Abstract
In 2014, there was a burst of studies on the molecular subtypes of bladder cancer in the published literature that was made possible by the advances in high-throughput technologies. Based on gene expression profiling, the major molecular classification subdivisions were basal and luminal subtypes, which resembled to those observed in breast cancers. These basal and luminal subtypes were further subdivided by TCGA into squamous, infiltrated, luminal-papillary, luminal/genomically unstable (GU), and neuronal/small cell carcinoma (SCC) subtypes. Recently, an international subtypes consensus project further expanded on the TCGA subtypes by defining a consensus molecular classification (CMC). A multidisciplinary team of experts generated CMC to overcome the difficulties of clinical applications due to several published bladder cancer molecular classifications with various nomenclatures and molecular features. It included six molecular subtypes with the addition of one more luminal subtype (luminal nonspecified) compared to the TCGA subtype classification. The initial research efforts have focused on the characterization of each subtype at the molecular and histopathologic levels, but more recent studies have examined their significance in terms of clinical utility, i.e., biomarkers that inform prognostication and/or to predict therapeutic responses to be tested in future clinical trials. This review provides an overview of recent investigations into the relationship between molecular subtypes and the clinical management of patients with bladder cancer.
Keywords: Molecular subtypes, bladder cancer, clinical application
Introduction
Bladder cancer is classified based on the depth of invasion as either non-muscle invasive (stages Ta, T1, and Tis) or muscle-invasive (stages T2, T3 and T4). Approximately 80% of bladder cancers are non-muscle invasive bladder cancer (NMIBCs) at diagnosis, and they have limited metastatic and/or lethal potential. However, NMIBCs have high rates of local recurrence, and a significant fraction (25–30%) progress to become muscle-invasive bladder cancer (MIBC). MIBCs are characterized by rapid progression to metastatic disease and are associated with higher mortality. Although this high-level heterogeneity is clinically significant, accumulating studies are demonstrating that NMIBCs and MIBCs are also highly heterogeneous with regard to clinical outcomes and treatment responses (1-6). Routine classification of bladder cancer has been mainly based on histopathologic status including tumor size, nodal status and distance metastasis (TNM classification). Though there are guidelines for the TNM classification system, histopathologic assessments are inherently subjective and are not robust enough to definitively characterize the underlying heterogeneity of bladder cancer.
Recent advances in genomics technologies have added new layers of understanding into the molecular heterogeneity of bladder cancers, most notably, by the identification of specific genetic alterations including DNA variations and molecular subtypes by gene expression profiling (2,5,6). With respect to the latter, in earlier studies, unsupervised cluster analyses of gene expression profiling data easily separated NMIBCs from MIBCs, demonstrating that the expression signatures of NMIBCs were distinct from MIBCs (7,8). Beyond this top-level division, several subsequent studies reported that NMIBCs and MIBCs can be further subdivided into molecular subtypes that exhibit distinct biological and possibly clinical properties. At the highest level, NMIBC and MIBC molecular subtypes can also be divided into two major subsets (basal-like and luminal-like) that share biomarkers with the well-characterized basal and luminal molecular subtypes that were observed in breast cancer (2,6).
Because of their shared molecular properties, the molecular subtypes of bladder cancer may have clinical value by improving the current diagnostic systems. For example, they can build on subjective histopathological assessment methods by adding objective scoring systems, and by identifying the underlying molecular heterogeneity that cannot be easily detected by microscopic assessments alone. The molecular subtypes may also contain therapeutic targets that are not determined by solely DNA mutational status. Overall, the value of the molecular subtypes will be determined by their clinical relevance. Therefore, this review provides a summary of the current understanding of the clinical relevance of the molecular subtypes of bladder cancer.
