Abstract
A subset of Spitz tumors is associated with copy number increase of chromosome 11p and activating mutations of HRAS. These aberrations have been reported to occur in association with desmoplastic Spitz nevi. Little is known to what extent 11p gains can also be found in non-desmoplastic tumors. To learn more about the spectrum of microscopic and cytogenetic changes that can be seen in Spitz lesions in association with 11p gains, we reviewed the clinical and pathologic features of 40 cases. Patient ages ranged from 3 to 75 years. The most common anatomic site was the head and neck region, followed by the upper extremities. Prominent desmoplasia was present in ten cases. Seven tumors lacked significant stromal fibrosis. Twenty tumors were mitotically active. Novel microscopic features encountered in a few cases include a tumor with a polypoid silhouette and papillomatous surface and rare atypical tumors with a deep bulbous growth pattern. Among 36 cases analyzed by single nucleotide polymorphism (SNP) array or comparative genomic hybridization (CGH), 28 tumors had gains of the entire or near-entire p arm of chromosome 11 with no other co-existing unbalanced genomic aberration. Eight cases had additional changes; six of these with one additional aberration per case, and two cases had several chromosomal aberrations. We also examined a subset of tumors by fluorescence in situ hybridization (FISH) for HRAS gene locus (11p15.5). All tumors were FISH-positive. In conclusion, we expand the spectrum of pathologic findings associated with Spitz tumors with 11p gains. This cytogenetic aberration is not restricted to desmoplastic Spitz nevi. It can also be seen in non-desmoplastic and papillomatous lesions, and atypical melanocytic tumors with a deep bulbous growth. We also document that in some Spitz tumors additional cytogenetic aberrations may be found, the significance of which remains to be determined.
Keywords: HRAS, Spitz nevus, 11p, atypical Spitz tumor, spitzoid, melanoma
INTRODUCTION
Spitz nevi can be confused with melanoma and vice versa[1–5]. Cytogenetic analysis has emerged as a useful adjunct method in the evaluation of spitzoid melanocytic neoplasms. A subset of Spitz nevi is associated with isolated gains of the p-arm of chromosome 11, HRAS gene amplification, and a high frequency of HRAS mutations. These lesions have been reported to display stromal desmoplasia, low cellularity, and variable nuclear pleomorphism[6, 7]. For the occasional differential diagnosis of a desmoplastic Spitz nevus versus melanoma, documentation of gain of 11p and/or the presence of an isolated HRAS aberration is useful to support a diagnosis of nevus over melanoma, since isolated gains of 11p or HRAS aberrations have not been found in desmoplastic melanomas, and are exceptionally rare in any kind of melanoma[8, 9].
As there is limited knowledge on the morphologic spectrum of Spitz lesions with 11p gains and we have encountered a few cases with unexpected microscopic features or additional cytogenetic aberrations, we wanted to review our experience with these lesions and detail the clinical, microscopic, and cytogenetic findings. To this end a total of 40 Spitz nevi and atypical Spitz tumors with cytogenetically confirmed gains of 11p were examined microscopically to record various histopathologic parameters. An additional goal of our study was to compare the sensitivity of FISH for 11p with CGH or SNP array results as whole genome cytogenetic testing may not be feasible when there is limited tumor tissue and entails higher cost and usually slower turnaround time.
MATERIALS AND METHODS
Case Selection
In the context of Institutional Review Board approved protocols, 40 cases of lesions classified as Spitz nevus or atypical Spitz tumor with gains of chromosome 11p were retrieved from Memorial Sloan Kettering Cancer Center (MSKCC) (n=35) and University of California San Francisco (n=5).
SNP array and CGH
Single nucleotide polymorphism (SNP) array (n=35) using the Affymetrix OncoScan assay (Affymetrix, Santa Clara, CA) as previously described[10] or array comparative genomic hybridization (CGH) (n=5) were used for genome-wide cytogenetic analysis.
FISH for melanoma
FISH analysis was performed on FFPE tissue sections using locus-specific probes and centromere probes (CEP) (Abbott Molecular, Inc., Des Plaines, IL). The four-color FISH panel consists of locus-specific probes targeting the RREB1 gene (6p25), MYB gene (6q23), and CCND1 gene (11q13) and the centromere of chromosome 6 (CEP6) probe, with additional probe sets for CDKN2A(9p21)/CEP9. Two cases with insufficient tissue for evaluation by SNP or CGH array were evaluated with all six aforementioned FISH probes. Three cases with loss of whole chromosome 9 by SNP array were examined by FISH for CDKN2A(9p21)/CEP9.
