We have come a long way in the last years of developing analytical strategies in metabolomics. We have seen huge progress in tackling multiplatform measurement, data analysis, data integration, and interpretation.1 Mass spectrometry (MS) is the unrivaled technology in the field. Following a divide and conquer strategy, successful approaches defined and addressed sub-omes individually. Recursively solving technical “subproblems” also with regard to the analytical tasks of quantification and identification allowed us to make significant progress. However, some of the challenges, as imposed by the metabolome’s complexity (molecules <1500 Da), are not entirely overcome to date.
Indeed, the physicochemical space occupied by this building block of life is vastly heterogeneous, spanning concentration ranges from (high) fM to mM2 and forming dynamic complex reaction networks. The complete scope of metabolic networks remains to be elucidated. This holds true for “simple” organisms such as bacteria with relatively small-sized metabolomes (in the hundreds) and even more for the human body metabolome considering that it consists of hundreds of different metabolome, depending on body fluids, cell type, status, and tissue. Next to the endogenous human pathways, numerous metabolites exist that are transformed and/or circulated upon the complex interplay with trillions of microbes constituting the “ecosystem” of the human body.3 Additionally, certain disease-specific metabolites (e.g. methylated amino acids) with biological function may occur, defining the so-called epi-metabolome.4 Damaged and repaired metabolites can be the result of enzymatic impairment.5 Finally, the human metabolome is highly dependent on nutrition and the surrounding environment. More than 200,000 food derived metabolites and 10,000 xenobiotics exist that are potentially circulating.6
Consequently, we have not yet reached the ultimate aim, which is to comprehensively identify and quantify all metabolites with one or at least a few analytical runs. Metabolome coverage, selectivity, sensitivity and throughput remain conflicting goals that we have to navigate.7 While this fact limits the pace of experimental assessment of metabolome inventories8 regarding different cell types and model organisms, there has been significant progress in customizing workflows, with the aim of providing a pragmatic base for informative metabolite measurements.9
The virtuous cycle of the global metabolomics workflow starts with discoveries by nontargeted analysis. Over the last few years, this analytical strategy has seen a tremendous impact across different metabolomics applications and beyond. At the same time analytical chemists, embracing the novel omics-type of measurement, have been and keep being challenged regarding quality control (QC), method standardization, and harmonization. Evidently, the computational methods for data processing and data analysis are by far more complex than in target analysis. Establishing metrics and guidelines for nontargeted analysis is not straightforward,10 especially compared to the well-established validation practice in target analysis. Experimental design and data quality11 are key to fully exploit the potential of nontargeted analysis with regard to e.g. biomarker discovery12,13 and beyond. The integration of reference materials in nontargeted workflows is still under debate. The complexity of omics-reference materials production, following stringent metrological criteria, results in high costs, which contradicts to the idea of affordable discoveries by large-scale studies. The authors assume that this lack of general acceptance has in turn reduced the pace of material development, and today we still have only a few biological matrix reference standards available. Finally, whether a discovery can be standardized might be debatable; however, a finding should be validated. In fact, a metabolomics experiment should not end with nontargeted methods, but the results should be validated both analytically and biologically.14
Thus, the final analytical step of our ideal virtuous metabolomics cycle includes targeted measurements using authentic standards. Typical sample numbers in metabolomics range from tens or hundreds up to a few thousand, depending on the study design.15 The more diverse the study cohort, the more samples must be analyzed in order to generate a meaningful hypothesis. Following the golden rules of step-wise discovery and stringent analytical validation is more demanding for large scale studies. Time spans between sampling, analysis, interpretation, discovery, and final validation together with the limited availability of authentic standards pose practical limitations towards this approach. A major aim of analytical development remains increasing throughput of measurement. Regarding compound annotation, de facto every current study accepts annotations with varying but defined degree of certainty. This holds true for metabolomics and lipidomics, where annotation is facilitated by rule-based MS data interpretation as enabled by the structural templates of lipids. It is common practice in both applications to report levels of annotation.16,17 However, estimating the proportion of potentially false assignments is still an exciting field of research.18 Finally, analytical validation should include the quantitative dimension of discoveries in nontargeted analysis. Despite significant progress in harmonization, standardization, and advanced statistical analysis,19 large scale multicenter studies remain challenging. Recent applications resort to small scale studies for hypothesis generation, followed by a (wide) targeted large scale study for hypothesis validation.20
Biological validation is dependent on the scope of the study. In metabolic phenotyping, biological probability checks are facilitated by massive joint efforts to deploy open-source metabolic atlases for a number of different organisms. Comparisons with both experimental data and predictions (reactions, rules, and enzymes) support the findings.21 The complexity of biological validation increases dramatically in the case of a hypothesized biological function. Then, validation of the generated hypothesis does not only address the mere presence/up- or downregulation of a certain metabolite/pathway, but the hypothesized biological function needs to be corroborated. For example, in functional metabolomics,22 cutting-edge multi-omics analysis23 together with biochemical assays unravels molecular functions and associated modulatory mechanisms of perturbed metabolism in relation to phenotype.
Undoubtedly, accepting multiple lines of evidence in nontargeted discoveries (with reported degree of confidence) has accelerated metabolomics research. The question to which degree analytical validation can be reduced or even entirely replaced by advanced computational methods and biological validation experiments needs to be addressed in the overwhelmingly interdisciplinary science of metabolomics. Reporting on the accurate assessment and the resulting degree of confidence alone is a minimum requirement.24 On the other hand, the ways of evidence besides strict analytical validation might promote the acceleration of the measurement step itself. High-throughput technologies proved to be fit-for-purpose in dedicated applications despite limited selectivity.25,26
This review will focus on recurring topics in MS-based metabolomics measurement (including lipids). We will emphasize the role of stable isotopes for both target and nontargeted analysis giving an overview on different standard materials derived from isotopically labeled biomass and strategies enabled by these materials. We will discuss the current state of the art of quantification, validation, and harmonization with respect to both metabolomics and lipidomics. We will include strategies enabling various ways of scientific evidence regarding the metabolite/lipid annotation task. Finally, we will survey the rationales of workflow design, which straddle coverage and throughput. Nearly five years have passed since Cajka and Fiehn published their review on the state of the art of metabolomics/lipidomics, proposing at the same time a vision of merging targeted and nontargeted analysis.9 Since then, many studies have realized the potential of simultaneous unanticipated discovery and quantification of a selected metabolite pool, a strategy enabled by high-resolution mass spectrometry (HRMS). We report on the progress of “merging” ideas. We think that lipidomics and metabolomics need to be integrated into one workflow. We will discuss the potential of chromatographic solutions as compared to recent high-throughput technologies for the simultaneous analysis of the two sub-omes, as a first key step.
Established Concepts of Quantification in Metabolomics/Lipidomics
In absolute accurate quantification, guidelines on bioanalytical method validation from the United States Food & Drug Administration (U.S. FDA)27 or European Medicines Agency (EMA)28 establish gold standards and metrological frames. However, application to omics-type analysis is challenged by the sheer number of analytes within one measurement, the lack of standards, and the need for an actual analyte-free matrix. In the following, we will give a brief tutorial summary on absolute quantification strategies currently established in the field of metabolomics and lipidomics. The term quantitative assessment in MS-based omics studies often refers to relative quantification of differences between sample groups, while here we refer to absolute quantification requiring proper standardization and analytical validation. A brief introduction will emphasize the need for standards and reference materials, in the form of both multi-mix standards and biological matrix material.
Recommended Absolute Quantification Approaches
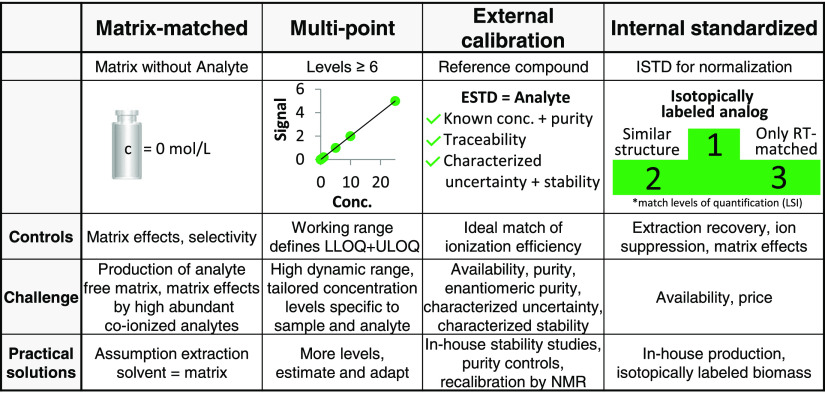
The method of highest metrological order in MS based analysis is isotope dilution established by matrix-matched multi-level external calibration with internal standardization. The internal standard (ISTD) added as early as possible in the analytical process and equilibration between sample and spike should be ensured prior to extraction. Multilevel calibration, preferred as the working range (given by the lower limit of quantification (LLOQ) and the upper limit of quantification (ULOQ)), is assessed and controlled along with the quantification exercise. This is not the case when isotope dilution is based on a single spike level (one-point calibration). Next to this gold standard, other external calibration strategies could meet the recommendations of widely accepted (bio-) analytical method validation guidelines, as well, as long as they properly employ internal standardization. As internal standards, either standards of similar structures or of matching retention time (RT) and thus co-ionization are commonly used. Spiking the same amount of ISTD to external calibrants and samples allows us to use ISTDs without certified concentration. Figure 1 dissects the calibration method into four major components and discusses their relevance. According to the guidelines, the analysis of biological matrix blanks is mandatory. The conceptualization of such a blank sample, i.e. a biological matrix free of endogenous metabolites, is challenging. Knockout experiments for specific metabolites, albeit tedious, offer a solution. However, most studies resort to simplifying approaches using extraction blanks or protein mixtures.
Figure 1.
Accurate absolute quantification according to the U.S. FDA guideline. Four requirements need to be fulfilled for calibration: 1, matrix-matched; 2, multipoint; 3, external standardization; 4, internal standardization. Additionally, their control point, the challenge, and a practical solution for omics-experiments are given. *The ranking of ISTD follows the levels of quantification of the Lipidomics Standards Initiative (LSI).29
The gold standard of quantification, if applied to -omics type of analysis requires a high number of external standards (ESTDs) and ISTDs, which are stable isotope labeled. Fully labeled standards are expensive but simplify data evaluation and validation. State of the art wide targeted assays in metabolomics implement hundreds of standards. In research practice, the need for fit-for-purpose methods has led to the implementation of alternative quantification strategies with the aim of reducing the overall number of standards, measurements, and costs involved. In lipidomics, calibration strategies resorting to few standards per lipid class have successfully been established as enabled by the structural templates of lipids.30 Moreover, recent development concerns the use of partial isotopic labeling for standard production.31Figure 2 provides an overview on established quantification methods in metabolomics/lipidomics compared to the gold standard (matrix-matched multi-point external calibration including internal standardization).
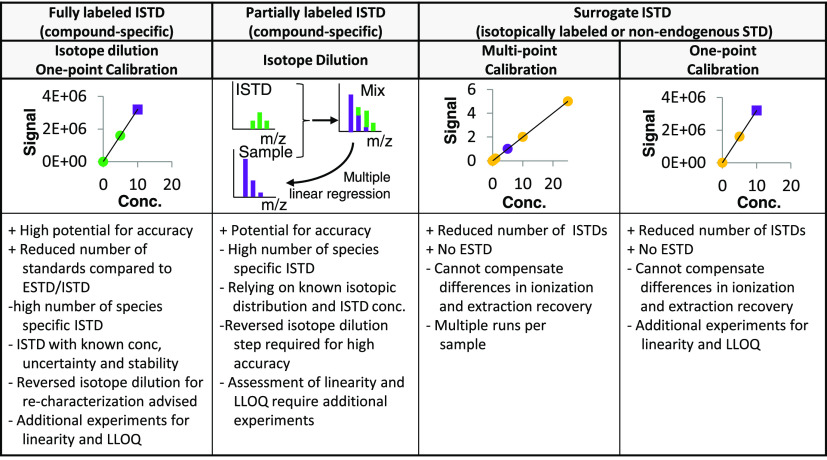
Figure 2.
Fit for purpose internal standard-based quantification strategies established in the field of metabolomics and lipidomics. Colors in the graphs symbolize values from the sample (purple), compound-specific standards (green), and surrogate standards (orange).
As previously mentioned, isotope dilution using a known amount of isotopically labeled ISTD with characterized concentration (traceable) offers a method of high metrological order. Both fully labeled and partially labeled ISTDs can be used. In the latter case, concentrations are calculated using multiple linear regressions. The “single spike” isotope dilution method is accurate, provided that (1) spike and sample are equilibrated upon extraction and (2) the blend ratio is within the linear dynamic range and (3) significantly different from the natural ratio. Thus, additional validation experiments are required. For highest metrological order, reversed isotope dilution experiments are necessary to characterize the spike with every experiment. These steps are mostly omitted in -omics measurements. The validation process is accelerated by kit solutions and commercial availability of ISTD mixtures with concentration levels tailored for specific applications. If no compound-specific calibrant is available, surrogate calibration is accomplished by structurally similar standards, either using isotopically labeled ISTD or non-endogenous ISTDs. Structurally similar standards are preferred over RT matched standards, which ensure co-ionization only. Surrogate internal standardization drastically reduced the number of necessary standards. It is executed as multi-point calibration32 or as one-point calibration.33 In lipidomics, surrogate calibration is accepted, provided lipid class co-ionization and the use of response factors34 is ensured. If lipid surrogate quantification is performed on the MS2 level, variations in signal intensities between the different fatty acyl chain fragments have to be mathematically corrected for. Schuhmann et al. recently published a model based on commercially available lipid standards to correct systematic errors (up to 60%) for common glycerophospholipids due to the differences in (1) the sn-1/2 positions of the glycerol backbone, (2) the length of the hydrocarbon chain, and (3) the number and location of double bonds.35
Stable Isotope Labeling
In contrast to radionuclides, isotopes have stable nuclei, hence representing a safe alternative for labeling approaches. The overall abundance of heavy stable isotopes in nature is low (<5%). Given the isotopic effect, i.e. the isotopic fractionation upon chemical reactions and biological processes, the natural abundance varies to a small degree, forming the basis for natural tracer studies in geochronology, ecology, archeology, or climatology. The low natural abundance facilitates the production of pure stable isotope labeled compounds, either via chemical synthesis or via in vivo synthesis.36 Stable isotopes and stable isotope labeling have a well-documented history in MS, which was exquisitely outlined for life sciences by Lehmann.37 In this review, we emphasize the pivotal role of stable isotope labeled biomass. Today, in vivo synthesized stable isotope labeled compounds have become essential tools for mass spectrometry-based identification or quantification in metabolomics (including lipidomics). The important application of supplied stable isotope tracers in metabolomics for flux and tracer studies is comprehensively covered elsewhere.38−41 Labeled biomass was used early on in quantitative omics workflows, e.g. amino acid labeling to monitor proteome changes upon system perturbation. Relative quantification in proteomics studies using cell culture based labeling42 was performed, but also successful labeling of higher organisms such as Caenorhabditis elegans, Drosophila melanogaster, and mice was reported.43−45 However, it is important to note that for higher organism complex nutrients and media composition are necessary so that in most cases only specific amino acids (SILAC approach) were labeled leading to amino acid labeling efficiencies of up to 98%.44 Only when fully labeled mircoorganisms such as Escherichia coli 98% enriched in 15N were fed to worms (C. elegans) or fruit flies (D. melanogaster), protein extracts with a labeling degree up to 94% were detectable.45 However, the smaller number of nitrogen atoms limits its use in metabolomics or lipidomics, thus carbon or deuterium labeling is preferred. Already in 2005, absolute quantification based on internal standardization by uniformly 13C-labeled yeast cell extracts was introduced, paving the way for absolute quantification of a high-numbered analyte panel.46 At that time no enrichment degrees were reported for metabolites or lipids. The use of labeled biomass for quantification tasks in metabolomics was facilitated by fully labeled E. coli grown in shaking flasks as pioneered by the group of Rabinowitz47 and further extended for eukaryotic uniformly labeled yeast grown in fermenters by Canelas et al.48
Enrichment Degree and Isotopologue Distribution
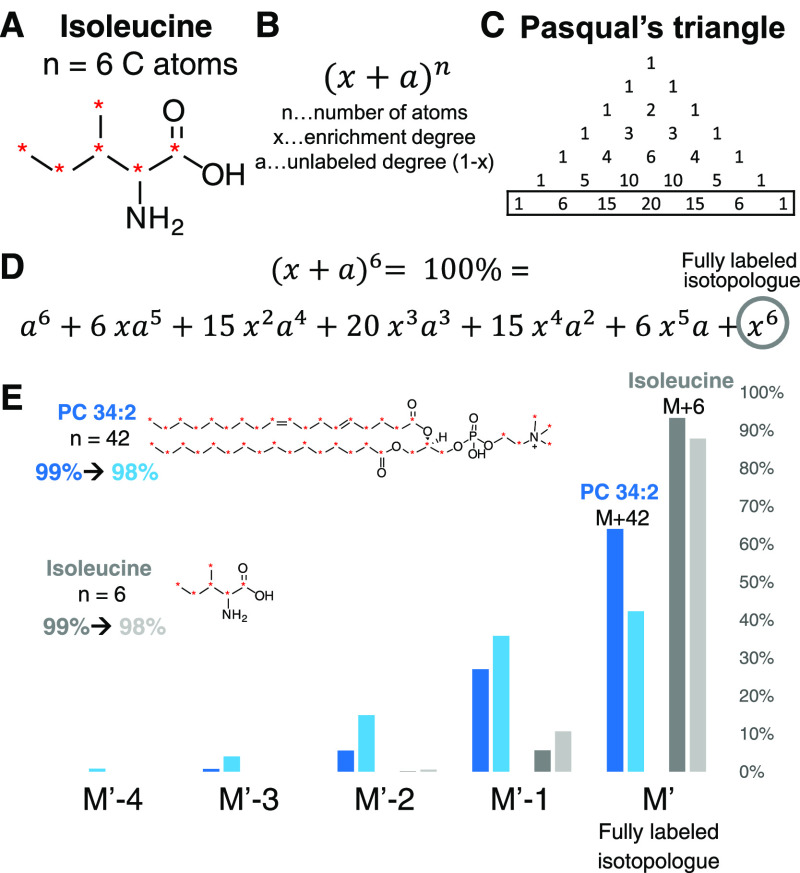
Isotopically labeled standards are characterized by the enrichment degree—often used interchangeably with the term labeling efficiency—which refers to the probability of finding a labeled atom at any possible label site. One has to be aware that the actual relative abundance of the heaviest isotopologue, i.e. the fully labeled isotopologue, is lower than the enrichment degree and depends on enrichment, the number of labeling sites, elemental composition, and mass resolution (see Figure 3 A–D). A simplified assumption of 100% abundance of the fully labeled isotopologue leads to errors in actual relative abundance in the mass spectra (Examples for leucine and phosphatidylcholine (PC) 34:2 can be found in Figure 3 E). This is relevant in absolute quantification relying on ISTDs with known concentration and especially crucial if the labeled compound is used as surrogate ISTD as e.g. often performed in lipidomics.33 In this case, either all isotopologues are summed up (after they have been checked for interferences) or the actual value is corrected e.g. similar to isotope correction Type 1 for natural unlabeled lipids.33 A useful tool for fast prediction of isotopologue distributions from molecular formulas is enviPat, which is available as website version and R package.49 Overall, in order to enable omics-type analysis, the known enrichment degree is of paramount importance. Spike materials of high enrichment degree (>99%) are preferred as they lead to more distinct isotopologue signals, reduced spectral overlay, and more straightforward data interpretation.
Figure 3.
Difference between enrichment degree and the relative isotopic abundance of a fully labeled isotopologue. (A) Isoleucine with 6 carbon atoms is used as an example. (B) Calculation of abundances for carbon as di-isotopic element is based on the binominal formula. Other elements with more than one isotope (e.g. H, N) influence the final abundance according to their natural abundance also based on a binominal formula. Polyisotopic elements (O) are based on polynomial terms. Usually, the contribution of H, N, and O to the overall difference is minimal (here 1–2%) but other elements must be considered (e.g. Cl, Br, S). (C) Determination of coefficients of a binominal formula for each term according to the n + 1 line in Pasqual’s triangle (for n = 6:1, 6, 15, 20, 15, 6, 1). (D) Binominal formula for n = 6. Each term is the relative abundance of the corresponding isotopologue without the consideration of other elemental isotopes. The last term corresponds to the fully labeled isotopologue. The sum of all isotopologues is always 100%. (E) Exemplarily, the effect of 1% enrichment difference (99%-darker color and 98%-lighter color) on the abundance is shown for PC 34:2 (n = 42, blue) and isoleucine (n = 6, grey). The bar chart shows the distribution from the fully labeled isotopologue (M′) until M′ – 4 for both molecules. The difference for the fully labeled isotopologue to 100% is already 12% for the 98% labeled isoleucine and 58% for PC 34:2. But even for a better enrichment (99%) the error for PC 34:2 is still 36%, highlighting the importance to consider the relative abundance for quantification workflows.50
Suite of In Vivo Synthesized Isotopically Labeled Materials
In the last decades, labeled organisms such as bacteria, yeast, or plants have been grown to create huge libraries of stable isotope labeled (13C, 15N, 34S, 2H) endogenous metabolites.51−53 Controlled growth conditions of E. coli or Pichia pastoris were particularly successful as enrichment degrees higher than 99% were achieved leading to the simultaneous production of hundreds of biologically relevant labeled metabolites covering the highly conserved primary metabolome.51,54−56 Some of the materials are already commercially available (such as labeled E. coli, yeast, and algae products; details on the materials can be found in Table 1).
Table 1. Overview on Labeled Biomass Materials.
| Organism | Kingdom | Isotope | Enrichment degree | Feed | Reference |
|---|---|---|---|---|---|
| Escherichia coli | Bacteria | 13C | >98% | Glucose | Mahieu and Patti 201757 |
| Escherichia coli | Bacteria | 15N | >98% | (NH4)2SO4 | Krüger et al. 200843 |
| Arthrospira platensis (Spirulina) | Bacteria | 13C | >97% | CO2 | Berthold et al. 199158 |
| Chlamydomonas reinhardtii (algae) | Bacteria | 13C | >98% | CO2 | Behrens et al. 199459 |
| Chlorella vulgaris (algae) | Bacteria | 13C | >98% | CO2 | Behrens et al. 199459 |
| Nannochloropsis oculata (algae) | Protist | 13C | >85% | CO2 | Doomun et al. 202060 |
| Pichia pastoris (yeast) | Fungi | 13C | >98% | Glucose | Neubauer et al. 201256 |
| Pichia pastoris (yeast) | Fungi | 34S | >95% | Na2SO4 | Hermann et al. 201661 |
| Saccharomyces cerevisiae (yeast) | Fungi | 15N | >94% | (NH4)2SO4 | Krüger et al. 200843 |
| Fusarium graminearum | Fungi | 13C | >99.5% | Glucose | Bueschl et al. 201452 |
| Arabidopsis thaliana | Planae | 13C | >95% | CO2 | Giavalisco et al. 200953 |
| Triticum durum (wheat) | Planae | 13C /15N | >96%/>95% | CO2/NO3 salts | Ćeranić et al. 202062 |
| Caenorhabditis elegans (worm) | Animalia | 15N | >98% | E. coli | Krüger et al. 200843 |
| Drosophila melanogaster (fly) | Animalia | 15N | >94% | S. cerevisiae | Krüger et al. 200843 |
| Rattus norvegicus domestica (rat) | Animalia | 15N | >94% | Spirulina | McClatchy et al. 200763 |
| Mus musculus (mice) | Animalia | 13C | 6–75% | Ralstonia eutropha | Dethloff et al. 201864 |
| Homo sapiens (HeLa cells) | Animalia | 2H | 0–5% | 5% D2O | Kim et al. 201931 |
| Homo sapiens (HCT116 cells) | Animalia | 13C | 0–99% | Glucose and AAs | Grankvist et al. 201865 |
The list of labeled organisms is further growing. For example, uniformly 13C labeled lipids derived from microalgae Nannochloropsis oculata were measured via MS/MS to calculate 13C enrichment for both the whole molecule and the different building blocks of a lipid.60 Such information can be useful to follow labeling of the head group versus fatty acids and might help to study lipid synthesis and remodeling processes. Advances in stable isotope labeling in plants using customized closed growth chambers enabled us to increase the enrichment degree to 96–98% for 13C and 95–99% for 15N adding a complex compound panel of primary and secondary metabolites.62 Still missing is a fully labeled mammalian organism. The complex feed or media and the resulting high costs limit the production to partial labeling approaches, which have been used successfully for relative quantification. For example, growing HeLa cells on a 5% deuterium oxide enriched medium together with a deconvolution algorithm facilitating classical isotopic dilution approaches enabled improved relative quantification for lipids.31 Even mice can be partially labeled (6–75% enrichment depending on the metabolite) for feeding a commercially available 13C-labeled bacterial diet (Ralstonia eutropha). This strategy was also applied for relative quantification, improving precision from 27% to less than 10%.64Table 1 summarizes the labeled biomass materials, used labeled isotopes, enrichment degree, feed, and literature.
Applications of Stable Isotope Labeled Biomass
Isotopically labeled biomass has three major applications in metabolomics and lipidomics, namely (1) credentialing by identification of biological metabolites using labeled and nonlabeled metabolite pairs, (2) validation of isotopologue distributions, and (3) standardization and normalization for quantification workflows.
Credentialing: Isotopically Labeled Biomass for Identification
Credentialing-type approaches involve the analysis of samples containing analytes in an unlabeled as well as a stable-isotope labeled form. Mixing of extracts from uniformly labeled organisms with those from unlabeled organisms allows us to distinguish metabolic features with biological origin from background contaminants by the occurrence of shifted m/z and MS/MS spectra and, in approaches implementing liquid chromatography (LC–MS), also matching RTs. An early application of comprehensive incorporation of stable isotope labeled biomass was published by Giavalisco et al.,53 who applied 13C labeling of Arabidopsis thaliana in order to recognize biological features and improve the molecular formula annotation of their flow injection (FI-) fourier-transform ion cyclotron resonance (FTICR) and reversed-phase (RP)-LC-FTICR analysis. The first open-source software MetExtract capable of automatizing assignment of LC-MS peaks originating from 13C labeled compounds to their endogenous counterparts was published by Bueschl et al.66 Later, other tools mostly relying on differential incorporation of isotopic labels into metabolites have been introduced, which simplify this type of analysis and include tracer analysis (MAVEN,67 mzMatch-ISO,68 X13CMS,69 isoMETLIN,70 george,71 and ALLocator72). The isotopic ratio outlier analysis (IROA) approach demonstrated the introduction of highly specific isotopologue patterns to further improve specificity and quantification capabilities using labeled organisms.73 In 2014, Mahieu et al.74 coined the term “credentialing” and further emphasized the importance of this type of approaches for the recognition of real biological features and the comparison and fine tuning of metabolomics workflows. Later they used stable isotope labeling combined with other feature grouping and noise removal approaches to show that the number of biological features in an E. coli extract can account for less than 5% of all features detected via nontargeted peak detection.57 MetExtract was later updated to MetExtract II to remove mismatches and group different ion-species as well as employ stable isotope patterns for the purpose of LC-MS peak detection, annotation/noise removal in fragmentation spectra, molecular formula elucidation, and isotopic tracer studies.75 This presented a significant step in harvesting the full potential of stable isotope labeling. In 2019, Wang et al. employed not only 13C but also 15N isotopically labeled organisms (Saccharomyces cerevisiae and E. coli).76 As in the original credentialing approach they combined stable isotope labeling with other noise reduction and feature grouping approaches in order to recognize biological features. Using this approach, they found a comparable number of biological features (only 4% of the peaks were annotated as apparent metabolites). Moreover, systematic annotation of peaks and discrimination of biological compounds (including isotopic variants) from adducts, fragments and MS artifacts was established. In fact, the correct identification of adducts was identified as a major bottleneck for elucidating the number of true sample molecules. In the following, the integration of stable isotope labeled buffers in LC-HRMS improved cost efficiency and introduced an universal stable isotope labeling approach for the corroboration and annotation of real chemical features to any kind of sample.77 The disadvantage of doubled measurement time is compensated by the comparable performance (for noise removal and annotation) to other credentialing approaches.
