Abstract
Recombinant viruses are the workhorse of modern neuroscience. Whether one would like to understand a neuron’s morphology, natural activity patterns, molecular composition, connectivity or behavioural and physiologic function, most studies begin with the injection of an engineered virus, often an adeno-associated virus or herpes simplex virus, among many other types. Recombinant viruses currently enable some combination of cell type-specific, circuit-selective, activity-dependent and spatiotemporally resolved transgene expression. Viruses are now used routinely to study the molecular and cellular functions of a gene within an identified cell type in the brain, and enable the application of optogenetics, chemogenetics, calcium imaging and related approaches. These advantageous properties of engineered viruses thus enable characterization of neuronal function at unprecedented resolution. However, each virus has specific advantages and disadvantages, which makes viral tool selection paramount for properly designing and executing experiments within the central nervous system. In the current Review, we discuss the key principles and uses of engineered viruses and highlight innovations that are needed moving forward.
A fundamental goal of neuroscience is to understand how the brain generates thought and action. The brain is the single most complex biological entity known to humankind, making it exceptionally difficult to study. One of the greatest barriers to understanding the CNS is its molecular and cellular heterogeneity — millions (in mice) to billions (in humans) of neurons are densely packed and interconnected, interacting with one another and with non-neuronal cell types, including several types of glia and endothelial cells. In order to understand how these diverse cell types together orchestrate thought and action, one first needs to access them.
Over the past century or so, Herculean efforts have been made to better understand how the nervous system’s structure gives rise to its function. These studies have often involved the use of a virus (Box 1). Wild-type viruses and, more recently, recombinant viruses (viral genomes with added ‘marker genes’, ‘reporter genes’ or other useful elements) have enabled unprecedented access to cells, by virtue of their activity, connectivity or expression of a marker gene. Recombinant viruses thus enable long-term, cell type-specific and spatiotemporally resolved genetic access to certain cell populations. Whether one is seeking to understand CNS function as it relates to molecules and excitability, cell types and circuits or behaviour and physiology, one typically starts today with the injection of an engineered virus. These recombinant viruses, in combination with the appropriate genetic tools, allow for uniquely sensitive, selective and modular approaches to neuroscientific studies.
Box 1 |. History of the use of viral vectors in neuroscience research.
Viruses were first employed to study nervous system connectivity, classically Papez’s use of rabies virus to delineate the limbic system in the 1930s121. This was followed several decades later by more widespread use of pseudorabies virus for anatomical tracing122. However, the toxicity of these vectors (leading to death within weeks) limited their application.
This changed in the 1990s with the introduction of several viruses that, upon injection into adult brain, expressed an encoded transgene. In 1993, three groups reported that adenovirus injected into rodent brain expressed LacZ in infected neurons and glia123–125. In 1994, AAV was used to express tyrosine hydroxylase in rat striatum126 and, in 1997, four groups reported the use of herpes simplex virus 1 (HSV-1)127–129 or lentivirus110 to express transgenes in the brain. These studies ushered in the modern era of viral-mediated gene transfer in neuroscience.
Carlezon et al.128 represented a crucial milestone: the first use of viruses to establish the causal relationship between a molecular adaptation in the brain with a specific functional end point. Previously, establishing causality required using mice where the manipulation of a gene occurred early in development and ubiquitously throughout brain and peripheral tissues. By contrast, viral-mediated gene transfer enabled far greater resolution in both space (targeting an injected brain region only) and time (targeting any point in an animal’s life).
The key features of HSVs used by Carlezon et al.128 were their diminished toxicity, due to their lack of wild-type HSV, purification from cellular debris and greatly increased ratios of amplicon (virus particles carrying the transgene) to helper virus (virus particles necessary to package the amplicon).
This advance led to the rapid application of viruses to establish the causal role of numerous classes of proteins in the brain130–133, including their role in specific neurons or glia, as well as to express Cre recombinase64,134–136 or tools for optogenetics, chemogenetics, fibre photometry, single-cell Ca2+ imaging and, most recently, CRISPR.
