Abstract
Background
Mounting evidence shows that microRNA-34a (miR-34a) is involved in cancer prognosis. Therefore, we summarize the predictive role of miR-34a for survival in patients with gastrointestinal cancers (GICs).
Methods
All eligible studies were found by searching PubMed, Web of Science and EMBASE, and survival results were extracted. Then, the hazard ratio (HR) with the corresponding 95% confidence interval (CI) was calculated to evaluate the prognostic role of miR-34a in GICs. The association between miR-34a expression and clinicopathological characteristics was estimated by odds ratios (ORs) and 95% CIs.
Results
A total of 20 studies were included in this meta-analysis. For overall survival (OS), lower miR-34a expression could probably predict poorer outcome in GICs, with a pooled HR of 1.86 (95% CI: 1.52–2.28, P < 0.01). For disease-free survival (DFS), progression-free survival (PFS), and recurrence-free survival (RFS), lower miR-34a expression was related to worse DFS/PFS/RFS with a pooled HR of 1.86 (95% CI: 1.31–2.63, P < 0.01). A significant relation of differentiation/TNM stage/lymphatic metastasis and the expression level of miR-34a was identified.
Conclusion
This meta-analysis revealed that lower miR-34a expression is significantly connected with worse OS and DFS/PFS/RFS in GIC patients. In addition, the miR-34a expression level is relatively lower in patients with lymph node metastasis than in patients without lymph node metastasis, and decreased miR-34a expression levels are linked to poor tumour differentiation and late TNM stage. MiR-34a may become a new factor for the prognosis prediction and progression of GICs.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-020-07751-y.
Keywords: microRNA-34a, Gastrointestinal cancer, Prognosis, Meta-analysis
Background
Gastrointestinal cancers (GICs) account for the majority of cancer-related deaths worldwide, especially in developing countries [1]. Previous studies have shown that stomach, oesophageal, liver, and colorectal cancers are commonly identified as the leading causes of cancer deaths [2]. Currently, common treatments for GICs include surgery, neoadjuvant chemoradiotherapy, adjuvant chemoradiotherapy and immunotherapy; however, the therapeutic effects are limited in patients at advanced stages. Therefore, there is an urgent need for early detection of GICs and recognition of high-risk patients with poor prognosis.
MicroRNAs (miRNAs) are small-molecule RNAs with a length of 19 to 25 nucleotides that regulate the posttranscriptional silencing of target genes by combining with the 3′-untranslated region (3′-UTR) of target messenger RNA [3]. MiRNAs participate in various biological processes, including cell multiplication, differentiation, apoptosis and cell cycle regulation [4]. Studies have reported that miRNAs are abnormally expressed in tumours and have strong diagnostic and prognostic values [5].
MicroRNA-34a (miR-34a), a member of the miR-34 family, has been verified to be abnormally expressed in various tumours, including oesophageal cancer (EC) [6], gastric cancer (GC) [7], colorectal cancer (CRC) [8], hepatocellular carcinoma (HCC) [9], pancreatic cancer (PC) [10], gallbladder cancer (GBC) [11], and other cancers [12]. Based on recent studies, miR-34a has been considered closely related to gastrointestinal cancer multiplication [13], invasion [14] and metastasis [15], which points to the important biological roles of miR-34a in cellular signalling pathways, such as the MAPK/Ras pathway [16], Wnt/β-Catenin pathway [17], PI3K/Akt pathway [18], SIRT1/p53 pathway [19], and FoxM1/c-Myc pathway [20]. However, the prognostic accuracy of miR-34a in GICs was inconsistent among these studies. Hu et al. [21], Hui et al. [22], and Yang et al. [23] reported that a low expression level of miR-34a predicted a worse survival rate in GIC patients. In contrast, Osawa et al. [24], Zhang et al. [25] and Mojin Wang et al. [26] found that GIC patients benefited from downregulated miR-34a expression. To assess the prognostic value of miR-34a in GICs systematically and to discuss the association between miR-34a expression and clinicopathological characteristics, we performed a meta-analysis on the basis of all published relevant studies.
Methods
Literature search
We searched the PubMed, Web of Science and Embase databases to identify relevant studies before January 1, 2020. The following keywords were used: ‘microRNA-34a’, ‘miR-34a’, ‘cancer’, ‘neoplasm’, ‘oesophageal’, ‘stomach’, ‘colorectal’, ‘colon’, ‘pancreatic’, ‘hepatocellular’, ‘liver’, ‘gallbladder’, ‘prognosis’, ‘survival’, ‘hazard ratio’, and ‘gastrointestinal’. These keywords were combined with ‘AND’ or ‘OR’. The results were limited to papers published in English.
Selection criteria
Studies were included based on the following conditions: (1) the diagnosis of GICs was confirmed by histopathology; (2) the expression of miR-34a in tissue or blood was measured and divided into high and low levels; and (3) the survival outcome was reported directly or survival data were provided from Kaplan-Meier survival curves. The exclusion criteria were as follows: (1) reviews, laboratory studies or letters; and (2) the lack of or inability to calculate key information about survival outcomes, such as the HR or 95% CI.
Data extraction and quality assessment
Two investigators (Yan-Ling Chen and Xiao-Lin Liu) independently extracted the data from all eligible references, including first author, publication time, country, tumour type, sample type, test method, TNM stage, follow-up time and cut-off value, HRs of miR-34a for OS and/or DFS, PFS, RFS, and 95% CIs. In addition, data on clinical characteristics were collected from the studies that reported such information. All eligible studies were retrospective. The Newcastle-Ottawa Scale (NOS) was used to assess the quality of each study. The range of scores is 0 to 9, and a score greater than 6 was considered high quality [27]. Any disagreement was finally resolved by discussion.
Statistical analysis
We used RevMan 5.3 (Cochrane Collaboration, Oxford, UK) and STATA 12.0 (StataCorp LP, College Station, TX, USA) to conduct the statistical analysis. The pooled HRs and corresponding 95% CIs were used to evaluate the prognostic value of low miR-34a expression in GICs. The heterogeneity among studies was calculated by Cochran’s Q test and Higgins’s I2 statistic. If P > 0.05 or I2 ≤ 50%, we considered no significant heterogeneity to exist, and a fixed-effect model was used; if P ≤ 0.05 or I2 > 50%, a random-effect model was used. Some studies did not provide the HRs and 95% CIs directly, and we obtained the key points and the relevant data from Kaplan-Meier survival curves by utilising Engauge Digitizer 4.1 software and then calculated the HR and corresponding 95% CI following Tierney’s method [28]. Publication bias was assessed by funnel plots and Egger’s test. In addition, we performed a sensitivity analysis by removing studies one by one to assess the influence of a single study. The association between miR-34a expression and clinicopathological characteristics was evaluated by the pooled OR and 95% CI.
