Abstract
Background
Bacterial degradation/transformation of steroids is widely investigated to create biotechnologically relevant strains for industrial application. The strain of Nocardioides simplex VKM Ac-2033D is well known mainly for its superior 3-ketosteroid Δ1-dehydrogenase activity towards various 3-oxosteroids and other important reactions of sterol degradation. However, its biocatalytic capacities and the molecular fundamentals of its activity towards natural sterols and synthetic steroids were not fully understood. In this study, a comparative investigation of the genome-wide transcriptome profiling of the N. simplex VKM Ac-2033D grown on phytosterol, or in the presence of cortisone 21-acetate was performed with RNA-seq.
Results
Although the gene patterns induced by phytosterol generally resemble the gene sets involved in phytosterol degradation pathways in mycolic acid rich actinobacteria such as Mycolicibacterium, Mycobacterium and Rhodococcus species, the differences in gene organization and previously unreported genes with high expression level were revealed. Transcription of the genes related to KstR- and KstR2-regulons was mainly enhanced in response to phytosterol, and the role in steroid catabolism is predicted for some dozens of the genes in N. simplex. New transcription factors binding motifs and new candidate transcription regulators of steroid catabolism were predicted in N. simplex.
Unlike phytosterol, cortisone 21-acetate does not provide induction of the genes with predicted KstR and KstR2 sites. Superior 3-ketosteroid-Δ1-dehydrogenase activity of N. simplex VKM Ac-2033D is due to the kstDs redundancy in the genome, with the highest expression level of the gene KR76_27125 orthologous to kstD2, in response to cortisone 21-acetate. The substrate spectrum of N. simplex 3-ketosteroid-Δ1-dehydrogenase was expanded in this study with progesterone and its 17α-hydroxylated and 11α,17α-dihydroxylated derivatives, that effectively were 1(2)-dehydrogenated in vivo by the whole cells of the N. simplex VKM Ac-2033D.
Conclusion
The results contribute to the knowledge of biocatalytic features and diversity of steroid modification capabilities of actinobacteria, defining targets for further bioengineering manipulations with the purpose of expansion of their biotechnological applications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12896-021-00668-9.
Keywords: Nocardioides simplex, Arthrobacter simplex, Pimelobacter simplex, Biocatalysts, Transcriptome, Phytosterol, Cortisone acetate, Progesterone
Key points
Role in phytosterol catabolism is predicted for some dozens of N. simplex genes
Four kstD genes are induced that provide superior 1(2)-dehydrogenase activity of the strain
Gene kstD2 KR76_27125 is highly over-expressed in response to AcC
Metabolism of cortisone acetate is not regulated by KstR-repressors
New candidate transcription regulators of steroid catabolism were predicted
Background
Steroids represent a huge class of specific organic molecules with many of them playing essential roles in all living systems. Diverse bacteria from different ecological niches have been evolved to degrade steroids as sources of carbon and energy, or detoxify exogenic steroids by their structural modifications. Among bacteria, actinobacteria excite a particular interest due to their comprehensive metabolic possibilities and high activity of the enzymatic systems towards various steroids.
Currently, degradation of sterols (such as cholesterol, or phytosterols) by actinobacteria is in the focus of intensive researches due to its exclusive role in pathogenicity of Mycobacterium tuberculosis, and established applications of the non-pathogenic species and their engineered derivatives in biotechnology for production of high-value steroids for the pharmaceutical industry.
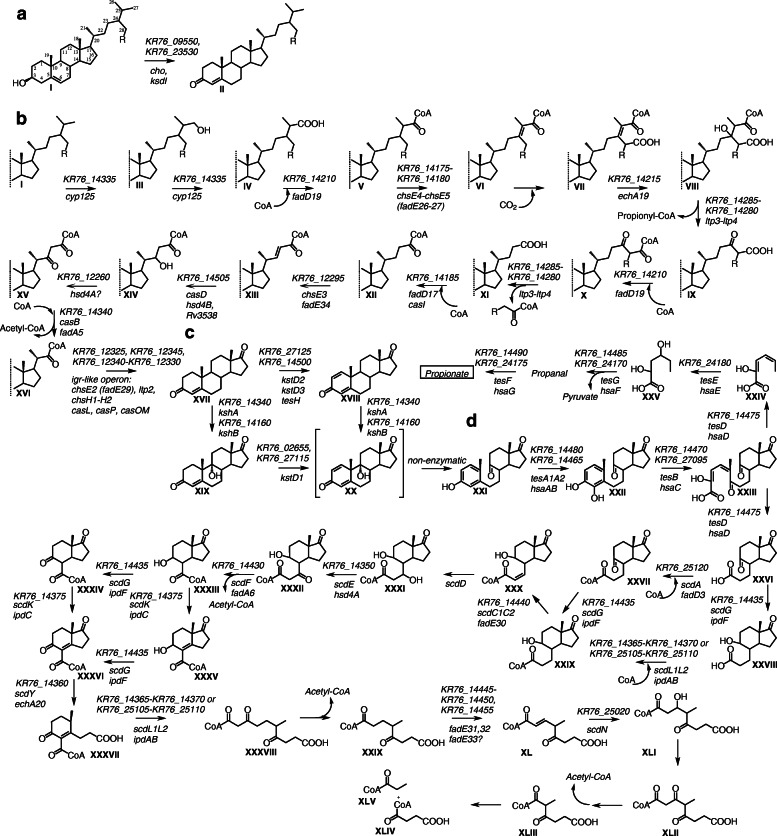
Sterol catabolism is a complicated, multi-step process that included degradation of side chain, rings A/B and rings C/D of steroid core oxidation (Fig. 1). This pathway was reported to be controlled by two TetR-type transcriptional repressors, KstR and KstR2 [1–4].
Fig. 1.
Putative scheme of phytosterol catabolism by N. simplex VKM Ac-2033D. “KstR” and “KstR2” in ovals mean regulons
Cholesterol catabolism has been intensively studied in the so-called mycolic acid rich actinobacteria such as pathogenic species: Mycobacterium tuberculosis [5], Rhodococcus strains [6, 7], as well as in the non-pathogenic species such as Mycolicibacterium smegmatis (basonym Mycobacterium smegmatis) mc2155 [4], Gordonia cholesterolivorans [8]. The roles of certain enzymes, mechanisms of their functioning and interactions with substrates were investigated. Cholesterol oxidases (ChOs) and/or 3β-hydroxysteroid dehydrogenases (3-HsDs) were shown to be responsible for modification of 3β-hydroxy-5-ene to 3-keto-4-ene- moiety that is considered as a first reaction of sterol degradation [4, 9]. Cytochrome P450 monooxygenases encoded by cyp125, cyp124 and cyp142 account for sterol hydroxylation at C-26(27) [10, 11], and aldol lyases encoded by ltp3 and ltp4 play role in the degradation of C24-branched chain sterols [12]. Acyl-CoA synthetase FadD19 catalyzes the formation of the cholestanoate CoA-thioester [13] that is further oxidized to C-17-ketosteroids by three successive cycles of β-oxidation [4, 14]. 9α-Hydroxylase (KshAB, consisting of KshA and KshB subunits) and 3-ketosteroid-∆1-dehydrogenase (KstD) [15, 16] are well-known as key enzymes of steroid core degradation whose cooperative action provides the ring B opening (Fig. 1). 3-Hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione (3-HSA) formed is further degraded by the enzymes encoded by hsaA, hsaB, hsaC, hsaD, hsaE, hsaF, hsaG [17–19]. The degradation of the last two rings (rings C/D) of the steroid core is catalyzed by the products of the genes of the so-called KstR2-regulon [20]. Unlike mycobacteria, rhodococci and other above-mentioned species related to Corynebacteriales order, much less is known on steroid catabolism in actinobacteria belonging to Propionibacteriales order such as representatives of Nocardioides genus and related genera. The capability of Arthrobacter simplex (syn. Nocardioides simplex) to degrade cholesterol has been reported more than 50 years ago and the major intermediate was identified as cholest-4-ene-3-one [21]. Since then mostly 3-ketosteroid-Δ1-dehydrogenases (KstDs) from A. simplex were studied, the gene coding for these enzymes were identified and the engineered strains were constructed to provide selective production of 1(2)-dehydrosteroids (e.g. [22, 23]).
Recent metagenome research revealed among others the presence of the potent steroid degraders belonging to Nocardiodaceae and Nocardioides in different habitats [24]. Dozens of genes associated with steroid catabolism were detected in Nocardioides dokdonensis FR1436 isolated from sand sediment [25]. Most the research on the molecular mechanisms of steroid catabolism have been provided by our team for the biotechnologically relevant strain of Nocardioides simplex VKM Ac-2033D [26–28]. The data obtained in [26] have been further used for the refolding in situ of a soluble cholesterol oxidase PsChO3 KR76_06800 (cholesterol oxidase ChoD) producing cholest-5-en-3-one [29], and for the rational design and protein engineering of cholesterol oxidase (PsChO) KR76_21605 producing 3-oxo-4-ene-steroid products thus allowing significant improvement of the enzyme substrate selectivity [30].
Superior 3-ketosteroid-Δ1-dehydrogenase (KstD) activity of Nocardioides simplex VKM Ac-2033D towards various 3-ketosteroids provides effective production of the pharmaceutical 1(2)-dehydroanalogs [31–34]. It is known that the 1(2)-dehydroanalogs often demonstrate higher therapeutic efficiency and lesser side effects as compared with the corresponding 1(2)-saturated steroid drugs [35, 36]. The N. simplex VKM Ac-2033D is also capable of hydrolyzing steroid esters such as 21-acetylated steroids, reduction of C17 and C20 carbonyl groups of androstanes and pregnanes, respectively [31–33] and able to grow on cholesterol [27].
High KstD activity of N. simplex VKM Ac-2033D had been earlier demonstrated towards cortisol [37] and its 6-methylated derivative [38], androst-4-ene-3,17-dione (AD) and its 6-aminomethylated derivatives [34], testosterone [34], androsta-4,9-diene-3,17-dione [39], 21-acetoxy-pregna-4(5),9(11),16(17)-triene-21-ol-3,20-dione and pregna-4,9(11)-diene-17α,21-diol-3,20-dione acetates [32, 33], 7α- and 7β-hydroxytestololactone [40], etc. (more information is given in the Supplementary Table S1). In this study, the substrate spectrum of steroids for bioconversion with N. simplex was expanded with progesterone (Pr) and its 17α-hydroxylated and 11α,17α-dihydroxylated derivatives (17α-OH-Pr and 11α,17α-di-OH-Pr).
The strain had originally been isolated from the Pamir highland soils [41] and identified firstly as Mycobacterium sp. 193 [42], then re-classified as Mycobacterium globiforme 193 [43], and later as Arthrobacter globiformis 193 based on its cultural and morphological properties [44].
In 1976, Prauser had proposed a new genus Nocardioides for the Gram-positive, aerobic, non-sporeforming species, with LL-2,6-diaminopimelic acid in their cell walls and lack of mycolic acids [45]. Accordingly, the coryneform bacteria differed from other Arthrobacter species in their cell wall composition such as previously reported Arthrobacter simplex had been re-classified as belonging to Nocardioides genus [46]. Currently, more than 90 validly published species are within this genus [47]. The strain Arthrobacter globiformis 193 had been re-classified as Nocardioides simplex VKM Ac-2033D in 2003 [31].
Intriguingly, Suzuki and Komagata proposed the genus Pimelobacter (pimele oil - inhabiting; bacterium - a rod) for the Arthrobacter species with LL-2,6-diaminopimelic acid instead of lysine in the peptidoglycan layer with a type strain Pimelobacter simplex (syn. Arthrobacter simplex) [48]. As a result of taxonomic re-evaluation, Pimelobacter simplex was later re-classified as N. simplex [49, 50].
This complicated story resulted in the confusing situation when Arthrobacter simplex, Pimelobacter simplex and Nocardioides simplex names are often used as synonyms in the current literature for similar actinobacteria. In our case, a complete genome sequence of N. simplex VKM Ac-2033D was presented to NCBI database, and indicated by NCBI database as Pimelobacter simplex VKM Ac-2033D [26] under accession no. CP009896, its detailed bioinformatics analysis was carried out in [27].
In our previous study, the putative genes and gene clusters related to the sterol uptake system, aliphatic side chain degradation, A/B- and C/D-ring opening and degradation systems were revealed in the genome of N. simplex VKM Ac-2033D [27]. Previously unknown steroid metabolism gene clusters have been predicted. Orthologous clusters were later discovered also in N. dokdonensis FR1436T [25].
