The Trithorax Cxxc1 molecule specifically regulates the chromatin accessibility of the genes that are downregulated during priming and reactivated in the later phase in differentiating CD4 T cells. The authors show Cxxc1 deletion induces CD4 T cell hyperactivation and enhanced allergic airway inflammation.
Abstract
Different dynamics of gene expression are observed during cell differentiation. In T cells, genes that are turned on early or turned off and stay off have been thoroughly studied. However, genes that are initially turned off but then turned on again after stimulation has ceased have not been defined; they are obviously important, especially in the context of acute versus chronic inflammation. Using the Th1/Th2 differentiation paradigm, we found that the Cxxc1 subunit of the Trithorax complex directs transcription of genes initially down-regulated by TCR stimulation but up-regulated again in a later phase. The late up-regulation of these genes was impaired either by prolonged TCR stimulation or Cxxc1 deficiency, which led to decreased expression of Trib3 and Klf2 in Th1 and Th2 cells, respectively. Loss of Cxxc1 resulted in enhanced pathogenicity in allergic airway inflammation in vivo. Thus, Cxxc1 plays essential roles in the establishment of a proper CD4+ T cell immune system via epigenetic control of a specific set of genes.
Graphical Abstract
Introduction
Differentiation from multipotent progenitor cells to committed cells, which is often accompanied by epigenetic changes, is regulated by priming signals and commitment factors, including environmental stimuli and localization (Gallusci et al., 2017; Nimmo et al., 2015; Weinberger et al., 2016), resulting in the induction of epigenetic variation in each of the committed cells, as illustrated in Fig. S1 A. In peripheral T cells, antigen stimulation via TCR/coreceptor signaling constitutes the priming signals, and environmental cytokines are essential commitment factors (Chen and Flies, 2013; Chi, 2012; Hu et al., 2011; O’Garra, 2000). APCs are important for T cells to recognize antigenic peptides on MHC molecules and induce TCR/coreceptor signaling (Eisenbarth, 2019; Rossjohn et al., 2015); however, cessation of TCR/coreceptor signaling through APC dissociation from T cells is another important commitment factor for differentiation into T helper (Th) cell subsets (Fig. 1 A, top; Bohineust et al., 2018).
Figure S1.
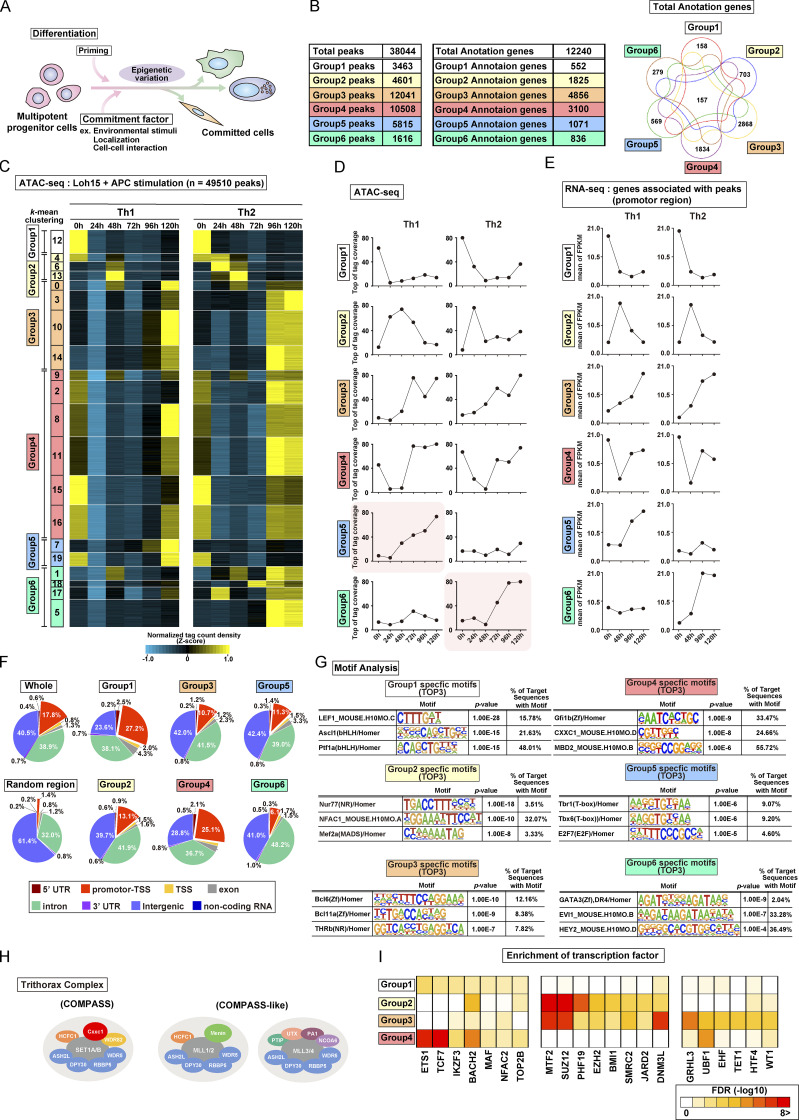
TCR/coreceptor signaling alters chromatin accessibility landscapes in naive CD4 T cells stimulated with antigenic peptide and APCs (related to Fig. 1). (A) A schematic illustration of differentiation from the multipotent progenitor cells to committed cells. The differentiation is regulated by priming signals and commitment factors including environmental stimuli, both of which are critical for the induction of epigenetic variation. (B) Th1 and Th2 cells were generated using plate-bound anti-TCRβ and a soluble anti-CD28 antibody stimulation system. The numbers of groups 1–6 regions (left) and groups 1–6 genes (middle) are shown. All genes that had at least one ATAC-seq peak in the promoter region were classified into the corresponding six groups while allowing overlap (right). (C–E) Th1 and Th2 cells were generated using the DO11.10 TCR Tg system; (i.e., naive CD4+ T cells were stimulated with OVA peptide presented by APCs (also see Materials and methods). The heatmap demonstrates the level of chromatin accessibility at 49,510 regulatory regions measured by ATAC-seq for naive cells (0 h) and the process of differentiation induced by Loh15 peptide plus APC stimulation (24 h, 48 h, 72 h, 96 h, and 120 h; C). Accessible regions were organized in groups by k-means clustering (k = 20). Clusters were assembled into six metaclusters (groups 1–6), depending on their accessibility patterns. The average tag count of each peak of ATAC-seq for differentiating Th1 or Th2 cells (D). The RNA expression of genes whose promoters (i.e., from 5 kb upstream to 2 kb downstream of the transcription start site) were associated with groups 1–6 regions (E). (F) The pie charts display the annotation of groups 1–6 regions. The results obtained with random regions of the genome and pooled data of groups 1–6 are shown as Random and Whole, respectively. (G) The top three motifs enriched in Fig. 1 F are shown with the DNA sequence, P value, and percentage. (H) A schematic representation of the Trithorax complex. Cxxc1 forms the Compass complex with SET1A/B that methylates H3K4 (left). MLL1/2-containing (middle) and MLL3/4-containing (right) Compass-like complexes are also shown. (I) Potential proteins binding to the promoters of the groups 1 (left), 2 (middle), and 3 (right) genes were predicted on the basis of transcription factor enrichment analysis. FPKM, fragments per kilobase of exon model per million mapped reads; UTR, untranslated region.
Figure 1.
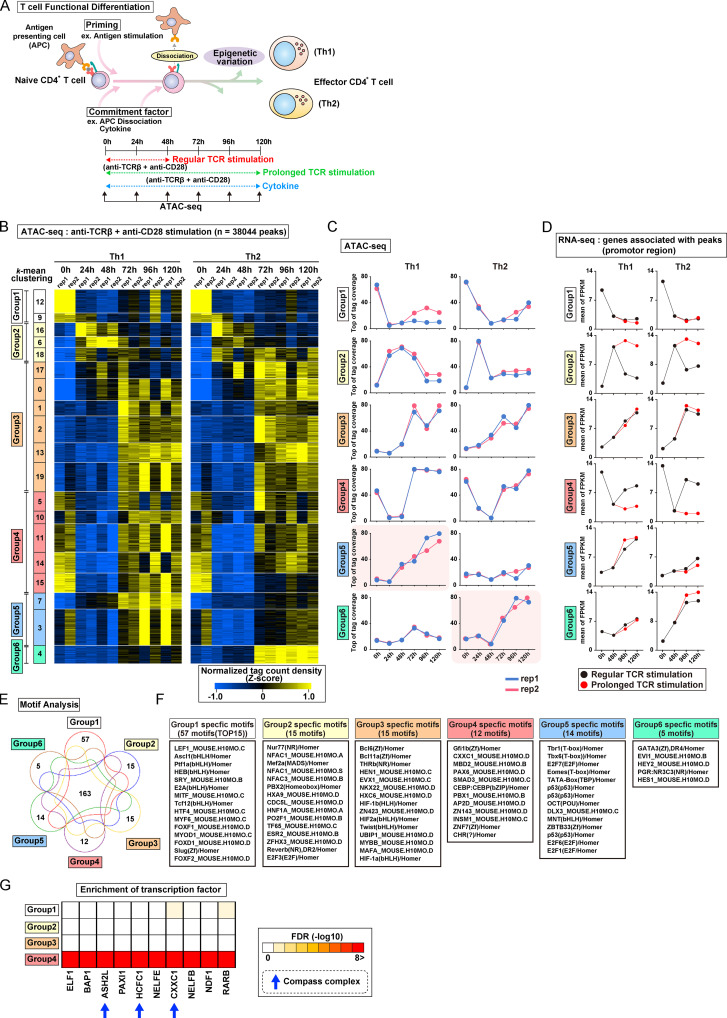
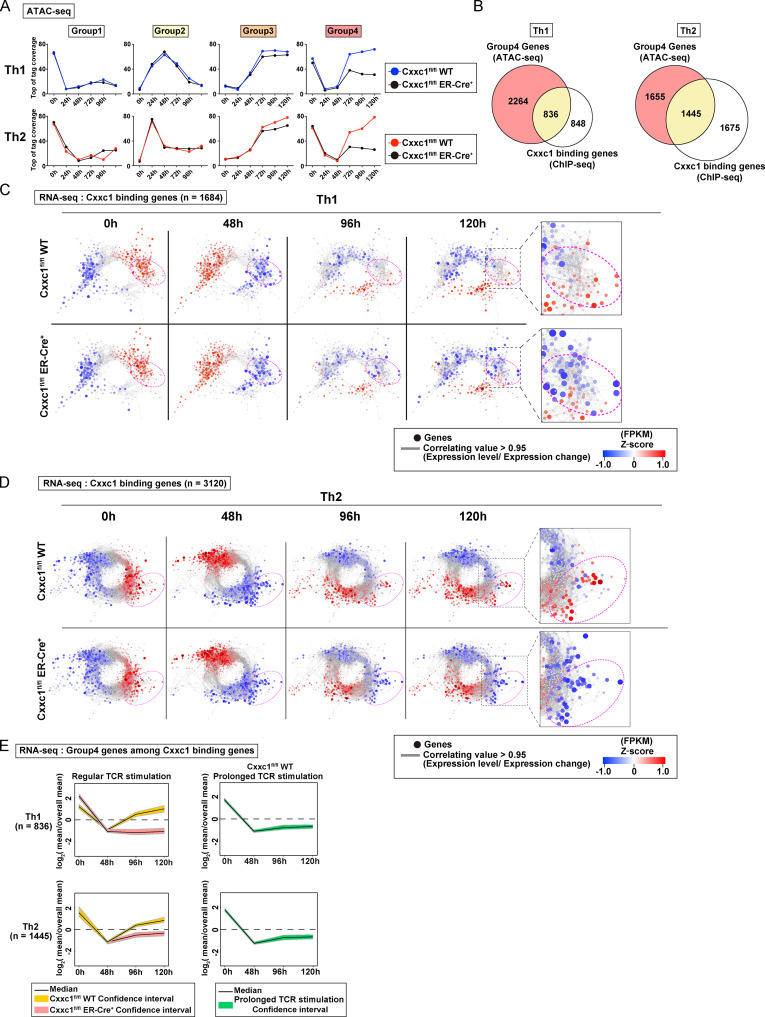
TCR/coreceptor signaling alters chromatin accessibility landscapes responsible for Th1 and Th2 cell differentiation. (A) In peripheral T cells, antigen stimulation via TCR/coreceptor signaling is recognized as the priming signal, while environmental cytokines are known as commitment factors. APCs are important for T cells to receive TCR/coreceptor signaling; however, the dissociation of APCs from T cells is considered one of the important commitment factors for differentiation into Th subsets. ATAC-seq was performed with differentiating Th1 and Th2 cells at the indicated time points. Th1 and Th2 cells that received prolonged TCR/coreceptor stimulation throughout the differentiation process (from 0 to 120 h; green dashed line) and those that received regular TCR/coreceptor stimulation (from 0 to 48 h; red dashed line) were prepared. (B) The heatmap demonstrates the level of chromatin accessibility at 38,044 regulatory regions measured by ATAC-seq. Results with two replicates are shown for each time point. Accessible regions were organized in groups by k-means clustering (k = 20). Clusters were assembled into six metaclusters (groups 1–6), depending on their accessibility patterns. (C) The average tag count of each peak of ATAC-seq in differentiating Th1 or Th2 cells. Results with two replicates are shown for each time point. (D) The RNA expression levels of genes whose promoters (i.e., from 5 kb upstream to 2 kb downstream of the transcription start site) have ATAC peaks as shown in C. The mean values of two replicates were used for plotting. FPKM, fragments per kilobase of exon per million reads mapped. (E) The Venn diagram displays the overlap of the enriched motifs among groups 1–6 regions. (F) A list of specific motifs found in each group. (G) Potential proteins binding to the promoters of the group 4 genes were predicted based on a transcription factor enrichment analysis.
Different dynamics of gene expression are observed during cell differentiation and often involve epigenetic changes (Briggs et al., 2018; Strober et al., 2019; White et al., 1999). Using T cell differentiation as a model system, genes that are turned on early by TCR/coreceptor stimulation have been thoroughly studied; they are controlled by nuclear translocation of preexisting transcription factors such as NFAT and NF-κB (Fisher et al., 2006; Hogan, 2017; Müller and Rao, 2010). Genes that are turned on and stay on or turned off and stay off (Hawkins et al., 2010; Henning et al., 2018; Semrau et al., 2017) tend to enforce lineage commitment and ensure the stability of cell lineages (Rosenbauer and Tenen, 2007; Yui and Rothenberg, 2014); the transcription factors include T-bet and GATA3 as lineage-determining stimuli for Th1 and Th2 cells, respectively (Ho et al., 2009; Nakayama et al., 2017; Szabo et al., 2000), and TCF1 (also known as Tcf7), which confers naive T cell stemness (Nish et al., 2017; Pace et al., 2018). However, transcription factors or epigenetic regulators that regulate genes that are initially turned off but then turned on again after TCR/coreceptor stimulation has ceased have not been defined; they are obviously important, particularly for the T cells, because they distinguish between transient and prolonged TCR/coreceptor stimulation, especially in the context of acute versus chronic viral infections and antitumor responses (Speiser et al., 2014).
