Abstract
The coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), presents an unprecedented challenge to global public health with researchers striving to find a possible therapeutic candidate that could limit the spread of the virus. In this context, the present study employed an in silico molecular interaction-based approach to estimate the inhibitory potential of the phytochemicals from ethnomedicinally relevant Indian plants including Justicia adhatoda, Ocimum sanctum and Swertia chirata, with reported antiviral activities against crucial SARS-CoV-2 proteins. SARS-CoV-2 proteins associated with host attachment and viral replication namely, spike protein, main protease enzyme Mpro and RNA-dependent RNA polymerase (RdRp) are promising druggable targets for COVID-19 therapeutic research. Extensive molecular docking of the phytocompounds at the binding pockets of the viral proteins revealed their promising inhibitory potential. Subsequent assessment of physicochemical features and potential toxicity of the compounds followed by robust molecular dynamics simulations and analysis of MM-PBSA energy scoring function revealed anisotine against SARS-CoV-2 spike and Mpro proteins and amarogentin against SARS-CoV-2 RdRp as potential inhibitors. It was interesting to note that these compounds displayed significantly higher binding energy scores against the respective SARS-CoV-2 proteins compared to the relevant drugs that are currently being targeted against them. Present research findings confer scopes to explore further the potential of these compounds in vitro and in vivo towards deployment as efficient SARS-CoV-2 inhibitors and development of novel effective therapeutics.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, Justicia adhatoda, Ocimum sanctum, Swertia chirata, phytocompounds, molecular docking, molecular dynamics simulations, therapeutic candidates
1. Introduction
Coronaviruses (CoVs) are non-segmented, positive-sense, single-stranded, enveloped RNA viruses that belong to the family Coronaviridae and potentiate the spread of human and veterinary important diseases (Wu & McGoogan, 2020). Zoonotic transmission of coronaviruses has occurred at least six times before 2019 and has included Human (H) CoV-229E, HCoV-OC43, HCoV-HKU1 and HCoV-NL63 that typically presented mild symptoms (Andersen et al., 2020). On the other hand, the outbreaks of SARS-CoV (severe acute respiratory syndrome) in 2003 and MERS-CoV (Middle East respiratory syndrome) in 2012 caused severe and life-threatening illness associated with the infection of the respiratory tract. The present coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected over 62 million people and claimed more than one million lives so far (updated on November 28, 2020) (https://www.worldometers.info/coronavirus/). The disease originated in Wuhan, China in December 2019 and has since caused widespread panic and imposed excessive strain on global public healthcare systems. The global scientific community has begun to amass a wealth of knowledge that depicts the complex genomic features (Wu & McGoogan, 2020) and the evolutionary enigma (Andersen et al., 2020) to understand the infectivity and epidemiology of SARS-CoV-2. While most studies related to COVID-19 are in nascent stages, a more in-depth research is required to enhance the knowledge about this virus (Balachandar et al., 2020). Several proteins encoded by the viral genome including the spike (S) glycoprotein, the main protease enzyme (Mpro) and the RNA-dependent RNA polymerase (RdRp) have attracted interest as promising druggable and vaccine targets given their pivotal roles in viral replication and infection (Tai et al., 2020).
Several methods like drug repurposing (Ciliberto & Cardone, 2020), administration of convalescent plasma transfusion (Shen et al., 2020) and usage of SARS-CoV and MERS-CoV antibodies (Huang et al., 2020) are presently being employed to combat the catastrophic COVID-19. Additionally, several drugs and vaccines are currently in clinical trials throughout the world. However, effective treatments specifically targeted to treat and cure COVID-19 associated pathologies, are yet to be achieved.
Biological diversity has afforded mankind an invaluable source of molecular entities that have, for centuries, been a resource in the production of traditional and formal pharmaceuticals (Kar et al., 2020). The process of drug discovery relies heavily upon phytochemicals as remedies to various ailments. Considering the importance of morphine, codeine and taxol, plants in particular have had an important role in drug discovery and development. Thus, plants may be an untapped reserve of diverse chemical constituents that could prove valuable in the development of drugs targeting COVID-19. Recently, our group reported inhibitory prospects of phytochemicals from Clerodendrum spp. against the replication and infection-associated proteins of SARS-CoV-2 based on molecular docking and molecular dynamics simulations (Kar et al., 2020). In India, Justicia adhatoda (syn. Adhatoda Vasica), Ocimum sanctum and Swertia chirata are frequently used to treat the symptoms associated with respiratory disorders involving flu, bronchitis and pneumonia (Chavan & Chowdhary, 2014; Ghoke et al., 2018; Verma et al., 2008). The crude extracts of the above mentioned plants have been reported to display inhibition of viral infection without any cytotoxic effects towards normal tissues (Chavan & Chowdhary, 2014; Ghoke et al., 2018; Verma et al., 2008). Several biologically active secondary metabolites with considerable antiviral and antimicrobial potential have been identified within the three plant species and have been depicted in Figure 1 (Astani et al., 2011; Benencia & Courrèges, 2000; Jha et al., 2012; Li et al., 2016; Pilau et al., 2011; Rao & Sinsheimer, 1974; Zheng & Lu, 1989). In the present study, a molecular docking-based approach was employed to determine the affinity of these compounds towards the spike, Mpro and RdRp proteins of SARS-CoV-2 as a means for the identification of possible drug leads. A comprehensive investigation of the physicochemical features of the screened phytocompounds was conducted in the light of the general rules of drug-likeness. Furthermore, robust molecular dynamics simulations followed by detailed analysis of molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) binding free energy of the protein-ligand complexes was conducted to profile the most promising inhibitory candidates against each of the SARS-CoV-2 proteins, targeted towards the development of effective therapeutics.
Figure 1.
The structures of the selected phytocompounds from the plants Justicia adhatoda, Ocimum sanctum and Swertia chirata employed for the present analysis. (A) Anisotine. (B) Adhatodine. (C) Vasicoline. (D) Beta-sitosterol. (E) Vasicolinone. (F) Vasicine. (G) Beta-carotene. (H) Eugenol. (I) Caryophyllene. (J) Carvacrol. (K) Cineole. (L) Amarogentin. (M) Mangiferin. (N) Beta-amyrin. (O) Swerchirin. (P) Swertianin. (Q) Swertiamarin.