Isotopically Labeled Biomass for Validated Isotopologue Distribution Elucidations
Another way to harvest stable-isotope labels in metabolomics is the investigation of differential incorporation of labels into organisms comprehensively reviewed elsewhere.38−40 However, we want to highlight the application of labeled biomass with controlled labeling pattern78−80 to validate isotope tracer analysis workflows.81 In the past, it was shown that 13C tracer and flux experiments demand dedicated validation tools. Spectral accuracy, i.e. an instrument’s ability to truly measure the fractional abundance of the different isotopologues, is crucial. Metabolite standards with natural isotopic pattern (as well as fully labeled standards) are not well suited to assess the accuracy of carbon isotopologue distribution in tracer studies. Due to the low natural abundance of 13C, heavy natural isotopologues are below the limit of detection. Using in vivo synthesis, tailored carbon isotopologue distribution of primary metabolites can be obtained, which serves as ideal reference. Isotopologue distribution of stable isotopologe-labeled compounds can be assessed with excellent precisions of <1% and trueness bias as small as 0.01–1%.
Isotopically Labeled Biomass for Quantification
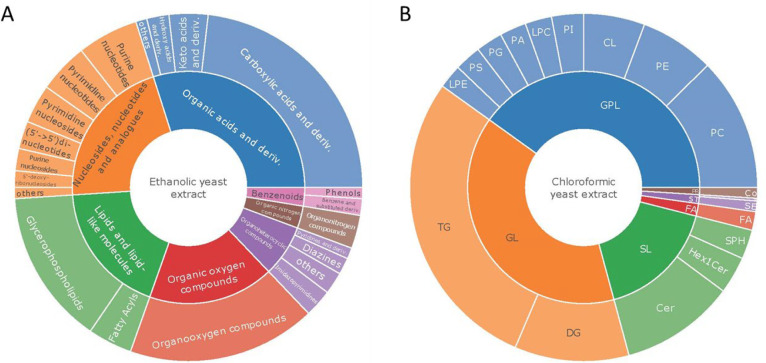
Starting in the 1980s, stable isotope-labeled ISTDs and isotope dilution approaches in combination with LC- and gas chromatography (GC)-MS/MS were used to improve quantification of small molecules.37 In metabolomics, internal standardization is widely adopted for absolute quantification, as the analytical process consists of multiple steps and requires normalization. Chemical synthesis of isotope labeled standards precludes omics-type of analysis, as hundreds of ISTDs are required to make isotopically labeled biomass a promising alternative. The cost-effective in vivo synthesized metabolites standards are characterized with respect to their isotope labeling degree but not their concentrations. Thus, normalization between samples (relative quantification) or internal standardization of external calibration (absolute quantification)47,48,54,56 is accomplished by spiking known amounts of labeled biomass into the samples. The benefits of these quantification workflows are well documented. Overall, improved analytical figures of merit (trueness, precision, and linearity) have been observed upon the integration of labeled yeast extracts.54,55,82 The use of HRMS together with stable isotope labeled standards supports workflows merging absolute quantification and nontargeted unanticipated discoveries (relative quantification and annotation) in one analytical run. This powerful strategy has been addressed in metabolomics and lipidomics.54,82 In lipidomics, only a slight decrease of identified lipids (∼10%) was observed in the presence of labeled biomass.82 This can be explained with ion competition in complex matrices when applying data-dependent fragmentation and can be further optimized by deep metabolite profiling or data-independent acquisition. Stable isotope labeled materials as an intermediate have to be chosen on the bases of sufficient metabolite/lipid class coverage and biomass availability/costs. Labeled yeast, e.g. P. pastoris, offers a reasonable compromise for quantitative studies, as it is an eukaryotic organism that can be easily cultivated under controlled conditions on a sole carbon source. Yeasts share a high metabolome and lipidome overlap with humans including the evolutionarily conserved primary metabolome, e.g. amino acids, nucleotides, organic acids, and metabolites of the central carbon metabolism. But also lipids are covered as shown by Natter et al.83 Wolrab et al. summarized the most frequently up- and downregulated lipids in oncology including the classes phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidylinositols (PI), phosphatidylserines (PS), lysophosphatidylcholines (LPC), lysophosphatidylethanolamines (LPE), lysophosphatidic acids (LPA), free fatty acids (FA), triacylglycerols (TG), diacylglycerols (DG), cholesterol esters (CE), sphingomyelins (SM), ceramides (Cer), monosialodihexosylganglioside (GM3), and sulfatides (SHexCer) in both tissue and body fluids,84 and except for CE, SM, GM3, and SHexCer, all of the listed classes are present in yeast. In the past, P. pastoris yeast extracts were successfully spiked to human plasma (including standard reference material (SRM) 1950 from the national institute of standards and technology (NIST), USA), different cell extracts, and yeast, either as ISTD based on ethanolic extracts or chloroform based lipidome isotope labeling of yeast (LILY) extracts, for metabolites and lipids, respectively (Figure 4A, B).
Figure 4.
Current in-house library of annotated metabolites and lipids found in Pichia pastoris (yeast). (A) Metabolite classes in ethanolic yeast extract85 classified using the ClassyFirer86 annotation system. (B) Lipid classes annotated in chloroformic yeast extract.87 GPL, glycerophospholipids; GL, glycerolipids; SL, sphingolipids; ST. sterols; PR, prenols; Hex1Cer, hexosyl ceramides; SPH, shingosine bases; SE, steryl esters; Co, coenzyme Q; PG, phosphatidylglycerols; PA, phosphatidic acids; CL, cardiolipins.
At the present state, a library of 206 metabolites for the ethanolic yeast extract covering the classes of (1) organic acids and derivatives, (2) nucleosides, nucleotides, and analogues, (3) lipids and lipid-like molecules, (4) organic oxygen compounds, (5) organoheterocyclic compounds, (6) organic nitrogen compounds, and (7) benzoids is established (Figure 4A). All of the identified metabolites were also present in The Human Metabolome Database (HMDB)88. This can be in part attributed to the human microbiome, but also to the evolutionary (inter-species) conservation of the primary metabolome. With regard to the yeast and human lipidome, major differences exist including a different sphingoid base—SPH 18:0;3 instead of SPH 18:1;2—as well as other sphingolipid classes (inositol phosphoceramide (IPC), mannosylinositol phosphoceramide (MIPC), and mannose-bis(inositolphospho)ceramide (M(IP)2C) instead of SM, ceramide 1-phosphates (CerP), and gangliosides. Yeasts also contain a smaller diversity of fatty acids with a maximum of three double bonds with a lack of higher polyunsaturated fatty acids (PUFA). Furthermore, no ether lipids (plasmanyl (ether bond), plasmenyl (vinyl bond)) are present and cholesterol is replaced by ergosterol in yeast. Overall, this leads to a list of 405 lipid species (Figure 4B) combining information from reports on LILY from chloroform extracts by RP-LCMS82 and an improved preparative supercritical fluid chromatography (SFC) workflow.87 Optimized extraction strategies and confirmation by authentic standards can further increase the metabolite and lipid list in yeast.89 Here, we want to emphasize the possibility of class or retention-time specific standardization if the target metabolite or lipid is not present in the yeast extract. By using these labeled compounds as class or retention-time specific ISTD if the target analyte is not present in the yeast extract,90 the list of possible analytes in a quantitative approach can further be enlarged and adapted to the sample of interest.
Harmonization and Reference Materials
Joint efforts toward harmonized metabolomics protocols and the definition of a minimum of quality requirements are of paramount importance. There is a vivid scientific community working toward harmonization to raise transparency and quality of published results.17,91−95 Standardized methods and reference materials provide benchmarks, paving the way to reproducibility and most importantly interassay commutability, with regard to both targeted and nontargeted analysis.
Reference Materials and Interlaboratory Comparisons
Certified reference materials represent the highest metrological order benchmarks enabling traceable and accurate quantification in metabolomics workflows. Certification requires an inherently long lead time, as composition and quantitative values are reported with characterized uncertainty and stability. Certified reference materials are provided by metrological institutions or by accredited material producers. While the application of (certified) reference materials in absolute quantification is well established, their integration for nontargeted metabolomics is emerging. A recent multi-platform study by hydrophilic interaction liquid chromatography (HILIC)/RP-LC HRMS96 demonstrated the power of using high-quality benchmarks in large-scale nontargeted metabolomics. Three pooled human plasma reference materials (Qstd3, 211 CHEAR, NIST SRM 1950) were repeatedly measured along with 3600 samples over a period of 17 months, providing a convincing strategy for data normalization and estimative concentration levels.
As the pace of standard production suitable for omics-type research in national metrological institutions is slow, international ring trials/interlaboratory initiatives drive standardization by offering measurement protocols and consensus values for biological matrix materials which can be distributed to the community. For the widely adopted NIST reference material human plasma SRM 1950, the number of consensus values assessed by international ring trials is continuously growing. Consensus values for 250 metabolites (amino acids, biogenic amines, acylcarnitines, glycerolipids, glycerophospholipids, cholesteryl esters, sphingolipids, hexoses) were assessed on the basis of the Biocrates AbsoluteIDQp400HR.97 Interlaboratory comparisons are of paramount importance in lipidomics, since reference materials are lacking. In 2017, an international ring trial provided consensus values for 339 lipids (from the major categories: fatty acids, glycerolipids, glycerophospholipids, sphingolipids, sterols) in SRM 1950.98 Recently, Triebl et al.99 further emphasized the need for reference samples by showing that lipidomics workflows continue to suffer from limitations associated with reproducibility and commutability of quantitative data from different platforms, even when isotopically labeled ISTDs were included. The authors compared direct infusion, HILIC, and RP-LC-MS workflows for lipid analysis showing that upon normalization to the reference sample SRM 1950, platform-dependent quantitative bias was successfully removed.99 The frequent use of SRM 1950 in both metabolomics and lipidomics studies96,97,100,101 highlights its key role as a reference point for merged workflows. Another recent interlaboratory study tested seven distinct materials including human urine pools from four SRMs and one research-grade test material (RGTM) provided by NIST.102 Untargeted analytical profiles for these materials were obtained using a variety of common metabolomics platforms (nuclear magnetic resonance (NMR), GC- and LC-MS), leading to the conclusion that all platforms were able to detect compositional differences despite some platform-dependent differences.
Community-Based Guidelines in Metabolomics
Community guidelines on how to report and perform metabolomics workflows form the basis of standardization. The metabolomics standardization initiative (MSI) of the metabolomics society91 has worked intensively on definitions and guidelines considering all steps of the targeted and nontargeted analytical process for many years. This includes defining the analytical task, sampling/analysis of data standards, data evaluation, and reporting.17,93
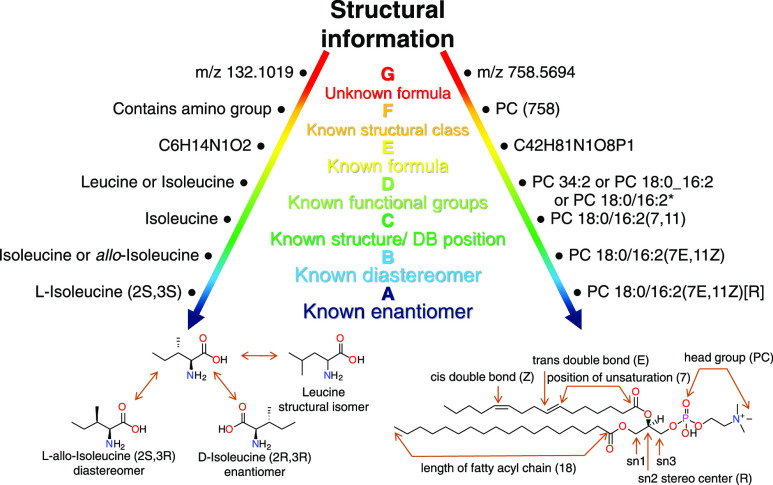
The metabolomics community is currently, revisiting the standards of metabolite reporting by the state of the art level of confidence scala94 (1–3) introducing new subclasses (A–F) for unambiguous metabolite identification such as cis/trans configuration information. In October 2020, a new guideline on lipid classification, nomenclature, and shorthand notation was published95 including major changes for the annotation of double bond equivalents and the number of oxygens as well as newly delineated oxygenated lipid species. Figure 5 shows the metabolite and lipid identification ranking according to the newly proposed guidelines of the metabolomics community.
Figure 5.
Metabolite (left) and lipid (right) identification according to the proposed guidelines of the metabolomics society (A–G) using the examples of leucine and a PC 18:0/16:2(7E,11Z)[R]. The lowest annotation level corresponds to known accurate mass information (G) followed by a known compound class (F), known compound sum formula (E), known functional moieties (D), known structure (isoleucine)/double bond position (PC 18:0/16:2(7,11) (C), known diastereomer (B), and the highest level to enantiomer-specific identification (A). *in lipidomics105 3 intermediate steps are distinguished at level D: sum of carbon and double bond number for all fatty acyl chains (PC 34:2)/known distribution (PC 18:0_16:2) and known position of the fatty acyl chains (PC 18:0/16:2).
Updated metabolomics repositories such as MetaboLights103 provide openness and transparency of reported data sets. These repositories will be essential for developing of community-based benchmark materials and will facilitate the development of accepted guidelines.
Instrument-dependent compound identification workflows complicate cross-platform evaluations and call for harmonization of reference libraries. A recent European interlaboratory study published harmonization guidelines for acquisition and processing of tandem MS data. Interestingly, they also revealed that under certain collision energies time of flights ((TOF)s) and Orbitrap fragmentation spectra are comparable.104
Quality Control and Benchmarking
QC and normalization strategies are essential for successful large-scale studies. Normalization can be performed by QC samples and data-driven or via ISTDs and is extensively summarized elsewhere.106−109 In large-scale metabolomics and lipidomics studies, the concept of a pooled sample for QC has gained worldwide acceptance, also allowing us to correct for intra- and interbatch variations and to accomplish MS/MS measurements required for annotation.109,110 However, the production of sufficient amounts of pooled samples can be problematic for multicenter studies in clinical metabolomics. Additionally, if only one sample pool including all sample groups is produced, dilution effects can mask low abundant metabolite signals. The production of QCs for each group represents an alternative; however, in some cases preparing a pooled sample is simply impossible. For example, in many large-scale investigations such as longitudinal clinical studies or population profiling, all samples are not available at the beginning of the analysis. Alternatively, multistandard mixes of metabolites and/or lipids are established reference samples, which can be either produced user-defined in the lab or ordered as commercially available stocks, e.g. LSMLS or MSMLS (from IROA) including 400 metabolites each 1 mg in well plates or 600 metabolites each 5 μg per well plate. Lipid-specific kits are also offered, e.g. AbsoluteIDQ (from Biocrates) including 180 or 400 lipids. More recently, lipid mixes with matrix-specific concentrations are commercially available e.g. SPLASH LIPIDOMIX (Avanti) products which include one deuterated ISTD of all major lipid classes at ratios relative to human plasma. Another possibility is to take deuterated standards from the UltimateSPLASH (Avanti) panel from different lipid classes to prepare a customized lipid mix. These valuable standard panels offer reference materials for streamlined validation protocols and accelerate harmonization. However, it should be emphasized that harmonization efforts enabled by reference standard mixes and kit-type of analysis will not replace certified reference materials which are fully traceable. Recently, the concept of a cheap and easily accessible biological benchmark material was proposed for metabolomics and lipidomics. The idea was resumed from proteomics, where HeLa cell extracts have become the gold standard for benchmarking instrument performance and proof-of-principle experiments upon introduction of new analytical methods.111−115 Yeast ethanolic extracts with a characterized metabolome, not only enabled testing for the chemical space and coverage upon method implementation and developments but also enabled in-house routines for instrumental performance tests with additional potential for batch to batch corrections in large scale nontargeted metabolomics studies. The benchmark material is obtained from P. pastoris from fully controlled fermentations, which can be easily reproduced in a lab with fermentor access.85 Additionally, these extracts are also commercially available in both endogenous and 13C-labeled formate. An open-source yeast metabolite and lipid library is established for the material. All reported compounds were reported in the human metabolome data base, showing once more that yeast is a cost-effective benchmark material for human metabolomics. 104 out of 206 metabolites were stable for several years when stored in aliquots at −80 °C.85
Nontargeted Data Analysis—Increasing Quality by Multiple Lines of Evidence
Nontargeted metabolomics workflows consist of key steps that need to be addressed individually with regard to standardization. The first step of a nontargeted experiment involves the analytical process aspects, discussed in several reviews.1,116,117 Data analysis constitutes the most time consuming and complex step of nontargeted experiments. Many tools and approaches are available for this process and have been summarized extensively.107,118−120 More specifically, data analysis follows stepwise data preprocessing, features table processing, statistical analysis (feature prioritization and biomarker elucidation), annotation, and biological contextualization like pathway mapping and integration with other omics data, all of which (with the exception of statistical analysis) are discussed in the following. We will emphasize the multiple strategies of corroborating nontargeted read-outs and deliberately focus on different aspects that improve quality.
Data Preprocessing
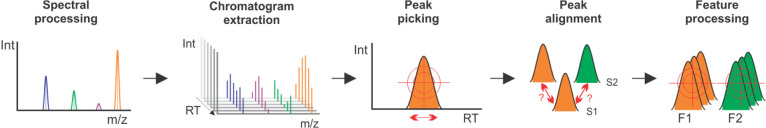
Data preprocessing (DPP) presents the first major challenge in nontargeted metabolomics since it facilitates the translation of raw data into the less complex format of so-called feature tables. While approaches enabling metabolomics DPP keep being improved, the general steps have remained unchanged across different tools (Figure 6) (with very few exceptions as in ref (121)). However, despite this fact and the development of different DPP parameter optimization tools122−124 it often suffers from extensive problems. Those include false negative and false positive reports of ion species as well as wrongly reported abundance values and other issues.125−128 It should be noted that data pre-processing is not challenging because it is hard to perform, but because it is hard to perform well. This point was laid out by Sindelar et al., who demonstrated why poor performance of data preprocessing could lead to much harder downstream data analysis.129 It is therefore essential to control the effectiveness of this process.
Figure 6.
General steps of nontargeted data preprocessing.
There are a number of advances we would like to highlight in this context. One recent R package, named patRoon, combines different data preprocessing and annotation algorithms into a single framework and thereby allows us to build pipelines in the R-environment.130 This increases flexibility in data processing choices considerably since it allows us to combine the strengths of many different tools and to compare them more easily. It is worth noting that patRoon supports any HRMS platform and supports algorithms from many widely used tools such as ProteoWizard,131 XCMS.132 Two other tools which should be noted here are NeatMS133 and MetaClean134, which are based on deep learning and machine learning, respectively. Both tools allow us to comprehensively assess the peak picking quality as conducted via different tools for experimental datasets.133 To the best of our knowledge these recently published works represent the only available tools to comprehensively assess peak picking quality for all picked peaks, which poses a significant advancement. However, RT alignment and false negatives are not considered in this approach which makes further development necessary. To address this need mzRAPP was introduced, a tool enabling reliability assessment of different nontargeted data preprocessing steps (under submission). It is based on automatically validated and extended benchmarks (starting from user supplied integration boundaries per molecular formula) and allows us to derive different performance metrics including the proportion of false negatives, affected isotopic ratios, and the number of alignment errors for nontargeted DPP of any experimental datasets. It is worth noting that the use of benchmark datasets in this context enables us to investigate the number of false negative peaks as they provide a so called “ground truth” as a reference point. While this also offers several other apparent advantages for the benchmarking of different DPP tools,135 benchmark datasets in metabolomics come with significant problems. First, their curation process requires extensive manual work and is hugely time intensive (although some do exist; e.g., ref (136)). This, in turn, implies that it is impractical to create benchmarks for different types of datasets (e.g. sample complexities or choices in instrumentation or acquisition mode such as RP-LC, HILIC, orbitrap MS, TOF MS) which might imply different needs for applied DPP software. Secondly, it can be problematic to consider benchmark datasets as “ground truth” without sufficient validation. mzRAPP tackles this problem by automatically applying a number of validation metrics to check the consistency of user supplied benchmark candidates.
Elimination of Redundancies and Noise from Feature Tables
As discussed, nontargeted data preprocessing of LC-HRMS data generally leads to aligned feature tables. Ideally, (when bioinformatic noise is not considered) rows in those feature tables correspond to chromatographic peaks with specific mz@RT values in different samples. Hence, each mz@RT value ideally reflects an ion species originating from a sample molecule eluting from the chromatographic dimension and being ionized in the electrospray. However, preprocessing workflows typically introduce significant numbers of bioinformatics noise-features into data sets. In this context, we would like to highlight three recently published tools allowing us to remove those noise features from datasets. MetProc allows us to remove features based on missing value structures in QC samples.137 Another tool called genuMet is solely relying on injection order to identify false positive features without relying on measured QC samples.138 Finally MS-CleanR has been added to the MS-DIAL139 workflow, allowing us to (among other things not discussed here) filter features based on blank signals, background drifts, unusual mass decimals and relative standard deviations (RSDs).140 Since all of those tools offer slightly different approaches, their compatibility for different data sets remains to be elucidated.
Over the last years, many papers and authors have discussed the challenge that the number of reported ion-species cannot be directly translated to the number of sample molecules.11 This is due to bioinformatic noise and because one molecule will form multiple ion species due to the presence of different isotopologues and adducts. It has been reported that a single metabolite can lead to more than 100 different ion species during the ionization process.141 More recently, it was also shown that adduct species differ significantly in HILIC compared to RP chromatography.142 The same work also highlighted the problem of in-source fragmentation, which poses a significant risk for wrong annotation.
Over the years a number of approaches have been developed to group those different ion-species in order to eliminate redundancies or even gain additional reliability for annotations. Many tools enabling this and other important data analysis steps are summarized elsewhere.120 An interesting experimental approach which has been shown to allow improved and simplified annotation of adducts has been to measure samples twice with different LC-MS buffer compositions (14NH3–acetate and 15NH3–formate buffer)77 (in fact, this approach has also been used by ref (142)). In both conducted studies this approach showed great potential for annotating adducts and eliminating noise. Unlike credentialing approaches74,143 this workflow is applicable to any samples even if it cannot be labeled via stable isotopes. However, as it requires two measurements for each sample, it dramatically increases measurement time and might not be applicable to small sample volumes. Nevertheless, this approach presents significant improvement in increased control over noise reduction and adduct annotation.
The Annotation Task
In metabolomics the term annotation refers to the assignment of molecular information to features. This information can involve details on contributing atoms (molecular formula e.g. C6H12O6), structural class (e.g. steroid), atomic connections (e.g. phenylalanine), relative stereochemistry (e.g. leucine or isoleucine) or chirality (e.g. d-leucine). Different approaches allow to collect evidence for the affiliation of a feature on any of those levels. In fact the Metabolite Identification Task Group of the Metabolomics Society has proposed reporting standards for different levels of identification depending on the nature of collected evidence (Figure 5 shows the proposed metabolite annotation). While those standards are defining specific types of evidence which have to be collected for a level to be reached (e.g. matching of acquired MS/MS scans against a mass spectral library), there are no consensus criteria for the necessary strength of collected evidence (e.g. what constitutes a valid spectral match). In this context one of the most discussed topics is the adaptation of a false discovery rate (FDR) for spectral matching as it is routinely applied in the proteomics field. Over recent years a range of different strategies allowing us to apply this idea also in metabolomics has been proposed or implemented.18,144−147 While their actual application is still scarce, they definitely pose a step toward increased reliability of annotations. Another point, which needs to be considered in this regard is the nature of reference spectra used for spectral matching. Until now, matching against experimental spectral libraries has been considered the gold standard for this kind of approach. Although spectral libraries have been growing to impressive sizes (e.g. recently METLIN reached more than 850 K standard spectra),148 a recent evaluation on available reference spectra from authentic chemical standards149 regarding the coverage of different MS spectral libraries in different genome scale metabolic models (GSMs) revealed that on average only <40% of metabolites in the models are represented. Meanwhile, in silico approaches MetFrag150 (a combinatorial fragmenter) and machine learning based methods such as CFM-ID151 (an in-silico fragmenter) and CSI:FingerID152 (a structure predictor) are more and more accepted. This is mainly due to their increased coverage of the molecular space since they do not rely on experimental fragmentation data but molecular structure databases such as PubChem.153 Indeed some of those can even go beyond that (e.g. in combination with tools like EMMF154).
The advantage is evident since such structure databases are many orders of magnitude larger than any spectral library. Indeed, this might lead to an improved FDR when using this kind of approach as compared to matching against a spectral library with less metabolic coverage. Another strategy worth mentioning involves the support of annotations utilizing reactivities of specific functional groups. Briefly, this involves the specific derivatization of functional groups (such as amines, carboxylic acids, alcohols, etc.) commonly referred to as sub-omes.155 Derivatization improves overall ionization efficiency and enables selective separation and enrichment using reversed-phase stationary phases. Moreover, the production of sample specific ISTDs is facilitated. Blends of sample derivatized with isotopically labeled reagent or unlabeled reagent, respectively, served for relative as well as absolute quantification.156 This also enables credentialing-type approaches (as discussed above).157 As a drawback, these approaches take considerable effort in terms of data analysis. Dedicated RT and spectral libraries for identification of derivatized molecules (available for some derivatization strategies such as dansylation158) are required. It should be noted that derivatization approaches reduce throughput and require dedicated validation, due to challenges arising from matrix effects and decreased stability.159 Hence, derivatization strategies can potentially bring many advantages, but require an extensive amount of work in order for validation and method development.
H/D exchange on the other hand is more straightforward in its application and can be included into existing data evaluation pipelines. Recently, there have been significant advancements in infrastructure for this type of analysis. For example, the software MetFrag supports H/D exchange data.160 Although H/D exchange only allows us to investigate acidic moieties, its potential for annotation has been shown in multiple studies.161,162
In cases where the annotation strategies discussed above fail to deliver the desired insight, novel approaches based upon complex bioinformatics algorithms fill the void. These innovations utilize molecular networking of fragmentation spectra (spectral similarity translated to biochemical and chemical substructures) or machine learning algorithms. In this context MS2LDA, which was initially published in 2016,163 associates specific fragments and/or neutral losses with chemical moieties and, thereby inspecting complex structural relationships between different unknown analytes. This algorithm has been further developed to now directly enable differential analysis of chemical substructures between different samples (such as investigations on the regulation of xenobiotic derivatives across different samples.164 More recently also feature based molecular networking, allowing us to consider the chromatographic and/or ion mobility dimension in this type of analysis, has been introduced.165 This way, isomers and in-source fragments can potentially be investigated. Another tool we wish to highlight here is CANOPUS166, which classifies features via their MS/MS spectra even when existing spectral libraries do not include MS/MS scans of the class in question.
