Highlights
-
•
A deep learning network facilitated dose calculation from CBCT.
-
•
A single network achieved CBCT-based dose calculation generating synthetic CT for head-and-neck, lung, and breast cancer patients with similar performance to a network specifically trained for each anatomical site.
-
•
Generation of synthetic-CT can be achieved within 10 s, facilitating online adaptive radiotherapy scenarios.
Keywords: Adaptive radiotherapy, Dose calculation, Image-guided radiotherapy, CBCT, Deep learning, Artificial intelligence, Image-to-image translation, Machine learning
Abstract
Background and purpose Adaptive radiotherapy based on cone-beam computed tomography (CBCT) requires high CT number accuracy to ensure accurate dose calculations. Recently, deep learning has been proposed for fast CBCT artefact corrections on single anatomical sites. This study investigated the feasibility of applying a single convolutional network to facilitate dose calculation based on CBCT for head-and-neck, lung and breast cancer patients.
Materials and Methods Ninety-nine patients diagnosed with head-and-neck, lung or breast cancer undergoing radiotherapy with CBCT-based position verification were included in this study. The CBCTs were registered to planning CT according to clinical procedures. Three cycle-consistent generative adversarial networks (cycle-GANs) were trained in an unpaired manner on 15 patients per anatomical site generating synthetic-CTs (sCTs). Another network was trained with all the anatomical sites together. Performances of all four networks were compared and evaluated for image similarity against rescan CT (rCT). Clinical plans were recalculated on rCT and sCT and analysed through voxel-based dose differences and -analysis.
Results A sCT was generated in 10 s. Image similarity was comparable between models trained on different anatomical sites and a single model for all sites. Mean dose differences were obtained in high-dose regions. Mean gamma (3%, 3 mm) pass-rates were achieved for all sites.
Conclusion Cycle-GAN reduced CBCT artefacts and increased similarity to CT, enabling sCT-based dose calculations. A single network achieved CBCT-based dose calculation generating synthetic CT for head-and-neck, lung, and breast cancer patients with similar performance to a network specifically trained for each anatomical site.
1. Introduction
In modern external beam image-guided radiotherapy (IGRT), cone-beam computed tomography (CBCT) plays a crucial role in accurate patient position verification [1]. Also, CBCT can facilitate online adaptive radiotherapy (ART) by visualising daily anatomical variations [2], without recurring to additional rescanning on CT.
CBCT image quality is inferior to that of CT in soft-tissue contrast and CT number consistency due to the presence of artefacts [3]. Therefore, CBCT is not sufficient to perform accurate dose calculations and patients need to be referred for a rescan CT (rCT) when anatomical differences are noted between daily images and the planning CT [4]. However, scheduling and acquiring an rCT adds logistic complexity and patient burden to the treatment. On the contrary, with ART based on CBCT these issues can be addressed. For example, by enabling accurate dose calculations on the daily CBCT images could eliminate the need for acquiring an rCT [2]. A prerequisite for online ART based on CBCT is that the CT number accuracy is sufficient to enable dose calculation.
Considerable literature has recently emerged proposing methods for how to correct CBCT imaging artefacts and increase image intensity consistency using: look-up table-based approaches [5], deformable image registration (DIR) of the planning CT to the daily anatomy on CBCT [6] and model- or Monte Carlo-based methods for scatter estimation and correction [7], [8]. Specifically, DIRs enabled accurate dose calculations for head-and-neck (HN) [9] but obtained lower dose accuracy in more complex anatomical regions with potential large inter-fraction changes such as lung [10] and pelvis [11]. Also, Monte Carlo-based methods were suitable for ART [12]. These techniques can be deployed on a time scale of minutes, which is not acceptable when aiming to use CBCT images for daily online dose evaluation or online pre-treatment adaptation.
Recently, deep learning has been proposed for fast CBCT artefact correction [13] as it can solve image-to-image translation problems within seconds [14], [15]. In this sense, previous work demonstrated the use of a two-dimensional (2D) U-net to improve CBCT image quality [13], [16], [17]. Moreover, it has been shown that converting CBCT with deep learning resulted in accurate dose calculation for prostate cancer patients [18], [19] and HN cancer patients [20], [21].
In this study, we investigated whether CBCTs converted to synthetic–CT (sCT) with a single convolutional network can be used as a surrogate of the daily anatomy for dose calculations of multiple anatomical regions. We employed a network to synthetise CT from CBCT of patients with HN, lung, or breast cancer. A single network trained for all the anatomical sites was compared to three networks trained per anatomical site. The performances of the single network was compared to the site-specific networks in terms of image similarity and dose calculation accuracy.