It is known that most bacterial enzyme systems involved in steroid metabolic pathways are mainly induced with different steroids, but not expressed constitutively [51]. In this study phytosterol and cortisone 21-acetate (AcC) were estimated as inducers. Phytosterol is a mixture of plant sterols with a branched side chain (with β-sitosterol as a main component) (Fig. 2). Unlike natural sterols such as cholesterol, or phytosterol, AcC is a synthetic corticosteroid ester which is often applied for induction of KstD activity in N. simplex and related strains [38, 52]. However, neither growth on AcC, no AcC bioconversion with N. simplex VKM Ac-2033D has been examined so far. The fundamental questions also are: whether AcC can be metabolized by N. simplex and whether the gene network that over-expressed in response to AcC corresponds to that of steroid degradation. To address these issues, we compared differential transcriptomes of N. simplex cultured with (and without) phytosterol, and with (and without) AcC.
Fig. 2.
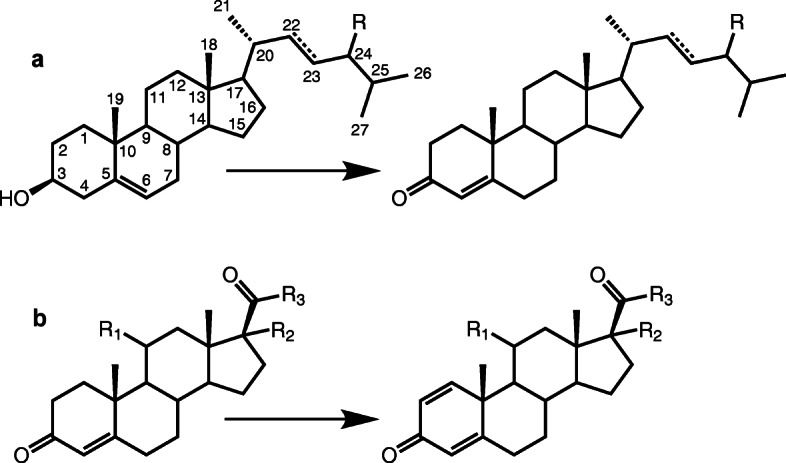
Major reactions of steroid bioconversion by N. simplex: a. Oxidation of 3-hydroxy group of phytosterol constituents: R = C2H5 β-sitosterol/stigmast-4-en-3-one (β-sitostenone); R = C2H5, Δ22,23stigmasterol/stigmasta-4,22-dien-3-one; R = CH3 campesterol/campest-4-en-3-one, R = CH3, brassicasterol/brassicast-4-en-3-one. b. R1 = H, R2 = H, R3 = CH3, Progesterone (Pr)/1(2)-dehydro-progesterone (DPr); R1 = H, R2 = OH, R3 = CH3, 17α-hydroxyprogesterone (17α-OH-Pr)/1(2)-dehydro-17α-hydroxyprogesterone (17α-OH-DPr); R1 = OH, R2 = OH, R3 = CH3, 11α,17α-dihydroxyprogesterone (11α,17α-di-OH-Pr)/1(2)-dehydro-11α,17α-dihydroxyprogesterone (11α,17α-di-OH-DPr); R1 = O, R2 = OH, R3 = CH2-O-C(O)-CH3 21-acetate of cortisone or cortisone acetate (AcC)/21-acetate of 1(2)-dehydro-cortisone or 21-acetate of prednisone (AcP)
Differentially expressed genes (DEG) sets in different induction conditions were compared and expression of the predicted steroid catabolic genes and their clusters was evaluated. The operons with highest expression level and new candidate transcription regulators motifs of steroid catabolism were predicted. The kstD gene with superior expression in response to AcC was revealed, and steroid conversion assay confirmed high level of KstD activity towards progesterone (a predicted preferable substrate for the product of gene kstD2) and progesterone derivatives. Moreover, the over-expressed genes encoding the enzymes related to the basic catalytic properties of the N. simplex VKM Ac-2033D were revealed.
Results
Steroid substrates and major bioconversion products
Structures of the steroid substrates used in this work and the bioconversion products detected are presented in Fig. 2. The corresponding 1(2)-dehydroderivatives were identified as major bioconversion products from Pr, 17α-OH-Pr and 11α,17α-di-OH-Pr and AcC. The main intermediates of phytosterol (comprising a mixture of β-sitosterol, stigmasterol, campesterol, brassicasterol) transformation by N. simplex cells were identified as corresponding phytostenones (stigmast-4-en-3-one (β-sitostenone), stigmasta-4,22-dien-3-one, campest-4-en-3-one, brassicast-4-en-3-one) (Supplementary Table S2 and S3). Earlier, campest-4-en-3-one, stigmast-4-en-3-one, and stigmasta-4,22-dien-3-one had been reported as intermediates at bioconversion of campesterol, β-sitosterol, stigmasterol, respectively, with Arthrobacter simplex IAM 1660 [21].
Growth and steroid bioconversion
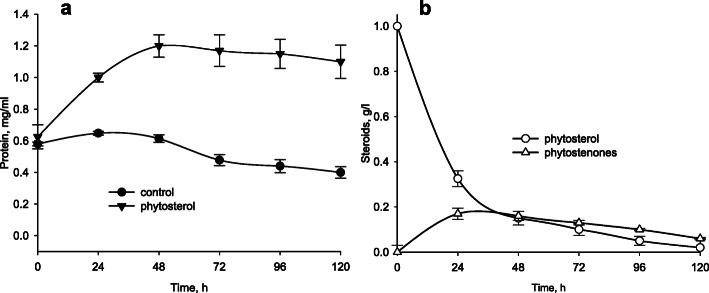
As shown in Fig. 3a, N. simplex VKM Ac-2033D can grow on phytosterol while no growth was observed in the mineral medium with only MCD (control). As shown in Fig. 3b, phytosterol was completely degraded by N. simplex in approx. 120 h via transit intermediates (phytostenones).
Fig. 3.
a: Growth of N. simplex VKM Ac-2033D on phytosterol. Medium without phytosterol was used in control. b: Phytosterol consumption and phytostenones accumulation during strain growth on phytosterol
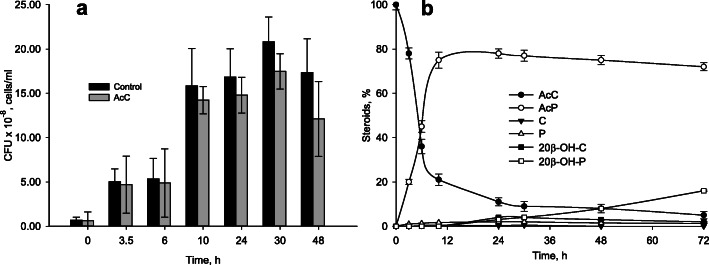
No growth difference was observed when N. simplex incubating with AcC in comparison with control (without AcC) (Fig. 4a). Maximum CFU number for AcC which was observed on 24 h was smaller than CFU number in the control: 17.4 × 108 ± 2 × 108 and 20.8 × 108 ± 2.8 × 108 cells/ml, respectively. Unlike phytosterols, AcC was not degraded by N. simplex, but converted into the mixture of steroids (21-acetate of prednisone, AcP (main product), prednisone (P), cortisone (C) and their 20-hydroxyderivatives (Fig. 4b)). Quantification of all steroids formed from AcC evidenced the absence of steroid core destruction. Expectedly, no AcP, or any other steroids were detected if no AcC was added (control) (data not shown).
Fig. 4.
a: Growth of N. simplex VKM Ac-2033D in the presence of AcC. Media without AcC was used as a blank. b: Dynamics of AcC bioconversion. Data are the average of three replicates. Abbreviations: AcC – cortisone 21-acetate; C - cortisone, 20β-OH-C - 20β-hydroxy cortisone, 20β-OH-P - 20β-hydroxy prednisone
High-throughput mRNA sequencing
Complete transcriptomes of the N. simplex cells grown in the presence, or absence of phytosterol, as well as in the presence, or absence of AcC were obtained. The total number of reads, the percentage of reads mapping to the rRNA genes, and links to SRA repository in NCBI for experimental variants are provided in the Table 1.
Table 1.
Sequencing statistics and links to reads
| Samples | Total number of reads | Percent of reads mapping to the rRNA genes | Link to the reads in SRA |
|---|---|---|---|
| Phytosterol, 1st replicate | 10,718,187 | 2 | https://www.ncbi.nlm.nih.gov/sra/SRX5542009 |
| Phytosterol, 2nd replicate | 10,138,962 | 2.9 | https://www.ncbi.nlm.nih.gov/sra/SRX5542008 |
| AcC, 1st replicate | 8,938,451 | 3 | https://www.ncbi.nlm.nih.gov/sra/SRX5542005 |
| AcC, 2nd replicate | 11,883,597 | 4.4 | https://www.ncbi.nlm.nih.gov/sra/SRX5542004 |
| Control, 1st replicate | 10,207,198 | 2.1 | https://www.ncbi.nlm.nih.gov/sra/SRX5542007 |
| Control, 2nd replicate | 12,237,275 | 5 | https://www.ncbi.nlm.nih.gov/sra/SRX5542006 |
Differentially expressed genes (DEG)
DEG analysis showed 91 up- and 42 down-regulated genes in phytosterol plus conditions, while 81 and 31 genes were up- and down regulated in response to AcC, respectively (Table 2). A gene was considered differentially expressed, if its expression increased or decreased more than threefold with q-value less or equal to 0.01.
Table 2.
Differentially expressed genes in gene clusters of N. simplex VKM Ac-2033D
| Cluster | Genes | Phytosterol | AcC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| I | Ave | Max | D | I | Ave | Max | D | ||
| А | KR76_14160-KR76_14505 | 31 | 10.94 | 31.24 | 0 | 0 | 0 | ||
| B | KR76_12190-KR76_12345 | 12 | 6.90 | 14.70 | 0 | 0 | 0 | ||
| C | KR76_25015- KR76_25200 | 14 | 15.10 | 59.00 | 0 | 0 | 0 | ||
| D | KR76_27035-KR76_27130 | 7 | 13.35 | 24.53 | 0 | 3 | 415.5 | 1207.7 | 0 |
| E | KR76_17995-KR76_18070 | 0 | 0.00 | 0 | 0 | 0 | |||
| Out of clusters | 27 | 7.06 | 12.44 | 42 | 79 | 32 | |||
| Total | 91 | 9.94 | 42 | 82 | 32 | ||||
I - the number of genes that significantly (more than 3 times with q-value < 0.01) increased their expression
Ave - the average level of induction among the genes that significantly increased expression in the cluster
Max - the maximum level of induction
D - the number of genes that significantly (more than 3 times with q-value < 0.01) reduced their expression
In phytosterol induced cells, maximum expression level (59-fold) was observed for gene KR76_25195 coding for ABC amino acid transporter; while the minimum level was indicated for KR76_00525, a putative two-component system sensor kinase, at 0.06 of phytosterol minus conditions. When inducing with AcC, maximum over-expression (1207-fold) was determined for the gene KR76_27125 encoded KstD; and the minimum level was shown by KR76_15900 RidA/YjgF/TdcF/RutC subgroup, at 0.03 of phytosterol minus conditions.
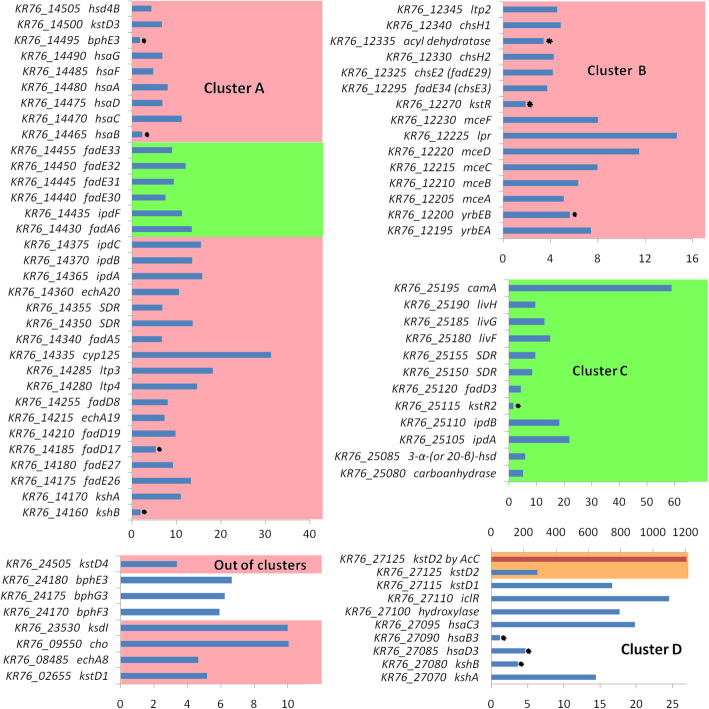
The steroid induced genes were combined into the operons using bioinformatics methods, supported by the data from the transcriptome analysis. The detailed information about these operons, predicted binding motifs of transcription factors and levels of gene expression is provided in Supplementary Table S4. Induced genes with homology to the known genes of steroid catabolism are depicted in Fig. 5.
