ABSTRACT
The genetic variations and dysbiosis of gut microbiota are associated with ASD. However, the role of the microbiota in the etiology of ASD in terms of host genetic susceptibility remains unclear. This study aims to systematically explore the interplay between host genetic variation and gut microbiota in ASD children. Whole-exon sequencing was applied to 26 ASD children and 26 matched controls to identify the single nucleotide variations (SNVs) in ASD. Our previous study revealed alteration in gut microbiota and disorder of metabolism activity in ASD for this cohort. Systematic bioinformatic analyses were further performed to identify associations between SNVs and gut microbiota, as well as their metabolites. The ASD SNVs were significantly enriched in genes associated with innate immune response, protein glycosylation process, and retrograde axonal transport. These SNVs were also correlated with the microbiome composition and a broad aspect of microbial functions, especially metabolism. Additionally, the abundance of metabolites involved in the metabolic network of neurotransmitters was inferred to be causally related to specific SNVs and microbes. Furthermore, our data suggested that the interaction of host genetics and gut microbes may play a crucial role in the immune and metabolism homeostasis of ASD. This study may provide valuable clues to investigate the interaction of host genetic variations and gut microbiota in the pathogenesis of ASD.
KEYWORDS: Autism, genetic variation, gut microbiota, metabolites, cytokine
Introduction
Autism Spectrum Disorder (ASD) has genetic risk factors as there is a much higher concordance rate for the disease in monozygotic twins than indizygotic ones1. Previous large-cohort studies have revealed a large set of polymorphisms conferring various levels of risk.2–4 However, the de novo mutations, common variants, and short nucleotide polymorphisms identified across numerous ASD cases altogether only account for approximately 50% of the cases.5,6 As such, many studies highlight the possibility of environmental risk factors and associated medical co-morbidities that may contribute to the core neurobehavioral symptoms of the disorder.7,8
ASD children suffer from a range of gastrointestinal (GI) symptoms, such as gaseousness, diarrhea, and constipation, with the prevalence shown to be anywhere from 9% to 91%.9 These associations of ASD with a greater prevalence of GI symptoms motivate explorations of the role of gut microbiome in ASD pathogenesis, which is emerging as a key regulator of intestinal physiology, neuroimmunity, and host behavior. Growing studies have reported dysbiosis of the gut microbiota in ASD individuals. For example, studies have observed an altered abundance of Sutterella, Desulfovibrio, Bacteroides vulgatus, Akkermansia, Prevotella, Coprococcus, and Firmicutes in ASD patients.10,11 The gnotobiotic animal and probiotic studies demonstrated that alteration of microbiota can directly cause behavioral abnormalities and neuropathological endophenotypes of ASD. The phenotype can be improved by transplantation of normal gut microbiome.12
Microbial communities influence human physiology through their metabolites, cellular and molecular components and provide crucial signals for the development and function of the immune system.13,14 Both metabolites and immunity disorders were observed in ASD children. In the ASD mice model, 5-aminovaleric acid (5AV) was significantly lower than in the normal controls (TD), and the administration of 5AV improved behaviors in ASD mice.15 Elevated levels of IL-17α, IL-4, and IL-10 have been detected in the serum of a subset of autistic children.16–18
Recent studies have shown the role of host genetics in shaping both the overall microbiome composition and the individual bacterial taxa. For instance, Knight et al.19 reported that Crohn’s risk variants located in the NOD2 gene were associated with changes in the abundance of Enterobacteriaceae. A genome-wide host genetics and microbiome association conducted in HMP healthy cohort validated the associations between a loss-of-function variant in the fucosyltransferase 2 (FUT2) and a variant conferring hypolactasia near the lactase (LCT) gene, with Bifidobacterium longum abundance in the stool.20 Also, our previous study has found that the genetic variation of the Jmjc domain of lysine demethylase 5 (KDM5) protein in drosophila can lead to compositional changes in the gut microbiota, overactivation of innate immunity pathway, and abnormal level of 5-HT, then the autism-like behaviors.21
Nevertheless, the detailed association study of host genetic variation, gut microbiota-metabolites, the immune system in ASD remains unknown. In this study, to investigate the nature and extent of host genome-microbiome interplay in ASD, a total of 26 ASD patients with constipation and 26 matched typically developed children (TD) were supplied to exon sequencing. The SNVs enriched in ASD were identified by comparing with the TDs, and removing SNVs with minor allele frequency (MAF) more than 5% as recorded in ExAc, 1000 Genome database, and the Chinese Millionome Database (CMDB). Then the fecal microbiome, fecal metabolites, and serum cytokines were characterized to dissect the association with the enriched SNVs. Besides, the causal relationship between host SNVs and gut microbiota was inferred. The association analysis of functional genetic variations, gut microbiota, metabolites, and cytokines can provide novel and insightful clues revealing the pathogenesis of ASD diseases.
Results
SNVs in ASD are related to the gut microbiome
A total of 567 exonic SNVs (Table S1) were identified in our ASD cohort using the matched TD cohort and the public databases as control (Figure 1). These SNVs shared the same mutation pattern (Figure 2(a)) and mutation signature (Figure 2(b)) with the previously reported ASD-related mutations collected in the AutismKB, a knowledgebase for the genetic evidence of autism spectrum disorder.22 We calculated the coordinates underlying variability in the host SNV genotype data using Principal Component Analysis (PCA). We then computed alpha diversity, a measure of within-sample microbial diversity (Simpson index within a sample). We found that the first coordinated of the functional SNVs (protein-altering SNVs, including non-synonymous, stop-gain, and stop-loss SNVs) was significantly correlated with alpha diversity (Figure 2(c), R = 0.45, P = .020) of the gut microbiome. The altered alpha diversity links to lots of human health disease,23,24 including autism.15 Our results suggested a possible role for host genetic variations in shaping it. Next, we looked for the association of host genetics with the overall composition of the gut microbiome. A marginally significant correlation was observed between the first principal coordinate of functional SNVs and microbiome PC1 (Figure 2(c), R = 0.37, P = .059). However, when considering the total enriched SNVs, no significant correlation between the host genetics and microbiome diversity or composition was detected (Figure S1). These correlations suggest a potential relationship between the functional SNVs and microbiome.
Figure 1.
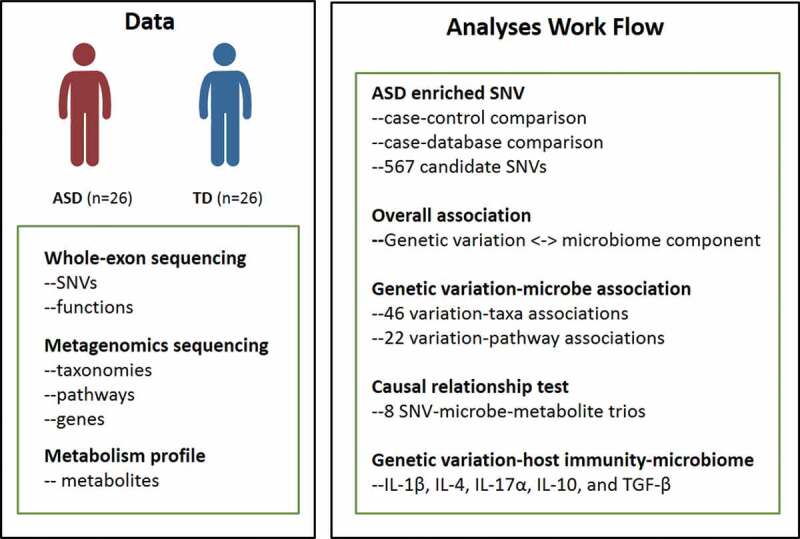
Schematic overview of the study
We performed whole exome sequencing of the host genome, whole genome shotgun sequencing and metabolite profile of fecal samples of 26 ASD individuals and 26 TDs. Associations were tested between genetic variations and microbes, as well as microbial genes, metabolites and pathways, and host immunity.
