Abstract
Background and purpose
Computed tomography (CT) radiomics of head and neck cancer (HNC) images is susceptible to dental implant artifacts. This work devised and validated an automated algorithm to detect CT metal artifacts and investigate their impact on subsequent radiomics analyses. A new method based on features from total variation, gradient directional distribution, and Hough transform was developed and evaluated.
Materials and methods
Two HNC datasets were analyzed: a training set of 131 patients for developing the detection algorithm and a testing set of 220 patients. Seven designated features were extracted from ROIs (regions of interest) and machine learning with random forests was used for building the artifact detection algorithm. Performance was assessed using the area under the receiver operating characteristics curve (AUC).
Results
The testing results of artifacts detection yielded a cross-validated AUC of 0.91 (95% CI: 0.89–0.94), and a test AUC of 0.89. External testing validation yielded an accuracy of 0.82. For radiomics model prediction, training with artifacts yielded an AUC of 0.64 (95% CI: 0.63–0.65), while training on images without artifacts improved the AUC to 0.75 (95% CI: 0.74–0.76). This was compared to visual inspection of artifacts (AUC = 0.71 [95% CI: 0.69–0.73]).
Conclusion
We developed a new method for automated and efficient detection of streak artifacts. We also showed that such streak artifacts in HNC CT images can worsen the performance of radiomics modeling.
Keywords: Artifact detection, Radiomics, Machine learning
1. Introduction
With recent advances in medical imaging technologies, contrast-enhanced computed tomography (CT), magnetic resonance (MR), and positron emission tomography (PET) imaging are being routinely acquired during the diagnosis, staging and radiotherapy treatment planning of head and neck cancers (HNC). The most widely used imaging modality for diagnosis and therapy is CT, which can assess the tissue/lesion density, shape and texture, and has been a good image-based data resource for patient’s outcome modeling (e.g., radiomics) [1]. A large number of imaging features can be extracted from CT images for such radiomics analysis. These features are widely explored in HNC CT image analysis (e.g., segmentation, predictive and prognostic biomarkers, etc). Aerts et al. found that the CT radiomic signature constructed from non-small cell lung cancer (NSCLC) patients preserved significant prognostic performance for head and neck squamous cell carcinoma (HNSCC). Significant associations were discovered between the radiomics features and gene-expression patterns [2]. Zhang et al. found that CT texture features such as primary mass entropy and histogram skewness were independent predictors of overall survival in a dataset of 72 HNSCC patients [3]. However, it is sometimes overlooked that the existence of metal artifacts in CT HNC images, due to dental implants, may corrupt the reliability and the precision of such radiomics analysis and may cause misleading results. Since metal artifacts in the original images can lead to changes in underlying texture features, which form the basis of radiomics analysis [4], [5].
Although large amounts of promising studies were carried out for CT-based HNC radiomic analysis, artifacts influence was not taken into account or at least wasn’t mentioned in these articles. Until recently, less attention was paid into this critical issue in CT HNC image analyses. Bogowicz et al. conducted studies aimed to predict tumor local control (LC) after radiochemotherapy of HNSCC and human papilloma virus (HPV) status using CT radiomics. In their study, contours were manually removed from artifact-affected slices. Scans with more than a half of the contoured slices affected by metal artifacts were not included in the analysis [6]. Elhalawani et al. also applied exclusion of slices with metal artifacts in their HPV prediction model [7]. For these studies that excluded the artifact-affected slices or patients, manual filtering was applied, which is a very time-consuming process. There are also some approaches proposed for the metal artifacts reduction (MAR) [8], [9], [10], [11], [12]. Yet, these methods are likely to introduce new artifacts to images, degrade their resolution, and influence the statistical distribution of the original images, rendering them detrimental to any subsequent radiomic analysis [13], [14]. In order to overcome these challenges, we proposed a novel method that enabled the classification of artifact-affected slices/ROIs using extracted features automatically and efficiently, which has the potential to simplify the preprocessing and make the radiomic signatures more reliable. We have applied our algorithm on an external dataset to investigate the impact of artifacts on radiomics modeling as well. Our current approach aims to flag images with artifacts that would allow building more robust radiomic models with artifact-free images.