FFPE sections (4 μm) generated from FFPE blocks of tumor specimens were pretreated by deparaffinizing in xylene and dehydrating in ethanol. Dual-color FISH was performed according to the Vysis protocol for FFPE sections (Abbott Molecular) with a few minor modifications. FISH analysis and signal capture were performed using fluorescence microscopes (Axio; Carl Zeiss AG, Jena, Germany) coupled with an ISIS FISH Imaging System (MetaSystems GmbH, Altlussheim, Germany). Thirty tumor cell interphase nuclei were evaluated for each specimen. Cutoff values for positive results are as follows: gains of RREB1 > 29%, or relative gains of RREB1/CEP 6 > 55%, or relative loss of MYB/CEP 6 > 40%, or gains of CCND1 in > 38%, or homozygous loss of CDKN2A (p16) > 33% of the cells examined.
FISH for 11p
FISH to evaluate selectively for 11p gains was performed in 22 cases (18 of which had 11p gains previously detected by genome-wide SNP-array) and 20 negative controls. Formalin-fixed paraffin-embedded tissue sections of 4 μm thickness with marked tumor areas were used for FISH analysis following standard protocols. Briefly, the tissue sections were de-paraffinized in Citrisolv solution (Fisher Scientific), followed by dehydration in 100% ethanol. After heating in 20% citrate buffer, the tissue sections were treated with pepsin (0.5mg/ml in 0.1N HCl) for 10–25 min at 40° C, followed by fixation in 10% formalin, and dehydration in a series of 70%, 85% and 100% ethanol. A BAC FISH probe of 720 kb flanking the HRAS gene locus (11p15.5) from Cytotest (Rockville, M.D., USA), in combination with a centromere specific probe CCP11 as internal control, was used to assess the copy level of HRAS. After applying the FISH probes to the tissue areas, both tissue and probes were co-denatured at 94° C for 7 minutes, and then incubated at 37° C overnight, followed by post-hybridization washing in 2xSSC/0.3% NP-40 at 77° C for 2 minutes. Tissue sections were counterstained with DAPI. Signal analysis was performed in combination with morphology correlation, and at least 100 interphase cells within the marked tumor area were evaluated and imaged using a Zeiss fluorescence microscope coupled with Metasystems ISIS software (Newton, MA). A cut -off value of 16% of cells was determined for gain of HRAS, with a ratio of HRAS/CCP11 over 2.
Next-Generation Sequencing
Two Spitz tumors with nodular deep pushing border/bulbous growth, one of which showed multiple genomic aberrations in addition to 11p gains by SNP array, were further analyzed by MSK-AmpliSeq, an amplicon-capture based next-generation sequencing assay used to identify specific mutations in 98 genes including RAS genes, ROS1, TERT, BRAF, BAP1, ALK, and others[11] against an unmatched normal sample.
RESULTS
Clinical and Morphologic Findings of Spitz nevi with +11p
Forty patients with Spitz nevi/tumors harboring chromosome 11p gains were identified. They included twenty-five female and fifteen male patients, ranging in age from 3 to 75 (mean = 32.2 years; median = 28 years). Seventeen lesions arose in the head and neck region, ten were located on the upper extremity, seven on the lower extremity, and six on the trunk.
The pathologic features are summarized in Table 1 and illustrated in Figures 1–4. The surface of twenty-six lesions (26/33) was raised, 3 of which were papillomatous, seven lesions were flat (7/33). The silhouette of the lesions was wedge-shaped in 25 cases (25/31) and broad plaque-like in six cases (6/31). All lesions were predominantly intradermal. Fifteen cases (15/40) had junctional melanocyte nests. None of the tumors were ulcerated. Three (3/31) lesions extended into the superficial subcutis. Melanocytes were present in variably-formed short fascicles or strands, nests, sheets, and single cells; with most lesions (31/40) showing a combination of growth patterns. In all 34 evaluable cases, a dispersed cell growth between collagen bundles was observed towards the periphery and depth of the lesions. However, two tumors showed in addition to single cell dispersion a bulbous nodular pushing border in part of their deep aspect. Variation in cellularity was noted within and across lesions, most showing higher cellularity towards the surface. Stromal desmoplasia was present in 33/40 cases. It was prominent (>50% of intervening stroma showing sclerotic changes) in ten cases (10/40). No significant stromal desmoplasia was seen in seven cases (7/40).
Table 1.
