Abstract
Malignant glioma is one of the deadliest types of cancer. Understanding how the cell of origin progressively evolves toward malignancy in greater detail could provide mechanistic insights and lead to novel concepts for tumor prevention and therapy. Previously we have identified oligodendrocyte precursor cell (OPC) as the cell of origin for glioma following the concurrent deletion of p53 and NF1 using a mouse genetic mosaic system that can reveal mutant cells prior to malignancy. In the current study, we set out to deconstruct the gliomagenic process in two aspects. First, we determined how the individual loss of p53 or NF1 contributes to aberrant behaviors of OPCs. Second, we determined how signaling aberrations in OPCs progressively change from pre-malignant to transformed stages. We found that while the deletion of NF1 leads to mutant OPC expansion through increased proliferation and decreased differentiation, the deletion of p53 impairs OPC senescence. Signaling analysis showed that, while PI3K and MEK pathways go through stepwise over-activation, mTOR signaling remains at the basal level in pre-transforming mutant OPCs but is abruptly up-regulated in tumor OPCs. Finally, inhibiting mTOR via pharmacological or genetic methods, led to a significant blockade of gliomagenesis but had little impact on pre-transforming mutant OPCs, suggesting that mTOR is necessary for final transformation but not early progression. In summary, our findings show that deconstructing the tumorigenic process reveals specific aberrations caused by individual gene mutations and altered signaling events at precise timing during tumor progression, which may shed light on tumor-prevention strategies.
Keywords: glioma, oligodendrocyte precursor cell, genetic mosaic mouse model, tumor initiation, mTOR
Introduction
Glioma is the most deadly type of brain cancer (1–3). Grade IV gliomas, also known as glioblastoma (GBM), are the most commonly diagnosed form of glioma. Primary GBM appears without evidence of progression from lower grades, while secondary GBM invariably progresses from lower grade tumors. GBM is incurable and gives patients a median survival time of only 15-month (4, 5). Sequencing of more than 600 GBM patient samples by The Cancer Genome Atlas (TCGA) project has revealed mutations in three critical pathways: p53, receptor tyrosine kinases (RTKs), and RB G1 checkpoint (6, 7). However, how these pathways transform which normal brain progenitor cells into malignant tumor cells remains unclear. To deconvolute this complex problem, it would be critical to first identify the cell of origin for glioma, then to study the impact of each mutated pathway on tumor initiation, progression, and transformation.
Unequivocal identification of the cell of origin is almost impossible with patient tumor samples due to significant alterations of cellular morphologies and marker gene expression in malignant tumor cells. Therefore, access to the pre-malignant stage holds the key to this problem. Since this is nearly impossible with human patients, mouse genetic models have been widely used to help pinpoint the glioma cell of origin in the brain. While conventional mouse models cannot reveal pre-malignant mutant cells, we have used a mouse genetic system termed MADM (Mosaic Analysis with Double Markers) to circumvent the problem (8). Through inter-chromosomal mitotic recombination, MADM generates mutant cells labeled with GFP and its sibling WT cells labeled with RFP from a heterozygous, colorless mother cell (Fig. 1A). The sparseness of mutant cells resulted from the low frequency of inter-chromosomal recombination not only closely mimics the clonal nature of human cancer, but also allows one to visualize mutant cells at previously inaccessible tumor initiation and early progression stages (illustrated in Fig. 1B). Most importantly, the definitive correlation between color and genotype greatly facilitates the revelation of tumor initiation based on the increased ratio of green-to-red cell numbers (G/R ratio). In our previous studies, we found that, after introducing p53/NF1 mutations into neural stem cells with MADM, cell lineage-specific analysis revealed a dramatic increase of G/R ratio at pre-malignant stage only in oligodendrocyte precursor cells (OPCs) but not in any other brain cell types (Fig. 1B, middle panel, and Fig. 1C for representative images of MADM-based analysis) (9). Along with other evidences, we pinpointed OPC as the cell-of-origin in this model, a finding supported by other studies and verified with human glioma samples (10–14).
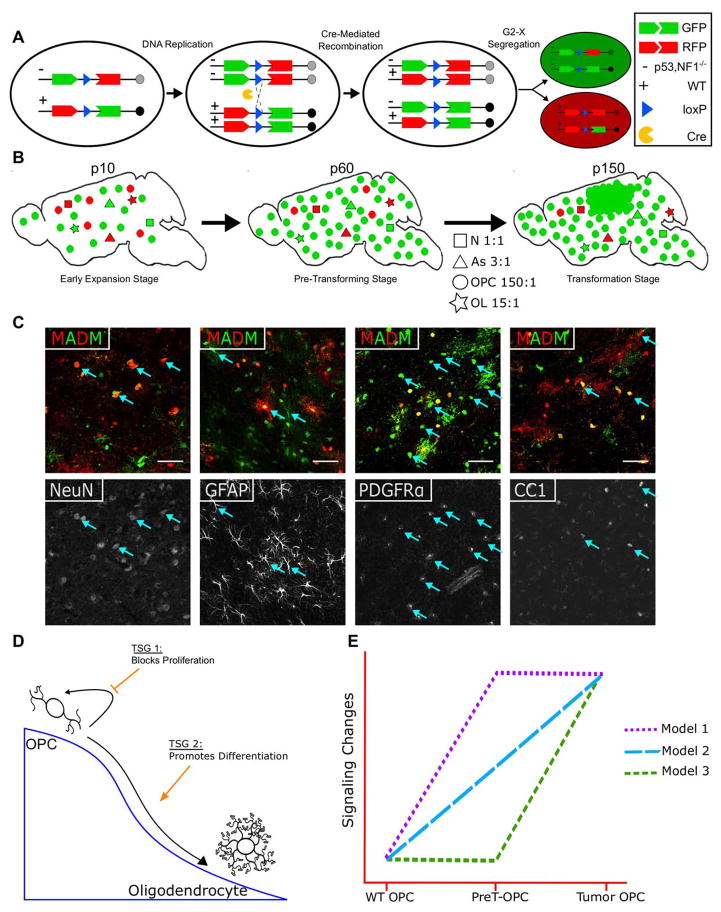
Fig. 1. Hypothetical framework of the current study.
(A) Schematic illustration of how MADM generates GFP+ mutant and sibling RFP+ WT cells from a heterozygous, colorless cell via Cre-Mediated inter-chromosomal mitotic recombination.
(B) In our previous study, upon the concurrent deletion of p53 and NF1, GFP+ mutant OPCs progressively outnumber RFP+ WT OPCs (as reflected by G/R ratio) while mutant neurons, astrocytes, and oligodendrocytes showed minimal expansion. By P150, glioma formed at full penetrance from GFP+ mutant OPCs in the MADM model, indicating that OPC is a cell of origin for glioma.
(C) Co-staining of cell type-specific markers with MADM labeling allows for cell-type specific analysis. When a neural stem cell Cre line is used, all neuroglial cells including neurons (NeuN), astrocytes (GFAP), OPCs (PDGFRα), and oligodendrocytes (CC1) can be readily identified to be labeled by MADM. Images were taken from a P60 brain.
(D) Schematic illustration of our first working hypothesis that two tumor suppressor genes (TSGs), p53 and NF1, may exert their tumor suppressing activities by inhibiting OPC proliferation or by promoting OPC differentiation, respectively.
(E) Schematic illustration of our second working hypothesis on progressive alterations of signaling pathways during gliomagenesis. Model 1: full activation in both PreT-OPCs and tumor OPCs. Model 2: gradual activation from PreT-OPCs to tumor OPCs. Model 3, minimal activation in Pre-T OPCs followed by abrupt activation in tumor OPCs.
While our previous data showed that increased proliferation and decreased differentiation of p53-null, NF1-null OPCs precedes gliomagenesis, it remains unclear which mutation leads to which phenotypic aberrations. Therefore, in the current study, we set out to deconstruct the contribution of individual tumor suppressor gene (TSG) mutation toward glioma initiation and progression, using the MADM system (Fig. 2A). We hypothesized that p53 mutation and NF1 mutation might play distinct roles to promote gliomagenesis: the deletion of one could promote proliferation while the deletion of the other could cripple differentiation, thereby cooperatively leading to tumor formation (Fig. 1D). The identification of OPCs as the glioma cell of origin also provides a unique opportunity to investigate signaling aberrations at distinct stages of tumor progression, since we can use the OPC-specific surface marker PDGFRα to purify both pre-transforming mutant OPCs (PreT-OPCs) and malignant tumor cells for signaling analysis (15–18). Since NF1 is known as a RasGAP (19–22), we focused our analysis on the downstream effectors of the Ras pathway to examine the route of progressive activation of these molecules from pre-malignant to malignant stages (Fig. 1E). Based on our findings, we hope to develop therapeutic strategies that are tailored to target malignant tumor cells and pre-transforming mutant cells in a precise fashion to achieve the best efficacy.