2. Materials and methods
2.1. Retrieval and refinement of protein and ligand structures
The high-resolution X-ray diffraction crystal structures of the receptor-binding domain (RBD) of SARS-CoV-2 spike protein (PDB ID: 6LZG Chain B; 2.50 Å resolution) (Wang et al., 2020), the SARS-CoV-2 Mpro (PDB ID: 6LU7 Chain A; 2.16 Å resolution) (Jin et al., 2020) and the SARS-CoV-2 RdRp (PDB ID: 7BV2 Chain A; 2.50 Å resolution) (Ciliberto & Cardone, 2020) were retrieved from PDB. The proteins were prepared by removing the inhibitor and water molecules prior to analysis. Polar hydrogen atoms and Kollman charges were added to the corresponding PDB files using AutoDock tools. The profiling of phytochemicals present in Justicia adhatoda, Ocimum sanctum and Swertia chirata was achieved by screening the Dr. Duke's Phytochemical and Ethnobotanical Databases (https://phytochem.nal.usda.gov/phytochem/search/list) and the respective structures were retrieved from NCBI PubChem (https://pubchem.ncbi.nlm.nih.gov/). The three-dimensional structures were generated through the Open Babel software (O’Boyle et al., 2011). Energy-optimization of the structures was achieved using the PRODRG server (Schüttelkopf & Van Aalten, 2004). Gromos 96 force field was applied for the process of energy minimization. Gasteiger charges were added to the corresponding ligand structures using AutoDock tools.
2.2. Molecular docking of the viral proteins with the phytocompounds
Molecular docking was achieved using the AutoDock Vina software (Trott & Olson, 2010) employing a grid-based docking method opting a rigid protein receptor and flexible ligand docking protocol (Naidoo et al., 2020). Briefly, after a thorough evaluation of information regarding the active site residues of each protein, the prepared ligands were docked at the active binding pockets of the RBD of SARS-CoV-2 spike protein, SARS-CoV-2 Mpro and SARS-CoV-2 RdRp (Andersen et al., 2020; Ciliberto & Cardone, 2020; Wang et al., 2020). For the SARS-CoV-2 spike protein, a grid box with center_x = −37.0, center_y = 31.5, center_z = 1.6, size_x = 42.0, size_y = 98.0 and size_z = 44.0 involving the active site residues was generated. Similarly for the SARS-CoV-2 Mpro a grid box with center_x =−10.9, center_y = 12.3, center_z =68.7, size_x = 19.3, size_y = 29.9 and size_z = 21.7 encompassing the active binding pocket was used. In case of SARS-CoV-2 RdRp a grid box with center_x = 91.6, center_y = 92.4, center_z = 103.8, size_x = 15.3, size_y = 17.6 and size_z = 18.9 was generated to involve the active site residues. Results returned from AutoDock Vina were validated using the DINC server (Antunes et al., 2017). Pymol software (version 1.7.4) was used to identify molecular interactions between the receptor-ligand complexes exhibiting the lowest binding scores and RMSD values < 2.0 Å. These were further validated using the Protein-Ligand Interaction profiler (Salentin et al., 2015). The compounds displaying binding energy scores ≤ −7.0 kcal/mol were selected for the next phases of analyses (Naidoo et al., 2020).
2.3. Assessment of physicochemical features and potential toxicity of the phytocompounds
The SwissADME and pkCSM servers were employed for an in-depth analysis of the physicochemical features of the selected phytochemicals (Daina et al., 2017; Pires et al., 2015). The Pan-Assay Interference Structures (PAINS) medicinal chemistry analysis was conducted employing the SwissADME server and was further validated using the FAF-Drugs4 (Lagorce et al., 2017). The toxicity parameters like mutagenicity (AMES mutagenesis) and cytotoxicity of the phytochemicals were assessed using the ProTox-II web-server (Banerjee et al., 2018) and validated with the vNN-ADMET server (Schyman et al., 2017).
2.4. Molecular dynamics simulations
Molecular dynamics (MD) simulations provide validations to the results of molecular docking and help to accurately assess the potential stability of a receptor-ligand complex (Kar et al., 2020; Naidoo et al., 2020). The complexes of the ligands displaying the best interaction (in terms of binding energy values) with each SARS-CoV-2 spike, Mpro and RdRp proteins were subjected to MD simulations for a timescale of 120 nanoseconds (ns) using the GROMACS software (version 2019) (Abraham et al., 2020) with GROMOS96 43a1 force field parameters (Chiu et al., 2009). The protein and ligand topologies were generated using the ‘pdb2gmx’ script and PRODRG server (Schüttelkopf & Van Aalten, 2004) respectively. The generated ligand topologies were rejoined to the processed protein structures. Counter ions (Na + cation and Cl- anion) were added to the protein-ligand complexes to make the system electrostatically neutral. The energy minimization of the protein-ligand complexes was achieved in multiple steps using steepest descent method. The entire systems were then progressively heated up to 300 K on a time scale of 100 ps (Khan et al., 2020). The equilibration steps were set with the constant pressure and temperature (NPT) ensemble (Umesh et al., 2020). The MD simulations were carried out at a standard temperature of 300 K and a pressure level of 1.013 bar (Umesh et al., 2020). The trajectories were estimated by analyzing the root mean square deviation (RMSD) and the root mean square fluctuation (RMSF) of the protein-ligand complexes for the timescale of 120 ns (Kar et al., 2020). The structures of the protein-ligand complexes generated after the MD simulations of 120 ns were used to analyze the interactions and assess whether the complexes suffered any change in interaction using the LIGPLOT software (Wallace et al., 1995).
2.5. Molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA)-based estimation of binding free energies
Molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method was opted for estimating the free energies of binding of the protein-ligand complexes to infer about their conformational stabilities (Bhardwaj et al., 2020). MM-PBSA based free energies of binding of the selected complexes were estimated employing the ‘g_mmpbsa’ script of GROMACS. The total free energy of binding of a protein-ligand complex involves the potential energy, polar and non-polar solvation energies and is calculated as:
where ΔGbind signifies the total energy of binding of the receptor-ligand complex, ΔGreceptor and ΔGligand represent the energies of the free receptor and unbound ligand respectively (Bhardwaj et al., 2020).
2.6. Statistical analysis
SPSS software package (version 17.0) was employed to perform the statistical test of significance at p < 0.01.
3. Results and discussion
3.1. Molecular docking of the phytocompounds with the receptor-binding domain of SARS-CoV-2 spike protein
The spike protein of SARS-CoV-2 attaches the human receptor angiotensin-converting enzyme 2 (ACE2) via the receptor-binding domain (RBD) and facilitates viral entry in host cells (Tai et al., 2020). Molecular docking of the selected phytocompounds was conducted at the active binding pocket of the RBD of SARS-CoV-2 spike protein with prior knowledge about the active site residues that bind with the human receptor ACE2 (Andersen et al., 2020; Tai et al., 2020). The detailed binding energy scores of the concerned phytochemicals have been provided in Table 1. The compounds anisotine, adhatodine, beta-carotene, eugenol, mangiferin and beta-amyrin were found to display encouraging binding energy scores of −7.8 (±0.01) kcal/mol, −7.0 ± (±0.02) kcal/mol, −7.2 (±0.01) kcal/mol, −7.3 (±0.03) kcal/mol, −7.5 (±0.03) kcal/mol and −7.1 (±0.01) kcal/mol, respectively, among the concerned phytocompounds (Table 1) with anisotine showing the most promising inhibitory potential in terms of binding energy score.
Table 1.