Spitz neoplasms with 11p gains.
| Case | Age | Gender | Site | Cytogenetics* | Surface | Silhouette | Epidermal changes | Paucicel | Growth pattern | Sclerosis | Borders | Cytomorph | Multinucl | Mit | Pigmt | Solar elast | Inflam |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12 | F | Jaw line | +11p, −8p (subset) | R/Pa | NE | Y | N | Ns, Fa | Mi | NE | E, Sp | Fo | 1 | It | NE | Pv |
| 2 | 18 | F | Back | +11 p | R/Pa | W | Y | N | S, Fa | Mi | I | E, Sp | A | 1 | Fo | A | Pv |
| 3 | 20 | M | Helix | +11p | R/Pa | NE | Y | N | Ns | Mi | I | E, Sp | A | 2 | Fo | NE | Pv |
| 4 | 41 | M | Back | +11p | Fl | W | Y | N | Fa | P | I | E, Sp | P | 1 | N | A | Pv |
| 5 | 11 | F | Helix | +11p | R | W | Y | N | Fa, SC | Mi | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 6 | 18 | M | UE | +11p | R | W | Y | N | Fa | P | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 7 | 66 | F | UE | +11p | Fl | Pl | N | Y | SC | A | I | E, Sp | Fo | 0 | N | Mi | Pv |
| 8 | 27 | M | Forehead | +11p | R | W | Y | N | Fa | Mi | I | E, Sp | Fo | 1 | N | Mi | Pv |
| 9 | 3 | M | Cheek | +11p | R | W | Y | N | S | Mi | I, No | E, Sp | P | 1 | Fo | A | Pv |
| 10 | 40 | F | UE | +11p, 18qLOH | NE | NE | Y | N | Fa | P | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 11 | 22 | F | LE | +11p | R | Pl | Y | N | Ns, SC | A | I | E | A | 0 | Fo | A | Pv |
| 12 | 75 | F | UE | +11p | F | Pl | Y | Y | Fa, SC | Mi | I | E, Sp | A | 0 | Fo | Mi | Pv |
| 13 | 28 | F | LE | +11p | R | W | Y | N | S | Mi | I | E, Sp | Fo | 1 | Fo | A | Pv |
| 14 | 61 | M | UE | +11 p | R | NE | Y | N | Ns, Fa | Mi | NE | E, Sp | A | 1 | Fo | Mi | Pv |
| 15 | 28 | F | Nose | +11p | R | W | Y | N | Fa, SC | A | I | E, Sp | Fo | 0 | N | Mi | Pv |
| 16 | 25 | F | UE | +11p | Fl | W | Y | N | Fa, SC | Mi | I | E, Sp | Fo | 0 | N | Mi | Pv |
| 17 | 40 | M | UE | +11p | Fl | Pl | Y | N | Ns, SC | Mi | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 18 | 14 | F | Cheek | +11p | R | W | Y | N | Ns, Fa | Mi | I | E, Sp | P | 0 | Fo | A | Pv |
| 19 | 72 | F | Back | +11p | Fl | Pl | Y | N | Fa, SC | Mi | I | E, Sp | A | 0 | N | A | Pv |
| 20 | 10 | M | Flank | +11p, +8p | R | W | Y | N | Fa, SC | P | I | E, Sp | P | 2 | Fo | A | Pv |
| 21 | 45 | F | UE | +11p | R | W | Y | N | Ns, SC | Mi | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 22 | 33 | F | UE | +11p | R | W | Y | Y | Ns, SC | P | I | E, Sp | Fo | 1 | N | A | Pv |
| 23 | 37 | M | LE | +11p | R | W | Y | N | S, Fa | Mi | I | E, Sp | P | 0 | Fo | A | BL |
| 24 | 30 | F | Back | +11p | R | W | Y | N | Ns, Fa | Mi | I | E, Sp | Fo | 1 | Fo | A | Pv |
| 25 | 30 | M | LE | +11p | NE | NE | Y | N | Ns, Fa | A | I | E, Sp | Fo | 1 | Fo | NE | In |
| 26 | 56 | M | LE | +11p | R | W | Y | N | Ns, Fa | A | I | E, Sp | Fo | 0 | Fo | A | Pv |
