Neurological symptoms are frequently observed in COVID-19. Here, we examine the neuroinvasive potential of SARS-CoV-2 and demonstrate infection of neurons in three separate approaches: mouse model, human brain organoid, and autopsy of COVID-19 patients.
Abstract
Although COVID-19 is considered to be primarily a respiratory disease, SARS-CoV-2 affects multiple organ systems including the central nervous system (CNS). Yet, there is no consensus on the consequences of CNS infections. Here, we used three independent approaches to probe the capacity of SARS-CoV-2 to infect the brain. First, using human brain organoids, we observed clear evidence of infection with accompanying metabolic changes in infected and neighboring neurons. However, no evidence for type I interferon responses was detected. We demonstrate that neuronal infection can be prevented by blocking ACE2 with antibodies or by administering cerebrospinal fluid from a COVID-19 patient. Second, using mice overexpressing human ACE2, we demonstrate SARS-CoV-2 neuroinvasion in vivo. Finally, in autopsies from patients who died of COVID-19, we detect SARS-CoV-2 in cortical neurons and note pathological features associated with infection with minimal immune cell infiltrates. These results provide evidence for the neuroinvasive capacity of SARS-CoV-2 and an unexpected consequence of direct infection of neurons by SARS-CoV-2.
Graphical Abstract
Introduction
As of September 2020, SARS-CoV-2 has infected >25 million people globally. While a majority of COVID-19 patients present with respiratory symptoms, neurological involvement, including impaired consciousness and headache, have been reported in patients (Mao et al., 2020). To date, human autopsy studies have identified viral RNA transcripts in brain tissues (Puelles et al., 2020; Solomon et al., 2020) and viral proteins in the endothelial cells within the olfactory bulb (Cantuti-Castelvetri et al., 2020) in people who succumbed to COVID-19. Several reports have come out indicating SARS-CoV-2 infection of the central nervous system (CNS) cells (Bullen et al., 2020; Jacob et al., 2020; Pellegrini et al., 2020; Ramani et al., 2020; Yang et al., 2020; Zhang et al., 2020). However, there are still many unknowns regarding the frequency or the consequences of neuroinvasion. Understanding the full extent of viral invasion is crucial to treating patients as we begin to try to figure out the long-term consequences of COVID-19, many of which are predicted to have possible CNS involvement (De Felice et al., 2020; Heneka et al., 2020; Pereira, 2020; Zhang et al., 2020).
Because the CNS is not the primary organ affected by SARS-CoV-2, studying neurological disease in COVID-19 patients systemically provides several challenges, including having only a subset of the population of patients with neuroinvasion, lacking technology to sample CNS tissues directly, and distinguishing direct neuroinvasion versus systemic viremia within the brain. Therefore, robust, reliable model systems are required to answer the questions underlying SARS-CoV-2 neuropathology. During the Zika virus (ZIKV) pandemic, several groups used human brain organoids to answer key questions regarding ZIKV neuroinvasion and its consequences (Garcez et al., 2016; Qian et al., 2016; Xu et al., 2019). Using a similarly well-characterized human brain organoid model (Lancaster and Knoblich, 2014; Lancaster et al., 2013), we test the infection capacity of SARS-CoV-2 in CNS tissue. Using single-cell RNA sequencing (RNA-seq), we uncover the transcriptional changes caused by SARS-CoV-2 infection of neurons.
Beyond the neuroinvasive potential of SARS-CoV-2, the question remains whether ACE2 is the main route of entry of SARS-CoV-2 into neuronal cells and what strategies might block viral infection. ACE2 expression in the CNS, and in neurons in particular, is still unclear (Xia and Lazartigues, 2008). Moreover, in addition to ACE2, other cofactors such as TMPRSS2 (Hoffmann et al., 2020) and Neuropilin-1 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020) seem to affect infection rates, but it is unclear if these are also required for neuronal infection. Thus, we used transcriptional profiling along with blocking antibody studies to demonstrate the requirement of ACE2 and SARS-CoV-2 spike protein to infect neurons.
To gain in vivo relevance for these findings, we examine the neuroinvasive potential of SARS-CoV-2 using mouse models of SARS-CoV-2, and observe vascular remodeling in infected regions, independent of vascular infection, providing a valuable tool that can be used to dissect out the consequences of CNS infection. Finally, by examining postmortem COVID-19 patient brain tissues, we provide evidence of neuroinvasion by SARS-CoV-2 and identify associations between infection and ischemic infarcts in localized brain regions.
Results
Modeling SARS-CoV-2 neuroinvasion and cellular death using human brain organoids
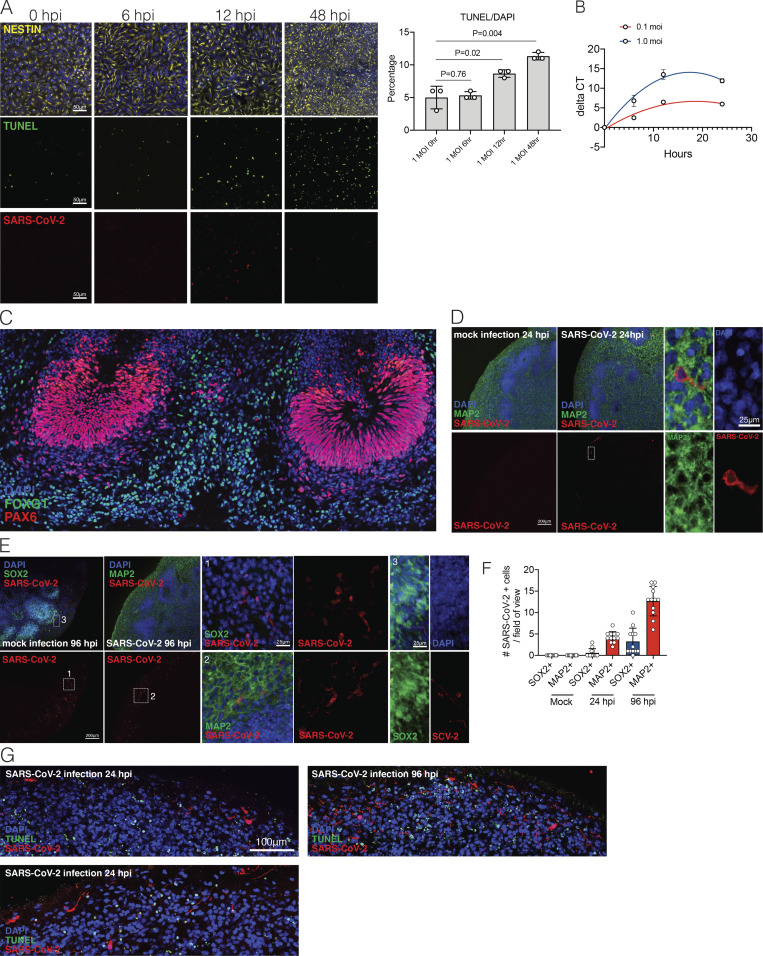
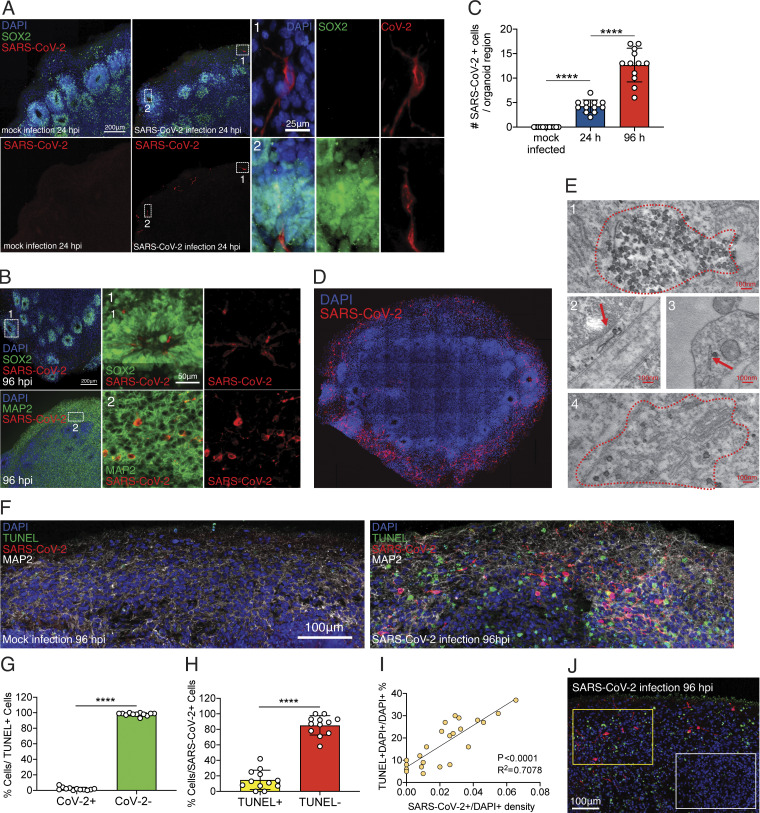
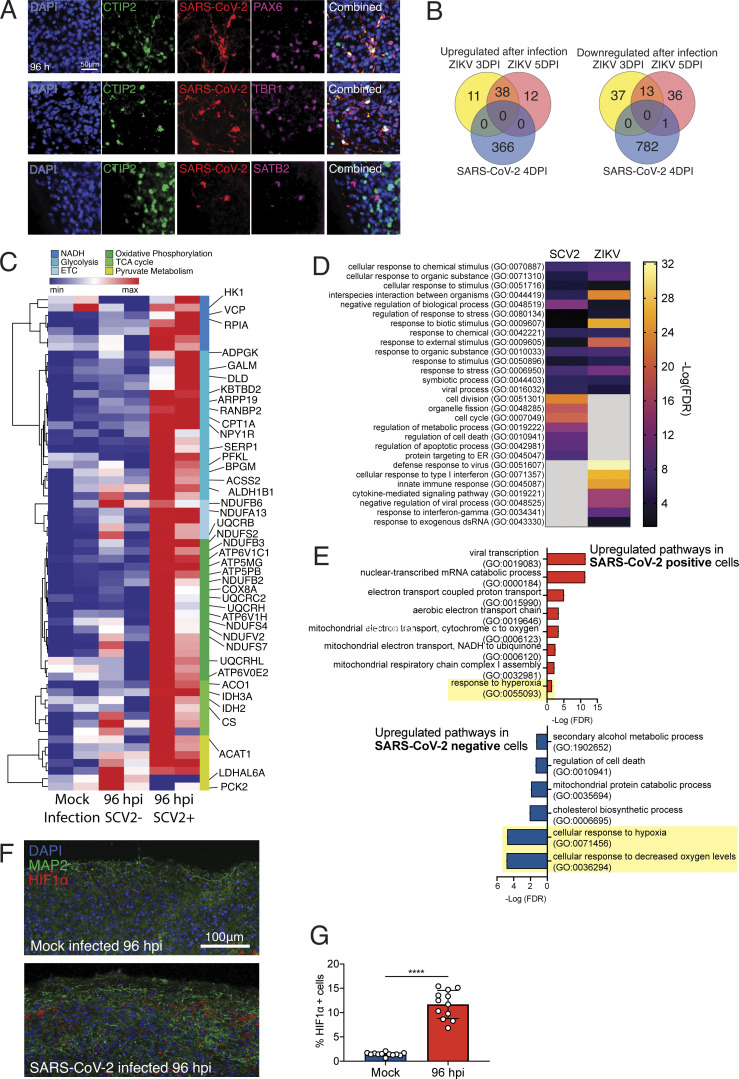
To dissect the mode and consequences of infection, we first established the neuroinvasive potential of SARS-CoV-2 in a human brain model system. We used human induced pluripotent stem cell (hiPSC) lines (Y1 and Y6), derived from healthy individuals (Fig. 1), to generate forebrain-specific human neural progenitor cells (hNPCs). In culture, we observed replication of SARS-CoV-2 in 2-wk-old hNPCs, with peak viral titers as early as 12 h postinfection (hpi; Fig. S1, A and B). In addition, Tdt-mediated dUTP-biotin nick end labeling (TUNEL) staining indicated increased cell death (Fig. S1 A). Next, we generated hiPSC-derived brain organoids (Amin and Paşca, 2018; Pellegrini et al., 2020; Qian et al., 2016; Velasco et al., 2019) to model the SARS-CoV-2 infection of neuronal cells in 3D. We confirmed a dorsal cortical identity of organoids by immunostaining for markers FOXG1, PAX6, and CTIP2 (Fig. S2 C). Similar to a recent report (Ramani et al., 2020), we observed infection of neuronal cells in 9-wk-old organoids as early as 24 hpi, with significantly increased number of SARS-CoV-2–positive cells at 96 hpi (Fig. 2, A–C; and Fig. S1 C). Although the majority of the SARS-CoV-2–infected cells were localized within MAP2-positive cellular fields of mature neurons (Fig. 2 B [2] and Fig. S1 C), we also observed infection of SOX2-positive neural stem cells with bipolar morphology and cells localized around the neural tube–like structures (Fig. 2 B [1]). We observed an increase in SARS-CoV-2–positive cells 96 hpi compared with 24 hpi (Fig. 2 C and Fig. S1, C–E). 96 hpi, we observed a widespread infection, mostly limited to the regions with high cortical cell density in the organoid (Fig. 2 D). Using electron microscopy, we visualized viral particles within the organoid (Fig. 2 E), with discrete regions of high-density virus accumulation (Fig. 2 E [1]) and other regions showing viral budding from ER-like structures (Fig. 2 E [2–4]), suggesting that SARS-CoV-2 is able to use the neuronal cell machinery to replicate. Organoid infection resulted in extensive neuronal cell death; strikingly, however, the majority of TUNEL-positive cells were SARS-CoV-2 negative (Fig. 2, F and G; and Fig. S1 F), and only ∼15% of the cells infected with SARS-CoV-2 were TUNEL positive (Fig. 2 H). Increased cell death was correlated with a higher density of SARS-CoV-2–positive cells (Fig. 2 I), which, however, were not overtly dying (Fig. 2, G and H). Even within a single plane, we noticed the high-density SARS-CoV-2 area to have more TUNEL-positive cells (Fig. 2 J, yellow box) compared with low-density SARS-CoV-2 regions (Fig. 2 J, white box). Together, these data indicated that SARS-CoV-2 can infect cells of neural origin and suggested that infected cells can promote death of nearby cells.