Classification of bladder cancer
NMIBC
NMIBCs are heterogeneous in terms of their propensities to progress to MIBC and their responses to current frontline intravesical therapies. The current standard of care for NMIBC is transurethral resection of bladder tumor (TURBT) with or without intravesical therapy with chemotherapy (mitomycin C, gemcitabine plus docetaxel) or Bacillus Calmette-Guérin (BCG), with the latter being the therapy of choice for high-risk NMIBC. Even though initially response rates are high (70%), recurrence is a major problem and many patients develop BCG unresponsive disease, for which the current guidelines recommend definitive therapy with radical cystectomy (RC) or trimodal therapy with maximal TURBT plus chemoradiation. Currently, there are no predictive biomarkers for the identification of the patients who will or will not respond to BCG, which has led researchers to perform whole-genome studies to characterize NMIBC heterogeneity. Therefore, NMIBCs have been and continue to be subjected to whole transcriptome mRNA expression profiling and molecular subtype analyses in an attempt to identify biomarkers that are associated with these variable clinical outcomes.
The group at Lund performed comprehensive analyses using combined NMIBC and MIBC datasets to report the existence of five molecular subtypes: squamous cell carcinoma-like (SCCL), urobasal (uro) B (uroB), infiltrated, genomically unstable (GU), and urobasal A (uroA) (5). SCCL and uroB expressed molecular markers shared by basal-like breast cancers, while uroA and GU expressed luminal markers. The vast majority of NMIBCs belonged to the uroA or GU subtypes, and low-grade Ta tumors were largely classified as uroA. Though uroA and uroB expressed luminal and basal biomarkers, respectively, both uroA and uroB displayed high FGFR3 and FGFR3 pathway gene expression. Interestingly, protein expression patterns of basal markers in uroA and uroB subtypes were confined only in the basal cell layer (9). These similarities between uroA and uroB led to the hypothesis that uroB might evolve from uroA. GU and infiltrated were mainly comprised of T1 and high-grade tumors, and the infiltrated group was enriched with immune and other stromal cells.
More recently, the UROMOL study, involving a cohort of 460 NMIBCs, was the largest multi-institutional transcriptional analysis project to stratify NMIBCs at the molecular level (2). The cohort was heterogeneous in terms of its content of low- and high-grade tumors, and the tumors also represented all NMIBC stages [Ta, T1 and carcinoma in situ (CIS)]. However, one weakness of the cohort was that it did not contain a large number of tumors from patients treated with BCG, and the study was therefore not powered to correlate molecular subtypes with BCG response. Unsupervised clustering of the RNAseq data identified three subtypes with distinct basal- and luminal-like characteristics based on cell cycle genes and differentiation markers. Class 1 tumors mainly consisted of Ta tumors and were associated with the best clinical prognoses. Class 2 tumors were enriched with mutations of TP53 and ERBB2 (HER2), and APOBEC-related mutational signatures, that led the investigators to propose the hypothesis that these tumors were the ones that progressed to luminal MIBCs. Class 3 displayed some basal-like features, and they expressed high levels of lncRNAs. These features resulted in a proposed pathway for class 3 tumors as dormant luminal tumors that can indirectly shift to become MIBCs, with the dormant state defined by histone and chromatin remodeling. These three subtypes are hypothesized to represent three different developmental pathways of NMIBC.
A third study reported two copy number subtypes: genomic subtype 1 (GS1) and genomic subtype 2 (GS2), based on unsupervised cluster analysis of low pass whole-genome sequencing and array-CGH data from 141 Ta tumors (10). GS1 showed no or few copy number alterations, while GS2 was characterized by the high-frequency deletion of chromosome 9q involving the region encoding TSC1. To investigate the similarity in gene expression between copy number based subtypes and mRNA-based subtypes, transcriptome analyses were performed on a subset of the tumors (GS1 =48 and GS2 =31). When tumors were assigned to the Lund molecular subtypes (5), the majority of tumors (40/48 GS1 and 24/31 GS2) belonged to the uroA subtype irrespective of their copy number status, indicating that Ta tumors displayed luminal subtype biology.
