Highlights
-
•
Automatic delineation software shows promising results in terms of time-saving.
-
•
Standard geometry measures do not have a high correlation with delineation time.
-
•
New evaluation measures were introduced: added path length (APL) and surface DSC.
-
•
(Added) path length showed the highest correlation with time-recordings.
-
•
This makes APL the most representative measure for clinical usefulness.
Keywords: Radiotherapy, Automatic delineation, Contouring time, Time-saving, Hausdorff distance, Dice similarity coefficient, Surface DSC, Added path length
Abstract
Background and purpose
In radiotherapy, automatic organ-at-risk segmentation algorithms allow faster delineation times, but clinically relevant contour evaluation remains challenging. Commonly used measures to assess automatic contours, such as volumetric Dice Similarity Coefficient (DSC) or Hausdorff distance, have shown to be good measures for geometric similarity, but do not always correlate with clinical applicability of the contours, or time needed to adjust them. This study aimed to evaluate the correlation of new and commonly used evaluation measures with time-saving during contouring.
Materials and methods
Twenty lung cancer patients were used to compare user-adjustments after atlas-based and deep-learning contouring with manual contouring. The absolute time needed (s) of adjusting the auto-contour compared to manual contouring was recorded, from this relative time-saving (%) was calculated. New evaluation measures (surface DSC and added path length, APL) and conventional evaluation measures (volumetric DSC and Hausdorff distance) were correlated with time-recordings and time-savings, quantified with the Pearson correlation coefficient, R.
Results
The highest correlation (R = 0.87) was found between APL and absolute adaption time. Lower correlations were found for APL with relative time-saving (R = −0.38), for surface DSC with absolute adaption time (R = −0.69) and relative time-saving (R = 0.57). Volumetric DSC and Hausdorff distance also showed lower correlation coefficients for absolute adaptation time (R = −0.32 and 0.64, respectively) and relative time-saving (R = 0.44 and −0.64, respectively).
Conclusion
Surface DSC and APL are better indicators for contour adaptation time and time-saving when using auto-segmentation and provide more clinically relevant and better quantitative measures for automatically-generated contour quality, compared to commonly-used geometry-based measures.
1. Introduction
Contouring of organs-at-risk (OARs) and target volumes is an important step in radiation treatment planning [1], [2]. However, the delineation quality and time spent on contouring strongly depend on the experience of the radiation oncologist or radiotherapy technician (RTT) [3], [4]. In the last decades, auto-segmentation algorithms have been developed, including atlas-based methods and deep-learning algorithms based on convolutional neural networks [5], [6]. These methods have the potential to reduce inter- and intra-observer variability and speed-up the contouring process. However, the majority of the automatically generated contours still require manual corrections to make them clinically acceptable, although many studies show time-savings compared to full manual contouring [7], [8], [9], [10], [11].
Multiple studies have been performed to evaluate auto-segmentation of OARs for different treatment sites [10], [11], [12], [13], [14], [15], [16]. Despite the promising results in terms of efficiency and consistency, it remains a challenge to objectively assess the automatically generated contours in terms of changes that still remain to be made. Often, the automatically generated contours are compared to manual contours. A large range of metrics can be used for this, as shown by Taha and Hanbury [17]. The most common measures are the volumetric Dice Similarity Coefficient (DSC) and the Hausdorff distance, which are good measures for the geometric quantification of contour similarities [9], [10], [15], [18], [19], [20], [21]. The volumetric DSC evaluates the intersection of two contour volumes relative to the union, and the Hausdorff distance represents the maximum nearest neighbor Euclidean distance between contours.
However, these measures have a low correlation to clinical contour quality or time needed to adjust the contours [9]. As auto-segmentation techniques are nowadays frequently introduced in clinical practice to reduce contouring time, it is desirable to estimate this time-saving. However, direct evaluation of the reduction in contouring time is, of itself, time-consuming. Thus, there is a need for quantitative evaluation measures that correlate with clinical practice (e.g. predict time-saving using automatically generated contours).
In this study, we evaluated existing standard quantitative geometric measures and propose a new evaluation measure: the path length of a contour that has to be added. In addition to the standard DSC and Hausdorff distance, the correlation with time-saving was also investigated for the surface DSC, a measure introduced by Nikolov et al. [22]. Compared to the volumetric DSC, the surface DSC compares two contours (i.e. surfaces) instead of volumes. Therefore, the surface DSC and added path length (APL) measures are hypothesized to be more clinically meaningful performance measures of segmentation, especially in terms of time-saving, compared to the Hausdorff distance and volumetric DSC. We evaluated these measures using atlas-based and deep-learning auto-segmentation methods for OARs in the thorax.
