Abstract
Microsatellite instability (MSI) is an important biomarker for predicting response to immune checkpoint inhibitor therapy, as emphasized by the recent checkpoint inhibitor approval for MSI-high (MSI-H) solid tumors. Herein, we describe and validate a novel method for determining MSI status from a next-generation sequencing comprehensive genomic profiling assay using formalin-fixed, paraffin-embedded samples. This method is 97% (65/67) concordant with current standards, PCR and immunohistochemistry. We further apply this method to >67,000 patient tumor samples to identify genes and pathways that are enriched in MSI-stable or MSI-H tumor groups. Data show that although rare in tumors other than colorectal and endometrial carcinomas, MSI-H samples are present in many tumor types. Furthermore, the large sample set revealed that MSI-H tumors selectively share alterations in genes across multiple common pathways, including WNT, phosphatidylinositol 3-kinase, and NOTCH. Last, MSI is sufficient, but not necessary, for a tumor to have elevated tumor mutation burden. Therefore, MSI can be determined from comprehensive genomic profiling with high accuracy, allowing for efficient MSI-H detection across all tumor types, especially those in which routine use of immunohistochemistry or PCR-based assays would be impractical because of a rare incidence of MSI. MSI-H tumors are enriched in alterations in specific signaling pathways, providing a rationale for investigating directed immune checkpoint inhibitor therapies in combination with pathway-targeted therapies.
Genomic microsatellite instability (MSI) is a condition characterized by variability in repetitive DNA sequences known as microsatellites. MSI can be caused by germline or somatic inactivation of genes in the DNA mismatch repair (MMR) pathway. The MMR pathway is primarily responsible for performing two functions associated with specific types of DNA damage: recognizing nucleotide mismatches or small insertions/deletions (indels) incorporated during replication, then excising and resynthesizing the correct DNA sequences to avoid propagating DNA damage.1, 2 Deficiencies in the MMR pathway lead to an accumulation of repeating indel units at microsatellite loci across the genome. This phenomenon occurs because small indels most commonly occur at microsatellite repetitive regions because of replication slippage at homopolymers.3 Thus, measuring the length variability of an individual's microsatellites compared with a standard can inform MSI status as well as infer MMR functional status.
MSI is clinically important as it confers an increased risk for colorectal, gastric, endometrial, and other cancers. MSI and MMR deficiency are most well known for their association with Lynch syndrome.4 Patients with MSI-high (MSI-H) colorectal carcinoma (CRC) and endometrial carcinoma have a better prognosis than those with MSI-stable (MSS) tumors; however, the prognostic value of MSI in other tumor types is controversial and requires further clarification.5, 6, 7, 8, 9, 10 In addition, recent studies have shown that MSI status is an important cancer biomarker for predicting therapeutic response to immune checkpoint inhibitors,11 presumably through its ability to confer increased probability of expressing immune reactive neoantigens caused by high genomic instability. The clinical importance of this biomarker is further underlined by the recent pan-solid tumor US Food and Drug Administration approval of pembrolizumab for MSI-H or mismatch repair–deficient tumors, an unprecedented approval from the agency using a biomarker-driven, tumor-type agnostic label.12, 13
MSI is clinically diagnosed as instability at two mononucleotide and three dinucleotide poly-A loci using a PCR-based test, according to the Bethesda guidelines.14, 15 However, many PCR-based tests require both tumor and matched normal sample, as the five loci are polymorphic in the human population. Instability at two of the five loci is sufficient to consider a tumor MSI-H. Alternatively, immunohistochemical (IHC) staining of tumor and nontumor nuclei for expression of the four clinically relevant MMR proteins [mutL homolog 1 (MLH1), mutS homolog 2 (MSH2), mutS homolog 6 (MSH6), PMS1 homolog 2, mismatch repair system component (PMS2)] is also used to evaluate MSI-H status.16 Absence of expression of one or more MMR proteins indicates an MSI-H tumor if the proteins are expressed in matched normal tissue. However, the IHC approach is not a direct phenotypic measurement of MSI, and it can miss cases in which MMR deficiency is due to inactivation of genes other than the four tested; in addition, deleterious variants may not result in loss of protein expression. Although PCR- or IHC-based MSI testing is routinely performed in CRC and increasingly in uterine endometrial carcinoma, traditional testing is impractical in tumor types in which MSI is rare, despite most other solid tumor types having an MSI-H patient population. This may hinder the pan-solid tumor adoption of checkpoint inhibitors.
Herein, we present a next-generation sequencing (NGS)–based MSI detection method that is routinely applied to clinical samples sequenced on a Comprehensive Genomic Profiling (CGP) assay. Although other NGS-based methods have been described in recent years,17, 18, 19 the method described herein is novel in that it uses principal component analysis (PCA) to generate an MSI score for stratification of MSI-H and MSS patients. This method is sensitive (97.0%; 95% CI, 89.6%–99.6%) and specific (positive predictive value > 95.0%) when compared with corresponding PCR and IHC assessments of the same tissue samples. The method does not require matched normal tissue and can be applied to NGS sequencing data if a sufficient number of homopolymers are captured on targeted or comprehensive cancer assays. Herein, we apply this novel method to >67,000 patient tumor samples spanning multiple disease types to investigate the genomic landscape of MSI-H in clinically advanced cancers, expanding on analyses of smaller sample sizes.20, 21 From this investigation, we demonstrate specific gene and pathway enrichment in MSI-H or MSS tumors as well as a correlation between tumor mutation burden (TMB) and MSI status.
Materials and Methods
Platform and Source of Samples
This study was reviewed and approved by the Western Institutional Review Board. Samples were submitted to a Clinical Laboratory Improvement Amendments–certified, New York State–accredited, and College of American Pathologists–accredited laboratory (Foundation Medicine, Inc., Cambridge, MA) for hybrid capture, followed by next-generation sequencing.22 For samples submitted for FoundationOne testing (n = 61,731), 395 cancer genes, 315 of which are reported clinically, were sequenced on DNA, covering 2.1 total MB. For samples submitted for FoundationOneHeme testing (n = 5913), the coding exons of 405 cancer-related genes (2.2 MB DNA) plus targeted RNA sequencing for 265 genes frequently involved in novel cancer-related fusions were sequenced.23 In addition to variant detection in these genes, TMB was assessed, as described by Chalmers et al.24 This study used the previously set cutoffs of <6 mutations/Mb for TMB-low, ≥6 and <20 mutations/Mb for TMB-intermediate, and ≥20 mutations/Mb for TMB-high.24 Disease ontologies were combined into similar groups for analysis (Supplemental Table S1). For the validation study, 30 CRC samples from Lifespan Academic Medical Center (Providence, RI) with prior IHC testing were sequenced, 40 CRC and uterine endometrial samples from Fox Chase Cancer Center (Philadelphia, PA) with prior PCR and/or IHC testing were sequenced, and five, CRC (n = 1), endometrial (n = 2), duodenum adenocarcinoma (n = 1), and gastroesophageal junction carcinoma (n = 1), samples from UC Davis (Sacramento, CA) with prior IHC and/or PCR testing were sequenced. Samples from the validation study were tested on the basis of tissue availability and patient consent. To determine limit of detection, five MSI-H tumor samples from the validation study were diluted to obtain different tumor content ratios. The limit of detection study also used two HapMap controls (Horizon Discovery, Cambridge, UK; GM24143). For all other analyses, clinical samples from the Foundation Medicine, Inc., database that underwent comprehensive genomic testing on FoundationOne or FoundationOneHeme were used.
NGS MSI Detection Method
Selection of Microsatellite Loci
An initial pool of 1880 mononucleotide homopolymers of 7 to 39 bp in repeat length, sequenced using the FoundationOne assay, such that no wet laboratory modifications of the assay were necessary, was established. A subset of the available loci was selected for inclusion in the PCA training. The selection criteria prioritized coverage and variability in observed allelic lengths at each microsatellite across all the training samples. A minimum of 250× median depth at each locus was enforced to ensure robust detection of variant alleles. In addition, microsatellites that did not show any variability in allelic lengths, compared with the hg19 reference genome and other genomes in the training data set, would not be informative to the PCA training and were removed. All selected loci were also required to be intronic and have an hg19 reference length of 10 to 20 bp because of the limitations of aligning 49-bp reads over a repetitive sequence >20 bp. The full list of mononucleotide homopolymers is provided in Supplemental Table S2.
