Abstract
Background
Cancer patients are at increased risk of death from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Cancer and its treatment affect many haematological and biochemical parameters, therefore we analysed these prior to and during coronavirus disease 2019 (COVID-19) and correlated them with outcome.
Patients and methods
Consecutive patients with cancer testing positive for SARS-CoV-2 in centres throughout the United Kingdom were identified and entered into a database following local governance approval. Clinical and longitudinal laboratory data were extracted from patient records. Data were analysed using Mann–Whitney U test, Fisher's exact test, Wilcoxon signed rank test, logistic regression, or linear regression for outcomes. Hierarchical clustering of heatmaps was performed using Ward's method.
Results
In total, 302 patients were included in three cohorts: Manchester (n = 67), Liverpool (n = 62), and UK (n = 173). In the entire cohort (N = 302), median age was 69 (range 19-93 years), including 163 males and 139 females; of these, 216 were diagnosed with a solid tumour and 86 with a haematological cancer. Preinfection lymphopaenia, neutropaenia and lactate dehydrogenase (LDH) were not associated with oxygen requirement (O2) or death. Lymphocyte count (P < 0.001), platelet count (P = 0.03), LDH (P < 0.0001) and albumin (P < 0.0001) significantly changed from preinfection to during infection. High rather than low neutrophils at day 0 (P = 0.007), higher maximal neutrophils during COVID-19 (P = 0.026) and higher neutrophil-to-lymphocyte ratio (NLR; P = 0.01) were associated with death. In multivariable analysis, age (P = 0.002), haematological cancer (P = 0.034), C-reactive protein (P = 0.004), NLR (P = 0.036) and albumin (P = 0.02) at day 0 were significant predictors of death. In the Manchester/Liverpool cohort 30 patients have restarted therapy following COVID-19, with no additional complications requiring readmission.
Conclusion
Preinfection biochemical/haematological parameters were not associated with worse outcome in cancer patients. Restarting treatment following COVID-19 was not associated with additional complications. Neutropaenia due to cancer/treatment is not associated with COVID-19 mortality. Cancer therapy, particularly in patients with solid tumours, need not be delayed or omitted due to concerns that treatment itself increases COVID-19 severity.
Key words: COVID-19, cancer, SARS-CoV-2
Highlights
-
•
Pre-infection haematological/biochemical characteristics are not associated with COVID-19 severity.
-
•
Significant haematological/biochemical changes occur upon infection with heterogeneity in response observed.
-
•
High not low neutrophils were associated with oxygen requirement and COVID-19 mortality – GCSF should be used with caution.
Age, haematological cancer, high neutrophil:lymphocyte ratio, CRP and low albumin were significant predictors of death in multivariable analysis.
No significant complications requiring readmission were seen upon restart of cancer therapy following diagnosis of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19) represents an unprecedented global health challenge. A diverse spectrum of clinicopathological syndromes have been reported, ranging from asymptomatic cases to multiorgan failure and death.1
Patients with cancer are at significantly increased risk of COVID-19 and severe complications, including need for invasive ventilation and death2,3 with reported fatality rates of 10%-30%,3, 4, 5 with a recent meta-analysis of 18 650 patients with COVID-19 and cancer reporting a probability of death of 25.6%.6 Older patients, those with multiple comorbidities and male sex appear to be at a particularly high risk for poor outcomes.2,4,5,7
Large-scale studies in the general population have identified a number of clinical and pathological factors associated with adverse outcomes in patients with confirmed COVID-19.8,9 In the general population, patients with neutrophilia, lymphopaenia, raised C-reactive protein (CRP) and lactate dehydrogenase (LDH) are significantly more likely to develop acute respiratory distress syndrome (ARDS).8,10 In addition to the clinical features of fever, dyspnoea and cough, commonly observed biochemical abnormalities in cancer patients with COVID-19 include lymphopaenia, high levels of CRP and hypoalbuminaemia; however, the prognostic significance of these derangements within an oncology population has not yet been established.11
In addition to the inherent immunosuppression of cancer and its therapies, malignancy itself can cause biochemical abnormalities such as raised LDH/CRP and low albumin. We aimed to understand whether these abnormalities were associated with worse outcomes for cancer patients during COVID-19 illness. We analysed changes in biochemical and haematological parameters both before and during infection to identify systemic changes which occur when patients with cancer become infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and correlated them with measures of severity.
Methods
Database approval
Institutional approval was obtained following local information governance processes for case-note review at each site in order to establish a database to support wider clinical decision-making. In line with Health Research Authority (UK) guidance, database creation to support public health surveillance and clinical decisions is exempt from Ethics Committee review, and anonymised data within a database can be used for research purposes if local governance approval is obtained.12 Data were fed into other registries such as the UK Cancer Coronavirus Monitoring Project (UKCCMP) and ESMO Co-Care registry.4 Patients with a cancer diagnosis testing PCR positive for SARS-CoV-2 were included. Asymptomatic patients screened prior to planned surgery were excluded from the database due to insufficient data; however, asymptomatic patients with nosocomial infection were included. Longitudinal data were available from two oncology centres (Manchester and Liverpool). Data on clinical and laboratory features before and during SARS-CoV-2 infection were extracted from patient records. All patient data within the database were pseudoanonymised with the key matching the study identity to patient identities kept at local National Health Service (NHS) sites. The data were anonymous to the study team performing the analysis.
Study design
A variety of measures were used to provide information regarding preinfection and during infection parameters. For a sample size calculation we used a pilot study of 52 patients from Manchester. Assuming a clinically meaningful difference between means of 25 mg/l, 1.5 × 109 g/l, 0.25 × 109 g/l and 1 g/l for CRP, neutrophils, lymphocytes and albumin, respectively, we estimated that the sample size of 136 patients was needed to achieve 80% power and with a 5% alpha.
To evaluate chronic preinfection states (CHRONIC), values were taken as the average results from the previous 6 months up to 14 days (the maximum incubation period of SARS-CoV-213) prior to COVID-19 diagnosis. We used the value closest to day −15 preinfection (IMMED) to provide information about the patient's state just prior to diagnosis of COVID-19. During infection, we assessed day 0 values defined as the date of SARS-CoV-2 PCR confirmation. Minimum and maximum values were the lowest and highest values, respectively, during infection defined as day 0-14. Oxygen requirement (O2) and death were used as measures of severity of infection. According to clinical assessments and national guidance,14 due to advanced cancer stage and frailty of the majority of patients, only a minority would have benefited from critical organ support and requirement for intensive care unit (ICU) was not used as an outcome measure.
