Abstract
Background and Purpose
Four-dimensional (4D) computed tomography (CT) is widely used in radiotherapy (RT) planning and remains the current standard for motion evaluation. We assess a 4D magnetic resonance imaging (MRI) sequence in terms of motion and image quality in a phantom, healthy volunteers and patients undergoing RT.
Materials and Methods
The 4D-MRI sequence is a prototype T1-weighted 3D gradient echo with radial acquisition with self-gating. The accuracy of the 4D-MRI respiratory sorting based method was assessed using a MRI-CT compatible respiratory simulation phantom. In volunteers, abdominal viscera were evaluated for artefact, noise, structure delineation and overall image quality using a previously published four-point scoring system. In patients undergoing abdominal RT, the tumour (or a surrogate) was utilized to assess the range of motion on both 4D-CT and 4D-MRI. Furthermore, imaging quality was evaluated for both 4D-CT and 4D-MRI.
Results
In phantom studies 4D-MRI demonstrated amplitude of motion error of less than 0.2 mm for five, seven and ten bins. 4D-MRI provided excellent image quality for liver, kidney and pancreas. In patients, the median amplitude of motion seen on 4D-CT and 4D-MRI was 11.2 mm (range 2.8–20.3 mm) and 10.1 mm (range 0.7–20.7 mm) respectively. The median difference in amplitude between 4D-CT and 4D-MRI was −0.6 mm (range −3.4–5.2 mm). 4D-MRI demonstrated superior edge detection (median score 3 versus 1) and overall image quality (median score 2 versus 1) compared to 4D-CT.
Conclusions
The prototype 4D-MRI sequence demonstrated promising results and may be used in abdominal targeting, motion gating, and towards implementing MRI-based adaptive RT.
Keywords: MRI; Motion management; 4D-MRI, abdominal radiotherapy; Four dimensional magnetic resonance imaging
1. Introduction
Four-dimensional (4D) computed tomography (CT) is widely used in radiotherapy (RT) and remains the current standard for motion evaluation and delineation of an internal target volume (ITV) as per ICRU62 [1]. Unfortunately, 4D-CT has additional radiation exposure to a three-dimensional (3D) planning CT alone, and has poor soft tissue contrast. 4D-CT inadequately models respiratory motion for approximately 30% of patients [2]. Magnetic resonance imaging (MRI) has emerged as a potential replacement for CT in the RT planning process [3], [4]. MRI has advantages of avoiding radiation exposure, and offering superior soft tissue delineation compared to CT, both are of enormous value in abdominal RT given the proximity of a number of organs at risk (OAR) and its potential use for adaptive replanning and real-time image guidance. The development of a clinically useful 4D-MRI sequence in abdominal radiotherapy has received much attention [5], [6], [7], [8].
Until recently, ITV formation using MRI has involved unacceptable trade-offs between spatial resolution, acquisition time and signal to noise ratio. The majority of attempts to provide motion information with MRI have included acquisition of dynamic 2D-MRI images across all phases of respiratory cycle [9], [10], [11], sorting and reconstructing k space data [7], [8], retrospective binning of 2D or 3D-MRI images [12], and use of motion vectors applied to 2D cine or 3D images [7], [13].
True 4D MRI has recently become possible thanks to efficient sampling schemes using radial projections which acquire successive data through centre of k-space.
In this paper we present a sequence utilising a stack of stars trajectory for use in abdominal radiotherapy planning. This sequence uses incremental projections taken at the ‘golden angle’ which provides an optimal distribution of the radial data. We compare motion information between 4D-CT and the 4D-MRI sequence in phantom and patients. Using a previously published scoring system, we assess the 4D-MRI sequence image quality in the parameters of noise, artefact, structure delineation and overall image quality for upper abdominal viscera in healthy volunteers. We also assess and compare imaging quality between 4D-CT and 4D-MRI in patients. There is little published literature comparing image quality and motion information of 4D-MRI acquired through a stack of stars trajectory, with 4D-CT.
2. Materials and methods
The prototype 4D-MRI sequence was tested in: (1) a respiratory simulation phantom; (2) ten healthy volunteers; and (3) ten patients undergoing abdominal RT.