Establishment of MSI Score and Cutoff
For each sample at each locus, every NGS read that fully spanned the repeat region was used to determine an allelic length, which allowed a distribution of allelic lengths to be obtained. For a sample that is MSI-H, variability of allelic length is expected to significantly increase while the mean allelic length often decreases due to the higher likelihood that polymerase slippage will result in deletion than insertion. Thus, for each sample at every locus, the mean and variance of the allelic length were sufficient to predict MSI and were recorded. To combine the separate mean and variance information for all the loci, PCA was used to project the multidimensional data into a corresponding number of PCs. The exemplar training data set was used, and the first principal component (PC1) explained 45% of the data variance, whereas PC2 and onwards explained no more than 5% each and were, therefore, discarded. The projection vector onto PC1 of the training data set was fixed, and the PC1 value was used as the MSI score. The cutoff of MSI-H versus MSS was established by comparing the MSI score with orthogonal MSI testing data.
Genomic Enrichment Assessment and Statistical Analysis
Gene or pathway enrichment was established using a 2 × 2 contingency table with Fisher exact testing. To be included in a comparison, a gene must have been altered in at least 25 samples, at least one member of a pathway must have been altered in at least 25 samples, and at least 25 samples must have been assigned the MSI status. Odds ratios, P values, and multiple hypothesis (Holm-Sidak) adjusted P values were obtained from Fisher exact test and used to determine significance (adjusted P < 0.05) and directionality of enrichment (odds ratio < 1 enriched in MSS, odds ratio > 1 enriched in MSI-H). TMB comparison between MSS and MSI-H samples used the Mann-Whitney-Wilcox test of significance to determine a P value. All statistical analyses were performed in python version 2.7.12 (Python Software Foundation, Wilmington, DE; www.python.org) using scipy stats package version 0.18.1 (SciPy, http://www.scipy.org) or R version 3.3.2 (The R Project for Statistical Computing, https://www.r-project.org) using built-in packages and RStudio version 1.0 (RStudio, Inc., Boston, MA).
Exclusion of Polymerase Slippage MMR-Related Variants
Insertion or deletion events occurring in a repetitive region (two or more trinucleotide or dinucleotide repeats or three or more single-nucleotide repeats) were considered potential MMR-related variants. For the gene enrichment analyses, unless otherwise noted, variants occurring in regions of likely polymerase slippage were considered MMR-related variants and were excluded.
Results
Development of an NGS-Based MSI Detection Algorithm
Herein, we present data from a separate exemplar training data set to illustrate how the development of the NGS-based MSI detection was conducted and to highlight key features of the method. The exemplar data set includes a mixture of 300 uterine endometrial and CRC cases characterized as MSI-H as well as 300 cases from a mixture of cancers characterized as MSS. Although the racial backgrounds are unknown, others have reported that rates of MSI-H are similar in African American, white, and Hispanic CRC patients.25
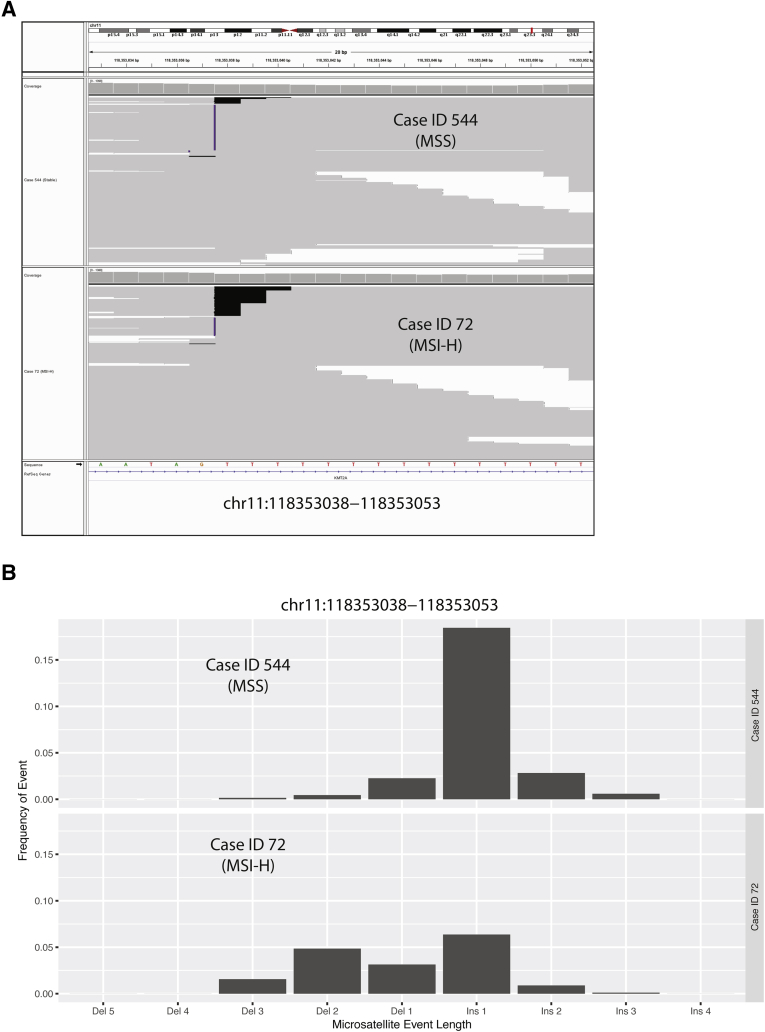
Genomic instability was measured at microsatellites in the cancer genome for tumors deficient in MMR. Figure 1A shows an Integrative Genomics Viewer version 2.3.57 snapshot of a representative locus to illustrate the types of NGS sequencing data that are generated for MSI-H versus MSS tumors.26, 27 Case 544 is a gastrointestinal stromal tumor previously characterized as MSS, and case 72 is a CRC previously characterized as MSI-H (Figure 1A). Cases were chosen to highlight extreme differences at a single loci to illustrate the differences underlying this method; however, the specific loci with variance will vary for each sample. For similar depth of coverage at chromosome 11:118,353,038 to 118,353,053 (Case 544 to approximately 596X and case 72 to approximately 749X), the MSI-H case visually had a higher frequency of deletion events. Homopolymer loci such as this often display differences between the reference genome and individuals, which is why this exemplar MSS case (Figure 1A) displays frequent apparent insertions at this locus. In contrast, the MSI-H case has greater variance at these loci. Figure 1B shows the frequency distribution of indel events at chromosome 11:118,353,038 to 118,353,053 for the same two cases presented in Figure 1A. The differences in distributions shown in Figure 1B suggest that NGS sequencing data from microsatellite regions provide some discriminating power between MSI-H and MSS tumors, as expected. The average microsatellite length and variance are captured for each microsatellite locus and are used in the NGS-based MSI algorithm.
Figure 1.
Development of next-generation sequencing (NGS)–based microsatellite instability (MSI) algorithm. Overview of the development of the NGS-based algorithm to determine MSI status. A: Integrative Genomics Viewer screenshot of one representative MSI-stable (MSS) and MSI-high (MSI-H) sample sorted for sequence reads with insertion (Ins)/deletion (Del; indel) events. Homopolymer shown is chromosome (Chr) 11:118,353,038 to 118,353,053, which is one location used in the MSI algorithm. B: Histogram summarizing frequency of the different length indel events observed at Chr11:118,353,038 to 118,353,053 for the two cases shown in A.
