Abstract
Cellular differentiation is caused by highly controlled modifications in the gene expression but rarely involves a change in the DNA sequence itself. Histone acetylation is a major epigenetic factor that adds an acetyl group to histone proteins, thus altering their interaction with DNA and nuclear proteins. Illumination of the histone acetylation during dentinogenesis is important for odontoblast differentiation and dentinogenesis. In the current study, we aimed to discover the roles and regulation of acetylation at histone 3 lysine 9 (H3K9ac) and H3K27ac during dentinogenesis. We first found that both of these modifications were enhanced during odontoblast differentiation and dentinogenesis. These modifications are dynamically catalyzed by histone acetyltransferases (HATs) and deacetylases (HDACs), among which HDAC3 was decreased while p300 increased during odontoblast differentiation. Moreover, overexpression of HDAC3 or knockdown p300 inhibited odontoblast differentiation in vitro, and inhibition of HDAC3 and p300 with trichostatin A or C646 regulated odontoblast differentiation. Taken together, the results of our present study suggest that histone acetylation is involved in dentinogenesis and coordinated expression of p300- and HDAC3-regulated odontoblast differentiation through upregulating histone acetylation.
Keywords: cell differentiation, dentinogenesis, HDAC3, histone acetylation, p300
1 |. INTRODUCTION
Multiple signaling pathways and transcriptional factors converge to induce cell proliferation, differentiation, and biomineralization during dentinogenesis. Signaling pathways, such as sonic hedgehog signaling, bare morphogenetic protein signaling, fibroblast growth factor signaling, Notch signaling, and Wnt/catenin signaling, are critical for tooth development.1 In our previous studies, KLF4, SP1, and QKI have been shown to be important factors that promote odontoblast differentiation.2–4 The appropriate temporal and spatial expression of these genes is vital to proper tooth development. And the specific gene expression patterns induced during cell differentiation are assumed to be regulated by several epigenetic factors such as microRNA (miRNA)-mediated posttranscriptional regulation and histone modification–mediated posttranslational modulation. Previously, we have shown that miRNAs such as miRNA-143, miRNA-145, and miRNA-338–3p could posttranscriptionally regulate KLF4, OSX, and RUNX25,6 and form a competing endogenous RNA-regulatory network governing odontoblastic differentiation.2,7 However, the functions of histone modification during odontoblastic differentiation are not completely understood.
Different histone modifications are associated with distinct biological functions.8–10 For instance, histone H3 and H4 are known as marks associated with active transcription.11 Distal regulatory regions, such as enhancers, are marked by the H3K27ac.12–14 Acetylation of H3K9 is also a mark of active transcription.15 The level of histone acetylation is catalyzed by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs).16,17 There are several families of HATs such as Gcn-related acetyltransferases, p300/cAMP response element-binding protein (CREB)-binding protein and p300/CREB binding protein-associated factor (P/CAF).18 HDACs have four classes: class I HDACs (HDAC1, −2, −3, and −8) are ubiquitously expressed, whereas class II HDACs (HDAC4, −5, −6, and −7) are highly expressed in certain tissues, such as brain, heart, and skeletal muscle.19 In addition, there are class III HDACs, also called Sirtuins, which include SIRT1, −2, −3, −4, −5, −6, and −7. Sirtuins differ in their subcellular localization and interact with transcriptional factors and nonhistone substrates.20 HDAC 11 is the only class IV HDAC and shows sequence similarity to class I and II HDACs. These two groups of enzymes can homeostatically maintain the proper level of acetylation modification. The balance of histone acetylation can be disturbed by histone deacetylase inhibitors (HDACi), which modify cellular processes, including cell cycle, angiogenesis, differentiation, apoptosis, and gene expression.21 Recent studies have demonstrated that HDACi stimulate odontoblast differentiation through upregulating odontoblastic-related genes such as Alp, Dmpl, Dspp, with increased mineralized nodules.22–25 Also, a previous report showed that p300 played a key role in regulating expression of key pluripotency genes in human dental pulp cells and in modifying odontogenic differentiation.26 On the basis of these, we hypothesized that histone acetylation might involve in odontoblast differentiation. Therefore, we sought to determine the role of histone acetylation during tooth development in this study, and we chose to study the role of H3K9ac and H3K27ac, which are two landmarks of active transcription. To this end, we investigated the expression of histone acetylases and found that the expression levels of p300 were increased while HDAC3 was decreased during odontoblast differentiation. Moreover, we found that p300 and HDAC3 themselves could regulate odontoblast differentiation. Thus, our data provided important insights into the regulation of dentinogenesis by histone acetylation.
2 |. MATERIALS AND METHODS
2.1 |. Animals and embryo collection
All mouse experiments were performed in accordance with the approval of Institutional Animal Care and Use Committees at the School and Hospital of Stomatology of Wuhan University (protocol no. S07917110D). The mandibles of Kunming mice at postnatal day 2 (PN 2) were dissected and then fixed individually in 4% paraformaldehyde at 4°C for 24 hours. The sample was decalcified for 2 days in 10% ethylenediaminetetraacetic acid and then processed for paraffin section for histological analysis. At least three mice were used for each study in histological analysis.