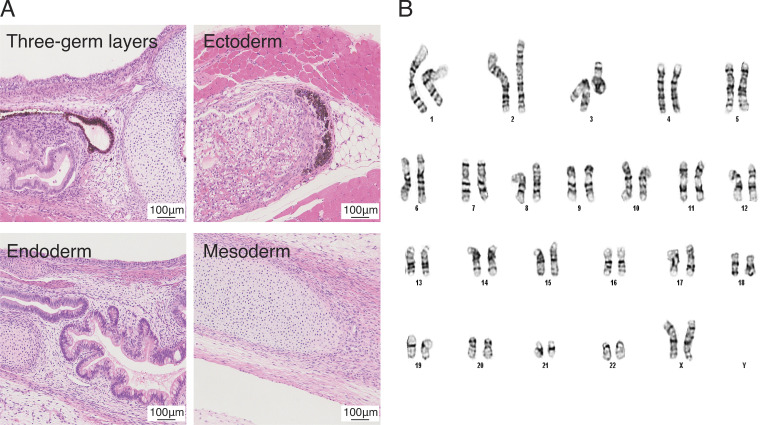
Figure 1.
Pluripotency of Y6 line. (A) Sample images of cell types of three germ layers in teratomas following transplantation to Rag2−/−GammaC−/− mice. Scale bar = 100 µm. (B) Sample image showing normal karyotype for Y6 line. Scale bars = 100 µm.
Figure S1.
SARS-CoV-2 infection in hPSC-derived NPCs and 9-wk organoids. (A) Representative images of immunostaining of hNPCs with TUNEL staining, anti-Nestin, and anti–SARS-CoV-2 nucleocapsid antibody 0, 6, 12, and 48 hpi. Scale bar = 50 µm. (B) RT-PCR amplification of COVID-genome from infected hNPC cells. (C) Images of FOXG1 and PAX6 staining for dorsal neuron characterization. (D) Sample images of immunostaining of 9-wk organoids with DAPI, anti-MAP2, and anti–SARS-CoV-2 antibody 24 hpi after SARS-CoV-2 or mock infection. Note the perinuclear and neuritic staining of SARS-CoV-2 in the MAP2+ cell. Dashed white box corresponds to SARS-CoV-2+ and MAP2+ single-cell in 24-hpi organoid. Scale bar = 200 µm for zoomed-out images and 25 µm for zoomed-in images. (E) Sample images of immunostaining of 9-wk organoids with DAPI, anti-MAP2, or anti-Sox2 and anti–SARS-CoV-2 antibody 96 hpi after SARS-CoV-2 or mock infection. Dashed white box 1 corresponds to “1 panel” showing magnified SARS-COV-2+/SOX2− cell in 96-hpi organoid. Dashed white box 2 corresponds to 2 panel showing SARS-COV-2+ and MAP2+ cell in 96-hpi organoid; 3 panel shows SOX2+/SARS-CoV-2+ cell in 96-hpi organoid. Scale bar = 200 µm for zoomed-out images and 25 µm for zoomed-in images. (F) Quantification of number of SARS-CoV-2+ cells/field of view that are either SOX2+ or MAP2+ in mock, 24-hpi, and 96-hpi organoids. (G) Organoids were stained with TUNEL to evaluate cell death 24 and 96 hpi. n = 4 organoids per condition, 12 cortical regions from two iPSC lines. Experiments were performed twice for reproducibility. Scale bar = 100 µm.
Figure S2.
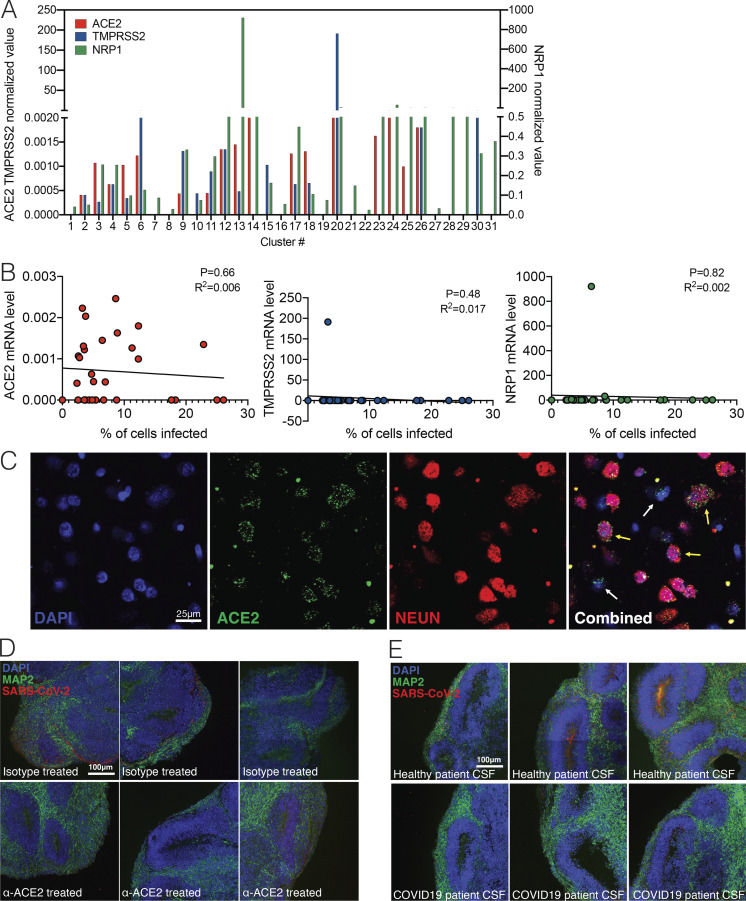
SARS-CoV-2 infection depends on hACE2. (A) Expression levels of ACE2, TMPRSS2, and NRP1 from single-cell RNA-seq data. (B) Correlation of ACE2, TMPRSS2, and NRP1 expression to percentage of cells infected in each cluster. (C) Imaging of postmortem COVID-19 patient brains stained with ACE2 and NEUN. Scale bar = 25 µm. (D) Uncropped images of ACE2 antibody–treated organoids shown in Fig. 3 B. Scale bar = 100 µm. (E) Uncropped images of CSF-treated organoids shown in Fig. 3 E. All experiments were performed with unique organoid, n = 4 per condition, with images from n = 12 cortical regions with two IPSC lines. Experiments were performed twice for reproducibility. Scale bar = 100 µm.
Figure 2.
SARS-CoV-2 infects human brain organoids and induces cell death. Human brain organoids were infected with SARS-CoV-2 and collected 24 or 96 hpi to analyze for different cellular markers. (A) Images of brain organoids looking at SARS-CoV-2 infection (in red) 24 hpi (see Fig. S2 C for additional images). Scale bar = 200 µm for zoomed-out images and 25 µm for zoomed-in images. (B) Images of brain organoids looking at SARS-CoV-2 infection (in red) 96 hpi (see Fig. S2 D for additional images). Scale bar = 200 µm for zoomed-out images and 50 µm for zoomed-in images. (C) Quantification of SARS-CoV-2–positive cells in a single microscope image of cortical region of organoids (A and B). (D) Tiled image of 96-hpi organoid. (E) Electron microscopy image of SARS-CoV-2 viral particles in brain organoids (see Fig. S3 for uncropped and additional images). Scale bar = 100 nm. (F) Organoids were stained with TUNEL to evaluate cell death at 96 hpi. Scale bar = 100 µm. (G) Quantification of SARS-CoV-2 and TUNEL double-positive (yellow) or SARS-CoV-2–negative, TUNEL-positive (green) cells over total TUNEL-positive cells. (H) Quantification of SARS-CoV-2 and TUNEL double-positive (yellow) or SARS-CoV-2–positive, TUNEL-negative (red) cells over total SARS-CoV-2–positive cells. (I) Correlation between the frequency of TUNEL-positive cells and presence SARS-CoV-2 in different regions of the organoid. (J) Representative image of TUNEL and SARS-CoV-2 staining showing high-density SARS-CoV-2 region (yellow box) and low-density SARS-CoV-2 region (white box) in the same plane. Scale bar = 100 µm. All experiments were performed with unique organoid, n = 4 per condition, from the same culturing batch, with images from n = 12 cortical regions with two IPSC lines, and Student’s t test was performed (****, P < 0.0001). Experiments were performed twice independently for reproducibility.
