Abstract
This review highlights recent efforts to detect bacteria, using engineered small molecules that are processed and incorporated similarly to their natural counterparts. There are both scientific and clinical justifications for these endeavors. The use of detectable, cell-wall targeted chemical probes has elucidated microbial behavior, with several fluorescent labeling methods in widespread laboratory use. Furthermore, many existing efforts including ours, focus on developing new imaging tools to study infection in clinical practice. The bacterial cell wall, a remarkably rich and complex structure, is an outstanding target for bacteria-specific detection. Several cell wall components are found in bacteria but not mammals, especially peptidoglycan, lipopolysaccharide, and teichoic acids. As this review highlights, the development of laboratory tools for fluorescence microscopy has vastly outstripped related positron emission tomography (PET) or single photon emission computed tomography (SPECT) radiotracer development. However, there is great synergy between these chemical strategies which both employ mimicry of endogenous substrates to incorporate detectable structures. As the field of bacteria-specific imaging grows, it will be important to understand the mechanisms involved in microbial incorporation of radionuclides. Additionally, we will highlight the clinical challenges motivating this imaging effort.
Keywords: infection, imaging, peptidoglycan, nuclear medicine, chemical biology
Graphical Abstract
A. Introduction and scope of review:
The last decade has showed remarkable progress in detecting and characterizing bacterial pathogens non-invasively. Cell-wall derived techniques abound in this area, are both driving our understanding of microbial behavior and inviting new human-compatible imaging technologies. In this review, we will focus on the bacteria-specific structures found in or proximal to the cell wall, including transporter proteins and membrane-bound components such as penicillin-binding proteins. In particular, we will discuss elements targeted for fluorescent probe and positron emission tomography (PET) and single photon enhanced computed tomography (SPECT) tracer development, with an emphasis on clinically relevant approaches and discoveries. This review will generally be limited to non-mycobacterial structures and components seen in multiple species of gram-positive and gram-negative pathogens.
We will begin by highlighting the clinical challenges motivating this imaging effort, and follow with a discussion addressing fluorescent versus PET/SPECT detection methods, with the latter focused on imaging human-relevant pathogens via tomographic techniques. In general, fluorescent tools that study bacteria and their behavior in vitro are much more highly evolved in the literature, with numerous elegant studies highlighting the incorporation of fluorescent D-amino acid analogues and other cell-wall specific structures. The review will then describe direct versus pre-targeted detection strategies. Steric effects frequently justify the incorporation of “clickable” moieties into bacterial structures, which can be subsequently detected via bio-orthogonal reactions with frequently bulky and structurally complex fluorescent probes. Finally, we will illustrate several small-molecules targeting cell wall structures that either bind cell-wall related proteins, or are metabolic precursors for the cell wall itself. This discussion will enumerate the basic differences between gram-negative and gram-positive cells which could potentially allow distinguishing between the two in key disease contexts. As the structural features of the bacterial cell wall are detailed, we will see that several components are likely bacteria-specific, whereas others are frequently found in fungi, mammals, and other organisms.
B. Clinical justifications:
Imaging studies are frequently used in the evaluation of infected patients, particularly important in identifying the presence and location of infection, and documenting response to antimicrobial therapy. These imaging approaches generally rely on structural changes, detecting abnormal tissue edema or fluid by standard radiography, computed tomography (CT), and magnetic resonance (MR). In contrast, the nuclear imaging techniques positron emission tomography (PET) and single photon emission computed tomography (SPECT) add biochemical and functional information. Current clinical methods include 2-deoxy-2-[18F]fluoro-D-glucose([18F]FDG)-PET, the [111In]WBC scan, and gallium-derived tools (67Ga and 68Ga for SPECT and PET respectively)1–6. The mechanisms of these tracers are beyond the scope of this review, but they generally image the host immune response to active infection rather than the bacteria themselves. Therefore, these tools may fail to differentiate active infection from sterile inflammation.
In response to this limitation, newer radiotracers have targeted bacteria-specific metabolism, by exploiting basic differences between mammalian and bacterial cells. For example, sorbitol-derived radiotracers are highly sensitive to Enterobacteriaceae but not mammalian cells7, while para-aminobenzoic acid (PABA)8,9 and trimethoprim-derived probes10 can detect the bacterial folate generating pathway. These and related approaches are expected to have an expanding clinical role in the next decade. Cell-wall and membrane-targeted agents also hold special promise, complementing the detection of cytoplasmic and nuclear targets. Targeted structures include both structural constituents of the cell wall (peptidoglycan, teichoic acids, LPS), and relevant cell-wall and membrane proteins especially transporters. The use of all pathogen-targeted imaging methods is somewhat complicated by the normal human microbiome11. However, the microbiome is generally well-sampled, easily accessible via the skin and aerodigestive tract. Many analytic tools investigating the microbiome are ex vivo, for example using the stool and/or biofluids such as sputum12–14. Therefore the described imaging methods are most helpful in settings where these traditional sampling methods fail. In particular, rapid methods to diagnose bacterial infection will be most helpful for triaging acutely ill patients. Depending on the radiotracers used, imaging studies can reveal (1) the location of infection (2) the type of organism i.e. gram-negative versus gram-positive (3) response to antimicrobial therapy and (4) the presence of resistant pathogens. We believe that new imaging tools are essential in the following clinical scenarios:
1. Infection of deeper, normally sterile spaces:
Successful diagnosis of infection involving the intervertebral discs and other joint spaces (hips, knee, etc.) is frequently difficult in clinical practice, even using magnetic resonance imaging (MRI). Percutaneous sampling of these spaces is frequently needed which can be insensitive, costly and potentially dangerous for the patient. For example, vertebral discitis-osteomyelitis represents a major diagnostic challenge even when advanced imaging is employed, highlighted in several reports15,16. Another example of a sterile region that is not easily accessed is the biliary tract and pancreas. It can be difficult to diagnose cholangitis (infection of the biliary system)17 and infected pancreatitis which may occur in the context of pancreatic pseudocysts18. The pancreas and biliary system are not easily sampled, and diagnostic tools compatible with full-body cross-sectional imaging are needed.
2. Infection that occurs in the presence of inflammatory mimics:
For several entities, imaging and other diagnostic studies cannot separate acute infection from sterile inflammation. This distinction is of critical importance for treatment, since opposite treatments are frequently used for these two entities. A patient who has an acute bacterial joint infection requires urgent antibiotic therapy, while the inflammation associated with rheumatoid arthritis or gout is treated with nonsteroidal anti-inflammatory drugs (NSAIDS) or corticosteroids. Other critical infections include those of the diabetic foot19, or P. aeruginosa pneumonias in cystic fibrosis patients20.
3. Infection in patients who cannot mount an immune response:
As discussed above, most nuclear medicine tools used in clinical practice image the host response to infection rather than the bacteria themselves. Both [18F]FDG and [111In]WBC scans identify immune cells trafficked to the area, while gallium-derived radiotracers sense transferrin21 which is upregulated in the acute phase of infection22. A large number of patients cannot mount an immune response, especially seen in human immunodeficiency virus (HIV), blood/bone marrow cancers, hereditary diseases and drug-related disorders. Frequently these patients can be afebrile, and have a normal or low white blood-cell count. Accurate methods to detect living bacteria would be critical in this population.
C. Fluorescence- the origin of nuclear imaging tools:
As highlighted by this review, many of the newer imaging methods (i.e. PET and SPECT) targeting the cell wall have been motivated by related fluorescence-based techniques. Fluorescent molecules have largely been studied in vitro and have contributed significantly to our understanding of bacterial behavior especially cell-wall remodeling. The advantages and disadvantages of fluorescent detection are highlighted in Table 1, with the clear benefits being (1) lower cost (2) diversity of chemical structures employed and (3) activatable or “tunable” signals. In contrast, PET and SPECT hold major advantages for in vivo imaging, with both techniques compatible with clinical translation. In contrast to fluorescence-based methods, which have a limited depth of penetration into tissues, PET and SPECT have been used in humans for decades to assess the metabolic activity of deeper, non-accessible human organs including the brain23–25. PET is highly sensitive, with the obtained spatial resolution benefitting from modern time-of-flight scanners26. Most academic centers in the United States have adopted PET as their primary nuclear imaging tool, based on both this high sensitivity and ease of radionuclide incorporation into small-molecules structures of interest including metabolites. This is particularly true for 11C and 18F radionuclei for which substitution likely induces fewer perturbations of biologic behavior. For 11C metabolites, structures can be generated that are chemically identical to those of their parent molecules27. This strategy is also a key feature of the most commonly used PET radiotracer [18F]FDG, which is a substrate for both glucose transporters (GLUTs) and hexokinase. Many radionuclides have been used to image infection, including 11C (PET, t1/2 =20 min), 68Ga(PET, t1/2 = 68 min), 18F(PET, t1/2 = 110 min), 99mTc (SPECT, t1/2 = 6 h), 64Cu (PET, t1/2 = 13 h), 111In (SPECT, t1/2 = 67 h), 201Tl (t1/2 = 73 h), 67Ga(SPECT, t1/2 = 78 h), 89Zr (PET, t1/2 = 78 h), 124I (PET, t1/2 = 4.2 d).
Table 1.
Comparison between optical imaging and radionuclide-based PET and SPECT imaging.
| OPTICAL | PET/SPECT | |
|---|---|---|
| In vitro analysis | Yes | Yes |
| In vivo analysis | Yes | Yes |
| Penetrating depth | ~cm | Full body |
| Activatable | Yes | No |
| Detected component | Fluorophore | Radionuclide |
| Detection method | Photons | Positrons/Gamma rays |
| Concentration | nM | fM-pM |
| Instrumentation | Smaller, more portable | Larger, PET/CT or PET/MR |
With respect to the bacterial cell wall, several recent studies have demonstrated the synergy of optical and nuclear imaging. Work published by Ning et al. from the Murthy group using maltohexose derivatives highlights the scope and limitations of both approaches for in vivo imaging. Figure 1 depicts both NIR-dye modified28 and 18F labeled29 maltohexose derivatives, both synthesized using click chemistry. Similarly, there is a rich history in the fluorescence literature describing the use of D-amino acid derived probes targeting the bacterial cell wall. Figure 2A shows the structures of two D-amino acid derived structures that use 7-hydroxycoumarin 3-carboxylic acid and 4-chloro-7-nitrobenzofuran to make HADA and NADA respectively, fluorescent sensors used for in vitro analysis30. In contrast Figure 2B depicts D-amino acid derived structures for PET namely D-[methyl-11C]methionine (D-[11C]met) and D-[3-11C]alanine (D-[11C]ala)31–33 which were used both in vitro and in vivo to study pathogenic bacteria. The radiosynthesis of D-[11C]ala from a glycine precursor using a cinchonidinium-derived chiral catalyst is depicted in Figure 2C.
Figure 1.