2.2 |. Cell culture and reagents
Mouse dental papilla mesenchymal cells (mDPCs) were isolated from the first molars of PN 0.5 from Kunming mice and then digested at 37°C for 1 hour in a solution of 4 mg/mL of dispase and 3 mg/mL collagenase type I. The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, UT) containing 10% fetal bovine serum (FBS) (HyClone), plus streptomycin (100 μg/mL), and penicillin (100U/mL) at 37°C in a humidified atmosphere of air containing 5% CO2. The medium was changed every 2 days. After reaching confluence, the cells were spread into a new dish.
For odontoblast differentiation, the cells were cultured in DMEM containing 10% FBS, antibiotics, 10nmol/L dexamethasone, 50 μg/mL ascorbic acid, and 10mmol/L sodium β-glycerophosphate (Sigma, St Louis, MO).
Trichostatin A (TSA) and C646 were used at a concentration of 20 nM and 10 μM, respectively. They were both purchased from Selleck (Shanghai, China). Cells were treated with TSA or C646 for 24 hours before being cultured with differentiation medium.
2.3 |. Alizarin red staining
Mineral nodules were assessed by alizarin red staining (Sigma), after the cells were induced at indicated times. In brief, 1% alizarin red was prepared in distilled water and adjusted to pH 5.5. The cells were washed with phosphate-buffered saline (PBS) three times and then fixed with 95% ice-cold ethanol for 20 minutes. After three times wash with PBS, alizarin red was applied to the cells at room temperature for 30 minutes. Then the cells were washed with distilled water. When the calcified nodules were quantified, 10% cylpyridinium chloride (Sigma) solution was added to the stained calcified nodule samples overnight, and the solution was collected. The OD value of the solutions was measured by a microplate reader (wavelength = 540 nm).
2.4 |. Transfection
Cells were seeded in six-well plates and transfected according to the manufacturer’s instructions. Specifically, small interfering RNA (siRNA) of p300 (Gene Pharma, Shanghai, China) and scrambled siRNA (Invitrogen, Carlsbad, CA) was used at a final concentration of 80 nmol/L using Lipofectamine 2000 (Invitrogen). The HDAC3 overexpression plasmid was transfected at a final concentration of 1 μg, which was purchased from Sangon Biotech (Shanghai, China). After 48 hours of transfection, RNA and protein lysates were collected for further analysis.
2.5 |. Immunohistochemistry
Sagittal sections of the mice mandibular incisor at PN 2 were cut into 5 μm thick sections. Then the slides were de-waxed and rehydrated. Immunostaining was proceeded to follow the Vectastain ABC Elite kit protocols (Vector labs, Burlingame, CA). To be precise, the slides were incubated for 15 minutes in 3% hydrogen peroxide and boiled for 5 minutes (121°C) in 10 mmol/L citrate buffer (pH 6.0) for antigen retrieval. The slides were blocked with bovine serum albumin (BSA) and then incubated with antibodies individually against H3K9ac (1:200, 9649S; CST, MA) or H3K27ac (1:200, ab4729; Abcam, Cambridge, UK) at 4°C overnight. After incubation with horseradish peroxide (HRP) secondary antibody, the slides were counterstained with hematoxylin, mounted, and photographed.
2.6 |. Cell immunofluorescence
Cells were dispersed into single-cell and seeded on coverslips. The slides of cells were carefully washed with PBS and fixed with 4% paraformaldehyde at room temperature for 15 minutes. They were permeabilized with 0.25% Triton X-100 for 5 minutes, and then washed with PBS three times. 2.5% BSA was used to block nonspecific binding for 1 hour. Cells were incubated with antibodies against H3K9ac or H3K27ac individually at 4°C overnight. The cells were incubated with Cy3-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 1 hour after they were washed with PBS three times. Cells on coverslips were detected and photographed by a fluorescence microscope (Leica, Wetzlar, Germany). Nuclei were labeled with 4’,6-diamidino-2-phenylindole (ZSGB-BIO), and cellular F-actin was stained with FITC-phalloidin (Yeasen, Shanghai, China).
2.7 |. Quantitative reverse transcriptase polymerase chain reaction analysis
Total RNA was extracted from cultured cells using the HP Total RNA Kit (Omega bio-tech, Norcross, GA). The complementary DNA (cDNA) was synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc, Rockford, IL). Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed on an ABI 7500 real-time PCR System (Applied Biosystems, California) with SYBR Premix Ex TaqTM (Roche). Glyceraldehyde 3-phosphate dehydrogenase was used as an internal reference. The primers for qRT-PCR are shown in Table 1.
TABLE 1.