Statistical analysis
Analysis was performed using SPSS version 21, GraphPad Prism 8.1.2 and Python Version 3.7. Differences between groups (O2 and death) were calculated using Wilcoxon signed rank test (for the change from preinfection to during infection), Mann–Whitney U test and Fisher's exact test, with the significance level P = 0.05. The effect size was calculated as common language effect size (CL),15 or odds ratio (OR) accordingly. Details regarding numbers of patients included in each analysis and the mean values of each group are presented in Supplementary Tables S1 and S2, available at https://doi.org/10.1016/j.esmoop.2020.100005. Heatmaps were grouped using hierarchical clustering according to Ward's method, based on values from day 0 to day 7. For clustering purposes only, the missing values within these days were imputed using linear interpolation, and filled forward after the latest value with only the known (not imputed) values shown on the heatmap. A multivariable analysis for O2 and death for features at day 0 was performed using logistic regression. For duration of hospitalisation in patients who died from COVID-19, linear regression was performed. Missing values in numeric features at day 0 (less than 10%) were imputed using Bayesian ridge regression. Before that, a logarithmic transformation was applied to highly skewed features. Missing values of cancer stage (<10%) were imputed using logistic regression trained on all remaining features. In all aforesaid tests P < 0.05 was considered statistically significant.
Results
Clinical characteristics
In the Manchester and Liverpool cohorts 67 and 62 patients were identified, respectively, with a further 173 patients identified in the UK cohort. Clinical features of the Manchester, Liverpool and UK cohorts are presented in Table 1 and Supplementary Tables S3-S5, available at https://doi.org/10.1016/j.esmoop.2020.100005. Across the entire cohort, median age was 69 years, range 19-93, with 163 males and 139 females. Two hundred and sixteen had a solid tumour and 86 a haematological cancer; 39% (117/302) of patients received chemotherapy within the 4 weeks preceding infection, 16% (48/302) received targeted therapy and 4.6% (14/302) immunotherapy [programmed cell death protein 1/programmed death-ligand 1/cytotoxic T lymphocyte antigen 4 (PD-1/PD-L1/CTLA-4) inhibitors]. The rest received radiotherapy, hormone treatment or no active treatment within the 4 weeks preceding infection. In Manchester and Liverpool, 99 patients presented to hospital with symptoms of COVID-19, whereas 30 patients were likely nosocomial infection cases.
Table 1.
Characteristics of the total cohort of cancer patients presenting with coronavirus 2019 (COVID-19)
| Characteristic | Number (percentage) |
|---|---|
| Median age (range) | 69 (19-93) |
| Sex | |
| Male | 163 (54.0) |
| Female | 139 (66.0) |
| Comorbidities | |
| Hypertension | 95 (31.5) |
| Chronic obstructive pulmonary disease | 38 (12.6) |
| Diabetes | 60 (19.9) |
| Cardiovascular disease | 68 (22.5) |
| Cancer type | |
| Haematological | 86 (28.5) |
| Solid | 216 (71.5) |
| Cancer stage | |
| Early stage | 96 (31.8) |
| Distant metastases | 184 (60.9) |
| Unknown | 22 (7.2) |
| Therapy within 4 weeks of infectiona | |
| Chemotherapy | 117 (38.7) |
| Targeted therapy | 48 (15.8) |
| Immune therapy | 14 (4.6) |
Can have more than one therapy.
Clinical outcomes
At time of data cut-off, we grouped patients according to worst outcome/maximal supportive therapy. In total, 13% (39/302) of patients were discharged within 24 hours, without further complications/admissions; 11% (34/302), already admitted due to complications from cancer and/or cancer therapy, were incidentally diagnosed during screening for SARS-CoV-2 infection; 15% (46/302) of patients were admitted but did not require oxygen therapy; ~23% (70/302) required oxygen and 2% (7/302) were admitted to ICU; 29% (87/302) died due to COVID-19 with a further 6% (17/304) dying from non-COVID-19 causes such as documented cancer progression. Two patients did not have a specified outcome. Of the patients in the Manchester and Liverpool cohorts still alive and on treatment, ~19% (10/52) continued throughout COVID-19 illness (either hormone treatment or radiotherapy). A further 58% (30/52) have restarted therapy [11 targeted therapy, 3 immunotherapy, 14 chemotherapy (including 4 on high-dose chemotherapy), 2 radiotherapy] at a median of 21 days following positive SARS-CoV-2 test. None of these patients have had further complications due to SARS-CoV-2, or readmission within 30-days.
Haematological features
To provide a detailed characterisation of the clinical course of COVID-19 in cancer patients attending hospital, we examined various haematological characteristics both preinfection and during infection.
Lymphocytes
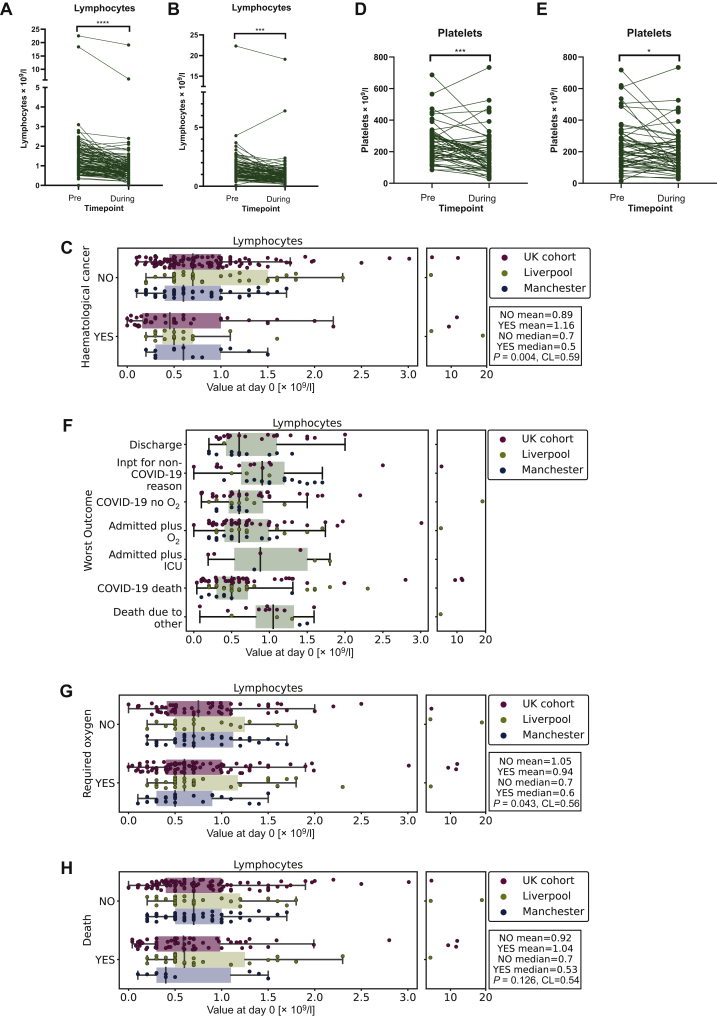
We observed lymphopaenia in 247/286 patients with data available at diagnosis of COVID-19 (day 0) in the entire cohort. Haematological parameters are often affected by cancer and its treatment, therefore in the Manchester and Liverpool cohort we examined whether COVID-19 infection affected these compared with each patient's long-term baseline (average over a 6-month period = CHRONIC) and immediate baseline (last test prior to infection = IMMED). The majority of patients (76/116 with tests available prior to infection) were lymphopaenic with 76/116 having CHRONIC lymphopaenia. We therefore examined whether lymphocyte counts were further reduced during COVID-19 through comparing CHRONIC and IMMED preinfection counts with their average lymphocyte counts from the first 7 days of diagnosed infection (Figure 1A and B). This revealed that while patients with cancer are generally lymphopaenic, infection with SARS-CoV-2 was significantly associated with a further decrease in counts (Figure 1A: mean 1.59 versus 0.99; P < 0.0001, CL = 0.72 and Figure 1B: 1.50 versus 0.99; P < 0.0001, CL = 0. 72). In patients with haematological cancer, preinfection lymphocyte count was not significantly different from solid tumours (Supplementary Figure S1A, available at https://doi.org/10.1016/j.esmoop.2020.100005; CHRONIC P = 0.18, CL = 0.58 and IMMED Figure 1B; P = 0.96, CL = 0.48); however, at day 0 of infection they had significantly lower counts compared with patients with nonhaematological malignancy (Figure 1C; P = 0.004, CL = 0.59). Furthermore, we found that 107/290 patients in the entire cohort had thrombocytopaenia (<150 × 109/l) at COVID-19 diagnosis. Again, in the Manchester cohort we compared patients' preinfection platelets (CHRONIC/IMMED) with the average counts during COVID-19 and found a significant reduction (CHRONIC mean 252 versus 199; P = 0.0008, CL = 0.67; Figure 1D; IMMED mean 236 versus 199; P = 0.03, CL = 0.58; Figure 1E).