| 27 | 7 | M | Nose | +11p | R | NE | Y | N | Ns, Fa | P | NE | E, Sp | P | 0 | Fo | NE | Pv |
| 28 | 41 | F | Nose | +11p | NE | NE | Y | N | Ns, Fa | Mi | NE | E, Sp | Fo | 1 | Fo | NE | Pv |
| 29 | 16 | F | Face | (**) | R | W | Y | N | Ns, Fa | P | I | E, Sp | P | 0 | N | A | Pv |
| 30 | 57 | M | Nose | (***) | NE | W | Y | Y | Fa, SC | P | I | E, Sp | Fo | 0 | N | Mo | Pv |
| 31 | 8 | F | Cheek | +11p, −6p, −9, 18pq, 19pLOH | R | W | Y | N | S | A | I, No | E, Sp | P | 3 | Fo | A | In |
| 32 | 28 | F | Cheek | (**) | NE | W | Y | N | Fa, SC | Mi | I | E, Sp | Fo | 1 | N | Mi | Pv |
| 33 | 33 | F | LE | +11p, +7q | NE | NE | Y | N | Ns, Fa | Mi | NE | E, Sp | A | 2 | N | NE | Pv |
| 34 | 18 | F | Eyebrow | +11p | R | W | Y | N | Ns, Fa | P | I | E, Sp | Fo | 1 | Fo | A | Pv |
| 35 | 19 | F | Nose | +11p, +7q | R | W | Y | N | Ns, Fa | Mi | I | E, Sp | P | 1 | N | A | Pv |
| 36 | 19 | F | Forehead | +11p | R | W | Y | N | Fa, SC | P | I | E, Sp | A | 0 | Fo | A | Pv |
| 37 | 59 | M | Cheek | (**) | NE | NE | Y | N | Fa, SC | Mi | NE | E, Sp | Fo | 0 | N | Mo | Pv |
| 38 | 66 | F | LE | +11p | Fl | Pl | Y | N | Fa, SC | Mi | I | E, Sp | A | 0 | Fo | A | Pv |
| 39 | 42 | F | UE | +11p, −9, 13pLOH | R | W | Y | N | Ns, Fa | Mi | I | E, Sp | A | 1 | Fo | A | Pv |
| 40 | 14 | M | Back | +11p, −9 | R | W | Y | N | Fa, SC | A | I | E, Sp | A | 3 | Fo | A | Pv |
Paucicel= paucicellular, Cytomorph= cytomorphology, Multinucl= multinucleation, Mit= dermal mitoses per mm2, Pigmt= pigment, Solar elast= solar elastosis, Inflam= inflammation, F= female, M= male, UE= upper extremity, LE= lower extremity, R=raised, Pa=papillomatous Fl=flat, NE= not evaluable, W=wedge-shaped, Pl= plaque-like, Y= yes, N= no, S= sheet-like, Ns= nests, Fa= short fascicles or strands, SC= single cells, Mi= mild, P= prominent, A= absent, I=infiltrative, No= nodular/bulbous, E= epithelioid, Sp= spindle, Mo=moderate, Pv=perivascular, In=interstitial, It= intermediate, Fo=focal, BL = band-like
SNP or CGH array was performed except where specified: (**) 11p gains detected by FISH and melanoma FISH assay performed, or (***) where only FISH for 11p was performed and detected gains
Figure 1.
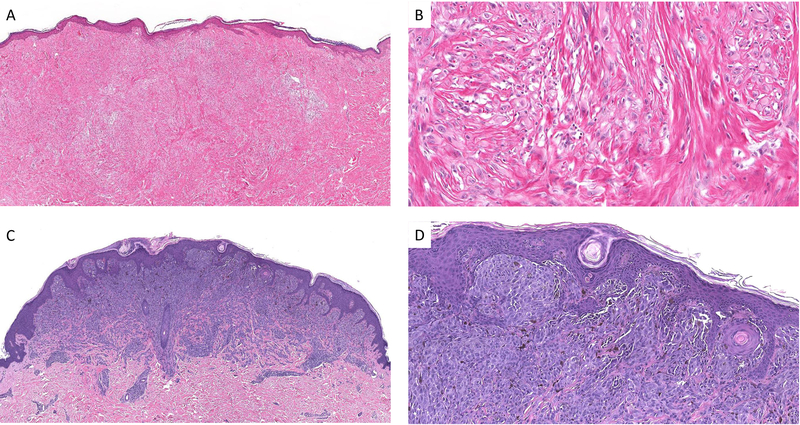
An example of a desmoplastic Spitz nevus with 11 p gain (case #4). The lesion is densely fibrotic with a pink appearance (A) Amelanotic spindle and epithelioid melanocytes are dispersed in a fibrotic stroma at low cell density (B). Example of a Spitz nevus with 11 p gain and only minimal sclerosis (case #2). There is a sharply circumscribed compound melanocytic proliferation with evidence of maturation (C). Nests of spindle and epithelioid melanocytes with focal melanin pigment are seen in the superficial aspect of the lesion (D).
Figure 4.
