoint blockers (ICBs) are increasingly applied to treat patients with advanced HCC. However, the overall survival (OS) of HCC patients is still unsatisfactory, and there is no confirmed immune-related and prognostic gene to identify patients who could clinically benefit from this treatment. The tumor microenvironment (TME) is known to be closely related to immunotherapy and plays a pivotal role in the recurrence and progression of HCC. Our aim is to explore TME-related genes and identify the prognostic value in HCC patients.
Methods
ESTIMATE, immune, and stromal scores were calculated for HCC patients based on RNA expression data from The Cancer Genome Atlas database. Differential expression analysis was performed to screen the differentially expressed genes (DEGs). A protein-protein interaction (PPI) network was constructed to identify the key DEGs. Univariate and multivariate Cox analyses were adopted to validate hub DEGs associated with clinical prognosis, and a single-sample gene set enrichment analysis (ssGSEA) algorithm was used to dissect the landscape of tumor-infiltrating cells (TIC) in HCC. Finally, the relationship between hub immune-related genes and TIC was explored through difference and correlation analyses.
Results
ESTIMATE, immune and stromal scores were all found to be associated with the OS of patients (P<0.05). A total of 1,112 DEGs were identified by comparing low and high score groups of immune and stromal scores. Most of DEGs were enriched in immune-related gene sets by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Additionally, the top 34 genes were included in the protein–protein interaction (PPI) network, and univariate Cox analysis focus on a novel prognosis-related gene cluster CXCL5/CXCL8 (P<0.001). Regarding the immune landscape of HCC, univariable Cox regression analysis showed six immune cells to be associated with OS. Finally, 21 immune cells were commonly determined between high and low expression of CXCL5/CXCL8, suggesting there is a close relationship between expression of CXCL5 and CXCL8 .
Conclusions
Our study has revealed that the immune-related gene cluster of CXCL5 /CXCL8 could be a promising prognostic indicator for HCC and a potential novel biomarker to guide the selection of HCC patients for ICB immunotherapy.
Keywords: Hepatocellular carcinoma (HCC), tumor microenvironment (TME), CXCL5, CXCL8
Introduction
Hepatocellular carcinoma (HCC) has become the sixth most common tumor globally (1,2). However, due to its occult symptoms, late-stage diagnosis rate, rapid progression, and early recurrence, it is ranked fourth of all cancers in terms of mortality. Apart from the known key risk factors, such as hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, alcohol consumption, and exposure to aflatoxin or aristolochic acid, other etiologies have also contributed to the increased incidence of HCC, including non-alcoholic fatty liver disease (NAFLD), diabetes (3), and hereditary hemochromatosis (4). Despite rapid advances in liver transplantation, surgical resection, and molecular targeted therapy, there are still several patients who do not have access to surgery, chemotherapy, or radiotherapy due to large medical costs or advanced stage, so these treatments have limited application in HCC (5).
In recent decades, immunotherapy has become a promising and revolutionary therapeutic alternative for patients with melanoma, non-small cell lung cancer, and other malignancies (6). The recent finding that immunotherapy is an effective treatment for HCC patients is a milestone in the history of this chronic and stubborn disease (7). However, only a small proportion of HCC patients can benefit from immunotherapy, possibly due to the complexity and heterogeneity of tumors and unknown changes to the tumor microenvironment (TME). The TME refers to the area surrounding a tumor, and comprises blood vessels, fibroblasts, signaling molecules, and the extracellular matrix (ECM) (8,9), among which immune cells and stromal cells are the main influential components (10). Accumulating evidence demonstrates that tumor-infiltrating immune cells (TICs) in the TME, including innate and adaptive immune cells, could serve as promising indicators for the outcomes (11) and therapeutic effects of cancer patients (12,13). A recent study reported that the immune score and stromal score of HCC could help to determine tumor purity and the ratio of TICs in the TME (14). Furthermore, some long non-coding RNAs (lncRNAs) have been confirmed to be associated with the proportion of liver TICs and self-renewal. For instance, both Linc00210 and LncMAPK6 can facilitate TIC self-renewal; the former can promote the self-renewal and propagation of liver TICs by the Wnt/β-catenin signaling pathway (15), while the latter mainly enhances the self-renewal of liver TICs through MAPK6 expression (16). These results suggested that both TICs and immune-related genes could regulate the initiation and progression of HCC. Thus, to explore novel immune-associated genes and prognosis-related TICs in HCC is urgently needed in order to gain a comprehensive understanding of the dynamic accommodation of immune and stromal components in the TME.
In our study, we used ESTIMATE and a single-sample gene set enrichment analysis (ssGSEA) algorithm to calculate the stromal score, immune score, and proportion of TICs in HCC samples obtained from The Cancer Genome Atlas (TCGA) database. By using a series of bioinformation methods, we for the first time identified an immune- and prognosis-related gene cluster CXCL5/CXCL8, which might be a potential marker for dynamic modulation of the TME in HCC.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-556).
Methods
Data collection
Transcriptome RNA sequencing (RNA-seq) data of 50 paracancerous and 374 cancerous tissues were downloaded from TCGA Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Fragments per kilobase of exon per million reads mapped (FPKM) was used to quantify the gene expression of HCC patients. The corresponding clinicopathological features of HCC patients, including age, sex, American Joint Committee on Cancer (AJCC) TNM stage, tumor differentiation grade, survival time, and survival state, were also obtained from the TCGA database. Only 369 patients had a detailed survival data and were selected for the survival analysis.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
ESTIMATE, immune, stromal scores of HCC patients and survival analysis
The ESTIMATE, immune, and stromal scores, which represent the ratio of the immune/stromal components, of each HCC sample were generated using the “estimate” package in R (17). These three scores have been demonstrated to have a positive correlation with the TME in cancer samples (18). OS was regarded as the primary prognostic endpoint of HCC patients and survival analysis was revealed by Kaplan-Meier (K-M) survival curves. The cutoff values of these three scores were set using “survival” and “survminer” in R. The log-rank test was used to compare the difference of K-M curves. P<0.05 was deemed to be statistically significant.
Correlation analysis of immune/stromal score with clinical stage and grade
Correlation analysis of immune/stromal score with the clinical stage and grade of HCC patients was performed using R package. Comparisons between two or more groups were performed using the Wilcoxon and Kruskal-Wallis tests. The difference plot and comparisons between groups were done using the “ggpubr” package in R.