Figure 1.
Longitudinal changes in lymphocytes and platelets.
(A) CHRONIC lymphocyte counts (day −170 to day −15) preinfection versus 7 days during infection in the Manchester cohort (∗∗∗∗P < 0.0001). (B) Lymphocyte count IMMED (last test pre-infection) versus 7 days during infection in the Manchester cohort (∗∗∗P < 0.001). (C) Boxplot of day 0 lymphocyte count (total cohort) grouped by diagnosis of haematological malignancy. (D) CHRONIC platelet counts preinfection versus 7 days during infection in the Manchester cohort (∗∗∗P = 0.0008). (E) Platelet count IMMED preinfection versus 7 days during infection in the Manchester cohort (∗P = 0.03). (F) Boxplot of lymphocyte count according to worst outcome in the entire cohort measured at day 0. Discharge, discharged within 24 hours; Inpt for non-COVID-19 reason, inpatient due to reason other than COVID-19 and outcome not altered by infection; COVID-19 no O2, admitted due to COVID-19 infection but did not require oxygen; admitted plus O2, admitted due to COVID-19 and required oxygen; admitted plus ICU, admitted due to COVID-19 and required intensive care; COVID-19 death, death due to other. (G) Boxplot of lymphocyte count grouped by oxygen requirement in the entire cohort measured at day 0. (H) Boxplot of lymphocyte count grouped by whether patient died in the entire cohort measured at day 0. COVID-19, coronavirus disease 2019; ICU, intensive care unit.
Next, we examined whether day 0 lymphocyte count was associated with different outcomes in the entire cohort (Figure 1F; Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100005). We observed that low day 0 lymphocyte count was significantly associated with O2 during COVID-19 (Figure 1F; P = 0.043, CL = 0.56) but not with death (Figure 1G; P = 0.126, CL = 0.54). Furthermore, minimum lymphocyte count during day 0-14 of infection was significantly associated with O2 (Supplementary Figure S1C, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.005, CL = 0.61) but not death (Supplementary Figure S1D, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.07, CL = 0.56) in the Manchester/Liverpool cohorts. As many patients were lymphopaenic due to cancer or cancer treatment prior to COVID-19, we assessed whether this affected outcome following infection. Critically, we did not find a significant association of O2 or death with average (Supplementary Figure S1E, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.26; CL = 0.56 and Supplementary Figure S1F, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.33; CL = 0.55 O2 and death, respectively) or day –15 lymphocyte count (Supplementary Figure S1G and S1H, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.47; CL = 0.52 and P = 0.79; CL = 0.49, O2 and death, respectively).
Neutrophils
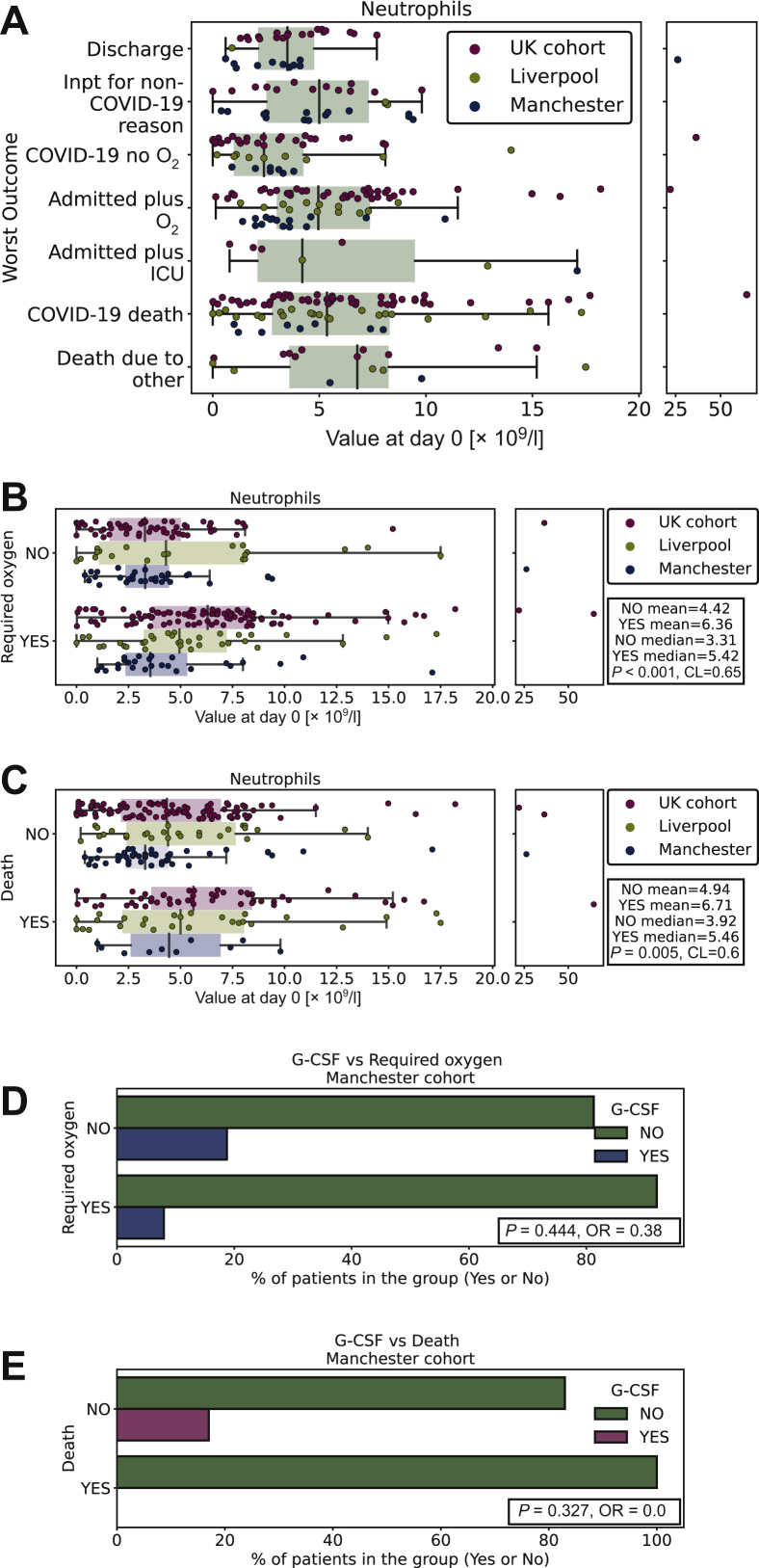
We examined whether neutrophil counts changed during SARS-CoV-2 infection. In the entire cohort with data available, 57/287 were neutropaenic at baseline (absolute neutrophil count <2 × 109/l) with 32 of these having neutrophils <1 × 109/l, and 20 patients <0.5 × 109/l. In the Manchester cohort, of the 32 patients presenting with neutropaenia at any point from day −14 to 14, 6 were on chemotherapy and were in treatment nadir, 2 were neutropaenic while taking palbociclib (CDK4/6 inhibitor) and 3 patients had a baseline neutropaenia due to haematological cancer. Although patients with haematological cancer did not have lower CHRONIC neutrophil counts compared with patients with solid malignancies (Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.1, CL = 0.66), they had lower neutrophil counts: both IMMED preinfection (Supplementary Figure S2B, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.002, CL = 0.68) and at day 0 (Supplementary Figure S2C, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.001, CL = 0.67). In Manchester, patients with neutropaenia were given granulocyte colony-stimulating factor (G-CSF). Longitudinal data for the 32 Manchester patients presenting with neutropaenia are shown in Supplementary Figure S2D, available at https://doi.org/10.1016/j.esmoop.2020.100005. Intriguingly, in these patients the drop in neutrophils occurred close to the time of SARS-CoV-2 test, which for the majority was when they presented with symptoms and rapidly improved even without G-CSF support, with a median neutropaenia duration of 3 days.
