Abstract
Background
Recently, autophagy was found related to several malignances.
Methods
To explore the diagnostic and prognostic values of autophagy in prostate cancer (PCa), we first identified differentially expressed autophagy-related genes (DEARGs) based on The Cancer Genome Atlas (TCGA) Prostate Adenocarcinoma (PRAD) dataset. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were applied to perform gene functional annotation. Then, multivariate logistic regression analysis was applied to construct the risk index (RI). Receiver operating characteristic (ROC), calibration and decision curve analysis (DCA) curves were performed to identify the effectiveness of RI. Next, multivariate Cox regression analyses were performed to construct the prognostic index (PI) and autophagy-clinical prognostic index (ACPI).
Results
We identified 16 DEARGs and functional annotation demonstrated the relevance of these genes to autophagy and revealed the association of these DEARGs with digestive system, drug resistance and apoptosis. Then, the RI was constructed based on 5 DEARGs and the area under the ROC curve (AUC) was 0.9858. Validation based on Gene Expression Omnibus (GEO) datasets suggested that the RI was effective. Next, 7 ARGs were identified associated with overall survival (OS) and the PI was developed composed of 3 ARGs. Finally, ACPI was constructed based on PI and the M stage.
Conclusions
This study provided potential models for predicting the risk and prognosis of PCa and indicated the molecular insights of autophagy in PCa. While no other dataset was applied to test the effectiveness of the PI and ACPI models attribute to the well prognosis of PCa.
Keywords: Prostate cancer (PCa), prognosis, autophagy, The Cancer Genome Atlas, gene signature
Introduction
Prostate cancer (PCa) is the most common malignancy in genitourinary system, leading to the eighth cause of death in general population. There were 1,276,106 estimated new cases and 358,989 estimated deaths globally in 2018 (1). Since the late of 1980s, prostate-specific antigen (PSA) has been the most important biomarker for PCa screening (2). In 2012, the United States Preventive Services Task Force (USPSTF) recommended against PSA-based screening for PCa regardless of age groups (3). However, the USPSTF reviewed this recommendation in 2018 and proposed the opportunity of PSA screening should be discussed in men younger than 70 years old (4).
With the increasing awareness of diseases and the popularization of PSA screening, more and more patients were diagnosed at early stage. Even though, PCa acts as the third leading cause of death in men (5) and it remains to be a significant public health burden (6). Nowadays, localized PCa is treated in a variety of ways, including watchful waiting, active surveillance, surgery or radiotherapy, while management for advanced PCa remains to be a tricky problem for clinicians. Although a large number of clinical and molecular studies on PCa have emerged, the diagnostic and prognostic biomarkers have not been fully investigated.
Autophagy is the process of transporting damaged, denatured or aged proteins and organelles to lysosomes for digestion and degradation (7). Meanwhile, it can inhibit tumor formation by limiting inflammation, removing toxic unfolded proteins, and removing damaged mitochondria that produce reactive oxygen species (DNA damaging). Consequently, it is an important biological process which plays a crucial role in survival, differentiation and homeostasis (8). Under normal physiological conditions, autophagy is conducive to maintaining the homeostasis of the cell. During metabolic stress, autophagy can prevent the accumulation of toxic or carcinogenic damage proteins and organelles, and inhibit cell canceration (9-11). However, autophagy provides more nutrition for cancer cells and promotes tumor growth once the cancer has formed (12-14). Therefore, autophagy shows two sides in the process of tumorigenesis and development (15).
Numerous programs have explored the role of autophagy in the development and progression of PCa. Lu et al. (16) found that autophagy induced by overexpression of DCTPP1 promoted tumor progression and predicted poor survival outcomes in PCa. Blessing et al. (17) demonstrated that transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor could promote the progression of PCa. On the contrary, Liu et al. (18) considered that SGK1 inhibition-induced autophagy could impaired PCa metastasis by reversing EMT because of the amplified antimetastatic effects. Hence, it is of great value to investigate the role of the entire subset of autophagy genes in the diagnosis and prognosis of PCa.
Attributed to the advent of the era of big data and the development of high-throughput sequencing, it is convenient to study the diagnostic and prognostic values of autophagy-related genes (ARGs) in PCa patients from public databases. In our study, we identified five diagnostic ARGs and developed a risk index (RI) which could predict the risk of PCa with high accuracy. In addition, three survival related ARGs were investigated and a prognostic model was constructed. Furthermore, we developed a prognostic model based on both the ARGs and clinical characteristics. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-498).
Methods
Data processing
TCGA database of Prostate Adenocarcinoma (PRAD) including gene expression profiles and related clinical information were downloaded and extracted from the Genomic Data Commons Data Portal (GDC, https://portal.gdc.cancer.gov). The data of mRNA expression were transformed to values in transcripts per million (TPM). ARGs were obtained from The Human Autophagy Database (HADb, http://www.autophagy.lu/index.html), which is an autophagy-dedicated database aiming at reserving human genes involved in autophagy. mRNA expression profiles of GSE32571, GSE46602, GSE70768 were downloaded from Gene Expression Omnibus (GEO). All cases with missing values in required variables were deleted. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differentially expressed ARGs and functional annotation
Genes exhibiting at least 2-fold changes along with an adjusted P value less than 0.05 were selected as significantly differentially expressed ARGs (DEARGs). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene ontology (GO) analysis of ARGs were carried out using Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov, version 6.8) online tools: functional annotation. Enriched GO terms and KEGG pathways were identified according to the cut-off criterion of P<0.05.