Single cell–level profiling of human brain organoids
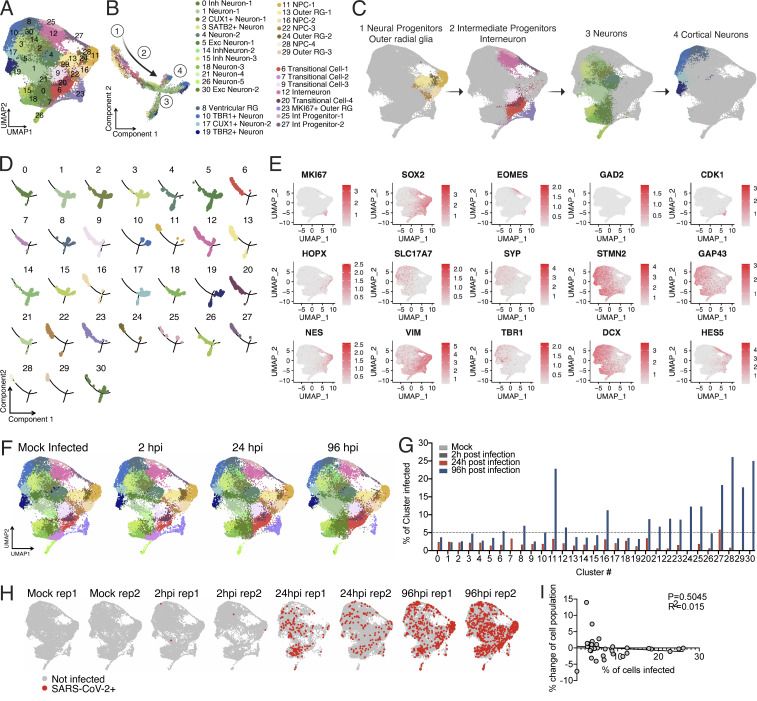
We hypothesized that the cellular heterogeneity of brain organoids may be leading to certain cells being more susceptible to infection and others to death, and that this system would provide an ideal platform to understand the cellular tropism of SARS-CoV-2 in the CNS and elucidate the consequences of SARS-CoV-2 neuroinvasion. To address these questions, we performed single-cell RNA-seq to dissect the cellular states and transcriptional changes occurring after SARS-CoV-2 infection in both infected and noninfected cells within the organoid. We analyzed 60-d-old organoids that were either mock infected or infected with SARS-CoV-2 and collected at 2, 24, and 96 hpi (n = 2 for each time point); 96,205 cells were used to create 31 distinct clusters (Fig. S3 A). Annotations from previous studies (Cakir et al., 2019; Kanton et al., 2019; Velasco et al., 2019) allowed us to characterize the identity of each cluster (Fig. S3, A–C). Monocle trajectory further classified these clusters into four major cellular states: neural progenitors/outer radial glia, intermediate progenitor/interneurons, neurons, and cortical neurons (Fig. S3, B–D). To confirm the cell-types we found in the single-cell sequencing, we used additional staining with dorsal cortical markers CTIP2, PAX6, and TBR1 and observed infection primarily overlapping with CTIP2-positive, PAX6-negative cells or CTIP2/TBR1 double-positive, Pax6-negative cells, which are neuronal cells with a deep-layer fate (layers 5/6). We also observed infection of SATB2-positive, CTIP2-negative cells or SATB2/CTIP2 double-positive cells, indicating that upper-layer cortical neurons are also susceptible to infection in the organoid model.
Figure S3.
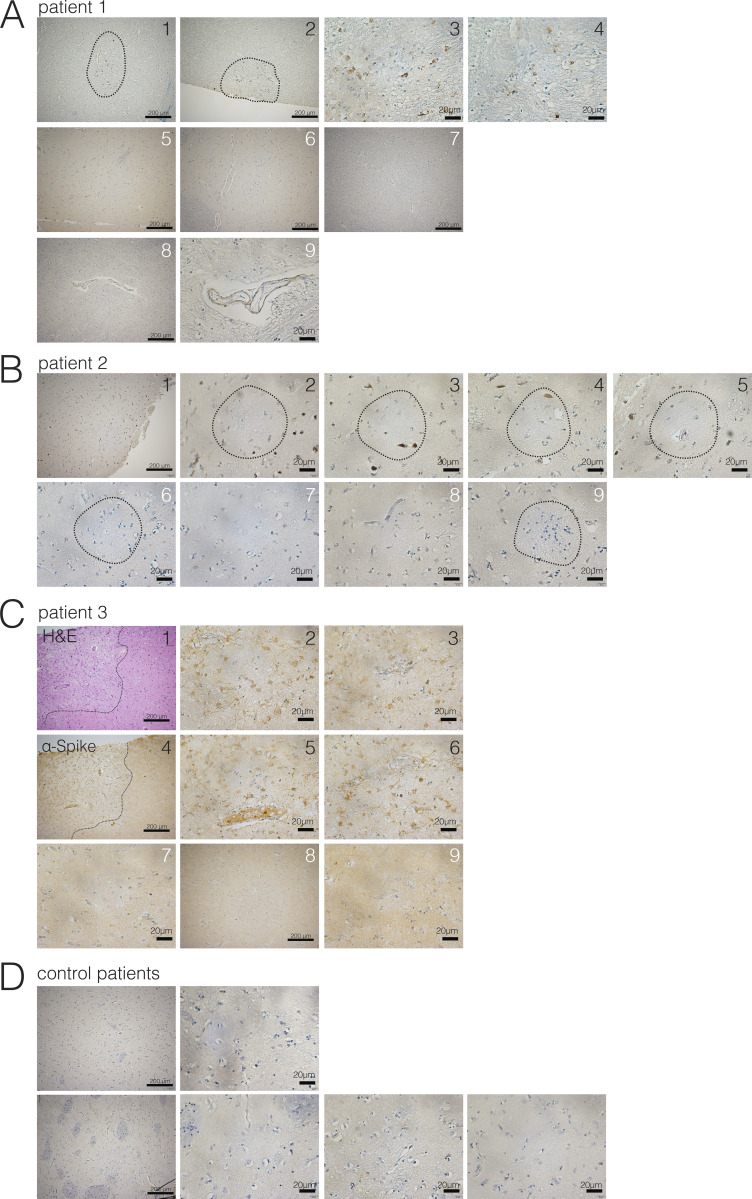
Evidence of neuroinvasion in postmortem COVID-19 patient brains (caudate). FFPE sections of brain tissue from COVID-19 patients were stained using H&E or anti–SARS-CoV-2-spike antibody. (A) Images of regions of the caudate of patient 1. White numbers indicate unaffected regions, and black numbers indicate regions with infected cells. Dotted lines around 1 and 2 indicate ischemic infarcts with virus staining. (B) Images of regions of the caudate of patient 2. White numbers indicate unaffected regions, and black numbers indicate regions with infected cells. Dotted circles indicate ischemic infarcts. (C) Images of regions of the caudate of patient 3. White numbers indicate unaffected regions, and black numbers indicate regions with infected cells. Dotted lines around 1 and 4 indicate ischemic infarcts with virus staining. Images 1, 4, and 5 are shown in Fig. 4. (D) Example images from control patient brains. Scale bar = 200 µm for zoomed-out images and 20 µm for zoomed-in images.
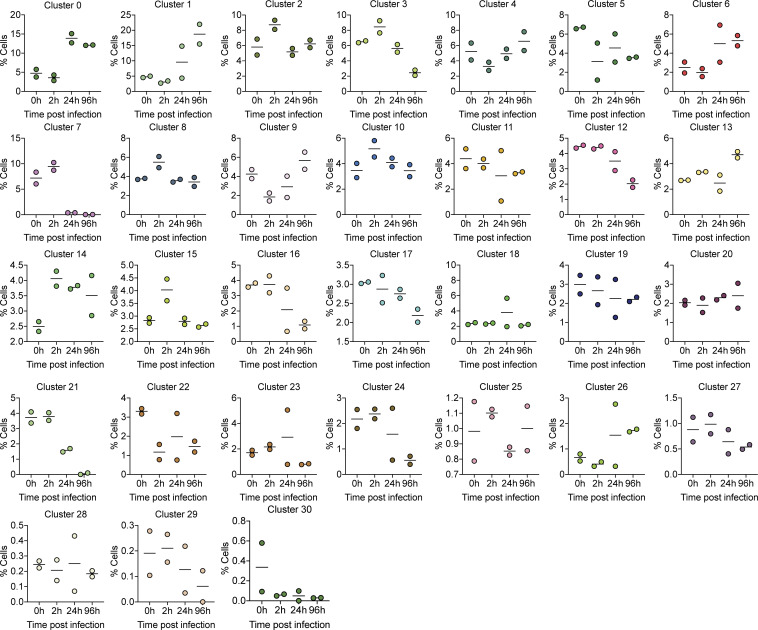
Comparing global UMAP clusters in specimens collected before and after infection, we were able to identify cellular population changes during infection (Fig. 4). With an added SARS-CoV-2 annotation, SARS-CoV-2 transcript reads were localized to a variety of cell clusters (Fig. 3 A), demonstrating the widespread infectivity of SARS-CoV-2 in neurons, radial glia, and neuronal progenitor cells. Several cell clusters showed large changes in their representation within the organoid, such as cluster 1 (from 5% to ∼20% of the population), or cluster 7 (from ∼10% to nearly 0%; Fig. 4). However, this was not uniformly seen with all infected clusters, as cells in cluster 11 showed high infection rates (Fig. 3 G) but little change in population representation (Fig. 4). This is consistent with findings from TUNEL staining (Fig. 2, E–G), which demonstrated minimal overlap between SARS-CoV-2–infected cells and those undergoing cell death. Alternatively, it is possible that infection resulted in differentiation or change in cell’s phenotype in populations such as cluster 7, which is characterized to be a transitional cell type.
Figure 4.
Frequency of each cell cluster from single-cell RNA-seq. Graphs display the percentage frequency of each cluster during a given infection status, 0, 2, 24, and 96 hpi.
Figure 3.
Single cell RNA-seq of SARS-CoV-2 infected organoids. (A) UMAP projection of cells from single cell sequencing. (B) Monocle trajectory analysis resulted in four distinct states of cells from the organoid. (C) The four major clusters consisted of the following: (1) Neural progenitor, outer radial glia like cells; (2) intermediate progenitor, interneurons; (3) neurons; and (4) cortical neurons. (D) Monocle projection of individual clusters. (E) Heatmap of commonly used genes for identification of cell subtypes in human brain organoids. (F) UMAP projection of organoids depending on infection status. (G) Percentage of infected cells in each cluster. (H) UMAP heatmap of SARS-CoV-2 transcript + cells separated by infection status. (I) Correlation between % change of cell population versus % of cells infected in each cluster. Single cell data were produced by two separate organoids (see F) to ensure reproducibility.
Next, we compared the changes in the transcriptome of SARS-CoV-2–infected organoids with another well-studied neurotropic virus: ZIKV. Differentially expressed genes (DEGs) after a brain organoid infection with ZIKV (Watanabe et al., 2017) showed almost no overlap with DEGs from SARS-CoV-2–infected organoids (Fig. 5 B). In both cases, the transcriptome showed evidence of viral infection and invasion (Fig. 5 D). However, unique processes were enriched in each of the infections. SARS-CoV-2–infected brain organoid up-regulated pathways related to cell division, organelle fission, and metabolic processes, while ZIKV showed enrichment in type I IFN pathways (Fig. 5 D). SARS-CoV-2 induced a unique transcriptional state within neurons compared with ZIKV, consistent with the fact that SARS-CoV-2 induces a moderate IFN-stimulated gene response in other tissues (Blanco-Melo et al., 2020) and previous reports of specific virus replication being controlled by alternative pathways by neurons (Daniels et al., 2019; Yordy et al., 2012).
Figure 5.
Neuronal cells undergo a unique metabolic response to SARS-CoV-2 infections. Brain organoids were infected with SARS-CoV-2 and sequenced with 10× single-cell sequencing strategies. (A) Validation of neuronal subtypes using CTIP2, PAX6, and TBR1 antibodies for confocal imaging. Scale bar = 50 µm. (B) DEGs from brain organoids infected with ZIKV (Watanabe et al., 2017) were compared with DEGs from SARS-CoV-2–infected organoids. (D) Enriched gene ontology terms (http://www.geneontology.org) for up-regulated genes from B. (E) Enriched gene ontology terms in SARS-CoV-2–infected cells (top) and SARS-CoV-2–negative bystander cells from 96-hpi organoids (bottom). (C) Heatmap of genes from metabolic pathways. (F) HIF1A staining of brain organoids that were mock infected versus 96 hpi. Scale bar = 100 µm. (G) Quantification of HIF1A-positive cells in SARS-CoV-2–infected organoids. Single-cell RNA-seq was performed in duplicate with one IPSC line (Y6) for reproducibility. HIF1A staining was performed with unique organoid, n = 4 per condition, from the same culturing batch, with images from n = 12 cortical regions with two IPSC lines, and Student’s t test was performed (****, P < 0.0001). Experiments were performed twice for reproducibility.
