Abstract
Radiology reports contain a radiologist’s interpretations of images, and these images frequently describe spatial relations. Important radiographic findings are mostly described in reference to an anatomical location through spatial prepositions. Such spatial relationships are also linked to various differential diagnoses and often described through uncertainty phrases. Structured representation of this clinically significant spatial information has the potential to be used in a variety of downstream clinical informatics applications. Our focus is to extract these spatial representations from the reports. For this, we first define a representation framework based on the Spatial Role Labeling (SpRL) scheme, which we refer to as Rad-SpRL. In Rad-SpRL, common radiological entities tied to spatial relations are encoded through four spatial roles: trajector, landmark, diagnosis, and hedge, all identified in relation to a spatial preposition (or spatial indicator). We annotated a total of 2,000 chest X-ray reports following Rad-SpRL. We then propose a deep learning-based natural language processing (NLP) method involving word and character-level encodings to first extract the spatial indicators followed by identifying the corresponding spatial roles. Specifically, we use a bidirectional long short-term memory (Bi-LSTM) conditional random field (CRF) neural network as the baseline model. Additionally, we incorporate contextualized word representations from pre-trained language models (BERT and XLNet) for extracting the spatial information. We evaluate both gold and predicted spatial indicators to extract the four types of spatial roles. The results are promising, with the highest average F1 measure for spatial indicator extraction being 91.29 (XLNet); the highest average overall F1 measure considering all the four spatial roles being 92.9 using gold indicators (XLNet); and 85.6 using predicted indicators (BERT pre-trained on MIMIC notes).
The corpus is available in Mendeley at http://dx.doi.org/10.17632/yhb26hfz8n.1 and https://github.com/krobertslab/datasets/blob/master/Rad-SpRL.xml.
Keywords: Spatial relations, Radiology report, NLP, Deep learning
1. Introduction
There has been a growing interest in automatically extracting useful information from unstructured reports in the medical domain. One of the most explored free text clinical report types for information extraction using NLP has been radiology reports, which contain a wealth of clinically significant patient information. Automatic recognition of important information such as actionable findings and their corresponding location and diagnoses facilitates the time-consuming process of manual review of the reports containing radiologists’ descriptions of imaging results. However, extracting such spatial information associated with radiographic findings has been less researched and forms the focus of our work.
Besides radiology-specific knowledge and experience, interpreting spatial relations from radiological images requires good spatial ability skills on the part of radiologists as it often involves mental visualization of complex 3D anatomical structures to describe the locations of radiographic findings. A few studies [1,2] have highlighted the possible requirement of these skills in prospective radiologists to perceive and understand the spatial relationships between different objects in radiology practice. These spatial interpretations from images are summarized in the corresponding free text reports. Thus, radiology reports have a high prevalence of spatial relations in the way radiologists describe radiographic findings and their association with anatomical structures. These spatial relations provide sufficient contextual information related to the findings. Moreover, some of these spatially-grounded findings demand immediate action by the physician ordering the imaging examination. Therefore, it is important to understand the spatial meanings from the unstructured reports and generate structured representations of the spatial relations for various downstream clinical applications. Such applications include easy visualization of the important actionable findings, predictive modeling, cohort retrieval, automated tracking of findings, and automatic generation of more complete annotations for associated images containing spatial and diagnosis-related information of findings.
However, spatial language understanding in the radiology domain has remained less explored, and often the language used for representing spatial relations is complex. We therefore aim to automatically extract important spatial information from radiology reports in this work.
In the general domain, earlier studies [3,4] have formulated and evaluated the spatial role labeling (SpRL) task for extracting spatial information from text by mapping language to a formal spatial representation. In the SpRL annotation scheme, an object of interest (trajector) is associated with a grounding location (landmark) through a preposition or spatial trigger (spatial indicator). For example, in the sentence, “The book is on the table”, the spatial preposition ‘on’ indicates the existence of a spatial relationship between the object ‘book’ (trajector) and its location ‘table’ (landmark).
In the medical domain, a limited number of studies have utilized the SpRL scheme. Kordjamshidi et al. [5] extracted relations between bacteria names and their locations from scientific text. Roberts et al.[6] utilized SpRL in the extraction of spatial relations between symptoms/disorders and anatomical structures from consumer-related texts. In this paper, we also construct similar spatial roles for radiology texts based on SpRL. For instance, in a radiology report sentence, “Mild streaky opacities are present in the left lung base”, the location of a clinical finding ‘opacities’ (trajector) has been described with respect to the anatomy ‘left lung base’ (landmark) using the spatial preposition ‘on’ (spatial indicator).
Moreover, radiologists oftentimes document potential diagnoses related to the clinical findings which are spatially grounded. Consider the following example:
Stable peripheral right lower lobe opacities seen between the anterior 7th and 8th right ribs which may represent pleural reaction or small pulmonary nodules.
Here, presence of a finding – ‘stable peripheral right lower lobe opacities’ at a specific location –’anterior 7th and 8th right ribs’ may elicit the radiologist to document two possible diagnoses – ‘pleural reaction’ and ‘small pulmonary nodules’. As the actual occurrence of a disorder is highly dependent on various patient factors such as other physical examinations, laboratory tests, and symptoms, the radiologists usually describe diagnoses with uncertainty phrases or hedges. For instance, in the example above, the hedge term ‘may represent’ is used to relate a finding and its corresponding body location with the most probable diagnoses.
In this paper, we propose a framework as a preliminary step to understand textual spatial semantics in chest X-ray reports. We define a basic spatial representation framework that extends SpRL for radiology (Rad-SpRL) involving interactions among common radiology entities. As most of the actionable clinical findings in all types of radiology reports are spatially located and represent a probable diagnosis, Rad-SpRL can potentially be extended to other report types. Consider the following sentence from a head CT report:
A well circumscribed hypodense 1 cm lesion is seen in the right cerebellar hemisphere consistent with prior stroke.
Here, the spatial preposition ‘in’ describes that the finding ‘lesion’ is located inside the anatomical structure ‘right cerebellar hemisphere’ which is also consistent with the diagnosis ‘stroke’. To evaluate this representation, we manually annotated a corpus of 2000 radiology reports (a subset of publicly available Open-i® chest X-ray reports [7]) using Rad-SpRL and applied deep learning models to identify the spatial roles.
Owing to the promising results of applying deep learning models in entity and relation extraction, we have adopted two classes of neural network models to investigate the automatic extraction of these relations. The first method is based on a bidirectional long short-term memory (Bi-LSTM) recurrent neural network with a conditional random field (CRF) layer as our baseline model to identify detailed spatial relationships, including diagnosis and hedging terms from the reports. The main intention behind using Bi-LSTM is that LSTM units work well for taking in long-distance dependencies in a sentence, and the bi-directional sequential architecture adds more benefits by considering both the right and left context of a word. This has been shown to achieve state-of-the-art results on the 2012–2013 SemEval datasets for SpRL [8]. We additionally incorporated character embeddings to better handle out-of-vocabulary, rare, and misspelled words.
The second method is based on transformer-based language models. Recent deep learning works have leveraged pre-trained language models and demonstrated improved performance in a variety of NLP tasks [9–11]. Some studies [10,12,13] have particularly focused on pre-training the language models on a clinical domain corpus – including clinical notes and biomedical literature – with the aim to generate enhanced contextual word representations to be used for fine-tuning various downstream clinical NLP models. Motivated by this, we utilize contextualized embedding models based on transformers by applying BERT- and XLNet-based models for extracting spatial relations.
Understanding spatial relations relies on the syntactic structure of a sentence as demonstrated in previous works where various syntactic features and rules based on lexico-syntactic patterns and syntactic parse trees were employed [14–17]. BERT [18], based on a deep bi-directional transformer architecture, encodes rich linguistic information in a hierarchical manner with syntactic features in the middle layers. Thus BERT captures structural or syntactic information about language [19,20]. The deeper architecture along with multi-headed self-attention helps in achieving better long-range dependencies while learning the contextualized representation of words. Moreover, a recent study [9] achieved the state-of-the-art results by applying BERT in the more general task of semantic role labeling, the class of NLP problems to which Rad-SpRL belongs. More recently, the XLNet model based on autoregressive pre-training outperformed BERT on multiple NLP tasks including reading comprehension and document ranking [21]. XLNet is also a transformer-based model, though larger than BERT in the total number of parameters. Inspired by all these, we fine-tune both the BERT and XLNet models on our annotated Rad-SpRL corpus.
Our work recognizes granular information about the interpreted diagnoses by identifying them in context to the same spatial preposition (e.g., in, of, within, around) connecting a clinical finding to an anatomical location. Thus, we extract detailed information about a finding, the body location where the finding is detected, possible diagnoses associated with the finding, and also any hedging term used by radiologists in interpreting these diagnoses. Additionally, the finding and the location terms contain their respective descriptors (e.g., the descriptor ‘mild streaky’ associated with the finding ‘opacities’).
The organization of the rest of the paper is as follows. Section 2 highlights the previous relevant studies on spatial representation frameworks, chest radiology, spatial relation extraction, and the Open-i dataset [7]. Section 3 describes our new corpus of 2000 chest X-ray reports annotated according to Rad-SpRL and the annotation process that produced this corpus. Section 4 includes a description of our automatic methods, including Bi-LSTM-CRF as well as BERT- and XLNet-based methods for extracting spatial information, plus the implementation and evaluation details. Results are summarized in Section 5, while Section 6 discusses the results and limitations of this work. Section 7 concludes the paper and provides directions for future work.