2. Materials and methods
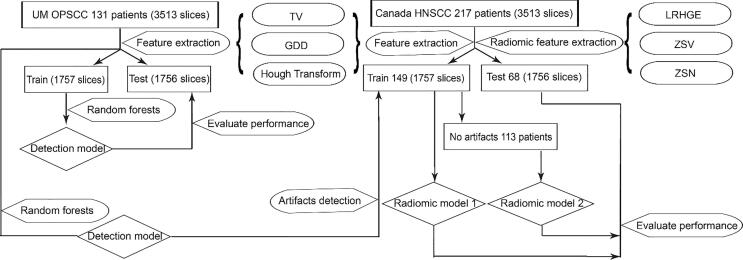
A total of 131 oropharyngeal squamous cell carcinoma patients (3513 slices, among which 360 slices had visually identified metal artifacts in the regions of interest [ROIs]), treated at the University of Michigan Department of Radiation Oncology, and a set of 220 head-and-neck squamous cell carcinoma patients (17956 slices) from a previously published dataset, treated at four hospitals in Canada were included in this study [15]. The two datasets will be referred to as UM data and the Canadian data, respectively. We determined the ground truth non-artifact slices by visually inspecting all slices in the UM data set and only looking at the tumor ROI. This means that if the tumor ROI on a given slice did not contain metal artifacts, it would be considered as a negative sample even if other parts of that same ROI contained artifacts, which will help save valuable data. For the Canadian data, due to the very large amount of slices, we visually determined if any ROI contained artifacts, as opposed to slice-by-slice. For this case, after we obtained predicted slice label, if for a ROI, there was at least one slice that was labeled to contain artifact, the whole ROI would be labeled as artifact-present. Example slice of the artifact-affected ROI was shown in Supplemental materials (SM) Fig. S1.
The UM data was randomly and equally split into training and test sets. Training set was used to train a random forests artifacts detection model (all hyper-parameters and parameters), then applied to the hold-out test set. The Canadian data was split by hospital: 148 patients from Hôpital général juif (HGJ) and Centre hospitalier universitaire de Sherbrooke (CHUS) were used as training set for the proposed radiomics model for distant metastases in Vallières et al study [15]. Seventy-two patients from Hôpital Maisonneuve-Rosemont (HMR) and Centre hospitalier de l’Université de Montréal (CHUM) combined were used as test set for evaluation. A brief workflow is shown in Fig. 1. More details about the datasets were given in SM Table S1.
Fig. 1.
Brief workflow for artifacts detection and impact on radiomic model performance.
2.1. Features design and extraction
2.1.1. Total variation based-feature
The concept of total variation (TV) was introduced first by Rudin et al. [16] for noise removal in image processing using first-order norms, since noisy images tended to have a high TV value compared with noise-free images. Similarly, metal artifacts led to an increased TV value relative to that of the regions of interest (ROIs) without artifacts, so the TV score could be taken as a measure of the artifacts. Below is the formula for calculating the TV (feature 1):
| (1) |
where is the intensity for pixel , are number of pixels along the two directions, is the number of pixels in the ROI. TV sums the absolute values of two-dimensional gradients for each pixel point of an image here we are referring to in-plane directions. Additionally, TV values were normalized by dividing by the number of pixels in ROIs to exclude the influence of image size.
2.1.2. Gradient direction distribution (GDD) based features
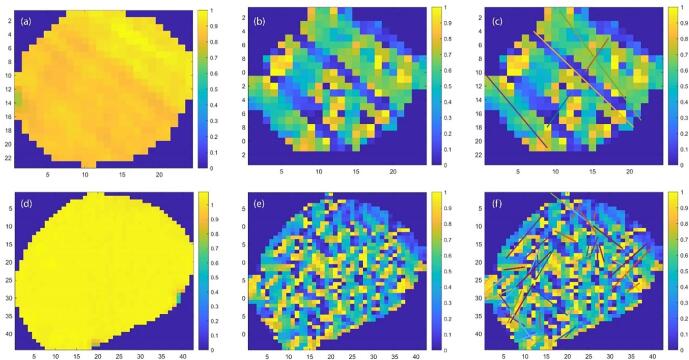
Compared with TV exacting gradient magnitude information, GDD features extracted gradient direction information. Some image pre-processing procedures were necessary prior to the extraction. The aim of pre-processing was to improve the image data quality by suppressing unwanted distortions and enhance the metal artifacts for preparation of the feature extraction. As shown in Fig. 2(a), the raw ROI images were noisy and the artifacts were hard to detect directly.