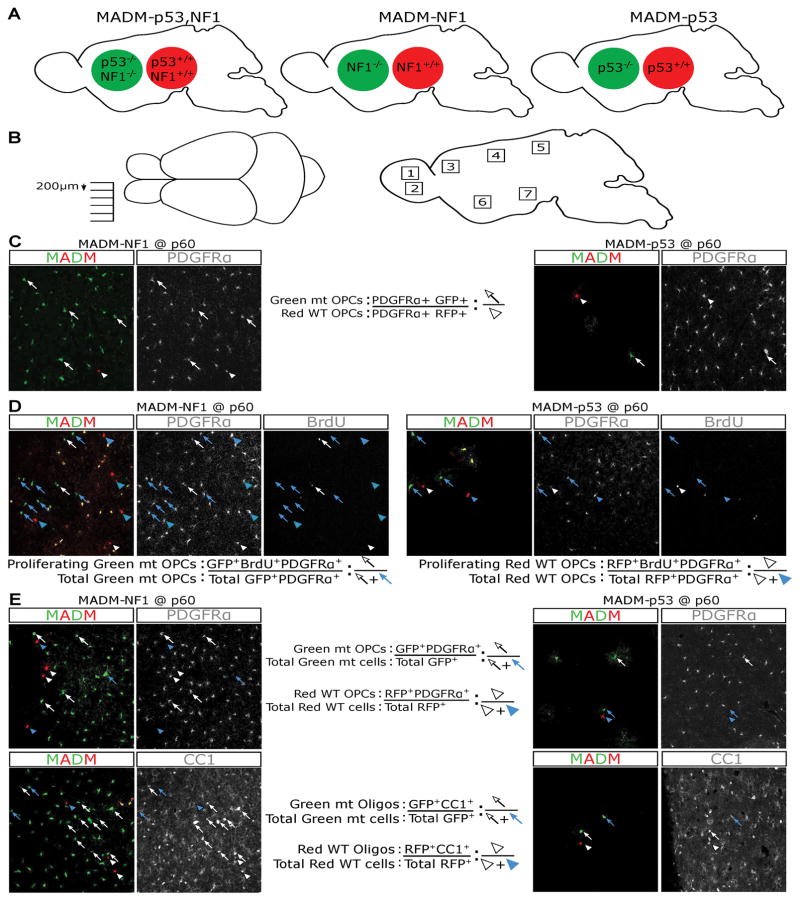
Fig. 2. MADM models used in this study and systematic sampling schemes and quantification methods for determining the effects of NF1 or p53 deletion in vivo.
(A) Simplified schematic to show MADM-p53,NF1 tumor model, MADM-p53 and MADM-NF1 models. GFP+ cells are null for specified genes while RFP+ cells are WT.
(B) To ensure systematic sampling, brains were cut mediolaterally and sections were collected every 200 μm. Seven sections were collected for each sample. Within each brain section, 7 distinct regions (dorsal and ventral olfactory bulb; anterior, middle, and posterior cortex; ventral striatum; hypothalamus) were imaged for quantifications to avoid biases due to potential regional heterogeneity of OPCs.
(C) To evaluate the extent of mutant OPC expansion (G/R ratio in Figure 2A), the total number of mutant OPCs (GFP+ PDGFRα+) and WT OPCs (RFP+ PDGFRα+) were counted and the sum of all 49 areas (7 sections per brain, 7 regions per section) was used to calculate the G/R ratio in each brain. Arrows point to mutant OPCs, and arrowheads point to WT OPCs.
(D) To assess the proliferative rate of OPCs, the total number of proliferating mutant OPCs (BrdU+, GFP+, PDGFRα+) was divided by the total number of mutant OPCs (GFP+ PDGFRα+); and an equivalent scheme was used for WT OPCs. The sum of all 49 areas was used to calculate the final percentage of proliferating OPCs in each populations. Arrows point to mutant OPCs, and arrowheads point to WT OPCs. Blue: non-proliferating OPCs; white: proliferating OPCs.
(E) To assess the differentiation potential of OPCs (the proportion of OPCs versus oligodendrocytes), brains were stained for either PDGFRα or CC-1. The total number of GFP+ PDGFRα+ cells was counted and divided by the total GFP+ cells to determine the relative proportion of OPCs within the mutant population; and an equivalent scheme was used to quantify the WT population. CC1 was used to determine the proportion of oligodendrocytes within both the WT and mutant populations with the same scheme. The sum of all 49 areas was used to calculate the final numbers. Arrows point to mutant OPCs, and arrowheads point to WT OPCs. Blue: OPCs; white: oligodendrocytes.
MATERIALS & METHODS
Mouse lines and genotyping
All animal procedures were based on animal care guidelines approved by the Institutional Animal Care and Use Committee. The following mouse strains were used to obtain experimental and control mice: TGML (stock no. 022977, JAX), GTML (stock no. 022976, JAX), hGFAP-Cre (stock no. 004600, JAX), NG2-Cre (stock no. 008533; JAX), NG2-CreER (stock no. 008538, JAX), p53KO (stock no. 002101; JAX), Rosa-tdTomato (stock no. 007908; JAX), neurofibromin 1 (NF1)Flox (strain no. 01XM4; NCI) and p53flox (strain no. 01XC2; NCI), mTORFlox (stock no. 011009, JAX). Mice used for single NF1 and p53 studies were TGML11-NF1Flox/GTML11; NG2-Cre and TGML11-p53KO/GTML11; NG2-Cre, respectively. Mice used for glioma generation and purification of OPC-like tumor cells were TGML11-p53KO, NF1Flox/GTML11; hGFAP-Cre. Mice used for purification of p53-null, NF1-null OPCs were p53KO, NF1flox/p53flox, NF1flox; hGFAP-Cre; Rosa-tdT. Wild type OPCs were generated from mice carrying no p53 or NF1 allele and no Cre. Mice used for mTOR studies were p53flox, NF1flox/p53flox, NF1flox; Rosa-tdT; mTORflox (homozygous and heterozygous); NG2-CreER. All genotyping was performed as previously described (9, 13).
BrdU injection for assessing proliferative rate of OPCs in vivo
BrdU was given by i.p. injection at 50 mg/kg. Mice were given one injection then brains were harvested after 3 hours (MADM P10), one injection per day for 4 consecutive days then brains were harvested 1-day after the last injection (MADM P60 and P240), or one injection per day for 7 consecutive days then brains were harvested 1-day after the last injection (CKO).
Tissue collection
To dissect brains, mice were transcardially perfused first with PBS supplemented with procaine and heparin (Sigma) to flush out the blood, followed by 4% (wt/vol) paraformaldehyde. Brains were then dissected out and post-fixed in 4% (wt/vol) paraformaldehyde overnight at 4°C. After post-fixing, brains were washed in PBS 3x at room temperature (RT), soaked in 30% (wt/vol) sucrose overnight at 4°C for cryoprotection, then embedded in OCT and stored at −80°C.
Immunostaining
Brains were cut as 25 μm-thick sections on glass slides. Brain sections were allowed to dry at RT for 1hr prior to staining. Slides with brain sections were washed 3 times with PBS for 10 minutes each, then incubated in blocking/permeabilization buffer (5% normal donkey serum/NDS in PBS with 0.3% triton X-100) for 20 minutes at RT. Primary antibody incubation was performed at 4°C overnight in the same blocking/permeabilization buffer. Secondary antibody incubation was performed for 2 to 4 hours at RT or 4°C overnight in blocking/permeabilization buffer. If necessary, slides were incubated in DAPI solution for 20 minutes as the last step before coverslipping. Secondary antibodies were conjugated to the Alexa Fluor® family dyes, purchased from Invitrogen. Primary antibodies are listed below:
| Antigen | Species | Dilution | Source | Catalog no. |
|---|---|---|---|---|
| APC-CC1 | Mouse | 1:100 | Millipore | OP80 |
| BrdU | Rat | 1:250 | AbD Serotec | OBT0030G |
| GFP | Chicken | 1:500 | Aves Lab | GFP-1020 |
| c-myc | Goat | 1:200 | Novus | NB600-338 |
| MBP | Mouse | 1:500 (TC) | Covance | SMI-99P |
| PDGFRa | Goat | 1:200 (1:400 TC) | R&D Systems | AF1062 |
| tdTomato | Rabbit | 1:100 | Clontech | 632496 |
For BrdU immunostaining, slides with brain sections were washed 3 times with PBS, post-fixed at RT with 4% PFA for 20 minutes, then treated with freshly prepared 2 N HCl prepared in PBS at 37° C for 30 minutes. After acid treatment, slides were washed 4 times with PBS for 7 minutes each and then general immunostaining procedures as described above were followed.
When Edu (5-ethynyl-2′-deoxyuridine, Invitrogen, cat. #10044) was used to assess the number of cells in the S phase, it was added to culture media for 3 hours. After EdU treatment, cells were fixed with 4% PFA for 20min at RT, permeabilized with 0.1% PBT, and stained with azide conjugated with Alexa Fluor® 647 (Invitrogen, cat. #A10277) through the Click-iT reaction.
For in vitro work, cells were grown on 12 mm glass coverslips coated with poly-D-lysine (Sigma). Cultured cells were fixed by 4% paraformaldehyde (PFA) for 30 minutes at room temperature, followed by 3 washes with PBS. Coverslips were allowed to air dry for at least 30 minutes to allow cells attach tightly for further staining. After incubating in blocking/permeabilization buffer (10% NDS in PBS with 0.1% triton X-100) for 30 minutes at RT, coverslips were incubated in primary antibody diluted in the blocking/permeabilization buffer for 1hr at RT, then appropriate secondary antibodies for 30 minutes at RT. If necessary, coverslips were incubated in 1 ug/ml DAPI solution (Sigma, cat. # 32670-25MG-F) for 20 minutes as the last step before mounting with anti-fade gel mounting media (EMS, cat. # 17985-10).
OPC culture
Wild type OPCs and p53-null, NF1-null PreT-OPCs were purified from P8 mice, and OPC-like tumor cells were purified from fresh glioma tissues through PDGFRA-immunopanning as previously described (9). All cells were genotyped for p53 and NF1 status using appropriate primer sets to confirm their purity and correct genotype. Cells were maintained in Neurobasal (NB) media (Gibco 21103-049) supplemented with B27 (50X, Gibco 17504-044), GlutaMAX (100X, Gibco 35050-061), Penicilin/Streptomycin (100X, Gibco 15140122), plus 10 ng/mL PDGF-AA (Peprotech 100-13A) for tumor OPC experiments. For all experiments in this paper, Tumor OPC cell lines used were between passages 2 to 10. All cells were kept in a 5% CO2 incubator at 37°C.