To understand the impact of SARS-CoV-2 infection on the brain organoid at the cellular level, we characterized cells that were infected with SARS-CoV-2 versus neighboring uninfected cells without the presence of SARS-CoV-2 transcript, in cluster 11, which comprised the highest number of infected cells 96 hpi. SARS-CoV-2–positive cells showed enrichment of genes corresponding to viral transcription, along with enrichment for metabolic processes including electron transport–coupled proton transport, cytochrome c to oxygen, and NADH to ubiquinone (Fig. 5 E, top panel). Conversely, SARS-CoV-2–negative cells showed a mitochondrial catabolic state with the up-regulation of alcohol metabolism, cholesterol synthesis, and regulation of cell death (Fig. 5 E, bottom panel). These two cell populations displayed antagonizing pathway enrichment, with the infected cells responding to hyperoxia and the bystander cells responding to hypoxia (Fig. 5 E, highlight in yellow). The hypermetabolic state is unique to the SARS-CoV-2–infected cells (Fig. 5 C) and highlights the ability of SARS-CoV-2 to hijack the host neuron machinery to replicate (Fig. 2 D). Finally, we confirmed that infection by SARS-CoV-2 induced a locally hypoxic environment in neuronal regions by staining for HIF1α (Fig. 5, F and G) in mock-infected and SARS-CoV-2–infected organoids. Together, these results indicate the potential of SARS-CoV-2 in manipulating host metabolic programming, which may create a resource-restricted environment for cells.
Host ACE2 receptor is required for infection of neurons
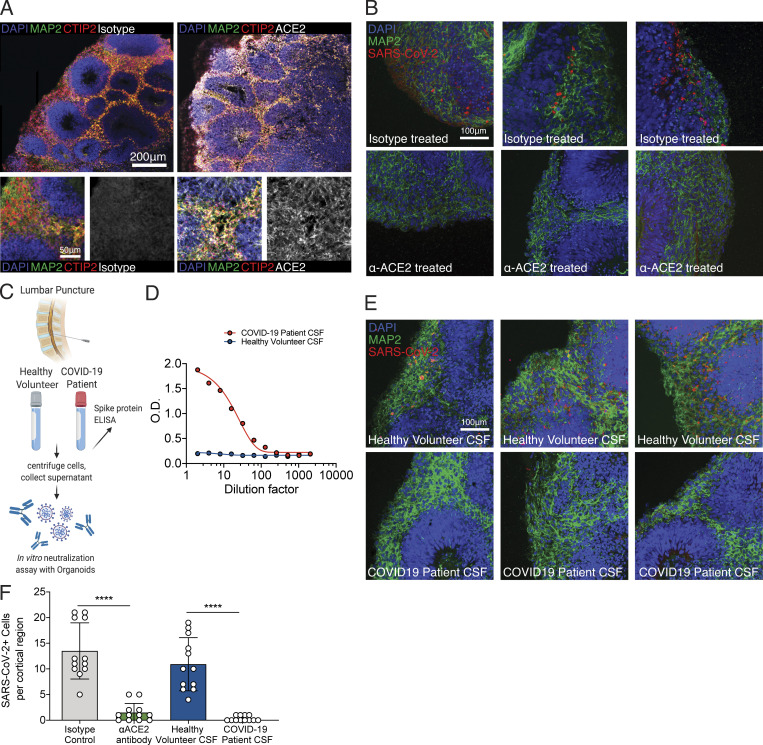
One of the ongoing questions regarding SARS-CoV-2 neuroinvasion is that mRNA levels of ACE2 appear to be very low in the CNS (Li et al., 2020; Qi et al., 2020; Sungnak et al., 2020). Indeed, our single-cell RNA-seq dataset demonstrated low levels of ACE2; it was, however, detectable in many clusters (Fig. S2 A). In addition, we did not observe a correlation between the percentage of cells infected in each cluster to ACE2, TMPRSS2, or Neuropilin-1 expression (Fig. S2 B). However, it remained possible that ACE2 protein may be expressed on the cell surface to promote viral entry. Consistent with this idea, we found widespread expression of ACE2 protein in both MAP2-positive neurons and cells in the neural tube-like structures of the organoids (Fig. 6 A), indicating that the mRNA level of ACE2 does not accurately reflect ACE2 protein expression. In addition, we used postmortem human brain tissue to stain for neurons and ACE2 and found that neurons in the cortical gray matter colocalized with ACE2 staining (Fig. S2 C, yellow arrow), and we found other cells in the vicinity that also stained for ACE2 (Fig. S2 C, white arrow). To test the requirement of ACE2 for SARS-CoV-2 infection, we incubated organoids with an anti-ACE2 blocking monoclonal antibody before infection with SARS-CoV-2. We detected significant inhibition of SARS-CoV-2 infection upon pretreatment with ACE2 antibody compared with isotype control, indicating the requirement of ACE2 for infection of brain organoids (Fig. 6, B and F; and Fig. S2 C).
Figure 6.
SARS-CoV-2 neural infection depends on ACE2 and can be neutralized by anti-spike antibodies found in CSF of COVID-19 patients. (A) Immunofluorescence staining of ACE2 in brain organoids. Scale bar = 200 µm for zoomed-out images and 50 µm for zoomed-in images. (B) Immunofluorescence staining of organoids preincubated with isotype antibodies (top row) or anti-ACE2 antibodies (bottom row) and infected with SARS-CoV-2. Scale bar = 100 µm. (C) Schematic showing collection of clinical lumbar puncture from patients with and without COVID-19 for assays shown in D–F. (D) Quantification of anti–SARS-CoV-2 spike antibodies present in CSF of healthy versus COVID-19 patient in limiting dilution using ELISA. (E) Immunofluorescence staining of organoids infected with SARS-CoV-2 preincubated with CSF from health patients (top row) or CSF from COVID-19 patients (bottom row). Scale bar = 100 µm. (F) Quantification of figures from C and E. All experiments were performed with unique organoid, n = 4 per condition, from the same culturing batch, with images from n = 12 cortical regions with two IPSC lines, and Student’s t test was performed (****, P < 0.0001). Experiments were performed twice for reproducibility.
Next, we investigated whether there are humoral antibody responses against SARS-CoV-2 in the CNS of infected patients, and whether antibodies present in the CNS can prevent infection of neurons. We analyzed cerebrospinal fluid (CSF) from a patient hospitalized with COVID-19 and acute encephalopathy (Fig. 6 C) and from a healthy control volunteer by performing ELISA against the spike protein of SARS-CoV-2 (Fig. 6 D). We detected IgG antibodies specific to the spike protein in the patient’s CSF even at 100× dilution (Fig. 6 D). Using this patient’s CSF, we performed a neutralization assay against SARS-CoV-2 and validated its efficacy in preventing brain organoid infection (Fig. 6 E). CSF-containing antiviral antibodies blocked SARS-CoV-2 infection in organoids (Fig. 6, E and F; and Fig. S2 D).
Mouse models of COVID-19 confirm the neuroinvasive potential of SARS-CoV-2
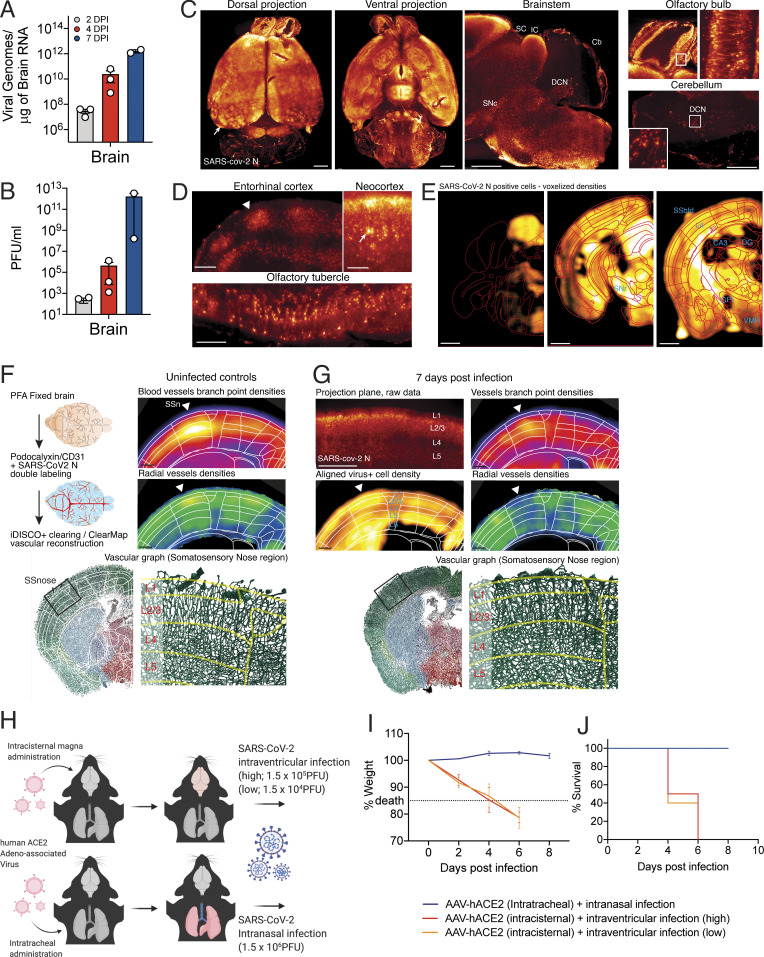
To examine the consequences of SARS-CoV-2 infection in a more physiologically complete system, we examined the neuroinvasive potential of SARS-CoV-2 in vivo by using transgenic mice expressing human ACE2 under the K18 promoter (K18-hACE2; McCray et al., 2007). Similar to previous reports of SARS-CoV showing neurotropism (McCray et al., 2007; Netland et al., 2008), we observed increasing viral titers in the brain of mice after intranasal administration of SARS-CoV-2 (Fig. 7, A and B). We next analyzed the distribution of the virus in the whole brain by immunolabeling against the nucleocapsid protein, clearing, and light sheet microscopy imaging using iDISCO+ (Renier et al., 2014; Fig. 7 and Video 1). 7 d after infection, the virus was widely present in neural cells throughout the forebrain (Fig. 7 C). The cortex was unevenly infected, as the infected cells were visible in columnar patches and in sensory regions, while the layer 4 was mostly devoid of infection (Fig. 7, C and D). We mapped the density of infected cells using ClearMap (Renier et al., 2016) and confirmed that most brain regions contained a high density of infected cells, with the notable exception of the cerebellum (Fig. 7 E). Other regions also contained a relatively low density of infected cells, for instance the dentate gyrus, the globus pallidus, and cortical layer 4. Of note, whole-brain analysis of the virus distribution did not detect the presence of the virus in the vascular endothelium. To explore the possibility that the viral expression by neural cells could indirectly affect the organization of the vascular network, we double-labeled brains for the nucleocapsid protein and the vascular endothelium with CD31 and Podocalyxin, and used ClearMap again to reconstruct the whole-brain vascular network 7 d after infection (Fig. 7 F; Kirst, 2020). We focused on the organization of the cortical vasculature. In infected brains, we focused on hot spots of infections represented as the number of detected infected cells and measured the density and orientation of all blood vessels. In many instances, the expression of the virus coincided with a disruption in the normal vascular topology expected in the cortex, with an important loss of the normal enrichment in radially oriented vessels in upper layers.
Figure 7.