Validation of DEGs and common DEGs between high-score and low-score subgroups
A total of 369 tumor tissue samples were stratified into high and low subgroups for immune score and stromal score using the cutoff values set by “survminer” package. Limma package was used to validate the DEGs between the high- and low-score samples. DEGs were identified by the Wilcoxon rank-sum test, and were defined as genes with a fold change (FC) >2 and false discovery rate (FDR) <0.05. These DEGs were then visualized in a volcano plot. After that, these DEGs were intersected with the “VennDiagram” package in R, and the common DEGs were selected for further Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
GO, KEGG pathway, and Gene Set Enrichment Analysis
GO and KEGG pathway enrichment analyses were performed with “ClusterProfiler” and “ggplot2” package. Enriched GO and KEGG terms were defined as those with both a P value and a q-value of <0.05. Gene Set Enrichment Analysis (GSEA) was conducted with 1,000 permutation numbers using the software (GSEA 4.10), which was downloaded from the GSEA website (https://www.gsea-msigdb.org/gsea/index.jsp). The C7.all.v7.1 gene se t (immunologic gene sets) and C2. curated gene sets (KEGG gene sets) were downloaded from MSigDB (Molecular Signatures Database) and were selected as the target gene sets. The cut-off criteria were set as |normalized enrichment score (NES)| >1.5 and nominal P value (NOM P value) <0.05.
Construction of a protein-protein interaction (PPI) network and Cox regression analysis
A PPI network was constructed using the STRING database (https://string-db.org/). For nodes, a score of >0.9 indicated a high confidence interaction . These relationship networks were analyzed using Cytoscape (3.61 version). The top 34 genes (with >10 nodes) in the complex network were retained and considered to be hub genes. Univariate Cox regression analysis was carried out using the ‘survival’ package in R. A P value <0.05 was considered as statistically significant.
Quantification of immune cells and their relation with hub genes
The TIC abundance profiles of 374 HCC patients were estimated with Immune Cell Abundance Identifier (ImmuCellAI; http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/) and an ssGSEA algorithm. Twenty-seven immune cells were evaluated based on TCGA RNA-seq data, including 12 T-cell subtypes and 15 other immune cell types [B cells, natural killer (NK) cells, monocytes, macrophages, neutrophils, dendritic cells (DCs), and eosinophils].
Statistical analysis
All statistical analysis and plots were performed with R (3.63 version; https://www.r-project.org/). P value less than 0.05 was statistically significant. Difference analysis between two groups was used Wilcoxon rank sum test and difference analysis between three and more groups was adopted Kruskal-Wallis test. Relation analysis were identified by Spearman correlation analysis. Survival analysis was visualized by K-M curves and determined by Log-rank test. Related R packages including “estimate”, “ggplot2”, “edgeR”, “ggpubr”, “ClusterProfiler”, “survival” and “survminer” were downloaded from the Lanzhou University Open Source Society.
Results
Relation of ESTIMATE scores with OS of HCC patients
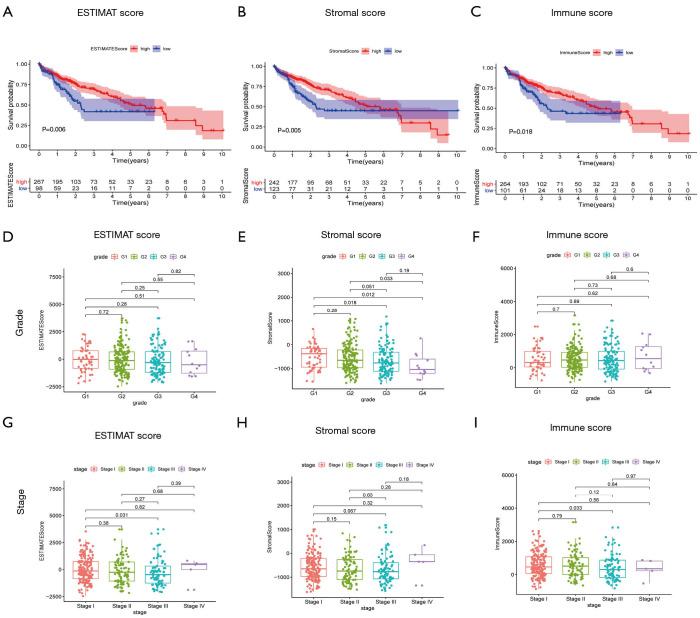
A cohort of 374 HCC patients with available expression data and 365 HCC patients with survival information available was analyzed. Clinicopathological characteristic data of HCC patients obtained from TCGA are shown in Table 1. Based on expression data, the immune and stromal scores of each patient were generated to determine the presence of immune and stromal cells, and to further to study their relationship with the prognosis of HCC. The ESTIMATE score is the sum of the immune and stromal scores, and these three scores represent distinct components of the TME in HCC. According to the ESTIMATE, immune, and stromal scores and their corresponding optimal cutoff points determined by “survminer” in R, HCC patients were stratified into two groups, the high- and low-score groups. Survival analysis showed that the high-score groups all displayed better outcomes than the low-score groups, demonstrating that the components of the TME strongly influence the occurrence and progression of HCC (Figure 1A,B,C).
Table 1. Clinicopathological characteristics of HCC patients in TCGA (n=371).
| Characteristics | n | % |
|---|---|---|
| Age [mean (SD)] | 59.66 | 13.35 |
| Sex (%) | 251 | |
| Male | 251 | 67.7 |
| Female | 120 | 32.3 |
| Grade (%) | ||
| G1 | 55 | 14.8 |
| G2 | 178 | 48.0 |
| G3 | 120 | 32.3 |
| G4 | 13 | 3.5 |
| Unknown | 5 | 1.3 |
| Stage (%) | ||
| Stage I | 174 | 46.9 |
| Stage II | 85 | 22.9 |
| Stage III | 84 | 22.6 |
| Stage IV | 4 | 1.1 |
| Unknown | 24 | 6.5 |
| T (%) | ||
| T1 | 184 | 49.6 |
| T2 | 92 | 24.8 |
| T3 | 79 | 21.3 |
| T4 | 14 | 3.8 |
| Unknown | 2 | 0.5 |
| M (%) | ||
| M0 | 269 | 72.5 |
| M1 | 3 | 0.8 |
| MX | 99 | 26.7 |
| N (%) | ||
| N0 | 253 | 68.2 |
| N1 | 4 | 1.1 |
| NX | 113 | 30.5 |
| Unknown | 1 | 0.3 |
HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
Figure 1.
