Abstract
Artificial intelligence (AI) and machine learning (ML) in medicine are currently areas of intense exploration, showing potential to automate human tasks and even perform tasks beyond human capabilities. Literacy and understanding of AI/ML methods are becoming increasingly important to researchers and clinicians. The first objective of this review is to provide the novice reader with literacy of AI/ML methods and provide a foundation for how one might conduct an ML study. We provide a technical overview of some of the most commonly used terms, techniques, and challenges in AI/ML studies, with reference to recent studies in cardiac electrophysiology to illustrate key points. The second objective of this review is to use examples from recent literature to discuss how AI and ML are changing clinical practice and research in cardiac electrophysiology, with emphasis on disease detection and diagnosis, prediction of patient outcomes, and novel characterization of disease. The final objective is to highlight important considerations and challenges for appropriate validation, adoption, and deployment of AI technologies into clinical practice.
Keywords: artificial intelligence, atrial fibrillation, cardiac electrophysiology, computers, diagnosis, machine learning
Artificial intelligence (AI) refers to machine-based data processing to achieve objectives that typically require human cognitive function (Table 1). In the modern era, AI has mined dense data and provided the potential to classify complex patterns and novel representations of data beyond direct human interpretation. Machine learning (ML) is a subdiscipline of AI and employs algorithms to learn patterns empirically from data. ML extends the range of traditional statistics because it is able to identify nonlinear relationships and high-order interactions between multiple variables that may be challenging for traditional statistics. Deep learning (DL) has emerged as a powerful ML approach that leverages large datasets and recent increases in computational power to make efficient decisions on complex data. The successes of ML and DL in diverse disciplines, ranging from language processing, gaming, computer vision, engineering, industrial, and scientific arenas, has led to an increasing public awareness of the promise of AI across multiple facets of life.
Table 1.
Glossary of Commonly Used Terms in Artificial Intelligence and Machine Learning Studies
| Term | Definition |
|---|---|
| AI | Machine-based data processing to achieve objectives that typically require human cognitive function. |
| ML | A subdiscipline of AI referring to the algorithms and statistical models used to learn patterns from data. |
| Supervised machine learning | Development of a model that can make successful predictions on new data, by first training the model to link input data to labeled outputs, then validating and testing the model using independent input data. |
| Unsupervised machine learning | Identification of natural patterns within complex input data, without training the model to link input data to labeled outputs. Does not require the input data to have corresponding labels, nor separate training and testing data. |
| Artificial neural network | Computational model loosely based on the neuron connections in biological neural networks. The network consists of layers of nodes that serve as artificial neurons, and connections between layers to serve as synapses. Any node within a layer is connected to every node in neighboring layers. |
| DL | ML built from artificial neural networks with many hidden layers. In contrast to artificial neural networks, DL does not require nodes from neighboring layers to be fully interconnected. Convolutional neural networks, which use convolutional filters to partially connect neighboring layers, are the most widely used DL networks, but multiple types of DL networks exist. |
| Feature | Quantifiable property of the input data. |
| Training | Fitting an ML model by learning the relationships between the features and the data labels. |
| Testing | Evaluating a trained ML model on new data. |
| Cross-validation | An unbiased technique used to evaluate the effect of different ML model parameters during training to develop an optimized model. |
| AUC | A common metric to evaluate the performance of ML classifiers, referring to area under the receiver operating characteristic curve. AUC signifies how well the classifier can distinguish between 2 classes, with 0.5 indicating random classification and 1 indicating perfect classification. Generally, interpretation of AUC values may be: <0.70 poor, ≥0.70 fair, ≥0.80 good, ≥0.90 excellent. |
| Overfitting | When a model learns noise and random fluctuations as relationships in the training data. A principal cause of poor performance in ML models. |
| Cluster analysis | An unsupervised ML method to identify partitions within the data that share similarity. |
| Dimensionality reduction | A method to reduce high-dimensional data into lower-dimensional representations while preserving relevant variation and structure present in the full-dimensional data. |
AI indicates artificial intelligence; AUC, area under the curve; DL, deep learning; and ML, machine learning.
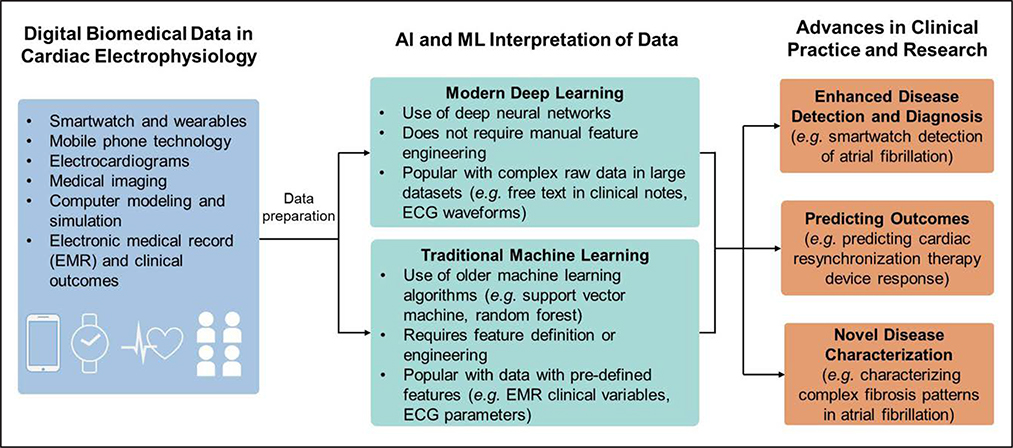
AI is not a new concept in cardiac electrophysiology with automated ECG interpretation existing since the 1970s.1 However, the relatively recent development of large electronic databases in which data have been labeled by experts, innovations in algorithms, software tools, and hardware capabilities are rapidly transforming the role of AI in cardiovascular imaging2 and cardiac electrophysiology. AI tools have shown promise in automating and assisting disease diagnosis, and tools are now being developed to enhance prediction of disease prognosis and response to therapeutics and provide novel characterization of health and disease (Figure 1).
Figure 1. Overview of artificial intelligence and machine learning in cardiac electrophysiology.
A broad overview of how increasing quantities of diverse digital data in cardiac electrophysiology are being interpreted by artificial intelligence methods to generate advances in clinical practice and research. EMR indicates electronic medical record.
We structure this review with 3 objectives. (1) We first provide novice readers with literacy of the technical concepts in AI and ML, as well as a basic foundation to conduct ML studies. Throughout this section, we provide brief relevant examples from cardiac electrophysiology to illustrate technical concepts. (2) We then provide current perspective of how recent AI studies have influenced the direction of clinical practice and research in cardiac electrophysiology, with an emphasis on clinical results rather than detailed technical methodology. (3) Lastly, we briefly discuss important considerations for adopting AI technologies into clinical practice for cardiac electrophysiology.
TECHNICAL OVERVIEW OF COMMON AI METHODS
Representing Data as Features
AI interpretation of data requires that input data are structured as feature vectors. A feature refers to a quantifiable property of the input data. Features are assembled into feature vectors to mathematically represent the input data and are subsequently computationally processed by ML algorithms. Features are inclusive of a wide variety of data. In their simplest form, features are minimally processed from the original data, such as clinical variables stored in the electronic medical record (EMR). In traditional ML studies that do not use DL, handcrafted features are engineered by humans to measure specific, relevant attributes of the data. For example, in early attempts to automate ECG interpretation, typical features consisted of coefficients of mathematical representations of ECG waveform morphology.3–5 Meanwhile, modern DL approaches, which are discussed in further detail in the DL section, are capable of automatically computing and selecting relevant features from raw input data, such as in recent studies using DL to interpret 12-lead ECG waveforms.6–10
Supervised ML
The objective of supervised ML is to train a model to relate input data, represented by feature vectors, to labeled (known) outcomes of interest. Classification refers to predicting outcome labels on new data. This can be done in a binary manner (eg, identifying normal versus abnormal ECG) or in a multiclass manner (eg, classifying the type of rhythm on ECG; normal versus atrial fibrillation versus atrioventricular block, etc). Regression refers to predicting a continuous outcome (eg, predicting the degree of ejection fraction improvement after heart failure intervention).
In supervised ML, training data consisting of input features and corresponding data labels (outputs) are provided to an ML algorithm. The ML algorithm then fits the ML model by learning the relationships between the features and the data labels, a process referred to as training. Once the model is trained, it is able to make predictions from new data, a process referred to as testing.
Traditional Supervised ML
Traditional supervised ML, which we use to refer to non-DL techniques, has existed for several decades. A variety of supervised ML algorithms can identify relationships between input data and output labels in linear and nonlinear manners. An overview of some common algorithms is provided in Table 2. In traditional supervised ML, beyond algorithm selection, the most critical component to model performance often lies in feature engineering and selection to represent the input data in the most relevant manner.
Table 2.
Overview of Common Supervised Machine Learning Classification Algorithms
| Algorithm | Description | Advantages and Disadvantages | Suitable Data in Cardiac Electrophysiology |
|---|---|---|---|
| Deep learning | Mimics human neuronal structure with many processing layers | Represents state-of-the-art performance with raw input data in complex tasks and does not require any feature engineering but often requires extremely large datasets, intensive computational power, significant processing time, and algorithms are difficult to interpret | Raw 12-lead ECG, imaging (CT, MRI, echocardiography), clinical text, or other diagnostic and monitoring data (telemetry, Holter/wearables, electrograms, electrophysiological studies) |
| Traditional supervised ML algorithms | |||
| Logistic regression | Linear combinations of log-odds | Transparent and fast but performs poorly with large numbers of variables and does not automatically capture interactions | Well-selected features obtained from the electronic medical record or manually engineered from processed data (eg, features extracted from ECG signal processing, specific measurements made on images, etc) |
| Support vector machine | Identifies hyperplane that separates classes, can use a linear or nonlinear kernel | Fast can be flexible with kernel adjustment but is difficult to interpret and features may need normalization and scaling | |
| Naïve Bayes | Bayes’ theorem of conditional probabilities | Fast, scalable, does not require large amounts of data, but is not directly interpretable and assumes independence between variables | |
| Random forest | Large ensembles of decision trees | Resistant to noise and overfitting even with large amounts of variables, flexible with continuous and categorical variables, capable of capturing variable interactions | High quantities of features obtained from the electronic medical record or manually engineered from processed diagnostic data (ECG, imaging, etc) |
CT indicates computed tomography; ML, machine learning; and MRI, magnetic resonance imaging.
A study using surface ECG to predict abnormal myocardial relaxation illustrates the basic use of traditional supervised ML.11 The ECG was the first signal processed from the time-voltage domain into spectrograms (energy spectra in the time-frequency domain) using the wavelet transform. Handcrafted features were extracted from these spectrograms. An ML model was trained using a random forest algorithm to use input features (ECG spectrogram metrics) to predict corresponding data labels (presence of impaired myocardial relaxation).