Fig. 5.
Induction of sterol catabolism genes by phytosterol and AcC in N. simplex VKM Ac-2033D. Blue bars – induction by phytosterols, red bar – induction by AcC. Pink color – predicted KstR regulon; green color – predicted KstR2 regulon; orange color – predicted Acin sites. The asterisks mark the changes from q-value over the threshold 0.01; the data for these genes are presented because they are related to steroid catabolism
Almost two-thirds of the genes that were induced by phytosterol locate in the “typical” steroid metabolism clusters A and B (Table 2), in the newly cluster C that was previously predicted in [27], as well as in the cluster D, which does not have predicted binding motifs for the transcription factors KstR and KstR2.
The only gene that was highly up-regulated both in the presence of phytosterol and AcC, was KR76_27125, kstD2 coding for KstD. The gene belongs to cluster D. Three genes with unknown functions were significantly down-regulated in phytosterol plus and AcC plus conditions (Supplementary Table S4). No cases of meaningful reverse gene behavior, when one inducer caused significant increase in expression, while another caused significant expression decrease, were detected.
Except for kstD2, no significant expression of the known genes putatively involved in the steroid degradation was observed in AcC plus conditions. No pronounced clustering of genes was observed either: the operons that enhanced expression were evenly distributed throughout the genome (Supplementary Table S4). A number of the AcC-induced genes could be combined into small clusters, specifically, KR76_09390 - KR76_09460; KR76_16215 - KR76_16555; KR76_24345 - KR76_24405; KR76_24685 - KR76_24800. These clusters included the genes related to various transporters, amino acid and organic acid metabolism enzymes, cytochromes, transcriptional regulators, dehydrogenases including 3-ketosteroid-Δ1-dehydrogenase etc.
Expression of steroid catabolism genes orthologs and identification of candidates for new transcription factor regulators of steroid catabolism genes
Our previous study [27] allowed prediction of the KstR and KstR2 binding sites in 71 and 29 operons, respectively. Of these, in phytosterol plus conditions putative KstR-regulated genes of 27 operons (38%) and putative KstR2-regulated genes of 10 operons (34%) increased their expression. In response to AcC, no cases of gene induction with the predicted KstR or KstR2 binding site were observed.
Transcription of nine operons was enhanced in the presence of phytosterol, though the genes do not have predicted binding sites to KstR, or KstR2. Five of them (in which a total of seven genes increased expression significantly) located in cluster D (KR76_27035-KR76_27130).
To identify candidate motifs for the binding of transcription factors, we used the tool FIMO from the MEME 4.10.0 suite [53]. The search was carried out on the sections 500 bps upstream to 50 bps downstream. Identification was carried out for the operons that (i) increased, (ii) decreased their expression in AcC plus conditions and (iii) down-regulated in phytosterol plus conditions. Several motifs were identified (Supplementary Table S5): three motifs for the operons that increased expression on AcC (Acin1, Acin2 and Acin3) and one motif for the operons that decreased expression on phytosterol (Sitdec1). Nothing was found before the operons that reduced expression on AcC. The operons with these sites are listed in the Supplementary Table S4.
Progesterone bioconversion
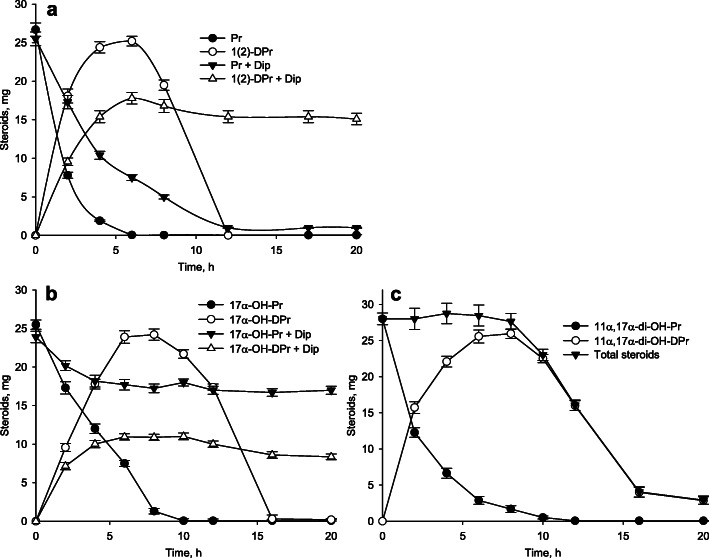
As reported earlier, preferable substrate for KstD2 from Rhodococcus ruber is progesterone [54]. The kstD2 from N. simplex has the moderate amino acid sequence similarity (69%) with that KstD2 from R. ruber. In this study, we tested KstD activity of N. simplex in vivo towards progesterone (Pr), and its hydroxylated derivatives, such as 17α-OH-Pr and 11α,17α-di-OH-Pr. As shown in Fig. 6, the corresponding 1(2)-dehydroderivatives were formed as major products from all three substrates. Their accumulation reached maximum level at 10 h, followed by the consequent degradation. Lower degradation was observed in the presence of α,α-dipyridyl (the inhibitor of iron containing enzymes).
Fig. 6.
a: Bioconversion of progesterone (Pr, 0.4 g/l) with and without α,α-dipyridyl (Dip). b: Bioconversion of 17α-hydroxy-progesterone (17α-OH-Pr, 1 g/l) with and without α,α-dipyridyl. c: Bioconversion of 11α,17α-di-hydroxy-progesterone (11α,17α-di-OH-Pr, 1 g/l) without α,α-dipyridyl. In all variants resting AcC-induced N. simplex VKM Ac-2033D cells were used
Discussion
Sterol degradation
The genetics of sterol (phytosterol and β-sitosterol) degradation by mycolic-acid rich actinobacteria belonging to Mycolicibacterium (syn. Mycobacterium) (e.g. M. smegmatis, M. neoaurum), Mycobacterium (e.g. M. tuberculosis), Rhodococcus, Gordonia and other genera have been studied intensively during past two decades, while little information is known for the representatives of Nocardioides and related actinobacteria. Similar to many other soil-dwelling bacteria [55] N. simplex could be able to utilize abundant plant sterols such as sitosterol which is a core component of plant membranes. Indeed, N. simplex VKM Ac-2033D grows on phytosterol as a sole source of carbon and energy as it was confirmed in this study. Phytostenones were detected as major intermediates at phytosterol degradation by the N. simplex VKM Ac-2033D cells.
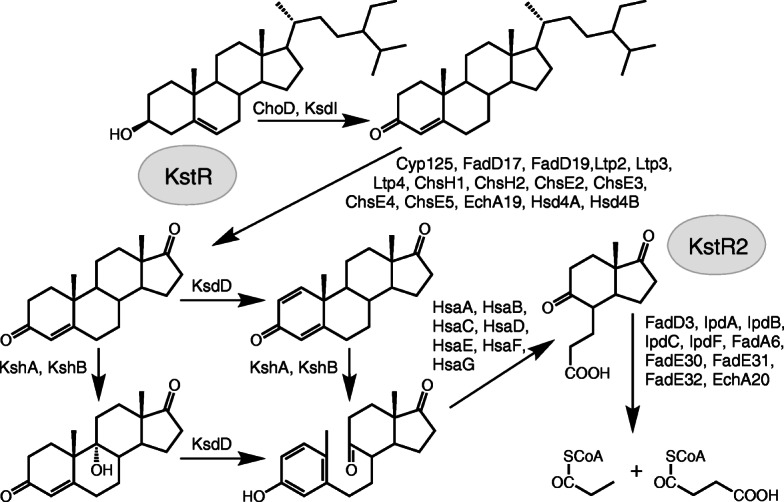
N. simplex is capable of full degradation of phytosterol and different other steroids without substituents in steroid core such as androstenedione, or progesterone (Supplementary Table S1). Genes whose products could play role in the reactions of steroid catabolism in N. simplex VKM Ac-2033D were predicted (Table 3, Fig. 7) based on their orthology with the known genes and new transcriptomic data.
Table 3.
N. simplex VKM Ac-2033D genes whose products taking part in steroid degradation, predicted
| Step | Enzyme annotation | M. tub | R. jostii | C. testosteroni | References | N. simplex | Cluster | Regulon | Phyt |
|---|---|---|---|---|---|---|---|---|---|
| Regulation | |||||||||
| TetR transcriptional repressor | kstR | [2] | KR76_12270 | B | KstR | ||||
| TetR transcriptional repressor | kstR2 | [2] | KR76_25115 | C | KstR2 | ||||
| A-ring activation | |||||||||
| I-II | cholesterol oxidase | cho | [56] | KR76_09550 | KstR | + | |||
| I-II | ∆5-∆4–3-ketosteroid isomerase | ksi | [57] | KR76_23530 | KstR | + | |||
| [57] | KR76_24125 | KstR | |||||||
| Cholesterol side chain degradation | |||||||||
| III-V | cytochrome P450 125 | cyp125 | [10, 58] | KR76_14335 | A | KstR | + | ||
| V-VI | 3-oxocholest-4-en-26-oate-CoA ligase | fadD19 | [59] | KR76_14210 | A | KstR | + | ||
| VI-VII | Acyl-coA dehydrogenases | chsE4–5 (fadE26–27) | [60] | KR76_14175-KR76_14180 | А | KstR | + | ||
| VII-VIII | 2-enoyl-CoA hydratase | echA19 | [6] | KR76_14215 | A | KstR | + | ||
|
IX-X; XI-XII |
aldol lyase | ltp3 | [12] | KR76_14285 | А | KstR | + | ||
| aldol lyase | ltp4 | KR76_14280 | А | KstR | + | ||||
| XII-XIII | steroid-22-oyl-CoA synthetase | fadD17 | casI | [61] | KR76_14185 | A | KstR | ||
| XIII-XIV | acyl-CoA dehydrogenase | chsE3 (fadE34) | [60, 62] | KR76_12295 | B | KstR | + | ||
| XIV-XV | 3α,7α,12α-trihydroxy-5β-chol-22-en-24-oyl-CoA hydratase |
hsd4B, Rv3538 |
casD | [4, 61, 63] | KR76_14505 | A | KstR | + | |
| XV-XVI | dehydrogenase | hsd4A | [4] | KR76_12260 | В | KstR | |||
| XIV-XVII | 3-ketoacyl-CoA thiolase | fadA5 | casB | [64] | KR76_14340 | А | KstR | + | |
|
XVII-XVIII; igr-operon |
7α,12α-dihydroxy-23,24-bisnorchola-1,4-dien-22-oyl-CoA dehydrogenase | chsE2 (fadE29) | casL | [65] | KR76_12325 | B | KstR | + | |
| acetyl-CoA acetyltransferase | chsH2 | casM | [66, 67] | KR76_12330 | B | KstR | + | ||
| acyl dehydratase | casN | [66] | KR76_12335 | B | KstR | ||||
| beta-hydroxyacyl-ACP dehydratase | chsH1 | casO | [66, 67] | KR76_12340 | B | KstR | + | ||
| 3-ketoacyl-CoA thiolase | ltp2 | casP | [66] | KR76_12345 | B | KstR | + | ||
| Opening of B-ring | |||||||||
| XVIII-XIX; XX-XXI |
Δ-3-ketosteroid dehydrogenase with preference for 9-OH-AD and testosterone |
kstD | kstD1 | [54, 68] | KR76_02655 | KstR | + | ||
| KR76_27115 | + | ||||||||
|
Δ-3-ketosteroid dehydrogenase with preference for progesterone |
kstD | kstD2 | [69] | KR76_27125 | D | Acin1,3 | + | ||
|
Δ-3-ketosteroid dehydrogenase with preference for saturated steroid substrates |
kstD | kstD3 | tesH | [54, 70] | KR76_14500 | A | KstR | + | |
| XVIII-XX; XIX-XXI | 3-ketosteroid 9α-hydroxylase, oxygenase subunit | kshA | [71] | KR76_14170 | A | KstR | + | ||
| KR76_27070 | + | ||||||||
| 3-ketosteroid 9α-hydroxylase, reductase subunit | kshB | KR76_14160 | A | KstR | |||||
| KR76_27080 | |||||||||
| eCleavage of A-ring | |||||||||
| XXII-XXIII | flavin-dependent monooxygenase | hsaA | tesA1A2 | [18, 72] | KR76_14480 | A | KstR | + | |
| hsaB | KR76_14465 | A | KstR | ||||||
| KR76_27090 | D | ||||||||
| XXIII-XXIV | extradiol dioxygenase | hsaC | tesB | [72, 73] | KR76_14470 | A | KstR | + | |
| KR76_27095 | D | + | |||||||
| XXIV-XXV | carbon–carbon hydrolase | hsaD | tesD | [19, 72] | KR76_14475 | A | KstR | + | |
| KR76_27085 | D | ||||||||
| XXV-XXVI | hydratase | hsaE | tesE | [17, 74] | KR76_14495 | A | KstR | ||
| KR76_24180 | + | ||||||||
| XXVI-Propanal | 4-hydroxy-2-oxovalerate aldolase | hsaF | tesG | KR76_14485 | A | KstR | + | ||
| KR76_24170 | + | ||||||||
| Propanal-Propionate | acetaldehyde dehydrogenase, acetylating | hsaG | tesF | KR76_14490 | A | KstR | + | ||