Binding energy scores and interaction profile of the phytocompounds with the receptor-binding domain (RBD) of SARS-CoV-2 spike protein.
| Plant | Ligands | Binding energy scores (kcal/mol) | Predicted Inhibition Constant (Ki) (μM) | Interacting residues |
|---|---|---|---|---|
| RBD of SARS-CoV-2 spike protein (6LZG) | ||||
| Justicia adhatoda | Anisotine | −7.8 ± 0.01 | 1.90 | Tyr495, Phe497, Tyr453, Tyr505*, Lys417ǂ |
| Adhatodine | −7.0 ± 0.02 | 7.35 | Tyr495, Phe497, Gln498, Asn501, Tyr505* | |
| Vasicoline | −6.5 ± 0.01 | 17.09 | Tyr495, Tyr505*, Arg403# | |
| Beta-sitosterol | −6.5 ± 0.03 | 17.09 | Tyr449, Leu452, Phe490 | |
| Vasicolinone | −6.2 ± 0.02 | 28.37 | Tyr495, Phe497, Tyr505* | |
| Vasicine | −5.6 ± 0.01 | 78.14 | Tyr495, Phe497, Tyr505, Gln493 | |
| Ocimum sanctum | Beta-carotene | −7.2 ± 0.01 | 5.24 | Lys378, Val407, Ala411, Asn437, Asn440, Val503 |
| Eugenol | −7.3 ± 0.03 | 4.42 | Tyr453, Tyr495, Tyr505 | |
| Caryophyllene | −6.8 ± 0.02 | 10.30 | Tyr453, Tyr495, Tyr505 | |
| Carvacrol | −5.9 ± 0.01 | 47.08 | Tyr505* | |
| Cineole | −5.8 ± 0.01 | 55.74 | Tyr453, Tyr505, Arg403 | |
| Swertia chirata | Amarogentin | −6.6 ± 0.02 | 14.44 | Tyr495, Arg403, Asn501, Tyr505* |
| Mangiferin | −7.5 ± 0.03 | 3.16 | Tyr505, Arg403#, Gly496, Asn501 | |
| Beta-amyrin | −7.1 ± 0.01 | 6.21 | Tyr495, Tyr505 | |
| Swerchirin | −5.7 ± 0.02 | 66.01 | Tyr495, Tyr505 | |
| Swertianin | −6.0 ± 0.01 | 39.77 | Tyr495, Arg403# | |
| Swertiamarin | −6.3 ± 0.02 | 23.96 | Tyr495, Phe497, Tyr505, Tyr453, Gln493, Gly496, Asn501, Gly502 |
Hydrophobic interactions are marked in italics, hydrogen bonds are highlighted in bold, π-cation interactions are displayed with #, π-stacking are displayed with * and salt bridges are displayed with ǂ.
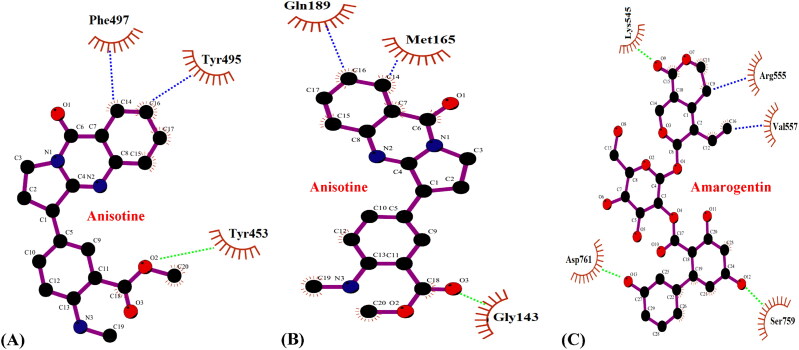
A detailed interaction profiling of the anisotine-RBD SARS-CoV-2 spike protein complex revealed that anisotine interacted with the residues Lys417, Tyr453, Tyr495, Phe497 and Tyr505 of the RBD of SARS-CoV-2 spike protein (Figure 2A and Table 1). Interestingly, the RBD of SARS-CoV-2 spike protein has been reported to attach the human ACE2 receptor with high affinity and involve the same residues Lys417, Tyr453, Tyr495 and Tyr505 in its interaction (Andersen et al., 2020; Ortega et al., 2020). Thus, anisotine might possess considerable potential to competitively bind with the RBD of the SARS-CoV-2 spike protein and limit host attachment via ACE2 receptor, however, further experiments are demanded to draw an inference.
Figure 2.
(A) Mode of interaction of anisotine with the receptor-binding domain (RBD) of SARS-CoV-2 spike protein. Yellow sphere represents the receptor-binding domain (RBD) of SARS-CoV-2 spike protein. Anisotine is represented as red sphere. Hydrogen bonds, π-stacking and salt bridges have been represented as blue, green dashed and yellow dashed lines respectively. (B) Mode of interaction of anisotine with SARS-CoV-2 main protease Mpro. Blue sphere represents SARS-CoV-2 main protease Mpro. Anisotine is represented as orange sphere. Hydrophobhic interactions and hydrogen bonds have been represented as grey dashed and blue lines respectively. (C) Mode of interaction of amarogentin with SARS-CoV-2 RdRp. Brass sphere indicates SARS-CoV-2 RdRp. Amarogentin is represented as green sphere. Hydrophobhic interactions and hydrogen bonds have been represented as grey dashed and blue lines respectively.
To provide an account of the potency of each compound, we calculated an inhibition constant associated with the binding energy score as per the scheme opted by Naidoo and colleagues (Naidoo et al., 2020). A compound that exhibits an inhibition constant (ki) within micromolar range (1-40 μM) is considered a hit or lead compound and should be considered for further drug development (Hughes et al., 2011). The compounds anisotine (1.90 μM), adhatodine (7.35 μM), beta-carotene (5.24 μM), eugenol (4.42 μM), mangiferin (3.16 μM) and beta-amyrin (6.21 μM) were predicted to be potent inhibitors and promising lead compounds based on our observations of inhibition constant values (Table 1).
Recently, our group (Kar et al., 2020) conducted an interaction profiling of the drug arbidol, that is commonly targeted against SARS-CoV-2 spike protein to inhibit viral attachment with human ACE2 receptor, and reported a binding energy score of −6.2 kcal/mol with the RBD of SARS-CoV-2 spike protein (Kar et al., 2020; Sanders et al., 2020). It was interesting to note that the compounds anisotine, adhatodine, beta-carotene, eugenol, mangiferin and beta-amyrin exhibited significantly higher binding energy scores (p < 0.01) (Table 1) with respect to the drug arbidol. Thus, the present findings offer scopes to further evaluate the inhibitory prospects of these compounds in vitro and in vivo towards the development of novel effective inhibitors against SARS-CoV-2 spike protein.