2. Material and methods
2.1. Imaging protocols
Ninety-nine patients diagnosed with HN (33), lung (33) or breast (33) cancer undergoing radiotherapy were retrospectively included in this study (Supplementary Material 1) in accordance to the guidelines of the local Medical Ethical Committee. Irradiations were performed between May 2016 and February 2019 on Agility linacs (Elekta AB, Sweden) with CBCT-based pre-treatment position verification.
An rCT was acquired in case anatomical variations were noted on the CBCT, including at least fourteen patients with rCT per site.
CBCTs were translated to apply clinical set-up corrections and resampled to the planning CT within the X-ray volumetric imaging (XVI, v5.0.2b72 Elekta AB, Sweden) system. CBCT acquisition occurred on ten different linacs. Imaging protocols and clinical registration procedures are detailed in Supplementary Material 2.
2.2. Image pre-processing
CT and CBCT were cropped to the size of the CBCT field of view (FOV) that was identified according to the following steps. CBCTs were thresholded at −999.9, obtaining a binary mask. In each transverse slice, morphological closure was performed, and a bounding box containing a circular mask with diameter of at least 26.9 cm was found for each transverse plane. The largest circular mask over the transverse slices was propagated for all the slices obtaining . CT and CBCT were cropped in the bounding box containing .
In addition to cropping, image intensity of CT and CBCT was clipped within the interval [-1024;3071] and linearly rescaled to [0;1].
2.3. Network architecture and training
To generate sCT from CBCT, a 2D cycle-generative adversarial network (cycle-GAN) was adopted [22]. Cycle-GANs enable unpaired training, which, compared to paired training, makes the network less sensitive to residual registration mismatch of CT and CBCT.
The network consisted of two cycles called “forward” and “backwards” during which GANs generated CT from CBCT and vice versa. Moreover, so-called “cycle-consistency” was enforced with an -norm such that after converting from CBCT to CT and vice versa, the original image should be obtained. The architecture (Supplementary Material 3) was based on the cycle-GAN provided by Zhu et al. [22], and it was implemented in Tensorflow (v1.3.0)1.
Three networks were trained separately on each anatomical site; another network was trained on all anatomical sites to investigate whether a single network may generalise for all anatomical sites together. To train, validate and test the network, the patient population for each anatomical was split into three datasets: 15 patients per site for training, 8 for validation and the remaining 10 for testing. For the single network, the data split in train/validation/test was maintained, e.g. same patient in each set, for the single network: 45 patients per site for training, 24 for validation and the remaining 30 for testing. The validation set was used to aid hyperparameter optimisation and to determine at which iteration the training should be stopped to avoid over-fitting (early-stopping), while the test set was used to evaluate the performance of the network.
Training of the cycle-GAN was performed in the transverse plane; for each iteration, random CT and CBCT slices of potentially different patients and different location in the cranio-caudal direction were supplied. A total of 3668 CBCT and 3668 CT slices were considered for training composed by 1606x2 slices for HN, 1046x2 for lung and 1016x2 for breast cancer patients.
To investigate the impact on dose calculation of a different CT, the patients included in the test set were selected among the patients with an rCT and with CBCT and rCT acquired with minimal time differences aiming at minimal antomical differences. Patients’ demographics were controlled to ensure data balancing in terms of the number of patients in the three sets. Also, we inspected the distribution of male/female, age, tumour staging and linac on which CBCT were acquired (Supplementary Material 1).
2.4. Image post-processing
First, the trained model was applied within of the pre-processed CBCTs (as described in 2.2). Then, the HU intensity range of [-1024;3071] was restored with a linear rescaling obtaining the so-called . were bi-linearly resampled from a matrix size of 256x256 to the CT matrix size within .
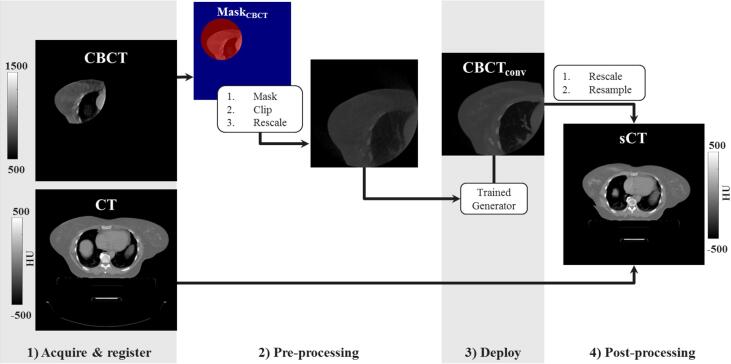
The CT FOV was about two-four times larger than the size of . However, to enable replanning, the volume covered by the treatment beams should be considered. To generate images with full CT FOV, the was substituted in the planning CT within the , obtaining the so-called synthetic–CT (sCT) (Fig. 1).