2. Materials and methods
2.1. Patient cohort and contouring
Patient CT imaging data, delineated contours, and time recordings were taken from the study by Lustberg et al. [7]. In short, the mid-ventilation phase of a 4D CT scan (resolution = 0.977 × 0.977 mm, slice spacing = 3 mm) of twenty consecutively treated stage I-III non-small cell lung cancer (NSCLC) patients was used to contour various OARs: the left lung, right lung, heart, spinal cord, esophagus, and mediastinum. The OARs were fully manually contoured using the clinically available treatment planning system ([TPS], Eclipse, version 11.0, Varian, Palo Alto, United States of America), except for the lungs which were segmented using auto-threshold options available in the TPS, followed by manual corrections. A commercial atlas-based and a prototype deep-learning contouring method (EmbraceCT and DLCexpert, respectively, from Mirada Medical Ltd., Oxford, United Kingdom) were used for auto-segmentation, followed by manual adjustments for fine-tuning to make them clinically acceptable. By using both atlas and deep-learning based contouring methods, variation in contouring quality is ensured [7]. For each method, the time required for manual contouring and contour adjustment was recorded for each patient and OAR individually. All contouring tasks were performed by one experienced RTT, aware of the time being recorded, to reduce inter-observer variability. This retrospective study was approved by the Institutional Review Board.
2.2. Clinical contour accuracy and quality measures
All contours were imported in Matlab R2018b (The MathWorks Inc., Natick, MA, USA) for further analysis. For each OAR, the similarity between automatically generated and user-adjusted contours was quantitatively assessed in terms of contour quality and time-saving. The volumetric DSC, the mean slice-wise Hausdorff distance (MSHD), the surface DSC and the new APL measure were used (Fig. 1). Although the surface DSC was previously introduced by Nikolov et al. [22], it has not been correlated to time-saving yet. The volumetric DSC is defined as the intersection of two contour volumes relative to the union in 3D. The MSHD is defined as the maximum nearest neighbor Euclidean distance between the surfaces of the two contours in one slice, calculated from the auto-contour towards the user-adjusted contour, and averaged over all slices. The surface DSC is defined as the intersection surface of the two contours normalized by the union of the two contours, in 3D. The path length of a contour that had to be added to meet the institutional guidelines for contouring was calculated by considering all manual adjustments in terms of pixels added, both expansion and shrinkage of the automatically generated contour.
Fig. 1.
Illustration of evaluation measures used in this study. The solid line represents the automatically generated contour, the dashed line the user-adjusted contour. A Volumetric DSC, defined as the union of two volumes (green volume region) normalized by the mean of the two volumes. B Hausdorff distance, defined as the maximum nearest neighbor Euclidean distance (green arrow). C Surface DSC, defined as the union of two contours (yellow contour region) normalized by the mean surface of the two contours. D Added path length (APL) (yellow contour region). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The absolute time needed (s) for adjusting the auto-contours and the relative time-saving (%) compared to manual contouring were measured per OAR. Pearson correlation coefficients (R) between these time measures and the evaluation measures were calculated using linear regression.
To evaluate whether the measures correlate to actual editing time, the similarity between each automatically generated and user-adjusted OAR contour was calculated, as this represents the actual amount of editing performed. Consequently, the tolerance parameter introduced by Nikolov et al. [22], representing inter-observer variation in segmentations, was set to 0 mm for both surface DSC and APL, because all edits should be analyzed.
In addition, this analysis was repeated for the automatically generated contours compared to the manual contours, as a manual “ground truth” would be used to evaluate auto-contouring in the absence of user-editing. Because both the automatic and manual contour might be considered clinically acceptable when considering a small difference, which would result in no adjustment of the automatic contour, a tolerance of 1 voxel (1 mm) was considered for this analysis. This tolerance was taken into account to avoid penalizing contours within clinically acceptable tolerance.
3. Results
In total 235 automatically generated contours were evaluated. The median time required to contour all OARs manually was 20 min per patient [range 16–25 min]. The total median relative time-saving when using auto-contouring compared to manual contouring was 40% [range −321% (i.e., more time was needed) to 93% (i.e., time was saved)] for the atlas-based method and 71% [range −50 to 94%] for DLC, demonstrating the contour quality difference between the two methods.