2. Related work
2.1. Spatial representation frameworks for text
Different representation frameworks have been proposed to encode spatial knowledge in textual data for various use cases. Among the early works, Hayward et al. investigated the structural similarities between visual and linguistic representations of space [22]. Mani et al. proposed SpatialML to represent geographical location information including geo-coordinates and orientation and annotated ACE English documents as per SpatialML [23]. This representation encodes the spatially-related entities through contextual information such as direction and distance as well as the actual physical connection between the related entities (using the Region Connection Calculus). However, this representation is specific to the geographical aspects of the spatial language. At the same time, Kordjamshidi et al. proposed Spatial Role Labeling (SpRL) that involves extracting spatial arguments of the spatial relations in a sentence [3]. This framework is an improvement over representations such as SpatialML and STM spatio-temporal markup [24] as this is more generalizable in terms of spatial language expressiveness and handles a greater number of spatial concepts (both static and dynamic). This has also been utilized on biomedical [5] and consumer health data [6] (described in Section 1). Later, Fasola and Mataric devised methods to represent dynamic spatial relations for facilitating interactive instruction of robots [25]. For text-to-scene generation, Chang et al. proposed a representation that converts an input text describing a scene to output a 3D scene by transforming the text to a set of constraints consisting of the objects and the spatial relations between them as well as by learning priors on how the objects occur in 3D scenes [26]. Kergosien et al. designed a framework to extract relevant spatial information from web textual data (newspaper articles) to annotate satellite images with additional meaningful information for use cases such as image annotation and land use planning [27]. Collell et al. used both visual and linguistic features to generate distributed spatial representations by feeding them into a neural network model that learns to predict 2D spatial arrangements of objects provided their instances and the relationship between them [28]. More recently, Ulinski et al. designed the SpatialNet framework to encode spatial language based on frame semantic principles and additionally proposed ways to incorporate external knowledge sources for disambiguating the spatial expressions [29]. All these highlight some important works relevant to spatial information representation in text.
2.2. Types of chest radiology entities extracted
Numerous studies have focused on extracting specific information such as clinical findings or imaging observations, differential diagnoses, and anatomical locations from chest-related reports. In Table 1, we compare the specific information types or the radiology entities extracted in the previous studies from chest radiology reports using NLP. We primarily pay attention to the clinically-important entities which are common across various types of radiology reports. We also do not take into account the cases where uncertainty and negation information were used to detect the presence or absence of a particular finding or a disease [30]. For example, Hedge is not considered as extracted in Table 1 when the uncertainty levels are classified into negative, uncertain or positive for each finding term extracted [31]. Further, in Table 1, we have not considered studies dealing with specific body locations (e.g., mammography reports containing breast imaging information, and head CT reports) as the entities of interest are usually very domain-specific such as ‘Clock face’, ‘Depth’, ‘BI-RADS category’ etc. in the case of mammography reports. We also do not take into account the works which focused on detecting a specific disease such as pneumothorax [32] or pulmonary lesion [33] from chest radiographs. Note that these works did not attempt to extract all the information types collectively, neither did they focus on identifying any association or relation among these entities. Our work aims to relate the isolated entities (e.g., findings, locations, probable diagnoses) from spatial context. We specifically extract findings whose associated anatomical locations are described through spatial expressions as well as identify the probable diagnoses associated with these spatially-located findings.
Table 1.
Comparison of our corpus with the studies extracting clinically-relevant radiological entities from chest X-ray and chest CT reports.
2.3. Relation extraction from radiology reports, including spatial relations
Friedman et al. [39] proposed a formal model (MedLEE) based on grammar rules to map clinical information in radiology reports, including central findings and their contextual information like body location, degree, and certainty modifiers into a structured format utilizing controlled vocabulary and synonym knowledge base. They also worked toward providing an interface for using MedLEE for different applications [40]. In another work, Friedman et al. [41] adapted MedLEE to generate the most specific Unified Medical Language System (UMLS) code based on a finding and its associated modifier information. Later, Sevenster et al. [42] built a reasoning engine to correlate clinical findings and body locations in radiology reports utilizing MedLEE. However, the major limitation of this work is the system’s poor recall. Yim et al. [43] worked on extracting relations containing tumor-specific information from radiology reports of hepatocellular carcinoma patients. A recent work by Steinkamp et al. [44] extracted facts representing clinical assertions and recognized contextual information such as location, image citation, and description of change over time related to a target entity (e.g., finding) identified for that fact. However, this system does not necessarily capture the related entities from a spatial perspective and does not identify all the fine-grained spatial information. Another work by Beatrice et al. [45] identified relations between observation entities with their location (deep/cortical) and recency (old/recent) modifiers from brain imaging reports. However, the location information includes two broad categories and is relevant to two specific observations (stroke and microbleed). In Table 2, we present the two works relevant to spatial information extraction from radiology reports. The main limitations of Rink et al. [16] are the usage of appendicitis-specific lexicons and the requirement of manual effort in crafting rules based on syntactic dependency patterns to identify the spatially-grounded inflammation description. Besides being domain-specific, another limitation of Roberts et al. [15] is that the study extracts only the location entities associated with an actionable finding and this required relying on heavy feature engineering.
Table 2.
Studies focusing on spatial relations in radiology reports.
2.4. Studies using Open-i X-ray report annotations
Open-i is a biomedical image search engine.1 One of its datasets is a public chest X-ray dataset containing 3955 de-identified radiology reports from the Indiana Network for Patient Care released by the National Library of Medicine [7]. (Hereafter referred to simply as the Open-i dataset.) We have presented an example of the manual annotation of a sample report in the Open-i dataset in Fig. 1 (the annotations are inspired by MeSH terms). Although most of the Open-i annotations embody the relationship between finding and location, there are, however, a few missing relations. For example, note that in Fig. 1 the Open-i manual annotations contain the normalized finding Pulmonary Emphysema corresponding to the phrase ‘emphysematous changes’ in the report, but do not annotate the associated location ‘right upper lobe’. The Open-i dataset has been used previously in many studies, presented in Table 3. However, most of the these studies focused on the extraction of only the disease/finding [30,46–49]. Two studies worked on automatically annotating both disease and disease descriptions (e.g., location, severity) [50,51] similar to the human annotations in Demner-Fushman et al. [7]. However, all these works ignored distinguishing diagnosis terms from findings (except for Peng et al. [47]), and annotating correlations between them. We describe annotation-specific limitations of each of these works in Table 3.
Fig. 1.
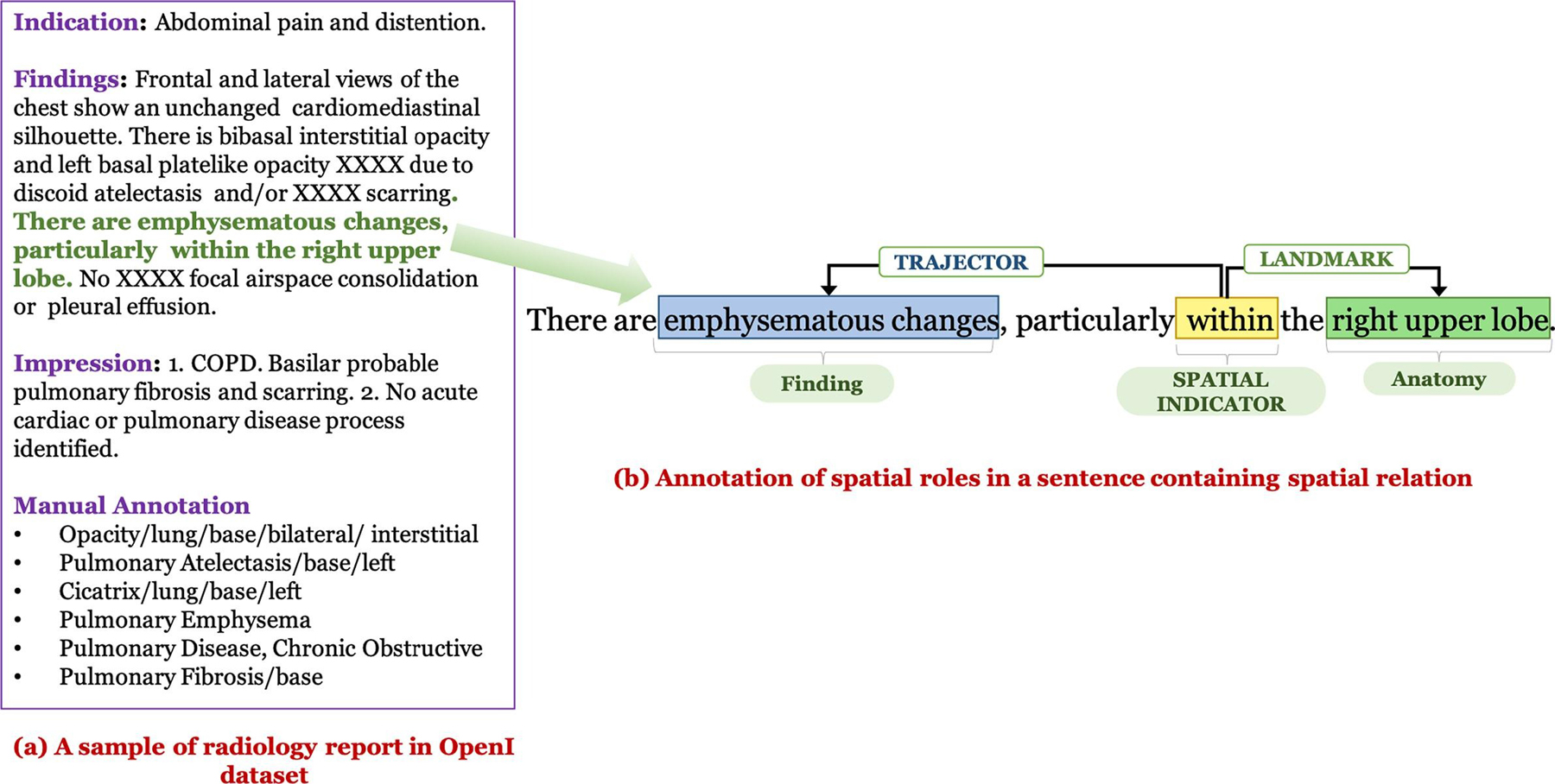
Examples of manual annotations: (a) Open-i annotations, (b) Our spatial relation annotations.
Table 3.