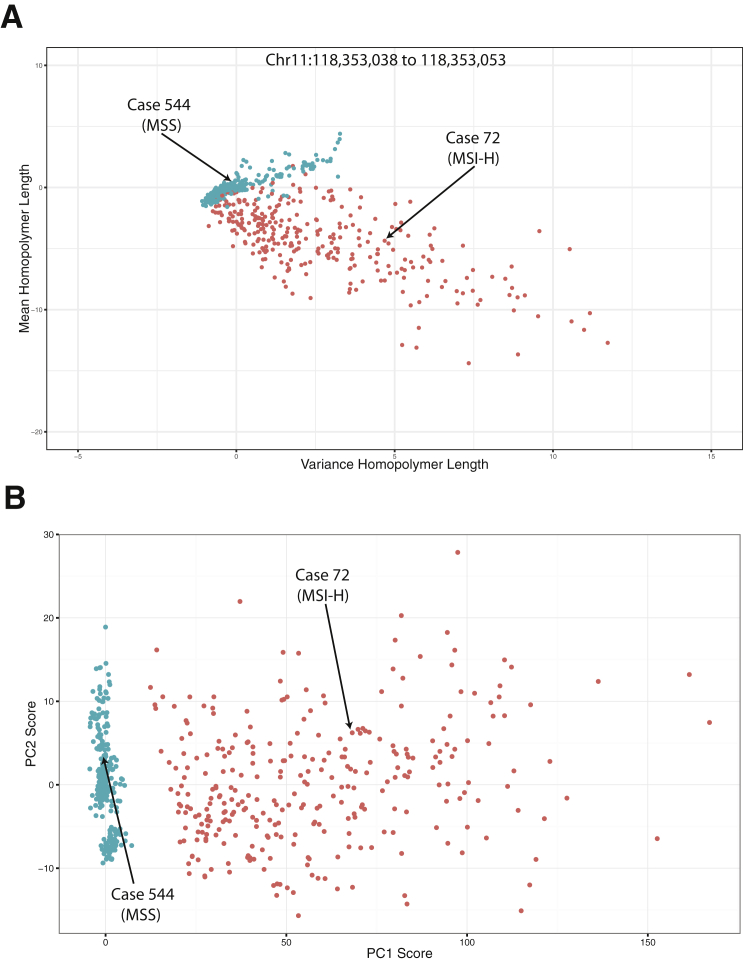
Looking more broadly at all 600 exemplar cases (Figure 2A), again at the chromosome 11:118,353,038 to 118,353,053 locus, illustrates the power of a single locus to stratify MSI-H and MSS cases using microsatellite mean repeated length and variance. The 300 MSI-H cases had a shorter microsatellite length but a larger variance than the 300 MSS cases. The two data points from the cases presented in Figure 1, are labeled for context. Some MSI-H cases were more divergent at this locus, whereas others had similar distributions to the MSS cases. These results suggest that although a single locus or even a low number of loci can separate some MSI-H cases from some MSS cases, there are also many for which this single locus is not sufficient to successfully segregate all MSI-H and MSS cases using this method without matched normal DNA.
Figure 2.
Development of next-generation sequencing (NGS)–based microsatellite instability (MSI) algorithm. Overview of the development of the NGS-based algorithm to determine MSI status. A: An example plot of normalized mean length of homopolymer and variance in length at one locus [chromosome (Chr) 11:118,353,038 to 118,353,053]. Normalization was achieved using the mean and SD values from the MSI-stable (MSS) training group. Each point represents a single sample. A total of 300 MSS (teal) and 300 MSI-high (MSI-H; orange) samples are shown to visualize separation of MSS and MSI-H cases based on mean and variance statistics. B: Principal component (PC) analysis scores plot, showing PC1 and PC2 scores, with PC1 separating the two groups (MS, teal; MSI-H, orange). Same 300 MSS and MSI-H cases as presented in Figure 1C.
Originally, microsatellite loci were selected for PCA that had high variability of mean length and variance among all the training samples. The same microsatellite loci were used for PCA in this exemplar data set. Figure 2B shows the PCA scores plot for PC1 versus PC2 in the 600-sample exemplar set. The final results showed that PC1, the NGS-based MSI score, was capable of efficiently separating the MSI-H and MSS cases along the PC1 axis. There was no need to extend beyond the first principal component, as it explained approximately 45% of the total data variance, whereas none of the other principal components explained >5% each. PCA ranges of the MSI score were manually assigned to MSI-H, MSI-intermediate, or MSS using unsupervised clustering of specimens for which gold standard MSI status was previously assessed. In the exemplar data, these ranges would be assigned MSS for a score < 7.5, MSI-intermediate for scores 7.5 to 12, and MSI-H for scores > 12, although these values are not directly interpretable and are relevant only to status assignments. MSI-low (MSI-L) calls were not made because there was no available training set with orthogonal PCR–determined MSI with an MSI-L score, but MSI-L is expected to significantly overlap with the MSI-intermediate category. Although MSI-H has been well studied as a biomarker predicting response to checkpoint inhibitor therapy, MSI-L and the likely similar MSI-intermediate have not been established as biomarkers for response and, therefore, the use of these statuses in clinical management of patients is not established.
NGS-Based MSI Detection Is Highly Concordant with Traditional Methods
Concordance was evaluated by comparing NGS-based MSI status for 73 CRC or uterine endometrial patient samples and an additional two non-CRC/endometrial (one gastroesophageal junction adenocarcinoma and one duodenum adenocarcinoma) samples, as the validation study, with prior standard-of-care PCR or IHC MSI testing results (Table 1). Successful NGS sequencing was achieved for 74 samples, of which 72 (24 MSI-H and 48 MSS) had definitive NGS MSI results. The remaining two samples had NGS MSI results of MSI-intermediate, which represents an equivocal region of the method, and were, therefore, excluded from the concordance analysis. Results were 97% (70/72) concordant with PCR/IHC-based MSI testing (Table 1) and 95% (70/74) concordant if MSI-intermediate samples were included and considered discordant. Comparison to IHC showed 100% concordance, although only five cases were identified as MSI-H in this data set. One of the discordant samples was negative by PCR but positive by NGS. On close inspection of the reported genomic results from the FoundationOne assay, a high number of indel variants were confirmed at homopolymers in gene coding regions (JAK1 2573delA, DNMT3A 171delC, and ARID1A 1015delG), indicating a possible false-negative result by PCR.
Table 1.
Concordance of NGS and Traditional MSI Testing Methods
| Variable | IHC |
PCR |
IHC and PCR |
||||
|---|---|---|---|---|---|---|---|
| MSI-H | MSS | MSI-H | MSS | MSI-H | MSS | ||
| NGS | MSI-H | 5 | 0 | 18 | 1 | 23 | 1 |
| MSS | 0 | 29 | 1 | 18 | 1 | 47 | |
Data are expressed as n. NGS-based MSI testing results were 97% concordant with the combined IHC and PCR result set. IHC test results were 100% concordant, and PCR test results were 95% concordant.
IHC, immunohistochemistry; MSI, microsatellite instability; MSI-H, MSI-high; MSS, MSI-stable; NGS, next-generation sequencing.
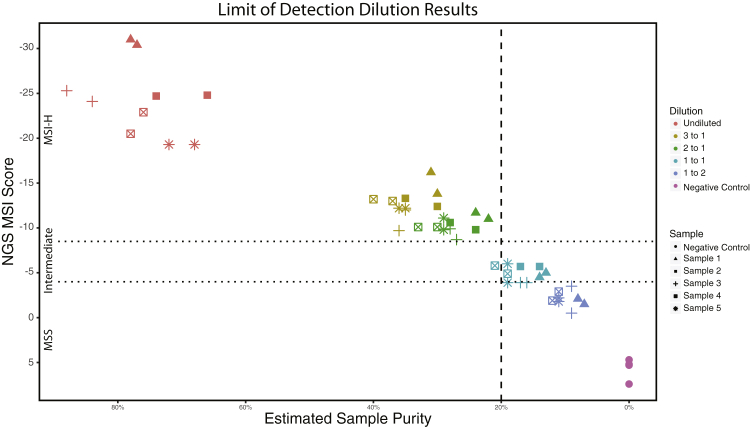
To assess the performance of the NGS MSI detection method at low sample purity, five MSI-H samples from the validation study were selected for dilution with a control sample (Horizon Discovery; GM24143). Samples were diluted to five different tumor/normal ratios (based on DNA quantity): undiluted, 3:1, 2:1, 1:1, and 1:2. Because of differences in relative DNA capture efficiencies between tumor and control samples, tumor purity was estimated computationally after sequencing (rather than from presequencing volumetric dilution ratios). Sequencing was performed for each dilution on both Illumina HiSeq2500 and HiSeq4000 platforms (Illumina, San Diego, CA). Figure 3 shows the calculated MSI score as a function of tumor purity. MSI-H status was unchanged for dilutions >20% but was diminished for dilutions <20% tumor purity with 0% (0/17) of cases reported as MSI-H. One of the 1:2 dilutions (sample 2) failed during laboratory processing and, thus, does not have a result. Figure 3 highlights that the PCA-based MSI score correlates with the tumor purity; however, above the 20% purity cutoff, the MSI status remains stable as tumor purity changes, despite the MSI raw score relationship to purity. Given the raw score is only used to determine the appropriate MSI status to assign, this indicates that tumor purity (>20%) does not alter the performance of this method.
Figure 3.
Limit of detection. Estimated sample purity of microsatellite instability–high (MSI-H) samples diluted to represent varying sample purities. Each sample (identified by shape) was diluted (identified by color) and next-generation sequencing (NGS) MSI score was assessed. Dotted lines represent cutoffs for MSI-stable (MSS; top line) and MSI-H (bottom line). Dashed line represents 20% cutoff for estimated tumor purity.