For human imaging, volunteers and patients were scanned in an abdominal RT planning position with a customized vacuum bag (BlueBAG, Elekta, Stockholm, Sweden) for immobilization and a flat wing board (MTWB09 Wingboard, CIVCO Medical Solutions, Orange City, IA) and arms above the head. All MR imaging was performed on the departmental RT dedicated 3.0 Tesla simulator wide bore MRI (Siemens MAGNETOM Skyra, Erlangen, Germany) with a flatbed insert (CIVCO Medical Solutions, Orange City, IA) using a 32-channel posterior in-table RF coil and 18-channel flexible RF coil array. Sequences included T2-w HASTE gated with phase navigation, breath hold T1-w volumetric interpolated breath hold examination (VIBE) and multiphasic (Arterial, venous and transitional phases) breath-hold T1-w VIBE enhanced with 0.1 ml/kg Gadobutrol (Gadovist, Bayer). Additionally, the new 4D-MRI sequence was acquired in all volunteers and patients. The 4D-MRI sequence is a respiratory phase-resolved 3D T1-w gradient echo (VIBE) radial acquisition with self-gating (Siemens, Erlangen, Germany). K-space sampling was performed using a stack-of-stars radial trajectory with golden angle increment [14] (radial encoding for read-out and Cartesian encoding for slices). The sequence used data from the centre of k-space to extract a respiratory motion surrogate signal, which permits self-gating [15], [16]. The self-gating respiratory signal was then used for retrospective binning of the k-space data to reconstruct the images in defined number of breathing states (respiratory phases). The RT treatment planning system (TPS) only permits contouring on 4D-CT in the axial plane. In order to directly compare 4D-CT and 4D-MRI, images were acquired in axial plane. The 4D-MRI sequence was acquired with resolution of 1.2 × 1.2 × 3.0 mm3, 9 degrees flip angle, TR/TE of 4.33/1.98 ms, 320 × 320 base resolution with 2000 radial views and 380 × 380 mm2 field of view.
The 4D-MRI sampling scheme followed a “stack-of-stars” trajectory with radial read-out and Cartesian encoding of the slices. All slices were encoded for a given azimuth angle and then the acquisition angle was rotated by 111.25 degree (golden angle) [14]. Thus, for every acquired angle or “radial views”, one readout was acquired in kz = 0 plane (k-space centreline). Thus with N radial views, N k-space centrelines were used to calculate a respiratory motion surrogate signal with N samples.
Based on the surrogate signal, the data from all radial views was equally divided into defined number of respiratory phases (number of bins) based on the respiratory amplitude where the data was acquired. For example, if the data was sorted into five bins, five respiratory amplitude bins could be resolved. The respiratory amplitude bin was defined based on the amplitude between end-exhale to end-inhale, not considering upward or downward side of the signal, i.e. hysteresis was not included in current version of the prototype. Each respiratory amplitude bin contained same amount of data. All the 4D-MRI reconstruction was performed on the scanner and all the images took on average 95 s to be reconstructed online. Gradient non-linearity such as subject induced distortions due to susceptibility and chemical shift effects were not accounted and corrected in this version of the work in progress (WIP) but has been planned for future version of the WIP release.
2.1. Phantom study
The accuracy of the 4D-MRI sequence was assessed using an MRI-CT compatible respiratory simulation phantom (QUASAR™, Modus Medical Devices Inc., London, Canada). The respiratory simulation platform was driven to move a 3 cm diameter sphere target filled with gadolinium doped water with various sinusoidal waveforms (10–15 mm amplitude, 10–20 breaths per minute) and two patient specific respiratory waveforms. The 4D-MRI acquisition time was 3 min in the phantom study. The phantom experiment was performed on a CT simulator (Brilliance™ CT, Philips Medical Systems, The Netherlands) using the gold standard 4D-CT technique that produces ten respiratory bins over a complete expiration to expiration cycle. The experiments were repeated using the 4D-MRI sequence with reconstruction of three, five, seven and ten bins, over expiration to inspiration. All 4D datasets were imported into image analysis software (MiM MaestroTM, Cleveland, USA) for target contouring. Four dimensional structure sets were created by propagating the manually drawn contours from one frame of 4D data series to other frames and compared to known values. Four dimensional structure sets were created using deformable propagation with manual editing to contour sphere on all phases.
2.2. Volunteer study
Ten healthy volunteers were scanned using the prototype 4D-MRI sequence using a stereotactic body radiation therapy (SBRT) positioning system (Omni V®, Bionix Radiation Therapy, Ohio, USA). The first two volunteers were scanned with 4D-MRI using three bins, five bins and ten bins. With information from phantom study and first two volunteers, an optimal number of bins was determined for use in the subsequent volunteers by two radiation oncologists and MRI radiographer. The optimal number of bins was determined for clinical use and provided an appropriate compromise between imaging quality and motion information. The volunteers were not given any respiratory coaching, and were instructed to breathe normally. Artefact, noise, structure delineation and image quality were graded on a previously published four-point scale [17] for 4D-MRI (see Supplementary Material) for liver, right kidney, duodenum and pancreas by two radiation oncologists and an MRI radiographer. Lower scores in this scoring system indicate a higher quality image. Scores were obtained by consensus. Artefact was defined as presence of signal wrap, ghosting and distortion that impairs the definition of structures on the MRI, or motion artefact resulting in poor reconstruction of the 4D segmented scan. Image noise refers to the visible reduction in signal-to-noise ratio. Structure delineation was a composite score assessing ability to delineate structure and is impacted by noise and artefact. An overall image quality score was also given.