Results
Literature search
A total of 1196 records were obtained in the beginning. A total of 825 studies were excluded because of duplication, and 282 records were excluded after screening the titles and abstracts. According to the selection criteria, 19 studies were finally identified as eligible, including 2 EC, 5 GC, 4 HCC, 4 PC, 3 CRC, and 1 GBC. Since one of the studies contained two different groups, 20 independent experiments were included for quantitative analysis. The flow diagram of the study selection is shown in Fig. 1.
Fig. 1.
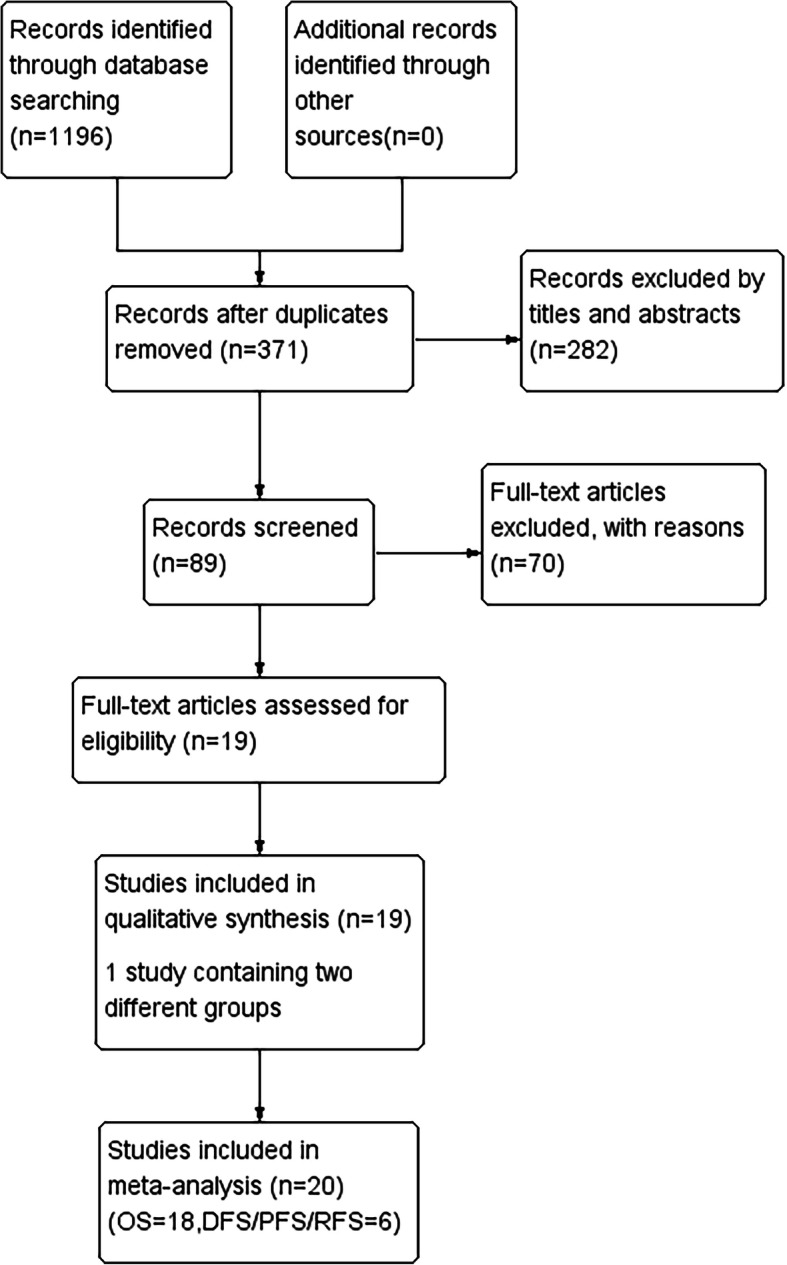
Flow diagram of the study selection process
Characteristics of the eligible studies
The main features of eligible studies are summarized in Table 1, and the summary of HRs and their 95% CIs are shown in Table 2. The eligible articles were published between 2011 and 2019, including 1691 participants with OS data and 676 participants with DFS/PFS/RFS data from China, America, Japan, Scotland, and Slovakia. The types of GICs included EC, GC, CRC, HCC, PC, and GBC. Quantitative real-time PCR (qRT-PCR) was extensively used in all studies to assess the expression of miR-34a. Tumour tissues were the most commonly used sample, except for Long L-M’s study [10] in which plasma samples were used. Among the included studies, 8 studies reported HRs and the corresponding 95% CIs directly, and the HRs and 95% CIs of the remaining 12 studies were calculated by Kaplan-Meier survival curves.
Table 1.