Fig. 1.
Schematic of the image workflow for applying the trained generator on a new 2D transverse slice of the CBCT of a breast cancer patient to create a sCT. After image acquisition, registration (1) and pre-processing (2) the trained network is deployed producing converted CBCT (, 3) which substituted the original CT within obtaining the so-called synthetic CT (sCT).
2.5. Image evaluation
Image similarity between sCT and rCT was compared to evaluate whether the single network trained with all the anatomical sites was comparable to the three networks trained per anatomical site. The appropriateness of the CBCT conversion on the test set was assessed performing an image and a dose comparison.
Similarities between the image intensity of the rCT and either the sCT, CBCT, or planning CT were calculated within in terms of mean absolute error (MAE) and mean error (ME) over the 30 patients in the test set. The rCT was considered as ground truth, and the metrics were calculated in terms of mean and range. Wilcoxon signed-rank tests were conducted between sCT/rCT and planning CT/rCT for MAE. Additional metrics, e.g. peak signal-to-noise ratio and structural similarity, are reported in Supplementary Material 4. The comparison planning CT/rCT was conducted to understand the impact of set-up position and anatomical differences.
2.6. Dose evaluation
For the patients in the test sets, clinical plans were recalculated on planning CT, sCT and rCT images in Monaco (v 5.11.02, Elekta AB, Sweden) using a Monte Carlo algorithm on a grid of 3 × 3 × 3 mm3 with 5% statistical uncertainty for volumetric-modulated arc therapy and 3% for intensity-modulated radiotherapy plans. Clinical contours, delineated by a radiation oncologist on the planning CT, were rigidly transferred to the sCT and rCT except for the body contour, which was automatically re-delineated. These contours were considered as volumes of interest (VOIs).
Dose distributions were analysed through voxel-wise relative dose differences ( and ) in the high dose region (dose of the prescribed dose, inside ). Also, 3D -analysis [23] with 3%,3 mm and 2%,2 mm criteria relative to dose on rCT for regions with a dose >10% of the prescription dose were performed. For all dose comparisons, a 15 mm cropping for planning CT, sCT and rCT in the proximity of body contour was performed to take account of dose build-up [24] in the proximity of the skin [25].
To investigate the impact of dose difference within VOIs, analysis of dose-volume histogram (DVH) points was performed on sCT and rCT. Maximum dose and mean dose were inferred from the DVH. OARs were considered for such analysis when they were present in at least four of the patients for each anatomical site: submandibular and parotid glands, spinal cord, larynx and brain stem for HN patients; lungs, heart, oesophagus, spinal cord and trachea for lung patients; lungs, heart, oesophagus, humerus and spinal cord for breast patients.
3. Results
Cycle-GANs required about eight days and five hours on a Tesla P100 GPU (NVIDIA Corporation) to train 200 epochs. As a result of the early stopping investigation, we opted for utilising 160000 iterations (100 epochs) for HN, 180000 (170 epochs) for lung, 180000 (160 epochs) for breast, and 360000 iterations (100 epochs) for the network trained on all the three sites combined. Generating sCT required 10 s for an entire CBCT volume (70 slices) on GPU and about 40 s on a Intel Xeon(R) CPU (E5-2690 v4 @ 2.60 GHz, with 56 threads).
3.1. Image comparison
The time between rCT and CBCT in the test set was on average ( [min; max]) [0;8] days and [8;67] days between CT and CBCT. Image similarity metrics over the test patients are reported in Table 1.
Table 1.
Overview of the image comparison. Image comparison calculated as mean () and range ([min;max]) of the test dataset (30 patients; 10 patients for each treatment site) compared to the reference dataset in terms of mean absolute error (MAE) and mean error (ME) between the test (Test) image minus the reference (Ref) image.
| Site | Head-and-Neck | Lung | Breast | ||||
|---|---|---|---|---|---|---|---|
| Test | Ref | MAE | ME | MAE | ME | MAE | ME |
| [HU] | [HU] | [HU] | [HU] | [HU] | [HU] | ||
| CBCT | rCT | 195 ± 20 [160;230] | −122 ± 33 [−183;−71] | 219 ± 44 [133;280] | 153 ± 48 [94;230] | 152 ± 40 [98;213] | 71 ± 37[7;115] |
| sCT single networka | rCT | 53 ± 12 [37;77] | −3 ± 7 [−15;10] | 83 ± 10 [72;104] | −2 ± 11 [−25;10] | 66 ± 18 [41;95] | −6 ± 13[−24;13] |
| sCT separate networksb | rCT | 51 ± 12 [35;74] | −6 ± 6 [−16;4] | 86 ± 9 [73;105] | −5 ± 14 [−28;10] | 67 ± 18 [41;98] | −5 ± 11[−18;14] |
| CT | rCT | 63 ± 17 [−40;90] | −18 ± 15 [−46;3] | 94 ± 23 [68;146] | 9 ± 22 [−33;36] | 63 ± 24 [40;115] | 8 ± 20[−14;54] |
a sCT obtained from a single network trained on all the anatomical sites.
b sCT obtained from three different networks trained on each anatomical site.
