Abstract
Background
Digestive processes in the rumen lead to the release of methyl-compounds, mainly methanol and methylamines, which are used by methyltrophic methanogens to form methane, an important agricultural greenhouse gas. Methylamines are produced from plant phosphatidylcholine degradation, by choline trimethylamine lyase, while methanol comes from demethoxylation of dietary pectins via pectin methylesterase activity. We have screened rumen metagenomic and metatranscriptomic datasets, metagenome assembled genomes, and the Hungate1000 genomes to identify organisms capable of producing methyl-compounds. We also describe the enrichment of pectin-degrading and methane-forming microbes from sheep rumen contents and the analysis of their genomes via metagenomic assembly.
Results
Screens of metagenomic data using the protein domains of choline trimethylamine lyase (CutC), and activator protein (CutD) found good matches only to Olsenella umbonata and to Caecibacter, while the Hungate1000 genomes and metagenome assembled genomes from the cattle rumen found bacteria within the phyla Actinobacteria, Firmicutes and Proteobacteria. The cutC and cutD genes clustered with genes that encode structural components of bacterial microcompartment proteins. Prevotella was the dominant genus encoding pectin methyl esterases, with smaller numbers of sequences identified from other fibre-degrading rumen bacteria. Some large pectin methyl esterases (> 2100 aa) were found to be encoded in Butyrivibrio genomes. The pectin-utilising, methane-producing consortium was composed of (i) a putative pectin-degrading bacterium (phylum Tenericutes, class Mollicutes), (ii) a galacturonate-using Sphaerochaeta sp. predicted to produce acetate, lactate, and ethanol, and (iii) a methylotrophic methanogen, Methanosphaera sp., with the ability to form methane via a primary ethanol-dependent, hydrogen-independent, methanogenesis pathway.
Conclusions
The main bacteria that produce methyl-compounds have been identified in ruminants. Their enzymatic activities can now be targeted with the aim of finding ways to reduce the supply of methyl-compound substrates to methanogens, and thereby limit methylotrophic methanogenesis in the rumen.
Keywords: Rumen, Bacterial, Methyl-compound, Methanol, Methylamines
Background
Methane (CH4) is an important greenhouse gas (GHG) accounting for ~ 14% of total global GHG emissions [1]. Approximately 40% of this comes from agriculture, with the single largest source being enteric fermentation in ruminants. Ruminants are important to the economies of many developed and developing countries, and finding ways to reduce CH4 emissions from ruminants is a challenge facing farmers worldwide [2]. As a consequence of the digestive processes in the rumen, by-products of fibre degradation and fermentation end-products, including hydrogen (H2), carbon dioxide (CO2), methanol, methylamines, and methylsulphides, are formed but not used by the host animal. Hydrogenotrophic and methylotrophic methanogens in the rumen are able to remove these end products by reducing them to CH4, which is eructated from the animal leading to atmospheric emissions of CH4 [3]. Hydrogenotrophic rumen methanogens mainly belong to the genus Methanobrevibacter, while the core rumen methylotrophic methanogens are from the genus Methanosphaera and the order Methanomassiliicoccales [3].
The main methyl-compounds found in the rumen are methanol and methylamines. Methanol is present from around 0.8 mM in the rumen of cattle fed hay and grain [4] to around 0.07 mM in Brahman steers fed Rhodes grass hay [5] and is thought to be derived from demethyoxylation of dietary pectins via the action of pectin methyl esterases (PMEs; EC3.1.1.11). Pectin is a significant component of the plant cell wall (PCW) after cellulose, hemicellulose and lignin, and is found in the middle lamellae that joins cells together. While research with environmental bacteria has stressed the importance of pectin degradation in the initiation of PCW breakdown [6], little is known about the organisms that carry out pectin degradation and methanol release in the rumen. The rumen bacterium Lachnospira multipara produces pectin lyase (PL) and PME activities [7, 8] and has been regarded as the primary pectin fermenter isolated from rumen contents of animals fed diets high in pectin [9]. During pectin fermentation by L. multipara, methanol is formed as a product of PME activity [10], and pectin fermentation can cross-feed methanol-utilising bacteria such as Eubacterium limosum as has been demonstrated with co-cultures of these species [11]. However, Lachnospira is not normally abundant in the rumen [12], and other more abundant genera with pectin-degrading ability, notably Butyrivibrio and Prevotella, are likely to be the major pectin degraders.
Mono-, di- and tri-methylamines are produced mainly as the end-product of plant phosphatidylcholine degradation [13] via choline. Methylamine has been measured at around 0.085 mM in the rumen fluid of dairy cows fed a cereal grain diet [14] and ranges from 0.0285 to 0.703 mM in the rumen of cows fed varying amounts of barley grain [15] and from 0.334 to 0.564 mM in Brahman steers on the tropical forage, Rhodes grass [5]. Very little is known about how methylamines are produced in the rumen. It has been shown that labelled choline dosed into the rumen was rapidly metabolized to trimethylamine (TMA) by rumen microorganisms and the labelled methyl groups ended up as CH4 [16, 17]. A more recent study found a negative relationship between rumen Methanomassiliicoccales populations and urinary trimethylamine-N-oxide (TMAO) concentration [18], thought to be due to Methanomassiliicoccales using TMA for methane formation in the rumen, and diverting it from being oxidised to TMAO in the liver. More is known about the metabolism of choline and TMA in the human gut, as the TMAO formed in the liver is correlated with atherosclerosis in animal models and is linked with cardiovascular risks in human clinical trials [19, 20]. The release of TMA from choline was reported in the human gut bacterium, Proteus mirabilis, mediated by the enzyme choline trimethylamine lyase (CTMAL; EC:4.3.99.4) [21]. Microbial choline TMA lyase was found to be an enzyme complex composed of a catalytic choline utilisation polypeptide CutC, and an associated activating protein CutD, encoded by adjacent genes within a gene cluster that also contains genes encoding bacterial microcompartment proteins [22]. This gene cluster was first described from the rumen sulphate reducing bacterium, Desulfovibrio desulfuricans, and confining this activity within a bacterial microcompartment is seen as a means to prevent the volatile and toxic acetaldehyde intermediate damaging other cellular processes [22]. Several other human gut bacteria with choline TMA lyase activity have been identified [23, 24] and gut metagenomes have been screened for TMA-producing catabolic genes [25].
In order to target ruminal CH3-compound formation as a means to reduce methanogenesis, the types of organisms producing CH3-compounds in the rumen and the enzymes involved need to be identified. Here, we report a survey of rumen-derived metagenomic and metatranscriptomic datasets [26] and rumen metagenome assembled genomes [27] to identify the genes encoding the production of CH3-compounds, and which organisms express these genes under conditions prevailing in the rumen. We also screen the Hungate1000 genomes [28] for the occurrences of these genes and examine their arrangement within each genomic context, to give additional insights into the potential physiological context and genetic regulation of processes leading to CH3-compound release. Furthermore, we describe an enrichment culture experiment using pectin to encourage the growth of methanol-forming microbes from sheep rumen contents, and report the identification and analysis of metagenome assembled genomes (MAGs) from this enrichment.