| KR76_24175 | + | ||||||||
| Ring D degradation | |||||||||
| XVII-XVIII | HIP-CoA synthetase | fadD3 | scdA | [75, 76] | KR76_25120 | С | KstR2 | + | |
| XVII-XIX | 5-Oxo HIC-CoA oxidase | ipdF | scdG | [20, 76] | KR76_14435 | A | KstR2 | + | |
| XXIX-XXX | COCHEA-CoA hydrolase, α-subunit | ipdA | scdL1L2 | [20, 76] | KR76_14365 | A | KstR2 | + | |
| KR76_25105 | С | KstR2 | + | ||||||
| COCHEA-CoA hydrolase, β-subunit | ipdB | KR76_14370 | A | KstR2 | + | ||||
| KR76_25110 | С | KstR2 | + | ||||||
| XXX-XXXI | acyl-CoA dehydrogenase | fadE30 | scdC1C2 | [76, 77] | KR76_14440 | A | KstR2 | + | |
| XXXI-XXXII | acyl-CoA hydrolase | scdD | [76] | ||||||
| XXXII-XXXIII |
dehydrogenase, oxidized a hydroxyl in propionate chain |
hsd4A | scdE | [76] | KR76_14350 | A | KstR2 | + | |
| XXXIII-XXXIV | acetyl-CoA acetyltransferase | fadA6 | scdF | [20, 76] | KR76_14430 | A | KstR2 | + | |
|
XXXIV-XXXV; XXXVI-XXXVII |
5-Oxo HIC-CoA oxidase | ipdF | scdG | [20, 76] | KR76_14435 | A | KstR2 | + | |
|
XXXIV-XXXVI; XXXV-XXXVII |
5-OH HIC-CoA reductase | ipdC | scdK | [20, 76] | KR76_14375 | A | KstR2 | + | |
| Ring C degradation | |||||||||
| XXXVII-XXXVIII | HIEC-CoA hydrolase | echA20 | scdY | [20, 76] | KR76_14360 | A | KstR2 | + | |
| XXXVIII-XXXIX | CoA transferase | ipdA | scdL1L2 | [20, 76] | KR76_14365 | A | KstR2 | + | |
| KR76_25105 | С | KstR2 | + | ||||||
| CoA transferase | ipdB | KR76_14370 | A | KstR2 | + | ||||
| KR76_25110 | С | KstR2 | + | ||||||
| XL-XLI | acyl-CoA dehydrogenases | fadE31, fadE32 | ro04591-ro04593 | [61, 78] |
KR76_14445 KR76_14450 |
A | KstR2 | + | |
| acyl-CoA dehydrogenases | fadE33 | KR76_14455 | A | KstR2 | + | ||||
| XLI-XLII | enoyl-CoA hydratase | echA13 | scdN | [20, 76] | KR76_25020 | С | KstR2 | + | |
Step – degradation step from Fig. 7
Enzyme annotation – function of enzyme
M. tub. – name of gene in M. tuberculosis investigations
R. jostii - name of gene (or locus_tag) in R. jostii investigations
C. testosteroni - name of gene in C. testosteroni investigations
N. simplex - locus_tag of ortholog that is candidate gene for this function. Functions are predicted by the annotation, location, predicted regulon that ortholog belongs to and gene expression changes
Cluster – cluster where gene is located in genome [27]
Regulon – repressor whose binding site is predicted in the promoter of operon that ortholog belongs to
Phyt – gene in N. simplex VKM Ac-2033D that is induced by phytolesterol (+)
Fig. 7.
Biochemical scheme of sterol catabolism in N. simplex VKM Ac-2033D. Genes coding respective proteins are denoted, locus-tags from N. simplex VKM Ac-2033D are predicted, see Table 3. a Modification of 3β-ol-5-ene to 3-keto-4-ene moiety in A-ring of steroid core; b degradation of the C24-branched sterol side chain to C19-steroids; c steroid core modifications; d steroid core degradation via 9(10)-seco pathway. I - sterols; II - stenones. R = H, campesterol, campestenone; R = CH3, sitosterol, stigmast-4-en-3-one (β-sitostenone); XVII - androst-4-ene-3,17-dione (AD), XVIII - androsta-1,4-diene-3,17-dione (ADD), XIX - 9α-hydroxy-AD, XX - unstable 9α-hydroxy-ADD, XXI - 3β-hydroxy-9,10-seco-androsta-1,3,5(10)- triene-9,17-dione (3βHSA), XXII - 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (3,4-DHSA), XXIII - 4,5-9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oic acid (4,9-DSHA), XXIV - 2-hydroxyhexa-2,4-dienoic acid (2-HHD), XXV - 4-hydroxy-2-oxohexanoic acid, XXVI - 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid (DOHNAA) or 3aα-H-4α-(3′-propanoate)-7aβ-methylhexahydro-1,5-indadione (HIP), XXVII - 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oyl-CoA (HIP-CoA) XXVIII - 9-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid or 3aα-H-4α(3′-propanoate)-5α-hydroxy-7aβ-methylhexahydro-1-indanone (5-OH-HIP), XXIX - 9-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oyl-CoA (5-OH-HIP-CoA), XXX 9-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrost-6-ene-5-oyl-CoA (HIPE-CoA), XXXI - 7,9-dihydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oyl-CoA, XXXII - 9-hydroxy-7,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oyl-CoA, XXXIII - 9-hydroxy-17-oxo-1,2,3,4,5,6,10,19-octa-norandrostan-7-oyl-CoA or 3aα-H-4α(carboxylCoA)-5α-hydroxy-7aβ-methylhexahydro-1-indanone (5-OH-HIC-CoA), XXXIV - 9,17-dioxo-1,2,3,4,5,6,10,19-octa-norandrostan-7-oyl-CoA, XXXV - 9-hydroxy-17-oxo-1,2,3,4,5,6,10,19-octa-norandrost-8(14)-en-7-oyl-CoA, XXXVI - 9,17-dioxo-1,2,3,4,5,6,10,19-octa-norandrost-8(14)-en-7-oyl-CoA or 7a-Methyl-1,5-dioxo-2,3,5,6,7,7a-hexahydro-1H-indene-4-carboxylic acid (HIEC-CoA), XXXVII - 9-oxo-1,2,3,4,5,6,10,19-octanor-13,17-secoandrost-8(14)-ene-7,17-dioic acid-CoA-ester or (R)-2-(2-carboxyethyl)-3-methyl-6-oxocyclohex-1-ene-1-carboxyl-CoA (COCHEA-CoA), XXXVIII - 6-methyl-3,7-dioxo-decane-1,10-dioic acid-CoA ester, XXXIX - 4-methyl-5-oxo-octane-1,8-dioic acid-CoA ester, XL - 4-methyl-5-oxo-oct-2-ene-1,8-dioic acid-CoA ester (MOODA-CoA), XLI - 3-hydroxy-4-methyl-5-o xo-octane-1,8-dioic acid-CoA ester, XLII - 4-methyl-3,5-dioxo-octane-1,8-dioic acid-CoA ester, XLIII - 2-methyl-3-oxo-hexane-1,6-dioic acid-CoA ester, XLIV – succinyl-CoA, XLV – propionyl-CoA. Adopted from: [6, 12, 14, 17, 20, 60, 67, 75, 76, 79]
Bacterial steroid degradation pathway was shown to be controlled by two TetR-type transcriptional repressors named KstR and KstR2 [1–4]. In Actinobacteria 3-oxocholest-4-en-26-oic acid and its derivatives may regulate the transcription factor KstR [58], while the effector of KstR2 is HIP-CoA [80]. The reciprocal BLAST with the corresponding regulators in M. tuberculosis confirmed the amino acid sequence similarity of KR76_12270 with KstR and KR76_25115 with KstR2 [27].
As shown for saprophytic mycobacteria, side chain oxidation and ring B modification are the independent processes [81]. The alkyl side chain of sterols is cleaved according to the mechanism of fatty acid β-oxidation, which is triggered by hydroxylation at C26(27). Further oxidation of the side chain occurs with sequential cleavage of two- and three-carbon fragments. Steroid core degradation seems to carry out through the 9(10)-seco pathway with the B ring opening (see below). After that step sequential destruction of the rings A and B remnants, as well as the rings C and D occurs (Table 3, Fig. 7).
We considered in more details the genes related to the B ring opening. As mentioned above, the only known mechanism of bacterial steroid core degradation is the 9(10)-seco pathway which comprises 1(2)-dehydrogenation catalyzed by KstDs and 9α-hydroxylation by 3-ketosteroid-9α-hydroxylase (KshAB) (Fig. 7). The former enzymes encoded by kstDs [82], while the Rieske 3-ketosteroid-9α-hydroxylase consists of KshA monooxygenase and KshB reductase subunits encoding by kshA and kshB, respectively [71].
There are three kshA orthologs in the genome of N. simplex [27]. Transcription of two of them was enhanced in phytosterol induced cells, specifically, KR76_14170 that located in cluster A (10.9-fold), and KR76_27070 in cluster D (14-fold), which has no predicted kstR binding sites (Table 3). Noteworthy, none of them increased expression in response to AcC.
Broadest substrate range without clear substrate preference was reported for KshA5 of Rhodococcus rhodochrous DSM 43269 [83]. The enzyme showed activity towards different Δ4, Δ1,4, 5α-H and 5β-H steroids, steroids with bulky aliphatic side chains and was found to be an only KshA enzyme of the DSM 43269 strain that exhibited activity towards 5α-androstane-17β-ol-3-one and cortisol [83]. The closest ortholog of kshA5 of R. rhodochrous DSM43269 is KR76_27070 in N. simplex VKM Ac-2033D (identity 62%). Its product could be involved in progesterone (and other steroids) degradation by N. simplex via 9α-hydroxylation of their 1(2)-dehydrogenated intermediates. Unlike R. rhodochrous DSM 43269, the N. simplex VKM Ac-2033D does not degrade cortisol (Supplementary Table S1). It is possible that the product of KR76_27070 is not active towards cortisol. The 3-keto-4-ene-steroid substrates substituted in positions 6, 7, 9, and 11 are considered more protected then unsubstituted 3-keto-4-ene-steroids from the action of the key enzymes involved in the degradation the steroid core; such ‘protected’ 3-keto-4-ene-steroids are widely used as the substrates in bioconversions with N. simplex [34, 37, 40].
The amino acid alignment and conserved amino acid residues [71] of expressed KshAs of N. simplex are shown in Supplementary Figure S1. N240 in the catalytic domain is replaced with D240. Similar replacement was earlier observed for the KshA of Mycolicibacterium sp. VKM Ac-1817D (syn. Mycobacterium sp. VKM Ac-1817D) [84]. Since N. simplex VKM Ac-2033D is not capable of degrading 11-functionalised steroids (e.g. cortisol, prednisolone, cortisone), this replacement might have altered the catalytic site in N. simplex.
It is well known that the number of oxygenase (KshA) and reductase (KshB) subunits of 9α-hydroxylase can differ [83]. For instance, five genes coding for KshA and two kshB genes were revealed in the genome of Mycobacterium sp. VKM Ac-1817D. Interestingly, when N. simplex VKM Ac-2033D growing on phytosterol, two kshAs and one kshB were up-regulated [84].
The domain structure of KshB of N. simplex is similar to KshB from R. erythropolis strain SQ1 [85] (Supplementary Fig. S2), with two replacements in a NAD binding domain of KR76_14160 that might indicate disturbed functionality of KR76_14160 consistent with its lower induction level (1.9-fold) (Table 4, Fig. 5).
Table 4.