Key Points in Traditional Supervised ML
Supervised ML algorithms are capable of learning linear and nonlinear relationships from labeled data.
Model success is most dependent on successful feature engineering.
Deep Learning
DL is a powerful subtype of ML and has shown state-of-the-art performance in speech recognition, computer vision and image/video processing, game-playing, and medical diagnosis. When compared with traditional supervised ML, the ultimate strength of DL is that it is a powerful, flexible way of representing complex raw input data that does not require manual feature engineering. For example, in the problem of automated ECG interpretation, early traditional supervised ML studies relied on human-defined ECG features. Meanwhile, a modern DL model learned patterns within raw ECGs to diagnose sinus rhythm and multiple other arrhythmias, with performance similar to cardiologists.12
DL is predated by and built from artificial neural networks, which are computational systems modeled after biological neuronal connections. Layers of nodes (processing elements to represent neurons) manipulate and transform the input data to create a data representation, which ultimately links the input data with outputs. Each node from one layer is connected to each node from the neighboring layers, termed fully connected layers. Layers between the input and output layer are termed hidden layers. The strength of connections between nodes from different layers, which represent synapses, are quantified by weights. As learning occurs, weights are iteratively adjusted to move network error (the cost function) in the direction of the steepest negative slope, a process termed gradient descent. Gradient descent ultimately identifies node connection weights that minimize the cost function, thus maximizing the network’s accuracy. Figure 2A depicts an artificial neural network from an early study predicting cardiovascular mortality from ECG features.13
Figure 2. Architectures of an artificial neural network vs deep learning in ECG interpretation.
A, An example of an artificial neural network used to predict whether or not a patient will experience cardiovascular mortality, using 4 clinical features and 132 resting ECG features (intervals and amplitudes of various ECG segments). These create a 1×136 feature vector input to the neural network, represented by neurons (x1, x2, x3,…x136). The input neurons are then connected to a single fully connected hidden layer of 70 neurons (h1, h2, h3,…h70), and then ultimately connected to the output node (y), which yields a prediction score of cardiovascular mortality. The black lines between nodes represent weights, which are iteratively adjusted during the training process to minimize output prediction error. B, An example of a deep learning convolutional neural network based on the network used to predict whether or not a patient has left ventricular dysfunction from the waveforms of a 10-s 12-lead ECG. The input is the entire 12-lead ECG signal, formatted as a 12×1024 sample matrix. This network first learns temporal features within each lead, by extracting feature maps via 6 iterations of 1-dimensional convolution in the temporal axis followed by 1-dimensional pooling. Next, the network learns how the temporal features are distributed across leads by spatial feature learning via convolution across the 12 ECG leads. The resulting feature maps are flattened and passed to 2 fully connected layers (ha,1, ha,2, ha,3,…ha,64) and (hb,1, hb,2, hb,3,…hb,32), which used the learned temporal and spatial features to classify whether or not the patient has left ventricular dysfunction, as predicted in output node y.
Compared to artificial neural networks with only a small number of layers, DL is defined by the use of neural networks with many successive hidden layers before a final output is generated. Another important distinction is that DL does not require the nodes from neighboring layers to be fully connected and offers different types of neural network architectures to manipulate data in varying ways. The ensuing processing complexity has required contemporary advances in computational software and hardware.
DL most commonly involves deep convolutional neural networks (CNNs), which typically process raw input data (eg, images, ECGs) to predict a categorical output. The functional building blocks of CNNs are convolutional layers. Each convolutional layer uses a set of convolution filters, which are mathematical operations that detect data features (eg, straight edges, curves in images), to construct feature maps. After each convolutional layer, a pooling layer subsamples the feature maps. Repeated layers of convolution and pooling learn higher-level features from the previous feature maps, creating hierarchical representations of the data (eg, learning how edges and curves construct more complex shapes). CNNs then employ fully connected layers at the end of the network to generate global data classifications from the feature representations learned from the convolutional layers. CNNs have become the primary network architecture to make advanced predictions from ECGs in recent studies.6–10,12,14,15 Figure 2B depicts one of these modern DL architectures used to predict left ventricular dysfunction from ECG.7
Fully convolutional networks are CNNs that consist only of convolutional layers and lack the fully connected layers at the end. Instead of making a categorical prediction from the input data, fully convolutional networks are able to use the learned features to make pixel-level predictions on the input image, which becomes especially useful for image segmentation. As an example, dual fully convolutional networks have been used to automatically segment the left atrial epicardium and endocardium in late gadolinium-enhanced magnetic resonance images in patients with atrial fibrillation (AF).16
Although CNNs are typically used to process data from a single point in time, recurrent neural networks are built to analyze the temporal sequence of data and are often useful in speech or text analysis. In recurrent layers, each neuron’s output is adjusted at each time step. A recent study used recurrent layers after a series of convolutional layers to analyze temporal patterns of 5-second ECG segments within longer ECG recordings, and subsequently classify AF from other rhythms in single-lead ECG recordings of variable length.14
Generative adversarial networks17 are DL architectures using 2 networks to compete with each other. A generator network creates new data, and a discriminator network assesses whether the generated data is similar to training data, with the intention of creating synthetic data that mimics the original data. Generative adversarial networks are well known in digital artwork and image creation. For example, a generative adversarial network generated artificial ECG data to address data scarcity in medical data analysis studies.18 Such use of generative adversarial networks has simultaneously led to concerns of developing realistic fraudulent data.
A final important concept in DL is transfer learning. Developing a strong DL model from scratch requires a very large, labeled training data set, and may take several days to train. However, in transfer learning, layers and weights (and thus learned data features) from an existing trained model are used as the starting point for a new model with a different, but related, task. The new model will already have access to the low-level features learned from the pretrained model, and the new model will then focus on relating these features to the new task. In image classification tasks, seminal DL models trained from large scale databases, such as AlexNet,19 VGG Net,20 GoogLeNet,21 and ResNet,22 among others, are available to use for transfer learning. In one instance, the authors used transfer learning from AlexNet to classify ECGs from the Massachusetts Institute of Technology–Beth Israel Hospital arrhythmia database.23 However, to date, most DL models in cardiac electrophysiology have been developed in-house, which is often a result of institutional ownership of the data required to develop the models. Transfer learning presents an intriguing opportunity to diversify the clinical applications of existing pretrained DL models. For example, consider the features learned by CNN trained to predict left ventricular dysfunction from ECG,7 and applying transfer learning to develop a new network to predict subtle signs of ischemia on ECG.
Key Points in DL
DL is well-suited for interpreting complex raw data and does not require manual feature engineering.
DL most commonly involves the use of CNNs for classification problems, but many other types of DL exist.
Model Development and Evaluation
In supervised ML, it is difficult to predict what type(s) of model will perform best, so model development and optimization are typically empirical. In traditional supervised ML, model development and optimization consists of selecting a learning algorithm, an optimal feature set, and tuning hyperparameters of the learning algorithm. In DL, model optimization includes testing different neural network architectures and tuning hyperparameters within the architectures. Some important hyperparameters in the DL training process are learning rate, batch size, and epoch size. Hyperparameters of the architecture itself, such as number of nodes, layers, and types of layers, activation functions, should also be considered. Then, hyperparameters within the layers can be adjusted (eg, filter sizes and number of filters in convolutional layers).
There are several ways to evaluate model performance throughout model development. Classification algorithms produce a continuous value rather than a binary or discrete output. Receiver operating characteristic curve analysis is used to compute area under the curve (AUC) based on classifier output. At specific operating points on the receiver operating characteristic curve, model accuracy, sensitivity (also called recall), specificity, positive predictive value (also called precision), negative predictive value, and the F1 score (2×precision*recall/[precision+recall]) can also quantify performance. Confusion matrices specify where classification errors occur. Regression performance can be assessed with mean absolute error, root mean-squared error, or R2 goodness of fit (Table 1).
Careful data partitioning is required during model development and evaluation. Data used to evaluate model adjustments should never be incorporated in the training process of that model. One common scheme for unbiased data partitioning is to use the training set to perform model adjustments over successive iterations of k-fold cross validation11,24–26 and evaluate the optimal model on a dedicated testing set. An alternative method uses 3 partitions: a training set, a hold-out validation set, and a testing set. The model is fit using data from the training set, and the effect of model adjustments are evaluated on the validation set. The optimal adjustments are then used to develop a final model that can undergo unbiased evaluation on the testing set. Compared with cross-validation, this approach reduces computational time but does not maximize the amount of data that may be used for model development. Given reduced computational burden, a validation set is often used in DL studies.7,8,10,12
Key Points in Model Development and Optimization
Model development and optimization is performed empirically and involves algorithm selection and hyperparameter adjustment.
Careful data partitioning into training, validation, and testing sets optimizes evaluation of model performance without bias.
Challenges in Conducting Supervised ML and DL Studies
The first limitation of supervised ML is that all data must be annotated with labels, which can be laborious, may limit the amount of data available for analysis, and may necessitate arbitrary dichotomization of labels that are intrinsically continuous. In cardiac electrophysiology studies, this may require annotating ECGs, segmenting images, and collecting clinical outcomes. Approaches have been proposed to reduce the need for labeling, such as heuristic pretraining to classify AF from smartwatch photoplethysmography tracings.27 Such strategies should be developed to enable the analysis of very large clinical data which would otherwise be difficult or impossible to label categorically. In addition to needing labeled data, the quantity of data should also be maximized. DL in particular often requires large annotated datasets to achieve strong performance (eg, 91 232 annotated ECGs trained a CNN to classify ECG rhythm).12 This may also require intense computational processing and advanced hardware capabilities through graphics processing units. As mentioned above, transfer learning can also be used to address this challenge in DL.23
Once data have been collected, a common issue in classification tasks is imbalanced class proportions. For example, in the study to automate ECG rhythm classification,12 some arrhythmias such as atrioventricular block were rarely present in comparison to sinus rhythm. Thus, the model could maintain a high overall accuracy by labeling all cases of atrioventricular block as sinus rhythm, without ever learning to detect atrioventricular block. Evaluating model performance with metrics, such as AUC and F1 score, is more helpful in this situation. The dataset can also be resampled to be more balanced by removing negative cases or replicating positive cases. The authors in this study oversampled rare rhythms in training to ensure that the model learned to identify these rhythms.12 Penalization for false negatives could also be introduced into a model, such as penalizing failure to detect life-threatening arrhythmias.
Another critical challenge in supervised ML is overfitting, which occurs when the model learns noise and random fluctuations as false relationships in the training data, resulting in high performance on the training data with lower performance on the testing data. Common causes of overfitting include insufficient training data, excessive noise in training data, or excessive model complexity. These can be combated by improving the training data (eg, increasing the quantity, diversity, or quality of data) or reducing model complexity (reducing model variance).