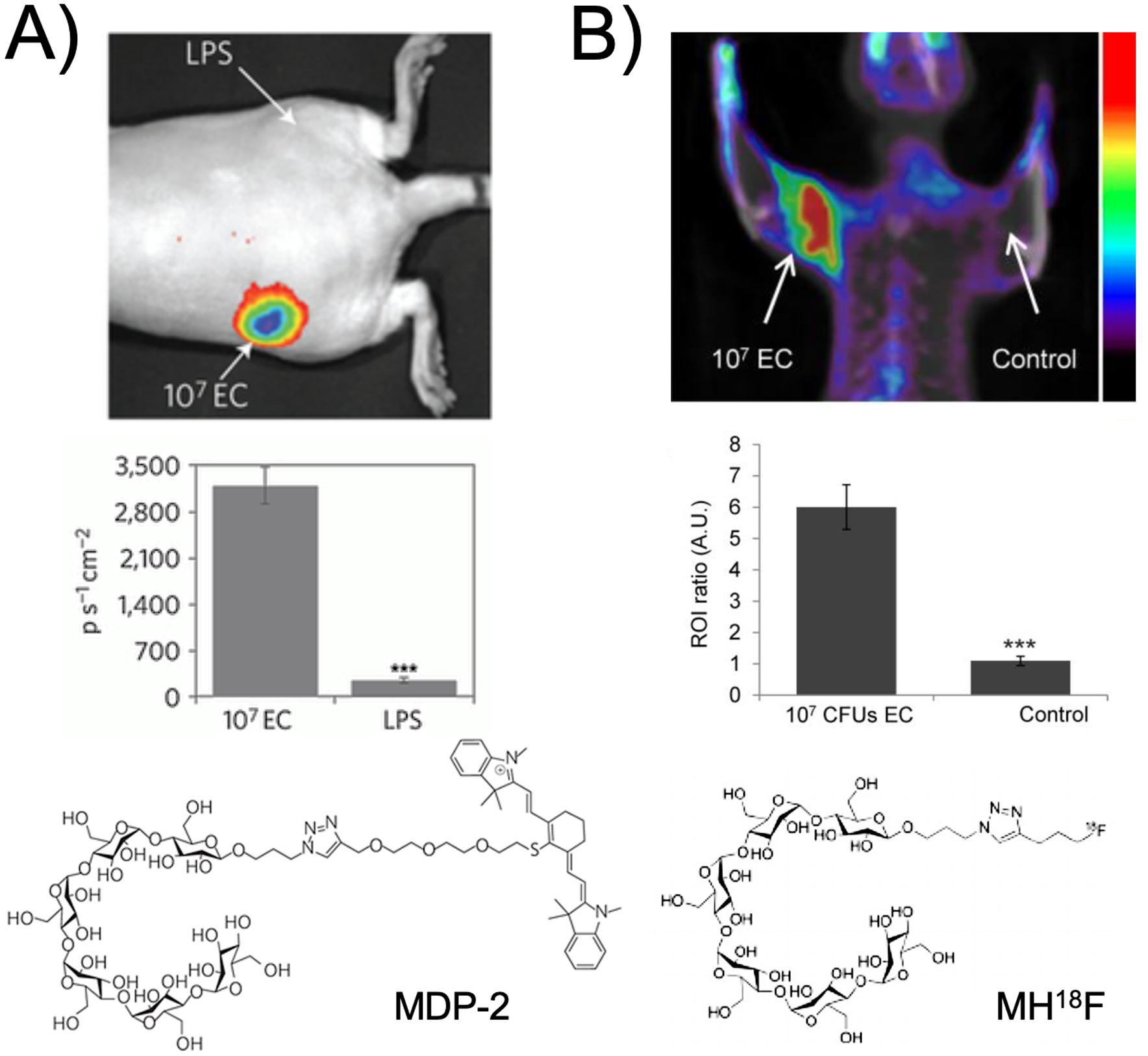
Maltodextrin transporter-targeted fluorescent and PET probes published from the Murthy group, highlighting the synergy between optical imaging and PET. (A) Data obtained using the MDP-2 NIR probe in a simple preclinical model of bacterial infection, in which fluorescent signal is seen in living E. coli but not in LPS-induced inflammation in the opposite flank (adapted from Ning et al. 2011). (B) Similar findings for the MH18F maltohexose-derived positron emission tomography tracer (adapted from Ning et al. 2014).
Figure 2.
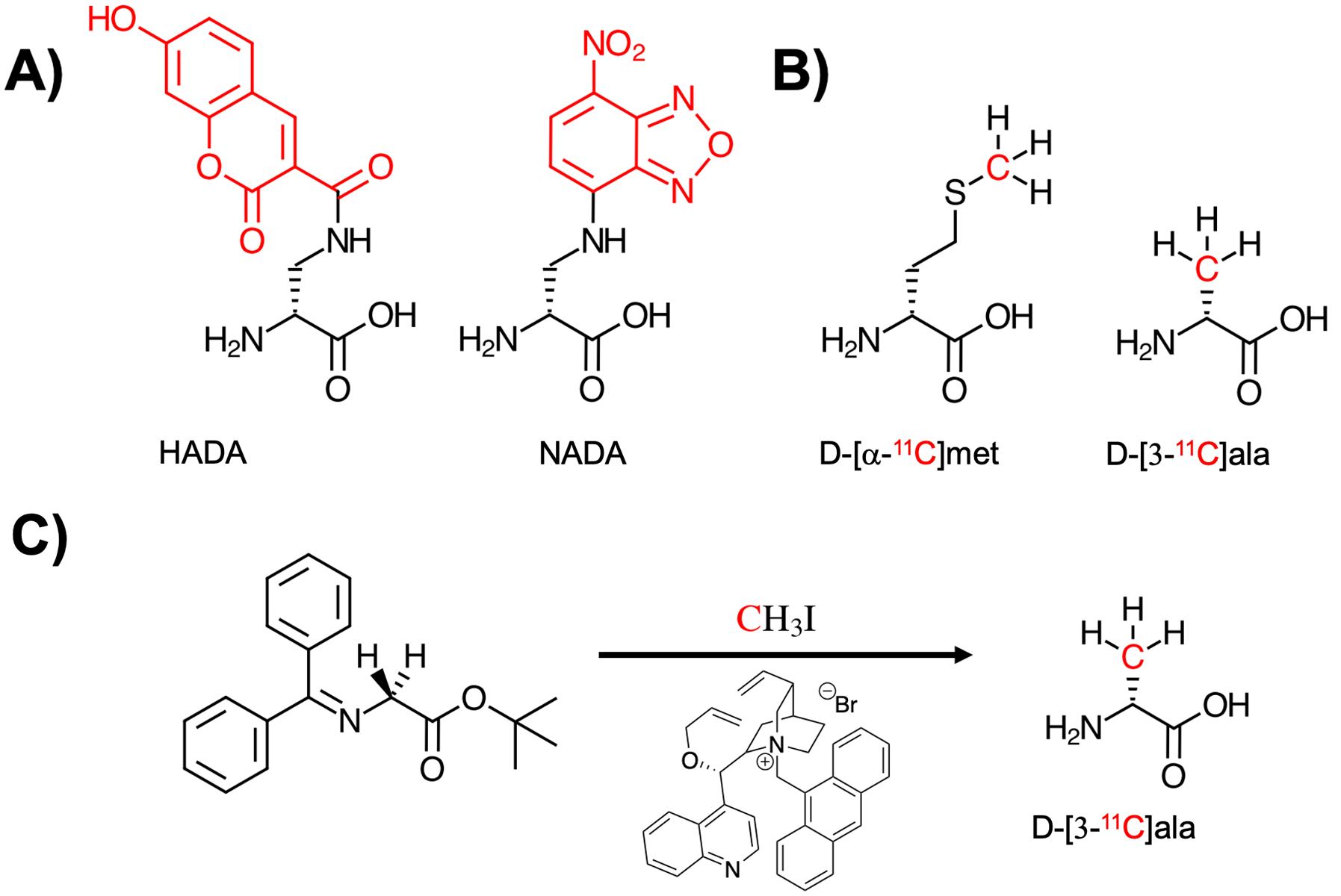
Comparison of D-amino acid derived structures modified for fluorescent detection and positron emission tomography (PET) imaging, with the sites of modification marked in red. (A) Two D-amino acid structures incorporating hydroxycoumarin and nitrobenzofuran-derived fluorophores, HADA and NADA respectively (Kuru et al. 2012). (B) Two 11C-labelled amino acids D-[α−11C]met (Neumann et al. 2017) and D-[3-11C]ala (Parker et al. 2020) with the 11C nucleus highlighted in red. (C) The enantioselective radiosynthesis of D-[3-11C]ala from an achiral glycine-derived precursor via reaction with 11C methyl iodide in the presence of a cinchonidinium-derived phase-transfer catalyst.
D. Direct versus “clickable” detection technologies:
Especially with respect to fluorescent D-amino acids (FDAAs), bacterial structures have shown marked promiscuity in incorporating fluorophore-modified small molecules. However, any significant deviations from canonical structures (i.e. D-alanine and D-glutamate) impart changes to biochemical behavior based on size, lipophilicity, and solvation properties34. Figure 3 shows the dramatic relationship between FDAA side-chain and bacterial accumulation with smaller linkers seemingly favored. Numerous tools for chemical biology have been developed to address this issue, by incorporating structures capable of detection into biologic systems. If visualization of FDAAs via muropeptide incorporation in the periplasm is considered “direct,” an indirect approach would be the incorporation of a chemically and biochemically inert D-amino acid into peptidoglycan, that is identified later via reaction with a second, fluorescent chemical moiety. This may be accomplished via a “bio-orthogonal” or “click” reaction whereby the two components are reactive with each-other, but otherwise stable in the biologic milieu35. The first methods used for click-chemistry dependent D-amino acid visualization36 is described in Figure 4A, while more advances in click chemistry are highlighted in Figures 4B/C including nitrone-based cycloaddition chemistry, where strained cyclooctyne-bearing fluorophores are reacted with nitrone derivatives of alanine and lysine and the corresponding adducts studied for bacterial incorporation37. In addition to the previously described steric advantages of using less bulky metabolites in biologic systems, another major consideration is pre-targeting. In other words, the modified cell-wall metabolite could be introduced into a biologic system over time if needed, with subsequent observation more dependent on the pharmacokinetics and reactivity of the fluorescent label. This mechanism of bacterial detection has been explored beyond D-amino acid utilization, with similar methods used to image N-acetyl muramic acid and KDO incorporation via generating a fluorescent product38. These cell-wall components are described more precisely later in the review. To our knowledge, a clickable, pre-targeted approach has not been described to image bacteria-specific structures using SPECT or PET. This is true despite the heavy use of bio-orthogonal methods to image antibodies and other biomolecules in nuclear imaging39–42. For in vivo imaging, click approaches need to overcome significant technical hurdles including addressing the use of copper for many reported in vitro methods43.
Figure 3.