Studies who have used Open-i manual annotations.
| Paper | How Open-i chest X-ray dataset is involved | Limitation (Radiology entities annotated/considered for model evaluation) |
|---|---|---|
| Demner-Fushman et al. [7] | Manually annotated or coded the collected reports with findings, diagnoses, body parts using MeSH terms supplemented by RadLex codes. Automatic annotation was also produced by the Medical Text Indexer (MTI). | This is a manual annotation process relying on MeSH terms and standard qualifier terms. The coded terms were not well-distinguished between findings and diagnoses. Moreover, the annotation lacks other information such as relation between findings and diagnoses. The automatic labeling does not include the related body parts for the labeled finding. (Positive Findings/Diagnoses and Body parts) |
| Shin et al. [50] | Trained CNNs using existing image annotations from Demner-Fushman et al. [7] and considered images labeled with a single disease using unique MeSH term combinations (this accounted for around 40 percent of the full Open-i dataset and 17 unique disease annotation patterns). Generated image annotations including disease as well as its contexts such as location, severity, and the affected organs by taking into account image/text contexts while training CNNs. | Although the annotation includes disease context and that way it generates different image captions based on severity/location contexts, it is limited to one major disease provided an image. (Findings/Diagnoses and their context such as location and severity) |
| Wang et al. (2017) [30] | Used text mining (DNorm [52] and MetaMap [53]) to label disease names using reports. Evaluated their image labeling method on Open-i reports using the key findings/disease names coded by human annotators as gold standard [7]. Note that additional datasets are also used. | Only used the available annotations for evaluating their proposed method. (Findings/Diagnoses) |
| Wang et al. (2018) [46] | Evaluated a text-image embedding auto-annotation framework on the Open-i dataset using the key findings/disease names coded by human annotators as the gold standard [7]. Additional datasets are also used. | Used the annotated Open-i dataset for evaluating proposed disease classification method for 14 diseases. (Findings/Diagnoses) |
| Peng et al. [47] | Defined rules utilizing universal dependency graphs to identify negation or uncertainty related to findings. Manually checked the annotations in Open-i and organized the findings into 14 domain-important and generic types of medical findings. | Used the Open-i dataset both for designing the patterns and testing. Although they mentioned that organizing the findings into fine-grained categories can facilitate in correlating findings with the diagnosis, the terms distinguished as diagnoses or body parts were not utilized in the study for showing any correlation. (Findings) |
| Daniels et al. [48] | Proposed a deep neural network that predicts one or more diagnoses given an image by jointly learning visual features and topics from report findings. | Used the Open-i dataset and their corresponding ‘findings’ annotations both for fine-tuning and evaluating the model. (Findings/Diagnoses) |
| Huang et al. [51] | Proposed a neural sequence-to-sequence model by leveraging “indication” information of the report which includes annotating the relationship between the positions where the finding term appears. They used the Open-i manual annotations as a reference annotation for evaluating the model. | Although this generated annotations for multiple diseases per image and also aimed to improve the results of Shin et al. [50] in annotating disease along with context such as location and severity, they did not annotate other useful contexts including spatial information of the finding as well as the associated diagnosis. (Findings/Diagnoses and their context such as location and severity) |
| Zech et al. [49] | To assess the generalizability of a deep learning model for screening pneumonia across 3 hospital systems. Used human-annotated pathology labels of the Open-i dataset for testing. | Used Open-i only for evaluation. (Findings/Diagnoses) |
| Candemir et al. [54] | Fine-tuned several deep CNN architectures to detect presence of cardiomegaly. Used Open-i dataset both for training and testing. | Manually annotated each Open-i image into one of the following severity categories: borderline, mild, moderate, severe, and non-classified using the corresponding reports having cardiomegaly. (Findings, specifically cardiomegaly and their severity levels) |
3. Proposed spatial relation annotation framework
3.1. Dataset for spatial relation annotation
A subset of 2000 reports from a total of 2470 non-normal reports as judged by two human annotators in Demner-Fushman et al. [7] was used to create our spatial relation corpus. This newly annotated chest X-ray corpus contains spatial relations between findings and body locations as well as the correlated probable diagnoses and the hedging terms used in qualifying the diagnoses. We have presented a simple comparison between the Open-i manual annotations and our spatial annotations of a sample report in Fig. 1. Note that we have not annotated other findings appearing in the report such as Opacity and Pulmonary Fibrosis as their corresponding body locations are not described through any spatial preposition.
3.2. Rad-SpRL
Our spatial representation framework (Rad-SpRL) consists of 4 spatial roles (trajector, landmark, hedge, and diagnosis) with respect to a spatial indicator. The spatial roles and the spatial indicator are defined as follows:
spatial indicator: term (usually a preposition, e.g., in, within, at, near) that triggers a spatial relation
trajector: object (finding, anatomical location) whose spatial position is being described
landmark: location of the trajector (may also be chained as a trajector to another landmark)
hedge: phrase indicating uncertainty (e.g., could be, may represent), generally in reference to the diagnosis and very rarely in the trajector
diagnosis: disease/clinical condition the radiologist associated with the finding
In most of the cases where a sentence contains spatial information, a finding (trajector) is usually detected at a particular body location (landmark) where the trajector term appears to the left of the spatial indicator and the landmark to its right. However, there are instances where a spatial preposition describes the body location (landmark) with its associated abnormality (trajector) and the trajector term appears to the right of the spatial indicator and landmark to the left (refer to example in Fig. 2(d)). We have presented a few specific examples to highlight how various spatial roles and spatial indicators are identified in sentences following the above definitions of Rad-SpRL in Fig. 2. Please note that we have considered disease/condition terms as diagnosis only when they are documented in conjunction with any spatially-located finding, or in other words are entirely probable diagnoses inferred from the finding. Also note that there is some ambiguity between a finding and a diagnosis, such that the same phrase may appear as a diagnosis in one relation while being a trajector in another. Our purpose here is not to formally distinguish between a finding and a diagnosis, but rather to identify the spatial relationships in radiology reports where the trajector is generally a finding (or artifact in the image) and the diagnosis is generally a well-understood disease term.
Fig. 2.
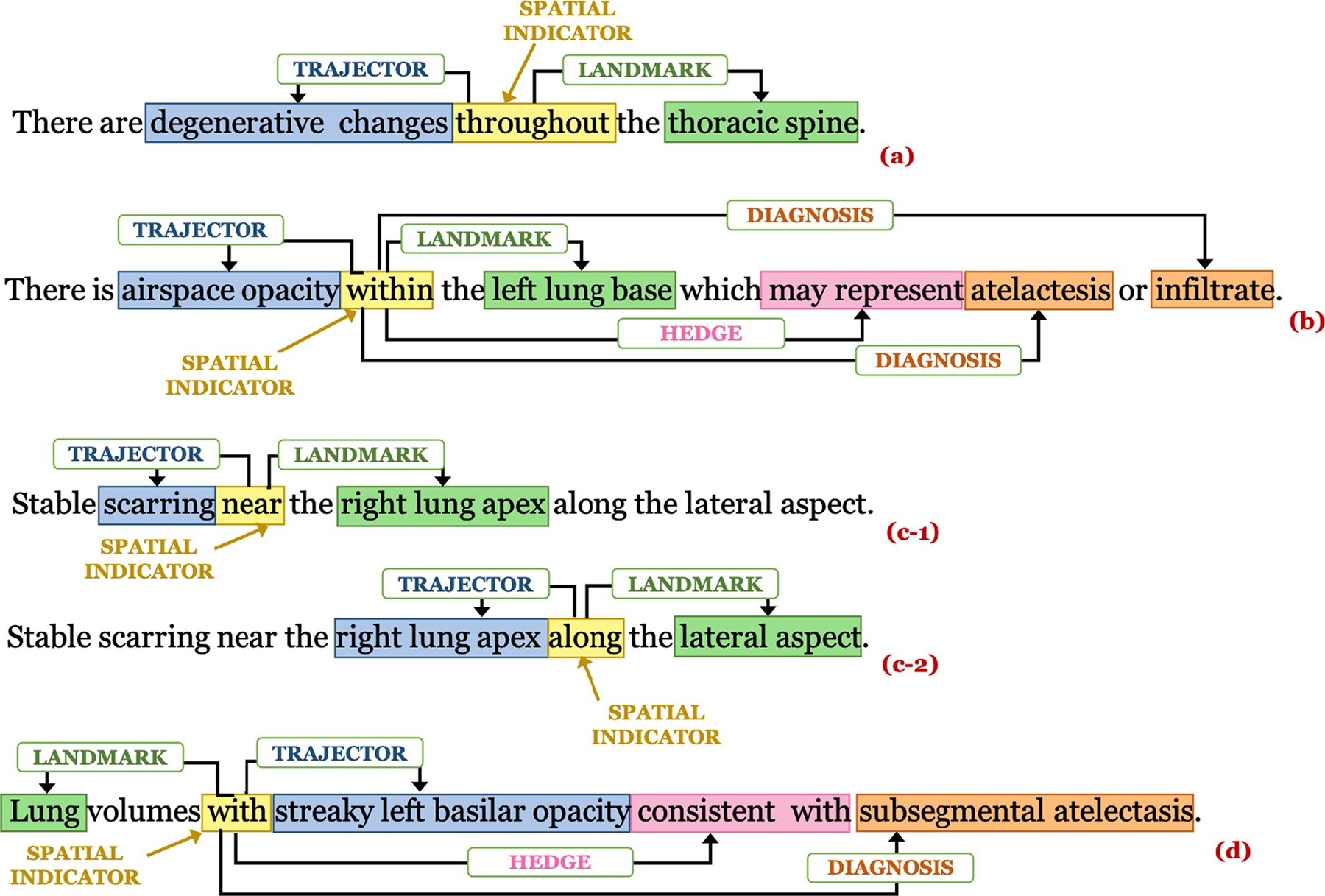
Examples of spatial role annotations: (a) Sentence having trajector and landmark, (b) Sentence having trajector, landmark, hedge, and diagnosis, (c-1) and (c-2) show the annotations of the same sentence containing 2 spatial indicators where the same entity right lung apex acts as a landmark in (c-1) and a trajector in (c-2), and (d) Sentence where a landmark is described with a trajector.
3.3. Annotation process
Two annotators (S.E.S., a medical librarian, and L.R., an MD) annotated the spatial roles for each identified spatial indicator in each of the 2000 reports independently. They also were the annotators that manually coded the findings/diagnoses available as part of the Open-i dataset [7]. The spatial relation annotations were conducted in two rounds and reconciled after each. The first round consisted of annotating the first 500 reports and the second round consisted of annotating the remaining 1500. Fig. 3 shows a sample annotated report from the corpus.