The main characteristics of included 20 studies
| Study | Year | Country | Type | Design | Sample | Num. | Stage | Cut-off | Follow-up | Test method | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hu et al. [21] | 2011 | America | EC | R | Tissue | 99 | I-IV | Median | > 250 | qRT-PCR | OS/DFS |
| Lin et al. [29] | 2015 | China | EC | R | Tissue | 111 | I-IV | Median | NR | qRT-PCR | OS |
| Osawa et al. [24] | 2011 | Japan | GC | R | Tissue | 37 | II-III | 70% | 60 | qRT-PCR | OS |
| Hui et al. [22] | 2015 | China | GC | R | Tissue | 76 | I-III | Mean | > 60 | qRT-PCR | OS |
| Wei et al. [30] | 2015 | China | GC | R | Tissue | 157 | I-IV | NR | > 100 | qRT-PCR | OS |
| Zhang et al. [31] | 2015 | China | GC | R | Tissue | 137 | I-IV | 2.44 | 68 | qRT-PCR | OS |
| Yang et al. [23] | 2015 | China | GC | R | Tissue | 50 | I-IV | Median | 60 | qRT-PCR | OS |
| Li et al. [32] | 2015 | China | HCC | R | Tissue | 114 | I-IV | ROC | 90 | qRT-PCR | OS/PFS |
| Yang et al. [33] | 2013 | China | HCC | R | Tissue | 30 | NR | Mean | 60 | qRT-PCR | OS |
| Cui et al. [34] | 2015 | China | HCC | R | Tissue | 120 | NR | Median | 60 | qRT-PCR | OS/RFS |
| Xu et al. [20] | 2015 | China | HCC | R | Tissue | 75 | I-IV | Median | 60 | qRT-PCR | OS/RFS |
| Ohuchida et al. [35] | 2011 | Japan | PC | R | Tissue | 90 | NR | NR | >100 | qRT-PCR | OS |
| Jamieson et al. [36] | 2012 | Scotland | PC | R | Tissue | 72 | NR | Median | 48 | qRT-PCR | OS |
| Long et al. [10] | 2016 | China | PC | R | plasma | 159 | I-IV | Mean | 24 | qRT-PCR | OS |
| Sun et al. [37] | 2018 | China | PC | R | Tissue | 139 | I-IV | Mean | 60 | qRT-PCR | OS |
| Zhang et al. [25] | 2017 | China | CRC | R | Tissue | 84 | I-IV | 2 | 36 | qRT-PCR | OS |
| Hasakova et al. [38] | 2019 | Slovakia | CRC | R | Tissue | 64 | I-IV | Median | 100 | qRT-PCR | OS |
| Gao et al. [39] | 2014 | China | CRC | R | Tissue | 205 | II-III | 0.307 | >80 | qRT-PCR | DFS |
| Gao et al. [39] | 2014 | China | CRC | R | Tissue | 63 | II-III | 0.307 | >80 | qRT-PCR | DFS |
| Jin et al. [11] | 2013 | China | GBC | R | Tissue | 77 | NR | Mean | 24 | qRT-PCR | OS |
Abbreviations: CRC colorectal cancer; DFS disease-free survival, EC esophageal cancer; GBC gallbladder cancer; GC gastric cancer; HCC hepatocellular carcinoma, NR no report, OS overall survival, PC pancreatic cancer, PFS progressive-free survival, qRT-PCR quantitative real-time PCR, R retrospective, RFS recurrence-free survival
Table 2.
Summary of HRs and their 95% CI
| Study | Year | Country | Tumor type | Outcome | HR | 95% CI | NOS |
|---|---|---|---|---|---|---|---|
| Hu et al. [21] | 2011 | America | EC |
OS DFS |
1.41 1.39 |
0.81–2.44 0.82–2.35 |
8 |
| Lin et al. [29] | 2015 | China | EC | OS | 3.182 | 1.273–10.184 | 6 |
| Osawa et al. [24] | 2011 | Japan | GC | OS | 0.2 | 0.06–0.68 | 6 |
| Hui et al. [22] | 2015 | China | GC | OS | 2.327 | 1.099–4.927 | 7 |
| Wei et al. [30] | 2015 | China | GC | OS | 2.31 | 0.13–40.12 | 8 |
| Zhang et al. [31] | 2015 | China | GC | OS | 1.33 | 1.14–1.61 | 8 |
| Yang et al. [23] | 2015 | China | GC | OS | 3.05 | 0.6–15.50 | 8 |
| Li et al. [32] | 2015 | China | HCC |
OS PFS |
1.81 1.22 |
1.03–3.18 0.92–1.62 |
6 |
| Yang et al. [33] | 2013 | China | HCC | OS | 3.54 | 1.67–7.52 | 7 |
| Cui et al. [34] | 2015 | China | HCC |
OS RFS |
1.44 1.49 |
1.13–1.72 1.15–1.79 |
8 |
| Xu et al. [20] | 2015 | China | HCC |
OS RFS |
1.96 1.96 |
1.04–3.57 1.10–3.45 |
8 |
| Ohuchida et al. [35] | 2011 | Japan | PC | OS | 2.92 | 1.303–6.295 | 8 |
| Jamieson et al. [36] | 2012 | Scotland | PC | OS | 6.67 | 2.684–16.573 | 8 |
| Long et al. [10] | 2016 | China | PC | OS | 1.88 | 1.35–2.64 | 8 |
| Sun et al. [37] | 2018 | China | PC | OS | 2.24 | 1.38–3.36 | 7 |
| Zhang et al. [25] | 2017 | China | CRC | OS | 1.76 | 1.01–3.05 | 6 |
| Hasakova et al. [38] | 2019 | Slovakia | CRC | OS | 1.34 | 0.65–2.75 | 8 |
| Gao et al. [39] | 2014 | China | CRC |
DFS DFS |
3.819 2.973 |
2.438–5.983 1.339–6.602 |
8 |
| Jin et al. [11] | 2013 | China | GBC | OS | 2.37 | 1.11–5.06 | 8 |
Abbreviations: 95%CI 95% confidence interval, CRC colorectal cancer, DFS disease-free survival, EC esophageal cancer, GBC gallbladder cancer, GC gastric cancer, HCC hepatocellular carcinoma, HR hazard ratio, OS overall survival, PC pancreatic cancer, PFS progressive-free survival, RFS recurrence-free survival
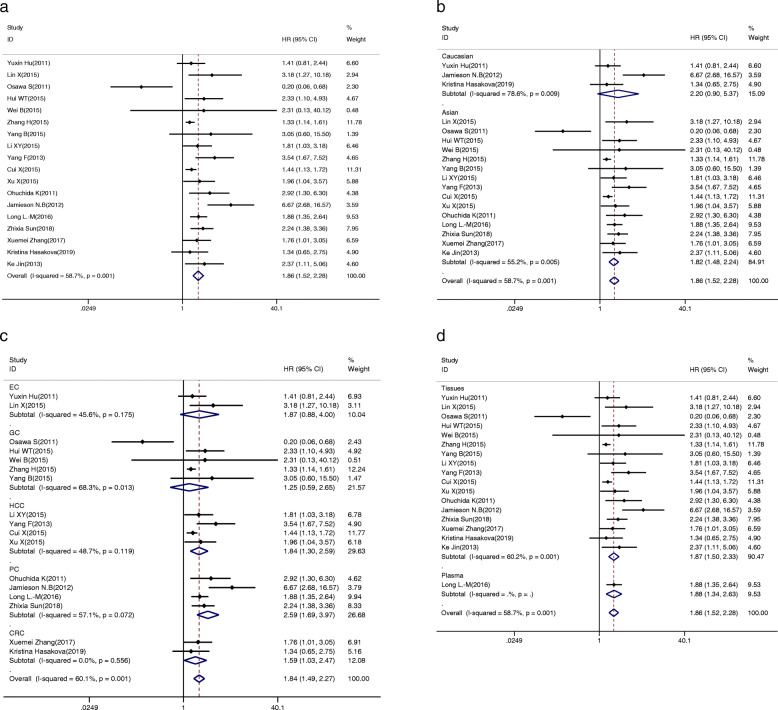
Overall survival is associated with miR-34a expression
We analysed the association between low expression of miR-34a and OS at first, and remarkable heterogeneity between studies was found (I2 = 58.7%, P = 0.001, Table 3). Therefore, the random-effect model was used to compute the pooled HR and corresponding 95% CI. The result showed that a lower expression level of miR-34a significantly predicted worse OS, with a pooled HR of 1.86 (95% CI: 1.52–2.28; Fig. 2a).