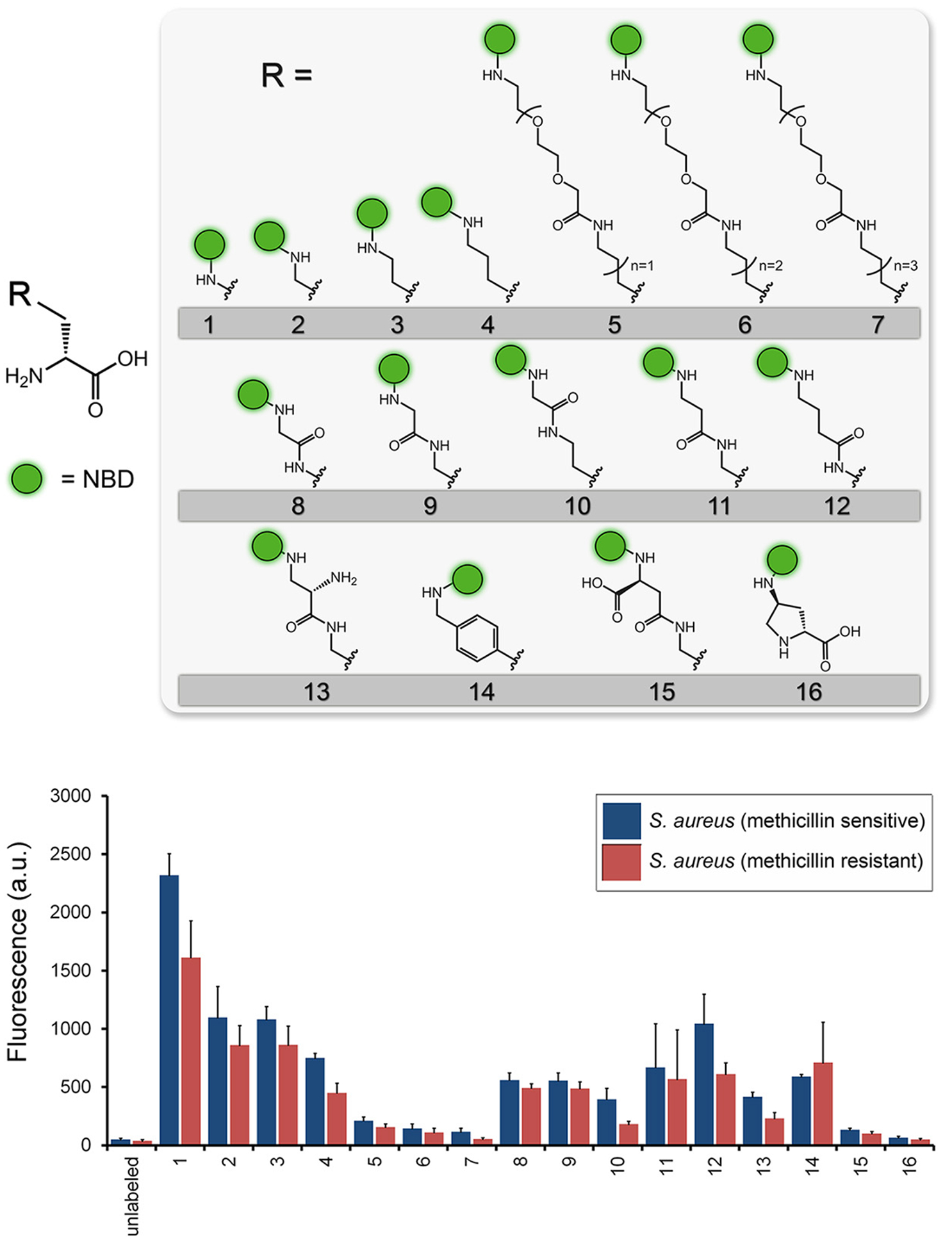
Use of D-amino acid derived structures to label peptidoglycan, via direct incorporation. This study employed linkers of various lengths suggesting the preference of S. aureus for smaller D-amino acid derived structures (adapted from Fura et al. 2015).
Figure 4.
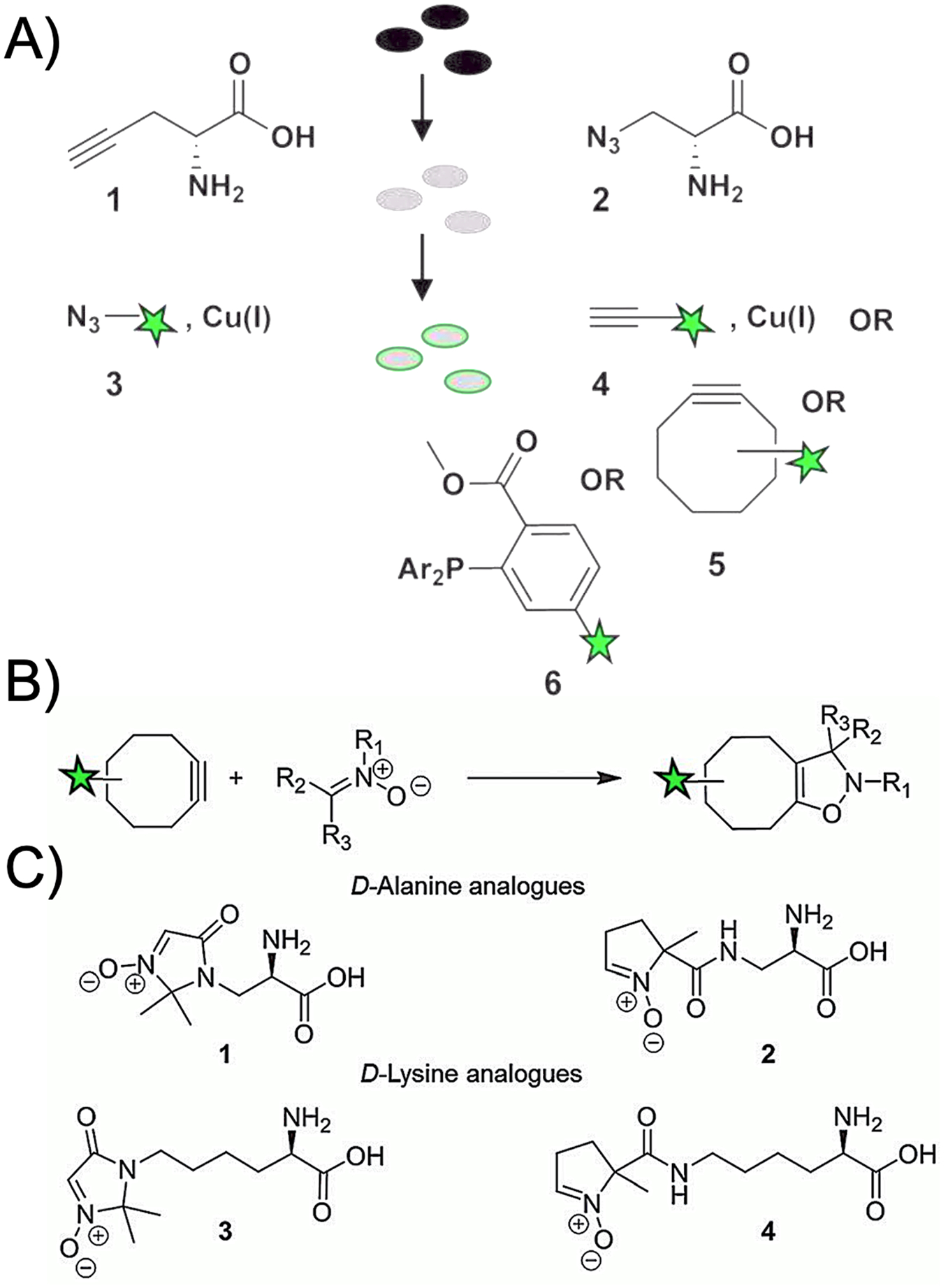
Bio-orthogonal chemistry developed to detect D-amino acids. (A) Chemistry developed by Siegrist et. al. used either an alkyne or azide containing side-chain with subsequent click reaction with a fluorescent moiety. (adapted from Siegrist et al. 2013). (B) A newer approach to bio-orthogonal chemistry using nitrone and strained alkyne cycloaddition chemistry. (C) Nitrone cycloadditions applied to D-alanine and D-lysine analogs (adapted from MacKenzie et al. 2015).
E. Targeting proteins proximal to the cell wall:
The first class of molecules we will consider are cell-wall specific proteins especially peptidoglycan-remodeling enzymes and bacterial-metabolite specific transporters. The most established approach to imaging cell-wall proteins is via modified antibiotic structures, with several recent in vivo methods targeting bacterial transport.
1. Antibiotic targets: fluorescent and radiolabeled sensors:
There is extensive literature regarding antibiotics, and their corresponding cell-wall protein targets as bacteria-specific imaging probes. Of note, antibiotics targeting the cell wall are not intrinsically fluorescent (as is the case for DNA-targeted antibiotics mithramycin, chlromamycin A3, and olivomycin), and thus conjugation to a fluorophore is required. Specifically, cell wall-targeted antibiotics that have been conjugated with fluorophores include β-lactams, vancomycin, ramoplanin, and polymyxin B44–47 meeting varying degrees of success in bacterial detection. Figure 5 contrasts two vancomycin-derived compounds that have been conjugated for visualization via SPECT and fluorescence45,48,49. Fluorescent conjugates have been used for imaging especially using NIR fluorophores50, but importantly they also may be turned “on” or “off” during binding, metabolism, and response to the microenvironment. Therefore these probes have been used to study antimicrobial resistance, mode of action, and toxicity; for an outstanding review see Stone et al. 201849. An important limitation in developing related PET and SPECT tracers is that much of this information is potentially lost in using a radionuclide that is always “on.” Other important limitations of imaging radiolabeled antibiotics via SPECT and PET are the signal depends on probe-protein affinity and the concentration of the target which may not be sufficient and there is no turnover benefit, i.e. the maximum signal is stoichiometric. These considerations suggest that imaging probes metabolized and incorporated by living, intact bacterial machinery have a significant advantage. Historically the most successful imaging probes are those that undergo biotransformation and incorporation for example FDG51, although this thinking is challenged by the dramatic successes of affinity-based imaging methods for example those targeting PSMA in humans52. Regardless, antibiotic-derived PET and SPECT tracers have historically shown limited in vivo success detecting bacterial infection53,54, although more recently reported trimethoprim analogues have shown outstanding data in preclinical models10. In other cases, labeled antimicrobial agents can be used to help us better understand therapeutic resistance, as is the case for [11C]rifampin55.
Figure 5.