Fig. 3.
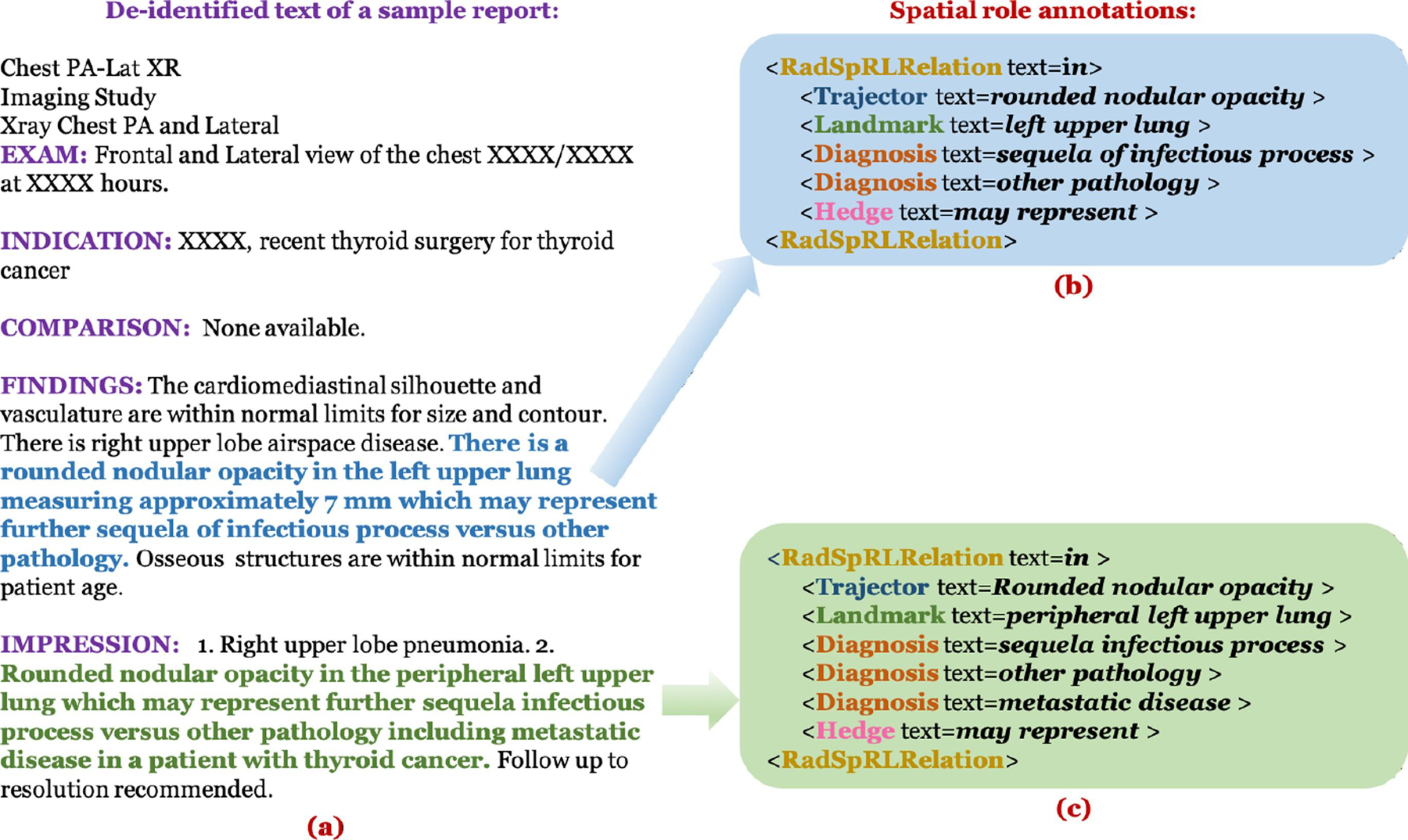
(a) Example of a de-identified report in our corpus, (b) Spatial role label annotations for the sentence represented by blue text in (a), and (c) Spatial role label annotations for the sentence represented by green text in (a). RadSpRLRelation indicates the text of the respective spatial indicators implying the existence of a spatial relation in both the sentences.
3.3.1. Annotation agreement
The inter-annotator agreement statistics for both spatial indicator and spatial roles are shown in Table 4. The Kappa (κ) agreement between the two annotators has been calculated for spatial indicator (as this is a binary classification task) whereas we report the overall F1 agreement for annotating the spatial role labels (as this is a role identification task). The Kappa agreement is high for spatial indicators in both annotation rounds. The F1 agreements for the 4 spatial roles are fairly low in the first round with much improvement in the second round. This is mainly because it is relatively easy and unambiguous to locate a spatial preposition in a sentence compared to identifying the spatial roles. All conflicts were reconciled with an NLP expert (K.R.) following each round of annotation. The moderate agreement rate for trajector and diagnosis roles was likely due to ambiguity in distinguishing the two roles in a sentence, especially when the language pattern is different from the usual. Consider the examples below:
Probably scarring in the left apex, although difficult to exclude a cavitary lesion.
There are irregular opacities in the left lung apex, that could represent a cavitary lesion in the left lung apex.
Table 4.
Annotator agreement.
| Number of Reports | Kappa (κ) | Overall F1 | |||
|---|---|---|---|---|---|
| spatial indicator | trajector | landmark | diagnosis | hedge | |
| First 500 | 0.88 | 0.44 | 0.50 | 0.25 | 0.49 |
| Remaining 1500 | 0.93 | 0.66 | 0.71 | 0.62 | 0.57 |
| Complete 2000 | 0.92 | 0.59 | 0.64 | 0.49 | 0.55 |
In the first example, ‘scarring’ was annotated as a trajector after reconciliation as its spatial location (‘left apex’) is described directly, although there is a higher chance of annotating it as a diagnosis since most of the probable diagnoses terms are usually preceded by a hedge term (‘Probably’ in this case). Similarly, ‘cavitary lesion’ is indirectly connected to the same body location (‘left apex’) and has been interpreted as an additional finding. So, ‘cavitary lesion’ was also annotated as a trajector and not as a diagnosis. In the second example, ‘cavitary lesion’ was annotated as a diagnosis in context to the first ‘in’ in the sentence, whereas the same term ‘cavitary lesion’ was annotated as a trajector when its role was identified in context to the second ‘in’. As previously noted, this difference where the same term can be both a trajector and diagnosis in different sentences is a consequence of focusing on explicitly representing the spatial language as described as well as the natural ambiguity between a finding and diagnosis in radiology. As a result, some downstream processing or interpretation is still required, which we leave to future work.
3.3.2. Annotation statistics
A total of 1962 spatial relations are annotated in our corpus of 2000 reports. Most of the trajector terms were findings. However, 176 out of 2293 terms annotated as trajectors were anatomical locations (example shown in Fig. 2(c–2)). 118 spatial indicators had more than one probable diagnosis, out of which 98 were associated with 2 diagnosis terms, 17 were associated with 3 diagnosis terms, and 3 had 4 associated diagnosis terms. There are 1052 reports containing at least one sentence triggering a spatial relation. In those reports, there are 1742 sentences each containing at least one spatial indicator (1522 sentences containing exactly one spatial indicator and remaining 220 containing more than one spatial indicator). We have highlighted some brief descriptive statistics of our corpus based on the reconciled version of the annotations in Table 5.
Table 5.
Descriptive statistics of the annotations.
| Parameter | Frequency |
|---|---|
| Average length of sentence containing spatial relation | 13 |
| spatial indicator | 1962 |
| trajector | 2293 |
| landmark | 2167 |
| diagnosis | 455 |
| hedge | 388 |
| Sentences containing at least 1 spatial indicator | 1742 |
| Maximum number of spatial indicator in any sentence | 4 |
| Spatial relations containing only trajector and landmark | 1589 |
| Spatial relations containing only trajector, landmark, and diagnosis | 9 |
| Spatial relations containing only trajector, landmark, and hedge | 70 |
| Spatial relations containing all 4 spatial roles | 304 |
| Spatial relations containing more than 1 diagnosis | 118 |
| Maximum diagnosis terms associated with any spatial relation | 4 |
4. Methods for spatial relation extraction
We apply Bi-LSTM CRF as the baseline model and additionally utilize two pre-trained transformer language models (BERT and XLNet) for extracting the spatial indicators in a sentence and consequently to extract the associated spatial roles for each spatial indicator. For spatial role extraction, we evaluate both the gold and the predicted spatial indicators in a sentence.
4.1. Baseline model
We formulate the spatial role extraction as a sequence labeling task. We utilize a Bi-LSTM CRF framework similar to the proposed architecture in Lample et al. [55] both for spatial indicator extraction and spatial role labeling. The CRF in the decoding layer takes into account the sequential information in the sentence while predicting the sequence labels related to any spatial role (trajector, landmark, diagnosis, and hedge). We utilize a Bi-LSTM that incorporates a character embedding (where each character is denoted ci,j) for each word wi in a sentence. Here, irepresents the word position and jstands for the position of the character in the word wi. For every word, this character embedding is then concatenated with the respective pre-trained word embedding . For extracting the spatial role labels, additionally a spatial indicator embedding is concatenated to the word and character embeddings to distinguish the indicators from non-indicator words. The final concatenated representation [; ; ] is fed into the final Bi-LSTM network with one hidden layer. The overall architecture is presented in Fig. 4.
Fig. 4.