Annotation in the Field of Lipidomics
The general annotation strategies applied for metabolomics are often not applicable in nontargeted lipidomics. This is reflected in a survey among lipidomics researchers from 2018167 revealing that 60% of all researchers rely mostly on manual (visual) annotation. Even though software tools are available and commonly applied (e.g. LDA;168 MS-DIAL,169 LIFS software tools170), manual annotation remains an integral part of lipid annotation highlighting the lack of adequate nontargeted analysis tools in lipidomics. Most available software tools are based on two approaches: library matching (MS-DIAL, LipidSearch, Greazy, LipidDex, etc.) and decision rule-based identification (LDA, LipidXplorer, LipidMatch, LipidHunter, etc.). Due to building blocks of lipids leading to a distinct MS2 pattern within the same class, decision rule sets based on well-defined fragments (fragment rules) and their intensity relationships (intensity rules) can be described for specific lipid classes.171,172 For library matching similar principles are applied as in standard metabolomics workflows using accurate mass, MS2 spectra, and scoring algorithm. Both experimental or in-silico databases are applied in lipidomics. Unfortunately, false discovery rate calculation is not possible up to now and a certain level of false assignments is state of the art in nontargeted lipidomics. Hence, it is of utmost importance to reliably estimate the proportion of potentially false assignments. Filtering of false positive annotations can be done by relative RT; the homologous lipid series of the same class depends on relative carbon number and/or relative double bond number.173 Using regressions models the so-called equivalent carbon number (ECN) model can be applied for manual annotation174 or RT prediction175 in order to exclude false positive hits and confirm lipids. Additionally, Kendrick mass plots can be used to identify homologous series in lipid data sets.176 The application of Bayesian statistics presents an interesting and promising direction and may overcome some limitations of hand-crafted rule sets.177 Excellent community-based resources provide guidelines (see ILS, LSI92) on criteria and characteristic fragments for MS/MS annotation. The LIPID MAPS178 and LSI website list continuously update information on manual inspection of MS/MS data reporting on obligatory fragment ions for unambiguous annotation of lipids. Still, only the minimum requirements have been defined (see ILS and LSI) so that openness and transparency of reported datasets remains inevitable to bring harmonization in lipidomics to the next level. As nontargeted lipidomics remains error-prone and still requires expert knowledge, comprehensive information on lipid annotations is essential. The periodicity of lipids offers further control points in lipid identification. In our opinion, lipidomics and metabolomics annotation have to be harmonized and is already possible using the proposed identification levels by the metabolomics society (Figure 5).
Retention Time and Cross Section as Orthogonal Parameter in Nontargeted Analysis
Retention Time for Annotation
Orthogonal data as chromatographic RTs are key to increase the confidence of MS-based compound annotations. So far, the poor reproducibility and commutability of experimental retention times across labs even when using reversed-phase chromatography only precluded the wide adoption of RT libraries179 for high quality annotation across labs. RT prediction from molecular structures is a currently very active area of research. The most relevant developments are summarized elsewhere.180 The most recent advances not covered in the review are provided by the software tools Retip181 and QSRR automator.182 Retip is a machine learning based tool which has been trained using more than 800 standard compounds for each, RP and HILIC chromatography. Retip was integrated into the MS-DIAL tool-box. QSRR automator has been published as a Python package and builds RT prediction models for in-house chromatographic methods.182 It is worth noting that RT prediction is not (yet) accurate enough to enable accurate identification of small molecules. However, it can be applied for the annotation of (miss-)annotated in-source fragments and allows reranking of positional isomers which can provide valuable insights.181
Collision Cross Section Value for Compound Annotation
The role of collision cross sections (CCS) obtained from ion mobility spectrometry (IMS) for confident compound annotation has been extensively discussed.183−186 The pace of generating CCS databases (both experimental and in-silico predicted) has been enormous.187−189 Currently there are two unified databases, CCS Compendium186 and AllCCS.190 Novel open-source software tools facilitate data evaluation.131,169
Seminal studies showed that interlaboratory reproducibility of CCS assessment outperforms191,192 reproducibility of chromatographic RTs. As a drawback, a CCS value correlates with the measured accurate mass of a molecule, while chromatographic retention offers an entirely orthogonal identifier. Due to the current limitations in ion mobility resolution, isomer separation of small primary metabolites is limited. In complex samples, only molecules exhibiting CCS differences in the low % range (typically 3%) are routinely resolved. The resolution is improved by novel advanced instrumental concepts.193,194 Recently, the potential of trapped IMS (TIMS) to separate lipid isomers was shown.195 The obtained resolving power allowed us to discriminate lipid species exhibiting CCS differences of <1% in complex biological mixtures. Several studies implemented ion mobility for structurally characterizing lipids with a high degree of specificity. Information on double bond position and geometry was obtained combining IMS with ozonolysis and Paternò–Büchi reaction.196,197
Navigating the Conflicting Goals of Metabolome Coverage and Throughput
Up to date metabolome analysis is the best fit for purpose compromise between coverage, selectivity, and throughput. High coverage implies a wide interrogation window with regard to both the chemical molecular dimension and the metabolite abundance dimension (8 orders of magnitude concentration difference). Major application areas of metabolomics such as, e.g., precision medicine envisage the measurement of large cohorts (thousands of samples) in regulated environments. The current transitory phase from small scale experiments to large scale studies, industry- and clinical applications, triggers exciting developments regarding streamlined workflows and tailored solutions with advanced throughput. As the field moves forward, economic considerations regarding cost effectiveness and automation of the complete workflow become more important. Miniaturization accommodates the analysis of small precious samples, bears the potential of increasing sensitivity, and reduces solvent consumption following the principle of green chemistry. We will discuss key aspects of current developments—from sample preparation to analysis, advancing automation, miniaturization, and throughput—and discuss the methods with regard to coverage and selectivity.
Sample Preparation
High-throughput sample preparation is still a bottleneck preventing exploitation of the full potential of high-throughput MS-based metabolomics. A recent review discusses the need for high-throughput technologies emphasizing the role of sample preparation.198 The state of the art of sample preparation strategies for all relevant sample matrices is comprehensively reviewed elsewhere.1 Protein precipitation upon dilution, liquid–liquid extraction, and solid phase extraction (SPE) are widely accepted methods in metabolomics and lipidomics analysis which can be adopted for robotic liquid handing systems (e.g. methyl tert-butyl ether (MTBE) extractions in lipidomics199). Further advancement of classical sample preparation strategies in metabolomics and lipidomics is driven by emerging application fields such as biotechnological large-scale enzyme activity screens and plate-based biomarker or drug screening and includes the development of miniaturized green sample pre-treatment (e.g. micro-liquid–liquid extraction, low volume SPE), offering favorable extraction kinetics, high preconcentration rates, and increased throughput. For example, implementation of a commercially available, fully automated SPE system using small volume SPE cartridges achieved a duty cycle of less than 15 seconds per sample preparation.200 Automated nondispersive micro-liquid–liquid extraction allows high-throughput through parallelization. Dispersive micro-liquid–liquid extraction ameliorates extraction kinetics, but severe limitations regarding automation of phase separation have been reported.198,201 Currently, solid-phase microextraction (SPME) and electromembrane extraction methods are “re-explored” for metabolomics, given their potential for fully automated parallel extraction in well-plate formats and enrichment through miniaturization.202−204
SPME is a nondestructive and nonexhaustive extraction showing great promise in probing and extraction of “tiny” metabolomes. While multianalyte quantification remains a challenge, low invasiveness of SPME and the nonexhaustive nature of extraction, together with recently developed extractive phases, make the technique particularly attractive for time-resolved or spatially resolved metabolomics fingerprinting.202 For example, a high-throughput time-course metabolomic analysis was achieved through multiple extraction of 96-well-plate cell cultures.205 Direct immersion (DI) in vivo sampling enabled time-resolved metabolic fingerprinting of animal brains206,207 and a method for the analysis of small molecules from semi-solid tissue relying on DI-SPME and desorption electrospray ionization, (DESI)-MS, has been proposed, promising space-resolved analysis of tissues.208 Non-exhaustive in vivo extraction followed by GC X GC qTOFMS analysis enabled real-time monitoring of apple metabolism during the process of ripening on the tree. The slim geometry of the extraction device avoided tissue wounding and oxidative degradation of analytes seen with conventional workflows relying on harvesting, metabolism quenching, and ex vivo extraction.209 However, the current selection of commercially available DI-SPME extractive devices is very narrow, limiting the wide adoption of this technique.202
Electromembrane extraction is a combination of partitioning-based liquid–liquid extraction and electrophoresis. Fundamentals of electromembrane extraction have been summarized in a review by Douin et al.204 Analytes move from a donor phase, usually an aqueous sample, through a water-immiscible organic layer acting as purification filter, into an aqueous (or optionally organic) acceptor phase. Mass transfer is driven by an electric field introduced between donor and acceptor phase via insertion of electrodes and application of direct current in the milliampere range, which speeds up the extraction process and enhances extraction yield compared to simple partitioning-based extraction. For optimized systems, selective analyte enrichment up to 100-fold and recoveries up to 100%204 and excellent cleanup potential have been reported (salt- and protein-removal,210 phospholipid-removal211). The technique holds high potential for point of care analysis as enabled by parallelization and downscaling of analysis as well as implementation into microfluidic chips (e.g. Hansen et al.212). However, the extraction principle is inherently limited to ionizable molecule species and is not suited for molecules prone to degradation by electrolysis, and electrolysis phenomena are aggravated with decreasing acceptor volume. Plus, electromembrane extraction is a selective extraction procedure,204 preventing the full scope of wide coverage metabolomics. On the other hand, high selectivity towards target analytes is a desirable feature for specialized routine application in regulated environments as it facilitates process validation.
Direct Analysis in Metabolomics and Lipidomics
Flow Injection-MS
Direct analysis has its undisputed role as a rapid first-pass metabolic fingerprinting method. It comes with a reduced analysis time of 2–5 min, thereby increasing the analytical throughput by one order of magnitude compared to typical LC-MS-based metabolomics. A recent review gives an excellent summary on successful applications and well-known limitations imposed by matrix effects and the occurrence of isomers and in-source fragments.213 Ion suppression and ion competition were studied in fundamental experiments using injections of 5 μL at flow rates <100 μL min–1, where ion competition was shown to be a major cause for limited sensitivity in orbitrap MS.214 As a consequence, sensitivity could be increased by optimizing data acquisition. The use of sequential narrow mass segments in trapping MS with fixed m/z windows or variable sample specific windows showed to be valid strategies for improving sensitivity and linear dynamic range.214
A recent study combined FI-HRMS with online fractionation improving the metabolome coverage and reducing matrix effects.215 The fully automated sequential fractionation was based on solid-phase extraction on complementary ion-exchange and reversed-phase chemistries. Fast and high coverage screening (3 min per polarity) was thoroughly validated for targeted analysis of 50 diagnostic and explorative biomarkers in plasma samples, including amino acids, amines, purines, sugars, acylcarnitines, organic acids, and fatty acids. The sensitivity of FI was significantly improved. LLOQ values comparable to conventional LC-MS/MS were reported. FI-HRMS for quantification of high abundant cholesterol and cholesterylester utilizing compound specific response factors proved to be fit for the purpose for cultured cells, tissue homogenates, and serum samples.34
IMS offers a rapid (millisecond-regime) post-ionization separation dimension,216 which makes it particularly attractive for FI analysis. Its benefit for both targeted and nontargeted metabolomics has been investigated.217,218 Compared to FI-MS alone, FI-IMS-MS offers improved linearity and reduced noise level. Nonetheless, ionization suppression due to matrix effects remains a major obstacle with detrimental impact on sensitivity, peak capacity, and consequently, coverage.219 It is therefore unlikely that IMS will render chromatographic separations obsolete in nontargeted analysis.
Ultimate Throughput–Duty Cycles of Seconds Per Sample
The cycle time of the sample transfer to the MS limits the throughput of FI-MS-based metabolomics. For example, the fastest commercially available SPE system offers a sample cycle time of 10 seconds, limited by the required SPE elution volumes.200 When used without SPE, the rate limiting step becomes the autosampler, enabling a duty cycle of 2.5 seconds per sample,220 a setting which was proposed for drug discovery and high-throughput MS targeted assays.
Duty cycles of seconds per sample are also realized in alternative ambient MS approaches. However, despite significant progress, large scale metabolomics studies have not yet been put into practice. Excellent duty cycles in the second-regime were, for example, obtained by immediate drop on demand technology combined with open port sampling interfaces (I-DOT-OPSI-MS).221 Recent studies on single cell metabolomics demonstrate the power of high throughput MS. Another emerging high-throughput-technique enables nanoliter-scale infusion MS at sampling rates of up to 6 Hz installing plate robotic handling.222 Acoustic droplet ejection (ADE) uses acoustic pulses to generate nanoliter-droplets directly from a microtiter plate in a contactless manner with high speed, precision, and accuracy. The potential areas of future applications are evident and range from high-throughput drug screening assays, plate based synthetic chemistry, and large-scale biotechnological studies addressing enzyme kinetics. Interfacing ADE with MS involved (1) acoustic mist ionization (AMI) coupled to MS222 or (2) acoustic ejection MS (AEMS) using an open port interface (OPI) with electrospray ionization (ESI).223,224 While the first approach integrated droplet generation and ionization, the latter configuration used ADE only for sample delivery for subsequent ionization by ESI. This way, matrix effects and adverse effects caused by contamination of MS transfer capillaries were reduced. Excellent analytical figures of merit were obtained upon injection of 25,000 samples (standards) revealing excellent RSD of 8.5% for peak intensity and full width at half maximum (177 ms), respectively. The peak width was in the order of 200 ms.224
Miniaturization–Nanoflow Direct Infusion
Miniaturization of direct analysis toward nanoflow proved to be particularly attractive because of the inherent features of nanoESI. Ionization at this flow regime is characterized by increased ionization efficiency. At the same time, differences in ionization efficiency for different molecules are significantly reduced as compared to ESI at higher flow rates.225 Shotgun lipidomics accomplished by chip based nanoESI orbitrap MS have become an essential tool of the trade for both lipid identification and quantification.35,226 A 50 min analysis time consuming only 10 μL of sample solution is theoretically possible.227 In practice, a 5–15 min run time ensures analysis at both polarities while applying data dependent acquisition (DDA) or data independent acquisition (DIA) approaches. Today, MS2 methods based on DIA (covering the whole mass range in 1 Da steps228) prevail over DDA (follows the intensity order226). Dedicated software solutions allow for noise filtering accelerating data processing.229 Typically, several hundred lipids are identified on a species level covering the abundant lipid classes. Several strategies enable increased coverage, by e.g. including derivatization.230 Pitfalls regarding lipid identification are summarized and curated by the LSI.29 Quantification is achieved by ISTDs. The lipid head group determines the ionization efficiency to a large extent, allowing us to minimize the number of calibrators to one or a few per class. Response factor corrections were introduced for the quantification of neutral lipids.34 Quantification on the molecular species level is complicated as the required MS2 level, different fatty acyl chain moieties show different responses (up to 60%), jeopardizing accuracy without correction.231
Chromatography—Key Steps Toward Coverage and Throughput
Miniaturization of Liquid Chromatography
In MS-based metabolomics, microscale and nanoscale separations have been developed with the aim of advancing small scale sample analysis, increasing sensitivity and thus coverage of low abundant analytes, and finally reducing costs by overall reduced reagent consumption. Miniaturized separation used with tailored low-diameter ESI-emitters offers unrivaled absolute detection limits (fmol on column). Combinations with large volume injection and online enrichment allows the analysis of very low analyte concentrations and very efficient sample use. However, the successful application of online-enrichment-nano-RP-LC faces limitations: Numerous primary analytes show poor retention on RP-LC, and sample volumes may be extremely limited, as in single cell analysis. In such cases, the full sensitivity potential of nano-RP-LC-MS platforms may be exploited by analyte derivatization, increasing RP-retention and ionization efficiency. A comprehensive summary of theory, common approaches, and over 20 of the most recent applications of nano-LC-MS in metabolomics and lipidomics investigation can be found elsewhere.232 Single cell analysis is an emerging application of small-scale metabolomics by nano-LC-MS. Recently, Nakatani et al. reported a method for derivatization-free targeted quantification of hydrophilic metabolites in single HeLa cells. Living single cells were sampled from culture using an in-house developed nano-pipette device, and the sampling capillary was directly connected to a sample loop line. The optimized nano-LC-MS/MS method based on a self-packed RP-LC column (pentafluorophenylpropyl Discovery HSF5, 0.1 × 180 mm, 3 μm) and multiple reaction monitoring yielded an average sensitivity increase of 26-fold compared to a conventional flow setup (2.1 x 150 mm) employing the same column chemistry. 18 relatively abundant hydrophilic metabolites (16 amino acids and 2 nucleic acid related metabolites) were detected and quantified in 22 single HeLa cells. Clustering in different groups was observed.233
Another emerging nano-LC-MS application is the in-depth, high-coverage analysis of the lipidome. With a recently published 110 min nano-LC-MS method, linear dynamic range and sensitivity could be substantially increased by 1–2 and 2–3 orders of magnitude, respectively, when compared to conventional high-performance LC (HPLC) (150 × 2.1 mm, 2.7 μm). The proposed workflow displayed excellent analytical figures of merit after careful optimization of sample reconstitution. Lipidome coverage was evaluated for the phospholipidome of S. cerevisiae and achieved increased lipid identification (436 phospholipids) compared to conventional-flow HPLC and a shotgun approach. Low abundant lipid species and isomers could be detected even when they were coeluting.234 When combined to a new data evaluation pipe-line, almost 900 lipid species in 26 lipid classes in S. cerevisiae were identified. The identification rate was increased by a factor of 4 compared to previous whole yeast lipidome shotgun studies.89 The high potential of this workflow for in-depth lipidome analysis is highlighted due to the detection of less common lipid classes like monomethyl-PE (MMPE) and dimethyl-PE (DMPE) and lipids with incorporated odd-chain and diunsaturated fatty acids.
For a long time, the development and wide adoption of microscale separations in metabolomics suffered from the fact that many stationary phase chemistries were not commercialized for the required column dimensions (1.5–0.5 mm inner diameter for micro-LC; 0.5–0.15 mm I.D. for capillary LC235). Micro-LC separations are more common, since ionization performance of ESI sources is compromised at the flow regime of capillary-LC (0.01–0.001 mL min–1). The sensitivity gain of micro-LC is moderate as compared to microbore-LC (3.2–1.5 mm I.D.). In a recently published study, the optimized microflow-LC-MS/MS improved sensitivity in a compound-dependent manner by 6- to 49-fold when compared to conventional microbore-LC-MS/MS.236
In metabolomics, the sensitivity gain provided by micro-LC has been exploited to design rapid separations for metabolic phenotyping. Throughput has been optimized at the expense of chromatographic performance, providing fit for purpose platforms with enhanced but not maximized sensitivity upon miniaturization.237−241 A systematic comparison to conventional HPLC methods in terms of LOD and LLOQ was beyond the scope of these studies. Short micro-ultraperformance (UPLC) type of separation utilizing RP materials with sub-2-μm particles (100% wettable, 1.0 × 50 mm, 1.7 μm) and separation times of 2.5 min was successfully applied in large scale phenotyping studies237 and recently explored in combination with ion mobility.238,239 A rapid micro-HILIC method utilizing sub-2-μm particles (Acquity UPLC BEH-Amide, 1.0 × 50 mm, 1.7 μm) addressed the analysis of polar metabolites in rat urine in less than 3.5 min. Comparison to conventional HILIC-MS demonstrated 4-fold reduction of analysis time, 75% reduction in solvent consumption, and 18-fold reduction of sample consumption, while providing sufficient retention of polar metabolites (e.g. hexoses, methylhistidine, kynurenic acid, creatinine), excellent reproducibility (RT RSDs between 0.31 and 6.3% over 134 sample injections), and excellent run-to-run reproducibility.240 Rapid lipidomic profiling of plasma by micro-LC-IMS-MS proved to be fit for the purpose for clustering plasma lipotypes as assessed in breast cancer patients and healthy controls indicating the suitability of micro-LC–IMS–MS as a rapid platform for large scale lipidomics screening.241 The combination of online SPE enrichment with micro-LC-MS was validated for targeted analysis of 13 steroid hormones from human plasma. After careful optimization, large volume injections obtained excellent LOQs in the sub-ng/mL range at high throughput (below 3 min per sample). Validation according to FDA guidelines showed the suitability for high-throughput analysis in a clinical routine laboratory.242 Cebo et al. introduced a validated approach based on offline mixed-mode SPE enrichment and micro-UHPLC-ESI-triple quadruple (QqQ)-MS/MS for the quantification of 42 oxylipins in plasma and platelets at reasonable throughput (13 min per sample).243 Limits of detection were between 2 and 250 fmol on column, offering comparable LODs to well established conventional LC approaches, but at significantly reduced solvent consumption.
Multidimensional Chromatography
Comprehensive two-dimensional chromatography is undoubtedly a powerful strategy for the separation of complex mixtures.244,245 In order to maximize the separation space and thus the peak capacity, orthogonality and compatibility of the two dimensions is essential. Each chromatographic peak of the first dimension requires sampling several times, efficient transfer, and rapid separation by a second dimension, in order to maintain the chromatographic resolution of the first dimension. The time spans between successive separations in the second dimension should be minimized requiring short separation and equilibration times. This is well established in GC X GC but still a technical challenge in 2D-liquid-chromatography. The wide suite of successful GC X GC applications in metabolomics has been summarized elsewhere.1,245 In LC X LC, method development is still regarded as a bottleneck. The experimental design regarding the two separation dimensions is not straightforward, as separation conditions influence and restrict each other.244 Solvent incompatibility with regard to mismatch of elution strength and immiscibility requires dilution of the sample upon transfer to the second dimension. Typical 2D-LC-MS designs involve HILIC and RP-LC. Long microLC/microbore LC columns (flow rate 10–50 μL/min) are employed as the first dimension, followed by short, thick columns (flow rate in the mL/min regime) in the second dimension. In the last years many new instrumental 2D-LC designs have been developed for metabolomics/lipidomics applications facilitating flexibility and universality.246 Elaborate constructions allow versatile modulation (i.e. sample collection and transfer) including active modulation with dilution conditions optimized over the separation time. Despite significant progress, the theoretical peak capacity in LC X LC-MS is hardly reached in practise, due to incomplete usage of separation space, still suboptimal cutting, and peak deterioration upon remaining solvent incompatibility.
Adoption of comprehensive LC X LC-MS in metabolomics and lipidomics has been limited mainly because comprehensive 2D-LC-MS metabolomics approaches developed up to date have largely suffered from incomplete usage of separation space in HILIC and RP-LC combinations and from severe sensitivity loss.247 The latter diminishes actual coverage in nontargeted screenings. Solvent evaporation interfaces as featured in SFC X RP-LC lipidomics might overcome this challenge. Recent reports on 2D-SFC-RP-MS, separating 370 lipids from 10 lipid classes of human plasma within 38 min, are promising.248 2D-LC-MS relying on heart-cutting strategies proved to be powerful in selected applications246 including the separation of secondary metabolites in plants and emerging targeted chiral metabolomics.249,250
Dual/Parallel Chromatography
Several column switching approaches have been introduced as elegant solutions for increased throughput and coverage within one analytical run. One successful configuration (Figure 7A) integrated serial orthogonal chromatography in order to transfer the poorly retained metabolites of the first dimension onto a second orthogonal column and enable two parallel separations subsequently. This configuration offered a valuable alternative to heart-cut chromatography.246 A simple six-port valve was installed between the two chromatographic columns enabling to transfer metabolites eluting from the first column onto the second column. Two independent separations were carried out by the switching of the valve. This setting was successfully employed for high coverage metabolome251 and lipidome analysis.252 For metabolome analysis, reversed-phase and porous graphitized carbon LC were combined. The method was validated by targeted absolute quantification of 80 primary metabolites in P. pastoris. Excellent RT stability (average 0.4%) even in the presence of a biological matrix was obtained. An interplatform comparison with GC- and LC-tandem-MS analyses showed the power of the method even with respect to sugar phosphate isomer quantification.251 The same separation concept combined HILIC and RP-LC for high coverage lipidome analysis.253 The void volume of the HILIC separation containing non-polar lipids was transferred to the RP column which enabled the on-line combination of HILIC with RP without any dilution in the second dimension. Rapid consecutive separation for polar lipids and class specific separation for nonpolar lipids was accomplished within one analytical run of only 15 min (including re-equilibration time, using stationary phases with sub-2-μm particles and UPLC).
Figure 7.
Practical setup solutions for sequential and parallel LC. (A) In valve position A, the void volume of the first column is transferred to the second column. Afterward, the valve is switched in position B and the sample is analyzed on both columns parallel.251,252 (B) In valve position A, the first extract is injected on the first column and analyzed. Meanwhile, the second column is equilibrated and the mobile phase is flushed into waste. After separation on the first column, the valve is switched to position B and the second extract is injected on the second column and analyzed while the first column is equilibrated.253 (C) In valve position A, the sample is loaded and divided into two sample loops equally. In valve position B, both parts of the sample are injected onto two orthogonal columns and analyzed.254
Figure 7 summarizes options for successful column switching technologies. Dual chromatography extends the separation space by fully automated consecutive or parallel execution of orthogonal chromatographic separations. Different configurations enabled orthogonal dual HILIC/RP-LC separation by parallel injection of two extracts from one biological specimen (Figure 7B) or of one sample extract (Figure 7C). While the latter configuration was proposed for simultaneous analysis of nonpolar and polar metabolites,254 the parallel injection of different sample extracts facilitated the development of merged metabolomics/lipidomics.253 The HRMS workflow integrated biphasic extractions, parallel injection, separation, and MS analysis providing the full scope of targeted and nontargeted metabolomics and lipidomics within one analytical run. Wang et al. proposed a dual chromatography approach for simultaneous lipidomics and metabolomics analysis implementing parallel HILIC and RP separation in a heart cutting 2D-LC configuration where parallel analysis was preceded by prefractionation on a first separation dimension.255 As a drawback, this method precluded the integration of biphasic extractions, since only one sample could be analysed. Recently, sample preparation and reconstitution were reoptimized in order to provide high coverage within one measurement solution.256
MS Platforms and Data Acquisition Strategies—Improving Coverage, Selectivity, and Reliability
Despite significant progress, cutting edge low-resolution tandem-MS outperforms latest generation HRMS for quantitative analysis, both in terms of sensitivity and linear dynamic range. Large scale metabolomics studies are often performed on triple quadrupole-MS based platforms profiting from increased robustness and high quantitative capabilities.257 Today, the implementation of multiple reaction monitoring (MRM) approaches is supported by significant computational resources. A library containing MRM transitions for more than 15,500 molecules is publicly available.258 Both experimentally assessed and in-silico generated MRM transitions are included. Dedicated software tools enable optimization of MRM transitions including collision energies using mass spectral libraries, such as METLIN and HMDB.259 Large-scale metabolomics as enabled by QqQ-MS was successfully applied in wide-targeted assays providing absolute quantification of a high coverage metabolite panel.1,97 Recently, hybrid MS approaches emerged which offer attractive ways bridging the concepts of targeted and nontargeted analysis.20 These workflows successfully exploit the power for accurate relative quantification by QqQ-MS, without omitting a discovery step. The “discovery” is realized through optimizing the MRM transitions based on a sample matrix representative for the large-scale study. This optimization/discovery step can be performed using low mass resolution MS only or integrating high mass resolution. In MS/MS analysis by HRMS, sensitivity and selectivity (and thus the coverage) are significantly influenced by the type of mass spectrometer, but also by the selected data acquisition strategy. DIA and DDA acquisition modes have their specific applications in both targeted quantification and nontargeted compound annotation.260−262 For both DIA and DDA, new tools for online/on-the-fly and offline scan-level control, fragmentation, and acquisition optimization are available to support automated mass spectrometer parameter choice.262−264
Toward MS-Based Multi-omics
Emerging multi-omics analysis led to significant efforts for methods integrating multiple omics layers for one sample. Significant progress relates to the experimental design of multi-omics measurement and data evaluation strategies. Multi-omics applications profit from global phenotype metabolomics data acquired at reasonable throughput. Sophisticated sample preparation protocols together with high coverage multi-platforms characterize the tools of the trade for multi-omics analysis. Cutting edge network analysis enables us to integrate MS-based datasets with genome, transcriptome, proteome, and metabolome information derived from orthogonal platforms.