The epigenetic regulation of gene expression is essential for the differentiation of every cell type from its precursor (Boland et al., 2014). In particular, histone modifications have been shown to play important roles in controlling the proper differentiation of immune cells and maintaining their functions (Ansel et al., 2006; Kanno et al., 2012; Nakayama and Yamashita, 2008). The Trithorax complex is one of the most fundamental epigenetic regulators and exerts its functions via the methylation of histone H3 lysine 4 (H3K4; Schuettengruber et al., 2011). In mammals, six H3K4 methylases have been identified, and four of them (mixed-lineage leukemia [MLL] 1–4) are included in the Compass-like complex, whereas the others (SET1A and B) are components of the Compass complex (Shilatifard, 2012). We have reported that MLL1 and menin, which are essential components of the Compass-like complex, are involved in maintenance of memory Th2 cell functions both in vitro and in vivo (Kuwahara et al., 2014; Onodera et al., 2010, 2017; Yamashita et al., 2006). Additionally, the lack of menin impairs Th17 cell differentiation and results in decreased IL-17A production (Watanabe et al., 2014). In contrast, the Compass complex methylates H3K4 in an H2B ubiquitination–dependent manner and possesses higher methyltransferase activity than the Compass-like complex (Kim et al., 2013; Wu et al., 2008). It has been proven that the Compass complex interacts with Cxxc1, and this interaction is required for its binding to ubiquitinated H2B. Clinically, mutations of the CXXC1 gene impair this interaction and are risk factors for colon cancer, lung cancer, and chronic lymphoblastic leukemia in humans (Bader et al., 2003; Houlston et al., 2010). Furthermore, Cxxc1 is reported to be highly expressed in CD4+CD8+ double-positive T cells in the thymus, and a lack of Cxxc1 results in impaired thymocyte development (Cao et al., 2016). Cxxc1 is also highly expressed in peripheral lymphoid tissues (Chun et al., 2014). However, the role of Cxxc1 in peripheral CD4+ T cells (Brown et al., 2017; Sha et al., 2018) still remains unclear.
In the present study, we show that the Cxxc1 subunit of the Trithorax complex directs transcription of genes initially turned off by TCR/coreceptor stimulation but then turned on again in a later phase. The late up-regulation of these genes was impaired both by prolonged TCR/coreceptor stimulation and Cxxc1 deficiency, and we show that these secondary Cxxc1-induced genes are required for the generation of functional immunocompetent Th1 and Th2 cells. For example, the Cxxc1-dependent up-regulation of Trib3 in Th1 cells was found to be required to prevent hyperactivation of the PI3K-Akt signaling, MAPK, and JAK-STAT cascades that are induced by TCR/coreceptor stimulation (Dong et al., 2002; O’Shea et al., 2015; Okkenhaug, 2013). Cxxc1 deficiency led to hyperproduction of IFNγ in Th1 cells. In contrast, Cxxc1-dependent up-regulation of Klf2, which prevents the hyperproduction of Th2 cytokines (IL-4, -5, and -13), was found to be important for the proper Th2 cell differentiation. Cxxc1-deficient cells caused enhanced pathogenicity in mouse models of allergic airway inflammation in vivo. Thus, our data indicate that Cxxc1 plays essential roles in the establishment of a proper immune system via epigenetic control of a specific set of genes during Th1/Th2 cell differentiation.
Results
TCR/coreceptor signaling alters chromatin accessibility landscapes critical for Th1 and Th2 cell differentiation
To analyze chromatin accessibility changes during the onset and cessation of Th cell differentiation, we used assay for transposase-accessible chromatin sequencing (ATAC-seq). Naive CD4+ T cells were stimulated with immobilized anti-TCR mAb and anti-CD28 mAb for 48 h under Th1 or Th2 cell differentiation culture conditions (Fig. 1 A, bottom). In most experiments, stimulation was interrupted at 48 h by removal of T cells from the wells (see Materials and methods). Based on accessibility patterns at the different time points (0, 24, 48, 72, 96, and 120 h after stimulation), the accessible regions were clustered into 20 groups and then into six major groups by k-means clustering (see heatmaps in Fig. 1 B and Fig. S1 B; genes in each of the major groups are listed in Table S1). Although we observed some variation between two replicates in a small group (Fig. 1 B), all of the major groups (1–6) exhibited little replicate variation (Fig. 1 C). Groups 1–4 displayed similar time courses of accessibility in both Th1 and Th2 cells, whereas groups 5 and 6 showed increasing accessibility, specifically in Th1 and Th2 cells, respectively. The 0, 24, and 48 h time points represent the “early phase,” which depends on TCR/coreceptor stimulation, whereas the 72, 96, and 120 h time points represent the “late phase” that follows the cessation of TCR/coreceptor stimulation at 48 h. The accessibility of group 1 regions decreased upon TCR/coreceptor stimulation and was not regained; group 2 regions transiently gained accessibility upon TCR/coreceptor stimulation but lost accessibility in the late phase; group 3 regions showed increased accessibility during the late phase; and group 4 regions showed decreased accessibility upon TCR/coreceptor stimulation and regained accessibility during the late phase. Similar results were obtained when naive CD4+ T cells from DO11.10 TCR transgenic (Tg) mice were stimulated with their cognate antigen Loh15 presented by APCs (Fig. S1, C and D).
To assess the relationship between chromatin accessibility and gene expression, we used RNA sequencing (RNA-seq) to classify genes that had one or more ATAC-seq peaks in the promoter region into the six groups defined by ATAC-seq while allowing overlap (Fig. S1 B). The time courses observed for gene expression were well correlated with the time course of changes in chromatin accessibility (Fig. 1 D, black lines; and Fig. S1 E). Genes in groups 1–4 were similarly regulated in differentiating Th1 and Th2 cells, whereas genes in groups 5 and 6 were expressed at higher levels in Th1 and Th2 cells, respectively, consistent with the increased accessibility of their promoter regions in these two differentiated cell types.
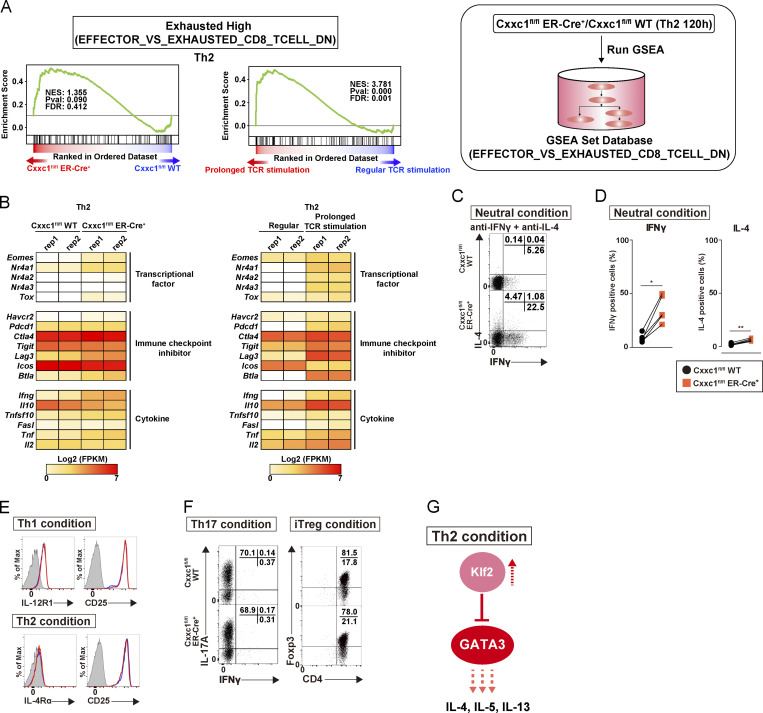
As our focus was on the transcriptional control mechanisms that accompanied the cessation of TCR/coreceptor signaling, which in our experimental system occurred at 48 h, we compared cells subjected to transient TCR/coreceptor stimulation from 0 to 48 h (our standard conditions) with Th1 or Th2 cells that received prolonged TCR/coreceptor stimulation throughout the differentiation process (from 0 to 120 h; green dashed line in Fig. 1 A). The cells that received prolonged TCR/coreceptor stimulation showed altered gene expression patterns of the group 2 and group 4 genes, with no significant effect on the expression patterns of genes in groups 1, 3, 5, and 6 (Fig. 1 D). We identified potential regulators of the dynamic expression changes in each group through motif analysis (Fig. 1, E and F). As expected, the T-box and GATA3 motifs were significantly enriched in regions accessible in groups 5 and 6, respectively, confirming the known involvement of T-bet and GATA3 in controlling chromatin accessibility in Th1 and Th2 cells (Ho et al., 2009; Nakayama et al., 2017; Szabo et al., 2000); likewise, the consensus binding motif for the TCF/lymphoid enhancer factor (LEF) family was significantly enriched in group 1 regions, which lose accessibility during differentiation of naive T cells, consistent with the loss of TCF1 and LEF1 expression over the same time period (Fig. 1 F and Fig. S1 G; Chen and Yu, 2015; Steinke et al., 2014). The NFAT family motif (also known as NFAC) was enriched in group 2 regions whose accessibility was strongly correlated with the NFAT activity driven by TCR/coreceptor stimulation (Hogan, 2017; Müller and Rao, 2010). In contrast, the top three motifs found in the group 4 regions were associated with the epigenetic regulators, which can also bind to DNA in a sequence-independent manner. To identify regulatory factors responsible for the observed patterns of chromatin accessibility in more detail, we next performed a transcription factor enrichment analysis based on published chromatin immunoprecipitation with sequencing (ChIP-seq) data (Kawakami et al., 2016). This analysis utilizes published chromatin immunoprecipitation with sequencing (ChIP-seq) data, thus allowing us to predict potential transcriptional regulators involved in the accessibility changes even if they do not directly bind DNA and so lack specific binding motifs in the peak. Notably, components of the Compass complex (Fig. S1 H), which is involved in H3K4 methylation (Nakayama et al., 2017; Onodera and Nakayama, 2015; Shilatifard, 2012; Tumes et al., 2017), were predicted to bind specifically to the promoters of group 4 genes (see blue arrows in Fig. 1 G and Fig. S1 I). We therefore focused on Cxxc1 and studied it in detail.
Cxxc1 specifically regulates a set of genes that are down-regulated during the early phase and reactivated during the late phase
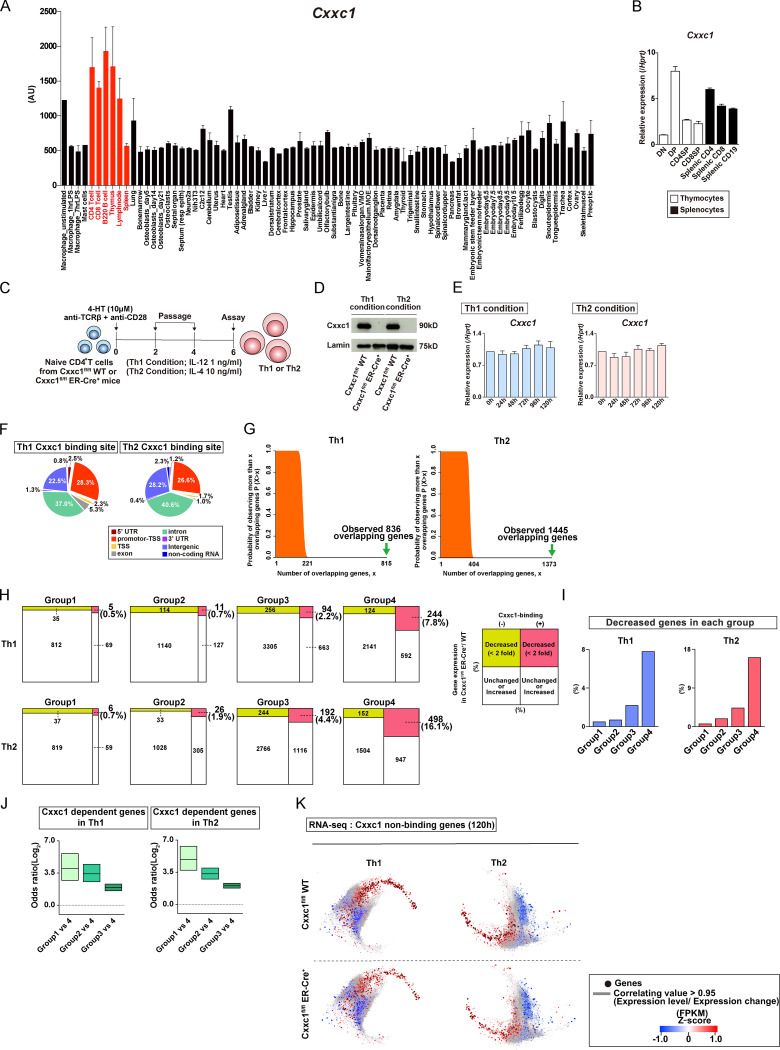
Cxxc1 is highly expressed in lymphocytes and the lymphoid tissues (ImmGen database [http://www.immgen.org]; Fig. S2 A). As previously reported, CD4+CD8+ double-positive cells exhibited the highest expression level of the Cxxc1 gene in the thymocytes (Fig. S2 B; Cao et al., 2016). Since CD4-cre–driven Cxxc1 deletion affected thymocyte development, we used Cxxc1fl/fl ERT2-Cre+ mice (KO mice) and deleted the Cxxc1 gene by 4-hydroxytamoxifen (4-HT) treatment for the analysis of peripheral CD4+ T cells (Fig. S2 C). Cxxc1fl/fl ERT2-Cre–negative mice (WT mice) were used as controls (hereafter ERT2-Cre is referred as to ER-Cre). 4-HT treatment resulted in the loss of Cxxc1 protein expression in both Th1 and Th2 cells (Fig. S2 D). In WT cells, Cxxc1 mRNA expression showed no significant change during Th1 or Th2 differentiation (Fig. S2 E).
Figure S2.
Cxxc1 specifically regulates the reactivation of the genes that are suppressed during the early phase (related to Fig. 2). (A) The gene expression profile of Cxxc1 in GeneAtlas DNA microarray (gntlm05381_a_at) datasets. (B) The relative expression of Cxxc1 (normalized to Hprt) in the indicated cells from the thymus and spleen is shown with SDs. (C) Experimental protocols to induce Th1 or Th2 cell differentiation from naive CD4+ T cells for Cxxc1fl/fl mice. To delete the Cxxc1 gene, 4-HT treatment was conducted on days 0 to 1. (D) Western blotting of Cxxc1 proteins with Th1 or Th2 cell lysates from WT or Cxxc1-deficient cells. (E) The Cxxc1 expression in Th1 and Th2 cells was analyzed by qRT-PCR. (F) The pie chart shows annotation of the Cxxc1 binding sites. (G) The overlap of group 4 and Cxxc1-bound genes in Th1 and Th2 cells was analyzed by hypergeometric analysis. Plots show the probability of observing (y axis) more than the indicated number of overlapping genes (x axis) if sets with 3,100 and 1,684 genes (left) or 3,100 and 3,120 genes (right) are sampled randomly (Martinez et al., 2015). (H and I) Genes that met the following criteria were defined as Cxxc1-dependent genes in each group: (1) directly bound by Cxxc1 or (2) more than twofold decrease in the mRNA expression in Cxxc1-deficient cells at 120 h compared with WT control. The numbers of Cxxc1-dependent genes (H) are shown with percentages (I) in each group in Th1 and Th2 cells. (J) Frequencies of Cxxc1-dependent genes in groups 1–3 were compared with those in group 4 in Th1 and Th2 cells using Fisher’s exact test. The odds ratio is shown for each comparison (group 1 versus group 4; group 2 versus group 4; and group 3 versus group 4). (K) A coregulation transcriptome network is shown for the Cxxc1 nonbinding genes. Node color indicates the gene expression level based on Z-scores. AU, arbitrary units; FPKM, fragments per kilobase of exon per million reads mapped.