As neutropaenia could result in impaired immune response to SARS-CoV-2, we assessed whether this influenced outcome. Low neutrophil count at diagnosis of COVID-19 (day 0) was not associated with increased infection severity in the entire cohort (Figure 2A). By contrast, we observed a significant association with O2 (Figure 2B; P < 0.001, CL = 0.65) and death (Figure 2C; P = 0.007, CL = 0.60) in patients with a higher neutrophil count at day 0. Comparison of minimum neutrophil count (Manchester/Liverpool cohorts) during COVID-19 was not significantly associated with O2 (Supplementary Figure S2E, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.38, CL = 0.54) or death (Supplementary Figure S2F, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.24, CL = 0.56). However, higher maximal neutrophil count (patients given G-CSF excluded, Manchester/Liverpool cohorts), although not significantly associated with O2 (Supplementary Figure S2G, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.086, CL = 0.59), was significantly associated with death (Supplementary Figure S2H, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.026, CL = 0.63). In addition, high neutrophil-to-lymphocyte ratio (NLR) at day 0 was found to be associated with O2 (Supplementary Figure S2I, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.001, CL = 0.67) and death (Supplementary Figure S2J, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.01, CL = 0.59), as was the rate of rise (O2: P = 0.003; CL = 0.75, Supplementary Figure S2K, available at https://doi.org/10.1016/j.esmoop.2020.100005; death: P = 0.004; CL = 0.81, Supplementary Figure S2L, available at https://doi.org/10.1016/j.esmoop.2020.100005). Critically, in patients treated with G-CSF, we did not observe increased numbers requiring oxygen or dying (O2: P = 0.44; OR = 0.38, Figure 2D; and death: P = 0.33; OR = 0.0, Figure 2E).
Figure 2.
Longitudinal changes in neutrophils.
(A) Boxplot of neutrophil count according to worst outcome in the entire cohort measured at day 0. Discharge, discharged within 24 hours; Inpt for non-COVID-19 reason, inpatient due to reason other than COVID-19 and outcome not altered by infection; COVID-19 no O2, admitted due to COVID-19 infection but did not require oxygen; admitted plus O2, admitted due to COVID-19 and required oxygen; admitted plus ICU, admitted due to COVID-19 and required intensive care; COVID-19 death, death due to other. (B) Boxplot of neutrophil count grouped by oxygen requirement in the entire cohort measured at day 0. (C) Boxplot of neutrophil count grouped by death in the entire cohort measured at day 0. (D) Boxplot of whether the given G-CSF grouped by oxygen requirement in the Manchester cohort measured at day 0. (E) Boxplot of whether the given G-CSF grouped by death in the Manchester cohort measured at day 0. COVID-19, coronavirus disease 2019; G-CSF, granulocyte colony-stimulating factor; ICU, intensive care unit.
Finally, as many cancer treatments and cancer itself can result in neutropaenia, we assessed whether low neutrophils preinfection were associated with outcome. Again, we did not find a significant association between O2 or death and CHRONIC average count preinfection (Supplementary Figure S2M and S2N, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.8; CL = 0.52 and P = 0.56; CL = 0.56, O2 and death, respectively) or IMMED (last value preinfection; Supplementary Figure S2O and S2P, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.55; CL = 0.53 and P = 0.5; CL = 0.54, O2 and death, respectively).
