Highlights
-
•
We developed an automatic method to segment cardiac sub-structures for radiotherapy planning CTs.
-
•
The Dice Similarity Coefficients and Average Surface Distance were up to 97% and < 11 mm, respectively.
-
•
The whole heart showed the absolute dose difference < 0.3 Gy whereas the coronary arteries showed < 2.3 Gy in breast radiotherapy simulations.
-
•
No notable improvement in our method beyond 10 atlases and using the manual guide points.
Keywords: Cardiac structures, Automatic segmentation, Deformation, Breast radiotherapy
Abstract
Background and purpose
We developed an automatic method to segment cardiac substructures given a radiotherapy planning CT images to support epidemiological studies or clinical trials looking at cardiac disease endpoints after radiotherapy.
Material and methods
We used a most-similar atlas selection algorithm and 3D deformation combined with 30 detailed cardiac atlases. We cross-validated our method within the atlas library by evaluating geometric comparison metrics and by comparing cardiac doses for simulated breast radiotherapy between manual and automatic contours. We analyzed the impact of the number of cardiac atlas in the library and the use of manual guide points on the performance of our method.
Results
The Dice Similarity Coefficients from the cross-validation reached up to 97% (whole heart) and 80% (chambers). The Average Surface Distance for the coronary arteries was less than 10.3 mm on average, with the best agreement (7.3 mm) in the left anterior descending artery (LAD). The dose comparison for simulated breast radiotherapy showed differences less than 0.06 Gy for the whole heart and atria, and 0.3 Gy for the ventricles. For the coronary arteries, the dose differences were 2.3 Gy (LAD) and 0.3 Gy (other arteries). The sensitivity analysis showed no notable improvement beyond ten atlases and the manual guide points does not significantly improve performance.
Conclusion
We developed an automated method to contour cardiac substructures for radiotherapy CTs. When combined with accurate dose calculation techniques, our method should be useful for cardiac dose reconstruction of a large number of patients in epidemiological studies or clinical trials.
1. Introduction
The adventitious irradiation of cardiac substructures during radiation therapy can lead to a variety of cardiac complications including pericarditis, myocardial fibrosis, and coronary artery disease (CAD) [1], [2], [3]. Several studies [4], [5], [6] have discussed radiation dose-volume predictors of acute and late cardiovascular effects in patients. However, the mechanism of radiation-induced cardiac toxicity and which cardiac structures are involved is not clearly understood and data showing the relationship between radiation dose to heart substructures and subsequent cardiac complications is scarce [6]. To fill this gap in knowledge it is important to consider dose to the cardiac substructures in risk analysis rather than using the mean absorbed dose to the whole heart as a proxy. Such an effort will require accurate segmentation and dose estimation for cardiac substructures and is the subject of this paper [7], [8].
The whole heart is routinely contoured on planning computed tomography (CT) images during radiotherapy whenever heart radiation toxicity is of concern due to its proximity to the planning target volume (e.g., breast or lung cancer radiotherapy). However, cardiac substructures are not routinely delineated due to limited image quality of treatment planning CTs and time constraints. Automated heart segmentation methods have been reported in the literature by various research groups [9], [10], [11], [12], [13], [14]. However, most of the existing studies focus on geometrical performance of their methods rather than dosimetric performance. Furthermore, most of the current atlas-based methods use a limited number of cardiac atlases (less than 15) and it is not clear how many cardiac atlases are optimal. It is also essential to check if manual guide points [15] would improve performance.
The purpose of the current study was to develop an automatic segmentation method for performing cardiac substructure dose calculations on a large number of patients as needed for epidemiological studies or clinical trials. We cross-validated our method within the atlas library by evaluating standard geometric comparison metrics and by comparing cardiac substructure doses for simulated breast radiotherapy between manual and automatic contours. We analyzed the impact of the number of cardiac atlas in the library and the use of manual guide points on the performance of our method. We also tested the performance of our method for a simulated breast cancer radiotherapy application.