Generic network vs site-specific networks
Considering the HN case, the network trained on sole HN patients resulted in an MAE of 51 ± 12 HU with a range of [35;74]; while, when considering the network trained on all the patients, MAE was 53 ± 12 HU with a range of [35;77]. Similar results were obtained for lung and breast cancer patients. In this sense, MAE between networks trained per separate anatomical site and the single network trained on all sites together were compatible in terms of range and with average values within one , with p > 0.35.
Accuracy of CT numbers
Similarity was higher between sCT and rCT compared to between CBCT and rCT, e.g. MAE decreased from 195 ± 20 (CBCT/rCT) to 53 ± 12 HU (sCT/rCT) for HN. All the similarity metrics calculated between sCT/rCT and CT/rCT can be considered equivalent, with no large differences when considering the range, for all three anatomical sites. The mean MAE and range for sCT/rCT were smaller, about 5–10 HU, compared to CT/rCT due to the reduced time between sCT/rCT, which resulted in less anatomical differences.
Fig. 2, Fig. 3, Fig. 4 show examples of CBCT and sCT obtained from the single network for a HN, lung and breast cancer patient. The network reduced scatter artefacts while retaining anatomical accuracy. For the lung patient shown in Fig. 3, atelectasis occurred between CT and rCT, which was conserved in the sCT.
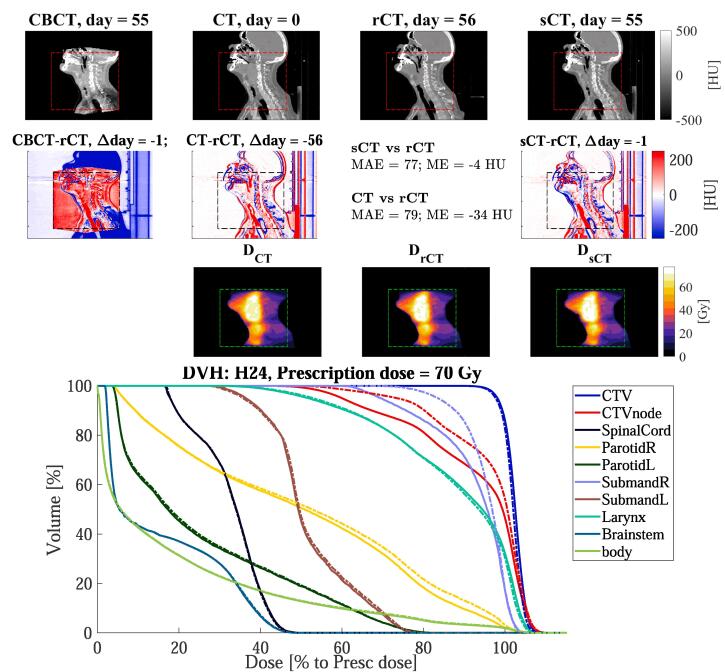
Fig. 2.
Sagittal views for the head-and-neck cancer patient H24 of: (1st row) CBCT (1st column), CT (2nd column), rescan CT (rCT, 3rd column) and synthetic CT (sCT, 4th column), along with (2nd row) the respective difference to rCT, and the doses (3rd row). The red, black, or green dotted rectangles indicate the position of . The days refer to the acquisition date relative of the planning CT. In the 4th row, the DVH is shown for target and OARs of sCT (solid lines) and rCT (dashed lines). Note that for the clinical target volume (CTV) of the node (CTVnode) and the right (R) submandibular, the DVH differed between rCT and sCT. This is due to anatomical differences between sCT and rCT.