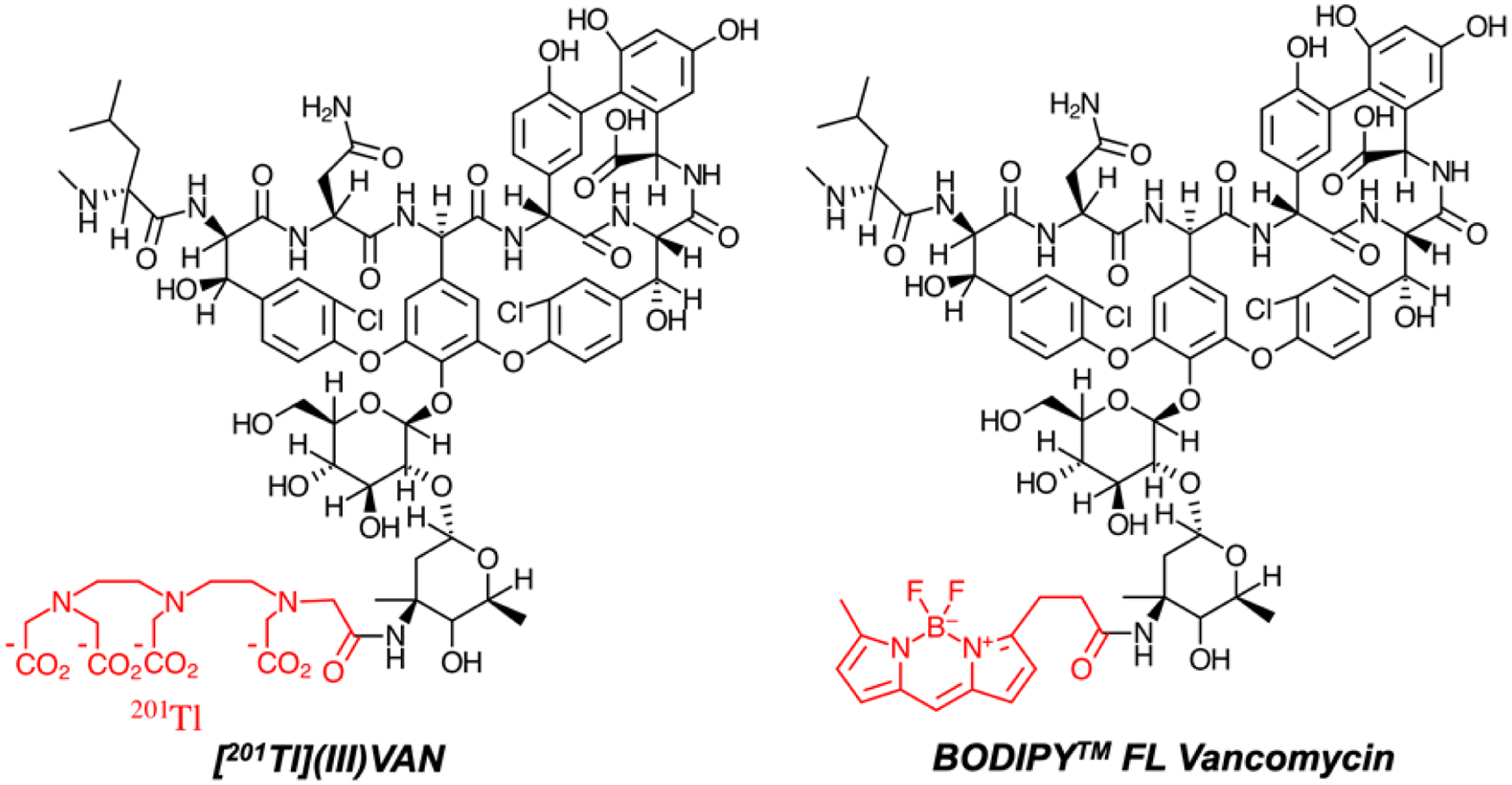
Vancomycin-derived conjugates used for single photon emission computed tomography (SPECT) and fluorescence imaging. (A) Modified 201Tl-containing derivative of vancomycin (Jalilian et al. 2008) (C) A commercially available BODIPY functionalized vancomycin.
2. Bacteria-specific metabolite transporters:
In the last decade, there has been a pronounced interest in imaging pathogens using bacteria-specific metabolic pathways. This is best accomplished using PET probes that are metabolized and incorporated by bacteria but not their mammalian hosts. Published approaches include radiolabeled versions of para-amino benzoic acid (PABA)8,9, trimethoprim10, sorbitol7,56,57, maltose/maltotriose/maltohexose29,58–60, arabinofuranoses61, and bacterial siderophores62. These technologies in particular the sorbitol-derived radiotracer 2-deoxy-2-[18F]fluorosorbitol ([18F]FDS) have recently been applied to patients suffering from bacterial infection. There are outstanding reviews of this emerging field published elsewhere63–65, but an important consideration is that these approaches likely rely heavily on bacteria-specific transport. The transporter used by [18F]FDS was incompletely characterized by Weinstein et al., but they did note that [18F]FDS uptake was outcompeted by concentrations of sorbitol above 40 mg/ mL7. As indicated above, several probes have also targeted the maltose-maltodextrin transport system66. For [18F]FDG, active transport of the probe (via GLUT 1,3,4) is very important and difficult to saturate. At our institution we determined that the concentration of glucose present in a patient-administered [18F]FDG sample was approximately 1 mM, reinforcing the need for a high transporter Km if competing unlabeled metabolites are present67.
F. Structure of gram-negative and gram-positive cell walls:
The basic differences between gram-negative and gram-positive bacteria are essential to consider in developing cell-wall targeted metabolic probes. The basic membrane structures are highlighted in Figure 6A, which shows the relative contribution of peptidoglycan, the strong and elastic polymer that both protects bacteria from attack, and contributes to cell-cell signaling and quorum sensing68,69. Of course it is the peptidoglycan component that renders bacteria gram-negative versus gram-positive, with the latter organisms better identified by Gram staining due to the higher concentration of peptidoglycan (90% by dry-weight versus 10% in gram-negatives). The basic structure of the peptidoglycan monomer is shown in Figure 6B indicating the sugar-derived components N-acetyl glucosamine and N-acetyl muramic acid as well as the muramic acid pentapeptide. Extensive data has shown that both pathogen classes can be detected using D-amino acid derived sensors, which may be of value in vivo in documenting the presence of infection versus other diseases32,33. However, for antibiotic selection in the acute setting, determining the presence of gram-negative or gram-positive pathogens would also be of high value. Therefore, structures found only in gram-negative or gram-positive organisms are of high interest. In gram-negatives these include lipopolysaccharide (LPS), which is composed of a lipid and polysaccharide composed of O-antigen, outer core and inner core, found in the outer membrane.59–61 In gram-positives, cell-wall teichoic acids are used. As discussed subsequently many of these components have been targeted by fluorescence methods and other detection methods.62–68
Figure 6.
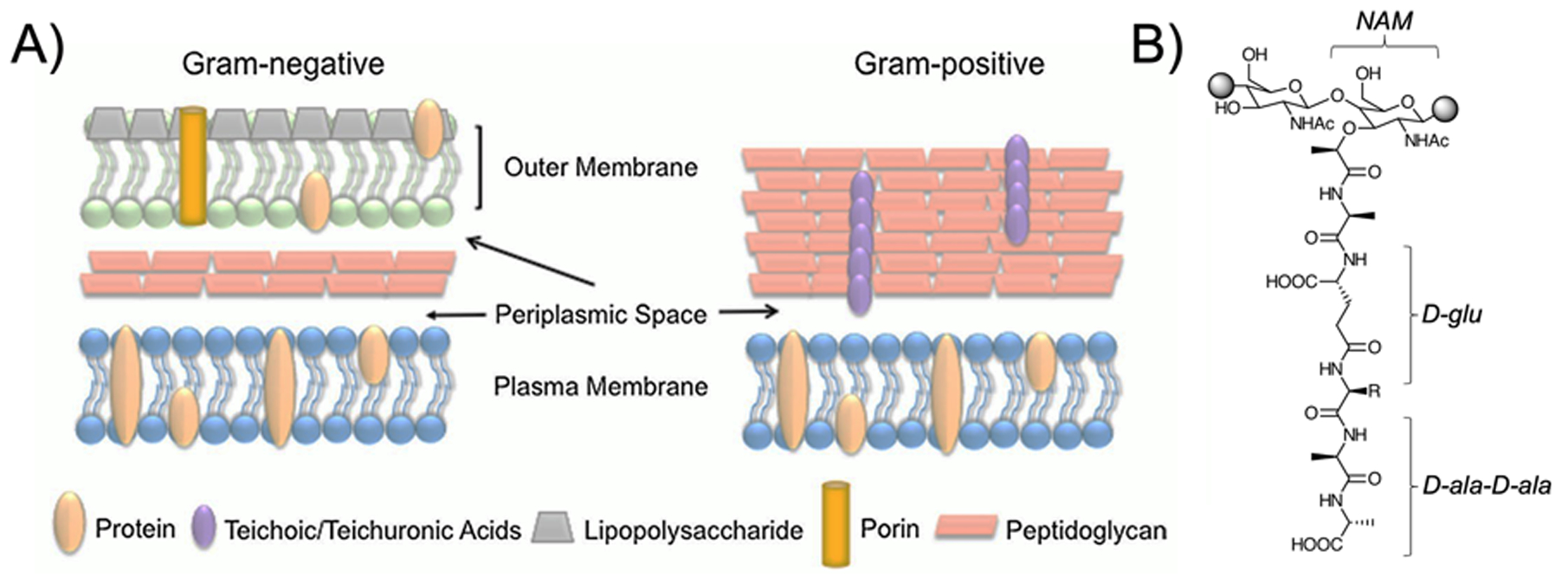
Targeted cell wall structures in fluorescent and SPECT/PET Probe development (. (A) Basic structures of gram-negative and gram-positive cell walls highlighting the location of peptidoglycan, lipopolysaccharide, teichoic acids, and transporters (adapted from Slavin et al. 2017)100 (B) Structure of the peptidoglycan monomer; both muropeptide D-amino acids and N-acetyl muramic acid from the sugar backbone have been targeted for detection.
G. Cell wall component-derived sensors:
Our group and others are particularly enthusiastic about small molecules that are readily incorporated into the bacterial cell wall, with structural modifications that allow subsequent detection. This is in contrast to previously described sensors that interface with membrane proteins, for example penicillin-binding proteins and sugar transporters. The data on D-amino acids is particularly robust, having been used by both fluorescence and PET to characterize living bacteria. Another peptidoglycan constituent is N-acetyl muramic acid which has been modified for bacterial detection via a bio-orthogonal approach. We will also briefly summarize gram-negative and gram-positive specific structures that have been targeted for imaging especially LPS.