2.3. Patient study
Patients undergoing abdominal RT including 4D-CT and a planning MRI were consented prospectively via an opt-out process for an additional ten-minute 4D-MRI sequence. Ethics approval was obtained through South Western Sydney Local Health District (SWSLHD) Human Research and Ethics Committee (HREC) (LNR/17/LPOOL/259). 4D-MRI was performed on the same day immediately following the planning CT and 4D-CT.
Clinical and demographic details were collected including gender, histology, primary site and site of RT from patient electronic medical records and RT TPS (Pinnacle3, Philips Medical Systems, The Netherlands). Each patient had tumour (or a surrogate within the region of interest (ROI) in the case of adjuvant RT) selected for the determination of amplitude of motion. Volumes were contoured by consensus in MiMTM by two radiation oncologists on maximal inspiratory and expiratory phases for both 4D-CT and 4D-MRI. The total distance between centroids was calculated using Euclidian distances for both modalities. Differences in amplitude of movement were recorded by subtracting 4D-MRI Euclidian distance from 4D-CT distance eg. A positive value would indicate that 4D-CT shows greater amplitude of movement. Artefact, noise, edge detection and image quality were graded on a four-point scale for both 4D-MRI and 4D-CT. The lower the score, the more superior the imaging quality for that particular parameter eg. Score of one superior to score of three. This scoring system has been utilised previously [17].
3. Results
3.1. Phantom study
For all phantom scans the mean acquisition time was 172 seconds (range 169–175 seconds). With the exception of the 4D-MRI sequence using three bins, the 4D-CT and 4D-MRI mapped amplitude of movement to within 0.2 mm of the ground truth. The difference between the three bin 4D-MRI and the ground truth ranged from 1.1 to 1.6 mm (see Supplementary Material). Outside of the three bin 4D-MRI, minimal impact of amplitude accuracy was seen with increased breaths per minute (See Fig. 1 and Supplementary Material). 4D-CT and 4D-MRI were comparable for motion detection for the five bin, seven bin and ten bin 4D-MRI, with difference being less then 0.1 mm for the amplitudes and BPM tested.
Fig. 1.
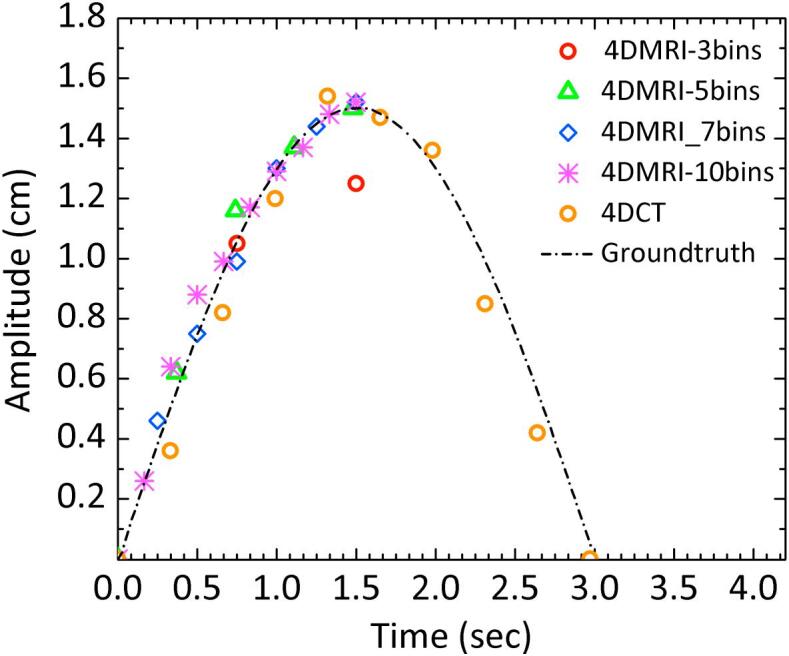
Plot of amplitude against time from the 4D-MRI and 4D-CT datasets acquired with the respiratory simulation phantom. The ground truth waveform (15 mm, 20 bpm) has also been shown for comparison.