Multi-omics Sample Preparation Strategies
Multi-omics sample preparation approaches have to deal with the challenge that preferred collection methods, storage techniques, required quantity, and choice of biological samples are not directly transferable from one omics field to the other, especially when quantification rather than profiling has to be performed.265 The metabolomics part of multi-omics studies is especially challenging as degradation, oxidation, or conversion of metabolites (including lipids) might occur during sample preparation. Moreover, the procedures have to be tailored for the two sub-omes.9 In the multi-omics setting, discovery studies call for minimal pretreatment in order to prevent the potential loss of metabolites.266 Multi-omics sample preparation strategies based on a single sample enable the true combination of multi-molecular information without influences of different sample aliquots. One-phase extractions coined as sample preparation for multi-omics technologies (SPOT) were successfully applied for the parallel analysis of the proteome and metabolome.267 Other multi-omics strategies involve filter tips such as cellulose based filter tips, which enable detergent-free single-pot metabolomics and proteomics by capturing the protein fraction, collecting the metabolite flow-through and the peptide fraction after tryspin digestion.268 For lipid sub-ome integration, extraction strategies involve different solvent mixtures with higher organic content, e.g. chloroform/MeOH269 or MTBE/MeOH.199 Two-phase extractions such as chloroform/MeOH/water270 or MTBE/MeOH/water226,253,271,272 proved to be particularly successful for global high coverage analysis as metabolites and lipids can be analyzed from a single sample at the same time separating the protein fraction. In terms of sample handling and automatization potential, MTBE/MeOH/water is preferable as the protein pellet is found at the vial bottom after phase induction and is not present in the intermediate phase between the polar and nonpolar phase. Such phase separation strategies reduce the sample amount needed and pave the way for direct-infusion and LC-MS based multi-omics (metabolomics, lipidomics, and proteomics) from a single sample.226,273 Additional lipidome coverage can be achieved using approaches such as (1) 3-phase extraction separating neutral lipids in the upper phase and glycerophospholipids in the middle organic phase274 or (2) the combination of two-phase liquid extraction with MTBE combined with SPE.271 In order to understand metabolite action on the subcellular level, experimental resolution of subcellular metabolism is needed. Recently, non-aqueous fractionation was successfully applied to resolve subcellular plant metabolism and the corresponding proteome.275 In this comprehensive approach, organic solvent mixtures and ultracentrifugation was applied to analyze proteins and primary metabolite in a four-compartment plant model comprising chloroplasts, cytosol, vacuole, and mitochondria.275 However, additional steps such as various liquid phases or subcellular analysis increase the sample preparation significantly so that scientists have to decide in each case at what time expense they aim to increase metabolite coverage.
Multiplatform Analysis Strategies for Metabolomics and Lipidomics
High coverage analysis in MS-based metabolomics is only achieved through multiplatform solutions involving orthogonal chromatographic separations integrating both HRMS and MS/MS methods. Despite significant progress in HRMS and its quantification capabilities, up-to-date tandem MS is the method of choice when aiming at the analysis of low abundant metabolites (e.g. bile acids or fatty acyl-CoA esters). Hydrophilic primary metabolites—the biological definition of metabolites involved in growth, development, and reproduction—are among the most evolutionarily conserved biomolecules. Multiple isomers and isomeric in-source fragments intrinsically challenge MS analysis.276 Examples are hexose phosphates, pentose phosphates, 3-phosphoglycerate/2-phosphoglycerat, citrate/isocitrate, homoserin/threonine, leucine/isoleucine, adenosine monophosphate/deoxyguanosine monophosphate, adenosine triphosphate/deoxyguanosine triphosphate, and alanine/sarcosine. As a consequence, common multiplatform workflows include chromatographic separations providing selectivity for primary metabolites, a prerequisite for accuracy regarding both identification and quantification, respectively. In LC, ion-pairing and HILIC are the methods of choice separating water-soluble central metabolites. A currently accepted protocol of wide targeted analysis relies on RP ion-pairing MS/MS analysis. The method covers 215 metabolites including amino acids, citric acid cycle intermediates, and other carboxylic acids, nucleobases, nucleosides, phosphosugars, and fatty acids.277 Sugar phosphate isomers are quantified based on distinct MS/MS fragmentation pattern278 (as no base-line separation is provided). There are several drawbacks associated with the use of ion-pairing reagents, such as MS system contamination, ion suppression effects limiting overall sensitivity, together with the fact that metabolites ionizing only in positive mode such as e.g. carnitines and S-adenosylmethionine cannot be measured. Generally, the use of ion-pairing reagents implies the establishment of a dedicated MS systems, often precluding the combination with HRMS. For HILIC separations, two stationary phases, i.e. the BEH amid phase and the polymeric zwitterionic phase,279 prevail, using both acidic and neutral/basic eluents. HILIC separations are versatile, but as a drawback there is no single experimental setting covering all relevant primary metabolites.280 When optimized properly the separation selectivity of phosphorylated carbohydrates is comparable to ion-pairing chromatography. Thus, GC remains the unrivaled separation method when addressing intermediates of glycolysis and pentose phosphate pathways. Wide coverage of the primary metabolome is established upon two-step derivatization procedures (ethoximation/methoximation followed by trimethylsilylation). Routine applications involve robotic just-in time derivatization. However, GC is not suitable for measuring the energy status of a given sample, as important nucleotides and cofactors are not covered. For this purpose, both ion-pairing281 chromatography or HILIC can be applied after careful consideration of sample preparation.280 Up to date, despite excellent separation power for polar and ionic metabolites, the application of capillary electrophoresis (CE) in metabolomics is limited to expert laboratories. The most recent CE developments were comprehensively summarized elsewhere.1,198 Examples of multiplatform combinations practice integrating the analysis of different extracts (tailored preparation for sub-ome analysis) followed by nontargeted assays (RP-LC-HRMS for lipidomics, GC-HRMS for primary metabolites,280 complementary HILIC-HRMS for metabolites not amenable to GC-MS, and targeted tandem mass spectrometric assays for low abundant metabolites such as bile acids,282 steroids, and oxylipins).283 Alternatively, the number of MS platforms can be reduced by replacing the combination of GC-HRMS/LC-HRMS by two complementary LC-HRMS methods (either two HILIC methods with acidic and basic eluents/positive and negative ionization or the combination of acidic RP-LC-HRMS and basic HILIC-HRMS).
In lipidomics, the majority of lipid classes can be covered by state of the art profiling approaches such as direct infusion MS or RP-MS. Multiplatform combinations in lipidomics often involve shotgun lipidomics for bulk lipid analysis in combination with LC-MS strategies enlarging the lipidome in terms of lower abundant lipids as recently shown for the platelet lipidome.284 An excellent review by Rustam and Reid summarizes the analytical challenges and advances in lipidomics including common MS-based review, chromatographic solutions, and possible combinations.285 The major challenge remains measuring high-abundant membrane lipids besides very low-abundant signal molecules across a huge polarity range (log P value from 5–35). Low concentrated lipid mediators are an important subclass often analyzed by RP-LC to cover different eicosanoide isomers.286 Sphingolipids analysis is of high interest as they are involved in signaling and protein sorting and are often suppressed by other membrane lipids.287 Extraction and analysis of glycosphingolipids subclasses such as gangliosides or sulfatides is challenging, and potential strategies are summarized in a recent workflow by Barrientos et al.288 If specific lipid sub-omes (e.g. sterols or prenols) are of interest, LIPID MAPS provides methods and protocols for LC-MS and GC-MS based analysis as starting points including a summary of available analytical standards (see resources section).178
Merging Metabolomics and Lipidomics
Monitoring the metabolic phenotype should always consider lipids due to their critical function in health and disease. Lipids make up 79% (90,678 lipids in 114,126 metabolites) of all listed metabolites in the HMDB 4.0 (accessed October 2020)88 highlighting the need to cover them in the analysis workflows. Especially when it comes to biomarker research the metabolome including the lipidome should be monitored to follow disease relevant changes as shown in recent studies on cancer prediction289,290 or cardiovascular disease.291 Cajka and Fiehn provide an excellent overview on the challenges and opportunities of merged metabolomics and lipidomics workflows.9 Here, we want to emphasize that global metabolite and lipid profiling in one analytical run is possible shown by two-phase MTBE extraction and fully automated parallel HILIC chromatography for metabolites and RP chromatography for lipids.253 The instrumental setup was realized by a HRMS, a dual-injection autosampler, and two positional 6-ports enabling simultaneous lipid and metabolite analysis in one analytical run of 32 min (Figure7B). Untargeted screening of human plasma samples resulted in >100 metabolite (organic acids, amino acids, nucleotides, acyl carnitines) and >380 lipid (phospholipids, sphingolipids, cholesteryl esters, di-and triglycerides). Stable-isotope labeled metabolites and lipids from yeast extracts further enabled us to merge targeted and nontargeted identification and are generally possible when labeled biomass is utilized. Moreover, LC-MS based proteomics, metabolomics, and lipidomics can be performed from a single sample which provides the starting-point for interesting multi-omics studies based on network analysis, e.g. protein-metabolite interactions in mesenchymal stem cell adipogenesis.283
Network Analysis and Visualization of Multi-omics Workflows
Multi-omics derived data sets including different sub-omes rely on appropriate data integration originating from several layers of information. The ultimate aim is to understand “the flow of information” underlying a certain phenotype. Here, we will consider relevant solutions—a selection of tools can be found in Table 2, which emerged to satisfy the need to facilitate downstream analysis of metabolomics data and generate or validate an underlying biological hypothesis. Visualization tools play a crucial role for biological interpretation of metabolomics data, and well-covered overviews can be found in several recent reviews.292−294 Such tools are required at the end of a metabolomics workflow pipeline and assume successful tackling of steps prior in the pipeline292,295 including construction of adept study design, biological experiment, sample preparation, identification, quantification, assessing QC standards, adjustment to batch effects, etc. The aspects and critical points of a metabolomics experiment from study design and sample preparation to data analysis and evaluation of various tools have been discussed in a comprehensive review.293
Table 2. Selected Tools for Data Analysis and Visualization, Metabolic Networking, and Databases.
| Name of the tool | Literature | Innovation |
|---|---|---|
| MetaboAnalyst | (Chong et al., 2019)328 | One-in-all metabolomics data analysis tool collection. |
| MetExplore | (Chazalviel et al., 2018; Cottret et al., 2018)303,304 | Visualization of metabolic networks and pathways, facilitates the analysis of omics data in biochemical context and pathway enrichment. |
| KEGG | (Kanehisa et al., 2017)329 | “Encyclopedia of genes and genomes”. Several model organisms. KEGG orthology for genes and proteins. |
| Reactome | (Bohler et al., 2016; Fabregat et al., 2018)309,330 | Knowledge base of biomolecular pathways: free, open-source, open-data, curated and peer-reviewed. |
| Cyc databases | (Caspi et al., 2020)311 | The “largest curated collection of metabolic pathways”. Many different model organisms. |
| Virtual Metabolic Human database | (Noronha et al., 2017) (Noronha et al., 2019)310,314 | Human and gut microbiome metabolism, 255 diseases, and also microbial genes, microbes. |
| WikiPathways | (Slenter et al., 2018)312 | Browsable, editable database curated by the research community |
| Chemical Similarity Enrichment Analysis (ChemRICH) | (Barupal and Fiehn, 2017)306 | Alternative to biochemical pathway mapping for metabolomic datasets. Not based on biochemistry directly but on structural similarity. The enrichment test is Kolmogorov–Smirnov test based (not hypergeometric test or Fisher exact test). |
| Metabox | (Wanichthanarak et al., 2017)308 | Metabolomics data analysis and interpretation toolbox for integration of proteomics and transcriptomics data. |
| Metscape | (Gao et al., 2010; Karnovsky et al., 2012)322,323 | Cytoscape plugin, metabolomics correlation networks and KEGG-based metabolic networks integrating gene expression and metabolomics. |
| PathBank | (Wishart et al., 2020)313 | Comprehensive, interactive database for metabolic pathways in 10 different model organisms. |
| OmicsNet | (Zhou and Xia, 2018)318 | Multi-omics data integration, biological networks (genes, proteins, microRNAs, transcription factors, metabolites) |
| GEM-Vis | (Buchweitz et al., 2020)324 | Visualization of time-course metabolomic data within the context of metabolic network maps. |
| FEMTO | (Nägele et al. 2016)302 | Integration of metabolomic time-series analysis and network information. |
Among the commonly applied strategies are uni- and multivariate statistical methods. Multivariate methods involve both several unsupervised methods like principal component analysis (PCA) or hierarchical clustering (HC) as well as supervised methods (partial least squares discriminant analysis (PLS-DA), orthogonal projections to latent structures discriminant analysis (oPLS-DA), (linear) discriminant analysis ((L)DA), (canonical) correspondence analysis (C)DA, random forests (RF), support vector machines (SVM), neural networks (NN), and feature selection strategies (recursive feature elimination, genetic algorithms, sparse models (Lasso, Elastic Net, sparse PLS)). Such methods are capable of capturing and pinpointing the unique metabolic fingerprints related to the underlying phenotype.119 While this is no concern with unsupervised techniques, a particular issue with supervised methods is the risk of overfitting to the labeled data.296 However, accompanying cross-validation can help avoid this issue.297
A powerful approach beyond the realm of statistics and machine learning algorithms is pathway analysis, which is taking advantage of established biological knowledge. In the simplest variation, metabolites of interest derived from a metabolomics experiment can be mapped on the pathways defined in a particular library. In the case of over-representation analysis (ORA), corresponding p-values can be obtained based on the metabolites list for the affected pathways. Extending the metabolite list with quantitative information (fold-change, intensity, and absolute amounts) can be further exploited by metabolite set enrichment analysis (MSEA). Even beyond this, considering the position of affected metabolites within pathways informs about the perturbation of the pathway and is a useful additional metric next to the enrichment, as it is capable of identifying subtle but consistent changes, whereby affected metabolites are ranked based on centrality measures thus contributing more to perturbation. Such a strategy constitutes the core of combined enrichment and topology analysis.298,299 Although pathway analysis is a powerful tool and there is great merit in identifying relevant pathways corresponding to a phenotype, it suffers from several limitations. First, definitions of pathways differ to a varying degrees across databases.300 Second, highly linked metabolites with a high number of possible biochemical reactions are also constituents of multiple pathways and pathways might overlap. Hence, it is challenging to explain changes over several pathways.301 In such a scenario, time-series analysis might support the identification of regulatory hubs within a metabolic network.302
A great way to comprise results and enable biological interpretation is the use of network-based approaches. Similar to pathway analysis, metabolic networking relies on reference databases for biochemical and signaling pathway information but constructs a single network where each metabolite is linked by all possible biochemical reactions. With the help of several strategies and options to extract a subnetwork capturing all relevant metabolites from the input metabolite list, they can represent a global and concise picture of metabolism.301 MetExplore303,304 allows metabolic network construction, exploration, and combination with omics data analysis. It can access several databases for multiple model organisms and allows collaboration in curation and annotation of metabolic networks. The interpretation of results is aided by metabolite set enrichment analysis (MSEA) and extraction of relevant subnetworks. Correlation-network construction does not require biochemical knowledge but is based on quantitative information. It can establish correlations and metabolites can be grouped based on the magnitude and sign of correlations, while the network visualization strategy lends itself well to identify the corresponding clusters and relationships among them.305 Furthermore, correlations can also be determined based on chemical similarity in a metabolite list (ChemRICH)306 or spectral similarity (MS2LDA).164 The full potential and perspectives of network-analysis in metabolomics data analysis and system biological approach for biological interpretation have been discussed in recent reviews including various possibilities and respective tools.294,305 A plethora of bioinformatics and data analysis tools was developed in the R ecosystem, also for the metabolomics community. These tools and their evolution have been extensively reviewed.120 However, the barrier of entry can be substantially higher to users with no programming skills or experience with command line tools. One response by R developers for this is to include a graphical user interface (GUI) within the package, which allows users to work within the comfort of their browser. Several web-based tools emerged in the last years, which function as a metabolomics data analysis toolbox and allow the visualization of metabolomics results via different modules and offer multiple solutions from the aforementioned options (MetaboAnalyst,307 MetaBox,308 MetExplore304). Such tools lower the barrier of entry with aesthetic and user-friendly GUI and example datasets. As a prime example, MetaboAnalyst and its equivalent R-based package MetaboAnalystR123 serve multiple functionalities in several modules ranging from metabolite identification, exploratory data analysis, pathway enrichment analysis, combined MSEA and topology analysis, and multi-omics integration, just to name a few.
Several pathway databases exist (KEGG, Reactome,309 Recon,310 Cyc,311 WikiPathways,312 PathBank,313 etc., Table 2) with a different focus, number of model organisms contained, and thus target audience, features, and applications. They have been reviewed extensively.293 Most of them provide a basic functionality to map metabolites from a list to their pathways, visualize, and some form of quantitative analysis (ORA, MSEA). As a unique feature Virtual Metabolic Human310,314 integrates the largest database of human and gut microbiome metabolism and presents a virtual human model with many possible pathological conditions.
The final integration of data from multiple omics-type experiments (like genomics, transcriptomics, and proteomics) complementing metabolomics studies315 depends on the possibility to combine multilayer information. MetaboAnalyst, Reactome, Recon, PaintOmics 3,316 the R-package mixOmics,317 and OmicsNet318,319 contain several modules for multi-omics data integration. A comprehensive review by Wörheide et al.320 discusses the various ways how to perform data-integration in multi-omics workflows. MetScape321−323 is a Cytoscape plugin to facilitate the visualization of correlation networks and metabolic networks based on metabolomics data. Metabolomics networks can also integrate transcriptomics data to inspect gene-metabolite connections, and subnetworks can be extracted. The new visualization technique GEM-Vis324 facilitates the visualization and exploration of time-course metabolomics experiments as metabolic network maps.
Although the field of metabolomics downstream data analysis and visualization clearly gained momentum with an increasing number of novel tools in the last years,120,293,325 there are many software examples which are not available anymore through the uniform resource locator (URL) originally referenced. This is by no means specific to metabolomics, but rather to bioinformatics software in general.326 In addition, some tools—though still available in online repositories—are not compatible with current R versions or require specific dependencies. Using virtual environments, virtual machines or containers are technical solutions to the problem of long-term software availability. Here it has to be mentioned that funding of scientific research is often not capable to encompass the full life cycle of software development as functional tools require maintenance even after their publication.327 Hence, sustainability of software solutions is of utmost importance as the challenge of growing data complexity increases our dependency on data interpretation pipelines.
Conclusions
Years of successful analytical development led to informative tailored methods. An optimal metabolomics workflow should cover the lipid dimension and has to find the right balance of coverage, throughput, and accuracy. State of the art workflows consist of complementary multiplatform modules which allow nontargeted discoveries and targeted absolute quantification. Only recently, measurement and data evaluation strategies of the two sub-ome specific disciplines metabolomics and lipidomics converge. Both high-resolution and low-resolution tandem MS are integral parts of multiplatform approaches. Up to date, coverage of low abundant metabolites (pM/low nM concentrations) is ensured by quadrupole-based tandem MS. While, there has been a paradigmatic shift in using HRMS for targeted absolute quantification, thereby enabling us to merge targeted and nontargeted approaches, typical limits of detection of HRMS workflows remain in the (low) nM range. The final measurement strategy depends on sample type and size, sampling frequency, the envisaged depth of the metabolomics/lipidomic profile, the different experimental conditions addressed, and finally, the type of information expected as outcome. Accurate quantification and identification are the prerequisite for correct biological interpretation, a bold argument which remains valid even in the context of powerful multi-omics data rich in analysis and network integration. While the gold standard validating the quantitative aspect in MS-based metabolomics will remain stringent analytical validation using standards and reference materials, the corroboration of the qualitative realm in metabolomics is currently revolutionized by bioinformatics tools. For example, in silico approaches are more and more accepted as an alternative to spectral library search. However, one should not forget that the development and validation of these tools is inherently linked to the availability of excellent community-based resources. Providing standards, reference materials, setting up and curating open-source data sets and experimental spectral libraries was and still is of paramount importance of the progress to the field.
Acknowledgments
This work was supported by the University of Vienna, the Faculty of Chemistry, and the Vienna Metabolomics Center (VIME; http://metabolomics.univie.ac.at/) of the University of Vienna. The authors thank Gerrit Hermann, Tim Causon, Stephan Hann, and Thomas Nägele for discussion, Martin Schaier for graphical abstract preparation, and all members of the Koellensperger group (University of Vienna) for continuous support.
Biographies
Evelyn Rampler is a senior scientist at the Department of Analytical Chemistry of the University of Vienna. She studied biotechnology and specialized in analytical chemistry during her Ph.D. studies at the University of Natural Resource and Life Sciences Vienna. During two independent post-doc phases at the Institute of Molecular Pathology and the University of Vienna as well as several research stays abroad (US, Germany, Australia), she gained expertise in LC-MS based proteomics, glycomics, and metabolomics. Currently, she leads projects on the development of novel LC-MS based metabolomics, (glyco)lipidomics, and multi-omics workflows for the application in different sample matrices (human plasma, plants, stem and tumor cells) and is part of the Board of the Austrian Proteomics & Metabolomics Association.
Yasin El Abiead is currently a Ph.D. candidate at the department of Analytical Chemistry at the University of Vienna where he has also obtained his M.Sc. degree as well as been a member of the Junior Board of the Austrian Proteomics & Metabolomics Association. His research deals with the intersection of analytical chemistry and bioinformatics workflows and the validation of nontargeted metabolomics experiments.
Harald Schoeny is currently a Ph.D. candidate at the Department of Analytical Chemistry of the University of Vienna where he has also obtained his M.Sc. degree. During his study he has spent several research stays abroad, e.g. at Royal Holloway, University of London, UK and at the Leibniz-Institut für Analytische Wissenschaften - ISAS, Germany. His research is focused on the development of novel lipid workflows and the optimization of quantitative lipidomics workflows toward a standardized and harmonized field.
Mate Rusz studied chemical engineering at the Technical University of Budapest (Hungary) and biochemistry at the University of Vienna. He is currently a Ph.D. candidate in metabolomics at the University of Vienna, where he investigates acquired resistance toward metal-based drugs and oxidative stress in preclinical cancer cell models with metabolomics.
Felina Hildebrand is currently a Ph.D. candidate in analytical chemistry at the University of Vienna. She obtained her M.Sc. degree in chemical biology at the Friedrich Schiller University Jena, Germany. Her main field of interest is the development of novel workflows in lipidomics and metabolomics focusing on sensitivity and high-throughput.
Veronika Fitz is a Ph.D. candidate in analytical chemistry at the University of Vienna, where she obtained M.Sc. degrees in Pharmacy and Pharmaceutical Quality Management. Her current research focus lies on the development of sensitive LC-MS workflows for quantitative multi-residue bioanalysis of small molecules.
Gunda Koellensperger was appointed as full professor for Analytical Chemistry by the University of Vienna in 2013. Following her studies at the Vienna University of Technology, she did her tenure at the University of Natural Resources and Life Sciences, Vienna. In 2011 she was guest professor at the Humboldt University, Berlin. From 2010–2014 she was key researcher of the Austrian Centre for Industrial Biotechnology. Since 2020 she has been head of the Institute of Analytical Chemistry. She is vice president of the Austrian Society of Analytical Chemistry, board member of the Austrian Society for Proteomics and Metabolomics, and vice chair of the Vienna Metabolomics Center (VIME). She is author of more than 160 papers and book chapters. Her current research covers methods in metabolomics/lipidomics and biotechnological production of standards for mass spectrometry and metallomics.
The authors declare no competing financial interest.