MSI-H Samples Occur at Low Frequency across Many Disease Types
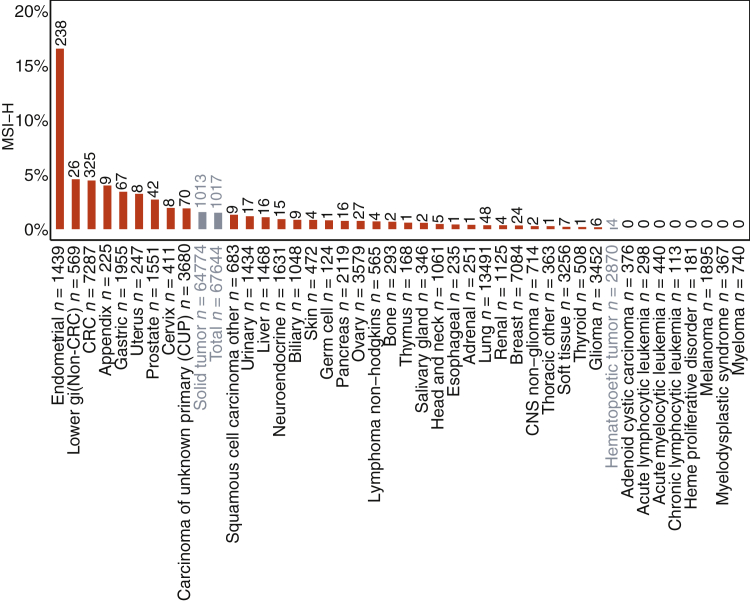
The distribution of MSI-H was determined in a large cohort of real-world cancers across a variety of cancer types. Overall, 1.5% of all tumors (solid and hematopoietic) were MSI-H (Figure 4), accounting for >1000 samples. Some cancer types (Supplemental Table S1) had higher rates of MSI-H (Figure 4), including uterine endometrial (16.5%), small bowel (4.6%), CRC (4.5%), appendix (4%), gastric (3.4%), uterine other (3.2%), prostate (2.7%), cervix (1.9%), and carcinoma of unknown primary (1.9%) (Figure 4). MSI-H tumors were rare or absent in some cancer types, like melanoma (0% of 1895 samples), lung (0.36% of 13,491 samples), and many hematopoietic malignancies. MSI-H percentages by disease ontology are included in Supplemental Table S3. These data highlight that MSI-H tumors are not equally distributed among cancers, but they occur at low frequency in many tumor types.
Figure 4.
Microsatellite instability–high (MSI-H) sample distribution across tumor types. Prevalence (%) of MSI-H in each disease group in order of frequency displayed with the number of MSI-H samples on top of each bar and the total number of samples assessed with the disease group label. Each disease group (defined in Supplemental Table S1) must have at least 100 total samples with an evaluated MSI status to be included. Disease groups in gray are super groups and encompass all solid tumors (solid tumor), all hematopoietic tumors (hematopoietic tumor), or all tumors profiled (total). CNS, central nervous system; CRC, colorectal carcinoma.
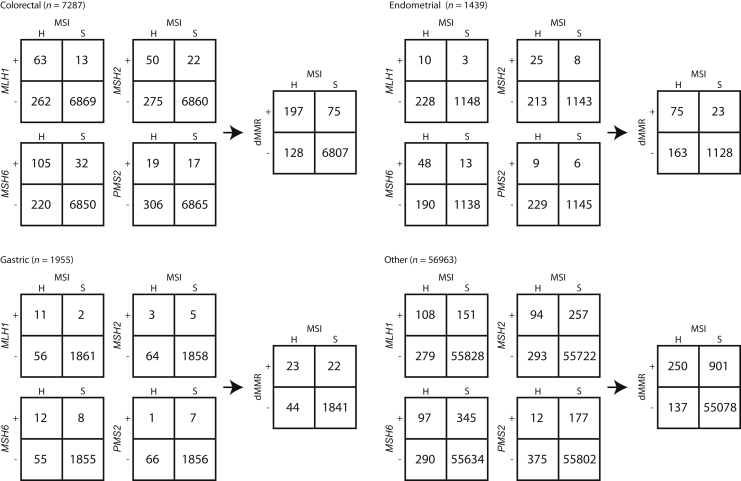
MMR-Related Genes Are Frequently Mutated in MSI-H Samples
Considering the known association between MMR deficiency and MSI, the frequency of deleterious mutations in the commonly tested MMR genes (MLH1, MSH2, MSH6, and PMS2) was investigated across different disease groups. The frequency of genomic deficiency of the individual MMR genes varied among disease groups but was generally enriched in MSI-H cases. Mutations in MMR genes were significantly associated with MSI status, except PMS2 in gastric cancer (Fisher exact test; P < 0.005) (Figure 5). Cases were identified within each disease group with genomic deficiencies in MMR genes that were classified as MSS. Conversely, cases were also identified that were MSI-H but had no detected MMR mutation. The overall frequency of these events was <4%, with the exception of endometrial cancer, which was approximately 13%.
Figure 5.
Frequency of deleterious mutations in mismatch repair (MMR) genes by microsatellite instability (MSI) status. There are 2 × 2 tables for sample counts of each gene (MLH1, MSH2, MSH6, and PMS2) mutation frequency. The vertical axis plus (+) indicates presumed deleterious variant(s) observed, and the vertical axis minus (−) indicates no deleterious variants observed. Samples are split by MSI status on the horizontal axis (H indicates high, and S indicates stable). Results are further segregated into disease categories: colorectal, uterine endometrial, gastric, and other, with other representing all other tumor types not already displayed. The MMR-deficient (dMMR) 2 × 2 in each disease group represents the combined results from the four gene-level 2 × 2 tables.
MSS samples with genomic alterations in MMR genes are likely explained by the maintenance of MMR function through the second, wild-type allele. Loss of heterozygosity at the MMR gene is a key factor needed for MMR deficiency, leading to the development of the MSI-H phenotype, and is not assessed in this analysis. In addition, several of the MSS cases with MMR mutations harbored inactivating mutations in the polymerase genes POLE/D and had extremely high tumor mutational burdens (average TMB > 30 mutations/Mb). These cases were likely genomically deficient for MMR but had not yet developed the MSI-H phenotype, as tumors with this biology are likely driven by deficiency of a separate repair pathway.28 The frequency of MSS samples with a genomic alteration in MMR genes (1.5%) is within the expected error for this method (97% concordance), suggesting a subset of these cases may be incorrectly assigned, as this method's concordance would suggest occurs in 2% of cases. The MSI-H cases without detected MMR alterations may exhibit other mechanisms of MMR deficiency, such as epigenetic silencing, which has been well described for MLH1 promoter and is not detected by this assay, or novel MMR variants of unknown significance.
Common Genomic Alterations across MSI-H Tumors
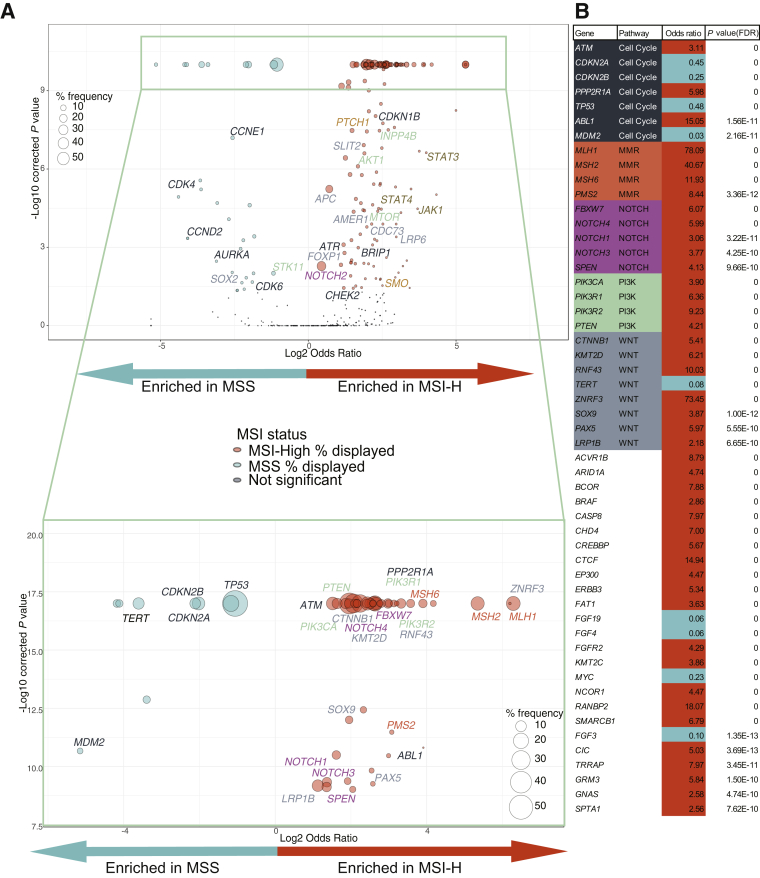
Because MSI-H tumors shared mutations in MMR-related genes, we hypothesized that MSI-H tumors may have additional alterations in common. Given the rarity of MSI-H tumors in hematological malignancies and sarcomas, the remaining analyses were focused on solid tumors. Gene enrichment analysis was performed in each disease group as well as across all solid tumors, specifically excluding polymerase slippage–related variants. The exclusion of polymerase slippage MMR-related variants allowed us to exclude variants that were most likely related to the high mutation rate of these tumors.