2. Methods and materials
2.1. Cardiac structure atlas
We created a library of cardiac structure atlases using 30 sets of contrast-enhanced diagnostic chest-abdomen-pelvis (CAP) CTs of adult female patients (age 31 ± 6.5 years and Body Mass Index 26 ± 5.9 kg/m2) obtained from the National Institutes of Health Clinical Center under an Internal Review Board (IRB)-exempt protocol. The scans were conducted on a Siemens SOMATOM Definition Flash scanner using standard imaging protocols. The imaging protocols varied slightly amongst the convenience sample of patients, but typical parameters were: 120 kVp, 0.33 s rotation time, 64 × 0.6 mm collimation, and pitch of 0.8. The following structures were manually segmented using the Eclipse treatment planning system (Varian Medical Systems, Palo Alto, CA) with reference to recognized cardiac delineation guidelines [16]: the whole heart (WH), the four heart chambers (left atrium (LA), right atrium (RA), left ventricle (LV), and right ventricle (RV)), and the four coronary arteries (left main coronary artery (LMCA), left anterior descending artery (LAD), left circumflex artery (LCX), and right coronary artery (RCA)). The contours were independently verified and edited by two practicing radiologists to ensure accuracy of the structure delineations. Fig. S1 shows a 3D rendering of an example heart atlas in perspective and left lateral views. Fig. S2 illustrates the distribution of volumes for each cardiac structure amongst the 30 hearts in the library. The mean volume (coefficient of variation) for the WH, LA, RA, LV, and RV was 577 cm3 (18%), 49 cm3 (29%), 57 cm3 (29%), 174 cm3 (20%), 100 cm3 (20%), respectively. For the arteries LMCA, LAD, LCX, and RCA these values were 0.2 cm3 (39%), 1.5 cm3 (18%), 1.0 cm3 (34%), 1.3 cm3 (38%), respectively.
2.2. Automatic segmentation method
Our method assumed, as a starting point, that we are provided with a patient CT having the WH segmented (DICOM-RT structure format) as is routine clinical practice during radiotherapy treatment planning. From this starting point, our method segmented the heart structures by a two-step process: selection of most-similar cardiac atlas followed by a 3D deformation of the most-similar atlas. First, the 30 cardiac atlases were linearly scaled with the origin point at the center of mass in the heart using the volume ratios between the WH of the given patient and each one of the 30 cardiac atlases. The algorithm selected the most similar atlas having the greatest Dice Similarity Coefficient (DSC) [17], a spatial overlap index between two objects ranging from zero (no overlap) to 100% (complete overlap), calculated between each of the 30 scaled atlases and the given WH. Next, non-rigid transformation between the patient WH contour (not CT images) and the selected atlas contour was performed by B-spline 3D deformation using elastix, an open-source software for rigid and non-rigid registration of image [18]. The contours of the WH and substructures of the selected atlas were transformed using the same transformation matrices. Finally, the resulting cardiac contours were written back into DICOM-RT structure format.
2.3. Leave-one-out cross-validation
To evaluate the performance of our automatic segmentation algorithm we conducted a leave-one-out cross-validation within the set of 30 cardiac atlases. One of the 30 atlases was used as a test case (true contours) and the remaining 29 atlases were used as the set of atlases from which the most similar atlas is selected. The accuracy of the automatic segmentation method was then quantified by calculating DSC or Average Surface Distance (ASD), the average of all distances between the surfaces of two objects. We repeated the comparison for each of the 30 atlases. For the WH and the four heart chambers, the DSC was calculated between manual and automatic segmentation. For the case of small structures such as the arteries, we opted instead to use the ASD.