Lentivirus production and cell infection
The viral packaging line HEK 293T was maintained in DMEM media with High Glucose (Gibco, 11995-065) supplemented with 10% fetal bovine serum (FBS - GIBCO, 16000-044). Lentivirus production was performed through calcium phosphate transfection of HEK 293T cell line with a 3rd generation packaging system (3 packaging plasmids + vector plasmid). 6–12 hours after transfection, HEK 293T cells were switched to NB media supplemented with B27, GlutaMAX and Penicilin/Streptomycin (without serum or growth factors). Media containing lentivirus were collected daily for 3–4 days, filtered through a 0.22 um pore syringe filter membrane, and supplemented with the appropriate growth factors for OPC culture before being added to the target cells. Two rounds of infection were performed to achieve the maximal viral transduction rate, with 24 hours for each round of infection. All cells were kept in a 5% CO2 incubator at 37°C.
Flow analysis of cell death and cell-cycle profile
Cells were trypsinized to detach from flasks. For cell death analysis, cells were washed 1 time with 5 mL of 1x PBS then treated with 1mg/mL propidium iodide (PI). Because live cells with intact plasma membrane will not take up PI, any PI positive cells were considered “dead” cells. To avoid including potential cell debris, we only considered the population within the top portion of the flow plot as dead cells (Fig. S3). For cell-cycle analysis, cells were washed 1 time with 1x PBS and centrifuged down at 300 g for 5 minutes. Cells were then suspended in 0.5 mL PBS and fixed in 70% ethanol for 2 hours. Following fixation, cells were centrifuged down at 300 g for 5 minutes and then washed 1 time with PBS. Cells were centrifuged down again, suspended in 1 mL PBS containing 1 mg/mL PI for 15 min at RT, and used for flow analysis.
Quantitative RT-PCR
Total RNA was extracted by TRI Reagent® (Sigma, cat. # T9494-200ML), then treated with DNAse I (New England Biolabs Inc., cat. # M0303). cDNA was synthesized using iScript Reverse Transcription Supermix (Bio-Rad, 170-8841). qPCR was performed with KAPA SYBR FAST ABI Prism Kit (KAPA Biosystems, KK4605) in an Applied Biosystems StepOnePlusTM Real-Time PCR System for 40 cycles (denaturation: 95°C, 3 seconds; annealing: 60°C, 30 seconds), followed by a default Melting Curve program. Beta-actin gene expression was used as an internal control for every sample. Ct values were measured within the geometric amplification phase, and triplicates were used to get the average value.
Western blotting
Cells were lysed in ice-cold lysis buffer 17 (R&D, 895943), supplemented with protease inhibitor cocktail tablets (Roche, 11836153001 – 1 tablet for 10 mL of Lysis buffer) and Halt Phosphatase Inhibitor (100X, Thermo Scientific, 1862495). Sample loading was adjusted according to the measurement of protein concentration by Pierce® BCA Protein Assay Kit (Thermo Scientific, cat. # 23227), and beta-actin antibody (Sigma, cat. # A5441) was used as an internal control for normalization. Samples were run in Mini-PROTEAN® TGXTM Gels (Bio-Rad, cat. # 456-9033), and transfer was performed with Nitrocellulose membranes (Amersham, cat. # RPN 2020 D). Membranes were blocked in 5% BSA solution for 1 hour at RT, then incubated with primary antibody (see list below) overnight at 4°C. After washing with TBST, membranes were incubated with secondary antibody for 1 hour at RT. Secondary antibodies used were all purchased from Li-Cor® or Jackson Immunology Laboratories. Detection was performed in an Odyssey Infrared Imaging System or Gel Doc.
| Antigen | Species | Dilution | Source | Catalog no. |
|---|---|---|---|---|
| Total 4EBP1 | Rabbit | 1:1000 | Cell Signaling | 9452 |
| p-4EBP1 | Rabbit | 1:500 | Cell Signaling | 2855 |
| Total AKT | Mouse | 1:1000 | Cell Signaling | 2920 |
| p-AKT | Rabbit | 1:500 | Cell Signaling | 9275 |
| Total ERK | Mouse | 1:1000 | Cell Signaling | 9107 |
| p-ERK | Rabbit | 1:500 | Cell Signaling | 4370 |
| Total 70S6K | Rabbit | 1:1000 | PMID # 17967879 | - |
| p-70S6K | Mouse | 1:500 | Cell Signaling | 9206 |
MTT assay
MTT solution (Invitrogen, M-6494) 5 mg/mL was added to cell media in a 1:10 dilution, and incubated in the dark at 37oC, 5% CO2 for 3 hours. Media was then aspirated and formazan crystals were dissolved in DMSO. Absorbance was read on a plate reader at 570 nm wavelength.
Drug assays
PreT-OPC and Tumor OPC cells were plated in 96-well plates at the same density, and then treated with pre-determined concentrations of Temsirolimus (Selleckchem, cat. # S1044), BEZ235 (Selleckchem, cat. # S1105), or PD0325901 (Selleckchem cat. # S1036) 72 hours after treatment, cell viability was accessed by MTT assay.
Quantifications
For all in vivo quantifications, images were systematically taken throughout the entire brain along rostrocaudal, dorsoventral and mediolateral axes. All quantifications are representative of >200 cells positive for each marker analyzed. For p53-null and WT brains, quantifications are representative of at least 50 cells/marker/mouse since labeling efficiency using MADM is significantly lower. For in vitro quantifications, >300 cells/marker were counted per data point.
Statistics
Data are shown as an average of 3 (in vitro) or >5 (in vivo) independent experiments ± standard deviation (SD) or standard error of the mean (SEM). Significance of multiple comparison in Fig. 3A, 4B–E, 5B,E,F, and S2B was determined by a 1-way ANOVA with Dunnett tests, while significance of pair wise comparison in Fig. 6B, 7B, and S1A,C was determined by student t test. Significance in Fig. 3B–C was determined by 2-way fixed effects ANOVA with replication, because we made the comparison between single-gene knockout and double-knockout to assess synergy or antagonism between the two single genotypes (p53 and/or NF1) in the double knockout.
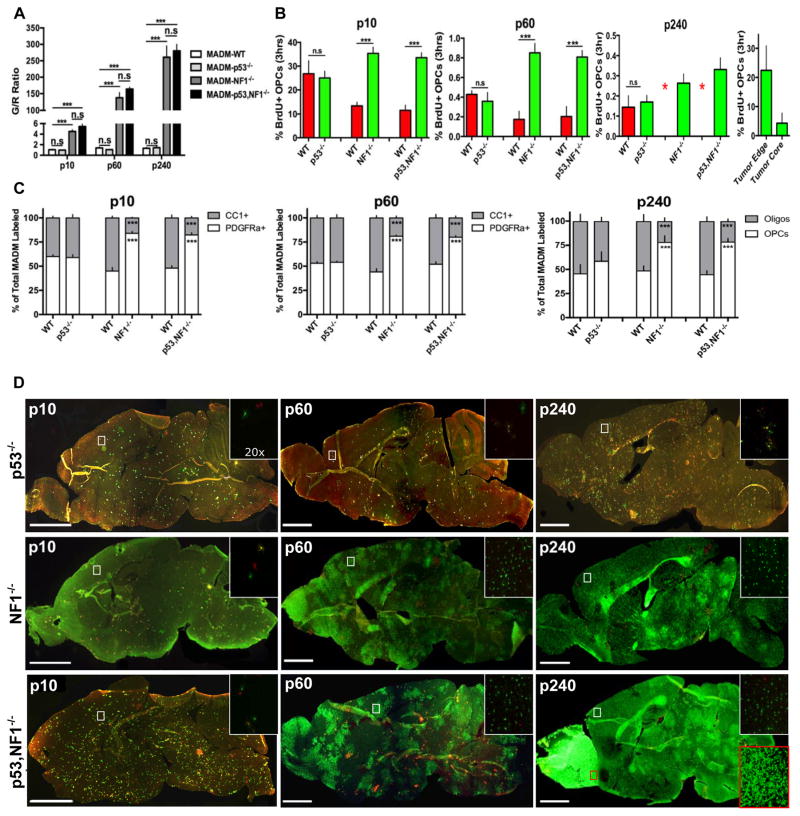
Fig. 3. Loss of NF1 but not p53 is sufficient for over-expansion, increased proliferation and hindered differentiation of mutant OPCs.
(A) Following the individual loss of either NF1 or p53, MADM labeled cells were quantified for the mutant to WT ratio (G/R ratio) at early (P10), pre-transforming (P60) and tumor stages (P240). The loss of NF1 led to massive over-expansion of mutant OPCs comparable to p53-null, NF1-null OPCs at all ages examined, but the loss of p53 had no effects. (n=10) 1-way ANOVA with Dunnett tests; Error Bar ± SEM (*** P < 0.001).