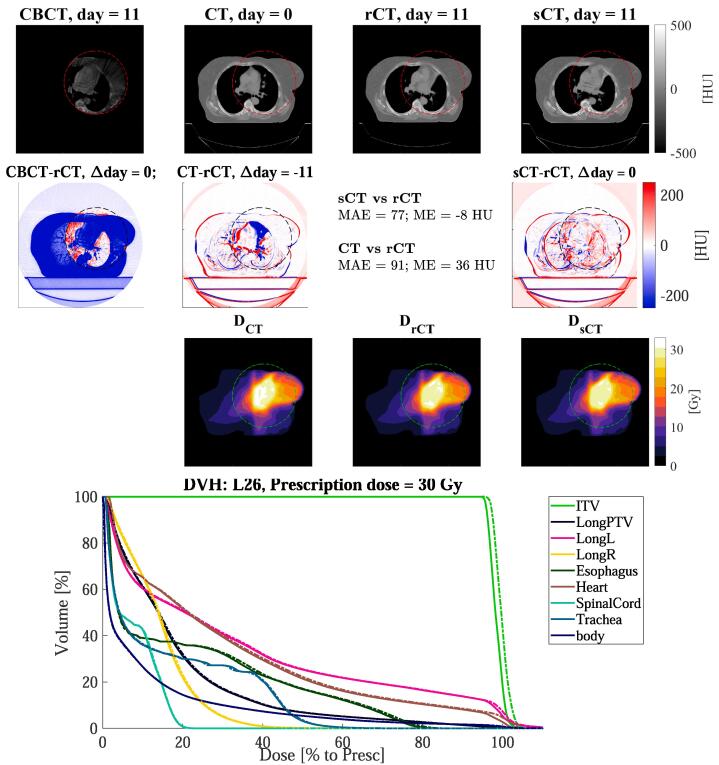
Fig. 3.
Axial views for the lung cancer patient L26 of: (1st row) CBCT (1st column), CT (2nd column), rescan CT (rCT, 3rd column) and synthetic CT (sCT, 4th column), along with (2nd row) the respective difference to rCT, and the doses (3rd row). The red, black, or green dotted rectangles indicate the position of . The days refer to the acquisition date relative of the planning CT. In the 4th row, the DVH is shown for target and OARs of sCT (solid lines) and rCT (dashed lines).
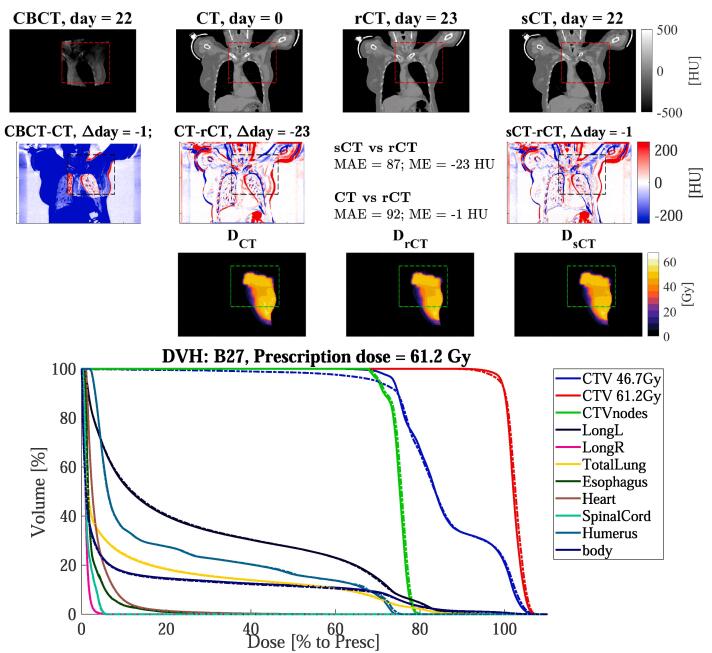
Fig. 4.
Coronal views for the breast cancer patient B27 of: (1st row) CBCT (1st column), CT (2nd column), rescan CT (rCT, 3rd column) and synthetic CT (sCT, 4th column), along with (2nd row) the respective difference to rCT, and the doses (3rd row). The red, black, or green dotted rectangles indicate the position of . The days refer to the acquisition date relative of the planning CT. In the 4th rows, the DVH is shown for target and OARs of sCT (solid lines) and rCT (dashed lines).
3.2. Dose comparison
Fig. 2, Fig. 3, Fig. 4 reports dose distributions calculated on CT, rCT and sCT along with their DVHs. Dose comparison was performed on the sCT generated with the network trained on all sites. We observed small differences between doses on rCT and sCT. On average over the ten patients for each site, dose differences between sCT/rCT () were lower than for CT/rCT (), e.g. in the high dose region (D) the maximum absolute mean differences were below 0.2% for and below 0.9% for (Table 2).
Table 2.