Construction of a RI based on ARGs
Multicollinearity of DEARGs were evaluated by liner regression analysis. Then DEARGs having no collinearity with other variables were subjected to a multivariate logistic regression analysis using the method of Forward Stepwise (Likelihood Ratio) to retain the genes that might be an independent indicator in monitoring. Then, a RI composed of these genes was developed. The formula of RI based on a linear combination of the relative expression level of genes multiplied regression coefficients, which represented the relative weight of genes in the multiple logistic analysis. Next, receiver operating characteristic (ROC) and decision curve analysis (DCA) curves were performed to identify the effectiveness and benefits of RI. In addition, goodness-of-fit of RI was evaluated using Hosmer and Lemeshow tests (H-L Tests) and visualized with calibration curves. GEO datasets were used to validation the RI.
Survival related ARGs and construction of the prognostic index and autophagy-clinical prognostic index (ACPI)
We first applied univariate Cox regression analyses to select the ARGs significantly associated with OS of patients with PCa. Then, a multivariate Cox regression analysis using the method of Forward Stepwise (Likelihood Ratio) was performed to screen the genes whose expression might be an independent factor related to prognosis. Subsequently, a prognostic index (PI) was constructed with a linear combination of the relative expression level of genes multiplied regression coefficients. Next, we separated PCa patients into high- and low-risk groups based on the median PI value to establish a survival curve plotted by Kaplan-Meier (K-M) method. Finally, we applied multivariate Cox regression analysis using the method of Forward Stepwise (Likelihood Ratio) to construct the ACPI. K-M method and ROC curve were used to assess ACPI.
Statistical analyses
All statistical analyses were performed on R software (version 3.6.1). DEARGs were identified utilizing Wilcoxon signed-rank test. Collinearities were defined as tolerances <0.1 or variance inflation factors (VIFs) >10. Multivariate logistic regression analysis was used to construct the RI model. The ROC curves and the corresponding area under the curve (AUC) were performed by the package of “pROC” in R. The DCA curves were plotted using the package of “rmda” in R. Univariate Cox regression analyses were used to evaluate the relationship between expression profiles and OS. The PI and ACPI model were developed according to the results of multivariate Cox proportional hazards regression model. Both the univariate and the multivariate Cox regression analyses were performed with the package of “survival”. Nomogram and calibration curves were visualized using package of “rms”. All analyses were two sided and P<0.05 was statistically significant.
Results
DEARGs
The flow chart of this study was exhibited in Figure 1. Gene expression profiles and corresponding clinical information of 52 normal tissues and 495 tumor tissues were extracted from TCGA. In total, we identified 16 DEARGs genes meeting the criteria of FDR <0.05 and |log2(Fold Change)| >1 (Figure 2A,B). Among them, 5 ARGs (ATG9B, BIRC5, CAMKK2, CDKN2A, NKX2-3) were significantly up regulated and 11 (BCL2, DNAJB2, FAM215A, HSPB8, ITGB4, ITPR1, NRG1, NRG2, SUPT20HL2, TMEM74, TP63) were down regulated. Furthermore, box plots of the expression pattern of these 16 DEARGs were visualized in Figure 2C.
Figure 1.

Flow chart of this study.
Figure 2.
Differentially expressed ARGs between PCa and normal prostate tissues. (A) The volcano plot for the 222 ARGs from the TCGA PRAD dataset. Red indicated high expression and green low expression. (B) The heatmap of DEARGs based on their expression levels. (C) The box plots of 16 DEARGs. Each red dot represents a tumor sample and blue a non-tumor sample. ARGs, auto-related genes; TCGA, The Cancer Genome Atlas; PRAD, prostate adenocarcinoma; DEARGs, differentially expressed autophagy-related genes.
Gene functional annotation
GO analysis and KEGG pathway enrichment were applied to perform gene functional annotation. Figure 3A exhibited the top 10 most significant GO terms for biological processes in GO analysis. We found that these DEARGs were notably associated with autophagy, process utilizing autophagic mechanism, digestive tract development, digestive system development, regulation of autophagy and so on. Besides, KEGG pathway analysis (Figure 3B) showed that these DEARGs were significantly related to autophagy-animal, platinum drug resistance, EGFR tyrosine kinase inhibitor resistance, apoptosis and apoptosis-multiple species. In a word, these results further demonstrated the relevance of these genes to autophagy, and at the same time, revealed the association of these DEARGs with digestive system, drug resistance and apoptosis. As shown in Figure 3C,D, the Z-scores of enriched pathways were mostly less than zero, indicating that most of the pathways were more likely to be decreased.
Figure 3.