(B) Following the individual loss of either NF1 or p53, the proliferative rate of MADM labeled cells was assessed at P10, P60 and P240. The loss of NF1 led to increased proliferation of mutant OPCs comparable to p53-null, NF1-null OPCs but the loss of p53 had no effects. At P240 WT OPC numbers were too low in MADM-NF1 and MADM-p53,NF1 models for the accurate quantification of proliferative rate (marked as red asterisks). Right panel: tumor cells at the edge of tumor mass showed comparable proliferative rate as perinatal OPCs. 2-way fixed effects ANOVA with replication; Error Bar ± SEM (*** P < 0.001, ** P < 0.01)
(C) Following the individual loss of either NF1 or p53, the relatively proportion of OPCs and oligodendrocytes among MADM-labeled cells were quantified at P10, P60 and P240. The loss of NF1 but not p53 was sufficient for the hindrance in OPC differentiation at all ages examined. 2-way fixed effects ANOVA with replication; Error Bar ± SEM (*** P < 0.001, ** P < 0.01)
(D) Individual loss of either p53 or NF1 alone was not sufficient for gliomagenesis even though NF1 loss led to massive expansion of cell number, increased proliferative rate, and hindered differentiation of OPCs. Insets show higher magnification images of boxed brain region. The red inset in the bottom right panel shows the tumor region in the MADM-p53,NF1 model.
Scale bar 2mm; Inset 50μm
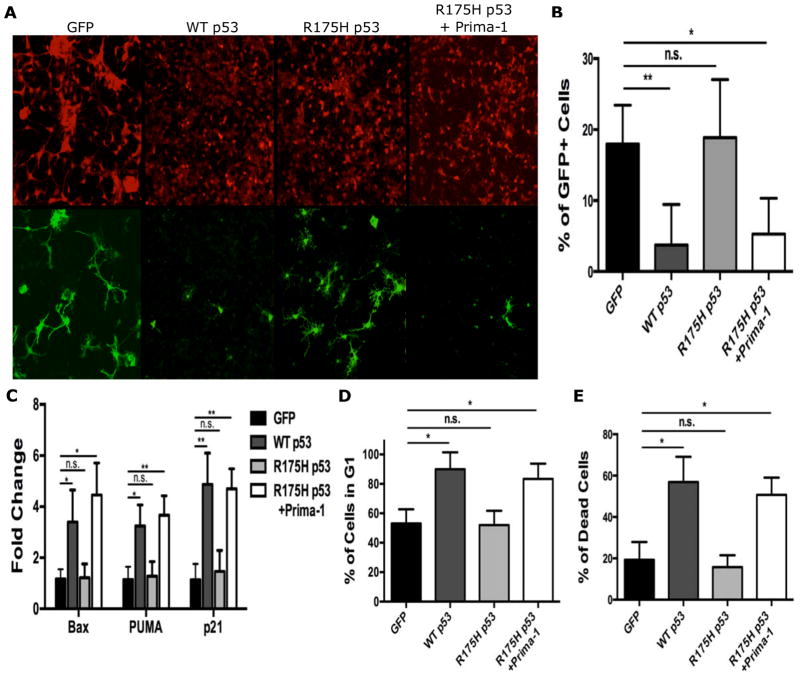
Fig. 4. Restoration of WT p53 function in tumor OPCs led to cell cycle arrest and cell death.
(A–B) 48 hours after the initial infection, the percent of GFP+ tumor OPCs infected with WT p53 decreased significantly in comparison to GFP control and mutant p53R175H-infected cells. With the functional restoration of p53R175H by PRIMA-1, the percentage of GFP+ tumor OPCs also significantly decreased. Scale bar 150μm. N=3
(C) 48 hours after infection or treatment with PRIMA-1, the transcriptional activity of known p53 targets were analyzed. Following the infection of WT p53 or the functional restoration of p53R175H with PRIMA-1, there was significant up regulation of Bax, PUMA, and p21 transcripts compared to GFP control. The infection of tumor OPCs with p53R175H alone had no significant effect on transcript levels of all three genes.
(D) 48 hours after infection, flow analysis of cell cycle profiling was performed on tumor OPCs. WT p53 and p53R175H-nfected cells treated with PRIMA-1 had a significant increase in the percentage of cells in G1 compared to GFP control and p53R175H-nfected cells without PRIMA-1.
(E) 48 hours after infection, flow analysis of cell death was performed on tumor OPCs. WT p53 and p53R175H-infected cells treated with PRIMA-1 had a significant increase in the percent of dead cells compared to GFP control and p53R175H-nfected cells without PRIMA-1. N=3
Statistics in B–E: 1-way ANOVA with Dunnett tests; Error Bar ± SD (* P < 0.05, ** P < 0.005)
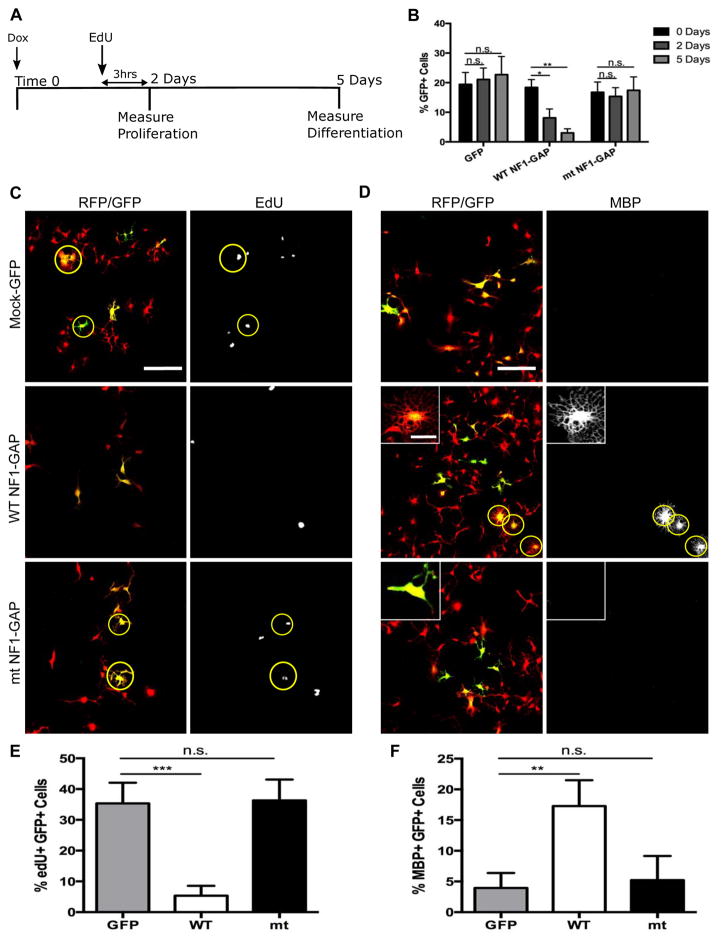
Fig. 5. Restoration of NF1-GAP function is sufficient to reduce proliferation and promote differentiation of tumor OPCs.
(A) Timeline schematic for determining the effect of NF1-GAP restoration in tumor OPCs. Cells were treated with Doxycycline to induce expression of GAP domain at Time 0. 2 days after the initial Doxycycline treatment, tumor OPCs were treated with EdU for 3 hours to measure the proliferative rate. 5 days after the initial Doxycycline treatment, tumor OPC differentiation was determined by immuno-staining cells for MBP, a marker for differentiated oligodendrocytes.
(B) 2 and 5 days after infection of tumor OPCs with WT NF1-GAP, the percent of GFP+ tumor OPCs significantly decreased at both time points compared to mock-GFP control and mutant NF1-GAP infected tumor OPCs. (n=3) Student t Test; Error Bar ± SD (* P < .05, ** P < 0.005)
(C) Following 2 days of NF1-GAP expression, tumor OPCs were pulsed with EdU for 3 hours to determine the proliferative rate. While mutant NF1-GAP infected tumor OPCs showed no significant change compared to mock-GFP control, WT NF1-GAP infected tumor OPCs had a significant reduction in the amount of EdU+ tumor OPCs.
(D) 5 days after restoration of WT NF1-GAP but not mock-GFP or mutant NF1-GAP, tumor OPCs developed highly elaborated oligodendrocyte-like morphology and started expressing oligodendrocyte marker MBP. Inset shows the morphological differences between WT and mutant NF1-GAP expressing cells (note the ramified processes and web-like appearance of MBP+ cell in the middle panel).
(E) Quantification of the percentage of EdU+, GFP+ tumor OPCs in mock-GFP, WT NF1-GAP, and mutant NF1-GAP infected cells.
(F) Quantification of the percentage of MBP+, GFP+ tumor OPCs in mock-GFP, WT NF1-GAP, and mutant NF1-GAP infected cells.
Statistics in B,E,F: 1-way ANOVA with Dunnett tests; Error Bar ± SD (** P < 0.005)
Scale bar, 100 μM; inset, 25 μM
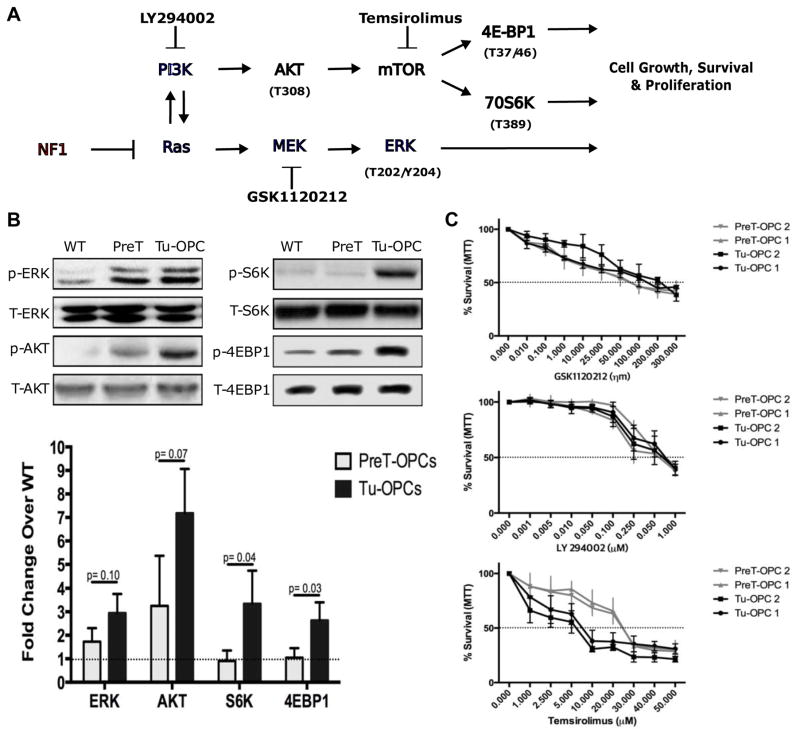
Fig. 6. Deletion of NF1 leads to differential activation of downstream Ras effectors in Pre-T and tumor OPCs.