MIBC
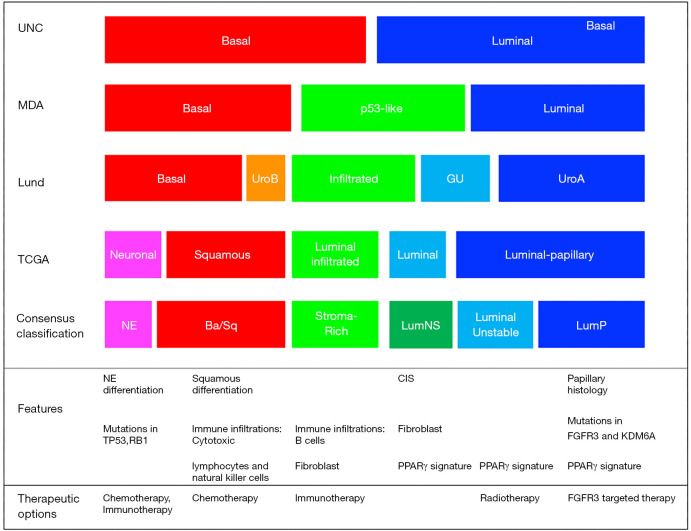
The comprehensive characterization of MIBC tumors identified basal and luminal molecular subtypes that were associated with distinct gene expression patterns and clinical features (4-6,11-13). Though several groups including Lund, UNC, MDA and TCGA reported various numbers (k=2-5) of molecular subtypes, their underlying biological characteristics were highly concordant (Figure 1). Basal/SCCL-like tumors were characterized by high expression of basal cell markers such as KRT5/6 and KRT14 and were typically enriched with squamous and sarcomatoid features. Luminal tumors were characterized by high-level expression of uroplakins, KRT20, ERBB2 and luminal differentiation markers such as forkhead box A1 (FOXA1), GATA-binding protein 3 (GATA3), tripartite motif-containing protein 24 (TRIM24) and peroxisome proliferator-activated receptor-γ (PPARγ). Basal and luminal biomarkers tended to be expressed in a mutually exclusive manner in the molecular subtypes, and many appeared to be excellent therapeutic targets. Luminal tumors could be further subdivided into infiltrated/p53-like, urobasal A/luminal papillary, and GU tumors. The infiltrated/p53-like subtype was enriched with cancer-associated fibroblasts (CAFs) and extracellular matrix (ECM) markers and exhibited low-level expression of cell cycle and proliferation markers. UrobasalA/luminal papillary tumors were enriched with activating FGFR3 mutations, FGFR3-related mRNA signatures and papillary histopathological features, while GU tumors were distinctive from uroA tumors by containing fewer FGFR3 mutations and higher tumor mutational burdens (TMBs) (4,5,14).
Figure 1.
Molecular subtype classification of bladder cancer. Associations among subtype classifications were assigned based on biological marker expressions. UNC, University of North Carolina; MDA, MD Anderson cancer center; NE, neuroendocrine.
Consensus molecular classifications (CMCs)
Even though the various reported molecular subtype classifications shared common characteristics, their distinct nomenclatures and various granularity of molecular features were major obstacles to their clinical application. To overcome these obstacles, a multidisciplinary team of experts was created to generate a consensus classification scheme utilizing six published approaches and transcriptomic profiles from 1,750 MIBCs obtained from 18 public datasets (15). Six consensus subtypes (luminal-papillary:LumP, luminal-nonspecified:LumNS, luminal unstable:LumU, stroma-rich, basal/squamous:Ba/Sq and neuroendocrine-like: NE-like) were identified by network-based analyses (16), then multivariable Cox models were used to determine the clinical outcomes correlations to the subtypes (15). Subtype calls from the consensus classification had significant associations with the calls generated from each of the original classification systems. The Ba/Sq subtype comprised of the largest sample size (35% of all samples) and was associated with squamous differentiation and poor survival outcomes. Also, it had a strong association with STAT3 and HIF1A regulon activities and frequent mutations of TP53 (61%) and RB1 (25%) (15). The NE-like tumors were highly related to small cell/neuroendocrine variant histology. In addition, they exhibited either mutation or deletion of TP53 (94%) and RB1 (94%) that were consistent with previous reports and were associated with the worst survival outcomes overall. LumP tumors represented the second largest population (24% of all samples) and were enriched with papillary histological morphology and activation of the FGFR3 transcriptional pathway. Tumors with LumNS displayed higher stromal infiltration than LumP tumors, while LumU tumors exhibited the highest cell cycle activity among the three luminal subtypes. All three luminal subtypes exhibited urothelial differentiation signatures and PPARG, ERBB2, and GATA3 regulon activities. Finally, stroma-rich tumors were mainly characterized by stromal infiltration.