3.2. Volunteer study
For all volunteers and patients the mean acquisition time was 305 seconds (range 263–314 seconds). The average reconstruction time was 95 s for all respiratory phases. After scanning the first two volunteers, it was apparent that the 4D-MRI sequence using three bins provided inadequate motion information. Increased radial streaking artefact seen with ten bins was also noted (see Fig. 2). The subsequent eight volunteers were scanned with 4D-MRI sequence using five bins. Duodenum had a median score of 3 for artefact, and 3 for noise. The median score for structure delineation was superior for liver (median score 1.5) compared to right kidney, pancreas and duodenum (median score 2, 2, and 2.5 respectively). The median overall image quality score for duodenum (median score 3) was inferior to liver, right kidney and pancreas which all had a median score of 2 (see Supplementary Material).
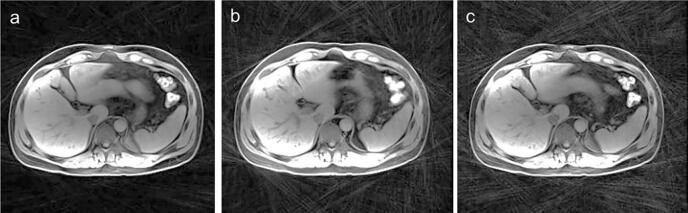
Fig. 2.
The radial streak artifact seen with three (a), five (b) and ten (c) bins. All images using 2000 radial views.
3.3. Patient study
Clincal and demographic details are provided in Table 1. The tumour was used to measure amplitude in all patients receiving definitive RT. All patients receiving definitive RT had the tumour used to measure amplitude, whilst for the two receiving adjuvant radiotherapy a surgical clip was used to measure motion at the region of interest.
Table 1.
Clinical and demographic details of ten patients scanned using both 4D-CT and 4D-MRI. Pt: Patient; Adeno: Adenocarcinoma.
| Patient | Age/Sex | Primary | Histology | Site of RT | ROI |
|---|---|---|---|---|---|
| 1 | 78M | Colorectal | Adeno | Liver | Surgical clip |
| 2 | 51F | Breast | Adeno | Liver | Tumour |
| 3 | 61F | Pancreas | Adeno | Pancreas | Tumour |
| 4 | 53M | Pancreas | Adeno | Pancreas | Tumour |
| 5 | 52F | Pancreas | Adeno | Pancreas | Tumour |
| 6 | 59F | Lung | Adeno | Right Adrenal | Tumour |
| 7 | 27M | Bile Ducts | Adeno | Bile Ducts | Surgical clip |
| 8 | 72F | Breast | Adeno | Liver | Tumour |
| 9 | 63F | Bowel | Adeno | Para-aortic node | Tumour |
| 10 | 67F | Breast | Adeno | Liver | Tumour |
The median amplitude of motion for 4D-CT and 4D-MRI was 11.2 mm (range 2.8 mm to 20.3 mm) and 10.1 mm (range 0.7 mm to 20.7 mm) respectively (see Table 2). The difference in motion between 4D-CT and 4D-MRI ranged from −3.4 to + 5.2 mm (median difference −0.6 mm). 4D-CT showed a greater amplitude of motion for five of the ten patients, and 4D-MRI showed greater amplitude of motion for five of ten patients.
Table 2.
Amplitude of tumour (or surrogate) motion in three directions, as well as total Euclidian Distance for 4D-CT and 4D-MRI. Difference in amplitude also shown, with “+” demonstrating greater movement on 4D-CT and “-“ demonstrating greater movement on 4D-MRI.
| Pt # | Centroid of Tumour (or ROI) movement (mm) |
Total Euclidian Distance (mm) |
Diff (mm) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 4D-CT |
4D-MRI |
||||||||
| ΔX | ΔY | ΔZ | ΔX | ΔY | ΔZ | 4D-CT | 4D-MRI | ||
| 1 | 3.6 | 4.5 | 11.5 | 4.7 | 1.3 | 8.6 | 12.9 | 9.9 | +3.0 |
| 2 | 3.6 | 3.9 | 19.6 | 5.3 | 5.9 | 19.1 | 20.3 | 20.7 | -0.4 |
| 3 | 2.0 | 1.7 | 1.0 | 3.1 | 0.7 | 2.8 | 2.8 | 4.2 | -1.4 |
| 4 | 6.1 | 4.9 | 24.3 | 0.4 | 0.6 | 20.3 | 25.5 | 20.3 | +5.2 |
| 5 | 4.7 | 4.4 | 7.0 | 1.4 | 0.6 | 10.1 | 9.5 | 10.2 | -0.7 |
| 6 | 0.2 | 1.2 | 3.5 | 0.1 | 0.0 | 0.7 | 3.7 | 0.7 | +3.0 |
| 7 | 2.5 | 2.4 | 3.8 | 1.9 | 1.7 | 8.0 | 5.1 | 8.4 | -3.3 |
| 8 | 3.1 | 6.3 | 12.6 | 2.9 | 9.0 | 15.1 | 14.4 | 17.8 | -3.4 |
| 9 | 1.3 | 5.4 | 6.2 | 1.8 | 1.9 | 6.3 | 8.3 | 6.8 | +1.5 |
| 10 | 0.1 | 9.1 | 11.0 | 2.1 | 6.0 | 15.5 | 14.3 | 16.8 | -2.5 |
| Median | 2.8 | 4.5 | 9.0 | 2.0 | 1.5 | 9.4 | 11.2 | 10.1 | -0.6 |
The median score for artefact with 4D-CT was 2 (range 1 to 4), and 4D-MRI was 2 (range 2 to 3). The median score for noise with 4D-CT and 4D-MRI was 2 (range 1 to 3). Median score for edge detection with 4D-CT was 3 (range 1 to 4) and for 4D-MRI was 1 (range 1 to 3). Median overall image quality for 4D-CT was 2 (range 1 to 4) and for 4D-MRI was 1 (range 1 to 3) (see Table 3)
Table 3.