An example of automatically generated and user-adjusted contours of the heart for two patients is shown in Fig. 2. The volumetric DSC of the automatically generated and user-adjusted contour was similar, but the MSHD, surface DSC, APL, and contouring time saved were deviating.
Fig. 2.
Example of transverse CT images of two patients showing the automatically generated (solid line) and user-adjusted (dashed line) contours of the heart. For both patients, the volumetric DSC of the automatically generated and user-adjusted contour is similar (94%), but the MSHD, surface DSC, and APL differ between these patients (0.92 cm vs. 1.34 cm, 44% vs. 68%, 678 cm vs. 409 cm, respectively). The contouring time needed to make both contours clinically acceptable for these patients also differed (3.6 min vs. 3.1 min and 6% vs. 20%, respectively). Manual contouring time for both contours was approximately 4 min.
When comparing the manual contouring time with the path length of the manual contour, a correlation coefficient of 0.90 was found (excluding the lungs, see Fig. S1, Supplementary Material). Correlation coefficients for all measures with respect to the absolute time needed for adjustment are shown in Fig. 3. A correlation coefficient of −0.32 was found for the volumetric DSC, 0.64 for the Hausdorff distance, −0.69 for the surface DSC, and 0.87 for the APL. Table S1 and S2 in the Supplementary Material show for each measure the correlation coefficients and linear fit parameters for all OARs separately. For the APL, the newly introduced measure, the correlation coefficients ranged between 0.44 and 0.88, with the right lung showing the lowest correlation. Almost all organs showed similar slopes for the APL, except for one outlier for the spinal cord, and the esophagus. The mediastinum had the highest APL (826 cm [range 290–2441 cm], compared to a median range of 24–413 cm for the other OARs).
Fig. 3.
All measures against absolute time needed to adjust the automatically generated contour, comparing the automatic to the adjusted contour. Atlas-based (circles) and deep-learning contouring (triangles) were combined. MSHD = Mean Slice-wise Hausdorff Distance.
Fig. 4 shows correlation coefficients for all measures with respect to the relative contouring time saved when using auto-segmentation. A correlation coefficient of 0.44 was found for the volumetric DSC, −0.64 for the Hausdorff distance, 0.57 for the surface DSC, and −0.38 for the APL. Supplementary Material Tables S3 and S4 show for each measure the correlation coefficients and linear fit parameters for all OARs separately. For the surface DSC, the correlation coefficients ranged between 0.50 and 0.92, with the right lung showing the lowest correlation. The esophagus had the lowest surface DSC with the most spread (63% [range 7–94%], compared to a median range of 66–97% for the other OARs). For the atlas-based method, the lungs show a different distribution of points compared to the other OARs. The slope of the linear fit is less steep, i.e. for a certain surface DSC, less contouring time is saved compared to the other OARs. If the atlas-based contoured lungs are removed from the total comparison, a correlation coefficient between the surface DSC and relative time saved of 0.76 was found.
Fig. 4.
All measures against relative contouring time saved after adjusting the automatically generated contour, comparing the automatic to the adjusted contour. Atlas-based (circles) and deep-learning contouring (triangles) were combined. MSHD = Mean Slice-wise Hausdorff Distance. One outlier (x = −321%) is not shown in these graphs.
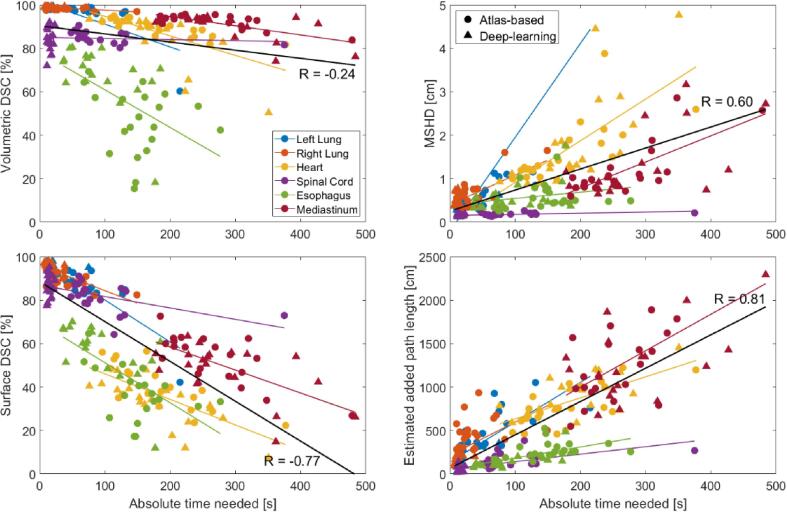
The results of the comparison of the automatic and manual contours, which represent expected edits, are shown in Fig. 5. For the absolute time needed for adjustment, a correlation coefficient of −0.24 was found for the volumetric DSC, 0.60 for the Hausdorff distance, −0.77 for the surface DSC, and 0.81 for the estimated APL. For the relative contouring time saved, a correlation coefficient of 0.37 was found for the volumetric DSC, −0.63 for the Hausdorff distance, 0.55 for the surface DSC, and −0.35 for the estimated APL (Fig. S2, Supplementary Material).