1. Peptidoglycan:
Not surprisingly, peptidoglycan has been investigated extensively as a way of detecting bacteria and differentiating them from mammalian cells. This is certainly true pathologically where Gram staining can identify peptidoglycan-rich bacteria such as S. aureus as well as identify their typical morphology. Numerous antibiotics are based on inhibition of peptidoglycan-processing enzymes, for example the penicillins, cephalosporins, and vancomycin70–72. More recently, microbe detection has become a critical focus for developing new chemistries. Many of these have employed D-amino acids which are incorporated into peptidoglycan via distinct pathways. Literature supports an intracellular, racemase-dependent assimilation of the canonical D-amino acids D-alanine and D-glutamine73,74, but a more permissive “swapping” of muropeptide DAAs in the periplasm whereby D-alanine can be substituted for an introduced substrate75. This mechanism has been used extensively by FDAAs in recent literature, via either direct incorporation of a FDAA or use of a bio-orthogonal DAA/ fluorescent detector pair. One exciting feature of fluorescence-based methods is the potential to build a full palette of frequency-specific molecules76.
More recently the use of DAA-derived sensors for PET has been explored, using 11C-labeled DAAs whose chemical structures match those of endogenous peptidoglycan substrates31,32. Numerous data have shown that while exogenous D-alanine and D-glutamine show the most avid incorporation into bacteria, other DAA show significant accumulation75,77. Since this is true for D-methionine, a radiosynthesis of D-[11C]met was developed, that showed accumulation of signal in living bacteria but not heat-killed inoculation31. This study set the stage for developing the more synthetically challenging DAA substrates like D-[11C]ala that show an order of magnitude enhancement of sensitivity to bacteria33. One limitation that has been inadequately addressed is the metabolic fate of D-amino acid derived structures in mammals. For example, mammals may convert D-amino acids to their corresponding L-amino acids via the activity of D-amino acid racemases78 or to their corresponding α-keto acids via D-amino acid oxidases79. For example, D-alanine itself can be converted to pyruvate, with rapid subsequent metabolism in bacteria and mammalian tissues. Developing D-amino acid sensors that are resistant to these and other host metabolic pathways represents a critical component of imaging probe development.
While D-amino acid-derived probes target peptidoglycan muropeptides, another approach to detecting the cell wall uses bacteria-specific sugars especially muramic acid and N-acetyl muramic acid. The sugar backbone of peptidoglycan is composed of this amine sugar as well as N-acetyl glucosamine making a repeating “NAM-NAG” structure. A recently published method uses a modified N-acetyl muramic acid that is incorporated into peptidoglycan, and subsequently detected using a bio-orthogonal reaction80 as highlighted in Figure 7. In contrast N-acetyl glucosamine is a substrate for bacterial, mammalian, and fungal metabolism and thus derived imaging methods derived from this structure would likely lack the desired bacterial specificity. In fungi, N-acetyl glucosamine molecules connected via β-(1,4) linkages form chitin, a primary component of the cell wall.81 In humans, N-acetyl glucosamine is used as a treatment for osteoarthritis and inflammatory bowel disease (IBD) including Crohn’s disease and ulcerative colitis.82 The first in vivo study using N-[18F]fluoroacetyl-D-glucosamine targeted tumors, based on the hypothesis that the hyaluronic acid concentration is a tumor biomarker.83 A more recent study developed a new radiosynthesis of this compound, that was used to image E. coli infection in rats.84 Although, N-[18F]fluoroacetyl-D-glucosamine demonstrated selectivity to live bacterial infection in comparison to sterile inflammation, FDG still outperformed this probe with 3-fold greater accumulation in the targeted region.
Figure 7.
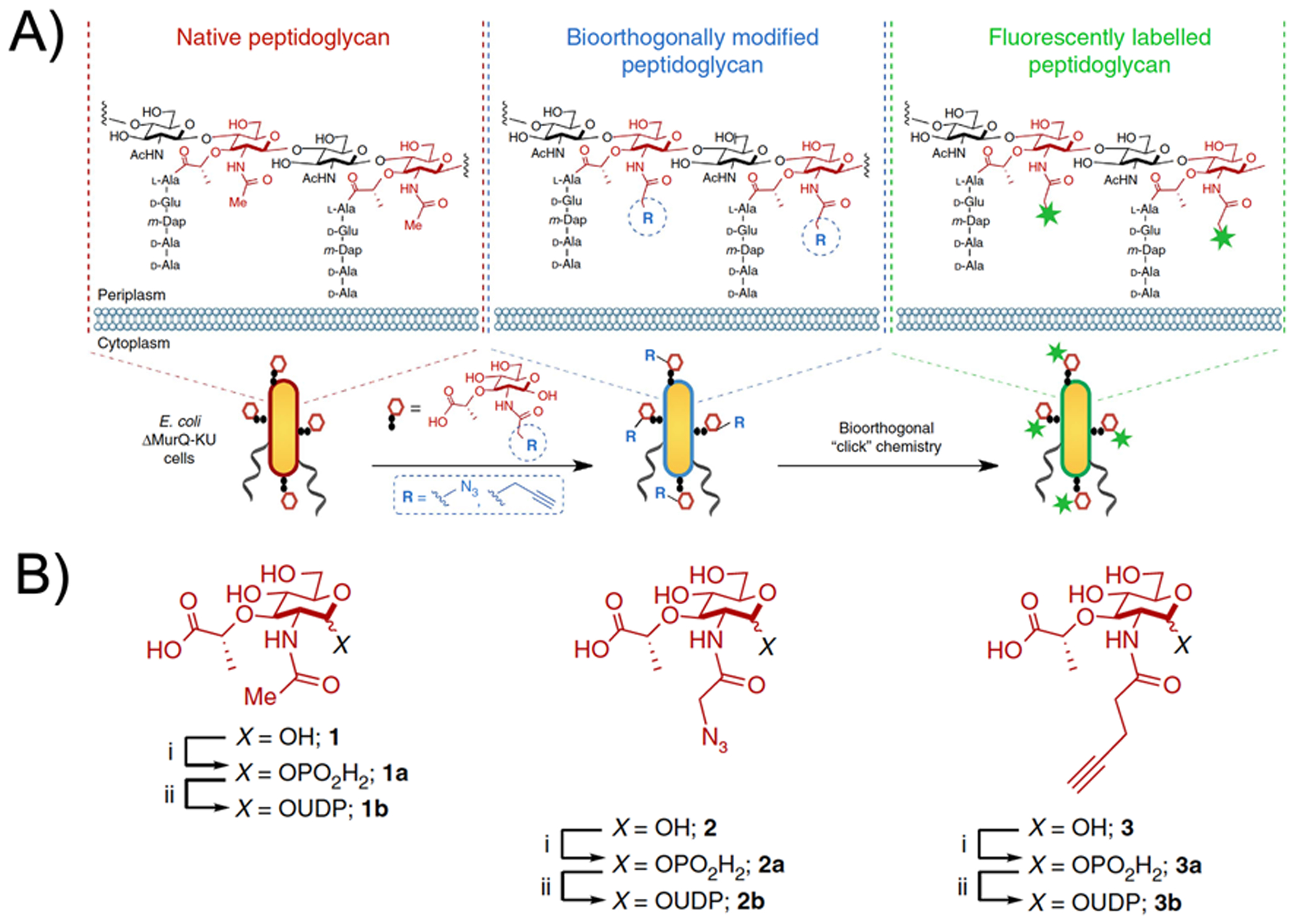
Use of bio-orthogonal chemistry to detect peptidoglycan incorporation of N-acetyl muramic acid (adapted from Liang et al. 2017). (A) The chemical structure of native peptidoglycan, with biorthogonal modified peptidoglycan and site(s) of fluorescent labelling indicated. (B) Structures of N-acetyl muramic acid monomer and its functionalized derivatives.
2. Gram-negative specific components:
The marked uptake of [18F]FDS in Enterobacteriaceae relative to gram-positive bacteria highlights the possibility of specifically imaging this class of bacteria.7 The accumulation of [18F]FDS appears to be transporter and kinase dependent, but there are also structural constituents of the gram-negative cell wall that might be targeted for specific detection of this class. For example, a major component of the outer membrane of bacteria are lipopolysaccharides (LPS) also known as “endotoxin.”85 LPS is composed of three 3 structural components. O-antigen, the outermost polysaccharide domain. The core domain, containing the sugar 3-deoxy-D-manno-oct-2-ulosonic acid (KDO). Lipid A is a phosphorylated glucosamine disaccharide connected to several fatty acids. In the last several years, a chemical strategy of in vitro sensing LPS has been reported and used based on the bacteria-specific KDO structure38,86,87. As highlighted in Figure 8, this labeling technique uses an azide-modified analogue of 3-deoxy-D-manno-octulosonic acid. Click chemistry can subsequently be used to detect metabolic incorporation of this moiety.
Figure 8.
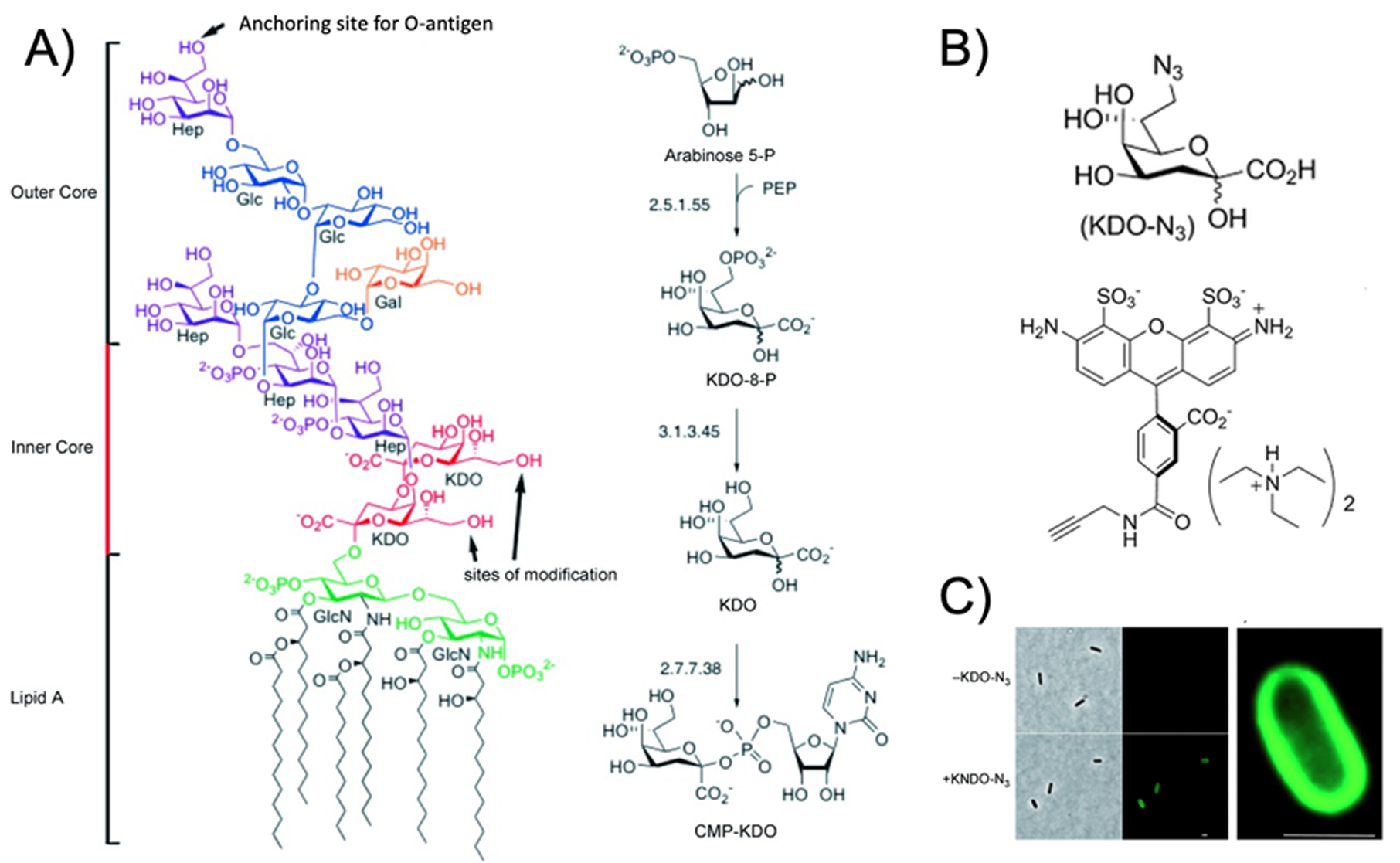
Use of bio-orthogonal chemistry to detect incorporation of 3-deoxy-D-manno-octulosonic acid (KDO) in gram-negative bacteria (adapted from Dumont et al. 2012). (A) The chemical structure of lipopolysaccharide with KDO content highlighted, with biorthogonal modified peptidoglycan and site(s) of fluorescent labelling indicated. (B) Structures of modified KDO (KDO-N3) and alkyne-bearing fluorescent dye used (C) Fluorescent microscopy showing detection of bacteria using KDO-N3 with magnified view showing localization of signal to the membrane.