Biochemical features
C-reactive protein
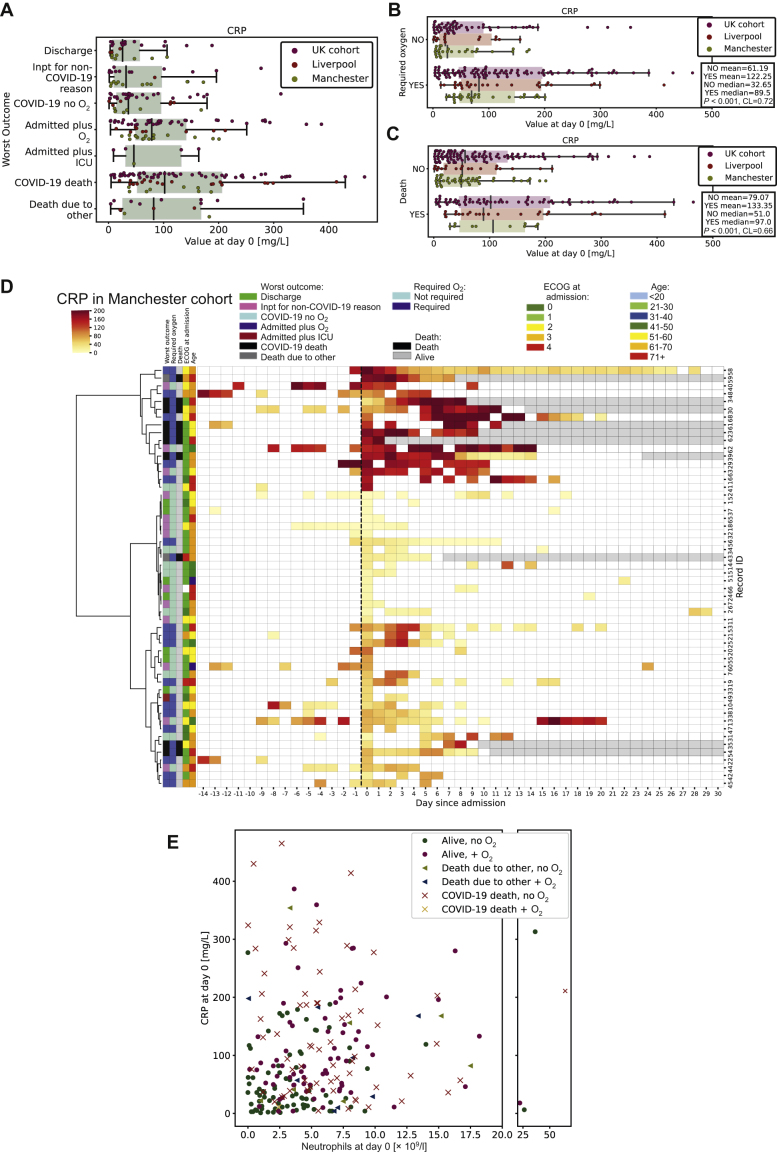
As CRP, an acute phase protein, is associated with inflammation,16 we examined this at day 0 according to worst outcome (Figure 3A). At diagnosis of COVID-19, in all cohorts, raised CRP was significantly associated with O2 (Figure 3B; P < 0.001, CL = 0.72) and death (Figure 3C; P < 0.001, CL = 0.66) from COVID-19. Critically, different CRP trajectories were observed when comparing patients longitudinally (Figure 3D). Patients not requiring oxygen clustered together and had low CRP levels, which remained flat over time. Those requiring oxygen had high day 0 but subsequently stable CRP levels, whereas those patients who died had high day 0 CRP or rapidly increasing CRP levels (Figure 3D). Furthermore, rate of CRP increase was significantly associated with O2 or death (Supplementary Figure S3A, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.013; CL = 0.71 and Supplementary Figure S3B, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.009; CL = 0.77, O2 and death respectively). These data suggest that high baseline CRP or rapidly rising CRP was associated with poor outcomes in COVID-19.
Figure 3.
Longitudinal changes in C-reactive protein.
(A) Boxplot of CRP according to worst outcome in the entire cohort measured at day 0. Discharge, discharged within 24 hours; Inpt for non-COVID-19 reason, inpatient due to reason other than COVID-19 and outcome not altered by infection; COVID-19 no O2, admitted due to COVID-19 infection but did not require oxygen; admitted plus O2, admitted due to COVID-19 and required oxygen; admitted plus ICU, admitted due to COVID-19 and required intensive care; COVID-19 death, death due to other. (B) Boxplot of CRP grouped by oxygen requirement in the entire cohort measured at day 0. (C) Boxplot of CRP grouped by whether patient died in the entire cohort measured at day 0. (D) Heatmap of the CRP level (darker red = higher CRP) against time (pre/post SARS-CoV-2-positive PCR test). Each patient record is represented by a row and each column by a timepoint. Hierarchical clustering is based on values from day 0 to 7. (E) Scatter graph of CRP versus neutrophil count measured at day 0 in the entire cohort. Blue, required oxygen; red, did not require oxygen; triangle, died from non-COVID-19 cause; cross, died due to COVID-19. COVID-19, coronavirus disease 2019; CRP, C-reactive protein; ECOG, Eastern Cooperative Oncology Group; ICU, intensive care unit; SARS-CoV, severe acute respiratory syndrome coronavirus 2.
As both high neutrophil count and CRP were associated with worse outcomes and may represent deranged inflammatory processes, we examined whether they correlated at day 0 in the entire cohort. We did not observe a correlation between CRP and neutrophil count. Intriguingly we found that although patients with low CRP and low neutrophil count had better outcomes following infection, patients with either a high CRP/low neutrophil count or a low CRP but high neutrophil count had worse outcomes (Figure 3E). This suggests heterogeneous inflammatory responses in patients with cancer and COVID-19, which can all result in oxygen requirement and death.