Genes enriched in MSI-H tumors included PIK3CA (38% of all MSI-H tumors); CTNNB1 (15%); and the MMR-related genes MLH1 (17%), MSH2 (15%), MSH6 (6%), and PMS2 (3%). Those enriched in MSS tumors included TP53 (57%), CDKN2A (22%), CDKN2B (14%), and TERT (11%) (Figure 6, Supplemental Tables S4 and S5, and Supplemental Figure S1). In addition, enrichment of genes that are not associated with CRC or endometrial MSI-H (from which 55% of MSI-H samples in this data set are associated with these two disease groups), including SLIT2, which is enriched in ovary (Supplemental Figure S1) MSI-H samples, and ZNRF3 and SOX9, which are both enriched in the overall data set (Figure 6), but not in either CRC (Supplemental Figure S1) or endometrial (Supplemental Figure S1) alone, was found. These MSI-H–enriched genes fell into common pathways, such as the phosphatidylinositol 3-kinase (PI3K) and WNT pathways. MMR and pathway-related genes with significantly enriched alterations are labeled in Figure 6 (Supplemental Table S6). For improved visualization, the most significant (P < 1 × 10−9) region of the plot is expanded in Figure 6A and capped at 1 × 10−17. Figure 6B provides odds ratios and adjusted P values for the genes displayed in Figure 6A.
Figure 6.
Gene enrichment in microsatellite instability–high (MSI-H) or MSI-stable (MSS) tumors. Pan-tumor enrichment of genes in MSI-H (log2 odds ratio > 1) or MSS (log2 odds ratio < 1). Circle sizes represent the frequency of gene alteration in either MSI-H tumors (orange) or MSS tumors (teal). Circles representing genes without significant enrichment (dark blue) are not scaled to size. Multiple hypothesis–corrected –log10 P values are on the y axis. A: All genes with at least 25 samples are displayed. Top panel: Pathway-related (Supplemental Table S6) or mismatch repair (MMR) related genes with P < 0.05 and P > 1e-09 are labeled. P values are capped at 1e-09 for visualization purposes. Bottom panel: Enlargement of highly significantly enriched genes (P < 1e-09). P values are capped at 1e-17 for visualization purposes. Pathway-related (Supplemental Table S6) or MMR-related genes are labeled. B: List of all genes with P < 1e-09 with odds ratio (>1 enriched in MSI-H, and <1 enriched in MSS) and multiple hypothesis–corrected P value. Odds ratios corresponding with MSI-H enrichment are coded in orange, and those with MSS are coded in teal. Genes included in pathway analyses are color coded, and the pathway is listed. FDR, false discovery rate; PI3K, phosphatidylinositol 3-kinase.
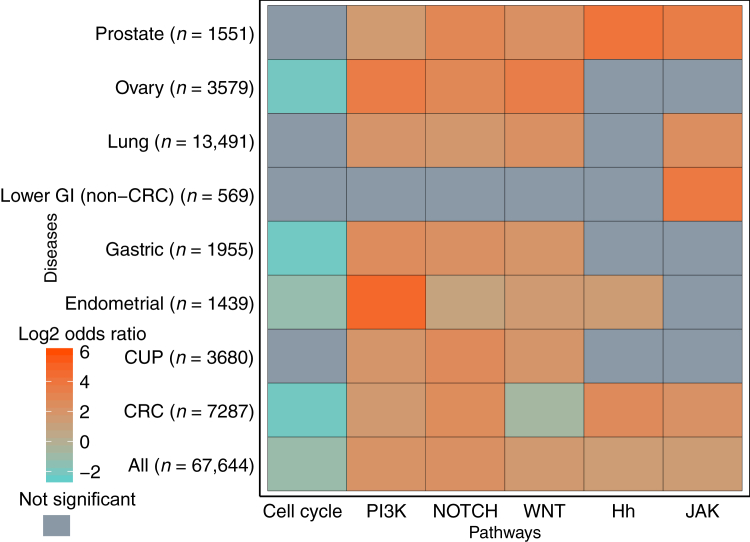
After identifying enrichment of alterations in specific MMR, PI3K, and WNT pathway genes in MSI-H tumors, the overall enrichment of six total pathways of interest in MSS versus MSI-H tumors was investigated (Supplemental Table S6). PI3K, NOTCH, and WNT pathways were generally enriched in MSI-H samples (Figure 7 and Supplemental Figure S2). Interestingly, Janus kinase/signal transducers and activators of transcription (JAK) and Hedgehog pathways were also enriched in MSI-H tumors across all disease groups when regions of polymerase slippage were included (Supplemental Figure S2 and Supplemental Table S7), but not when polymerase slippage regions were excluded (Figure 7 and Supplemental Table S8). Therefore, the genes in these pathways included in this targeted sequencing panel could be specific targets of MMR deficiency because of the frequency of homopolymer regions within JAK- and Hedgehog-related genes. Alterations in PI3K, WNT, and NOTCH pathways were enriched in almost all disease groups, even when MMR-related variants were excluded (Figure 7 and Supplemental Table S8). These pathways may not simply be frequent targets of MMR deficiency, but rather they may be necessary for MSI-H tumor survival.
Figure 7.
Pathway enrichment with mismatch repair polymerase slippage regions removed. Enrichment of various pathways (Supplemental Table S6 for genes included in each pathway) in indicated disease types. Log2 odds ratio displayed by color: orange indicates enrichment in microsatellite instability–high, and teal indicates enrichment in MSI-stable. Gray indicates nonsignificant multiple hypothesis–corrected P values for enrichment (P ≥ 0.05). CRC, colorectal carcinoma; CUP, carcinoma of unknown primary; GI, gastrointestinal; Hh, Hedgehog; JAK, Janus kinase/signal transducers and activators of transcription; PI3K, phosphatidylinositol 3-kinase.
In contrast, alterations in genes involved in the cell cycle are consistently enriched in MSS tumors regardless of the tumor type. The commonality of cell cycle mutations across tumor types is expected given the inclusion of common tumor suppressor mutations, such as TP53, in the pathway.
MSI-H Correlates with High Tumor Mutational Burden
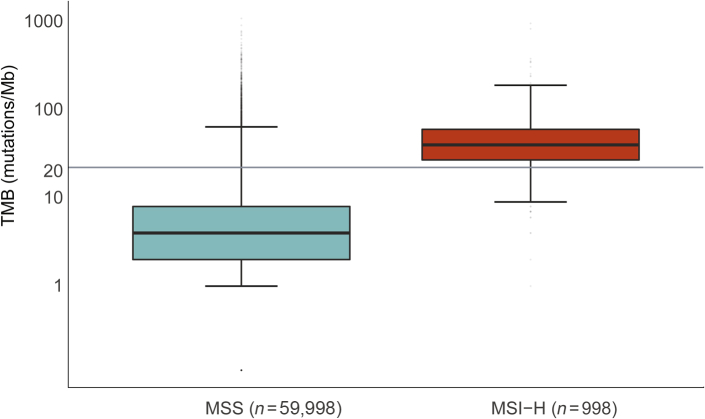
As a biomarker for response to immunotherapy, MSI is a surrogate for high TMB, which is, in turn, a surrogate for production of neoantigens to which the immune system can mount a response. In 59,998 MSS and 998 MSI-H samples across all tumor types, MSI-H tumors had a significantly increased median TMB (36 mutations/Mb) compared with MSS samples (3.6 mutations/Mb). The first quartile of MSI-H samples is above the TMB-high cutoff of 20 mutations/Mb, whereas the third quartile of MSS samples remains below this cutoff line (Figure 8). Although 83% of MSI-H tumors have high TMB, 6.6% of MSS tumors also have high TMB. This highlights the fact that, although MSI-H may be sufficient in most cases to predict high TMB, MSI-H is not necessary for high TMB.