3. Gram-positive specific components:
As previously discussed, strategies targeting peptidoglycan should inherently produce higher signal in gram-positive organisms, although recent PET experiments highlight that this is not always the case32. Teichoic acids are polymers of sugar alcohols (ribitol, glycerol) linked to carbohydrates via phosphodiester bonds88. They are found within the cell wall of several important gram-positive pathogens including the genera Staphylococcus and Streptococcus. Teichoic acids can be either tethered to components of peptidoglycan more superficially (in particular D-alanine and muramic acid) or anchored to the lipid membrane and referred to as lipoteichoic acids (LTA’s). Several components involved in the biosynthesis of teichoic acids are the “Tar” enzymes: TarA, TarB, TarF, TarK, TarL, TarO.89 Understanding the roles and expressions of these enzymes would be crucial in targeting teichoic acid components for gram-positive detection via a chemical biology approach, which to our knowledge has not been reported. Recent papers have described fluorescence microscopy and force microscopy imaging of cell wall teichoic acids, concluding that the distribution of wall teichoic acids affects cell morphology, elongation, and division.90 In addition, an antibody-based imaging method has been explored to detect gram-positive organisms. Specifically, an 89Zr-labeled antibody (anti-LTA mAb) specific for LTA ([89Zr]SAC55) was synthesized, that exhibited specific binding in vitro (2 fold increase over background) to LTA-expressing bacteria.91 The potential of this method was highlighted via in vivo studies that showed statistically significant distinction of infection over sterile implant sites. Finally, a series of structures have been reported that are sensitive to the glycoprotein structure of the gram-positive cell surface, but whose precise mechanism of detection has not been elucidated. These include boronic acid-containing probes for example the recent BODIPY derivative BacGO92, hexidium93, and wheat-germ agglutinin94.
H. Species-specific cell wall structures:
Detecting a single species can frequently be of high clinical interest, especially in a suspected case of tuberculosis. Imaging specific to M. tuberculosis would be very important because this disease is so difficult to diagnose and treat. The complex cell wall of mycobacteria95 contains numerous possibilities to sense this bacteria. For example, several reports have described sensing trehalose glycolipids, with related strategies explored for fluorescence and nuclear imaging96,97. Other examples of potentially species-specific structures are siderophores and their associated membrane receptors98,99.
I. Conclusions:
Sensors of bacterial cell wall components have taken a central role in elucidating microbial behavior, and are expected to impact diagnostic imaging tools in the near future. As highlighted in this review, there is tremendous synergy between fluorescence-based tools that are primarily used in vitro, and radiotracers developed for in vivo PET and SPECT imaging. Several of the structural components found in gram-positive and gram-negative bacterial cell walls are not seen in humans or other pathogens, representing a strong foundation for bacteria-specific imaging. Patient-compatible tracers based on this premise will join a complement of innovative imaging methods bound for clinical use. In the field of bacteria-specific imaging, the diversity of approaches considered represents a major strength moving forward, with the potential to benefit many acutely ill patients.
Synopsis:
Cell-wall targeted probes, labeled for both fluorescence microscopy and nuclear imaging can improve our understanding of microbial behavior in vitro and in vivo.
Funding sources:
Grant sponsors NIH R01EB024014, NIH R01EB025985, DOD A132172. UCSF Resource Allocation Program.
Bibliography:
- (1).Treglia G Diagnostic Performance of 18F-FDG PET/CT in Infectious and Inflammatory Diseases according to Published Meta-Analyses. Contrast Media Mol. Imaging 2019, 2019, 3018349 DOI: 10.1155/2019/3018349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Vaidyanathan S; Patel CN; Scarsbrook AF; Chowdhury FU FDG PET/CT in infection and inflammation--current and emerging clinical applications. Clin Radiol 2015, 70, 787–800 DOI: 10.1016/j.crad.2015.03.010. [DOI] [PubMed] [Google Scholar]
- (3).Magnuson JE; Brown ML; Hauser MF; Berquist TH; Fitzgerald RH; Klee GG In-111-labeled leukocyte scintigraphy in suspected orthopedic prosthesis infection: comparison with other imaging modalities. Radiology 1988, 168, 235–239 DOI: 10.1148/radiology.168.1.3380966. [DOI] [PubMed] [Google Scholar]
- (4).Bar-Shalom R; Yefremov N; Guralnik L; Keidar Z; Engel A; Nitecki S; Israel O SPECT/CT using 67Ga and 111In-labeled leukocyte scintigraphy for diagnosis of infection. J. Nucl. Med 2006, 47, 587–594. [PubMed] [Google Scholar]
- (5).Erba PA; Leo G; Sollini M; Tascini C; Boni R; Berchiolli RN; Menichetti F; Ferrari M; Lazzeri E; Mariani G Radiolabelled leucocyte scintigraphy versus conventional radiological imaging for the management of late, low-grade vascular prosthesis infections. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 357–368 DOI: 10.1007/s00259-013-2582-9. [DOI] [PubMed] [Google Scholar]
- (6).Kumar V; Boddeti DK (68)Ga-radiopharmaceuticals for PET imaging of infection and inflammation. Recent Results Cancer Res 2013, 194, 189–219 DOI: 10.1007/978-3-642-27994-2_11. [DOI] [PubMed] [Google Scholar]
- (7).Weinstein EA; Ordonez AA; DeMarco VP; Murawski AM; Pokkali S; MacDonald EM; Klunk M; Mease RC; Pomper MG; Jain SK Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci Transl Med 2014, 6, 259ra146 DOI: 10.1126/scitranslmed.3009815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mutch CA; Ordonez AA; Qin H; Parker M; Bambarger LE; Villanueva-Meyer JE; Blecha J; Carroll V; Taglang C; Flavell R; et al. [11C]Para-Aminobenzoic Acid: A Positron Emission Tomography Tracer Targeting Bacteria-Specific Metabolism. ACS Infect. Dis 2018, 4, 1067–1072 DOI: 10.1021/acsinfecdis.8b00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhang Z; Ordonez AA; Wang H; Li Y; Gogarty KR; Weinstein EA; Daryaee F; Merino J; Yoon GE; Kalinda AS; et al. Positron Emission Tomography Imaging with 2-[18F]F- p-Aminobenzoic Acid Detects Staphylococcus aureus Infections and Monitors Drug Response. ACS Infect. Dis 2018, 4, 1635–1644 DOI: 10.1021/acsinfecdis.8b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sellmyer MA; Lee I; Hou C; Weng C-C; Li S; Lieberman BP; Zeng C; Mankoff DA; Mach RH Bacterial infection imaging with [18F]fluoropropyl-trimethoprim. Proc. Natl. Acad. Sci. USA 2017, 114, 8372–8377 DOI: 10.1073/pnas.1703109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gilbert JA; Blaser MJ; Caporaso JG; Jansson JK; Lynch SV; Knight R Current understanding of the human microbiome. Nat. Med 2018, 24, 392–400 DOI: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wright EK; Kamm MA; Teo SM; Inouye M; Wagner J; Kirkwood CD Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflamm. Bowel Dis 2015, 21, 1219–1228 DOI: 10.1097/MIB.0000000000000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mahdavinia M; Keshavarzian A; Tobin MC; Landay AL; Schleimer RP A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin. Exp. Allergy 2016, 46, 21–41 DOI: 10.1111/cea.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Haldar K; Bafadhel M; Lau K; Berg A; Kwambana B; Kebadze T; Ramsheh MY; Barker B; Haldar P; Johnston S; et al. Microbiome balance in sputum determined by PCR stratifies COPD exacerbations and shows potential for selective use of antibiotics. PLoS One 2017, 12, e0182833 DOI: 10.1371/journal.pone.0182833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Dumont RA; Keen NN; Bloomer CW; Schwartz BS; Talbott J; Clark AJ; Wilson DM; Chin CT Clinical Utility of Diffusion-Weighted Imaging in Spinal Infections. Clin Neuroradiol 2018, 1–8 DOI: 10.1007/s00062-018-0681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Talbott JF; Shah VN; Uzelac A; Narvid J; Dumont RA; Chin CT; Wilson DM Imaging-Based Approach to Extradural Infections of the Spine. Semin Ultrasound CT MR 2018, 39, 570–586 DOI: 10.1053/j.sult.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zimmer V; Lammert F Acute Bacterial Cholangitis. Viszeralmedizin 2015, 31, 166–172 DOI: 10.1159/000430965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Aghdassi AA; Mayerle J; Kraft M; Sielenkämper AW; Heidecke C-D; Lerch MM Pancreatic pseudocysts--when and how to treat? HPB (Oxford) 2006, 8, 432–441 DOI: 10.1080/13651820600748012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sanverdi SE; Ergen BF; Oznur A Current challenges in imaging of the diabetic foot. Diabet. Foot Ankle 2012, 3 DOI: 10.3402/dfa.v3i0.18754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bhagirath AY; Li Y; Somayajula D; Dadashi M; Badr S; Duan K Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med 2016, 16, 174 DOI: 10.1186/s12890-016-0339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Weiner RE The mechanism of 67Ga localization in malignant disease. Nucl Med Biol 1996, 23, 745–751 DOI: 10.1016/0969-8051(96)00119-9. [DOI] [PubMed] [Google Scholar]
- (22).Goldsmith SJ; Vallabhajosula S Clinically proven radiopharmaceuticals for infection imaging: mechanisms and applications. Semin Nucl Med 2009, 39, 2–10 DOI: 10.1053/j.semnuclmed.2008.08.002. [DOI] [PubMed] [Google Scholar]