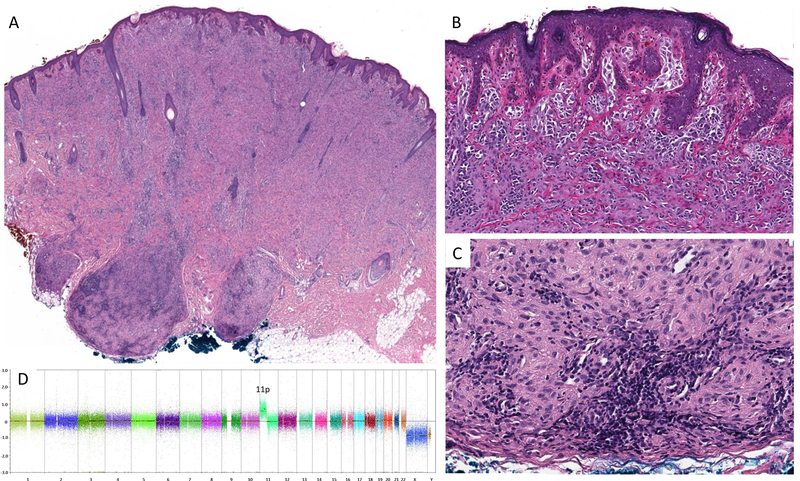
Silhouette of the tumor from the cheek of a 3 year-old boy (Case #9). The tumor shows two small bulbous projections at its base (A). Superficial portion of nests of plump spindle and epithelioid cells (B). Aggregates of epithelioid melanocytes in the deep bulbous portion of the tumor are associated with lymphocytes (C). No cytogenetic aberrations other than an isolated gain of 11p were found in this tumor (D).
Cytologically, the tumors were characterized by a mixture of plump spindle and epithelioid melanocytes. Multinucleation was a frequent finding, identified in 29/40 cases as either a prominent (9/40) or focal (20/40) feature. Mitotic figures were observed in 20/40 cases, with a mitotic rate of 1/mm2 in 15/40 cases, 2 mitoses/mm2 found in 3/40 cases, and 3 mitoses/mm2 in 2/40 cases. The tumors were usually pauci-melanotic (26/40), with melanin pigment, if present, limited to the superficial dermis and seen in less than 10% of the melanocytes. Only one case (1/40) showed melanin pigment in ~30% of tumor cells. Pigment was also present in melanophages. Thirteen cases (13/40) were completely amelanotic.
Associated epidermal changes from slight acanthosis and rete ridge elongation to verrucous hyperplasia were present in thirty-nine cases (39/40); Kamino bodies were conspicuous in 3/40. Solar elastosis was present in 9 cases (9/34, 7 mild and 2 moderate). An associated mild lymphocytic inflammatory infiltrate with a predominant perivascular pattern was noted in thirty-seven cases (37/40); a denser band-like or interstitial inflammatory infiltrate was present in three cases (3/40).
Clinical follow-up was very limited because most cases were seen in consultation and reported a Spitz nevi. Follow-up was available on 3 of 40 patients, ranging from 10 – 24 months. The cases with follow-up included the two microscopically most atypical tumors with bulbous growth (cases 9 and 31). None of the tumors have recurred.
Cytogenetic Findings
Genome-wide cytogenetic evaluation by SNP or CGH array was performed on 36 cases. Gains of the entire or near-entire p-arm of chromosome 11 (including the 11p15.5 HRAS gene locus) was the only aberration detected in 28 cases. Eight cases showed additional aberrations; six of which carried a single additional abnormality, which included segmental gains in chromosome 7q (n=2), loss of chromosome 8p in a minor subset of tumor cells (n=1), segmental gains in chromosome 8p (n=1), copy-neutral loss of heterozygosity (CN-LOH) in a segment of chromosome 18q (n=1), and loss of chromosome 9 (n=1). One case showed two additional aberrations, loss of chromosome 9 and CN-LOH in a segment of chromosome 13p. One case showed low level segmental loss of chromosome 6p, loss of chromosome 9, segmental loss of chromosome 18, and CN-LOH in a segment of chromosome 19p. FISH for 9p (including CDKN2A locus) was performed in the three cases with chromosome 9 loss, none of these cases reached the cutoffs for homozygous deletion of CDKN2A associated to melanoma.
In four cases there was insufficient material for SNP or CGH array, and gains of 11p were detected by FISH. Two of these cases were negative for the melanoma FISH assay. One case had gains in 11q (CCND1) as the only abnormality detected by a melanoma FISH assay performed at an outside institution which in conjunction with the gains in 11p and centromere 11 subsequently detected was interpreted as chromosome 11 polysomy.
Fluorescence In Situ Hybridization for 11p
FISH for 11p (including HRAS gene locus) was performed on 22 cases, 18 of which had 11p gains previously documented by SNP-array (described above and in Table 1), and on 20 negative controls. FISH detected 11p gains in all cases (22/22) and was negative in the control group (0/20). An example of a positive test result is illustrated in Fig. 5.
Figure 5.