Figure 8.
Microsatellite instability–high (MSI-H) is sufficient but not necessary for high tumor mutation burden (TMB). Spectrum of TMB in MSI-stable (MSS) and MSI-H samples. The y axis is TMB (mutations/Mb) plotted on a logarithmic scale to better display extreme outliers. Horizontal lines in each box indicates the median TMB, whereas the lower and upper bounds of the box represent 25th and 75th percentiles, respectively. Whiskers extend to extreme values no more than 1.5 times the interquartile range beyond the box. Points beyond whiskers are considered outliers and are plotted individually. P < 0.001 (calculated from a Mann-Whitney-Wilcox test).
Discussion
Herein, we provide validation for a novel method of calling MSI using a targeted hybrid capture NGS-based approach. This method has excellent concordance with existing methods and maintains performance specifications in specimens with at least 20% tumor content. Applying this method across our database of 67,644 patient tumor samples, it was shown that 1.5% of all tumors were MSI-H, whereas individual tumor types ranged from 0% to 16.5% MSI-H. This data set may be biased because of an enrichment for late-stage advanced disease, but the genomic findings are still informative. On investigation of the genomic landscape of these samples, previous reports that MSI-H samples are associated with genomic deficiency in MMR genes were confirmed.20, 21 Some gene alterations are enriched in MSI-H or MSS across multiple indications, and these correlated to pathway enrichment in MSI-H or MSS tumors. Finally, MSI-H is a sufficient, but not necessary, biomarker for high TMB.
MSI can be effectively determined from an NGS-based panel approach, allowing efficient testing for MSI while also screening for additional relevant genomic information. Using the described method, MSI status can be reliably determined regardless of the specific baits on a targeted NGS panel, as long as there is sufficient genomic coverage. Similar to PCR, the method assesses the effect of mismatch repair–deficient and not the cause; however, the validation study did not include samples annotated as having MLH1 promoter methylation or MSI-L samples. This is a limitation of this study. Validation results show that the method is sensitive and specific, with high concordance to traditional methods, enabling confident NGS-based assessment of MSI status without the requirement for matched normal tissue. Concordance, sensitivity, and specificity favorably compare with other recent NGS-based MSI methods. The method described herein is 97% concordant, with 95% sensitivity and 98% specificity, similar to comparable methods from Nowak et al29 (97% concordance, 91% sensitivity, and 98% specificity) and the mSINGS method (98% concordance, 98% sensitivity, and 98% specificity).18
In light of growing evidence that immunotherapy improves quality and duration of patient lives and the recent pan-cancer approval of pembrolizumab for MSI-H cancers, many physicians and patients will want to determine whether immune checkpoint inhibitor therapy could be an option. For some diseases, like uterine endometrial, where a relatively large percentage of patients are MSI-H (16.5%), existing methods for detecting MSI, such as PCR or IHC, may be practical. However, MSI-H is rare in most disease types (Figure 4), so routine testing for MSI alone is not realistic, despite the potential value to those rare patients. Determining the MSI status in combination with other potentially actionable alterations from a single tumor specimen is a logical approach that is cost-effective and spares tissue. For example, in lung cancer, where several genomic alterations are biomarkers for response to approved targeted therapies, but for which the MSI-H frequency is low (0.36%), this type of CGP approach could provide multiple valuable results from a single tumor specimen.
As expected, alterations in MMR genes MLH1, MSH2, MSH6, and PMS2 were enriched in MSI-H tumors (Figure 6 and Supplemental Table S4). MSI-H cases with no detected genomic alterations in MMR genes were also identified. This is consistent with the expectation that there may be other mechanisms of MMR deficiency, such as epigenetic silencing of MMR genes. In contrast, cases with genomic alterations in MMR genes that were MSS were also identified. These genomically deficient MSS cases fall broadly into two categories, the first being tumors with ultra-high mutation burdens resulting from gene alterations in other repair pathways, such as polymerase E or D (POLE/D). One potential explanation is that these cases have acquired MMR mutations incidentally and have not had sufficient time to develop the MSI-H phenotype. The second category would include tumors carrying MMR mutations that have not yet acquired biallelic inactivation at the MMR gene locus and are, thus, still MMR proficient.30
It is possible that because of the higher mutational burden in MSI-H tumors (Figure 8), any given pathway or gene would be enriched in MSI-H tumors. To conservatively decrease the noise associated with high mutational burden when performing enrichment analysis, polymerase slippage regions were removed in Figures 6 and 7 and Supplemental Figure S1. By broadly removing variants in polymerase slippage regions, variants that do not appear to be a direct result of MMR deficiency were specifically studied. Although MMR deficiency may be an initiating event in MSI-H tumors, the subsequent events that led to transformation appear to cluster in certain pathways. Some of this clustering could be driven by an enrichment of polymerase slippage regions and, thus, is likely a direct consequence of MSI, like RNF43, which, in addition to being identified herein, was found using the more focused MSMutSig method as having multiple microsatellite hotspot loci within the gene.31 However, other pathways enriched in MSI-H cases, such as NOTCH, do not depend on variants in polymerase slippage regions, as determined by our broadly conservative exclusion of variants in these regions. This is highly suggestive of positive selection of these pathways in MSI-H tumors, rather than incidental enrichment caused by frequent regions of polymerase slippage. The enrichment of pathways independent of polymerase slippage–related variants suggests that, if these pathways are truly selected for, they may be necessary for tumor survival.
Regardless of the mechanism, it is clear that MMR deficiency can lead to both microsatellite instability as well as pathogenic variants in other common cancer pathways. Some of these pathways, such as PI3K and NOTCH, are strongly enriched across MSI-H samples of all tumor types, suggesting molecular commonalities among MSI-H tumors regardless of tumor site of origin. Although a few signaling pathways were studied (cell cycle, NOTCH, JAK/STAT, Hedgehog, WNT, and PI3K) (Supplemental Table S6), the analysis was not exhaustive and does not exclude the possibility that other pathways may be enriched in MSI-H or MSS tumors.
One of the most important topics in immunotherapy is understanding not only what makes a patient likely to respond to checkpoint inhibition, but also what may make a patient less likely to respond. Some emerging factors that may impair response are still being explored. For example, disrupted JAK/STAT signaling, which was enriched in MSI-H tumors, may mediate resistance to programmed death-ligand 1 (PD-L1) blockade by disabling interferon-γ receptor signaling.32, 33, 34 In addition, activation of the WNT signaling pathway in the tumor may enhance immune evasion, a necessary survival mechanism in many MSI-H tumors that are likely to express neoantigens that confer susceptibility to clearance by the immune system. Multiple studies suggest differing mechanisms of WNT-related immune evasion, including WNT-triggered tumor activation leading to T-cell exclusion from the tumor or suppression of dendritic cell recruitment and a subsequent decrease in tumor-associated T cells.35, 36, 37 Herein, we show that WNT pathway activation is enriched in many MSI-H tumors (Figure 7), notably except CRC, even when MMR deficiency-related variants are excluded (Supplemental Figure S2), suggesting a possible selective pressure for WNT activation in the context of MSI-H. This finding supports further investigation into WNT pathway activation as a potential resistance mechanism to immune checkpoint inhibitor therapy. In general, pathway enrichment studies such as this can inform combination therapies that may warrant investigation (eg, combining immune checkpoint inhibitors and targeted pathway inhibitors). More important, in addition to the overall enrichment of pathways in MSI-H tumors (Figure 7), each tumor group was also analyzed separately, to avoid biases caused by the high number of samples from certain disease types (CRC and endometrial specifically).