Statistics of the dose comparison of the thirty patients in the test set for the sCT trained on all the anatomical site together. The values are reported as percentage mean and range [min; max].
| sCT vs rCT | CT vs rCT | |||||
|---|---|---|---|---|---|---|
| Sites | b | c | b | c | ||
| [%] | [%] | [%] | [%] | [%] | [%] | |
| Head-and-neck | ||||||
| Lung | ||||||
| Breast | ||||||
a on dose >90% of the prescribed dose.
b Pass rates of on dose >10% of the prescribed dose.
c Pass rates of on dose >10% of the prescribed dose.
d on dose >90% of the prescribed dose.
The mean gamma pass rates with the 2%, 2 mm criteria were and higher for sCT/rCT compared to CT/rCT for all VOIs, which is in line with the dose differences observed. The mean and maximum doses on the sCT differed on average 0.5% compared to the rCT. Images of the patients with dose differences in VOIs were inspected on a single-case basis, as reported in the Supplementary Material 5. DVH points differences were except for left lung and spinal cord of two lung patients (L25, −2.1% and L27, 3.8%), the heart of a breast patient (B31, −5.6%), and the oesophagus of two breast patients (B30, 3.1% and B31, 2.3%). For one lung case (L25, Fig. S4), dose differences to lung and spinal cord of about 2–3% were observed due to anatomical differences of the lung. Also, residual artefacts characterised by inhomogeneous CT numbers seem to be present along the craniocaudal direction in the lungs for sCT; it appears that for this case the CBCT artefacts were not fully corrected by the network within the lungs. Also for the other lung case (L27, Fig. S5), anatomical differences were observed in the lung. In addition, the CBCTs were characterised by severe scatter artefacts due to obesity. On sCT, the spinal cord was not entirely recovered, possibly resulting in local differences. Besides, the spinal cord is located in a low-dose region, which may be highlighted when considering metrics as voxel-wise relative differences.
4. Discussion
The cycle-GAN increased the accuracy of CT numbers compared to CBCT, enabling sCT-based calculations for HN, lung and breast cancer patients. Our main finding is that a single network trained on all the three sites performed similarly to three networks trained on each anatomical site, as justified by the results from the image comparison.
When investigating the accuracy of CT numbers on sCT calculating image similarity to rescan CT, we found that HU values were comparable to values observed between CT and rCT. We observed a slight improvement in performance for HN compared to lung and breast cancer patients. The network was trained with higher amount of slices for HN (1606) compared to lung (1046) and breast (1016). We hypothesise that this data imbalancing may have resulted in relatively increased performances for HN cancer patients. Also, the use of immobilisation masks for HN case may increase the reproducibility of patient set-up or reduce motion artefacts in the images (both for CT and CBCT) [26]. Though variations in the CBCT imaging protocol were reported, e.g. kV, mAs and linac where the images were acquired, we did not observe any effect on the quality of sCT. It may be of interest to investigate the robustness of the method against variations of acquisition settings, as performed by Maier et al. [17].
In terms of dose calculation accuracy, we compared sCT to rCT, achieving excellent results for all the anatomical sites. We observed that the largest dose differences were in low-dose regions, which are more sensitive to statistical differences due to the low amount of events in the Monte Carlo dose calculations. Previous work with deep learning was performed only on single anatomies, e.g. prostate [13], [18], [27], [19], HN [20], [28] and lung [16] patients. For HN patients, similar findings were reported by Liang et al. [20], where also a cycle-GAN was utilised achieving a mean () 2%, 2 mm pass-rates of 98.4 ± 1.7% compared to 97.8% of this work. Also, Li et al. used a 2D U-net with residual convolutional units obtaining mean DVH point difference [28]. In our study, similar mean differences () were achieved, which demonstrates the high sCT quality resulting from our approach. For lung patients, Xie et al. applied patch-based residual learning on lung patients obtaining a conspicuous correction of cupping and streaking artefacts [16]. However, they did not perform any dose calculations making it difficult to compare the dose calculation accuracy of the studies.
Repositioning inevitably occurred between planning CT, CBCT and rCT. To further minimise anatomical and set-up differences, we could have used DIR to increase the similarity of CBCT/sCT and CT/rCT. However, we opted against it for the following reasons: (i) since we were trying to reproduce the dose derived by CT-based calculations, we did not want to modify CT or rCT further; (ii) residual deformation errors should be thoroughly evaluated [29], and this was deemed out of the scope of this investigation; (iii) recurring to using solely translation mimics the set-up procedure that is currently performed clinically at the linacs, and we aimed at observing the impact of dose evaluation in a comparable setting.