Table 3.
Association between miR-34a expression levels and overall survivals
| No.of studies | No.of patients | Pooled HR (95%CI) | Meta regression p-value | Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| Fixed | Random | I2 | p-value | ||||
| Overall | 18 | 1691 | 1.600 (1.44–1.77) | 1.86 (1.52–2.28) | 58.7% | 0.001 | |
| Ethnicity | 0.806 | ||||||
| Asian | 15 | 1456 | 1.58 (1.42–1.76) | 1.82 (1.48–2.24) | 55.2% | 0.005 | |
| Caucasian | 3 | 235 | 1.86 (1.25–2.76) | 2.20 (0.90–5.37) | 78.6% | 0.009 | |
| Sample Size | 0.979 | ||||||
| ≥ 100 | 7 | 937 | 1.51 (1.34–1.69) | 1.61 (1.35–1.92) | 36.1% | 0.153 | |
| <100 | 11 | 754 | 1.98 (1.59–2.48) | 2.00 (1.37–2.93) | 63.2% | 0.002 | |
| NOS Scores | 0.978 | ||||||
| ≥ 8 | 11 | 1100 | 1.53 (1.36–1.71) | 1.75 (1.42–2.16) | 49.5% | 0.031 | |
| < 8 | 7 | 591 | 2.00 (1.56–2.55) | 1.87 (1.20–2.93) | 65.8% | 0.008 | |
| Specimen | 0.933 | ||||||
| tissue | 17 | 1532 | 1.57 (1.41–1.75) | 1.87 (1.50–2.33) | 60.2% | 0.001 | |
| plasma | 1 | 159 | 1.88 (1.34–2.63) | 1.88 (1.34–2.63) | – | – | |
| Cancer Types | 0.494 | ||||||
| EC | 2 | 210 | 1.69 (1.04–2.74) | 1.87 (0.88–4.00) | 45.6% | 0.175 | |
| GC | 5 | 457 | 1.33 (1.13–1.57) | 1.25 (0.59–2.65) | 68.3% | 0.013 | |
| HCC | 4 | 339 | 1.60 (1.33–1.92) | 1.84 (1.30–2.59) | 48.7% | 0.119 | |
| PC | 4 | 460 | 2.27 (1.77–2.89) | 2.59 (1.69–3.97) | 57.1% | 0.072 | |
| CRC | 2 | 148 | 1.59 (1.03–2.47) | 1.59 (1.03–2.47) | 0.0% | 0.556 | |
| GBC | 1 | 77 | 2.37 (1.11–5.06) | 2.37 (1.11–5.06) | – | – | |
Abbreviations: 95%CI 95% confidence interval, CRC colorectal cancer, EC esophageal cancer, GBC gallbladder cancer, GC gastric cancer, HCC hepatocellular carcinoma, HR hazard ratio, NOS Newcastle-Ottawa Scale, PC pancreatic cancer
Fig. 2.
The association between miR-34a expression levels and (a) overall survival; subgroup analyses of (b) ethnicity (Asian and Caucasian), c cancer type (EC, GC, HCC, PC, CRC), and (d) specimen (plasma and tissues)
To explicate the heterogeneity in OS, subgroup analysis was conducted by ethnicity (Asian and Caucasian), sample capacity (≥100 and < 100), NOS scores (≥8 and < 8), specimen (plasma and tissue) and tumour types (EC, GC, CRC, HCC and PC). As a result, homogeneity was achieved in the CRC group (I2 = 0.00%, P = 0.556; Table 3) and the correlation was obvious (HR = 1.59, 95% CI:1.03–2.47, Fig. 2c). Additionally, there were significant correlations between the expression level of miR-34a and OS in Asian populations (HR = 1.82, 95% CI: 1.48–2.24, Fig. 2b); a sample capacity greater than or equal to 100 (HR = 1.61, 95% CI: 1.35–1.92, Supplementary Fig. 1A) or less than 100 (HR = 2.00, 95% CI: 1.37–2.93, Supplementary Fig. 1A); NOS scores equal to or greater than 8 (HR = 1.75, 95% CI: 1.42–2.16, Supplementary Fig. 1B) or less than 8 (HR = 1.87, 95% CI: 1.20–2.93, Supplementary Fig. 1B); specimens removed the plasma (HR = 1.87, 95% CI: 1.50–2.33, Fig. 2d), HCC (HR = 1.84, 95% CI: 1.30–2.59, Fig. 2c), and PC (HR = 2.59, 95% CI:1.69–3.97, Fig. 2c) by the random-effect model. As shown in Table 3, the significance disappeared in Caucasian and EC groups when the fixed-effect model was transformed into the random-effect model. Moreover, the heterogeneities were still evident among subgroups, except for the CRC group. Ultimately, to analyse heterogeneity, meta regression was performed, but it was unable to explain the variation in HRs (p = 0.806 for ethnicity, p = 0.979 for sample capacity, p = 0.978 for NOS scores, p = 0.933 for specimen, and p = 0.494 for cancer types, Table 3). Moreover, the sensitivity analysis was performed to assess the contribution of each study, and no study seemed to make a difference to the pooled results (Supplementary Fig. 2A). In addition, publication bias was evaluated by funnel plots and Egger’s test. As shown in Supplementary Fig. 2B, the funnel plots showed no obvious asymmetry, and Egger’s test revealed that no significant publication bias existed (P = 0.058).