Figure 2.
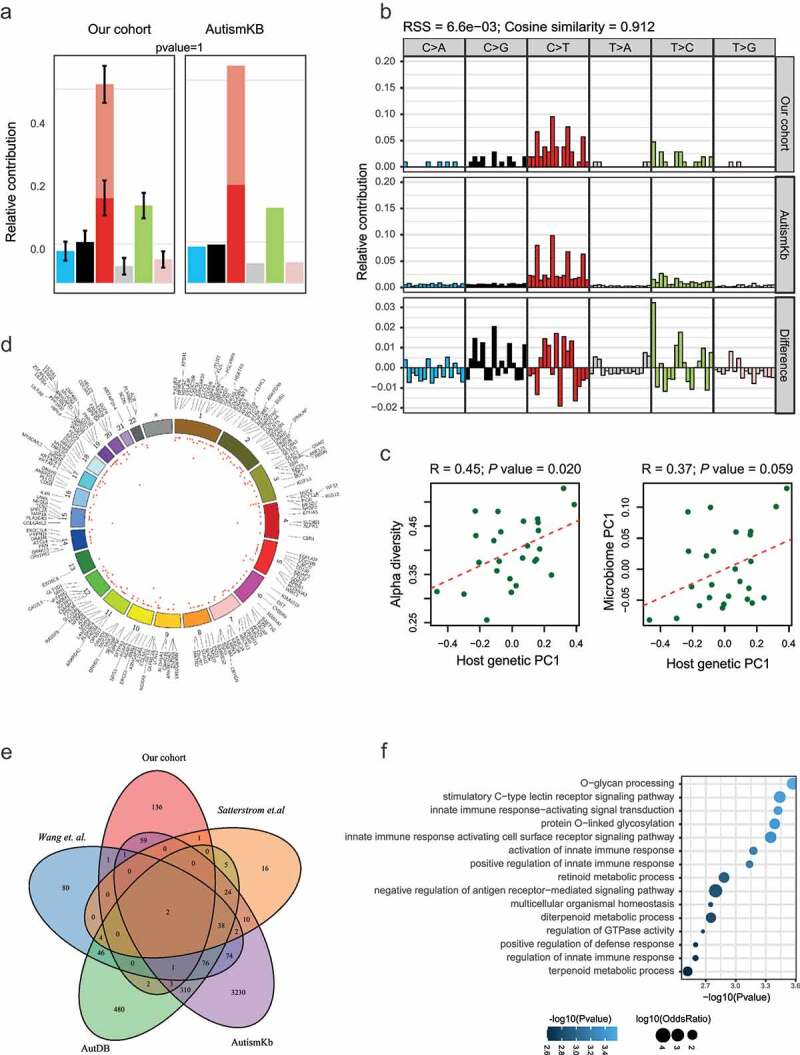
SNVs enriched in ASD children. (A-B) The comparison of the mutation pattern (A) and mutation signature(B) between our identified SNVs and mutations collected from the AutismKb database. (C) Correlation of the first PC of host genetic data (x-axis) and alpha diversity (left panel) or first PC of the stool microbiome (right panel). (D) Circos plot of the functional SNVs enriched in ASD. The height of the points represents the number of mutant samples. Genes mapped by SNVs were labeled outside the circle. (E) Overlap of genes with the ASD enriched SNVs in our studies with those collected from other studies. (F) Functional annotation of genes with functional SNVs
The functional SNVs were mapped to 206 genes (Figure 2(d)), including 70 previously reported genes,4,22,25,26 such as MUC4, SETD5, and ANKRD1127-29 (Figure 2(e)). Functional enrichment analysis on these genes revealed a significant enrichment on the protein glycosylation process, innate immune response, and retinoid metabolic process (figure 2(f)). The mucins encoded by MUC family genes are O-glycosylated proteins that play an essential role in forming protective mucous barriers on epithelial surfaces,30 and mucins are components of innate and adaptive immune responses to mucosal infection.31 Increasing evidence has shown that variants in genes encoding glycosylated extracellular proteins or enzymes (glycogene) involved in glycosylation may contribute to the etiology and pathogenesis of ASD.32 For examples, the mucin encoding gene MUC4 and the glycosyltransferase B3GALNT2.33 Studies in mouse models of congenital disorders of glycosylation (CDGs) and behavioral phenotypes observed in CDG patients support the idea that glycogene variants either cause or contribute to the development of idiopathic ASD.32 Retinoids are vitamin A and its derivatives. The retinoid signaling plays a significant role in regulating brain functions, including neuronal differentiation, neurite growth, patterning of the anteroposterior axis of the neural tube, neurotransmitter release, and long term potentiation.34 Defective retinoid signaling has been evidenced in the pathology of Alzheimer’s disease,35 and Vitamin A deficiencies were reported to exacerbate symptoms in children with ASD.36
SNVs correlated with bacteria taxa
Systematic screening for associations between each functional SNVs and abundance of each species was conducted using standard linear regression after adjusting for age and sex for confounding factors. There were 46 candidate associations identified (Figure 3, Table S2). Among the results, a variation in the glycosyltransferase B3GALNT2 (chr1:235659576, c.C155T, p.A52V) was associated with the increased abundance of Catenibacterium mitsuokai in a subset of ASD children. A variation of PGLYRP4 (rs148195147, c.G602A, p.R201Q) was related to Faecalibacterium prausnitzii. The PGLYRP4 is a part of the innate immune system and encodes a known antibacterial protein that recognizes peptidoglycan, a ubiquitous component of bacterial cell walls. The deficiency of PGLYRP4 leads to changes in the gut microbiota.37 Other associations involved by the immunity-related genes include CD180 variation (chr5:66479217, c.C1454A, p.T485N) associated with Bacteroides dorei and Clostridium sp. CAG:7, RAET1G variation (rs4870111, c.G668A, p.R223K) associated with two Bacteroides species and Catenibacterium mitsuokai, and RTN4 variation associated with Hungatella hathewayi. CD180 is a TLR-associated molecule that is mainly expressed on macrophages, DCs, and B cells, which can also sense bacterial LPS to trigger the growth of dormant multiplemyeloma cells.38 Both RAET1G and RTN4 mutations were reported in ASD cohorts.22 RAET1G encodes a member of the major histocompatibility complex (MHC) class I family of proteins. It is one of the ligands of natural killer group 2, member D (NKG2D) receptor, which functions as an activating receptor in innate and adaptive immunity.39 RTN4 is necessary for immune responses triggered by nucleic acid-sensing TLRs.40 Besides, RTN4 is also an inhibitor of axonal regeneration in the CNS.
Figure 3.