Fig. 2.
(a) Original ROI with artifacts; (b) corresponding Gradient direction map of the ROI; (c) detected lines by modified Hough transform; (d)–(f) are similar with (a), (b), (c), while without artifacts.
We cropped and resized the original ROIs so that they had comparable size to provide a good estimate of the distribution. First, we resized ROIs to 25 × 25 pixels (median size of ROI slices in UM dataset), then cropped outer pixels to remove any edge effects, such that all images had the same size of 16 × 16.
With size-modified ROIs, the gradient direction of each pixel point in the ROIs was approximated using the Sobel operator [17]. The direction ranged from −180° to 180° counterclockwise from the positive x-axis. In Fig. 2(b) and (e), the gradient direction map of the ROIs was plotted. In these plots, the streak artifacts were more pronounced than in the original images. Histograms provided useful information about image statistics, from which we could extract discriminant features to help with artifacts detection. For ROIs with artifacts there should be a dominant direction of the gradient orientation distribution, while the ROIs without artifacts would tend to have more uniform distribution. We extracted the maximum gradient direction percentage (feature 2) from the histogram of angles for the gradient with 36 bins (bin width of 10°):
| (2) |
where is the bin index. Due to varying tumor shapes, even though the bounding ROI box was modified, there were still some non-tumor parts of the original images included in the analyzed region. In order to remove shape effects, another complementary feature was calculated (feature 3):
| (3) |
2.1.3. Grey-scale Hough transform based features
Conventional Hough transform is a well-known method for line detection [18], [19], [20]. However, conventional Hough transform (CHT) requires input images to be edge-enhanced binary images, which are obtained by edge detection algorithms followed by thresholding or thinning. This will lead to loss of information and it requires selecting a threshold (harder for rich contrast, blurry, and band structure images). In our case, the artifacts were not obvious, and they were dispersed band structures. We used an extended Hough transform algorithm that dealt specifically with grey-scale images and avoided the thresholding process encountered in the conventional Hough transform [21]. Keck et al. proposed the use of direct output from the edge operator [22]. Thus, there was no threshold used to suppress the edges. Instead, the intensities in the edge image was considered to be the weighting coefficient for the Hough transform – grey-scale Hough transform (GSHT). However, the traditional edge operators performed poorly in the ROIs in our application, since the lines in our case were dispersed with relatively gradually changing intensities. Instead of applying the edge operators, we input the gradient direction map for GSHT. Subsequently, we applied a local maxima filter to the obtained Hough map. The modified GSHT algorithm was summarized in SM Table S2.
We used the GSHT as feature extractor instead of directly line detection because of dispersive characteristic of the artifacts. As shown in Fig. 2(f), if there were too many lines detected, it indicated high noisiness of the image and also absence of artifacts. Hence, the number of lines detected was a distinctive feature (feature 4). Since most of the artifact lines extended through the whole tumor, the ratio of the line length detected over the length of the tumor along that same direction should be close to one for artifact lines (feature 5).
| (4) |
For tumors with artifacts, the length of detected lines should be relatively large. Thus, the number of lines with ratios larger than a priori threshold was another distinct feature:
| (5) |
The threshold for large lines we used here was empirically found to be 0.6 using our training data. For ROIs with artifacts, the lines detected were expected to have similar orientations, while those false lines had directions without any regular pattern. We counted the number of lines that had similar orientations with the maximum ratio line in one ROI. Here, we defined similar orientation as angle difference smaller than (20) using prior knowledge.
| (6) |
where D represents the direction of a line.
After designed these seven features, we did principal component analysis (PCA) for these features to explain the variance in the data.
2.2. Random forests artifacts detection classifier construction
In summary, we have devised 7 features for the automatic detection of artifacts which were summarized in Table 1.
Table 1.