References
- González-Riano C.; Dudzik D.; Garcia A.; Gil-de-la-Fuente A.; Gradillas A.; Godzien J.; López-Gonzálvez Á.; Rey-Stolle F.; Rojo D.; Ruperez F. J.; Saiz J.; Barbas C. Recent Developments along the Analytical Process for Metabolomics Workflows. Anal. Chem. 2020, 92 (1), 203–226. 10.1021/acs.analchem.9b04553. [DOI] [PubMed] [Google Scholar]
- Dunn W. B.; Ellis D. I. Metabolomics: Current Analytical Platforms and Methodologies. TrAC, Trends Anal. Chem. 2005, 24 (4), 285–294. 10.1016/j.trac.2004.11.021. [DOI] [Google Scholar]
- Van Treuren W.; Dodd D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu. Rev. Pathol.: Mech. Dis. 2020, 15 (1), 345–369. 10.1146/annurev-pathol-020117-043559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter M. R.; Cajka T.; Fiehn O. Epimetabolites: Discovering Metabolism beyond Building and Burning. Curr. Opin. Chem. Biol. 2017, 36, 70–76. 10.1016/j.cbpa.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma-Ortiz C.; Jeffryes J. G.; Cooper A. J. L.; Niehaus T. D.; Thamm A. M. K.; Frelin O.; Aunins T.; Fiehn O.; de Crécy-Lagard V.; Henry C. S.; Hanson A. D. ‘Nothing of Chemistry Disappears in Biology’: The Top 30 Damage-Prone Endogenous Metabolites. Biochem. Soc. Trans. 2016, 44 (3), 961–971. 10.1042/BST20160073. [DOI] [PubMed] [Google Scholar]
- Bloszies C. S.; Fiehn O.. Using Untargeted Metabolomics for Detecting Exposome Compounds. Curr. Opin. Toxicol. April 2018, 8, 87–92 10.1016/j.cotox.2018.03.002. [DOI] [Google Scholar]
- Ortmayr K.; Causon T. J.; Hann S.; Koellensperger G. Increasing Selectivity and Coverage in LC-MS Based Metabolome Analysis. TrAC, Trends Anal. Chem. 2016, 82, 358–366. 10.1016/j.trac.2016.06.011. [DOI] [Google Scholar]
- Viant M. R.; Kurland I. J.; Jones M. R.; Dunn W. B. How Close Are We to Complete Annotation of Metabolomes?. Curr. Opin. Chem. Biol. 2017, 36, 64–69. 10.1016/j.cbpa.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Fiehn O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- Broadhurst D.; Goodacre R.; Reinke S. N.; Kuligowski J.; Wilson I. D.; Lewis M. R.; Dunn W. B. Guidelines and Considerations for the Use of System Suitability and Quality Control Samples in Mass Spectrometry Assays Applied in Untargeted Clinical Metabolomic Studies. Metabolomics 2018, 14 (6), 72. 10.1007/s11306-018-1367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E. S.; Patti G. J. Perspectives on Data Analysis in Metabolomics: Points of Agreement and Disagreement from the 2018 ASMS Fall Workshop. J. Am. Soc. Mass Spectrom. 2019, 30 (10), 2031–2036. 10.1007/s13361-019-02295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Broadhurst D. I.; Wilson M.; Wishart D. S. Translational Biomarker Discovery in Clinical Metabolomics: An Introductory Tutorial. Metabolomics 2013, 9 (2), 280–299. 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage E. G.; Barbas C. Metabolomics in Cancer Biomarker Discovery: Current Trends and Future Perspectives. J. Pharm. Biomed. Anal. 2014, 87, 1–11. 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- Nash W. J.; Dunn W. B. From Mass to Metabolite in Human Untargeted Metabolomics: Recent Advances in Annotation of Metabolites Applying Liquid Chromatography-Mass Spectrometry Data. TrAC, Trends Anal. Chem. 2019, 120, 115324. 10.1016/j.trac.2018.11.022. [DOI] [Google Scholar]
- Dunn W. B.; Lin W.; Broadhurst D.; Begley P.; Brown M.; Zelena E.; Vaughan A. A.; Halsall A.; Harding N.; Knowles J. D.; Francis-McIntyre S.; Tseng A.; Ellis D. I.; O’Hagan S.; Aarons G.; Benjamin B.; Chew-Graham S.; Moseley C.; Potter P.; Winder C. L.; Potts C.; Thornton P.; McWhirter C.; Zubair M.; Pan M.; Burns A.; Cruickshank J. K.; Jayson G. C.; Purandare N.; Wu F. C. W.; Finn J. D.; Haselden J. N.; Nicholls A. W.; Wilson I. D.; Goodacre R.; Kell D. B. Molecular Phenotyping of a UK Population: Defining the Human Serum Metabolome. Metabolomics 2015, 11 (1), 9–26. 10.1007/s11306-014-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaleckis R.; Meister I.; Zhang P.; Wheelock C. E. Challenges, Progress and Promises of Metabolite Annotation for LC–MS-Based Metabolomics. Curr. Opin. Biotechnol. 2019, 55, 44–50. 10.1016/j.copbio.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Sumner L. W.; Samuel T.; Noble R.; Gmbh S. D.; Barrett D.; Beale M. H.; Hardy N. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3 (3), 211–221. 10.1007/s11306-007-0082-2.Proposed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheubert K.; Hufsky F.; Petras D.; Wang M.; Nothias L.-F.; Dührkop K.; Bandeira N.; Dorrestein P. C.; Böcker S. Significance Estimation for Large Scale Metabolomics Annotations by Spectral Matching. Nat. Commun. 2017, 8 (1), 1494. 10.1038/s41467-017-01318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W. R.; Grapov D.; Fahrmann J. F.; DeFelice B.; Rom W. N.; Pass H. I.; Kim K.; Nguyen U. T.; Taylor S. L.; Gandara D. R.; Kelly K.; Fiehn O.; Miyamoto S. Metabolomic Markers of Altered Nucleotide Metabolism in Early Stage Adenocarcinoma. Cancer Prev. Res. 2015, 8 (5), 410–418. 10.1158/1940-6207.CAPR-14-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.; Zhong F.; Zhu J. Bridging Targeted and Untargeted Mass Spectrometry-Based Metabolomics via Hybrid Approaches. Metabolites 2020, 10 (9), 348. 10.3390/metabo10090348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H.; Nakabayashi R.; Mori T.; Yamada Y.; Takahashi M.; Rai A.; Sugiyama R.; Yamamoto H.; Nakaya T.; Yamazaki M.; Kooke R.; Bac-Molenaar J. A.; Oztolan-Erol N.; Keurentjes J. J. B.; Arita M.; Saito K. A Cheminformatics Approach to Characterize Metabolomes in Stable-Isotope-Labeled Organisms. Nat. Methods 2019, 16 (4), 295–298. 10.1038/s41592-019-0358-2. [DOI] [PubMed] [Google Scholar]
- Hu L.; Liu J.; Zhang W.; Wang T.; Zhang N.; Lee Y. H.; Lu H. FUNCTIONAL METABOLOMICS DECIPHER BIOCHEMICAL FUNCTIONS AND ASSOCIATED MECHANISMS UNDERLIE SMALL-MOLECULE METABOLISM. Mass Spectrom. Rev. 2020, 39 (5–6), 417–433. 10.1002/mas.21611. [DOI] [PubMed] [Google Scholar]
- Cavill R.; Jennen D.; Kleinjans J.; Briedé J. J. Transcriptomic and Metabolomic Data Integration. Briefings Bioinf. 2016, 17 (5), 891–901. 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- Schymanski E. L.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H. P.; Hollender J. Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol. 2014, 48 (4), 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Ortmayr K.; Dubuis S.; Zampieri M. Metabolic Profiling of Cancer Cells Reveals Genome-Wide Crosstalk between Transcriptional Regulators and Metabolism. Nat. Commun. 2019, 10 (1), 1–13. 10.1038/s41467-019-09695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M.; Enke T.; Chubukov V.; Ricci V.; Piddock L.; Sauer U. Metabolic Constraints on the Evolution of Antibiotic Resistance. Mol. Syst. Biol. 2017, 13 (3), 917. 10.15252/msb.20167028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidance for Industry Bioanalytical Method Validation Guidance for Industry Bioanalytical Method Validation. Biopharm. Fed. Regist. 2001, (No. 100), 66. [Google Scholar]
- Wharf C.; Kingdom U.. Guideline on Bioanalytical Method Validation; 2012; Vol. 44. [Google Scholar]
- Lipidomics Standards Initiative. https://lipidomics-standards-initiative.org.
- Liebisch G.; Ekroos K.; Hermansson M.; Ejsing C. S.; Gallego S. F.; Sprenger R. R.; Neess D.; Pauling J. K.; Færgeman N. J.; Ejsing C. S.; Sandra K.; Sandra P.; Xiao J. F.; Zhou B.; Ressom H. W.; Harkewicz R.; Dennis E. A.; Husen P.; Tarasov K.; Katafiasz M.; Sokol E.; Vogt J.; Baumgart J.; Nitsch R.; Ekroos K.; Ejsing C. S.; Manuscript A.; Human T.; Lipidome P.; Ovčačíková M.; Lísa M.; Cífková E.; Holčapek M.; Hayes R.; Ahmed A.; Edge T.; Zhang H.; Marat A. L.; Haucke V.; Majerus P. W.; York J. D.; Kiyonami R.; Peake D.; Drew D.; Sun X.; Liu X.; Miller K.; Ball S.; Holčapek M.; Lísa M.; Ulmer C. Z.; Koelmel J. P.; Ragland J. M.; Garrett T. J.; Bowden J. A.; Wörmer L.; Lipp J. S.; Schröder J. M.; Hinrichs K. U.; Müller C.; Kanawati B.; Rock T. M.; Forcisi S.; Moritz F.; Schmitt-Kopplin P.; James P. F.; Perugini M. A.; O’Hair R. A. J.; Lísa M.; Holčapek M.; Gesta S.; Tseng Y. H.; Kahn C. R.; Li Q.; Bozek K.; Xu C.; Guo Y.; Sun J.; Paabo S.; Sherwood C. C.; Hot P. R.; Ely J. J.; Li Y.; Willmitzer L.; Giavalisco P.; Khaitovich P.; Darby R. A.; Cartwright S. P.; Dilworth M. V.; Bill R. M.; Lehmann W.; Almeida R.; Pauling J. K.; Sokol E.; Hannibal-bach H. K.; Ejsing C. S. A TIMELINE OF STABLE ISOTOPES AND MASS SPECTROMETRY IN THE LIFE SCIENCES. J. Chromatogr. A 2017, 17 (2), 242–256. 10.1016/j.cell.2007.10.004. [DOI] [Google Scholar]
- Kim J.; Kang D.; Lee S. K.; Kim T. Y. Deuterium Oxide Labeling for Global Omics Relative Quantification: Application to Lipidomics. Anal. Chem. 2019, 91 (14), 8853–8863. 10.1021/acs.analchem.9b00086. [DOI] [PubMed] [Google Scholar]
- Drotleff B.; Illison J.; Schlotterbeck J.; Lukowski R.; Lämmerhofer M. Comprehensive Lipidomics of Mouse Plasma Using Class-Specific Surrogate Calibrants and SWATH Acquisition for Large-Scale Lipid Quantification in Untargeted Analysis. Anal. Chim. Acta 2019, 1086, 90–102. 10.1016/j.aca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- Wang M.; Wang C.; Han X. SELECTION OF INTERNAL STANDARDS FOR ACCURATE QUANTIFICATION OF COMPLEX LIPID SPECIES IN BIOLOGICAL EXTRACTS BY ELECTROSPRAY IONIZATION MASS SPECTROMETRY—WHAT, HOW AND WHY?. Mass Spectrom. Proteins Pept. Methods Mol. Biol. (Clifton, N.J.) 2017, 36, 693–714. 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höring M.; Ejsing C. S.; Hermansson M.; Liebisch G. Quantification of Cholesterol and Cholesteryl Ester by Direct Flow Injection High-Resolution Fourier Transform Mass Spectrometry Utilizing Species-Specific Response Factors. Anal. Chem. 2019, 91 (5), 3459–3466. 10.1021/acs.analchem.8b05013. [DOI] [PubMed] [Google Scholar]
- Schuhmann K.; Moon H. K.; Thomas H.; Ackerman J. M.; Groessl M.; Wagner N.; Kellmann M.; Henry I.; Nadler A.; Shevchenko A. Quantitative Fragmentation Model for Bottom-Up Shotgun Lipidomics. Anal. Chem. 2019, 91 (18), 12085–12093. 10.1021/acs.analchem.9b03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachleder V.; Vítová M.; Hlavová M.; Moudříková Š.; Mojzeš P.; Heumann H.; Becher J. R.; Bišová K. Stable Isotope Compounds - Production, Detection, and Application. Biotechnol. Adv. 2018, 36 (3), 784–797. 10.1016/j.biotechadv.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Lehmann W. D. A Timeline of Stable Isotopes and Mass Spectrometry in the Life Sciences. Mass Spectrom. Rev. 2017, 36 (1), 58–85. 10.1002/mas.21497. [DOI] [PubMed] [Google Scholar]
- Jang C.; Chen L.; Rabinowitz J. D. Metabolomics and Isotope Tracing. Cell 2018, 173 (4), 822–837. 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U. Metabolic Networks in Motion: 13 C-based Flux Analysis. Mol. Syst. Biol. 2006, 2 (1), 62. 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N.; Saghatelian A.; Patti G. J. Defining the Metabolome: Size, Flux, and Regulation. Mol. Cell 2015, 58 (4), 699–706. 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triebl A.; Wenk M. R.. Analytical Considerations of Stable Isotope Labelling in Lipidomics. Biomolecules 2018, 8 ( (4), ), 151. 10.3390/biom8040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. E.; Mann M. Mass Spectrometry–Based Proteomics Turns Quantitative. Nat. Chem. Biol. 2005, 1 (5), 252–262. 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Krüger M.; Moser M.; Ussar S.; Thievessen I.; Luber C. A.; Forner F.; Schmidt S.; Zanivan S.; Fässler R.; Mann M. SILAC Mouse for Quantitative Proteomics Uncovers Kindlin-3 as an Essential Factor for Red Blood Cell Function. Cell 2008, 134 (2), 353–364. 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Sury M. D.; Chen J. X.; Selbach M. The SILAC Fly Allows for Accurate Protein Quantification in Vivo. Mol. Cell. Proteomics 2010, 9 (10), 2173–2183. 10.1074/mcp.M110.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijgsveld J.; Ketting R. F.; Mahmoudi T.; Johansen J.; Artal-Sanz M.; Verrijzer C. P.; Plasterk R. H. A.; Heck A. J. R. Metabolic Labeling of C. Elegans and D. Melanogaster for Quantitative Proteomics. Nat. Biotechnol. 2003, 21 (8), 927–931. 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- Wu L.; Mashego M. R.; Van Dam J. C.; Proell A. M.; Vinke J. L.; Ras C.; Van Winden W. A.; Van Gulik W. M.; Heijnen J. J. Quantitative Analysis of the Microbial Metabolome by Isotope Dilution Mass Spectrometry Using Uniformly 13C-Labeled Cell Extracts as Internal Standards. Anal. Biochem. 2005, 336 (2), 164–171. 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Bennett B. D.; Yuan J.; Kimball E. H.; Rabinowitz J. D. Absolute Quantitation of Intracellular Metabolite Concentrations by an Isotope Ratio-Based Approach. Nat. Protoc. 2008, 3 (8), 1299–1311. 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canelas A. B.; Ten Pierick A.; Ras C.; Seifar R. M.; Van Dam J. C.; Van Gulik W. M.; Heijnen J. J. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Anal. Chem. 2009, 81 (17), 7379–7389. 10.1021/ac900999t. [DOI] [PubMed] [Google Scholar]
- Loos M.; Gerber C.; Corona F.; Hollender J.; Singer H. Accelerated Isotope Fine Structure Calculation Using Pruned Transition Trees. Anal. Chem. 2015, 87 (11), 5738–5744. 10.1021/acs.analchem.5b00941. [DOI] [PubMed] [Google Scholar]
- Gross J. H.Mass Spectrometry—A Textbook; Springer, 2011. [Google Scholar]
- Weiner M.; Tröndle J.; Schmideder A.; Albermann C.; Binder K.; Sprenger G. A.; Weuster-Botz D. Parallelized Small-Scale Production of Uniformly 13C-Labeled Cell Extract for Quantitative Metabolome Analysis. Anal. Biochem. 2015, 478, 134–140. 10.1016/j.ab.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Lemmens M.; Adam G.; Wiesenberger G.; Maschietto V.; Marocco A.; Strauss J.; Bo S.; Schuhmacher R. Metabolomics 2014, 10, 754–769. 10.1007/s11306-013-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P.; Köhl K.; Hummel J.; Seiwert B.; Willmitzer L. 13C Isotope-Labeled Metabolomes Allowing for Improved Compound Annotation and Relative Quantification in Liquid Chromatography-Mass Spectrometry-Based Metabolomic Research. Anal. Chem. 2009, 81 (15), 6546–6551. 10.1021/ac900979e. [DOI] [PubMed] [Google Scholar]
- Schwaiger M.; Rampler E.; Hermann G.; Miklos W.; Berger W.; Koellensperger G. Anion-Exchange Chromatography Coupled to High-Resolution Mass Spectrometry: A Powerful Tool for Merging Targeted and Non-Targeted Metabolomics. Anal. Chem. 2017, 89 (14), 7667–7674. 10.1021/acs.analchem.7b01624. [DOI] [PubMed] [Google Scholar]
- Rampler E.; Coman C.; Hermann G.; Sickmann A.; Ahrends R.; Koellensperger G. LILY-Lipidome Isotope Labeling of Yeast: In Vivo Synthesis of 13 C Labeled Reference Lipids for Quantification by Mass Spectrometry. Analyst 2017, 142 (11), 1891–1899. 10.1039/C7AN00107J. [DOI] [PubMed] [Google Scholar]
- Neubauer S.; Haberhauer-Troyer C.; Klavins K.; Russmayer H.; Steiger M. G.; Gasser B.; Sauer M.; Mattanovich D.; Hann S.; Koellensperger G. U13C Cell Extract of Pichia Pastoris - A Powerful Tool for Evaluation of Sample Preparation in Metabolomics. J. Sep. Sci. 2012, 35 (22), 3091–3105. 10.1002/jssc.201200447. [DOI] [PubMed] [Google Scholar]
- Mahieu N. G.; Patti G. J. Systems-Level Annotation of a Metabolomics Data Set Reduces 25 000 Features to Fewer than 1000 Unique Metabolites. Anal. Chem. 2017, 89 (19), 10397–10406. 10.1021/acs.analchem.7b02380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold H. K.; Hachey D. L.; Reeds P. J.; Thomas O. P.; Hoeksema S.; Klein P. D. Uniformly 13C-Labeled Algal Protein Used to Determine Amino Acid Essentiality in Vivo. Proc. Natl. Acad. Sci. U. S. A. 1991, 88 (18), 8091–8095. 10.1073/pnas.88.18.8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens P. W.; Sicotte V. J.; Delente J. Microalgae as a Source of Stable Isotopically Labeled Compounds. J. Appl. Phycol. 1994, 6 (2), 113–121. 10.1007/BF02186065. [DOI] [Google Scholar]
- Elahee Doomun S. N.; Nie S.; Loke S.; Kowalski G. M.; Beech P. L.; Callahan D. L. Ultrahigh-Resolution Mass Spectrometry Method for Resolving 13 C-Enrichment Patterns in a Microalgal Lipidome. J. Am. Soc. Mass Spectrom. 2020, 31 (8), 1763–1772. 10.1021/jasms.0c00192. [DOI] [PubMed] [Google Scholar]
- Hermann G.; Møller L. H.; Gammelgaard B.; Hohlweg J.; Mattanovich D.; Hann S.; Koellensperger G. In Vivo Synthesized 34 S Enriched Amino Acid Standards for Species Specific Isotope Dilution of Proteins. J. Anal. At. Spectrom. 2016, 31 (9), 1830–1835. 10.1039/C6JA00039H. [DOI] [Google Scholar]
- Ćeranić A.; Doppler M.; Büschl C.; Parich A.; Xu K.; Koutnik A.; Bürstmayr H.; Lemmens M.; Schuhmacher R. Preparation of Uniformly Labelled 13C- And 15N-Plants Using Customised Growth Chambers. Plant Methods 2020, 16 (1), 1–15. 10.1186/s13007-020-00590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy D. B.; Dong M. Q.; Wu C. C.; Venable J. D.; Yates J. R. 15N Metabolic Labeling of Mammalian Tissue with Slow Protein Turnover. J. Proteome Res. 2007, 6 (5), 2005–2010. 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethloff F.; Bueschl C.; Heumann H.; Schuhmacher R.; Turck C. W. Partially 13C-Labeled Mouse Tissue as Reference for LC-MS Based Untargeted Metabolomics. Anal. Biochem. 2018, 556 (March), 63–69. 10.1016/j.ab.2018.06.023. [DOI] [PubMed] [Google Scholar]
- Grankvist N.; Watrous J. D.; Lagerborg K. A.; Lyutvinskiy Y.; Jain M.; Nilsson R. Profiling the Metabolism of Human Cells by Deep 13C Labeling. Cell Chem. Biol. 2018, 25 (11), 1419–1427.e4. 10.1016/j.chembiol.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Berthiller F.; Lirk G.; Winkler S.; Krska R.; Schuhmacher R. Metextract: A New Software Tool for the Automated Comprehensive Extraction of Metabolite-Derived Lc/Ms Signals in Metabolomics Research. Bioinformatics 2012, 28 (5), 736–738. 10.1093/bioinformatics/bts012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E.; Vastag L.; Rabinowitz J. D. Metabolomic Analysis and Visualization Engine for LC - MS Data. Anal. Chem. 2010, 82 (23), 9818–9826. 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkathukalam A.; Jankevics A.; Creek D. J.; Achcar F.; Barrett M. P.; Breitling R. MzMatch-ISO: An R Tool for the Annotation and Relative Quantification of Isotope-Labelled Mass Spectrometry Data. Bioinformatics 2013, 29 (2), 281–283. 10.1093/bioinformatics/bts674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Chen Y. J.; Cho K.; Nikolskiy I.; Crawford P. A.; Patti G. J. X13CMS: Global Tracking of Isotopic Labels in Untargeted Metabolomics. Anal. Chem. 2014, 86 (3), 1632–1639. 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C.; Montenegro-Burke J. R.; Domingo-Almenara X.; Palermo A.; Warth B.; Hermann G.; Koellensperger G.; Huan T.; Uritboonthai W.; Aisporna A. E.; Wolan D. W.; Spilker M. E.; Benton H. P.; Siuzdak G. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90 (5), 3156–3164. 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellades J.; Navarro M.; Samino S.; Garcia-Ramirez M.; Hernandez C.; Simo R.; Vinaixa M.; Yanes O. GeoRge: A Computational Tool to Detect the Presence of Stable Isotope Labeling in LC/MS-Based Untargeted Metabolomics. Anal. Chem. 2016, 88 (1), 621–628. 10.1021/acs.analchem.5b03628. [DOI] [PubMed] [Google Scholar]
- Kessler N.; Walter F.; Persicke M.; Albaum S. P.; Kalinowski J.; Goesmann A.; Niehaus K.; Nattkemper T. W. ALLocator: An Interactive Web Platform for the Analysis of Metabolomic LC-ESI-MS Datasets, Enabling Semi-Automated, User-Revised Compound Annotation and Mass Isotopomer Ratio Analysis. PLoS One 2014, 9 (11), e113909. 10.1371/journal.pone.0113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong F. A.; Beecher C. Addressing the Current Bottlenecks of Metabolomics: Isotopic Ratio Outlier Analysis, an Isotopic-Labeling Technique for Accurate Biochemical Profiling. Bioanalysis 2012, 4 (18), 2303–2314. 10.4155/bio.12.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahieu N. G.; Huang X.; Chen Y. J.; Patti G. J. Credentialing Features: A Platform to Benchmark and Optimize Untargeted Metabolomic Methods. Anal. Chem. 2014, 86 (19), 9583–9589. 10.1021/ac503092d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueschl C.; Kluger B.; Neumann N. K. N.; Doppler M.; Maschietto V.; Thallinger G. G.; Meng-Reiterer J.; Krska R.; Schuhmacher R. MetExtract II: A Software Suite for Stable Isotope-Assisted Untargeted Metabolomics. Anal. Chem. 2017, 89 (17), 9518–9526. 10.1021/acs.analchem.7b02518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Xing X.; Chen L.; Yang L.; Su X.; Rabitz H.; Lu W.; Rabinowitz J. D. Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal. Chem. 2019, 91 (3), 1838–1846. 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.; Xing X.; Wang L.; Chen L.; Zhang S.; McReynolds M. R.; Rabinowitz J. D. Improved Annotation of Untargeted Metabolomics Data through Buffer Modifications That Shift Adduct Mass and Intensity. Anal. Chem. 2020, 92 (17), 11573–11581. 10.1021/acs.analchem.0c00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuillet M.; Bellvert F.; Cahoreau E.; Letisse F.; Millard P.; Portais J. C. Methodology for the Validation of Isotopic Analyses by Mass Spectrometry in Stable-Isotope Labeling Experiments. Anal. Chem. 2018, 90 (3), 1852–1860. 10.1021/acs.analchem.7b03886. [DOI] [PubMed] [Google Scholar]
- Millard P.; Massou S.; Portais J. C.; Létisse F. Isotopic Studies of Metabolic Systems by Mass Spectrometry: Using Pascal’s Triangle to Produce Biological Standards with Fully Controlled Labeling Patterns. Anal. Chem. 2014, 86 (20), 10288–10295. 10.1021/ac502490g. [DOI] [PubMed] [Google Scholar]
- Mairinger T.; Hann S. Implementation of Data-Dependent Isotopologue Fragmentation in 13C-Based Metabolic Flux Analysis. Anal. Bioanal. Chem. 2017, 409 (15), 3713–3718. 10.1007/s00216-017-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger-Haber M.; Hermann G.; El Abiead Y.; Rampler E.; Wernisch S.; Sas K.; Pennathur S.; Koellensperger G. Proposing a Validation Scheme for 13C Metabolite Tracer Studies in High-Resolution Mass Spectrometry. Anal. Bioanal. Chem. 2019, 411 (14), 3103–3113. 10.1007/s00216-019-01773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampler E.; Criscuolo A.; Zeller M.; El Abiead Y.; Schoeny H.; Hermann G.; Sokol E.; Cook K.; Peake D.; Delanghe B.; Koellensperger G. A. Novel Lipidomics Workflow for Improved Human Plasma Identification and Quantification Using RPLC-MSn Methods and Isotope Dilution Strategies. Anal. Chem. 2018, 90 (11), 6494–6501. 10.1021/acs.analchem.7b05382. [DOI] [PubMed] [Google Scholar]
- Natter K.; Kohlwein S. D. Yeast and Cancer Cells - Common Principles in Lipid Metabolism. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2013, 1831 (2), 314–326. 10.1016/j.bbalip.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolrab D.; Jirásko R.; Chocholoušková M.; Peterka O.; Holčapek M. Oncolipidomics: Mass Spectrometric Quantitation of Lipids in Cancer Research. TrAC, Trends Anal. Chem. 2019, 120, 115480. 10.1016/j.trac.2019.04.012. [DOI] [Google Scholar]
- Rampler E.; Hermann G.; Grabmann G.; Abiead Y. El; Schoeny H.; Baumgartinger C.; Köcher T.; Koellensperger G.. Benchmarking Non-Targeted Metabolomics Using Yeast Derived Libraries. bioRxiv 2020, 2020.10.06.319160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou Feunang Y.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminf. 2016, 8 (1), 1–20. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeny H.; Rampler E.; Hermann G.; Grienke U.; Rollinger J. M.; Koellensperger G. Preparative Supercritical Fluid Chromatography for Lipid Class Fractionation—a Novel Strategy in High-Resolution Mass Spectrometry Based Lipidomics. Anal. Bioanal. Chem. 2020, 412 (10), 2365–2374. 10.1007/s00216-020-02463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Marcu A.; Guo A. C.; Liang K.; Vázquez-Fresno R.; Sajed T.; Johnson D.; Li C.; Karu N.; Sayeeda Z.; Lo E.; Assempour N.; Berjanskii M.; Singhal S.; Arndt D.; Liang Y.; Badran H.; Grant J.; Serra-Cayuela A.; Liu Y.; Mandal R.; Neveu V.; Pon A.; Knox C.; Wilson M.; Manach C.; Scalbert A. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46 (D1), D608–D617. 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danne-Rasche N.; Rubenzucker S.; Ahrends R. Uncovering the Complexity of the Yeast Lipidome by Means of NLC/NSI-MS/MS. Anal. Chim. Acta 2020, 1140, 199–209. 10.1016/j.aca.2020.10.012. [DOI] [PubMed] [Google Scholar]
- Schoeny H.; Rampler E.; Abiead Y. El; Hildebrand F.; Zach O.; Hermann G.; Koellensperger G.. A Combined Flow Injection/Reversed Phase Chromatography-High Resolution Mass Spectrometry Workflow for Accurate Absolute Lipid Quantification with 13 C-Internal Standards. bioRxiv 2020, 2020.11.04.367987 10.1101/2020.11.04.367987. [DOI] [PubMed] [Google Scholar]
- Metabolomics Society: Standardization in Metabolomics Experiments. http://metabolomicssociety.org/resources/metabolomics-standards (accessed 2020-07-14).