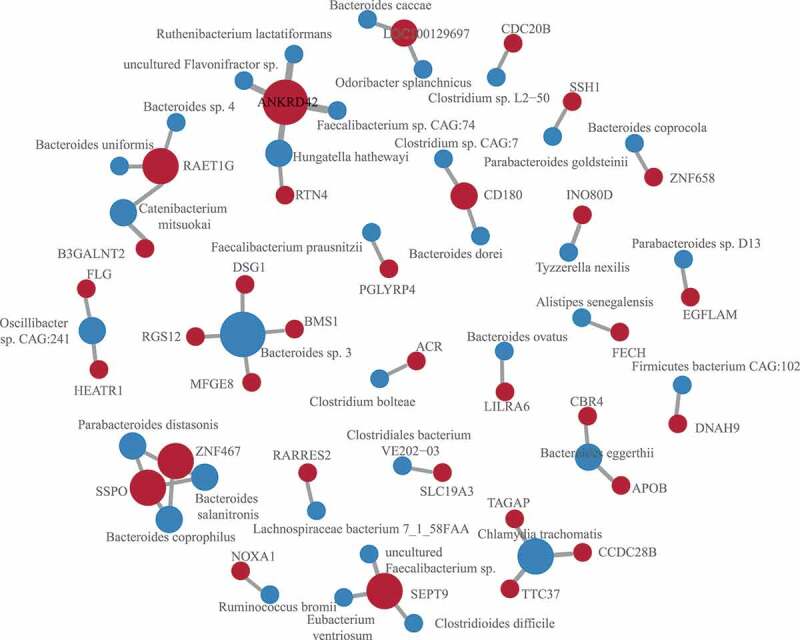
SNVs associated with microbes
SNV-microbe associations identified by regression. The SNVs were mapped to genes. The size of the node is proportional to the number of associations involved by the gene or microbe.
Also, APOB (Apolipoprotein B, rs13306187, c.G4163A, p.R1388H) and RARRES2 (Retinoic Acid Receptor Responder 2, rs147597725, c.G440A, p.S147N), two genes involved in the retinoid metabolic process, were related to the abundance of Lachnospiraceae bacterium 7_1_58FAA and Bacteroides eggerthii, respectively. Apolipoprotein B was an innate barrier against bacterial infection,41 and novel APOB mutations were observed in autism.42 The Bacteroides eggerthii was reported to enhance colitis in mice.43 Studies have reported that the Bacteroidetes/Bacteroidales were among the key taxa related to vitamin A in ASD children.44 Other associations include the thiamine transporters SLC19A3 (rs117864472, c.A1132G, p.I378V) and Clostridiales bacterium VE202-03, among others. The thiamine deficiency was observed in Alzheimer’s disease and contribute to synapse and neural circuit defects.45
Previous studies have demonstrated that ASD genetic risk can act in part through expression quantitative trait loci,46 namely, eQTL, and epigenetic regulation,47 such as meQTL. Accordingly, we explored the association between microbes and the eQTL or meQTL SNVs. We mapped the total SNVs to the publicly available eQTL48 and meQTL47 maps for multiple tissues. Thirty SNVs in our ASD cohort were the eQTL or meQTL loci of neuropsychiatric disorders (Figure 4(a)). However, a limited number of these loci were associated with microbes (Figure 4(a)). An exonic variation in the non-coding RNA LOC285819 (rs34104395), which has an eQTL effect on BTN3A2 in both colon and brain tissue, was associated with Clostridium sp. L2-50. BTN3A2 is a member of the immunoglobulin superfamily, residing in the juxta-telomeric region of the major histocompatability class 1 locus. Increased expression of BTN3A2 might confer risk for schizophrenia by altering excitatory synaptic function. Previous studies have reported the influence of MHC gene variation in shaping gut microbiome composition in both mice49 and human.50 This observation suggested a possible genetic regulation of gut microbiota mediated by the host immune system. Besides, the eQTL loci (rs75634125) of MRPS22 (Mitochondrial Ribosomal Protein S22) was associated with the abundance of Roseburia faecis. The MRPS22 mutation was reported to be associated with the mitochondrial disorder.51
Figure 4.

The association between QTL SNVs and microbial pathways, the causal relationship between SNVs, microbes, and metabolites
(A) The association between eQTL/meQTL loci and microbes, and the genes putatively regulated by these loci. The inner labels are ASD SNVs that mapped to the eQTL or meQTL map, and the outermost labels represent genes regulated by the eQTL or meQTL loci. The blue links represent the associations between ASD SNVs and the gut microbes. (B) The associations between SNVs (gene) and microbial pathways (KEGG pathway, levele 3).
Host SNVs associated with microbial pathways and microbiome gene abundance
To assess how and to what extent host SNVs impact the functions of microbiome, associations between SNVs and microbial pathways and genes were tested. We identified 22 associations between 13 host genes and 20 microbial pathways (Figure 4b, Table S3). Several variations were associated with metabolism pathways, including variations located in MUC3A (chr7:100550525,c.C1106T, p.P369L; chr7:100550534, c.C1115G, p.S372 C) and ANKRD20A2/3 (Ankyrin Repeat Domain 20 Family Member A1, rs201420500, c.C2456A, p.S819Y) that are associated with lipid metabolism pathways, and LAMTOR1 variation (rs146341570, c.C374T, p.P125L) that associated with lysine degradation. Notably, variations in PLCG2 (rs201080992, c.C579G, p.H193Q) and ALDH1A (rs8187929, c.A529T, p.I177F) were associated with the pathway of Xenobiotics biodegradation and metabolism (caprolactam degradation and naphthalene degradation pathways). ALDH1A1 is a multifunctional enzyme with dehydrogenase, esterase, and anti-oxidant activities. Moreover, coding variation in PLGG2 implicated microglial-mediated innate immunity in Alzheimer’s disease.52 Toxin exposure has been epidemiologically demonstrated as one of the main etiological factors of ASD, and deficiencies in toxicant-degradation pathways were reported in ASD.53 These results implicated the possible genetic factors that were underlying the microbial detoxification deficiency. Besides, a couple of gene variations were associated with the immune system. For example, variation in an ASD-related gene SSH1 (rs117900986, c.A1862G, p.N621S) was associated with the human immune disease. SSH1 encodes proteins dephosphorylate and activate the actin-binding/depolymerizing factor cofilin. It is required by NOD1 to detect bacterial-induced changes in actin dynamics leading to NF-κB activation and innate immune responses.54 Other associations were observed between variation within NOXA1 (rs201388549, c.C526T, p.R176W), an activator of Nox2-based NADPH oxidase,55 and the carbohydrate digestion and absorption, and variation of ANKRD42 (rs14346246, c.G1547A, p.G516D) with the wnt signaling pathway.
Furthermore, compared to the total annotated genes, the host-SNVs-associated genes were enriched in multiple metabolism pathways in the KEGG annotation (Table S4), including carbohydrate metabolism, amino acid metabolism, metabolism of cofactors, nucleotide metabolism and vitamins, and biosynthesis of other secondary metabolites. Additionally, enriched pathways include membrane transport (Figure S2A). Also, these microbial genes were enriched in cell motility for the eggNOG annotation (Figure S2B).