Relationship of scores with patient outcome, clinical stage, and grade. (A,B,C) Correlation of ESTIMATE score, stromal score, and immune score with outcome of HCC patients examined by survival analysis. (A) Kaplan-Meier survival analysis for the high- and low-ESTIMATE score groups of HCC patients (P=0.006). (B) Survival analysis for stromal score (P=0.005) and (C) immune score (P=0.018). (D,E,F) Association of ESTIMATE score, stromal score, and immune score with clinicopathological stage of HCC patients. (G,H,I) Distribution of ESTIMATE, stromal, and immune scores with clinical stage. ESTIMATE, Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data; HCC, hepatocellular carcinoma.
Correlation of ESTIMATE scores with clinicopathological information of HCC patients
To further clarify the role of the TME in HCC, we analyzed the relationships of the three scores with clinicopathological information. As shown in Figure 1, with the advance of clinicopathological grade, the ESTIMATE score and Stromal score had significantly declined while Immune score remained unchanged (Figure 1D,E,F). Interestingly, as the tumor staged advanced, the ESTIMATE score and Immune score had a decreased trend while StromalScore had no changes (Figure 1G,H,I). Together, these results suggested that the ESTIMATE, Stromal, and Immune scores had a negative association with tumor stage and grade, demonstrating that immune and stromal components played an important role in the progression of HCC.
Determination of DEGs shared by samples with high and low immune and stromal scores
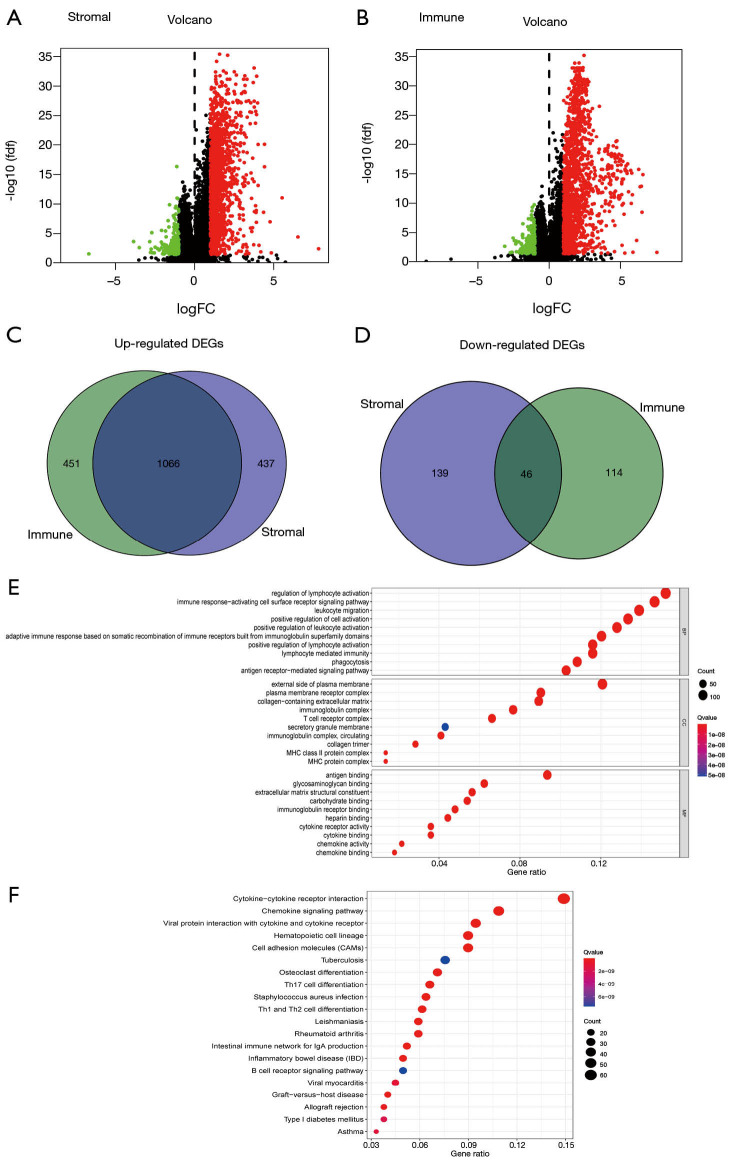
To find the genes determined by immune and stromal score, different analysis was performed between samples with high and low immune/stromal scores. Using the same cutoff value validated by survival analysis, the HCC patients were divided into high or low stromal/immune score groups. 1,688 DEGs were obtained from samples with high and low stromal scores, among which 1,503 genes were up-regulated and 185 genes were down-regulated (Figure 2A). Similarly, 1,677 DEGs were obtained from samples with high and low immune scores ; 160 genes were down-regulated and 1,517 genes were up-regulated (Figure 2B). The DEGs were visualized in a volcano diagram, with red points representing up-regulated genes and blue representing down-regulated genes (Figure 2A,B). Next, DEGs between low score and high score of stromal and immune score were intersected, and the results showed that the expression of 1,066 genes was increased (Figure 2C) and the expression of 46 genes was decreased in HCC (Figure 2D). These 1,112 DEGs (up- and down-regulated genes) were selected for KEGG enrichment analysis.
Figure 2.
Identification of common DEGs and function enrichments. (A,B) Volcano diagram displays the DEGs for stromal (A) and immune score (B). Red points represent up-regulated genes while blue points represent down-regulated genes. P value <0.05 and fold change >2 as significance threshold. (C,D) Common up-regulated and down-regulated genes shared by stromal (C) and immune score (D) are shown in Venn plots. (E,F) GO and KEGG pathway enrichment analysis of common DEGs; P<0.05 was considered statistically significance. DEGs, differentially expressed genes; GO, gene oncology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Common DEGs were enriched in immune-related gene sets
The GO enrichment analysis showed the common DEGs to mainly be enriched in immune-related gene sets, such as those involved in lymphocyte regulation, leukocyte activation, the immune response-activating cell surface receptor signaling pathway, leukocyte migration, and lymphocyte-mediated immunity (Figure 2E). Likewise, the KEGG enrichment analysis revealed that these DEGs were mainly enriched in cytokine and cytokine receptor interaction, the chemokine signaling pathway, and T-cell differentiation, all which are associated with the TME (Figure 2F). Overall, these results indicated that these DEGs obtained through analysis of the stromal and immune scores of HCC tissue samples were strongly associated with the TME and TICs.