Differentially expressed genes encoding KstDs and KshA/B
| Locus_tag | Ortholog in Mtb | Localization | Product | q-value Phyt/Ca | Phyt/C | q-value AcC/Cb | AcC/C |
|---|---|---|---|---|---|---|---|
| KR76_14170 | kshA | Cluster А | 3-ketosteroid-9α-hydroxylase | 2.24E-55 | 10.9 | 1 | 1.3 |
| KR76_27070 | kshA | Cluster D | 3-ketosteroid-9α-hydroxylase | 3.15E-59 | 14.4 | 1 | 1 |
| KR76_14160 | kshB | Cluster А | 3-ketosteroid-9α-hydroxylase | 0.7 | 1.9 | 1 | 1.2 |
| KR76_27080 | kshB | Cluster D | 3-ketosteroid-9α-hydroxylase | 1 | 3.7 | 1 | 1 |
| KR76_02655 | kstD1 | Out of clusters | 3-ketosteroid-Δ1-dehydrogenase | 1.01E-07 | 5.2 | 1 | 1.4 |
| KR76_27115 | kstD1 | Cluster D | 3-ketosteroid-Δ1-dehydrogenase | 1.4E-131 | 16.7 | 1 | 1.2 |
| KR76_27125 | kstD2 | Cluster D | 3-ketosteroid-Δ1-dehydrogenase | 1.44E-14 | 6.4 | 0 | 1207.7 |
| KR76_01140 | kstD3 | Out of clusters | 3-ketosteroid-Δ1-dehydrogenase | 1 | 0.3 | 1 | 0.3 |
| KR76_14500 | kstD3 | Cluster А | 3-ketosteroid-Δ1-dehydrogenase | 1.12E-12 | 6.7 | 1 | 1 |
aPhyt/C – expression changes on Phytosterol with respect to control
bAcC/C – expression changes on AcC with respect to control
With regard to KstDs, five candidate kstD genes were earlier revealed in the genome of N. simplex that are divided into three orthogroups, specifically, kstD1, kstD2 and kstD3 [27].
KstD1 of Rhodococcus erythropolis PR4 and R. ruber shows substrate preference for 9-OH-AD and testosterone [54]. The orthologs of kstD1 are KR76_02655 and KR76_27115 which increased their expression in phytosterol plus conditions.
KstD2 of R. ruber showed maximum preference for progesterone [54], while KstD2 of Gordonia neofelifaecis NRRL B-59395 transformed progesterone and cholest-4-en-3-one [69]. We tested KstD activity of N. simplex in vivo towards Pr, and its hydroxylated derivatives, such as 17α-OH-Pr and 11α,17α-di-OH-Pr. The corresponding 1(2)-dehydroderivatives were formed (Fig. 6). Lower degradation was observed in the presence of α,α-dipyridyl (the inhibitor of the iron containing enzymes, such as KshA). We suppose that progesterone bioconversion might be carried out by the kstD2 KR76_27125 product. Amino acid sequence of KstD2 KR76_27125 of N. simplex differ from consensus KstD2 [86] in some residues (Supplementary Fig. S3). Role of such substitution is not clear at the moment.
KstD3 of R. ruber preferred saturated steroid substrates (such as 5α-androstane-17β-ol-3-one) followed by progesterone [54]; KstD3 of Gordonia neofelifaecis showed higher activity to 16α,17α-epoxyprogesterone > AD > cholest-4-en-3-one > progesterone [69]. The expression of kstD3 ortholog KR76_14500 in N. simplex was increased on phytosterol 6.7-fold and was not increased in response to AcC.
Thus, effective 1(2)-dehydrogenation of 3-ketosteroids by N. simplex VKM Ac-2033D (Supplementary Table S1) is due to the presence of five kstDs, of which four genes are over-expressed in response to phytosterol and one (kstD2) is induced with AcC, but its transcription level is highest among all genes (more than 1200-fold).
Therefore, the genome-wide transcriptome analysis confirmed the transcriptional activation of the gene clusters related to the 9(10)-seco pathway of steroid catabolism in phytosterol induced N. simplex cells.
Cortisone 21-acetate conversion
Unlike phytosterol, AcC could not be utilized by N. simplex VKM Ac-2033D as a carbon and energy source (Fig. 4a), but was converted mainly to its 1(2)-dehydroanalog - AcP (Fig. 4b). Probably, the presence of 11-oxo group in the molecule of AcC (or 11β-hydroxyl group in cortisol) hinders the action of 9α-hydroxylase (KshAB). These results are in accordance with the previously reported N. simplex activity towards cortisol and its derivatives [34, 37], and correlate with the data presented in Table 4 demonstrating quite different gene expression under phytosterol and AcC induction.
Along with the KstD activity, slight deacetylase and 20β-reductase activities were observed to form cortisone (C), prednisone (P), and 20β-reduced derivatives (Fig. 4b).
As shown earlier, efficiency of the 1(2)-dehydrogenation by N. simplex VKM Ac-2033D did not depend on the presence of acetyl group in the steroid inducer [31]. The order of deacetylation and 1(2)-dehydrogenation reactions can differ depending on the induction conditions (Fig. 8). In the AcC induced cells, 1(2)-dehydrogenation preceded deacetylation that coincides with the highest level of kstD2 expression (Table 4), while in the absence of AcC induction, the reverse order of reactions was observed (data not shown).
Fig. 8.
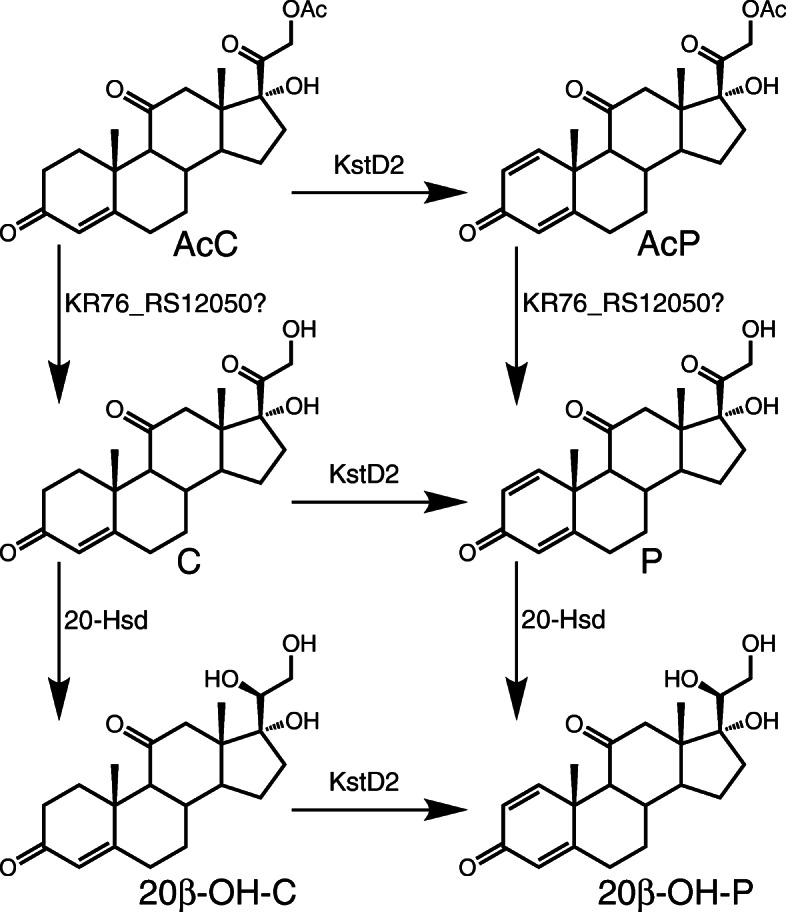
N. simplex VKM Ac-2033D AcC bioconversion scheme. Designations: AcP – 21-acetate of 1(2)-dehydro-cortisone or 21-acetate of prednisone; C – cortisone; P – prednisone; 20β-OH-C - 20β-hydroxy cortisone; 20β-OH-P - 20β-hydroxy prednisone
Both cytosol-dissolved and membrane-associated constitutive steroid esterases were earlier reported for N. simplex [32]. Among the genes whose transcription is enhanced in response to AcC, no inducible esterases were detected. However, the analysis of the complete transcriptome revealed KR76_12050 acyl-CoA thioesterase II with a high level of constitutive expression. Suggesting that the product of this gene might be responsible for 21-deacetylation we check a domain composition of the corresponding protein using InterProScan [87]. Since no membrane-bound domains were detected in this protein, the cytosolic enzymes are most likely responsible for the hydrolysis of 21-acetyl group in AcC.
Searching of the genes related to 20-reductase activity was carried out with protein BLAST. In N. simplex genome, three homologs of fabG3 coding for 3α,20β-hydroxysteroid dehydrogenase [88] were revealed. Of these, expression of KR76_13560 is greatly enhanced in response to AcC (13.7-fold) (Supplementary Table S4). Product of this gene is similar (amino acid sequence) by 56% with the cortisone-20β-reductase from Streptomyces hydrogenans [89].
FabG of Bacillus megaterium was identified as the enzyme with 20β-HSD activity [90] that reduce the β-ketoacyl group to a β-hydroxy group. Only one of homologs in N. simplex VKM Ac-2033D, namely, KR76_14350, increased its expression in the presence of phytosterol (Supplementary Table S4). None of these genes was induced by AcC.
Probably, the product of the gene KR76_13560 may be responsible for the 20β-reducing activity of N. simplex. As shown earlier by Medentsev et al. [91], steroid 1(2)-dehydrogenation by Arthrobacter grobiformis (syn. N. simplex) did not depend on the level of NAD(P) H, but the resulting redox equivalents transferred to the respiratory chain to generate transmembrane potential (ΔμH+). Hypothetically, accumulated reduced equivalents may stimulate constitutive NADH-dependent 20β-HSD activity.
Transcription factors binding motifs
Totally, about one third part of the operons was induced in which the predicted binding motifs for KstR and KstR2 were found earlier [27]. We have three possible explanations for such a low percentage. i) Some of the operons may be repressed by other transcription factors aside from KstR and KstR2. Even after KstR and KstR2 unbind, those other factors might continue to repress the operons. ii) Some of the predicted KstR and KstR2 binding sites could be false positive predictions. iii) If two operons are directed outward (“← →”), the intergenic region between them is small and a KstR or KstR2 site is present there, we supposed that it regulates both operons, while in fact it would regulate one of them. The relative contributions of these three effects were not estimated in this study.
Search for motifs for the binding of candidate factors revealed several recurring motifs before operons that strengthened, or weakened their expression.
Two sites (Acin1, Acin3) were found before KR76_27125, kstD2, with a maximum observed level of induction (Fig. 5). On the other side of these sites the transcriptional regulator gene KR76_27130 belonging to the TetR family is located whose expression was induced 10-fold. The increased expression of this gene and the absence of the predicted binding sites for the transcription factors KstR and KstR2 in this area suggest that KR76_27130 could be involved in the regulation of kstD2 induction and that the predicted motifs belong to it. Recently, the TetR family transcriptional factor KR76_27130 was confirmed to be a steroid catabolism regulator named SRTF1 (Steroid Responsive Transcriptional Factor 1) [92]. Its binding site coincided with the predicted Acin1 motif.
Other genes annotated as transcription factors that induced by AcC, specifically KR76_27330 (transcriptional regulator, MerR family) and KR76_26400 (transcriptional regulator, PadR family), both are located in an operon with predicted Acin3 site; and KR76_24405 (putative transcriptional regulator) is in an operon with the predicted Acin1 site. However, the found Acin and Sitdec1 motifs are hardly enough to explain the change in the expression of all the operons upon the addition of phytosterol, or AcC. It seems that there would be another transcription factor with a small number of motifs that could not be detected by MEME tools. Post-transcriptional regulation of mRNA from these operons also cannot be fully excluded.
Nevertheless, the calculations and predictions indicate the most likely candidate genes that can serve as a target for future biotechnological research.
In the future, it is planned to expand the experiments by studying of a range of new inducers and increasing the number of replicates. New inducers will clarify the role of genes that are homologous to already known steroid catabolism genes but do not have binding motifs with its confirmed regulators. It is interesting to use as inducers substances that are intermediates at various stages of steroid catabolism, for example, androstenedione or androstendiendione. This will make it possible to determine the complexes of genes that are activated at later stages and to compare the catabolism of steroids in N. simplex VKM Ac-2033D with catabolism in more studied organisms. Knockout and proteomic experiments are also promising, confirming the role of calculated transcription factors.
Conclusion
The gene network involved in phytosterol degradation by N. simplex VKM Ac-2033D is generally similar to those described for mycobacteria and related actinobacteria, but differs mainly by the existence of a cluster that has its own system of regulation.
In this study, we have generated experimental and computational evidences for the functionality of the clusters A, B, C, D those were identified by the bioinformatics analysis of the genome [27]. While the clusters A and B have been previously described for other bacteria, the clusters C and D were detected only by the analyzing of the N. simplex VKM Ac-2033D genome. Probably, the cluster D has its own previously unreported regulation system. We predicted a role in phytosterol catabolism for some dozens of genes of N. simplex VKM Ac-2033D; the genes taking part in AcC bioconversion were also suggested.