Extracted features.
| Extraction Method | Total variation | GDD | Modified grey-scale Hough transform | ||||
|---|---|---|---|---|---|---|---|
| Feature index | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Name | Total variation | Maximum gradient direction | Ratio of pixels inside ROIs | Number of lines detected | Maximum ratio of detected lines | Number of lines larger than a certain threshold | Number of lines with similar orientation with the longest line detected |
Tree-based methods are commonly used in machine learning to build predictive models by partitioning the feature space into a set of rectangles. We randomly split UM data into training and testing sets (with equal samples). Random forests were implemented on the training set to construct the detection model. A Bayesian optimizer was used to optimize the 5-fold cross-validated loss objective function to tune the hyper-parameters (minimum leaf size and number of trees used) to control the tree depth. Then, we fixed the hyper-parameters to re-train the model. The trained model was applied to the testing set, with 10 times 5-fold cross-validation to provide the confidence interval for the training results. Feature importance was computed as well. Slice level artifacts detection model was trained and tested on the training and testing sets of UM data, while ROI level detection was trained on the UM data and tested both on the testing sets of UM data and externally on the Canadian data. Furthermore, based on the variance explained using principal component analysis (PCA), we tried different models using all the 7 features and less features to examine whether we could simplify the features we used and still obtain a generalizable model.
2.3. Evaluation of impact of artifacts in tumor ROIs on radiomic prediction performance
For all the 148 train and 72 test ROIs, we implemented the feature extraction method described above. A random forests classifier (that classifies the presence of metal artifacts in each slice) using all the samples of UM data (131 patients, 3513 slices) was constructed, and applied to the Canadian data (220 patients, 344 ROIs, 17,956 slices) to obtain the predicted labels for whether or not one slice has artifacts and then determine if the ROI contains artifacts. The ground truth for these data are visually determined. Radiomic models for distant metastases were built on three sets of data: (1). all 148 train samples; (2). samples without artifacts based on the algorithm; (3) samples without artifacts based on visual detection. The three models were further tested on test set (72 patients containing no metal artifacts). For the model construction detail please refer to the paper [15]. The prediction results were presented by plotting the receiver operating characteristic (ROC) curve and calculating the corresponding area under the curve (AUC). The clinical patient characteristics were evaluated for patients without artifacts and all the patients in Canadian data to make sure the subgroup (without artifacts) clinical characteristics were not biased. For categorical variables, Pearson’s Chi-squared test was carried out, and for continuous variables, pairwise t-test was used to check if there was significant bias or deviance for the subgroup compared to the whole set.
3. Results
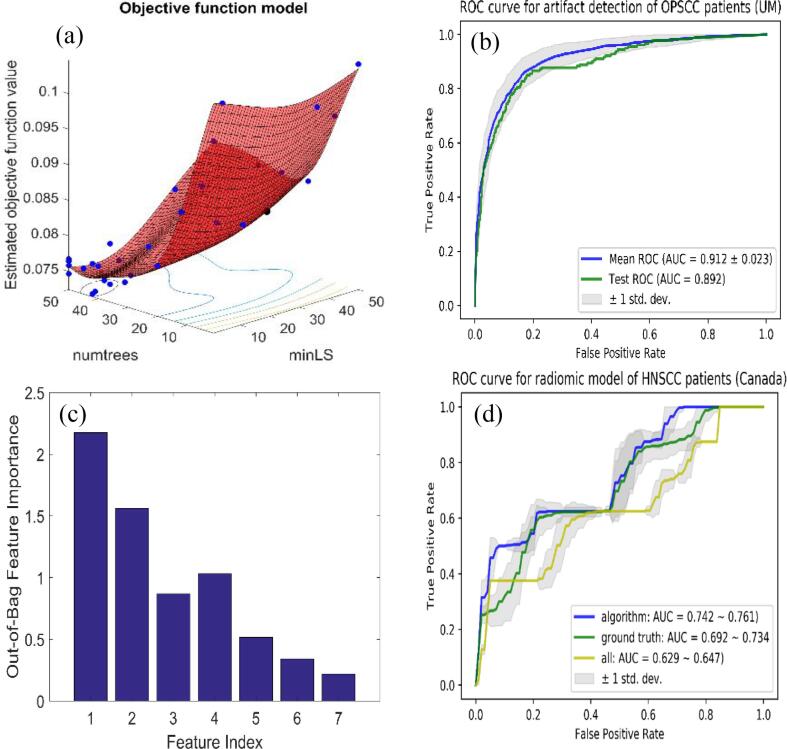
Fig. 3(a) showed the training of objective function in terms of number of trees and minimum leaf size. The optimal values for these two parameters were 42 trees and 9 minimum number of leaf node observations. Fig. 3(b) showed that the training AUC achieved 0.91 (95% CI: 0.89–0.94), testing 0.89. The out-of-bag feature importance, measured by bootstrapping technique in random forests algorithm, was also calculated and presented in Fig. 3(c) [23]. The ranking showed that first four important features were total variation, max GDD, number of lines detected by Hough transform and ratio of valid pixels in the images and contributed to 99% of the unexplained variation using PCA. Since the first 4 features could explain most of the variance, we examined the models using less features. The slice level AUC on UM data saturated after 4 features (∼0.90), details are summarized in SM Table 3. The confusion matrix for ROI level performance on UM and Canadian data using different features (4–7) was shown in Table 3. The results for 5–7 features were the same for UM (combined in the table), with an accuracy of 0.70/0.77, specificity of 0.66/0.83, sensitivity of 0.74/0.71, F1 score 0.73/0.77 for 4 feature and 5–7 feature models, respectively, as shown in Table 3. Since a 4-feature model didn’t perform well for ROI level classification, we tested only 5 and 7 feature models on Canadian data. The confusion matrix and corresponding metric results were shown in Table 2, Table 3 as well, with an accuracy of 0.79/0.82, specificity of 0.80/0.88, sensitivity of 0.77/0.70, F1 score 0.69/0.71 for 5 feature and 7 feature models, respectively. After checking the clinical characteristics, we found that none of the characteristics showed significant deviance from the original dataset.