GO and KEGG pathway analysis of DEARGs. (A,B) Bubble plots of biological process GO terms and KEGG pathway analysis for DEARGs. (C,D) Circle plots of biological process GO terms and KEGG pathway analysis for DEARGs. The outer circle of circle plots showed a scatter plot for each term of the logFC of the assigned genes. Red circles represented up-regulation, and blue ones down-regulation. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEARGs, differentially expressed autophagy-related genes.
Construction of RI
The tolerances of all the DEARGs were greater than 0.1 and VIFs less than 10, indicating there existed no multicollinearity within these genes (Table S1). Multivariate logistic regression analysis results were shown in Table 1. The formula of RI was as followed: RI = [3.320 × exp(ATG9B)] + [−0.005 × exp(DNAJB1)] + [−0.009 × exp(HSPB8)] + [1.895 × exp(NKX2-3)] + [−0.054 × exp(TP63)] [exp(ARG) represented the expression value of ARG]. The model was visualized in a nomogram (Figure S1). The classification accuracy in was 96.5%, with a positive predictive value (PPV) of 97.4% and a negative prediction value (NPV) of 85.7%. It was noticed that ATG9B and NKX2-3 had greater influences on the index. What’s more, the coefficient of TP63, HSPB8 and DNAJB1 were negative, indicating that expression of NKX2-3 and ATG9B were associated with an increased risk of PCa, while expression of TP63, HSPB8 and DNAJB1 were related to a decreased PCa risk.
Table 1. Uni- and multivariate logistic regression analysis of DEARGs.
| Genes | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | ||
| TP63 | 0.913 | 0.895–0.931 | <1×10–4 | 0.948 | 0.920–0.976 | 3.85×10–4 | |
| HSPB8 | 0.985 | 0.981–0.988 | <1×10–4 | 0.991 | 0.985–0.996 | 1.06×10–3 | |
| ATG9B | 18.527 | 7.184–47.782 | <1×10–4 | 27.663 | 5.088–150.415 | 1.22×10–4 | |
| NKX2_3 | 35.315 | 9.692–128.672 | <1×10–4 | 6.655 | 1.810–24.467 | 4.33×10–3 | |
| DNAJB1 | 0.998 | 0.997–0.999 | 3.74×10–4 | 0.995 | 0.991–0.999 | 6.00×10–3 | |
| CAMKK2 | 1.034 | 1.024–1.044 | <1×10–4 | 1.43×10–1 | |||
| ITPR1 | 0.969 | 0.959–0.980 | <1×10–4 | 7.58×10–1 | |||
| TMEM74 | 0.003 | 0.001–0.021 | <1×10–4 | 8.66×10–1 | |||
| NRG1 | 0.359 | 0.264–0.489 | <1×10–4 | 2.71×10–1 | |||
| NRG2 | 0.424 | 0.349–0.515 | <1×10–4 | 3.23×10–1 | |||
| ITGB4 | 0.972 | 0.964–0.981 | <1×10–4 | 5.95×10–2 | |||
| BCL2 | 0.904 | 0.878–0.930 | <1×10–4 | 8.05×10–1 | |||
| CDKN2A | 2.262 | 1.616–3.164 | <1×10–4 | 9.23×10–1 | |||
| BIRC5 | 1.882 | 1.502–2.359 | <1×10–4 | 9.51×10–1 | |||
| SUPT20HL2 | 0.000 | 0.000–0.001 | <1×10–4 | 3.01×10–1 | |||
| FAM215A | 0.001 | 0.000–0.011 | <1×10–4 | 6.73×10–1 | |||
DEARGs, differentially expressed auto-related genes; OR, odds ratio; CI, confidence interview.
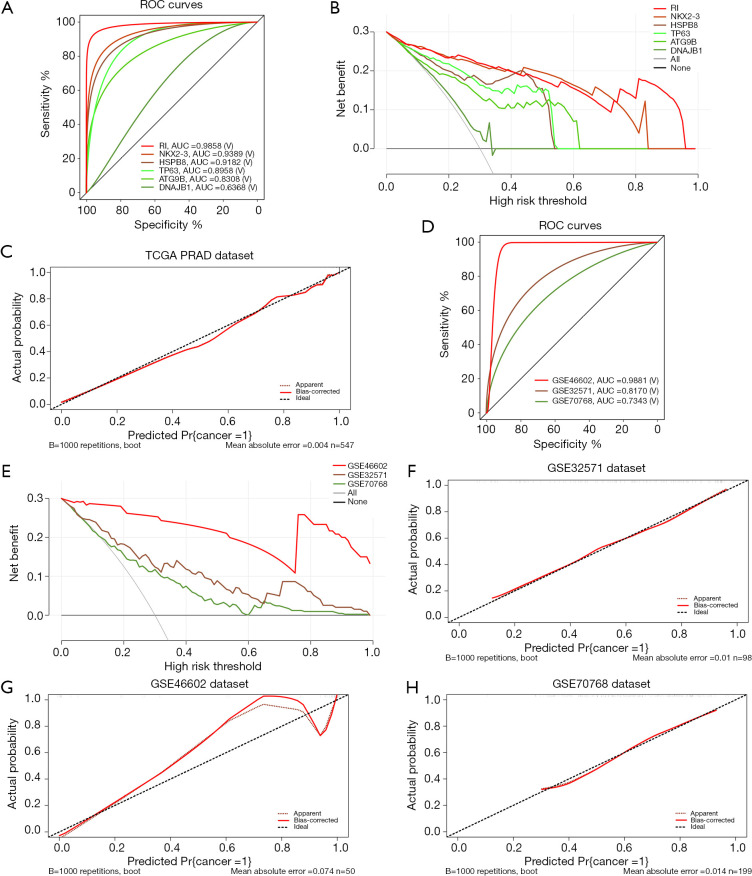
To evaluate how well these 5 diagnostic ARGs and the RI predicted the risk of PCa, we then performed ROC curve analysis based on TCGA database (Figure 4A). The AUCs of ATG9B, DNAJB1, HSPB8, NKX2-3, TP63 and the RI were 0.8308, 0.6368, 0.9182, 0.9389, 0.8958, 0.9858, respectively, implying a high prediction performance of PCa. Also, the DCA curve (Figure 4B) suggested that the RI model was beneficial. What’s more, the goodness-of-fit is well with a P value of 9.87×10−1 in the H-L Test (Figure 4C, Table S2).