The causal relationship between SNV, microbiome and its metabolites
One of the critical mechanisms by which microbes regulate host physiology is through their metabolites.15,56 To decipher how the host SNVs impact microbiome and their metabolites in ASD, a causal inference test (CIT) proposed by Millstein et al.57 was applied to infer the causal relationship between SNV, microbes and their metabolites. Focusing on the SNVs associated with specific microbes identified in the previous section, 8 SNV-microbe-metabolite trios, comprising 4 SNV-microbe associations detected before and 8 metabolites (Figure 5(a), Table S5), were identified.
Figure 5.
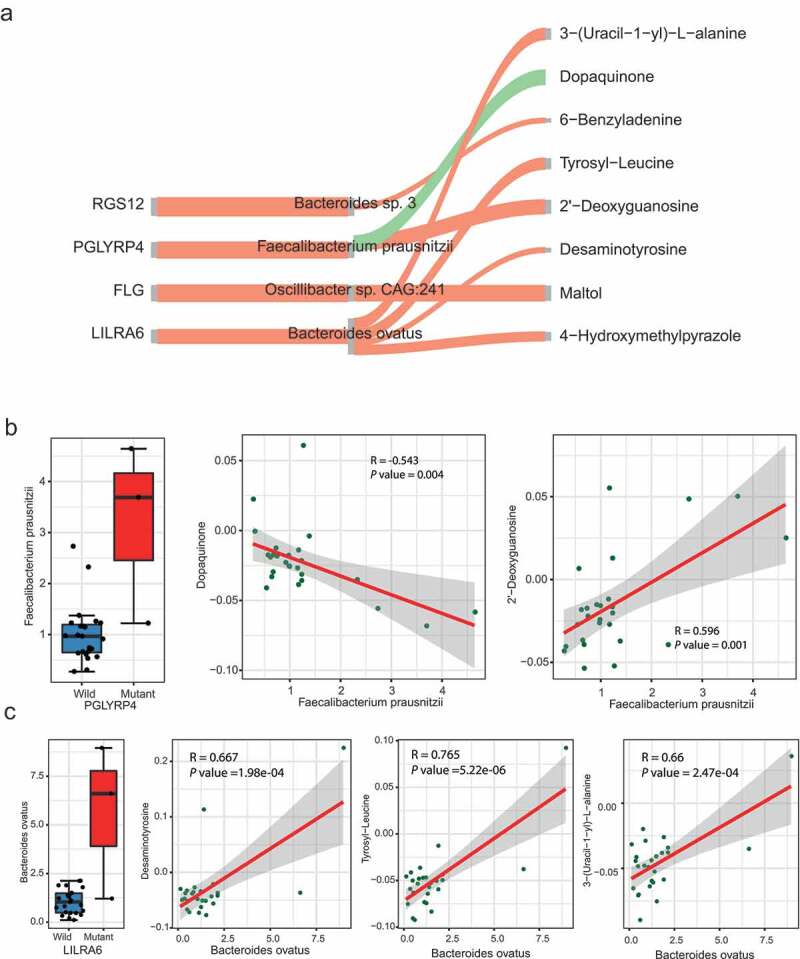
The causal relationship between SNVs, microbes, and metabolites
(A) The causal relationship of SNV-microbe-metabolite trios inferred by causal inference test. The red and green links represent positive and negative assoications, respectively.(B) The abundance of Faecalibacterium prausnitzi related to PGLYRP4a SNV status, and the correlation between Faecalibacterium prausnitzi and Dopaquinone and 2ʹ-Deoxyguanosine.(C) The abundance of Bacteroides ovatus related to SNV status of LILRA6 and the correlation between Bacteroides ovatus and Desaminotyrosine, Tyrosyl-Leucine, and 3-(Uracil-1- yl)-L- alanine.
Three metabolites in the putative causal relationship network were involved in tryptophan and tyrosine metabolism, i.e. Tyrosyl-Leucine, Dopaquinone, and Desaminotyrosine. The level of Dopaquinone was causally related to the variation of PGLYRP4 (rs148195147, c.G602A, p.R201Q) and the abundance of Faecalibacterium prausnitzii (Figure 5(b)). Also, the Faecalibacterium prausnitzii is associated with the level of the 2ʹ-Deoxyguanosine, a metabolite involved in the folate biosynthesis pathway. A variation located in LILRA6 (rs56257556, c.A931 G, p.N311D) was associated with Desaminotyrosine, Tyrosyl-Leucine, and an Alanine metabolites 3-(Uracil-1-yl)-L-alanine mediated by Bacteroides ovatus (Figure 5(c)). LILRA6 is a leukocyte immunoglobulin-like receptor and plays roles in the pathways of the innate immune system and class I MHC mediated antigen processing and presentation. The de novo mutation of LILRA6 was previously observed in ASD.58
Association of cytokines with host genetics and microbiota
Emerging research suggests that immune dysfunction is a risk factor contributing to the neurodevelopmental defects observed in ASD.59 We then tested the levels of seven cytokines in the blood serum (Table S6). The level of IL-1β, IL-4, IL-17α, IL-10, and TGF-β was significantly higher in ASD children compared with TDs (Figure 6(a)), consistent with the previous studies.8,17,18 Several studies have suggested that host genetic can reshape the gut microbiome and its metabolites through aberrant immune activation.8 The association between genetic variation and the differing cytokines was tested. The level of all the five differentially expressed cytokines was associated with SNVs (Figure 6(b), Table S7). Including known ASD-associated genes MUC2, MUC4, SETD5, MUC5B, INO80D, SEPT9, PGLYRP4, DST, and CHD6, among others. Besides, all the five cytokines were correlated with microbes and metabolites (Figure 6(c-d), Table S8-9). These results indicated that the interaction of gut microbes-metabolites and host genetics might play essential roles in the aberrant immune activation in ASD children.
Figure 6.