- Liebisch G.; Ahrends R.; Arita M.; Arita M.; Bowden J. A.; Ejsing C. S.; Griffiths W. J.; Holčapek M.; Köfeler H.; Mitchell T. W.; Wenk M. R.; Ekroos K.. Lipidomics Needs More Standardization. Nat. Metab. 2019, 1, 745–747. 10.1038/s42255-019-0094-z. [DOI] [PubMed] [Google Scholar]
- Sansone S.-A.; Fan T.; Goodacre R.; Griffin J. L.; Hardy N. W.; Kaddurah-Daouk R.; Kristal B. S.; Lindon J.; Mendes P.; Morrison N.; Nikolau B.; Robertson D.; Sumner L. W.; Taylor C.; van der Werf M.; van Ommen B.; Fiehn O. The Metabolomics Standards Initiative. Nat. Biotechnol. 2007, 25 (8), 846–848. 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- Salek R. M.; Steinbeck C.; Viant M. R.; Goodacre R.; Dunn W. B. The Role of Reporting Standards for Metabolite Annotation and Identification in Metabolomic Studies. GigaScience 2013, 2 (1), 2047–217X-2–13. 10.1186/2047-217X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Fahy E.; Aoki J.; Dennis E. A.; Durand T.; Ejsing C.; Fedorova M.; Feussner I.; Griffiths W. J.; Koefeler H.; Merrill A. H.; Murphy R. C.; O’Donnell V. B.; Oskolkova O. V.; Subramaniam S.; Wakelam M.; Spener F. Update on LIPID MAPS Classification, Nomenclature and Shorthand Notation for MS-Derived Lipid Structures. J. Lipid Res. 2020, 1–53. 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. H.; Nellis M.; Uppal K.; Ma C.; Tran V.; Liang Y.; Walker D. I.; Jones D. P. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem. 2020, 92 (13), 8836–8844. 10.1021/acs.analchem.0c00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. W.; Adams K. J.; Adamski J.; Asad Y.; Borts D.; Bowden J. A.; Byram G.; Dang V.; Dunn W. B.; Fernandez F.; Fiehn O.; Gaul D. A.; Hühmer A. F. R.; Kalli A.; Koal T.; Koeniger S.; Mandal R.; Meier F.; Naser F. J.; O’Neil D.; Pal A.; Patti G. J.; Pham-Tuan H.; Prehn C.; Raynaud F. I.; Shen T.; Southam A. D.; St. John-Williams L.; Sulek K.; Vasilopoulou C. G.; Viant M.; Winder C. L.; Wishart D.; Zhang L.; Zheng J.; Moseley M. A.. International Ring Trial of a High Resolution Targeted Metabolomics and Lipidomics Platform for Serum and Plasma Analysis. Anal. Chem. 2019, 91, 14407. 10.1021/acs.analchem.9b02908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J. A.; Heckert A.; Ulmer C. Z.; Jones C. M.; Koelmel J. P.; Abdullah L.; Ahonen L.; Alnouti Y.; Armando A. M.; Asara J. M.; Bamba T.; Barr J. R.; Bergquist J.; Borchers C. H.; Brandsma J.; Breitkopf S. B.; Cajka T.; Cazenave-Gassiot A.; Checa A.; Cinel M. A.; Colas R. A.; Cremers S.; Dennis E. A.; Evans J. E.; Fauland A.; Fiehn O.; Gardner M. S.; Garrett T. J.; Gotlinger K. H.; Han J.; Huang Y.; Neo A. H.; Hyötyläinen T.; Izumi Y.; Jiang H.; Jiang H.; Jiang J.; Kachman M.; Kiyonami R.; Klavins K.; Klose C.; Köfeler H. C.; Kolmert J.; Koal T.; Koster G.; Kuklenyik Z.; Kurland I. J.; Leadley M.; Lin K.; Maddipati K. R.; McDougall D.; Meikle P. J.; Mellett N. A.; Monnin C.; Moseley M. A.; Nandakumar R.; Oresic M.; Patterson R.; Peake D.; Pierce J. S.; Post M.; Postle A. D.; Pugh R.; Qiu Y.; Quehenberger O.; Ramrup P.; Rees J.; Rembiesa B.; Reynaud D.; Roth M. R.; Sales S.; Schuhmann K.; Schwartzman M. L.; Serhan C. N.; Shevchenko A.; Somerville S. E.; St. John-Williams L.; Surma M. A.; Takeda H.; Thakare R.; Thompson J. W.; Torta F.; Triebl A.; Trötzmüller M.; Ubhayasekera S. J. K.; Vuckovic D.; Weir J. M.; Welti R.; Wenk M. R.; Wheelock C. E.; Yao L.; Yuan M.; Zhao X. H.; Zhou S. Harmonizing Lipidomics: NIST Interlaboratory Comparison Exercise for Lipidomics Using SRM 1950–Metabolites in Frozen Human Plasma. J. Lipid Res. 2017, 58 (12), 2275–2288. 10.1194/jlr.M079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triebl A.; Burla B.; Selvalatchmanan J.; Oh J.; Tan S. H.; Chan M. Y.; Mellet N. A.; Meikle P. J.; Torta F.; Wenk M. R. Shared Reference Materials Harmonize Lipidomics across MS-Based Detection Platforms and Laboratories. J. Lipid Res. 2020, 61 (1), 105–115. 10.1194/jlr.D119000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolrab D.; Chocholoušková M.; Jirásko R.; Peterka O.; Holčapek M. Validation of Lipidomic Analysis of Human Plasma and Serum by Supercritical Fluid Chromatography–Mass Spectrometry and Hydrophilic Interaction Liquid Chromatography–Mass Spectrometry. Anal. Bioanal. Chem. 2020, 412 (10), 2375–2388. 10.1007/s00216-020-02473-3. [DOI] [PubMed] [Google Scholar]
- Galvez L.; Rusz M.; Schwaiger-Haber M.; El Abiead Y.; Hermann G.; Jungwirth U.; Berger W.; Keppler B. K.; Jakupec M. A.; Koellensperger G. Preclinical Studies on Metal Based Anticancer Drugs as Enabled by Integrated Metallomics and Metabolomics. Metallomics 2019, 11 (10), 1716–1728. 10.1039/C9MT00141G. [DOI] [PubMed] [Google Scholar]
- Bearden D. W.; Sheen D. A.; Simón-Manso Y.; Benner B. A.; Rocha W. F. C.; Blonder N.; Lippa K. A.; Beger R. D.; Schnackenberg L. K.; Sun J.; Mehta K. Y.; Cheema A. K.; Gu H.; Marupaka R.; Nagana Gowda G. A.; Raftery D. Metabolomics Test Materials for Quality Control: A Study of a Urine Materials Suite. Metabolites 2019, 9 (11), 270. 10.3390/metabo9110270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K.; Cochrane K.; Nainala V. C.; Williams M.; Chang J.; Jayaseelan K. V.; O’Donovan C. MetaboLights: A Resource Evolving in Response to the Needs of Its Scientific Community. Nucleic Acids Res. 2019, 48 (1), 440–444. 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberacher H.; Sasse M.; Antignac J. P.; Guitton Y.; Debrauwer L.; Jamin E. L.; Schulze T.; Krauss M.; Covaci A.; Caballero-Casero N.; Rousseau K.; Damont A.; Fenaille F.; Lamoree M.; Schymanski E. L. A European Proposal for Quality Control and Quality Assurance of Tandem Mass Spectral Libraries. Environ. Sci. Eur. 2020, 32 (1), 43. 10.1186/s12302-020-00314-9. [DOI] [Google Scholar]
- Liebisch G.; Vizcaíno J. A.; Köfeler H.; Trötzmüller M.; Griffiths W. J.; Schmitz G.; Spener F.; Wakelam M. J. O. Shorthand Notation for Lipid Structures Derived from Mass Spectrometry. J. Lipid Res. 2013, 54 (6), 1523–1530. 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Delgado P.; Dudzik D.; Miguel V.; Lamas S.; Barbas C. Data-Dependent Normalization Strategies for Untargeted Metabolomics—a Case Study. Anal. Bioanal. Chem. 2020, 412 (24), 6391–6405. 10.1007/s00216-020-02594-9. [DOI] [PubMed] [Google Scholar]
- Misra B. B. Data Normalization Strategies in Metabolomics: Current Challenges, Approaches, and Tools. Eur. J. Mass Spectrom. 2020, 26 (3), 165–174. 10.1177/1469066720918446. [DOI] [PubMed] [Google Scholar]
- Vu T.; Riekeberg E.; Qiu Y.; Powers R. Comparing Normalization Methods and the Impact of Noise. Metabolomics 2018, 14 (8), 1–10. 10.1007/s11306-018-1400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S.; Kind T.; Cajka T.; Hazen S. L.; Tang W. H. W.; Kaddurah-Daouk R.; Irvin M. R.; Arnett D. K.; Barupal D. K.; Fiehn O. Systematic Error Removal Using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91 (5), 3590–3596. 10.1021/acs.analchem.8b05592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudzik D.; Barbas-Bernardos C.; García A.; Barbas C. Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics. a Review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. 10.1016/j.jpba.2017.07.044. [DOI] [PubMed] [Google Scholar]
- Altelaar A. F. M. F. M.; Frese C. K.; Preisinger C.; Hennrich M. L.; Schram A. W.; Timmers H. T. M. T. M.; Heck A. J. R.; Mohammed S. Benchmarking Stable Isotope Labeling Based Quantitative Proteomics. J. Proteomics 2013, 88, 14–26. 10.1016/j.jprot.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Ahn N. G.; Shabb J. B.; Old W. M.; Resing K. A. Achieving In-Depth Proteomics Profiling by Mass Spectrometry. ACS Chem. Biol. 2007, 2 (1), 39–52. 10.1021/cb600357d. [DOI] [PubMed] [Google Scholar]
- Navarro P.; Kuharev J.; Gillet L. C.; Bernhardt O. M.; MacLean B.; Röst H. L.; Tate S. A.; Tsou C. C.; Reiter L.; Distler U.; Rosenberger G.; Perez-Riverol Y.; Nesvizhskii A. I.; Aebersold R.; Tenzer S. A Multicenter Study Benchmarks Software Tools for Label-Free Proteome Quantification. Nat. Biotechnol. 2016, 34 (11), 1130–1136. 10.1038/nbt.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelstrup C. D.; Bekker-Jensen D. B.; Arrey T. N.; Hogrebe A.; Harder A.; Olsen J. V. Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J. Proteome Res. 2018, 17 (1), 727–738. 10.1021/acs.jproteome.7b00602. [DOI] [PubMed] [Google Scholar]
- Köcher T.; Pichler P.; Swart R.; Mechtler K. Analysis of Protein Mixtures from Whole-Cell Extracts by Single-Run Nanolc-Ms/Ms Using Ultralong Gradients. Nat. Protoc. 2012, 7 (5), 882–890. 10.1038/nprot.2012.036. [DOI] [PubMed] [Google Scholar]
- Segers K.; Declerck S.; Mangelings D.; Heyden Y. Vander; Eeckhaut A. Van. Analytical Techniques for Metabolomic Studies: A Review. Bioanalysis 2019, 11 (24), 2297–2318. 10.4155/bio-2019-0014. [DOI] [PubMed] [Google Scholar]
- Pezzatti J.; Boccard J.; Codesido S.; Gagnebin Y.; Joshi A.; Picard D.; González-Ruiz V.; Rudaz S. Implementation of Liquid Chromatography–High Resolution Mass Spectrometry Methods for Untargeted Metabolomic Analyses of Biological Samples: A Tutorial. Anal. Chim. Acta 2020, 1105, 28–44. 10.1016/j.aca.2019.12.062. [DOI] [PubMed] [Google Scholar]
- O’Shea K.; Misra B. B. Software Tools, Databases and Resources in Metabolomics: Updates from 2018 to 2019. Metabolomics 2020, 16 (3), 36. 10.1007/s11306-020-01657-3. [DOI] [PubMed] [Google Scholar]
- Liebal U. W.; Phan A. N. T.; Sudhakar M.; Raman K.; Blank L. M. Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 2020, 10 (6), 243. 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanstrup J.; Broeckling C.; Helmus R.; Hoffmann N.; Mathé E.; Naake T.; Nicolotti L.; Peters K.; Rainer J.; Salek R.; Schulze T.; Schymanski E.; Stravs M.; Thévenot E.; Treutler H.; Weber R.; Willighagen E.; Witting M.; Neumann S. The MetaRbolomics Toolbox in Bioconductor and Beyond. Metabolites 2019, 9 (10), 200. 10.3390/metabo9100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochategui E.; Jaumot J.; Tauler R. ROIMCR: A Powerful Analysis Strategy for LC-MS Metabolomic Datasets. BMC Bioinf. 2019, 20 (1), 256. 10.1186/s12859-019-2848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C.; Kujawinski E. B. AutoTuner: High Fidelity and Robust Parameter Selection for Metabolomics Data Processing. Anal. Chem. 2020, 92 (8), 5724–5732. 10.1021/acs.analchem.9b04804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z.; Chong J.; Li S.; Xia J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10 (5), 186. 10.3390/metabo10050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmiger M. P.; Poetzsch M.; Steuer A. E.; Kraemer T. Parameter Optimization for Feature and Hit Generation in a General Unknown Screening Method - Proof of Concept Study Using a Design of Experiment Approach for a High Resolution Mass Spectrometry Procedure after Data Independent Acquisition. Anal. Chem. 2018, 90 (5), 3531–3536. 10.1021/acs.analchem.7b05387. [DOI] [PubMed] [Google Scholar]
- Hemmer S.; Manier S. K.; Fischmann S.; Westphal F.; Wagmann L.; Meyer M. R. Comparison of Three Untargeted Data Processing Workflows for Evaluating LC-HRMS Metabolomics Data. Metabolites 2020, 10 (9), 378. 10.3390/metabo10090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Lu Y.; Guo Y.; Cao H.; Wang Q.; Shui W. Comprehensive Evaluation of Untargeted Metabolomics Data Processing Software in Feature Detection, Quantification and Discriminating Marker Selection. Anal. Chim. Acta 2018, 1029, 50–57. 10.1016/j.aca.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Züllig T.; Zandl-Lang M.; Trötzmüller M.; Hartler J.; Plecko B.; Köfeler H. C. A Metabolomics Workflow for Analyzing Complex Biological Samples Using a Combined Method of Untargeted and Target-List Based Approaches. Metabolites 2020, 10 (9), 342. 10.3390/metabo10090342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L.; Wang J.; Page D.; Asthana S.; Zetterberg H.; Carlsson C.; Okonkwo O. C.; Li L. Comparative Evaluation of MS-Based Metabolomics Software and Its Application to Preclinical Alzheimer’s Disease. Sci. Rep. 2018, 8 (1), 1–10. 10.1038/s41598-018-27031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindelar M.; Patti G. J. Chemical Discovery in the Era of Metabolomics. J. Am. Chem. Soc. 2020, 142 (20), 9097–9105. 10.1021/jacs.9b13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus R.; Laak T. Ter; de Voogt P.; van Wezel A.; Schymanski E.. Patroon: Open Source Software Platform for Environmental Mass Spectrometry Based Non-Target Screening. 2020. 10.21203/rs.3.rs-36675/v1. [DOI] [PMC free article] [PubMed]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M.-Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. A Cross-Platform Toolkit for Mass Spectrometry and Proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; Want E. J.; O’Maille G.; Abagyan R.; Siuzdak G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78 (3), 779–787. 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Gloaguen Y.; Kirwan J.; Beule D.. Deep Learning Assisted Peak Curation for Large Scale LC-MS Metabolomics. bioRxiv 2020, 2020.08.09.242727 10.1101/2020.08.09.242727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetnik K.; Petrick L.; Pandey G. MetaClean: A Machine Learning-Based Classifier for Reduced False Positive Peak Detection in Untargeted LC–MS Metabolomics Data. Metabolomics 2020, 16 (11), 117. 10.1007/s11306-020-01738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangul S.; Martin L. S.; Hill B. L.; Lam A. K.-M.; Distler M. G.; Zelikovsky A.; Eskin E.; Flint J. Systematic Benchmarking of Omics Computational Tools. Nat. Commun. 2019, 10 (1), 1393. 10.1038/s41467-019-09406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller E.; Huber C.; Beckers L.-M.; Brack W.; Krauss M.; Schulze T. A Data Set of 255,000 Randomly Selected and Manually Classified Extracted Ion Chromatograms for Evaluation of Peak Detection Methods. Metabolites 2020, 10 (4), 162. 10.3390/metabo10040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin M. D.; Cao L.; Deik A. A.; Clish C. B.; Hu F. B.; Martínez-González M. A.; Razquin C.; Bullo M.; Corella D.; Gómez-Gracia E.; Fiol M.; Estruch R.; Lapetra J.; Fitó M.; Arós F.; Serra-Majem L.; Ros E.; Liang L. MetProc: Separating Measurement Artifacts from True Metabolites in an Untargeted Metabolomics Experiment. J. Proteome Res. 2019, 18 (3), 1446–1450. 10.1021/acs.jproteome.8b00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Clish C.; Hu F. B.; Martínez-González M. A.; Razquin C.; Bullo-Bonet M.; Corella D.; Gómez-Gracia E.; Fiol M.; Estruch R.; Lapetra J.; Fitó M.; Arós F.; Serra-Majem L.; Ros E.; Liang L.. GenuMet: Distinguish Genuine Untargeted Metabolic Features without Quality Control Samples. bioRxiv 2019, 837260. 10.1101/837260. [DOI] [Google Scholar]
- Tsugawa H.; Cajka T.; Kind T.; Ma Y.; Higgins B.; Ikeda K.; Kanazawa M.; Vandergheynst J.; Fiehn O.; Arita M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12 (6), 523–526. 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisier-Vannier O.; Chervin J.; Cabanac G.; Puech V.; Fournier S.; Durand V.; Amiel A.; André O.; Benamar O. A.; Dumas B.; Tsugawa H.; Marti G. MS-CleanR: A Feature-Filtering Workflow for Untargeted LC–MS Based Metabolomics. Anal. Chem. 2020, 92 (14), 9971–9981. 10.1021/acs.analchem.0c01594. [DOI] [PubMed] [Google Scholar]
- Mahieu N. G.; Spalding J. L.; Gelman S. J.; Patti G. J. Defining and Detecting Complex Peak Relationships in Mass Spectral Data: The Mz.Unity Algorithm. Anal. Chem. 2016, 88 (18), 9037–9046. 10.1021/acs.analchem.6b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.; Chiles E.; Maimouni S.; Wondisford F. E.; Zong W. X.; Song C. In-Source CID Ramping and Covariant Ion Analysis of Hydrophilic Interaction Chromatography Metabolomics. Anal. Chem. 2020, 92 (7), 4829–4837. 10.1021/acs.analchem.9b04181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Xing X.; Chen L.; Yang L.; Su X.; Rabitz H.; Lu W.; Rabinowitz J. D. Peak Annotation and Verification Engine for Untargeted LC-MS Metabolomics. Anal. Chem. 2019, 91 (3), 1838–1846. 10.1021/acs.analchem.8b03132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Jones D. R.; Shaw T. I.; Cho J. H.; Wang Y.; Tan H.; Xie B.; Zhou S.; Li Y.; Peng J. Target-Decoy-Based False Discovery Rate Estimation for Large-Scale Metabolite Identification. J. Proteome Res. 2018, 17 (7), 2328–2334. 10.1021/acs.jproteome.8b00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Liu B.; Zheng H.; Xiao X.; Li Z.; Luan E.; Li W.; Yang Y.; Wang Y.; Long Q.; Song J.; Zhang G. XY-Meta: A High-Efficiency Search Engine for Large-Scale Metabolome Annotation with Accurate FDR Estimation. Anal. Chem. 2020, 92 (8), 5701–5707. 10.1021/acs.analchem.9b03355. [DOI] [PubMed] [Google Scholar]
- Martano G.; Leone M.; D’oro P.; Matafora V.; Cattaneo A.; Masseroli M.; Bachi A. SMfinder: Small Molecules Finder for Metabolomics and Lipidomics Analysis. Anal. Chem. 2020, 92 (13), 8874–8882. 10.1021/acs.analchem.0c00585. [DOI] [PubMed] [Google Scholar]
- Reisdorph N. A.; Walmsley S.; Reisdorph R.. A Perspective and Framework for Developing Sample Type Specific Databases for LC/MS-Based Clinical Metabolomics. Metabolites 2020, 10 ( (1), ), 8. 10.3390/metabo10010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J.; Guijas C.; Benton H. P.; Warth B.; Siuzdak G. METLIN MS2Molecular Standards Database: A Broad Chemical and Biological Resource. Nat. Methods 2020, 17, 953–954. 10.1038/s41592-020-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frainay C.; Schymanski E. L.; Neumann S.; Merlet B.; Salek R. M.; Jourdan F.; Yanes O. Mind the Gap: Mapping Mass Spectral Databases in Genome-Scale Metabolic Networks Reveals Poorly Covered Areas. Metabolites 2018, 8 (3), 51. 10.3390/metabo8030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttkies C.; Schymanski E. L.; Wolf S.; Hollender J.; Neumann S. MetFrag Relaunched: Incorporating Strategies beyond in Silico Fragmentation. J. Cheminf. 2016, 8 (1), 3. 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y.; Pon A.; Karu N.; Zheng J.; Li C.; Arndt D.; Gautam M.; Allen F.; Wishart D. S.. Cfm-Id 3.0: Significantly Improved Esi-Ms/Ms Prediction and Compound Identification. Metabolites 2019, 9 ( (4), ), 72. 10.3390/metabo9040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K.; Shen H.; Meusel M.; Rousu J.; Böcker S. Searching Molecular Structure Databases with Tandem Mass Spectra Using CSI:FingerID. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (41), 12580–12585. 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W.; Beck J.; Brister J. R.; Bolton E. E.; Canese K.; Comeau D. C.; Funk K.; Ketter A.; Kim S.; Kimchi A.; Kitts P. A.; Kuznetsov A.; Lathrop S.; Lu Z.; McGarvey K.; Madden T. L.; Murphy T. D.; O’Leary N.; Phan L.; Schneider V. A.; Thibaud-Nissen F.; Trawick B. W.; Pruitt K. D.; Ostell J. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48 (D1), D9–D16. 10.1093/nar/gkz899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour N.; Alden N.; Menon R.; Jayaraman A.; Lee K.; Hassoun S.. Biological Filtering and Substrate Promiscuity Prediction for Annotating Untargeted Metabolomics. Metabolites 2020, 10 ( (4), ), 160. 10.3390/metabo10040160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Li L. Chemical Derivatization in LC-MS Based Metabolomics Study. TrAC, Trends Anal. Chem. 2020, 131, 115988. 10.1016/j.trac.2020.115988. [DOI] [Google Scholar]
- Guo K.; Li L. Differential 12C-/13C-Isotope Dansylation Labeling and Fast Liquid Chromatography/Mass Spectrometry for Absolute and Relative Quantification of the Metabolome. Anal. Chem. 2009, 81 (10), 3919–3932. 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- Guo K.; Bamforth F.; Li L. Qualitative Metabolome Analysis of Human Cerebrospinal Fluid by 13C-/ 12C-Isotope Dansylation Labeling Combined with Liquid Chromatography Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2011, 22 (2), 339–347. 10.1007/s13361-010-0033-4. [DOI] [PubMed] [Google Scholar]
- Huan T.; Wu Y.; Tang C.; Lin G.; Li L. DnsID in MyCompoundID for Rapid Identification of Dansylated Amine- and Phenol-Containing Metabolites in LC-MS-Based Metabolomics. Anal. Chem. 2015, 87 (19), 9838–9845. 10.1021/acs.analchem.5b02282. [DOI] [PubMed] [Google Scholar]
- Han W.; Li L. Matrix Effect on Chemical Isotope Labeling and Its Implication in Metabolomic Sample Preparation for Quantitative Metabolomics. Metabolomics 2015, 11 (6), 1733–1742. 10.1007/s11306-015-0826-3. [DOI] [Google Scholar]
- Ruttkies C.; Schymanski E. L.; Strehmel N.; Hollender J.; Neumann S.; Williams A. J.; Krauss M. Supporting Non-Target Identification by Adding Hydrogen Deuterium Exchange MS/MS Capabilities to MetFrag. Anal. Bioanal. Chem. 2019, 411 (19), 4683–4700. 10.1007/s00216-019-01885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R. P.; Garg A.; Putlur S. P.; Wagh S.; Kumar V.; Rao V.; Singh S.; Mandlekar S.; Desikan S. Practical and Economical Implementation of Online H/D Exchange in LC-MS. Anal. Chem. 2013, 85 (22), 10904–10912. 10.1021/ac402339s. [DOI] [PubMed] [Google Scholar]
- Kostyukevich Y.; Acter T.; Zherebker A.; Ahmed A.; Kim S.; Nikolaev E. Hydrogen/Deuterium Exchange in Mass Spectrometry. Mass Spectrom. Rev. 2018, 37 (6), 811–853. 10.1002/mas.21565. [DOI] [PubMed] [Google Scholar]
- Hooft J. J. J. van der; Wandy J.; Barrett M. P.; Burgess K. E. V; Rogers S. Topic Modeling for Untargeted Substructure Exploration in Metabolomics. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (48), 13738–13743. 10.1073/pnas.1608041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Hooft J. J. J.; Wandy J.; Young F.; Padmanabhan S.; Gerasimidis K.; Burgess K. E. V.; Barrett M. P.; Rogers S. Unsupervised Discovery and Comparison of Structural Families Across Multiple Samples in Untargeted Metabolomics. Anal. Chem. 2017, 89 (14), 7569–7577. 10.1021/acs.analchem.7b01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothias L.-F.; Petras D.; Schmid R.; Dührkop K.; Rainer J.; Sarvepalli A.; Protsyuk I.; Ernst M.; Tsugawa H.; Fleischauer M.; Aicheler F.; Aksenov A. A.; Alka O.; Allard P.-M.; Barsch A.; Cachet X.; Caraballo-Rodriguez A. M.; Da Silva R. R.; Dang T.; Garg N.; Gauglitz J. M.; Gurevich A.; Isaac G.; Jarmusch A. K.; Kameník Z.; Kang K. Bin; Kessler N.; Koester I.; Korf A.; Le Gouellec A.; Ludwig M.; Martin H C.; McCall L.-I.; McSayles J.; Meyer S. W.; Mohimani H.; Morsy M.; Moyne O.; Neumann S.; Neuweger H.; Nguyen N. H.; Nothias-Esposito M.; Paolini J.; Phelan V. V.; Pluskal T.; Quinn R. A.; Rogers S.; Shrestha B.; Tripathi A.; van der Hooft J. J. J.; Vargas F.; Weldon K. C.; Witting M.; Yang H.; Zhang Z.; Zubeil F.; Kohlbacher O.; Böcker S.; Alexandrov T.; Bandeira N.; Wang M.; Dorrestein P. C. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17 (9), 905–908. 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K.; Nothias L. F.; Fleischauer M.; Ludwig M.; Hoffmann M.; Rousu J.; Dorrestein P.; Böcker S.. Classes for the Masses: Systematic Classification of Unknowns Using Fragmentation Spectra. bioRxiv 2020, 2020.04.17.046672. 10.1101/2020.04.17.046672. [DOI] [Google Scholar]
- Bowden J. A.; Ulmer C. Z.; Jones C. M.; Koelmel J. P.; Yost R. A. NIST Lipidomics Workflow Questionnaire: An Assessment of Community-Wide Methodologies and Perspectives. Metabolomics 2018, 14 (5), 53. 10.1007/s11306-018-1340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartler J.; Armando A. M.; Trötzmüller M.; Dennis E. A.; Köfeler H. C.; Quehenberger O.. Automated Annotation of Sphingolipids Including Accurate Identification of Hydroxylation Sites Using MS n Data. Anal. Chem. 2020, acs.analchem.0c03016. 10.1021/acs.analchem.0c03016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H.; Ikeda K.; Takahashi M.; Satoh A.; Mori Y.; Uchino H.; Okahashi N.; Yamada Y.; Tada I.; Bonini P.; Higashi Y.; Okazaki Y.; Zhou Z.; Zhu Z. J.; Koelmel J.; Cajka T.; Fiehn O.; Saito K.; Arita M.; Arita M.. A Lipidome Atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159. 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- LIFS. https://lifs.isas.de/tools.html (accessed 2020-11-06).