Scores of Artefact (A), Noise (N), Edge Detection (ED) and overall Image Quality (IQ) in ten patients undergoing 4D-CT and 4D-MRI during abdominal radiotherapy planning. Note two patients (patient 1 and 7) received adjuvant treatment so tumour edge detection was not applicable (NA). Note that lower scores indicate a superior image score.
| Patient | 4D-CT |
4D-MRI |
||||||
|---|---|---|---|---|---|---|---|---|
| A | N | ED | IQ | A | N | ED | IQ | |
| 1 | 1 | 2 | NA | 1 | 2 | 1 | NA | 2 |
| 2 | 1 | 2 | 3 | 2 | 2 | 2 | 1 | 1 |
| 3 | 1 | 2 | 3 | 1 | 2 | 1 | 1 | 1 |
| 4 | 4 | 2 | 3 | 4 | 3 | 3 | 3 | 3 |
| 5 | 1 | 1 | 3 | 1 | 2 | 2 | 3 | 1 |
| 6 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 |
| 7 | 2 | 3 | NA | 4 | 2 | 2 | NA | 1 |
| 8 | 3 | 3 | 3 | 3 | 2 | 2 | 1 | 1 |
| 9 | 4 | 2 | 2 | 4 | 2 | 2 | 1 | 2 |
| 10 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 |
| Median | 2 | 2 | 3 | 2 | 2 | 2 | 1 | 1 |
4. Discussion
Here we present phantom, volunteer and clinical data of a prototype T1 weighted 4D-MRI sequence. This study is unique in that directly compared motion and quality information between 4D-CT and 4D-MRI. In more than half of the patients undergoing abdominal radiotherapy, 4D MRI showed superior edge detection and image quality indicating potential benefits of 4DMRI. The amplitude on 4D-CT and 4D-MRI were similar. This is further evidence that 4D-MRI may supersede 4D-CT in motion mapping during the planning process.
Many attempts to provide motion information with MRI have included acquisition of dynamic 2D MRI images across all phases of respiratory cycle in two orthogonal planes [10], [11]. Slice stacking has been utilised with variable success [9] but can have significant stepping artefact, may not account for tumour deformation and triggering efficiency can be problematic. Respiratory amplitude-based triggering systems can acquire image slices at chosen phases of respiration using internal and external signals [18], [19], [20]. Internal MRI signal has been shown to be superior then external signals in the setting of pancreatic tumours [6]. Respiratory based sorting has its limitations including incomplete binning leading to gaps in data for which methods of compensation exist [11], [19].
Radial acquisition with self-gating and retrospective sorting have shown encouraging results [5], [7]. In 2005, Larson et al introduced a free-breathing self-gated MRI cine technique [21]. Stemkens et al used a stack of stars trajectory to generate a respiratory correlated 4D-MRI with linear phase binning [7]. Several authors have also demonstrated the feasibility of an online constructed 4D-MRI using a stack of stars trajectory similar to the sequence presented here [7], [22]. Online 4D-MRI using “pre beam” 4D-MRI and online 2D-cine has shown encouraging results [23]. While the existent literature on 4D-MRI is broad, the unique aspects of this paper are the comparison of 4D-MRI to the current gold standard 4D-CT in terms of amplitude and imaging quality.
In the phantom study, the five bin, seven bin and ten bin 4D-MRI demonstrated very similar motion (less than 0.2 mm difference) to both the 4D-CT and the ground truth (see Fig. 1 and Table 2). This result is encouraging and suggests that this 4DMRI sequence may adequately map motion at the amplitudes and BPM tested.