Figure 4.
Assessment and validation of the RI. (A) ROC curves for the 5 independent diagnostic genes (ATG9B, DNAJB1, HSPB8, NKX2-3, TP63) and the RI based on TCGA PRAD dataset. (B) DCA curves for the 5 independent diagnostic genes (ATG9B, DNAJB1, HSPB8, NKX2-3, TP63) and the RI based on TCGA PRAD dataset. (C) Calibration curves of the RI in TCGA PRAD dataset. (D) ROC curves and AUC for the RI based on the 4 datasets (GSE32571, GSE46602 and GSE70768). (E) DCA curves for the RI based on the 3 GEO datasets. (F,G,H) Calibration curves of the RI in GSE 32571, GSE46602 and GSE70768. RI, risk index; TCGA, The Cancer Genome Atlas; PRAD, prostate adenocarcinoma; ROC, receiver operating characteristic; AUC, area under the ROC curve; DCA, decision curve analysis; GEO, Gene Expression Omnibus.
In addition, we tested the effectiveness and benefits of the RI in GEO datasets. As shown in Figure 4D, the AUCs of the ROC curves were 0.8170, 0.9881 and 0.7343 for GSE32571, GSE46602 and GSE70768, respectively, indicating a high diagnostic value in all the datasets. What’s more, the RI showed good benefits in all the datasets (Figure 4E). Finally, H-L Tests and the calibration curves showed well goodness-of-fit of the RI in these GEO datasets (Figure 4F,G,H and Table S2).
Identification of survival related ARGs and construction of PI
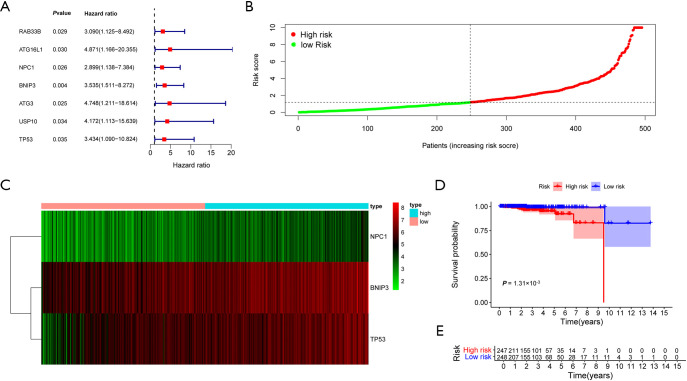
Using univariate Cox regression analyses, we identified 7 ARGs (RAB33B, ATG16L1, NPC1, BNIP3, ATG3, USP10, TP53) associated with PCa OS based on the data extracted from TCGA database (Figure 5A). Functional annotations of these genes were shown in Figure S2. Also, there existed no multicollinearity within these genes (Table S3). Further multivariate Cox regression analysis retained 3 survival related genes to construct the PI model: PI = [1.337 × exp(BNIP3)] + [0.861 × exp(NPC1)] + [1.079 × exp(TP53)]. Figure 5B,C showed the PI distribution of patients and the heatmap of the three genes’ expression profiles in the TCGA PRAD dataset. Furthermore, the K-M plot were developed to analyze the different survival time between the high- and low-risk groups based on the PI (Figure 5D). The results showed that patients in the high-risk group suffered significantly worse prognose than those in the low-risk group (P=1.31×10−3). Figure 5E exhibited the number of patients in high- and low-risk groups.
Figure 5.
Survival related ARGs and the PI. (A) Forest plot of the 7-survival related ARGs. (B) The PI distribution of patients in the TCGA PRAD dataset. (C) The heatmap of the 3 key genes’ expression profiles. (D) Kaplan–Meier plot compared the OS between patients in the high-risk group and those in the low-risk group. (E) The number of patients in different risk groups stratified by survival years. ARGs, auto-related genes; PI, prognostic index; TCGA, The Cancer Genome Atlas; PRAD, prostate adenocarcinoma; OS, overall survival.