Oligonucleotide primer sequences used in the qRT-PCR
| Primer | Sequence 5′- 3′ |
|---|---|
| DSPP | Forward: GTGGGATCATCAGCCAGTCAG |
| Reverse: TGCCTTTGTTGGGACCTTCA | |
| DMP1 | Forward: ACCACAATACTGAATCTGAAAGCTC |
| Reverse: TGCTGTCCGTGTGGTCACTA | |
| ALP | Forward: CTGATGTGGAGTATGA |
| Reverse: TGTATCTCGGTTTGAA | |
| GAPDH | Forward: TGTGTCCGTCGTGGATCTGA |
| Reverse: TTGCTGTTGAAGTCGCAGGAG | |
| HDAC3 | Forward: TTGAAGATGCTGAACCATGC |
| Reverse: TGGCCTGCTGTAGTTCTCCT | |
| p300 | Forward: GAAGAACAGCCAAGCACCTC |
| Reverse: CGGTAAAGTGCCTCCAATGT |
Abbreviations: GAPDH, glyceraldehyde 3-phosphate dehydrogenase; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
2.8 |. Western blot analysis
Cells were harvested and lysed in lysis buffer and centrifuged for 15 minutes at 13 000 rpm. Total proteins were measured using the BCA Protein Assay Kit (Thermo). Equal amounts of proteins were loaded onto a 10% polyacrylamide gel and transferred to polyvinylidene fluoride membranes (Roche Diagnostics GmbH, Mannheim, Germany). The membranes were blocked with 5% nonfat milk for 1 hour at room temperature followed by incubation with the primary antibodies: DMP1 (a kind gift from Dr. Chunlin Qin),27 DSP (sc-18328; Santa Cruz Biotechnology, Santa Cruz, CA), HDAC3 (ab32369; Abcam), p300 (05–257; EMD Millipore, Darmstadt, Germany) H3K9ac or H3K27ac. ACTIN (AC028; ABclonal Biotechnology Co, Ltd) was used as an internal reference.
Then the membranes were incubated with HRP-labeled IgG (Ebioscience, San Diego, CA) at room temperature for 1 hour. Enhanced chemiluminescence solution (Merck Millipore, Darmstadt, Germany) was used for detection.
2.9 |. Statistical analysis
All data are presented as the mean ± standard deviation. Data analyses were performed by the Student t-test (two-tailed). P < .05 was considered statistically significant. Experiments in this study were repeated independently at least three times.
3 |. RESULTS
3.1 |. Acetylated H3K9 and H3K27 correlated with dentinogenesis
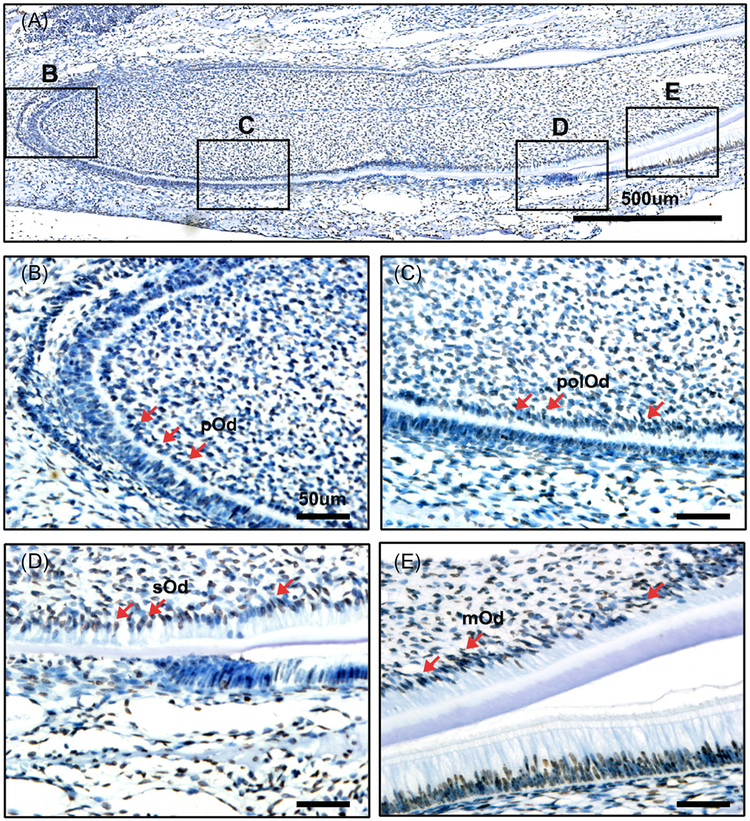
To detect the correlation of histone acetylation and dentinogenesis, we detected the expression of acetylated H3K9 and H3K27 in vivo. Immunohistochemistry was performed in lower incisors of PN 2 mouse for all stages of odontoblasts present in one slide.28 Acetylated H3K9 and H3K27 showed extensive expression in tooth, and they were upregulated during differentiation (Figures 1A and 2A). Both H3K9ac and H3K27ac showed a low level in the pre-odontoblasts (Figures 1B,C and 2B,C). Then they were intensely expressed in the secretary and mature odontoblasts (Figures 1D,E and 2 D,E). Taken together, these results suggest the strong correlation of acetylated H3K9 and H3K27 with dentinogenesis.
FIGURE 1.
Immunohistochemical detection of acetylated H3K9 in the PN2 mouse lower incisor. A, Immunohistochemical detection of acetylated H3K9 during dentinogenesis. B, The expression of acetylated H3K9 was rarely detected at preodontoblasts. C, Acetylated H3K9 was weakly detected in the polarized odontoblasts. D, Acetylated H3K9 was highly expressed in the secretory odontoblasts. E, The expression of acetylated H3K9 was strongly localized in mature odontoblasts. mOd, mature odontoblasts; pOd, preodontoblast; polOd, polarizing odontoblasts; sOd, secretory odontoblasts. Scale bar: A, 500 μm; B-E, 50 μm
FIGURE 2.