In traditional ML approaches, model complexity is often caused by high-dimensional feature spaces, requiring a large quantity of training data to adequately represent the variability in feature combinations.28 The optimal number of features is problem-dependent, but a 10:1 ratio of data samples to features is a general starting point. Regularization techniques penalize the model based on magnitude of feature weights to emphasize small feature weights and reduce model variance. The feature space can also be reduced before training by feature selection.29 As an alternative, dimensionality reduction, discussed later, can reduce the data requirement and problem complexity, albeit at the cost of reduced interpretability of lower-dimensional feature sets.
Several techniques in DL can reduce model complexity and resist overfitting. Networks can be simplified by reducing the number of nodes or layers. Weight regularization within the network helps constrain the magnitude of feature weights. Adding a dropout layer to the network drops outputs from a random sample of nodes from the previous layer, enforcing the network not to rely on a few nodes, and instead learn features from other nodes to make more robust decisions. Lastly, in DL, early stopping can be used to stop training a network once the error on the validation set rises, prohibiting the network from learning more noise. Comparable performance in training and validation datasets helps confirm that overfitting is avoided.
Finally, interpreting supervised ML results requires thoughtful consideration of bias. If the study data is poorly reflective of other datasets, the model will generalize poorly to external datasets. This effect can be considered in study design. For example, the ECG rhythm classification model was validated on an external set of ECGs from PhysioNet to assess whether the model was generalizable to other ECG data sources.12
Key Points in Challenges of Supervised ML
Large quantities of annotated data are needed.
Recognition and mitigation of overfitting is imperative.
Brief Guide to Conducting a Supervised ML Study
Decide on classification or regression
Prepare the input data and annotate the desired corresponding outcomes
- Decide on a traditional ML approach versus DL
- If using traditional ML, engineer and extract features
- If using DL, format the input data to be compatible with a DL architecture
- Partition data
- Training, validation, and testing, or
- Training (cross-validation), and testing
Optimize model parameters on the validation set or with cross-validation
- Test the optimal model
- Train the model on the training set, evaluate on the testing set
- Calculate performance metrics
Continue to externally validate the model on additional datasets
Unsupervised ML
Unsupervised ML is relatively underdeveloped compared with supervised ML but offers intriguing possibilities. In contrast to supervised ML, unsupervised ML does not train a model to predict labels from input data. Unsupervised ML instead quantifies natural patterns within input data, blind to the labels of interest. Parsing out these patterns reveals underlying structure to complex data which may help to identify relevant subgroups. For example, unsupervised ML of clinical data combined with complex echocardiographic left ventricular strain and volume traces was used to identify different phenotypes of heart failure patients in the MADIT-CRT trial (Multicenter Automatic Defibrillator Implantation Trial With Cardiac Resynchronization Therapy), and the resulting phenotype groups experienced significantly different treatment effect from cardiac resynchronization therapy (CRT).30
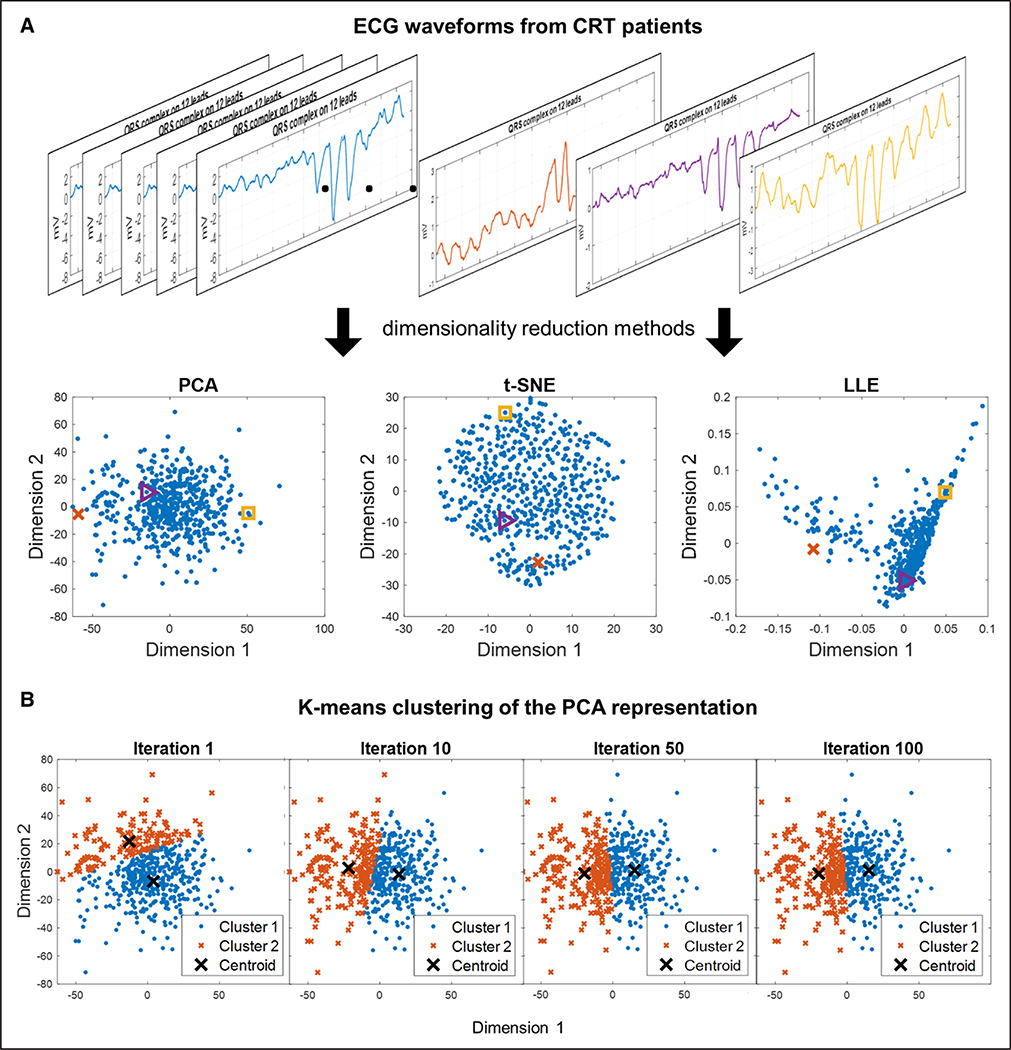
Cluster analysis is an unsupervised technique often used to identify subgroups from complex input data. Hierarchical agglomerative clustering is a popular clustering approach that identifies groups of data that are similar to each other. It starts by identifying small clusters of similarity, then sequentially creating larger clusters by merging small clusters in a bottom-up approach. Hierarchical clustering results are typically presented with dendrograms and heat maps, which can help provide visual interpretation of high-dimensional data. An example of this approach is provided by authors who used hierarchical clustering of large quantities of clinical variables identified novel clinical phenotypes of AF.31 As an alternative to hierarchical clustering, k-means clustering identifies a prespecified number k clusters within the data such that every data point belonging to a given cluster is closer to that cluster’s centroid than all other cluster centroids (Figure 3). For example, k-means clustering was used to identify 4 different heart failure phenotype groups in the aforementioned analysis of the MADIT-CRT trial.30
Figure 3. Unsupervised machine learning: dimensionality reduction and k-means clustering.
Demonstration of an unsupervised machine learning approach to identify 2 subgroups of cardiac resynchronization therapy (CRT) patients based on ECG QRS complex waveforms.75 A, Visualization of different dimensionality reduction techniques to reduce ECG QRS waveforms from 539 CRT patients into data points in a 2-dimensional representation. Principal components analysis (PCA) is a common linear dimensionality reduction method. Other pictured techniques are nonlinear dimensionality reduction techniques: t-distributed stochastic neighbor embedding (t-SNE), and locally linear embedding (LLE). Colors of the example ECG waveforms and corresponding colors in the scatterplots indicate where each example waveform is projected in the 2-dimensional representations. B, Demonstration of k-means clustering with k=2 on the PCA representation of the ECG waveforms to create 2 clusters. The k-means algorithm is as follows: (1) k centroids are created at random locations, (2) each data point is then assigned to the nearest centroid, (3) the locations of the centroids are then updated to represent the mean location of the data points assigned to the centroid. Steps 2 and 3 are repeated over many iterations until the centroids no longer update. The centroid location and cluster assignment change over several iterations until reaching convergence.
Dimensionality reduction is an unsupervised technique that yields representations of input data that may facilitate clustering (Figure 3). High-dimensional data is reduced into lower-dimensional representations while preserving the relevant variation and structure within the full-dimensional data. An intuitive example of dimensionality reduction is representing Earth (3-dimensional) using a map (2-dimensional). Many linear and nonlinear dimensionality reduction algorithms exist. In the previously mentioned example identifying heart failure phenogroups of patients in MADIT-CRT, multikernel dimensionality reduction simplified a 1682-dimensional feature vector of complex echocardiography traces and clinical parameters into a 2-dimensional representation to facilitate k-means clustering.30
Challenges in Conducting Unsupervised ML Studies
Like supervised ML, unsupervised ML studies may require extensive processing to prepare useful input data. The patterns that unsupervised ML uncovers also may not be the patterns that are best related to the outcome of interest, and a supervised ML approach may be more optimal. When using cluster analysis, it can be nebulous to decide how many clusters are needed. For example, although 4 heart failure phenogroups were identified from patients in the MADIT-CRT study,30 the authors ran their algorithm using 3 to 8 clusters and chose the cluster number that maximized statistical significance of the treatment effect among the clusters. Additionally, some nonlinear dimensionality reduction and clustering processes use distributions specific to the analyzed data which cannot be directly applied to new data.
Key Points in Unsupervised ML
Unsupervised ML does not involve training a model but instead finds underlying structure in data.
Common strategies are dimensionality reduction to simplify complex data or cluster analysis to identify relevant groups.
Unsupervised ML is relatively underdeveloped compared with supervised ML.
Brief Guide to Designing an Unsupervised ML Study
Choose clinical outcome(s) of interest (eg, event-free survival).
Choose how to represent the input data with a feature vector (eg, handcrafted features, raw signals, or dimensionality reduction methods to alter the data representation).
Cluster analysis on chosen input data representation to identify subgroups.
Compare outcomes in the subgroups identified by the cluster analysis.
Interpret the input data representation and differences between subgroups.
Continue to assess if unsupervised learning representations and clusters generalize to new cohorts.
Appropriate Use of ML
ML and DL approaches offer great potential, but to most appropriately use ML, it is useful to consider (1) if ML/DL yields better results than simpler traditional regression, and (2) when reduced interpretability in ML/DL approaches is and is not a concern.