2.4. Dosimetric performance for simulated breast cancer treatment
We also studied the dosimetric performance of our automatic segmentation algorithm when applied to left breast radiotherapy plans simulated for the 30 patients that were used for the cardiac atlas development. Because the selected patients were not CT scanned for purposes of breast radiotherapy planning, the posture of the patients, and subsequently their breast shape, was not exactly as one would expect for breast radiotherapy patients. For example, unlike typical breast patients, the female patients used in this study did not have any immobilization devices such as a breast board. Despite the differences, these patients were deemed suitable for this study because our primary purpose was to compare radiation dose between manually- and automatically-segmented contours—not to quantify typical dose from breast treatments. Using the Eclipse system, typical tangential fields (lateral + medial) treatment plans were created by a clinical medical physicist using 6 MV, 16 MV, or combination of the two photon energies to have the 95% isodose surface cover the entire left breast tissue as homogeneously as possible depending on the breast shape and size, while limiting the hotspots below 115% of the 50 Gy prescription dose. Due to the aforementioned limitation, a relatively simple wedge-pair treatment planning technique was used. Dose to the cardiac structures was calculated by the treatment planning system using the Analytical Anisoptric Algorithm (AAA) and compared between the manual and automatic contours.
2.5. Sensitivity analysis
We performed two sensitivity analyses to evaluate the impact of (1) the number of heart atlases employed in the algorithm and (2) the use of manually-defined guide points on the performance of our automatic segmentation method. First, we tested the hypothesis that the inclusion of more atlas sets would improve the accuracy of our automatic segmentation method. We randomly selected ten atlases from the 30-atlas library as test cases (ground truth). We applied our segmentation algorithm to the WH contour for these test cases and selected the most-similar atlas from a library containing 1, 5, 10, 15, or 20 atlases (randomly selected from the remaining 20 atlases). We calculated DSC and ASD for each case.
Second, we tested the hypothesis that the performance of our method would improve by using manually-placed guide points to aid the selection of the most similar heart atlas from our library. We manually placed nine guide points at key anatomical locations within the heart for each of the 30 cardiac atlases as follows (Fig. S3): apex midpoint (ApexMid), aortic valve (AorticValve), pulmonary valve (PulmValve), junction of LAD and left coronary artery (JunctLADLCA), junction of aorta and RCA (JunctAortaRCA), junction of coronary sinus and RA (CorSinus), junction of right lower lobe