Abstract
We demonstrated a general method to evaluate systematic errors related to Magnetic Resonance (MR) imaging sequences in marker-based co-registration of MR and Computed Tomography (CT) images, and investigated the effect of MR image quality in the co-registration process using clinical MR and CT protocols for stereotactic ablative body radiotherapy (SABR) planning of the liver. Small systematic errors (under 1.6 mm) were detected, unlikely to be a clinical risk to liver SABR. The least favourable marker configuration was found to be a co-planar arrangement parallel to the transaxial image plane.
Keywords: Registration, Fiducial markers, Stereotactic radiotherapy, Liver
1. Introduction
The fusion of magnetic resonance (MR) and computed tomography (CT) images combines the superior soft tissue contrast of MRI with the CT-based electron density information required for Radiotherapy (RT) planning [1]. MR is the diagnostic imaging modality of choice for liver tumours and aids radiotherapy target delineation, which can be challenging particularly for tumours difficult to visualise on CT such as liver metastases [2], [3]. Optimal MR-CT co-registration is therefore essential to the accuracy of MR derived target volume (TV) delineation and the definition of the disease extent. For mobile anatomical structures, metallic markers inserted around the TV may be used for accurate patient set up before treatment and to enable the MR-CT co-registration during RT planning [4], [5]. Markers may also be used to track tumour motion in real time during X-Ray guided RT delivery thus mitigating the effects of physiological motion [6], [7], [8], [9]. In X-Ray Guided Stereotactic Ablative Body Radiotherapy (SABR) of non-resectable metastatic liver disease, a minimum of three non-colinear markers is used to track tumour motion during RT treatment.
There are practical constraints in placing markers. In livers, access to the tumour is limited by the surrounding organs and ribcage [10]. Liver CT images are acquired relatively rapidly in exhale breath hold. However, there are practical constraints to the spatial resolution of MRI images acquired within a breath-hold. Typical breath-holds for liver examinations last for 10–20 s and require 2D or 3D techniques characterised by thicker slices (at least 4 mm) and data truncation to enable the entire liver volume to be examined; the nominal in-plane spatial resolution is rarely achieved in practice in MRI. In MRI markers are characterised by susceptibility-related signal loss, which depends on the MRI pulse sequence properties and the orientation of the marker in relation to the main magnetic field and to the image plane [11]. Both the receiver bandwidth and the frequency encoding direction have an effect on the depiction of the signal void around the marker and the signal loss pattern is not necessarily symmetric in relation to the marker position [12].
Clinical studies of CT-MR co-registration have considered different error sources, including marker migration and tissue deformations [10], [13], and different methods have been proposed to improve registration accuracy [14], [15]. A previous multi-institutional study reported considerable uncertainties employing MR-CT deformable registration for liver cancer [16]. Automated and semi-automated segmentation of internal structures may improve registration accuracy and can potentially facilitate tumour delineation in SABR of the liver [17]. In the liver, marker group deformations and rotations have been observed; they can be significant in the vicinity of the tumour, close to high dose gradients and this could compromise target coverage [4]. In contrast, clinical prostate studies have demonstrated smaller discrepancies between marker midpoints [12], [18], suggesting greater accuracy with a smaller, more rigid organ.
In clinical studies, the compromise between time and spatial resolution in MRI impacts on image quality, but the effect of the latter on the registration accuracy cannot be investigated independently from clinical factors such as motion artifacts and marker migrations. We hypothesise that data truncation artifacts and the low resolution of MRI datasets (compared to CT) are detrimental to the co-registration process. In this work we assess the accuracy of the CT-MR co-registration by implanting markers in gel test objects to be scanned (MR and CT) with clinical liver protocols. By using homogenous gel test objects, the effect of marker image quality on CT-MR registration is assessed separately. This registration is challenging, as it cannot be guided by heterogeneous tissue structure and anatomical borders in the vicinity of the lesion. The methods used in this study therefore aim to evaluate the limitations of breath hold MR sequences used for liver SABR under a variety of scenarios including those which are expected to be least favourable.