Results
Identification of genes encoding production of mono-, di- and tri-methylamines
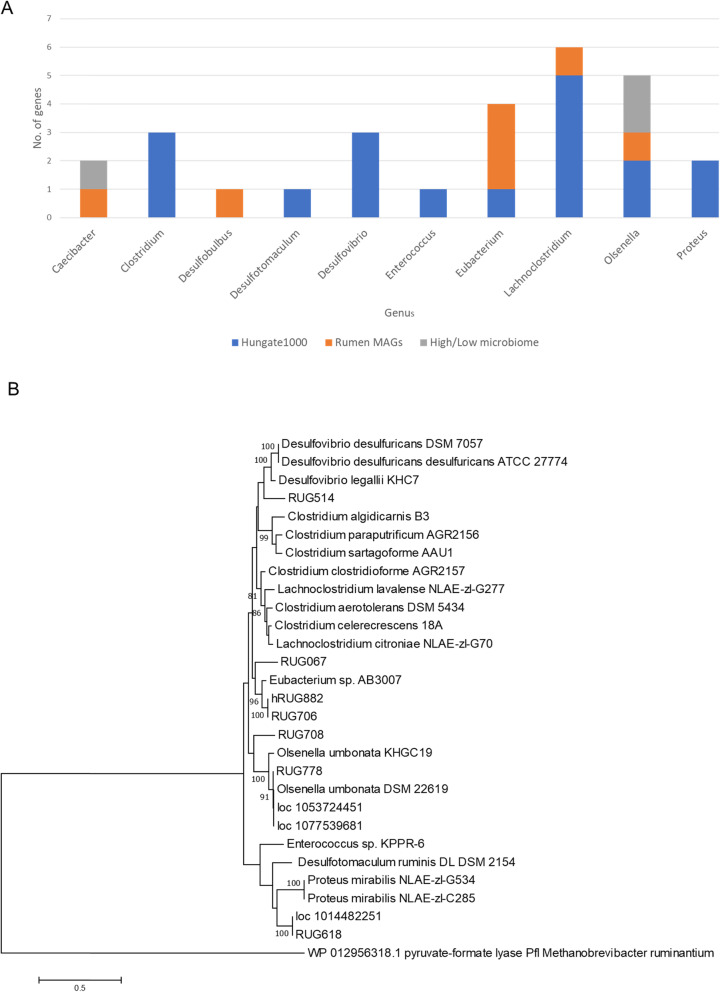
The presence of genes encoding choline TMA lyase and the associated choline TMA lyase activator in rumen metagenome datasets was determined using the HMM models for CutC and CutD [25]. Analyses against the combined assembly of metagenome and metatranscriptome reads derived from rumen contents of sheep selected for differences in CH4 yield (11,801,660 ORFs) [26] revealed good matches for both CutC and CutD from Olsenella umbonata (Actinobacteria, Coriobacteriaceae, two hits) and Caecibacter (Firmicutes, Veillonellaceae, one hit), but to no other organisms. (Figure 1a, Additional file 1: Table S1A = CutC MG&MT sheet). CutC transcript abundances were low in the sheep metatranscriptome dataset, suggesting low level expression of these genes in the rumen of these animals. The contigs were quite short in the combined assembly so it was not possible to get an indication of the genome context from these data. However, examination of the SPADES re-assembled metagenomes from the same study has provided additional information on the genome context for these genes (Additional file 2: Figure S1A). Analysis against the predicted ORFs of 913 cattle rumen MAGs) [27] indicated that just seven MAGs contained a putative CutC gene (Fig. 1a).
Fig. 1.
Choline TMA lyase (cutC) gene abundance and diversity in a combined rumen metagenome and metatranscriptome dataset, metagenome-assembled genomes and Hungate1000 bacterial genomes (a), and a phylogenetic tree showing the relationships of CutC protein sequences from all these sources (b)
The Hungate1000 Collection genomes were also screened for CutC and/or CutD domains (Table 1) and a phylogenetic tree of CutC sequences retrieved from rumen genome and metagenome/metatranscriptome sources is shown in Fig. 1b. In all cases the CutC and CutD genes were part of a larger cluster which included genes for the structural components of bacterial microcompartment proteins (Additional file 2: Figure S1B&C). A total of 18 bacterial strains were identified, 10 of rumen origin and 8 from faeces. None of these bacterial genera are regarded as abundant or prevalent members of the rumen microbiome based on results from the Global Rumen Census study [12]. The abundance of CutC sequences identified from Hungate1000 Collection genomes were assessed in the high and low methane yield sheep metagenome and metatranscriptome datasets (Additional file 1: Table S1A = CutC MG&MT table). The CutC from Olsenella umbonata DSM 22619 was most abundant in the metagenome dataset, followed by Eubacterium sp. AB3007 and Desulfovibrio legallii KHC7. CutC transcripts from Hungate1000 Collection genomes were mainly from D. desulfuricans subsp. desulfuricans ATCC 27774, D. legallii KHC7 and O. umbonata DSM 22619. Genes encoding CutC also include two non-specific Pfam domains (Pfam01228: glycine radical and Pfam02901: pyruvate formate lyase-like), but a further search using these domains did not find additional examples of choline TMA lyase.
Table 1.
Choline TMA lyase gene occurrence in rumen microbial datasets
| Rumen bacteria | Phylum | Strain | IMG gene ID | Locus Tag | aa | Origin |
|---|---|---|---|---|---|---|
| Hungate1000 [28] | ||||||
| Rumen bacteria | ||||||
| Olsenella umbonata | Actinobacteria | A2 (DSM 22619) | 2608576534 | Ga0059087_11836 | 847 | sheep |
| Olsenella umbonata | Actinobacteria | KHGC19 | 2663665377 | Ga0104367_1620 | 847 | cow |
| [Eubacterium] sp. | Firmicutes | AB3007 | 2561437736 | P156DRAFT_0760 | 847 | cow |
| Enterococcus sp. | Firmicutes | KPPR-6 | 2608578278 | Ga0059091_105198 | 857 | cow |
| Lachnoclostridium aerotolerans | Firmicutes | X8A62 (DSM 5434) | 2558962511 | T546DRAFT_01546 | 848 | sheep |
| Clostridium sartagoforme | Firmicutes | AAU1 | 2543441078 | A500_11264 | 846 | buffalo |
| Desulfotomaculum ruminis | Firmicutes | DL (DSM 2154) | 650891141 | Desru_2090 | 847 | sheep |
| Desulfovibrio legallii | Proteobacteria | KHC7 | 2654384186 | Ga0104380_103103 | 848 | cow |
| Desulfovibrio desulfuricans | Proteobacteria | G11 (DSM 7057) | 2595153453 | IE73DRAFT_02008 | 848 | cow |
| Desulfovibrio desulfuricans subsp. desulfuricans | Proteobacteria | Mb (ATCC 27774) | 643582164 | Ddes_1357 | 848 | sheep |
| Faecal bacteria | ||||||
| Clostridium algidicarnis | Firmicutes | B3 | 2562235685 | BV55DRAFT_1892 | 847 | cow |
| Clostridium paraputrificum | Firmicutes | AGR2156 | 2525457652 | G594DRAFT_02041 | 846 | calf |
| Lachnoclostridium citroniae | Firmicutes | NLAE-zl-G70 | 2624603054 | Ga0070263_11089 | 855 | goat |
| Lachnoclostridium clostridioforme | Firmicutes | AGR2157 | 2525451452 | G614DRAFT_00825 | 853 | calf |
| Lachnoclostridium lavalense | Firmicutes | NLAE-zl-G277 | 2624766256 | Ga0070262_11929 | 846 | goat |
| Lachnoclostridium celerecrescens | Firmicutes | 18A (DSM 5628) | 2596304492 | H171DRAFT_4337 | 847 | cow |
| Proteus mirabilis | Proteobacteria | NLAE-zl-G534 | 2624727893 | Ga0066901_10954 | 1142 | goat |
| Proteus mirabilis | Proteobacteria | NLAE-zl-C285 | 2656994829 | Ga0104388_10857 | 1142 | cow |
| Rumen metagenome assembled genomes (MAGs [27]) | ||||||
| hRUG882 | Firmicutes | N/A | N/A | NODE_2309_length_24057 _cov_25.2708_2 | 857 | cow |
| RUG067 | Firmicutes | N/A | N/A | k87_66874274_20 | 872 | cow |
| RUG514 | Proteobacteria | N/A | N/A | k87_1124619_4 | 811 | cow |
| RUG618 | Firmicutes | N/A | N/A | k87_25912225_4 | 855 | cow |
| RUG706 | Firmicutes | N/A | N/A | scaffold_2105_5 | 857 | cow |
| RUG708 | Firmicutes | N/A | N/A | k87_13261599_27 | 860 | cow |
| RUG778 | Actinobacteria | N/A | N/A | scaffold_14892_2 | 848 | cow |
| High/Low microbiome [26] | ||||||
| NZ sheep rumen microbiome combined assembly | Actinobacteria | N/A | 37167 | loc_1053724451 | 799 | sheep |
| NZ sheep rumen microbiome combined assembly | Actinobacteria | N/A | 37167 | loc_1077539681 | 398 | sheep |
| NZ sheep rumen microbiome combined assembly | Firmicutes | N/A | 37167 | loc_1014482251 | 394 | sheep |
Identification of genes encoding production of methanol