(A) Schematic illustration of the Ras signaling pathways, with a focus on PI3K and MEK/ERK. Phosphorylation sites for activation through specific pathways are shown. Also included are known inhibitors for some of the signaling proteins.
(B) Western blots shows that while the deletion of NF1 leads to increased levels of phosphorylated (p) ERK, pAKT, pS6K, and p4EBP1 in tumor OPCs. In Pre-T OPCs, only pERK and pAKT are elevated in comparison to WT OPCs, but pS6K and p4EBP1 show no significant changes. Bottom graph shows the quantification of the level of pERK, pAKT, pS6K and p4EBP1 in WT OPCs, PreT-OPCs, and tumor OPCs. Student t test; Error Bar ± SD
(C) PreT-OPCs and tumor OPCs were treated with a MEK inhibitor (GSK1120212), PI3K inhibitor (LY294002), or mTOR inhibitor (Temsirolimus) and assayed 48 hours later for survival with MTT. Dashed line represents IC50 value. (n=3)
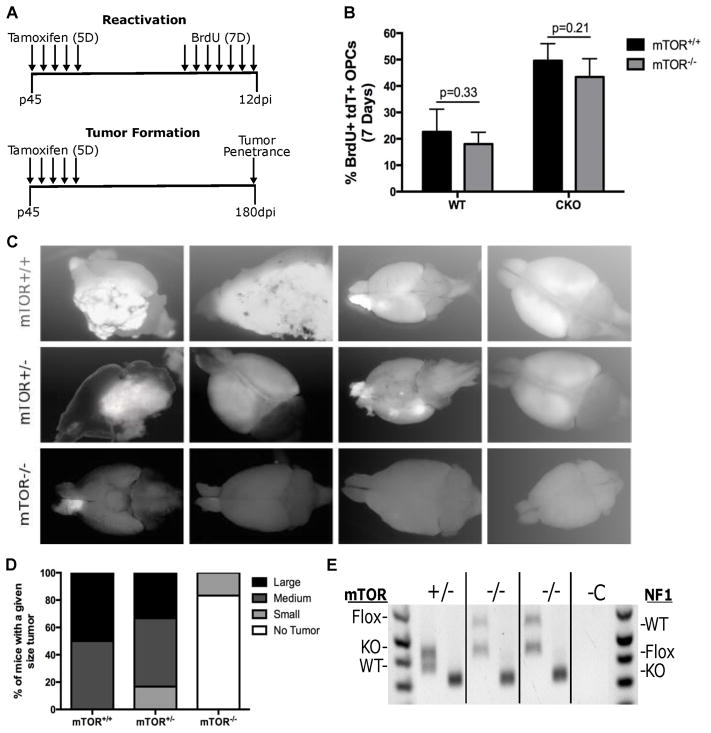
Fig. 7. mTOR deletion blocked gliomagenesis.
(A) Top schematic shows the experimental design for testing the effect of mTOR deletion on PreT-OPC reactivation. Mice were treated for 5 days with Tamoxifen to induce cre-mediated recombination and then treated for 7 days with BrdU to label dividing OPCs. Bottom schematic shows the experimental design for testing the effect of mTOR deletion on gliomagenesis. Following 5 days of Tamoxifen treatment, mice were kept for 180 days to determine the tumor penetrance.
(B) Following 7 days of BrdU injections, mice were examined for the percent of dividing OPCs. In both WT and glioma model mice, mTOR-KO led to no significant differences of OPC reactivation. (n=4 WT, n=5 CKO) Student t Test; Error Bar ± SD
(C) Glioma model mice (p53-null,NF1-null) with either mTOR-WT, mTOR-het, and mTOR-null status were dissected at P240 to determine the tumor penetrance. While mTOR-WT CKO mice always had large tumors, mTOR-null CKO mice had either none or occasionally very small tumors. The images are representative of the average tumor size in mTOR-WT, mTOR-het, and mTOR-null glioma mice. More than 10 mice were examined in each group.
(D) Semi-quantitative representation of tumor penetrance and sizes in glioma model mice (p53-null,NF1-null) with either mTOR-WT, mTOR-het, and mTOR-null status.
(E) Genotyping of tumor OPCs purified from mTOR-null brains. For each genotype (indicated at top), the left bands represent mTOR PCR and the right band represents NF1 PCR. While Cre-mediated recombination of the NF1-flox alleles was complete (only KO band was detected) in all tumor cells, the recombination of mTOR-flox alleles was incomplete in both mTOR-null tumors (evidenced by the presence of the mTOR-flox band). We are confident that the mTOR-flox allele in tumor cells came from “escaper cells” rather than contaminated non-tumor cells because of the absence of mTOR-flox band in the +/− lane and our confirmation of cell purity based on PDGFRα staining after immunopanning of tumor OPCs.
Results
Establishment of MADM-based models for dissecting the individual roles of p53 and NF1 loss in glioma initiation and progression
To investigate the individual roles of p53 and NF1 in gliomagenesis, we first established two MADM-based models in which sporadic GFP+ OPCs are either p53-null (MADM-p53) or NF1-null (MADM-NF1), respectively (Fig. 2A). Along with the original MADM tumor model in which GFP+ OPCs are null for both p53 and NF1 (MADM-p53, NF1) and MADM mice with no mutations (MADM-WT) as controls, we examined four parameters of OPC biology in all these mice: 1) the overall expansion of mutant cells based on the ratio between GFP+ mutant cells and RFP+ WT cells (G/R ratio); 2) the proliferative rate; 3) the differentiating capability; 4) the capacity for malignant transformation of GFP+ mutant OPCs. Since MADM-labeled mutant OPCs are detectable within hours of their generation, it enabled us to perform phenotypic analysis from pre-malignant to malignant stages, specifically postnatal day 10 (P10) when normal OPCs proliferate rapidly; P60 when both WT and mutant OPCs are mostly quiescent, and P240 when tumors start forming in the MADM-p53,NF1 model as previously shown (9).
To analyze these parameters in a systematic fashion, we sectioned each brain sagitally at 20 μm thickness, performed immuno-fluorescent staining on 7 brain sections that are 200 μm apart medialaterally (Fig. 2B left), and quantified cell numbers on 7 distinct areas per section (Fig. 2B right). To assess the effect of gene deletion on mutant OPC expansion based on G/R ratio, the total number of GFP+ OPCs were divided by the total number of RFP+ OPCs (Fig. 2C). Next, to assess the effect of gene deletion on OPC proliferation, the number of dividing mutant OPCs (BrdU+ GFP+ PDGFRa+) were divided by the total number of mutant OPCs (GFP+ PDGFRa+) (Fig. 2D). Proliferative rate of WT OPCs was calculated with a similar method by focusing the analysis on RFP+ PDGFRa+ cells (Fig. 2D). Last, to quantify the effect of gene deletion on the differentiation potential of mutant and WT cells, MADM brains were stained with markers for both OPCs (PDGFRa) and oligodendrocytes (CC1), and then the composition of each population among total mutant (GFP+) or WT (RFP+) cells was calculated (Fig. 2E) (23). In summary, we expect that, using RFP+ WT sibling cells produced along with GFP+ mutant cells in the MADM model as an internal control, we should be able to detect even the subtlest phenotypes in mutant cells.
Individual deletion of p53 or NF1 led to distinct phenotypes in OPCs but was insufficient for malignant transformation
To assess how single gene deletion contributes to the previously reported pre-malignant expansion of OPCs in the MADM-p53,NF1 model (24), we systematically quantified the G/R ratio throughout the brains of MADM-p53 and MADM-NF1 models at the aforementioned time points (Fig. 2C, see Materials and Methods for detailed procedures). While we expected that deletion of p53 or NF1 alone would each partially contribute to OPC expansion, we were surprised to find that the single deletion of NF1 resulted in a G/R ratio that phenocopied the concurrent deletion of p53 and NF1 at all time points that we examined (Fig. 3A), and that the single deletion of p53 had a G/R ratio of approximately 1, comparable to MADM-WT mice (Fig. 3A). This indicates that NF1 deletion is solely responsible for the pre-malignant expansion of mutant OPCs.