The central role that viruses occupy within neuroscientific investigations means that selection of the appropriate recombinant virus is paramount. Many factors come into play when choosing the right virus, including delivery modality, tropism (specificity to infect a given cell type), toxicity (impact on cell health and survival) and packaging (ease of producing sufficient particles), among many others. Tailoring the appropriate viral strategy to the desired biological question is thus essential for a successful experimental outcome.
Several types of recombinant viruses are available for conducting basic neuroscience research, each of them serving different purposes through their diverse properties. Animal viruses are classified in several different ways, such as their Linnaean taxonomy, genomic architecture (as in the Baltimore classification scheme), genome size and capsid symmetry, and whether the virus particle is naked or enveloped1,2. However, such classifications do not per se inform one about key viral properties for use in vivo. To carry out a successful experiment, the neuroscientist needs to be aware of several critical considerations, such as the limits for a given genome length that can be packaged in a particle (the theoretical payload capacity), the time to maximal expression of genes in the viral genome (including experimental transgenes), the onset and persistence of such expression, whether the viral genome integrates into the host genome and so on. These properties help to determine the feasibility and likelihood of success for different viral strategies used in the CNS. These considerations and caveats are outlined in Table 1.
Table 1 |.
Key properties of viruses used in neuroscience studies
| Family genome architecture and BC | Virus (abbreviation) | Genome size and maximum payload | Onset and duration of expression | Integrating or non-integrating | Directionality of spreada | Toxicity | Tropism | Types of experiments | Refs |
|---|---|---|---|---|---|---|---|---|---|
| Adenoviridae (linear dsDNA, BC I) | Canine adenovirus 2 (CAV-2) | Size: 32 kb | Onset: days to week(s) | Non-integrating | Retrograde | Limited | Preferentially infects nerve terminals (over soma) | Projection specificity; long-term studies | 24,97 |
| Payload: ~30 kb | Duration: months | ||||||||
| Herpesviridae (linear dsDNA, BC I) | Herpes simplex virus (HSV) | Size: ~152 kb | Onset: short (hours) | Non-integrating | Local: standard | Dependent on strain, replication competence and purity of vector preparation | Multiple strains capable of infecting CNS, many specific for neurons | Projection and cell type specificity; rapid expression; short-term studies; circuit and molecular perturbation; large payloads | 8,12,54, 98–102 |
| Payload: 30+ kbb | Duration: standard, 5–7 days; LT-HSV, weeks to months | Anterograde: HSV-H129 | |||||||
| Retrograde: LT-HSV | |||||||||
| Pseudorabies virus (PRV) | Size: ~142 kb | Onset: short (hours) | Non-integrating | Bidirectional: Becker | Dependent on strain and replication competence | Preferentially infects nerve terminals (over soma) | Projection specificity; polysynaptic tracing; large payloads | 13,58, 103,104 | |
| Payload: 30+ kbb | Duration: variable | Retrograde: Bartha | |||||||
| Parvoviridae (sssDNA, BC II) | Adeno-associated virus (AAV) | Size: ~4.7 kb | Onset: 1–2 weeks | Rare integration (largely episomal) | Local: AAV1, AAV2, AAV5, AAV8, AAV9 | Minimal | Many serotypes infect CNS; primarily infects soma | Projection and cell type specificity; long-term studies; circuit and molecular perturbation | 10,30, 59–61, 105–107 |