Fig. 5.
All measures against absolute time needed to adjust the automatically generated contour, comparing the automatic to the manual contour. Atlas-based (circles) and deep-learning contouring (triangles) were combined. MSHD = Mean Slice-wise Hausdorff Distance.
4. Discussion
In this study, the surface DSC and APL measures were found to be better indicators of time-saving, and thus clinical applicability and quality of automatically-generated contours, compared to the standard geometry measures volumetric DSC and MSHD. Even though the MSHD shows the highest correlation for relative time-saving when including all OARs, MSHD gives no information about the amount of contour that is adjusted, only how far the adjusted contour is from the original one.
A number of studies evaluated auto-segmentation of OARs [10], [11], [12], [13], [14], [15], [16]. The most commonly-used measures, the volumetric DSC and the Hausdorff distance, were found to be good measures for the geometric quantification of contour similarities [9], [10], [15], [18], [19], [20]. Next to these measures, the average surface distance (ASD) is also frequently used, leading to similar correlation coefficients as the Hausdorff distance as used in this study (see Fig. S3–S4, Supplementary Material). Van der Veen et al. found a 33% shorter delineation time using automated delineation compared to manual contouring [10]. Gooding et al. show time-savings for atlas-based auto-contouring of the same OARs as in this study ranging from 12% (esophagus) to 77% (right lung), and obtained scores for the volumetric DSC ranging from 0.46 (esophagus) to 0.98 (lungs) [9]. Yang et al. also show volumetric DSC ranging from 0.55 (esophagus) to 0.98 (lungs) [23]. These values are comparable to the values in our study, indicating that the auto-contouring algorithms used in our study perform equally well to those in literature. However, it remains challenging to relate the quality of the automatically generated contour in terms of changes that still need to be made with a measure showing clinical applicability. In this study, two clinically useful measures were found to correlate well with time-recordings and time-savings, showing these can be used to estimate the quality of automatically generated contours in terms of applicability and usefulness.
When comparing automatically generated and user-adjusted contours, the surface DSC and APL calculations are based on adaptations of the auto-contours. However, in a typical investigation, the auto-contour would normally be compared with a “ground truth” manual contour. Comparable results were found (Fig. 3, Fig. 5), indicating that both surface DSC and APL can be used in this way to estimate editing time. The higher correlation observed with APL may suggest that this measure is a more effective tool in such an assessment. However, a fixed tolerance of 1 mm was used in this study and the clinical relevance of this tolerance will most likely differ per OAR, which may slightly change the results observed here.
The use of the evaluation measures surface DSC and APL in clinical commissioning allows for time-saving estimations without elaborate manual timing by experts. A correlation between path length adjustments and time-saving only needs to be determined in a small group (e.g. 5–10 patients) from which then an estimation on a larger population can be made. Further evaluation could include more treatment sites. However, we investigated multiple OARs in the thorax and found that especially for APL the correlation and time-saving did not depend on OAR size or location in the thorax (see Fig. 3 and Supplementary Material). Although individual OAR analysis showed that an OAR may have a slightly higher correlation for one of the traditional measures, the APL shows the highest correlation when combining all OARs, which ensures that the whole range of contours and adaptations is included. This indicates that the correlation for APL is robust and does not depend as much on the specific OAR as for the other measures.
Generally, time-saving depends on three main aspects; whether the boundary of an OAR is visible, the volume of the OAR, and the delineation tool(s) used for manual contouring and editing (e.g. pen or brush, the diameter of the brush, and the ability to interpolate structures, axial contouring or contouring in 3D). Both for automatic and manual segmentation, boundary visibility hampers easy delineation. This is indicated by, for example for the esophagus, a low surface DSC and longer contouring times, indicating that more was adjusted and consequently, more time was needed. The low contrast also impairs the visual identification and precise localization of the esophagus. However, the esophagus is volumetrically a relatively small organ, and the absolute time needed to adjust the contour is low compared to adjusting a bigger organ by the same percentage. Furthermore, the DSC measures are normalized by volume or surface, resulting in low values and outlier results for the esophagus. Similarly, if editing slice-by-slice, the number of slices that an OAR covers also has to be taken into account when considering the absolute editing time needed (e.g. the heart covers fewer slices than the esophagus). The APL measure takes these issues into account.