pulmonary vein and LA (RLowPulmVein), moderator band (ModBand), and bifurcation of pulmonary artery (Bifurcation). Our segmentation method was then revised to select the most similar atlas having the smallest root-mean-square-error (RMSE) between the two sets of guide points (after linear scaling and B-spline 3D deformation). We then repeated the same leave-one-out cross-validation described in Section 2.3 but this time using the new guide point-based atlas selection method. We then compared DSC and ASD from this new approach to the values from the original method, the WH volume-based selection described in Section 2.2.
3. Results
3.1. Automatic segmentation of cardiac structures
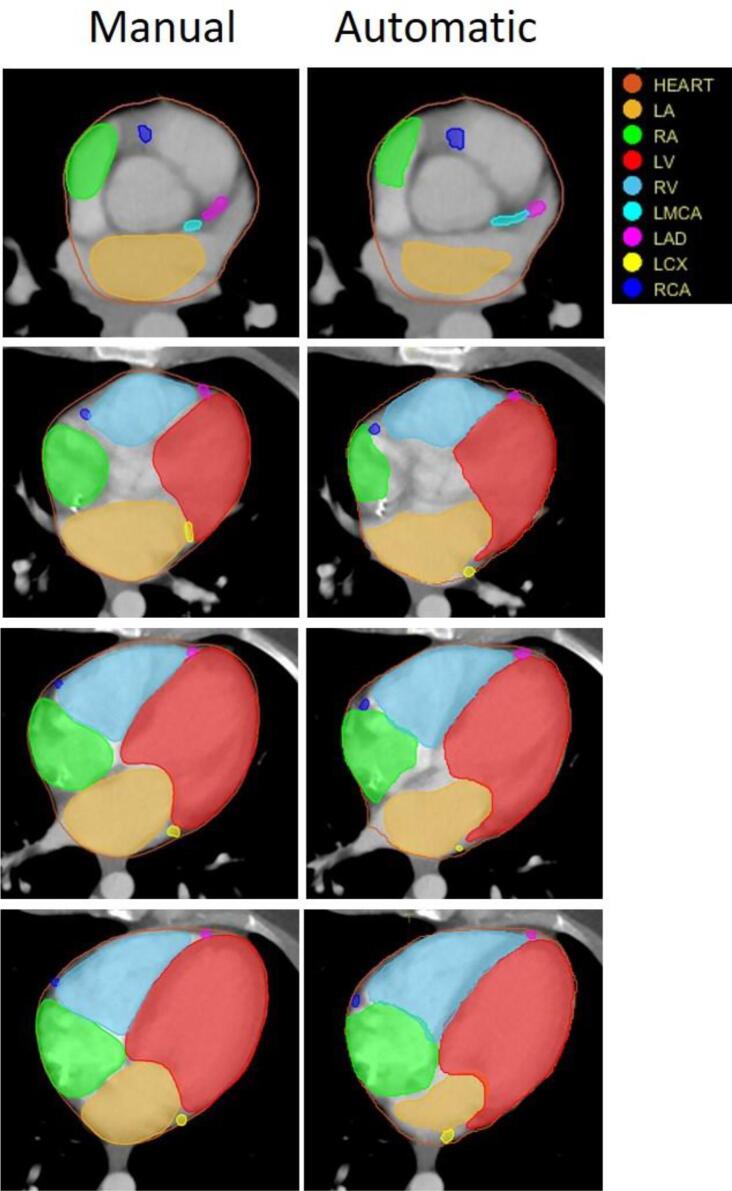
Fig. 1 shows transverse cross-sectional views of the manually-segmented (left column) and automatically-segmented (right column) cardiac structures at four different z-axis positions (inferior-superior) for an example atlas. The four chambers and four arteries are shown. Total processing time per case was less than ten minutes without manual intervention.
Fig. 1.
Transverse cross sectional views of manually-drawn (left) and automatically-generated (right) heart substructures at four different z-axis positions (inferior-superior) from the top to the bottom of the heart.
3.2. Geometric and dosimetric performance
On average, the DSC for the WH was 97%, while an average of about 70% was achieved for the four chambers (Table 1). The WH had the best ASD, 1.0 mm, and the manual and automatic segmentation of the four chambers matched within about 4.5 mm on average. The ASD for the four coronary arteries was, on average, less than 10.3 mm, with the LAD showing the best agreement with an average of 7.3 mm. Comparing doses for manual and automatically drawn contours, the differences for the WH and atria were less than 0.06 Gy and for the ventricles less than 0.3 Gy, on average. For the coronary arteries the differences were less than 0.3 Gy (LMCA, LCX, and RCA) and 2.3 Gy (LAD), on average (Table 2). However, individual performance varied and in worst case was 0.2 Gy for the whole heart, 0.2 Gy for the RA, 2.1 Gy for the RV, 0.7 Gy for LCX, and 16.9 Gy for LAD. Median dose difference for the LAD was 1 Gy.
Table 1.
Mean and standard deviation (SD) of the DSC (%) and ASD (mm) for the cardiac structures derived by manual and automatic segmentation.
| Cardiac Structure | DSC (%) |
ASD (mm) |
||
|---|---|---|---|---|
| Mean (SD) | Min – Max | Mean (SD) | Min – Max | |
| WH | 97 (1) | 96–98 | 1.0 (0.2) | 0.7–1.5 |
| LA | 67 (9) | 49–84 | 4.4 (1.5) | 2.3–10.8 |
| RA | 65 (10) | 44–78 | 4.7 (1.7) | 2.7–11.0 |
| LV | 80 (6) | 65–90 | 4.3 (1.5) | 2.2–9.1 |
| RV | 69 (8) | 51–81 | 4.7 (1.2) | 2.7–7.5 |
| LMCA | NA | NA | 8.4 (5.6) | 2.9–24.8 |
| LAD | NA | NA | 7.3 (2.5) | 4.2–13.4 |
| LCX | NA | NA | 9.2 (4.4) | 3.2–22.3 |
| RCA | NA | NA | 10.3 (4.4) | 4.7–24.6 |
Table 2.