Albumin
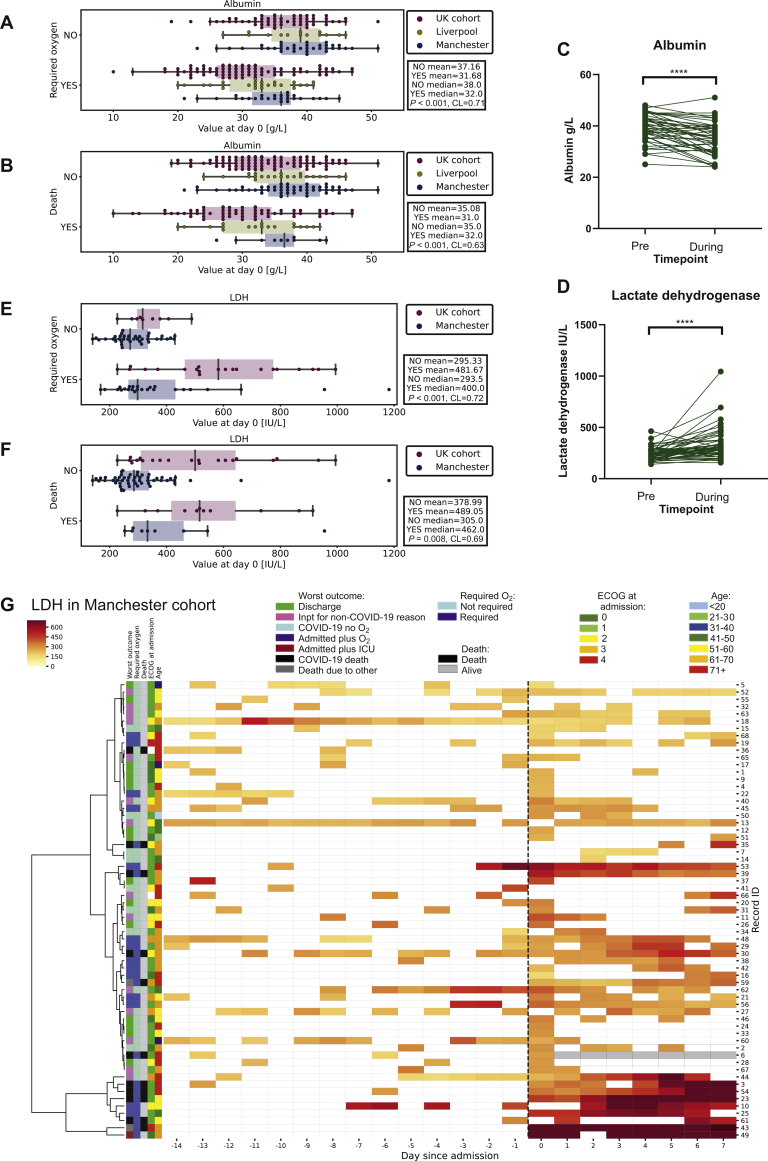
We found that that lower albumin at time of COVID-19 diagnosis was associated with both O2 (Figure 4A; P < 0.001, CL = 0.71) and death due to COVID-19 (Figure 4B; P < 0.001, CL = 0.63). In the Manchester cohort, albumin levels significantly dropped from pre-COVID-19 infection (IMMED) to the average 7 days during infection (mean 39 versus 35; P < 0.0001; Figure 4C, CL = 0.71), suggesting that low albumin was associated with an acute phase response to the viral illness rather than systemic effects from tumour burden.
Figure 4.
Longitudinal changes in albumin and LDH.
(A) Boxplot of albumin grouped by oxygen requirement in the entire cohort measured at day 0. (B) Boxplot of albumin grouped by whether patient died in the entire cohort measured at day 0. (C) Albumin IMMED (last test preinfection) versus 7 days during infection in the Manchester cohort (∗∗∗∗P < 0.0001). (D) LDH IMMED (last test preinfection) versus 7 days during infection in the Manchester cohort (∗∗∗∗P < 0.0001). (E) Boxplot of LDH grouped by oxygen requirement in the Manchester and UK cohorts measured at day 0. (F) Boxplot of LDH grouped by whether patient died in the Manchester and UK cohorts measured at day 0. (G) Heatmap of the LDH level (darker red = higher LDH) against time (pre/post SARS-CoV-2-positive PCR test). Each patient record is represented by a row and each column a timepoint. Hierarchical clustering is based on values from day 0 to 7. LDH, lactate dehydrogenase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Lactate dehydrogenase
Raised LDH has been associated with increased COVID-19 severity, in addition to increased tumour burden and aggressive clinical course in cancer patients.17 We therefore assessed whether LDH changed during COVID-19 compared with the potential effect of cancer burden causing a raised LDH prior to infection. In the Manchester cohort, LDH significantly rose from pre-COVID-19 infection (IMMED) to the average 7 days during infection (mean 242 versus 336; P < 0.0001, CL = 0.73, Figure 4D). Preinfection LDH (day –15) was not associated with worse outcome (Supplementary Figure S4A, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.51, CL = 0.55 and Supplementary Figure S4B, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.56, CL = 0.56 for O2 and death, respectively).
We observed that higher LDH at the time of COVID-19 diagnosis was associated with both O2 (Figure 4E; P < 0.001, CL = 0.72) and death due to COVID-19 (Figure 4F; P = 0.008, CL = 0.69). In addition, similar to CRP, longitudinal analysis of LDH revealed a high day 0 value, which was more sustained during COVID-19 in patients requiring oxygen or who died compared with those who had a mild disease course (Figure 4G). Although patients did not cluster together in LDH trajectories as clearly as CRP measurements (Figure 4G), rate of LDH rise was also found to be significantly associated with O2 and death (Supplementary Figure S4D, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.001, CL = 0.78 and Supplementary Figure S4E, available at https://doi.org/10.1016/j.esmoop.2020.100005; P = 0.002, CL = 0.83 for O2 and death, respectively).
Multivariable analyses
Finally, we evaluated both the day 0 haematological and biochemical parameters together with clinical factors such as age, sex, comorbidities, haematological/solid cancer, and systemic treatment within the preceding 4 weeks (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100005). In the univariable analysis of clinical features, only age (P < 0.001) and total number of comorbidities (P < 0.001) were significant for O2, whereas age (P < 0.001), total number of comorbidities (P < 0.01), male sex (P = 0.047) and haematological cancer (P = 0.02) were significantly associated with death. In multivariable analysis of all day 0 haematological, biochemical and clinical features presented in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100005, age (P = 0.002), haematological cancer (P = 0.034), CRP (P = 0.004), NLR (P = 0.036) and albumin (P = 0.02) remained significant predictors of death. In addition, multivariable analysis of time to death from hospital admission (n = 81 patients) revealed that higher neutrophils (P = 0.027) and advanced cancer stage (P = 0.042) were associated with earlier death (Supplementary Table S6, available at https://doi.org/10.1016/j.esmoop.2020.100005).
Discussion
Although numerous studies have examined COVID-19 features associated with severity of infection, many of these have been in noncancer populations and thus may not represent the specific characteristics of patients with cancer. The two largest series to date, in almost 2000 patients with cancer, have reported that age, male sex, Eastern Cooperative Oncology Group (ECOG) performance status, smoking status and presence of other comorbidities such as hypertension were significantly associated with COVID-19 mortality.4,5 In addition, the COVID-19 and Cancer Consortium (CCC19) study showed that active cancer was associated with increased mortality, with patients with progressive disease having the worst outcomes.5 Age was a significant independent risk factor in both cohorts,4,5 which again we validated further as a significant predictor of mortality in our cohort. Notably, neither study showed that type of cancer treatment received prior to infection was associated with increased severity of illness.
Analyses of large data sets have shown that patients with cancer have more severe outcomes when infected with COVID-19 compared with noncancer populations.3,9 Our study provides further insight into preinfection characteristics of patients with cancer and changes during COVID-19. We observed that both LDH and albumin significantly changed during infection with SARS-CoV-2 compared with levels immediately preinfection. Low albumin at presentation in particular was significantly associated with poor outcome and this remained significant in multivariable analysis. In addition, rate of LDH rise was associated with increased severity, although this should be interpreted with caution, as fewer data points were available in those with better outcomes. LDH is a nonspecific marker of cell damage and death, is widely distributed throughout the body18 and increases in a number of inflammatory processes including ARDS.18 Albumin is an acute phase protein that typically decreases during inflammation due to capillary leakage.19,20 Both are affected by inflammatory processes associated with cancer, with high LDH also reflecting high turnover of tumour cells and low albumin also associated with poor nutritional state and losses through processes such as accumulation of ascites.17 However, in our cohort, levels of either LDH or albumin preinfection were not associated with COVID-19 severity. Thus, it appears that changes seen were associated with response to SARS-CoV-2 itself, rather than pre-existing inflammatory processes associated with cancer.