Notably, ATAC-seq analysis of control and Cxxc1-deficient Th1 and Th2 cells showed that the chromatin accessibility of group 4 regions was significantly reduced in both cell types, whereas the accessibility of regions in other groups was not affected (Fig. 2 A). As expected, because group 4 regions lost accessibility early but regained accessibility beginning at 72 h, Cxxc1 deficiency had little impact on the accessibility of group 4 regions at 0, 24, and 48 h but strongly diminished chromatin accessibility at 72, 96, and 120 h during Th1 or Th2 cell differentiation (Fig. 2 A), reinforcing the idea that the late reacquisition of accessibility in group 4 genes involved Cxxc1 and the Compass complex.
Figure 2.
Cxxc1 specifically regulates the expression of genes that are suppressed during the early phase and reactivated during the late phase. (A) The average tag count of each peak of ATAC-seq for differentiating Th1 or Th2 cells. Data from Cxxc1fl/fl WT or Cxxc1-deficient (Cxxc1fl/fl ER-Cre+) cells are shown for each group. WT data are adapted from Fig. 1 D. (B) The Venn diagrams display the overlap between the genes belonging to group 4 and Cxxc1-bound genes in Th1 and Th2 cells. (C and D) Coregulation transcriptome networks created based on two RNA-seq datasets (i.e., in naive [0 h] and the differentiation process [48 h] of WT cells) are shown. All Cxxc1-bound genes in Th1 (C; n = 1,684) and Th2 (D; n = 3,120) cells are shown. Dashed ellipsis (added manually) indicates genes down-regulated in the absence of Cxxc1. The node color indicates the gene expression levels at each time point (0, 48, 96, and 120 h) based on Z-scores. FPKM, fragments per kilobase of exon per million reads mapped. (E) A time-course analysis of the expression of Cxxc1-bound group 4 genes in WT, Cxxc1-deficient (left), and WT cells that received prolonged TCR/coreceptor stimulation (right). Lines denote the median, and shaded areas represent the 95% confidence intervals.
ChIP-seq analysis of Cxxc1 in WT cells allowed us to identify Cxxc1-binding genes with at least one Cxxc1 peak in the promoter region (Fig. 2 B and Fig. S2 F). In both Th1 (n = 836) and Th2 (n = 1,445) cells, approximately half of the promoters that bound Cxxc1 overlapped with the promoters of group 4 genes, defined by early loss and later increase in accessibility, and this overlap frequency was significantly higher than for randomly selected genes (Fig. S2 G; Acquaviva et al., 2013; Martinez et al., 2015; Thomson et al., 2010). A set of these Cxxc1-binding genes was down-regulated at 48 h and up-regulated in WT but not Cxxc1-deficient cells at 96 or 120 h in both Th1 and Th2 cells (indicated by dashed ellipses in Fig. 2, C and D), consistent with the results of ATAC-seq analysis. In brief, 244 and 498 genes were defined as Cxxc1-dependent group 4 genes (promoters bound by Cxxc1 and expression down-regulated more than twofold in the absence of Cxxc1) in Th1 and Th2 cells, respectively (Fig. S2 H). In contrast, the effect of Cxxc1 deletion on mRNA expression was limited in the Cxxc1-binding groups 1–3 genes (Fig. S2, I and J). Similarly, genes whose promoters were not bound by Cxxc1 showed no obvious changes in mRNA expression in Cxxc1-deficient cells (Fig. S2 K). To confirm the correlation between accessibility and gene expression changes in the Cxxc1-binding group 4 genes, we plotted mRNA expression of these genes in WT and Cxxc1-deficient cells with 95% confidence intervals. The up-regulation of group 4 gene expression at 96 and 120 h observed in WT cells was significantly attenuated in the absence of Cxxc1 (Fig. 2 E). Highlighting the connection between Cxxc1 involvement and the recovery phase after cessation of TCR/coreceptor signaling, prolonged TCR/coreceptor stimulation inhibited the late up-regulation of group 4 genes, indicating that cessation of TCR/coreceptor stimulation is needed for reactivation of Cxxc1-binding group 4 genes. Thus, Cxxc1 preferentially regulates a set of genes whose expression is down-regulated by TCR/coreceptor signaling in the early phase but reactivated after removal of the TCR/coreceptor signal in the late phase in a manner that correlates with chromatin remodeling: the early loss of chromatin accessibility upon TCR/coreceptor stimulation followed by its reversal after the stimulus is removed.
Loss of Cxxc1 resulted in the enhanced expression of Th1 and Th2 cytokines in CD4+ T cells via distinct mechanisms
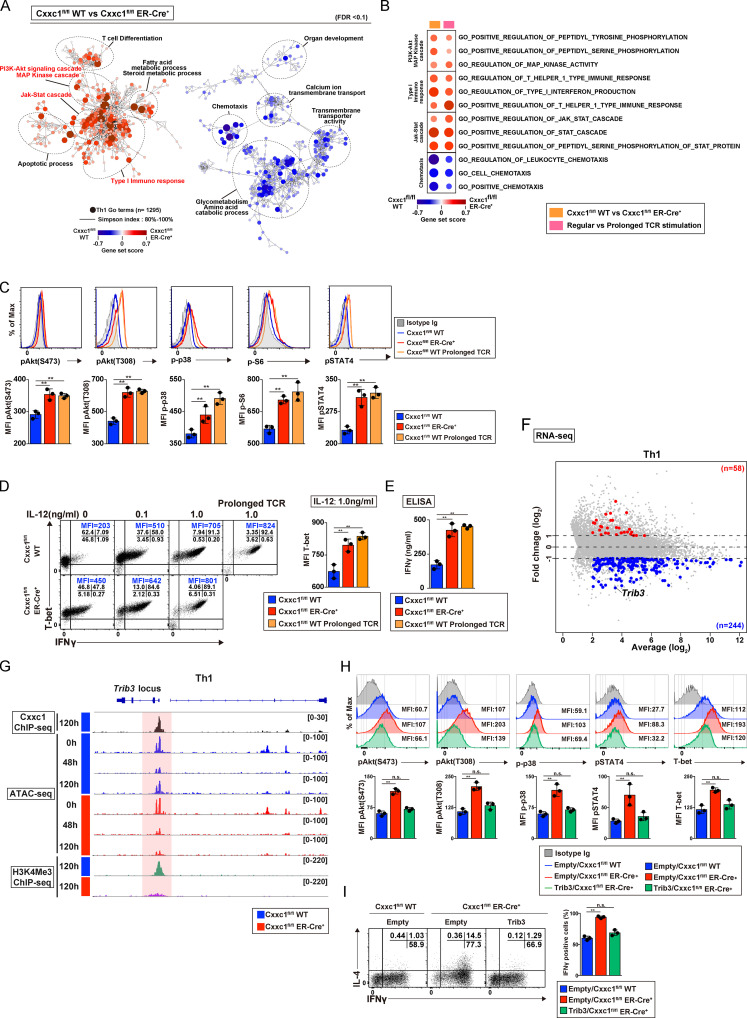
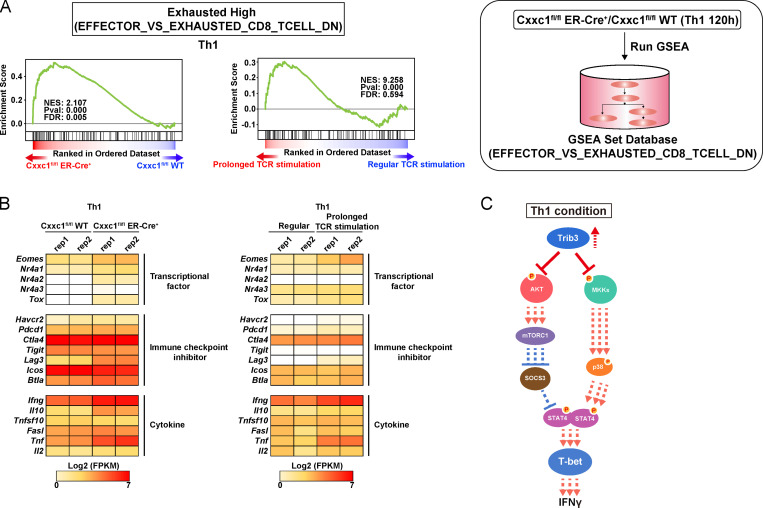
To understand the mechanism of action of Cxxc1, we performed a pathway analysis of the RNA-seq data from Th1 cells (Fig. 3 A). PI3K/Akt signaling, MAPK, and JAK-STAT cascades were significantly hyperactivated in Th1 cells in the absence of Cxxc1. The type I immune response pathway was also found to be activated in the absence of Cxxc1, and, more interestingly, gene set enrichment analysis (GSEA) revealed that Cxxc1-deficient cells exhibited “exhausted” signatures (up-regulation of the Eomes, Nr4a1, Nr4a3, Tox, and Lag3 genes) that reported in CD8+ T cells (Fig. S3, A and B; Crawford et al., 2014; Miller et al., 2019; Wherry et al., 2007). A similar pattern of activation of these cascades and the exhausted signatures was observed in Th1 cells that received prolonged TCR/coreceptor stimulation (Fig. 3 B; and Fig. S3, A and B). In fact, lack of Cxxc1 resulted in the enhanced phosphorylation of Akt, p38, S6 kinase, and STAT4 (Fig. 3 C). Consequently, IFNγ production and T-bet expression were significantly increased under Cxxc1-deficient conditions compared with WT controls (Fig. 3, D and E). A similar increase was observed in Th1 cells that received prolonged TCR/coreceptor stimulation (Fig. 3, C–E). To identify genes whose expression levels are dysregulated in the absence of Cxxc1, which might be responsible for the hyperproduction of IFNγ in Th1 cells, we focused on nuclear proteins encoded by the 244 Cxxc1-dependent group 4 genes shown in Fig. S2 H. Among these candidate genes, we found that expression of the Trib3 gene, which is reported to inhibit the phosphorylation of Akt or MAPK (Eyers et al., 2017), was markedly decreased in the absence of Cxxc1 (Fig. 3 F). Moreover, Cxxc1 deletion abrogated the late increase in chromatin accessibility at the Trib3 gene locus that occurred between the 48 and 120 h time points (Fig. 3 G, ATAC-seq tracks) and led to a striking decrease in H3K4me3 levels at the Trib3 promoter at 120 h (Fig. 3 G, H3K4me3 ChIP-seq tracks). Direct binding of Cxxc1 to the Trib3 promoter was detected by ChIP-seq, suggesting that Cxxc1 directly regulates the expression of Trib3 (Fig. 3 G). Finally, retroviral expression of Trib3 normalized the PI3K-Akt and MAPK pathways in Cxxc1-deficient cells (Fig. 3 H) and repressed IFNγ hyperproduction (Fig. 3 I) in differentiating Cxxc1-deficient Th1 cells, indicating that decreased expression of Trib3 is responsible for this set of phenotypes associated with Cxxc1 deficiency (Fig. S3 C).
Figure 3.
Loss of Cxxc1 results in the enhanced production of Th1 cytokines via dysregulation of Trib3 expression in CD4+ T cells. (A) The pathways that were changed in Cxxc1-deficient Th1 cells compared with WT controls are shown. Dashed ellipses (added manually) indicate groups of similar GO terms. The node color indicates the gene set score. (B) The pathways changed in both Cxxc1-deficient Th1 cells and Th1 cells that received prolonged TCR/coreceptor stimulation are shown. (C) Representative staining profiles of phosphorylated (p)-AKT (T308), p-AKT (S473), p-p38, p-S6, and p-STAT4 in WT (with or without prolonged TCR/coreceptor stimulation) and Cxxc1-deficient Th1 cells are shown. The bar graphs show mean fluorescence intensity (MFI) with SDs from three independent experiments (**, P < 0.01). (D) Naive CD4+ T cells from the indicated mice were cultured under Th1 cell–inducing conditions with the indicated concentrations of IL-12. The cultured cells were restimulated with PMA plus ionomycin for 4 h. The intracellular staining profiles of IFNγ and T-bet were analyzed by flow cytometry. The bar graph shows the T-bet MFI with SDs from three independent experiments (**, P < 0.01). (E) The protein expression of IFNγ from WT Th1 cells, Cxxc1-deficient Th1 cells, or Th1 cells that received prolonged TCR/coreceptor stimulation. IFNγ secreted by Th1 cells stimulated with anti-TCRβ for 24 h was measured by ELISA. Data from three independent experiments are shown as the mean values and SDs (**, P < 0.01). (F) The MA plot depicts 10,298 genes in WT versus Cxxc1-deficient Th1 cells based on RNA-seq. The blue and red points indicate the Cxxc1-dependent group 4 genes that were down-regulated or up-regulated, respectively (greater than twofold) in the absence of Cxxc1. (G) A genome browser view of ChIP-seq signals in Th1 cells for 3xFlag-Cxxc1 and H3K4Me3 is shown for the Trib3 gene. Retroviral transduction of 3xFlag-Cxxc1 into Th1 cells was performed before the ChIP-seq analysis. ATAC-seq signals of naive (0 h) and differentiation process (48 h and 120 h) of WT or Cxxc1-deficient cells are also shown. (H) Flow cytometry of Cxxc1-deficient Th1 cells, which were transduced with retroviral vector encoding Trib3. Representative profiles of p-AKT (T308), p-AKT (S473), p-p38, p-STAT4, and T-bet expression are shown. Bar graphs show data from three independent experiments with mean values and SDs (**, P < 0.01). (I) Intracellular staining profiles of IFNγ and IL-4 in the indicated cells restimulated with PMA plus ionomycin for 4 h are shown. The bar graph shows the percentage of IFNγ-positive cells from three independent experiments with mean values and SDs (**, P < 0.01). n.s., not significant.
Figure S3.
Loss of Cxxc1 results in the enhanced expression of Th1 cytokines in CD4+ T cells (related to Fig. 3). (A) GSEA was performed with the gene sets down-regulated (DN) in comparison of effector CD8+ T cells with exhausted CD8+ T cells in GSEA Molecular Signatures Database. The genes were rank ordered on the basis of relative expression in Cxxc1-deficient (red) versus WT (blue) Th1 cells (left) or prolonged (red) versus regular TCR/coreceptor stimulation (blue; right). The normalized enrichment score with P value and FDR are also shown. (B) The heatmap shows the RNA expression of the genes that are reported to be dysregulated in exhausted CD8+ T cells. Genes encoding transcription factors, inhibitory receptors, and effector molecules were analyzed in WT versus Cxxc1-deficient (left) or in regular versus prolonged TCR/coreceptor stimulation (right). (C) A schematic representation of downstream signaling controlled by Cxxc1 in Th1 cells. Cxxc1 controls the IFNγ expression via Trib3-dependent phosphorylation of the Akt-mTORC1-T-bet axis and mitogen-activated protein kinase kinase (MKK)–p38–T-bet axis. Cxxc1 deletion results in hyperproduction of IFNγ due to a lack of suppression of Akt and MKKs. FPKM, fragments per kilobase of exon per million reads mapped.