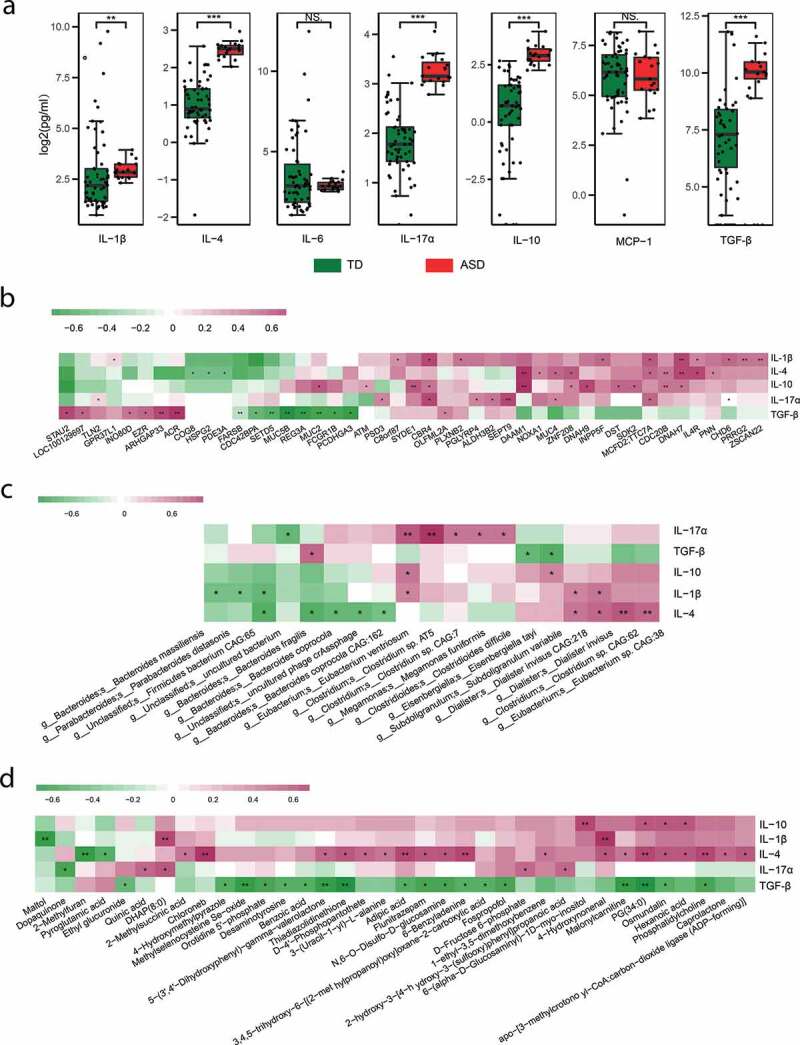
Cytokines in ASD and their association with gene, microbes, and metabolites
(A) The level of in cytokines in ASD and control samples, Wilcoxon test, ***fdr<0.001, **fdr<0.01, and *fdr<0.05.(B) The heatmap of associations between SNV (gene) and cytokines, * linear regression, p-value<0.05, ** p-value<0.01.(C) The heatmap of associations between microbes and cytokines, Spearman correlation, * p-value<0.05, ** p-value<0.01.(d) The heatmap of associations between metabolites and cytokines, Spearman correlation, * p-value<0.05, ** p-value<0.01.
Discussion
In this study, we first investigated the interplay between host and gut microbial in terms of host genetic, microbial composition, microbial gene abundance, microbial metabolites, and host immunity in ASD. We provided new insight into the interaction between host genetic variations and gut microbiota in the pathogenesis of ASD (Figure 7).
Figure 7.
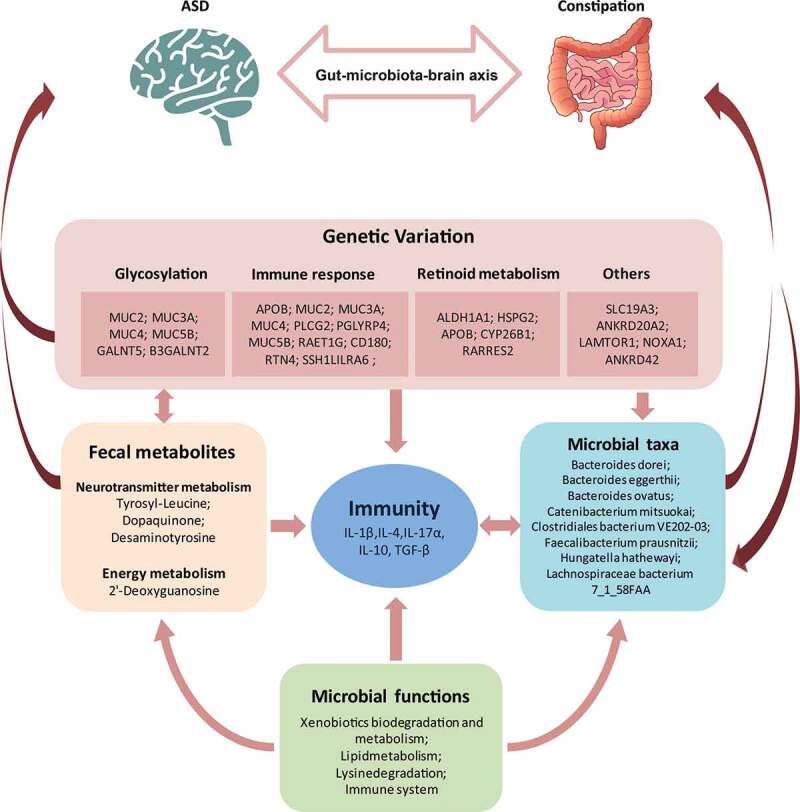
The summary of the interplay among host genetics, gut microbes, fecal metabolites, and cytokines
The link between ASD and the gut microbiome has been highlighted by many recent studies.12,15,60,61,62 Given that ASD is a disease influenced by both genetic and environmental factors, it is of interest to study the microbiome in the context of host genetic variation. Here, we jointly analyzed the SNV profile and the taxonomic composition of the fecal microbiome as well as its metabolites, and additionally, the host cytokines in ASD. We found that the composition of the microbiome is correlated with functional SNVs. The functional SNVs located in genes involved in glycosylation process, innate immune response, and retinoid metabolic process were significantly enriched in the presence of ASD, indicating that the abnormal function of proteins that participated in these biological processes may be involved in the pathogenesis of ASD. Consistent with that, the subjects recruited in this study all had symptoms of constipation, the SNVs identified in the samples were enriched in the genes associated with gut functions, e.g. the mucin gene (MUC2, MUC3A, MUC4, and MUC5B). The mucins can act as the protective barrier or luminal sensor for gut immunity and regulate gut microbiota.63 Also, the increased gastric mucin was observed in the treatment of chronic constipation with Lubiprostone.64 More importantly, the mucin gene mutation was reported in multiple ASD cohorts.42,65,66 The association analysis between functional SNVs with microbes in this current study revealed the involvement of SNVs within MUC family genes in the interaction between gut microbiota of constipated ASD.
Immune dysfunction is a viable risk factor contributing to the neurodevelopmental deficits observed in ASD,59 and many autism susceptibility genes are related to immune/infection pathways.67 On the other hand, microbes influence the activation of peripheral immune cells and contributes to the etiopathogenesis of neurobehavioral and neurodegenerative disorders, such as ASD, anxiety depression, Alzheimer’s disease and Parkinson’s disease.68 In this study, we found several associations between the immune-related gene variations and gut microbial taxa. For example, the variations located in PGLYRP4, APOB, CD180, RAET1G, and LILRA6 were associated with multiple species, mainly the Bacteroides, altered abundance of which was reported in multiple ASD studies.69 These observations implicated a scenario where the host genetic shaped the gut microbes and indicated an interplay between immunity and microbes in the context of ASD. However, for the other type of SNVs (intergenic, upstream/downstream, intronic SNVs, and non-coding exonic SNVs), thought tens of them were mapped to eQTL or meQTL loci, a limited number of these SNVs were associated with the abundance of microbes (Figure 4(a)). There were two possibilities regarding this observation. First, the interplay between genetic variations and gut microbiota abundance in ASD might occur mostly through the loss-of-function (LoF) gene variations. Second, another possible explanation is that due to the small sample size of our study, there was not enough statistical power to detect indirect associations between SNVs and microbes, which were mediated by the eQTL or meQTL genes. In this case, by increasing the sample size, we may expect an increased number of associations between nonfunctional SNVs and gut microbes being observed.