To determine the presence of genes for PMEs in rumen metagenome datasets, the HMM model for Pfam01095 (Pectinesterase) was used to search against the combined assembly of metagenome and metatranscriptome reads screened from rumen contents of sheep described above [26]. Using the HMM default settings, a total of 2414 hits were retrieved which were analysed using BLAST searches (Fig. 2; Additional file 1: Table S1B = PME MG sheet). The sequences of the top BLAST hits were almost entirely (2398) of bacterial origin. Of the bacterial sequences, 1012 (42%) gave a top BLAST hit to a rumen isolate from the Hungate 1000 Collection. Prevotella was the dominant genus with 475 sequences that gave top BLAST hits to rumen isolates, along with Ruminococcus (171), Bacteroides (147), Butyrivibrio (49), Fibrobacter (39), Lachnospira (19), Oribacterium (19), as well as unclassified Lachnospiraceae (19) and Erysipelotrichaceae (14). Only 63 of the 2414 BLAST hits (2.6%) were derived from ‘uncultured’ organisms. Of these, 61 matched to the same sequence (AEF12641) which encodes a 1501 aa protein, annotated as being from an uncultured Prevotella from a bovine rumen sample. This protein shows ~ 70% aa identity with PMEs from the rumen Prevotella strains TF2–5 and BPI-148. Many hits (115) show > 90% aa identity to PMEs from rumen bacterial isolates, the best matches (> 99% aa identity) were to Prevotella bryantii (4 different PMEs), Lachnospira multipara (3 different PMEs), Ruminococcus sp., Prevotella sp., Butyrivibrio sp. and Oribacterium sp. The largest PMEs detected (> 2100 aa) were predominantly from Butyrivibrio spp. An analysis of PME transcript abundance also indicated that PMEs from Prevotella spp. were the most highly expressed (Additional file 1: Table S1C=PME MT sheet).
Fig. 2.
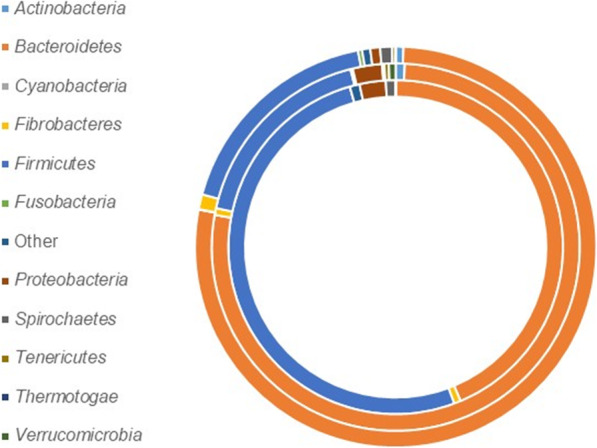
Abundance (%) and diversity of genes encoding pectin methyl esterase (PME; PF01095)-domain containing proteins in a combined rumen metagenome and metatranscriptome dataset (outer circle; n = 2414), metagenome-assembled genomes (middle circle; n = 505) and Hungate1000 bacterial genomes (inner circle; n = 315)
Similar results were obtained from a BLAST search analysis of the predicted ORFs from 913 cattle rumen MAGs [27]. This indicated the presence of 505 putative PME genes of bacterial origin (Additional file 1: Table S1D = PME RUG). Of these genes 146 (29%) gave a top BLAST hit to a rumen isolate from the Hungate 1000 Collection, with Prevotella again being the dominant genus. Only 5 ORFs derived from ‘uncultured’ organisms, and of these, 4 matched to the same sequence (AEF12641) that was observed in the combined assembly analysis. The largest PMEs detected were from Butyrivibrio spp.
Bacterial isolates from the human and pig gut microbiomes, and sequences from human microbiome metagenome studies, also made up significant numbers of the top BLAST hits retrieved from this analysis. Members of the genus Prevotella again provided the greatest number of sequences, and many of these sequences also give BLAST matches to rumen Prevotella isolates. This indicates, as previously reported [29], that more cultures are needed to capture the full diversity of rumen Prevotella species. Overall, 1394 sequences (58%) from the combined assembly and 245 ORFs (49%) from the MAGs gave a best match to sequences from members of the genus Prevotella. Further examination of these Prevotella results showed that 583 sequences (24%) from the combined assembly and 94 ORFs (19%) from the MAGs match to a PME of 324–330 aa, usually containing a signal peptide sequence at the N-terminus. Prevotella belongs to the phylum Bacteroidetes, and polysaccharide utilization is a characteristic feature within this group of organisms. The genes encoding polysaccharide breakdown are usually organised within polysaccharide utilization loci (PULs), which are defined as co-located genes organized around a susCD gene pair. PULs are thought to coordinate the breakdown of complex glycans via the carbohydrate-degrading enzymes co-located within the PUL. PULs are catalogued in the CAZy PUL database (PULDB) [30] which has recently been updated to include the Hungate1000 Collection genomes. Using the PULDB, the genomic context of the PME encoding genes was examined and most of the PME genes (including those encoding proteins of 324–330 aa) were found outside of PULs in the rumen Prevotella, even though they encode numerous examples of PULs in their genomes (ranging from 14 in P. albensis, up to 38 in Prevotella sp. strain KH1P2). However, several P. bryantii strains (B14, C21a, FB3001, KHPX14), P. ruminicola strains (D31d, Ga6B6, KHT3 AGR2160,) and Prevotella sp. strains (P6B1, P6B4, RM4, TC2–28, BPI-34, TF2–5) had one to three CE8 genes located within PULs. For example, in P. bryantii C21a two CE8 genes (G638DRAFT_00481, G638DRAFT_00861) were found in PULs 2 and 10 where they are co-located with genes for glycoside hydrolases and polysaccharide lyases suggesting that in this bacterium, pectin breakdown is a coordinated process.
The Hungate1000 Collection reference genome set was searched using information from the CAZy (http://www.cazy.org/) database (carbohydrate esterase family 8, CE8) and the protein domain specific for PME (Pfam01095), with the results shown in Additional file 3: Table S2. A total of 315 genes encoding PMEs were found in 159 microbial strains with up to six different PME-encoding genes found in a single strain. Strains belonging to the phylum Bacteroidetes showed the highest prevalence of PME genes. Many of the predicted PMEs contained signal peptide sequences, indicating a cell-surface or extracellular location. In addition, several genes encoded large multi-domain proteins, the most commonly associated domains included pectate lyases (Pfams 00544 and 09492), hydrolases (lipases/esterases Pfams 07859 and 13,472) and putative cell surface-binding components (Pfams 01473, 13,149 and 13,205).
Pectin enrichment culture from sheep rumen contents
While the above analyses focused on individual organisms and highlighted the detection of their genes in metagenomic and metatranscriptomic rumen datasets, a complementary aspect of the current study was to investigate the inter-relationships between members of the rumen microbial community that provide methylotrophic substrates for methanogenesis. To achieve this we carried out an enrichment experiment using homogalacturonan pectin (methyloxylated polygalacturonic acid) as a potential source of methanol, which in turn would act as the substrate for methanogenesis. A pectin-utilising, methane-producing enrichment was established and DNA extracted from the resulting microbial consortium was sequenced (BioProject accession: PRJNA365034).