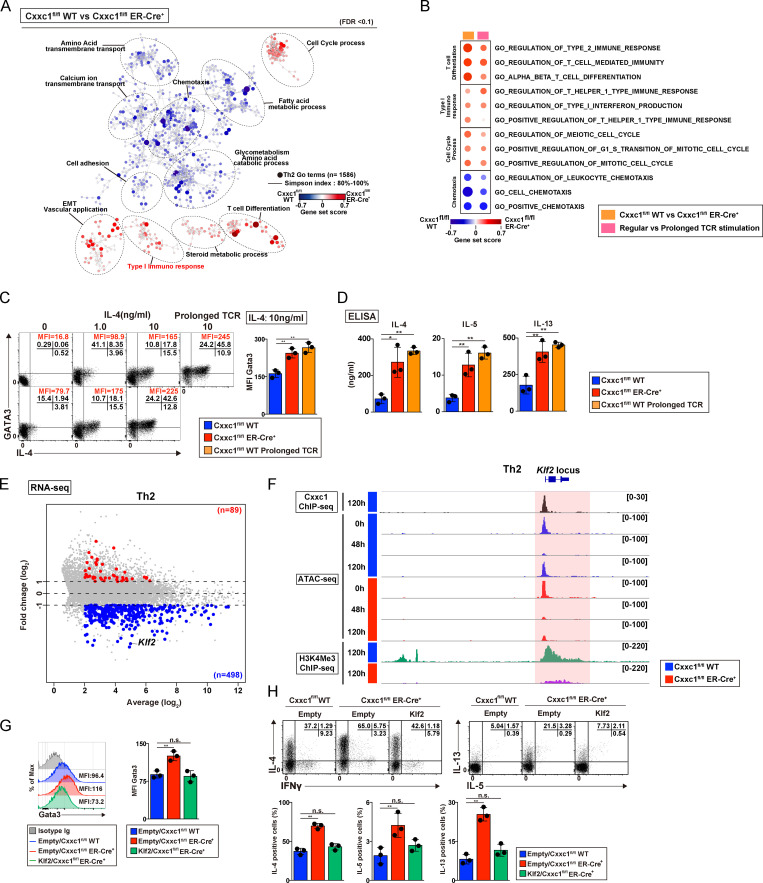
Although the hyperactivation of PI3K-Akt signaling, MAPK, and JAK-STAT cascades was not evident (Fig. 4, A and B), cytokine hyperproduction was also observed in Th2 cells (Fig. 4, B and C). Interestingly, GSEA revealed that Cxxc1 deficiency or prolonged TCR/coreceptor stimulation induced exhausted signatures (up-regulation of the Eomes, Nr4a1, Nr4a2, Nr4a3, Tox, Lag3, and Btla genes) in Th2 cells as well as Th1 cells (Fig. S4, A and B). To identify genes that are dysregulated in the absence of Cxxc1, which might be responsible for the hyperproduction of IL-4, -5, and -13 in Th2 cells, we focused on nuclear proteins encoded by the 498 Cxxc1-dependent group 4 genes shown in Fig. S2 H. Among these candidate genes, we focused on the Klf2 gene, which is reported to inhibit the hyperproduction of Th2 cytokines (Fig. 4 E; Weinreich et al., 2009). Moreover, Cxxc1 deletion abrogated the late increase in chromatin accessibility at the Klf2 gene locus that occurred between the 48 and 120 h time points (Fig. 4 F, ATAC-seq tracks) and led to a striking decrease in H3K4me3 levels at the Klf2 promoter at 120 h (Fig. 4 F, H3K4me3 ChIP-seq tracks). Direct binding of Cxxc1 to the Klf2 promoter was detected by ChIP-seq, suggesting that Cxxc1 directly regulates the expression of Klf2 (Fig. 4 F). Finally, retroviral expression of Klf2 normalized GATA3 protein expression (Fig. 4 G) and repressed cytokine hyperproduction (Fig. 4 H) in Cxxc1-deficient Th2 cells, indicating that decreased expression of Klf2 is responsible for this set of phenotypes associated with Cxxc1 deficiency (Fig. S4 G).
Figure 4.
Loss of Cxxc1 results in the enhanced expression of Th2 cytokines in CD4+ T cells. (A) The pathways that were changed in Cxxc1-deficient Th2 cells in comparison to WT control cells are shown. Dashed ellipses (added manually) indicate groups of similar GO terms. The node color indicates the gene set score. EMT, epithelial-to-mesenchymal transition. (B) The pathways changed in both Cxxc1-deficient Th2 cells and Th2 cells that received prolonged TCR/coreceptor stimulation are shown. (C) The representative profiles of IL-4 and GATA3 are shown. Naive CD4+ T cells from the indicated mice were cultured under Th2 cell–inducing conditions with the indicated concentrations of IL-4. The cultured cells were restimulated with PMA plus ionomycin for 4 h. A bar graph shows GATA3 mean fluorescence intensity (MFI) with SDs from three independent experiments (**, P < 0.01). (D) The protein expression of the indicated cytokines from WT Th2 cells, Cxxc1-deficient Th2 cells, or Th2 cells that received prolonged TCR/coreceptor stimulation. The indicated cytokines secreted by Th2 cells stimulated with anti-TCRβ for 24 h were measured by ELISA. Data from three independent experiments are shown with mean values and SDs (*, P < 0.05; **, P < 0.01). (E) The MA plot depicts 10,235 genes in WT versus Cxxc1-deficient Th2 cells based on RNA-seq. The blue and red points indicate Cxxc1-dependent group 4 genes that were down-regulated and up-regulated (greater than twofold) in the absence of Cxxc1, respectively. (F) A genome browser view of ChIP-seq signals in Th2 cells for 3xFlag-Cxxc1 and H3K4Me3 is shown for the Trib3 gene. Retroviral transduction of 3xFlag-Cxxc1 into Th2 cells was performed before ChIP-seq analysis. ATAC-seq signals of naive (0 h) and differentiation process (48 h and 120 h) of WT or Cxxc1-deficient cells are also shown. (G) Representative flow cytometry histograms of the GATA3 expression are shown (left). A bar graph shows GATA3 MFI with SDs from three independent experiments (**, P < 0.01; right). (H) Representative profiles of IFNγ versus IL-4 or IL-5 versus IL-13 in the indicated cells restimulated with PMA plus ionomycin for 4 h are shown (top). The percentages of IL-4–, IL-5–, and IL-13–positive CD4+ T cells are shown with the mean values and SDs (**, P < 0.01; bottom). n.s., not significant.
Figure S4.
Loss of Cxxc1 results in the enhanced expression of Th2 cytokines in CD4+ T cells (related to Fig. 4). (A) GSEA was performed as shown in Fig. 3 A. The genes were rank ordered based on relative expression in Cxxc1-deficient (red) versus WT (blue) Th2 cells (left) or prolonged (red) versus regular TCR/coreceptor stimulation (blue; right). (B) The heatmap shows the RNA expression of the genes that are reported to be dysregulated in exhausted CD8+ T cells. Genes encoding transcription factors, inhibitory receptors, and effector molecules were analyzed in WT versus Cxxc1-deficient (left) or regular versus prolonged TCR/coreceptor stimulation (right). FPKM, fragments per kilobase of exon per million reads mapped. (C and D) Naive CD4+ T cells from WT or Cxxc1-deficient mice were cultured under neutral conditions (in the presence of anti-IFNγ and anti-IL-4 Abs and IL-2). The cultured cells were restimulated with PMA plus ionomycin for 4 h. Intracellular staining profiles of IFNγ and IL-4 were analyzed by flow cytometry. Data from five independent experiments are shown (*, P < 0.05; **, P < 0.01). (E) Cell surface staining of the indicated markers on CD4+ T cells from WT or Cxxc1-deficient mice cultured under Th1 (top) or Th2 (bottom) cell-inducing conditions. Staining with a control antibody is indicated by the gray shaded area. 20,000 events are displayed for each sample. (F) Naive CD4+ T cells from WT or Cxxc1-deficient mice were cultured under Th17 or iT reg cell–inducing conditions. The cultured Th17 cells were restimulated with PMA plus ionomycin for 4 h, and intracellular staining profiles of IFNγ and IL-17A were analyzed by flow cytometry (left). For iT reg cells, the Foxp3 expression was analyzed together with the CD4 expression by flow cytometry (right). Three independent experiments were performed with similar results. (G) A schematic representation of downstream signaling controlled by Cxxc1 in Th2 cells. Cxxc1 controls the Th2 cytokine expression via Klf2-dependent suppression of the GATA3. Thus, Cxxc1 deficiency results in the down-regulation of the Klf2 expression, hyperactivation of the GATA3, and hyperproduction of Th2 cytokines.
Increased production of IFNγ and IL-4 was also observed in Cxxc1-deficient cells when cultured under neutral conditions where no polarizing cytokine, IL-4, or IL-12 was added (Fig. S4, C and D). The increased expression of T-bet and GATA3 appeared to be a cell-intrinsic effect because the expression of cytokine receptors, which induce Th1 or Th2 differentiation, was not changed (Fig. S4 E). In contrast to Th1 and Th2 cells, lack of Cxxc1 had little effect on induced regulatory T (iT reg) and Th17 cell differentiation (Fig. S4 F). Taken together, Cxxc1-mediated up-regulation of Trib3 and Klf2 is required for the proper differentiation of Th1 and Th2 cells, respectively.
Cxxc1 prevents the exacerbation of airway inflammation induced by antigen-specific CD4+ T cells in vivo
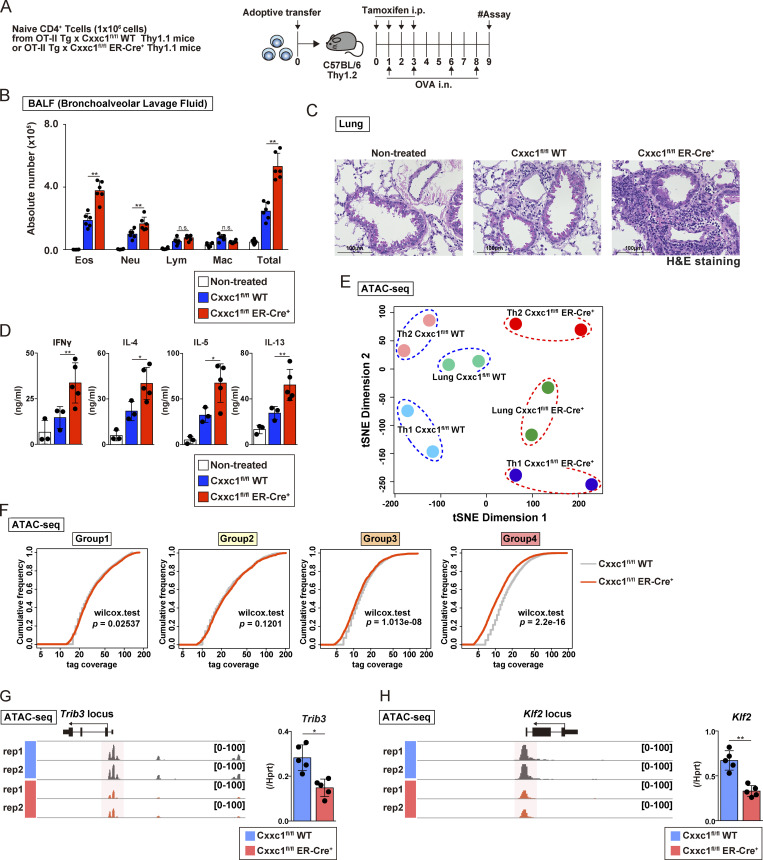
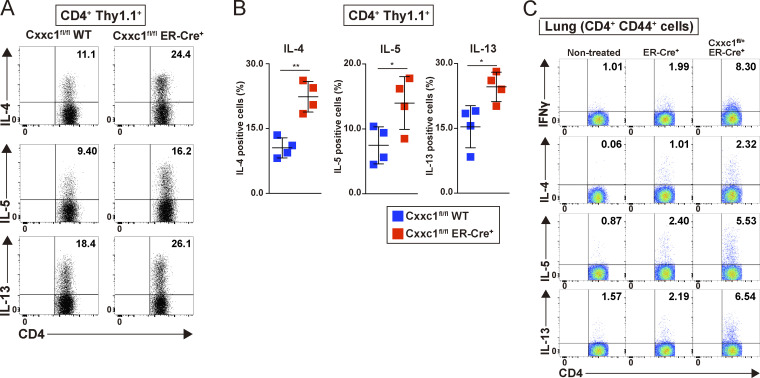
Finally, we examined the impact of the loss of Cxxc1 on inflammation pathology in vivo. Because Cxxc1 deletion resulted in increased IFNγ production in cultured T cells, we used mouse models of experimental airway inflammation induced by antigen-specific CD4+ T cells, where we expected inflammation to be exacerbated by the absence of Cxxc1. First, the same numbers of naive CD4+ T cells isolated from OT-II Tg Cxxc1fl/fl Thy1.1 (WT) and OT-II Tg Cxxc1fl/fl ER-Cre+ Thy1.1 (KO) mice were transferred into Thy1.2+ C57BL/6 recipient mice, which were treated with tamoxifen (by i.p. injection) and OVA (i.n. administration; Fig. 5 A). The numbers of eosinophils and neutrophils in the bronchoalveolar lavage (BAL) fluid were significantly increased in the group that received Cxxc1-deficient CD4+ T cells (Fig. 5 B), indicating that inflammation mediated by both Th1 and Th2 cells was enhanced since IFNγ-producing Th1 and IL-5–producing Th2 cells are known to recruit neutrophils and eosinophils, respectively (Israel and Reddel, 2017). Consistent with these findings, the deletion of Cxxc1 in CD4+ T cells resulted in the enhanced infiltration of mononuclear cells around the peribronchiolar and perivascular regions of the lungs (Fig. 5 C), as well as increased concentrations of IFNγ, IL-4, IL-5, and IL-13 in the BAL fluid were also observed (Fig. 5 D). At the same time, a dramatic elevation in the proportion of IL-4–, IL-5–, and IL-13–producing CD4+Thy1.1+ cells was observed in the lung in the absence of Cxxc1 (Fig. S5, A and B). Two-dimensional visualization of the ATAC-seq data from CD4+Thy1.1+ cells in the lung using t-distributed stochastic neighbor embedding (t-SNE) revealed that Cxxc1 deficiency had similar effects on chromatin accessibility in the lung CD4+ T cells as it did in in vitro cultured Th1 and Th2 cells (Fig. 5 E), with both lung and in vitro differentiated Cxxc1-deficient T cell subsets separated from the corresponding WT subsets in t-SNE dimension 1. The same analysis also revealed that CD4+ T cells activated in vivo exhibited epigenetic phenotypes intermediate between Th1 and Th2 (t-SNE dimension 2), consistent with the mixed Th1/Th2 signatures of airway inflammation that we observed in Fig. 5 B. Consistent with our observations in vitro (see Fig. 2), the genes in group 4 were most strongly affected by Cxxc1 deletion in vivo (Fig. 5 F). The Trib3 and Klf2 genes, which we showed were responsible for hyperactivation of Th1 and Th2 cells, respectively, showed reduced accessibility, and their expression was decreased in the absence of Cxxc1 in vivo (Fig. 5, G and H).
Figure 5.