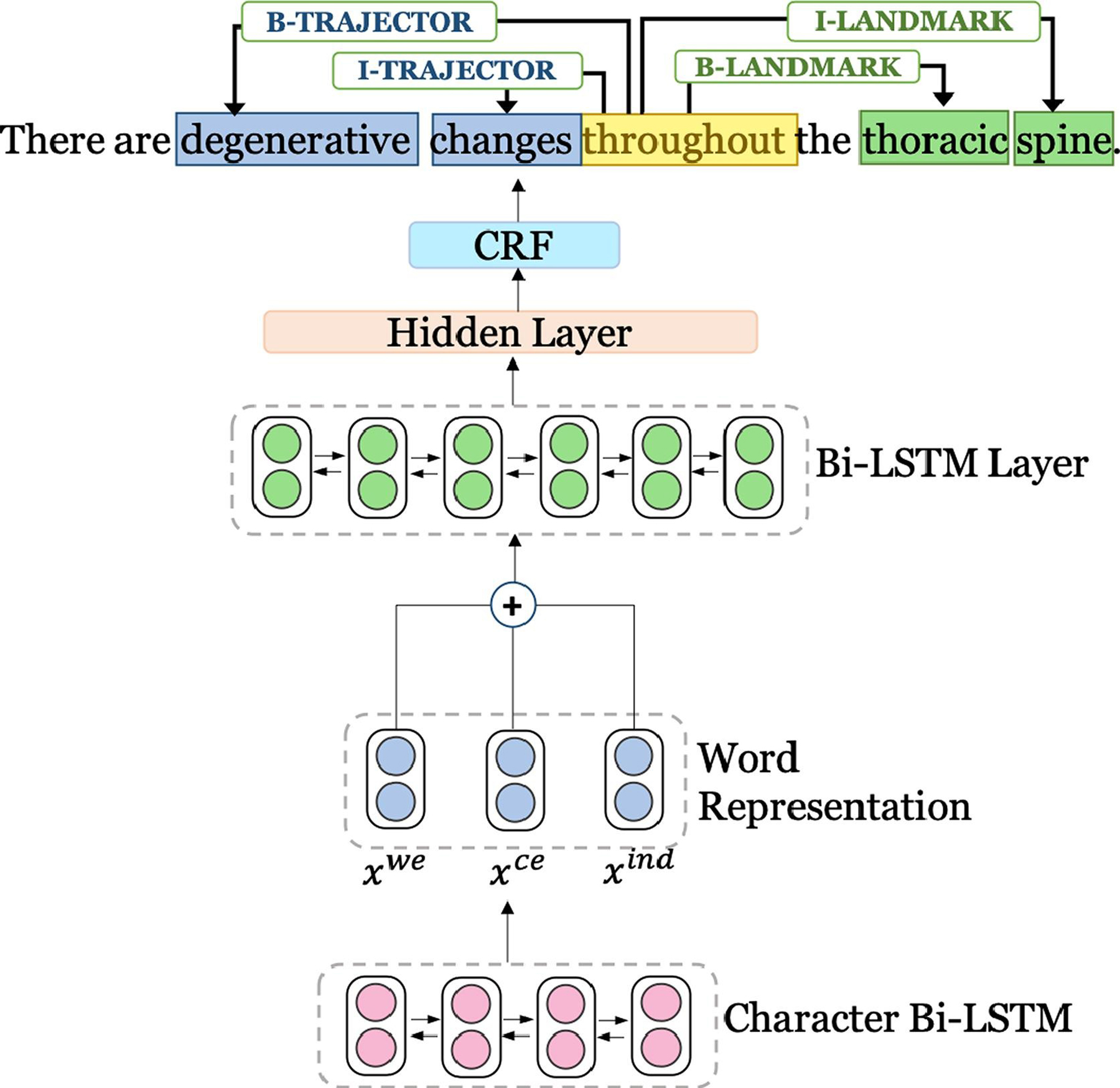
Baseline model architecture. For each word, a character representation is fed into the input layer of the Bi-LSTM network. For each word, xwe represents pre-trained word embeddings, xce represents character embeddings, and xind represents indicator embeddings. The final predictions for the spatial role labels in a sentence are made combining the Bi-LSTM’s final score and CRF score.
4.2. BERT and XLNet-based models
First, we fine-tune BERT for extracting the spatial indicators in a sentence and second, we apply the fine-tuned model for labeling the four spatial roles provided the spatial indicator in a sentence. In this work, we represent a sentence obtained after WordPiece tokenization as [[CLS] sentence [SEP]] for constructing a single input sequence following the original BERT paper [18], where [CLS] is a symbol added at the beginning of each input sentence and [SEP] is a separator token for separating sentences. The input sequences are then fed into the BERT model to generate contextual representations. For spatial role labeling, we mask the spatial indicator term with an identifier ‘$spin$’ to better encode the positional information of the specific spatial indicator in a sentence for which the spatial roles are annotated. The contextual BERT representation corresponding to each word in the sequence [[CLS] sentence [SEP]] is then concatenated with a spatial indicator embedding similar to the baseline Bi-LSTM CRF model. The concatenated representation is fed into a simple linear classification layer for predicting the final labels for each token. The model architecture is illustrated in Fig. 5.
Fig. 5.
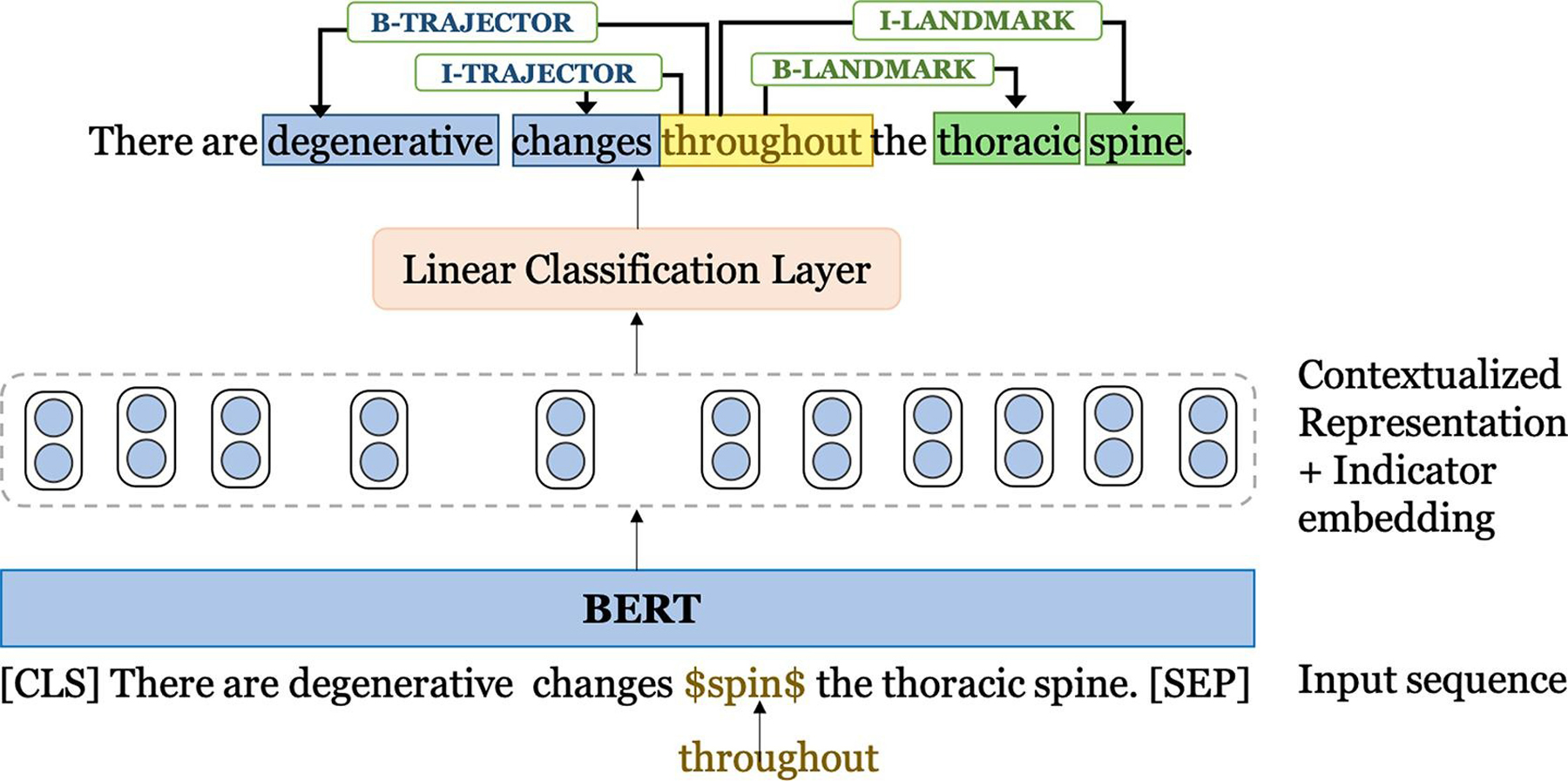
BERT-based model.
To fine-tune BERT for spatial role labeling for the Rad-SpRL corpus, we initialize the model with the publicly available pre-trained checkpoints of the BERT large model (BERTlarge). We also initialize the model parameters obtained by pre-training BERT on medical corpus (MIMIC-III clinical notes). We have adopted these pre-trained parameters from a previous work [10] where clinical domain embedding models were pre-trained on MIMIC-III clinical notes, referred to as BERTlarge (MIMIC), after initiating from the BERTlarge released checkpoint. Owing to the best performance of BERTlarge (MIMIC) on clinical concept extraction for four benchmark datasets [10], we initiate our model with the pre-trained parameters of BERTlarge (MIMIC) to fine-tune on our spatial role labeling task.
For XLNet, the model input is similar to BERT and we feed [sentence [SEP] [CLS]] into the model. We have utilized a similar simple architecture as BERT for fine-tuning XLNet on Rad-SpRL. However, we have initialized the model with the released pre-trained model parameters (XLNetlarge) for fine-tuning as experimenting with the MIMIC pre-trained parameters has yet to result in further performance improvement.
4.3. Pre-processing
4.3.1. spatial indicator extraction
We preprocess the Rad-SpRL dataset to generate input sequence for the models. We follow Beginning (B), Inside (I), and Outside (O) tagging scheme to label the words in a sentence. The input to the models consists of the sequence of words and the corresponding BIO tags. The following example shows how a sentence containing two spatial indicators is tagged.