The delineation tool used might also affect the similarity measure that would be most appropriate, e.g. the APL assumes drawing lines. If you would use a brush tool without auto-filling, then the added area might be more meaningful. The diameter of the brush also influences the time needed to draw a contour part. When using the surface DSC as a delineation time measure, it has to be considered that this measure will give the same result when slices are added or deleted, although it can be assumed that adding contours takes much longer than deleting a whole slice using a “cut” tool. Deleting a slice can typically be done in one click with an appropriate tool in most contouring software. The APL takes this into account implicitly, as only drawn path pixels are included in the calculation. However, when using the paintbrush to fully delete a contour, the surface DSC may be more appropriate.
When analyzing relative time-saving, it is assumed that the manual contouring is being done fully manually. This is not always the case, which results in a lower correlation. For example, for the surface DSC, the lungs show a less steep linear fit compared to the other OARs, especially for contours of lower quality (i.e. atlas-based contours). This is because for the lungs the manual contouring is typically done using auto-threshold options available in the TPS, which is already a time-efficient procedure. Therefore, small manual adaptations of e.g. blood vessels or bronchi in the auto-segmented lung contours take relatively a lot of time compared to the auto-threshold contouring. In a couple of cases, a collapsed lung was present, which was not correctly contoured using atlas-based segmentation. The higher correlation of 0.76 that was found when removing the lungs from the total comparison indicates that it is better to investigate the semi-automatic contouring of organs such as the lungs separately.
A study by Gooding et al. showed that it is difficult for a clinical observer to determine whether a contour was automatically generated or manually delineated if the automated contour is at a certain quality level [9]. It was found that the inability to judge the source of a contour correctly indicates that the quality of the contour is sufficient, which results in a reduced need for editing and therefore a greater time-saving. Being able to evaluate the time-saving is an important aspect of the clinical applicability of auto-segmentation techniques. Ideally, a technician should be able to score the quality of the auto-segmented contour before starting the editing process in clinical practice by visual inspection to determine if they should edit the auto-contour or start from scratch.
Application of the new path length measure in clinical practice implicates that a technician should perform this quality assessment by considering the path length that needs adjustment instead of estimating the extent of volume overlap, which is usually the current way experts score the usefulness of contours. Better visual estimation of the time required for editing may improve adoption of auto-segmentation techniques in clinical practice, by reducing frustration with editing auto-contours that do not save time, and fully utilizing those that do.
In general, when an observer would expect to change an automatically generated contour by more than ~40%, it is expected that no time would be saved, which follows from the results of the surface DSC. This also means that if APL is estimated to be more off than a certain threshold, manual contouring would be recommended. Delineation should hence be judged this way and not by volumetric overlap (Fig. 2). Further investigation could include evaluation of the inter-observer variability for the new measures by including more observers.
To conclude, two recently introduced/new evaluation measures have been evaluated: the surface DSC and the APL. Compared to the standard measures Hausdorff distance and volumetric DSC, these measures are better indicators for the clinical delineation time saved and absolute time needed using software-generated contours. They may provide additional objectively quantifiable surrogates for assessing time-saving and clinical applicability and quality of automatically generated contours in the delineation process.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2019.12.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rasch C.R.N., Duppen J.C., Steenbakkers R.J., Baseman D., Eng T.Y., Fuller C.D. Human-computer interaction in radiotherapy target volume delineation: a prospective, multi-institutional comparison of user input devices. J Digit Imaging. 2011;24:794–803. doi: 10.1007/s10278-010-9341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harari P.M., Song S., Tomé W.A. Emphasizing conformal avoidance versus target definition for IMRT planning in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77:950–958. doi: 10.1016/j.ijrobp.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinod S.K., Jameson M.G., Min M., Holloway L.C. Uncertainties in volume delineation in radiation oncology: a systematic review and recommendations for future studies. Radiother Oncol. 2016;121:169–179. doi: 10.1016/j.radonc.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Schick K., Sisson T., Frantzis J., Khoo E., Middleton M. An assessment of OAR delineation by the radiation therapist. Radiography. 2011;17:183–187. doi: 10.1016/j.radi.2011.01.003. [DOI] [Google Scholar]