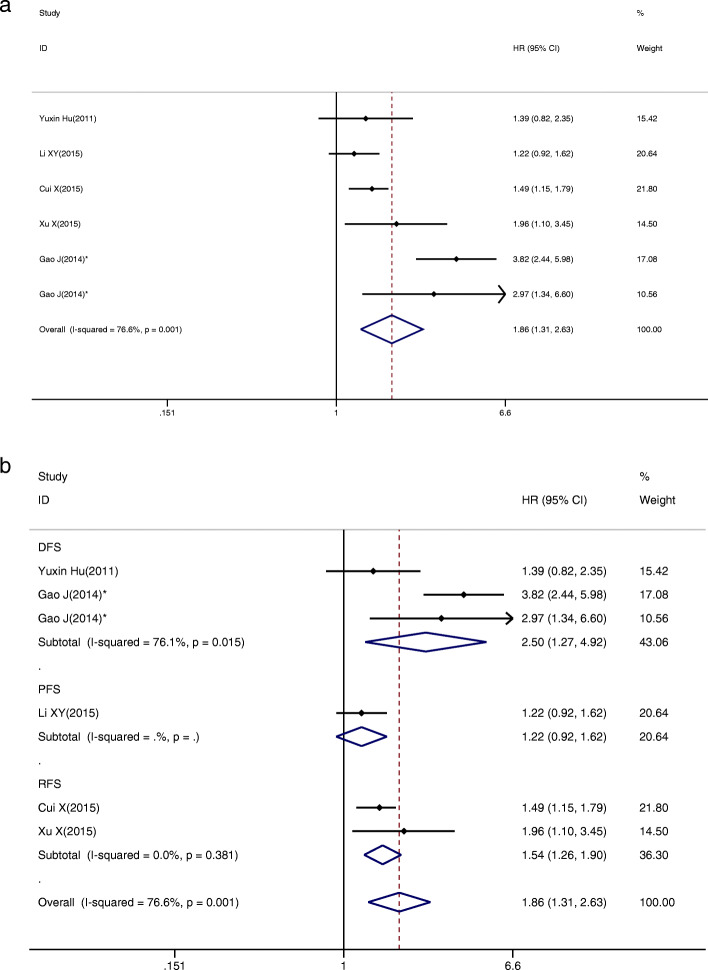
Tumour progression is associated with miR-34a expression
To evaluate the association between miR-34a expression and DFS/PFS/RFS, 6 studies were included in this analysis, and the data revealed that low miR-34a expression predicted a worse outcome with a combined HR of 1.86 (95% CI: 1.31–2.63) via a random-effect model (P = 0.001, I2 = 76.6%; Fig. 3a). To explain the heterogeneity, we performed subgroup analysis by DFS, PFS and RFS, showing a significant correlation with the expression of miR-34a (HR = 2.50, 95% CI: 1.27–4.92 for DFS; HR = 1.54, 95% CI: 1.26–1.90 for RFS; Fig. 3b). Moreover, homogeneity was achieved in the RFS group. Then, the sensitivity analysis was performed by removing studies one by one to assess the influence of a single study. As shown in Supplementary Fig. 2C, the stability of the entire study was not influenced by individual studies. Finally, funnel plots and Egger’s test were implemented to evaluate publication bias. The funnel plot was roughly symmetric (Supplementary Fig. 2D), and the P value of Egger’s test was 0.909. Therefore, no evidence for significant publication bias existed.
Fig. 3.
The association between miR-34a expression levels and (a) DFS/PFS/RFS; b subgroup analyses of DFS/PFS/RFS. Gao J*, study containing two different groups
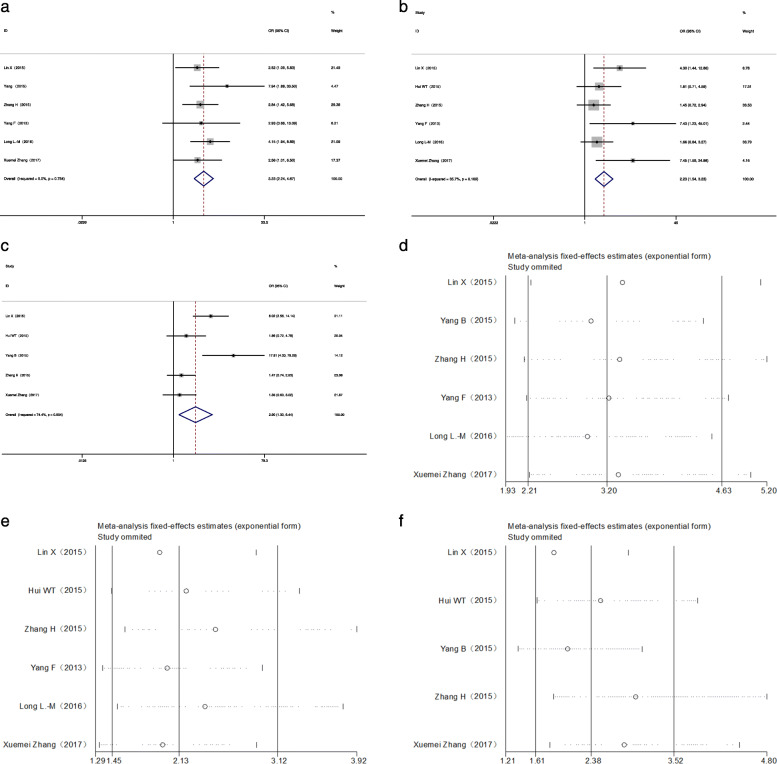
Correlation between miR-34a levels and clinicopathological features in GICs
For obtaining relevant statistics to evaluate the relation between miR-34a expression levels and different clinicopathological characteristics, seven studies containing 647 patients with GICs were screened out. As shown in Table 4, we observed a significant association between the expression level of miR-34a and lymphatic metastasis (OR = 3.231, 95% CI: 2.237–4.666; Fig. 4a) and differentiation degree (OR = 2.228, 95% CI: 1.538–3.228; Fig. 4b) via the fixed-effect model, as well as TNM stage (OR = 2.896, 95% CI: 1.302–6.442; Fig. 4c) via the random-effect model. There was no significant correlation identified between miR-34a level and tumour size (OR = 0.736, CI: 0.460–1.177). In addition, the expression level of miR-34a was unaffected by gender (OR = 0.776, 95% CI: 0.566–1.065). The heterogeneity disappeared in the gender group (I2 = 0.00%, P = 0.888), lymphatic metastasis group (I2 = 0.00%, P = 0.754), medium level of the tumour size group (I2 = 20.5%, P = 0.284), and differentiation degree group (I2 = 35.7%, P = 0.169), but it was obvious in the TNM stage group (I2 = 74.4%, P = 0.004). Sensitivity analysis was applied to assess the stability, including lymphatic metastasis (Fig. 4d), differentiation degree (Fig. 4e) and TNM stage (Fig. 4f), suggesting no study had significant impact on the results.
Table 4.