| Payload: ~4.7 kb | Duration: months | Anterograde: AAV1, AAV9 | |||||||
| Retrograde: AAVrgc | |||||||||
| Retroviridae (ssRNA-RT, BC VI) | Lentivirus (LV) | Size: ~9.8 kb (based on HIV-1) | Onset: days to weeks | Integrating | Dependent on pseudotype | Limited | VSV-G coat: local transduction; RbV-G coat: terminals and soma | Larger payloads than accommodated by AAV | 10,98, 108–111 |
 |
Payload: 8 kb | Duration: months | |||||||
| Rhabdoviridae ((−) ssRNA, BC V) | Rabies virus (RbV) | Size: ~12 kb | Onset: short (~2 days) | Non-integrating | Retro | Dependent on strain and replication competence | G coat: neurons and terminals; EnvA coat: TVA+ cells | Projection specificity; monosynaptic/ polysynaptic tracing | 11,56, 77,112 |
| Payload: 3.7+ kb | Duration: months | ||||||||
| Vesicular stomatitis virus (VSV) | Size: ~11 kb | Onset: short (hours) | Non-integrating | Dependent on pseudotype: | High cytotoxicity | Broadly infective, but dependent on pseudotype | Viral tracing; rapid expression | 113–115 | |
| Payload: 6 kb | Duration: toxicity-limited | Anterograde: VSV-G? | |||||||
| Retrograde: RbV-G | |||||||||
| Togaviridae ((+) ssRNA, BC IV) | Sindbis virus | Size: ~12 kb | Onset: short (hours) | Non-integrating | Local | Variable | Local infection for neurotropic strain used in mapping | Barcode-based tracing | 116–120 |
| Payload: ~6 kb? | Duration: toxicity-limited |
AAVrg, retrograde-tracing variant of AAV; BC, Baltimore classification; dsDNA, double-stranded DNA; EnvA, avian ASLV type A envelope protein; HSV, herpes simplex virus; HSV-H129, anterograde-tracing variant of HSV; LT-HSV, long-term HSV; RbV-G, rabies glycoprotein, protein G; ssRNA, single-stranded RNA; TVA, avian tumour virus receptor A; VSV-G, VSV glycoprotein, protein G.
More information on infection pattern (circuit-specific, monosynaptic or polysynaptic) is shown in FIG. 2.
Based on HSV; for amplicons, this number is theoretically 152 kb.
Among others, with lower efficiency.
In the current Review, we discuss the state of the art for viral methods available in the neuroscientist’s toolbox. We describe key considerations for choosing the experimentally appropriate recombinant virus and discuss unresolved questions for targeting cell types throughout the mammalian nervous system.
Virology principles for neuroscientists
Although many recombinant viruses are available for experimentation, there are certain universal properties that define each of these vectors and their utility for a given study. In this section, we define the key principles of virology, restricted to the properties most important to the neuroscientist; for a deeper dive into virology, we recommend the excellent Principles of Virology3 (as well as Addgene’s viral vector guides). We address the key question of what one needs to know about recombinant viruses to design and execute the optimized neuroscience experiment. We focus, in particular, on six key concepts and offer some examples of how specific viruses match up. These concepts are packaging and payload, delivery, tropism, access, infectivity and toxicity, and transgene expression (FIG. 1). Gaining a better understanding of each of these concepts (and how they define a given recombinant virus) will lead to a new lens through which one can design and analyse a neuroscientific study.
Fig. 1 |. Key principles for viral-mediated gene transfer in neuroscience.