2. Materials and methods
Patients scheduled for liver SABR at our institution had at least two pairs of linked fiducial markers (FlexiMarc G/T™, FM-1.0-2-20-GT-18-20, CIVCO) inserted before undertaking MR and CT examinations for RT planning. Each marker’s diameter is 1 mm and each pair of markers is separated by a 20 mm titanium rod and was inserted along a different path under local anaesthetic using CT guidance by an interventional radiologist. The proximity and arrangement of the markers with respect to the lesion are determining factors in the accuracy of tumour tracking [4], [10]. Radiologists aimed to place the markers proximal to the treatment site in a non-colinear orientation, approximately centred on the tumour, but without contacting the lesion. The latter prevents the deposit of cancerous cells along the needle tract during removal. This resulted in inter-marker separations as low as 20–40 mm for small lesions.
In a clinical setting, transaxial MR images were acquired during an exhale breath-hold (Skyra 3.0T, Siemens, Erlangen, Germany) and registered to an exhale breath hold CT (LightSpeed RT16, GE Medical Systems). T1-weighted (T1w), T2-weighted (T2w) and T2*-weighted (T2*w) images with 4 mm slice thickness required truncation of the acquisition matrix to be acquired within a breath-hold; MR datasets were thus anisotropic, with higher in-plane resolution in relatively thick slices. Both T1w and T2w images were used for visualisation of tumours, and T2*w images were used to enable visualisation of small markers by emphasising susceptibility-related signal loss. MR sequence parameters are provided in Table 1. Helical exhale breath hold CT examinations had 1.25 mm slice thickness and 1.0 × 1.0 mm2 in-plane resolution. Rigid MR-CT co-registration was undertaken using automatic and manual rigid-body registration (i.e. rotation and translation only) (Eclipse, Varian Medical Systems, Inc. Palo Alto, CA).
Table 1.
MR sequence parameters.
| MR sequences | T1w (3D Dixon-VIBE) |
T2w (2D fast spin-echo) |
T2*w (2D spoiled gradient echo) |
|---|---|---|---|
| TR/TE (ms) | 5.87/2.47 | 1600/96 | 230/4.92 |
| Voxel size (mm3) | 1.5 × 1.5 × 4 | 1.2 × 1.2 × 4 | 1.5 × 1.5 × 4 |
| Reconstruction/acquisition matrix | 256 × 192/ 256 × 144 |
320 × 240/ 320 × 194 |
256 × 192/ 256 × 154 |
| % Sampling | 75 | 81 | 80 |
| Pixel bandwidth (Hz/px) | 1030 | 710 | 1395 |
| Readout gradient direction | R/L | R/L | R/L |
| Parallel imaging factor | 2 | 2 | 2 |
2.1. Test object development and imaging
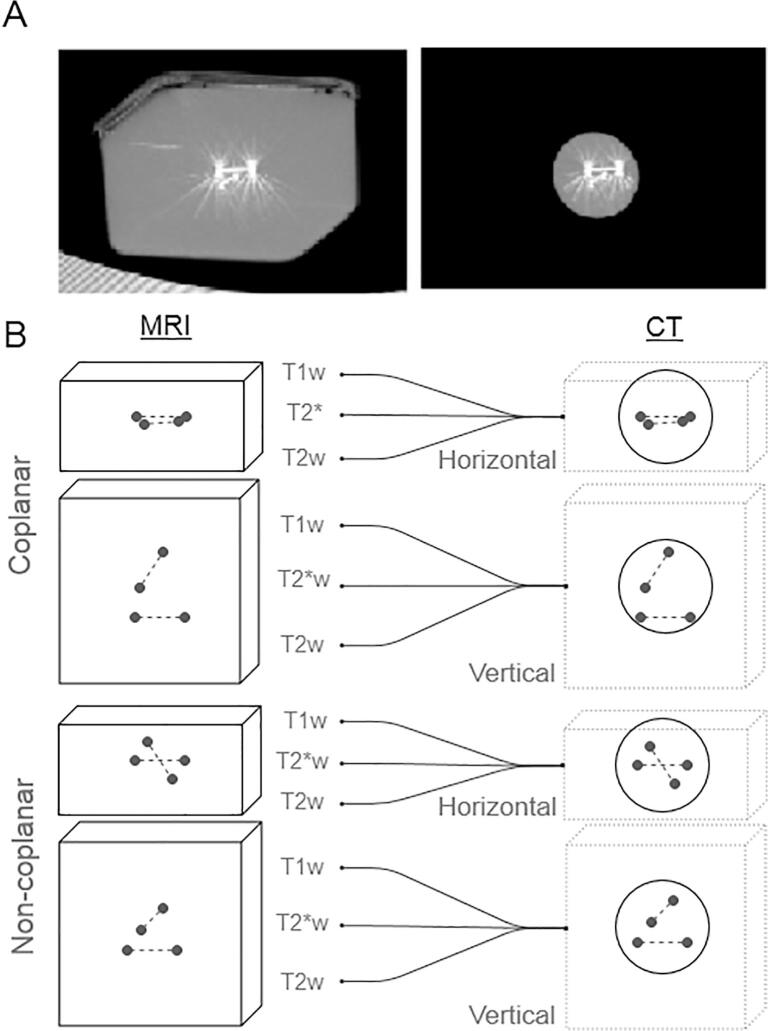
In order to investigate the accuracy of CT-MR co-registration, two test objects were built by suspending two linked pairs of markers in porcine gelatine (100 g/L) inside a 14 cm × 14 cm × 8 cm plastic container. The position of the markers was chosen to represent the most challenging clinical situations. Τhe most unfavourable marker configurations were chosen by limiting the furthest distance between any two markers to the 20–30 mm range. This range represents the typical minimum marker separation in the treatment of metastatic liver tumours using stereotactic radiosurgery at our institution.