Loss of the Cxxc1 enhances the airway inflammation in vivo. (A) An experimental protocol of allergic airway inflammation. In brief, the same numbers of naive CD4+ T cells from Cxxc1fl/fl (WT) or Cxxc1fl/fl ER-Cre+ (KO) mouse cells were transferred into C57BL/6 recipient mice that were treated with tamoxifen (by i.p. injection) and OVA (by i.n. injection). Mice that did not undergo cell transfer were used as controls. (B) The cell numbers of eosinophils (Eos), neutrophils (Neu), lymphocytes (Lym), and macrophages (Mac) in the BAL fluid are shown as the mean values with SDs (nontreated, n = 5; Cxxc1fl/fl [WT], n = 6; Cxxc1fl/fl [WT] ER-Cre+, n = 6; *, P < 0.05). (C) Images of H&E-stained lung specimens. Scale bars, 1 mm. (D) The indicated cytokines in the BAL fluid were measured by ELISA (nontreated, n = 3; Cxxc1fl/fl [WT], n = 3; Cxxc1fl/fl ER-Cre+, n = 6; *, P < 0.05; **, P < 0.01). (E) t-SNE plot visualization of Cxxc1fl/fl WT or Cxxc1-deficient (Cxxc1fl/fl ER-Cre+) cells based on ATAC-seq. Antigen-specific CD4+ T cells collected from the inflamed lung were referred to as lung Cxxc1fl/fl WT or Cxxc1fl/fl ER-Cre+. Data of Th1 and Th2 cells generated in vitro are adapted from Fig. 2 A and are also shown. (F) Cumulative distribution function plots of the normalized tag counts in ATAC-seq peaks in groups 1–4 are shown for Cxxc1fl/fl WT and Cxxc1-deficient (Cxxc1fl/fl ER-Cre+) cells. In each group, the P value was calculated by the Wilcoxon rank-sum test. (G and H) A genome browser view of the ATAC-seq signals in the indicated gene locus is shown (left). The mRNA expression levels in WT or Cxxc1-deficient cells were measured by qPCR (right). n.s, not significant.
Figure S5.
Loss of Cxxc1 enhances airway inflammation in vivo (related to Figs. 5 and 6). (A) Airway inflammation was induced based on the allergic inflammation model shown in Fig. 5 A. Intracellular staining profiles of IL-4, IL-5, and IL-13 in CD4+ T cells adaptively transferred (CD4+Thy1.1+) in the lung. (B) Percentages of IL-4–, IL-5–, and IL-13–positive CD4+ T cells are shown with mean values and SDs (Cxxc1fl/fl [WT] ER-Cre−, n = 4; Cxxc1fl/fl ER-Cre+, n = 4; *, P < 0.05; **, P < 0.01). (C) Airway inflammation was induced based on the allergic inflammation model shown in Fig. 6 D. Intracellular staining profiles of IFNγ, IL-4, IL-5, and IL-13 in activated CD4+ T cells (CD4+ CD44+) in the lung.
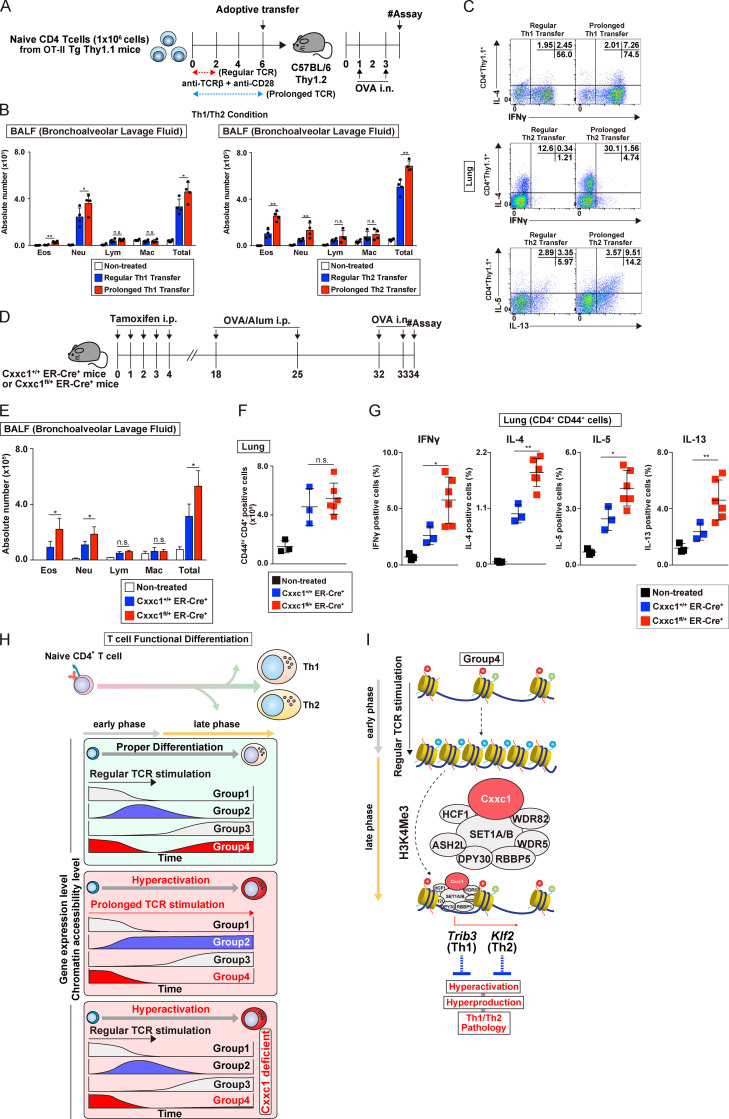
Next, we examined whether prolonged TCR/coreceptor stimulation of CD4+ T cells could affect airway inflammation. Consequently, we found that CD4+ T cells that received prolonged TCR/coreceptor stimulation before being transferred enhanced airway inflammation in vivo (Fig. 6, A–C). Finally, we confirmed our results using a system independent of cell transfer, in which Cxxc1fl/+ ERT2-Cre+ mice were treated with tamoxifen in vivo, immunized, and challenged by the i.n. administration of OVA (Fig. 6, D–G; and Fig. S5 C). Taken together, our in vivo results indicate that Cxxc1 plays an important role in repressing the dysregulation of antigen-specific Th1 and Th2 cells and the resulting pathology in airway inflammation, and they strengthen the connection between Cxxc1 expression and the late gene up-regulation once TCR/coreceptor stimulation is stopped.
Figure 6.
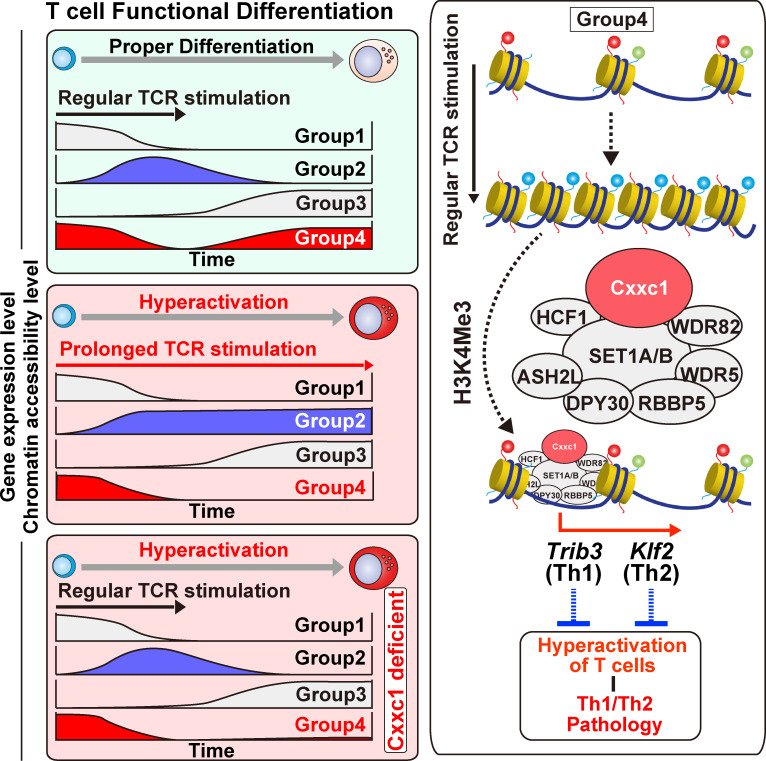
Prolonged TCR/coreceptor stimulation enhances airway inflammation in vivo. (A) A schematic protocol of the allergic airway. In brief, the same numbers of Th1 or Th2 cells with or without prolonged TCR/coreceptor stimulation were transferred into C57BL/6 recipient mice, which were treated with OVA (by i.n. injection). Mice that did not undergo cell transfer were used as controls. (B) The cell numbers of eosinophils (Eos), neutrophils (Neu), lymphocytes (Lym), and macrophages (Mac) in the BAL fluid are shown as the mean values with SDs (nontreated, n = 4; Th WT, n = 4; Th with prolonged TCR/coreceptor stimulation, n = 4) are shown (*, P < 0.05; **, P < 0.01). (C) Intracellular staining profiles of IFNγ, IL-4, IL-5, and IL-13 in transferred CD4+ T cells (CD4+ Thy1.1+) in the lung. (D) An experimental protocol of allergic airway inflammation in which Cxxc1+/+ ER-Cre+ (WT) and Cxxc1fl/+ ER-Cre+ mice were treated with tamoxifen in vivo and immunized with OVA/Alum (i.p. injection). At 1 wk after immunization, these mice were challenged with OVA (i.n. administration) and analyzed. Mice without immunization were used as controls. (E) The cell numbers of eosinophils, neutrophils, lymphocytes, and macrophages in the BAL fluid are shown as the mean values with SDs (nontreated, n = 3; Cxxc1+/+ ER-Cre+, n = 3; Cxxc1fl/+ ER-Cre+, n = 6; *, P < 0.05). (F) The numbers of activated CD4+ T cells in the lung were detected by cell surface staining of CD4 and CD44. (G) The indicated cytokine-producing cells among activated CD4+ T cells (CD4+CD44+) in the lung. (nontreated, n = 3; Cxxc1+/+ ER-Cre+, n = 3; Cxxc1fl/+ ER-Cre+, n = 6; *, P < 0.05; **, P < 0.01). (H) A schematic representation of the Cxxc1-dependent epigenetic regulation of Th subset differentiation. Both onset and cessation of the TCR/coreceptor stimulation are important for gene classification (top). Expression of group 1 genes decreases upon receiving TCR/coreceptor stimulation (i.e., early phase) and never increases after cessation of the TCR/coreceptor stimulation (i.e., late phase). Group 2 genes are turned on during early phase and turned off during late phase. Group 3 genes are silenced during early phase but up-regulated during late phase. Group 4 genes, which we focused on in the present study, exhibit decreased and increased expression before and after cessation of the TCR/coreceptor stimulation, respectively. Prolonged TCR/coreceptor stimulation specifically affects expression of the groups 2 and 4 genes (middle). Without cessation of the TCR/coreceptor stimulation, group 2 and group 4 genes are constitutively active and silenced, respectively. Group 4 genes are also dysregulated in the absence of Cxxc1, resulting in decreased expression during late phase (bottom). (I) Among the group 4 genes, the chromatin structure was closed after TCR/coreceptor stimulation in the early phase and reopened after the cessation of TCR/coreceptor stimulation (later phase). Cxxc1-containing Compass complex controls the reopening of the group 4 genes, probably via its H3K4me3 function. Cxxc1-containing Compass complex binds to the Trib3 gene in developing Th1 cells and inhibits hyperproduction of IFNγ. In developing Th2 cells, Cxxc1 binds to the Klf2 gene and inhibits the hyperproduction of Th2 cytokines. n.s., not significant.
Discussion
This study identifies the epigenetic machinery that governs dynamic transcriptional changes induced by TCR/coreceptor stimulation in early differentiating CD4+ T cells to become fully immunocompetent Th1 and Th2 cells. The Trithorax Cxxc1 molecule in the Compass complex was found to regulate selectively the expression of a set of genes (group 4) that are down-regulated by the TCR/coreceptor stimulation in the “early phase” and are reactivated in the “late phase” after the cessation of TCR/coreceptor stimulation (see Fig. 6 H). In the absence of Cxxc1, negative regulators such as Trib3 and Klf2 (Weinreich et al., 2009) fail to be re-up-regulated in the late phase, resulting in the hyperactivation of lineage-specifying transcription factors and the hyperproduction of Th1 and Th2 cytokines (see Fig. 6 I). Cxxc1 deficiency caused enhanced pathology in a mouse model of allergic airway inflammation in vivo. Prolonged TCR/coreceptor stimulation also induced a similar phenotype in vivo and in vitro, indicating that both the cessation of the TCR/coreceptor stimulation and Cxxc1-containing Trithorax Compass complex play essential roles in the establishment of a functional immune system in the CD4+ T cell–mediated immunity. The current study identified a set of Cxxc1 target genes and revealed for the first time that the functional consequences of Trithorax Compass complex–mediated epigenetic regulation.
The Cxxc1 target genes (e.g., Trib3 and Klf2) may function as gatekeepers; they are needed to be turned off in the early phase and turned on again in the late phase to accomplish proper CD4+ T cell differentiation. The expression of these genes may inversely correlate with cell proliferation; they should be turned off when T cells start to proliferate (Conley et al., 2016; Man and Kallies, 2015; Smith-Garvin et al., 2009) but need to be turned on again to repress proliferation (Conley et al., 2016; Daniels and Teixeiro, 2015; Man and Kallies, 2015). Similar mechanisms have been reported in myeloid cells (Rosenbauer and Tenen, 2007) and epidermal keratinocytes (Watt et al., 2008). In both cases, prolonged proliferation is proposed to be associated with the increased risk of oncogenesis, and each cell type has a fail-safe device that protects differentiating cells from uncontrolled proliferation. A previous study reported an interesting phenomenon in the thymus, named “coreceptor reversal,” in which TCR-signaled CD4+CD8+ double-positive thymocytes initially turn off and then reinitiate the expression of CD8 even when differentiating into CD8+ single-positive T cells (Brugnera et al., 2000). The initial termination of the CD8 expression generates CD4+CD8low intermediate thymocytes whose destination depends on whether TCR signaling persists or not. If TCR signaling persists, intermediate thymocytes differentiate into CD4+ single-positive T cells; however, if TCR signaling ceases, intermediate thymocytes undergo IL-7–dependent coreceptor reversal and differentiate into CD8+ single-positive T cells. The expression pattern of CD8 during the development of CD8+ single-positive T cells in the thymus seems similar to that of group 4 genes in peripheral CD4+ T cells. Although it remains unclear how Cxxc1 is involved in the termination and reinitiation of the CD8 expression, the blocked thymocyte development observed in the absence of Cxxc1 might be associated with impaired coreceptor reversal (Cao et al., 2016).
We previously reported that the menin/MLL1-containing Trithorax Compass–like complex is involved in the maintenance of the GATA3 expression in developed Th2 cells, such as memory Th2 cells, and that the lack of this complex resulted in decreased expression of GATA3 and Th2 cytokines (Yamashita et al., 2006; Onodera et al., 2010, 2017, 2018; Kuwahara et al., 2014). Interestingly, however, the current study revealed that Cxxc1 deficiency resulted in the hyperactivation of the master transcription factors (T-bet in Th1 and GATA3 in Th2) and the subsequent hyperproduction of Th1 and Th2 cytokines (IFNγ for Th1 and IL-4, IL-5, and IL-13 for Th2). We identified that Cxxc1 indirectly controls the expression of GATA3 through direct binding and regulation of the Klf2 expression. These results indicate that these two Trithorax complexes (i.e., menin/MLL1-containing Compass-like and Cxxc1/Setd1-containing Compass complex) mediate H3K4me3 (Piunti and Shilatifard, 2016; Shilatifard, 2012), but they appear to have specific target genes and a distinct regulatory function in the differentiation of CD4+ Th1 and Th2 cells. Thus, the current study revealed the dynamic regulation of CD4+ T cell differentiation by two distinct Trithorax complexes.