The first two volunteers were scanned with three bins, five bins and ten bins. This revealed that three bins demonstrated reduced amplitude compared to five bins, whilst ten bins showed increased radial streaking artefact. For this reason the subsequent eight volunteers and ten patients were scanned with five bins. These findings were consistent with the phantom data which showed that three bin 4D-MRI showed an inaccurate reduction in amplitude of movement as compared to five bins and ten bins. Image quality was similar across all bins of 4D-MRI. As expected, a small amount of artefact and noise was present in all organs, but this did not impact imaging quality significantly. Structure delineation was best for liver, followed by right kidney, pancreas and duodenum (see Table 4). Overall image quality of each organ was high with the exception of duodenum, which was high in some cases but poor in the majority of cases.
Table 4.
Scores for Artefact (A), Noise (N), Structure Delineation (SD) and overall Image Quality (IQ) for liver, right kidney, duodenum and pancreas in ten healthy volunteers. Note that lower scores indicate a superior image score.
| Volunteer | Liver |
Right Kidney |
Duodenum |
Pancreas |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | N | SD | IQ | A | N | SD | IQ | A | N | SD | IQ | A | N | SD | IQ | |
| 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 4 | 3 | 2 | 2 | 2 |
| 2 | 3 | 2 | 1 | 2 | 2 | 2 | 1 | 1 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| 3 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 2 | 2 | 1 | 2 |
| 4 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 |
| 5 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| 6 | 2 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 1 |
| 7 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2 |
| 8 | 3 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 |
| 9 | 2 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 |
| 10 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 |
| Median | 2 | 2 | 1.5 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 2.5 | 3 | 2 | 2 | 2 | 2 |
In the ten upper abdominal RT patients studied, 4D-MRI corroborated tumour motion amplitude information from the current gold standard 4D-CT (see Table 2). The median difference in motion between 4D-CT and 4D-MRI was −0.6 mm. Of note, several patients demonstrated varying movements in different planes. This was most apparent in patient 4, where the difference in tumour motion was greater than 3 mm in x, y and z planes. The possible reasons for these variations are a difference in breathing pattern between CT and MRI for individual patients, difference in reconstruction and lastly contouring variation between CT and MRI. Furthermore, 4D-MRI scored similarly for artefact and noise to 4D-CT, but had a greater accuracy for edge detection and better overall image quality (see Table 3) as expected with its superior soft tissue contrast.
There were some notable findings in this small cohort of patients. Image quality in the sagittal and coronal planes on the TPS (Pinnacle3, Philips Medical Systems, The Netherlands) is poor due to the voxel size of 1.2 × 1.2 × 3mm, with preference given to axial plane (1.2 × 1.2 mm). Signal variation and discontinuities were present (see Supplementary Material) due to absence of coil correction. Routine scans using surface coils require intensity correction and this is best obtained using a patient specific pre-scan adjustment obtained from the more uniform body coil (e.g. prescan normalisation (Siemens), PURE(GE)). We will incorporate Prescan Normalize in our future protocols. The benefits of improved delineation with 4D-MRI over 4D-CT were not apparent on ROI delineation for patients receiving adjuvant RT. For these patients, clips were easier visualised on 4D-CT compared to 4D-MRI. This potentially accounts for the different movement seen in the different planes for patient one and patient seven. Nonetheless, motion information from these contoured ROIs was still of value and appeared to be effective surrogates of target position in the planning process, and would still be beneficial during image guided RT. Pancreas contouring was difficult on 4D-CT and 4D-MRI in isolation, as images were often limited by poor tissue contrast, artefact from biliary stents and difficult to define tumours. Two of the three patients with liver metastasis had excellent tissue delineation on 4D-MRI (see Fig. 3), even without the presence of contrast. For these patients contouring volumes was easy on all phases of 4D-MRI, but not 4D-CT. The third patient with breast metastasis (patient ten) had poor tumour definition on both 4D-CT and 4D-MRI due to significant background liver cirrhosis. Potential improvements in tumour detection for liver metastases on 4D-MRI may be possible by using intravenous contrast that is not as effected by the time taken for image acquisition (e.g. disodium gadoxetate).
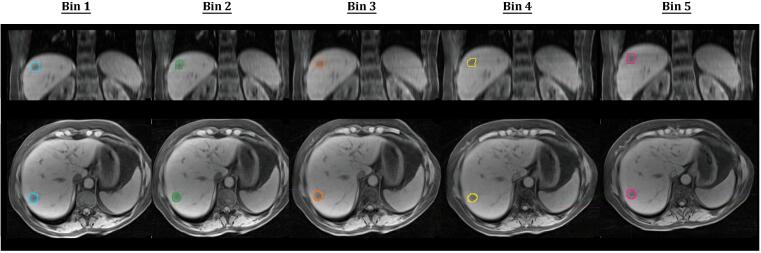
Fig. 3.