Construction of ACPI and the relationships between ARGs and clinical characteristics
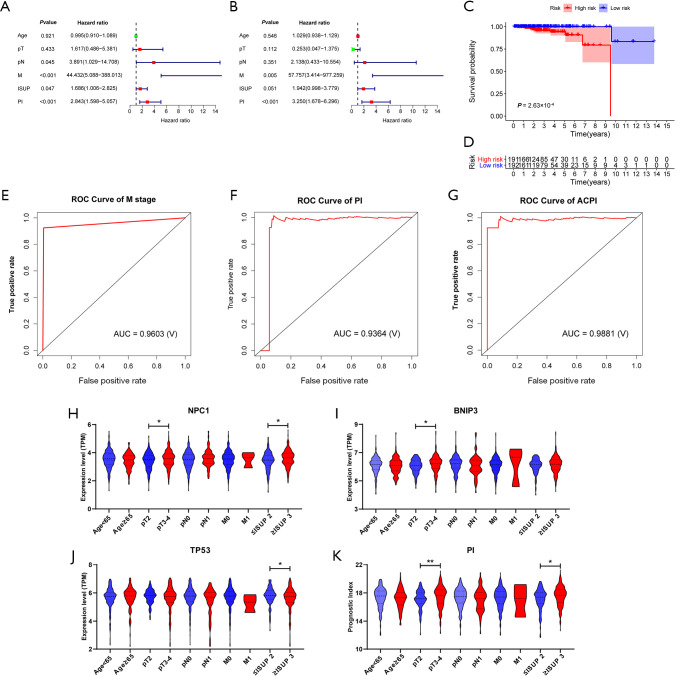
Univariate Cox regression analyses detected that pathological N stage, clinical M stage, International Society of Urological Pathology (ISUP) grade along with PI were significantly associated with PCa OS (Figure 6A). Then, clinical M stage and PI were retained after multivariate Cox regression analysis (Figure 6B). The ACPI was developed as followed: ACPI = (4.457 × PI) + 1.112 (if there existed metastasis). The K-M plot suggested that this model could significantly separate bad prognostic patients from well ones (Figure 6C, P=2.63×10−4). Figure 6D exhibited the number of patients in high- and low-risk groups. In addition, the AUCs of ROC curves for clinical M stage, PI and ACPI were 0.9603, 0.9364 and 0.9881, respectively, indicating a high prediction performance for PCa prognosis (Figure 6E,F,G).
Figure 6.
ACPI and relationship between 3 prognostic genes and clinical characteristics. (A) Forest plot of the univariate Cox regression analyses for the relationship between genetic or clinical factors and OS. (B) Forest plot of the multivariate Cox regression analyses for the relationship between genetic or clinical factors and OS. (C) Kaplan-Meier plot compared the OS between patients in the high-risk group and those in the low-risk group separated by the ACPI. (D) The number of patients in different risk groups stratified by survival years. (E,F,G) Survival prediction ROC curves of clinical M stage, PI and ACPI. (H,I,J) The relationship between three prognostic genes (NPC1, BNIP3 and TP53) and clinical characteristics. *, P<0.05; **, P<0.01; ***, P<0.001. Student’s t-test for normal distribution data and Mann-Whitney U test for non-normal distribution data. (K) The relationship between PI and clinical characteristics. *, P<0.05; **, P<0.01; ***, P<0.001. Student’s t-test for normal distribution data and Mann–Whitney U test for non-normal distribution data. PI, prognostic index; ACPI, autophagy-clinical prognostic index; ROC, receiver operating characteristic; AUC, area under the ROC curve; TPM, transcripts per million; OS, overall survival.
Furthermore, we investigated relationship of the three prognostic genes (NPC1, BNIP3, TP53) and PI with clinical characteristic. As Figure 6H,I,J showed, elevated expressions of NPC1 and BNIP3 were dramatically associated with advanced pathological T stages. In addition, overexpression of NPC1 was significantly related to high ISUP grades. On the contrary, elevated TP53 expression was detected in patients with low ISUP grades. Finally, we observed higher PI value occurred in advanced pathological T stages and high ISUP grades.
Discussion
On the whole, the incidence and mortality of PCa are high in general population (ranking the third and the eighth, respectively) (1). For patients with advanced or metastatic diseases, there are no further satisfying treatment strategies, especially in castration resistant prostate cancer (CRPC) patients. Thus, it is very important to find out the potential biomarkers related to the diagnosis and prognosis of PCa. Roles of autophagy in tumorigenesis, progression, and therapeutic resistance has been reported in various cancers (9), such as glioma (19,20), breast cancer (21), hepatocellular carcinoma (22) and so on. However, most studies only limited to a signal ARG. In this study, we identified essential prognostic and diagnostic ARGs and further established predictive models.
A total of 16 DEARGs were identified between of 52 normal tissues and 495 PCa tissues. Subsequently, GO analysis and KEGG analysis were performed. GO analysis showed that these DEARGs were notably associated with autophagy, process utilizing autophagic mechanism, digestive tract/system development, regulation of autophagy and so on. Additionally, KEGG pathway analysis revealed that these DEARGs were significantly related to autophagy-animal, platinum drug resistance, EGFR tyrosine kinase inhibitor resistance and so on. Moreover, a total of 5 ARGs (ATG9B, DNAJB1, HSPB8, NKX2-3, TP63) were recognized to be significantly related to the risk of PCa. Then, we identified 3 prognostic ARGs (BNIP3, NPC1, TP53) closely associated with OS of patients with PCa and developed a predictive model eventually.