Another interesting feature is the phenotype of Cxxc1-deficient cells, which is similar to that caused by the lack of Ezh2. Ezh2 is a member of a repressive Polycomb complex that mediates H3K27me3 and a counterpart of the Trithorax complex mediating H3K4me3 (Schuettengruber et al., 2017; Tumes et al., 2013). Ezh2 is known to control the expression of Th1 and Th2 cytokines via direct binding to the genes encoding master transcription factors (T-bet and GATA3), while Cxxc1 indirectly regulates the expression of these master transcription factors by controlling the expression of Trib3 in Th1 cells and Klf2 in Th2 cells. Moreover, Cxxc1 deficiency was not associated with cellular senescence, which is frequently observed in the absence of the Polycomb proteins and characterized by the increased expression of the granzyme family proteins (Chuang et al., 2014; Nagel et al., 2010; Tumes et al., 2013). In fact, the expression of granzyme family proteins was down-regulated in Cxxc1-deficient cells.
The present study explored the molecular basis for why the cessation of TCR/coreceptor stimulation is required for the proper differentiation of Th1 and Th2 cells. It is well known that the prolonged TCR/coreceptor stimulation causes exhaustion of CD8+ cytotoxic T cells (Wherry and Kurachi, 2015). The exhausted CD8+ T cells are characterized by the expression of inhibitory receptors, including PD-1, TIM3, and LAG3, and also hyperactivation of transcription factors Eomes, TOX, and IRF-4 (Hope et al., 2019; McLane et al., 2019; Xia et al., 2019). T cell exhaustion is one of the major mechanisms of immune dysfunction in the tumor microenvironment, and reinvigoration of the exhausted CD8+ T cells by blocking PD-1–PD-L1 interactions has been highlighted as a potent therapeutic strategy in cancer immunotherapy (McLane et al., 2019; Wherry and Kurachi, 2015). On the other hand, CD4+ T cells at the site of autoimmune inflammation have been shown to receive prolonged TCR/coreceptor signaling; however, the functional characterization of these CD4+ T cells has not yet been achieved (Bending et al., 2018). In the periphery, prolonged TCR/coreceptor stimulation in chronic infection appeared to induce the same effects on both CD8+ and CD4+ T cells (Crawford et al., 2014) to some extent. In the present study, CD4+ T cells that received prolonged TCR/coreceptor stimulation or that lacked Cxxc1 exhibited similar gene expression signatures to those of exhausted CD8+ T cells. For example, Eomes, one of the exhaustion signature genes (Buggert et al., 2014; McLane et al., 2019), was up-regulated in Cxxc1-deficient CD4+ T cells. In addition, Tbx21, which is known to be transiently up-regulated in progenitor of exhausted CD8+ T cells (McLane et al., 2019), was found to be increased in Cxxc1-deficient Th1 cells in the present study. Similarly, the Nr4a family and Tox were up-regulated. However, the expression of most of the inhibitory molecules was intact in the absence of Cxxc1, suggesting that Cxxc1 is not involved in the regulation by these inhibitory receptors. Only the Lag3 gene showed increased expression levels in both Th1 and Th2 conditions. For two genes encoding effector molecules (i.e., Ifng and Tnf), Cxxc1 deficiency had an opposite effect (up-regulation) to that observed in exhausted CD8+ T cells. Taken together, Cxxc1 deficiency in CD4+ T cells increased the expression of the transcription factors associated with CD8+ T cell exhaustion. Cxxc1 deficiency had little effect on most of the inhibitory molecule gene expression except Lag3. Among some effector molecule genes, which are down-regulated in exhausted CD8+ T cells, Ifng and Tnf were up-regulated in Cxxc1-deficient CD4+ T cells.
In summary, we found that the cessation of TCR/coreceptor signaling in the later phase of T cell activation is a critical commitment factor that induces the epigenetic changes important for the proper functional differentiation of CD4+ T cells. The target specificity of the Trithorax Compass complex Cxxc1 molecule was identified. Furthermore, two unique Cxxc1-controlled signaling cascades downstream of TCR in Th1 and Th2 cells were identified. Thus, the Trithorax Compass complex appears to be central to the epigenetic variation in the differentiating CD4+ T cells to become immunocompetent Th1 and Th2 cells.
Materials and methods
Mice
C57BL/6 mice were purchased from CLEA. Cxxc1fl/fl mice were established by Haruhiko Koseki (Brown et al., 2017) and backcrossed at Chiba University to a C57BL/6 background 10 times. Cxxc1fl/fl mice were crossed with Rosa26-Cre-ERT mice (TaconicArtemis), OT-II TCR Tg mice, and Thy1.1 mice (The Jackson Laboratory). OT-II TCR Tg mice express TCRs specific for residues 323–339 of the OVA protein. All mice used in this study were maintained under specific pathogen–free conditions and ranged from 6 to 8 wk of age. The protocols of all of the animal experiments were approved by the Chiba University animal committee. All mice used in this study were female. All animal care was performed in accordance with the guidelines of Chiba University. The research proposals were reviewed by the ethics committee for animals at Chiba University (registration numbers 1-254 and 1-300).
Antibodies
The antibodies used for the ChIP assay were anti–trimethyl-histone H3K4 (AR-0169; LP Bio) and anti-Flag mAb (M2; Sigma-Aldrich). The antibodies used for the immunoblot analyses were anti-Cgbp (35; Santa Cruz Biotechnology) and anti-lamin (H-110; Santa Cruz Biotechnology). The antibodies used for cytoplasmic and cell surface staining were as follows: BV421-conjugated anti-IFNγ mAb (XMG1.2; BioLegend), allophycocyanin-conjugated anti–IL-4 mAb (11B11; BioLegend), PE-conjugated anti–IL-5 mAb (TRFK5; BioLegend), Alexa Fluor 488–conjugated anti–IL-13 mAb (eBio13A; eBioscience), allophycocyanin-conjugated anti–IL-17A mAb (TC11-18H10.1; BioLegend), Alexa Fluor 647–conjugated anti-Foxp3 mAb (MF23; BioLegend), Alexa Fluor 647–conjugated anti-GATA3 (560068; BD PharMingen), PE-conjugated anti-GATA3 (4B10; BD PharMingen), BV421-conjugated anti-CD4 mAb (GK1.5; BioLegend), FITC-conjugated anti-CD90.1 mAb (HIS51; BD PharMingen), PE-conjugated anti-CD212 mAb (114; BD PharMingen), PE-conjugated anti-CD124 mAb (mIL4R-M1; BD PharMingen), allophycocyanin-conjugated anti-CD25 mAb (3C7; BioLegend), FITC-conjugated anti-CD44 mAb (IM7; BD PharMingen), PE-conjugated anti-Akt (pT308) mAb (J1-223.371; BD PharMingen), PE-conjugated anti-Akt (pS473; M89-61; BD PharMingen), Alexa Fluor 647–conjugated anti-p38 (pT180/Y182) mAb (36/p38 [pT180/Y182]; BD PharMingen), Alexa Fluor 647–conjugated anti-Stat4 (pY693) mAb (38/p-Stat4; BD PharMingen), Alexa Fluor 647–conjugated anti-S6 (pS235/pS236) mAb (N7-548; BD PharMingen), and Alexa Fluor 647–conjugated anti-Irf4 mAb (IRF4.3E4; BioLegend). The antibodies used for cell culturing were anti–TCR mAb (H57-597; homemade), anti-CD28 mAb (37.51; BioLegend), anti–IL-4 mAb (11B11; homemade), and anti-IFNγ mAb (R46A2; homemade).
Reagents
Mouse IL-12 was purchased from BD PharMingen, and recombinant mouse IL-4, IL-6, IL-1β, and recombinant human TGF-β1 were purchased from PeproTech. The OVA peptide (residues 323–339; ISQAVHAAHAEINEAGR) was synthesized by BEX. The primer was purchased from Sigma-Aldrich. The Universal Probe Library was purchased from Roche. The sequences of the Hprt primers and Universal Probe Library number used are as follows: sense primer, 5′-CCTGGTTCATCATGGCTAATC-3′; antisense primer, 5′-TCCTCCTCAGACCGCTTTT-3′; and Universal Probe Library no. 95. The sequences of the Cxxc1 primers and Universal Probe Library number used are as follows: sense primer, 5′-TCTGTGAGCGGAGATATGGA-3′; antisense primer, 5′-TCCCCATTCTCAGACTTGCT-3′; and Universal Probe Library no. 85. The sequences of the Trib3 primers and Universal Probe Library number used are as follows: sense primer, 5′-TCATCTGATCCAGCTCACG-3′; antisense primer, 5′-CCTTTGTCTTCAGCAACTGT-3′; and Universal Probe Library no. 67. The sequences of the Klf2 primers and Universal Probe Library number used are as follows: sense primer, 5′-TAGTGGCGGGTAAGCTCGT-3′; antisense primer, 5′-CTAAAGGCGCATCTGCGTA-3′; and Universal Probe Library no. 48.
In vitro Th1/Th2/Th17/iT reg cell differentiation
Splenic naive CD4 T cells (CD44loCD62Lhi) from C57BL/6, Cxxc1fl/fl (WT), or Cxxc1fl/fl ER-Cre+ (KO) mice were purified using a magnetic cell sorter (autoMACS; Miltenyi Biotec), yielding a purity of >98%. Purified naive CD4+ T cells were stimulated with immobilized anti-TCRβ mAb (H57-597; 10 µg/ml) in the presence of IL-2 (25 U/ml), IL-12 (10 U/ml; BD PharMingen), anti–IL-4 mAb (11B11; 5% vol/vol), and CD28 mAb (37.51; 1 µg/ml) for Th1 cell differentiation; IL-2 (25 U/ml), IL-4 (100 U/ml; PeproTech), anti-IFNγ mAb (R46A2; 5% vol/vol), and CD28 mAb (37.51; 1 µg/ml) for Th2 cell differentiation; TGF-β1 (2.5 ng/ml; PeproTech), IL-6 (25 ng/ml; PeproTech), anti–IL-4 mAb (11B11; 5% vol/vol), anti-IFNγ mAb (R46A2; 5% vol/vol), anti–IL-2 mAb (JES6-1A12; 2.5 µg/ml; BD PharMingen), and CD28 mAb (37.51; 1 µg/ml) for Th17 cell differentiation; and IL-2 (50 U/ml), TGF-β1 (10 ng/ml; PeproTech), anti-IFNγ mAb (R46A2; 5% vol/vol), anti–IL-4 mAb (11B11; 5% vol/vol), and anti-CD28 mAb (37.51; 1 µg/ml) for iT reg cell differentiation. The anti-TCR mAb and anti-CD28 mAb stimulation was stopped at 48 h, after which the cells were cultured with the above-described cytokines. The cells are referred to as Th1 or Th2 cells with prolonged TCR/coreceptor stimulation if the anti-TCR mAb and anti-CD28 mAb stimulation was not stopped at 48 h.
In vitro Th1/Th2 cell differentiation in DO11.10 TCR Tg system
Splenic naive CD4+ T cells (CD44loCD62Lhi) from DO11.10 OVA-specific TCR Tg mice were purified using a magnetic cell sorter (autoMACS; Miltenyi Biotec), yielding a purity of >98%. Purified naive CD4+ T cells were stimulated with OVA peptides (Loh15; 0.3 µM) plus irradiated (3,500 rad) Thy1.2-depleted splenic cells (i.e., APCs) in the presence of IL-2 (25 U/ml), IL-12 (10 U/ml; BD PharMingen), and anti–IL-4 mAb (11B11; 5% vol/vol) for Th1 cell differentiation, IL-2 (25 U/ml), IL-4 (100 U/ml; PeproTech), and anti-IFNγ mAb (R46A2; 5% vol/vol) for Th2 cell differentiation.
Immunofluorescence staining and flow cytometry
For intracellular staining of cytokines, cultured Th cells were restimulated with PMA (50 ng/ml) plus ionomycin (500 nM) for 4 h in the presence of 2 µM monensin (Sigma-Aldrich). Cells were fixed with 4% (wt/vol) paraformaldehyde for 10 min at room temperature and permeabilized in a permeabilizing solution (50 mM NaCl, 5 mM EDTA, 0.02% NaN3, pH 7.5) containing 0.5% Triton X-100 for 10 min on ice. After blocking with 3% (wt/vol) BSA in PBS for 15 min, cells were incubated on ice for 30 min with appropriate staining antibodies. Cells were washed with FACS buffer at the end of each step. Flow cytometry was performed on a FACSVerse (BD Biosciences) or FACSCanto II instrument (BD Biosciences), and the results were analyzed with the FlowJo software program (FlowJo, LLC).
Phosphoflow
Cells were fixed for 10 min at 37°C with Lyse/Fix Buffer (BD Biosciences) and permeabilized for 30 min on ice with Perm Buffer III (BD Biosciences) before staining for CD4 and phosphorylated Akt (pT308), Akt (pS473), p38 (pT180/Y182), Stat4 (pY693), or S6 (pS235/pS236).
Measurement of cytokines by ELISA
The purified Th1 or Th2 cells were restimulated with immobilized anti-TCRβ mAb (clone H57-597; 2.5 mg/ml) and anti-CD28 mAb (clone 37.51; 1 mg/ml) for 24 h. The IFNγ, IL-4, and IL-5 production in cell culture supernatants was analyzed by a standard sandwich ELISA protocol using the following Ab pairs: clone R4-6A2 and clone XMG1.2-biotin (BD Biosciences) for IFNγ, clone BVD4-1D11 and clone BVD6-24G2-biotin (BD Biosciences) for IL-4, and clone TRFK5 and clone TRFK4-biotin (BD Biosciences) for IL-5. IL-13 was measured using a DuoSet mouse IL-13 ELISA set (R&D Systems).
Quantitative PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen), and cDNA was synthesized using oligo(dT) primers and Superscript II RT (Invitrogen). Quantitative RT-PCR was performed using a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) via the comparative threshold cycle method. The primers and TaqMan probes used for the amplification and detection of the indicated genes were purchased from Roche. The expression of the target genes was normalized to the Hprt or Actb signals.
Immunoblotting
Cytoplasmic extracts and nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo Fisher Scientific). The antibodies used for the immunoblot analyses were anti-Cgbp (35; Santa Cruz Biotechnology) and anti-lamin (H-110; Santa Cruz Biotechnology).