In less complex problems, advanced ML algorithms often do not enhance performance beyond traditional regression with well-selected variables, a finding suggested in predicting CRT response from clinical variables,24 as well as predicting risk of AF.32 Advanced ML/DL is also unlikely to overcome limitations of the data itself, such as small datasets or poor outcome acquisition, and in such scenarios, traditional regression is a better-suited approach. Advanced ML is most appropriate when studying large datasets with large numbers of features where complex pattern recognition may be important, such as interpreting subtle patterns in ECG waveforms from hundreds of thousands of patients.
Another major limitation in ML, and especially DL, is its black box nature: difficulty in interpreting the contribution of features to model output.33 Poor interpretability is generally acceptable in performance-based tasks without immediate clinical ramifications, such as automated image segmentation. However, interpretability is especially important when trusting an AI model for clinical decision-making. For example, a clinician may feel uncomfortable with diagnosing a patient with disease detected by a DL ECG model that did not provide any interpretable rationale for that classification. This concern becomes even more important when considering that DL models are susceptible to adversarial attacks to input data that cause erroneous model output. A recent study demonstrated the ability to create adversarial examples, which are small perturbations imperceptible to humans, to ECG waveforms that were provided to a DL ECG rhythm classification model.34 Despite being invisible to human experts, these adversarial examples reduced the DL ECG rhythm classification accuracy from 88% to 26%.
One solution to poor interpretability is to develop inherently interpretable models,33 via cleverly designed features that reflect and quantify relevant biology. Another alternative is the development of glass box models, which return some explainability to ML model output. In traditional ML, some algorithms can provide variable importance metrics to assist interpreting the model. Several approaches are also being investigated to better interpret DL models.35 For example, a DL model to identify cardiac rhythm devices from chest radiographs was also able to highlight relevant aspects of the image that might otherwise pass unnoticed to help educate the observer.36
A recent study illustrates prudent use of ML models with different levels of interpretability when attempting to automate disease diagnosis from ECG.26 First, the investigators used uninterpretable DL to automatically segment the ECG waveforms, a purely performance-based task. However, they performed disease detection using an interpretable supervised ML classifier with variable importance metrics. Unlike an uninterpretable DL ECG model, this classifier could reveal which segments of the ECG waveform were most important in making the classification, which could be reexamined and corroborated by a clinician. An interpretable framework is important to consider when using ML to generate clinical decisions or make new disease insights, and perhaps human interpretation augmented by ML model output may ultimately perform best.
Key Points in Appropriate Use of ML
In many scenarios, simpler models perform as well as complex models.
Implications of poor model interpretability should be considered.
AI and ML in Clinical Cardiac Electrophysiology: Where Are We Now?
To provide pragmatic perspective, we discuss recent applications of AI to transform clinical cardiac electrophysiology and the implications this has on disease diagnosis, clinical outcomes prediction models, and novel characterization of disease. A summary of recent studies is provided in Table in the Data Supplement.
Disease Detection and Diagnosis
The development of mobile and wearable technology and AI is rapidly altering the landscape of disease detection and diagnosis in cardiac electrophysiology.
Mobile Technology to Detect Arrhythmia
Wearable photoplethysmographic sensors have transformed possibilities for AF screening by enabling long-term, passive assessment of pulse rate and regularity to detect an irregular pulse consistent with AF. In the Apple Heart Study,37 the photoplethysmographic monitoring algorithm of the Apple Watch was evaluated in 419 297 participants, with 2161 participants receiving an irregular pulse notification when 5 out of 6 photoplethysmographic tachograms suggest AF. In the 450 participants who subsequently wore an external heart monitor for ≈1 week, AF was identified in 34% of participants, and positive predictive value between the photoplethysmographic and a concurrent external monitor was 84%.37 In a similar study in China,38 AF screening using photoplethysmographic monitoring technology in Huawei wristband and wristwatches was assessed in 187 912 individuals. Two hundred sixty-two individuals received notification for potential AF and had effective follow-up with clinical evaluation and ECG monitoring. Eighty-seven percent of these individuals were confirmed to have AF, and positive predictive value of the photoplethysmographic signals was 92%.
Beyond photoplethysmographic pulse detection, Kardiaband and Apple Watch Series 4–5 are cleared by the Food and Drug Administration to use wearable ECG recording capabilities for on-demand ECG confirmation of photoplethysmographic-based detection of AF. On Apple Watch Series 4–5, irregular rhythm detection on photoplethysmographic prompts the user to record a single-lead ECG using a sensor of the digital crown. The Kardiaband algorithm uses photoplethysmographic and pedometer sensors on an Apple Watch Series 2 or 3 to continually assess heart rate and activity level. Discordances in these measures prompt the wearer to record a modified lead I ECG by placing their thumb on the proprietary sensor embedded in the watchband. The performance of the algorithm was validated on 24 patients with implantable cardiac monitors and a history of paroxysmal AF. Episode sensitivity for AF episodes ≥1 hour was 97.5% with a duration sensitivity of 97.7%.39 The Kardiaband-coupled app for ECG interpretation algorithm was assessed separately in 100 patients undergoing cardioversion. Compared with ECG, the Kardiaband app for AF detection algorithm interpreted AF with 93% sensitivity, 84% specificity, and a K coefficient of 0.77.40 False positives or unclassified readings for both Kardiaband and Apple Watch Series 4 are seen most frequently in the presence of bundle branch blocks, frequent ectopy, junctional rhythm, or rates outside the Food and Drug Administration–mandated range of programmed range of 50 to 150 bpm for Apple Watch and 50 to 100 BPM for Kardiaband.
In addition to passive photoplethysmographic recordings on smartwatches, active assessment of contact-free facial and fingertip photoplethysmographic measurements using smartphone cameras has also shown potential for AF screening and diagnosis across a series of studies.41–45 A meta-analysis of studies to date found that AF was diagnosed with a combined sensitivity and specificity of 94% and 96%.46
Expanding the Use of the 12-Lead ECG
The obvious application of AI to ECG interpretation is the rapid, reliable, and automated determination of ECG diagnosis. In this realm, automated cardiologist-level classification of 12 different rhythms has been obtained through DL of single-lead ECGs.12 However, AI interpretation of the ECG can also confer information about disease not typically diagnosed on ECG. These 2 capabilities represent complementary application of AI to medicine - scaling our current workflow and insuring quality but also adding value to a routine medical test.
A recent series of studies have investigated DL interpretation of raw ECG waveforms to expand the utility of the ECG in several arenas. One example is determining serum potassium. Extreme potassium concentration perturbations have well-described ECG manifestations, but more subtle potassium changes may be detectable by DL of the ECG and a CNN identified hyperkalemia with an AUC of 0.85 to 0.88.10 This approach may have implications for outpatient titration of medications that disrupt potassium homeostasis or renal function, or for altering dialysis schedules. Such work may yield a bloodless assay for serum electrolyte concentrations.
Similarly, although myocardial diseases causing poor function are often detectable on the ECG, the ECG itself is not a good screening test for asymptomatic left ventricular dysfunction—a condition affecting up to 2% to 5% of the adult population. However, a CNN trained using ECG and echocardiography pairs could reliably detect left ventricular dysfunction (AUC, 0.93).7 This network performed well in a subsequent validation study performed at the same institution,47 and is currently being tested in a prospective, cluster-randomized clinical trial (EAGLE [ECG AI-Guided Screening for Low Ejection Fraction], URL: https://www.clinicaltrials.gov; Unique identifier: NCT04000087).48
DL of sinus rhythm ECGs also identified patients with paroxysmal AF. A CNN was trained to recognize ECG patterns to diagnose AF while in sinus rhythm in those patients who had both rhythms at different times.9 The network performed well (AUC, 0.87) and when multiple sinus rhythm ECGs were considered together, the model improved to an AUC of 0.90. This approach may help identify patients who benefit from longitudinal screening for AF or patients who may benefit from anticoagulation after a stroke of undetermined source.
Most recently, a CNN used ECGs to predict sex (AUC, 0.97) and estimate age (average error of 6.9±5.6 years).6 Patients whose CNN-predicted age exceeded their actual age by >7 years had a higher proportion of low ejection fraction, hypertension, and coronary disease, suggesting that the CNN interpretation of age based on the ECG may be a mechanism to measure overall health. This finding was further bolstered by a study using a CNN to predict 1-year mortality from ECGs (AUC, 0.88), even among ECGs that were interpreted to be normal by physicians (AUC, 0.85).49
Predictive and Prognostic Models for Response to Therapy
The Precision Medicine Initiative in 2015 raised expectations for understanding individual variation in prevention and treatment of disease. The Centers for Medicare and Medicaid Services mandated the use of evidence-based patient decision aids in primary prevention implantable cardioverter-defibrillator implant50 and left atrial appendage closure51 to foster shared decision-making with patients. Meeting these objectives necessitates models tailored to individual patients and methods to present these models. ML algorithms that use diverse data from the EMR may become central to this process.
Improving CRT Response Prediction
Several recent studies demonstrated early-stage attempts at developing models to answer this need in CRT. One study25 developed a random forest model to predict a composite end point of heart failure event or death following CRT using 45 commonly available baseline variables. The model differentiated outcomes (AUC, 0.74) better than current clinical discriminators of bundle branch block morphology and QRS duration. Another study24 predicted echocardiographic response to CRT in a retrospective cohort from 2 institutions. The authors evaluated a variety of ML algorithms and clinical variable sets and found that an ML model created with a naive Bayes classifier and only 9 variables performed better than other models with wider feature sets. The ML model outperformed current guidelines in predicting response and improved discrimination of event-free survival, although improvement was modest—AUC for ML model 0.70 versus 0.65 for guidelines. A third study52 sought to predict 1- to 5-year all-cause mortality in CRT patients using 33 clinical variables to train random forest classifiers to generate the SEMMELWEIS-CRT score, and again found that ML classifiers predicted mortality (AUC range, 0.77–0.80) better than preexisting clinical risk scores (AUC range, 0.53–0.74). The final CRT study sought to predict CRT response using clinical variables in combination with 2-word features extracted from clinical free note text via natural language processing.53 A model trained using a gradient boosting classifier predicted patients with reduced CRT benefit (defined as <0% improvement in left ventricular ejection fraction or death by 18 months) with an AUC of 0.75 and was able to identify 26% of the patient population who experienced reduced CRT benefit with a precision of 0.79 and accuracy of 0.65. These studies begin to build a framework for using ML models to predict patient outcomes, but significant work remains in clinical adoption, which we discuss in the Validation and Translation section.
Novel Characterization of Disease
AI opens possibilities to provide novel characterization of disease processes and phenotypes between individuals and potentially to develop novel granular classifications that enable personalized medicine, which may ultimately assist with enhanced disease diagnosis and prediction of patient outcomes.