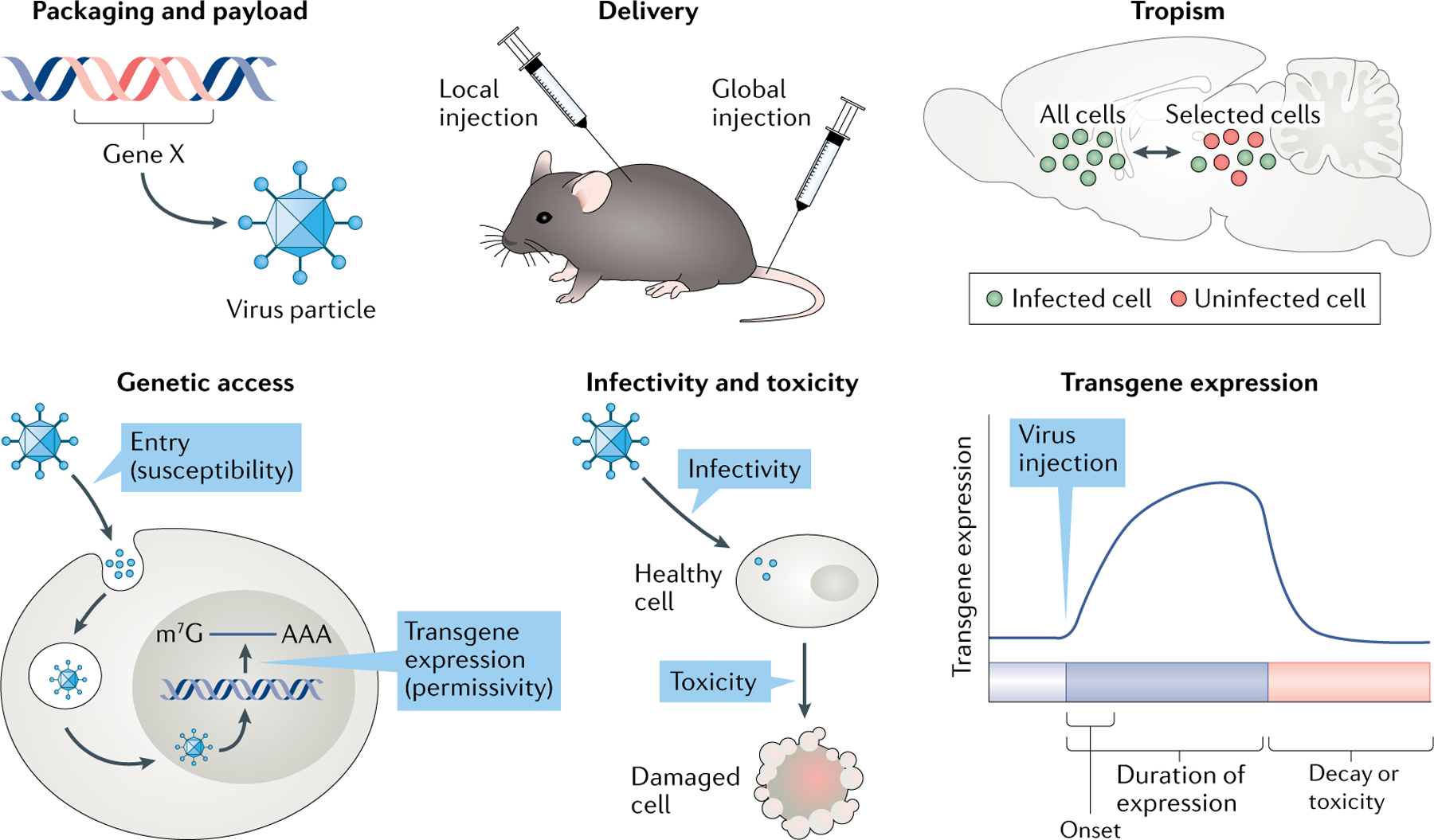
Schematic demonstrating six key principles essential for the neuroscientist: viral packaging limit (how much nucleic acid a virus particle can carry) and payload (the length and type of genomic material that can be successfully packaged into a virus particle), delivery methods (local versus global injections), tropism (specificity of a virus for a given cell type(s)), access (ability of a virus to enter a cell type and express its gene product(s)), infectivity and toxicity (how efficiently a virus infects a cell and how harmful it is to the cell), and transgene expression dynamics (time course of onset and persistence of transgene expression). These principles play a key role in determining a neuroscientist’s choice of virus by weighing the advantages and disadvantages of a given virus. AAA, 3′ poly(A) tail for mRNA; Gene X, a transgene being packaged into a virus particle; m7G, 7-methylguanosine (5′ cap for mRNA).