Immunohistochemical detection of acetylated H3K27 in the PN2 mouse lower incisor. A, Immunohistochemical detection of acetylated H3K27 during tooth development. B, Acetylated H3K27 was rarely detected at preodontoblasts. C, Weak detection of acetylated H3K27 was observed in the polarized odontoblasts. D, Acetylated H3K27 was highly expressed in the secretory odontoblasts. E, The expression of acetylated H3K27 was strongly localized in mature odontoblasts. Scale bar: A, 500 μm; B-E, 50 μm
3.2 |. Expression of acetylated H3K9 and H3K27 during odontoblast differentiation
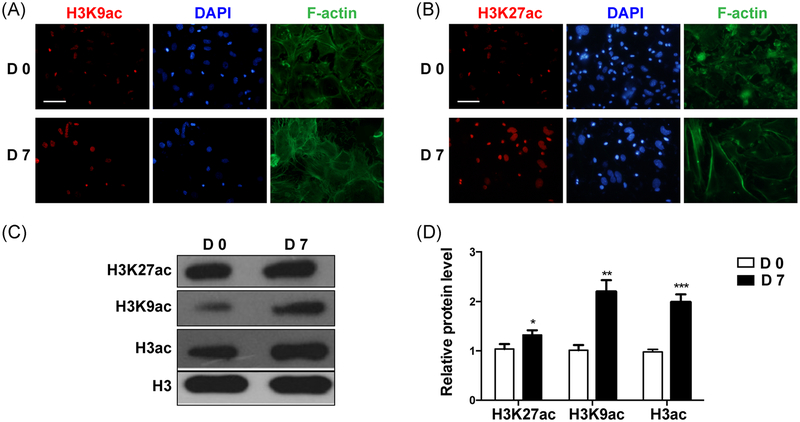
mDPCs are known as progenitors of odontoblasts, and they have the capacity to differentiate into odontoblasts after induction in vitro. Thus, it is an ideal in vitro model to study dentinogenesis.29 To elucidate the expression of H3K9ac and H3K27ac during odontoblast differentiation, mDPCs were cultured in the odontoblastic medium for 7 days. Immunofluorescent staining showed an upregulation of H3K9ac and H3K27ac expression in mDPCs cells after induction (Figure 3A,B). The protein level of H3K9ac and H3K27ac (Figure 3C,D) increased obviously on day 7. These results are in accordance with the results of in vivo dentinogenesis, which further verified that histone acetylation was involved during dentinogenesis.
FIGURE 3.
The expression of acetylated H3K9 and H3K27 during odontoblast differentiation of the mDPCs. mDPCs were cultured with odontoblastic differentiation medium for 7 days. Immunofluorescence was performed to detect the expression levels of acetylated H3K9 (A) and H3K27 (B); the nucleus was labeled with DAPI. The protein levels of acetylated H3K9 and H3K27 (C, D) were detected. The D0 group was set as the control. All data were presented as mean ± standard deviation (SD) and were based on three independent experiments. DAPI, 4’,6-diamidino-2-phenylindole; mDPCs, mouse dental papilla mesenchymal cells. *P < .05, **P < .01, ***P < .001
3.3 |. Expression of histone acetylase and histone deacetylases during odontoblast differentiation of mDPCs
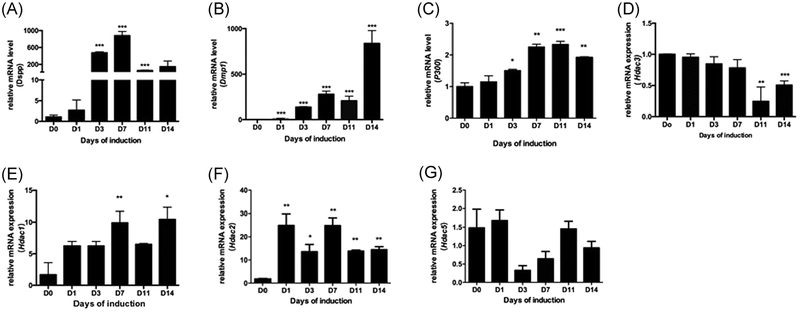
Since histone acetylation was increased during odontoblast differentiation and acetylated states are determined by the balance between HATs and HDACs, we wanted to know which histone acetylase was responsible for the upregulation of histone acetylation. Hence, we detected the expression of histone acetylase during odontoblast differentiation by qRT-PCR. mDPCs were cultured in odontoblastic medium for 14 days, and then the expression levels of odontoblastic-related marker genes were measured by qRT-PCR. We found that the messenger RNA (mRNA) level of Dspp (Figure 4A) and Dmpl (Figure 4B) increased significantly on day 3, and continued to rise until day 14. Also, the mRNA level of p300 (Figure 4C) increased on day 7 and kept rising until day 11, which were in accordance with the expression of odontoblastic-related genes Dspp and Dmpl. In contrast, the mRNA level of Hdac3 (Figure 4D) decreased on day 11 and downregulated until day 14, which showed a different trend with the expression of odontoblastic-related gene Dspp and Dmpl (results similar to Figure 4C,D have been published elsewhere). But other HDAC family members showed no obvious change (Figure 4E–G). These findings indicated that the coordinated expression of p300 and Hdac3 might be responsible for upregulated histone acetylation during differentiation.