- Hartler J.; Triebl A.; Ziegl A.; Trötzmüller M.; Rechberger G. N.; Zeleznik O. A.; Zierler K. A.; Torta F.; Cazenave-Gassiot A.; Wenk M. R.; Fauland A.; Wheelock C. E.; Armando A. M.; Quehenberger O.; Zhang Q.; Wakelam M. J. O. O.; Haemmerle G.; Spener F.; Köfeler H. C.; Thallinger G. G. Deciphering Lipid Structures Based on Platform-Independent Decision Rules. Nat. Methods 2017, 14 (12), 1171–1174. 10.1038/nmeth.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzenboeck L.; Troppmair N.; Schlachter S.; Koellensperger G.; Hartler J.; Rampler E. Chasing the Major Sphingolipids on Earth: Automated Annotation of Plant Glycosyl Inositol Phospho Ceramides by Glycolipidomics. Metabolites 2020, 10 (9), 1–20. 10.3390/metabo10090375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovčačíková M.; Lísa M.; Cífková E.; Holčapek M. Retention Behavior of Lipids in Reversed-Phase Ultrahigh-Performance Liquid Chromatography–Electrospray Ionization Mass Spectrometry. J. Chromatogr. A 2016, 1450, 76–85. 10.1016/j.chroma.2016.04.082. [DOI] [PubMed] [Google Scholar]
- Lísa M.; Holčapek M. Triacylglycerols Profiling in Plant Oils Important in Food Industry, Dietetics and Cosmetics Using High-Performance Liquid Chromatography–Atmospheric Pressure Chemical Ionization Mass Spectrometry. J. Chromatogr. A 2008, 1198–1199, 115–130. 10.1016/j.chroma.2008.05.037. [DOI] [PubMed] [Google Scholar]
- Aicheler F.; Li J.; Hoene M.; Lehmann R.; Xu G.; Kohlbacher O. Retention Time Prediction Improves Identification in Nontargeted Lipidomics Approaches. Anal. Chem. 2015, 87 (15), 7698–7704. 10.1021/acs.analchem.5b01139. [DOI] [PubMed] [Google Scholar]
- Korf A.; Vosse C.; Schmid R.; Helmer P. O.; Jeck V.; Hayen H. Three-Dimensional Kendrick Mass Plots as a Tool for Graphical Lipid Identification. Rapid Commun. Mass Spectrom. 2018, 32 (12), 981–991. 10.1002/rcm.8117. [DOI] [PubMed] [Google Scholar]
- Koelmel J. P.; Li X.; Stow S. M.; Sartain M. J.; Murali A.; Kemperman R.; Tsugawa H.; Takahashi M.; Vasiliou V.; Bowden J. A.; Yost R. A.; Garrett T. J.; Kitagawa N. Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using a Rapid and User-Friendly Software. Metabolites 2020, 10 (3), 101. 10.3390/metabo10030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIPID MAPS® Lipidomics Gateway. https://www.lipidmaps.org (accessed 2020-03-15).
- Domingo-Almenara X.; Guijas C.; Billings E.; Montenegro-Burke J. R.; Uritboonthai W.; Aisporna A. E.; Chen E.; Benton H. P.; Siuzdak G. The METLIN Small Molecule Dataset for Machine Learning-Based Retention Time Prediction. Nat. Commun. 2019, 10 (1), 1–9. 10.1038/s41467-019-13680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting M.; Böcker S. Current Status of Retention Time Prediction in Metabolite Identification. J. Sep. Sci. 2020, 43 (9–10), 1746–1754. 10.1002/jssc.202000060. [DOI] [PubMed] [Google Scholar]
- Bonini P.; Kind T.; Tsugawa H.; Barupal D. K.; Fiehn O. Retip: Retention Time Prediction for Compound Annotation in Untargeted Metabolomics. Anal. Chem. 2020, 92 (11), 7515–7522. 10.1021/acs.analchem.9b05765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor B. C.; Catrow J. L.; Maschek J. A.; Cox J. E. QSRR Automator: A Tool for Automating Retention Time Prediction in Lipidomics and Metabolomics. Metabolites 2020, 10 (6), 237. 10.3390/metabo10060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglia G.; Williams J. P.; Menikarachchi L.; Thompson J. W.; Tyldesley-Worster R.; Halldórsson S.; Rolfsson O.; Moseley A.; Grant D.; Langridge J.; Palsson B. O.; Astarita G. Ion Mobility Derived Collision Cross Sections to Support Metabolomics Applications. Anal. Chem. 2014, 86 (8), 3985–3993. 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Atri V.; Causon T.; Hernandez-Alba O.; Mutabazi A.; Veuthey J.-L.; Cianferani S.; Guillarme D. Adding a New Separation Dimension to MS and LC-MS: What Is the Utility of Ion Mobility Spectrometry?. J. Sep. Sci. 2018, 41 (1), 20–67. 10.1002/jssc.201700919. [DOI] [PubMed] [Google Scholar]
- Mairinger T.; Causon T. J.; Hann S. The Potential of Ion Mobility–Mass Spectrometry for Non-Targeted Metabolomics. Curr. Opin. Chem. Biol. 2018, 42, 9–15. 10.1016/j.cbpa.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Picache J. A.; Rose B. S.; Balinski A.; Leaptrot K. L.; Sherrod S. D.; May J. C.; McLean J. A. Collision Cross Section Compendium to Annotate and Predict Multi-Omic Compound Identities. Chem. Sci. 2019, 10 (4), 983–993. 10.1039/C8SC04396E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Mesa M.; Le Bizec B.; Monteau F.; García-Campaña A. M.; Dervilly-Pinel G. Collision Cross Section (CCS) Database: An Additional Measure to Characterize Steroids. Anal. Chem. 2018, 90 (7), 4616–4625. 10.1021/acs.analchem.7b05117. [DOI] [PubMed] [Google Scholar]
- Nichols C. M.; Dodds J. N.; Rose B. S.; Picache J. A.; Morris C. B.; Codreanu S. G.; May J. C.; Sherrod S. D.; McLean J. A. Untargeted Molecular Discovery in Primary Metabolism: Collision Cross Section as a Molecular Descriptor in Ion Mobility-Mass Spectrometry. Anal. Chem. 2018, 90 (24), 14484–14492. 10.1021/acs.analchem.8b04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. H.; Cho J. H.; Xu L. Breaking down Structural Diversity for Comprehensive Prediction of Ion-Neutral Collision Cross Sections. Anal. Chem. 2020, 92 (6), 4548–4557. 10.1021/acs.analchem.9b05772. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Luo M.; Chen X.; Yin Y.; Xiong X.; Wang R.; Zhu Z. J. Ion Mobility Collision Cross-Section Atlas for Known and Unknown Metabolite Annotation in Untargeted Metabolomics. Nat. Commun. 2020, 11 (1), 1–13. 10.1038/s41467-020-18171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow S. M.; Causon T. J.; Zheng X.; Kurulugama R. T.; Mairinger T.; May J. C.; Rennie E. E.; Baker E. S.; Smith R. D.; McLean J. A.; Hann S.; Fjeldsted J. C. An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Anal. Chem. 2017, 89 (17), 9048–9055. 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causon T. J.; Ivanova-Petropulos V.; Petrusheva D.; Bogeva E.; Hann S. Fingerprinting of Traditionally Produced Red Wines Using Liquid Chromatography Combined with Drift Tube Ion Mobility-Mass Spectrometry. Anal. Chim. Acta 2019, 1052, 179–189. 10.1016/j.aca.2018.11.040. [DOI] [PubMed] [Google Scholar]
- Wojcik R.; Nagy G.; Attah I. K.; Webb I. K.; Garimella S. V. B.; Weitz K. K.; Hollerbach A.; Monroe M. E.; Ligare M. R.; Nielson F. F.; Norheim R. V.; Renslow R. S.; Metz T. O.; Ibrahim Y. M.; Smith R. D. SLIM Ultrahigh Resolution Ion Mobility Spectrometry Separations of Isotopologues and Isotopomers Reveal Mobility Shifts Due to Mass Distribution Changes. Anal. Chem. 2019, 91 (18), 11952–11962. 10.1021/acs.analchem.9b02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K.; Ujma J.; Wildgoose J.; Pringle S.; Richardson K.; Langridge D.; Green M. A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91 (13), 8564–8573. 10.1021/acs.analchem.9b01838. [DOI] [PubMed] [Google Scholar]
- Fouque K. J. D.; Ramirez C. E.; Lewis R. L.; Koelmel J. P.; Garrett T. J.; Yost R. A.; Fernandez-Lima F. Effective Liquid Chromatography-Trapped Ion Mobility Spectrometry-Mass Spectrometry Separation of Isomeric Lipid Species. Anal. Chem. 2019, 91 (8), 5021–5027. 10.1021/acs.analchem.8b04979. [DOI] [PubMed] [Google Scholar]
- Xie X.; Xia Y. Analysis of Conjugated Fatty Acid Isomers by the Paternò-Büchi Reaction and Trapped Ion Mobility Mass Spectrometry. Anal. Chem. 2019, 91 (11), 7173–7180. 10.1021/acs.analchem.9b00374. [DOI] [PubMed] [Google Scholar]
- Poad B. L. J.; Zheng X.; Mitchell T. W.; Smith R. D.; Baker E. S.; Blanksby S. J. Online Ozonolysis Combined with Ion Mobility-Mass Spectrometry Provides a New Platform for Lipid Isomer Analyses. Anal. Chem. 2018, 90 (2), 1292–1300. 10.1021/acs.analchem.7b04091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miggiels P.; Wouters B.; van Westen G. J. P.; Dubbelman A.-C.; Hankemeier T. Novel Technologies for Metabolomics: More for Less. TrAC, Trends Anal. Chem. 2019, 120, 115323. 10.1016/j.trac.2018.11.021. [DOI] [Google Scholar]
- Matyash V.; Liebisch G.; Kurzchalia T. V.; Shevchenko A.; Schwudke D. Lipid Extraction by Methyl- Tert -Butyl Ether for High-Throughput Lipidomics. J. Lipid Res. 2008, 49 (5), 1137–1146. 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Romm M.; Zheng X.; Zink E. M.; Kim Y. M.; Burnum-Johnson K. E.; Orton D. J.; Apffel A.; Ibrahim Y. M.; Monroe M. E.; Moore R. J.; Smith J. N.; Ma J.; Renslow R. S.; Thomas D. G.; Blackwell A. E.; Swinford G.; Sausen J.; Kurulugama R. T.; Eno N.; Darland E.; Stafford G.; Fjeldsted J.; Metz T. O.; Teeguarden J. G.; Smith R. D.; Baker E. S. SPE-IMS-MS: An Automated Platform for Sub-Sixty Second Surveillance of Endogenous Metabolites and Xenobiotics in Biofluids. Clin. Mass Spectrom 2016, 2, 1–10. 10.1016/j.clinms.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexovič M.; Horstkotte B.; Šrámková I.; Solich P.; Sabo J. Automation of Dispersive Liquid–Liquid Microextraction and Related Techniques. Approaches Based on Flow, Batch, Flow-Batch and in-Syringe Modes. TrAC, Trends Anal. Chem. 2017, 86, 39–55. 10.1016/j.trac.2016.10.003. [DOI] [Google Scholar]
- Reyes-Garcés N.; Gionfriddo E. Recent Developments and Applications of Solid Phase Microextraction as a Sample Preparation Approach for Mass-Spectrometry-Based Metabolomics and Lipidomics. TrAC, Trends Anal. Chem. 2019, 113, 172–181. 10.1016/j.trac.2019.01.009. [DOI] [Google Scholar]
- Filipiak W.; Bojko B.. SPME in Clinical, Pharmaceutical, and Biotechnological Research – How Far Are We from Daily Practice? TrAC - Trends Anal. Chem. June 1, 2019, 115, 203–213. 10.1016/j.trac.2019.02.029. [DOI] [Google Scholar]
- Drouin N.; Kubáň P.; Rudaz S.; Pedersen-Bjergaard S.; Schappler J. Electromembrane Extraction: Overview of the Last Decade. TrAC, Trends Anal. Chem. 2019, 113, 357–363. 10.1016/j.trac.2018.10.024. [DOI] [Google Scholar]
- Jaroch K.; Boyaci E.; Pawliszyn J.; Bojko B. The Use of Solid Phase Microextraction for Metabolomic Analysis of Non-Small Cell Lung Carcinoma Cell Line (A549) after Administration of Combretastatin A4. Sci. Rep. 2019, 9 (1), 1–9. 10.1038/s41598-018-36481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani S. A.; Lendor S.; Boyaci E.; Pawliszyn J.; Womelsdorf T. Multineuromodulator Measurements across Fronto-Striatal Network Areas of the Behaving Macaque Using Solid-Phase Microextraction. J. Neurophysiol. 2019, 122 (4), 1649–1660. 10.1152/jn.00321.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Garcés N.; Diwan M.; Boyacl E.; Gómez-Ríos G. A.; Bojko B.; Nobrega J. N.; Bambico F. R.; Hamani C.; Pawliszyn J. In Vivo Brain Sampling Using a Microextraction Probe Reveals Metabolic Changes in Rodents after Deep Brain Stimulation. Anal. Chem. 2019, 91 (15), 9875–9884. 10.1021/acs.analchem.9b01540. [DOI] [PubMed] [Google Scholar]
- Lendor S.; Gómez-Ríos G. A.; Boyacl E.; Vander Heide H.; Pawliszyn J. Space-Resolved Tissue Analysis by Solid-Phase Microextraction Coupled to High-Resolution Mass Spectrometry via Desorption Electrospray Ionization. Anal. Chem. 2019, 91 (15), 10141–10148. 10.1021/acs.analchem.9b02157. [DOI] [PubMed] [Google Scholar]
- Risticevic S.; Souza-Silva E. A.; Gionfriddo E.; DeEll J. R.; Cochran J.; Hopkins W. S.; Pawliszyn J.. Application of in Vivo Solid Phase Microextraction (SPME) in Capturing Metabolome of Apple (Malus ×domestica Borkh.) Fruit. Sci. Rep. 2020, 10 ( (1), ), 10.1038/s41598-020-63817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubáň P.; Boček P. Capillary Electrophoresis with Capacitively Coupled Contactless Conductivity Detection: A Universal Tool for the Determination of Supported Liquid Membrane Selectivity in Electromembrane Extraction of Complex Samples. J. Chromatogr. A 2012, 1267, 96–101. 10.1016/j.chroma.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Vardal L.; Gjelstad A.; Huang C.; Oiestad E. L.; Pedersen-Bjergaard S. Efficient Discrimination and Removal of Phospholipids during Electromembrane Extraction from Human Plasma Samples. Bioanalysis 2017, 9 (8), 631–641. 10.4155/bio-2017-0027. [DOI] [PubMed] [Google Scholar]
- Hansen F. A.; Sticker D.; Kutter J. P.; Petersen N. J.; Pedersen-Bjergaard S. Nanoliter-Scale Electromembrane Extraction and Enrichment in a Microfluidic Chip. Anal. Chem. 2018, 90 (15), 9322–9329. 10.1021/acs.analchem.8b01936. [DOI] [PubMed] [Google Scholar]
- González-Domínguez R.; Sayago A.; Fernández-Recamales Á. Direct Infusion Mass Spectrometry for Metabolomic Phenotyping of Diseases. Bioanalysis 2017, 9 (1), 131–148. 10.4155/bio-2016-0202. [DOI] [PubMed] [Google Scholar]
- Sarvin B.; Lagziel S.; Sarvin N.; Mukha D.; Kumar P.; Aizenshtein E.; Shlomi T. Fast and Sensitive Flow-Injection Mass Spectrometry Metabolomics by Analyzing Sample-Specific Ion Distributions. Nat. Commun. 2020, 11 (1), 1–11. 10.1038/s41467-020-17026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan T.; Dubbelman A.-C.; Duisters K.; Kindt A.; Harms A. C.; Hankemeier T. High-Throughput Fractionation Coupled to Mass Spectrometry for Improved Quantitation in Metabolomics. Anal. Chem. 2020, 92 (21), 14330–14338. 10.1021/acs.analchem.0c01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds J. N.; Baker E. S. Ion Mobility Spectrometry: Fundamental Concepts, Instrumentation, Applications, and the Road Ahead. J. Am. Soc. Mass Spectrom. 2019, 30 (11), 2185–2195. 10.1007/s13361-019-02288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthias F.; Wang Y.; Alhajji E.; Rieul B.; Moussa F.; Benoist J. F.; Maître P. Identification and Quantification of Amino Acids and Related Compounds Based on Differential Mobility Spectrometry. Analyst 2020, 145 (14), 4889–4900. 10.1039/D0AN00377H. [DOI] [PubMed] [Google Scholar]
- Zang X.; Monge M. E.; Gaul D. A.; Fernández F. M. Flow Injection-Traveling-Wave Ion Mobility-Mass Spectrometry for Prostate-Cancer Metabolomics. Anal. Chem. 2018, 90 (22), 13767–13774. 10.1021/acs.analchem.8b04259. [DOI] [PubMed] [Google Scholar]
- Wernisch S.; Pennathur S. Application of Differential Mobility-Mass Spectrometry for Untargeted Human Plasma Metabolomic Analysis. Anal. Bioanal. Chem. 2019, 411 (24), 6297–6308. 10.1007/s00216-019-01719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T.; Ozbal C.; Holstein M.; Winter M.; Buettner F. H.; Thamm S.; Bischoff D.; Luippold A. H. RapidFire BLAZE-Mode Is Boosting ESI-MS Toward High-Throughput-Screening. SLAS Technol. 2019, 24 (4), 386–393. 10.1177/2472630318822449. [DOI] [PubMed] [Google Scholar]
- Van Berkel G. J.; Kertesz V.; Boeltz H. Immediate Drop on Demand Technology (I-DOT) Coupled with Mass Spectrometry via an Open Port Sampling Interface. Bioanalysis 2017, 9 (21), 1667–1679. 10.4155/bio-2017-0104. [DOI] [PubMed] [Google Scholar]
- Sinclair I.; Bachman M.; Addison D.; Rohman M.; Murray D. C.; Davies G.; Mouchet E.; Tonge M. E.; Stearns R. G.; Ghislain L.; Datwani S. S.; Majlof L.; Hall E.; Jones G. R.; Hoyes E.; Olechno J.; Ellson R. N.; Barran P. E.; Pringle S. D.; Morris M. R.; Wingfield J. Acoustic Mist Ionization Platform for Direct and Contactless Ultrahigh-Throughput Mass Spectrometry Analysis of Liquid Samples. Anal. Chem. 2019, 91 (6), 3790–3794. 10.1021/acs.analchem.9b00142. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Liu C.; Hua W.; Ghislain L.; Liu J.; Aschenbrenner L.; Noell S.; Dirico K.; Lanyon L.; Steppan C.; Arnold D.; Covey T.; Datwani S.; Troutman M.. Acoustic Ejection Mass Spectrometry for High-Throughput Analysis. bioRxiv 2020, 2020.01.28.923938. 10.1101/2020.01.28.923938. [DOI] [PubMed] [Google Scholar]
- Häbe T. T.; Liu C.; Covey T. R.; Simon R. P.; Reindl W.; Büttner F. H.; Winter M.; Bischoff D.; Luippold A. H.; Runge F. Ultrahigh-Throughput ESI-MS: Sampling Pushed to Six Samples per Second by Acoustic Ejection Mass Spectrometry. Anal. Chem. 2020, 92 (18), 12242–12249. 10.1021/acs.analchem.0c01632. [DOI] [PubMed] [Google Scholar]
- Schmidt A.; Karas M.; Dülcks T. Effect of Different Solution Flow Rates on Analyte Ion Signals in Nano-ESI MS, or: When Does ESI Turn into Nano-ESI?. J. Am. Soc. Mass Spectrom. 2003, 14 (5), 492–500. 10.1016/S1044-0305(03)00128-4. [DOI] [PubMed] [Google Scholar]
- Coman C.; Solari F. A.; Hentschel A.; Sickmann A.; Zahedi R. P.; Ahrends R. Simultaneous Metabolite, Protein, Lipid Extraction (SIMPLEX): A Combinatorial Multimolecular Omics Approach for Systems Biology. Mol. Cell. Proteomics 2016, 15 (4), 1453–1466. 10.1074/mcp.M115.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann K.; Almeida R.; Baumert M.; Herzog R.; Bornstein S. R.; Shevchenko A. Shotgun Lipidomics on a LTQ Orbitrap Mass Spectrometer by Successive Switching between Acquisition Polarity Modes. J. Mass Spectrom. 2012, 47 (1), 96–104. 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- Simons B.; Kauhanen D.; Sylvänne T.; Tarasov K.; Duchoslav E.; Ekroos K. Shotgun Lipidomics by Sequential Precursor Ion Fragmentation on a Hybrid Quadrupole Time-of-Flight Mass Spectrometer. Metabolites 2012, 2 (1), 195–213. 10.3390/metabo2010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann K.; Thomas H.; Ackerman J. M.; Nagornov K. O.; Tsybin Y. O.; Shevchenko A. Intensity-Independent Noise Filtering in FT MS and FT MS/MS Spectra for Shotgun Lipidomics. Anal. Chem. 2017, 89 (13), 7046–7052. 10.1021/acs.analchem.7b00794. [DOI] [PubMed] [Google Scholar]
- Hu C.; Wang C.; He L.; Han X.. Novel Strategies for Enhancing Shotgun Lipidomics for Comprehensive Analysis of Cellular Lipidomes. TrAC - Trends Anal. Chem. November 1, 2019, 120, 115330. 10.1016/j.trac.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann K.; Moon H. K.; Thomas H.; Ackerman J. M.; Groessl M.; Wagner N.; Kellmann M.; Henry I.; Nadler A.; Shevchenko A. Quantitative Fragmentation Model for Bottom-Up Shotgun Lipidomics. Anal. Chem. 2019, 91 (18), 12085–12093. 10.1021/acs.analchem.9b03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders K. L.; Edwards J. L. Nano-Liquid Chromatography-Mass Spectrometry and Recent Applications in Omics Investigations. Anal. Methods 2020, 12 (36), 4404–4417. 10.1039/D0AY01194K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani K.; Izumi Y.; Hata K.; Bamba T.. An Analytical System for Single-Cell Metabolomics of Typical Mammalian Cells Based on Highly Sensitive Nano-Liquid Chromatography Tandem Mass Spectrometry. Mass Spectrom.. 2020, 9 ( (1), ), A0080. 10.5702/massspectrometry.A0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danne-Rasche N.; Coman C.; Ahrends R. Nano-LC/NSI MS Refines Lipidomics by Enhancing Lipid Coverage, Measurement Sensitivity, and Linear Dynamic Range. Anal. Chem. 2018, 90 (13), 8093–8101. 10.1021/acs.analchem.8b01275. [DOI] [PubMed] [Google Scholar]
- Vasconcelos Soares Maciel E.; de Toffoli A. L.; Sobieski E.; Domingues Nazário C. E.; Lanças F. M. Miniaturized Liquid Chromatography Focusing on Analytical Columns and Mass Spectrometry: A Review. Anal. Chim. Acta 2020, 1103, 11–31. 10.1016/j.aca.2019.12.064. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Shou W.; Ogura T.; Li S.; Weller H. Optimization of Microflow LC-MS/MS and Its Utility in Quantitative Discovery Bioanalysis. Bioanalysis 2019, 11 (11), 1117–1127. 10.4155/bio-2019-0076. [DOI] [PubMed] [Google Scholar]
- Gray N.; Adesina-Georgiadis K.; Chekmeneva E.; Plumb R. S.; Wilson I. D.; Nicholson J. K. Development of a Rapid Microbore Metabolic Profiling Ultraperformance Liquid Chromatography-Mass Spectrometry Approach for High-Throughput Phenotyping Studies. Anal. Chem. 2016, 88 (11), 5742–5751. 10.1021/acs.analchem.6b00038. [DOI] [PubMed] [Google Scholar]
- Nye L. C.; Williams J. P.; Munjoma N. C.; Letertre M. P. M.; Coen M.; Bouwmeester R.; Martens L.; Swann J. R.; Nicholson J. K.; Plumb R. S.; McCullagh M.; Gethings L. A.; Lai S.; Langridge J. I.; Vissers J. P. C.; Wilson I. D. A Comparison of Collision Cross Section Values Obtained via Travelling Wave Ion Mobility-Mass Spectrometry and Ultra High Performance Liquid Chromatography-Ion Mobility-Mass Spectrometry: Application to the Characterisation of Metabolites in Rat Urine. J. Chromatogr. A 2019, 1602, 386–396. 10.1016/j.chroma.2019.06.056. [DOI] [PubMed] [Google Scholar]
- Letertre M.; Munjoma N. C.; Slade S. E.; Plumb R. S.; Swann J.; Coen M.; Nicholson J. K.; Wilson I. D. Metabolic Phenotyping Using UPLC–MS and Rapid Microbore UPLC–IM–MS: Determination of the Effect of Different Dietary Regimes on the Urinary Metabolome of the Rat. Chromatographia 2020, 83 (7), 853–861. 10.1007/s10337-020-03900-4. [DOI] [Google Scholar]
- King A. M.; Mullin L. G.; Wilson I. D.; Coen M.; Rainville P. D.; Plumb R. S.; Gethings L. A.; Maker G.; Trengove R. Development of a Rapid Profiling Method for the Analysis of Polar Analytes in Urine Using HILIC–MS and Ion Mobility Enabled HILIC–MS. Metabolomics 2019, 15 (2), 17. 10.1007/s11306-019-1474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M.; Trengove R. D.; Mullin L. G.; Rainville P. D.; Isaac G.; Plumb R. S.; Gethings L. A.; Wilson I. D. Rapid Profiling Method for the Analysis of Lipids in Human Plasma Using Ion Mobility Enabled-Reversed Phase-Ultra High Performance Liquid Chromatography/Mass Spectrometry. J. Chromatogr. A 2020, 1611, 460597. 10.1016/j.chroma.2019.460597. [DOI] [PubMed] [Google Scholar]
- Márta Z.; Bobály B.; Fekete J.; Magda B.; Imre T.; Mészáros K. V.; Bálint M.; Szabó P. T. Simultaneous Determination of Thirteen Different Steroid Hormones Using Micro UHPLC-MS/MS with on-Line SPE System. J. Pharm. Biomed. Anal. 2018, 150, 258–267. 10.1016/j.jpba.2017.12.014. [DOI] [PubMed] [Google Scholar]
- Cebo M.; Fu X.; Gawaz M.; Chatterjee M.; Lämmerhofer M. Micro-UHPLC-MS/MS Method for Analysis of Oxylipins in Plasma and Platelets. J. Pharm. Biomed. Anal. 2020, 189, 113426. 10.1016/j.jpba.2020.113426. [DOI] [PubMed] [Google Scholar]
- Stoll D. R.; Carr P. W. Two-Dimensional Liquid Chromatography: A State of the Art Tutorial. Anal. Chem. 2017, 89 (1), 519–531. 10.1021/acs.analchem.6b03506. [DOI] [PubMed] [Google Scholar]
- Haggarty J.; Burgess K. E. Recent Advances in Liquid and Gas Chromatography Methodology for Extending Coverage of the Metabolome. Curr. Opin. Biotechnol. 2017, 43, 77–85. 10.1016/j.copbio.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Lv W.; Shi X.; Wang S.; Xu G. Multidimensional Liquid Chromatography-Mass Spectrometry for Metabolomic and Lipidomic Analyses. TrAC, Trends Anal. Chem. 2019, 120, 115302. 10.1016/j.trac.2018.11.001. [DOI] [Google Scholar]