In Test Object A, the markers were co-planar; in Test Object B, the markers were non-coplanar (Fig. 1). Both Test Objects were scanned using the clinical liver MR and CT protocols described above, in two different positions (vertical and horizontal). The vertical Test Object orientation places the co-planar arrangement of markers parallel to the image plane (transaxial).
Fig. 1.
A. CT Images of the coplanar marker configuration before (left) and after (right) processing to remove the external container shape. The vertical test object orientation makes the arrangement of co-planar markers parallel to the image plane (transaxial) B. A total of N = 12 combinations of test object configuration and MR sequence were registered with CT.
Fully sampled MR datasets were also acquired to obtain a ‘gold standard’ for the CT-MR registrations (100% sampling in the phase and readout directions). These fully sampled images are not degraded by data truncation artifacts, but cannot be acquired within the duration of a clinical breath-hold scan. All images were transferred to the Treatment Planning System (TPS) for CT-MR co-registration.
2.2. CT-MR co-registration and data analysis
‘Gold-standard’ CT-MR registrations were performed by a RT expert using the fully sampled MR datasets and were guided by the external shape of the Test Object (Fig. 1A). The same CT datasets were then duplicated and edited to remove any information related to the external Test Object shape which could contribute towards co-registration: a 46 mm diameter spherical volume containing the markers only was defined and the image intensity was assigned to zero elsewhere (Fig. 1A). The edited CT dataset was then registered to the obtained MR Test Object images by a different TPS user. This second registration is based on marker information only. In total 12 registrations were performed independently (Fig. 1B): two different Test Objects (A and B) in two different Test Object orientations (Vertical and Horizontal), and 3 different pulse sequences.
For each registration the transformation coordinates were extracted from the exported DICOM registration object, using in-house software (python, v2.7, and pydicom package v0.9.9). The accuracy of each marker-based registration, Rmarker, was then quantified by the offset d in each marker position from the position associated with the gold standard registration, RGoldStd:
where pGoldStd and pMarker are the marker coordinate vectors for each registration:
Here pCT is the marker position vector in the original CT image, as found manually by point selection in the TPS.
One sample t-tests were performed to test the null hypothesis that the population mean offset is zero on each component of the displacement d (along the slice selection, readout and phase encoding directions). One-way analysis of variance (ANOVA) was used to find any significance in the difference between the three sequence results. Two sample t-tests were performed to test the null hypothesis that the coplanar and non-coplanar test objects share the same mean offset, and that there is no dependence on the test object imaging orientation. A test significance limit of 5% is used for all tests.