Importantly, distinct immune microenvironments were also associated with each subtype. Immune infiltration was observed mostly in the Ba/Sq and stroma-rich tumors. Ba/Sq tumors contained gene expression signatures associated with cytotoxic lymphocytes and natural killer cells, while the stroma-rich tumors exhibited overexpression of B-cell markers (15). Since each consensus subtype contained distinct biological properties, clinical characteristics, and therapeutic targets, a single-sample classifier was developed and made available online that can assign tumors to the consensus class labels (15). This tool will be utilized for the development of predictive biomarkers in future clinical trials.
Bladder cancer variants
About 90–95% of bladder cancers exhibit conventional urothelial carcinoma histopathological features, but about 5–10% of tumors contain histological variants (i.e., neuroendocrine, sarcomatoid, squamous, papillary, etc.) (17). Because identifying variant histologies is somewhat subjective, efforts are underway to develop more robust methods to classify these tumors objectively, and approaches based on mRNA expression profiling hold significant promise.
Neuroendocrine bladder carcinomas (NEBC) are rare and associated with rapid progression and high metastatic potential. Based on tissue morphology, NEBCs are subdivided into small cell carcinoma (SCC) and large cell carcinoma. SCCs are the majority of NEBC since the incidence of large cell carcinoma is infrequent. A majority of NEBCs are mixed with conventional urothelial bladder cancer (18,19). To objectively diagnose these highly-aggressive tumors, multiple studies have reported on the development of molecular classifiers for NEBCs. The Lund group was the first to propose the existence of an NEBC subtype (CC3-2) (20). Using Consensus Cluster analysis, about 7% of tumors expressed conventional neuroendocrine markers (SOX2 and ENO2), and they were also characterized by a highly proliferative phenotype. In TCGA’s final molecular classification study, it also reported on the existence of a “neuronal” subtype (n=20 out of 408 tumors), characterized by the expression of neuronal differentiation and development genes. Inactivating DNA alterations affecting TP53 and RB1 are typical of neuroendocrine urothelial bladder cancers, with half of TCGA’s neuronal tumors containing mutations in TP53 and/or RB1. Most (17/20) tumors in TCGA’s neuronal subtype did not exhibit neuroendocrine histology, suggesting that molecular classifiers might identify tumors with neuroendocrine biology that would be missed by conventional histopathology. Finally, we have analyzed the mutational and gene expression profiles of fairly large cohorts of SCCs and identified that SCCs have the distinctive mutational landscape and gene expression patterns compared to conventional urothelial carcinoma (PMID:32061548)
Sarcomatoid carcinoma is another rare bladder cancer histologic subtype with aggressive clinical features. It usually occurs in older males and is associated with advanced stage at presentation. Sarcomatoid carcinoma was originally named carcinosarcoma because it has both carcinomatous and sarcomatous characteristics (21). Sarcomatoid carcinomas expressed markers that were shared by the basal molecular subtype in conventional bladder cancers and were subdivided into “epithelial” and “mesenchymal” subsets, with the latter being more clinically aggressive (22). More specifically, the mesenchymal tumors were characterized by the epithelial-to-mesenchymal transition (EMT) and displayed loss of adherence genes (CDH1, Claudins, etc.) and overexpression of EMT transcriptional factor (SNAI2) (22). Also, about half of sarcomatoid carcinomas displayed significant immune cell infiltration, suggesting that patients with these tumors might benefit from immune checkpoint blockade and/or other immune-based therapy.
Micropapillary carcinoma is the third aggressive bladder cancer variant that exhibits the unique microscopic feature of the small infiltrating area of tumor cells residing in empty spaces (23). A majority of micropapillary carcinoma expressed characteristics of MDA’s luminal molecular subtype with enrichment of PPARg-associated gene expression signatures and suppression of the downstream of p63 pathways (23). Other work demonstrated that the biomarker profiles of micropapillary cancers are identical to those observed in the Lund GU and consensus luminal unstable subtypes (15). Based on these properties, it appears that micropapillary carcinomas progress from conventional luminal unstable tumors. Some micropapillary carcinomas expressed biomarkers shared by MDA’s p53-like subtype, and these tumors were particularly aggressive and associated with shorter survival outcomes (23). Though the molecular mechanisms that underlie the emergence of micropapillary histology are not well characterized, downregulation of miR-296 with upregulation of its targets and activation of the molecular co-chaperone, RUVBL1, may be involved (23).