Computational Modeling and ML to Study AF
The explosion of mapping and imaging in AF patients provides increasingly detailed data that could be used by AI to classify AF and personalize therapy for patients. Several approaches have been reported. Recent studies show a role for multiscale computer modeling to identify specific ablation targets in individuals.54 In studies of computer models derived from magnetic resonance images (MRI) of left atrial geometry in AF patients, ML of spatial atrial fibrosis patterns predicted sites of AF drivers that were unaffected by ablation.55–57 In separate work, ML has been shown to potentially clarify the controversial field of AF mapping.58,59 A major unmet need is to reduce ambiguity in mapped AF patterns because current AF mapping systems require operator interpretation by automatically identifying ablation targets. Deep CNNs were recently trained on 175 000 AF maps from 35 patients to identify potential sites for ablation, including termination sites of persistent AF, and provided accuracy of 95%.60 Another recent study used a cellular automaton model to simulate the ability of ablation lesions to eliminate swirling and sometimes meandering vortices of fibrillatory activity.49 The authors found that ablating approximately one-third of the grid area eliminated currently apparent vortices, although residual wavelets continued to propagate. Conversely, simultaneous electrical stimulation at electrodes distributed throughout the atria was able to terminate AF.49 Recent work has extended ML to locating re-entrant drivers in cellular automaton models.50
Image-Based Characterization of AF for Ablation
Integrating imaging with cellular computational modeling provided important insight into the relationship between atrial fibrosis and AF mechanisms. Late gadolinium-enhanced MRI is an important imaging modality to visualize fibrosis and objectively assess scar.61 An important subsequent processing task is segmentation of relevant anatomic structures62 and fibrotic regions on MRI, a laborious and intensive task with interobserver variability when performed manually. DL has been able to generate automated segmentation of atrial fibrosis,63,64 as well as the left atrial epicardium and endocardium in MRI.16 Extension of this work has yielded a computational framework to estimate 3-dimensional atrial wall thickness,65 which may further yield improved characterization of atrial remodeling and subsequently impact clinical management of AF. ML will likely continue to play a role in better characterizing relevant fibrosis patterns for clinical outcomes. This will also be relevant outside of AF, such as detecting for amyloidosis versus scar and has been used to analyze spatial scar patterns and risk for ventricular arrhythmia.66
Meanwhile, novel image characterization of left atrial morphology has been useful in predicting ablation outcomes in AF. AF induces morphological changes of the left atrium, which can manifest as changes in atrial volume and shape. Recent studies have developed image-based features from advanced imaging to predict AF recurrence using radiographic features. On cardiac MRI of AF patients who received catheter ablation, quantitative assessment of left atrial shape via particle-based modeling generated 19 shape features that were used to predict time to AF recurrence.67 The authors identified 3 features via feature selection and found that adding these shape features into a Cox regression model of clinical parameters and left atrial fibrosis increased the model’s concordance index from 0.68 to 0.72. Visualized shapes showed that a round left atrial shape with a shorter, laterally rotated appendage was associated with recurrence. Another study used statistical shape models from MRIs to identify salient image variations across the study cohort, then used an ML classifier to distinguish patients with AF recurrence versus nonrecurrence with an AUC of 0.71.68 Alternative to traditional ML approaches, DL has been useful to quantify dynamic shape features in other cardiac investigations.69 Although DL of left atrial shape features in AF has not yet been published, it is likely a fruitful area of future investigation.
Clinical Phenotyping of AF
AF is a heterogeneous multifactorial disorder with diverse phenotypic expression, and unsupervised clustering may improve phenotypic classification of AF to aid clinical evaluation and management. In an analysis of 9749 patients with AF from the United States, cluster analysis using 60 clinical characteristics identified 4 primary cluster phenotypes31: (1) AF with limited risk factors, (2) younger AF patients with comorbid behavioral disorders, (3) AF patients with tachycardia-bradycardia with device implantation due to sinus node dysfunction, and (4) AF with atherosclerotic vascular disease. The cluster phenotypes were validated in a separate United States cohort. Interestingly, the clusters were not driven by left atrial size nor type of AF but rather by comorbid illness. Similar to other cluster analyses in cardiovascular disorders, the cluster phenotypes had distinct associations with cardiovascular outcomes. More recently, a cluster analysis in a Japanese population with AF found different cluster phenotypes, including younger patients with paroxysmal AF, (2) persistent AF with left atrial enlargement, and (3) AF with atherosclerotic vascular disease.70 Thus, regional variation may be evident and important in defining AF phenotypes, or perhaps resulting cluster differences could be an effect of data differences between registries.
Cluster phenotyping not only enables better description of AF but also may guide therapy. Certain risks and outcomes inherent to certain cluster phenotypes can be targeted with specific pharmacological, behavioral, or catheter-based therapies. Beyond clinical management, cluster phenotyping may also facilitate clinical investigation. For example, when designing a clinical trial for a novel investigational therapy, targeting a specific cluster phenotype may be more relevant than targeting a broad population.
Phenotyping Heart Failure Among CRT Candidates
Integration of complex imaging data with standard clinical variables for unsupervised phenotyping has also been demonstrated on heart failure patients in the MADIT-CRT trial.30 To expand the utility of echocardiography beyond standard single measurements, the authors used multiple kernel learning (unsupervised dimensionality reduction) integrating cycle-wide left ventricular strain and volume traces with clinical data of 1106 heart failure patients randomized to receive CRT or implantable cardioverter-defibrillator. This approach integrated complex regional patterns of cardiac function (1632 echocardiographic data points per cardiac cycle) with extensive clinical parameters. Four phenogroups were identified by k-means clustering with significantly different clinical and echocardiographic characteristics. Two phenogroups were associated with the highest proportion of clinical characteristics known to be predictive of volumetric response to CRT and had substantially better treatment effect from CRT. The algorithm outperformed independent analyses of clinical parameters or complex echocardiographic descriptors alone. The results suggest that unsupervised ML may yield novel, interpretable, and clinically meaningful phenotyping of heterogeneous patient cohorts that may also aid in patient selection for therapy.
Validation and Translation
There is an abundance of AI/ML studies in clinical research, but relatively few have been adopted into clinical practice. In this section, we address proper practice to validate and translate AI toward meaningful improvements to clinical practice.
Importance of Generalizability
As in other fields of medicine, before widespread use of an AI model, it is important to assess whether AI-based models are generalizable to data external to model development. For example, hospital-specific biases hampered the reliability of a generalized DL model used to detect pneumonia on chest x-ray.71 Nearly all AI studies have been retrospective and use data from limited numbers of institutions or high-quality clinical trial data that may not be generalizable to other cohorts. In AI models in cardiac electrophysiology, it should be assessed if the patient population is representative of other patient populations and if the AI model is robust to variation in data collection and processing techniques (eg, imaging scanners, imaging sequences, ECG acquisition equipment and techniques, interobserver differences in data collection protocol, feature computation definitions).
Validation, Translation, and Adoption
AI has been implemented in many industries, and successful implementation improves decision-making, decreases resource utilization, and provides cost savings. But the application of AI in medicine is more complex than in the business, commercial, or technical sectors. First, before clinical deployment, a fixed AI model must be selected for Food and Drug Administration scrutiny, which is important to consider given the possibility of AI models that may continuously change as more training data becomes available over time. Additionally, for implementation of AI in cardiac electrophysiology, the potential for catastrophic error (eg, missed diagnosis of a life-threatening arrhythmia), susceptibility to adversarial attack,34 the need to explain model outputs (eg, selecting patients for CRT implantation), the importance of maintaining patient privacy (eg, could AI identify patients from anonymous ECG waveforms?), and potential subsequent legal ramifications are also crucial factors that should be considered.72 Many AI studies have been retrospective proof-of-concept studies, but developing trust in the practicality and the benefits of AI in a real-world setting will require prospective clinical studies. Prospective studies can focus on AI outcomes data and patient experience compared with traditional decision models, which may help define what constitutes successful AI. The prospective EAGLE trial,48 the Apple Heart Study,37 and the Huawei Heart study38 are among the few early examples in cardiac electrophysiology.
Furthermore, for clinical adoption, AI tools need to better immersed in the clinical ecosystem. Currently, many AI models exist as inaccessible research tools at specific institutions. In a modestly more accessible format, 2 of the CRT prediction studies provided an online calculator for clinicians to use with patients,24,52 but these calculators are still not integrated with the EMR. Even if the model itself is accessible, the data required as input for the AI model must be able to easily pass from the clinical data environment to the AI model. Tools that use data beyond clinical EMR variables become even more challenging. For example, for optimal clinical adoption of the DL ECG interpretation tools, a digital ECG acquired in clinic must be stored at that institution’s data server. The digital ECG waveforms must be in a compatible format and then be securely exported to the AI model, which subsequently provides an interpretation. The clinical provider can then use this interpretation to guide clinical management of the patient. Without a fluid ecosystem, this may prove to be a cumbersome process for practical clinical use.
Lastly, reimbursement for AI technologies may occur in the future and would likely introduce cost savings. A cost-benefit analysis of AI utilized in imaging revealed the low cost of a graphics processing unit, the ability to process thousands of images per second, and the potential imaging capacity of millions of images per day, creating a low-cost imaging reading output.73 One could envision such a system for cardiologist-level ECG diagnosis, but this model must take into consideration the need for human input and contextual reasoning. With the emphasis on value-based care, improving efficiency and realizing the maximum benefit from AI models may improve a hospital’s financial performance, potentially decreasing operating costs.
Gaps, Needs, and Future Directions
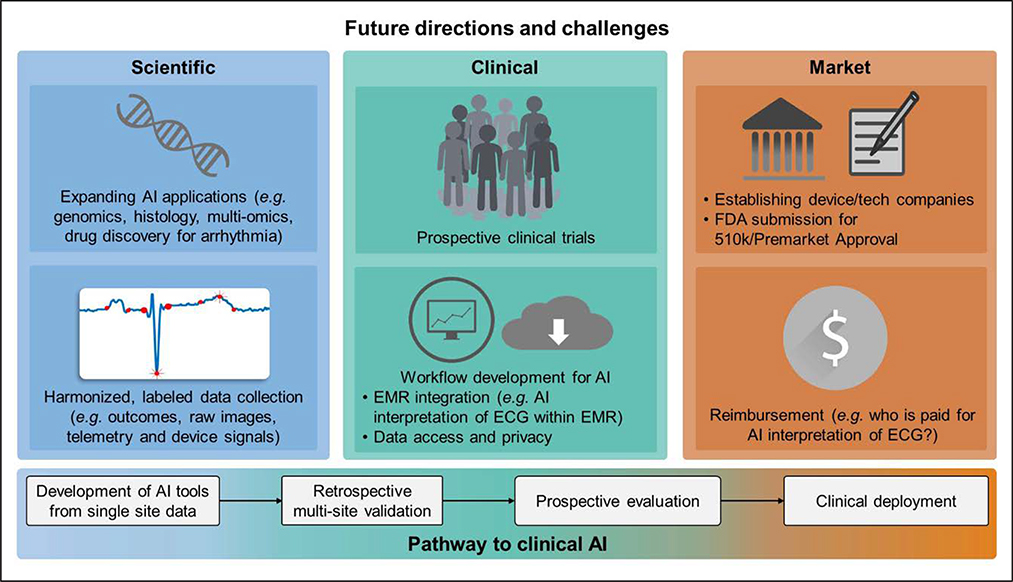
To date, AI in cardiac electrophysiology has shown great preliminary promise, but significant needs remain in basic and translational research, an institution-level improvement in data collection and harmonization practices, and clinical validation and practical implementation (Figure 4). From a research standpoint, opportunities remain to investigate AI in more basic science applications: genomic and proteomic data, histological characterization, or drug discovery in understanding and treating arrhythmia. Additionally, most recent AI studies exist in isolation. An advantage of ML techniques is the capability to fuse different types of data. For instance, building off from many AF studies discussed in the article, one could imagine using AI to integrate the DL interpretation of ECG waveform data with patient-specific fibrosis patterns from MRI with clinical variable cluster phenotypes from the EMR. Similar efforts for data fusion in multiomic studies on a more basic science level may help relate biological understanding of diseases such as AF with their heterogeneous clinical phenotypes.