The main limitation of this study is deemed to be the cohort size: ten patients per anatomical sites in the test set may be considered as a low number. Before clinical implementation, a study including a larger number of patients should be initiated, paying particular attention to the data variability and data balancing among anatomical sites. Besides, we did not adapt the contours of targets and OARs, which is necessary to investigate the clinical impact of replanning thoroughly. Notwithstanding the relatively limited sample, this work offers valuable insights into the generalisation capability of a single cycle-GAN, and, in general, it shows that a single neural network can convert CBCTs of multiple sites. This study was a single-center, and a next study should investigate the feasibility of applying the same, or a re-trained model, in a multi-center setting to ensure the robustness of the model.
Specifically, we showed that a single cycle-GAN can be utilised for multiple anatomical sites as HN, lung and breast. This finding has important implications for simplifying the training of a convolutional network since a single network may be adopted for different anatomical sites. To fully understand whether a single network may facilitate CBCT-based dose calculations for the whole body, we are currently performing a novel study including additional anatomical areas, e.g. pelvis, lower abdomen and brain.
In this work, we proved that a single cycle-GAN can convert CBCTs into CTs, resulting in sCTs that have sufficient quality to enable dose recalculation. Also, the conversion occurred in a matter of seconds, which is line with the sCT generation time reported by other deep learning approaches for lung [16], prostate [13], [18], [27], [19] and HN [28], [20]. We foresee the speed of conversion as an important step toward online ART. Besides, in conventional non-adaptive radiotherapy, this methodology could be used to evaluate the dosimetric impact of anatomical differences occurring during treatment, supporting the decision to perform a rescan CT or not.
A single cycle-GAN was successfully trained to synthesise CT from CBCT using unpaired training data. The resulted sCT resembled a diagnostic quality planning CT and featured the anatomy of the CBCT. In terms of dose calculation accuracy, good results were obtained for all the anatomical sites. In general, the proposed approach enables considerably fast image conversion, and it may facilitate online adaptive radiotherapy treatments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Quadro P5000 GPU used for prototyping this research. The study was conducted in accordance to the guidelines of the local Medical Ethical Committee.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.phro.2020.04.002.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Boda-Heggemann J., Lohr F., Wenz F., Flentje M., Guckenberger M. kV cone-beam CT-based IGRT. Strahlen Onkol. 2011;187:284–291. doi: 10.1007/s00066-011-2236-4. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q.J., Li T., Wu Q., Yin F.F. Adaptive Radiation Therapy. Cancer J. 2011;17:182–189. doi: 10.1097/PPO.0b013e31821da9d8. [DOI] [PubMed] [Google Scholar]
- 3.Schulze R., Heil U., Groß D., Bruellmann D., Dranischnikow E., Schwanecke U. Artefacts in CBCT: a review. Dentomax Radiol. 2011;40:265–273. doi: 10.1259/dmfr/30642039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramella S., Fiore M., Silipigni S., Zappa M.C., Jaus M., Alberti A.M. Local control and toxicity of adaptive radiotherapy using weekly CT imaging: results from the LARTIA trial in stage III NSCLC. J Thor Oncol. 2017;12:1122–1130. doi: 10.1016/J.JTHO.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Dunlop A., McQuaid D., Nill S., Murray J., Poludniowski G., Hansen V.N. Comparison of CT number calibration techniques for CBCT-based dose calculation. Strahl Onkol. 2015;191:970–978. doi: 10.1007/s00066-015-0890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhen X., Gu X., Yan H., Zhou L., Jia X., Jiang S.B. CT to cone-beam CT deformable registration with simultaneous intensity correction. Phys Med Biol. 2012;57:6807–6826. doi: 10.1088/0031-9155/57/21/6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bootsma G.J., Verhaegen F., Jaffray D.A. Efficient scatter distribution estimation and correction in CBCT using concurrent Monte Carlo fitting. Med Phys. 2014;42:54–68. doi: 10.1118/1.4903260. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W., Vernekohl D., Zhu J., Wang L., Xing L. A model-based scatter artifacts correction for cone beam CT. Med Phys. 2016;43:1736–1753. doi: 10.1118/1.4943796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peroni M., Ciardo D., Spadea M.F., Riboldi M., Comi S., Alterio D. Automatic segmentation and online virtualCT in head-and-neck adaptive radiation therapy. Int J Radiat Oncol Biol Phys. 2012;84:e427–e433. doi: 10.1016/J.IJROBP.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Veiga C., Janssens G., Teng C.L., Baudier T., Hotoiu L., McClelland J.R. First clinical investigation of cone beam computed tomography and deformable registration for adaptive proton therapy for lung cancer. Int J Radiat Oncol Biol Phys. 2016;95:549–559. doi: 10.1016/J.IJROBP.2016.01.055. [DOI] [PubMed] [Google Scholar]