4.3.2. Spatial role labeling
For each spatial indicator in a sentence, we create an instance or sample of the sentence. For each instance, we tag all the spatial roles (trajector/landmark/diagnosis/hedge) as well as the spatial indicator. Creating separate sentence instance for each spatial indicator helps in dealing with cases where the same word can be both a trajector and a landmark in context of two different spatial indicators in the sentence (example shown in (c-1) and (c-2) in Fig. 2). Also, annotating only the roles associated with a single spatial indicator provides the model unambiguous information about the position of the specific indicator term to which these roles are associated. We again follow the BIO tagging scheme. The input to the final model consists of words and the corresponding B, I, O labels for a set of sentences. However, in the case of applying BERT and XLNet, the input sentence is tokenized by WordPiece and SentencePiece tokenizers before feeding into the BERT and XLNet encoders, respectively. The following example shows the tagged words for the sentence – “Minimal degenerative changes of the thoracic spine”.
4.4. Experimental settings and evaluation
We use pre-trained medical domain MIMIC-III word embeddings of 300 dimensions2 in our Bi-LSTM experiments. The character and the indicator embeddings are initialized randomly and altered during training. The dimensions of character and indicator embeddings are 100 and 5 respectively. The model is implemented using TensorFlow [56], and the hyperparameters are chosen based on the validation set. LSTM hidden size is set at 500, dropout rate at 0.5, learning rate at 0.01, and learning rate decay at 0.99. We use the Adam optimizer and train the model for a maximum of 20 epochs.
For fine-tuning BERT, both for BERTlarge and BERTlarge (MIMIC), we largely followed the standard BERT parameters, including setting the maximum sequence length at 128, learning rate at 2e-5, and using the cased version of the models. Additionally, we set the number of training epochs at 4 based on the performance of the models on the validation set. For BERTlarge (MIMIC), we initialize the model parameters pre-trained on MIMIC after 320000 steps. For XLNet, the maximum sequence length and learning rates are the same as used for BERT, casing is also preserved, and the number of training steps is set at 2500 based on the validation set performance. In both BERT and XLNet, the dimension of indicator embedding is set at 5.
First, we perform 10-fold cross validation (CV) – with data splits at the report level – to evaluate the performance of the three models for spatial indicator extraction. The training, validation, and test sets are split in the ratio of 80%, 10%, and 10% respectively. There are a total of 1742 sentences with at least one spatial indicator and 31779 sentences without any indicator in the dataset. To ensure that the performance of the models is not impacted due to the imbalance in the number of sentences with and without spatial indicators, we additionally run both the Bi-LSTM CRF and the BERTlarge (MIMIC) models by randomly undersampling the negative sentences (i.e., sentences without an indicator) while training. We experiment using different number of negative instances such that #negative sentences after undersampling = n * #positive sentences in each train and validation sets, where n = 1,2,3,4,5,6). We found that the performance of both the models (average F1 score of a 10-fold CV) improves as n is increased from 1 through 3 and starts to decline 4 onwards. Therefore, we select the value of n as 3 for conducting all our experiments. However, to evaluate the performance of the models on the full original dataset, we include all sentences in the reports of the test sets so that we get a more realistic sense of how well the models perform.
To better assess the generalizability of the models, we randomize the fold creation 5 times and conduct 10-fold cross validation for each fold variation. We then report the average Precision, Recall, and F1 measures across 50 (5*10) different instantiations for each model. We also include the 95% confidence intervals of the average F1 measures.
Second, we evaluate the performance of the three models in extracting the spatial roles in context to a spatial indicator. We use the same fold settings and the same training, development, and test splits as in the spatial indicator extraction for spatial role labeling. For training and validation, we utilize only the sentences containing a gold spatial indicator in the sentence. However, for testing, we experiment providing both the gold and the predicted spatial indicators (i.e., the output of the first model). The same trained model weights are used in predicting the roles using gold and predicted indicators. We report the average Precision, Recall, and F1 measures of each of the 4 spatial roles across 50 instantiations for each model. We also calculate the overall measures of the three metrics considering all the roles collectively. We report the 95% confidence intervals of the average overall F1 measures. Exact match is performed for evaluating the performance on the test set.
5. Results
The average results of the 10-fold CV across 5 different runs with fold variation are shown in Table 6 for spatial indicator extraction on the Rad-SpRL corpus. Note that we test the models on all sentences (both with and without indicator). We see that either the recall or precision is higher than 90% for Bi-LSTM CRF, BERTlarge, and XLNet models. BERTlarge (MIMIC) had better balance in precision and recall (both higher than 90%). The highest F1 score is obtained by XLNetlarge, which is 91.29.
Table 6.
spatial indicator extraction results: Average Precision, Recall, and F1 measures of 10-fold CV across 5 different fold variations. CI - 95% confidence intervals of the average F1 measures across 50 iterations.
| Models | Precision | Recall | F1 (CI) |
|---|---|---|---|
| Bi-LSTM CRF | 84.73 | 92.38 | 88.33 (± 0.56) |
| BERTlarge | 94.07 | 83.54 | 87.85 (± 2.49) |
| BERTlarge (MIMIC) | 90.69 | 91.60 | 91.08 (± 3.68) |
| XLNetlarge | 88.62 | 94.40 | 91. 29 (± 0.70) |
For spatial role extraction, we report the average performance metric values of the 10-fold CV across 5 different fold variations, both considering the gold and the predicted spatial indicators in sentences of the test sets. Note that the test sets for each of the 50 different runs of the models are same for both indicator and role extraction. We create a separate instance of a sentence for each of the predicted spatial indicators (in case multiple indicators are extracted by a model). When extracting the spatial roles using the predicted spatial indicators, we take into account all the spatial roles predicted for the false positive spatial indicators in calculating the precision loss, and consider the spatial roles predicted for the false negative spatial indicators in assessing the recall loss. This provides a more realistic end-to-end evaluation of the models.
The results using gold and predicted indicators are presented in Table 7 and Table 8, respectively. We note that contextualized word representations help in improving spatial role extraction except for BERTlarge, which performed slightly inferior to the baseline model (Bi-LSTM CRF) when the predicted spatial indicators are used (see Table 8). XLNet performed the best (highest average overall F1 score of 92.9) in extracting the spatial roles when gold indicators are used, however, its performance is comparable to BERTlarge (MIMIC) when predicted indicators are used (85.4 for XLNet and 85.6 for BERT with the same confidence interval). For trajector, the highest average F1 for the end-to-end evaluation is 85.7, whereas for landmark the highest average F1 is 89.3, both obtained by BERTlarge (MIMIC) (Table 8). For all the models, the average F1 measures for diagnosis and hedge are comparatively lower than trajector and landmark, with the highest values being 79.0 and 78.6, respectively. Although the highest overall F1 is achieved by BERTlarge (MIMIC) for the end-to-end evaluation, XLNet performed better in extracting the diagnosis and hedge roles.
Table 7.
Spatial role extraction results using gold spatial indicators: Average Precision (P%), Recall (R%), and F1 measures of 10-fold CV across 5 different fold variations. CI - 95% confidence intervals of the average F1 measures across 50 iterations. BLSTM-C - Bi-LSTM CRF, BERT-L - BERTlarge, BERT-LM - BERTlarge (MIMIC), XLNet-L - XLNetlarge.
| Models |
trajector |
landmark |
diagnosis |
hedge |
overall |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 (CI) | |
| BLSTM-C | 88.8 | 87.3 | 88.0 | 94.1 | 89.9 | 91.9 | 76.6 | 75.0 | 75.2 | 78.4 | 76.3 | 77.0 | 89.0 | 86.4 | 87.6 (±0.55) |
| BERT-L | 89.7 | 91.8 | 90.7 | 95.4 | 96.1 | 95.8 | 72.7 | 85.5 | 78.4 | 72.8 | 84.1 | 77.8 | 88.8 | 92.4 | 90.5 (±0.42) |
| BERT-LM | 91.2 | 93.1 | 92.1 | 95.6 | 96.6 | 96.1 | 72.3 | 83.9 | 77.4 | 75.0 | 86.1 | 80.1 | 89.5 | 93.3 | 91.4 (±0.54) |
| XLNet-L | 92.8 | 94.1 | 93.5 | 96.1 | 96.8 | 96.4 | 78.6 | 88.0 | 82.8 | 79.6 | 88.6 | 83.7 | 91.6 | 94.2 | 92.9 (±0.38) |
Table 8.
Spatial role extraction results using predicted spatial indicators: Average Precision (P%), Recall (R%), and F1 measures of 10-fold CV across 5 different fold variations. CI - 95% confidence intervals of the average F1 measures across 50 iterations. BLSTM-C - Bi-LSTM CRF, BERT-L - BERTlarge, BERT-LM - BERTlarge (MIMIC), XLNet-L - XLNetlarge.
| Models |
trajector |
landmark |
diagnosis |
hedge |
overall |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 (CI) | |
| BLSTM-C | 77.7 | 83.9 | 80.6 | 83.0 | 86.6 | 84.7 | 72.3 | 72.2 | 71.7 | 71.5 | 72.5 | 71.6 | 78.8 | 83.1 | 80.8 (±0.76) |
| BERT-L | 85.7 | 73.9 | 78.1 | 90.0 | 77.2 | 82.8 | 73.1 | 75.3 | 73.8 | 71.4 | 71.8 | 71.1 | 85.1 | 75.2 | 79.3 (±2.16) |
| BERT-LM | 85.8 | 85.9 | 85.7 | 89.6 | 89.2 | 89.3 | 72.5 | 83.0 | 77.3 | 72.0 | 81.6 | 76.3 | 84.8 | 86.6 | 85.6 (±0.65) |
| XLNet-L | 82.3 | 88.9 | 85.3 | 86.0 | 90.9 | 88.2 | 73.8 | 85.6 | 79.0 | 73.2 | 85.3 | 78.6 | 82.1 | 89.1 | 85.4 (±0.65) |
6. Discussion
In this paper, we extract the four spatial roles with respect to a spatial indicator in a sentence following the Rad-SpRL annotation scheme. This includes identifying the probable diagnoses with associated hedges in reference to a spatial relation between any finding and its associated location. The results in Table 7 and Table 8 demonstrate that the models achieve promising results in extracting the spatial roles from the Rad-SpRL corpus. We observe that incorporating contextualized word representations by fine-tuning BERT (pre-trained on MIMIC) and XLNet models on the Rad-SpRL dataset performs better than a Bi-LSTM CRF network in extracting the spatial indicators as well as the spatial roles. Thus, BERTlarge (MIMIC) and XLNetlarge are currently the best performing models. However, more work is needed to determine which between these two models is more robust in extracting spatial information from chest X-ray reports. We also note that the average F1 measures are high for trajector and landmark roles and are comparatively low for diagnosis and hedge. The reason behind this can be attributed to the lesser number of diagnosis and hedge terms in the dataset (5–6 times less than both trajector and landmark terms) and greater distance between the spatial indicator and the diagnosis/hedge terms compared to the trajector/landmark terms.