Therapeutic implications of molecular subtypes
Chemotherapy
Neoadjuvant cisplatin-based combination chemotherapy (NAC) with M-VAC (methotrexate, vinblastine, doxorubicin, and cisplatin) or GC (gemcitabine and cisplatin) followed by RC is the standard frontline treatment option for bladder cancer. However, major pathological responses (≤pT1 at cystectomy) are observed in only 50–60% of patients and response durations can be short with eventual disease progression in many patients. Therefore, a top priority of ongoing clinical research is to identify biomarkers that can be used to predict NAC responses. To achieve this, the “coexpression extrapolation” (CoXEN) algorithm was developed using NCI 60 cell line data to predict chemosensitivity to either frontline therapies for MIBC (24). To attempt to validate it, a two-arm Phase II clinical trial was designed by the Southwest Oncology Group (S1314), which completed accrual in late 2018 and is currently being evaluated. A total of 237 cisplatin-eligible patients were randomized to receive either MVAC (every 14 days for 4 cycles) or GC (every 21 days for 4 cycles). When the COXEN algorithm was applied to the gene expression data from the trial samples, the results demonstrated there was no relationship between the COXEN MVAC score and response, but the CoXEN GC score was loosely associated with downstaging in both the GC arm and the combined cohort of GC and MVAC treated tumors. The potential relationship between the GC and MVAC COXEN scores and survival outcomes will be evaluated after the data become more mature.
Gene expression data have also been studied for prognostic markers to identify optimal treatment with better survival outcomes. Several studies, including ours, have shown that chemotherapeutic response varies across the molecular subtypes of bladder cancer (3,12,13,25). In early work we showed that the infiltrated/p53-like tumors were chemoresistant, whereas basal tumors were associated with the most survival benefit in patients treated with NAC (3,13). To build upon this finding, a larger retrospective cohort of TUR specimens used a single-sample subtype classifier, where molecular subtypes were shown to have different phenotypic responses after NAC (25). The conclusions of this study partially confirmed the earlier work by showing that basal tumors were associated with the most survival benefit from NAC. However, there was an uncoupling between survival outcomes and pathologic responses, since none of the published subtype classifiers revealed an association between the molecular subtypes and pathologic responses (13,25). One speculation for the explanation of this uncoupling is sample selection bias that may cause that basal tumors with poor survival outcomes were enriched in non-chemotherapy cohort while basal tumors with better survival outcomes might be schewed in published NAC cohorts (3,25). An attempt to validate these observations is also underway with the whole transcriptome expression data that were generated in the CoXEN trial. These results will determine whether molecular subtype classifiers should be used with other approaches to select patients with basal tumors for treatment with NAC.
Recent retrospective studies have demonstrated that inactivating DNA alterations in genes involved in DNA repair are associated with response, and in particular, defects in ERCC2 were correlated with complete pathological response in patients with MIBC treated with NAC (26). In parallel, another group correlated alterations of one or more of three DNA repair-associated genes (ATM, RB1, and FANCC) with pathologic responses and survival outcomes in a separate clinical trial cohort (27). These DNA biomarkers were also built into the biomarker studies within the CoXEN clinical trial, and they are currently undergoing prospective evaluation. Furthermore, open clinical trials (NCT02177695) are examining whether patients whose tumors contain these alterations can be managed by active surveillance rather than definitive surgery.