Fig. 3.
(a) Optimization of hyper-parameters for random forests: number of trees (41) and minimum leaf size (17); (b) ROC curve for test data, with AUC of 0.89; (c) Out-of-bag feature importance; (c) Radiomic model test results for distant metastases using: all train samples (148 patients, yellow); samples filtered by our artifacts detection algorithm (107 patients, blue) and samples filtered visually (100 patients, green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Performance for UM and Canadian data of ROI artifacts.
| Feature # | Accuracy | Specificity | Sensitivity | F1 score | |
|---|---|---|---|---|---|
| UM | 4 features | 0.70 | 0.66 | 0.74 | 0.73 |
| 5–7 features | 0.77 | 0.83 | 0.71 | 0.77 | |
| Canada | 5 features | 0.79 | 0.80 | 0.77 | 0.69 |
| 7 features | 0.82 | 0.88 | 0.70 | 0.71 | |
Table 2.
Confusion matrices for UM and Canadian data of ROI artifacts.
| UM (4 features/5–7 features) | Positive | Negative |
|---|---|---|
| Predicted positive | 26/25 | 10/5 |
| Predicted negative | 9/10 | 19/24 |
| Canada (5 features/7 features) | Positive | Negative |
| Predicted positive | 81/73 | 47/29 |
| Predicted negative | 24/32 | 192/210 |
The results for the radiomic models were shown in Fig. 3(d). Radiomic model constructed using samples without artifacts, either filtered by our algorithm or visually, yielded a substantially better performance than using the original training set, which included 32% (48/148) artifacts patients. The AUC were 0.64 (95% CI: 0.63–0.65), 0.71 (95% CI: 0.69–0.73), and 0.75 (95% CI: 0.74–0.76) for the radiomic models trained on all train samples, samples excluding artifacts affected ones by our algorithm and by visual detection, respectively.
4. Discussion
In this study, a set of features extracted from total variation, gradient direction distribution and grey-scale Hough transform algorithm was designed. UM testing AUC of 0.89 showed that the proposed approach was able to accurately classify slices with metal artifacts. The robustness of these features was further validated by relatively good performance on external Canadian data. Confusion matrix for external validation was used since the slice-by-slice labels in Canadian data were not obtained due to the large sample size (17,596 slices). A ROI would be labeled positive, if one or more slices contained artifacts.
Though first 4 features explained most of the variance and slice level AUC saturated after 4 features, adding feature 5 increased the ROI level performance (accuracy, specificity, and F1 score) for UM data, due to less false positive, and similar true positive cases. This was reasonable since the last few features were mainly designed to regularize or reduce the false positive classification. Similar trend was captured in the Canadian data as well, the specificity increased from 0.80 to 0.88 with more features. While, the decrease of sensitivity (0.77–0.70) for 7 features-model was due to less true positives. In all, 5 features-model was comparable for slice-by-slice detection to the 7 features-model. For the ROI level detection, 5 features-model resulted in less artifacts dataset, while 7 features-model tended to reserve more samples but with more artifacts cases as well. The slice level model generalizability was not harmed by adding more features, probably because random forests algorithm is able to select the most robust features for the task.