Comparison of the mean (standard deviation) cardiac structure doses from manual and automatic segmentations.
| Cardiac Structures | Manual Dose (Gy) |
Automatic Dose (Gy) |
Absolute Dose Difference (Gy) |
||||
|---|---|---|---|---|---|---|---|
| Mean (SD) | Min – Max | Mean (SD) | Min – Max | Mean (SD) | Median | Max | |
| WH | 2.4 (1.2) | 1.1–5.9 | 2.5 (1.2) | 1.1–6.1 | 0.05 (0.05) | 0.03 | 0.19 |
| LA | 1.2 (0.3) | 0.8–2.0 | 1.2 (0.3) | 0.8–1.9 | 0.06 (0.04) | 0.05 | 0.14 |
| RA | 1.1 (0.4) | 0.5–1.8 | 1.0 (0.4) | 0.5–1.8 | 0.06 (0.05) | 0.05 | 0.19 |
| LV | 3.3 (2.0) | 1.5–9.8 | 3.1 (1.7) | 1.4–8.0 | 0.3 (0.6) | 0.1 | 2.6 |
| RV | 2.1 (1.0) | 0.9–5.8 | 2.2 (1.1) | 0.9–5.8 | 0.3 (0.4) | 0.1 | 2.1 |
| LMCA | 1.8 (0.5) | 1.0–2.7 | 1.7 (0.5) | 1.0–3.0 | 0.3 (0.2) | 0.3 | 0.7 |
| LAD | 8.8 (8.6) | 2.4–33.1 | 8.2 (6.4) | 2.7–25.3 | 2.3 (3.6) | 1.0 | 16.9 |
| LCX | 1.8 (0.4) | 1.2–2.8 | 1.8 (0.5) | 1.2–3.2 | 0.2 (0.2) | 0.1 | 0.7 |
| RCA | 1.3 (0.5) | 0.6–2.1 | 1.3 (0.5) | 0.5–2.6 | 0.2 (0.1) | 0.1 | 0.5 |
3.3. Sensitivity analysis
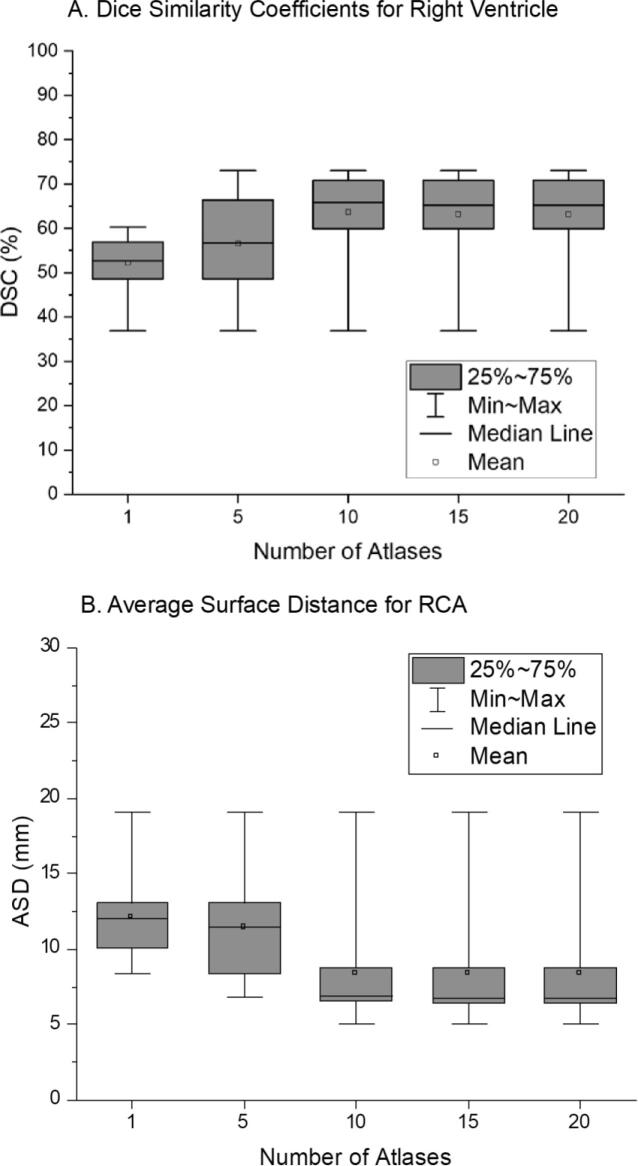
We observed that the average DSC for the four chambers improved as the number of atlases increased, but the performance plateaued for libraries containing more than ten atlases. The average DSC (Fig. 2A) of the ten-atlas method is 22% greater than that of the one-atlas method and two-sample t-testing found this difference to be statistically significant (p = 0.0104). Average ASD overall decreased as the number of atlases increased. The average ASD for the right coronary artery (RCA) (Fig. 2B) for the ten-atlas method was significantly lower by 30% (p = 0.0316) than that for the one-atlas method. For both DSC and ASD, no notable improvement was observed beyond the ten atlases. Therefore we conclude that results in Table 1, Table 2 would likely not be improved by adding more heart atlases to the library, suggesting that, within the uncertainty of our method, 10 unique hearts geometries may form a sufficient set to cover most patients.
Fig. 2.
(a) Dice similarity coefficients (DSC) for right ventricle (RV) and (b) average surface distance (ASD) for right coronary artery (RCA) for when the number of atlases in the library is increased from 1 to 20.