Excellent tumour edge detection for patient 2 is shown on all five bins of the 4D-MRI. Bin 1: end-inspiratory; Bin 5: end-expiratory.
The inaccuracy of surrogates for tumour position such as the diaphragm and stent has been demonstrated previously [24], [25]. Respiratory bellows have been demonstrated to be an inaccurate predictor of upper abdominal viscera movement [6]. Therefore, newer techniques that provide soft tissue contrast and facilitate accurate motion detection are required. We intend to explore the benefit of 4D-CT over 4D-MRI in a larger cohort of patients, and also investigate the value of MRI specific contrast agents; particularly in liver stereotactic RT. Recruitment to a larger study is ongoing. We intend to map duodenal position throughout the respiratory cycle using 4D-MRI, and believe this can have enormous implication for duodenum planning risk volume particularly in pancreas SBRT [6]. A study looking at 4D-MRI in patients undergoing pancreas SBRT has opened for recruitment.
These results using 4D-MRI offer insight into the benefits of online imaging in the context of an MRI-Linac. Prior to any online imaging within a MRI-linac, a comparison of 4D-CT and 4D-MRI is paramount. Clinical work assessing the feasibility and quality of 4D-MRI at the time of simulation is important prior to any clinical transition to a MRI-Linac. Information obtained at the point of simulation by 4D-MRI may stratify patients into those that will need MR guidance, margin adaptation or other tools for motion management.
Conflicts of Interest
Dr Li Pan is employed by Siemens Healthineers.
Acknowledgements:
The authors wish to thank Drs. Himanshu Bhat and Robert Grimm for assistance in developing the 4D-MRI prototype sequence and Dr. Benjamin Schmitt for ongoing input into this research project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2018.09.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Landberg T., Chavaudra J., Dobbs J., Gerard J.P., Hanks G., Horiot J.C. Report 62. J Int Commission Radiation Units Measurements. 1999 [Google Scholar]
- 2.Rietzel E., Chen G.T. Improving retrospective sorting of 4D computed tomography data. Med Phys. 2006;33:377–379. doi: 10.1118/1.2150780. [DOI] [PubMed] [Google Scholar]
- 3.Liney G.P., Moerland M.A. Magnetic resonance imaging acquisition techniques for radiotherapy planning. Semin Radiat Oncol. 2014;24:160–168. doi: 10.1016/j.semradonc.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Raaymakers B., Jürgenliemk-Schulz I., Bol G., Glitzner M., Kotte A., van Asselen B. First patients treated with a 1.5 T MRI-Linac: clinical proof of concept of a high-precision, high-field MRI guided radiotherapy treatment. Phys Med Biol. 2017;62:L41. doi: 10.1088/1361-6560/aa9517. [DOI] [PubMed] [Google Scholar]
- 5.Deng Z., Pang J., Yang W., Yue Y., Sharif B., Tuli R. Four-dimensional MRI using three-dimensional radial sampling with respiratory self-gating to characterize temporal phase-resolved respiratory motion in the abdomen. Magn Reson Med. 2016;75:1574–1585. doi: 10.1002/mrm.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stemkens B., Tijssen R.H., de Senneville B.D., Heerkens H.D., van Vulpen M., Lagendijk J.J. Optimizing 4-dimensional magnetic resonance imaging data sampling for respiratory motion analysis of pancreatic tumors. Int J Radiat Oncol Biol Phys. 2015;91:571–578. doi: 10.1016/j.ijrobp.2014.10.050. [DOI] [PubMed] [Google Scholar]
- 7.Stemkens B., Tijssen R.H., de Senneville B.D., Lagendijk J.J., van den Berg C.A. Image-driven, model-based 3D abdominal motion estimation for MR-guided radiotherapy. Phys Med Biol. 2016;61:5335. doi: 10.1088/0031-9155/61/14/5335. [DOI] [PubMed] [Google Scholar]
- 8.Rank C.M., Heußer T., Buzan M.T., Wetscherek A., Freitag M.T., Dinkel J. 4D respiratory motion-compensated image reconstruction of free-breathing radial MR data with very high undersampling. Magn Reson Med. 2017;77:1170–1183. doi: 10.1002/mrm.26206. [DOI] [PubMed] [Google Scholar]
