Abstract
Medicago ruthenica is a well-known high-quality forage due to its good palatability and strong tolerance to drought, cold and saline-alkali stress. Here, the complete chloroplast genome sequence of M. ruthenica was reported. The chloroplast genome is 126,939 bp in length. This chloroplast genome has no inverted repeat (IR) regions, which is very common in the family Fabaceae. The M. ruthenica chloroplast genome encodes 107 genes, including 73 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis result strongly suggested that M. ruthenica is a distinct lineage in Medicago, being sister to highly supported clade composed of three species (M. hybrida, M. papillosa and M. sativa).
Keywords: Medicago ruthenica, chloroplast genome, phylogenetic analysis, Fabaceae
Medicago ruthenica is a perennial plant, which is one of the core species of Section Pialcarpae (Small and Jomphe 1989). It is considered to be a relic species of Tertiary flora in the Palearctic (Campbell et al. 1997). It is widely distributed in Mongolia, Korea, Russia (Siberia, Far East) and high latitude and cold regions in northern China (Hao and Shi 2006). Its habitat is mostly in extremely cold areas and saline-alkali lands with little rain, little snow or no snow cover in winter (Balabaev 1934). It has strong tolerance to drought, cold and saline-alkali stress (Balabaev 1934). At the same time, M. ruthenica is a perennial wild legume species, has high-crude protein content and good palatability (Huang, et al., 2007). For these reasons, M. ruthenica is considered to be a high-quality perennial leguminous forage resource with domestication potential, which is expected to be cultivated and utilized in areas where M. sativa and other alfalfa species cannot survive the winter. However, no studies on the plastome of M. ruthenica have been published. In this study, the complete chloroplast genome of M. ruthenica (Genbank accession number: MN901635) was sequenced on the Illumina NavoSeq Platform (Illumina, San Diego, CA, USA), which will provide genetic and genomic information to promote its ecological restoration and systematics research of Fabaceae.
In this study, M. ruthenica were collected from Mari village, Xunhua County, Haidong City, Qinghai Province, China (35.68°N, 102.35°E). The fresh and young leaves were dried immediately by silica gels. The complete chloroplast genome of M. ruthenica was extracted from the dried leaves (about 0.2 g) with a modified CTAB method. The voucher specimen was kept in Herbarium of the Northwest Institute of Plateau Biology, Northwest Institute of Plateau Biology, Chinese Academy of Sciences (HNWP, XIE2019015). Genome sequencing was performed using the Illumina NovaSeq Platform at Genepioneer Biotechnologies Inc., Nanjing, China. The trimmed reads were mainly assembled by SPAdes v3.10.1 (Bankevich et al. 2012). Then PCGs, rRNAs and tRNAs were annotated by prodigal v2.6.3, hmmer v3.1b2 and aragorn v1.2.38, respectively.
The complete chloroplast genome of M. ruthenica has an atypical chloroplast genome structure with a length of 126,939 bp. This chloroplast genome has no inverted repeat (IR) regions, which is very common in the family Fabaceae. The GC content of the whole chloroplast genome is 34.12%. A total of 107 functional genes were annotated, including 73 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Fourteen of them contain one intron and 1 of them contains two introns.
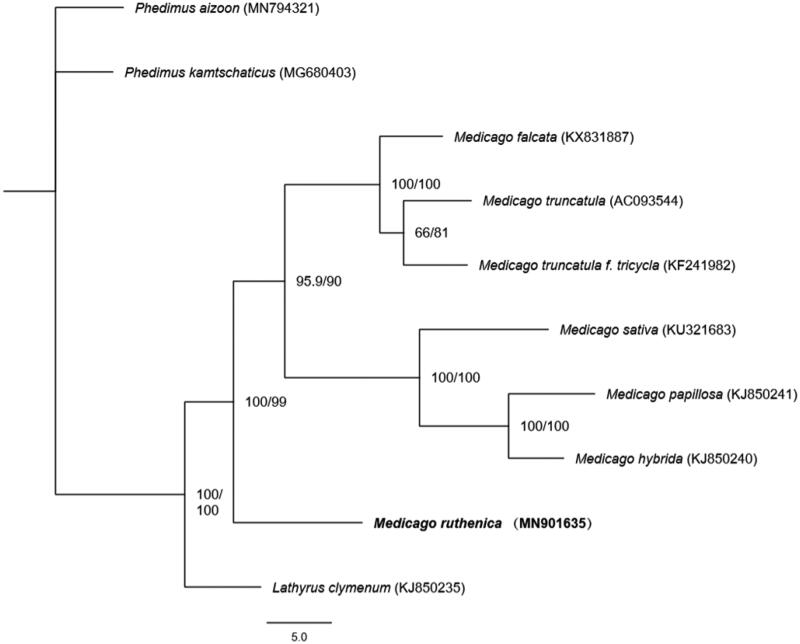
Based on the complete chloroplast genomes assembled here and downloaded from GenBank, phylogenetic relationships of 8 Fabaceae species (7 and 1 from Medicago and Lathyrus, respectively) with two species from Crassulaceae as outgroups were resolved by the mean of maximum likelihood with 1000 bootstrap repeats (model: F81 + F + I + G4). After aligned using MAFFT (Katoh and Standley 2013), the maximum likelihood tree (Figure 1) was built using W-IQ-TREE (Trifinopoulos et al. 2016). The phylogenetic tree showed that M. ruthenica was a distinct lineage in Medicago.
Figure 1.
The maximum likelihood tree based on 10 complete chloroplast genome sequences. Support values written on the branches: SH-aLRT support (%)/ultrafast bootstrap support (%).
Funding Statement
This research was supported by the Natural Science Foundation of Qinghai Province [2017-ZJ-706], National Natural Science Foundation of China [31760622], National International Science and Technology Cooperation Special Project [2015DFG31870].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov, reference number MN901635.
References
- Balabaev GA. 1934. Yellow lucernes of Siberia, Medicago ruthenica (L.) Lebd. and M. platycarpos (L.). Lebd Bull App Bot Genet Plant Breed Serv. 7:13–123. [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikow AS, Lesin VN, Nikloenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5) :455-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell TA, Bao G, Xia ZL.. 1997. Agronomic evaluation of Medicago ruthenica collected in Inner Mongolia. Crop Sci. 37(2):599–604. [Google Scholar]
- Hao JH, Shi FL.. 2006. Study on drought resistance of Melilotoides ruthenica accessions. Chin J Grassland. 28(3):39-43 [Google Scholar]
- Huang YX, Zhou DW, Yue XQ, Yang JY.. 2007. Research progress of Medicago ruthenica. Pratacultural Science. 24(12):34–39. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E, Jomphe M.. 1989. A synopsis of the genus Medicago (Leguminosae). Can J Bot. 67(11):3260–3294. [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ.. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank of National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov, reference number MN901635.