We did not observed statistically significant differences in DSC (Table S1) or ASD (Table S2) when using the guide point-based selection method. The DSC for the four chambers with the guide points was highly variable and was sometimes slightly worse (average −2% ± 9%). The ASD was slightly improved for the arteries (average 1.2 ± 5.2 mm), but differences were not significant. The guide point-based selection method showed borderline significant improvement in the accuracy of mean dose against the WH volume-based selection method only for LMCA (p = 0.049) (Table S3).
4. Discussion
The purpose of the current study was to develop an automatic method to segment cardiac substructures given a radiotherapy planning CT to support epidemiological studies or clinical trials looking at cardiac disease endpoints after radiotherapy. Ultimately our aim is to apply this method to help identify the cardiac substructures most responsible for late adverse health effects observed in some radiotherapy patients. Such information could be used to optimize radiotherapy plans to reduce cardiac toxicity, especially when relatively high heart doses may be involved such as is sometimes the case for patients treated for breast cancer, Wilms tumor, Hodgkin’s lymphoma, and lung cancer.
A fundamental challenge of this study (and patient organ segmentation studies in general) is that ground truth anatomy is not known; rather the gold standard segmentation is typically taken as that delineated by a trained observer on the best quality images available. However, the heart is not static within the patient. Many previous studies have shown that manual segmentation suffers from significant inter- and intra-observer variability [11], [19]. Furthermore, it should be noted that it takes significant time and patience to produce high-quality manual contours, even for a trained observer. A key strength of our automatic segmentation method is that it takes advantage of a large library of 30 detailed cardiac atlases developed and revised with great effort and care from patient images over the course of months—effort well beyond what would be feasible as part of routine clinical practice. The auto-segmented hearts resulting from our method, after applying linear scaling and 3D deformation, retain the aesthetic features of the detailed atlases in our library (e.g. no skipped CT slices and smooth interpolation between CT slices). The final segmented heart structures are written into the original DICOM-RT STRUCTURE file. Total processing time per case was less than ten minutes without manual intervention, and with additional effort can likely be made to run faster (speed was not the focus of the current study).