Taking into account the relatively low F1 measure for diagnosis and hedge, we performed a brief analysis of the errors. On average, the best performing BERTlarge (MIMIC) model in the end-to-end evaluation (shown in Table 8) misses around 10% of the gold annotated diagnosis terms, misclassifies 1% of the terms as trajectors, and misidentifies the beginning of around 2.6% of the diagnosis terms as inside. Some of the diagnosis terms that are misclassified as trajectors include ‘bronchovascular crowding’, ‘edema’, ‘pulmonary fibrosis’, ‘atelectasis’, and ‘scarring’. This is mainly because of the different ways certain common radiographic findings are also described as differential diagnoses. For example, in the sentence – “Low lung volumes with bibasilar opacities may represent bronchovascular crowding.”, the diagnosis ‘bronchovascular crowding’ is falsely classified as a trajector. This might be because there are instances in the dataset where ‘bronchovascular crowding’ appears as trajector (e.g., in the sentence – “There are low lung volumes with bronchovascular crowding as a result.”), as often a diagnosis term itself appears in a spatial relationship. The main reason for the errors associated with incorrect starting boundary of a predicted diagnosis term is that sometimes an extra adjacent term to the left of the actual diagnosis term is predicted by the model. For example, in “Increasing prominence of the superior mediastinum may be secondary to enlarging thyroid mass.”, the model outputs ‘enlarging thyroid mass’ as the predicted diagnosis instead of the annotated ‘thyroid mass’. For hedge, one of the major contributing factors of incorrect predictions of gold terms is that the BERTlarge (MIMIC) model misses around 14% of the gold annotated hedge terms. Most of these missed terms (e.g.,’questionable’, ‘suggestion of’, ‘appears’, ‘alternatively’) occur very infrequently in the dataset. Another challenge could be the variety of ways the hedging terms are used and positioned in a sentence to suggest any finding or differential diagnosis. Future work should attempt to improve the models to better handle complex description of sentences.
In this work, we have considered both positive and negative spatial relations as our focus was on identifying the spatial relationship itself, not the presence or absence of the condition to which the relation refers. We aim to differentiate the negated relations in future. Future work should also be directed toward building an end-to-end system based on neural joint learning models [57,58] that would extract both spatial indicator and the spatial roles together, reducing discongruencies between predicted roles. This work extracts single word prepositional spatial expressions. We further aim to consider non-prepositional spatial expressions as spatial indicators (e.g., verbs such as ‘demonstrates’, ‘shows’ etc.) that indicate the presence of any spatial relation between finding and body location. Additionally, we will address expressions containing multiple words (e.g., ‘projects in’, ‘projecting through’, ‘projected over’) in our future work, although such expressions occur rarely in this dataset to describe the location of findings. We will also investigate the performance of our proposed systems for extracting the spatial roles when the spatial indicators are non-prepositional or multi-word expressions. Besides radiographic findings, we also intend to extend the Rad-SpRL framework to extract other important and common spatially-grounded radiology entities such as medical devices from the reports. The multi-word spatial expressions we described above such as ‘projects over’ and ‘extends below’ are more common in describing the location of devices that we aim to extract in the future. The following example illustrates a sample sentence where the medical device ‘Right IJ venous catheter’ acts as the trajector in reference to its associated location ‘proximal SVC’ that acts as the landmark:
Right IJ venous catheter terminates at the proximal SVC.
Apart from diagnoses and hedging terms, we additionally aim to extract other important contextual information related to spatial relations across different imaging modalities in our later work. Another limitation of this study is that intersentence spatial relations are not covered, although the frequency of such cases are rare in the Rad-SpRL corpus. We also aim to evaluate the generalizability of our sequence labeling methods in extracting the spatial roles from datasets across institutions. For standardization of the extracted spatial roles, we further aim to normalize them utilizing the existing radiology lexicons such as RadLex [59] codes. From a method perspective, we plan to apply some alternative deep learning methods such as highway networks [60] and tree-based LSTMs [58] to further improve the performance of spatial role extraction from the Rad-SpRL corpus.
7. Conclusion
This paper proposes a spatial representation framework in radiology (Rad-SpRL). It provides a detailed description of the annotation scheme used for extracting spatial information from radiology reports. This consists of annotating four radiology-specific spatial roles in a dataset of 2000 chest X-ray reports. The spatial roles are annotated in the context of a spatial indicator which denotes the presence of a spatial relation between clinical findings and body locations. It additionally identifies probable diagnoses and hedging terms associated with the spatially-related finding-location. For this, we first employ a Bi-LSTM CRF model as the baseline model to automatically extract the spatial indicators and the spatial roles from our annotated Rad-SpRL corpus. We then experiment with BERT and XLNet-based models. The models achieve satisfactory performance with the highest average F1 measure of 91.29 for extracting spatial indicators and F1 measures of 85.7, 89.3, 79.0, and 78.6 for identifying trajector, landmark, diagnosis, and hedge roles, respectively using the predicted indicators. In the future, we aim to evaluate the models on much larger datasets, extend the annotation framework to capture more fine-grained spatial information, and adopt joint learning models for extracting the spatial indicator and the spatial roles jointly.
Acknowledgements
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering (NIBIB: R21EB029575), the U.S. National Library of Medicine (NLM: R00LM012104), the Patient-Centered Outcomes Research Institute (PCORI: ME-2018C1-10963) and the Cancer Prevention Research Institute of Texas (CPRIT: RP160015).
Footnotes
Declaration of Competing Interest
None.
References
- [1].Birchall D, Spatial ability in radiologists: a necessary prerequisite? Br. J. Radiol 88 (1049) (2015) 6–8, 10.1259/bjr.20140511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Corry C, The future of recruitment and selection in radiology. Is there a role for assessment of basic visuospatial skills? Clin. Radiol 66 (5) (2011) 481–483, 10.1016/j.crad.2010.12.003. [DOI] [PubMed] [Google Scholar]
- [3].Kordjamshidi P, Otterlo MV, Moens M-F, Spatial Role Labeling: Task Definition and Annotation Scheme, in: Proceedings of the Language Resources & Evaluation Conference, 2010, pp. 413–420. [Google Scholar]
- [4].Kordjamshidi P, Rahgooy T, Manzoor U, Spatial Language Understanding with Multimodal Graphs using Declarative Learning based Programming, Proceedings of the 2nd Workshop on Structured Prediction for Natural Language Processing, 2017, pp. 33–43, 10.18653/v1/w17-4306. [DOI] [Google Scholar]
- [5].Kordjamshidi P, Roth D, Moens M-F, Structured learning for spatial information extraction from biomedical text: Bacteria biotopes, BMC Bioinform 16 (1) (2015) 1–15, 10.1186/s12859-015-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roberts K, Rodriguez L, Shooshan S, Demner-Fushman D, Automatic Extraction and Post-coordination of Spatial Relations in Consumer Language, 2015 AMIA Annual Symposium Proceedings, 2015, pp. 1083–1092. [PMC free article] [PubMed] [Google Scholar]
- [7].Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ, Preparing a collection of radiology examinations for distribution and retrieval, J. Am. Med. Inform. Assoc 23 (2) (2016) 304–310, 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ramrakhiyani N, Palshikar G, Varma V, A Simple Neural Approach to Spatial Role Labelling, in: Advances in Information Retrieval, 2019, pp. 102–108. doi: 10.1007/978-3-030-15719-7_13. [DOI] [Google Scholar]
- [9].Shi P, Lin J, Simple BERT Models for Relation Extraction and Semantic Role LabelingarXiv:1904.05255 [Google Scholar]
- [10].Si Y, Wang J, Xu H, Roberts K, Enhancing clinical concept extraction with contextual embeddings, J. Am. Med. Inform. Assoc (2019) 1–8, 10.1093/jamia/ocz096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lan Z, Chen M, Goodman S, Gimpel K, Sharma P, Soricut R, ALBERT: A Lite BERT for Self-supervised Learning of Language Representations (2019) 1–16arXiv:1909.11942. [Google Scholar]
- [12].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J, BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics (2019) 1–8, 10.1093/bioinformatics/btz682 arXiv:arXiv:1901.08746v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang K, Altosaar J, Ranganath R, ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission (2019) 1–19arXiv:1904.05342. [Google Scholar]
- [14].Kordjamshidi P, Moens M-F, Global machine learning for spatial ontology population, J. Web Semant 30 (2015) 3–21, 10.1016/j.websem.2014.06.001. [DOI] [Google Scholar]
- [15].Roberts K, Rink B, Harabagiu SM, Scheuermann RH, Toomay S, Browning T, Bosler T, Peshock R, A machine learning approach for identifying anatomical locations of actionable findings in radiology reports, 2012 AMIA Annual Symposium Proceedings, 2012, pp. 779–788. [PMC free article] [PubMed] [Google Scholar]
- [16].Rink B, Roberts K, Harabagiu S, Scheuermann RH, Toomay S, Browning T, Bosler T, Peshock R, Extracting actionable findings of appendicitis from radiology reports using natural language processing, 2013 AMIA Joint Summits on Translational Science Proceedings, 2013, p. 221. [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang C, Zhang X, Jiang W, Shen Q, Zhang S, Rule-based extraction of spatial relations in natural language text, in: 2009 International Conference on Computational Intelligence and Software Engineering, no. 40971231, IEEE, 2009, pp. 1–4. doi: 10.1109/CISE.2009.5363900. [DOI] [Google Scholar]