Figure 4. Future directions for artificial intelligence (AI) in cardiac electrophysiology.
On the bottom is a typical pathway from development of AI tools to their clinical deployment. Progress thus far in electrophysiology has largely been achieved through the retrospective studies component of this pathway, with early prospective studies just beginning. There is currently significant need to evaluate and develop existing AI technologies toward clinical deployment, and potential clinical and market challenges to do so are outlined. Future scientific efforts to develop new AI tools are also outlined. EMR indicates electronic medical record; and FDA, Food and Drug Administration.
In addition, an institution-level focus on improved data collection would only facilitate and strengthen future AI research. Currently, most AI studies are performed by extracting data independently from within the institution. However, this makes it challenging to combine data across institutions. A concerted effort in collecting and labeling clinical data and outcomes and harmonizing data across institutions would be immensely beneficial to AI. This data collection should move beyond clinical variables in the EMR. In cardiac electrophysiology, data from imaging, 12-lead ECG, Holter and wearables, telemetry, and electrophysiological studies could also be collected and harmonized. Storing harmonized data in repositories that are made more accessible to researchers would also rapidly advance AI in this field. There already exist some publically available data repositories for electrophysiological data, such as the ECG databases available from PhysioNet,74 which have led to great development in AI technologies with ECG, but there is significant room for growth in increasing availability in other data with clinical outcomes.
Lastly, as the potential for AI is further expanded, the next challenge is developing a framework for incorporating AI into clinical practice. As discussed above, the clinical safety, reliability, and benefit of AI models need to be more rigorously validated in prospective studies. Mechanisms to smoothly integrate AI models into clinical practice while considering needs of data access and transfer and patient privacy are needed. Meanwhile, market needs of commercial establishment, Food and Drug Administration approval, and reimbursement mechanisms must be addressed.
Conclusions
Modern processing hardware, innovation in software algorithms, and collection of large electronic datasets have placed AI in a position to alter the landscape of biomedical research and clinical practice, with AI showing promise both to perform expert-level tasks and extend capabilities beyond human cognition. In cardiac electrophysiology, we have begun to see how AI is changing traditional mechanisms to detect and diagnose disease, predict patient outcomes, and understand and characterize disease processes. Significant work remains to better understand the capabilities, pitfalls, and appropriate deployment of AI in order for it to be integrated clinically. Leaders in the field will need to thoughtfully consider the appropriate application of AI and its implications. Meanwhile, researchers and clinicians alike would be empowered to have literacy in AI and ML, providing them with the ability to interpret the output of such methods as they continue to be adopted into modern use.
Supplementary Material
Acknowledgments
Sources of Funding
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number R01-HL111314, the American Heart Association Atrial Fibrillation Strategically Focused Research Network grant, the National Institutes of Health National Center for Research Resources for Case Western Reserve University and Cleveland Clinic Clinical and Translational Science Award under award number UL1-RR024989, Center of Excellence in Cardiovascular Translational Functional Genomics, Heart & Vascular Institute and Lerner Research Institute funds, Tomsich Atrial Fibrillation Research Fund, Heart & Vascular Institute and Lerner Research Institute Philanthropy funds, National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, R01 CA220581-01A1, and 1U01 CA239055-01, the National Institute for Biomedical Imaging and Bioengineering under award number 1R43EB028736-01, the National Center for Research Resources under award number 1 C06 RR12463-01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Department of Defense (DOD) Breast Cancer Research Program Breakthrough Level 1 Award W81XWH-19-1-0668, the DOD Prostate Cancer Idea Development Award (W81XWH-15-1-0558), the DOD Lung Cancer Investigator-Initiated Translational Research Award (W81XWH-18-1-0440), the DOD Peer Reviewed Cancer Research Program (W81XWH-16-1-0329), the Ohio Third Frontier Technology Validation Fund, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering, the British Heart Foundation (Project, Programme, and Centre of Research Excellence Grants), and the National Institute for Health Research Biomedical Research Centre, United Kingdom. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the US Department of Veterans Affairs, the Department of Defense, or the United States Government.
Disclosures
Dr Chung serves on the steering committee for and has spoken at conferences for EPIC Alliance, a forum for networking and mentoring of women in cardiac electrophysiology sponsored by Biotronik, but declines honoraria from device companies. Dr Madabhushi is an equity holder in Elucid Bioimaging and Inspirata; is a scientific advisory consultant for Inspirata; has been a scientific advisory board member for Inspirata, AstraZeneca, and Merck; and has sponsored research agreements with Philips and Inspirata. His technology has been licensed to Elucid Bioimaging and Inspirata. He is involved in National Institutes of Health grants with Path-Core and Inspirata. Mayo Clinic has licensed the underlying technology to EKO, a maker of digital stethoscopes with embedded ECG electrodes. Mayo Clinic may receive financial benefit from the use of this technology, but at no point will Mayo Clinic benefit financially from its use for the care of patients at Mayo Clinic. Drs Friedman, Kapa, Noseworthy, and Attia may also receive financial benefit from this agreement. Dr Narayan reports consulting from Beyondai Inc, TDK Inc, Up to Date, Abbott Laboratories, and American College of Cardiology Foundation, and intellectual property rights from the University of California Regents and Stanford University. Dr Passman receives research support, consulting fees, and speaking fees from Medtronic, and research support from AliveCor. Dr Perez reports grants from Apple Inc as well as personal fees from Apple Inc and Boehringer-Ingelheim. Dr Peters reports consulting fees from Google. Dr Piccini receives grants for clinical research from Abbott, American Heart Association, Boston Scientific, Gilead, Janssen Pharmaceuticals, National Heart, Lung, and Blood Institute, and Philips; and serves as a consultant to Abbott, Allergan, ARCA Biopharma, Biotronik, Boston Scientific, Johnson & Johnson, LivaNova, Medtronic, Milestone, Oliver Wyman Health, Sanofi, Philips, and Up-to-Date. Dr Tarakji reports consulting and advisory board fees from Medtronic, AliveCor, and Boston Scientific. Dr Trayanova is a founder of CardioSolv, holds an equity ownership interest in the company and acts as its Chief Scientific Officer. Dr Turakhia reports grants from Apple Inc as well as grants from Janssen Inc, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, American Heart Association, and SentreHeart; personal fees from Medtronic Inc, Abbott, Precision Health Economics, iBeat Inc, iRhythm, Novartis, Biotronik, Sanofi-Aventis, and Pfizer; other support from AliveCor; and grants and personal fees from Cardiva Medical. Dr Wang reports honoraria/consultant fees from Janssen, St. Jude Medical, Amgen, Medtronic; fellowship support from Biosense-Webster, Boston Scientific, Medtronic, St. Jude Medical; clinical studies from Medtronic, Siemens, Cardiofocus, ARCA Biopharma; and stock options from Vytronus. The other authors report no conflicts.
Nonstandard Abbreviations and Acronyms
- AF
atrial fibrillation
- AI
artificial intelligence
- AUC
area under the curve
- CNN
convolutional neural network
- DL
deep learning
- EMR
electronic medical record
- MADIT-CRT
Multicenter Automatic Defibrillator Implantation Trial With Cardiac Resynchronization Therapy
- ML
machine learning
- MRI
magnetic resonance image
- ROC
receiver operating characteristic
Footnotes
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCEP.119.007952.
Contributor Information
Albert K. Feeny, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, OH.
Mina K. Chung, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, OH; Department of Cardiovascular Medicine, Cleveland Clinic, OH.
Anant Madabhushi, Department of Biomedical Engineering, Case Western Reserve University, OH; Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, OH.
Zachi I. Attia, Department of Cardiovascular Medicine, Mayo Clinic College of Medicine, Rochester, MN.
Maja Cikes, Department of Cardiovascular Diseases, University of Zagreb School of Medicine & University Hospital Center Zagreb, Croatia.
Marjan Firouznia, Department of Biomedical Engineering, Case Western Reserve University, OH.
Paul A. Friedman, Department of Cardiovascular Medicine, Mayo Clinic College of Medicine, Rochester, MN.
Matthew M. Kalscheur, Division of Cardiovascular Medicine, Department of Medicine, School of Medicine & Public Health, University of Wisconsin; William S. Middleton Veterans Hospital, Madison, WI.
Suraj Kapa, Department of Cardiovascular Medicine, Mayo Clinic College of Medicine, Rochester, MN.
Sanjiv M. Narayan, Division of Cardiovascular Medicine, Stanford University, CA; Veterans Affairs Palo Alto Health Care System, CA.
Peter A. Noseworthy, Department of Cardiovascular Medicine, Mayo Clinic College of Medicine, Rochester, MN.
Rod S. Passman, Division of Cardiology, Northwestern University, Feinberg School of Medicine, Chicago, IL.
Marco V. Perez, Division of Cardiovascular Medicine, Stanford University, CA; Veterans Affairs Palo Alto Health Care System, CA.
Nicholas S. Peters, National Heart Lung Institute & Centre for Cardiac Engineering, Imperial College London, United Kingdom.
Jonathan P. Piccini, Duke Clinical Research Institute, Duke University Medical Center, Durham, NC.
Khaldoun G. Tarakji, Department of Cardiovascular Medicine, Cleveland Clinic, OH.
Suma A. Thomas, Department of Cardiovascular Medicine, Cleveland Clinic, OH.
Natalia A. Trayanova, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD.
Mintu P. Turakhia, Division of Cardiovascular Medicine, Stanford University, CA; Veterans Affairs Palo Alto Health Care System, CA; Center for Digital Health, Stanford University School of Medicine, CA.