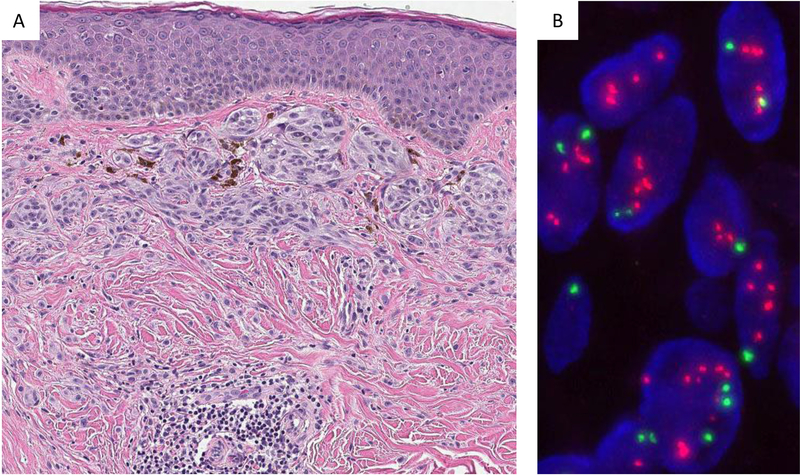
FISH analysis for HRAS. Spitz nevus with partial dermal sclerosis (Case #17) (A). Analysis of the tumor cells by FISH (B) reveals amplification/gain of HRAS with a signal pattern of numerous and clustered HRAS signals (orange-red). CCP11 labelled in green serves as internal control to assess the HRAS/CCP11 ratio. CCP = centromeric chromosome probe of chromosome 11
Mutational Analysis by Next-generation Sequencing
Mutational analysis was performed on both tumors displaying a deep bulbous growth pattern. Both tumors carried an HRASQ61K mutation. No other mutations were detected. The panel of genes analyzed included TERT promoter sequences and no TERT promoter mutations were identified.
DISCUSSION
A distinct spectrum of genomic aberrations has been identified in Spitz tumors[6, 12]. Most often genomic rearrangements are found involving various receptor tyrosine kinases, such as ALK, ROS1, NTRK1, NTRK3, RET, or MET, and the serine-threonine kinases BRAF or MAP3K8[13, 14]. Historically the first unique genetic aberration associated with a Spitz nevus, was the detection of gains of chromosome 11p with increased copy number or mutations in HRAS. These aberrations were reported to be typical of some desmoplastic Spitz nevi[6, 7]. Indeed, most of the Spitz lesions with 11p gains in our cohort showed some morphologic features associated with Spitz nevi/tumors with gains of 11p. We found 11p gains also in papillomatous compound Spitz nevi and Spitz nevi without desmoplasia. The lesions varied from exophytic to flat with wedge-shaped to plaque-like distribution of lesional melanocytes. Three lesions were densely inflamed. Two atypical tumors displayed a deep bulbous growth pattern.
Twenty percent of the cases in this cohort are over 50 years of age and the oldest patient was 75 years old. As desmoplastic Spitz nevi can occur in the elderly and frequently arise in the head and neck region, the microscopic findings on a partial biopsy sample may raise concerns about desmoplastic melanoma. Although epithelioid cytomorphology and abundant, pale cytoplasm, and positive immunoreactivity for Melan A can already favor a nevus in this context, the detection of an isolated gain of 11p is evidence in support of a desmoplastic Spitz nevus as it has not yet been reported to occur in desmoplastic melanomas[12, 15].
As whole genome cytogenetic assessment by SNP array or CGH can be problematic or not feasible due to limited tumor tissue in small samples or pauci-cellular lesions, or because dense associated inflammation; similarly to others[6, 12] we sought to evaluate the use of FISH for 11p (including the HRAS locus). To this end, we tested several cases by FISH in parallel with SNP array demonstrating there was consistency in the test results, which confirms that FISH for 11p is a valuable alternative for cases when more comprehensive cytogenetic testing is not possible due to high cost, lack of availability, limited amount of tissue, or low cellularity of the lesion.
Gains of 11p have not been reported as a frequent aberration associated to malignant melanoma[12, 15, 16], and isolated gains of 11p are exceptionally rare in melanoma. This likely reflects the fact that HRAS is an initiating mutation in only a small fraction of melanomas. Nonetheless, the sole detection of gains of 11p by FISH should not be construed as proof of benignity or as sufficient evidence to definitively exclude melanoma. As observed in 8 cases of our cohort, co-existence of 11p gains with additional cytogenetic aberrations is possible. When several additional aberrations are detected in association with atypical microscopic findings (e.g., mitoses, bulbous growth), the differential is then between an “atypical Spitz tumor” or “Spitz melanocytoma” and Spitz melanoma. In the one case with several genomic aberrations (case #31) the possibility of a Spitz melanoma was considered and discussed as the occurrence of Spitz melanomas with HRAS aberrations has recently been reported[17]. However, based on the constellation of microscopic findings in context with absence of additional mutations, in particular TERT promoter mutations, and lack of staining for PRAME, a “high grade Spitz melanocytoma” or “severely atypical Spitz tumor” was favored, acknowledging uncertainty about the biologic potential of the neoplasm. While no recurrence has been found on preliminary follow-up for any of our atypical Spitz tumors with 11p gains, long term follow-up studies are needed to determine the biologic significance of additional cytogenetic aberrations and/or the deep bulbous growth pattern. It is of interest that one lesion with bulbous growth had no unbalanced genomic aberration other than an isolated gain of 11p (case 9).