In the association analysis between host SNVs and the gut microbial pathways and genes, we observed notable associations between the SNVs and immune system and metabolic pathways. Metabolites act as messengers of information between the intestinal microbiota and host cells. The presence of metabolites, which depends on the microbial metabolic activity, thus impacts host development, health, and pathogenesis.70 For example, amino acids serve as precursors for many neuroactive molecules, such as serotonin, and GABA. The amino acid metabolism were reported to be different between TD and ASD individuals.71,72 The association between host SNVs with microbial genes related to amino acid metabolism suggested a mechanism through which ASD host genetic variations influence neurodevelopment and behavior in a microbe-mediated way. The additional causal inference test also identified multiple genetic variations associated with the level of metabolites involved in the neurotransmitter metabolism. This result provides a possible mechanism underlying the genetic susceptibility of ASD. To identify variations with higher accuracy, we only considered variations detected in more than two samples. Considering the small sample size of our study, some rare or less common variations which may also contribute to the pathogenesis of ASD will be filtered. Thus, it is necessary to recruit more patients to capture such variations linking to ASD.
Taken together, the current study provides important clues to explore the mechanism of ASD from the perspective of genetic and environmental interaction. Many studies describe gut dysbiosis, disorders of metabolism activity, and immune dysfunction, as a change in ASD. However, little is known about whether these changes are a cause or consequence of an altered pathological state. Therefore, further animal experiments are needed to understand exactly how the candidate genetic variants influence the interplay between microbiota, metabolism, and the immune system in pathology of ASD.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Affiliated Yixing Hospital of Jiangsu University (Ethics NO, 2016055). All participants and their legal guardians were provided a written informed consent upon enrollment. Once the consent forms were signed, we screened them for eligibility criteria and sent questionnaires and sample collection kits to participants who meet the study eligibility criteria.
Study subject recruitment
The children with ASD were recruited as we described in our previous study73. Briefly, ASD children were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition. Each participant underwent physical, neurological, and behavioral examinations. Children were excluded from the study if they were previously diagnosed with a genetic condition such as tuberous sclerosis, significant sensory impairment, clinically evident inflammatory conditions, celiac disease, and other physiological conditions (i.e. depressive disorder, schizophrenia, and bipolar disorder). Participants that are on anti-inflammatory or anti-oxidant drugs, or take antibiotics, probiotics, prebiotics, and antifungal medications in 3 months before the feces collection were excluded. The Rome IV criteria for functional constipation were used for evaluating GI problems. The typically developed children (TD) for the fecal metabolism and cytokines measure were recruited from kindergartens through pediatricians and under physical, neurological, and behavioral examinations as the ASD group, subjects with any pathological conditions, including GI problems were excluded (Table S10).
Exon sequencing and data process
Peripheral blood samples were collected from the ASD and TD group for extracting DNA. Exome capture was performed using the Agilent SureSelect Human All Exon V6 r2 kit followed by Illumina paired-end sequencing.
The raw reads were quality-controlled using FastQC v0.11.2 and then mapped to the human reference genome (hg19) using bwa v0.7.17.74 PCR duplicates were removed using Picard tools v2.18.12. SNVs were identified using FreeBayes v1.2.075 and filtered using bcftools v1.9. SNVs with base quality < 30, coverage < 30, with less than 3 reads mapped to the alternative alleles or located within 5bp nearby a gap were removed.
SNVs presented in the TD groups were removed. Then, SNVs were annotated by the annovar software to extract allele frequencies from the public database Allele frequency of these SNVs in the normal population was extracted from the 1000 Genome project,76 the Exome Aggregation Consortium (ExAC) database,77 and the Chines MillionomeI database (CMDB),78 a large-scale Chinese genomics database including comprehensive variation and their allele frequency information from 141,431 unrelated healthy Chinese individuals. SNVs with MAF < 5% in these public databases and detected in more than two ASD samples were considered as candidates. GO enrichment analysis of genes with SNVs was conducted in R package.
Fecal sample collection
Fecal samples of each participant were obtained at the hospital or home and collected as directed in the instruction provided and frozen immediately until shipment. Samples were shipped using dry ice overnight to Nanjing Medical University and stored at −80°C until extraction.
Metagenomic sequencing and preprocess
About 2 μg DNA per sample was prepared for library construction and then sequenced on the Illumina HiseqX platform. Low quality or contaminated human reads were removed, and the high-quality reads were assembled into contigs using SOAPdenovo (v2.04).79 MetaGeneMark (v2.10)80 was used to predict genes from the assembled contigs. CD-HIT v4.5.881 was used to generate a non-redundant gene catalog. Abundances of the genes were computed by aligning high-quality reads to the reference gene database. For taxonomic identity and functional assignment of unigenes, reads were aligned to the NCBI NR database (e-value≤1e-5) using DIAMOND (v0.9.9).82 The LCA algorithm83 was used to conduct annotation. The detailed methods and results have been presented by our other study.
Gene functional annotations
The unigenes were functionally annotated by mapping to different functional protein databases with BLAST software. Predicted unigenes were assigned to the Kyoto encyclopedia of genes and genomes (KEGG), the evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG), and carbohydrate-active enzymes database (CAZy) database using DIAMOND (v0.9.9). The abundances of each functional annotation were the sum of the abundance of annotation of each functional level.
Metabonomics analysis and data process
For each ASD and TDs, about 50 mg feces were mixed with 1000 μL of extract solvent, and samples were transferred to UHPLC-QE Orbitrap/MS analysis. The differential analysis was performed using the ropls package(v1.12.0), and metabolites were with VIP > 1, P-value (T-test) < 0.05 and FC>1.5 were defined as significant different in abundance. The detailed methods have been presented in our previous study.
Association analysis
Association test was performed using standard linear regression after adjusting for established confounding factors (age and sex). p-values were then corrected for multiple tests using the Benjamini-Hochberg method.
Causal inference analysis
The SNV-microbe-metabolite trios were assessed using the causal inference test (CIT)57 to test the possibility that SNV causally influences microbe abundance and then the microbe metabolite. The enriched SNVs and their associated microbes, and the differential metabolites between ASD and TD group were subjected to the test. Briefly, the CIT has statistical tests for four conditions, all of which must be met for the microbe -mediated causal classification: (1) SNV and microbe are associated; (2) SNV is associated with microbe after adjusting for metabolite; (3) microbe is associated with metabolite after adjusting for SNV; and (4) SNV is independent of metabolite after adjusting for microbe. The CIT p-value was defined as the maximum of the component test p-values, and multivariate linear regression was used in the four-component tests.
Cytokine assay in healthy control and ASD patients
We measured cytokines in blood serum of the ASD cohort and an independent TD group including IL-1ß, IL-4, IL-6, IL-10, IL-17α, MCP-1, and TCG-β using BD™ CBA Flex Set (Table S6). Then the level of them was log-transformed, and the Mann-Whitney U test was applied to test the difference between ASD and TDs. These p-values were then corrected for multiple tests using the Benjamini-Hochberg method and cytokines with FDR<0.05 were considered as differentially expressed.
Supplementary Material
Funding Statement
This work was supported by NSFC [grant 81671983 and 81871628], Natural science funding [BK20161572] from Jiangsu province and starting package from NJMU (Xingyin Liu). Starting funding for the team of gut microbiota research in NJMU (Xingyin Liu, Faming Zhang, Yankai Xia, and Chuan Su).
Data availability
All metagenomic raw data have been deposited in GEO under accession number GSE113540.