Identification of hub gene cluster CXCL5/CXCL8 using PPI network and correlation analysis
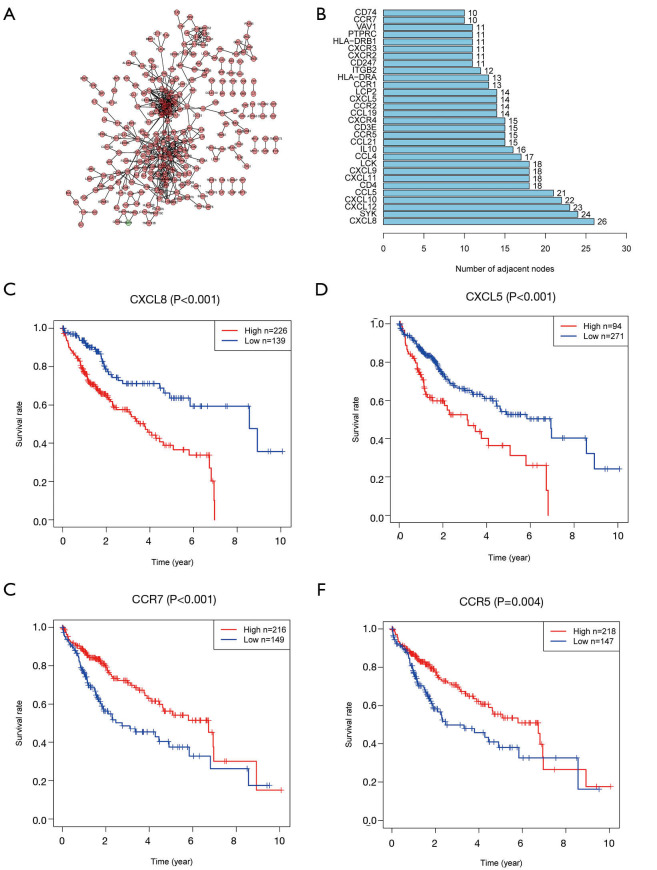
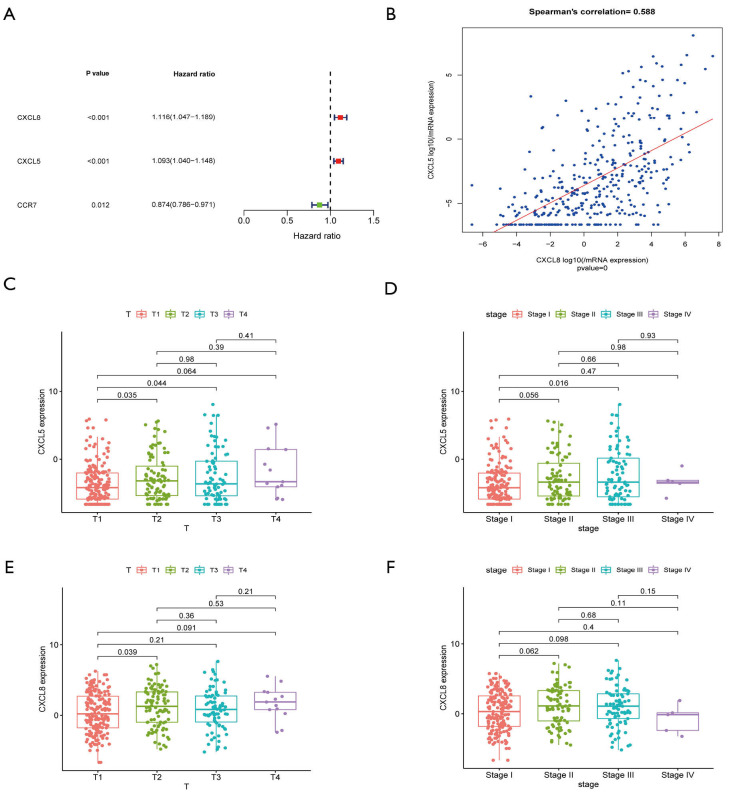
To further mine the hub immune-related genes, we constructed a PPI network and visualized the results with Cytoscape. A total of 554 PPIs was determined (Figure 3A). The top 34 genes (>10 nodes) are shown in the bar plot (Figure 3B). Survival analysis using the K-M method revealed 17 immune-related genes to have a strong relevance to survival time. The top two clusters ranked by P value were selected for further analysis. The CXCL5/CXCL8 cluster was found to be negatively related with OS (Figure 3C,D), whereas CCR5 and CCR7 were benign prognostic indicators (Figure 3E,F). Multivariate Cox regression analysis was utilized to further identify the predominant genes among the 17 genes from the survival analysis of HCC patients. After the analysis, only CXCL5, CXCL8, and CCR7 were retained, with CXCL5/CXCL8 having a damaging effect and CCR7 having a protective effect (Figure 4A). Correlation analysis of gene expression demonstrated that a strongly positive relationship existed between CXCL5 and CXCL8 (Figure 4B, Spearman’s correlation =0.588; Figure S1A, combined score =0.942, Figure S1B,C). Taken together, these results suggest that CXCL5/CXCL8 have a highly positive interaction and might represent a prognostic cluster in the TME in HCC.
Figure 3.
PPI networks and survival analysis of hub genes. (A) Protein-protein interaction networks were built with interaction confidence >0.9. (B) The top genes with more than 10 nodes are shown in a bar plot. (C,D,E,F) The top five genes ranked by P value were CXCL8, CXCL5, CCR5, HLA-DRB1, and CCR7. Correlation of two gene clusters (CXCL5/CXCL8, CCR5/CCR7) with survival are shown in K-M curves (all P<0.01). PPI, protein-protein interaction; CXCL8, C-X-C motif chemokine ligand 8; CXCL5, C-X-C motif chemokine ligand 5; CCR5, cytokine C-C motif receptor 5; CCR7, cytokine C-C motif receptor 7; K-M curves, Kaplan-Meier curve.
Figure 4.