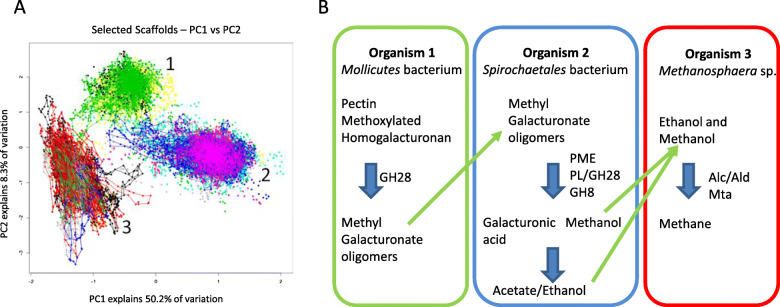
The consortium metagenome sequences assembled into 107 contigs and MetaBAT analysis grouped the 24 largest scaffolds into three bins, each of which represent uncultivated members of the rumen microbiome (Fig. 3a; Additional file 4: Table S3). The assembled genome of Organism 1 consisted of three contigs, with a combined size of 1.46 Mb and a GC content of ~ 38%. CheckM analysis indicated the assembled genome was 99.39% complete with 0% contamination. The 16S and 23 rRNA genes did not show a close relationship with any cultivated organisms, the closest matches being to members of the family Erysipelotrichaceae. Examination of the gene complement of Organism 1 identified a small number of genes encoding carbohydrate active enzymes (CAZymes), including members of glycoside hydrolase families GH10, GH32, GH43, GH53 and GH65, which indicates an ability to ferment plant polysaccharides. It also encodes genes for tandem signal peptide-containing polygalacturonases (GH28) which show weak homology (~ 40% nucleotide identity) to metagenome assembled genomes from environmental Tenericutes [31]. The second of these polygalacturonases contains a CBM32 domain (Pfam00754) which has been shown to mediate binding to polygalacturonate [32].
Fig. 3.
a: Tetranucleotide plot of the 24 scaffolds defining the 3 organisms recovered from a pectin-utilising, methane-producing enrichment culture. b: Predicted metabolic relationship between the three organisms enabling pectin conversion to methane
Organism 2 had 11 contigs associated with its assembled genome, giving a size of 3.61 Mb with a GC content of ~ 52%. CheckM analysis indicated 97.13% genome completeness with 0% contamination. The 16S rRNA gene found on one contig gives top BLAST hits to members of the genus Sphaerochaeta at ~ 91% identity, placing this organism in the phylum Spirochaetes. This organism appears to share the key features that distinguish Sphaerochaeta from most Spirochaetes, namely the lack of motility and non-spiral morphology; analysis of the genome indicated the absence of motility and chemotaxis genes, while examination of the enrichment culture by phase contrast microscopy did not show the presence of organisms with helical morphology characteristic of other members of the Spirochaetes phylum. The genome of Organism 2 also encodes numerous carbohydrate metabolism and fermentation genes [33], including a PME, a pectate lyase/polygalacturonase and six GH88 family unsaturated glucuronyl hydrolases predicted to mediate homogalacturonan metabolism. The PME, the pectate lyase/polygalacturonase and three of the GH88 proteins show homology (~ 62–84% aa identity) with a Spirochaetales MAG from activated sludge. None of the predicted proteins have signal peptide sequences indicating they function intracellularly. However, a large number of ABC carbohydrate transporters were identified, including 52 substrate-binding proteins identified as belonging to COG1653, which is frequently associated with the uptake of oligosaccharides. A pectinesterase gene with a best BLAST match to Sphaerochaeta coccoides DSM 17374 was also identified from one of the cattle rumen MAGs (RUG703).
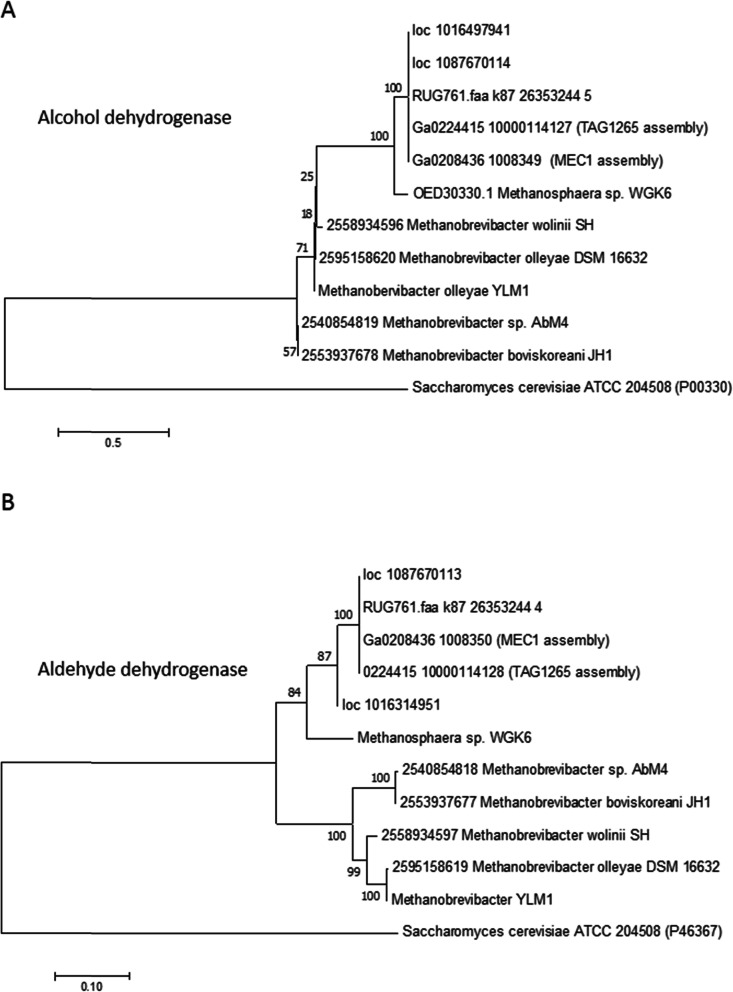
Ten contigs were associated with a third organism predicting a genome size of 2.0 Mb and a GC content of ~ 30%. CheckM analyses indicated that the assembled genome was 97.6% complete with 0% contamination. The 16S rRNA gene of Organism 3 gave a top hit to the type strain of Methanosphaera stadtmanae at 97% identity. Members of the genus Methanosphaera are methylotrophic methanogens [34], but although they are known to be present in the rumen from community profiling [3] only a few rumen isolates are available for study [35]. The assembled genome encodes the genes required for producing methane from methanol, but not from methylamines, and like M. stadtmanae DSM3091, lacks the genes for molybdopterin biosynthesis which suggests that it may be unable to reduce CO2 to methane due to the lack of this co-factor. Unlike M. stadtmanae, Organism 3 encodes a pair of genes encoding putative alcohol and aldehyde dehydrogenases which cluster with similar genes from Methanosphaera sp. WGK6 isolated from the wallaby gut [36], Methanosphaera sp. metagenome assembled genome from cattle (RUG761, [27]) and sheep (TAG1265, [35]), and more distantly with similar genes from the genome of the rumen methanogens, Methanobrevibacter sp. AbM4 [37] and Mbb. boviskoreani [38] (Fig. 4). Overall, the results from the analysis of the assembled genomes (Additional file 4: Table S3) show that these three organisms are likely to act together to convert pectin to methane (Fig. 3b).
Fig. 4.