Our analysis suggests that the bioconversion of cortisone acetate by N. simplex is not regulated by the KstR-repressor.
The 3-ketosteroid-Δ1-dehydrogenase activity of N. simplex VKM Ac-2033D is firstly shown for the progesterone structure. The extremely high level of 1(2)-dehydrogenation of 3-ketosteroids is explained by the multiplicity and high level of induction of 3-ketosteroid-Δ1-dehydrogenases (kstD2 KR76_27125 in the case of AcC). A number of genes that can be responsible for the increased 20β-reducing activity have been found, however, in the case of different inducers, transcription of different genes is enhanced.
The knowledge of the genome organization, the functionality of the existing sterol catabolism genes and their possible involvement in steroid oxidation process contributes to our understanding of steroid bioconversion by actinobacteria, as well as to the expansion of the application field of the N. simplex VKM Ac-2033D in biotechnology.
Materials and methods
Reagents
Phytosterol (total sterols content – 95.47%; β-sitosterol – 42.39%, stigmasterol – 26.08%, campesterol – 23.48%, brassicasterol – 3.52%) was obtained from Jiangsu Spring Fruit Biological Products Co., Ltd., China. Pregn-4-ene-3,20-dione (progesterone, Pr), pregn-4-en-17α-ol-3,20-dione (17α-hydroxy-progesterone, 17α-OH-Pr), pregn-4-ene-11α,17α-diol-3,20-dione (11α,17α-dihydroxy-progesterone, 11α,17α-di-OH-Pr) were purchased from Sigma (USA). 21-Acetoxy-pregn-4-ene-17α,21-diol-3,11,20-trione (21-acetate of cortisone, cortisone 21-acetate, AcC) was purchased from Steraloids (USA), yeast extract from Difco (USA), soya peptone from HiMedia (India), randomly methylated β-cyclodextrin (MCD) from Wacker Chemie (Germany). Other materials and solvents were of analytical grade and purchased from domestic commercial suppliers.
Microorganism
A strain of Nocardioides simplex VKM Ac-2033D was obtained from the All-Russian Collection of Microorganisms (VKM). For RNA extraction, N. simplex VKM Ac-2033D was cultured as described in [27] on the mineral medium (control) supplemented with (g/l): yeast extract - 1.0, glycerol - 5.0, MCD - 3.46, and the same medium supplemented with phytosterol (0.5 g/L), or AcC (0.2 g/L).
Cultivation was carried out aerobically on a rotary shaker (200 rpm) at 30 °C for 16 h. For steroid bioconversion experiments, the cells were harvested by centrifuge (4 °C, 8000×g, 30 min) and washed with sterile saline as described in [31].
Growth estimations
Growth on phytosterol, or AcC as a sole source of carbon was estimated as described in [27] on the mineral medium supplemeted with MCD 17 (g/L) (control), phytosterol (1 g/L), or AcC (1 g/L).
Because of low phytosterol and AcC solubilities, optical or dry weight measurements were inaccurate, and the biomass grown upon phytosterol plus or AcC plus conditions was assayed by protein content or CFU estimation, respectively. The protein concentration was determined by the Lowry method with preliminary alkaline hydrolysis of biomass by adding 0.9 ml of 0.5 N NaOH to a 0.1 ml sample. For colony-forming units (CFU) counting, some broth samples were serially diluted with saline under vigorous agitation and plated on the solid LB medium. The experiments were carried out in no less than three replicates.
Bioconversion of steroids by the resting cells, isolation and identification of steroids
Progesterone, 17α-hydroxy-progesterone, and 11α,17α-dihydroxy-progesterone (0.4, 1.0, and 1.0 g/l, respectively) were tested as the substrates for bioconversion by resting whole cells of N. simplex (0.4, 0.2 and 0.2 g/l, respectively). Biomass was prepared as described above (in 2.2). Bioconversions were carried out in 0.02 М Na-phosphate buffer, рН 7.0, aerobically (200 rpm) at 30 °C in 750-ml shake flasks containing 100 ml buffered media with MCD (3.5–8.0 g/l). Concentration of α,α-dipyridyl when added was 0.4 g/l. The experiments were carried out in triplicates.
Steroid metabolites were isolated by preparative TLC [34]. Chromatographic purity of the compounds was monitored by TLC. For TLC, DC-Fertigfolien ALUGRAM® SIL G / UV25410 × 20 cm plates (Macherey-Nagel, Germany) were used. Steroids were developed in benzene:acetone 3:1 (v/v) or 4:1 (v/v) and visualized under UV-light (254 nm) in the chemiscope CN-6 (VilberLourmat, Germany).
HPLC analyses of steroid bioconversions were performed on a 5 μm C18 reverse phase column Symmetry 4.6 × 250 mm with a guard column Symmetry C18, 5 μm, 3.9 × 20 mm (Waters, Ireland). Mobile phase consisted of CH3CN:H2O:CH3COOH (40:60:0.01, v/v); at 50 °C, with a flow rate of 1 ml min− 1 and UV detection at 240 nm were used. Peaks were compared to authentic progesterone (P), 17α-hydroxy-progesterone (17α-OH-P), and 11α,17α-dihydroxy-progesterone (11α,17α-di-OH-P) (an external standard calibration method).
The structure of some steroids was confirmed by mass spectrometry (MS) and 1H-NMR spectroscopy. MS spectra of the reported steroids were recorded on a Finnigan LCQ Advantage MAX quadrupole ion trap mass spectrometer (Thermo Electron San. Jose, USA) in positive ions [M + H]+, at evaporator temperature 350 °C, capillar – 170 °C. MS/MS spectra were obtained using normalized collision energy (Normolized Collision EnergyTM) ranging from 20 to 40%. Collection and processing of data were performed using the Xcalibur software. Proton nuclear magnetic resonance (1H-NMR) spectra were recorded at 400 MHz with a Bruker Avance 400 spectrometer. Chemical shifts were measured relative to a solvent signal.
Isolation of mRNA and high-throughput sequencing
For RNA isolation, the cells were taken in the exponential growth phase (16 h), harvested by centrifuge at 8000×g for 10 min and immediately ground in a porcelain mortar under liquid nitrogen. Total RNA was isolated using Qiagen RNeasy mini kit according to the instructions of the supplier (Qiagen, Netherlands). The resulting mixture was treated with DNAse I. The ribosomal RNA was depleted using Ribo-Zero rRNA Removal Kit (Bacteria) according to the protocols of the supplier (Epicentre, USA).
Sample preparation of mRNA for high-throughput sequencing was made with TruSeq RNA Sample prep kit v.2 according to the protocols of the manufacturer (Illumina, USA). Sequencing was conducted on HiSeq 2000 (50-nucleotide single-read run) according to the protocols of the manufacturer (Illumina, USA). The experiments were carried out in duplicate.
Real-time PCR validation was performed with using the AriaMx Realtime PCR system (Agilent, Richardson, TX, USA) with an M-439 kit (Eva Green I) (Syntol, Moscow, Russian Federation). The nucleotide sequences of primers used in this study for target and reference genes are listed in Supplementary Table S6. The amplification was performed as follows: 95 °C 5 min (1 cycle), 95 °C 10 s, 54 °C 15 s, 72 °C 30 s (40 cycles). Relative gene expression levels were calculated using the double delta Cq method [93].
Computational analyses
The differential expression analysis was performed by Rockhopper 2.03 [94]. Rockhopper does not require the mapping of a whole read (mapping part of a read is enough). It was thus unnecessary to trim the reads before processing, and the raw reads were not trimmed. The reads were mapped by Rockhopper in the strand specific mode, with the seed length of 20 bp, allowing at most 1 mismatch between the mapped part of a read and the genome. The options “compute operons” (−y) and “identify transcript boundaries” (−t) were switched on. A gene was considered differentially expressed between the conditions phytosterol plus and phytosterol minus or AcC plus and AcC minus, if its expression increased or decreased more than threefold with q-value less or equal to 0.01.
To check the percentage of reads corresponding to rRNA, the reads were mapped to the rRNA genes of N. simplex by CLC Assembly Cell 4.2 (https://www.qiagenbioinformatics.com/products/clc-assembly-cell/); the reads were required to map with at least 98% sequence similarity.
The operon predictions made by Rockhopper were supplemented with the operon predictions made by FgenesB [95]. M. tuberculosis H37Rv was used as the “closest organism” in FgenesB. If Rockhopper and FgenesB provided different predictions for a particular operon, we chose the shortest prediction.
The predictions of KstR and KstR2 binding sites were taken from [27]. The search for membrane-associated domains in the protein corresponding to the gene with the locus tag KR76_12050 was performed by the InterProScan [87] web server (https://www.ebi.ac.uk/interpro/interproscan.html) on August 5, 2018.
Alignments of the KshA, KshB and KstD proteins were done with CLC Genomics Workbench, v. 7.5.1.
Genes whose products could play role in the biochemical reactions were predicted based on the annotation, homology with the known enzymes (using orthogroups), gene expression changes, location and predicted regulons to which the orthologs belong.
Searching of the genes related to 20-reductase activity was carried out with protein BLAST for fabG3 coding for 3α,20β-hydroxysteroid dehydrogenase [88] and fabG of Bacillus megaterium [90].
Search of transcription factors binding motifs
To find out binding motifs of transcription factors that may be responsible for the change in operon expression, the following protocol was used:
Regions spanning from 500 bps upstream to 50 bps downstream of start codons of the first genes in the operons that changed their expression were taken.
In these regions, potential binding motifs were de novo predicted by MEME 4.10.0 [96]. The minimum allowed motif width was set to 8 bps, the maximum to 50 bps. Any number of motifs was allowed in each of the regions (the option “-mod anr”). Only motifs with e-value, less or equal to 0.1 were considered further.
Potentially, the found motifs may correspond to low complexity sequences, or promoters. To identify and discard such motifs, we took the regions spanning from 500 bps upstream to 50 bps downstream of start codons of all N. simplex genes and searched for the motifs corresponding to all the motifs found in the previous paragraph. The search was done by the tool FIMO from the MEME 4.10.0 suite [53] with the q-value threshold for reported motifs of 0.05. If a motif had 1000 sites or more, it was deemed unrelated to a transcription factor.
If a motif had less than 1000 sites, it was considered potentially corresponding to some transcription factor.
This protocol was applied separately three times: for the operons that (i) increased, or (ii) decreased their expression in AcC conditions, and (iii) decreased their expression in phytosterol conditions.
Supplementary Information
Additional file 1: Supplementary Table S1. Steroid bioconversion by N. simplex VKM Ac-2033D. Supplementary Table S2.MS-characteristics of the steroid substrates and N. simplex bioconversion products. Supplementary Table S3. 1H-NMR spectra. Supplementary Table S5. New candidate motifs for the binding of transcription factors for steroid catabolism regulation in N. simplex VKM Ac-2033D. Supplementary Table S6. Real-time qPCR.
Additional file 2: Supplementary Table S4. DEGs and operons.
Additional file 3: Supplementary Figure S1-S3. Alignments of KshA, KshB, KstD [pdf]. Supplementary Figure S1. KshA alignment. Supplementary Figure S2. KshB alignment. Supplementary Figure S3. KstD alignment.
Acknowledgements
This work has been performed in the frames of ERA CoBioTech program on the Project: Synthetic biology for industrial production of steroids (Acronim: Syntheroids) and was supported exclusively by the Ministry of Science and Higher Education of Russian Federation (unique project identifier RFMEFI58818X0008). Authors are very grateful to Maria Logacheva (Moscow State University, Moscow) for the high-throughput sequencing.
Abbreviations
- MCD
methyl-β-cyclodextrin
- AcC
cortisone 21-acetate
- KstD
3-ketosteroid-Δ1-dehydrogenase
- DEG
differentially expressed gene(s)
Authors’ contributions
VS designed research, made sequencing libraries, analyzed the data; MS provided bioinformatics analysis; VF performed microbiological works; MD administrated and coordinated the project; EB isolated RNA; VF and AS conducted biochemical and chemical analyses; VS, MS, VF, MD wrote the manuscript; EB, VF and AS draw figures. All authors read and approved the manuscript.
Funding
ERA CoBioTech program on the Project: Synthetic biology for industrial production of steroids (Acronim: Syntheroids); Ministry of Science and Higher Education of Russian Federation (unique project identifier RFMEFI58818X0008).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files as Supplementary Table S3. Reads are in https://www.ncbi.nlm.nih.gov/sra/SRX5542009, https://www.ncbi.nlm.nih.gov/sra/SRX5542008, https://www.ncbi.nlm.nih.gov/sra/SRX5542005, https://www.ncbi.nlm.nih.gov/sra/SRX5542004, https://www.ncbi.nlm.nih.gov/sra/SRX5542007, https://www.ncbi.nlm.nih.gov/sra/SRX5542006.