FIGURE 4.
Expression pattern of Hdac3 and p300 during odontoblast differentiation of the mDPCs. mDPCs were cultured with odontoblast differentiation medium over 14 days. mRNAs were prepared on days 0, 1, 3, 7, 11, and 14 after induction. qRT-PCR was performed to detect the expression levels of Dspp (A) and Dmp1 (B), p300 (C) and Hdac3 (D), GAPDH was used as the internal reference. E-G, The expression pattern of HDAC family members were detected. The D0 group was set as the control. All data were presented as mean ± standard deviation (SD) and were based on three independent experiments. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; mDPCs, mouse dental papilla mesenchymal cells; mRNA, messenger RNA; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction. *P < .05, **P < .01, ***P < .001
3.4 |. Overexpression of Hdac3 in mDPCs
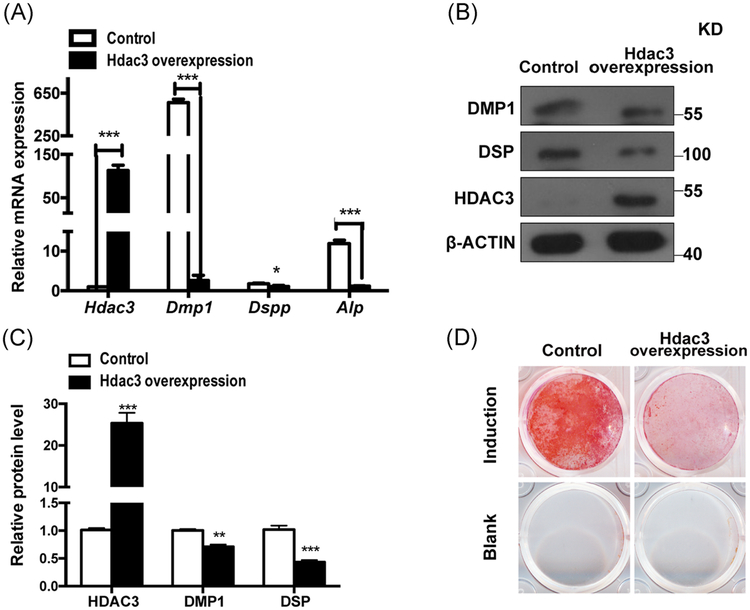
Then we explored whether these two histone acetylases were able to regulate odontoblast differentiation and dentinogenesis. To test the function of Hdac3 directly, we performed in vitro differentiation assays with mDPCs for 7 days. Then RNA and protein were obtained, qRT-PCR and immunoblots were performed for Hdac3, Dmpl, Dspp, Alp, and normalized to Actin. Mineralization was determined by alizarin red staining. As Hdac3 was downregulated during odontoblast differentiation, we overexpressed Hdac3 in mDPCs and then the cells were inducted for 7 days. We found that Dmpl, Dspp and Alp were notably downregulated in Hdac3 overexpression group (Figure 5A,B). Cells in Hdac3 overexpression group showed significantly decreased mineral deposition capacity (Figure 5C,D).
FIGURE 5.
Overexpression of Hdac3 inhibited the odontoblast differentiation of the mDPCs. mDPCs were infected with either control or Hdac3 overexpression plasmid. Then the cells were treated with odontoblastic induction medium. RNA and protein were obtained after 7 day’s induction. qRT-PCR (A) and immunoblots (B) were performed for Hdac3, Dmp1, Dspp, Alp, and normalized to Actin. C, Normalized protein levels were shown in the chart. D, Mineralization was determined by alizarin red staining. All data were based on three independent experiments. mDPCs, mouse dental papilla mesenchymal cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction. *P < .05, **P < .01, ***P < .001
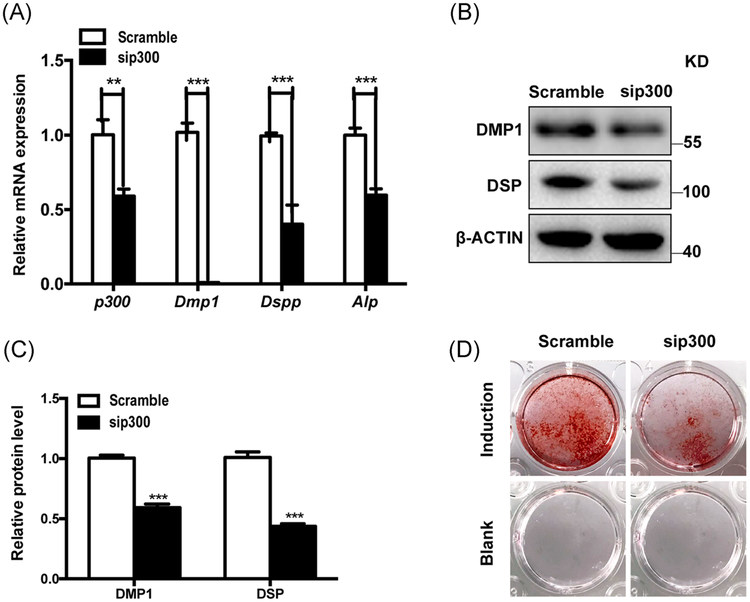
3.5 |. Knockdown of p300 in mDPCs
Since p300 was upregulated during odontoblast differentiation, sip300 transduction was applied in mDPCs. The scramble siRNA was used as the control group. Our results demonstrated that Dmpl, Dspp, and Alp were markedly downregulated in the p300-knockdown groups (Figure 6A,B). When cultured under induction medium for 7 days, the p300-knockdown groups showed significantly decreased mineral deposition ability (Figure 6C,D).