Disclosure
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Muhle R, Trentacoste SV, Rapin I.. The genetics of autism. Pediatrics. 2004;113:e472–16. doi: 10.1542/peds.113.5.e472. [DOI] [PubMed] [Google Scholar]
- 2.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J, Park CY, Theesfeld CL, Wong AK, Yuan Y, Scheckel C, Fak JJ, Funk J, Yao K, Tajima Y, et al. Whole-genome deep-learning analysis identifies contribution of non-coding mutations to autism risk. Nat Genet. 2019;51:973. doi: 10.1038/s41588-019-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Guo H, Xiong B, Stessman HAF, Wu H, Coe BP, Turner TN, Liu Y, Zhao W, Hoekzema K, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, Levy D, Allen J, Ye K, Ronemus M, Lee Y-H, Yamrom B, Wigler M. Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci U S A. 2015;112:E5600–5607. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goyal DK, Miyan JA. Neuro-immune abnormalities in autism and their relationship with the environment: a variable insult model for autism. Front Endocrinol. 2014;5:29. doi: 10.3389/fendo.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson BJ, Dovgan K, Takahashi N, Beversdorf DQ. The relationship among gastrointestinal symptoms, problem behaviors, and internalizing symptoms in children and adolescents with autism spectrum disorder. Front Psychiatry. 2019;10:194. doi: 10.3389/fpsyt.2019.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dan Z, Mao X, Liu Q, Guo M, Zhuang Y, Liu Z, Chen K, Chen J, Xu R, Tang J, et al. Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes. 202020; 11:1246–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zurita MF, Cárdenas PA, Sandoval ME, Peña MC, Fornasini M, Flores N, Monaco MH, Berding K, Donovan SM, Kuntz T, et al. Analysis of gut microbiome, nutrition and immune status in autism spectrum disorder: a case-control study in ecuador. Gut Microbes. 2020; 11:453–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. The microbiota modulates gut physiology and behavioral abnormalities associated with autism. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinolo MAR, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. 2011;22:849–855. doi: 10.1016/j.jnutbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Sharon G, Cruz NJ, Kang D-W, Gandal MJ, Wang B, Kim Y-M, Zink EM, Casey CP, Taylor BC, Lane CJ, et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in Mice. Cell. 2019;177(1600–1618.e17):1600–1618.e17. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Ayadhi LY, Mostafa GA. Elevated serum levels of interleukin-17A in children with autism. J Neuroinflammation. 2012;9:158. doi: 10.1186/1742-2094-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krakowiak P, Goines PE, Tancredi DJ, Ashwood P, Hansen RL, Hertz-Picciotto I, Van de Water J. Neonatal cytokine profiles associated with autism spectrum disorder. Biol Psychiatry. 2017;81:442–451. doi: 10.1016/j.biopsych.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molloy CA, Morrow AL, Meinzen-Derr J, Schleifer K, Dienger K, Manning-Courtney P, Altaye M, Wills-Karp M. Elevated cytokine levels in children with autism spectrum disorder. J Neuroimmunol. 2006;172:198–205. doi: 10.1016/j.jneuroim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolde R, Franzosa EA, Rahnavard G, Hall AB, Vlamakis H, Stevens C, Daly MJ, Xavier RJ, Huttenhower C. Host genetic variation and its microbiome interactions within the human microbiome project. Genome Med. 2018;10:6. doi: 10.1186/s13073-018-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, Mao J-H, Secombe J, Dan Z, Chen J-H, et al. Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host&Microbe. 2019;25:537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang C, Li J, Wu Q, Yang X, Huang AY, Zhang J, Ye AY, Dou Y, Yan L, Zhou W, et al. AutismKB 2.0: a knowledgebase for the genetic evidence of autism spectrum disorder. Database; 2018; 2018:bay106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. [DOI] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An J-Y, Peng M, Collins R, Grove J, Klei L, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereanu W, Larsen EC, Das I, Estévez MA, Sarkar AA, Spring-Pearson S, Kollu R, Basu SN, Banerjee-Basu S. AutDB: a platform to decode the genetic architecture of autism. Nucleic Acids Res. 2018;46:D1049–54. doi: 10.1093/nar/gkx1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ka M, Kim W-Y. ANKRD11 associated with intellectual disability and autism regulates dendrite differentiation via the BDNF/TrkB signaling pathway. Neurobiol Dis. 2018;111:138–152. doi: 10.1016/j.nbd.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee Y-H, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes IR, Cruz ACP, Ferrasa A, Phan D, Herai RH, Muotri AR. Genetic variations on SETD5 underlying autistic conditions. Dev Neurobiol. 2018;78:500–518. doi: 10.1002/dneu.22584. [DOI] [PubMed] [Google Scholar]
- 30.Bergstrom KSB, Mucin-type XL. O-glycans and their roles in intestinal homeostasis. Glycobiology. 2013;23:1026–1037. doi: 10.1093/glycob/cwt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dwyer CA, Esko JD. Glycan susceptibility factors in autism spectrum disorders. Mol Aspects Med. 2016;51:104–114. doi: 10.1016/j.mam.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Zwaag B, Franke L, Poot M, Hochstenbach R, Spierenburg HA, Vorstman JAS, van Daalen E, de Jonge MV, Verbeek NE, Brilstra EH, et al. Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS ONE. 2009;4:e5324. doi: 10.1371/journal.pone.0005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Sodhi RK, Singh N. Retinoids as potential targets for Alzheimer’s disease. Pharmacol Biochem Behav. 2014;120:117–123. doi: 10.1016/j.pbb.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 36.Guo M, Zhu J, Yang T, Lai X, Lei Y, Chen J, Vitamin LT. A and vitamin D deficiencies exacerbate symptoms in children with autism spectrum disorders. Nutr Neurosci. 2019;22:637–647. doi: 10.1080/1028415X.2017.1423268. [DOI] [PubMed] [Google Scholar]
- 37.Dabrowski AN, Shrivastav A, Conrad C, Komma K, Weigel M, Dietert K, Gruber AD, Bertrams W, Wilhelm J, Schmeck B, et al. Peptidoglycan recognition protein 4 limits bacterial clearance and inflammation in lungs by control of the gut microbiota. Front Immunol. 2019;10:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi J, Kuroda Y, Koyama D, Osada N, Izumi T, Yasui H, Kawase T, Ichinohe T, Furukawa Y. Myeloma cells are activated in bone marrow microenvironment by the CD180/MD-1 complex, which senses lipopolysaccharide. Cancer Res. 2018;78:1766–1778. doi: 10.1158/0008-5472.CAN-17-2446. [DOI] [PubMed] [Google Scholar]
- 39.Eagle RA, Flack G, Warford A, Martínez-Borra J, Jafferji I, Traherne JA, Ohashi M, Boyle LH, Barrow AD, Caillat-Zucman S, et al. Cellular expression, trafficking, and function of two isoforms of human ULBP5/RAET1G. PloS One. 2009;4:e4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura T, Endo S, Inui M, Saitoh S-I, Miyake K, Takai T. Endoplasmic protein Nogo-B (RTN4-B) interacts with GRAMD4 and regulates TLR9-mediated innate immune responses. J Immunol. 2015;194:5426–5436. doi: 10.4049/jimmunol.1402006. [DOI] [PubMed] [Google Scholar]