Paul J. Wang, Division of Cardiovascular Medicine, Stanford University, CA; Veterans Affairs Palo Alto Health Care System, CA.
REFERENCES
- 1.Nygårds ME, Hulting J. An automated system for ECG monitoring. Comput Biomed Res. 1979;12:181–202. doi: 10.1016/0010-4809(79)90015-6 [DOI] [PubMed] [Google Scholar]
- 2.Dey D, Slomka PJ, Leeson P, Comaniciu D, Shrestha S, Sengupta PP, Marwick TH. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol. 2019;73:1317–1335. doi: 10.1016/j.jacc.2018.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martis RJ, Acharya UR, Min LC. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomedical Signal Processing and Control 2013;8:437–448. [Google Scholar]
- 4.Zhao Q, Zhang L. ECG Feature extraction and classification using wavelet transform and support vector machines. International Conference on Neural Networks and Brain 2005;2:1089–1092. doi: 10.1109/ICNNB.2005.1614807 [DOI] [Google Scholar]
- 5.Lagerholm M, Peterson C, Braccini G, Edenbrandt L, Sörnmo L. Clustering ECG complexes using hermite functions and self-organizing maps. IEEE Trans Biomed Eng. 2000;47:838–848. doi: 10.1109/10.846677 [DOI] [PubMed] [Google Scholar]
- 6.Attia ZI, Friedman PA, Noseworthy PA, Lopez-Jimenez F, Ladewig DJ, Satam G, Pellikka PA, Munger TM, Asirvatham SJ, Scott CG, et al. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ Arrhythm Electrophysiol. 2019;12:e007284. doi: 10.1161/CIRCEP.119.007284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med. 2019;25:70–74. doi: 10.1038/s41591-018-0240-2 [DOI] [PubMed] [Google Scholar]
- 8.Ko WY, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, Demuth SJ, Ackerman MJ, Gersh BJ, Arruda-Olson AM, et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol. 2020;75:722–733. doi: 10.1016/j.jacc.2019.12.030 [DOI] [PubMed] [Google Scholar]
- 9.Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–867. doi: 10.1016/S0140-6736(19)31721-0 [DOI] [PubMed] [Google Scholar]
- 10.Galloway CD, Valys AV, Shreibati JB, Treiman DL, Petterson FL, Gundotra VP, Albert DE, Attia ZI, Carter RE, Asirvatham SJ, et al. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 2019;4:428–436. doi: 10.1001/jamacardio.2019.0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta PP, Kulkarni H, Narula J. Prediction of abnormal myocardial relaxation from signal processed surface ECG. J Am Coll Cardiol. 2018;71:1650–1660. doi: 10.1016/j.jacc.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 12.Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez MV, Dewey FE, Tan SY, Myers J, Froelicher VF. Added value of a resting ECG neural network that predicts cardiovascular mortality. Ann Noninvasive Electrocardiol. 2009;14:26–34. doi: 10.1111/j.1542-474X.2008.00270.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Z, Nash MP, Cheng E, Fedorov VV, Stiles MK, Zhao J. ECG signal classification for the detection of cardiac arrhythmias using a convolutional recurrent neural network. Physiol Meas. 2018;39:094006. doi: 10.1088/1361-6579/aad9ed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adam M, Gertych A, Tan RS. A deep convolutional neural network model to classify heartbeats. Comput Biol Med. 2017;89:389–396. doi: 10.1016/j.compbiomed.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 16.Xiong Z, Fedorov VV, Fu X, Cheng E, Macleod R, Zhao J. Fully automatic left atrium segmentation from late gadolinium enhanced magnetic resonance imaging using a dual fully convolutional neural network. IEEE Trans Med Imaging. 2019;38:515–524. doi: 10.1109/TMI.2018.2866845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial networks. arXiv:1406.2661 [cs, stat] 2014; http://arxiv.org/abs/1406.2661. Accessed March 1, 2020. [Google Scholar]
- 18.Zhu F, Twan WH, Tseng LC, Peng SH, Hwang JS. First detection of white spot syndrome virus (WSSV) in the mud shrimp Austinogebia edulis in Taiwan. Sci Rep. 2019;9:18572. doi: 10.1038/s41598-019-54837-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krizhevsky A, Sutskever I, Hinton GE. In Pereira F, Burges CJC, Bottou L, Weinberger KQ, ed. Advances in Neural Information Processing Systems 25. Curran Associates Inc;2012:1097–1105. [Google Scholar]
- 20.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 [cs]. 2015;http://arxiv.org/abs/1409.1556. [Google Scholar]
- 21.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going Deeper with Convolutions. In Computer Vision Pattern Recognition (CVPR) 2015. http://arxiv.org/abs/1409.4842. [Google Scholar]
- 22.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. arXiv:1512.03385 [cs]. 2015;http://arxiv.org/abs/1512.03385. [Google Scholar]
- 23.Isin A, Ozdalili S. Cardiac arrhythmia detection using deep learning. Procedia Computer Sci. 2017;120:268–275. [Google Scholar]
- 24.Feeny AK, Rickard J, Patel D, Toro S, Trulock KM, Park CJ, LaBarbera MA, Varma N, Niebauer MJ, Sinha S, et al. Machine learning prediction of response to cardiac resynchronization therapy: improvement versus current guidelines. Circ Arrhythm Electrophysiol. 2019;12:e007316. doi: 10.1161/CIRCEP.119.007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalscheur MM, Kipp RT, Tattersall MC, Mei C, Buhr KA, DeMets DL, Field ME, Eckhardt LL, Page CD. Machine learning algorithm predicts cardiac resynchronization therapy outcomes: lessons from the COMPANION trial. Circ Arrhythm Electrophysiol. 2018;11:e005499. doi: 10.1161/CIRCEP.117.005499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tison GH, Zhang J, Delling FN, Deo RC. Automated and interpretable patient ECG profiles for disease detection, tracking, and discovery. Circ Cardiovasc Qual Outcomes. 2019;12:e005289. doi: 10.1161/CIRCOUTCOMES.118.005289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tison GH, Sanchez JM, Ballinger B, Singh A, Olgin JE, Pletcher MJ, Vittinghoff E, Lee ES, Fan SM, Gladstone RA, et al. Passive detection of atrial fibrillation using a commercially available smartwatch. JAMA Cardiol. 2018;3:409–416. doi: 10.1001/jamacardio.2018.0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verleysen M, François D. The Curse of Dimensionality in Data Mining and Time Series Prediction In: Cabestany J, Prieto A, Sandoval F, eds. Computational Intelligence and Bioinspired Systems. Berlin, Heidelberg: Springer; 2005:758–770. [Google Scholar]
- 29.Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344 [DOI] [PubMed] [Google Scholar]
- 30.Cikes M, Sanchez-Martinez S, Claggett B, Duchateau N, Piella G, Butakoff C, Pouleur AC, Knappe D, Biering-Sørensen T, Kutyifa V, et al. Machine learning-based phenogrouping in heart failure to identify responders to cardiac resynchronization therapy. Eur J Heart Fail. 2019;21:74–85. doi: 10.1002/ejhf.1333 [DOI] [PubMed] [Google Scholar]
- 31.Inohara T, Shrader P, Pieper K, Blanco RG, Thomas L, Singer DE, Freeman JV, Allen LA, Fonarow GC, Gersh B, et al. Association of of atrial fibrillation clinical phenotypes with treatment patterns and outcomes: a multicenter registry study. JAMA Cardiol. 2018;3:54–63. doi: 10.1001/jamacardio.2017.4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bundy JD, Heckbert SR, Chen LY, Lloyd-Jones DM, Greenland P. Evaluation of risk prediction models of atrial fibrillation (from the Multi-Ethnic Study of Atherosclerosis [MESA]). Am J Cardiol 2020;125:55–62. doi: 10.1016/j.amjcard.2019.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudin C Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X, Hu Y, Foschini L, Chinitz L, Jankelson L, Ranganath R. Deep learning models for electrocardiograms are susceptible to adversarial attack. Nat Med. 2020;26:360–363. doi: 10.1038/s41591-020-0791-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montavon G, Samek W, Müller K-R. Methods for interpreting and understanding deep neural networks. Digital Signal Processing. 2018;73:1–15. [Google Scholar]
- 36.Howard JP, Fisher L, Shun-Shin MJ, Keene D, Arnold AD, Ahmad Y, Cook CM, Moon JC, Manisty CH, Whinnett ZI, et al. Cardiac rhythm device identification using neural networks. JACC Clin Electrophysiol. 2019;5:576–586. doi: 10.1016/j.jacep.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez MV, Mahaffey KW, Hedlin H, Rumsfeld JS, Garcia A, Ferris T, Balasubramanian V, Russo AM, Rajmane A, Cheung L, et al. ; Apple Heart Study Investigators. Large-scale assessment of a smartwatch to identify atrial fibrillation. N Engl J Med. 2019;381:1909–1917. doi: 10.1056/NEJMoa1901183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y, Wang H, Zhang H, Liu T, Liang Z, Xia Y, Yan L, Xing Y, Shi H, Li S, et al. ; MAFA II Investigators. Mobile photoplethysmographic technology to detect atrial fibrillation. J Am Coll Cardiol. 2019;74:2365–2375. doi: 10.1016/j.jacc.2019.08.019 [DOI] [PubMed] [Google Scholar]
- 39.Wasserlauf J, You C, Patel R, Valys A, Albert D, Passman R. Smartwatch performance for the detection and quantification of atrial fibrillation. Circ Arrhythm Electrophysiol. 2019;12:e006834. doi: 10.1161/CIRCEP.118.006834 [DOI] [PubMed] [Google Scholar]
- 40.Bumgarner JM, Lambert CT, Hussein AA, Cantillon DJ, Baranowski B, Wolski K, Lindsay BD, Wazni OM, Tarakji KG. Smartwatch algorithm for automated detection of atrial fibrillation. J Am Coll Cardiol. 2018;71:2381–2388. doi: 10.1016/j.jacc.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 41.Yan BP, Lai WHS, Chan CKY, Chan SCH, Chan LH, Lam KM, Lau HW, Ng CM, Tai LY, Yip KW, et al. Contact-free screening of atrial fibrillation by a smartphone using facial pulsatile photoplethysmographic signals. J Am Heart Assoc. 2018;7:e008585. doi: 10.1161/JAHA.118.008585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan PH, Wong CK, Poh YC, Pun L, Leung WWC, Wong YF, Wong MMY, Poh MZ, Chu DWS, Siu CW. Diagnostic performance of a smartphone-based photoplethysmographic application for atrial fibrillation screening in a primary care setting. J Am Heart Assoc. 2016;5:e003428. doi: 10.1161/JAHA.116.003428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brasier N, Raichle CJ, Dörr M, Becke A, Nohturfft V, Weber S, Bulacher F, Salomon L, Noah T, Birkemeyer R, et al. Detection of atrial fibrillation with a smartphone camera: first prospective, international, two-centre, clinical validation study (DETECT AF PRO). Europace. 2019;21:41–47. doi: 10.1093/europace/euy176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verbrugge FH, Proesmans T, Vijgen J, Mullens W, Rivero-Ayerza M, Van Herendael H, Vandervoort P, Nuyens D. Atrial fibrillation screening with photo-plethysmography through a smartphone camera. Europace. 2019;21:1167–1175. doi: 10.1093/europace/euz119 [DOI] [PubMed] [Google Scholar]
- 45.Rozen G, Vaid J, Hosseini SM, Kaadan MI, Rafael A, Roka A, Poh YC, Poh MZ, Heist EK, Ruskin JN. Diagnostic accuracy of a novel mobile phone application for the detection and monitoring of atrial fibrillation. Am J Cardiol. 2018;121:1187–1191. doi: 10.1016/j.amjcard.2018.01.035 [DOI] [PubMed] [Google Scholar]
- 46.O’Sullivan JW, Grigg S, Crawford W, Turakhia MP, Perez M, Ingelsson E, Wheeler MT, Ioannidis JPA, Ashley EA. Accuracy of smartphone camera applications for detecting atrial fibrillation: a systematic review and meta-analysis. JAMA Netw Open. 2020;3:e202064. doi: 10.1001/jamanetworkopen.2020.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attia ZI, Kapa S, Yao X, Lopez-Jimenez F, Mohan TL, Pellikka PA, Carter RE, Shah ND, Friedman PA, Noseworthy PA. Prospective validation of a deep learning electrocardiogram algorithm for the detection of left ventricular systolic dysfunction. J Cardiovasc Electrophysiol. 2019;30:668–674. doi: 10.1111/jce.13889 [DOI] [PubMed] [Google Scholar]
- 48.Yao X, McCoy RG, Friedman PA, Shah ND, Barry BA, Behnken EM, Inselman JW, Attia ZI, Noseworthy PA. ECG AI-Guided Screening for Low Ejection Fraction (EAGLE): rationale and design of a pragmatic cluster randomized trial. Am Heart J. 2020;219:31–36. doi: 10.1016/j.ahj.2019.10.007 [DOI] [PubMed] [Google Scholar]
- 49.Raghunath S, Ulloa Cerna AE, Jing L, vanMaanen DP, Stough J, Hartzel DN, Leader JB, Kirchner HL, Stumpe MC, Hafez A, et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat Med. 2020;26:886–891. doi: 10.1038/s41591-020-0870-z [DOI] [PubMed] [Google Scholar]
- 50.Decision Memo for Implantable Defibrillators (CAG-00157R3). https://www.cms.gov/medicare-coverage-database/details/nca-decision-memo.aspx?NCAId=148. Accessed November 15, 2019.