Validation of the prognosis-related immune gene cluster and its relevance with clinical stage. (A) Multivariate Cox regression analysis of the 34 hub genes from PPI network with P<0.05 as the filter threshold. (B) Correlation of gene expression of CXCL5 and CXCL8 displayed by scatter plot, and Spearman’s coefficient was applied to analyze the correlation. (C,D) The relationship of CXCL5 gene expression with clinicopathologic stage and T stage. (E,F) The correlation of CXCL8 gene expression with AJCC stage and T stage. (C,D,E,F) Kruskal-Wallis rank sum test was considered as the statistical test. PPI, protein-protein interaction; CXCL5, C-X-C motif chemokine ligand 5; CXCL8, C-X-C motif chemokine ligand 8.
Association of CXCL5/CXCL8 with the pathology and prognosis of HCC
Apart from HCC in the TCGA database, data obtained from HCCDB: Integrative Molecular Database of Hepatocellular Carcinoma, also confirmed that the outcomes of HCC patients with a high expression of CXCL5/CXCL8 were worse than those of patients with a low expression (Figure S1D,E,F). Regarding the correlation of CXCL5/CXCL8 with the clinical pathology of HCC, the gene cluster was found to be positively related to T classification and stage (Figure 4C,D,E,F).
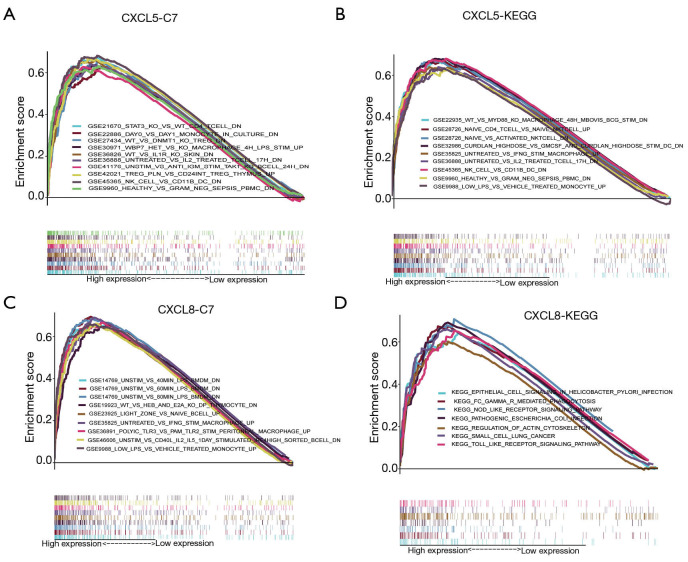
Based on median gene expression of CXCL5 and CXCL8, all HCC patients were divided into high and low expression groups. We found that the high expression of CXCL5 group was mainly enriched in immune cell related gene sets including CD4 T cells gene sets, NK cells gene sets, monocytes gene sets, and interleukin-2 gene sets (Figure 5A,B). Likewise, the high expression of CXCL8 group was mainly mapped to gene sets relating to Lipopolysaccharides (LPS), TRL3, B cells, monocytes, and the KEGG toll-like receptor signaling pathway (Figure 5C,D).
Figure 5.
GSEA analysis of CXCL5/CXCL8 cluster with KEGG and C7 immune pathway. (A,B) The enriched gene sets in C7 (immunologic gene sets) and KEGG (B) in HCC samples with high CXCL5 expression compared with the median expression of CXCL5. (C,D) Enriched gene sets in C7 and KEGG pathway in HCC samples with high CXCL8 expression. GSEA, Gene Set Enrichment Analysis; CXCL5, C-X-C motif chemokine ligand 5; CXCL8, C-X-C motif chemokine ligand 8; KEGG, Kyoto Encyclopedia of Genes and Genomes; HCC, hepatocellular carcinoma.
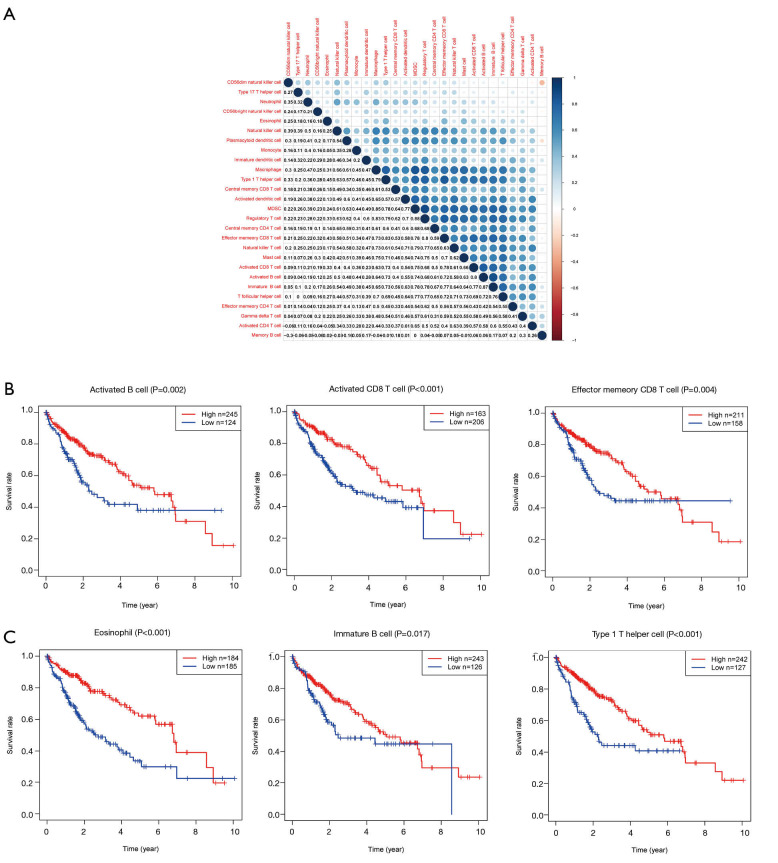
Six prognosis-related immune cells were identified by Cox regression analysis
Given that the expression of CXCL5 and CXCL8 was closely associated with patient outcomes, and enriched in immune-related gene sets including the regulation of multiple immune cells, 27 immune cell fractions were generated by using an ssGSEA algorithm to further confirm the correlation between CXCL5/CXCL8 and TME. The results of correlation analysis of these 27 immune cell fractions are displayed in a heat map (Figure 6A), and show that these immune cells were positively correlated with each other, suggesting that these cells had a synergistic effect in the HCC samples. Univariable Cox regression analysis indicated that there were six prognosis-related TICs including activated B cells, immature B cells, activated CD8 T cells, effector memory CD8 T cells, T helper 1 cells, and eosinophils (Table 2). Survival analysis of the six TICs displayed that patients with a high fraction of immune cells had a longer survival time than those with a low fraction (P<0.05; Figure 6B,C), which was consistent with the above conclusion that patients with a high immune score had better outcomes.