- 41.Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. Apolipoprotein B is an innate barrier against invasive staphylococcus aureus infection. Cell Host Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional, and chromatin genes disrupted in autism. Nature. 2014;515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dziarski R, Park SY, Kashyap DR, Dowd SE, Pglyrp-Regulated Gut GD, Mizoguchi E. Microflora prevotella falsenii, parabacteroides distasonis and bacteroides eggerthii enhance and alistipes finegoldii attenuates colitis in Mice. PloS One. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Liu X, Xiong X-Q, Yang T, Cui T, Hou N-L, Lai X, Liu S, Guo M, Liang X-H, et al. Effect of vitamin A supplementation on gut microbiota in children with autism spectrum disorders - a pilot study. BMC Microbiol. 2017;17:204. doi: 10.1186/s12866-017-1096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Q, Liu H, Sang S, Chen L, Zhao Y, Wang Y, Zhong C. Thiamine deficiency contributes to synapse and neural circuit defects. Biol Res. 2018;51:35. doi: 10.1186/s40659-018-0184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamazon ER, Badner JA, Cheng L, Zhang C, Zhang D, Cox NJ, Gershon ES, Kelsoe JR, Greenwood TA, Nievergelt CM, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrews SV, Ellis SE, Bakulski KM, Sheppard B, Croen LA, Hertz-Picciotto I, Newschaffer CJ, Feinberg AP, Arking DE, Ladd-Acosta C, et al. Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat Commun. 2017;8:1011. doi: 10.1038/s41467-017-00868-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, Bell R, Ajami NJ, Petrosino JF, Morrison L, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun. 2015;6:8642. doi: 10.1038/ncomms9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell JT, Roesch LFW, Ördberg M, Ilonen J, Atkinson MA, Schatz DA, Triplett EW, Ludvigsson J. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat Commun. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kılıç M, K-K O, Kılıç E, Yüksel D, Demirci H, MŞ S, Yücel-Yılmaz D, Özgül RK. A patient with mitochondrial disorder due to a novel mutation in MRPS22. Metab Brain Dis. 2017;32:1389–1393. doi: 10.1007/s11011-017-0074-5. [DOI] [PubMed] [Google Scholar]
- 52.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang M, Chu Y, Meng Q, Ding R, Shi X, Wang Z, He Y, Zhang J, Liu J, Zhang J, et al. A quasi-paired cohort strategy reveals the impaired detoxifying function of microbes in the gut of autistic children. Sci Adv. 2020; 6:eaba3760. [DOI] [PMC free article] [PubMed]
- 54.Bielig H, Lautz K, Braun PR, Menning M, Machuy N, Brügmann C, Barisic S, Eisler SA, Andree M, Zurek B, et al. The cofilin phosphatase slingshot homolog 1 (SSH1) Links NOD1 signaling to actin remodeling. PLoS Pathog. 2014;10:e1004351. doi: 10.1371/journal.ppat.1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawano M, Miyamoto K, Kaito Y, Sumimoto H, Tamura M.. Noxa1 as a moderate activator of Nox2-based NADPH oxidase. Arch Biochem Biophys. 2012;519:1–7. [DOI] [PubMed] [Google Scholar]
- 56.Liu H, Chen X, Hu X, Niu H, Tian R, Wang H, Pang H, Jiang L, Qiu B, Chen X, et al. Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome. 2019;7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner TN, Coe BP, Dickel DE, Hoekzema K, Nelson BJ, Zody MC, Kronenberg ZN, Hormozdiari F, Raja A, Pennacchio LA, et al. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–722.e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meltzer A. The role of the immune system in autism spectrum disorder. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2017;42:284–298. doi: 10.1038/npp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malkki H. Neurodevelopmental disorders: human gut microbiota alleviate behavioural symptoms in a mouse model of autism spectrum disorder. Nat Rev Neurol. 2014;10:60. [DOI] [PubMed] [Google Scholar]
- 61.:Wang M, Wan J, Rong H, He F, Wang H, Zhou J, Cai C, Wang Y, Xu R, Yin Z, et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019;4:e00321-18.. doi: 10.1128/mSystems.00321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li N, Yang J, Zhang J, Liang C, Wang Y, Chen B, Zhao C, Wang J, Zhang G, Zhao D, et al. Correlation of Gut Microbiome Between ASD Children and Mothers and Potential Biomarkers for Risk Assessment. Genomics Proteomics Bioinformatics 2019;17:26–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang C-S, Kao C-Y. Current understanding of the gut microbiota shaping mechanisms. J Biomed Sci. 2019;26:59. doi: 10.1186/s12929-019-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Majewski M, Sarosiek I, Wallner G, Edlavitch SA, Sarosiek J. Stimulation of mucin, mucus, and viscosity during lubiprostone in patients with chronic constipation may potentially lead to increase of lubrication. Clin Transl Gastroenterol. 2014;5:e66. doi: 10.1038/ctg.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D’Gama A, Kim SN, Hill RS, Goldberg AP, et al. Rates, distribution, and implications of post-zygotic mosaic mutations in autism spectrum disorder. Nat Neurosci. 2017;20:1217–1224. doi: 10.1038/nn.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen R, Davis LK, Guter S, Wei Q, Jacob S, Potter MH, Cox NJ, Cook EH, Sutcliffe JS, Li B. Leveraging blood serotonin as an endophenotype to identify de novo and rare variants involved in autism. Mol Autism. 2017;8:14. doi: 10.1186/s13229-017-0130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carter CJ. Autism genes and the leukocyte transcriptome in autistic toddlers relate to pathogen interactomes, infection and the immune system. A role for excess neurotrophic sAPPα and reduced antimicrobial Aβ. Neurochem Int. 2019;126:36–58. [DOI] [PubMed] [Google Scholar]
- 68.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu F, Li J, Wu F, Zheng H, Peng Q, Zhou H. Altered composition and function of intestinal microbiota in autism spectrum disorders: a systematic review. Transl Psychiatry. 2019;9:43. doi: 10.1038/s41398-019-0389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sittipo P, Shim J, Lee YK. Microbial metabolites determine host health and the status of some diseases. Int J Mol Sci. 2019;20:5296. doi: 10.3390/ijms20215296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aldred S, Moore KM, Fitzgerald M, Waring RH, Aldred S, Moore KM, Fitzgerald M, Waring RH . Plasma amino acid levels in children with autism and their families. J Autism Dev Disord. 2003;33:93–97. doi: 10.1023/A:1022238706604. [DOI] [PubMed] [Google Scholar]
- 72.Evans C, Dunstan RH, Rothkirch T, Roberts TK, Reichelt KL, Cosford R, Deed G, Ellis LB, Sparkes DL. Altered amino acid excretion in children with autism. Nutr Neurosci. 2008;11:9–17. doi: 10.1179/147683008X301360. [DOI] [PubMed] [Google Scholar]
- 73.Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinforma Oxf Engl. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv 2012;1207:3907 [Google Scholar]
- 75.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al . A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu S, Huang S, Chen F, Zhao L, Yuan Y, Francis SS, Fang L, Li Z, Lin L, Liu R, et al. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and chinese population history. Cell. 2018;175:347–359.e14. doi: 10.1016/j.cell.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 78.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinforma Oxf Engl. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 82.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All metagenomic raw data have been deposited in GEO under accession number GSE113540.