3.2. Molecular docking of the phytocompounds with SARS-CoV-2 mpro
SARS-CoV-2 replication is arrested by proper inhibition of its main protease enzyme Mpro that plays a crucial role in the proteolytic cleavage of the viral polyprotein orf1ab (Jin et al., 2020). A robust molecular docking of the selected phytocompounds revealed interesting facts regarding their inhibitory potential. A detailed account of the binding energy scores has been provided in Table 2. The compound anisotine was noted to exhibit the highest binding energy score of −8.4 (± 0.02) kcal/mol (Table 2). Furthermore, the phytocompounds adhatodine, vasicoline, vasicolinone, beta-carotene, eugenol, caryophyllene, amarogentin and mangiferin also displayed decent binding potential with binding energy values of −7.9 (±0.01) kcal/mol, −7.4 (±0.01) kcal/mol, −7.3 (±0.01) kcal/mol, −7.8 (±0.01) kcal/mol, −7.6 (±0.02) kcal/mol, −7.1 (±0.01) kcal/mol, −8.0 (±0.01) kcal/mol and −7.8 (±0.02) kcal/mol respectively (Table 2).
Table 2.
Binding energy scores and interaction profile of the phytocompounds with SARS-CoV-2 main protease Mpro.
| Plant | Ligands | Binding energy scores (kcal/mol) | Predicted Inhibition Constant (Ki) (μM) | Interacting residues |
|---|---|---|---|---|
| SARS-CoV-2 protease Mpro (6LU7) | ||||
| Justicia adhatoda | Anisotine | −8.4 ± 0.02 | 0.70 | Met165, Gln189, Gly143, His41* |
| Adhatodine | −7.9 ± 0.01 | 1.60 | His41, Met165, Gln189, Thr26 | |
| Vasicoline | −7.4 ± 0.01 | 3.74 | Met165, Glu166, Gln189 | |
| Beta-sitosterol | −6.7 ± 0.02 | 12.20 | Met165, Pro168 | |
| Vasicolinone | −7.3 ± 0.01 | 4.42 | Met165, Glu166, Gly143, Ser144, Cys145 | |
| Vasicine | −5.8 ± 0.03 | 55.74 | Met165, Gln189 | |
| Ocimum sanctum | Beta-carotene | −7.8 ± 0.01 | 1.90 | Thr25, Thr26, Met165, Pro168, Gln189, Ala191 |
| Eugenol | −7.6 ± 0.02 | 2.67 | Met49, Met165, Gln189, Glu166, His41* | |
| Caryophyllene | −7.1 ± 0.01 | 6.21 | Pro168, Gln189 | |
| Carvacrol | −5.8 ± 0.02 | 55.75 | Thr25 | |
| Cineole | −5.3 ± 0.01 | 129.69 | Thr25, Leu27, Gly143 | |
| Swertia chirata | Amarogentin | −8.0 ± 0.01 | 1.36 | Met165, Glu166, His41 |
| Mangiferin | −7.8 ± 0.02 | 1.90 | Met165, Gln189, Leu141, Gly143, Ser144, His164, Glu166, Thr190 | |
| Beta-amyrin | −6.9 ± 0.01 | 8.70 | Asn142, Met165, Glu166, Gln189 | |
| Swerchirin | −6.8 ± 0.01 | 10.30 | Met165, Gln189, His164, Glu166, His41* | |
| Swertianin | −6.4 ± 0.03 | 20.24 | Met165 | |
| Swertiamarin | −6.8 ± 0.01 | 10.30 | Met49, Asn142, Gly143, Leu141, Ser144, His163, Glu166 |
Hydrophobic interactions are marked in italics, hydrogen bonds are highlighted in bold and π-stacking are displayed with*.
An extensive scrutiny revealed that anisotine interacted with SARS-CoV-2 Mpro involving the residues His41, Gly143, Met165 and Gln189 at the active binding pocket (Jin et al., 2020) (Figure 2B and Table 2). His41 along with Cys145 forms the catalytic dyad at the active centre of SARS-CoV-2 Mpro and plays a major role in its enzymatic activity and facilitates the cleavage and processing of the large viral polyprotein orf1ab (Jin et al., 2020, Kar et al., 2020). Gly143 is an important canonical oxyanion hole residue that is located in a cleft between domains I and II of SARS-CoV-2 Mpro along with the His41-Cys145 catalytic dyad and helps in enzymatic activity (Gurung et al., 2020). Met165 is an important residue at the hydrophobic substrate binding S2 subsite of SARS-CoV-2 Mpro (Jin et al., 2020; Xue et al., 2008). The residue Gln189 links the domains II and III of SARS-CoV-2 Mpro and plays a key role in its enzymatic activity (Jin et al., 2020). Interaction of anisotine with these crucial residues at the active substrate binding site of SARS-CoV-2 Mpro promises to provide useful information about the inhibitory potential of the phytochemical.
It was evident on estimation of inhibition constant (ki) that the compounds adhatodine (1.60 μM), vasicoline (3.74 μM), vasicolinone (4.42 μM), beta-carotene (1.90 μM), eugenol (2.67 μM), caryophyllene (6.21 μM), amarogentin (1.36 μM) and mangiferin (1.90 μM) returned calculated ki values within the proposed threshold (1-40 μM) to be considered as potential lead compounds (Hughes et al., 2011) (Table 2). However, anisotine in particular displayed the most promising Ki value of 0.70 μM and was inferred to convey most potent biological activity (Table 2).
The binding efficacies of the phytocompounds were further compared with the drugs lopinavir and ritonavir that are currently being used to inhibit SARS-CoV-2 Mpro activity (Sanders et al., 2020). The drugs lopinavir and ritonavir have been reported to show binding energy scores of −8.1 kcal/mol and −7.5 kcal/mol with SARS-CoV-2 Mpro (Kar et al., 2020). It was striking to observe that anisotine exhibited significantly higher (p < 0.01) binding potential with SARS-CoV-2 Mpro than both the drugs. Thus, the present observations provide opportunities to further evaluate the inhibitory efficacy of anisotine in vitro and in vivo to deploy it as an effective therapeutic against SARS-CoV-2 Mpro.
3.3. Molecular docking of the phytochemicals with SARS-CoV-2 RdRp
The catalytic component nsp12, in association with nsp7 and nsp8, of the SARS-CoV-2 RdRp mediates viral replication (Yin et al., 2020). Inhibition of RdRp activity in SARS-CoV-2 arrests its growth and proliferation in host cells (Yin et al., 2020). The binding energy scores of the phytocompounds at the active binding pocket of the SARS-CoV-2 RdRp (Yin et al., 2020) have been detailed in Table 3. Among the concerned compounds, only beta-carotene and amarogentin displayed promising binding energy scores of −7.1 (±0.01) kcal/mol and −7.4 (±0.01) kcal/mol respectively with SARS-CoV-2 RdRp (Table 3). The compound amarogentin displayed a calculated inhibition constant of 3.74 μM (Table 3) whereas, beta-carotene returned a calculated inhibition constant of 6.21 μM, both of which were noted to fall within the proposed threshold (1-40 μM) to be considered as potential lead compounds (Hughes et al., 2011) (Table 3).
Table 3.