Figure 6.
Correlation between tumor-infiltrating immune cells and survival analysis of outcome-associated TICs. (A) Correlation between 27 different TICs and the value in each box represents the correlation coefficient between two types of immune cells. (B,C) Survival analysis for these six prognosis-related immune cells identified by univariate Cox analysis. TICs, tumor-infiltrating immune cells.
Table 2. Univariate Cox regression analysis of TICs.
| Immune cells | KM | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| Activated B cell | 0.008814 | 0.006805 | 0.000128 | 0.361176 | 0.013796 |
| Activated CD8 T cell | 0.0261 | 0.010087 | 0.000348 | 0.29215 | 0.007441 |
| Effector memory CD8 T cell | 0.01096 | 0.001177 | 1.69E-05 | 0.081823 | 0.001829 |
| Eosinophil | 1.44E-06 | 1.15E-05 | 1.75E-08 | 0.007619 | 0.000599 |
| Immature B cell | 0.398619 | 0.006532 | 6.42E-05 | 0.664926 | 0.032927 |
| Type 1 T helper cell | 0.085154 | 5.66E-06 | 3.11E-09 | 0.010309 | 0.001608 |
TICs, tumor-infiltrating immune cells.
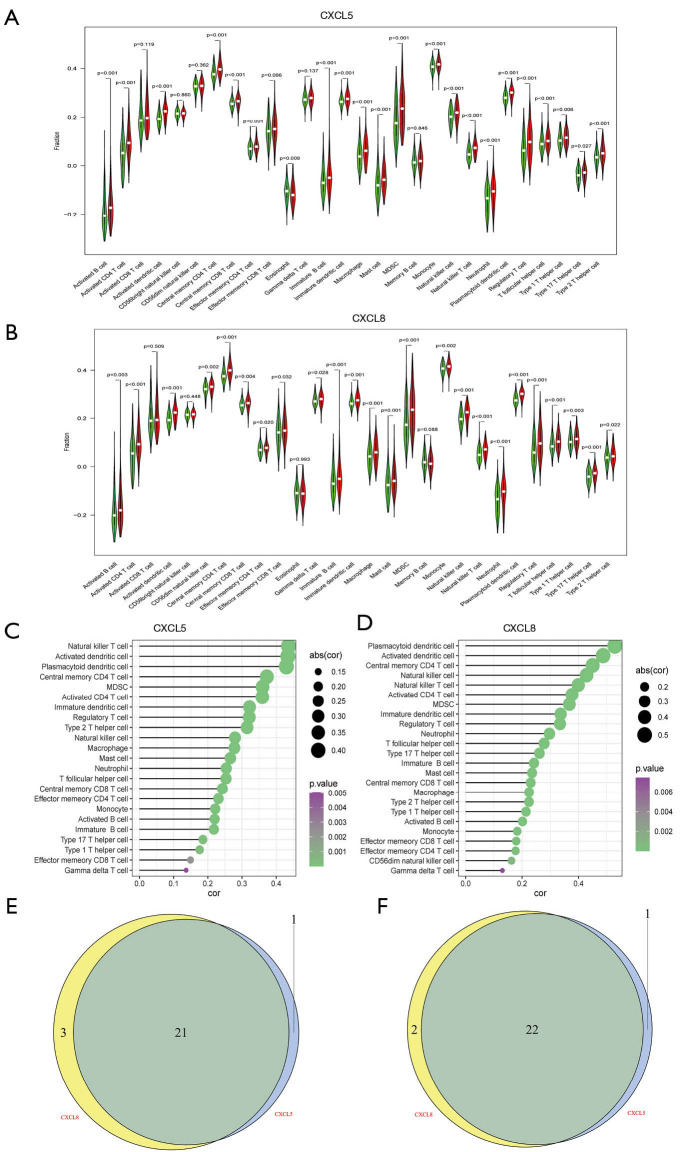
CXCL5/CXCL8-related TICs identified by difference analysis and correlation analysis
To confirm the relationship of CXCL5/CXCL8 with TICs, difference and correlation analyses were performed separately. As shown in Figure 7A,B, most TICs (22 for CXCL5 and 24 for CXCL8) were differentially infiltrated in the low- and high-groups (Figure 7C,D). Among the differentially infiltrated immune cells, 21 kinds of TIC were shared by CXCL5 and CXCL8 (Figure 7E). Likewise, correlation analysis showed that 23 TICs had an association with CXCL5, while 24 TICs were related to CXCL8, which supported that CXCL5/CXCL8 had a significant influence on immune cells and the cluster might be a promising target for the TME in HCC .
Figure 7.
Correlation of TICs with CXCL5/CXCL8 cluster. (A) Violin plot showing the different scores of immune cells between high and low CXCL5 gene expression compared to the median expression. (B) Violin plot displaying the different scores of TICs between high and low CXCL8 gene expression. (C,D) Correlation of CXCL5 (C) and CXCL8 (D) with 27 types of immune cells. P<0.05 was statistically significant. (E) Common different immune cells shared by CXCL5 and CXCL8. (F) Venn plot displaying 22 common kinds of TICs by correlation analysis of CXCL5/CXCL8 . TICs, tumor-infiltrating immune cells; CXCL5, C-X-C motif chemokine ligand 5; CXCL8, C-X-C motif chemokine ligand 8.
Discussion
Researches on tumorigenesis, heterogeneity, and drug resistance in HCC have seen significant attention and effort invested into the exploration of the TME. Tumors depend on the TME to sustain their proliferation, invasion, and metastasis (19). In brief, the TME consists of recruited immune cells, resident stromal cells and non-cellular components which could interact with cancer cells. Furthermore, as the tumor grows, infiltrating immune cells may be associated with immunity disruption (19). A number of studies have shed light on immune targets, especially on immune checkpoint blockers (ICBs) (7,20,21); however, ICBs, which may contribute to the heterogeneity and complexity of the TME in HCC, have not demonstrated an ideal response for HCC patients. However, ICBs still represent a novel therapeutic advancement for HCC patients, especially for those with advanced disease (22). Numerous studies have shown that immune infiltration, which is a signature of tumor heterogeneity and the TME, showed a better response to ICBs. According to a previous study that investigated several malignancies, the existence of gene indicators for CD8 T cells and T helper 1 cells contributes to preferable outcomes and prognosis (23). In the present study, we identified tumor immune landscape-related genes that are correlated with the prognostic outcomes of HCC patients and the regulation of TIC using data from the TCGA database. A series of bioinformatics analyses revealed that the novel gene cluster CXCL5/CXCL8 might be a potential biomarker for TME remodeling, prognostic prediction, and immunotherapy in HCC.