Autophagy related gene 9b (ATG9B), plays a key role in the regulation of lysosomal degradation pathway. Studies have identified that mutations in this gene and disruption of the autophagy process were tightly related to multiple cancers. Previous study (23) discussed that HPV 16E6/E7 could activate autophagy via ATG9B in cervical cancers. Deficiency of ATG9B could suppress autophagy and promote hepatocyte apoptosis in hepatocarcinogenesis (24). DnaJ heat shock protein family (Hsp40) member B1 (DNAJB1) has been reported to be linked with many cellular processes, including the viral infection (25), endoplasmic reticulum stress (26) and proteasome pathway (27). In lung cancer, Park et al. (28) proposed that DNAJB1 enhanced the cell proliferation by suppressing the stabilization of mitogen-inducible gene 6. Heat shock protein B8 (HSPB8) is a member of common small heart shock protein family. Shen et al. (29) found that HSPB8 promoted cell growth and was related to a poor prognosis in patients with gastric cancer. NK2 homeobox 3 (NKX2-3) is located on 10q24.2 and encodes homeodomain-containing transcription factor. Li et al. (30) reported that NKX2-3 expression levels was a valuable biomarker to predict the response effects of FOLFOX4 chemotherapy in patients with primary advanced colorectal cancer. TP63 is a famous tumor suppressor gene and plays an important role in regulating cell cycle, apoptosis and maintaining genomic stability.
Bcl-2/adenovirus E1B 19 kD protein-interacting protein 3 (BNIP3) is a member protein of the Bcl-2 family which contains only the BH3 domain (31). After cellular stress, BNIP3 evolves into a homopolymer on the outer membrane of mitochondria, which induces apoptosis, necrosis and autophagy (32). Previous studies have indicated that upregulation of BNIP3 was related to tumor progression and poor survival outcomes in many different cancer types. Giatromanolaki et al. (33) found that BNIP3 over-expression was tightly linked with hypoxia-regulated protein expression and indicated poor survival outcomes in non-small cell lung cancer (NSCLC). Tan et al. (34) discovered that BNIP3 expression was up-regulated in breast cancer patients with ductal carcinoma in suit (DCIS) and invasive carcinoma. Furthermore, poor survival outcomes and an increased risk of recurrence were found in DCIS when compared with invasive carcinoma, which suggested a pivotal switch from a cell death to survival function induced by BNIP3. Sington et al. (35) thought that BNIP3 could act as a biomarker in follicular lymphoma with more aggressive behavior, and be of great value in the distinction from reactive lymphadenitis. In our study, we found that overexpression of BNIP3 was related to advanced pathological T stage of PCa patients, thus may contribute to a worse prognosis.
TP53, is a tumor suppressor gene, located on the 17p13 with 11 exons. It plays an important role in cell cycle, DNA repair and apoptosis to maintain the genetic stability (36) by encoding the p53 protein (a 53-kD nuclear phosphoprotein). Because TP53 is one of the important tumor suppressor genes (37), there have been a large number of studies on its role in tumorigenesis, development and prognosis. A relationship between elevated TP53 expression and lower ISUP grades was detected in this study. Zang et al. (38) demonstrated that TP53 mutation may represent poor prognosis in patients with advanced NSCLC, and the prognostic value differed in various mutated exons. Similar conclusions were drawn in many other cancer types, such as non-Hodgkin Lymphomas (39), chronic lymphocytic leukemia (40) and so on. Previous study (41) has proven that p53 tumor suppressor protein could regulate cell death in PCa cells by bridging the autophagic and apoptotic signaling pathways. A relationship between elevated TP53 expression and lower ISUP grades was detected in this study.
Niemann-Pick type C1 (NPC1) maps to chromosome 18q11.2 with 28 exons. The transmembrane glycoprotein encoded by NPC1 resides in the limiting membrane of endosomes and lysosomes, and mediates intracellular cholesterol transport through the binding of cholesterol to its N-terminal domain (42). Li and his colleagues (43) putted forwards that the NPC1 N-terminal domain transferred cholesterol to a detected cavity on the other side of the protein taking advantage of flexibility of polyproline junction. Kagedal et al. (44) found that NPC1 was over-expressed in patients with Alzheimer’s disease (AD), which could be explained by the perturbed cholesterol homeostasis in AD. The results of this study suggested that overexpression of NPC1 was associated with advanced pathological T stages and high ISUP grades, acting as a tumor promoting factor.
Recently, many diagnostic and prognostic ARGs were identified and predictive models were established for cancer patients based on the shared public databases and advanced bioinformatics analysis. Previous study (45) identified and validated a PI in bladder cancer patients using prognostic ARGs. Wang and his teammates (46) developed a nomogram for glioma patients by combining autophagy signature and clinical outcomes.