Overall analysis of miR-34a expression association with clinicopathologic characteristics
| Clinicopathological characteristics | Num. of studies | Num. of patients | Pooled OR (95%CI) | Heterogeneity | ||
|---|---|---|---|---|---|---|
| Fixed | Random | I2 | p-value | |||
| Gender (male vs. female) | 7 | 647 | 0.776 (0.566–1.065) | 0.777 (0.565–1.067) | 0.0% | 0.888 |
| Tumor Size (≤5 vs > 5 cm) | 3 | 326 | 0.736 (0.460–1.177) | 0.284 (0.433–1.288) | 20.5% | 0.284 |
| Lymphatic Metastasis (YESvs.NO) | 6 | 571 | 3.231 (2.237–4.666) | 3.200 (2.210–4.635) | 0.0% | 0.754 |
| TNM stage (III + IV vs. I + II) | 5 | 458 | 2.468 (1.698–3.588) | 2.896 (1.302–6.442) | 74.4% | 0.004 |
| Differentiation (poor vs. others) | 6 | 597 | 2.228 (1.538–3.228) | 2.373 (1.430–3.938) | 35.7% | 0.169 |
Abbreviations: 95%CI 95% confidence interval, Fixed, fixed effects model, OR Odds ratio, Random, random pooling model
Fig. 4.
The association between miR-34a expression levels and (a) lymphatic metastasis, (b) tumour differentiation degree, and (c) TNM stage; sensitivity analyses for ORs of clinicopathological characteristics, such as (d) lymphatic metastasis, (e) tumour differentiation degree, and (f) TNM stage
Discussion
In the last few decades, miRNAs have attracted increasing interest among investigators as potential biomarkers for cancer diagnosis and prognosis. Many clinical trials have demonstrated that miRNAs play a pivotal role in tumour development by regulating the expression of target genes and tumour suppressors or directly performing their functions as oncogenes or tumour suppressors [40, 41]. MiR-34a expression is transcriptionally controlled by p53, a vital tumour suppressor pathway, which is disrupted in cancer frequently. It has been reported that miR-34a influenced tumour biological activities by targeting several genes or signal pathways, such as CCND1 in EC [42], PDGFR in GC [18], HMGB1 in CRC [43], and XIST in PC [37]. Recently, a systematic review has summarized numerous studies that reported the diagnostic and prognostic value of miR-34a in GICs [44]. However, among these studies, two opposing views were presented on whether patients could benefit from the high expression of miR-34a. Hao Wu et al. [7], Milad Asadi et al. [45] and Yan Zhou et al. [46] showed that the downregulation of miR-34a was linked to a poor prognosis in GIC patients, while Hiyoshi et al. [8], and Mojin Wang [26] reported that patients benefited from downregulated miR-34a. The prognostic value of miR-34a in GICs has been illustrated in many studies, but the particular prognostic role of miR-34a in GICs remains unclear. As far as we know, this is the most comprehensive meta-analysis exploring the clinical value of miR-34a in patients with GICs.
This meta-analysis discussed 20 papers and contained 2367 patients in total. Among these studies, 18 studies including 1691 patients provided the relevant OS statistics. By the random-effect model, the results showed that the decreased miR-34a expression was associated with poorer outcomes in patients with GICs. To explain the potential sources of heterogeneity, subgroup analyses were performed. As a result, homogeneity was reached in the CRC group, and the OS of the CRC group was found to be greatly associated with miR-34a expression levels. Though the expression level of miR-34a in CRC patients remains controversial, there are several potential mechanisms that suggest how low expression of miR-34a could induce an unfavourable outcome of CRC. MiR-34a expression is governed by p53 and could inhibit recurrence of CRC by inhibiting cell growth, migration and invasion and inducing cell apoptosis and cell cycle arrest in a p53-dependent manner [39]. Moreover, it has been reported that miR-34a served a key role in suppressing CRC metastasis by targeting and regulating Notch signalling [25] and the FMNL2 and E2F pathways [47, 48]. In addition, Jiang et al. showed that miR-34a suppressed tumour formation caused by loss of Apc and controlled intestinal stem cell and secretory cell homeostasis by downregulation of multiple target mRNAs, such as Pdgfra, Pdgfrb, and Axl [49].
As shown in Table 3, the associations between miR-34a expression levels and OS were also significant in other subgroups. In the included studies, mir-34a showed a low expression level in both tumour tissue and blood, except for Osawa’s study [24], in which tissue samples were used. Subgroup analysis showed that the miR-34a level from tissue (HR = 1.87, 95% CI: 1.50–2.33) and plasma (HR = 1.88, 95% CI: 1.34–2.63) were of equal importance in prognostic value. Since there was only one study included based on plasma samples, the conclusion remained unclear until now and required further verification. In addition, subgroup analysis of tumour types showed a closer relationship between a low miR-34a level and poor OS in patients with PC (HR = 2.59, 95% CI:1.69–3.97). Empirically, HR > 2 is considered strongly predictive [50]. As for the possible mechanism, Long et al. reported that miR-34a significantly inhibited the tumour growth of PC tumours by suppressing Notch1, Notch2 and Notch4 expression [10]. Tang et al. found that the EMT program activator Snail1 and the proliferation regulator Notch1 were both targets of miR-34a [51]. Overexpression of miR-34a suppressed the expression of Snail1, which in turn upregulated E-cadherin. Moreover, the HDAC inhibitor Vorinostat (SAHA) inhibited the expression of EMT inducers Zeb-1, Snail, and Slug by upregulating the expression of miR-34a, thereby attenuating the migration and invasion of PC cells [52]. Since the heterogeneities within the subgroups were still significant, meta regression was performed to illustrate the influence of different factors, including ethnicity, sample capacity, specimen, NOS scores and tumour classification, but there was no factor that significantly affected the variation in HR. The analysis of tumour progression and miR-34a expression revealed that low miR-34a expression seemed to predict a worse outcome, especially in DFS (HR = 2.50, 95% CI: 1.27–4.92). According to our research, we could infer that the decreased expression level of miR-34a was closely related to worse prognosis in patients with GICs. However, for the EC and GC groups, the results were still not stable and required more comprehensive studies to further research the miR-34a prognostic value in GICs.
To evaluate the association between miR-34a and the clinical characteristics, seven articles including 647 patients were included. Significant relations were observed between miR-34a expression levels and differentiation/TNM stage/lymphatic metastasis by a fixed- or random-effect model. Sensitivity analysis indicated that no study had a significant impact on the results. Based on the findings above, we could suggest that patients with decreased miR-34a expression are more likely to develop lymphatic metastasis, and decreased miR-34a expression level is linked to poor tumour differentiation and late TNM stage.