FIGURE 6.
Downregulation of p300 inhibited the odontoblast differentiation of the mDPCs. mDPCs were infected with either scramble or sip300. Then the cells were treated with odontoblastic induction medium. RNA and protein were obtained after 7 day’s induction. qRT-PCR (A) and immunoblots (B) were performed for p300, Dmp1, Dspp, Alp, and normalized to Actin. C, Normalized protein levels were shown in the chart. D, Mineralization was determined by alizarin red staining. All data were based on three independent experiments. mDPCs, mouse dental papilla mesenchymal cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction. *P < .05, **P < .01, ***P < .001
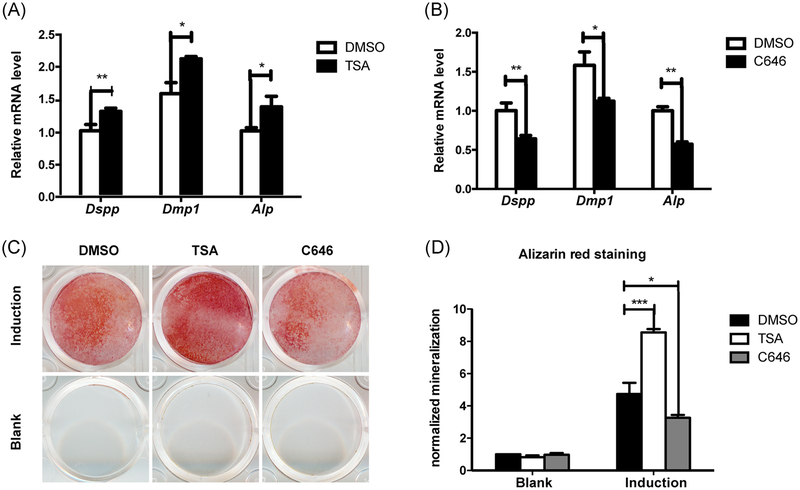
3.6 |. Inhibitors of p300 and HDAC3 in mDPCs
Then we took a further step by using inhibitors of p300 and HDAC3 to study the function of these two histone acetylases. When cells were stimulated with TSA, which blocks the activity of HDACs for 24 hours before induction, the mRNA levels of Dspp, Dmpl, and Alp were significantly increased after 7 days of induction (Figure 7A). As indicated by alizarin red staining, calcium nodules were increased in the TSA group on day 9 after induction (Figure 7C,D). C646 is a selective inhibitor of p300 and CBP. It decreased the mRNA levels of Dspp, Dmpl and Alp (Figure 7B), and the mineralization nodular was also decreased (Figure 7C,D). These findings demonstrated that p300 and HDAC3 regulated odontoblast differentiation.
FIGURE 7.
Inhibitors of HDAC3 and p300 could regulate the odontoblast differentiation of the mDPCs. mDPCs were treated with either TSA or C646 for 24 hours, 0.1% DMSO was used as controls, then the cells were treated with odontoblastic induction medium. RNA was obtained after 7 day’s induction. qRT-PCR (A, B) were performed for Dmp1, Dspp, Alp, and normalized to Actin. C, Mineralization was determined by alizarin red staining. D, Normalized mineralization was shown in the chart. All data were based on three independent experiments. mDPCs, mouse dental papilla mesenchymal cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction. DMSO, dimethyl sulfoxide; mDPCs, mouse dental papilla mesenchymal cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; TSA, trichostatin A. *P < .05, **P < .01, ***P < .001
Overall, the coordinated expression of p300 and HDAC3 could upregulate histone acetylation during dentinogenesis. Moreover, p300 and HDAC3 could regulate odontoblast differentiation.
4 |. DISCUSSION
Dentinogenesis, including odontoblast differentiation, relies on the coordinated expression of regulatory genes, including transcription and growth factors. Gene expression is influenced by epigenetic status, including DNA methylation and histone modifications. DNA methylation has been discovered to regulate odontoblast differentiation (unpublished data). However, the involvement of histone modifications in controlling the differentiation of odontoblasts has not been fully explored. In this study, we have demonstrated histone acetylation as an important regulator of dentinogenesis and odontoblast differentiation. We further showed that histone acetylases p300 and HDAC3 could regulate odontoblast differentiation in vitro.