- (23).Slough C; Masters SC; Hurley RA; Taber KH Clinical positron emission tomography (PET) neuroimaging: advantages and limitations as a diagnostic tool. J. Neuropsychiatry Clin. Neurosci 2016, 28, A4, 67–71 DOI: 10.1176/appi.neuropsych.16030044. [DOI] [PubMed] [Google Scholar]
- (24).Raji CA; Tarzwell R; Pavel D; Schneider H; Uszler M; Thornton J; van Lierop M; Cohen P; Amen DG; Henderson T Clinical utility of SPECT neuroimaging in the diagnosis and treatment of traumatic brain injury: a systematic review. PLoS One 2014, 9, e91088 DOI: 10.1371/journal.pone.0091088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Assessment of brain SPECT. Report of the Therapeutics and Technology Assessment Subcommittee of the American Academy of Neurology. Neurology 1996, 46, 278–285. [DOI] [PubMed] [Google Scholar]
- (26).Ullah MN; Pratiwi E; Cheon J; Choi H; Yeom JY Instrumentation for Time-of-Flight Positron Emission Tomography. Nucl. Med. Mol. Imaging 2016, 50, 112–122 DOI: 10.1007/s13139-016-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Neumann K; Flavell R; Wilson DM Exploring metabolism in vivo using endogenous 11C metabolic tracers. Semin Nucl Med 2017, 47, 461–473 DOI: 10.1053/j.semnuclmed.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ning X; Lee S; Wang Z; Kim D; Stubblefield B; Gilbert E; Murthy N Maltodextrin-based imaging probes detect bacteria in vivo with high sensitivity and specificity. Nat. Mater 2011, 10, 602–607 DOI: 10.1038/nmat3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ning X; Seo W; Lee S; Takemiya K; Rafi M; Feng X; Weiss D; Wang X; Williams L; Camp VM; et al. PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem. Int. Ed. Engl 2014, 53, 14096–14101 DOI: 10.1002/anie.201408533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kuru E; Hughes HV; Brown PJ; Hall E; Tekkam S; Cava F; de Pedro MA; Brun YV; VanNieuwenhze MS In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem. Int. Ed. Engl 2012, 51, 12519–12523 DOI: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Neumann KD; Villanueva-Meyer JE; Mutch CA; Flavell RR; Blecha JE; Kwak T; Sriram R; VanBrocklin HF; Rosenberg OS; Ohliger MA; et al. Imaging Active Infection in vivo Using D-Amino Acid Derived PET Radiotracers. Sci. Rep 2017, 7, 7903 DOI: 10.1038/s41598-017-08415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stewart MN; Parker MFL; Jivan S; Luu JM; Huynh TL; Schulte B; Seo Y; Blecha JE; Villanueva-Meyer JE; Flavell RR; et al. High Enantiomeric Excess In-Loop Synthesis of d-[methyl-11C]Methionine for Use as a Diagnostic Positron Emission Tomography Radiotracer in Bacterial Infection. ACS Infect. Dis 2019. DOI: 10.1021/acsinfecdis.9b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Parker MFL; Luu JM; Schulte B; Huynh TL; Stewart MN; Sriram R; Yu MA; Jivan S; Turnbaugh PJ; Flavell RR; et al. Sensing Living Bacteria in Vivo Usingd -Alanine-Derived11 C Radiotracers. ACS Cent. Sci 2020. DOI: 10.1021/acscentsci.9b00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Fura JM; Kearns D; Pires MM D-Amino Acid Probes for Penicillin Binding Protein-based Bacterial Surface Labeling. J. Biol. Chem 2015, 290, 30540–30550 DOI: 10.1074/jbc.M115.683342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Thirumurugan P; Matosiuk D; Jozwiak K Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev 2013, 113, 4905–4979 DOI: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- (36).Siegrist MS; Whiteside S; Jewett JC; Aditham A; Cava F; Bertozzi CR (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol 2013, 8, 500–505 DOI: 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).MacKenzie DA; Sherratt AR; Chigrinova M; Kell AJ; Pezacki JP Bioorthogonal labelling of living bacteria using unnatural amino acids containing nitrones and a nitrone derivative of vancomycin. Chem. Commun. (Camb) 2015, 51, 12501–12504 DOI: 10.1039/c5cc04901f. [DOI] [PubMed] [Google Scholar]
- (38).Dumont A; Malleron A; Awwad M; Dukan S; Vauzeilles B Click-mediated labeling of bacterial membranes through metabolic modification of the lipopolysaccharide inner core. Angew. Chem. Int. Ed. Engl 2012, 51, 3143–3146 DOI: 10.1002/anie.201108127. [DOI] [PubMed] [Google Scholar]
- (39).Hou S; Choi J-S; Garcia MA; Xing Y; Chen K-J; Chen Y-M; Jiang ZK; Ro T; Wu L; Stout DB; et al. Pretargeted Positron Emission Tomography Imaging That Employs Supramolecular Nanoparticles with in Vivo Bioorthogonal Chemistry. ACS Nano 2016, 10, 1417–1424 DOI: 10.1021/acsnano.5b06860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Pretze M; Pietzsch D; Mamat C Recent trends in bioorthogonal click-radiolabeling reactions using fluorine-18. Molecules 2013, 18, 8618–8665 DOI: 10.3390/molecules18078618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Billaud EMF; Belderbos S; Cleeren F; Maes W; Van de Wouwer M; Koole M; Verbruggen A; Himmelreich U; Geukens N; Bormans G Pretargeted PET Imaging Using a Bioorthogonal 18F-Labeled trans-Cyclooctene in an Ovarian Carcinoma Model. Bioconjug Chem 2017, 28, 2915–2920 DOI: 10.1021/acs.bioconjchem.7b00635. [DOI] [PubMed] [Google Scholar]
- (42).Peplow M Click chemistry targets antibody-drug conjugates for the clinic. Nat. Biotechnol 2019. DOI: 10.1038/d41587-019-00017-4. [DOI] [PubMed] [Google Scholar]
- (43).Kim E; Koo H Biomedical applications of copper-free click chemistry: in vitro, in vivo, and ex vivo. Chem. Sci 2019, 10, 7835–7851 DOI: 10.1039/c9sc03368h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhao G; Meier TI; Kahl SD; Gee KR; Blaszczak LC BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother 1999, 43, 1124–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Tiyanont K; Doan T; Lazarus MB; Fang X; Rudner DZ; Walker S Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. USA 2006, 103, 11033–11038 DOI: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Daniel RA; Errington J Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 2003, 113, 767–776 DOI: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- (47).Newton BA A fluorescent derivative of polymyxin: its preparation and use in studying the site of action of the antibiotic. J. Gen. Microbiol 1955, 12, 226–236 DOI: 10.1099/00221287-12-2-226. [DOI] [PubMed] [Google Scholar]
- (48).Jalilian AR; Hosseini MA; Majdabadi…, A. Evaluation of [201 Tl](III) Vancomycin in normal rats. Nuclear Medicine … 2008. [PubMed] [Google Scholar]
- (49).Stone MRL; Butler MS; Phetsang W; Cooper MA; Blaskovich MAT Fluorescent antibiotics: new research tools to fight antibiotic resistance. Trends Biotechnol 2018, 36, 523–536 DOI: 10.1016/j.tibtech.2018.01.004. [DOI] [PubMed] [Google Scholar]
- (50).Mills B; Bradley M; Dhaliwal K Optical imaging of bacterial infections. Clin. Transl. Imaging 2016, 4, 163–174 DOI: 10.1007/s40336-016-0180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Gambhir SS Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer 2002, 2, 683–693 DOI: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- (52).Hope TA; Aggarwal R; Chee B; Tao D; Greene KL; Cooperberg MR; Feng F; Chang A; Ryan CJ; Small EJ; et al. Impact of 68Ga-PSMA-11 PET on Management in Patients with Biochemically Recurrent Prostate Cancer. J. Nucl. Med 2017, 58, 1956–1961 DOI: 10.2967/jnumed.117.192476. [DOI] [PubMed] [Google Scholar]
- (53).Auletta S; Baldoni D; Varani M; Galli F; Hajar IA; Duatti A; Ferro-Flores G; Trampuz A; Signore A Comparison of 99mTc-UBI 29–41, 99mTc-ciprofloxacin, 99mTc-ciprofloxacin dithiocarbamate and 111In-biotin for targeting experimental Staphylococcus aureus and Escherichia coli foreign-body infections: an ex-vivo study. Q. J. Nucl. Med. Mol. Imaging 2019, 63, 37–47 DOI: 10.23736/S1824-4785.17.02975-2. [DOI] [PubMed] [Google Scholar]
- (54).Langer O; Brunner M; Zeitlinger M; Ziegler S; Müller U; Dobrozemsky G; Lackner E; Joukhadar C; Mitterhauser M; Wadsak W; et al. In vitro and in vivo evaluation of [18F]ciprofloxacin for the imaging of bacterial infections with PET. Eur. J. Nucl. Med. Mol. Imaging 2005, 32, 143–150 DOI: 10.1007/s00259-004-1646-2. [DOI] [PubMed] [Google Scholar]
- (55).Ordonez AA; Wang H; Magombedze G; Ruiz-Bedoya CA; Srivastava S; Chen A; Tucker EW; Urbanowski ME; Pieterse L; Fabian Cardozo E; et al. Dynamic imaging in patients with tuberculosis reveals heterogeneous drug exposures in pulmonary lesions. Nat. Med 2020. DOI: 10.1038/s41591-020-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Li J; Zheng H; Fodah RA; Warawa J; Ng CK Validation of 2-(18)F-fluorodeoxysorbitol ((18)F-FDS) as a potential radiopharmaceutical for imaging bacterial infection in the lung. J. Nucl. Med 2017, 59, 134–139 DOI: 10.2967/jnumed.117.195420. [DOI] [PubMed] [Google Scholar]
- (57).Yao S; Xing H; Zhu W; Wu Z; Zhang Y; Ma Y; Liu Y; Huo L; Zhu Z; Li Z; et al. Infection Imaging With (18)F-FDS and First-in-Human Evaluation. Nucl Med Biol 2016, 43, 206–214 DOI: 10.1016/j.nucmedbio.2015.11.008. [DOI] [PubMed] [Google Scholar]