Ethics approval and consent to participate
not applicable.
Consent for publication
not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Victoria Yu Shtratnikova, Email: vtosha@yandex.ru.
Mikhail I. Sсhelkunov, Email: shelkmike@gmail.com
Victoria V. Fokina, Email: 2vvfokina@gmail.com
Eugeny Y. Bragin, Email: bragory@yandex.ru
Andrey A. Shutov, Email: w___w@rambler.ru
Marina V. Donova, Email: donova@ibpm.pushchino.ru
References
- 1.Kendall SL, Withers M, Soffair CN, Moreland NJ, Gurcha S, Sidders B, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis: transcriptional repressor controlling a large lipid metabolism regulon in mycobacteria. Mol Microbiol. 2007;65:684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kendall SL, Burgess P, Balhana R, Withers M, ten Bokum A, Lott JS, et al. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology. 2010;156:1362–1371. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhía I, Galan B, Medrano FJ, Garcia JL. Characterization of the KstR-dependent promoter of the gene for the first step of the cholesterol degradative pathway in Mycobacterium smegmatis. Microbiology. 2011;157:2670–2680. doi: 10.1099/mic.0.049213-0. [DOI] [PubMed] [Google Scholar]
- 4.Uhía I, Galán B, Kendall SL, Stoker NG, García JL. Cholesterol metabolism in Mycobacterium smegmatis: cholesterol pathway. Environ Microbiol Rep. 2012;4:168–182. doi: 10.1111/j.1758-2229.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- 5.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. Plos Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A. 2007;104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLeod MP, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drzyzga O, Fernández de las Heras L, Morales V, Navarro Llorens JM, Perera J. Cholesterol degradation by Gordonia cholesterolivorans. Appl Environ Microbiol. 2011;77:4802–10. [DOI] [PMC free article] [PubMed]
- 9.Ivashina TV, Nikolayeva VM, Dovbnya DV, Donova MV. Cholesterol oxidase ChoD is not a critical enzyme accounting for oxidation of sterols to 3-keto-4-ene steroids in fast-growing Mycobacterium sp. VKM ac-1815D. J Steroid Biochem Mol Biol. 2012;129:47–53. doi: 10.1016/j.jsbmb.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Capyk JK, Kalscheuer R, Stewart GR, Liu J, Kwon H, Zhao R, et al. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids. J Biol Chem. 2009;284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston JB, Ouellet H, Ortiz de Montellano PR, et al. J Biol Chem. 2010;285:36352–36360. doi: 10.1074/jbc.M110.161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilbrink MH, van der Geize R, Dijkhuizen L. Molecular characterization of ltp3 and ltp4, essential for C24-branched chain sterol-side-chain degradation in Rhodococcus rhodochrous DSM 43269. Microbiology. 2012;158(Pt_12):3054–3062. doi: 10.1099/mic.0.059501-0. [DOI] [PubMed] [Google Scholar]
- 13.Casabon I, Swain K, Crowe AM, Eltis LD, Mohn WW. Actinobacterial acyl coenzyme a synthetases involved in steroid side-chain catabolism. J Bacteriol. 2014;196:579–587. doi: 10.1128/JB.01012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesbitt NM, Yang X, Fontán P, Kolesnikova I, Smith I, Sampson NS, et al. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect Immun. 2010;78:275–282. doi: 10.1128/IAI.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capyk JK, Casabon I, Gruninger R, Strynadka NC, Eltis LD. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J Biol Chem. 2011;286:40717–40724. doi: 10.1074/jbc.M111.289975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bragin EY, Shtratnikova VY, Schelkunov MI, Dovbnya DV, Donova MV. Genome-wide response on phytosterol in 9-hydroxyandrostenedione-producing strain of Mycobacterium sp. VKM ac-1817D. BMC Biotechnol. 2019;19:39. doi: 10.1186/s12896-019-0533-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carere J, McKenna SE, Kimber MS, Seah SYK. Characterization of an aldolase-dehydrogenase complex from the cholesterol degradation pathway of Mycobacterium tuberculosis. Biochemistry. 2013;52:3502–3511. doi: 10.1021/bi400351h. [DOI] [PubMed] [Google Scholar]
- 18.Dresen C, Lin LY-C, D’Angelo I, Tocheva EI, Strynadka N, Eltis LD. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J Biol Chem. 2010;285:22264–22275. doi: 10.1074/jbc.M109.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lack NA, Yam KC, Lowe ED, Horsman GP, Owen RL, Sim E, et al. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J Biol Chem. 2010;285:434–443. doi: 10.1074/jbc.M109.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowe AM, Casabon I, Brown KL, Liu J, Lian J, Rogalski JC, et al. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria. mBio. 2017;8. 10.1128/mBio.00321-17. [DOI] [PMC free article] [PubMed]
- 21.Nagasawa M, Bae M, Tamura G, Arima K. Microbial transformation of sterols. Part II: cleavage of sterol side chains by microorganisms. Agric Biol Chem. 1969;33:1644–1650. doi: 10.1271/bbb1961.33.1644. [DOI] [Google Scholar]
- 22.Choi K-P, Murooka Y, Molnár I. Secretory overproduction of Arthrobacter simplex 3-ketosteroid-delta-1-dehydrogenase by Streptomyces lividans with a multi-copy shuttle vector. Appl Microbiol Biotechnol. 1995;43:1044–1049. doi: 10.1007/BF00166923. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Tian Y, Wang J, Li Y, Wang H, Mao S, et al. Construction of engineered Arthrobacter simplex with improved performance for cortisone acetate biotransformation. Appl Microbiol Biotechnol. 2013;97:9503–9514. doi: 10.1007/s00253-013-5172-7. [DOI] [PubMed] [Google Scholar]
- 24.Holert J, Cardenas E, Bergstrand LH, Zaikova E, Hahn AS, Hallam SJ, et al. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio. 2018;9:e02345–e02317. doi: 10.1128/mBio.02345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak MJ, Kwon SK, Kim JF. Complete genome sequence of the sand-sediment actinobacterium Nocardioides dokdonensis FR1436T. Stand Genomic Sci. 2017;12. 10.1186/s40793-017-0257-z. [DOI] [PMC free article] [PubMed]
- 26.Shtratnikova VY, Schelkunov MI, Pekov YA, Fokina VV, Logacheva MD, Sokolov SL, et al. Complete genome sequence of steroid-transforming Nocardioides simplex VKM Ac-2033D. Genome Announc. 2015;3:e01406–14. [DOI] [PMC free article] [PubMed]
- 27.Shtratnikova VY, Schelkunov MI, Fokina VV, Pekov YA, Ivashina T, Donova MV. Genome-wide bioinformatics analysis of steroid metabolism-associated genes in Nocardioides simplex VKM Ac-2033D. Curr Genet. 2016;62:643–56. [DOI] [PubMed]
- 28.Shtratnikova VY, Schelkunov MI, Fokina VV, Bragin EY, Lobastova TG, Shutov AA, et al. Genome-wide transcriptome profiling provides insight on cholesterol and lithocholate degradation mechanisms in Nocardioides simplex VKM Ac-2033D. Genes. 2020;11:1229. [DOI] [PMC free article] [PubMed]
- 29.Qin H-M, Wang J-W, Guo Q, Li S, Xu P, Zhu Z, et al. Refolding of a novel cholesterol oxidase from Pimelobacter simplex reveals dehydrogenation activity. Protein Expr Purif. 2017;139:1–7. doi: 10.1016/j.pep.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Qin H-M, Zhu Z, Ma Z, Xu P, Guo Q, Li S, et al. Rational design of cholesterol oxidase for efficient bioresolution of cholestane skeleton substrates. Sci Rep. 2017;7. 10.1038/s41598-017-16768-6. [DOI] [PMC free article] [PubMed]
- 31.Fokina VV, Sukhodolskaya GV, Baskunov BP, Turchin KF, Grinenko GS, Donova MV. Microbial conversion of pregna-4,9(11)-diene-17α,21-diol-3,20-dione acetates by Nocardioides simplex VKM Ac-2033D. Steroids. 2003;68:415–21. [DOI] [PubMed]
- 32.Fokina VV, Donova MV. 21-Acetoxy-pregna-4(5),9(11),16(17)-triene-21-ol-3,20-dione conversion by Nocardioides simplex VKM Ac-2033D. J Steroid Biochem Mol Biol. 2003;87:319–25. [DOI] [PubMed]
- 33.Fokina VV, Sukhodol’skaya GV, Gulevskaya SA, Gavrish EY, Evtushenko LI, Donova MV. The 1(2)-dehydrogenation of steroid substrates by Nocardioides simplex VKM Ac-2033D. Microbiology. 2003;72:24–9. [PubMed]
- 34.Sukhodolskaya G, Fokina V, Shutov A, Nikolayeva V, Savinova T, Grishin Y, et al. Bioconversion of 6-(N-methyl-N-phenyl) aminomethyl androstane steroids by Nocardioides simplex. Steroids. 2017;118:9–16. doi: 10.1016/j.steroids.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Mutafova B, Mutafov S. Microbial transformations of plant origin compounds as a step in preparation of highly valuable pharmaceuticals. J Drug Metab Toxicol. 2016;7:204–215. doi: 10.4172/2157-7609.1000204. [DOI] [Google Scholar]
- 36.Costa S, Zappaterra F, Summa D, Semeraro B, Fantin G. Δ1-dehydrogenation and C20 reduction of cortisone and hydrocortisone catalyzed by Rhodococcus strains. Molecules. 2020;25:2192. doi: 10.3390/molecules25092192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sukhodolskaya GV, Donova MV, Nikolaeva VM, Koshcheyenko KA, Dovbnya DV, Khomutov SM, et al. Method of the producing 1(2)-dehydroderivatives of 4-delta-3-ketosteroids. RU Patent 2156302. 2000. [Google Scholar]
- 38.Fokina VV, Karpov AV, Sidorov IA, Andrjushina VA, Arinbasarova AY. The influence of β-cyclodextrin on the kinetics of 1-en-dehydrogenation of 6α-methylhydrocortisone by Arthrobacter globiformis cells. Appl Microbiol Biotechnol. 1997;47:645–649. doi: 10.1007/s002530050989. [DOI] [Google Scholar]
- 39.Sukhodol’skaja GV, Savinova TS, Fokina VV, Shutov AA, Nikolaeva VM, Lukashev NV, et al. Microbiological method of producing 1,2-dehydrogenated derivatives of delta-4-3-keto-steroids of androstane family in aqueous organic media. RU Patent 2447154. 2012. [Google Scholar]
- 40.Lobastova TG, Khomutov SM, Shutov AA, Donova MV. Microbiological synthesis of stereoisomeric 7(α/β)-hydroxytestololactones and 7(α/β)-hydroxytestolactones. Appl Microbiol Biotechnol. 2019;103:4967–4976. doi: 10.1007/s00253-019-09828-6. [DOI] [PubMed] [Google Scholar]
- 41.Zvyagintseva IS, Skryabin GK. Dehydrogenation of steroids by mycobacteria. Biol Bull Acad Sci USSR. 1964;4:525–32. [Google Scholar]
- 42.Krassilnikov N, Skryabin G, Aseeva I, Korsunskaya L. The 1,2-dehydrogenation of hydrocortisone by Mycobacterium sp. 193 cells. Dokl Biol Sci Sect. 1959;128:1063–1065. [Google Scholar]
- 43.Lestrovaya N, Nazaruk M, Skryabin G. The dehydrogenation and reduction of the a ring of delta-4-3-ketosteroids by cell-free extracts of Mycobacterium globiforme 193. Dokl Biol Sci Sect. 1965;163:768–770. [PubMed] [Google Scholar]
- 44.Arinbasarova AY, Koshcheyenko KA. Covalent binding of cells with activated silica gel. Prikl Biokhim Mikrobiol. 1980;16:854–861. [Google Scholar]
- 45.Prauser H. Nocardioides, a new genus of the order Actinomycetales. Int J Syst Bacteriol. 1976;26:58–65. doi: 10.1099/00207713-26-1-58. [DOI] [Google Scholar]
- 46.O’Donnell AG, Goodfellow M, Minnikin DE. Lipids in the classification of Nocardioides: reclassification of Arthrobacter simplex (Jensen) Lochhead in the genus Nocardioides (Prauser) emend. O’Donnell et al. as Nocardioides simplex comb. nov. Arch Microbiol. 1982;133:323–329. doi: 10.1007/BF00521299. [DOI] [PubMed] [Google Scholar]
- 47.Goodfellow M, Whitman WB, Bergey DH. Bergey’s manual of systematic bacteriology. Vol. 5. The Actinobacteria. 2. Ed. New York: Springer; 2012. [Google Scholar]
- 48.Suzuki K-I, Komagata K. Pimelobacter gen. Nov., a new genus of coryneform bacteria with LL-diaminopimelic acid in the cell wall. J Gen Appl Microbiol. 1983;29:59–71. doi: 10.2323/jgam.29.59. [DOI] [Google Scholar]
- 49.Collins MD, Dorsch M, Stackebrandt E. Transfer of Pimelobacter tumescens to Terrabacter gen. Nov. as Terrabacter tumescens comb. nov. and of Pimelobacter jensenii to Nocardioides as Nocardioides jensenii comb. nov. Int J Syst Bacteriol. 1989;39:1–6. doi: 10.1099/00207713-39-1-1. [DOI] [Google Scholar]