Next, to determine the effect of single NF1 or p53 deletion on OPC proliferation, we quantitatively measured the proliferative rate of OPCs via BrdU labeling at P10, P60, and P240 (Fig. 2D). First, we noticed that the proliferative rate of normal OPCs dramatically decreases after early development, from ~25% within a 3 hr time window at P10, to ~0.4% at P60, then to ~0.1% at P240 (Fig. 3B), which matches well with previous independent studies (25, 26). When gliomas form in our model, the proliferative rate of tumor OPCs returns to the level of neonatal stage, especially at the tumor edge (Fig. 3B, right most panel). Furthermore, while p53-null OPCs showed no significant difference in proliferative rate when compared to WT OPCs, NF1-null OPCs proliferated at significantly higher rate than WT counterparts in the MADM-NF1 model, comparable to p53-null,NF1-null OPCs (Fig. 3B). Interestingly, in both MADM-NF1 and MADM-p53,NF1 models, the proliferative rate of WT cells appeared to be suppressed to a level lower than those in MADM-WT or MADM-p53 brains, which eventually became undetectable at P240 (Fig. 3B, red asterisk). This suggests a possible non-cell autonomous suppression from NF1-null or p53-null,NF1-null OPCs toward their WT counterparts in the same brain. These findings demonstrated that NF1 deletion but not p53 deletion led to enhanced proliferation of mutant OPCs.
To evaluate the differentiating potential of OPCs in aforementioned four mouse models, we immunostained brain sections with specific markers for un-differentiated OPCs (PDGFRα) and differentiated oligodendrocytes (CC1), and quantified the percentage of each cell types within either the mutant (p53-null, NF1-null, or p53-null,NF1-null) or WT populations (Fig. 2E). While p53 deletion had little effect, the deletion of NF1 led to a nearly 1.5-fold increase (p<0.001) in OPC composition and a 50% reduction in oligodendrocyte composition (p<0.001) (Fig. 3C), replicating the observations in the MADM-p53,NF1 model. These data suggest that NF1 deletion but not p53 deletion led to a decrease in differentiation of OPCs. It is worth noting that, while the change of relative composition between OPC and oligodendrocyte could be alternatively explained by a shift in apoptotic rate in one or both cell types, decreased differentiation due to NF1 loss should be the more likely interpretation because data below showed that restoring NF1 activity enhanced differentiation capacity of transformed OPCs (Fig. 5D,F).
Our data so far have shown that, while NF1-null OPCs recapitulated p53-null,NF1-null OPCs in every aspect including cell number expansion, increased proliferation and compromised differentiation capacity, p53 deletion seemed to have no clear effect on OPCs. However, when we dissected mice of MADM-p53 and MADM-NF1 models at the tumor latency age of the MADM-p53,NF1 model, we found no tumors in both MADM-p53 and MADM-NF1 models. This was surprising since all 18 MADM-NF1 mice had massive expansion of mutant OPCs in the brain (Fig. 3D). This finding suggests that the over-expansion of mutant OPCs caused by NF1 deletion is not sufficient for malignant transformation, and that p53 plays a critical gate-keeping role to prevent tumor formation.
One unanswered question however, was what effect p53 deletion had on mutant OPCs, since our in vivo analysis revealed no detectable difference in OPC numbers, proliferation, and differentiation. Based on known p53 functions, we hypothesized that p53 could exert tumor-suppressing activity through promoting apoptosis and/or senescence in NF1-null OPCs. To test this hypothesis, we first compared the expression level of known p53-target genes involved in apoptosis and senescence between NF1-null and p53-null,NF1-null OPCs. We found that NF1-null OPCs exhibited significantly higher expression level of p21, Bax, or PUMA than p53-null,NF1-null OPCs did (Fig. S1A). Second, we used β-Gal to assess cellular senescence (27, 28) in cultured cells and found increased senescence in NF1-null OPCs in comparison to p53-null,NF1-null OPCs (Fig. S1B,C). This finding concurs with previous work showing that over-active Ras signaling in OPCs leads to p53-dependent senescence (29). All these data suggest that deletion of p53 most likely promotes gliomagenesis by allowing OPCs to escape apoptosis and senescence triggered by oncogenic stresses induced by mutations such as NF1 deletion.
Restoration of p53 in tumor OPCs caused massive cell-death and cell-cycle arrest
Having shown that deletion of p53 is vital for OPC transformation, we next wanted to determine if the restoration of p53 in p53-null,NF1-null tumor OPCs could have anti-tumor effects. While tumor OPCs infected with adenovirus encoding GFP control or p53-expressing viruses had similar rates of infection initially (Fig. S2A), after 48 hours the number of tumor OPCs transduced with p53-expressing virus were 3-fold lower when compared to those infected with the control virus (Fig. 4A,B), suggesting that the expression of p53 led to cell cycle arrest or cell death in tumor OPCs. To determine if p53 activity is required for the reduction of cell number, tumor OPCs were also infected with the mutant form of p53R175H that is commonly found in gliomas. While initial infection rates between GFP-virus and p53R175H-virus were similar (Fig. S2A), the percentage of p53R175H-expressing virus infected cells remain indistinguishable from control virus-infected cells after 48 hours (Fig. 4A,B). However, the treatment of the compound PRIMA-1 previously reported to restore DNA binding activity of p53R175H (30) led to a significant decrease of cell numbers in p53R175H-expressing cells, suggesting that restoring p53 activity in tumor OPCs has significant anti-tumor effects.
To confirm the restoration of p53 function, we analyzed expression levels of known transcriptional targets of p53. 18 hours post-infection Bax, PUMA, and p21 transcript levels significantly increased in cells over-expressing WT p53 (p<0.05) or p53R175H+PRIMA-1 (p<0.05) but not those over-expressing GFP or p53R175H (Fig. 4C). To test whether the loss of tumor OPCs infected with WT p53 was due to cell cycle arrest or cell death, we used FACS to analyze the cell cycle profiles based on DNA content and to quantify cell death based on propidium Iodide incorporation. 48 hours post-infection, both WT p53 and p53R175H+PRIMA-1 infected OPCs showed not only G1 arrest (Fig. 4D) but also increased cell death (Fig. 4E & S3A) in comparison to GFP- or p53R175H-expressing cells, suggesting that p53 deterred tumor OPCs through both cell cycle arrest and augmented cell death.
Finally, to confirm the human relevance of our findings, we repeated the experiments in p53-null human glioma cell line, LNZ-308. Cells were infected with GFP, WT p53, p53R175H, and p53R175H+PRIMA-1 and assayed for cell survival 48 hours after infection. Similar to mouse tumor OPCs, LNZ-308 cells exhibited a 3-fold increase in cell death in both WT p53 and p53R175H+PRIMA-1 infected cells when compared to either GFP control or p53R175H infected cells (Fig. S2B). These results demonstrated that restoring p53 activity could be a promising therapeutic strategy for glioma treatment.
Restoration of NF1 activity inhibits tumor OPC proliferation while promoting differentiation
MAMD-based analysis showed that the deletion of NF1 in OPCs led to increased proliferation and decreased differentiation long before malignant transformation, it would be interesting to determine whether the restoration of NF1 activity in tumor cells could reverse these effects. Because full-length NF1 cannot be readily packaged into viral vectors, we focused on its GAP (GTPase activating protein) domain since previous studies have shown that turning off Ras through its GAP activity is NF1 protein’s main function. Considering possible strong tumor-inhibiting activity of NF1-GAP, we constructed lentiviral vectors with a dual-promoter system to allow the verification of infection rate via GFP expression prior to inducible expression of NF1-GAP (Fig. S4A).
First we confirmed that the infection rate was equivalent between the control and NF1-GAP infected tumor OPCs (Fig. S4B). 48 hours after infection, doxycycline (Dox) was added to induce the expression of NF1-GAP (Fig. 5A, time 0) and then incubated for 2 days to assess proliferation and for 5 days to assess differentiation (Fig. 5A). It became readily evident that the number of NF1-GAP-infected tumor OPCs gradually decreased after 2- and 5-days of Dox treatment, while the number of mock-infected cells remained steady (Fig. 5B). Using EdU labeling as the S phase marker, we found that WT NF1-GAP expressing tumor OPCs had a much lower proliferative rate in comparison to mock-infected cells 2-days post-Dox treatment (Fig. 5C & E). Additionally, 5-days post-Dox treatment, while mock-infected Tumor OPCs had little morphological changes, NF1-GAP infected OPCs showed greatly increased ramifications, indicative of oligodendrocyte differentiation. To confirm this finding, we stained these cells with oligodendrocyte marker MBP and found a significant increase of MBP+ cells when NF1-GAP was over-expressed in tumor OPCs (Fig. 5D & F), suggesting NF1 restoration in glioma cells could promote their differentiation into oligodendrocytes. To firmly establish that the GAP activity is responsible for our observations, we also infected tumor OPCs with a mutant form of NF1-GAP in which the Arginine critical for enhancing the GTPase activity of Ras has been replaced with Proline, resulting in a loss of GAP activity. Contrary to the findings with WT NF1-GAP, the abundance of mutant NF1-GAP-infected tumor OPCs after 2- and 5-days of Dox induction remained the same as mock-infected cells (Fig. 5B). Additionally, mutant NF1-GAP-infected tumor OPCs showed no significant difference in proliferative or differentiating capacity compared to mock-infected cells (Fig. 5C–F). Taken together, the restoration of WT NF1-GAP is sufficient to inhibit proliferation as well as to promote differentiation of tumor OPCs, most likely through shutting down Ras activity.