Packaging and payload.
What can and cannot we put into a virus? The first key consideration for recombinant virus selection is the packaging and payload. Viral genomes are composed of DNA or RNA, and these genomes are packaged into special particles that can open up and deliver the genome to a cell. Each virus particle has a unique capacity to carry a defined size of nucleic acid molecule. This is the so-called ‘packaging limit’ and is set by the particular virus particle whose size is encoded by the viral genome. Virus particles are currently incapable of cell type-selective delivery of dyes, small molecules and the like to a cell; however, there are instances in which viral particles have been engineered with capsid modifications or ‘loaded’ to potentially co-deliver a non-genetic payload (albeit with reduced cell selectivity)4. Furthermore, a viral genome’s nucleic acid composition is a key initial determinant for virus selection. Given the currently limited toolkit available for manipulating RNA in vivo, RNA-based viruses are typically not used for ‘molecular logic operations’ (for example, Cre or Flp recombinase-On), unless they are of Baltimore classification VI5, single-stranded RNA reverse transcriptase (RT; Table 1) or used in combination with DNA-based vectors6. By contrast, DNA-based viruses such as adeno-associated virus (AAV) and herpes simplex virus (HSV) are frequently used for Cre- or Flp-dependent gene expression7–9.
Virus particles from different virus families differ substantially in the size of the genome nucleic acid that can be packaged, as well as how much of the original viral genome can be removed (without significantly influencing infectivity); these limitations establish how much new nucleic acid information can be added (their payload capacities). Some viruses, such as AAV or rabies virus (RbV), have limited payload capacities in the order of ~3.5–5 kb (refs10,11), whereas that of alphaherpes-viruses (for example, HSV and pseudorabies virus (PRV)) may exceed tens or, theoretically, even hundreds of kilobases12,13 (Table 1). Whereas size limitations may be overcome via creative methods, such as split proteins14 or split inteins15, or brute force using viral co-delivery6, the endogenous capacity of a virus particle to package nucleic acid is an important limitation.
A practical limitation is producing sufficiently high concentrations of infectious virus particles for experimental use: some of the engineered viruses do not grow well. Importantly, the number of infectious particles needed for a given experimental system is determined empirically. The readout of infection varies with the vector, the transgene that is delivered and expressed, and the targeted cell or animal. Delivery of a transgene and achieving sufficient expression in a high percentage of targeted cells within a given brain region often require injection of very large numbers of particles, meaning that a high concentration (titre) of virus is needed. High titres of virus, for example AAV (up to 1014 particles/ml) and HSV (up to 108 particles/ml), are now used routinely in neuroscience, which enables the infection of a high percentage of targeted cells and, therefore, superior linking of transgene expression to a downstream functional effect.
High-titre viral production enables not only quantitatively superior infections but qualitatively novel studies as well. For example, the development of PHP capsid variants of AAV, which are capable of targeting several tissues in tandem with viral production at a very high titre, has enabled infection of (and thus access to) a high percentage of distributed cell types (both neuronal and non-neuronal) across both the CNS and the peripheral nervous system. Of note, PHP-based vectors have enabled efficient transduction of myriad peripheral nervous system cell types in dorsal root ganglia, cardiac ganglia and enteric neurons16–19. Still, in contrast to numerous other commonly used vectors in neuroscience, producing high titres of other viruses, such as lentiviral vectors, remains a challenge; in the case of lentivirus, this is primarily due to the fragility of the recombinant virus particles.
Delivery.
How do we get a virus into the nervous system? Viral delivery of new genes into the CNS begins with an injection of infectious particles either directly into the CNS or via a peripheral route. Depending on the goals of the experiment, delivery may include a combination of different recombinant viruses injected in one or more locations, inside or outside the CNS. A simple and important dichotomy to consider is that of a ‘local’ injection versus a ‘global’ injection. A local injection requires surgery, often stereotaxic, to a defined anatomic locus within the CNS or periphery, whereas a global injection is given peripherally, often via the blood-stream, and relies on transgenic/biophysical methods or promoter-specific elements to drive cell type-specific gene expression in the CNS17,18,20,21.