In this study, the ESTIMATE, immune, and stromal scores were preliminarily calculated to determine the proportions of immune and stromal components in tumorous tissues, and showed a positive correlation with the prognosis of HCC patients. Consistently, subsequent analysis of the ESTIMATE, immune and stromal scores of HCC patients and their outcomes revealed that patients with high scores had a significantly better prognosis than those with low scores (P=0.05). Then, correlation analysis was conducted to mine the hub genes determined by the immune and stromal scores: 1,688 DEGs were obtained for stromal scores while 1,677 genes were obtained for immune scores. Next, function annotations illustrated that these DEGs were mainly mapped to cytokine, cytokine receptor and chemokine signaling pathways, which were closely connected with the chemotaxis of neutrophils and the immune landscape. Taken together, all these results indicated that these shared DEGs play a major role in the TME and TICs, and are closely related to immune regulation and TME modification.
In order to identify the hub genes, we constructed a PPI network (Figure 3A) and performed survival analysis. Seventeen genes with a relatively low P-value were obtained, among which were two gene clusters from the same family, cytokine C-X-C motif ligand 5/8 (CXCL5/CXCL8) and cytokine C-C motif receptor (CCR5/CCR7). The CXCL5/CXCL8 cluster was negatively correlated with OS, while the CCR5/CCR7 cluster contributed to benign OS of HCC patients. It is worth noting that only CXCL5/CXCL8 might be a prognostic cluster of the TME in HCC, as suggested by the results of univariate Cox regression analysis and correlation analysis of gene expression. Besides, an ssGSEA algorithm and univariable Cox regression analysis were utilized to build 27 profiles of immune cell types and to screen six prognosis-related immune cells, respectively. This was followed by a thorough investigation of the relationship between CXCL5 and CXCL8. Interestingly, both could interact with certain immune cells, most of which were the same for the two genes, according to a Venn diagram (Figure 7). Therefore, we may speculate that CXCL5/CXCL8 could be the signature of the TME and prognosis in HCC, and that targeting this cluster might serve as a therapeutic alternative to or improve the response of ICBs in HCC.
Proinflammatory cytokine CXCL5, a member of the CXC-type chemokine family, facilitates tumorigenesis and development. A previous study suggested that CXCL5 could serve as a promising prognosis-related biomarker (24), while another described that it could mediate the ratio of inflammation, and accelerate metastasis and the growth of prostate tumors in bone (25). Moreover, CXCL8 is an essential multifunctional cytokine that promotes tumor metastasis and invasion (26). Congruously, we have confirmed that HCC patients with a higher expression of CXCL5/CXCL8 always had a worse outcome than those with a low expression. CXCL5 had a strong association with immune-related gene sets including the pathways related to CD4 T cells, NK cells, monocytes, and interleukin-2, while CXCL8 was mapped to the pathways related to LPS, TRL3, B cells, monocytes, and the KEGG toll-like receptor signaling pathway. Previous studies found that a higher CXCL5 expression resulted in a higher number of M2 macrophage fragments in pancreatic ductal adenocarcinoma patients 27 and increased neutrophil accumulation in various cancers (27-29), which agrees with our conclusion (Figure 7). Subsequent survival analysis also confirmed that patients with a high immune score had a better outcome, due to having more immune cell infiltration. Notably, type 17 T (Th17) helper cells have shown a positive correlation with CXCL5 and CXCL8 (P=0.027 and 0.001; Figure 7A,B). There has been some evidence that Th17 cells/interleukin-17 (IL-17) can induce CXCL5 to recruit neutrophils in kidney tubular cells (30). One possible mechanism of this may be that IL-17 can induce CXCL5 by activating the nuclear factor kappa-B (NF-κB) signaling pathways (31). Alternatively, another study showed that when cultured under the condition of Th17 polarization, IL-8 (encoded by CXCL8) single producing cell’s accounted for a high proportion and CXCL8 was upregulated by activating aryl hydrocarbon receptor (AHR) (32). Moreover, Th17 can promote the expression of CXCL8 by enhancing the gene expression level of tumor necrosis factor (TNF)-α, which can induce the expression of neutrophil chemokine s, such as CXCL8 (33). Furthermore, TNF-α can regulate the inward flow of Th17 cells, tumor-associated macrophages, and tumor-associated neutrophils (34). Accordingly, Th17 can produce cytokines for tumor promotion (35).
In addition, targeting IL-8 by antibody (ABX-IL8) could be a potential therapeutic strategy to control angiogenesis, growth, and metastasis of melanoma (36). Nordy potently inhibited CXCR4 ligand SDF-1-induced production of IL-8 and vascular endothelial cell growth factor in malignant glioma cells (37). Therefore, the application of inflammatory factors and their receptors is expected to become a new tumor treatment strategy and method.
There are several drawbacks to our study. Limited by its retrospective nature, this study lacks data from prospective real-world studies. Second, basic experimental researches were not performed to expand on the results. Third, the relationship between CXCL5, CXCL8, and Th17 remains to be explored and verified.
In conclusion, our study discovered a novel prognostic gene cluster comprising CXCL5 and CXCL8. This cluster is not only closely related to the prognosis of HCC patients, but it might orchestrate the tumor immune landscape dynamically. Furthermore, the CXCL5/CXCL8 cluster could also be a potential biomarker for response to ICBs and a therapeutic target to alter the TME in HCC patients.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81672751) and the Key Research and Development Program of Shaanxi province (2019SF-010).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-556
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-556). The authors have no conflicts of interest to declare.