Immunotherapy
In 2016, the US FDA’s approval of atezolizumab for the treatment of locally advanced or metastatic bladder cancer based on the results of the IMvigor-210 clinical trial was a breakthrough for the bladder cancer management, since no new therapies had been approved for decades. Subsequent to the approval of atezolizumab, four other immune checkpoint inhibitors targeting either PD-L1 (atezolizumab, durvalumab, and avelumab) or PD-1 (nivolumab and pembrolizumab) were approved in the advanced/metastatic setting (28). More recently, the clinical efficacy of atezolizumab was also assessed in the neoadjuvant setting within the context of the ABACUS trial (29). Biomarker studies revealed that in the advanced/metastatic setting, responses were linked to stromal expression of PDL1, enrichment with IFNg and CD8+T effector cell gene expression signatures, and high neoantigen or tumor mutation burdens (TMBs), whereas high TMBs did not correlate with responses in the neoadjuvant setting (29,30). Instead, the quality of the immune infiltrate as measured by dual CD8 and granzyme B immunohistochemistry may predict response in the neoadjuvant setting (29). Conversely, TGFb pathway activation was implicated in resistance in both settings, and preclinical functional studies demonstrated that TGFβ activation shaped the tumor microenvironment to limit T cell infiltration. However, the relationship between response and molecular subtype membership is less clear. In IMvigor-210, TCGA [2014] cluster II tumors (which were similar to p53-like/luminal infiltrated tumors) were associated with the highest response rates, whereas cluster I (luminal papillary) tumors were found to be resistant (30). Subsequent studies failed to confirm the relationship between infiltration and response. For example, data from the Checkmate 275 clinical trial of nivolumab demonstrated that responses were enriched in basal molecular subtype and correlated with an interferon-g-signature that was also enriched in these tumors (31), but the association between the luminal papillary subtype and resistance was consistently observed (14). Finally, analysis of the complete ImVigor210 trial data revealed that the Lund GU subtype was associated with response, consistent with the observation that the GU subtype is characterized by the high TMBs. Recently, the TCGA team applied their final molecular subtype (k=5) taxonomy to the IMvigor 210 trial data using a novel single-patient subtype classifier, and they showed that the neuroendocrine-like (NE-like) and luminal subtypes were both associated with high response rates (32). Though Lund GU tumors and TCGA NE-like tumors showed the responses to immunotherapy respectively, the TCGA NE-like tumors might be classified as Lund GU tumors since GU tumors in earlier Lund classification were similar to the NE-like subtypes in later Lund classification (14,20,32). However, the opposite pattern of NE-like tumor’s response to immunotherapy was observed with the Decipher classifier developed by GenomeDx (25,33). Therefore, the investigation into the relationships between molecular subtype membership and immunotherapy response/resistance is still ongoing.
FGFR3 targeted therapy
Since the majority of NMIBCs and approximately 15–20% of MIBCs contain activating FGFR3 mutations, FGFR3 is an attractive therapeutic target in bladder cancer. In MIBC, tumors with mutations or over-expression of FGFR3 are enriched in the luminal-papillary molecular subtype (6,13,15). When altered by an activating mutation, FGFR3, a receptor tyrosine kinase, can activate signaling pathways to induce higher rates of cell proliferation and FGFR3 S249C, the most common mutation form in bladder cancer, creates an abnormal cysteine disulfide bridge that leads to ligand-independent constitutive dimerization and activation of receptor (34). Mutations are also found within the transmembrane domain (ex, Y373C) or kinase domain (ex, K652E), which also leads to constitutive receptor activation (34,35). Several small molecule receptor tyrosine kinase inhibitors have been developed to target FGFR3 (36,37). Among them, erdafitinib, a tyrosine kinase inhibitor targeting FGFR1-4, exhibited the strongest antitumor activity in bladder cancer (36). In a recently-completed Phase II clinical trial, 99 patients with locally-advanced or metastatic bladder cancer with tumors containing FGFR3 alterations that progressed after chemotherapy were treated with a median of five cycles of erdafitinib. Of these patients, 40% displayed complete or partial responses and no treatment-related death was reported. Based on this observation, the US FDA approved the drug for the treatment of metastatic bladder cancer patients with FGFR3 alterations in 2019. Since FGFR3 pathway activation and FGFR3 mutations are dominant features of luminal-papillary tumors, there are ongoing studies to investigate whether there is a direct relationship between response to erdafitinib and the luminal papillary subtype.