- Xu M.; Legradi J.; Leonards P. Evaluation of LC-MS and LC×LC-MS in Analysis of Zebrafish Embryo Samples for Comprehensive Lipid Profiling. Anal. Bioanal. Chem. 2020, 412 (18), 4313–4325. 10.1007/s00216-020-02661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Nie H.; Zhao F.; Song S.; Meng Y.; Bai Y.; Liu H. A Novel Online Two-Dimensional Supercritical Fluid Chromatography/Reversed Phase Liquid Chromatography–Mass Spectrometry Method for Lipid Profiling. Anal. Bioanal. Chem. 2020, 412 (10), 2225–2235. 10.1007/s00216-019-02242-x. [DOI] [PubMed] [Google Scholar]
- Koga R.; Miyoshi Y.; Sato Y.; Mita M.; Konno R.; Lindner W.; Hamase K. Enantioselective Determination of Citrulline and Ornithine in the Urine of D-Amino Acid Oxidase Deficient Mice Using a Two-Dimensional High-Performance Liquid Chromatographic System. J. Chromatogr. A 2016, 1467, 312–317. 10.1016/j.chroma.2016.07.053. [DOI] [PubMed] [Google Scholar]
- Kimura T.; Hamase K.; Miyoshi Y.; Yamamoto R.; Yasuda K.; Mita M.; Rakugi H.; Hayashi T.; Isaka Y. Chiral Amino Acid Metabolomics for Novel Biomarker Screening in the Prognosis of Chronic Kidney Disease. Sci. Rep. 2016, 6 (1), 1–7. 10.1038/srep26137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmayr K.; Hann S.; Koellensperger G. Complementing Reversed-Phase Selectivity with Porous Graphitized Carbon to Increase the Metabolome Coverage in an on-Line Two-Dimensional LC-MS Setup for Metabolomics. Analyst 2015, 140 (10), 3465–3473. 10.1039/C5AN00206K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampler E.; Schoeny H.; Mitic B. M.; El Abiead Y.; Schwaiger M.; Koellensperger G.. Simultaneous Non-Polar and Polar Lipid Analysis by on-Line Combination of HILIC, RP and High Resolution MS. Analyst 2018, 143, 1250. 10.1039/C7AN01984J. [DOI] [PubMed] [Google Scholar]
- Schwaiger M.; Schoeny H.; El Abiead Y.; Hermann G.; Rampler E.; Koellensperger G.. Merging Metabolomics and Lipidomics into One Analytical Run. Analyst 2019, 144 ( (1), ), 220. 10.1039/C8AN01219A. [DOI] [PubMed] [Google Scholar]
- Klavins K.; Drexler H.; Hann S.; Koellensperger G. Quantitative Metabolite Profiling Utilizing Parallel Column Analysis for Simultaneous Reversed-Phase and Hydrophilic Interaction Liquid Chromatography Separations Combined with Tandem Mass Spectrometry. Anal. Chem. 2014, 86 (9), 4145–4150. 10.1021/ac5003454. [DOI] [PubMed] [Google Scholar]
- Wang S.; Zhou L.; Wang Z.; Shi X.; Xu G. Simultaneous Metabolomics and Lipidomics Analysis Based on Novel Heart-Cutting Two-Dimensional Liquid Chromatography-Mass Spectrometry. Anal. Chim. Acta 2017, 966, 34–40. 10.1016/j.aca.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Lv W.; Wang L.; Xuan Q.; Zhao X.; Liu X.; Shi X.; Xu G. Pseudotargeted Method Based on Parallel Column Two-Dimensional Liquid Chromatography-Mass Spectrometry for Broad Coverage of Metabolome and Lipidome. Anal. Chem. 2020, 92 (8), 6043–6050. 10.1021/acs.analchem.0c00372. [DOI] [PubMed] [Google Scholar]
- Chen S.; Kong H.; Lu X.; Li Y.; Yin P.; Zeng Z.; Xu G. Pseudotargeted Metabolomics Method and Its Application in Serum Biomarker Discovery for Hepatocellular Carcinoma Based on Ultra High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry. Anal. Chem. 2013, 85 (17), 8326–8333. 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- Domingo-Almenara X.; Montenegro-Burke J. R.; Ivanisevic J.; Thomas A.; Sidibé J.; Teav T.; Guijas C.; Aisporna A. E.; Rinehart D.; Hoang L.; Nordström A.; Gómez-Romero M.; Whiley L.; Lewis M. R.; Nicholson J. K.; Benton H. P.; Siuzdak G. XCMS-MRM and METLIN-MRM: A Cloud Library and Public Resource for Targeted Analysis of Small Molecules. Nat. Methods 2018, 15 (9), 681–684. 10.1038/s41592-018-0110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.; Leclerc É.; Hamedpour V.; Kubota R.; Takizawa S. Y.; Sakai Y.; Minami T. Simplest Chemosensor Array for Phosphorylated Saccharides. Anal. Chem. 2019, 91 (24), 15570–15576. 10.1021/acs.analchem.9b03578. [DOI] [PubMed] [Google Scholar]
- Guo J.; Huan T. Comparison of Full-Scan, Data-Dependent, and Data-Independent Acquisition Modes in Liquid Chromatography-Mass Spectrometry Based Untargeted Metabolomics. Anal. Chem. 2020, 92 (12), 8072–8080. 10.1021/acs.analchem.9b05135. [DOI] [PubMed] [Google Scholar]
- Jian Guo T. H. Evaluation of Significant Features Discovered from Different Data Acquisition Modes in Mass Spectrometry-Based Untargeted Metabolomics. Sci. Total Environ. 2020, 703, 135577. 10.1016/j.scitotenv.2019.135577. [DOI] [PubMed] [Google Scholar]
- Barbier Saint Hilaire P.; Rousseau K.; Seyer A.; Dechaumet S.; Damont A.; Junot C.; Fenaille F. Comparative Evaluation of Data Dependent and Data Independent Acquisition Workflows Implemented on an Orbitrap Fusion for Untargeted Metabolomics. Metabolites 2020, 10 (4), 158. 10.3390/metabo10040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelmel J. P.; Kroeger N. M.; Gill E. L.; Ulmer C. Z.; Bowden J. A.; Patterson R. E.; Yost R. A.; Garrett T. J. Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J. Am. Soc. Mass Spectrom. 2017, 28 (5), 908–917. 10.1007/s13361-017-1608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandy J.; Davies V.; Van Der Hooft J. J. J.; Weidt S.; Daly R.; Rogers S.. In Silico Optimization of Mass Spectrometry Fragmentation Strategies in Metabolomics. Metabolites 2019, 9 ( (10), ), 219. 10.3390/metabo9100219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinu F. R.; Beale D. J.; Paten A. M.; Kouremenos K.; Swarup S.; Schirra H. J.; Wishart D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9 (4), 76. 10.3390/metabo9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raterink R.-J.; Lindenburg P. W.; Vreeken R. J.; Ramautar R.; Hankemeier T. Recent Developments in Sample-Pretreatment Techniques for Mass Spectrometry-Based Metabolomics. TrAC, Trends Anal. Chem. 2014, 61, 157–167. 10.1016/j.trac.2014.06.003. [DOI] [Google Scholar]
- Gutierrez D. B.; Gant-Branum R. L.; Romer C. E.; Farrow M. A.; Allen J. L.; Dahal N.; Nei Y.-W. W.; Codreanu S. G.; Jordan A. T.; Palmer L. D.; Sherrod S. D.; McLean J. A.; Skaar E. P.; Norris J. L.; Caprioli R. M. An Integrated, High-Throughput Strategy for Multiomic Systems Level Analysis. J. Proteome Res. 2018, 17 (10), 3396–3408. 10.1021/acs.jproteome.8b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zougman A.; Wilson J. P.; Roberts L. D.; Banks R. E. Detergent-Free Simultaneous Sample Preparation Method for Proteomics and Metabolomics. J. Proteome Res. 2020, 19 (7), 2838–2844. 10.1021/acs.jproteome.9b00662. [DOI] [PubMed] [Google Scholar]
- Folch J.; Lees M.; Sloane Stanley G. H. H. A Simple Method for Isolation and Purification of Total Lipides from Animal Tissue. J. Biol. Chem. 1952, 226, 497–509. [PubMed] [Google Scholar]
- Weckwerth W.; Wenzel K.; Fiehn O. Process for the Integrated Extraction, Identification and Quantification of Metabolites, Proteins and RNA to Reveal Their Co-Regulation in Biochemical Networks. Proteomics 2004, 4 (1), 78–83. 10.1002/pmic.200200500. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Cruickshank C.; Armstrong M.; Mahaffey S.; Reisdorph R.; Reisdorph N. New Sample Preparation Approach for Mass Spectrometry-Based Profiling of Plasma Results in Improved Coverage of Metabolome. J. Chromatogr. A 2013, 1300, 217–226. 10.1016/j.chroma.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Gonzalez F. J.; Xie C.; Jin Y.; Feng C.; Xue L.; Lu D.; Zhou Z.; Wu C. A Systemic Workflow for Profiling Metabolome and Lipidome in Tissue. J. Chromatogr. A 2019, 1589, 105–115. 10.1016/j.chroma.2018.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampler E.; Egger D.; Schoeny H.; Rusz M.; Pacheco M. P.; Marino G.; Kasper C.; Naegele T.; Koellensperger G. The Power of LC-MS Based Multiomics: Exploring Adipogenic Differentiation of Human Mesenchymal Stem/Stromal Cells. Molecules 2019, 24 (19), 3615. 10.3390/molecules24193615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale G.; Martin S. A.; Mitsche M. A.; Thompson B. M.; Eckert K. M.; McDonald J. G. Three-Phase Liquid Extraction: A Simple and Fast Method for Lipidomic Workflows. J. Lipid Res. 2019, 60 (3), 694–706. 10.1194/jlr.D090795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtauer L.; Küstner L.; Weckwerth W.; Heyer A. G.; Nägele T. Resolving Subcellular Plant Metabolism. Plant J. 2019, 100 (3), 438–455. 10.1111/tpj.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. F.; Lu W.; Rabinowitz J. D. Avoiding Misannotation of In-Source Fragmentation Products as Cellular Metabolites in Liquid Chromatography-Mass Spectrometry-Based Metabolomics. Anal. Chem. 2015, 87 (4), 2273–2281. 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain M.The Agilent Metabolomics Dynamic MRM Database and Method Author Technical Overview. [Google Scholar]
- Buescher J. M.; Moco S.; Sauer U.; Zamboni N. Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectrometry Method for Fast and Robust Quantification of Anionic and Aromatic Metabolites. Anal. Chem. 2010, 82 (11), 4403–4412. 10.1021/ac100101d. [DOI] [PubMed] [Google Scholar]
- Spalding J. L.; Naser F. J.; Mahieu N. G.; Johnson S. L.; Patti G. J. Trace Phosphate Improves ZIC-PHILIC Peak Shape, Sensitivity, and Coverage for Untargeted Metabolomics. J. Proteome Res. 2018, 17 (10), 3537–3546. 10.1021/acs.jproteome.8b00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.; Su X.; Klein M. S.; Lewis I. A.; Fiehn O.; Rabinowitz J. D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86 (1), 277–304. 10.1146/annurev-biochem-061516-044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.; Deja S.; Kucejova B.; Duarte J. A. G.; McDonald J. G.; Burgess S. C. Targeted Determination of Tissue Energy Status by LC-MS/MS. Anal. Chem. 2019, 91 (9), 5881–5887. 10.1021/acs.analchem.9b00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.; Chen X.; Wang Z.; Wang D.; Zhang J. L. Global Profiling and Identification of Bile Acids by Multi-Dimensional Data Mining to Reveal a Way of Eliminating Abnormal Bile Acids. Anal. Chim. Acta 2020, 1132, 74–82. 10.1016/j.aca.2020.07.067. [DOI] [PubMed] [Google Scholar]
- Griffiths W. J.; Abdel-Khalik J.; Crick P. J.; Yutuc E.; Wang Y. New Methods for Analysis of Oxysterols and Related Compounds by LC–MS. J. Steroid Biochem. Mol. Biol. 2016, 162, 4–26. 10.1016/j.jsbmb.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Peng B.; Geue S.; Coman C.; Münzer P.; Kopczynski D.; Has C.; Hoffmann N.; Manke M. C.; Lang F.; Sickmann A.; Gawaz M.; Borst O.; Ahrends R. Identification of Key Lipids Critical for Platelet Activation by Comprehensive Analysis of the Platelet Lipidome. Blood 2018, 132 (5), e1–e12. 10.1182/blood-2017-12-822890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustam Y. H.; Reid G. E. Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem. 2018, 90 (1), 374–397. 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- Coras R.; Kavanaugh A.; Boyd T.; Huynh Q.; Pedersen B.; Armando A. M.; Dahlberg-Wright S.; Marsal S.; Jain M.; Paravar T.; Quehenberger O.; Guma M. Pro- and Anti-Inflammatory Eicosanoids in Psoriatic Arthritis. Metabolomics 2019, 15 (4), 65–65. 10.1007/s11306-019-1527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Weintraub S. T.; Coman C.; Ponnaiyan S.; Sharma R.; Tews B.; Winter D.; Ahrends R. A Comprehensive High-Resolution Targeted Workflow for the Deep Profiling of Sphingolipids. Anal. Chem. 2017, 89 (22), 12480–12487. 10.1021/acs.analchem.7b03576. [DOI] [PubMed] [Google Scholar]
- Barrientos R. C.; Zhang Q.. Recent Advances in the Mass Spectrometric Analysis of Glycosphingolipidome – A Review. Anal. Chim. Acta 2020, 1132, 134. 10.1016/j.aca.2020.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage E. G.; Southam A. D. Monitoring Cancer Prognosis, Diagnosis and Treatment Efficacy Using Metabolomics and Lipidomics. Metabolomics 2016, 12 (10), 1–15. 10.1007/s11306-016-1093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyoğlu D.; Idle J. R. Metabolomic and Lipidomic Biomarkers for Premalignant Liver Disease Diagnosis and Therapy. Metabolites 2020, 10 (2), 50. 10.3390/metabo10020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkuchen A.; Carvalho V. M.; Venturini G.; Salgueiro J. S.; Freitas L. S.; Dellavance A.; Martins F. C.; Lorenzi-Filho G.; Cardozo K. H. M.; Drager L. F. Metabolomic and Lipidomic Profile in Men with Obstructive Sleep Apnoea: Implications for Diagnosis and Biomarkers of Cardiovascular Risk. Sci. Rep. 2018, 8 (1), 11270. 10.1038/s41598-018-29727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambiaghi A.; Ferrario M.; Masseroli M. Analysis of Metabolomic Data: Tools, Current Strategies and Future Challenges for Omics Data Integration. Brief. Bioinform. 2017, 18 (3), 498–510. 10.1093/bib/bbw031. [DOI] [PubMed] [Google Scholar]
- Ivanisevic J.; Want E. J.. From Samples to Insights into Metabolism: Uncovering Biologically Relevant Information in LC- HRMS Metabolomics Data. Metabolites 2019, 9 ( (12), ) 10.3390/metabo9120308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza L. P. De; Alseekh S.; Brotman Y.; Fernie A. R. Network-Based Strategies in Metabolomics Data Analysis and Interpretation: From Molecular Networking to Biological Interpretation. Expert Rev. Proteomics 2020, 17 (4), 243–255. 10.1080/14789450.2020.1766975. [DOI] [PubMed] [Google Scholar]
- Brown M.; Dunn W. B.; Ellis D. I.; Goodacre R.; Handl J.; Knowles J. D.; O’Hagan S.; Spasić I.; Kell D. B. A Metabolome Pipeline: From Concept to Data to Knowledge. Metabolomics 2005, 1 (1), 39–51. 10.1007/s11306-005-1106-4. [DOI] [Google Scholar]
- Steuer A. E.; Brockbals L.; Kraemer T.. Metabolomic Strategies in Biomarker Research–New Approach for Indirect Identification of Drug Consumption and Sample Manipulation in Clinical and Forensic Toxicology? Front. Chem. 2019, 7, 10.3389/fchem.2019.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst D.; Kell D. Statistical strategies for avoiding false discoveries in metabolomics and related experiments.. Metabolites 2006, 10.1007/s11306-006-0037-z. [DOI] [Google Scholar]
- Ma J.; Shojaie A.; Michailidis G. A Comparative Study of Topology-Based Pathway Enrichment Analysis Methods. BMC Bioinf. 2019, 20 (1), 546. 10.1186/s12859-019-3146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Wang S.; Dai E.; Zhou S.; Liu D.; Liu H.; Meng Q.; Jiang B.; Jiang W. Pathway Enrichment Analysis Approach Based on Topological Structure and Updated Annotation of Pathway. Briefings Bioinf. 2019, 20 (1), 168–177. 10.1093/bib/bbx091. [DOI] [PubMed] [Google Scholar]
- Ginsburg H. Caveat Emptor: Limitations of the Automated Reconstruction of Metabolic Pathways in Plasmodium. Trends Parasitol. 2009, 25 (1), 37–43. 10.1016/j.pt.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Frainay C.; Jourdan F. Computational Methods to Identify Metabolic Sub-Networks Based on Metabolomic Profiles. Briefings Bioinf. 2017, 18 (1), 43–56. 10.1093/bib/bbv115. [DOI] [PubMed] [Google Scholar]
- Nägele T.; Fürtauer L.; Nagler M.; Weiszmann J.; Weckwerth W. A Strategy for Functional Interpretation of Metabolomic Time Series Data in Context of Metabolic Network Information. Front. Mol. Biosci. 2016, 3, 6. 10.3389/fmolb.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazalviel M.; Frainay C.; Poupin N.; Vinson F.; Merlet B.; Gloaguen Y.; Cottret L.; Jourdan F. MetExploreViz: Web Component for Interactive Metabolic Network Visualization. Bioinformatics 2018, 34 (2), 312–313. 10.1093/bioinformatics/btx588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottret L.; Frainay C.; Chazalviel M.; Cabanettes F.; Gloaguen Y.; Camenen E.; Merlet B.; Heux S.; Portais J.-C.; Poupin N.; Vinson F.; Jourdan F. MetExplore: Collaborative Edition and Exploration of Metabolic Networks. Nucleic Acids Res. 2018, 46 (1), 495–502. 10.1093/nar/gky301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato A.; Tenori L.; Cascante M.; De Atauri Carulla P. R.; Martins dos Santos V. A. P.; Saccenti E. From Correlation to Causation: Analysis of Metabolomics Data Using Systems Biology Approaches. Metabolomics 2018, 14 (4), 37. 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barupal D. K.; Fiehn O. Chemical Similarity Enrichment Analysis (ChemRICH) as Alternative to Biochemical Pathway Mapping for Metabolomic Datasets. Sci. Rep. 2017, 7 (1), 1–11. 10.1038/s41598-017-15231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.; Soufan O.; Li C.; Caraus I.; Li S.; Bourque G.; Wishart D. S.; Xia J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucleic Acids Res. 2018, 46 (W1), W486–W494. 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanichthanarak K.; Fan S.; Grapov D.; Barupal D. K.; Fiehn O. Metabox: A Toolbox for Metabolomic Data Analysis, Interpretation and Integrative Exploration. PLoS One 2017, 12 (1), e0171046. 10.1371/journal.pone.0171046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A.; Korninger F.; Viteri G.; Sidiropoulos K.; Marin-Garcia P.; Ping P.; Wu G.; Stein L.; D’Eustachio P.; Hermjakob H. Reactome Graph Database: Efficient Access to Complex Pathway Data. PLoS Comput. Biol. 2018, 14 (1), e1005968. 10.1371/journal.pcbi.1005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha A.; Daníelsdóttir A. D.; Gawron P.; Jóhannsson F.; Jónsdóttir S.; Jarlsson S.; Gunnarsson J. P.; Brynjólfsson S.; Schneider R.; Thiele I.; Fleming R. M. T. ReconMap: An Interactive Visualization of Human Metabolism. Bioinformatics 2016, 33 (4), btw667. 10.1093/bioinformatics/btw667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R.; Billington R.; Keseler I. M.; Kothari A.; Krummenacker M.; Midford P. E.; Ong W. K.; Paley S.; Subhraveti P.; Karp P. D. The MetaCyc Database of Metabolic Pathways and Enzymes - a 2019 Update. Nucleic Acids Res. 2020, 48 (D1), D445–D453. 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slenter D. N.; Kutmon M.; Hanspers K.; Riutta A.; Windsor J.; Nunes N.; Mélius J.; Cirillo E.; Coort S. L.; Digles D.; Ehrhart F.; Giesbertz P.; Kalafati M.; Martens M.; Miller R.; Nishida K.; Rieswijk L.; Waagmeester A.; Eijssen L. M. T.; Evelo C. T.; Pico A. R.; Willighagen E. L. WikiPathways: A Multifaceted Pathway Database Bridging Metabolomics to Other Omics Research. Nucleic Acids Res. 2018, 46 (1), 661–667. 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Li C.; Marcu A.; Badran H.; Pon A.; Budinski Z.; Patron J.; Lipton D.; Cao X.; Oler E.; Li K.; Paccoud M.; Hong C.; Guo A. C.; Chan C.; Wei W.; Ramirez-Gaona M. PathBank: A Comprehensive Pathway Database for Model Organisms. Nucleic Acids Res. 2020, 48 (1), 470–478. 10.1093/nar/gkz861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha A.; Modamio J.; Jarosz Y.; Guerard E.; Sompairac N.; Preciat G.; Daníelsdóttir A. D.; Krecke M.; Merten D.; Haraldsdóttir H. S.; Heinken A.; Heirendt L.; Magnúsdóttir S.; Ravcheev D. A.; Sahoo S.; Gawron P.; Friscioni L.; Garcia B.; Prendergast M.; Puente A.; Rodrigues M.; Roy A.; Rouquaya M.; Wiltgen L.; Žagare A.; John E.; Krueger M.; Kuperstein I.; Zinovyev A.; Schneider R.; Fleming R. M. T.; Thiele I. The Virtual Metabolic Human Database: Integrating Human and Gut Microbiome Metabolism with Nutrition and Disease. Nucleic Acids Res. 2019, 47 (1), 614–624. 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher J. M.; Driggers E. M.. Integration of Omics: More than the Sum of Its Parts. Cancer Metab. 2016, 4, 10.1186/s40170-016-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-de-Diego R.; Tarazona S.; Martínez-Mira C.; Balzano-Nogueira L.; Furió-Tarí P.; Pappas G. J.; Conesa A. PaintOmics 3: A Web Resource for the Pathway Analysis and Visualization of Multi-Omics Data. Nucleic Acids Res. 2018, 46 (1), 503–509. 10.1093/nar/gky466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F.; Gautier B.; Singh A.; Lê Cao K.-A. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13 (11), e1005752. 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G.; Xia J. OmicsNet: A Web-Based Tool for Creation and Visual Analysis of Biological Networks in 3D Space. Nucleic Acids Res. 2018, 46 (W1), W514–W522. 10.1093/nar/gky510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G.; Xia J. Using OmicsNet for Network Integration and 3D Visualization. Curr. Protoc. Bioinforma. 2019, 65 (1), 1–26. 10.1002/cpbi.69. [DOI] [PubMed] [Google Scholar]
- Wörheide M. A.; Krumsiek J.; Kastenmüller G.; Arnold M. Multi-Omics Integration in Biomedical Research – A Metabolomics-Centric Review. Anal. Chim. Acta 2021, 1141, 144–162. 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S.; Duren W.; Evans C. R.; Burant C. F.; Michailidis G.; Karnovsky A. Sparse Network Modeling and Metscape-Based Visualization Methods for the Analysis of Large-Scale Metabolomics Data. Bioinformatics 2017, 33 (10), 1545–1553. 10.1093/bioinformatics/btx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.; Tarcea V. G.; Karnovsky A.; Mirel B. R.; Weymouth T. E.; Beecher C. W.; Cavalcoli J. D.; Athey B. D.; Omenn G. S.; Burant C. F.; Jagadish H. V. Metscape: A Cytoscape Plug-in for Visualizing and Interpreting Metabolomic Data in the Context of Human Metabolic Networks. Bioinformatics 2010, 26 (7), 971–973. 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky A.; Weymouth T.; Hull T.; Glenn Tarcea V.; Scardoni G.; Laudanna C.; Sartor M. A.; Stringer K. A.; Jagadish H. V.; Burant C.; Athey B.; Omenn G. S. Metscape 2 Bioinformatics Tool for the Analysis and Visualization of Metabolomics and Gene Expression Data. Bioinformatics 2012, 28 (3), 373–380. 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchweitz L. F.; Yurkovich J. T.; Blessing C.; Kohler V.; Schwarzkopf F.; King Z. A.; Yang L.; Jóhannsson F.; Sigurjónsson Ó. E.; Rolfsson Ó.; Heinrich J.; Dräger A. Visualizing Metabolic Network Dynamics through Time-Series Metabolomic Data. BMC Bioinf. 2020, 21 (1), 130–130. 10.1186/s12859-020-3415-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra B. B. New Tools and Resources in Metabolomics: 2016–2017. Electrophoresis 2018, 39 (7), 909–923. 10.1002/elps.201700441. [DOI] [PubMed] [Google Scholar]
- Mangul S.; Martin L. S.; Eskin E.; Blekhman R. Improving the Usability and Archival Stability of Bioinformatics Software. Genome Biol. 2019, 20 (1), 47. 10.1186/s13059-019-1649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser J. Funding for Key Data Resources in Jeopardy. Science (Washington, DC, U. S.) 2016, 351 (6268), 14–14. 10.1126/science.351.6268.14. [DOI] [PubMed] [Google Scholar]
- Chong J.; Soufan O.; Li C.; Caraus I.; Li S.; Bourque G.; Wishart D. S.; Xia J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucleic Acids Res. 2018, 46 (W1), W486–W494. 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45 (1), 353–361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohler A.; Wu G.; Kutmon M.; Pradhana L. A.; Coort S. L.; Hanspers K.; Haw R.; Pico A. R.; Evelo C. T. Reactome from a WikiPathways Perspective. PLoS Comput. Biol. 2016, 12 (5), e1004941. 10.1371/journal.pcbi.1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]