Phylogenetic analysis of alcohol dehydrogenase (a) and aldehyde dehydrogenase (b) genes from rumen methanogen genomes and rumen MAGs. Both trees were constructed with the Jones-Taylor Thornton (JTT) model. Saccharomyces cerevisiae ATCC 204508 was used as the outgroup. Numbers represent the relative frequency of branch clustering based on 1000 bootstrap runs, bootstrap values < 50% are removed. Rumen MAGs; MEC1, Organism 3 (Methanosphaera sp.) from the pectin enrichment culture in this study; TAG1265, metagenome assembled Methanosphaera sp. sequences from low methane yield sheep datasets [35]; RUG761, metagenome assembled Methanosphaera sp. sequences from cattle [27]
Discussion
Current rumen manipulation strategies targeting CH4 mitigation are focused on direct inhibition of methanogens, targeting their essential functions via small molecule inhibitors and antimicrobial peptides or surface proteins through methanogen-targeted vaccines [39]. There has been little exploration of the opportunities around manipulating the supply of substrates to methanogens. Methylotrophic methanogens in the rumen appear to be limited by the availability of CH3-compounds. The energy available from the reduction of methanol to CH4 (CH3OH + H2 → CH4 + H2O) is − 112.5 kJ/mole, compared to − 131 kJ/mole for the reduction of CO2 (CO2 + 4 H2 → CH4 + 2 H2O) [40] but reflecting reaction stoichiometries, methylotrophs require only 1 mole of H2 per mole of CH4, whereas hydrogenotrophs require 4 H2 per mole of CH4. This means that methylotrophs have a lower H2 threshold, and when the energy requirement for ATP biosynthesis is considered, methylotrophs always have a greater net free energy change than hydrogenotrophs under conditions prevailing in the rumen. However, despite this thermodynamic advantage, the hydrogenotrophic Methanobrevibacter spp. are the main methanogens making up 75–78% of the methanogenic archaea in the rumen, [3, 12]. This suggests that the growth of methylotrophic methanogens is governed by the availability of CH3-compounds rather than the dissolved H2 concentration. Nevertheless, methanogens capable of methylotrophic methanogenesis represent around 22–25% of the methanogens in the rumen and reducing their supply of CH3-compound substrates in the rumen offers an opportunity to target these methanogens to reduce CH4 formation.
Recent work on a global analysis of rumen microbial communities from ruminant species and microbiome characterisation studies [12, 26–28] have provided large datasets which can be used to identify the major rumen bacteria involved in releasing CH3-compounds from plant material, and the genes encoding these activities. Our screens for ruminal TMA production revealed surprisingly few genes and organisms involved in this process. A total of 18 bacterial strains were identified using the CutC/D HMM models, and they belong to the same three phyla (Actinobacteria, Firmicutes and Proteobacteria) that were identified in studies on TMA metabolism in the human gut [22, 25]. Overall it appears that TMA lyase and choline TMA lyase activator genes are rare in the rumen. None of the seven bacterial genera detected with these genes would be regarded as abundant or prevalent members of the rumen microbiome based on results from the Global Rumen Census study [12]. The metagenome/metatranscriptome dataset, indicate that Olsenella and Caecibacter are the main methylamine producers in sheep, while MAG-derived sequences indicate that organisms related to Olsenella, Caecibacter, and Eubacterium are likely to be important in cattle.
We used the pectinesterase Pfam (PF01095) (EC 3.1.1.11) to screen the rumen microbiome datasets for signatures of the methanol-producing enzyme, PME. Pectinesterase is commonly found in plants where it plays an important role in fruit ripening, but it is also found in plant pathogens where it is involved in the de-esterification of pectin into pectate and methanol during the breakdown of plant material. In the rumen, many organisms are involved in pectin degradation, and our screens identified the majority of the pectinesterase-containing organisms belonged to the genus Prevotella. The metagenome sequences were short (average of 253 aa) compared to the predicted full length of the PME proteins, which meant that it was not possible to get much genome context around these metagenomic and metatranscriptomic hits. The majority of the metagenome-derived PMEs were most similar to PMEs found in Prevotella genomes from the Hungate1000 Collection or reported from other gut environments. PME expression in Prevotella has been reported previously as part of a study investigating carbohydrate esterase activities involved in hemicellulose degradation [41]. The expression of P. ruminicola 23 pectin esterases, Pec E1 and Pec E2, were analysed during growth on differing carbohydrates; Pec E2 was found to be more than 2× upregulated on xylo-oligosaccharides derived from corn fiber relative to glucose, suggesting a potential role for this enzyme in hemicellulose degradation.
From our preliminary analysis it appears that Prevotella are the main providers of methanol in the rumen since they make up the bulk of the PME sequences. The particular prevalence of Prevotella PMEs in the 324–330 aa size range suggests that these enzymatic activities are significant contributors. From genomic analyses, it is likely that Prevotella bryantii, Bacteroides sp. KHT7, and Lachnospira multipara are specialised pectin degraders, while Prevotella ruminicola and other Prevotella, Butyrivibrio, and Oribacterium species are generalist bacteria with the ability to degrade pectin. Interestingly, the celluloytic bacteria Fibrobacter succinogenes and Ruminococcus spp. encode PMEs but are not able to use pectin for growth, and may therefore be using these activities for clearing away pectins to allow access to their primary substrate, cellulose.
The results of the pectin enrichment experiment add another dimension to this study, and showed the potential importance of members of the rumen microbiota distinct from those highlighted by analysis of individual genomes and metagenomes. Three genomes were assembled from the metagenome sequence of the pectin-enriched consortium and the analysis shows that the three organisms encoding these genomes likely act together to convert pectin to methane (Fig. 1). The 16S rRNA gene of Organism 1 was not closely associated with any cultivated organism, but the absence of genes involved in peptidoglycan biosynthesis in its genome, coupled with the predicted small genome size, strongly suggest that this organism is a member of the class Mollicutes in the phylum Tenericutes. There have been few studies of the rumen members of this bacterial group but they are characterised as having a fermentative metabolism and occurring in association with other rumen inhabitants [42]. The presence of CAZYmes GH10, GH32, GH43, GH53 and GH65, indicates a general ability to breakdown plant polysaccharides, while the presence of extracellular GH28 polygalacturonases with CBM32 polygalacturonate binding domains suggest some degree of pectin degradation ability. However, Organism 1 is probably unable to utilize the major products of homogalacturonan degradation as it does not encode a pectin methyl esterase or any of the enzymes from the galacturonate utilization pathway. Like the polysaccharide-degrading activities of other rumen bacteria [43, 44], Organism 1 may use its pectin-degrading activity to clear away pectin from plant cell walls and allow access to its preferred substrate, probably hemicelluloses.
In contrast, Organism 2 (Sphaerochaeta sp.) has the complete complement of genes encoding the enzymes necessary for galacturonate utilization, although it does not encode extracellular enzymes involved in this process. It has a well-developed uptake system for the products of pectin degradation, and likely transports the pectin degradation products of Organisms 1 to act as substrates for its growth. The PME encoded by this Sphaerochaeta sp. may act upon methoxylated oligogalacturonides to release methanol as a prelude to further depolymerisation and fermentation. The metabolic profile of the Sphaerochaeta sp. indicates that acetate, lactate, and ethanol would be also be formed from fermentation of pectin-derived substrates. These compounds are potential energy and carbon sources for Organism 3, the methylotrophic methanogen Methanosphaera sp., which has the gene complement required for producing methane from methanol. Furthermore, this Methanosphaera sp. has genes encoding putative alcohol and aldehyde dehydrogenases; in other methanogens these genes have been shown to allow ethanol to be used as a source of reducing power for methane production and growth in Methanosphaera sp. WGK6 [36], Methanobrevibacter sp. AbM4 [37, 45] and Mbb. ruminantium [39]. The strong similarities among these genes lead us to predict that the Methanosphaera sp. RUG761 [27] and Mbb. boviskoreani [38] both share the same ethanol-dependent methanogenesis capability.