ChIP assay
ChIP experiments for H3K4me3, Flag-tag, and control Ab were performed using Dynabeads (Invitrogen) as previously described (Onodera et al., 2015). In brief, 1–10 × 106 cells were fixed with 1% paraformaldehyde at 37°C for 10 min. Cells were sedimented, washed, and lysed with SDS lysis buffer (50 mM Tris-HCl, 1% SDS, 10 mM EDTA, 1 mM PMSF, 1 mg/ml aprotinin, and 1 mg/ml leupeptin). The lysates were sonicated to reduce the DNA lengths to between 200 and 1,000 bp. The soluble fraction was diluted in ChIP dilution buffer and incubated with Ab conjugated with Dynabeads proteins A and G overnight at 4°C. The immune complexes were then captured using a magnet and washed with low-salt, high-salt, LiCl, and Tris-EDTA wash buffer. Enriched chromatin fragments were eluted with elution buffer (0.1 M NaHCO3 containing 1% SDS). The eluted material was incubated at 65°C for 6 h to reverse the formaldehyde cross-links and treated with RNase A (10 mg/ml) and proteinase K (40 mg/ml). DNA was extracted with a QIAquick PCR purification kit (Qiagen). The total input DNA (cellular DNA without immunoprecipitation) was purified in parallel. Real-time qPCR was performed using a StepOnePlus Real-Time PCR System via the comparative cycle threshold method with TaqMan probes and primers. To calculate the enrichment of each protein to a particular target DNA, we divided the values obtained for each target by the amount of the corresponding target in the input fraction. All of the results are expressed as the percentage of input DNA.
ATAC-seq
ATAC-seq was performed as previously described with minor modification (Buenrostro et al., 2013; Shih et al., 2016). 50,000 cells were pelleted and washed with 50 µl of 1× PBS, followed by treatment with 50 µl of lysis buffer. The nuclei were resuspended in 40 µl of Tagment DNA enzyme buffer with 2 µl of Tn5 transposase (15027865, 15027866; Illumina) to tag and fragmentize the accessible chromatin. The reaction was incubated at 37°C with shaking at 300 revolutions per minute for 30 min. The fragmentized DNAs were purified using a QIAGEN MinElute kit and amplified with 11 or 12 PCR cycles, based on the amplification curve. The libraries were sequenced for 50 cycles (paired-end reads) on an Illumina HiSeq 1500.
ChIP-seq and Illumina sequencing
Ab-specific immunoprecipitates and total input DNA samples were prepared using a SMARTer ThruPLEX DNA-Seq Kit (TaKaRa). Adapter-ligated DNA was recovered using AMPure XP Beads (Beckman Coulter). This DNA was then amplified by 15 PCR cycles and again recovered using AMPure XP Beads. The libraries were sequenced for 50 cycles (paired-end reads) on an Illumina HiSeq 1500.
The analysis of ATAC-seq and ChIP-seq data
The ATAC-seq and ChIP-seq data were mapped to the mouse genome build NCBI37/mm9 using Bowtie (v2.4.2.6; http://bowtie-bio.sourceforge.net/index.shtml). MACS2 (Feng et al., 2011) was used for peak calling and the visualization of binding, with the parameters set as follows: window_size = 300, gap_size = 300, and false discovery rate (FDR) = 0.01. HOMER tag directories, which were created by the HOMER platform (makeTagDirectory; Heinz et al., 2010) from the aligned Sequence Alignment/Map (SAM) formats using SAMtools (Li et al., 2009). When ATAC and ChIP peaks were annotated to the promoter of the closest gene, HOMER annotatePeaks.pl (mm9 genome build) was used. BigWig files were generated from the aligned SAM or Browser Extensible Data (BED) file formats using SAMtools, BEDtools (Quinlan and Hall, 2010), and deepTools 2.0 (https://deeptools.readthedocs.io/en/develop/index.html). The tag count density was normalized as tags per 10 million reads in the original library. K-means clustering and Z-scoring were performed using Gene Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). K-means clustering first identified 20 groups. Groups with similar accessible patterns were merged (An et al., 2014). Tag coverage was created with deepTools 2.0 from the BiGWig formats. ATAC and ChIP region annotation was generated using the Cis-regulatory Element Annotation System (Shin et al., 2009). The Integrative Genomics Viewer software program (https://software.broadinstitute.org/software/igv/) was used for the visualization of BiGWig files. A motif enrichment analysis was performed with the findMotifsGenome.pl command in the HOMER package using a 200-bp window. A transcription factor enrichment analysis was performed using the wPGSA dataset (http://wpgsa.org/). T-SNE was performed using R (https://www.r-project.org/).
RNA-seq
RNA-seq was performed as previously described (Onodera et al., 2015). Total cellular RNA was extracted with TRIzol reagent (Invitrogen). For cDNA library construction, we used a TruSeq RNA Sample Prep Kit version 2 (Illumina) in accordance with the manufacturer’s protocol. Sequencing of library fragments was performed on an Illumina HiSeq 1500 System. For the data analyses, read sequences (50 bp) were aligned to the mm10 mouse reference genome (University of California, Santa Cruz; December 2011) using the Bowtie 2 and TopHat (version 1.3.2) software programs. Fragments per kilobase of exon per million mapped reads for each gene were calculated using the Cufflinks (version 2.0.2) software program. Down-regulated genes were selected with the following criteria: (1) absolute fragments per kilobase of exon per million mapped reads ≥1 in WT cells and (2) a ≤0.5-fold change in the expression level.
The analysis and graphic display of RNA-seq data
For the visualization of the coregulation network (Ichikawa et al., 2019), the first neighbor genes were determined using the following criteria: (1) correlation >0.95 and (2) ratio of norm 0.75–1.10. The network graph of genes was visualized using Cytoscape (http://www.cytoscape.org/). Gene Ontology (GO) enrichment analysis was performed with 10,899 terms using the R and GSA packages and c5.all.v5.2.symbols.gmt as a GO term database (http://software.broadinstitute.org/gsea/msigdb) with FDR <0.10 and P < 0.01 as the cutoff values. A network of enriched GO terms was generated by the programming language Julia. Connection of the nodes was determined with the cutoff value of the Simpson coefficient set at ≥0.5. The network graph was visualized using Cytoscape and the force-directed layout with the Simpson coefficient as a weight for the length of the spring (Ichikawa et al., 2019; Morimoto et al., 2018). An MA plot was constructed by using R. GSEA was performed with genes in Th1 and Th2 cells using Python (https://www.python.org/) and c7.all.v7.1.symbols.gmt as a GO term database.
Retroviral vectors and infection
The pMX–internal ribosome entry site–human nerve growth factor receptor plasmid was generated as previously described (Shinnakasu et al., 2006). The infected cells were enriched by autoMACS with anti–human nerve growth factor receptor (clone C40-1457; BD PharMingen) and were subjected to a qRT-PCR assay.
Airway inflammation model induced by the adoptive transfer of naive CD4+ T cells
Naive CD4 T cells from Cxxc1fl/fl Thy1.1 or Cxxc1fl/fl ER-Cre+ Thy1.1 OVA-specific OT-II TCR Tg mice were administered i.v. through the tail vein to C57BL/6 mice (1 × 106 cells per mouse; day 0). After cell transfer, OVA was administered i.n. on days 1, 3, 6, and 8, and tamoxifen was administered i.p. on days 0, 1, 2, and 3. Assays were conducted on day 9.
Airway inflammation model in mice immunized with OVA
Tamoxifen was administered i.p. into Cxxc1+/+ ER-Cre+ or Cxxc1fl/+ ER-Cre+ mice before immunization with OVA. These mice were immunized with OVA (i.p.) on days 18 and 25 and challenged by OVA (i.n.) on days 32 and 33. Assays were conducted on day 34.
Airway inflammation model induced by the adoptive transfer of Th1 or Th2 cells with or without prolonged TCR/coreceptor stimulation
Th1 or Th2 cells were generated from OVA-specific OT-II TCR Tg mice with the anti-TCR mAb and anti-CD28 mAb stimulation in vitro. Th1 or Th2 cells that received prolonged TCR/coreceptor stimulation as described above were also prepared. Th1 or Th2 cells with or without prolonged TCR/coreceptor stimulation were administered i.v. through the tail vein to C57BL/6 mice (1 × 106 cells per mouse; day 0). After cell transfer, OVA was administered i.n. on days 1 and 3. Assays were conducted on day 4.
Collection of BAL fluid
The collection of BAL fluid was performed 24 h after the last OVA challenge as described previously (Onodera et al., 2017). A total of 100,000 viable BAL cells were cytocentrifuged onto slides by a Cytospin 4 (Thermo Fisher Scientific) and stained with a Diff-Quik Stain Kit (Sysmex). 200 leukocytes were counted on each slide. The cell types were identified using morphological criteria.
Lung histology and immunohistochemistry
Mice were sacrificed 24 h after the last OVA challenge, and the lungs were fixed in 10% (vol/vol) formalin. The samples were sectioned and stained with H&E, and the pathological changes were examined under a light microscope.
Statistical analysis
Data were analyzed using the GraphPad Prism software program (version 6). Comparisons between two groups were performed using the Mann-Whitney U test (two tailed), a two-tailed paired t test, or a two-way ANOVA with Bonferroni’s multiple comparisons test. Fisher’s exact test was used to evaluate whether fold enrichment of the proportion of the Cxxc1-dependent genes in group 4 was statistically significant compared with other groups. P values <0.05 were considered to indicate statistical significance. The overlap of group 4 and Cxxc1-bound genes in Th1 and Th2 cells was analyzed by a hypergeometric analysis. P values of cumulative distribution function plots of the normalized tag counts in ATAC-seq peaks in groups 1–4 were calculated by the Wilcoxon rank-sum test.
Data availability statement
The raw and analyzed data reported in this paper are available under Gene Expression Omnibus accession no. GSE159505.
Code availability
The code that supported the findings of this study is available from the corresponding author upon reasonable request.
Online supplemental material
Fig. S1 shows that TCR/coreceptor signaling alters chromatin accessibility landscapes in naive CD4 T cells stimulated with antigenic peptide and APCs (related to Fig. 1). Fig. S2 shows that Cxxc1 specifically regulates the reactivation of the genes that are suppressed during the early phase (related to Fig. 2). Fig. S3 shows that loss of Cxxc1 results in the enhanced expression of Th1 cytokines in CD4+ T cells (related to Fig. 3). Fig. S4 shows that loss of Cxxc1 results in the enhanced expression of Th2 cytokines in CD4+ T cells (related to Fig. 4). Fig. S5 shows that loss of Cxxc1 enhances airway inflammation in vivo (related to Figs. 5 and 6). Table S1 lists the genes in groups 1–6.
Supplementary Material
lists the genes in groups 1–6.
Acknowledgments
We are grateful to Dr. Patrick Hogan (La Jolla Institute for Immunology) for critical reading of and for providing valuable suggestions on the manuscript. We thank Miki Kato, Toshihiro Ito, and Kaoru Sugaya for their excellent technical assistance.
This work was supported by the following grants: Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research (S) 26221305, JP19H05650, (B) 20H03685, (C) 17K08876, 18K07164, and 19K16683; Practical Research Project for Allergic Diseases and Immunology (Research on Allergic Diseases and Immunology) from the Japan Agency for Medical Research and Development (AMED) (JP20ek0410060 and JP19ek0410045); AMED-PRIME, AMED (JP20gm6110005); AMED-CREST, AMED (JP20gm1210003); the Mochida Memorial Foundation for Medical and Pharmaceutical Research; the MSD Life Science Foundation; the Naito Foundation; and the Takeda Science Foundation.
Author contributions: Conceptualization and experiment design: M. Kiuchi, A. Onodera, and T. Nakayama. Experiment performance: M. Kiuchi, A. Onodera, K. Hirahara, K. Kokubo, T. Ichikawa, Y. Morimoto, H. Koseki, E. Kawakami, and T. Nakayama. Performed ATAC-seq: N. Takayama and K. Eto. Data analysis and interpretation: M. Kiuchi, A. Onodera, K. Hirahara, K. Kokubo, T. Ichikawa, Y. Morimoto, E. Kawakami, and T. Nakayama. Writing, reviewing, and editing: M. Kiuchi, A. Onodera, K. Hirahara, and T. Nakayama.