AUC around 0.90 suggested the probability of correctly ranking a positive – negative pair was 0.90. In general, it is pretty good performance for a classification task. To the best of our knowledge, we did not find literature implementing this kind of metal artifacts detection, thus it was hard to compare how good the accuracy of 0.82 was. However, the UM data ROI level accuracy was 0.77, with the slice level AUC 0.90. Thus, we could infer that the slice level AUC for external data was probably comparable. In addition, the artifact-free subset filtered by our algorithm showed improvement of performance, which also proved the goodness of this level of classification accuracy for radiomics modeling. Based on the context that researchers usually remove the slices affected when building models not the ROIs, our technique should be applicable and meaningful. Another thing to notice was the lower UM ROI accuracy, which could be due to the different artifacts proportion, with UM having 55% and Canada having 31% artifact-affected ROIs. Hence, it made sense that the specificity as well as the accuracy would be lower, with comparable sensitivity for UM data.
Leijenaar, et al. tested a radiomics signature derived from non-small cell lung cancer (NSCLC) patients on an external dataset of oropharyngeal squamous cell carcinoma (OPSCC) patients (n = 542) [24]. They visually identified ROIs with artifacts, and resulted in a subset of 275 patients with artifacts. Their radiomics signature was validated on all the data, subset of patients with and without artifacts within the delineated tumor regions. They found that the features preserved discriminative value on both with and without artifacts subsets, however, they still suggested that there was an influence of CT artifacts on the model fit, which indicated a need for remodeling excluding samples with artifacts. This was consistent with our finding. Their research focused on validating the radiomics signature on head and neck tumor ROIs with and without artifacts to see the robustness of the features. We investigated the influence of presence of artifacts for the model construction and corresponding test performance on artifact-free data.
Another study related to ours is the one by Ger et al. [5]. They investigated metal artifacts caused by dental fillings and beam-hardening artifacts caused by bone. They found at least 73% of feature values were affected by the streak artifacts. And almost all features were robust with removal of up to 50% of the original GTV. In summary, they showed that metal artifacts affect radiomic feature values, suggesting that regions containing such artifacts should not be included in radiomics data set. Their research provided further support for the necessity of removing artifact-affected images before radiomics modeling.
We were also interested in understanding the nature of the misclassified cases. Some examples of both false negatives and false positives were shown in SM Fig. S2. The main challenge we met with in this detection task was the subtleness of the metal artifacts or small signal-to-noise ratio (SNR) of the ROIs. A lot of the misses were the cases with artifacts that were subtle and hard to detect. The false positive cases were some slices with line-like structures inside while not being a true artifact.
Finally, one thing to point out is that if the radiomic features are from 3D ROIs, we might have to remove the artifact-affected patients. Given the fact that around 30–50% of patients have metal artifacts, the radiomic models developed in this way might be suitable for not affected patients only. However, if we extract features from 2D slices, then we can remove the affected slices without excluding the patient. While, we do acknowledge that the artifact classification can be more beneficial to develop radiomics models which are more robust against the streak artifacts, which is out of our scope for this study.
In conclusion, we have developed a new method for CT artifacts detection in tumor regions for head and neck patients; achieved UM test dataset prediction AUC of 0.89 using random forests algorithm and investigated the impact of presence of artifacts for head and neck CT images using internal and external datasets. We recommend using the proposed automatic algorithm to filter samples before CT head and neck radiomics analysis.
Conflict of interest
We hereby, confirm that all authors have contributed to this paper’s conception and writing, and they all confirm that this submitted work is free of conflict of interest.