(English Language Editor: J. Reynolds)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Jindal A, Thadi A, Shailubhai K. Hepatocellular Carcinoma: Etiology and Current and Future Drugs. J Clin Exp Hepatol 2019;9:221-32. 10.1016/j.jceh.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pocha C, Xie C. Hepatocellular carcinoma in alcoholic and non-alcoholic fatty liver disease—one of a kind or two different enemies? Transl Gastroenterol Hepatol 2019;4:72. 10.21037/tgh.2019.09.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fattovich G, Stroffolini T, Zagni I, et al. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 2004;127:S35-50. 10.1053/j.gastro.2004.09.014 [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Zhong BY, Zhu HD, et al. Embolotherapy of unresectable hepatocellular carcinoma: Eastern perspective. Chin Clin Oncol 2019;8:60. 10.21037/cco.2019.11.01 [DOI] [PubMed] [Google Scholar]
- 6.Martinez P, Peters S, Stammers T, et al. Immunotherapy for the First-Line Treatment of Patients with Metastatic Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:2691-8. 10.1158/1078-0432.CCR-18-3904 [DOI] [PubMed] [Google Scholar]
- 7.El Dika I, Khalil DN, Abou-Alfa GK. Immune checkpoint inhibitors for hepatocellular carcinoma. Cancer 2019;125:3312-9. 10.1002/cncr.32076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74-80. 10.1126/science.aaa6204 [DOI] [PubMed] [Google Scholar]
- 9.Spill F, Reynolds DS, Kamm RD, et al. Impact of the physical microenvironment on tumor progression and metastasis. Curr Opin Biotechnol 2016;40:41-8. 10.1016/j.copbio.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21:309-22. 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 11.Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res 2019;79:4557-66. 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014-22. 10.1038/ni.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tovar V, Cornella H, Moeini A, et al. Tumour initiating cells and IGF/FGF signalling contribute to sorafenib resistance in hepatocellular carcinoma. Gut 2017;66:530-40. 10.1136/gutjnl-2015-309501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge PL, Li SF, Wang WW, et al. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging (Albany NY) 2020;12:5479-99. 10.18632/aging.102971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu X, Zhu X, Qin F, et al. Linc00210 drives Wnt/β-catenin signaling activation and liver tumor progression through CTNNBIP1-dependent manner. Mol Cancer 2018;17:73. 10.1186/s12943-018-0783-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Huang G, Jiang H, He Y, et al. LncMAPK6 drives MAPK6 expression and liver TIC self-renewal. J Exp Clin Cancer Res 2018;37:105. 10.1186/s13046-018-0770-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Heinemann J. Cluster Analysis of Untargeted Metabolomic Experiments. Methods Mol Biol 2019;1859:275-85. 10.1007/978-1-4939-8757-3_16 [DOI] [PubMed] [Google Scholar]
- 18.Bi KW, Wei XG, Qin XX, et al. BTK Has Potential to Be a Prognostic Factor for Lung Adenocarcinoma and an Indicator for Tumor Microenvironment Remodeling: A Study Based on TCGA Data Mining. Front Oncol 2020;10:424. 10.3389/fonc.2020.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Lin Y, Zhang J, et al. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38:447. 10.1186/s13046-019-1412-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H, Sun G, Chen H, et al. Trends in the treatment of advanced hepatocellular carcinoma: immune checkpoint blockade immunotherapy and related combination therapies. Am J Cancer Res 2019;9:1536-45. [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng Z, Yang B, Liao ZY. Current progress and prospect of immune checkpoint inhibitors in hepatocellular carcinoma. Oncol Lett 2020;20:45. 10.3892/ol.2020.11909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 2015;125:3384-91. 10.1172/JCI80011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu B, Fan H, Lv X, et al. Prognostic significance of CXCL5 expression in cancer patients: a meta-analysis. Cancer Cell Int 2018;18:68. 10.1186/s12935-018-0562-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roca H, Jones JD, Purica MC, et al. Apoptosis-induced CXCL5 accelerates inflammation and growth of prostate tumor metastases in bone. J Clin Invest 2018;128:248-66. 10.1172/JCI92466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev 2016;31:61-71. 10.1016/j.cytogfr.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Liu Q, Peng J, et al. CXCL5 overexpression predicts a poor prognosis in pancreatic ductal adenocarcinoma and is correlated with immune cell infiltration. J Cancer 2020;11:2371-81. 10.7150/jca.40517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soler-Cardona A, Forsthuber A, Lipp K, et al. CXCL5 Facilitates Melanoma Cell-Neutrophil Interaction and Lymph Node Metastasis. J Invest Dermatol 2018;138:1627-35. 10.1016/j.jid.2018.01.035 [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, Zhou J, Tang D, et al. Neutrophil infiltration mediated by CXCL5 accumulation in the laryngeal squamous cell carcinoma microenvironment: A mechanism by which tumour cells escape immune surveillance. Clin Immunol 2017;175:34-40. 10.1016/j.clim.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 30.Disteldorf EM, Krebs CF, Paust HJ, et al. CXCL5 drives neutrophil recruitment in TH17-mediated GN. J Am Soc Nephrol 2015;26:55-66. 10.1681/ASN.2013101061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia J, Xu X, Huang P, et al. The potential of CXCL5 as a target for liver cancer - what do we know so far? Expert Opin Ther Targets 2015;19:141-6. 10.1517/14728222.2014.993317 [DOI] [PubMed] [Google Scholar]
- 32.Gasch M, Goroll T, Bauer M, et al. Generation of IL-8 and IL-9 producing CD4+ T cells is affected by Th17 polarizing conditions and AHR ligands. Mediators Inflamm 2014;2014:182549. 10.1155/2014/182549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, Wang G, Tao Y, et al. The proinflammatory effect and molecular mechanism of IL- 17 in the intestinal epithelial cell line HT-29. J buon 2015;20:120-7. [PubMed] [Google Scholar]
- 34.Ben-Baruch A. The Tumor-Promoting Flow of Cells Into, Within and Out of the Tumor Site: Regulation by the Inflammatory Axis of TNFα and Chemokines. Cancer Microenviron 2012;5:151-64. 10.1007/s12307-011-0094-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883-99. 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Huang R, Chen L, et al. Identification of interleukin-8 as estrogen receptor-regulated factor involved in breast cancer invasion and angiogenesis by protein arrays. Int J Cancer 2004;109:507-15. 10.1002/ijc.11724 [DOI] [PubMed] [Google Scholar]
- 37.Ping YF, Yao XH, Chen JH, et al. The anti-cancer compound Nordy inhibits CXCR4-mediated production of IL-8 and VEGF by malignant human glioma cells. J Neurooncol 2007;84:21-9. 10.1007/s11060-007-9349-8 [DOI] [PubMed] [Google Scholar]