Signaling analysis in pre-transforming and tumor OPCs revealed progressive activation of ERK and AKT but abrupt activation of mTOR from pre-malignant to malignant stages
Our findings above showed that, even though pre-transforming mutant OPCs (PreT-OPCs) had increased proliferative capacity when compared to WT OPCs, tumor OPCs proliferate at an exceedingly faster rate. Since NF1 deletion and the subsequent Ras activation appears to be responsible for the heightened proliferative rate, we decided to examine whether a few signaling transducers in the Ras pathway (Fig. 6A) are activated to a different level from pre-transformation to malignant stages of gliomagenesis. First, we purified WT, PreT-OPCs that are p53-null,NF1-null, and tumor OPCs with the immunopanning method. Then we analyzed the levels of phosphorylated (p) ERK, AKT, S6K, and 4EBP1 by western blot (Fig. 6B). Interestingly, while the activation of ERK and AKT followed a progressive fashion, i.e. moderately increased in PreT-OPCs then further increased in tumor OPCs (model 2 in Fig. 1E), mTOR activation revealed by pS6K and p4EBP1 was rather abrupt. i.e. minimal activation in PreT-OPCs followed by sudden increase in tumor OPCs (model 3 in Fig. 1E). Therefore, over-active ERK and AKT are most likely responsible for the pre-malignant expansion of PreT-OPCs, while elevated mTOR activity could be specifically involved in final transformation.
It should be noted that, because WT OPCs used in this assay were purified from neonatal brains thus may have higher mTOR activity than adult OPCs, mTOR activity in PreT-OPCs could be slightly elevated and still play a role in progression toward malignancy. Nevertheless, we predicted that the responsiveness of PreT-OPCs and tumor OPCs toward mTOR inhibitor would be quite different. To test this hypothesis, we treated PreT-OPCS and tumor OPCs with mTORC1 inhibitor Temsirolimus, and also MEK and AKT inhibitors as controls. Data from MTT assays showed that, while the survival curve of PreT-OPCs and tumor OPCs mostly overlapped when treated with MEK and AKT inhibitors, tumor OPCs were much more sensitive to Temsirolimus than PreT-OPCs were, exactly as we predicted (Fig. 6C). This further demonstrated that elevated mTOR activity is critical in tumor OPCs and could be a driver event for malignant transformation, but is less critical during the pre-malignant stage.
mTOR is critical for the transformation of reactivated Pre-Transforming OPCs
Our data above showed that the deletion of NF1 leads to gradual activation of both ERK and AKT from pre-malignant to tumor stages yet mTOR signaling seems to be specifically activated at the tumor stage. Since previous studies have shown that pathways altered at the late stage of tumorigenesis increase malignancy (31–33) and that both AKT and ERK pathways converge on mTOR (34), we genetically inactivated mTOR to assess its impact on gliomagenesis.
Our previous studies demonstrated that the inactivation of p53 and NF1 through OPC-specific NG2-CreER in quiescent adult OPCs led to cell cycle re-entry (reactivation) at 12-day post injection (12 dpi) of Tamoxifen and subsequently transform into glioma at 180 dpi (13). In the current study, we introduced floxed alleles of mTOR into the previous model to determine the necessity of mTOR in OPC reactivation and transformation (Fig. 7A). Following the daily Tamoxifen administration at P45 for 5 consecutive days, mice were injected with BrdU for 7 days to label proliferating cells. In WT animals, the deletion of mTOR had no significant impact on OPC proliferation compared to control brains, suggesting that mTOR is not critical for baseline level proliferation of adult OPCs (Fig. 7B, columns on the left side). In the glioma model, the augmented proliferative rate of PreT-OPCs in comparison to WT OPCs was indistinguishable between mTOR WT and mTOR deletion mice (Fig. 7B, columns on the right side), indicating that mTOR activity is not needed for the reactivation of PreT-OPCs. To determine the effect of mTOR deletion on OPC transformation, mice were examined at 180 dpi for both tumor penetrance and sizes. Compared to control mice that normally form large tumors by 180 dpi, mTOR−/− mice rarely had tumors (2/10) and two tumors that formed were very small in size (Fig. 7C,D). Furthermore, after we purified tumor OPCs using immunopanning from those two small tumors in mTOR−/− mice, genotyping result showed that one of the floxed mTOR alleles was still intact, suggesting that they are “escaper cells” due to the inefficiency of NG2-CreER (Fig. 7E). Taken together, these data clearly demonstrated the absolute requirement of mTOR activity for malignant transformation. Therefore, while mTOR is not necessary for initial reactivation of PreT-OPCs, it is critical for their final transformation, making it an excellent target for glioma treatment.
Discussion
Gliomagenesis involves multiple pathway mutations (6, 7, 35) and goes through a long latency. Until this study, it remained unknown when and how mutations in both p53 and Ras pathways contribute to glioma formation, due to the inaccessibility to the pre-malignant stage with human samples or conventional mouse models. Such hurdle is readily circumvented using the MADM model, owing to its unequivocal fluorescent protein labeling of mutant and sibling WT cells. In the current study, we first used MADM to clearly distinguish tumor-promoting functions of p53 and NF1 at the pre-malignant stages in vivo. Then we demonstrated that the restoration of p53 or NF1 activity in fully-transformed p53-null,NF1-null glioma cells could still revert the biological aberrations caused by their loss. Next we examined the temporal sequence of downstream signaling protein activation upon NF1 loss by comparing their activities in Pre-T and tumor OPCs, and observed two distinct patterns: progressive and abrupt activation. Finally, targeting mTOR that is abruptly activated at the malignant stage successfully blocked glioma formation. Overall, our study demonstrated how fine dissection of tumorigenic process could shed light on rationally designed therapeutic strategies to prevent and treat glioma effectively.
Because others and we previously identified OPCs as a cell of origin for glioma, in our current study we focused on the roles of NF1 and p53 in OPCs but not other cell types that respond minimally to these mutations. During normal embryonic development, OPCs originate from neural stem cells, migrate tangentially throughout the brain, continue to proliferate after birth, and differentiate into oligodendrocytes by the weaning age (36). Adult OPCs remain in a quiescent state and only re-enter cell cycle at a low frequency to replenish the OPC population (26, 37, 38). Based on the progenitor nature of OPCs, it is conceivable that either increased proliferation or decreased differentiation of OPCs, or both, could lead to tumor initiation. We found that, while the loss of NF1 leads to mutant OPC expansion due to increased proliferation and compromised differentiation, loss of p53 contributes to gliomagenesis specifically by alleviating NF1-loss induced senescence/apoptosis thus allowing further progression toward final transformation, as seen in other tumor types (39–41). Of course, it remains possible that p53 exert other tumor-suppressing functions since recent reports have shown that p53 could block tumor formation independent of cell-cycle arrest, senescence, and apoptosis (42, 43). Regardless of how p53 exerts its tumor suppressor function, it is clear that p53 deletion is needed for the progression of NF1-null OPCs to tumor OPCs.
While some oncogenic mutations are only required for the initial transformation, other gene mutations are critical for sustaining tumor cells even after the malignant transformation, known as oncogene addiction (44, 45). To test whether p53 and NF1 pathways are critical for sustaining glioma cells, we restored their expression in tumor OPCs to determine if it could be therapeutically efficacious. The restoration of p53 in both muse tumor OPCs and human glioma cells led to senescence and cell death, suggesting that the restoration of p53 activity in glioma may prove effective in the clinics once it is pharmacologically feasible, a notion in line with recent work demonstrating that p53 restoration is efficacious in treating astrocytomas (46). Similarly, we also found that the restoration of NF1’s GAP activity (47, 48) could greatly inhibit proliferation and promote differentiation of tumor OPCs, demonstrating the promise to therapeutically target the Ras signaling axis to treat glioma patients. It should be noted that cellular differentiation cannot be measured with conventional survival assays such as MTT. Therefore, it would be wise to broaden assay formats to assess therapeutic efficacy beyond proliferation or cell death. Of course, pinpointing the cell of origin and identifying its differentiation markers would be the prerequisites for developing robust differentiation assays.
Since tumor OPCs proliferate much faster than Pre-T OPCs and NF1-loss/Ras activation is solely responsible for OPC proliferation, we reasoned that the difference could be caused by varied activities of Ras pathway effectors at pre-malignant versus malignant stages. Interestingly, we found that, while both ERK and AKT undergo partial activation at pre-malignant then full activation at malignant stages, mTOR remains at the basal level in Pre-T OPCs and only becomes highly activated in tumor OPCs. This suggests that, while partially activated ERK and AKT are likely involved in the pre-malignant progression, fully activated ERK, AKT, and mTOR are critical for malignant transformation. Considering the unique status of mTOR, we chose to target it genetically to see whether this signaling node is critical for OPC transformation. As expected, following mTOR deletion, PreT-OPC reactivation is unaffected but malignant transformation is completely blocked. This finding, while exciting, poses an apparent contradiction with our previous work showing that mTOR inhibitor temsirolimus could inhibit Pre-T OPC reactivation (13). One conceivable explanation is that, since mTOR KO is restricted to OPCs while the drug works on all cell types, the inhibition of OPC reactivation by the drug could go through non-cell autonomous mechanisms. Another plausible explanation is that temsirolimus can only inhibit mTORC1 while mTOR KO inactivates both mTORC1 and mTORC2. Nevertheless, combined results from both studies clearly pointed to the potential benefit of treating glioma patients with mTOR inhibitors. Clinically, while past trials with mTOR inhibitors for glioma patients showed limited efficacy, further studies demonstrated that most of these failures were caused by the inability of the drugs to effectively penetrate the blood-brain barrier (BBB) or to adequately inhibit their targets (49, 50). Encouragingly, very recently a new class of mTOR inhibitor that can effectively penetrate BBB and inhibit mTOR kinase activity was found to have significant anti-tumor effects on glioma cells (51). Our data echo well with these findings and the existing knowledge of mTOR’s critical role in OPC development (52), thus provide strong rationales for further testing of mTOR inhibitors in glioma patients.