We found that our auto-segmentation results are comparable to those from previous studies [9], [11]. The DSC from the cross-validation was as high as 97% and 70% for the whole heart and four chambers, respectively. The ASD for the LAD among the coronary arteries showed the best agreement, 7.3 mm; however, these differences still translated into quite a large dose discrepancy (mean: 2.3 Gy). It was previously shown that there is a large inter-observer variability of 6.1 ± 6.4 mm for manual LAD contouring [11]. The length of LAD often significantly affects dose calculation, especially for left breast cancer patient. The determination of top and bottom of LAD is critical for contouring because mid and distal LAD could be close to the in-field radiation [20].
Our hypothesis that the greater number of atlases and manually-placed guide points would improve the performance of the automatic segmentation method was not supported by the results from our sensitivity analysis. The use of guide point-based selection method slightly reduced DSC for the four chambers; however, ASD was slightly improved for the four arteries, but not significantly. Dose comparison between manual and automatic segmentation methods showed borderline significant improvement only for the LMCA. Indeed, there is uncertainty in the placement of the anatomical guide points and single-point landmarks are not optimal for long, cylindrical arteries.
We acknowledge a couple of limitations in our method. First, our method assumes that the WH segmentation is provided as a starting point. Therefore, the method cannot be used for radiotherapy patients when the WH is not contoured in treatment planning. However, the WH is manually contoured in most of radiation treatments where cardiac dose is of concern. Second, our segmentation process fully relies on the accuracy of the given WH contour. One cannot expect accurate segmentation of the cardiac substructures if the quality of the WH contour is poor. In those cases, manual correction of the WH contour is required before our segmentation algorithm is applied. Third, we tested the performance of our algorithm only for breast cancer radiotherapy in the current study but plan to extend the investigation to other types of radiotherapy. Lastly, we used diagnostic CT images for simulating breast cancer radiotherapy. Even though the treatment fields are carefully created to closely mimic typical breast radiation therapy, the cardiac dose data calculated from the performance evaluation do not necessarily represent actual breast radiotherapy patients.
In summary, we developed an automatic method to segment cardiac structures on radiotherapy CT images when the WH is already contoured. We confirmed the excellent geometric and dosimetric performance of our method from cross-validation and breast radiotherapy simulations.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by the intramural research program of the National Institutes of Health (NIH), National Cancer Institute, Division of Cancer Epidemiology and Genetics. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors thank Mr. Christopher Pelletier for his contribution to the conceptual design of the cardiac structure segmentation method using guide points.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2019.11.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Heidenreich P.A., Hancock S.L., Vagelos R.H., Lee B.K., Schnittger I. Diastolic dysfunction after mediastinal irradiation. Am Heart J. 2005;150:977–982. doi: 10.1016/j.ahj.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Schultz-Hector S., Trott K.-R. Radiation-induced cardiovascular diseases: is the epidemiologic evidence compatible with the radiobiologic data? Int J Radiat Oncol Biol Phys. 2007;67:10–18. doi: 10.1016/j.ijrobp.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 3.Stewart J.R., Fajardo L.F., Gillette S.M., Constine L.S. Radiation injury to the heart. Int J Radiat Oncol Biol Phys. 1995;31:1205–1211. doi: 10.1016/0360-3016(94)00656-6. [DOI] [PubMed] [Google Scholar]
- 4.Darby S.C., Cutter D.J., Boerma M., Constine L.S., Fajardo L.F., Kodama K. Radiation-related heart disease: current knowledge and future prospects. Int J Radiat Oncol Biol Phys. 2010;76:656–665. doi: 10.1016/j.ijrobp.2009.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veinot J.P., Edwards W.D. Pathology of radiation-induced heart disease: a surgical and autopsy study of 27 cases. Hum Pathol. 1996;27:766–773. doi: 10.1016/S0046-8177(96)90447-5. [DOI] [PubMed] [Google Scholar]
- 6.Gagliardi G., Constine L.S., Moiseenko V., Correa C., Pierce L.J., Allen A.M. Radiation dose-volume effects in the heart. Int J Radiat Oncol Biol Phys. 2010;76:S77–S85. doi: 10.1016/j.ijrobp.2009.04.093. [DOI] [PubMed] [Google Scholar]
- 7.NCRP. Second primary cancers and cardiovascular disease after radiotherapy. Bethesda, MD: 2012.