Conclusions
The work reported here has elucidated the main CH3-compound-forming pathways in the rumen and has identified the main bacteria that are involved. The ability to form methanol from methoxylated pectin via PME activity is widespread among rumen bacteria, but is most prevalent among members of the genus Prevotella. TMA release from plant-derived choline via TMA lyase activity is restricted to a much narrower spectrum of bacteria, principally Olsenella and Caecibacter in the ovine rumen and Olsenella, Caecibacter, and Eubacterium in the bovine rumen. The pectin enrichment experiment using sheep rumen contents has provided a unique insight into a specific example of a pectin-utilising and methane-forming consortium. As the techniques for assembling genomes from metagenomic sequencing data continue to improve, it is likely that more investigation of enrichment cultures and synthetic consortia will elucidate the complex relationships and inter-dependencies that occur in CH3-compound formation in the rumen. The screening work now allows the main CH3-compound-forming bacteria to be targeted specifically with the aim of finding ways to reduce their growth and/or enzymatic activities. By using such microbiological interventions we aim to reduce the supply of CH3-compound substrates to methanogens and thereby limit the amount of methane formed from by methylotrophic methanogens in the rumen.
Methods
Identification of TMA forming potential in rumen microbiome datasets
The Hidden Markov Model (HMM) profiles of CutC and CutD was kindly provided by Rath et al. [25]. The HMMER software package [46] using default cutoffs for CutD and a score cutoff of > 1500 for CutC was used to identify potential cut genes in the Hungate1000 Collection genomes [28], the rumen metagenome assembled genomes (MAGs) dataset [27] and the combined assembly of the High/Low dataset [26] and re-assembled (using SPADES) metagenome data of rumen microbial communities from low MY sheep (tags 1283, 1265, 1435, 1449 at 2 time points) used in the combined assembly of the High/Low dataset above. For phylogenetic alignment of the CutC genes, protein sequences were aligned using MUSCLE [47]. Maximum likelihood trees were constructed in MEGA7 [48] using the Le Gascuel 2008 method [49]. Statistical support for the tree was obtained by bootstrapping 100 iterations and the pyruvate-formate lyase gene from Methanobrevibacter ruminantium M1 (WP_012956318.1) [39] was used as the outgroup. A taxonomic classification of the CutC genes identified from the Rumen MAGs and the High/Low combined assembly datasets were assigned using the top blast hit result against the NCBI non-redundant (nr) protein database. An E-value cutoff of less than 1e-5 was used.
Identification of potential pectinesterase (PME) activity in rumen microbiome datasets
The Hidden Markov Model (HMM) profile of PF01095 (PME domain) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/), and HMMER software was used to detect the presence of PME genes using default cutoffs against the three datasets described above [26–28]. The taxonomy of the PME genes identified from the High/Low dataset were assigned using the top BLAST hit result against the NCBI non-redundant (nr) protein database, using an E-value cutoff of less than 1e-5.
Read mapping to identified CutC and PME genes
Metagenomic and metatranscriptome reads of each of the high/low microbiome samples (see Additional file 5: Table S4A and as described previously [50]) were mapped to the identified rumen CutC (n = 18) and PME-containing genes (n = 2730) from the Hungate1000 and the high/low combined assembly using BBmap (https://sourceforge.net/projects/bbmap) with an ID cut-off of 98% sequence similarity. Results were summarised using Samtools version 1.9 [51], see Additional file 5: Table S4B. Read counts were normalised using reads per kilobase per million (RPKM).
Pectin enrichment culture from sheep rumen contents
A pectin enrichment of the microbiota from sheep rumen contents was set up to assess the types of organisms capable of mediating pectin degradation coupled to methylotrophic methanogenesis. Rumen contents from sheep grazing a ryegrass-white clover pasture, were collected and filtered through 335 μm nylon mesh into Oakridge tubes which had been flushed with O2-free CO2. The tubes were centrifuged at low speed (200 x g) for 15 min at room temperature and the supernatant transferred to fresh tubes flushed with O2-free CO2. The tubes were centrifuged at 28,000 x g for 30 min at room temperature, the supernatant discarded, and the cell pellet was re-suspended in 5 mL of anaerobic RM02 base medium [52], then the volume taken up to 50 mL using the same medium. The tubes were centrifuged again at 28,000 x g for 30 min at room temperature, the supernatant discarded, and the cell pellet was re-suspended in 5 mL of anaerobic RM02 base medium under a stream of O2-free CO2. The re-suspended cells were 10-fold serially diluted into RM02 medium containing 1% pectin (Sigma apple pectin, poly-D-galacturonic acid methyl ester) and incubated at 39 °C. The gas composition of the headspace of each enrichment tube was monitored daily using gas chromatography [39] and when methane appeared, an aliquot of the culture was observed using phase contrast and fluorescence microscopy. The methane-producing enrichment tubes were dominated by fluorescent cocci, along with other non-fluorescent cells. Aliquots of methane-positive cultures were plated onto agar plates of RM02 medium containing 1% pectin inside an anaerobic chamber (Coy Laboratory Products, 96% CO2:4% H2 atmosphere) and incubated anaerobically in air-tight gas canisters at 39 °C until colonies formed. Single colonies were picked from plates inside the anaerobic chamber into fresh RM02-pectin broth medium and assessed for culture purity by PCR amplification using bacterial- and archaeal-specific 16S rRNA gene primers. One of the single-colony sub-cultures, designated MEC1, was found to contain a limited microbial diversity by phase contrast and fluorescent microscopy, and according to the 16S rRNA gene sequences retrieved from this culture, was dominated by two organisms; a methanogen associated with the genus Methanosphaera sp. and a bacterium affiliated with the family Sphaerochaetaceae.
Metagenome sequencing and assembly of the pectin enrichment culture
Community genomic DNA was extracted from the limited diversity MEC1 metagenome and submitted for sequencing as part of the Hungate1000 project at the Joint Genome Institute [28]. Sequencing used Illumina HiSeq 2500-1 TB technology and the metagenome sequences were assembled into 107 contigs using SPAdes V 3.11.1 [53]. The 26 largest contigs, ranging in size from 1.49 kb to 796 Kb, were sorted into 3 bins using MetaBAT [54]. Each bin had a scaffold which contained an almost full length 16S rRNA gene sequence allowing their preliminary taxonomic identification (Additional file 4: Table S3). Genomes were annotated by the DOE–JGI genome annotation pipeline [55–58]. CheckM analysis [59] of the three assembled genomes was conducted to estimate their completeness and degree of contamination. The evolutionary relationship of the alcohol dehydrogenase and aldehyde dehydrogenase genes from Organism 3 (Methanosphaera sp.) MAG with similar genes from rumen methanogens were inferred using the Neighbor-Joining method [60]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [61]. The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the JTT matrix-based method [62] and the units are the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA7 [48].
Supplementary information
Additional file 1: Table S1. A. CutC genes in the combined metagenome/metatranscriptome and Hungate1000 datasets. B. Metagenome abundance of PME genes identified from the combined metagenome/metatranscriptome and Hungate1000 datasets. C. Metatranscriptome abundance of PME transcripts of genes identified from the combined metagenome/metatranscriptome and Hungate1000 datasets. D. Top Blast hit against the 505 Pfam01095 containing ORFs identified from the rumen MAG dataset (n = 913).
Additional file 2: Figure S2. Choline trimethylamine lyase and bacterial microcompartment gene synteny in SPADES re-assembled metagenomes of low MY sheep (A), and in bacterial genomes of rumen (B) or ruminant faecal origin (C) in the Hungate1000 Collection.
Additional file 3: Table S2. Pectin methyl esterase genes (Pfam01095) identified in the Hungate 1000 collection reference genome set
Additional file 4: Table S3. Contigs and genome sizes of the three organisms identified from the MEC1 limited diversity metagenome.
Additional file 5: Table S4. A Overview of samples analysed in this study. B Results of the mapping of the high/low sample reads to the identified rumen CutC and PME-containing genes from the Hungate1000 and the high/low combined assembly.