- 5.Sharp G., Fritscher K.D., Pekar V., Peroni M., Shusharina N., Veeraraghavan H. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41:1–13. doi: 10.1118/1.4871620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardenas C.E., Yang J., Anderson B.M., Court L.E., Brock K.B. Advances in auto-segmentation. Semin Radiat Oncol. 2019;29:185–197. doi: 10.1016/j.semradonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Lustberg T., van Soest J., Gooding M., Peressutti D., Aljabar P., van der Stoep J. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126:312–317. doi: 10.1016/j.radonc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Young A.V., Wortham A., Wernick I., Evans A., Ennis R.D. Atlas-based segmentation improves consistency and decreases time required for contouring postoperative endometrial cancer nodal volumes. Int J Radiat Oncol Biol Phys. 2011;79:943–947. doi: 10.1016/j.ijrobp.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 9.Gooding M.J., Smith A.J., Tariq M., Aljabar P., Peressutti D., van der Stoep J. Comparative evaluation of autocontouring in clinical practice: a practical method using the Turing test. Med Phys. 2018;45:5105–5115. doi: 10.1002/mp.13200. [DOI] [PubMed] [Google Scholar]
- 10.Van der Veen J., Willems S., Deschuymer S., Robben D., Crijns W., Maes F. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol. 2019;138:68–74. doi: 10.1016/j.radonc.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Reed V.K., Woodward W.A., Zhang L., Strom E.A., Perkins G.H., Tereffe W. Automatic segmentation of whole breast using atlas approach and deformable image registration. Int J Radiat Oncol Biol Phys. 2009;73:1493–1500. doi: 10.1016/j.ijrobp.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stapleford L.J., Lawson J.D., Perkins C., Edelman S., Davis L., McDonald M.W. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2010;77:959–966. doi: 10.1016/j.ijrobp.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 13.van Baardwijk A., Bosmans G., Boersma L., Buijsen J., Wanders S., Hochstenbag M. PET-CT-based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int J Radiat Oncol Biol Phys. 2007;68:771–778. doi: 10.1016/j.ijrobp.2006.12.067. [DOI] [PubMed] [Google Scholar]
- 14.Kim J., Han J., Ailawadi S., Baker J., Hsia A., Xu Z. SU-F-J-113: Multi-atlas based automatic organ segmentation for lung radiotherapy planning. Med Phys. 2016;43:3433. doi: 10.1118/1.4956021. [DOI] [Google Scholar]
- 15.Heimann T., Van Ginneken B., Styner M.A., Arzhaeva Y., Aurich V., Bauer C. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging. 2009;28:1251–1265. doi: 10.1109/TMI.2009.2013851. [DOI] [PubMed] [Google Scholar]
- 16.Lim J.Y., Leech M. Use of auto-segmentation in the delineation of target volumes and organs at risk in head and neck. Acta Oncol. 2016;55:799–806. doi: 10.3109/0284186X.2016.1173723. [DOI] [PubMed] [Google Scholar]
- 17.Taha A.A., Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging. 2015;15:1–28. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allozi R., Li X.A., White J., Apte A., Tai A., Michalski J.M. Tools for consensus analysis of experts’ contours for radiotherapy structure definitions. Radiother Oncol. 2010;97:572–578. doi: 10.1016/j.radonc.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huttenlocher D.P., Klanderman G.A., Rucklidge W.A. Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell. 1993;15:850–863. doi: 10.1109/34.232073. [DOI] [Google Scholar]
- 20.Bueno G., Déniz O., Salido J., Carrascosa C., Delgado J.M. A geodesic deformable model for automatic segmentation of image sequences applied to radiation therapy. Int J Comput Assist Radiol Surg. 2011;6:341–350. doi: 10.1007/s11548-010-0513-9. [DOI] [PubMed] [Google Scholar]
- 21.Deeley M.A., Chen A., Datteri R., Noble J.H., Cmelak A.J., Donnelly E.F. Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: a multi-expert study. Phys Med Biol. 2011;56:4557–4577. doi: 10.1088/0031-9155/56/14/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolov S, Blackwell S, Mendes R, De Fauw J, Meyer C, Hughes C. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. ArXiv. 2018:180904430–180904431. [Google Scholar]
- 23.Yang J., Veeraraghavan H., Armato S.G., Farahani K., Kirby J.S., Kalpathy-Kramer J. Autosegmentation for thoracic radiation treatment planning: a grand challenge at AAPM 2017. Med Phys. 2018;45:4568–4581. doi: 10.1002/mp.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.