3. Results
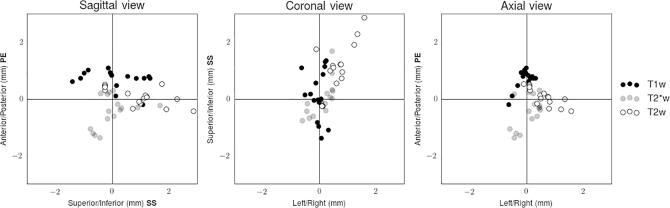
Table 2 lists the mean offset and standard deviation in marker-based registration for each breath-hold MR sequence (upper part) and each test object configuration (lower part), respectively. The mean absolute value of the offset across all registrations was found to be 1.2 ± 0.6 mm (mean ± standard deviation). The mean absolute displacement ranges from 0.9 to 1.4 mm for different sequences, and from 0.6 to 1.6 mm for different Test Objects and positions. Table 2 shows in bold type the measurements where one-sample t-tests rejected the null hypothesis that the population mean is zero, indicating there are sequence dependent systematic shifts along the phase encoding gradient for the T1w (p = 5.5 × 10−7) and T2*w (p = 5.3 × 10−3) sequences, and along the readout (p = 2.0 × 10−4) and slice selection (p = 8.2 × 10−4) gradients for the T2w sequence. Fig. 2 shows these data graphically for each MR sequence. The pulse sequence with the highest nominal in-plane spatial resolution (T2w) produced the largest displacement, and analysis of variance (one-way ANOVA) indicates significant differences between the sequences (p < 0.05 for all directions).
Table 2.
Marker displacements between gold standard registrations and marker-based registrations.
| Gold-std. registrations: Fully sampled MR Marker-based registrations: Breath-hold MR | Mean offset and standard deviation |
|||
|---|---|---|---|---|
| Phase Enc. (A/P) | Slice Sel. (S/I) | Readout (R/L) | ||
| MR sequence (N = 16) |
T1w | 0.7 ± 0.3 | 0.1 ± 0.9 | 0.0 ± 0.3 |
| T2*w | −0.4 ± 0.5 | 0.0 ± 0.6 | 0.1 ± 0.4 | |
| T2w | 0.1 ± 0.3 | 1.0 ± 0.9 | 0.6 ± 0.5 | |
| One way ANOVA result | p < 0.001 | p = 2.8 × 10−3 | p < 0.001 | |
| Marker arrangement (N = 24) |
Coplanar | 0.3 ± 0.5 | 0.6 ± 0.9 | 0.3 ± 0.5 |
| Non-coplanar | −0.1 ± 0.7 | 0.2 ± 0.9 | 0.1 ± 0.4 | |
| 2 sample t-test result | p = 0.04 | p = 0.13 | p = 0.07 | |
| Test object orientation (N = 24) |
Horizontal | 0.1 ± 0.7 | 0.1 ± 0.6 | 0.0 ± 0.4 |
| Vertical | 0.1 ± 0.5 | 0.7 ± 1.1 | 0.4 ± 0.4 | |
| 2 sample t-test result | p = 0.94 | p = 0.02 | p < 0.001 | |
Results are listed with the p-value from the relevant statistical significance test performed. Here N is the number of measurements. Values shown in bold indicate results which reject the null hypothesis of a population mean equal to zero using a one-sample t-test (p < 0.05).
Fig. 2.
Sagittal, coronal and axial views of the vector offset for each MR sequence. The phase encoding (PE), frequency encoding (FE) and slice selection (SS) gradient directions are indicated in bold on the axis labels.
The coplanar marker arrangement was associated with statistically significant non-zero offsets in all directions, although the two-sample t-test results only indicate a significant difference to non-coplanar registration in the phase encoding direction. As expected, the arrangement with the coplanar markers parallel to the Transaxial plane (vertical Test Object position) was associated with larger absolute displacements (1.6 ± 1.5 mm). There are statistically significant registration differences associated with the test object orientation in the slice selection (p = 6.7 × 10−3) and readout direction (p = 4.6 × 10−5) only.