Though this meta-analysis revealed that miR-34a was a promising biomarker of GICs, several potential limitations of this study should be considered. First, the number of included studies was limited; the current sample size was too small to explain the real relationship between miR-34a expression level and prognosis of GICs. Subgroup analyses were also affected by the relative lack of studies; for example, there was only one article related to PFS. The significance of this study lies in larger sample size experiments for further identification of the correlation between miR-34a and prognosis of GICs. Second, patients were all Asian and Caucasian, and the lack of data from other regions might have resulted in ethnic bias. Third, the cut-off values among studies were different, and we did not have absolute criteria to assess whether the expression of miR-34a was low or not, thus impacting the statistical power of the analysis. Finally, several HRs and 95% CIs were calculated according to the data extracted from survival curves, so it is difficult to exclude the influence of confounding bias.
Conclusion
In conclusion, our study demonstrates that lower miR-34a expression is significantly associated with poorer OS and DFS/PFS/RFS and may be a novel prognostic biomarker in GICs. Moreover, the miR-34a expression level is relatively lower in patients with lymph node metastasis, and a decreased expression level of miR-34a is related to poor tumour differentiation and late TNM stage. Further multicentre prospective clinical studies are needed to validate the association between miR-34a and the prognosis of GICs.
Supplementary Information
Additional file 1: Supplementary Figure 1. The association between miR-34a expression levels and (A) sample size (≥100 and < 100) and (B) NOS scores (≥8 and < 8).
Additional file 2: Supplementary Figure 2. Sensitivity analysis for the HR of (A) OS; (C) DFS/PFS/RFS; publication bias evaluation for (B) OS; (D) DFS/PFS/RFS.
Acknowledgements
We would like to thank all the people who helped us in this study.
Abbreviations
- 95% CI
95% confidence interval
- CRC
Colorectal cancer
- DFS
Disease-free survival
- EC
Oesophageal cancer
- GBC
Gallbladder cancer
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- HR
Hazard ratio
- NOS
Newcastle-Ottawa Scale
- NR
No report
- OR
Odds ratio
- OS
Overall survival
- PC
Pancreatic cancer
- PFS
Progression-free survival
- qRT-PCR
Quantitative real-time PCR
- R
Retrospective
- RFS
Recurrence-free survival
Authors’ contributions
Study design: YC. Data collection: YC and XL. Data analysis: YC, XL and LL. Manuscript composition: YC. Manuscript revision: XL. Table Drafting: YC and LL. Figure Drafting: YC and XL. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81900508), and the Natural Science Foundation of Jiangsu Province (BK20190172). The funders have no role in the study design, data analysis and manuscript composition.
Availability of data and materials
The authors declare that all data used or analysed during the current study are available on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiao-Lin Liu, Email: lxl55@foxmail.com.
Ling Li, Email: liling3243@sina.com.
References
- 1.Shams AZ, Haug U. Strategies for prevention of gastrointestinal cancers in developing countries: a systematic review. J Glob Health. 2017;7(2):020405. doi: 10.7189/jogh.07.020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Song X, Zhu J, Li M, Ji Y, Wu F, Chen Y, Cui X, Hu J, Wang L, et al. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J Oncol. 2017;51(1):378–388. doi: 10.3892/ijo.2017.4015. [DOI] [PubMed] [Google Scholar]
- 7.Wu H, Huang M, Liu Y, Shu Y, Liu P. Luteolin induces apoptosis by up-regulating miR-34a in human gastric Cancer cells. Technol Cancer Res Treat. 2015;14(6):747–755. doi: 10.7785/tcrt.2012.500434. [DOI] [PubMed] [Google Scholar]
- 8.Hiyoshi Y, Schetter AJ, Okayama H, Inamura K, Anami K, Nguyen GH, Horikawa I, Hawkes JE, Bowman ED, Leung SY, et al. Increased microRNA-34b and -34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancer. PLoS One. 2015;10(4):e0124899. doi: 10.1371/journal.pone.0124899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun TY, Xie HJ, Li Z, Kong LF, Gou XN, Li DJ, Shi YJ. Ding YZ: miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am J Transl Res. 2017;9(1):103–114. [PMC free article] [PubMed] [Google Scholar]
- 10.Long LM, Zhan JK, Wang HQ, Li S, Chen YY, Liu YS. The clinical significance of miR-34a in pancreatic ductal carcinoma and associated molecular and cellular mechanisms. Pathobiology. 2017;84(1):38–48. doi: 10.1159/000447302. [DOI] [PubMed] [Google Scholar]
- 11.Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao H, Li C, Chen Y. Zhao J: miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol. 2014;35(2):1503–1510. doi: 10.1007/s13277-013-1207-z. [DOI] [PubMed] [Google Scholar]
- 12.Gao H, Zhao H, Xiang W. Expression level of human miR-34a correlates with glioma grade and prognosis. J Neuro-Oncol. 2013;113(2):221–228. doi: 10.1007/s11060-013-1119-1. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, Zhou S, Liu J, Zhu J, Xue J, Gu L. Chen Y: miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathol Res Pract. 2016;212(5):444–449. doi: 10.1016/j.prp.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Chu J, Li H, Xing Y, Jia J, Sheng J, Yang L, Sun K, Qu Y, Zhang Y, Yin H, et al. LncRNA MNX1-AS1 promotes progression of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis. Biomed Pharmacother. 2019;116:109029. doi: 10.1016/j.biopha.2019.109029. [DOI] [PubMed] [Google Scholar]
- 15.Yoon JH, Choi WS, Kim O, Choi BJ, Nam SW, Lee JY, Park WS. Gastrokine 1 inhibits gastric cancer cell migration and invasion by downregulating RhoA expression. Gastric Cancer. 2017;20(2):274–285. doi: 10.1007/s10120-016-0617-1. [DOI] [PubMed] [Google Scholar]
- 16.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y. Zheng X: miR-34a inhibits migration and invasion by down-regulation of c-met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275(1):44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 17.Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107(Pt B):2620–2629. doi: 10.1016/j.ijbiomac.2017.10.154. [DOI] [PubMed] [Google Scholar]