Binding energy scores and interaction profile of the phytocompounds with SARS-CoV-2 RdRp.
| Plant | Ligands | Binding energy scores (kcal/mol) | Predicted Inhibition Constant (Ki) (μM) | Interacting residues |
|---|---|---|---|---|
| SARS-CoV-2 RdRp (7BV2) | ||||
| Justicia adhatoda | Anisotine | −5.9 ± 0.02 | 47.08 | Arg555, Val557, Asp760 |
| Adhatodine | −6.2 ± 0.03 | 28.37 | Arg555†, Asp623, Lys545 | |
| Vasicoline | −5.9 ± 0.01 | 47.08 | Lys545, Arg555 | |
| Beta-sitosterol | −5.8 ± 0.01 | 55.74 | Asp623, Thr687, Ala688, Tyr689, Arg553, Arg624 | |
| Vasicolinone | −6.1 ± 0.02 | 33.59 | Lys545, Arg555, Val557 | |
| Vasicine | −4.7 ± 0.01 | 357.23 | Arg555, Val557, Asn691 | |
| Ocimum sanctum | Beta-carotene | −7.1 ± 0.01 | 6.21 | Lys545, Val557 |
| Eugenol | −3.9 ± 0.02 | 1379.37 | Asp623, Thr680, Thr687, Asn691 | |
| Caryophyllene | −4.9 ± 0.01 | 254.89 | Tyr619, Lys551†, Arg553† | |
| Carvacrol | −4.0 ± 0.03 | 1165.04 | Arg555, Val557, Asp623 | |
| Cineole | −3.8 ± 0.01 | 1633.14 | Asp623, Thr687 | |
| Swertia chirata | Amarogentin | −7.4 ± 0.01 | 3.74 | Arg555, Val557, Lys545, Ser759, Asp761 |
| Mangiferin | −6.7 ± 0.01 | 12.19 | Lys545, Ile548, Arg555, Asp623, Thr680, Thr687, Asn691 | |
| Beta-amyrin | −6.7 ± 0.03 | 12.19 | Asp623, Thr680, Asn691 | |
| Swerchirin | −5.7 ± 0.01 | 65.00 | Arg555, Thr556, Asp623, Asn691, Ser759 | |
| Swertianin | −5.5 ± 0.02 | 92.52 | Arg555, Asp623, Arg553, Thr556, Arg624, Asn691, Ser759 | |
| Swertiamarin | −6.6 ± 0.02 | 14.43 | Ser682, Asp760, Arg553, Thr556, Asn691, Ser759 |
Hydrophobic interactions are marked in italics, hydrogen bonds are highlighted in bold and salt bridges are displayed with †.
The compound amarogentin was observed to interact with the residues Lys545, Arg555, Val557, Ser759 and Asp761 of SARS-CoV-2 RdRp at the active binding site (Yin et al., 2020) (Figure 2C and Table 3). Interestingly, the antiviral drug remdesivir has been reported to interact with the important active site residues Lys545 and Arg555 and inhibit SARS-CoV-2 RdRp activity (Yin et al., 2020). Furthermore, the residues Ser759 and Asp761 located at the active catalytic site of SARS-CoV-2 RdRp have been suggested to be crucial for the enzymatic activity of SARS-CoV-2 RdRp (Yin et al., 2020). Interaction of amarogentin with these important residues associated with proper enzymatic functioning of SARS-CoV-2 RdRp might provide meaningful insights about the inhibitory potential of the phytocompound.
Remdesivir and favipiravir are antiviral drugs that inhibit viral RdRp activity and arrest viral replication and are presently in clinical trials as possible drug candidates against COVID-19 (Sanders et al., 2020). The inhibitory potential of remdesivir on SARS-CoV-2 RdRp activity has also been demonstrated by Yin et al. (2020). Remdesivir and favipiravir have been reported to show binding energy scores of −6.3 kcal/mol and −3.6 kcal/mol respectively with SARS-CoV-2 RdRp based on molecular docking investigations (Kar et al., 2020). Interestingly, both the compounds amarogentin and beta-carotene displayed significantly higher binding energy scores (p < 0.01) than the drugs remdesivir and favipiravir as evident from the present analysis (Table 3). The present findings generate further possibilities to validate the inhibitory prospects of these compounds against SARS-CoV-2 RdRp in vitro and in vivo towards proposing them as effective inhibitors.
3.4. Physicochemical and drug-like features of the compounds
The physicochemical properties of the promising inhibitory candidates (binding energy score ≤ −7.0 kcal/mol) were assessed in order to evaluate them in accordance with Lipinski’s rule of five which predicts the oral bioavailability and membrane permeability of a drug candidate in terms of the pharmacokinetic properties of a concerned compound (Lipinski et al., 2001). Proper evaluation of the physicochemical features of a therapeutic candidate in accordance with the general rules of drug-likeness is a crucial step in the early stages of drug development (Lipinski et al., 2001). Lipinski's rule of five proposes that a drug candidate with the logarithm of partial coefficient (log P) ≤ 5, molecular weight (MW) ≤ 500 Dalton, number of hydrogen bond acceptors ≤ 10 and number of hydrogen bond donors ≤ 5 exhibits decent gastrointestinal absorption and enhanced permeability across membranes (Lipinski et al., 2001). Extensive analysis revealed that the physicochemical features of the compounds anisotine, adhatodine, vasicoline, vasicolinone, eugenol and caryophyllene (Table 4) were in accordance with Lipinski’s rule of five (RO5). However, the compounds beta-carotene (molecular weight > 500 Dalton; log p > 5), amarogentin (molecular weight > 500 Dalton; number of hydrogen bond acceptors > 10; number of hydrogen bond donors > 5), mangiferin (number of hydrogen bond acceptors > 10) and beta-amyrin (log p > 5) had slight violations from the Lipinski's rule of five (Table 4) despite showing promise based on their binding energy scores against the viral proteins. In this pretext, it is worth mentioning that most of the antiviral drugs like lopinavir (molecular weight > 500 dalton), ritonavir (molecular weight > 500 dalton) and remdesivir (molecular weight > 500 dalton), that are presently in clinical trials against COVID-19, have been reported to display slight violations to the Lipinski’s rule of five (RO5) (Kar et al., 2020). Indeed, highly sophisticated drug delivery systems including nanotechnology and the inhibition of efflux pumps have been developed to enhance oral bioavailability, while modifications to chemical and physical properties, molecular size reductions and salt formations are some techniques that have been employed to improve the bioavailability of a drug candidate (Fasinu et al., 2011).
Table 4.