Although H3K9ac and H3K27ac were widely expressed in the mesenchyme and epithelium, they had stronger staining in the secretory and mature odontoblasts. Thus, we investigated the expression of H3K9ac and H3K27ac in different stages during odontoblast differentiation. The in vitro study confirmed that histone acetylation was involved in dentinogenesis. It has been reported that the coordinated expression of four H3K9 methyltransferases in dental mesenchyme played important roles in tooth development.30 Therefore, we explored whether there was a coordinated expression of histone acetylases. We found that p300 was consistent with the spatiotemporal expression patterns of Dspp and Dmpl, while Hdac3 was the opposite during odontoblast differentiation. p300 was first considered a transcriptional adapter for many DNA-binding activators. Then, studies demonstrated that p300 per se was a histone acetytransferase.31 Substantial research has studied the involvement of p300 in the regulation of transcription in many cell lines. Studies showed that p300 could increase Runx2 acetylation and enhance the stability and transcriptional activation of Runx2, thus promoting osteoblast differentiation. It was reported that p300 was recruited to the promoter regions of OCN and DSPP and increased the acetylation of lysine 9 of histone H3, thus regulating the odontogenic differentiation of human dental pulp cells.26 And knockdown of p300 decreases the odontogenic differentiation potentials.32 In our study, p300 could also promote odontoblast differentiation of mouse dental papilla cells. HDAC3 belongs to class I HDACs and participates in many biological processes, such as cell cycle, cell proliferation, and development.33–35 Many studies have shown that HDAC3 could regulate osteoblasts and osteoclasts differentiation, thus contributing to skeletal development and bone homeostasis. Some demonstrated that HDAC3 facilitated osteoblastic differentiation; conversely, others found that HDAC3 accelerated osteoclastic differentiation.36–40 However, little is known about the role HDAC3 playing in odontoblast differentiation. Our results demonstrated that overexpressed HDAC3 hindered odontoblast differentiation. When TSA was used to inhibit HDAC3, it promoted odontoblast differentiation. A study demonstrated that TSA promoted rat adipose–derived stem cell osteogenic differentiation by altering the epigenetic modifications, with increased acetylated histone H3K9 recruitment onto the Runx2 promoter. Our results about TSA are consistent with these findings. Since TSA had the FDA’s permission in clinical therapy, it’s role in promoting odontoblast differentiation may give some hints to restorative dentistry.21
In conclusion, this study illustrated the expression patterns of H3K9ac and H3K27ac during dentinogenesis and odontoblast differentiation. Using in vivo mice mandibular incisor from which we could observe different differentiation stage of odontoblasts and in vitro odontoblastic differentiation assay, we demonstrated that H3K9ac and H3K27ac were upregulated during dentinogenesis and odontoblast differentiation. We then found that histone acetylases p300 and HDAC3 showed a unique expression pattern during differentiation. We also confirmed that histone acetylases p300 and HDAC3 could regulate odontoblast differentiation themselves through modulating histone acetylation. Coordinated expression of p300 and HDAC3 upregulates histone acetylation to regulate dentinogenesis.
ACKNOWLEDGMENTS
We thank Prof. Chunlin Qin from the Texas A&M Health Science Center for generously providing the antibody for Dmpl. The study was funded by grants from National Natural Science Foundation of China (No. 81271099,81420108011) to Prof. Zhi Chen, Young Elite Scientist Sponsorship Program by CAST (No. 2017QNRC001), Natural Science Foundation of Hubei Province (No. 2017CFB515) and National Natural Science Foundation of China (No. 81771057) to Dr Huan Liu, and National Natural Science Foundation of China (No. 81802973) to Dr Qiuhui Li.
Abbreviations:
- H3K9ac
acetylated lysine 9 at histone H3
- H3K27aca
cetylated lysine 27 at histone H3
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
REFERENCES
- 1.Jussila M, Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harbor Perspect Biol. 2012;4(4):a008425 10.1101/cshperspect.a008425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Lin C, Zhang J, et al. Quaking promotes the odontoblastic differentiation of human dental pulp stem cells. J Cell Physiol. 2018;233(9):7292–7304. 10.1002/jcp.26561 [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Liu H, Sun Q, Yuan G, Zhang L, Chen Z. KLF4 promoted odontoblastic differentiation of mouse dental papilla cells via regulation of DMP1. J Cell Physiol. 2013;228(10):2076–2085. 10.1002/jcp.24377 [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Lin H, Liu H, Zhang L, Yuan G, Chen Z. SP1 promotes the odontoblastic differentiation of dental papilla cells. Dev Growth Differ. 2015;57(5):400–407. 10.1111/dgd.12221 [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Lin H, Zhang L, et al. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem. 2013;288(13):9261–9271. 10.1074/jbc.M112.433730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Q, Liu H, Lin H, Yuan G, Zhang L, Chen Z. MicroRNA-338–3p promotes differentiation of mDPC6T into odontoblast-like cells by targeting Runx2. Mol Cell Biochem. 2013;377(1–2):143–149. 10.1007/s11010-013-1580-3 [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Liu H, Lin H, et al. Sp1 is a competitive endogenous RNA of Klf4 during odontoblast differentiation. Int J Biochem Cell Biol. 2017;85:159–165. 10.1016/j.biocel.2017.02.008 [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10(11):805–811. 10.1038/nrg2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. 10.1038/cr.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286(21):18347–18353. 10.1074/jbc.R110.205286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19(6):541–549. 10.1016/j.gde.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell. 2011;44(3):410–423. 10.1016/j.molcel.2011.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gates LA, Shi J, Rohira AD, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017;292(35):14456–14472. 10.1074/jbc.M117.802074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20(8):615–626. [DOI] [PubMed] [Google Scholar]