4. Discussion
CT-MR co-registration errors associated with MR image quality cannot be assessed independently in clinical studies, as the co-registration is necessarily affected by motion, deformation and marker migration. In this work we considered various challenging marker configurations and the effect of MRI sequence selection on the accuracy of marker-based CT-MR registrations in liver SABR planning. We considered all three types of MR contrast used in liver imaging (T1W, T2W and T2*W images), with parameters widely used in clinical practice: transaxial anisotropic datasets characterised by lower spatial resolution in the superior/inferior direction and data truncation to keep the examination within a breath-hold. It is important to stress that many different strategies can be used to keep the total MR data acquisition time within a 10–20 s breath-hold, using different degrees of data truncation, half-Fourier imaging and parallel imaging in 2D and 3D. As demonstrated by Jonsson et al. [12], the signal loss surrounding a marker is not necessarily symmetric in relation to the marker position and therefore assessment of clinical MR images cannot rule out systematic errors. Hence it is important to have a baseline assessment of MR-CT co-registration prior to considering other practical and clinical factors such as marker-group deformation and marker migration.
In this work we employed test objects constructed using a homogenous gel substrate, to represent the most challenging marker configurations for CT-MR registration. Co-registrations were performed by expert RT users with SABR experience and access to MR physics support. Despite having considered challenging marker placements, we found only relatively small displacements between the gold-standard registration and the marker-only registrations (1.2 ± 0.6 mm mean absolute displacement). There were significant differences between the sequences considered (p < 0.05), with T2*w imaging producing the smallest errors as the signal loss surrounding the marker is emphasised.
Patient studies reported MR-CT co-registration errors associated with liver deformation [16], [17], fiducial marker migration [10] and tumour spatial differences in MRI and CT [3]. Osorio et al reported liver deformations in the range of 2.8–10.7 mm after rigid MR-CT registration, which dropped to a range of 1.3–1.9 mm after non-rigid registration suggesting that liver can be considerably deformed especially after abdominal compression used during CT scan [17]. Smaller deformations were found when CT and MR scans were performed under more similar conditions [3], [16], [20]. Brock et al reported uncertainties between 3.9 mm and 6.5 mm employing MR-CT deformable registration for liver cancer [16], and a mean registration accuracy of under 3.4 mm was found by Elhawary et al. [20]. A close arrangement of the markers to the tumour would decrease the set up systematic error to 0.6 mm for liver compression and 0.4 mm for free-breathing [4]. The systematic errors we measured with our test object are smaller than most of the co-registration errors reported in clinical situations, and are only comparable to the results of Wunderink et al. [4] for the close arrangement of markers, suggesting that the systematic errors we detected are a small component of the overall CT-MR registration error.
The principal aspect of our work is to highlight sequence-dependent systematic displacements, which are typically sub-millimetre (Fig. 2). The largest systematic displacements were along the phase encoding direction with the T1w and T2*w sequences, which exhibited a submillimetre systematic offset in opposite directions. The most likely cause of this displacement is the users’ interpretation of the truncation artifacts (Gibbs), which will appear asymmetric when the markers are oblique to the slice considered. The displacement direction may be associated with different patterns of data truncation in different sequences. Additionally, the markers appear displaced along the slice selection direction with the T2w sequence and susceptibility-related slice warping cannot be ruled out. MRI artifacts dependent on receiver bandwidth can give rise to asymmetries arising in the readout encoding direction [12] and displacements are expected to be more pronounced using sequences with a lower bandwidth. However, the displacement along the readout direction is unexpected with sequences employing readout gradient reversals, and this may require further investigation.
Clinical marker placement is also a potential limiting factor in co-registration accuracy. During insertion, clinicians aim to minimise the potential for tissue distortion of the collective marker constellation by achieving minimal inter-marker distances [10], [19]. This however, has the corresponding effect of also potentially limiting the achievable co-registration accuracy. Intercostal marker insertions are also prone to result in marker alignment at small angles to the transaxial plane. Insertion protocols aim to avoid this, but nevertheless a coplanar marker arrangement represents a potential clinical scenario. Our work demonstrated clearly that a co-planar marker arrangement is less favourable.
Additionally, liver deformations contribute considerably to the geometric stability of the implanted fiducial markers and larger intermarker distances would increase registration errors [10]. Therefore, marker-based MR-CT registration is often combined with soft tissue alignment. In this sense, the relatively small errors reported by our work truly represent a very difficult scenario because (i) the maximum intermarker distance is small for our test objects and (ii) the registration does not include any information that is not marker-based. Therefore errors associated with MR image quality are very unlikely to exceed the values we measured.