Physicochemical properties and drug-likeness features of the phytocompounds.
| Phytocompound | Molecular weight (Dalton) | Number of hydrogen bond acceptor | Number of hydrogen bond donor | log P | Medicinal chemistry (PAINS) |
Mutagenicity (AMES mutagenesis) |
Cytotoxicity |
|---|---|---|---|---|---|---|---|
| Anisotine | 349.38 | 4 | 1 | 2.76 | 0 alert | No | No |
| Adhatodine | 335.40 | 3 | 1 | 3.54 | 0 alert | Yes | No |
| Vasicoline | 291.39 | 1 | 0 | 3.78 | 0 alert | Yes | No |
| Vasicolinone | 305.37 | 2 | 0 | 2.99 | 0 alert | Yes | No |
| Beta-carotene | 536.87 | 0 | 0 | 12.60 | 0 alert | Yes | No |
| Eugenol | 164.20 | 2 | 1 | 2.12 | 0 alert | No | No |
| Caryophyllene | 204.35 | 0 | 0 | 4.72 | 0 alert | No | No |
| Amarogentin | 586.54 | 13 | 6 | 1.05 | 0 alert | No | No |
| Mangiferin | 422.34 | 11 | 8 | −0.71 | 1 alert | Yes | No |
| Beta-amyrin | 426.72 | 1 | 1 | 8.1 | 0 alert | No | No |
Log P- Logarithm of partial coefficient; PAINS- Pan-Assay Interference Structures.
Pan-assay interference structures (PAINS) are a prominent source of false positive signals in the drug-discovery process that include fluorescence of small molecules, redox reactivity and covalent modifications of target proteins (Baell & Holloway, 2010; Gilberg et al., 2017). Pan-assay interference compounds are responsible for activity artifacts in biological screening assays owing to their tendencies of forming colloidal aggregation and chemically reactive nature in assay conditions (Gilberg et al., 2017). Thus, proper identification of these compounds with structural alerts and eliminating them from further phases of drug discovery process is an imperative step (Gilberg et al., 2017). All the compounds, except mangiferin, were predicted not to possess any structural alerts associated with false positive signals (Table 4).
Preclinical toxicity assessment of a novel therapeutic candidate is one of the most important steps in the domain of successful drug development (Kar et al., 2020; Naidoo et al., 2020). Mutagenicity refers to the adverse consequences that occur due to exposure to chemicals inducing genetic mutations (Kar et al., 2020; Naidoo et al., 2020). In silico mutagenicity assessment, in correlation with Ames mutagenicity dataset, revealed that the compounds anisotine, eugenol, caryophyllene, amarogentin and beta-amyrin were non-mutagenic whereas, the compounds adhatodine, vasicoline, vasicolinone, beta-carotene and mangiferin were predicted to be potential mutagens (Table 4). It was inferred from in silico estimation of potential cytotoxicity of the compounds that all of them were non-cytotoxic (Table 4). Thus, a thorough analysis of the molecular docking results followed by a subsequent assessment of physicochemical features and toxicity indices established anisotine and amarogentin as safe and effective candidates for further analyses.
3.5. MD simulations of the protein-ligand complexes
The compound anisotine displayed the best interaction results in terms of binding energy scores with the SARS-CoV-2 spike and Mpro proteins (Tables 1 and 2 respectively), and the phytochemical amarogentin exhibited the most encouraging result with SARS-CoV-2 RdRp (Table 3), among the selected phytocompounds. Furthermore, the compounds were predicted to be non-toxic (Table 4). Accordingly, molecular dynamics (MD) simulations of the respective protein-ligand complexes were performed for a timescale of 120 ns to validate the results of molecular docking and assess the conformational stability of the complexes. A thorough RMSD analysis along the timescale of 120 ns revealed that all the complexes were conformationally stable (Figure 3A–C). The average RMSD values of the anisotine-RBD SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes were found to be 1.8 Å, 1.3 Å and 1.6 Å respectively (Figure 3A–C). The anisotine-RBD SARS-CoV-2 spike protein complex experienced initial fluctuations in RMSD values of Cα atoms and attained stability after 84 ns and remained in equilibrium thereafter (Figure 3A). The anisotine-SARS-CoV-2 Mpro complex initially displayed fluctuations in RMSD values of Cα atoms. However, it attained stability after 88 ns and was noted to be in equilibrium after that (Figure 3B). The amarogentin-SARS-CoV-2 RdRp complex displayed significant dynamicity in RMSD values of Cα atoms until 100 ns after which it attained considerable stability and remained in equilibrium (Figure 3C).
Figure 3.
RMSD analysis of the complexes along the timescale of 120 ns. (A) anisotine-RBD SARS-CoV-2 spike protein (red). (B) anisotine-SARS-CoV-2 Mpro(blue). (C) amarogentin-SARS-CoV-2 RdRp (green).
Analysis of root mean square fluctuation (RMSF) of the residues of a protein-ligand complex is effective in addressing local changes along a protein chain and deciphering its conformational stability (Kar et al., 2020; Naidoo et al., 2020). Extensive analysis revealed that the average RMSF values of the anisotine-RBD SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes were 1.3 Å, 2.3 Å and 1.9 Å respectively (Figure 4A–C). The peaks in the RMSF plot signify the residues which experience major fluctuations during the MD simulations (Kar et al., 2020; Naidoo et al., 2020). It was evident from our analysis that the residues in the active binding pockets of the SARS-CoV-2 proteins that interacted with the respective compounds (Tables 1–3) were relatively stable (Figure 4A–C) throughout the course of the MD simulations for 120 ns and thus, reflected the conformation stability of the respective complexes.
Figure 4.
RMSF analysis of the complexes (A) anisotine-RBD SARS-CoV-2 spike protein (red). (B) anisotine-SARS-CoV-2 Mpro (blue). (C) amarogentin-SARS-CoV-2 RdRp (green).
The structures of the protein-ligand complexes generated after the MD simulations of 120 ns were used to analyze the interaction and assess whether the complexes suffered any change in interaction. A detailed investigation of the anisotine-RBD SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes revealed that all of them were conformationally stable and did not suffer any change in hydrogen bonds and hydrophobic interactions after the stipulated timescale of 120 ns (Figure 5A–C), thus, supporting our observations of molecular docking and inferences on the binding potential of the compounds with the concerned SARS-CoV-2 proteins.
Figure 5.
Interaction profile (Ligplot image) of the complexes after the molecular dynamics simulations for a timescale of 120 ns. (A) anisotine-RBD SARS-CoV-2 spike protein. (B) anisotine-SARS-CoV-2 Mpro. (C) amarogentin-SARS-CoV-2 RdRp. Hydrophobic interactions and hydrogen bonds have been marked as blue dashed and green dashed lines respectively.
3.6. Analysis of MM-PBSA free energies of binding of the protein-ligand complexes
The anisotine-RBD SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes were selected for further estimation of the free energies of binding using the molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) method. It has been suggested that MM-PBSA provides accurate estimates of free energies of binding of protein-ligand complexes and a more negative value indicates stronger binding (Bhardwaj et al., 2020). The detailed information regarding the electrostatic energy, SASA (Solvent Accessible Surface Areas) energy, van der Waals energy, polar solvation energy and final binding energy from 20 ns to 120 ns with 20 ns interval has been detailed in Table 5. It was evident that all forms of energy, except the polar solvation energy, favourably contributed to the interactions of the compounds with the viral proteins (Table 5). The anisotine-RBD SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes displayed final binding energy scores of −39.39 ± 1.29 kcal/mol, −42.44 ± 1.27 kcal/mol and −37.38 ± 0.79 kcal/mol respectively (Table 5) which implied considerable conformational stability of the complexes after the MD simulations of 120 ns.