Acknowledgements
The authors thank Silke Rath, Benjamin Heidrich, Dietmar Pieper and Marius Vital of the Microbial Interactions and Processes Research Group, Helmholtz Centre for Infection Research, in Braunschweig, Germany for making the cutC/D- cntA (two-component Rieske-type oxygenase/reductase) Hidden Markov Model [25] available for use in this project. We also acknowledge the assistance of Alan McCulloch of the AgResearch Technology and Digital Design group for computing support.
Abbreviations
- CO2
Carbon dioxide
- CTMAL
Choline trimethylamine lyase
- CutC
Choline trimethylamine lyase
- CutD
Choline trimethylamine lyase activator protein
- GH
Glycosyl hydrolase family
- H2
Hydrogen
- HMM
Hidden Markov Model
- MAG(s)
Metagenome Assembled Genome(s)
- PCW
Plant cell wall
- PL
Pectin lyase
- PMEs
Pectin methyl esterases
- PUL
Polysaccharide utilization loci
- PULDB
CAZy PUL database
- TMA
Trimethylamine
- TMAO
Trimethylamine-N-oxide
Authors’ contributions
The following authors contributed to the study design: GA, BK, SL, GC, SK, RM. Data collection and experimental procedures were conducted by: JK, PS, BK, SL, GA. Data analysis and interpretation: BK, SL, RS, GA, SM, CG. The manuscript was prepared by: BK, GA, SL, SM, GC, CG. All authors read and approved the final manuscript.
Funding
This work was funded by the New Zealand Government to support the objectives of the Livestock Research Group of the Global Research Alliance on Agricultural Greenhouse Gases.
Availability of data and materials
The metagenome and metatranscriptome datasets used in this study are accessible at the National Centre for Biotechnology Information Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) accession number SRA075938, BioProject number PRJNA202380, plus additional 16S rRNA gene amplicon sequence data under the SRA experiment accession numbers: SRX1079958 - SRX1079985. The Hungate1000 genomes are available from Joint Genome Institute’s Integrated Microbial Genomes and Microbiome Samples (IMG/M) which can be accessed at http://genome.jgi.doe.gov/. The raw sequence data and assembled genomes and proteomes from the 913 rumen uncultured genomes (RUG) and HiC rumen uncultured genomes (hRUG) are available the European Nucleotide Archive under project PRJEB21624. The SPADES assemblies of low methane yield sheep rumen microbial communities from New Zealand can be accessed via their IMG Database Project IDs: Sheep Tag 1265 (Gp0054682; Gp0053989), Sheep Tag 1283 (Gp0054684, Gp0054469); Sheep Tag 1435 (Gp0053990, Gp0054493), Sheep Tag 1494 (Gp0054822, Gp0054568).
Ethics approval and consent to participate
The collection of rumen contents from sheep was carried out under the approval of the AgResearch Ltd. Animal Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
William J. Kelly, Email: bill.kelly.donvis@gmail.com
Sinead C. Leahy, Email: sinead.leahy@agresearch.co.nz
Janine Kamke, Email: Janine.Kamke@horizons.govt.nz.
Priya Soni, Email: priya.soni@agresearch.co.nz.
Satoshi Koike, Email: skoike7@anim.agr.hokudai.ac.jp.
Roderick Mackie, Email: r-mackie@illinois.edu.
Rekha Seshadri, Email: rseshadri@lbl.gov.
Gregory M. Cook, Email: greg.cook@otago.ac.nz
Sergio E. Morales, Email: sergio.morales@otago.ac.nz
Chris Greening, Email: chris.greening@monash.edu.
Graeme T. Attwood, Email: graeme.attwood@agresearch.co.nz
Supplementary information
Supplementary information accompanies this paper at 10.1186/s42523-019-0016-0.
References
- 1.Reisinger A, Clark H. How much do direct livestock emissions actually contribute to global warming? Glob Change Biol. 2018;24:1749–1761. doi: 10.1111/gcb.13975. [DOI] [PubMed] [Google Scholar]
- 2.Wollenberg E, Richards M, Smith P, Havlik P, Obersteiner M, Tubiello F, et al. Reducing emissions from agriculture to meet the 2 °C target. Glob Change Biol. 2016;22:3859–3864. doi: 10.1111/gcb.13340. [DOI] [PubMed] [Google Scholar]
- 3.Seedorf H, Kittelmann S, Janssen PH. Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle. Appl Environ Microbiol. 2015;81:986–995. doi: 10.1128/AEM.03018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vantcheva ZM, Prodhan K, Hemken RW. Rumen methanol in vivo and in vitro. J Dairy Sci. 1970;53:1511–1514. doi: 10.3168/jds.S0022-0302(70)86424-4. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Fernandez G, Duval S, Kindermann M, Schirra HJ, Denman SE, McSweeney CS. 3-NOP vs. halogenated compound: Methane production, ruminal fermentation and microbial community response in forage fed cattle. Front Microbiol. 2018;9:1582. doi: 10.3389/fmicb.2018.01582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung D, Pattathil S, Biswal AK, Hahn MG, Mohnen D, Westpheling J. Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol Biofuels. 2014;7:147. doi: 10.1186/s13068-014-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silley P. A note on the pectinolytic enzymes of Lachnospira multiparus. J Appl Bacteriol. 1985;58:145–150. doi: 10.1111/j.1365-2672.1985.tb01441.x. [DOI] [Google Scholar]
- 8.Silley P. The production and properties of a crude pectin lyase from Lachnospira multiparus. Lett Appl Microbiol. 1986;2:29–31. doi: 10.1111/j.1472-765X.1986.tb01509.x. [DOI] [Google Scholar]
- 9.Bryant MP, Barrentine BF, Sykes JF, Robinson IM, Shawver CV, Williams LW. Predominant bacteria in the rumen of cattle on bloat-provoking ladino clover pasture. J Dairy Sci. 1960;43:1435–1444. doi: 10.3168/jds.S0022-0302(60)90346-5. [DOI] [Google Scholar]
- 10.Duskova D, Marounek M. Fermentation of pectin and glucose, and activity of pectin-degrading enzymes in the rumen bacterium Lachnospira multiparus. Lett Appl Microbiol. 2001;33:159–163. doi: 10.1046/j.1472-765x.2001.00970.x. [DOI] [PubMed] [Google Scholar]
- 11.Rode LM, Sharak-Genther BR, Bryant MP. Syntrophic association of methanol and CO2-H2-utilising species of Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl Environ Microbiol. 1981;42:20–22. doi: 10.1128/aem.42.1.20-22.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson G, Cox F, Ganesh S, Jonker J, Young W. Global Rumen Census Collaborators, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson RMC, Hemington NL. Digestion of grass lipids and pigments in the sheep rumen. Brit J Nutr. 1974;32:327–340. doi: 10.1079/BJN19740086. [DOI] [PubMed] [Google Scholar]
- 14.Ametaj BN, Zebeli Q, Saleem F, Psychogios NG, Lewis M, Dunn SM, et al. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics. 2010;6:583–594. doi: 10.1007/s11306-010-0227-6. [DOI] [Google Scholar]
- 15.Bovine Rumen Metabolome Database; http://www.rumendb.ca. Accessed 29 June 2018.