- 50.Yoon J-H, Park Y-H. The genus Nocardioides. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes. New York, NY: Springer New York; 2006. pp. 1099–1113. [Google Scholar]
- 51.Rohman A, Dijkstra BW. The role and mechanism of microbial 3-ketosteroid Δ1-dehydrogenases in steroid breakdown. J Steroid Biochem Mol Biol. 2019;191:105366. doi: 10.1016/j.jsbmb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 52.Catroux G, Fournier J-C, Blachère H. Importance de la forme cristalline de l’acétate de cortisone pour sa déshydrogénation en C-1 par Arthrobacter simplex. Can J Biochem. 1968;46:537–542. doi: 10.1139/o68-083. [DOI] [PubMed] [Google Scholar]
- 53.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinforma Oxf Engl. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guevara G, Fernández de las Heras L, Perera J, Navarro Llorens JM. Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isozymes in Rhodococcus ruber strain Chol-4. Microb Cell Factories. 2017;16. 10.1186/s12934-017-0657-1. [DOI] [PMC free article] [PubMed]
- 55.Bergstrand LH, Cardenas E, Holert J, Van Hamme JD, Mohn WW. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio. 2016;7:e00166. doi: 10.1128/mBio.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devi S, Kanwar SS. Cholesterol oxidase: source, properties and applications. Insights Enzyme Res. 2017;01:5–17. [Google Scholar]
- 57.Cho H-S, Choi G, Choi KY, Oh B-H. Crystal structure and enzyme mechanism of Δ5-3-ketosteroid isomerase from Pseudomonas testosteroni. Biochemistry. 1998;37:8325–8330. doi: 10.1021/bi9801614. [DOI] [PubMed] [Google Scholar]
- 58.García-Fernández E, Medrano FJ, Galán B, García JL. Deciphering the transcriptional regulation of cholesterol catabolic pathway in Mycobacteria: identification of the inducer of KstR repressor. J Biol Chem. 2014;289:17576–17588. doi: 10.1074/jbc.M113.545715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilbrink MH, Petrusma M, Dijkhuizen L, van der Geize R. FadD19 of Rhodococcus rhodochrous DSM43269, a steroid-coenzyme a ligase essential for degradation of C-24 branched sterol side chains. Appl Environ Microbiol. 2011;77:4455–4464. doi: 10.1128/AEM.00380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, Lu R, Guja KE, Wipperman MF, St Clair JR, Bonds AC, et al. Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 α2β2 acyl-CoA dehydrogenase initiates β-oxidation of 3-oxo-cholest-4-en-26-oyl CoA. ACS Infect Dis. 2015;1:110–125. doi: 10.1021/id500033m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, et al. Gene cluster encoding cholate catabolism in Rhodococcus spp. J Bacteriol. 2012;194:6712–6719. doi: 10.1128/JB.01169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruprecht A, Maddox J, Stirling AJ, Visaggio N, Seah SYK. Characterization of novel acyl coenzyme a dehydrogenases involved in bacterial steroid degradation. J Bacteriol. 2015;197:1360–1367. doi: 10.1128/JB.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol. 2014;49:269–293. doi: 10.3109/10409238.2014.895700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schaefer CM, Lu R, Nesbitt NM, Schiebel J, Sampson NS, Kisker C. FadA5 a thiolase from Mycobacterium tuberculosis: a steroid-binding pocket reveals the potential for drug development against tuberculosis. Structure. 2015;23:21–33. doi: 10.1016/j.str.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas ST, Sampson NS. Mycobacterium tuberculosis utilizes a unique heterotetrameric structure for dehydrogenation of the cholesterol side chain. Biochemistry. 2013;52:2895–2904. doi: 10.1021/bi4002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J Biol Chem. 2011;286:43668–43678. doi: 10.1074/jbc.M111.313643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang M, Guja KE, Thomas ST, Garcia-Diaz M, Sampson NS. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem Biol. 2014;9:2632–2645. doi: 10.1021/cb500232h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horinouchi M, Hayashi T, Kudo T. Steroid degradation in Comamonas testosteroni. J Steroid Biochem Mol Biol. 2012;129:4–14. doi: 10.1016/j.jsbmb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Q, Ren Y, He J, Cheng S, Yuan J, Ge F, et al. Multiplicity of 3-ketosteroid Δ1-dehydrogenase enzymes in Gordonia neofelifaecis NRRL B-59395 with preferences for different steroids. Ann Microbiol. 2015;65:1961–1971. doi: 10.1007/s13213-015-1034-0. [DOI] [Google Scholar]
- 70.Horinouchi M, Kurita T, Yamamoto T, Hatori E, Hayashi T, Kudo T. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem Biophys Res Commun. 2004;324:597–604. doi: 10.1016/j.bbrc.2004.09.096. [DOI] [PubMed] [Google Scholar]
- 71.Capyk JK, D’Angelo I, Strynadka NC, Eltis LD. Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J Biol Chem. 2009;284:9937–9946. doi: 10.1074/jbc.M900719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Horinouchi M, Koshino H, Malon M, Hirota H, Hayashi T. Steroid degradation in Comamonas testosteroni TA441: identification of metabolites and the genes involved in the reactions necessary before D-ring cleavage. Appl Environ Microbiol. 2018;84. 10.1128/AEM.01324-18. [DOI] [PMC free article] [PubMed]
- 73.Yam KC, D’Angelo I, Kalscheuer R, Zhu H, Wang J-X, Snieckus V, et al. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000344. doi: 10.1371/journal.ppat.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horinouchi M, Hayashi T, Koshino H, Kurita T, Kudo T. Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl Environ Microbiol. 2005;71:5275–5281. doi: 10.1128/AEM.71.9.5275-5281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casabon I, Crowe AM, Liu J, Eltis LD. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria: role of FadD3 in cholesterol catabolism. Mol Microbiol. 2013;87:269–283. doi: 10.1111/mmi.12095. [DOI] [PubMed] [Google Scholar]
- 76.Horinouchi M, Koshino H, Malon M, Hirota H, Hayashi T. Steroid degradation in Comamonas testosteroni TA441: identification of the entire β-oxidation cycle of the cleaved B ring. Appl Environ Microbiol. 2019;85. 10.1128/AEM.01204-19. [DOI] [PMC free article] [PubMed]
- 77.van der Geize R, Grommen AWF, Hessels GI, Jacobs AAC, Dijkhuizen L. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. Plos Pathog. 2011;7:e1002181. doi: 10.1371/journal.ppat.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wipperman MF, Yang M, Thomas ST, Sampson NS. Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an α2β2 heterotetrameric acyl coenzyme a dehydrogenase family. J Bacteriol. 2013;195:4331–4341. doi: 10.1128/JB.00502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujimoto Y, Chen CS, Gopalan AS, Sih CJ. Microbial degradation of the phytosterol side chain. II. Incorporation of [14C]-NaHCO3 onto the C-28 position. J Am Chem Soc. 1982;104:4720–4722. doi: 10.1021/ja00381a056. [DOI] [Google Scholar]
- 80.Casabon I, Zhu S-H, Otani H, Liu J, Mohn WW, Eltis LD. Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol Microbiol. 2013;89:1201–1212. doi: 10.1111/mmi.12340. [DOI] [PubMed] [Google Scholar]
- 81.Dovbnya DV, Egorova OV, Donova MV. Microbial side-chain degradation of ergosterol and its 3-substituted derivatives: a new route for obtaining of deltanoids. Steroids. 2010;75:653–658. doi: 10.1016/j.steroids.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 82.Itagaki E, Matushita H, Hatta T. Steroid transhydrogenase activity of 3-ketosteroid-delta 1-dehydrogenase from Nocardia corallina. J Biochem (Tokyo) 1990;108:122–127. doi: 10.1093/oxfordjournals.jbchem.a123150. [DOI] [PubMed] [Google Scholar]
- 83.Petrusma M, Hessels G, Dijkhuizen L, van der Geize R. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J Bacteriol. 2011;193:3931–3940. doi: 10.1128/JB.00274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bragin EY, Shtratnikova VY, Dovbnya DV, Schelkunov MI, Pekov YA, Malakho SG, et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. strains. J Steroid Biochem Mol Biol. 2013;138:41–53. doi: 10.1016/j.jsbmb.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 85.van der Geize R, Hessels GI, van Gerwen R, van der Meijden P, Dijkhuizen L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9alpha-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol Microbiol. 2002;45:1007–1018. doi: 10.1046/j.1365-2958.2002.03069.x. [DOI] [PubMed] [Google Scholar]
- 86.van der Geize R, Hessels GI, Dijkhuizen L. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ1-dehydrogenase isoenzyme. Microbiology. 2002;148:3285–3292. doi: 10.1099/00221287-148-10-3285. [DOI] [PubMed] [Google Scholar]
- 87.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang JK, Park MS, Waldo GS, Suh SW. Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis. Proc Natl Acad Sci. 2003;100:455–460. doi: 10.1073/pnas.0137017100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghosh D, Wawrzak Z, Weeks CM, Duax WL, Erman M. The refined three-dimensional structure of 3α,20β-hydroxysteroid dehydrogenase and possible roles of the residues conserved in short-chain dehydrogenases. Structure. 1994;2:629–640. doi: 10.1016/S0969-2126(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 90.Gerber A, Milhim M, Hartz P, Zapp J, Bernhardt R. Genetic engineering of Bacillus megaterium for high-yield production of the major teleost progestogens 17α,20β-di- and 17α,20β,21α-trihydroxy-4-pregnen-3-one. Metab Eng. 2016;36:19–27. doi: 10.1016/j.ymben.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Medentsev AG, Arinbasarova AY, Koshcheyenko KA, Akimenko VK, Skryabin GK. Regulation of 3-ketosteroid-1-en-dehydrogenase activity of Arthrobacter globiformis cells by a respiratory chain. J Steroid Biochem. 1985;23:365–368. doi: 10.1016/0022-4731(85)90418-2. [DOI] [PubMed] [Google Scholar]
- 92.Grazon C, Baer RC, Kuzmanović U, Nguyen T, Chen M, Zamani M, et al. A progesterone biosensor derived from microbial screening. Nat Commun. 2020;11. 10.1038/s41467-020-14942-5. [DOI] [PMC free article] [PubMed]
- 93.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods San Diego Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 94.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Solovyev V, Salamov A. Metagenomics and its applications in agriculture, biomedicine and environmental studies. Hauppauge: Nova Science Publisher’s; 2011. Automatic annotation of microbial genomes and metagenomic sequences; pp. 61–78. [Google Scholar]
- 96.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table S1. Steroid bioconversion by N. simplex VKM Ac-2033D. Supplementary Table S2.MS-characteristics of the steroid substrates and N. simplex bioconversion products. Supplementary Table S3. 1H-NMR spectra. Supplementary Table S5. New candidate motifs for the binding of transcription factors for steroid catabolism regulation in N. simplex VKM Ac-2033D. Supplementary Table S6. Real-time qPCR.
Additional file 2: Supplementary Table S4. DEGs and operons.
Additional file 3: Supplementary Figure S1-S3. Alignments of KshA, KshB, KstD [pdf]. Supplementary Figure S1. KshA alignment. Supplementary Figure S2. KshB alignment. Supplementary Figure S3. KstD alignment.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files as Supplementary Table S3. Reads are in https://www.ncbi.nlm.nih.gov/sra/SRX5542009, https://www.ncbi.nlm.nih.gov/sra/SRX5542008, https://www.ncbi.nlm.nih.gov/sra/SRX5542005, https://www.ncbi.nlm.nih.gov/sra/SRX5542004, https://www.ncbi.nlm.nih.gov/sra/SRX5542007, https://www.ncbi.nlm.nih.gov/sra/SRX5542006.