SARS-CoV-2 replicates efficiently in the brain of mice and can cause CNS-specific lethality. (A–C) Mice expressing human ACE2 under the K18 promoter (K18-hACE2) were infected with SARS-CoV-2 intranasally, and brains of the mice were collected on days 2, 4, and 7 hpi for qPCR (A) or plaque assay (B). (C–E) iDISCO+ whole brain immunolabeling against the nucleocapsid protein of SARS-CoV-2 7 d after an intranasal infection, shown as 300-µm projection planes. (C) Dorsal, ventral, and sagittal projections showing widespread distribution of the virus in the forebrain with patches of high viral density in the cortex (arrow). The virus is not detected in the cerebellum, except for the pial meninges and DCNs. (D) 300-µm-deep projection planes in the cortex showing cortical patches of viral expression (arrowhead), reduced infection of the cells in layer 4, and expression in pyramidal neurons (arrow). (E) ClearMap analysis of the infected cells distribution (n = 3), registered to the Allen Brain Atlas, showing wide distribution of the virus across brain regions, with a few regions with lower densities, among which are dentate gyrus (DG), globus pallidus internal segment (GPi), CA3 hippocampal region, cortical layer 4, and ventromedial hypothalamus (VMH). (F and G) Analysis of the vascular network using ClearMap and iDISCO+ 7 d after intranasal infection by mapping of the vascular network with colabeling of the N protein. Planes at the level of the nose somatosensory cortex are shown. (F) Control uninfected brains. Branch point densities (top panel) peak in controls at layer 4. The density of radially oriented vessels (middle panel) peaks in layers 1, 2, and 3 while decreasing in layers 4, 5, and 6. (G) Brain 7 d after infection. Expression of the N viral protein by neural cells is shown at the level of the nose somatosensory cortex (300-µm projection plane and mapped densities). While branch point densities of vessels still show a peak in layer 4, the normal radial organization of the vessels is not measured in the nose region (arrowhead). Representative render of the vascular graph shows a decrease in vessel orientations in control layers 2 and 3. (H) Schematic of experiment for I and J. Adeno-associated viruses coding for human ACE2 (AAV-hACE2) were injected into the cisterna magna or intratracheally to induce brain-specific or lung-specific expression of hACE2. Brain hACE2–expressing mice were infected with SARS-CoV-2 intraventricularly, and lung hACE2–expressing mice were infected with SARS-CoV-2 intranasally. (I and J) Weight loss curve (I) and survival curve (J) of mice infected with SARS-CoV-2 in the lung (blue) and the brain (red and orange; blue, n = 10; red, n = 4; orange, n = 4). Experiments were performed twice for reproducibility. Scale bars = 1 mm (A and C), 200 µm (B), and 500 µm (D and E). CB, cerebellum; DCN, deep cerebellar nuclei; IC, inferior colliculus; SC, superior colliculus; SN, substantia nigra (reticulata or compacta); SS, somatosensory cortex, nose of barrel field.
Video 1.
Whole-brain view of SARS-CoV-2 infection. iDISCO+ whole-brain immunolabeling against the nucleocapsid protein of SARS-CoV2 7 d after intranasal infection.
To dissect the consequences of SARS-CoV-2 infection in the CNS versus the respiratory system, we directed expression of human ACE2 using an adeno-associated virus vector (AAV-hACE2; Israelow et al., 2020) to the lungs via intratracheal delivery or to the brain via intracisternal delivery (Fig. 7 H). Mice were then infected with SARS-CoV-2 either intranasally or intraventricularly. Intranasally infected mice showed signs of lung pathology (Israelow et al., 2020) but no weight loss or death (Fig. 7, I and J). However, intraventricular administration of SARS-CoV-2 resulted in weight loss and death, even at the challenge virus dose 100-fold lower than that used for intranasal infection (Fig. 7, I and J). Altogether, these results highlight the neuroreplicative potential and lethal consequences of SARS-CoV-2 CNS infection in mice.
Evidence of SARS-CoV-2 neuroinvasion in COVID-19 patient brain autopsies
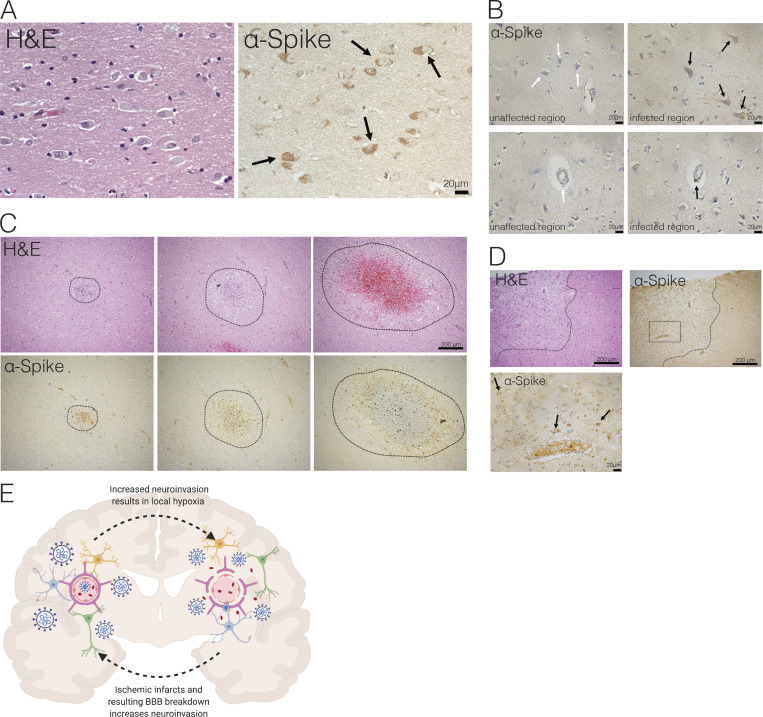
Finally, to determine whether there is evidence of SARS-CoV-2 CNS infection in COVID-19 patients, we examined formalin-fixed, paraffin-embedded (FFPE) sections of three patients who died after suffering from severe COVID-19–related complications. All patients had been admitted to the intensive care unit, they were sedated and ventilated due to respiratory failure (days 3, 10, and 18 for patients 3, 1, and 2, respectively), and their difficulty to be weaned from mechanical ventilation indicated the severity and highly pathogenic nature of the disease course (Table 1). We sampled several regions of the brain and stained with H&E and for SARS-CoV-2 spike protein with a validated protocol and antibody we have used previously (Hosier et al., 2020). While in control slides, there was no positive immunohistochemical staining for the SARS-CoV-2 spike protein, samples from COVID-19 patients showed distinct and specific spike protein staining, albeit to different degrees (Fig. 8 and Fig. S3). Neuropathological evaluations identified staining with anti-spike antibodies in cortical neurons (Fig. 8 A, Fig. 8 B [upper panel], and Fig. 8 D [black arrows]), along with signal in endothelial cells (Fig. 8 B, lower panel). In patient 1, medium spiny neurons in the basal ganglia did not show staining, however, similar to substantia nigra neurons in the midbrain, which were also negative for the SARS-CoV-2 spike protein (Fig. S4). The cellular staining pattern showed diffuse cytoplasmic and perinuclear staining, along with concentrated regions within the cells, consistent with our findings using electron microscopy in the organoids, which demonstrated localized hotspots within the infected cell. Tissue samples from patients with cells that were lipofuscin laden or contained microinfarcts but without COVID-19 showed no reactivity to the SARS-CoV-2 antibody, signifying the specificity of the antibody used. Patient 3, who failed to regain consciousness after cessation of sedation, was diagnosed with severe global encephalopathy and was noted to have multiple diffuse microbleeds, as demonstrated by diagnostic imaging (magnetic resonance imaging). Upon histological examination, we found multiple microscopic ischemic infarcts in the subcortical white matter, ranging from acute to subacute, and with focal hemorrhagic conversion (Fig. 8 C and Fig. S5 A). These varying stages of infarction indicated a temporal sequence of continued ischemic events. Most infarcts showed signs of tissue damage and localized cell death, and positive viral staining was present predominantly around the edges of the infarct and to a lesser degree within the center. At the hyperacute stage of infarction, viral proteins were present in endothelium. These infarcts are present predominantly in subcortical white matter, not within the cortex. In other samples, we found suspected viral staining in locally ischemic regions (Fig. 8 D and Fig. S5, B and C). Remarkably, all the regions of positive viral staining showed no lymphocyte or leukocyte infiltration. This is in contrast to other neurotropic viruses (ZIKV, rabies virus, and herpes virus) in which the infection is typically accompanied by large number of immune cell infiltrates, including T cells. These findings suggest that, although SARS-CoV-2 has neurotropic properties and can infect neurons in patients, it did not invoke an immune response typical of other neurotropic virus in this particular case.
Table 1. Clinical characteristics of COVID-19 patients.
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Gender | Male | Male | Female |
| Age (yr) | 63 | 58 | 49 |
| Medical history | Hypertension (no treatment) | Obesity | Renal transplantation (immunosuppressive therapy) |
| Hypertension | Type 1 diabetes | ||
| Sleep apnea syndrome | Hypertension | ||
| Pneumonia (2017) | Pulmonary embolism (2003) | ||
| Onset of illness | Day 0: Dyspnea and fever | Day 0: Diarrhea, asthenia | Day 0: Fever, dyspnea, cough, sputum |
| Day 16: Chills and dyspnea | |||
| SARS-CoV-2 PCR test positive | No SARS-CoV-2 PCR test | Day 1: Nasal SARS-CoV-2 PCR test positive | |
| Emergency department admission | Day 10: Worsening dyspnea → patient sedated and ventilated | ||
| ICU admission | Day 10: Acute respiratory distress and left myocardial failure | Day 18: Acute respiratory distress and fever → patient sedated and ventilated | Day 3: Dyspnea; mechanical ventilation |
| Day 5: Moderate acute respiratory distress syndrome; kidney failure | |||
| Medical course | Day 14: Difficulty waking the patient after cessation of sedation = areactive encephalopathy | Resuscitated hypoxic cardiac arrest due to aspiration during initial care | Difficult ventilator withdrawal |
| Compressive pneumothorax; refractory cardiac arrests | Day 16: Bacterial pneumoniae; transfer to the rehabilitation service | ||
| No SARS-CoV-2 PCR test during hospitalization | Day 19: Acute respiratory distress and kidney failure → transfer to ICU | ||
| Brain imaging | Day 32: Brain MRI; multiple diffuse micro bleedings (subcortical and deep regions) | No brain imaging | No brain imaging |
| Death | Day 43 | Day 19 (after 6–12 h of hospital medical care) | Day 27 |
| Postmortem SARS-CoV-2 PCR test on lungs | Positive | Positive | Positive |
ICU, intensive care unit; MRI, magnetic resonance imaging.
Figure 8.
Evidence of neuroinvasion in postmortem COVID-19 patient brains. FFPE sections of brain tissue from COVID-19 patients stained with H&E and anti–SARS-CoV-2-spike antibody. (A) Image of cortical neurons positive for SARS-CoV-2 (black arrows). Scale bar = 20 µm. (B) Images of unaffected regions (left) and infected regions (right) demonstrating infection of neurons (top row) and microvasculature (bottom row). Scale bar = 20 µm. (C) Ischemic infarcts found at different stages stained with H&E (top row) and SARS-CoV-2-spike antibody (bottom row). (D) Ischemic region (outlined with dotted line) with positive staining focused around ischemic infarct. Bottom image shows zoomed-in image indicated by dotted box in top image, and black arrows indicate infected neurons in the region. (E) Schematic of hypothesized consequences of SARS-CoV-2 neuroinvasion.
Figure S4.
Evidence of neuroinvasion in postmortem COVID-19 patient brains (Brodmann area 6 and midbrain). FFPE sections of brain tissue from COVID-19 patients were stained using anti–SARS-CoV-2-spike antibody. (A) Images of regions of Brodmann area 6 of patient 1. Images are also shown in Fig. 4. (B) Images of regions of patient 2. (C) Images of regions of the caudate of patient 3. (D) Example image from control patient brains. Scale bar = 200 µm for zoomed-out images and 20 µm for zoomed-in images.
Figure S5.
Evidence of SARS-CoV-2 neuroinvasion–associated ischemic infarcts. FFPE sections of brain tissue from COVID-19 patients were stained using H&E or anti–SARS-CoV-2-spike antibody. (A) H&E images of ischemic infarcts at different stages (1, earliest, to 9, latest). (B) SARS-CoV-2–stained images of ischemic infarcts at different stages (1, earliest, to 9, latest). Each number corresponds to H&E image in A. (C) Images of SARS-CoV-2–positive regions in brains of COVID-19 patients. (D) Example image from control patient brain. Scale bar = 200 µm for zoomed-out images and 20 µm for zoomed-in images.