Although particles delivered via local viral injection into the CNS parenchyma are initially restricted to the injection site (because they are too large to diffuse), particles can remain local or spread broadly depending on several parameters intrinsic to the virus. These key properties include the capacity of the virus particles to move anterogradely or retrogradely (between synaptically connected neurons), tropism for different cellular compartments (for example, soma versus nerve terminals) and replication competence, among others (that is, charge). An informative comparison is that of AAV, canine adenovirus (CAV) and PRV. All three of these viruses use local injections to achieve vastly different goals. Most AAVs infect a neuron’s soma to express a gene product, whereas CAV and PRV are preferentially taken up by nerve terminals and retrogradely transported from projection areas to cell soma13,22–24 (FIG. 2). CAV has been engineered to be replication incompetent by removal of many essential viral genes (for example, the genome is ‘gutted’ and thus encodes fewer cytotoxic elements), and accordingly is relatively non-toxic to neurons. The particles infect axons and move retrogradely long distances to the cell bodies where they deliver their payload to the neuron’s nucleus. As the genomes cannot replicate, no new virus particles are made. These engineered CAV vectors enable projection-specific delivery of gene payloads25,26. Replication-competent PRV (derived from the attenuated Bartha strain) is capable of infecting and replicating in a polysynaptic circuit moving specifically from postsynaptic cell to presynaptic cell until the animal is euthanized or succumbs to infection. AAV and CAV are thus used for longer-term genetic manipulations, whereas PRV’s utility is often restricted to shorter-term circuit-mapping studies20,24,27. Critically, as is clear from the examples of CAV and PRV, a local injection can ultimately result in the delivery of a gene payload distanced millimetres to centimetres away from the initial target.