- 16.Broad TE, Dawson RM. Role of choline in the nutrition of the rumen protozoon Entodinium caudatum. J Gen Microbiol. 1976;92:391–397. doi: 10.1099/00221287-92-2-391. [DOI] [PubMed] [Google Scholar]
- 17.Neill AR, Grime DW, Dawson RM. Conversion of choline methyl groups through trimethylamine into methane in the rumen. Biochem J. 1978;170:529–535. doi: 10.1042/bj1700529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgavi DP, Rathahao-Paris E, Popova M, Boccard J, Nielsen KF, Boudra H. Rumen microbial communities influence metabolic phenotypes in lambs. Front Microbiol. 2015. 10.3389/fmicb.2015.01060. [DOI] [PMC free article] [PubMed]
- 19.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol. 2018;16:171–181. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A. 2012;109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-del Campo A, Bodeaa S, Hamera HA, Marksa JA, Haiserb HJ, Turnbaugh PJ, et al. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio. 2015;6:e00042–e00015. doi: 10.1128/mBio.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability. MBio. 2015;6:e02481–e02414. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rath S, Heidrich B, Pieper DH, Vital M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome. 2017;5:54. doi: 10.1186/s40168-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S, et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014;24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, et al. Assembly of 913 microbial genomes from metagenomics sequencing of the cow rumen. Nat Commun. 2018. 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed]
- 28.Seshadri R, Leahy SC, Attwood GT, Teh KH, Lambie SC, Cookson AL, Eloe-Fadrosh EA, et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat Biotechnol. 2018;36:359–367. doi: 10.1038/nbt.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creevey CJ, Kelly WJ, Henderson G, Leahy SC. Determining the culturability of the rumen bacterial microbiome. Microb Biotechnol. 2014;7:467–479. doi: 10.1111/1751-7915.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terrapon N, Lombard V, Drula E, Lapébie P, Al-Masaudi S, Gilbert HJ, et al. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 2018;46(D1):D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 32.Abbott DW, Hrynuik S, Boraston AB. Identification and characterization of a novel periplasmic polygalacturonic acid binding protein from Yersinia enterolitica. J Mol Biol. 2007;367:1023–1033. doi: 10.1016/j.jmb.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 33.Caro-Quintero A, Ritalahti KM, Cusick KD, Löffler FE, Konstantinidis KT. The chimeric genome of Sphaerochaeta: nonspiral spirochetes that break with the prevalent dogma in spirochete biology. MBio. 2012;15:e00025–e00012. doi: 10.1128/mBio.00025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fricke W, Seedorf H, Henne A, Krüer M, Liesegang H, Hedderich R, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoedt EC, Parks DH, Volmer JG, Rosewarne CP, Denman SE, McSweeney CS, Muir JG, Gibson PR, Cuív PÓ, Hugenholtz P, Tyson GW, Morrison M. Culture- and metagenomics-enabled analyses of the Methanosphaera genus reveals their monophyletic origin and differentiation according to genome size. ISME J. 2018;12:2942–2953. doi: 10.1038/s41396-018-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoedt EC, Cuív PÓ, Evans PN, Smith WJ, McSweeney CS, Denman SE, et al. Differences down-under: alcohol-fueled methanogenesis by archaea present in Australian macropodids. ISME J. 2016;10:2376–2388. doi: 10.1038/ismej.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leahy SC, Kelly WJ, Li D, Li Y, Altermann E, Lambie SC, et al. The complete genome sequence of Methanobrevibacter sp. AbM4. Stand Genomic Sci. 2013;8:215–227. doi: 10.4056/sigs.3977691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J-H, Rhee M-S, Kumar S, Lee G-H, Chang D-H, Kim D-S, et al. Genome sequence of Methanobrevibacter sp. strain JH1, isolated from rumen of Korean native cattle. Genome Announc. 2013. 10.1128/genomeA.00002-13. [DOI] [PMC free article] [PubMed]
- 39.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, et al. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One. 2010. 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed]
- 40.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 41.Kabel MA, Yeoman CJ, Han Y, Dodd D, Abbas CA, de Bont JA, Morrison M, Cann IK, Mackie RI. Biochemical characterization and relative expression levels of multiple carbohydrate esterases of the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate. Appl Environ Microbiol. 2011;77:5671–5681. doi: 10.1128/AEM.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joblin KN, Naylor GE. The ruminal mycoplasmas: a review. J Appl Anim Res. 2002;21:161–179. doi: 10.1080/09712119.2002.9706368. [DOI] [Google Scholar]
- 43.Berg Miller ME, Antonopoulos DA, Rincon MT, Band M, Bari A, Akraiko T, et al. Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One. 2009. 10.1371/journal.pone.0006650. [DOI] [PMC free article] [PubMed]
- 44.Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, Deneke J, et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS One. 2011. 10.1371/journal.pone.0018814. [DOI] [PMC free article] [PubMed]
- 45.Weimar MR, Cheung J, Dey D, McSweeney C, Morrison M, Kobayashi Y, et al. Development of multiwell-plate methods using pure cultures of methanogens to identify new inhibitors for suppressing ruminant methane emissions. Appl Environ Microbiol. 2017;83:e00396–e00317. doi: 10.1128/AEM.00396-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. Accelerated profile HMM searches. PLOS Comp Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 50.Kamke J, Kittelmann S, Soni P, Li Y, Tavendale M, Ganesh S, Janssen PH, Shi W, Froula J, Rubin EM, Attwood GT. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome. 2016;4:56. doi: 10.1186/s40168-016-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G. Durbin R, and 1000 genome project data processing subgroup, the sequence alignment/map (SAM) format and SAMtools. Bioinfo. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kenters N, Henderson G, Jeyanathan J, Kittelmann S, Janssen PH. Isolation of previously uncultured rumen bacteria by dilution to extinction using a new liquid culture medium. J Microbiol Meth. 2011;84:52–60. doi: 10.1016/j.mimet.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peer J. 2015. 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed]
- 55.Tripp HJ, Sutton G, White O, Wortman J, Pati A, Mikhailova N, et al. Toward a standard in structural genome annotation for prokaryotes. Stand Genomic Sci. 2015;10:45. doi: 10.1186/s40793-015-0034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X, et al., editors. Research in Computational Molecular Biology. RECOMB 2013. Lecture Notes in Computer Science, vol 7821. Berlin: Springer; 2013. pp. 158–170. [Google Scholar]
- 57.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K, et al. The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4) Stand Genomic Sci. 2016;11:17. doi: 10.1186/s40793-016-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen I-M A, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D1:D507–D1:D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 61.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolut. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 62.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comp Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. A. CutC genes in the combined metagenome/metatranscriptome and Hungate1000 datasets. B. Metagenome abundance of PME genes identified from the combined metagenome/metatranscriptome and Hungate1000 datasets. C. Metatranscriptome abundance of PME transcripts of genes identified from the combined metagenome/metatranscriptome and Hungate1000 datasets. D. Top Blast hit against the 505 Pfam01095 containing ORFs identified from the rumen MAG dataset (n = 913).
Additional file 2: Figure S2. Choline trimethylamine lyase and bacterial microcompartment gene synteny in SPADES re-assembled metagenomes of low MY sheep (A), and in bacterial genomes of rumen (B) or ruminant faecal origin (C) in the Hungate1000 Collection.
Additional file 3: Table S2. Pectin methyl esterase genes (Pfam01095) identified in the Hungate 1000 collection reference genome set
Additional file 4: Table S3. Contigs and genome sizes of the three organisms identified from the MEC1 limited diversity metagenome.
Additional file 5: Table S4. A Overview of samples analysed in this study. B Results of the mapping of the high/low sample reads to the identified rumen CutC and PME-containing genes from the Hungate1000 and the high/low combined assembly.
Data Availability Statement
The metagenome and metatranscriptome datasets used in this study are accessible at the National Centre for Biotechnology Information Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) accession number SRA075938, BioProject number PRJNA202380, plus additional 16S rRNA gene amplicon sequence data under the SRA experiment accession numbers: SRX1079958 - SRX1079985. The Hungate1000 genomes are available from Joint Genome Institute’s Integrated Microbial Genomes and Microbiome Samples (IMG/M) which can be accessed at http://genome.jgi.doe.gov/. The raw sequence data and assembled genomes and proteomes from the 913 rumen uncultured genomes (RUG) and HiC rumen uncultured genomes (hRUG) are available the European Nucleotide Archive under project PRJEB21624. The SPADES assemblies of low methane yield sheep rumen microbial communities from New Zealand can be accessed via their IMG Database Project IDs: Sheep Tag 1265 (Gp0054682; Gp0053989), Sheep Tag 1283 (Gp0054684, Gp0054469); Sheep Tag 1435 (Gp0053990, Gp0054493), Sheep Tag 1494 (Gp0054822, Gp0054568).