References
- Acquaviva, L., Székvölgyi L., Dichtl B., Dichtl B.S., de La Roche Saint André C., Nicolas A., and Géli V.. 2013. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 339:215–218. 10.1126/science.1225739 [DOI] [PubMed] [Google Scholar]
- An, X., Schulz V.P., Li J., Wu K., Liu J., Xue F., Hu J., Mohandas N., and Gallagher P.G.. 2014. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 123:3466–3477. 10.1182/blood-2014-01-548305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansel, K.M., Djuretic I., Tanasa B., and Rao A.. 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24:607–656. 10.1146/annurev.immunol.23.021704.115821 [DOI] [PubMed] [Google Scholar]
- Bader, S., Walker M., McQueen H.A., Sellar R., Oei E., Wopereis S., Zhu Y., Peter A., Bird A.P., and Harrison D.J.. 2003. MBD1, MBD2 and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers. Oncogene. 22:3506–3510. 10.1038/sj.onc.1206574 [DOI] [PubMed] [Google Scholar]
- Bending, D., Prieto Martín P., Paduraru A., Ducker C., Marzaganov E., Laviron M., Kitano S., Miyachi H., Crompton T., and Ono M.. 2018. A timer for analyzing temporally dynamic changes in transcription during differentiation in vivo. J. Cell Biol. 217:2931–2950. 10.1083/jcb.201711048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohineust, A., Garcia Z., Beuneu H., Lemaître F., and Bousso P.. 2018. Termination of T cell priming relies on a phase of unresponsiveness promoting disengagement from APCs and T cell division. J. Exp. Med. 215:1481–1492. 10.1084/jem.20171708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, M.J., Nazor K.L., and Loring J.F.. 2014. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 115:311–324. 10.1161/CIRCRESAHA.115.301517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, J.A., Weinreb C., Wagner D.E., Megason S., Peshkin L., Kirschner M.W., and Klein A.M.. 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science. 360:eaar5780. 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.A., Di Cerbo V., Feldmann A., Ahn J., Ito S., Blackledge N.P., Nakayama M., McClellan M., Dimitrova E., Turberfield A.H., et al. 2017. The SET1 complex selects actively transcribed target genes via multivalent interaction with CpG island chromatin. Cell Rep. 20:2313–2327. 10.1016/j.celrep.2017.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera, E., Bhandoola A., Cibotti R., Yu Q., Guinter T.I., Yamashita Y., Sharrow S.O., and Singer A.. 2000. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 13:59–71. 10.1016/S1074-7613(00)00008-X [DOI] [PubMed] [Google Scholar]
- Buenrostro, J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert, M., Tauriainen J., Yamamoto T., Frederiksen J., Ivarsson M.A., Michaëlsson J., Lund O., Hejdeman B., Jansson M., Sönnerborg A., et al. 2014. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 10:e1004251. 10.1371/journal.ppat.1004251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W., Guo J., Wen X., Miao L., Lin F., Xu G., Ma R., Yin S., Hui Z., Chen T., et al. 2016. CXXC finger protein 1 is critical for T-cell intrathymic development through regulating H3K4 trimethylation. Nat. Commun. 7:11687. 10.1038/ncomms11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., and Flies D.B.. 2013. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 13:227–242. 10.1038/nri3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Yu D.. 2015. TCF-1 at the Tfh and Th1 Divergence. Trends Immunol. 36:758–760. 10.1016/j.it.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Chi, H. 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12:325–338. 10.1038/nri3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.H., Wang W.J., Li C.F., Ko C.Y., Chou Y.H., Chuu C.P., Cheng T.L., and Wang J.M.. 2014. The combination of the prodrugs perforin-CEBPD and perforin-granzyme B efficiently enhances the activation of caspase signaling and kills prostate cancer. Cell Death Dis. 5:e1220. 10.1038/cddis.2014.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, K.T., Li B., Dobrota E., Tate C., Lee J.H., Khan S., Haneline L., HogenEsch H., and Skalnik D.G.. 2014. The epigenetic regulator CXXC finger protein 1 is essential for murine hematopoiesis. PLoS One. 9:e113745. 10.1371/journal.pone.0113745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, J.M., Gallagher M.P., and Berg L.J.. 2016. T cells and gene regulation: the switching on and turning up of genes after T cell receptor stimulation in CD8 T cells. Front. Immunol. 7:76. 10.3389/fimmu.2016.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., and Wherry E.J.. 2014. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 40:289–302. 10.1016/j.immuni.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, M.A., and Teixeiro E.. 2015. TCR signaling in T cell memory. Front. Immunol. 6:617. 10.3389/fimmu.2015.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C., Davis R.J., and Flavell R.A.. 2002. MAP kinases in the immune response. Annu. Rev. Immunol. 20:55–72. 10.1146/annurev.immunol.20.091301.131133 [DOI] [PubMed] [Google Scholar]
- Eisenbarth, S.C. 2019. Dendritic cell subsets in T cell programming: location dictates function. Nat. Rev. Immunol. 19:89–103. 10.1038/s41577-018-0088-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers, P.A., Keeshan K., and Kannan N.. 2017. Tribbles in the 21st century: the evolving roles of Tribbles pseudokinases in biology and disease. Trends Cell Biol. 27:284–298. 10.1016/j.tcb.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., Liu T., and Zhang Y.. 2011. Using MACS to identify peaks from ChIP-Seq data. Curr. Protoc. Bioinformatics. 34:2.14.1–2.14.14. 10.1002/0471250953.bi0214s34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, W.G., Yang P.C., Medikonduri R.K., and Jafri M.S.. 2006. NFAT and NFkappaB activation in T lymphocytes: a model of differential activation of gene expression. Ann. Biomed. Eng. 34:1712–1728. 10.1007/s10439-006-9179-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci, P., Dai Z., Génard M., Gauffretau A., Leblanc-Fournier N., Richard-Molard C., Vile D., and Brunel-Muguet S.. 2017. Epigenetics for plant improvement: current knowledge and modeling avenues. Trends Plant Sci. 22:610–623. 10.1016/j.tplants.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Hawkins, R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S., et al. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 6:479–491. 10.1016/j.stem.2010.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 38(4):576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning, A.N., Klebanoff C.A., and Restifo N.P.. 2018. Silencing stemness in T cell differentiation. Science. 359:163–164. 10.1126/science.aar5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, I.C., Tai T.S., and Pai S.Y.. 2009. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 9:125–135. 10.1038/nri2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, P.G. 2017. Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium. 63:66–69. 10.1016/j.ceca.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, J.L., Stairiker C.J., Bae E.A., Otero D.C., and Bradley L.M.. 2019. Striking a balance—cellular and molecular drivers of memory T cell development and responses to chronic stimulation. Front. Immunol. 10:1595. 10.3389/fimmu.2019.01595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston, R.S., Cheadle J., Dobbins S.E., Tenesa A., Jones A.M., Howarth K., Spain S.L., Broderick P., Domingo E., Farrington S., et al. COINB Collaborative Group . 2010. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet. 42:973–977. 10.1038/ng.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, W., Troutman T.D., Edukulla R., and Pasare C.. 2011. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 35:1010–1022. 10.1016/j.immuni.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa, T., Hirahara K., Kokubo K., Kiuchi M., Aoki A., Morimoto Y., Kumagai J., Onodera A., Mato N., Tumes D.J., et al. 2019. CD103hi Treg cells constrain lung fibrosis induced by CD103lo tissue-resident pathogenic CD4 T cells. Nat. Immunol. 20:1469–1480. 10.1038/s41590-019-0494-y [DOI] [PubMed] [Google Scholar]
- Israel, E., and Reddel H.K.. 2017. Severe and difficult-to-treat asthma in adults. N. Engl. J. Med. 377:965–976. 10.1056/NEJMra1608969 [DOI] [PubMed] [Google Scholar]
- Kanno, Y., Vahedi G., Hirahara K., Singleton K., and O’Shea J.J.. 2012. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu. Rev. Immunol. 30:707–731. 10.1146/annurev-immunol-020711-075058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, E., Nakaoka S., Ohta T., and Kitano H.. 2016. Weighted enrichment method for prediction of transcription regulators from transcriptome and global chromatin immunoprecipitation data. Nucleic Acids Res. 44:5010–5021. 10.1093/nar/gkw355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kim J.A., McGinty R.K., Nguyen U.T., Muir T.W., Allis C.D., and Roeder R.G.. 2013. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell. 49:1121–1133. 10.1016/j.molcel.2013.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara, M., Suzuki J., Tofukuji S., Yamada T., Kanoh M., Matsumoto A., Maruyama S., Kometani K., Kurosaki T., Ohara O., et al. 2014. The Menin-Bach2 axis is critical for regulating CD4 T-cell senescence and cytokine homeostasis. Nat. Commun. 5:3555. 10.1038/ncomms4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, K., and Kallies A.. 2015. Synchronizing transcriptional control of T cell metabolism and function. Nat. Rev. Immunol. 15:574–584. 10.1038/nri3874 [DOI] [PubMed] [Google Scholar]
- Martinez, G.J., Pereira R.M., Äijö T., Kim E.Y., Marangoni F., Pipkin M.E., Togher S., Heissmeyer V., Zhang Y.C., Crotty S., et al. 2015. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. 42:265–278. 10.1016/j.immuni.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane, L.M., Abdel-Hakeem M.S., and Wherry E.J.. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Miller, B.C., Sen D.R., Al Abosy R., Bi K., Virkud Y.V., LaFleur M.W., Yates K.B., Lako A., Felt K., Naik G.S., et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20:326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, Y., Hirahara K., Kiuchi M., Wada T., Ichikawa T., Kanno T., Okano M., Kokubo K., Onodera A., Sakurai D., et al. 2018. Amphiregulin-producing pathogenic memory T helper 2 cells instruct eosinophils to secrete osteopontin and facilitate airway fibrosis. Immunity. 49:134–150.e6. 10.1016/j.immuni.2018.04.023 [DOI] [PubMed] [Google Scholar]
- Müller, M.R., and Rao A.. 2010. NFAT, immunity and cancer: a transcription factor comes of age. Nat. Rev. Immunol. 10:645–656. 10.1038/nri2818 [DOI] [PubMed] [Google Scholar]
- Nagel, S., Venturini L., Marquez V.E., Meyer C., Kaufmann M., Scherr M., MacLeod R.A., and Drexler H.G.. 2010. Polycomb repressor complex 2 regulates HOXA9 and HOXA10, activating ID2 in NK/T-cell lines. Mol. Cancer. 9:151. 10.1186/1476-4598-9-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, T., and Yamashita M.. 2008. Initiation and maintenance of Th2 cell identity. Curr. Opin. Immunol. 20:265–271. 10.1016/j.coi.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Nakayama, T., Hirahara K., Onodera A., Endo Y., Hosokawa H., Shinoda K., Tumes D.J., and Okamoto Y.. 2017. Th2 cells in health and disease. Annu. Rev. Immunol. 35:53–84. 10.1146/annurev-immunol-051116-052350 [DOI] [PubMed] [Google Scholar]
- Nimmo, R.A., May G.E., and Enver T.. 2015. Primed and ready: understanding lineage commitment through single cell analysis. Trends Cell Biol. 25:459–467. 10.1016/j.tcb.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Nish, S.A., Zens K.D., Kratchmarov R., Lin W.W., Adams W.C., Chen Y.H., Yen B., Rothman N.J., Bhandoola A., Xue H.H., et al. 2017. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J. Exp. Med. 214:39–47. 10.1084/jem.20161046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra, A. 2000. Commitment factors for T helper cells. Curr. Biol. 10:R492–R494. 10.1016/S0960-9822(00)00556-X [DOI] [PubMed] [Google Scholar]
- O’Shea, J.J., Schwartz D.M., Villarino A.V., Gadina M., McInnes I.B., and Laurence A.. 2015. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu. Rev. Med. 66:311–328. 10.1146/annurev-med-051113-024537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug, K. 2013. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu. Rev. Immunol. 31:675–704. 10.1146/annurev-immunol-032712-095946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera, A., and Nakayama T.. 2015. Epigenetics of T cells regulated by Polycomb/Trithorax molecules. Trends Mol. Med. 21:330–340. 10.1016/j.molmed.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Onodera, A., Yamashita M., Endo Y., Kuwahara M., Tofukuji S., Hosokawa H., Kanai A., Suzuki Y., and Nakayama T.. 2010. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J. Exp. Med. 207:2493–2506. 10.1084/jem.20100760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera, A., Tumes D.J., Watanabe Y., Hirahara K., Kaneda A., Sugiyama F., Suzuki Y., and Nakayama T.. 2015. Spatial interplay between Polycomb and Trithorax complexes controls transcriptional activity in T lymphocytes. Mol. Cell. Biol. 35:3841–3853. 10.1128/MCB.00677-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera, A., Kiuchi M., Kokubo K., Kato M., Ogino T., Horiuchi S., Kanai U., Hirahara K., and Nakayama T.. 2017. Menin controls the memory Th2 cell function by maintaining the epigenetic integrity of Th2 cells. J. Immunol. 199:1153–1162. 10.4049/jimmunol.1602129 [DOI] [PubMed] [Google Scholar]
- Onodera, A., Kokubo K., and Nakayama T.. 2018. Epigenetic and transcriptional regulation in the induction, maintenance, heterogeneity, and recall-response of effector and memory Th2 cells. Front. Immunol. 9:2929. 10.3389/fimmu.2018.02929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, L., Goudot C., Zueva E., Gueguen P., Burgdorf N., Waterfall J.J., Quivy J.P., Almouzni G., and Amigorena S.. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science. 359:177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Piunti, A., and Shilatifard A.. 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 352:aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Quinlan, A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer, F., and Tenen D.G.. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117. 10.1038/nri2024 [DOI] [PubMed] [Google Scholar]
- Rossjohn, J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., and McCluskey J.. 2015. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33:169–200. 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- Schuettengruber, B., Martinez A.M., Iovino N., and Cavalli G.. 2011. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12:799–814. 10.1038/nrm3230 [DOI] [PubMed] [Google Scholar]
- Schuettengruber, B., Bourbon H.M., Di Croce L., and Cavalli G.. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 171:34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Semrau, S., Goldmann J.E., Soumillon M., Mikkelsen T.S., Jaenisch R., and van Oudenaarden A.. 2017. Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat. Commun. 8:1096. 10.1038/s41467-017-01076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, Q.Q., Dai X.X., Jiang J.C., Yu C., Jiang Y., Liu J., Ou X.H., Zhang S.Y., and Fan H.Y.. 2018. CFP1 coordinates histone H3 lysine-4 trimethylation and meiotic cell cycle progression in mouse oocytes. Nat. Commun. 9:3477. 10.1038/s41467-018-05930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, H.Y., Sciumè G., Mikami Y., Guo L., Sun H.W., Brooks S.R., Urban J.F. Jr., Davis F.P., Kanno Y., and O’Shea J.J.. 2016. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell. 165:1120–1133. 10.1016/j.cell.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard, A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81:65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H., Liu T., Manrai A.K., and Liu X.S.. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics. 25(19):2605–2606. 10.1093/bioinformatics/btp479 [DOI] [PubMed] [Google Scholar]
- Shinnakasu, R., Yamashita M., Shinoda K., Endo Y., Hosokawa H., Hasegawa A., Ikemizu S., and Nakayama T.. 2006. Critical YxKxHxxxRP motif in the C-terminal region of GATA3 for its DNA binding and function. J. Immunol. 177:5801–5810. 10.4049/jimmunol.177.9.5801 [DOI] [PubMed] [Google Scholar]
- Smith-Garvin, J.E., Koretzky G.A., and Jordan M.S.. 2009. T cell activation. Annu. Rev. Immunol. 27:591–619. 10.1146/annurev.immunol.021908.132706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser, D.E., Utzschneider D.T., Oberle S.G., Münz C., Romero P., and Zehn D.. 2014. T cell differentiation in chronic infection and cancer: functional adaptation or exhaustion? Nat. Rev. Immunol. 14:768–774. 10.1038/nri3740 [DOI] [PubMed] [Google Scholar]
- Steinke, F.C., Yu S., Zhou X., He B., Yang W., Zhou B., Kawamoto H., Zhu J., Tan K., and Xue H.H.. 2014. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15:646–656. 10.1038/ni.2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober, B.J., Elorbany R., Rhodes K., Krishnan N., Tayeb K., Battle A., and Gilad Y.. 2019. Dynamic genetic regulation of gene expression during cellular differentiation. Science. 364:1287–1290. 10.1126/science.aaw0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Thomson, J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A.R., Deaton A., Andrews R., James K.D., et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 464:1082–1086. 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumes, D.J., Onodera A., Suzuki A., Shinoda K., Endo Y., Iwamura C., Hosokawa H., Koseki H., Tokoyoda K., Suzuki Y., et al. 2013. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 39:819–832. 10.1016/j.immuni.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Tumes, D.J., Papadopoulos M., Endo Y., Onodera A., Hirahara K., and Nakayama T.. 2017. Epigenetic regulation of T-helper cell differentiation, memory, and plasticity in allergic asthma. Immunol. Rev. 278:8–19. 10.1111/imr.12560 [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Onodera A., Kanai U., Ichikawa T., Obata-Ninomiya K., Wada T., Kiuchi M., Iwamura C., Tumes D.J., Shinoda K., et al. 2014. Trithorax complex component Menin controls differentiation and maintenance of T helper 17 cells. Proc. Natl. Acad. Sci. USA. 111:12829–12834. 10.1073/pnas.1321245111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, F.M., Frye M., and Benitah S.A.. 2008. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat. Rev. Cancer. 8:234–242. 10.1038/nrc2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger, L., Ayyash M., Novershtern N., and Hanna J.H.. 2016. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17:155–169. 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- Weinreich, M.A., Takada K., Skon C., Reiner S.L., Jameson S.C., and Hogquist K.A.. 2009. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 31:122–130. 10.1016/j.immuni.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., and Kurachi M.. 2015. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15:486–499. 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- White, K.P., Rifkin S.A., Hurban P., and Hogness D.S.. 1999. Microarray analysis of Drosophila development during metamorphosis. Science. 286:2179–2184. 10.1126/science.286.5447.2179 [DOI] [PubMed] [Google Scholar]
- Wu, M., Wang P.F., Lee J.S., Martin-Brown S., Florens L., Washburn M., and Shilatifard A.. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28:7337–7344. 10.1128/MCB.00976-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, A., Zhang Y., Xu J., Yin T., and Lu X.J.. 2019. T cell dysfunction in cancer immunity and immunotherapy. Front. Immunol. 10:1719. 10.3389/fimmu.2019.01719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, M., Hirahara K., Shinnakasu R., Hosokawa H., Norikane S., Kimura M.Y., Hasegawa A., and Nakayama T.. 2006. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 24:611–622. 10.1016/j.immuni.2006.03.017 [DOI] [PubMed] [Google Scholar]
- Yui, M.A., and Rothenberg E.V.. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14:529–545. 10.1038/nri3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the genes in groups 1–6.
Data Availability Statement
The raw and analyzed data reported in this paper are available under Gene Expression Omnibus accession no. GSE159505.