Table 5.
Details of binding free energies (± standard deviation) for anisotine-SARS-CoV-2 spike protein, anisotine-SARS-CoV-2 Mpro and amarogentin-SARS-CoV-2 RdRp complexes calculated using MM-PBSA method from 20 to 120 ns with 20 ns interval.
|
Protein-ligand complexes |
Time (ns) |
van der Waals energy (kcal/mol) |
SASA energy (kcal/mol) |
Electrostatic energy (kcal/mol) |
Polar solvation energy (kcal/mol) |
Binding energy (kcal/mol) |
|---|---|---|---|---|---|---|
| Anisotine-SARS-CoV-2 spike protein | 20 | −37.80 ± 2.71 | −3.41 ± 0.63 | −8.09 ± 0.55 | 15.39 ± 1.43 | −33.91 ± 2.46 |
| 40 | −40.73 ± 2.53 | −3.59 ± 0.38 | −8.38 ± 0.49 | 16.06 ± 1.35 | −36.64 ± 2.05 | |
| 60 | −42.03 ± 2.39 | −3.70 ± 0.29 | −8.64 ± 0.41 | 16.51 ± 1.29 | −37.86 ± 1.80 | |
| 80 | −42.67 ± 2.19 | −3.76 ± 0.21 | −8.83 ± 0.32 | 16.83 ± 1.17 | −38.43 ± 1.55 | |
| 100 | −43.38 ± 1.76 | −3.81 ± 0.17 | −8.93 ± 0.21 | 17.15 ± 1.04 | −38.97 ± 1.10 | |
| 120 | −43.88 ± 1.77 | −3.85 ± 0.19 | −9.05 ± 0.15 | 17.39 ± 0.82 | −39.39 ± 1.29 | |
| Anisotine-SARS-CoV-2 Mpro | 20 | −40.71 ± 2.45 | −3.68 ± 0.59 | −8.71 ± 0.71 | 16.57 ± 1.37 | −36.53 ± 2.38 |
| 40 | −43.86 ± 2.34 | −3.87 ± 0.47 | −9.02 ± 0.63 | 17.30 ± 1.26 | −39.45 ± 2.18 | |
| 60 | −45.26 ± 2.23 | −3.98 ± 0.35 | −9.30 ± 0.49 | 17.78 ± 1.14 | −40.76 ± 1.93 | |
| 80 | −45.95 ± 2.17 | −4.05 ± 0.31 | −9.51 ± 0.38 | 18.13 ± 1.08 | −41.38 ± 1.78 | |
| 100 | −46.72 ± 1.90 | −4.11 ± 0.27 | −9.61 ± 0.31 | 18.47 ± 1.01 | −41.97 ± 1.47 | |
| 120 | −47.26 ± 1.82 | −4.15 ± 0.14 | −9.75 ± 0.22 | 18.72 ± 0.91 | −42.44 ± 1.27 | |
| Amarogentin-SARS-CoV-2 RdRp | 20 | −35.87 ± 2.63 | −3.24 ± 0.65 | −7.68 ± 0.72 | 14.60 ± 1.47 | −32.19 ± 2.53 |
| 40 | −38.64 ± 2.45 | −3.41 ± 0.47 | −7.95 ± 0.59 | 15.24 ± 1.43 | −34.76 ± 2.08 | |
| 60 | −39.87 ± 2.36 | −3.51 ± 0.32 | −8.20 ± 0.47 | 15.66 ± 1.31 | −35.92 ± 1.84 | |
| 80 | −40.48 ± 2.08 | −3.57 ± 0.21 | −8.38 ± 0.41 | 15.97 ± 1.28 | −36.46 ± 1.42 | |
| 100 | −41.16 ± 1.79 | −3.62 ± 0.12 | −8.47 ± 0.32 | 16.27 ± 1.19 | −36.98 ± 1.04 | |
| 120 | −41.63 ± 1.53 | −3.66 ± 0.05 | −8.59 ± 0.23 | 16.50 ± 1.02 | −37.38 ± 0.79 |
4. Conclusion
COVID-19 pandemic has been a global calamity with its rapid transmission and infectivity. Specific drugs and vaccines targeted against SARS-CoV-2, the causal agent of COVID-19, has been the dire need of the hour. The present analysis, based on robust molecular docking and molecular dynamics simulations, proposes specific compounds from common Indian medicinal plants Justicia adhatoda and Swertia chirata namely, anisotine against SARS-CoV-2 spike and Mpro proteins and amarogentin against SARS-CoV-2 RdRp as potential inhibitors, with further in vitro and in vivo experiments demanded to draw a definite conclusion. Present research findings offer ample scopes to exploit further the potential of these compounds as successful inhibitors of SARS-CoV-2 and decipher associated mechanisms of inhibition towards the development of novel effective therapeutics against COVID-19.