Discussion
We examined the potential for SARS-CoV-2 to infect neural tissues of both mice and human origin and demonstrate potential consequences of its neuroinvasion. Our results suggest that neurological symptoms associated with COVID-19 may be related to consequences of direct viral invasion of the CNS. Specifically, our work experimentally demonstrates that the brain is a site for high replicative potential for SARS-CoV-2. We further show that SARS-CoV-2 causes significant neuronal death in human brain organoids. Using electron microscopy, we identified viral particles budding from the ER, indicating the virus’s ability to use the neuron cell machinery to replicate. Similar to neuronal loss observed in patient autopsies (Solomon et al., 2020), we noticed large numbers of cells dying in the organoid; however, this neuronal death did not colocalize directly with virus infection. Single-cell RNA-seq of the infected organoids showed metabolic changes in neurons without IFN or IFN-stimulated gene signatures, indicating that the neuroinvasive consequence of SARS-CoV-2 is unique compared with other neurotropic viruses such as ZIKV. Closer examination showed diverging metabolic changes in infected versus neighboring cells, suggesting that the infected cells can cause local changes to their microenvironment, affecting survival of nearby cells. It is possible that viral infection induces locally hypoxic regions, which aids in lowering the threshold for tissue damage in the context of an already oxygen-deprived state.
While ACE2 expression levels in the human brain are still being investigated, we showed that ACE2 is expressed at the protein level and is functionally required for SARS-CoV-2 infection in human brain organoids. Further, we detected robust antiviral antibody presence in the CSF of a COVID-19 patient who presented with acute neurological symptoms. This finding suggests that, at least in some patients with COVID-19 and neurological symptoms, there is robust antibody response against the virus within the CSF. In the in vivo setting of the CNS with vasculature and immune cells, neuronal death could have cascading downstream effects in causing and amplifying CNS inflammation.
Although our rodent model does not use endogenous ACE2 expression, it has been previously reported that even mouse-adapted SARS-CoV is still neurotropic in wild-type mice, and SARS-CoV-2 is neurotropic in mice with hACE2 expression from the endogenous locus (Roberts et al., 2007; Sun et al., 2020). Using mouse models, we demonstrate for the first time that SARS-CoV-2 neuroinvasion in mice can have significant remodeling of brain vasculature, providing a potential link between the hypoxia and what we see in both the human organoid and the patient brains.
Similar to previous reports of acute hypoxic ischemic damage without microthrombi in postmortem brain of COVID-19 patients (Solomon et al., 2020), we also found presence of ischemic damage and microinfarcts in postmortem brain samples of COVID-19 patients. In our study, we observed evidence of SARS-CoV-2 infection within the regions of micro–ischemic infarcts, suggesting the possibility of neuroinvasion-associated ischemia and vascular anomalies, consistent with what we observed in mice. However, a limitation of our study is that autopsy samples from only a small number of patients were examined, providing a snapshot of case reports from several patients rather than a generalizable phenomenon. Future studies are needed to examine whether there are other cases of neuroinvasion in the CNS, and the predisposition for such infection. Although we are unable to determine the exact relationship between neuroinvasion and ischemic infarcts, we pose a possible hypothesis from our findings in the patients, mice, and infections of human brain organoids: that SARS-CoV-2 neuroinvasion may cause locally hypoxic regions and disturbance of vasculature, and the disruption of brain vasculature can make vulnerable ischemic infarcts and regions more susceptible to viral invasion (Fig. 8 E). Our findings expand the utility of human brain organoids, beyond modeling fetal brains, and highlight the importance of using a variety of approaches to best model physiology of the human brain.
In future studies, identifying the route of SARS-CoV-2 invasion into the brain, in addition to determining the sequence of infection in different cell types in the CNS, will help validate the temporal relationship between SARS-CoV-2 and ischemic infarcts in patients. It may be through the nasal cavity–to-CNS connection through the cribriform plate, olfactory epithelium and nerve, or viremia, but regardless, the brain should be considered a SARS-CoV-2–susceptible organ system upon respiratory exposure (Baig and Sanders, 2020; Coolen et al., 2020).
Altogether, our study provides clear demonstration that neurons can become a target of SARS-CoV-2 infection, with devastating consequences of localized ischemia in the brain and cell death, highlighting SARS-CoV-2 neurotropism and guiding rational approaches to treatment of patients with neuronal disorders.
Materials and methods
All procedures were performed in a biosafety level 3 (BSL3)/animal BSL3 facility (for SARS-CoV-2–related work) with approval from the Yale Environmental Health and Safety committee (#20-19 and #18-16).
Mice
6- to 12-wk-old mixed-sex C57Bl/6 (B6J) mice purchased from Jackson Laboratory and B6.Cg-Tg(K18-ACE2)2Prlmn/J(K18-hACE2) mice (gift from Jackson Laboratory) were subsequently bred and housed at Yale University. Experiments with wild-type mice transduced with AAV-hACE2 were performed with littermate controls. Experiments performed with B6.Cg-Tg(K18-ACE2)2Prlmn/J(K18-hACE2) were from a single shipment batch of mice from Jackson Laboratory. All procedures used in this study (sex and age matched) complied with federal guidelines and the institutional policies of the Yale School of Medicine Animal Care and Use Committee.
AAV infection (intratracheal and intracisternal magna injection)
Adeno-associated virus 9 encoding hACE2 (AAV-CMV-hACE2) were purchased from Vector Biolabs.
Intratracheal injection
Animals were anaesthetized using a mixture of ketamine (50 mg/kg) and xylazine (5 mg/kg), injected i.p. The rostral neck was shaved and disinfected. A 5-mm incision was made, the salivary glands were retracted, and the trachea was visualized. Using a 500-µl insulin syringe, a 50-µl bolus injection of 1011 genome copies (GC) of AAV-CMV-hACE2 was injected into the trachea. The incision was closed with VetBond skin glue. Following intramuscular administration of analgesic (meloxicam and buprenorphine, 1 mg/kg), animals were placed in a heated cage until full recovery.
Intracisternal magna injection
Mice were anesthetized using ketamine and xylazine, and the dorsal neck was shaved and sterilized. A 2-cm incision was made at the base of the skull, and the dorsal neck muscles were separated using forceps. After visualization of the cisterna magna, a Hamilton syringe with a 15°, 33-gauge needle was used to puncture the dura. 3 µl of AAV9 (3 × 1012 viral particles/mouse) or mRNA (4–5 µg) was administered per mouse at a rate of 1 µl/min. Upon completion of the injection, the needle was left in to prevent backflow for an additional 3 min. The skin was stapled and disinfected, and the same postoperative procedures were performed as for intratracheal injections.
Generation of SARS-CoV-2 virus
To generate SARS-CoV-2 viral stocks, Huh7.5 cells were inoculated with SARS-CoV-2 isolate USA-WA1/2020 (#NR-52281; BEI Resources) to generate a P1 stock. To generate a working stock, VeroE6 cells were infected at multiplicity of infection 0.01 for 4 d. Supernatant was clarified by centrifugation (450 g for 5 min) and filtered through a 0.45-µm filter. To concentrate virus, 1 volume of cold (4°C) 4× PEG-it Virus Precipitation Solution (40% [wt/vol] PEG-8000 and 1.2 M NaCl) was added to 3 volumes of virus-containing supernatant. The solution was mixed by inverting the tubes several times and then incubated at 4°C overnight. The precipitated virus was harvested by centrifugation at 1,500 g for 60 min at 4°C. The pelleted virus was then resuspended in PBS and aliquoted for storage at −80°C. Virus titer was determined by plaque assay using Vero E6 cells.
SARS-CoV-2 infection of organoids
Brain organoids in low-adhesion plates were infected with SARS-CoV-2 at multiplicity of infection 1.
SARS-CoV-2 infection (intranasal)
Mice were anesthetized using 30% vol/vol isoflurane diluted in propylene glycol. With a pipette, 50 µl of SARS-CoV-2 (3 × 107 PFU/ml) was delivered intranasally.
SARS-CoV-2 infection (intraventricular)
Animals were anaesthetized using a mixture of ketamine (50 mg/kg) and xylazine (5 mg/kg) injected i.p. After sterilization of the scalp with alcohol and betadine, a midline scalp incision was made to expose the coronal and sagittal sutures, and a burr holes were drilled 1 mm lateral to the sagittal suture and 0.5 mm posterior to the bregma. A 10-µl Hamilton syringe loaded with virus and inserted into the burr hole at a depth of 2 mm from the surface of the brain and left to equilibrate for 1 min before infusion. Once the infusion was finished, the syringe was left in place for another minute before removal. Bone wax was used to fill the burr hole, and skin was stapled and cleaned. Following intramuscular administration of analgesic (meloxicam and buprenorphine, 1 mg/kg), animals were placed in a heated cage until full recovery. For the high condition, 5 µl of SARS-CoV-2 (3 × 107 PFU/ml) was used, and for the low condition, 5 µl of SARS-CoV-2 (3 × 106 PFU/ml).
Sample staining and iDISCO+ clearing
Whole-brain vasculature staining was performed following the iDISCO+ protocol previously described (Renier et al., 2016), with minimal modifications. All steps of the protocol were done at room temperature with gentle shaking unless otherwise specified. All buffers were supplemented with 0.01% sodium azide (Sigma-Aldrich) to prevent bacterial and fungal growth. Brains were dehydrated in an increasing series of methanol (Sigma-Aldrich) dilutions in water (washes of 1 h in methanol 20, 40, 60, 80, and 100%). An additional wash of 2 h in methanol 100% was done to remove residual water. Once dehydrated, samples were incubated overnight in a solution containing a 66% dichloromethane (Sigma-Aldrich) in methanol and washed twice in methanol 100% (4 h each wash). Samples were then bleached overnight at 4°C in methanol containing 5% hydrogen peroxide (Sigma-Aldrich). Rehydration was done by incubating the samples in methanol 60, 40, and 20% (1 h each wash). After methanol pretreatment, samples were washed in PBS twice, for 15 min and 1 h, in PBS containing 0.2% Triton X-100 (Sigma-Aldrich) and further permeabilized by 24-h incubation at 37°C in Permeabilization Solution (20% DMSO [Sigma-Aldrich] and 2.3% glycine [Sigma-Aldrich] in PBS-T).
To start the immunostaining, samples were first blocked with 0.2% gelatin (Sigma-Aldrich) in PBS-T for 24 h at 37°C, and the same blocking buffer was used to prepare antibody solutions. A combination of primary antibodies targeting different components of the vessel’s walls were used to achieve continuous immunostaining. Antibodies to podocalyxin and CD31 were combined with antibodies against the nucleocapsid (N) from GeneTex. Primary antibodies were incubated for 10 d at 37°C with gentle shaking, then washed in PBS-T (twice for 1 h and then overnight) and finally newly incubated for 10 d with secondary antibodies. Secondary antibodies conjugated to Alexa Fluor 647 were used to detect podocalyxin and CD31, while the NUCLEOCAPSID protein was stained with a secondary antibody conjugated to Alexa Fluor 555. After immunostaining, the samples were washed in PBS-T (twice for 1 h and then overnight), dehydrated in a methanol/water increasing concentration series (20, 40, 60, 80, and 100%, 1 h each, and then methanol 100% overnight), followed by a wash in 66% dichloromethane and 33% methanol for 3 h. Methanol was washed out with two final washes in dichloromethane 100% (15 min each), and finally the samples were cleared and stored in dibenzyl ether (Sigma-Aldrich) until light sheet imaging.