- 18.Peng Y, Guo JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34(3):e00112. doi: 10.1042/BSR20140020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Lai M, Du G, Shi R, Yao J, Yang G, Wei Y, Zhang D, Xu Z, Zhang R, Li Y, et al. MiR-34a inhibits migration and invasion by regulating the SIRT1/p53 pathway in human SW480 cells. Mol Med Rep. 2015;11(5):3301–3307. doi: 10.3892/mmr.2015.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L, Wan Y, Dong Y, Qu K, Liu C. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget. 2015;6(6):3988–4004. doi: 10.18632/oncotarget.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Correa AM, Hoque A, Guan B, Ye F, Huang J, Swisher SG, Wu TT, Ajani JA, Xu X-C. Prognostic significance of differentially expressed miRNAs in esophageal cancer. Int J Cancer. 2011;128(1):132–143. doi: 10.1002/ijc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hui W-T, Ma X-B, Zan Y, Wang X-J, Dong L. Prognostic significance of MiR-34a expression in patients with gastric Cancer after radical Gastrectomy. Chin Med J. 2015;128(19):2632–2637. doi: 10.4103/0366-6999.166019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B, Huang J, Liu H, Guo W. Li G: miR-335 directly, while miR-34a indirectly modulate survivin expression and regulate growth, apoptosis, and invasion of gastric cancer cells. Tumour Biol. 2016;37(2):1771–1779. doi: 10.1007/s13277-015-3951-8. [DOI] [PubMed] [Google Scholar]
- 24.Osawa S, Shimada Y, Sekine S, Okumura T, Nagata T, Fukuoka J, Tsukada K. MicroRNA profiling of gastric cancer patients from formalin-fixed paraffin-embedded samples. Oncol Lett. 2011;2(4):613–619. doi: 10.3892/ol.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Ai F, Li X, Tian L, Wang X, Shen S, Liu F. MicroRNA-34a suppresses colorectal cancer metastasis by regulating notch signaling. Oncol Lett. 2017;14(2):2325–2333. doi: 10.3892/ol.2017.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Zhang P, Li Y, Liu G, Zhou B, Zhan L, Zhou Z, Sun X. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microRNA-155 and microRNA-200c overexpression in human colorectal cancer. Med Oncol. 2012;29(5):3113–3118. doi: 10.1007/s12032-012-0241-9. [DOI] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. EurJ Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 28.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, Xu XY, Chen QS, Huang C. Clinical significance of microRNA-34a in esophageal squamous cell carcinoma. Genet Mol Res. 2015;14(4):17684–17691. doi: 10.4238/2015.December.21.41. [DOI] [PubMed] [Google Scholar]
- 30.Wei B, Huang QY, Huang SR, Mai W, Zhong XG. MicroRNA-34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol Med Rep. 2015;12(4):5255–5261. doi: 10.3892/mmr.2015.4110. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Li S, Yang J, Liu S, Gong X, Yu X. The prognostic value of miR-34a expression in completely resected gastric cancer: tumor recurrence and overall survival. Int J Clin Exp Med. 2015;8(2):2635–2641. [PMC free article] [PubMed] [Google Scholar]
- 32.Li X-Y, Wen J-Y, Jia C-C, Wang T-T, Li X, Dong M, Lin QU, Chen Z-H, Ma X-K, Wei LI, et al. MicroRNA-34a-5p enhances sensitivity to chemotherapy by targeting AXL in hepatocellular carcinoma MHCC-97L cells. Oncol Lett. 2015;10(5):2691–2698. doi: 10.3892/ol.2015.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F, Li Q-J, Gong Z-B, Zhou L, You N, Wang S, Li X-L, Li J-J, An J-Z, Wang D-S, et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13(1):77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 34.Cui X, Wu Y, Wang Z, Liu X, Wang S, Qin C. MicroRNA-34a expression is predictive of recurrence after radiofrequency ablation in early hepatocellular carcinoma. Tumour Biol. 2015;36(5):3887–3893. doi: 10.1007/s13277-014-3031-5. [DOI] [PubMed] [Google Scholar]
- 35.Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, Ueda J, Nagai E, Hashizume M, Tanaka M. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18(8):2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamieson NB, Morran DC, Morton JP, Ali A, Dickson EJ, Carter CR, Sansom OJ, Evans TRJ, McKay CJ, Oien KA. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18(2):534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 37.Sun Z, Zhang B, Cui T. Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep. 2018;39(4):1591–1600. doi: 10.3892/or.2018.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasakova K, Reis R, Vician M, Zeman M, Herichova I. Expression of miR-34a-5p is up-regulated in human colorectal cancer and correlates with survival and clock gene PER2 expression. PLoS One. 2019;14(10):e0224396. doi: 10.1371/journal.pone.0224396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34(31):4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 40.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 43.Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. Downregulation of HMGB1 by miR-34a is sufficient to suppress proliferation, migration and invasion of human cervical and colorectal cancer cells. Tumour Biol. 2016;37(10):13155–13166. doi: 10.1007/s13277-016-5261-1. [DOI] [PubMed] [Google Scholar]
- 44.Kong J, Wang W. A systemic review on the regulatory roles of miR-34a in gastrointestinal Cancer. OncoTargets Ther. 2020;13:2855–2872. doi: 10.2147/OTT.S234549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miR-34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2019;50(2):304–307. doi: 10.1007/s12029-018-0060-0. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Ding BZ, Lin YP, Wang HB. MiR-34a, as a suppressor, enhance the susceptibility of gastric cancer cell to luteolin by directly targeting HK1. Gene. 2018;644:56–65. doi: 10.1016/j.gene.2017.10.046. [DOI] [PubMed] [Google Scholar]
- 47.Lu G, Sun Y, An S, Xin S, Ren X, Zhang D, Wu P, Liao W, Ding Y, Liang L. MicroRNA-34a targets FMNL2 and E2F5 and suppresses the progression of colorectal cancer. Exp Mol Pathol. 2015;99(1):173–179. doi: 10.1016/j.yexmp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang L. Hermeking H: miR-34a and miR-34b/c suppress intestinal tumorigenesis. Cancer Res. 2017;77(10):2746–2758. doi: 10.1158/0008-5472.CAN-16-2183. [DOI] [PubMed] [Google Scholar]
- 50.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6(4):375–392. doi: 10.1023/A:1014778713034. [DOI] [PubMed] [Google Scholar]
- 51.Tang Y, Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the notch signaling pathway. Sci Rep. 2017;7:38232. doi: 10.1038/srep38232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6(8):e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. The association between miR-34a expression levels and (A) sample size (≥100 and < 100) and (B) NOS scores (≥8 and < 8).
Additional file 2: Supplementary Figure 2. Sensitivity analysis for the HR of (A) OS; (C) DFS/PFS/RFS; publication bias evaluation for (B) OS; (D) DFS/PFS/RFS.
Data Availability Statement
The authors declare that all data used or analysed during the current study are available on reasonable request.