- 11.Kurz C., Kamp F., Park Y.K., Zöllner C., Rit S., Hansen D. Investigating deformable image registration and scatter correction for CBCT-based dose calculation in adaptive IMPT. Med Phys. 2016;43:5635–5646. doi: 10.1118/1.4962933. [DOI] [PubMed] [Google Scholar]
- 12.Jia X., Yan H., Gu X., Jiang S.B. Fast monte carlo simulation for patient-specific ct/cbct imaging dose calculation. Phys Med Biol. 2012;57:577. doi: 10.1088/0031-9155/57/3/577. [DOI] [PubMed] [Google Scholar]
- 13.Kida S., Nakamoto T., Nakano M., Nawa K., Haga A., Kotoku J. Cone beam computed tomography image quality improvement using a deep convolutional neural network. Cureus. 2018;10 doi: 10.7759/cureus.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–1419. doi: 10.1002/mp.12155. [DOI] [PubMed] [Google Scholar]
- 15.Maspero M., Savenije M.H.F., Dinkla A.M., Seevinck P.R., Intven M.P.W., Jurgenliemk-Schulz I.M. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol. 2018;63:185001. doi: 10.1088/1361-6560/aada6d. [DOI] [PubMed] [Google Scholar]
- 16.Xie S., Yang C., Zhang Z., Li H. Scatter artifacts removal using learning-based method for CBCT in IGRT system. IEEE Access. 2018;6:78031–78037. doi: 10.1109/ACCESS.2018.2884704. [DOI] [Google Scholar]
- 17.Maier J., Eulig E., Vöth T., Knaup M., Kuntz J., Sawall S. Real-time scatter estimation for medical CT using the deep scatter estimation: method and robustness analysis with respect to different anatomies, dose levels, tube voltages, and data truncation. Med Phys. 2019;46:238–249. doi: 10.1002/mp.13274. [DOI] [PubMed] [Google Scholar]
- 18.Hansen D.C., Landry G., Kamp F., Li M., Belka C., Parodi K. ScatterNet: a convolutional neural network for cone-beam CT intensity correction. Med Phys. 2018;45:4916–4926. doi: 10.1002/mp.13175. [DOI] [PubMed] [Google Scholar]
- 19.Kurz C., Maspero M., Savenije M.H., Landry G., Kamp F., Pinto M. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab4d8c. [DOI] [PubMed] [Google Scholar]
- 20.Liang X., Chen L., Nguyen D., Zhou Z., Gu X., Yang M. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64:125002. doi: 10.1088/1361-6560/ab22f9. [DOI] [PubMed] [Google Scholar]
- 21.Harms J., Lei Y., Wang T., Zhang R., Zhou J., Tang X. Paired cycle-GAN based image correction for quantitative cone-beam CT. Med Phys. 2019 doi: 10.1002/mp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J.Y., Park T., Isola P., Efros A.A. 2017 IEEE International Conference on Computer Vision (ICCV) IEEE; Venice: 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks; pp. 2242–2251. [DOI] [Google Scholar]
- 23.Low D.A. Gamma dose distribution evaluation tool. J Phys. 2010;250:012071. doi: 10.1088/1742-6596/250/1/012071. [DOI] [Google Scholar]
- 24.Velkley D., Manson D., Purdy J., Oliver G., Jr Build-up region of megavoltage photon radiation sources. Med Phys. 1975;2:14–19. doi: 10.1118/1.594158. [DOI] [PubMed] [Google Scholar]
- 25.Maspero M., Seevinck P.R., Schubert G., Hoesl M.A.U., van Asselen B., Viergever M.A. Quantification of confounding factors in MRI-based dose calculations as applied to prostate IMRT. Phys Med Biol. 2017;62:948–965. doi: 10.1088/1361-6560/AA4FE7. [DOI] [PubMed] [Google Scholar]
- 26.van Lin E., van der Vight L., Huizenga H., Kaanders J., Visser A. Set-up improvement in head and neck radiotherapy using a 3D off-line EPID-based correction protocol and a customised head and neck support. Radiother Oncol. 2003;68:137–148. doi: 10.1016/S0167-8140(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 27.Landry G., Hansen D., Kamp F., Li M., Hoyle B., Weller J. Comparing Unet training with three different datasets to correct CBCT images for prostate radiotherapy dose calculations. Phys Med Biol. 2019;64:035011. doi: 10.1088/1361-6560/aaf496. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Zhu J., Liu Z., Teng J., Xie Q., Zhang L. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab2770. [DOI] [PubMed] [Google Scholar]
- 29.Paganelli C., Meschini G., Molinelli S., Riboldi M., Baroni G. Patient-specific validation of deformable image registration in radiation therapy: overview and caveats. Med Phys. 2018;45:e908–e922. doi: 10.1002/mp.13162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.