Challenges and future directions
The introduction of molecular subtype classifications represented a major advance in our understanding of the heterogeneity of bladder cancer. Sophisticated high-throughput technologies and tools for computational biology added additional layers to the understanding of biological events that lead to tumor development and progression and that represent attractive diagnostic and therapeutic targets. Recent analyses revealed that molecular subtypes have prognostic value. However, it is still unclear whether molecular subtyping can inform the selection of patients for particular treatments. Ongoing studies within the context of the CoXEN trial are exploring whether molecular subtyping can inform the selection of patients for treatment with neoadjuvant cisplatin-based chemotherapy, and a similar strategy will be required to determine the value of molecular subtyping for the selection of patients for treatment with other therapies. Furthermore, due to the intra-tumoral heterogeneity, molecular subtype calls may not be stable, especially in basal tumors that are potential candidates for chemotherapy treatment (38). Also, the diversity of the gene expression profiling platforms that were used for the generation of classifiers may introduce difficulties in comparing datasets leading to inaccurate subtype calls. In addition, the majority of current results linking therapeutic responses to molecular subtypes are from retrospective studies. Additional prospective studies are needed to determine whether the molecular subtype can predict the response to therapies.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by grant from DoD (W81XWH-19-1-0148).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Ja Hyeon Ku, Ho Kyung Seo, Seok Ho Kang) for the series “Muscle-Invasive Bladder Cancer” published in Translational Andrology and Urology. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-2019-mibc-12). The series “Muscle-Invasive Bladder Cancer” was commissioned by the editorial office without any funding or sponsorship. Dr. DJM reports grants from Astra-Zeneca, grants from Ranier Pharmaceuticals, personal fees from Ranier Pharmaceuticals, personal fees from Jassen, personal fees from H3 Biomedicine, outside the submitted work. The authors have no other conflicts of interest to declare.
References
- 1.Choi W, Ochoa A, McConkey DJ, et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur Urol 2017;72:354-65. 10.1016/j.eururo.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27-42. 10.1016/j.ccell.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 3.McConkey DJ, Choi W, Shen Y, et al. A Prognostic Gene Expression Signature in the Molecular Classification of Chemotherapy-naive Urothelial Cancer is Predictive of Clinical Outcomes from Neoadjuvant Chemotherapy: A Phase 2 Trial of Dose-dense Methotrexate, Vinblastine, Doxorubicin, and Cisplatin with Bevacizumab in Urothelial Cancer. Eur Urol 2016;69:855-62. 10.1016/j.eururo.2015.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjödahl G, Eriksson P, Liedberg F, et al. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J Pathol 2017;242:113-25. 10.1002/path.4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjödahl G, Lauss M, Lovgren K, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012;18:3377-86. 10.1158/1078-0432.CCR-12-0077-T [DOI] [PubMed] [Google Scholar]
- 6.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540-56.e25. 10.1016/j.cell.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res 2005;11:4044-55. 10.1158/1078-0432.CCR-04-2409 [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Carbayo M, Socci ND, Lozano J, et al. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol 2006;24:778-89. 10.1200/JCO.2005.03.2375 [DOI] [PubMed] [Google Scholar]
- 9.Sjödahl G, Lovgren K, Lauss M, et al. Toward a molecular pathologic classification of urothelial carcinoma. Am J Pathol 2013;183:681-91. 10.1016/j.ajpath.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 10.Hurst CD, Alder O, Platt FM, et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017;32:701-15.e7. 10.1016/j.ccell.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110-5. 10.1073/pnas.1318376111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi W, Czerniak B, Ochoa A, et al. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nat Rev Urol 2014;11:400-10. 10.1038/nrurol.2014.129 [DOI] [PubMed] [Google Scholar]