Acknowledgments
PK would like to thank University of North Bengal, India for providing fellowship. VK acknowledges DST-SERB Startup Research Scheme (File No. SRG/2019/001279) and Lovely Professional University, India for the infrastructure support. BV would like to thank Bharathiar University, India and Project funded and supported by MHRD-RUSA 2.0 – BEICH 2.0 (Ref No.BU/RUSA/BEICH/2019/65) for providing the necessary infrastructure facility. AR acknowledges Lovely Professional University, India for the infrastructure support. The Mangosuthu University of Technology, South Africa is acknowledged by DN and AA for infrastructure facility. MDS would like to thank Indian Council of Medical Research (ICMR) (File No. 2018-2786/CMB/Adhoc-BMS) for providing the necessary infrastructure support.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Abraham, P. K., Srihansa, T., Krupanidhi, S., Ayyagari, V. S., & Venkateswarulu, T. C. (2020). Design of multi-epitope vaccine candidate against SARS-CoV-2: A in-silico study. Journal of Biomolecular Structure & Dynamics, 1–9. 10.1080/07391102.2020.1770127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes, D. A., Moll, M., Devaurs, D., Jackson, K. R., Lizee, G., & Kavraki, L. E. (2017). DINC 2.0: A new protein-peptide docking webserver using an incremental approach. Cancer Research, 77(21), e55–e57. 10.1158/0008-5472.CAN-17-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astani, A., Reichling, J., & Schnitzler, P. (2011). Screening for antiviral activities of isolated compounds from essential oils. Evidence-Based Complementary and Alternative Medicine, 2011, 253643. 10.1093/ecam/nep187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell, J. B., & Holloway, G. A. (2010). New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry, 53(7), 2719–2740. 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- Balachandar, V., Mahalaxmi, I., Kaavya, J., Vivekanandhan, G., Ajithkumar, S., Arul, N., Singaravelu, G., Senthil Kumar, N., & Mohana Dev, S. (2020). COVID-19: Emerging protective measures. European Review for Medical and Pharmacological Sciences, 24(6), 3422–3425. 10.26355/eurrev_202003_20713 [DOI] [PubMed] [Google Scholar]
- Banerjee, P., Eckert, A. O., Schrey, A. K., & Preissner, R. (2018). ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Research, 46(W1), W257–263. 10.1093/nar/gky318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benencia, F., & CourrègGes, M. C. (2000). In vitro and in vivo activity of eugenol on human herpesvirus. Phytotherapy Research, 14(7), 495–500. [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan, R., & Chowdhary, A. (2014). In vitro inhibitory activity of Justicia adhatoda extracts against influenza virus infection and hemagglutination. International Journal of Pharmaceutical Sciences Review and Research, 25, 231–236. [Google Scholar]
- Chiu, S. W., Pandit, S. A., Scott, H. L., & Jakobsson, E. (2009). An improved united atom force field for simulation of mixed lipid bilayers. The Journal of Physical Chemistry B, 113(9), 2748–2763. 10.1021/jp807056c [DOI] [PubMed] [Google Scholar]
- Ciliberto, G., & Cardone, L. (2020). Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discovery Today, 25(6), 946–948. 10.1016/j.drudis.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasinu, P., Pillay, V., Ndesendo, V. M., Du Toit, L. C., & Choonara, Y. E. (2011). Diverse approaches for the enhancement of oral drug bioavailability. Biopharmaceutics & Drug Disposition, 32(4), 185–209. 10.1002/bdd.750 [DOI] [PubMed] [Google Scholar]
- Ghoke, S. S., Sood, R., Kumar, N., Pateriya, A. K., Bhatia, S., Mishra, A., Dixit, R., Singh, V. K., Desai, D. N., Kulkarni, D. D., Dimri, U., & Singh, V. P. (2018). Evaluation of antiviral activity of Ocimum sanctum and Acacia arabica leaves extracts against H9N2 virus using embryonated chicken egg model. BMC Complementary and Alternative Medicine, 18(1), 174–183. 10.1186/s12906-018-2238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilberg, E., Stumpfe, D., & Bajorath, J. (2017). Activity profiles of analog series containing pan assay interference compounds. RSC Advances, 7(57), 35638–35647. 10.1039/C7RA06736D [DOI] [Google Scholar]
- Gurung, A. B., Ali, M. A., Lee, J., Farah, M. A., & Al-Anazi, K. M. (2020). Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sciences, 255, 117831. 10.1016/j.lfs.2020.117831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J., Rees, S., Kalindjian, S., & Philpott, K. (2011). Principles of early drug discovery. British Journal of Pharmacology, 162(6), 1239–1249. 10.1111/j.1476-5381.2010.01127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, D. K., Panda, L., Lavanya, P., Ramaiah, S., & Anbarasu, A. (2012). Detection and confirmation of alkaloids in leaves of Justicia adhatoda and bioinformatics approach to elicit its anti-tuberculosis activity. Applied Biochemistry and Biotechnology, 168(5), 980–990. 10.1007/s12010-012-9834-1 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kar, P., Sharma, N. R., Singh, B., Sen, A., & Roy, A. (2020). Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: An in silico investigation. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1780947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., … Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce, D., Bouslama, L., Becot, J., Miteva, M. A., & Villoutreix, B. O. (2017). FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics, 33(22), 3658–3660. 10.1093/bioinformatics/btx491 [DOI] [PubMed] [Google Scholar]
- Li, Y., Lai, Y., Wang, Y., Liu, N., Zhang, F., & Xu, P. (2016). 1, 8-Cineol protect against influenza-virus-induced pneumonia in mice. Inflammation, 39(4), 1582–1593. 10.1007/s10753-016-0394-3 [DOI] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 46(1-3), 3–26. 10.1016/S0169-409X(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Naidoo, D., Roy, A., Kar, P., Mutanda, T., & Anandraj, A. (2020). Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: An in silico analysis. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1794972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3, 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, J. T., Serrano, M. L., Pujol, F. H., & Rangel, H. R. (2020). Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI Journal, 19, 410–417. 10.17179/excli2020-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilau, M. R., Alves, S. H., Weiblen, R., Arenhart, S., Cueto, A. P., & Lovato, L. T. (2011). Antiviral activity of the Lippia graveolens (Mexican oregano) essential oil and its main compound carvacrol against human and animal viruses. Brazilian Journal of Microbiology, 42(4), 1616–1624. 10.1590/S1517-838220110004000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using Graph-Based Signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, G. S., & Sinsheimer, J. E. (1974). Antiviral activity of triterpenoid saponins containing acylated beta-amyrin aglycones. Journal of Pharmaceutical Sciences, 63(3), 471–473. 10.1002/jps.2600630341 [DOI] [PubMed] [Google Scholar]
- Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F., & Schroeder, M. (2015). PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, J. M., Monogue, M. L., Jodlowski, T. Z., & Cutrell, J. B. (2020). Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. JAMA, 323(18), 1824–1836. 10.1001/jama.2020.6019 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf, A. W., & Van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Schyman, P., Liu, R., Desai, V., & Wallqvist, A. (2017). vNN web server for ADMET predictions. Frontiers in Pharmacology, 8, 889. 10.3389/fphar.2017.00889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C., Wang, Z., Zhao, F., Yang, Y., Li, J., Yuan, J., Wang, F., Li, D., Yang, M., Xing, L., Wei, J., Xiao, H., Yang, Y., Qu, J., Qing, L., Chen, L., Xu, Z., Peng, L., Li, Y., … Liu, L. (2020). Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA, 323(16), 1582–1589. 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y., & Du, L. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol, 17(6), 613–620. 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh, K. D., Selvaraj, C., Singh, S. K., & Dubey, V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, H., Patil, P. R., Kolhapure, R. M., & Gopalkrishna, V. (2008). Antiviral activity of the Indian medicinal plant extract, Swertia chirata against herpes simplex viruses: A study by in-vitro and molecular approach. Indian Journal of Medical Microbiology, 26(4), 322–326. 10.4103/0255-0857.43561 [DOI] [PubMed] [Google Scholar]
- Wallace, A. C., Laskowski, R. A., & Thornton, J. M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8(2), 127–134. 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., Lu, G., Qiao, C., Hu, Y., Yuen, K.-Y., Wang, Q., Zhou, H., Yan, J., & Qi, J. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell, 181(4), 894–904. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., & McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA, 323(13), 1239–1242. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W., Mao, C., Luan, X., Shen, D.-D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y.-C., Tian, G., Jiang, H.-W., Tao, S.-C., Shen, J., Jiang, Y., Jiang, H., Xu, Y., … Xu, H. E. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science, 368(6498), 1499–1504. doi: 10.1101/2020.04.08.032763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. S., & Lu, Z. Y. (1989). Antiviral effect of mangiferin and isomangiferin on herpes simplex virus. Zhongguo Yao li Xue Bao = Acta Pharmacologica Sinica, 10(1), 85–90. [PubMed] [Google Scholar]