- 51.Decision Memo for Percutaneous Left Atrial Appendage (LAA) Closure Therapy (CAG-00445N). https://www.cms.gov/medicare-coverage-data-base/details/nca-decision-memo.aspx?NCAId=281. Accessed November 15, 2019.
- 52.Tokodi M, Schwertner WR, Kovács A, Tősér Z, Staub L, Sárkány A, Lakatos BK, Behon A, Boros AM, Perge P, et al. Machine learning-based mortality prediction of patients undergoing cardiac resynchronization therapy: the SEMMELWEIS-CRT score. Eur Heart J. 2020;41:1747–1756. doi: 10.1093/eurheartj/ehz902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu SY, Santus E, Forsyth AW, Malhotra D, Haimson J, Chatterjee NA, Kramer DB, Barzilay R, Tulsky JA, Lindvall C. Can machine learning improve patient selection for cardiac resynchronization therapy? PLoS One. 2019;14:e0222397. doi: 10.1371/journal.pone.0222397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cantwell CD, Mohamied Y, Tzortzis KN, Garasto S, Houston C, Chowdhury RA, Ng FS, Bharath AA, Peters NS. Rethinking multiscale cardiac electrophysiology with machine learning and predictive modelling. Comput Biol Med. 2019;104:339–351. doi: 10.1016/j.compbiomed.2018.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali RL, Hakim JB, Boyle PM, Zahid S, Sivasambu B, Marine JE, Calkins H, Trayanova NA, Spragg DD. Arrhythmogenic propensity of the fibrotic substrate after atrial fibrillation ablation: a longitudinal study using magnetic resonance imaging-based atrial models. Cardiovasc Res. 2019;115:1757–1765. doi: 10.1093/cvr/cvz083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zahid S, Cochet H, Boyle PM, Schwarz EL, Whyte KN, Vigmond EJ, Dubois R, Hocini M, Haïssaguerre M, Jaïs P, et al. Patient-derived models link re-entrant driver localization in atrial fibrillation to fibrosis spatial pattern. Cardiovasc Res. 2016;110:443–454. doi: 10.1093/cvr/cvw073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyle PM, Zghaib T, Zahid S, Ali RL, Deng D, Franceschi WH, Hakim JB, Murphy MJ, Prakosa A, Zimmerman SL, et al. Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat Biomed Eng. 2019;3:870–879. doi: 10.1038/s41551-019-0437-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramirez FD, Birnie DH, Nair GM, Szczotka A, Redpath CJ, Sadek MM, Nery PB. Efficacy and safety of driver-guided catheter ablation for atrial fibrillation: a systematic review and meta-analysis. J Cardiovasc Electrophysiol. 2017;28:1371–1378. doi: 10.1111/jce.13313 [DOI] [PubMed] [Google Scholar]
- 59.Baykaner T, Rogers AJ, Meckler GL, Zaman J, Navara R, Rodrigo M, Alhusseini M, Kowalewski CAB, Viswanathan MN, Narayan SM, et al. Clinical implications of ablation of drivers for atrial fibrillation: a systematic review and meta-analysis. Circ Arrhythm Electrophysiol. 2018;11:e006119. doi: 10.1161/CIRCEP.117.006119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alhusseini M, Abuzaid F, Clopton P, Rogers A, Rodrigo M, Baykaner T, Wang P, Rappel W-J, Narayan S. Machine learning identifies sites where ablation terminates persistent atrial fibrillation. J Am Coll Cardiol. 2019;73:S1,301. [Google Scholar]
- 61.Marrouche NF, Wilber D, Hindricks G, Jais P, Akoum N, Marchlinski F, Kholmovski E, Burgon N, Hu N, Mont L, et al. Association of atrial tissue fibrosis identified by delayed enhancement MRI and atrial fibrillation catheter ablation: the DECAAF study. JAMA. 2014;311:498–506. doi: 10.1001/jama.2014.3 [DOI] [PubMed] [Google Scholar]
- 62.Zhuang X, Li L, Payer C, Štern D, Urschler M, Heinrich MP, Oster J, Wang C, Smedby Ö, Bian C, et al. Evaluation of algorithms for multi-modality whole heart segmentation: an open-access grand challenge. Med Image Anal. 2019;58:101537. doi: 10.1016/j.media.2019.101537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang G, Zhuang X, Khan H, Haldar S, Nyktari E, Ye X, Slabaugh G, Wong T, Mohiaddin R, Keegan J, et al. Segmenting Atrial Fibrosis from Late Gadolinium-Enhanced Cardiac MRI by Deep-Learned Features with Stacked Sparse Auto-Encoders In Valdés Hernández M, González-Castro V, eds. Medical Image Understanding and Analysis Springer International Publishing; 2017:195–206 [Google Scholar]
- 64.Yang G, Chen J, Gao Z, Zhang H, Ni H, Angelini E, Mohiaddin R, Wong T, Keegan J, Firmin D. Multiview sequential learning and dilated residual learning for a fully automatic delineation of the left atrium and pulmonary veins from late gadolinium-enhanced cardiac MRI Images. Conf Proc IEEE Eng Med Biol Soc 2018;2018:1123–1127. doi: 10.1109/EMBC.2018.8512550 [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Xiong Z, Nalar A, Hansen BJ, Kharche S, Seemann G, Loewe A, Fedorov VV, Zhao J. A robust computational framework for estimating 3D Bi-Atrial chamber wall thickness. Comput Biol Med. 2019;114:103444. doi: 10.1016/j.compbiomed.2019.103444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okada DR, Miller J, Chrispin J, Prakosa A, Trayanova N, Jones S, Maggioni M, Wu KC. Substrate spatial complexity analysis for the prediction of ventricular arrhythmias in patients with ischemic cardiomyopathy. Circ Arrhythm Electrophysiol 2020;13:e007975. doi: 10.1161/CIRCEP.119.007975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bieging ET, Morris A, Wilson BD, McGann CJ, Marrouche NF, Cates J. Left atrial shape predicts recurrence after atrial fibrillation catheter ablation. J Cardiovasc Electrophysiol. 2018;29:966–972. doi: 10.1111/jce.13641 [DOI] [PubMed] [Google Scholar]
- 68.Varela M, Bisbal F, Zacur E, Berruezo A, Aslanidi OV, Mont L, Lamata P. Novel computational analysis of left atrial anatomy improves prediction of atrial fibrillation recurrence after ablation. Front Physiol. 2017;8:68. doi: 10.3389/fphys.2017.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bello GA, Dawes TJW, Duan J, Biffi C, de Marvao A, Howard LSGE, Gibbs JSR, Wilkins MR, Cook SA, Rueckert D, et al. Deep learning cardiac motion analysis for human survival prediction. Nat Mach Intell. 2019;1:95–104. doi: 10.1038/s42256-019-0019-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inohara T, Piccini JP, Mahaffey KW, Kimura T, Katsumata Y, Tanimoto K, Inagawa K, Ikemura N, Ueda I, Fukuda K, et al. A cluster analysis of the Japanese multicenter outpatient registry of patients with atrial fibrillation. Am J Cardiol. 2019;124:871–878. doi: 10.1016/j.amjcard.2019.05.071 [DOI] [PubMed] [Google Scholar]
- 71.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30–36. doi: 10.1038/s41591-018-0307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beam AL, Kohane IS. Translating artificial intelligence into clinical care. JAMA. 2016;316:2368–2369. doi: 10.1001/jama.2016.17217 [DOI] [PubMed] [Google Scholar]
- 74.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. doi: 10.1161/01.cir.101.23.e215 [DOI] [PubMed] [Google Scholar]
- 75.Feeny AK, Rickard J, Trulock KM, Patel D, Toro S, Moennich LA, Varma N, Niebauer MJ, Gorodeski EZ, Grimm RA, et al. Machine learning of 12-lead QRS waveform patterns to identify cardiac resynchronization therapy patients with differential outcomes. Circ Arrhythm Electrophysiol 2020;13:e008210. doi: 10.1161/CIRCEP.119.008210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.