- 9.von Siebenthal M., Szekely G., Gamper U., Boesiger P., Lomax A., Cattin P. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- 10.Park S., Farah R., Shea S.M., Tryggestad E.J., Hales R., Lee J. Simultaneous tumor and surrogate motion tracking with dynamic MRI for radiation therapy planning. Phys Med Biol. 2017;63 doi: 10.1088/1361-6560/aaa20b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tryggestad E., Flammang A., Han-Oh S., Hales R., Herman J., McNutt T. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys. 2013;40:051909. doi: 10.1118/1.4800808. [DOI] [PubMed] [Google Scholar]
- 12.Celicanin Z., Bieri O., Preiswerk F., Cattin P., Scheffler K., Santini F. Simultaneous acquisition of image and navigator slices using CAIPIRINHA for 4D MRI. Magn Reson Med. 2015;73:669–676. doi: 10.1002/mrm.25134. [DOI] [PubMed] [Google Scholar]
- 13.Freedman J.N., Collins D.J., Bainbridge H., Rank C.M., Nill S., Kachelrieß M. T2-Weighted 4D magnetic resonance imaging for application in magnetic resonance-guided radiotherapy treatment planning. Invest Radiol. 2017;52:563–573. doi: 10.1097/RLI.0000000000000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkelmann S., Schaeffter T., Koehler T., Eggers H., Doessel O. An optimal radial profile order based on the Golden Ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007;26:68–76. doi: 10.1109/TMI.2006.885337. [DOI] [PubMed] [Google Scholar]
- 15.Grimm R, Bauer S, Kiefer B, Hornegger J. Block K: Optimal channel selection for respiratory self-gating signals. In: Proc. 21st Annual Meeting ISMRM, Salt Lake City, Utah, USA, 2013, p. 3749.
- 16.Grimm R., Block K., Hutter J., Forman C., Hintze C., Kiefer B. International Society for Magnetic Resonance in Medicine; 2012. Self-gating reconstructions of motion and perfusion for free-breathing T1-weighted DCEMRI of the thorax using 3D stack-of-stars GRE imaging, Proceedings of the 20th scientific meeting. [Google Scholar]
- 17.Kumar S., Rai R., Stemmer A., Josan S., Holloway L., Vinod S. Feasibility of free breathing Lung MRI for Radiotherapy using non-Cartesian k-space acquisition schemes. Br J Radiol. 2017;90:20170037. doi: 10.1259/bjr.20170037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Wei J., Olek D., Kadbi M., Tyagi N., Zakian K. Direct Comparison of Respiration-Correlated Four-Dimensional Magnetic Resonance Imaging Reconstructed Using Concurrent Internal Navigator and External Bellows. Int J Radiat Oncol Biol Phys. 2017;97:596–605. doi: 10.1016/j.ijrobp.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Yin F.F., Czito B.G., Bashir M.R., Cai J. T2-weighted four dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys. 2015;42:4460–4471. doi: 10.1118/1.4923168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui C., Wen Z., Stemkens B., Tijssen R., Van Den Berg C., Hwang K.-P. 4D MR imaging using robust internal respiratory signal. Phys Med Biol. 2016;61:3472. doi: 10.1088/0031-9155/61/9/3472. [DOI] [PubMed] [Google Scholar]
- 21.Larson A.C., Kellman P., Arai A., Hirsch G.A., McVeigh E., Li D. Preliminary investigation of respiratory self-gating for free-breathing segmented cine MRI. Magn Reson Med. 2005;53:159–168. doi: 10.1002/mrm.20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mickevicius N.J., Paulson E.S. Investigation of undersampling and reconstruction algorithm dependence on respiratory correlated 4D-MRI for online MR-guided radiation therapy. Phys Med Biol. 2017;62:2910. doi: 10.1088/1361-6560/aa54f2. [DOI] [PubMed] [Google Scholar]
- 23.Kontaxis C., Bol G.H., Stemkens B., Glitzner M., Prins F.M., Kerkmeijer L.G.W. Towards fast online intrafraction replanning for free-breathing stereotactic body radiation therapy with the MR-linac. Phys Med Biol. 2017;62:7233–7248. doi: 10.1088/1361-6560/aa82ae. [DOI] [PubMed] [Google Scholar]
- 24.Feng M., Balter J.M., Normolle D., Adusumilli S., Cao Y., Chenevert T.L. Characterization of pancreatic tumor motion using cine MRI: surrogates for tumor position should be used with caution. Int J Radiat Oncol Biol Phys. 2009;74:884–891. doi: 10.1016/j.ijrobp.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann M., Pichler B., Schölkopf B., Beyer T. Towards quantitative PET/MRI: a review of MR-based attenuation correction techniques. Eur J Nucl Med Mol Imaging. 2009;36:93–104. doi: 10.1007/s00259-008-1007-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.