Consistent with previous reports,3,21,22 we found that CRP was associated with increased COVID-19 severity and this remained significant in multivariable analysis. Rate of CRP rise was associated with increased severity of illness, thus patients with high and rising CRPs should be monitored closely, with early discussion about escalation of care. Intriguingly, CRP is not typically associated with viral infections, suggesting a specific underlying inflammatory biology related to the host response to SARS-CoV-2.23 Emerging preclinical data have implicated a deranged inflammatory cascade with increase in interleukin-6 (IL-6), IL-10, monocyte-chemoattractant protein-1 and interferon gamma-induced protein 10 associated with worse outcome.24,25 Notably, we observed that CRP was not correlated with neutrophil count and that response to SARS-CoV-2 was heterogeneous, with some patients with severe outcomes having increased CRP and others predominantly a neutrophilia, although patients with both high CRP and high neutrophil count commonly did poorly. Thus patients with higher neutrophils should be monitored even if CRP is low. Further understanding of this heterogeneous response may provide insight into personalising immunomodulatory therapies according to the predominant mechanism of immune dysregulation.
Many patients and clinicians are concerned about the potential impact of cancer treatments on the immune system resulting in increased severity of infection. Lower lymphocytes and neutrophils prior to or during infection were not associated with increased COVID-19 mortality, although lower lymphocytes were associated with oxygen requirement. In addition, high NLR was significantly associated with death. Lymphopaenia (<1 × 109/l) has been associated with increased severity of COVID-19 (with and without cancer) in a meta-analysis of 4911 patients.21 The observation that many patients were already lymphopaenic prior to infection may explain the lack of a significant difference in COVID-19 mortality of day 0 lymphocytes in our population. However, this may explain why cancer patients as a group are more susceptible to more severe COVID-19 as lymphocytes were not only low prior to diagnosis, but also they significantly dropped during COVID-19 infection with the majority being lymphopaenic at day 0 (247/286). In addition, patients with haematological cancer had lower lymphocyte counts at day 0 compared with those with solid tumours and in multivariate analysis they were more likely to die (P = 0.034; OR 2.0; 95% CI 1.05-3.79). This is supported by data from the UKCCMP that patients with haematological cancer have worse outcomes to COVID-19.26 Further comparisons of preinfection lymphocyte counts of cancer patients versus noncancer patients, changes during SARS-CoV-2 infection and correlation with viral load would be required to definitively test this hypothesis.
By contrast, and consistent with other noncancer cohorts,24 patients with higher neutrophil counts had poorer outcomes following presentation of COVID-19. Although this was not significant in multivariate analysis, higher neutrophil counts at day 0 were significantly associated with decreased time from admission to death. Of note, neutrophil counts associated with severe outcome at day 0 (and even maximal counts during illness) were the upper quartile of normal or not much higher than normal range. Although neutrophil counts were increased by G-CSF, it was not associated with severe outcome, potentially due to the timing of neutrophilia or immaturity of neutrophils. However, another study has shown that G-CSF was associated with worse outcome in 16 patients.27 Thus, use of G-CSF for neutropaenia during SARS-CoV-2 infection for patients at risk from serious infectious complications from neutropaenic sepsis as per American Society of Clinical Oncology (ASCO) guidelines could still be considered but should be used with caution.28 Neutrophils are associated with inflammation and increase in response to inflammatory cytokines such as IL-6, which have been shown to be raised during COVID-19.23,29 Neutrophilia has also been observed to be a poor prognostic marker in noncancer patients and aberrant neutrophil extracellular trap formation may contribute to organ damage in COVID-19.21,30 A neutrophil activation signature has been associated with mortality in patients with COVID-19 and critically, preceded severe illness, suggesting neutrophil activation as important in the pathogenesis of severe outcome.31 Furthermore, high neutrophil-to-T-cell ratio has been associated with increased COVID-19 severity21,24 and we observed worse outcomes in patients with higher NLRs. Taken together, our data reveal that patients with higher neutrophil counts or high NLRs at admission should be monitored carefully.
Others have shown no impact of cancer treatment type on severity of COVID-19.4,5 We did not observe further complications in patients who resumed therapy (including high-dose chemotherapy) following SARS-CoV-2 infection, although the numbers were small (n = 30). However, we still observed high mortality rates in our cohorts and cancer diagnosis is an independent risk factor for poor outcome.3,9 Other aspects of cancer, such as the effect of disease on frailty or performance status may therefore be more important. Critically, many cancer patients with late-stage disease and consequently poor overall prognosis are not always suitable candidates for intensive care, which may also affect outcomes. Large-scale analyses of all patients undergoing cancer treatments and comparisons of clinicopathological features with noncancer populations will enable better identification of those at highest risk of COVID-19 infection and adverse outcome. Our data suggest that low neutrophil counts resulting from cancer or its treatment are not associated with increased severity of COVID-19, particularly in patients with solid tumours and that restarting therapy in patients is safe.
Data were limited to routine tests available, thus other inflammatory markers such as D-dimers have not been described. In addition, more detailed analysis of immune cell subsets and their functional states would improve understanding of the biology of the different responses of cancer patients to SARS-CoV-2, although they will not be able to capture preinfection states. Furthermore, due to the retrospective nature of this study, data were not available from each patient for all parameters. LDH was only routinely collected for the Manchester cohort and some hospitals within the UK cohort (described in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100005). Further prospective studies are therefore needed to validate our findings fully.
Through longitudinal dissection of biochemical and haematological parameters in patients with cancer, we show that the features described here prior to infection do not appear to be associated with worse outcomes in patients presenting to hospital. However, COVID-19 results in heterogeneous inflammatory changes from baseline, which are associated with severe outcomes for cancer patients. High CRP, LDH and low albumin were associated with oxygen requirement and death from COVID-19. Critically, neutrophilia and not low neutrophils was associated with severity of COVID-19. Taken together, our data and those of large cohorts4,5 provide evidence that immunosuppression in terms of neutrophil counts seen in patients due to cancer and its treatments is not associated with worse outcome when infected by SARS-CoV-2, particularly in patients with solid tumours. However, differences in other immune cell subsets may still contribute to more severe illness. Our study adds to the understanding of the dynamic changes that occur in cancer patients infected with SARS-CoV-2 admitted to hospital and how these are associated with outcome.