The WNT pathway has been investigated previously in CRC as it relates to MSI status, but because of low sample numbers, consensus in the field has not been reached.38, 39, 40 A recent publication from Grasso et al41 provides a larger number of MSI-H samples studied (179) and suggests WNT pathway alterations are found equally across all CRCs, regardless of MSI status. Herein, evidence is provided using a large number of samples (325 MSI-H and 6887 MSS samples) that the WNT pathway is enriched in MSS CRC (Figure 7) in contrast to all other disease groups investigated. This discrepancy in our findings and those previously published41 is likely due to differences in WNT pathway gene definitions [eg, in this study, PTEN alterations are included as part of the PI3K pathway (Supplemental Table S6), whereas Grasso et al41 include it as part of the WNT pathway]. WNT pathway enrichment in MSS CRC appears to be driven primarily by APC, which itself is enriched in MSS CRC, whereas CTNNB1 is enriched in MSI-H CRC (Supplemental Figure S1A). The enrichment of APC variants in MSS tumors and the enrichment of CTNNB1 variants in MSI-H tumors are consistent with published observations.36, 38, 41
Response to immune checkpoint inhibitor therapy is predicted to be directly related to the number of neoantigens properly presented to the T cells.11 Although MSI-H is not a direct measure of neoantigen burden, MSI-H samples have a stochastically higher likelihood of neoantigen generation because of the increased overall mutational burden induced by MMR deficiency.28 Furthermore, results from NGS-based MSI methods are being successfully used in the clinic.42, 43, 44 It was confirmed that MSI-H samples have a high median TMB (36 mutations/Mb), whereas the median TMB for MSS samples is low (3.6 mutations/Mb), similar to a previously shown correlation between MSI-H and TMB-H.17 Also, there exist many TMB-high tumors that are MSS, in which a high TMB, and likely high neoantigen burden, is induced by other causes, such as UV exposure. Specifically, in melanoma, 1880 samples are MSS, 15 are MSI-intermediate, and 0 are MSI-H (Figure 4), despite a median TMB of 12.6 mutations/Mb. The increased TMB in melanoma is likely due primarily to UV exposure and not MMR deficiency; however, melanoma patients often have robust responses to immune checkpoint inhibitor therapy.45 This supports that, although MSI-H can be used as a biomarker for patients who are more likely to respond well to immune checkpoint inhibitor therapy, this approach will exclude other patients with high neoantigen burden unrelated to MMR deficiency who are also likely to respond. Future studies should continue to explore the best combinations of biomarkers that predict a positive response to immune checkpoint inhibitor therapy.
Acknowledgments
We thank Fox Chase Cancer Center, Lifespan Academic Medical Center, and UC Davis for collaboration and for providing samples used in validation of our method; the engineers and operations staff of Foundation Medicine, Inc., for supporting this research; and Dan Spirtz for helpful manuscript comments.
Footnotes
Supported by Foundation Medicine, Inc., and Genentech, Inc.
S.E.T. and K.G. contributed equally to this work.
Disclosures: S.E.T., K.G., E.S., D.A.F., P.J.S., G.M.F., V.A.M., J.S.R., R.J.H., J.P.G., and J.X.S. receive salary and have ownership interest (including patents) in Foundation Medicine, Inc.; S.L.M., S.A.H., and P.S.H. receive salary and have ownership interest in Genentech, Inc.; J.P.G. is on a speaker bureau for AstraZeneca and Bristol Myer Squib.
Samples used in this study are part of routine clinical testing, and raw data are not available for public release.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2019.06.011.
Supplemental Data
Gene enrichment in microsatellite instability–high (MSI-H) or MSI-stable (MSS) tumors. Enrichment of genes in MSI-H (log2 odds ratio > 1) or MSS (log2 odds ratio < 1). Circle sizes represent the frequency of gene alteration in either MSI-H tumors (orange) or MSS tumors (teal). Circles representing genes without significant enrichment (dark blue) are not scaled to size. Multiple hypotheses–corrected –log10 P values are displayed on the y axis. All genes with at least 25 samples are displayed. Pathway-related (Supplemental Table S6) or mismatch repair related genes with P < 0.05 are labeled. P values are capped at 1e-17 for visualization purposes. Colorectal carcinoma (CRC; A), carcinoma of unknown primary (B), endometrial (C), ovary (D), gastric (E), lower gastrointestinal (GI; non-CRC; F), lung (G), and prostrate (H) tumors are displayed.
Pathway enrichment in microsatellite instability–high (MSI-H) or MSI-stable (MSS) tumors. Enrichment of various pathways in multiple disease types, including variants occurring in regions of polymerase slippage. Log2 of odds ratio displayed by color: orange indicates enrichment in MSI-H, and teal indicates enrichment in MSS. Gray indicates nonsignificant P values for enrichment (P ≥ 0.05). CRC, colorectal carcinoma; CUP, carcinoma of unknown primary; GI, gastrointestinal; Hh, Hedgehog; JAK, Janus kinase/signal transducers and activators of transcription; PI3K, phosphatidylinositol 3-kinase.
References
- 1.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 2.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 3.Shah S.N., Hile S.E., Eckert K.A. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70:431–435. doi: 10.1158/0008-5472.CAN-09-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch H.T., Snyder C.L., Shaw T.G., Heinen C.D., Hitchins M.P. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 5.Sinicrope F.A., Foster N.R., Thibodeau S.N., Marsoni S., Monges G., Labianca R., Kim G.P., Yothers G., Allegra C., Moore M.J., Gallinger S., Sargent D.J. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelloo E., Bosse T., Nout R.A., MacKay H.J., Church D.N., Nijman H.W., Leary A., Edmondson R.J., Powell M.E., Crosbie E.J., Kitchener H.C., Mileshkin L., Pollock P.M., Smit V.T., Creutzberg C.L. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer: a TransPORTEC initiative. Mod Pathol. 2015;28:836–844. doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 7.Arabi H., Guan H., Kumar S., Cote M., Bandyopadhyay S., Bryant C., Shah J., Abdul-Karim F.W., Munkarah A.R., Ali-Fehmi R. Impact of microsatellite instability (MSI) on survival in high grade endometrial carcinoma. Gynecol Oncol. 2009;113:153–158. doi: 10.1016/j.ygyno.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Zighelboim I., Goodfellow P.J., Gao F., Gibb R.K., Powell M.A., Rader J.S., Mutch D.G. Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. J Clin Oncol. 2007;25:2042–2048. doi: 10.1200/JCO.2006.08.2107. [DOI] [PubMed] [Google Scholar]
- 9.Black D., Soslow R.A., Levine D.A., Tornos C., Chen S.C., Hummer A.J., Bogomolniy F., Olvera N., Barakat R.R., Boyd J. Clinicopathologic significance of defective DNA mismatch repair in endometrial carcinoma. J Clin Oncol. 2006;24:1745–1753. doi: 10.1200/JCO.2005.04.1574. [DOI] [PubMed] [Google Scholar]
- 10.Mackay H.J., Gallinger S., Tsao M.S., McLachlin C.M., Tu D., Keiser K., Eisenhauer E.A., Oza A.M. Prognostic value of microsatellite instability (MSI) and PTEN expression in women with endometrial cancer: results from studies of the NCIC Clinical Trials Group (NCIC CTG) Eur J Cancer. 2010;46:1365–1373. doi: 10.1016/j.ejca.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 11.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., Biedrzycki B., Donehower R.C., Zaheer A., Fisher G.A., Crocenzi T.S., Lee J.J., Duffy S.M., Goldberg R.M., de la Chapelle A., Koshiji M., Bhaijee F., Huebner T., Hruban R.H., Wood L.D., Cuka N., Pardoll D.M., Papadopoulos N., Kinzler K.W., Zhou S., Cornish T.C., Taube J.M., Anders R.A., Eshleman J.R., Vogelstein B., Diaz L.A., Jr. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark A. FDA News Release; Silver Spring, MD: 2017. FDA approves first cancer treatment for any solid tumor with a specific genetic feature. [Google Scholar]