In summary, our current study demonstrated the importance of dissecting tumorigenic process in multiple aspects. First, identifying the cell of origin allows the examination of specific functions of oncogenic mutations in a particular cell type that has its unique signaling context. Second, studying the contribution of individual mutations from pre-malignant to malignant stages with the MADM system sheds light on novel therapeutic strategies that target specific biological processes such as OPC differentiation in this case. Third, restoration of individual TSGs in tumor cells can pinpoint which biological aberrations are critical for sustaining tumor cells, which should be ideal therapeutic targets. Fourth, comparing signaling aberrations between pre-malignant and malignant stages could help one identify transformation-specific events that may carry exceptional therapeutic efficacy. Finally, our work clearly demonstrated great values in using genetic tools to test therapeutic effects of a particular candidate gene, which completely avoid problems commonly associated with pharmacological compounds such as chemical instability, incomplete target inhibition, general toxicity, off-target effects, which would cloud the conclusion. Once a target is validated with genetic method, drug development efforts then could move forward confidently. Such a two-step drug discovery process could prove ethically, medically, and financially advantageous over direct compound screening in the long-term.
Supplementary Material
Main Points.
NF1 and p53 play distinct roles during the pre-transforming stages of gliomagenesis that can be targeted for therapies. Furthermore, there is differential activation of downstream effectors of Ras signaling in pre-malignant mutant cells that are critical for transformation.
Acknowledgments
We thank Ying Jiang for technical support; David Brautigan and Kevin Janes for critical comments on the manuscript. This work is supported by a grant from the National Institutes of Health/National Cancer Institute (R01-CA136495 to H.Z.). P.P.G is partly supported by Research Supplements to Promote Diversity in Health-Related Research associated to R01-CA136495.
Abbreviations used in the manuscript
- PreT-OPCs
Pre-transforming mutant OPCs
- MADM-WT
MADM brains with no mutations
- MADM-NF1
MADM brains with GFP+ NF1-null OPCs
- MADM-p53
MADM brains with GFP+ p53-null OPCs
- MADM-p53
NF1, MADM brains with GFP+ p53, NF1-null OPCs
References
- 1.Agnihotri S, et al. Glioblastoma, a Brief Review of History, Molecular Genetics, Animal Models and Novel Therapeutic Strategies. Arch Immunol Ther Exp (Warsz) 2013;61(1):25–41. doi: 10.1007/s00005-012-0203-0. [DOI] [PubMed] [Google Scholar]
- 2.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From Molecular Pathology to Targeted Treatment. Annu Rev Pathol Mech Dis. 2014;9(1):1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 3.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;(26):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen P, Kesari S. Malignant Gliomas in Adults. N Engl J Med. 2008;359(17):1850–1850. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 5.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 6.Brennan CW, et al. The Somatic Genomic Landscape of Glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121(3):479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindberg N, Kastemar M, Olofsson T, Smits a, Uhrbom L. Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene. 2009;28(23):2266–2275. doi: 10.1038/onc.2009.76. [DOI] [PubMed] [Google Scholar]
- 11.Lindberg N, et al. Oncogenic signaling is dominant to cell of origin and dictates astrocytic or oligodendroglial tumor development from oligodendrocyte precursor cells. J Neurosci. 2014;34(44):14644–51. doi: 10.1523/JNEUROSCI.2977-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assanah M, et al. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galvao RP, et al. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proc Natl Acad Sci. 2014;111(40):E4214–E4223. doi: 10.1073/pnas.1414389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson AI, et al. Article Non-Stem Cell Origin for Oligodendroglioma. Cancer Cell. 2010;18(6):669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cadinu D, et al. Comparative proteomic analysis reveals characteristic molecular changes accompanying the transformation of nonmalignant to cancer lung cells. EuPA Open Proteomics. 2014;3:1–12. [Google Scholar]
- 16.Watson AL, et al. Canonical Wnt/ -catenin Signaling Drives Human Schwann Cell Transformation, Progression, and Tumor Maintenance. Cancer Discov. 2013;3(6):674–689. doi: 10.1158/2159-8290.CD-13-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maglietta R, et al. Molecular pathways undergoing dramatic transcriptomic changes during tumor development in the human colon. BM. 2012;12(608):1–16. doi: 10.1186/1471-2407-12-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorgoulis V, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 19.Klose A, et al. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet. 1998;7(8):1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- 20.Shin J, et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis Model Mech. 2012;5(6):881–894. doi: 10.1242/dmm.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffzek K, et al. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 1998;17(15):4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morcos P, Thapar N, Tusneem N, Stacey D, Tamanoi F. Identification of neurofibromin mutants that exhibit allele specificity or increased Ras affinity resulting in suppression of activated ras alleles. Mol Cell Biol. 1996;16(5):2496–2503. doi: 10.1128/mcb.16.5.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimou L, Gallo V. NG2-Glia and Their Functions in the Central Nervous System. Glia. 2015:1429–1451. doi: 10.1002/glia.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, et al. Mosaic Analysis with Double Markers Reveals Tumor Cell of Origin in Glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolt CC, et al. SoxD Proteins Influence Multiple Stages of Oligodendrocyte Development and Modulate SoxE Protein Function. Dev Cell. 2006;11(5):697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Young KM, et al. Oligodendrocyte dynamics in the healthy adult CNS: Evidence for myelin remodeling. Neuron. 2013;77(5):873–885. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WB, et al. Indicator Expression Directed by Regulatory Sequences of the Glial Fibrillary Acidic Protein ( GFAP ) Gene: In Vivo Comparison of Distinct GFAP-Lac2 Transgenes. Glia. 1995;184:174–184. doi: 10.1002/glia.440130304. [DOI] [PubMed] [Google Scholar]
- 28.Pherson SWMC, Heuss ND, Roehrich H, Gregerson DS. Bystander Killing of Neurons by Cytotoxic T Cells Specific for a Glial Antigen. Glia. 2006 Oct;466:457–466. doi: 10.1002/glia.20298. 2005. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd AC, Raff MC. Lack of Replicative Senescence in Cultured Rat Oligodendrocyte Precursor Cells. Science (80- ) 2001 Feb;291:868–872. doi: 10.1126/science.1056780. [DOI] [PubMed] [Google Scholar]
- 30.Lambert JMR, et al. PRIMA-1 Reactivates Mutant p53 by Covalent Binding to the Core Domain. Cancer Cell. 2009;15(5):376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Fearon EF, Vogelstein B. for Colorectal Tumorigenesis. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 32.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156(6):1821–5. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlomm T, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21:1371–1378. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 34.Kaul A, Toonen JA, Cimino PJ, Gianino SM, Gutmann DH. AKT- or MEK-mediated mTOR inhibition suppresses Nf1 optic glioma growth. Neuro Oncol. 2015;17(6):843–853. doi: 10.1093/neuonc/nou329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charles NA, Holland EC, Gilbertson R, Glass R. The Brain Tumor Microenvironment. Glia. 2011 Mar;1180:1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 36.Guo F, Lang J, Sohn J, Hammond E, Chang M. Canonical Wnt Signaling in the Oligodendroglial Lineage – Puzzles Remain. Glia. 2015:1671–1693. doi: 10.1002/glia.22813. [DOI] [PubMed] [Google Scholar]
- 37.Psachoulia K, Jamen F, Young KM, Richardson WD. Glia Biology: Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol. 2009;5(3):57–67. doi: 10.1017/S1740925X09990354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolswijk G, Noble M. Identification of an adult-specific glial progenitor cell. Development. 1989;400(105):387–400. doi: 10.1242/dev.105.2.387. [DOI] [PubMed] [Google Scholar]
- 39.Weiss MB, et al. Deletion of p53 in human mammary epithelial cells causes chromosomal instability and altered therapeutic response. Oncogene. 2010;29(33):4715–4724. doi: 10.1038/onc.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine AJ. p53, the Cellular Gatekeeper for Growth and Division. Cell. 1997 Feb 7;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 41.Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71(11):4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valente LJ, et al. P53 Efficiently Suppresses Tumor Development in the Complete Absence of Its Cell-Cycle Inhibitory and Proapoptotic Effectors p21, Puma, and Noxa. Cell Rep. 2013;3(5):1339–1345. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell – Intrinsic and Host-Dependent Mechanisms. Genes Cancer. 2010 Jun;6:597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torti D, Trusolino L. Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. Embo Mol Med. 2011;11(v):623–636. doi: 10.1002/emmm.201100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shchors K, et al. Using a preclinical mouse model of high-grade astrocytoma to optimize p53 restoration therapy. Proc Natl Acad Sci U S A. 2013;110(16):E1480–9. doi: 10.1073/pnas.1219142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corral T, Jiménez M, Hernández-Muñoz I, De Castro IP, Pellicer A. NF1 Modulates the Effects of Ras Oncogenes: Evidence of Other NF1 Function Besides Its GAP Activity. J Cell Physiol. 2003;197(2):214–224. doi: 10.1002/jcp.10349. [DOI] [PubMed] [Google Scholar]
- 48.Ismat Fa, Xu J, Min ML, Epstein Ja. The neurofibromin GAP-related domain rescues endothelial but not neural crest development in Nf1−/− mice. J Clin Invest. 2006;116(9):2378–2384. doi: 10.1172/JCI28341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cloughesy TF, et al. Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5(1):0139–0151. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrik-Outmezguine VS, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534(7606):272–276. doi: 10.1038/nature17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan Q, et al. A Kinase Inhibitor Targeted to mTORC1 Drives Regression in Glioblastoma. Cancer Cell. 2017;31(3):424–435. doi: 10.1016/j.ccell.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guardiola-Diaz HM, Ishii A, Bansal R. Erk1/2 MAPK and mTOR signaling sequentially regulates progression through distinct stages of oligodendrocyte differentiation. Glia. 2012;60(3):476–486. doi: 10.1002/glia.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.