- [18].Devlin J, Chang M-W, Lee K, Toutanova K, BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding, in: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Minneapolis, Minnesota, 2019, pp. 4171–4186. doi: 10.18653/v1/N19-1423. [DOI] [Google Scholar]
- [19].Clark K, Khandelwal U, Levy O, Manning CD, What Does BERT Look at? An Analysis of BERT’s Attention, in: Proceedings of the Second BlackboxNLP Workshop on Analyzing and Interpreting Neural Networks for NLP, Association for Computational Linguistics, 2019, pp. 276–286. doi: 10.18653/v1/W19-4828. [DOI] [Google Scholar]
- [20].Jawahar G, Sagot B, Seddah D, What Does BERT Learn about the Structure of Language? Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, 2019, pp. 3651–3657, 10.18653/v1/p19-1356. [DOI] [Google Scholar]
- [21].Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, Le QV, XLNet: Generalized Autoregressive Pretraining for Language UnderstandingarXiv:1906. 08237 [Google Scholar]
- [22].Hayward WG, Tarr MJ, Spatial language and spatial representation, Cognition 55 (1) (1995) 39–84, 10.1016/0010-0277(94)00643-Y. [DOI] [PubMed] [Google Scholar]
- [23].Mani I, Doran C, Harris D, Hitzeman J, Quimby R, Richer J, Wellner B, Mardis S, Clancy S, SpatialML: annotation scheme, resources, and evaluation, Lang. Resources Eval 44 (3) (2010) 263–280, 10.1007/s10579-010-9121-0. [DOI] [Google Scholar]
- [24].Pustejovsky J, Moszkowicz JL, Integrating Motion Predicate Classes with Spatial and Temporal Annotations, in: Coling 2008: Companion Volume: Posters, Coling 2008 Organizing Committee, 2008, pp. 95–98. URL <https://www.aclweb.org/anthology/C08-2024>. [Google Scholar]
- [25].Fasola J, Mataric MJ, Using semantic fields to model dynamic spatial relations in a robot architecture for natural language instruction of service robots, in: 2013 IEEE/RSJ International Conference on Intelligent Robots and Systems, 2013, pp. 143–150. 10.1109/IROS.2013.6696345. [DOI] [Google Scholar]
- [26].Chang A, Savva M, Manning CD, Learning Spatial Knowledge for Text to 3D Scene Generation, in: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Association for Computational Linguistics, 2014, pp. 2028–2038. 10.3115/v1/D14-1217. [DOI] [Google Scholar]
- [27].Kergosien E, Alatrista-Salas H, Gaio M, Güttler FN, Roche M, Teisseire M, When textual information becomes spatial information compatible with satellite images, in: 2015 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K), Vol. 01, 2015, pp. 301–306. [Google Scholar]
- [28].Collell G, Moens M-F, Learning representations specialized in spatial knowledge: leveraging language and vision, Trans. Assoc. Comput. Linguist 6 (2018) 133–144, 10.1162/tacl_a_00010. [DOI] [Google Scholar]
- [29].Ulinski M, Coyne B, Hirschberg J, SpatialNet: A Declarative Resource for Spatial Relations, in: Proceedings of the Combined Workshop on Spatial Language Understanding (SpLU) and Grounded Communication for Robotics (RoboNLP), Association for Computational Linguistics, 2019, pp. 61–70. 10.18653/v1/W19-1607. [DOI] [Google Scholar]
- [30].Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM, ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 3462–3471, 10.1109/CVPR.2017.369 arXiv:arXiv:1705.02315v5. [DOI] [Google Scholar]
- [31].Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, Ng AY, CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert ComparisonarXiv:1901.07031 [Google Scholar]
- [32].Wang Y, Sun L, Jin Q, Enhanced Diagnosis of Pneumothorax with an Improved Real-time Augmentation for Imbalanced Chest X-rays Data Based on DCNN, IEEE/ACM Trans. Comput. Biol. Bioinf 14 (8) (2019) 1, 10.1109/TCBB.2019.2911947. [DOI] [PubMed] [Google Scholar]
- [33].Pesce E, Withey SJ, Ypsilantis P-P, Bakewell R, Goh V, Montana G, Learning to detect chest radiographs containing lung nodules using visual attention networks, Med. Image Anal 53 (2019) 26–38, 10.1016/j.media.2018.12.007 arXiv:1712.00996. [DOI] [PubMed] [Google Scholar]
- [34].Hassanpour S, Langlotz CP, Information extraction from multi-institutional radiology reports, Artif. Intell. Med 66 (2016) 29–39, 10.1016/j.artmed.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cornegruta S, Bakewell R, Withey S, Montana G, Modelling Radiological Language with Bidirectional Long Short-Term Memory NetworksarXiv:1609.08409 [Google Scholar]
- [36].Bustos A, Pertusa A, Salinas J-M, de la Iglesia-Vayá M, PadChest: A large chest x-ray image dataset with multi-label annotated reportsarXiv:1901.07441 [DOI] [PubMed] [Google Scholar]
- [37].Hassanpour S, Bay G, Langlotz CP, Characterization of change and significance for clinical findings in radiology reports through natural language processing, J. Digit. Imaging 30 (3) (2017) 314–322, 10.1007/s10278-016-9931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G, Automated triaging of adult chest radiographs with deep artificial neural networks, Radiology 291 (1) (2019) 196–202, 10.1148/radiol.2018180921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB, A general natural-language text processor for clinical radiology, Journal of the American Medical Informatics Association: JAMIA 1 (2) (1994. Mar-Apr) 161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Friedman C, Johnson SB, Forman B, Starren J, Architectural requirements for a multipurpose natural language processor in the clinical environment, Proceedings of the Annual Symposium on Computer Application in Medical Care, 1995, pp. 347–351. [PMC free article] [PubMed] [Google Scholar]
- [41].Friedman C, Shagina L, Lussier Y, Hripcsak G, Automated encoding of clinical documents based on natural language processing, J. Am. Med. Inform. Assoc.: JAMIA 11 (5) (2004) 392–402, 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sevenster M, Van Ommering R, Qian Y, Automatically correlating clinical findings and body locations in radiology reports using MedLEE, J. Digit. Imaging 25 (2) (2012) 240–249, 10.1007/s10278-011-9411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yim W-W, Denman T, Kwan SW, Yetisgen M, Tumor information extraction in radiology reports for hepatocellular carcinoma patients, 2016 AMIA Joint Summits on Translational Science Proceedings, 2016, pp. 455–464. [PMC free article] [PubMed] [Google Scholar]
- [44].Steinkamp JM, Chambers C, Lalevic D, Zafar HM, Cook TS, Toward complete structured information extraction from radiology reports using machine learning, J. Digit. Imaging 32 (4) (2019) 554–564, 10.1007/s10278-019-00234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Alex B, Grover C, Tobin R, Sudlow C, Mair G, Whiteley W, Text mining brain imaging reports, J. Biomed. Semant 10 (1) (2019) 23, 10.1186/s13326-019-0211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang X, Peng Y, Lu L, Lu Z, Summers RM, TieNet: Text-Image Embedding Network for Common Thorax Disease Classification and Reporting in Chest X-Rays, The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2018, pp. 9049–9058, 10.1109/CVPR.2018.00943. [DOI] [Google Scholar]
- [47].Peng Y, Wang X, Lu L, Bagheri M, Summers R, Lu Z, NegBio: a high-performance tool for negation and uncertainty detection in radiology reports, 2018 AMIA Joint Summits on Translational Science Proceedings. 2018, pp. 188–196. [PMC free article] [PubMed] [Google Scholar]
- [48].Daniels ZA, Metaxas DN, Exploiting Visual and Report-Based Information for Chest X-Ray Analysis by Jointly Learning Visual Classifiers and Topic Models, in: IEEE 16th International Symposium on Biomedical Imaging (ISBI), 2019. [Google Scholar]
- [49].Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK, Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study, PLoS Med 15 (11) (2018) 1–17, 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shin H-C, Roberts K, Lu L, Demner-Fushman D, Yao J, Summers RM, Learning to Read Chest X-Rays: Recurrent Neural Cascade Model for Automated Image Annotation, The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 2497–2506. [Google Scholar]
- [51].Huang X, Fang Y, Lu M, Yao Y, Li M, An Annotation Model on End-to-End Chest Radiology Reports, in: IEEE Access, Vol. 7, IEEE, 2019. doi: 10.1109/access.2019.2917922. [DOI] [Google Scholar]
- [52].Leaman R, Khare R, Lu Z, Challenges in clinical natural language processing for automated disorder normalization, J. Biomed. Inform 57 (2015) 28–37, 10.1016/j.jbi.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aronson AR, Lang F-M, An overview of MetaMap: historical perspective and recent advances, J. Am. Med. Inform. Assoc 17 (3) (2010) 229–236, 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Candemir S, Rajaraman S, Thoma G, Antani S, Deep learning for grading cardiomegaly severity in chest x-rays: An investigation, in: IEEE Life Sciences Conference (LSC), IEEE, 2018, pp. 109–113. doi: 10.1109/LSC.2018.8572113. [DOI] [Google Scholar]
- [55].Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C, Neural Architectures for Named Entity Recognition, in: Proceedings of NAACL-HLT, 2016, pp. 260–270. arXiv:1603.01360. [Google Scholar]
- [56].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, et al. , Tensorflow: Large-scale machine learning on heterogeneous distributed systems, arXiv preprint arXiv:1603.04467 [Google Scholar]
- [57].Li F, Zhang M, Fu G, Ji D, A neural joint model for entity and relation extraction from biomedical text, BMC Bioinform 18 (1) (2017) 198, 10.1186/s12859-017-1609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Miwa M, Bansal M, End-to-End Relation Extraction using LSTMs on Sequences and Tree Structures, in: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, 2016, pp. 1105–1116. arXiv:arXiv:1601.00770v3, doi: 10.18653/v1/P16-1105. [DOI] [Google Scholar]
- [59].Langlotz CP, RadLex: a new method for indexing online educational materials, Radiographics 26 (6) (2006) 1595–1597, 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- [60].Srivastava RK, Greff K, Schmidhuber J, Highway NetWorks: Training Very Deep Networks, in: NIPS’15 Proceedings of the 28th International Conference on Neural Information Processing Systems, 2015, pp. 2377–2385. arXiv:1507.06228. [Google Scholar]