A full understanding of any systematic displacements is clearly desirable, but it requires knowledge of many pulse sequence parameters not easily available to the user. However, the assessment method presented here is straightforward and generally applicable to marker-based registration. It applies a gold standard CT-MR registration guided by the test object shape and employs a separate edited CT dataset for marker-only registration, thus providing considerable insight into a difficult clinical problem.
In a clinical setting, the visibility of the tumour has to be considered alongside marker visibility. It may be necessary to include more than one MR dataset in the planning process. A complex picture may thus emerge, with MR images based on different contrast mechanism originating from different breath-holds being registered to each other and to CT. Although small, different systematic errors associated with different datasets can thus be combined, depending on the characteristics of a particular workflow. A method to consider systematic errors associated with image quality alone may prove invaluable.
In conclusion, the error in MR-CT registration was found not to exceed 1.6 mm for Test Objects designed to represent very challenging clinical situations and to act as a worst-case scenario for marker configuration. Considering the CTV to PTV margins employed and all other motion and deformation uncertainties, the marker-based registration is unlikely to be a clinical risk in SABR planning for liver cancer. Our approach to assess the effect of image quality on marker-based registration is general, and the results can be extended to other clinical applications where marker-based registration is employed and the effect of MR image quality needs to be assessed.
Conflict of interest
None declared.
Acknowledgements
CRUK and EPSRC support to the Cancer Imaging Centre at ICR and RMH in association with MRC and Department of Health C1060/A10334, C1060/A16464 and NHS funding to the NIHR Biomedical Research Centre and the Clinical Research Facility in Imaging.
Contributor Information
Thomas Woolcot, Email: Thomas.Woolcot@bsuh.nhs.uk.
Evanthia Kousi, Email: Eva.Kousi@icr.ac.uk.
Emma Wells, Email: emma.wells@rmh.nhs.uk.
Katharine Aitken, Email: katharine.aitken@rmh.nhs.uk.
Helen Taylor, Email: Helen.Taylor@rmh.nhs.uk.
Maria A. Schmidt, Email: Maria.Schmidt@icr.ac.uk.
References
- 1.Schmidt M.A., Payne G. Radiotherapy planning using MRI. Phys Med Biol. 2015;60:323–361. doi: 10.1088/0031-9155/60/22/R323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pech M., Mohnike K., Wieners G., Bialek E., Dudeck O., Seidensticker M. Radiotherapy of liver metastases. comparison of target volumes and dose-volume histograms employing CT- or MRI-based treatment planning. Strahlenther Onkol. 2008;184:256–261. doi: 10.1007/s00066-008-1849-8. [DOI] [PubMed] [Google Scholar]
- 3.Voroney J.P., Brock K.K., Eccles C., Haider M., Dawson L.A. Prospective comparison of computed tomography and magnetic resonance imaging for liver cancer delineation using deformable image registration. Int J Radiat Oncol Biol Phys. 2006;66:780–791. doi: 10.1016/j.ijrobp.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 4.Wunderink W., Méndez Romero A., Seppenwoolde Y., de Boer H., Levendag P., Heijmen B. Potentials and limitations of guiding liver stereotactic body radiation therapy set-up on liver-implanted fiducial markers. Int J Radiat Oncol Biol Phys. 2010;77:1573–1583. doi: 10.1016/j.ijrobp.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 5.Nair J.V., Szanto J., Vandervoort E., Henderson E., Avruch L., Malone S. Feasibility, detectability and clinical experience with platinum fiducial seeds for MRI/CT fusion and real-time tumor tracking during CyberKnife stereotactic ablative radiotherapy. J Radiosurg SBRT. 2015;3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 6.Calcerrada Diaz-Santos N., Blasco Amaro J.A., Cardiel G.A., Andradas Aragones E. The safety and efficacy of robotic image-guided radiosurgery system treatment for intra- and extracranial lesions: a systematic review of the literature. Radiother Oncol. 