In our study, we identified ARGs associated with the diagnosis and prognosis of patients with PCa. Furthermore, predictive models with perfect accuracy and benefits were developed based on these ARGs to predict the risk and prognosis of PCa. However, some limitations could not be ignored. Firstly, expression profiles and clinical data of this study were respectively downloaded from public databases. Prospective and multicenter studies are needed in the future to validate our findings. Secondly, lack of basic experiments to explore the molecular mechanisms through which ARGs modulate the development and progression of PCa. Thirdly, there was no other dataset could be applied to test the effectiveness of the PI and ACPI models attribute to the well prognosis of PCa, also, limited by the follow-up time of TCGA database, there might exist some bias in the survival analysis.
Conclusions
In conclusion, this study identified ARGs and developed potential models for predicting the risk and prognosis of PCa. Further prospective studies and basic experiments are needed to validate our findings.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: National Natural Science Foundation of China (No. 81702520), Medical Research Project of Jiangsu Provincial Health and Family Planning Commission (No. H2018052), Research Project of Jiangsu Cancer Hospital (No. ZN201602), and the young talents program of Jiangsu Cancer Hospital (No. 2017YQL-04).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-498
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-498). XL serves as an unpaid Section Editor of Translational Andrology and Urology from Oct 2019 to Sep 2020. The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Stamey TA, Yang N, Hay AR, et al. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med 1987;317:909-16. 10.1056/NEJM198710083171501 [DOI] [PubMed] [Google Scholar]
- 3.Moyer VA, Force USPST. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2012;157:120-34. 10.7326/0003-4819-157-2-201207170-00459 [DOI] [PubMed] [Google Scholar]
- 4.US Preventive Services Task Force, Grossman DC, Curry SJ, et al. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018;319:1901-13. 10.1001/jama.2018.3710 [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 6.Kearns JT, Holt SK, Wright JL, et al. PSA screening, prostate biopsy, and treatment of prostate cancer in the years surrounding the USPSTF recommendation against prostate cancer screening. Cancer 2018;124:2733-9. 10.1002/cncr.31337 [DOI] [PubMed] [Google Scholar]
- 7.Macintosh RL, Ryan KM. Autophagy in tumour cell death. Semin Cancer Biol 2013;23:344-51. 10.1016/j.semcancer.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 8.Rabinowitz JD, White E. Autophagy and metabolism. Science 2010;330:1344-8. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White E, Karp C, Strohecker AM, et al. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 2010;22:212-7. 10.1016/j.ceb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007;21:1367-81. 10.1101/gad.1545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy 2007;3:28-31. 10.4161/auto.3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorin S, Hamai A, Mehrpour M, et al. Autophagy regulation and its role in cancer. Semin Cancer Biol 2013;23:361-79. 10.1016/j.semcancer.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 13.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006;10:51-64. 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo JY, Chen HY, Mathew R, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011;25:460-70. 10.1101/gad.2016311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 2009;15:5308-16. 10.1158/1078-0432.CCR-07-5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Dong W, He H, et al. Autophagy induced by overexpression of DCTPP1 promotes tumor progression and predicts poor clinical outcome in prostate cancer. Int J Biol Macromol 2018;118:599-609. 10.1016/j.ijbiomac.2018.06.005 [DOI] [PubMed] [Google Scholar]
- 17.Blessing AM, Rajapakshe K, Reddy Bollu L, et al. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy 2017;13:506-21. 10.1080/15548627.2016.1268300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Wang X, Wang Y, et al. SGK1 inhibition-induced autophagy impairs prostate cancer metastasis by reversing EMT. J Exp Clin Cancer Res 2018;37:73. 10.1186/s13046-018-0743-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang KH, Yan MD, Yao CJ, et al. Honokiol-induced apoptosis and autophagy in glioblastoma multiforme cells. Oncol Lett 2013;6:1435-8. 10.3892/ol.2013.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Lu X, Wang N, et al. Autophagy-related gene expression is an independent prognostic indicator of glioma. Oncotarget 2017;8:60987-1000. 10.18632/oncotarget.17719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Zhang J, Huang Y, et al. Cancer-Associated Fibroblasts Autophagy Enhances Progression of Triple-Negative Breast Cancer Cells. Med Sci Monit 2017;23:3904-12. 10.12659/MSM.902870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen DP, Ning WR, Li XF, et al. Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma. Autophagy 2018;14:1335-46. 10.1080/15548627.2018.1474994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tingting C, Shizhou Y, Songfa Z, et al. Human papillomavirus 16E6/E7 activates autophagy via Atg9B and LAMP1 in cervical cancer cells. Cancer Med 2019;8:4404-16. 10.1002/cam4.