- 8.Aznar M.C., Korreman S.-S., Pedersen A.N., Persson G.F., Josipovic M., Specht L. Evaluation of dose to cardiac structures during breast irradiation. Brit J Radiol. 2011;84:743–746. doi: 10.1259/bjr/12497075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirisli H.A., Schaap M., Klein S., Neefjes L.A., Weustink A.C., Walsum T.V. Fully automatic cardiac segmentation from 3D CTA data: a multi-atlas based approach. Int Soc Opt Photonics. 2010:762305. doi: 10.1117/12.838370. [DOI] [Google Scholar]
- 10.Ecabert O., Peters J., Schramm H., Lorenz C., von Berg J., Walker M.J. Automatic model-based segmentation of the heart in CT images. IEEE Trans Med Imaging. 2008;27:1189–1201. doi: 10.1109/TMI.2008.918330. [DOI] [PubMed] [Google Scholar]
- 11.Zhou R., Liao Z., Pan T., Milgrom S.A., Pinnix C.C., Shi A. Cardiac atlas development and validation for automatic segmentation of cardiac substructures. Radiother Oncol. 2017:1–6. doi: 10.1016/j.radonc.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Dijk-Peters F.B.J., Sijtsema N.M., Kierkels R.G.J., Vliegenthart R., Langendijk J.A., Maduro J.H. Validation of a multi-atlas based auto-segmentation of the heart in breast cancer patients. Radiother Oncol. 2015;115:S132–S133. doi: 10.1016/S0167-8140(15)40257-9. [DOI] [Google Scholar]
- 13.Eldesoky A.R., Yates E.S., Nyeng T.B., Thomsen M.S., Nielsen H.M., Poortmans P. Internal and external validation of an ESTRO delineation guideline – dependent automated segmentation tool for loco-regional radiation therapy of early breast cancer. Radiother Oncol. 2016;121:424–430. doi: 10.1016/j.radonc.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Kaderka R., Gillespie E.F., Mundt R.C., Bryant A.K., Sanudo-Thomas C.B., Harrison A.L. Geometric and dosimetric evaluation of atlas based auto-segmentation of cardiac structures in breast cancer patients. Radiother Oncol. 2018 doi: 10.1016/j.radonc.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Auger D.A., Zhong X., Epstein F.H., Meintjes E.M., Spottiswoode B.S. Semi-automated left ventricular segmentation based on a guide point model approach for 3D cine DENSE cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2014;16:8. doi: 10.1186/1532-429X-16-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng M., Moran J.M., Koelling T., Chughtai A., Chan J.L., Freedman L. Development and validation of a heart atlas to study cardiac exposure to radiation following treatment for breast cancer. Int J Radiat Oncol Biol Phys. 2011;79:10–18. doi: 10.1016/j.ijrobp.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dice L.R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- 18.Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 19.van den Bogaard V.A.B., Ta B.D.P., van der Schaaf A., Bouma A.B., Middag A.M.H., Bantema-Joppe E.J. Validation and modification of a prediction model for acute cardiac events in patients with breast cancer treated with radiotherapy based on three-dimensional dose distributions to cardiac substructures. J Clin Oncol. 2017;35:1171–1178. doi: 10.1200/JCO.2016.69.8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson G., Holmberg L., Garmo H., Duvernoy O., Sjögren I., Lagerqvist B. Distribution of coronary artery stenosis after radiation for breast cancer. J Clin Oncol. 2012;30:380–386. doi: 10.1200/JCO.2011.34.5900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.