Based on our experience and preliminary follow-up, we believe that an additional isolated aberration, such as that of a gain of chromosome 7q, is still in keeping with an indolent neoplasm. Although gains of chromosome 7q can be associated with BRAF gene fusions[18], we did not test for that event (HRAS mutation or copy number change, and BRAF fusions are as a rule mutually exclusive). When the microscopic features of a Spitz lesion with gains of 11p and 7q fit with a nevus, the lesion may still be designated as a Spitz “nevus”.
Significant limitations of this study include the use of a restricted sequencing panel that covers only 98 genes and limited clinical follow-up. The relatively narrow sequence panel implies that untested mutations could have been present in some Spitz tumors. The limited clinical follow-up is in large part related to the fact that most cases were seen in consultation from other pathologists and the majority of patients did not need to return for follow-up visits since they received a benign tumor diagnosis. We specifically reached out to obtain follow-up on the patients with the most atypical microscopic and/or cytogenetic findings. However, their tumor diagnoses were only established in the last 1 – 2 years, which precludes long term clinical follow-up.
In summary, we expand herein the morphologic and cytogenetic spectrum of Spitz lesions with gains of 11p to include non-desmoplastic nevi, papillomatous nevi, and atypical neoplasms with bulbous growth and/or additional cytogenetic aberrations. We also show that in cases with limited tumor tissue material FISH for 11p is a sensitive test to support the impression of a Spitz nevus with 11p gains. However, at least a subset of Spitz tumors with gains of 11p may bear additional cytogenetic aberrations, currently of unknown diagnostic or prognostic value, which require broader genomic analysis for detection. The scarcity of chromosome 11p gains in melanomas as a whole suggests that the large majority of such lesions are likely benign.
Figure 2.
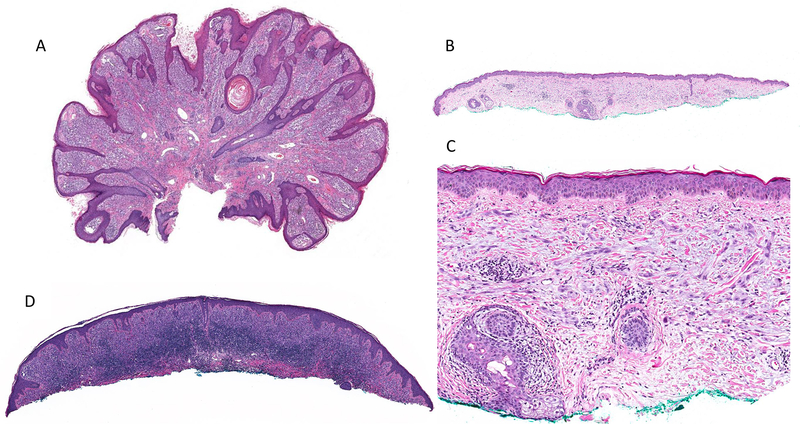
Low-power image showing case #3 with a markedly papillomatous surface with hyperplastic epidermis, high tumor cellularity, and sparse perivascular lymphocytic infiltrate (A). Case #7 is an example of a pauci-cellular tumor with flat surface, plaque-like distribution of melanocytes in the dermis, and no associated epidermal changes (B) composed of epithelioid and spindle cells without significant stromal desmoplasia (C). Case #23 showing a dense associated band-like lymphocytic infiltrate (D).
Figure 3.