2008;89:245–253. doi: 10.1016/j.radonc.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 7.Poels Kenneth, Dhont Jennifer, Verellen Dirk, Blanck Oliver, Ernst Floris, Vandemeulebroucke Jef. A comparison of two clinical correlation models used for real-time tumor tracking of semi-periodic motion: a focus on geometrical accuracy in lung and liver cancer patients. Radiother Oncol. 2015;115:419–442. doi: 10.1016/j.radonc.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Xu Zhiyuan, Ellis Scott, Lee Cheng-Chia, Starke Robert M., Schlesinger David, Vance Mary Lee. Silent corticotroph adenomas after stereotactic radiosurgery: a case control study. Int J Radiat Oncol Biol Phys. 2014;90:903–910. doi: 10.1016/j.ijrobp.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Poulsen Per Rugaard, Worm Esben S., Petersen Jørgen B.B., Grau Cai, Fledelius Walther, Høyer Morten. Kilovoltage intrafraction motion monitoring and target dose reconstruction for stereotactic volumetric modulated arc therapy of tumors in the liver. Radiother Oncol. 2014;111:424–430. doi: 10.1016/j.radonc.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Seppenwoolde Y., Wunderink W., Wunderink-van Veen S.R., Storchi P., Méndez Romero A., Heijmen B.J.M. Treatment precision of image-guided liver SBRT using implanted fiducial markers depends on marker–tumour distance. Phys Med Biol. 2011;56:5445–5468. doi: 10.1088/0031-9155/56/17/001. [DOI] [PubMed] [Google Scholar]
- 11.Callaghan P.T. Clarendon Press; Oxford: 1991. Principles of nuclear magnetic resonance microscopy; pp. 208–227. [Google Scholar]
- 12.Jonsson J.H., Garpebring A., Karlsson M.G., Nyholm T. Internal fiducial markers and susceptibility effects in MRI-simulation and measurement of spatial accuracy. Int J Radiat Biol Phys. 2012;82:1612–1618. doi: 10.1016/j.ijrobp.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Nichol A.M., Brock K.K., Lockwood G.A., Moseley D.J., Rosewall T., Warde P.R. A magnetic resonance imaging study of prostate deformation relative to implanted gold fiducial markers. Int J Radiat Oncol Biol Phys. 2007;67:48–56. doi: 10.1016/j.ijrobp.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Lee C.W.C., Tublin M.E., Chapman B.E. Registration of MR and CT images of the liver: comparison of voxel similarity and surface based registration algorithms. Comput Methods Programs Biomed. 2005;78:101–114. doi: 10.1016/j.cmpb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Tang S., Chen Y.W., Xu R., Wang Y., Morikawa S., Kurumi Y. MR-CT image registration in liver cancer treatment with an open configuration MR scanner. In: Pluim J.P.W., Likar B., Gerritsen F.A., editors. WBIR 2006, LNCS 4057. © Springer-Verlag Berlin Heidelberg; 2006. pp. 289–296. [Google Scholar]
- 16.Brock K.K. Results of a multi-institution deformable registration accuracy study (MIDRAS) Int J Radiat Oncol Biol Phys. 2010;76(2):583–596. doi: 10.1016/j.ijrobp.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 17.Vasquez Osorio E.M., Hoogeman M.S., Méndez Romero A., Wielopolski P., Zolnay A., Heijmen B.J. Accurate CT/MR vessel-guided nonrigid registration of largely deformed livers. Med Phys. 2012;39:2463–2477. doi: 10.1118/1.3701779. [DOI] [PubMed] [Google Scholar]
- 18.Kapanen M., Collan J., Beule A., Seppala T., Saarilahti K., Tenhunen M. Commissioning of MRI-only based treatment planning procedure for external beam radiotherapy of prostate. Magn Reson Med. 2013;70:127–135. doi: 10.1002/mrm.24459. [DOI] [PubMed] [Google Scholar]
- 19.Park J.C., Park S.H., Kim J.H., Yoon S.M., Song S.Y., Liu Z. Liver motion during cone beam computed tomography guided stereotactic body radiation therapy. Med Phys. 2012;39:6431–6442. doi: 10.1118/1.4754658. [DOI] [PubMed] [Google Scholar]
- 20.Elhawary H., Oguro S., Tuncali K., Morrison P.R., Tatli S., Shyn P.B. Multimodality non-rigid image-registration for planning, targeting and monitoring during CT-guided percutaneous liver tumor cryoablation. Acad Radiol. 2010;17:1334–1344. doi: 10.1016/j.acra.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]