- 13.Le D.T., Durham J.N., Smith K.N., Wang H., Bartlett B.R., Aulakh L.K. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boland C.R., Thibodeau S.N., Hamilton S.R., Sidransky D., Eshleman J.R., Burt R.W., Meltzer S.J., Rodriguez-Bigas M.A., Fodde R., Ranzani G.N., Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 15.Murphy K.M., Zhang S., Geiger T., Hafez M.J., Bacher J., Berg K.D., Eshleman J.R. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8:305–311. doi: 10.2353/jmoldx.2006.050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Rüschoff J., Fishel R., Lindor N.M., Burgart L.J., Hamelin R., Hamilton S.R., Hiatt R.A., Jass J., Lindblom A., Lynch H.T., Peltomaki P., Ramsey S.D., Rodriguez-Bigas M.A., Vasen H.F., Hawk E.T., Barrett J.C., Freedman A. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middha S., Zhang L., Nafa K., Jayakumaran G., Wong D., Kim H.R., Sadowska J., Berger M.F., Delair D.F., Shia J., Stadler Z., Klimstra D.S., Ladanyi M., Zehir A., Hechtman J.F. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precision Oncol. 2017;1:1–17. doi: 10.1200/PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salipante S.J., Scroggins S.M., Hampel H.L., Turner E.H., Pritchard C.C. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 19.Kautto E.A., Bonneville R., Miya J., Yu L., Krook M.A., Reeser J.W., Roychowdhury S. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. 2017;8:7452–7463. doi: 10.18632/oncotarget.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes-Ciriano I., Lee S., Park W.-Y., Kim T.-M., Park P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8:15180. doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonneville R., Krook M.A., Kautto E.A., Miya J., Wing M.R., Chen H.-Z., Reeser J.W., Yu L., Roychowdhury S. Landscape of microsatellite instability across 39 cancer types. JCO Precision Oncol. 2017;1:1–15. doi: 10.1200/PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J., Abdel-Wahab O., Nahas M.K., Wang K., Rampal R.K., Intlekofer A.M. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J., Huang F., He Y., Sun J., Tabori U., Kennedy M., Lieber D.S., Roels S., White J., Otto G.A., Ross J.S., Garraway L., Miller V.A., Stephens P.J., Frampton G.M. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashktorab H., Ahuja S., Kannan L., Llor X., Ellis N.A., Xicola R.M., Laiyemo A.O., Carethers J.M., Brim H., Nouraie M. A meta-analysis of MSI frequency and race in colorectal cancer. Oncotarget. 2016;7:34546–34557. doi: 10.18632/oncotarget.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell B.B., Light N., Fabrizio D., Zatzman M., Fuligni F., de Borja R. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak J.A., Yurgelun M.B., Bruce J.L., Rojas-Rudilla V., Hall D.L., Shivdasani P., Garcia E.P., Agoston A.T., Srivastava A., Ogino S., Kuo F.C., Lindeman N.I., Dong F. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19:84–91. doi: 10.1016/j.jmoldx.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang E., Chung D.C. Hereditary colon cancer: Lynch syndrome. Gut Liver. 2010;4:151–160. doi: 10.5009/gnl.2010.4.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruvka Y.E., Mouw K.W., Karlic R., Parasuraman P., Kamburov A., Polak P., Haradhvala N.J., Hess J.M., Rheinbay E., Brody Y., Koren A., Braunstein L.Z., D'Andrea A., Lawrence M.S., Bass A., Bernards A., Michor F., Getz G. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35:951. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin D.S., Zaretsky J.M., Escuin-Ordinas H., Garcia-Diaz A., Hu-Lieskovan S., Kalbasi A., Grasso C.S., Hugo W., Sandoval S., Torrejon D.Y., Palaskas N., Rodriguez G.A., Parisi G., Azhdam A., Chmielowski B., Cherry G., Seja E., Berent-Maoz B., Shintaku I.P., Le D.T., Pardoll D.M., Diaz L.A., Tumeh P.C., Graeber T.G., Lo R.S., Comin-Anduix B., Ribas A. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2016;7:188–201. doi: 10.1158/2159-8290.CD-16-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sveen A., Johannessen B., Tengs T., Danielsen S.A., Eilertsen I.A., Lind G.E., Berg K.C.G., Leithe E., Meza-Zepeda L.A., Domingo E., Myklebost O., Kerr D., Tomlinson I., Nesbakken A., Skotheim R.I., Lothe R.A. Multilevel genomics of colorectal cancers with microsatellite instability—clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med. 2017;9:46. doi: 10.1186/s13073-017-0434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albacker L.A., Wu J., Smith P., Warmuth M., Stephens P.J., Zhu P., Yu L., Chmielecki J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS One. 2017;12:e0176181. doi: 10.1371/journal.pone.0176181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pai S.G., Carneiro B.A., Mota J.M., Costa R., Leite C.A., Barroso-Sousa R., Kaplan J.B., Chae Y.K., Giles F.J. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. 2017;10:101. doi: 10.1186/s13045-017-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luke J.J., Bao R., Spranger S., Sweis R.F., Gajewski T. Correlation of WNT/β-catenin pathway activation with immune exclusion across most human cancers. J Clin Oncol. 2016;34:3004–3083. [Google Scholar]
- 37.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 38.Suraweera N., Robinson J., Volikos E., Guenther T., Talbot I., Tomlinson I., Silver A. Mutations within Wnt pathway genes in sporadic colorectal cancers and cell lines. Int J Cancer. 2006;119:1837–1842. doi: 10.1002/ijc.22046. [DOI] [PubMed] [Google Scholar]
- 39.Shitoh K., Furukawa T., Kojima M., Konishi F., Miyaki M., Tsukamoto T., Nagai H. Frequent activation of the beta-catenin-Tcf signaling pathway in nonfamilial colorectal carcinomas with microsatellite instability. Genes Chromosomes Cancer. 2001;30:32–37. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1065>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 40.Mirabelli-Primdahl L., Gryfe R., Kim H., Millar A., Luceri C., Dale D., Holowaty E., Bapat B., Gallinger S., Redston M. Beta-catenin mutations are specific for colorectal carcinomas with microsatellite instability but occur in endometrial carcinomas irrespective of mutator pathway. Cancer Res. 1999;59:3346–3351. [PubMed] [Google Scholar]
- 41.Grasso C.S., Giannakis M., Wells D.K., Hamada T., Mu X.J., Quist M. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–749. doi: 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharabi A., Kim S.S., Kato S., Sanders P.D., Patel S.P., Sanghvi P., Weihe E., Kurzrock R. Exceptional response to nivolumab and stereotactic body radiation therapy (SBRT) in neuroendocrine cervical carcinoma with high tumor mutational burden: management considerations from the Center for Personalized Cancer Therapy at UC San Diego Moores Cancer Center. Oncologist. 2017;22:631–637. doi: 10.1634/theoncologist.2016-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong J., Cho M., Sy M., Salgia R., Fakih M. Molecular profiling of metastatic colorectal tumors using next-generation sequencing: a single-institution experience. Oncotarget. 2017;8:42198–42213. doi: 10.18632/oncotarget.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santin A.D., Bellone S., Buza N., Choi J., Schwartz P.E., Schlessinger J., Lifton R.P. Regression of chemotherapy-resistant polymerase epsilon (POLE) ultra-mutated and MSH6 hyper-mutated endometrial tumors with nivolumab. Clin Cancer Res. 2016;22:5682–5687. doi: 10.1158/1078-0432.CCR-16-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodi F.S., O'Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C., Akerley W., van den Eertwegh A.J., Lutzky J., Lorigan P., Vaubel J.M., Linette G.P., Hogg D., Ottensmeier C.H., Lebbe C., Peschel C., Quirt I., Clark J.I., Wolchok J.D., Weber J.S., Tian J., Yellin M.J., Nichol G.M., Hoos A., Urba W.J. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene enrichment in microsatellite instability–high (MSI-H) or MSI-stable (MSS) tumors. Enrichment of genes in MSI-H (log2 odds ratio > 1) or MSS (log2 odds ratio < 1). Circle sizes represent the frequency of gene alteration in either MSI-H tumors (orange) or MSS tumors (teal). Circles representing genes without significant enrichment (dark blue) are not scaled to size. Multiple hypotheses–corrected –log10 P values are displayed on the y axis. All genes with at least 25 samples are displayed. Pathway-related (Supplemental Table S6) or mismatch repair related genes with P < 0.05 are labeled. P values are capped at 1e-17 for visualization purposes. Colorectal carcinoma (CRC; A), carcinoma of unknown primary (B), endometrial (C), ovary (D), gastric (E), lower gastrointestinal (GI; non-CRC; F), lung (G), and prostrate (H) tumors are displayed.
Pathway enrichment in microsatellite instability–high (MSI-H) or MSI-stable (MSS) tumors. Enrichment of various pathways in multiple disease types, including variants occurring in regions of polymerase slippage. Log2 of odds ratio displayed by color: orange indicates enrichment in MSI-H, and teal indicates enrichment in MSS. Gray indicates nonsignificant P values for enrichment (P ≥ 0.05). CRC, colorectal carcinoma; CUP, carcinoma of unknown primary; GI, gastrointestinal; Hh, Hedgehog; JAK, Janus kinase/signal transducers and activators of transcription; PI3K, phosphatidylinositol 3-kinase.