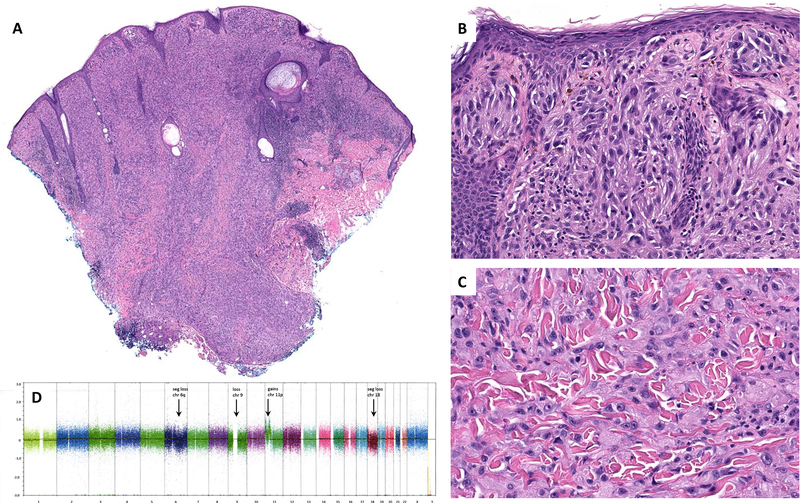
Low-power image of case #31 showing a raised surface, sheet-like growth pattern of tumor cells and nodular/bulbous pushing borders that extend into superficial subcutis (A). Nests of plump spindle and epithelioid melanocytes are present at the dermoepidermal junction and in the superficial dermis (B). Interstitial pattern of predominantly epithelioid melanocytes in the deep dermis (C). SNP array (D) showed gains in 11p and low level segmental loss in 6q, loss of chromosomes 9, segmental loss in chromosome 18, and CN-LOH of a segment of chromosome 19. seg = segmental; chr = chromosome
ACKNOWLEDGEMENTS
The authors wish to thank Maria Sanchez, Yesenia Gonzalez, and Fernando Garcia for their help retrieving archival material.
Disclosures: Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748. The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- [1].Barnhill RL. The Spitzoid lesion: rethinking Spitz tumors, atypical variants, ‘Spitzoid melanoma’ and risk assessment. Mod Pathol. 2006; 19 Suppl 2:S21–33. [DOI] [PubMed] [Google Scholar]
- [2].Barnhill RL, Argenyi ZB, From L, et al. Atypical Spitz nevi/tumors: lack of consensus for diagnosis, discrimination from melanoma, and prediction of outcome. Hum Pathol. 1999; 30:513–20. [DOI] [PubMed] [Google Scholar]
- [3].Cerroni L, Barnhill R, Elder D, et al. Melanocytic tumors of uncertain malignant potential: results of a tutorial held at the XXIX Symposium of the International Society of Dermatopathology in Graz, October 2008. Am J Surg Pathol. 2010; 34:314–26. [DOI] [PubMed] [Google Scholar]
- [4].Gerami P, Busam K, Cochran A, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol. 2014; 38:934–40. [DOI] [PubMed] [Google Scholar]
- [5].Harms KL, Lowe L, Fullen DR, et al. Atypical Spitz Tumors: A Diagnostic Challenge. Arch Pathol Lab Med. 2015; 139:1263–70. [DOI] [PubMed] [Google Scholar]
- [6].Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000; 157:967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].van Engen-van Grunsven AC, van Dijk MC, Ruiter DJ, et al. HRAS-mutated Spitz tumors: A subtype of Spitz tumors with distinct features. Am J Surg Pathol. 2010; 34:1436–41. [DOI] [PubMed] [Google Scholar]
- [8].Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell. 2015; 161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kodaz H, Kostek O, Hacioglu M, et al. Frequency of RAS mutations (KRAS, NRAS, HRAS) in human solid cancer. EJMO. 2017; 1:1–7. [Google Scholar]
- [10].Wang L, Rao M, Fang Y, et al. A genome-wide high-resolution array-CGH analysis of cutaneous melanoma and comparison of array-CGH to FISH in diagnostic evaluation. J Mol Diagn. 2013; 15:581–91. [DOI] [PubMed] [Google Scholar]
- [11].Chandramohan R, Sadowska J, Reilly CO, et al. MSK-AmpliSeq: A custom 94 gene AmpliSeq cancer panel with a custom paired tumor-normal analysis pipeline results in improved sensitivity and specificity [abstract]. In: proceedings of the Association for Molecular Pathology Annual Meeting 2015. Austin, TX. J Mol Diagn. 2015; 17:753–859. [Google Scholar]
- [12].Bastian BC, Wesselmann U, Pinkel D, et al. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999; 113:1065–9. [DOI] [PubMed] [Google Scholar]
- [13].Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014; 5:3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yeh I, Botton T, Talevich E, et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun. 2015; 6:7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998; 58:2170–5. [PubMed] [Google Scholar]
- [16].Namiki T, Yanagawa S, Izumo T, et al. Genomic alterations in primary cutaneous melanomas detected by metaphase comparative genomic hybridization with laser capture or manual microdissection: 6p gains may predict poor outcome. Cancer Genet Cytogenet. 2005; 157:1–11. [DOI] [PubMed] [Google Scholar]
- [17].Raghavan SS, Peternel S, Mully TW, et al. Spitz melanoma is a distinct subset of spitzoid melanoma. Mod Pathol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013; 26:845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]