Acknowledgement
This work was supported by the National Institutes of Health (NIH) via grants R37CA222215 and R01CA233487.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2019.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Coroller T.P., Grossmann P., Hou Y., Velazquez E.R., Leijenaar R.T.H., Hermann G. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H., Graham C.M., Elci O., Griswold M.E., Zhang X., Khan M.A. Locally advanced squamous cell carcinoma of the head and neck: CT texture and histogram analysis allow independent prediction of overall survival in patients treated with induction chemotherapy. Radiology. 2013;269:801–809. doi: 10.1148/radiol.13130110. [DOI] [PubMed] [Google Scholar]
- 4.Yu H., Scalera J., Khalid M., Touret A.-S., Bloch N., Li B. Texture analysis as a radiomic marker for differentiating renal tumors. Abdom Radiol. 2017;42:2470–2478. doi: 10.1007/s00261-017-1144-1. [DOI] [PubMed] [Google Scholar]
- 5.Ger R.B., Craft D.F., Mackin D.S., Zhou S., Layman R.R., Jones A.K. Practical guidelines for handling head and neck computed tomography artifacts for quantitative image analysis. Comput Med Imag Graph. 2018;69:134–139. doi: 10.1016/j.compmedimag.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogowicz M., Riesterer O., Ikenberg K., Stieb S., Moch H., Studer G. Computed tomography radiomics predicts HPV status and local tumor control after definitive radiochemotherapy in head and neck squamous cell carcinoma. Int J Radiation Oncol Biol Phys. 2017;99:921–928. doi: 10.1016/j.ijrobp.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Head MACC, Group NQIW. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Scientific reports. 2018; 8. [DOI] [PMC free article] [PubMed]
- 8.Abdoli M., Dierckx R.A., Zaidi H. Metal artifact reduction strategies for improved attenuation correction in hybrid PET/CT imaging. Med Phys. 2012;39:3343–3360. doi: 10.1118/1.4709599. [DOI] [PubMed] [Google Scholar]
- 9.Boas F.E., Fleischmann D. Evaluation of two iterative techniques for reducing metal artifacts in computed tomography. Radiology. 2011;259:894–902. doi: 10.1148/radiol.11101782. [DOI] [PubMed] [Google Scholar]
- 10.Joemai R.M., de Bruin P.W., Veldkamp W.J., Geleijns J. Metal artifact reduction for CT: development, implementation, and clinical comparison of a generic and a scanner-specific technique. Med Phys. 2012;39:1125–1132. doi: 10.1118/1.3679863. [DOI] [PubMed] [Google Scholar]
- 11.Xu C., Verhaegen F., Laurendeau D., Enger S.A., Beaulieu L. An algorithm for efficient metal artifact reductions in permanent seed implants. Med Phys. 2011;38:47–56. doi: 10.1118/1.3519988. [DOI] [PubMed] [Google Scholar]
- 12.Yazdi M., Lari M.A., Bernier G., Beaulieu L. An opposite view data replacement approach for reducing artifacts due to metallic dental objects. Med Phys. 2011;38:2275–2281. doi: 10.1118/1.3566016. [DOI] [PubMed] [Google Scholar]
- 13.Katsura M., Sato J., Akahane M., Kunimatsu A., Abe O. Current and novel techniques for metal artifact reduction at CT: practical guide for radiologists. Radio Graphics. 2018;38:450–461. doi: 10.1148/rg.2018170102. [DOI] [PubMed] [Google Scholar]
- 14.Gjesteby L., De Man B., Jin Y., Paganetti H., Verburg J., Giantsoudi D. Metal artifact reduction in CT: where are we after four decades? IEEE Access. 2016;4:5826–5849. [Google Scholar]
- 15.Vallières M., Kay-Rivest E., Perrin L.J., Liem X., Furstoss C., Aerts H.J. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7:10117. doi: 10.1038/s41598-017-10371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudin L.I., Osher S., Fatemi E. Nonlinear total variation based noise removal algorithms. Phys D. 1992;60:259–268. [Google Scholar]
- 17.Sobel I. An Isotropic 3 3 Image Gradient Operator, 2014.
- 18.Vc H.P. Method and means for recognizing complex patterns. Google Patents. 1962 [Google Scholar]
- 19.Duda R., Hart P. Use ofthe Hough transform to detectlinesand curvesin pictures. Commun ACM. 1972;15 [Google Scholar]
- 20.Ballard D.H. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recogn. 1981;13:111–122. [Google Scholar]
- 21.Peng T. Detect lines in grayscale image using Hough Transform. version 1.0 ed: mathworks; 2005.
- 22.Ruwwe C., Zölzer U., Hough Duprat O. transform with weighting edge-maps. Visualization Imag Image Process. 2005 [Google Scholar]
- 23.James G., Witten D., Hastie T., Tibshirani R. Springer; 2013. An introduction to statistical learning. [Google Scholar]
- 24.Leijenaar R.T., Carvalho S., Hoebers F.J., Aerts H.J., Van Elmpt W.J., Huang S.H. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54:1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.