Acknowledgements
The authors thank Clare Griffin for administrative support, Digital ECMT team for insights into data analysis and Manchester and Liverpool ECMC.
Funding
RJL is supported by the National Institute for Health Research as a Clinical Lecturer. TB is supported by the National Institute for Health Research as an academic clinical fellow. UTK is an MRC Clinical Training Fellow based at the University of Liverpool supported by the North West England Medical Research Council Fellowship Scheme in Clinical Pharmacology and Therapeutics, which is funded by the Medical Research Council [grant number MR/N025989/1]. Funding for COVID-19 work has been provided by The Christie Charitable fund [grant number 1049751].
Disclosure
RJL speaker fees BMS and Astrazeneca, MR honoraria from Astellas Pharma, speaker fees MSD and Servier. CW consultancy and speaker fees Pfizer, Amgen, Novartis, AA conference fee Merck, spouse shares in Astrazeneca. TR financial support to attend educational workshops from Amgen and Daiichi-Sankyo. JT is now working at Astra Zeneca. CD, outside of this scope of work, has received research funding from AstraZeneca, Astex Pharmaceuticals, Bioven, Amgen, Carrick Therapeutics, Merck AG, Taiho Oncology, Clearbridge Biomedics, Angle PLC, Menarini Diagnostics, GSK, Bayer, Boehringer Ingelheim, Roche, BMS, Novartis, Celgene, Thermofisher. CD is on advisory boards for, and has received consultancy fees/honoraria from, AstraZeneca, Biocartis and Merck KGaA. The remaining authors have no conflicts of interest to declare.
Supplementary data
References
- 1.Guan W., Ni Z., Hu Y. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang W., Guan W., Chen R. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol. 2020;21:335–337. doi: 10.1016/S1470-2045(20)30096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta V., Goel S., Kabarriti R. Case fatality rate of cancer patients with COVID-19 in a New York Hospital System. Cancer Discov. 2020;10(7):935–976. doi: 10.1158/2159-8290.CD-20-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee L.Y.W., Cazier J.B., Starkey T., Turnbull C.D., Kerr R., Middleton G. COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet. 2020;395:1919–1926. doi: 10.1016/S0140-6736(20)31173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuderer N.M., Choueiri T.K., Shah D.P. Clinical impact of COVID-19 on patients with cancer (CCC19): a cohort study. Lancet. 2020;395:1907–1918. doi: 10.1016/S0140-6736(20)31187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saini K.S., Tagliamento M., Lambertini M. Mortality in patients with cancer and coronavirus disease 2019: a systematic review and pooled analysis of 52 studies. Eur J Cancer. 2020;139:43–50. doi: 10.1016/j.ejca.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai M., Liu D., Liu M. Patients with cancer appear more vulnerable to SARS-COV-2: a multi-center study during the COVID-19 outbreak. Cancer Discov. 2020;10 doi: 10.1158/2159-8290.CD-20-0422. CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C., Chen X., Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williamson E.J., Walker A.J., Bhaskaran K. Factors associated with COVID-19 death in 17 million patients using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wynants L., Van Calster B., Bonten M.M.J. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Zhu F., Xie L. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann Oncol. 2020;31:894–901. doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Health Research Authority Guidance for Using Patient Data – Health Research Authority. https://www.hra.nhs.uk/covid-19-research/guidance-using-patient-data/ Available at.
- 13.Lauer S.A., Grantz K.H., Bi Q. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Institute of Clinical Excellence . NICE; London, UK: 2020. Admission to Critical Care. COVID-19 Rapid Guideline: Critical Care in Adults NICE Guidance. [Google Scholar]
- 15.McGraw K.O., Wong S.P. A common language effect size statistic. Psychol Bull. 1992;111(2):361–365. [Google Scholar]
- 16.Sproston N.R., Ashworth J.J. Role of C-reactive protein at sites of inflammation and infection. Front Immunol. 2018;9:754. doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrelli F., Cabiddu M., Coinu A. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta Oncol. 2015;54:961–970. doi: 10.3109/0284186X.2015.1043026. [DOI] [PubMed] [Google Scholar]
- 18.Drent M., Cobben N.A.M., Henderson R.F., Wouters E.F.M., van Dieijen-Visser M. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur Respir J. 1996;9(8):1736–1742. doi: 10.1183/09031936.96.09081736. [DOI] [PubMed] [Google Scholar]
- 19.Soeters P.B., Wolfe R.R., Shenkin A. Hypoalbuminemia: pathogenesis and clinical significance. J Parenter Enter Nutr. 2019;43(2):181–193. doi: 10.1002/jpen.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain S., Gautam V., Naseem S. Acute-phase proteins: as diagnostic tool. J Pharm Bioallied Sci. 2011;3:118–127. doi: 10.4103/0975-7406.76489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng X., Li S., Sun Q. Immune-inflammatory parameters in COVID-19 cases: a systematic review and meta-analysis. Front Med. 2020;7:301. doi: 10.3389/fmed.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albiges L., Foulon S., Bayle A. Determinants of the outcomes of patients with cancer infected with SARS-CoV-2: results from the Gustave Roussy cohort. Nat Cancer. 2020;1:965–975. doi: 10.1038/s43018-020-00120-5. [DOI] [PubMed] [Google Scholar]
- 23.Vabret N., Britton G.J., Gruber C. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann E.R., Menon M., Knight S.B. Longitudinal immune profiling reveals distinct features of COVID-19 pathogenesis. Sci Immunol. 2020;5(51):eabd6197. doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing A.G., Lorenc A., Del Barrio I.D.M. A consensus Covid-19 immune signature combines immuno-protection with discrete sepsis-like traits associated with poor prognosis. medRxiv. 2020 2020.06.08.20125112. [Google Scholar]
- 26.Lee L.Y.W., Cazier J.B., Starkey T. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: a prospective cohort study. Lancet Oncol. 2020;21(10):1309–1325. doi: 10.1016/S1470-2045(20)30442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morjaria S., Zhang A., Kaltsas A. The effect of neutropenia and filgrastim (G-CSF) in cancer patients with COVID-19 infection. medRxiv. 2020 doi: 10.1093/cid/ciab534. 2020.08.13.20174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith T.J., Bohlke K., Lyman G.H. Recommendations for the use of WBC growth factors: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. 2015;33(28):3199–3212. doi: 10.1200/JCO.2015.62.3488. [DOI] [PubMed] [Google Scholar]
- 29.Hashizume M., Higuchi Y., Uchiyama Y., Mihara M. IL-6 plays an essential role in neutrophilia under inflammation. Cytokine. 2011;54(1):92–99. doi: 10.1016/j.cyto.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meizlish M.L., Pine A.B., Bishai J.D. A neutrophil activation signature predicts critical illness and mortality in COVID-19. medRxiv. 2020 doi: 10.1182/bloodadvances.2020003568. 2020.09.01.20183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.