- (58).Gowrishankar G; Namavari M; Jouannot EB; Hoehne A; Reeves R; Hardy J; Gambhir SS Investigation of 6-[18F]-fluoromaltose as a novel PET tracer for imaging bacterial infection. PLoS One 2014, 9, e107951 DOI: 10.1371/journal.pone.0107951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Namavari M; Gowrishankar G; Hoehne A; Jouannot E; Gambhir SS Synthesis of [18F]-labelled maltose derivatives as PET tracers for imaging bacterial infection. Mol. Imaging Biol 2015, 17, 168–176 DOI: 10.1007/s11307-014-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Gowrishankar G; Hardy J; Wardak M; Namavari M; Reeves RE; Neofytou E; Srinivasan A; Wu JC; Contag CH; Gambhir SS Specific Imaging of Bacterial Infection Using 6″−18F-Fluoromaltotriose: A Second-Generation PET Tracer Targeting the Maltodextrin Transporter in Bacteria. J. Nucl. Med 2017, 58, 1679–1684 DOI: 10.2967/jnumed.117.191452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kalita M; Parker MFL; Luu JM; Stewart MN; Blecha JE; VanBrocklin HF; Evans MJ; Flavell RR; Rosenberg OS; Ohliger MA; et al. Arabinofuranose-derived positron-emission tomography radiotracers for detection of pathogenic microorganisms. J. Labelled Comp. Radiopharm 2020. DOI: 10.1002/jlcr.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Petrik M; Umlaufova E; Raclavsky V; Palyzova A; Havlicek V; Haas H; Novy Z; Dolezal D; Hajduch M; Decristoforo C Imaging of Pseudomonas aeruginosa infection with Ga-68 labelled pyoverdine for positron emission tomography. Sci. Rep 2018, 8, 15698 DOI: 10.1038/s41598-018-33895-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Auletta S; Varani M; Horvat R; Galli F; Signore A; Hess S PET radiopharmaceuticals for specific bacteria imaging: A systematic review. J Clin Med 2019, 8 DOI: 10.3390/jcm8020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Ordonez AA; Sellmyer MA; Gowrishankar G; Ruiz-Bedoya CA; Tucker EW; Palestro CJ; Hammoud DA; Jain SK Molecular imaging of bacterial infections: Overcoming the barriers to clinical translation. Sci Transl Med 2019, 11 DOI: 10.1126/scitranslmed.aax8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Lawal I; Zeevaart J; Ebenhan T; Ankrah A; Vorster M; Kruger HG; Govender T; Sathekge M Metabolic imaging of infection. J. Nucl. Med 2017, 58, 1727–1732 DOI: 10.2967/jnumed.117.191635. [DOI] [PubMed] [Google Scholar]
- (66).Boos W; Shuman H Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev 1998, 62, 204–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Carroll VN; Truillet C; Shen B; Flavell RR; Shao X; Evans MJ; VanBrocklin HF; Scott PJH; Chin FT; Wilson DM [(11)C]Ascorbic and [(11)C]dehydroascorbic acid, an endogenous redox pair for sensing reactive oxygen species using positron emission tomography. Chem. Commun. (Camb) 2016, 52, 4888–4890 DOI: 10.1039/c6cc00895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Bugg TD; Walsh CT Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep 1992, 9, 199–215. [DOI] [PubMed] [Google Scholar]
- (69).Lam H; Oh D-C; Cava F; Takacs CN; Clardy J; de Pedro MA; Waldor MK D-amino acids govern stationary phase cell wall remodeling in bacteria. Science (80-. ) 2009, 325, 1552–1555 DOI: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Lobanovska M; Pilla G Penicillin’s discovery and antibiotic resistance: lessons for the future? Yale J Biol Med 2017, 90, 135–145. [PMC free article] [PubMed] [Google Scholar]
- (71).Kalman D; Barriere SL Review of the pharmacology, pharmacokinetics, and clinical use of cephalosporins. Tex Heart Inst J 1990, 17, 203–215. [PMC free article] [PubMed] [Google Scholar]
- (72).Levine DP Vancomycin: a history. Clin. Infect. Dis 2006, 42 Suppl 1, S5–12 DOI: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- (73).Wei Y; Qiu W; Zhou X-D; Zheng X; Zhang K-K; Wang S-D; Li Y-Q; Cheng L; Li J-Y; Xu X; et al. Alanine racemase is essential for the growth and interspecies competitiveness of Streptococcus mutans. Int. J. Oral Sci 2016, 8, 231–238 DOI: 10.1038/ijos.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Azam MA; Jayaram U Inhibitors of alanine racemase enzyme: a review. J Enzyme Inhib Med Chem 2016, 31, 517–526 DOI: 10.3109/14756366.2015.1050010. [DOI] [PubMed] [Google Scholar]
- (75).Caparrós M; Pisabarro AG; de Pedro MA Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol 1992, 174, 5549–5559 DOI: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Hsu Y-P; Rittichier J; Kuru E; Yablonowski J; Pasciak E; Tekkam S; Hall E; Murphy B; Lee TK; Garner EC; et al. Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem. Sci 2017, 8, 6313–6321 DOI: 10.1039/c7sc01800b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Caparrós M; Arán V; de Pedro MA Incorporation of S-[3H]methyl-D-cysteine into the peptidoglycan of ether-treated cells of Escherichia coli. FEMS Microbiol. Lett 1992, 72, 139–146 DOI: 10.1016/0378-1097(92)90519-t. [DOI] [PubMed] [Google Scholar]
- (78).Yoshimura T; Esaki N Amino Acid Racemases: Functions and Mechanisms. J. Biosci. Bioeng 2003, 96, 103–109 DOI: 10.1263/jbb.96.103. [DOI] [PubMed] [Google Scholar]
- (79).Kawazoe T; Park HK; Iwana S; Tsuge H Human D‐amino acid oxidase: an update and review. The Chemical … 2007. [DOI] [PubMed] [Google Scholar]
- (80).Liang H; DeMeester KE; Hou C-W; Parent MA; Caplan JL; Grimes CL Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat Commun 2017, 8, 15015 DOI: 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zargar V; Asghari M; Dashti A A review on chitin and chitosan polymers: structure, chemistry, solubility, derivatives, and applications. ChemBioEng Reviews 2015. [Google Scholar]
- (82).Salvatore S; Heuschkel R; Tomlin S; Davies SE; Edwards S; Walker-Smith JA; French I; Murch SH A pilot study of N-acetyl glucosamine, a nutritional substrate for glycosaminoglycan synthesis, in paediatric chronic inflammatory bowel disease. Aliment. Pharmacol. Ther 2000, 14, 1567–1579. [DOI] [PubMed] [Google Scholar]
- (83).Fujiwara T; Kubota K; Sato T; Matsuzawa T; Tada M; Iwata R; Itoh M; Hatazawa J; Sato K; Fukuda H N-[18F]fluoroacetyl-D-glucosamine: a potential agent for cancer diagnosis. J. Nucl. Med 1990, 31, 1654–1658. [PubMed] [Google Scholar]
- (84).Martínez ME; Kiyono Y; Noriki S; Inai K; Mandap KS; Kobayashi M; Mori T; Tokunaga Y; Tiwari VN; Okazawa H; et al. New radiosynthesis of 2-deoxy-2-[(18)F]fluoroacetamido-D-glucopyranose and its evaluation as a bacterial infections imaging agent. Nucl Med Biol 2011, 38, 807–817 DOI: 10.1016/j.nucmedbio.2011.02.006. [DOI] [PubMed] [Google Scholar]
- (85).Raetz CRH; Whitfield C Lipopolysaccharide endotoxins. Annu. Rev. Biochem 2002, 71, 635–700 DOI: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Nilsson I; Grove K; Dovala D; Uehara T; Lapointe G; Six DA Molecular characterization and verification of azido 3,8-dideoxy-D-manno-oct-2-ulosonic acid incorporation into bacterial lipopolysaccharide. J. Biol. Chem 2017. DOI: 10.1074/jbc.M117.814962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Fugier E; Dumont A; Malleron A; Poquet E; Mas Pons J; Baron A; Vauzeilles B; Dukan S Rapid and Specific Enrichment of Culturable Gram Negative Bacteria Using Non-Lethal Copper-Free Click Chemistry Coupled with Magnetic Beads Separation. PLoS One 2015, 10, e0127700 DOI: 10.1371/journal.pone.0127700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Brown S; Santa Maria JP; Walker S Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol 2013, 67, 313–336 DOI: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Swoboda JG; Campbell J; Meredith TC; Walker S Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 2010, 11, 35–45 DOI: 10.1002/cbic.200900557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Andre G; Deghorain M; Bron PA; van Swam II; Kleerebezem M; Hols P; Dufrene YF Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem. Biol 2011, 6, 366–376 DOI: 10.1021/cb1003509. [DOI] [PubMed] [Google Scholar]
- (91).Pickett JE; Thompson JM; Sadowska A; Tkaczyk C; Sellman BR; Minola A; Corti D; Lanzavecchia A; Miller LS; Thorek DL Molecularly specific detection of bacterial lipoteichoic acid for diagnosis of prosthetic joint infection of the bone. Bone Res 2018, 6, 13 DOI: 10.1038/s41413-018-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Kwon H-Y; Liu X; Choi EG; Lee JY; Choi S-Y; Kim J-Y; Wang L; Park S-J; Kim B; Lee Y-A; et al. Development of universal fluorescent Gram-positive bacteria probe. Angew. Chem. Int. Ed. Engl 2019. DOI: 10.1002/anie.201902537. [DOI] [PubMed] [Google Scholar]
- (93).Mason DJ; Shanmuganathan S; Mortimer FC; Gant VA A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl. Environ. Microbiol 1998, 64, 2681–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Cash HL; Whitham CV; Behrendt CL; Hooper LV Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (80-. ) 2006, 313, 1126–1130 DOI: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Alderwick LJ; Harrison J; Lloyd GS; Birch HL The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan. Cold Spring Harb. Perspect. Med 2015, 5, a021113 DOI: 10.1101/cshperspect.a021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Kamariza M; Shieh P; Bertozzi CR Imaging mycobacterial trehalose glycolipids; 2018; Vol. 598, pp. 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Rundell SR; Wagar ZL; Meints LM; Olson CD; O’Neill MK; Piligian BF; Poston AW; Hood RJ; Woodruff PJ; Swarts BM Deoxyfluoro-d-trehalose (FDTre) analogues as potential PET probes for imaging mycobacterial infection. Org. Biomol. Chem 2016, 14, 8598–8609 DOI: 10.1039/c6ob01734g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol. Med 2016, 22, 1077–1090 DOI: 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Petrik M; Zhai C; Haas H; Decristoforo C Siderophores for molecular imaging applications. Clin. Transl. Imaging 2017, 5, 15–27 DOI: 10.1007/s40336-016-0211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Slavin YN; Asnis J; Häfeli UO; Bach H Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnology 2017, 15, 65 DOI: 10.1186/s12951-017-0308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]