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang N, Tan HY, Li S, et al. Atg9b Deficiency Suppresses Autophagy and Potentiates Endoplasmic Reticulum Stress-Associated Hepatocyte Apoptosis in Hepatocarcinogenesis. Theranostics 2017;7:2325-38. 10.7150/thno.18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batra J, Tripathi S, Kumar A, et al. Human Heat shock protein 40 (Hsp40/DnaJB1) promotes influenza A virus replication by assisting nuclear import of viral ribonucleoproteins. Sci Rep 2016;6:19063. 10.1038/srep19063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenna S, Farina AG, Martyanov V, et al. Increased expression of endoplasmic reticulum stress and unfolded protein response genes in peripheral blood mononuclear cells from patients with limited cutaneous systemic sclerosis and pulmonary arterial hypertension. Arthritis Rheum 2013;65:1357-66. 10.1002/art.37891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamazaki S, Uchiumi A, Katagata Y. Hsp40 regulates the amount of keratin proteins via ubiquitin-proteasome pathway in cultured human cells. Int J Mol Med 2012;29:165-8. [DOI] [PubMed] [Google Scholar]
- 28.Park SY, Choi HK, Seo JS, et al. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim Biophys Acta 2015;1853:2722-30. 10.1016/j.bbamcr.2015.07.024 [DOI] [PubMed] [Google Scholar]
- 29.Shen J, Li M, Min L. HSPB8 promotes cancer cell growth by activating the ERKCREB pathway and is indicative of a poor prognosis in gastric cancer patients. Oncol Rep 2018;39:2978-86. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Lu X, Chi P, et al. Identification of Nkx2-3 and TGFB1I1 expression levels as potential biomarkers to predict the effects of FOLFOX4 chemotherapy. Cancer Biol Ther 2012;13:443-9. 10.4161/cbt.19298 [DOI] [PubMed] [Google Scholar]
- 31.Chung LY, Tang SJ, Wu YC, et al. Platinum-based combination chemotherapy triggers cancer cell death through induction of BNIP3 and ROS, but not autophagy. J Cell Mol Med 2020;24:1993-2003. 10.1111/jcmm.14898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chinnadurai G, Vijayalingam S, Gibson SB. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene 2008;27 Suppl 1:S114-27. 10.1038/onc.2009.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giatromanolaki A, Koukourakis MI, Sowter HM, et al. BNIP3 expression is linked with hypoxia-regulated protein expression and with poor prognosis in non-small cell lung cancer. Clin Cancer Res 2004;10:5566-71. 10.1158/1078-0432.CCR-04-0076 [DOI] [PubMed] [Google Scholar]
- 34.Tan EY, Campo L, Han C, et al. BNIP3 as a progression marker in primary human breast cancer; opposing functions in in situ versus invasive cancer. Clin Cancer Res 2007;13:467-74. 10.1158/1078-0432.CCR-06-1466 [DOI] [PubMed] [Google Scholar]
- 35.Sington J, Giatromanolaki A, Campo L, et al. BNIP3 expression in follicular lymphoma. Histopathology 2007;50:555-60. 10.1111/j.1365-2559.2007.02657.x [DOI] [PubMed] [Google Scholar]
- 36.Glück S, Ross JS, Royce M, et al. TP53 genomics predict higher clinical and pathologic tumor response in operable early-stage breast cancer treated with docetaxel-capecitabine +/- trastuzumab. Breast Cancer Res Treat 2012;132:781-91. 10.1007/s10549-011-1412-7 [DOI] [PubMed] [Google Scholar]
- 37.Moriya M, Slade N, Brdar B, et al. TP53 Mutational signature for aristolochic acid: an environmental carcinogen. Int J Cancer 2011;129:1532-6. 10.1002/ijc.26077 [DOI] [PubMed] [Google Scholar]
- 38.Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70-5. 10.1016/j.lungcan.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 39.Xu P, Liu X, Ouyang J, et al. TP53 mutation predicts the poor prognosis of non-Hodgkin lymphomas: Evidence from a meta-analysis. PLoS One 2017;12:e0174809. 10.1371/journal.pone.0174809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SY, Jung DC, Oh YT, et al. Prostate Cancer: PI-RADS Version 2 Helps Preoperatively Predict Clinically Significant Cancers. Radiology 2016;280:108-16. 10.1148/radiol.16151133 [DOI] [PubMed] [Google Scholar]
- 41.Ringer L, Sirajuddin P, Tricoli L, et al. The induction of the p53 tumor suppressor protein bridges the apoptotic and autophagic signaling pathways to regulate cell death in prostate cancer cells. Oncotarget 2014;5:10678-91. 10.18632/oncotarget.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu XH, Jiang N, Yao PB, et al. NPC1, intracellular cholesterol trafficking and atherosclerosis. Clin Chim Acta 2014;429:69-75. 10.1016/j.cca.2013.11.026 [DOI] [PubMed] [Google Scholar]
- 43.Li X, Lu F, Trinh MN, et al. 3.3 A structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport. Proc Natl Acad Sci U S A 2017;114:9116-21. 10.1073/pnas.1711716114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kågedal K, Kim WS, Appelqvist H, et al. Increased expression of the lysosomal cholesterol transporter NPC1 in Alzheimer's disease. Biochim Biophys Acta 2010;1801:831-8. 10.1016/j.bbalip.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 45.Wang SS, Chen G, Li SH, et al. Identification and validation of an individualized autophagy-clinical prognostic index in bladder cancer patients. Onco Targets Ther 2019;12:3695-712. 10.2147/OTT.S197676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Gao L, Guo X, et al. Development and validation of a nomogram with an autophagy-related gene signature for predicting survival in patients with glioblastoma. Aging (Albany NY) 2019;11:12246-69. 10.18632/aging.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as