Light sheet imaging
We imaged with a 4× 0.35-NA objective cropped elongated field of view (600 × 2,200 µm) covering the narrow waist of the light sheet at 1.63 µm/pixel of lateral resolution, 1.6-µm spacing. A reference channel for registration to the annotated atlas using the sample autofluorescence was acquired at 5 µm/pixel. The acquisitions were done on a LaVision Ultramicroscope II equipped with infinity-corrected objectives. The microscope was installed on an active vibration filtration device, itself put on a marble compressed-air table. Imaging was done with the following filters: 595 nm/40 mW for Alexa Fluor 555, and 680 nm/30 mW for Alexa Fluor 647. The microscope was equipped with the following laser lines: OBIS-561 nm, 100 mW, OBIS-639 nm, 70 mW, and used the second-generation LaVision beam combiner. The images were acquired with an Andor complementary metal oxide semiconductor (CMOS) sNEO camera. Main acquisitions were done with the LVMI-Fluor 4×/0.3 WD6 LaVision Biotec objective. The brain was positioned in sagittal orientation, cortex side facing the light sheet. A field of view of 400 × 1,300 pixels was cropped at the center of the camera sensor. The light sheet numerical aperture was set to NA 0.1. Beam width was set to the maximum. Only the center-left light sheet was used. Laser powers were set to 100% (639 nm) or 10% (561 nm). Tile overlaps were set to 10%. The acquisition routine was set to the following: (1) Z-drive → save ome.tif stack; (2) filter change → Z-drive → save ome.tif stack; (3) change x position → repeat 1,2 12 times; and (4) change y position → repeat 1,2,3 6 times. At the end of the acquisition, the objective is changed to a MI PLAN 1.1×/0.1 for the reference scan at 488 nm excitation (tissue autofluorescence). The field of view is cropped to the size of the brain, the z-steps are set to 6 µm, and light sheet numerical aperture is set to 0.03 NA.
Viral RNA analysis
At indicated time points, mice were euthanized in 100% isoflurane. Brain tissue was placed in a bead homogenizer tube with 1 ml of PBS and 2% FBS. After homogenization, 250 µl of this mixture was placed in 750 µl Trizol LS (Invitrogen), and RNA was extracted with an RNeasy mini kit (Qiagen) per manufacturer protocol. To quantify SARS-CoV-2 RNA levels, real-time quantitative PCR (RT-qPCR) was performed using Luna Universal Probe Onestep RT-qPCR kits (New England Biolabs) with 1 µg of RNA and the Centers for Disease Control and Prevention real-time RT-PCR primer/probe sets for 2019-nCoV_N1.
Viral titer
Brain homogenates were centrifuged at 3,900 g for 10 min, and supernatant was taken for plaque assays. Supernatant at limiting dilutions was incubated on Vero E6 cells in MEM-supplemented NaHCO3, 4% FBS, and 0.6% Avicel RC-581. Plaques were resolved 48 hpi by fixing in 10% formaldehyde for 1 h followed by staining for 1 h in 0.5% crystal violet in 20% ethanol.
Stem cell culture
Human Y6 and Y1 iPS lines were obtained from Yale Stem Cell Center and New York Cell Stem Foundation, respectively. Cells were verified as being pluripotent, having normal karyotype, mycoplasma free, and cultured in feeder-free conditions on Matrigel-coated plates with mTeSR Plus culture medium (Stem Cell Technologies) and passaged using ReLeSR (Stem Cell Technologies).
Teratoma formation
106 Y6 iPSC cells were collected by collagenase treatment and resuspended in 100 ml of DMEM/F12, collagen, and Matrigel mix (2:1:1 ratio). Cells were injected i.m. into immunodeficient Rag2−/−GammaC−/− mice. After 8 wk, teratomas were harvested, fixed, and subjected to paraffin embedding and H&E staining.
G-band staining for karyotype analysis
Y6 iPSC small clumps were seeded on glass slides precoated with Matrigel and fed with mTESR medium for 3 d. Media were then switched to DMEM basal medium supplemented with 10% FBS for another 3 d, and the slides were transferred to Yale Cytogenetic Laboratory for G-band staining.
Neural progenitor cell culture
Y6 iPS lines were differentiated to NPCs on Matrigel-coated plates using the monolayer protocol of the StemDiff SMADI Neural Induction kit (#08581; Stem Cell Technologies) for two passages and then maintained in StemDiff Neural Progenitor media (#05833; Stem Cell Technologies). 12-d NPCs were used for all experiments.
Cerebral organoid culture
For preparation of embryoid bodies, 9,000 single cells were seeded in each well of low-attachment 96-well U-bottom plates and 10 µM Y-27632 ROCK inhibitor for 1 d. Cerebral organoids were generated following exactly the previously established protocol (Lancaster and Knoblich, 2014; Lancaster et al., 2013), using an orbital shaker for agitation.
Immunostaining
Brains of infected mice and organoids were collected and fixed in 4% paraformaldehyde (PFA). Samples were then dehydrated in a 30% sucrose solution. Cryostat sections were blocked in 0.1 M Tris-HCl buffer with 0.3% Triton and 1% FBS before staining. Slides were stained for IBA-1 (nb100-1028; Novus Biologicals), GFAP (ab4674, [Abcam] and rabbit anti–SARS-CoV-2 nucleocapsid [GeneTex]). Slides were mounted with Prolong Gold Antifade reagent (Thermo Fisher Scientific). All slides were analyzed by fluorescence microscopy (BX51; Olympus).
Organoids were fixed in 4% PFA in a BSL3 facility then moved to 30% sucrose solution at 4°C for ≥24 h. Organoids were embedded in tissue freezing medium, cut into 20-µm sections using a cryostat, and mounted on slides. After blocking and permeabilization with 0.25% Triton X-100 and 4% donkey serum, sections were incubated overnight with primary antibody: ms anti-Pax6 (1:300, #561462; BD PharMingen), rb anti-ACE2 (1:500, ab15348; Abcam), ms anti-Sox2 (1:500, sc-365823; Santa Cruz), rat anti-CTIP2 (1:500, ab18465; Abcam), rb anti-TBR1 (1:500, ab31940; Abcam), ms anti-MAP2 (1:500, MAB3418; Millipore), rb anti–SARS-CoV-2 nucleocapsid (1:250, GTX635679; GeneTex), and rb anti-HIF1 α antibody (GTX127309; Genetex). TUNEL assay was performed using the Click-It Plus TUNEL Assay for In Situ Apoptosis Detection (C10617; Thermo Fisher Scientific) per the manufacturer’s instructions. Alexa Fluor 488, 555, and 647 antibodies were applied for 1 h at room temperature (1:500) after three 10-min PBS washes. To mark nuclei, DAPI (1:3,000) was added to the secondary antibody incubation. Slides were then washed three times in PBS and then mounted with VectaShield Anti-Fade Mounting Medium. Images were acquired using an LSM 880 confocal microscope (Carl Zeiss) and prepared using Fiji (National Institutes of Health).
NPCs on coverslips were fixed in 4% PFA for 15 min at room temperature in BSL3 conditions and washed three times with PBS. After blocking and permeabilization with 0.25% Triton X-100 and 10% donkey serum, coverslips were incubated overnight with primary antibody as described above. TUNEL assay was performed as described above. Alexa Fluor 488, 555, and 647 antibodies were applied for 1 h at room temperature (1:500) after three 10-min PBS washes. Coverslips were then washed three times in PBS and then mounted on slides with Prolong Diamond Anti-Fade Mountant with DAPI (Thermo Fisher Scientific). Images were acquired using an LSM 880 confocal microscope (Carl Zeiss) and prepared using Fiji.
Human FFPE sections were heated for 30 min at 60°C and treated with xylenes followed by rehydration in decreasing concentrations of ethanol (100, 90, 80, and 70%). Antigen retrieval was performed using a pressure cooker (Biocare Decloaking Chamber) by boiling in sodium citrate (pH 6.0) for 20 min at 115°C. After blocking and permeabilization with 0.25% Triton X-100 and 4% donkey serum, sections were incubated overnight with primary antibody: rb anti-ACE2 (1:500, ab15348; Abcam) and ms anti-NeuN (1:500, ab104224; Abcam). Alexa Fluor 488 and 555 antibodies were applied for 1 h at room temperature (1:500) after three 10-min PBS washes. To mark nuclei, DAPI (1:3,000) was added to the secondary antibody incubation. Slides were then washed three times in PBS to remove detergent and treated with TrueBlack Lipofuscin Autofluorescence Quencher (Biotium) according to the manufacturer’s instructions. Sections were mounted with VectaShield mounting medium. Images were acquired using an LSM 880 confocal microscope (Carl Zeiss) and prepared using Fiji.
Electron microscopy
Organoids were fixed using 2.5% glutaraldehyde in 0.1 M phosphate buffer, osmicated in 1% osmium tetroxide, and dehydrated in ethanol. During dehydration, 1% uranyl acetate was added to the 70% ethanol to enhance ultrastructural membrane contrast. After dehydration, the organoids were embedded in Durcupan, and 70-nm sections were cut on a Leica ultramicrotome, collected on Formvar-coated single-slot grids, and imaged on a Tecnai 12 Biotwin electron microscope (FEI).
Single-cell RNA-seq
Organoids were collected, and single-cell suspensions were made using a papain dissociation system (Worthington Biochemical Corp.). Single-cell suspensions were loaded onto the Chromium Controller (10x Genomics) for droplet formation. Single-cell RNA-seq libraries were prepared using the Chromium Single Cell 3′ Reagent Kit (10x Genomics). Samples were sequenced on the NovaSeq.
R v3.4.2 (R Core Team) was used for all statistical analysis. Sequencing results were demultiplexed into Fastq files using Cell Ranger (v3.0.2; 10x Genomics) mkfastq function. Samples were aligned to GRCh38 (10x Genome). The count matrix was generated using the count function with default settings. Matrices were loaded into Seurat v3.1.5 for downstream analysis. Cells with <200 unique molecular identifiers or high mitochondrial content were discarded. Using FindIntegrationAnchors and IntegrateData functions, all libraries were integrated into a single matrix. Principal component values that were statistically significant were identified, and a cutoff point was determined using the inflection point after using the PCElbowPlot function. Clusters were determined using the RunUMAP, FindNeighbors, and FindClusters functions on Seurat. Cells were considered infected if transcripts aligned to viral ORF1ab, Surface glycoprotein (S), ORF3a, Envelope protein (E), Membrane glycoprotein (M), ORF6, ORF7a, ORF8, Nucleocapsid phosphoprotein (N), or ORF10. DEGs from the FindMarkers function were used to perform PANTHER-GO statistical overrepresentation tests for up-regulated and down-regulated genes in each condition shown in Fig. 5 B. Gene lists for Fig. 5 C were obtained from gene set enrichment analysis databases (https://gsea-msigdb.org/gsea/index.jsp).
Immunohistochemistry
Paraffin sections were heated for 30 min at 60°C and treated with xylenes, followed by rehydration in decreasing concentrations of ethanol (100, 90, 80, and 70%). Antigen retrieval was performed by boiling in sodium citrate (pH 6.0) for 15 min, and peroxidase activity was blocked with hydrogen peroxide for 10 min. Blocking was performed in 2.5% normal horse serum (Vector Laboratories) and incubated in primary antibody overnight at 4°C. Mouse anti–SARS-CoV-2 spike antibody (clone 1A9; GeneTex GTX632604) was used at a dilution of 1:400. Secondary antibody and detection reagents from the VECTASTAIN Elite ABC-HRP Kit (PK-7200; Vector Laboratories) were used according to manufacturer instructions. Sections were counterstained with Hematoxylin QS (H-3404; Vector Laboratories), dehydrated in increasing concentrations of ethanol, cleared with xylenes, and mounted with VectaMount permanent mounting medium (H-5000; Vector Laboratories).
ACE2 blocking
For ACE2 blocking assays, Organoids were preincubated with anti-ACE2 Antibodies (AF933; R&D Systems) or isotype antibodies at a concentration of 100 µg/ml for 1 h at 4°C. Organoids were then infected with SARS-CoV-2 as described above.
CSF neutralization assay
Excess CSF was obtained from a hospitalized patient with COVID-19 who underwent clinical lumbar puncture and from a healthy control volunteer. CSF was centrifuged to isolate cell-free supernatant, which was used for ELISA and neutralization assays as follows.