- 17.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4(10):944–947. 10.1038/sj.embor.embor941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrozza MJ, Utley RT, Workman JL, Côté J. The diverse functions of histone acetyltransferase complexes. TIG. 2003;19(6):321–329. 10.1016/S0168-9525(03)00115-X [DOI] [PubMed] [Google Scholar]
- 19.de Ruijte AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. 10.1042/BJ20021321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao Y, Yang Y, Zhu WG. Sirtuins: nodes connecting aging, metabolism and tumorigenesis. Curr Pharm Des. 2014;20(11):1614–1624. [DOI] [PubMed] [Google Scholar]
- 21.Duncan HF, Smith AJ, Fleming GJ, Cooper PR. HDACi: cellular effects, opportunities for restorative dentistry. J Dent Res. 2011;90(12):1377–1388. 10.1177/0022034511406919 [DOI] [PubMed] [Google Scholar]
- 22.Jin H, Park JY, Choi H, Choung PH. HDAC inhibitor trichostatin A promotes proliferation and odontoblast differentiation of human dental pulp stem cells. Tissue Eng Part A. 2013;19(5–6):613–624. 10.1089/ten.TEA.2012.0163 [DOI] [PubMed] [Google Scholar]
- 23.Kwon A, Park HJ, Baek K, et al. Suberoylanilide hydroxamic acid enhances odontoblast differentiation. J Dent Res. 2012;91(5):506–512. 10.1177/0022034512443367 [DOI] [PubMed] [Google Scholar]
- 24.Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors induced differentiation and accelerated mineralization of pulp-derived cells. J Endod. 2012;38(3):339–345. 10.1016/j.joen.2011.12.014 [DOI] [PubMed] [Google Scholar]
- 25.Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors epigenetically promote reparative events in primary dental pulp cells. Exp Cell Res. 2013;319(10):1534–1543. 10.1016/j.yexcr.2013.02.022 [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Liu H, Ning Y, Xu Q. The histone acetyltransferase p300 regulates the expression of pluripotency factors and odontogenic differentiation of human dental pulp cells. PLoS One. 2014;9(7):102117 10.1371/journal.pone.0102117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin C, Brunn JC, Cook RG, et al. Evidence for the proteolytic processing of dentin matrix protein 1: identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;278(36):34700–34708. 10.1074/jbc.M305315200 [DOI] [PubMed] [Google Scholar]
- 28.Ruch JV, Lesot H, Begue-Kirn C. Odontoblast differentiation. Int J Dev Biol. 1995;39(1):51–68. [PubMed] [Google Scholar]
- 29.Bègue-Kirn C, Ruch JV, Ridall AL, Butler WT. Comparative analysis of mouse DSP and DPP expression in odontoblasts, preameloblasts, and experimentally induced odontoblast-like cells. Eur J Oral Sci. 1998;106(Suppl 1):254–259. [DOI] [PubMed] [Google Scholar]
- 30.Kamiunten T, Ideno H, Shimada A, et al. Coordinated expression of H3K9 histone methyltransferases during tooth development in mice. Histochem Cell Biol. 2015;143(3):259–266. 10.1007/s00418-014-1284-0 [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–959. 10.1016/S0092-8674(00)82001-2 [DOI] [PubMed] [Google Scholar]
- 32.Liu HJ, Wang T, Li QM, Guan XY, Xu Q. Knock-down of p300 decreases the proliferation and odontogenic differentiation potentiality of HDPCs. Int Endontic J. 2015;48(10):976–985. 10.1111/iej.12392 [DOI] [PubMed] [Google Scholar]
- 33.Li S, Li M, Liu X, et al. Genetic and chemical screenings identify HDAC3 as a key regulator in hepatic differentiation of human pluripotent stem cells. Stem Cell Reports. 2018;11:22–31. 10.1016/j.stemcr.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castelo-Branco G, Lilja T, Wallenborg K, et al. Neural stem cell differentiation is dictated by distinct actions of nuclear receptor corepressors and histone deacetylases. Stem Cell Reports. 2014;3(3):502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh N, Gupta M, Trivedi CM, Singh MK, Li L, Epstein JA. Murine craniofacial development requires Hdac3-mediated repression of Msx gene expression. Dev Biol. 2013;377(2):333–344. 10.1016/j.ydbio.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feigenson M, Shull LC, Taylor EL, et al. Histone deacetylase 3 deletion in mesenchymal progenitor cells hinders long bone development. J Bone Miner Res. 2017;32(12):2453–2465. 10.1002/jbmr.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hesse E, Saito H, Kiviranta R, et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol. 2010;191(7):1271–1283. 10.1083/jcb.201009107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham L, Kaiser B, Romsa A, et al. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J Biol Chem. 2011;286(14):12056–12065. 10.1074/jbc.M110.216853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razidlo DF, Whitney TJ, Casper ME, et al. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS One. 2010;5(7):e11492 10.1371/journal.pone.0011492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with Runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279(40):41998–42007. 10.1074/jbc.M403702200 [DOI] [PubMed] [Google Scholar]