- 13.Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152-65. 10.1016/j.ccr.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544-8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamoun A, de Reynies A, Allory Y, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020;77:420-33. 10.1016/j.eururo.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantor AF, Hartge P, Hoover RN, et al. Epidemiological characteristics of squamous cell carcinoma and adenocarcinoma of the bladder. Cancer Res 1988;48:3853-5. [PubMed] [Google Scholar]
- 18.Blomjous CE, Vos W, De Voogt HJ, et al. Small cell carcinoma of the urinary bladder. A clinicopathologic, morphometric, immunohistochemical, and ultrastructural study of 18 cases. Cancer 1989;64:1347-57. [DOI] [PubMed] [Google Scholar]
- 19.Choong NW, Quevedo JF, Kaur JS. Small cell carcinoma of the urinary bladder. The Mayo Clinic experience. Cancer 2005;103:1172-8. 10.1002/cncr.20903 [DOI] [PubMed] [Google Scholar]
- 20.Aine M, Eriksson P, Liedberg F, et al. Biological determinants of bladder cancer gene expression subtypes. Sci Rep 2015;5:10957. 10.1038/srep10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dent ED, Jr. Carcinosarcoma (collision tumor) of the urinary bladder. J Urol 1955;74:104-8. 10.1016/S0022-5347(17)67250-1 [DOI] [PubMed] [Google Scholar]
- 22.Guo CC, Majewski T, Zhang L, et al. Dysregulation of EMT Drives the Progression to Clinically Aggressive Sarcomatoid Bladder Cancer. Cell Rep 2019;27:1781-93.e4. 10.1016/j.celrep.2019.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo CC, Dadhania V, Zhang L, et al. Gene Expression Profile of the Clinically Aggressive Micropapillary Variant of Bladder Cancer. Eur Urol 2016;70:611-20. 10.1016/j.eururo.2016.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JK, Havaleshko DM, Cho H, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A 2007;104:13086-91. 10.1073/pnas.0610292104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiler R, Ashab HAD, Erho N, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol 2017;72:544-54. 10.1016/j.eururo.2017.03.030 [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Mouw KW, Polak P, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 2016;48:600-6. 10.1038/ng.3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anari F, O'Neill J, Choi W, et al. Neoadjuvant Dose-dense Gemcitabine and Cisplatin in Muscle-invasive Bladder Cancer: Results of a Phase 2 Trial. Eur Urol Oncol 2018;1:54-60. 10.1016/j.euo.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghatalia P, Zibelman M, Geynisman DM, et al. Approved checkpoint inhibitors in bladder cancer: which drug should be used when? Ther Adv Med Oncol 2018;10:1758835918788310. 10.1177/1758835918788310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powles T, Kockx M, Rodriguez-Vida A, et al. Clinical efficacy and biomarker analysis of neoadjuvant atezolizumab in operable urothelial carcinoma in the ABACUS trial. Nat Med 2019;25:1706-14. 10.1038/s41591-019-0628-7 [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. 10.1016/S0140-6736(16)00561-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma P, Retz M, Siefker-Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 2017;18:312-22. 10.1016/S1470-2045(17)30065-7 [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Kwiatkowski D, McConkey DJ, et al. The Cancer Genome Atlas Expression Subtypes Stratify Response to Checkpoint Inhibition in Advanced Urothelial Cancer and Identify a Subset of Patients with High Survival Probability. Eur Urol 2019;75:961-4. 10.1016/j.eururo.2019.02.017 [DOI] [PubMed] [Google Scholar]
- 33.Batista da Costa J, Gibb EA, Bivalacqua TJ, et al. Molecular Characterization of Neuroendocrine-like Bladder Cancer. Clin Cancer Res 2019;25:3908-20. 10.1158/1078-0432.CCR-18-3558 [DOI] [PubMed] [Google Scholar]
- 34.Naski MC, Wang Q, Xu J, et al. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet 1996;13:233-7. 10.1038/ng0696-233 [DOI] [PubMed] [Google Scholar]
- 35.Munro NP, Knowles MA. Fibroblast growth factors and their receptors in transitional cell carcinoma. J Urol 2003;169:675-82. 10.1016/S0022-5347(05)63991-2 [DOI] [PubMed] [Google Scholar]
- 36.Loriot Y, Necchi A, Park SH, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med 2019;381:338-48. 10.1056/NEJMoa1817323 [DOI] [PubMed] [Google Scholar]
- 37.Lamont FR, Tomlinson DC, Cooper PA, et al. Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br J Cancer 2011;104:75-82. 10.1038/sj.bjc.6606016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sjödahl G, Eriksson P, Lovgren K, et al. Discordant molecular subtype classification in the basal-squamous subtype of bladder tumors and matched lymph-node metastases. Mod Pathol 2018;31:1869-81. 10.1038/s41379-018-0096-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as