ELISA
ELISAs were performed as previously reported (Israelow et al., 2020). In short, Triton X-100 and RNase A were added to serum samples at final concentrations of 0.5% and 0.5 mg/ml, respectively, and incubated at room temperature for 3 h before use to reduce risk from any potential virus in serum. 96-Well MaxiSorp plates (#442404; Thermo Fisher Scientific) were coated with 50 µl/well of recombinant SARS-CoV-2 S1 protein (100 µg, #S1N-C52H3; ACROBiosystems) at a concentration of 2 µg/ml in PBS and incubated overnight at 4°C. The coating buffer was removed, and plates were incubated for 1 h at room temperature with 200 µl of blocking solution (PBS with 0.1% Tween-20 and 3% milk powder). Serum was diluted 1:50 in dilution solution (PBS with 0.1% Tween-20 and 1% milk powder), and 100 µl of diluted serum was added for 2 h at room temperature. Plates were washed three times with PBS-T (PBS with 0.1% Tween-20), and 50 µl of mouse IgG-specific secondary antibody (1:10,000, #405306; BioLegend) diluted in dilution solution added to each well. After 1 h of incubation at room temperature, plates were washed three times with PBS-T. Samples were developed with 100 µl of TMB Substrate Reagent Set (#555214; BD Biosciences), and the reaction was stopped after 15 min by the addition of 2 N sulfuric acid.
Neutralization assay
Virus for infection was preincubated with 500 µl of healthy or COVID-19 CSF at 37°C for 1 h before infection of organoids. The organoid culture was supplemented with an additional 500 µl of CSF after infection until it was collected for imaging.
Human subjects
Research participants were enrolled at Yale University through Human Investigation Committee Protocols HIC#2000027690 and HIC #1502015318. The Institutional Review Board at Yale approved the protocols, and informed consent was obtained from all participants. Postmortem COVID-19 brain tissues were obtained from COVITIS Biobank (Assistance Publique Hopitaux de Paris, Paris, France).
Statistical analysis
No statistical methods were used to predetermine sample size. The investigators were not blinded during experiments and outcome assessment, but outcome assessment was additionally evaluated by animal technicians and vets blinded to the study. Survival curves were analyzed using a log-rank (Mantel–Cox) test. For other data, normally distributed continuous variable comparisons used a two-tailed unpaired Student’s t test or paired Student’s t test, with Prism software.
Data availability
Data files from single-cell RNA-seq can be found at Sequence Read Archive accession no. PRJNA682590.
Online supplemental material
Fig. S1 shows additional infection of 9-wk organoids with SARS-CoV-2 in addition to infection of hPSC-derived NPCs. Fig. S2 describes in more detail the hACE2 dependence of the virus in infecting neural cells. Fig. S3, Fig. S4, and Fig. S5 expand on the findings in human postmortem samples and show more examples of various regions of infected and noninfected brains. Video 1 shows a whole-brain view of SARS-CoV-2 infection.
Acknowledgments
We thank the Yale Environmental Health and Safety department for allowing safe working environments with the SARS-CoV-2 virus. We also thank the patient donors, and clinicians who helped with the collection of CSF for neutralization assays. We also thank the members of the Yale Center for Genome Analysis who have helped with all aspects of sequencing. Finally, we thank the members of the Iwasaki laboratory for insightful discussions regarding the project.
This study was supported by National Institutes of Health grants R01AI157488 (A. Iwasaki, S.F. Farhadian), R01NS111242 (A. Iwasaki), T32GM007205 (Medical Scientist Training Program training grant), F30CA239444 (E. Song), 2T32AI007517 (B. Israelow), and K23MH118999 (S.F. Farhadian); the Women’s Health Research at Yale University Pilot Project Program (A. Iwasaki and A. Ring); Fast Grant from Emergent Ventures at the Mercatus Center (A. Iwasaki, E. Song, and C.B. Wilen); the Mathers Foundation (A. Ring, C.B. Wilen, and A. Iwasaki); and the Ludwig Family Foundation (A. Iwasaki, A. Ring, and C.B. Wilen). A. Iwasaki is an investigator of the Howard Hughes Medical Institute.
Author contributions: E. Song, C. Zhang, K. Bilguvar, and A. Iwasaki planned the project and analyzed data. E. Song, C. Zhang, and A. Iwasaki wrote the manuscript. E. Song, C. Zhang, B. Israelow, A. Lu-Culligan, A.V. Prado, and S. Skriabine performed experiments. P. Lu, O-E. Weizman, F. Liu, Y. Dai, K. Szigeti-Buck, Y. Yasumoto, G. Wang, J. Heltke, E. Ng, J. Wheeler, and M.M. Alfajaro assisted with experiments. E. Levavasseur, S.A.J. Kazmi, K. Zhang, C. Castaldi, B. Fontes, D. Van Dijk, S. Mane, M. Gunel, A. Ring, C.B. Wilen, T.L. Horvath, I. Plu, N.G. Ravindra, S. Haik, J-L. Thomas, A. Huttner, D. Seilhean, A. Louvi, S.F. Farhadian, and K. Bilguvar provided expertise and materials for analysis of data. A.V. Prado, S. Skriabine, and N. Renier performed iDISCO+ imaging and analysis of mice. E. Levavasseur, S.A.J. Kazmi, K. Zhang, I. Plu, S. Haik, J-L. Thomas, A. Huttner, and D. Seilhean provided human samples and analysis of images. A. Iwasaki and K. Bilguvar supervised the project and secured funding.
References
- Amin, N.D., and Paşca S.P.. 2018. Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron. 100:389–405. 10.1016/j.neuron.2018.10.007 [DOI] [PubMed] [Google Scholar]
- Baig, A.M., and Sanders E.C.. 2020. Potential neuroinvasive pathways of SARS-CoV-2: Deciphering the spectrum of neurological deficit seen in coronavirus disease-2019 (COVID-19). J. Med. Virol. 92:1845–1857. 10.1002/jmv.26105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo, D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. 2020. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 181:1036–1045.e9. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen, C.K., Hogberg H.T., Bahadirli-Talbott A., Bishai W.R., Hartung T., Keuthan C., Looney M.M., Pekosz A., Romero J.C., Sillé F.C.M., et al. 2020. Infectability of human BrainSphere neurons suggests neurotropism of SARS-CoV-2. ALTEX. 37:665–671. 10.14573/altex.2006111 [DOI] [PubMed] [Google Scholar]
- Cakir, B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.J., Chapeton K., Patterson B., Yuan Y., He C.S., et al. 2019. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 16:1169–1175. 10.1038/s41592-019-0586-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri, L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Kallio K., Kaya T., Anastasina M., Smura T., et al. 2020. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 370:856–860. 10.1126/science.abd2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen, T., Lolli V., Sadeghi N., Rovai A., Trotta N., Taccone F.S., Creteur J., Henrard S., Goffard J.C., Dewitte O., et al. 2020. Early postmortem brain MRI findings in COVID-19 non-survivors. Neurology. 95:e2016–e2027. 10.1212/WNL.0000000000010116 [DOI] [PubMed] [Google Scholar]
- Daly, J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. 2020. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 370:861–865. 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, B.P., Kofman S.B., Smith J.R., Norris G.T., Snyder A.G., Kolb J.P., Gao X., Locasale J.W., Martinez J., Gale M. Jr., et al. 2019. The Nucleotide Sensor ZBP1 and Kinase RIPK3 Induce the Enzyme IRG1 to Promote an Antiviral Metabolic State in Neurons. Immunity. 50:64–76.e4. 10.1016/j.immuni.2018.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice, F.G., Tovar-Moll F., Moll J., Munoz D.P., and Ferreira S.T.. 2020. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and the Central Nervous System. Trends Neurosci. 43:355–357. 10.1016/j.tins.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez, P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., and Rehen S.K.. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science. 352:816–818. 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- Heneka, M.T., Golenbock D., Latz E., Morgan D., and Brown R.. 2020. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimers Res. Ther. 12:69. 10.1186/s13195-020-00640-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. 2020. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181:271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosier, H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., et al. 2020. SARS-CoV-2 infection of the placenta. J. Clin. Invest. 130:4947–4953. 10.1172/JCI139569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow, B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. 2020. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 217:e20201241. 10.1084/jem.20201241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, F., Pather S.R., Huang W.-K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M., et al. 2020. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell. 27:937–950.e9. 10.1016/j.stem.2020.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton, S., Boyle M.J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D., et al. 2019. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 574:418–422. 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- Kirst, C. 2020. Mapping whole brain structure and activity at cellular resolution: iDISCO+ and ClearMap. J. Biomol. Tech. In press. PMC.7424992 [Google Scholar]
- Lancaster, M.A., and Knoblich J.A.. 2014. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9:2329–2340. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., and Knoblich J.A.. 2013. Cerebral organoids model human brain development and microcephaly. Nature. 501:373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.Y., Li L., Zhang Y., and Wang X.S.. 2020. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 9:45. 10.1186/s40249-020-00662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., et al. 2020. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 77:683–690. 10.1001/jamaneurol.2020.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray, P.B. Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81:813–821. 10.1128/JVI.02012-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland, J., Meyerholz D.K., Moore S., Cassell M., and Perlman S.. 2008. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 82:7264–7275. 10.1128/JVI.00737-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, L., Bonfio C., Chadwick J., Begum F., Skehel M., and Lancaster M.A.. 2020. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 369:eaaz5626. 10.1126/science.aaz5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, A. 2020. Long-Term Neurological Threats of COVID-19: A Call to Update the Thinking About the Outcomes of the Coronavirus Pandemic. Front. Neurol. 11:308. 10.3389/fneur.2020.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles, V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. 2020. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 383:590–592. 10.1056/NEJMc2011400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, F., Qian S., Zhang S., and Zhang Z.. 2020. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526:135–140. 10.1016/j.bbrc.2020.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. 2016. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 165:1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani, A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A., et al. 2020. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 3:e106230. 10.15252/embj.2020106230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier, N., Adams E.L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A.E., Kadiri L., Umadevi Venkataraju K., Zhou Y., et al. 2016. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell. 165:1789–1802. 10.1016/j.cell.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier, N., Wu Z., Simon D.J., Yang J., Ariel P., and Tessier-Lavigne M.. 2014. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 159:896–910. 10.1016/j.cell.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Roberts, A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., et al. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. 10.1371/journal.ppat.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, I.H., Normandin E., Bhattacharyya S., Mukerji S.S., Keller K., Ali A.S., Adams G., Hornick J.L., Padera R.F. Jr., and Sabeti P.. 2020. Neuropathological Features of Covid-19. N. Engl. J. Med. 383:989–992. 10.1056/NEJMc2019373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S.H., Chen Q., Gu H.J., Yang G., Wang Y.X., Huang X.Y., Liu S.S., Zhang N.N., Li X.F., Xiong R., et al. 2020. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe. 28:124–133.e4. 10.1016/j.chom.2020.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak, W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. HCA Lung Biological Network . 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26:681–687. 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco, S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. 2019. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 570:523–527. 10.1038/s41586-019-1289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D., et al. 2017. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 21:517–532. 10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., and Lazartigues E.. 2008. Angiotensin-converting enzyme 2 in the brain: properties and future directions. J. Neurochem. 107:1482–1494. 10.1111/j.1471-4159.2008.05723.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.P., Qiu Y., Zhang B., Chen G., Chen Q., Wang M., Mo F., Xu J., Wu J., Zhang R.R., et al. 2019. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 29:265–273. 10.1038/s41422-019-0152-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffre F., et al. 2020. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 27:125–136. 10.1016/j.stem.2020.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy, B., Iijima N., Huttner A., Leib D., and Iwasaki A.. 2012. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 12:334–345. 10.1016/j.chom.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Li J., Liu H., Han N., Ju J., Kou Y., Chen L., Jiang M., Pan F., Zheng Y., et al. 2020. Long-term bone and lung consequences associated with hospital-acquired severe acute respiratory syndrome: a 15-year follow-up from a prospective cohort study. Bone Res. 8:8. 10.1038/s41413-020-0084-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data files from single-cell RNA-seq can be found at Sequence Read Archive accession no. PRJNA682590.