Fig. 2 |. Viral strategies for accessing neurons by virtue of their connectivity.
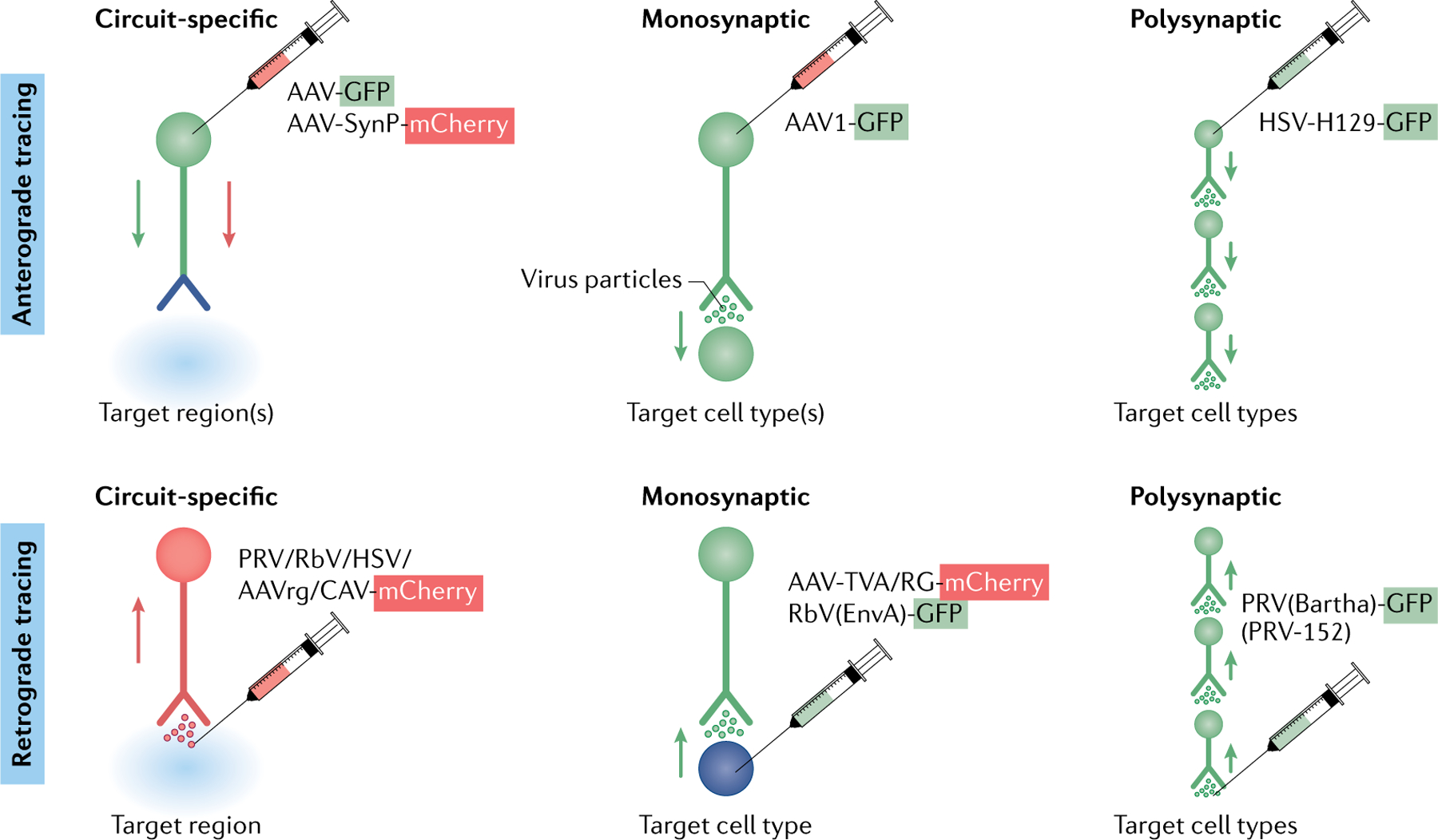
Viral tracing strategies commonly used to gain circuit-specific genetic access to cell types throughout the central nervous system. Tracing methods are commonly divided into anterograde (top) and retrograde (bottom) approaches, which often start by the injection of virus (indicated by a syringe) at a presynaptic soma or at the terminal or postsynaptic soma, respectively. Circuit-specific viral tracing methods most often utilize ‘gutted’ constructs (that is, they express few or no viral genes), which enable access to the cell type with limited toxicity. Monosynaptic and polysynaptic methods for accessing cell types are often, but not always, toxic to infected cells due to the frequent use of live, replication-competent viruses, which are capable of jumping one (monosynaptic) or more (polysynaptic) synapses. Green cell compartments are positive for GFP (green fluorescent protein), and red cell compartments are positive for mCherry (a monomeric, red fluorescent protein). Blue cell compartments (nerve terminals for circuit-specific anterograde tracing and postsynaptic soma in monosynaptic retrograde tracing) are positive for both GFP and mCherry. For circuit-specific anterograde tracing, AAV-GFP and AAV-SynP-mCherry, respectively, help to label the whole cell and the nerve terminals. Note that there is currently no way to access cells that are a given number of synapses upstream or downstream of a target cell type. AAV, adeno-associated virus (AAV1 is a variant of AAV); AAVrg, retrograde-tracing variant of AAV; CAV, canine adenovirus; EnvA, avian ASLV type A envelope protein (cognate ligand for TVA); HSV, herpes simplex virus; HSV-H129, anterograde-tracing variant of HSV; PRV, pseudorabies virus; RbV, rabies virus; RG, rabies G protein; SynP, a synaptophysin fusion construct (can target fluorophores to the presynaptic nerve terminal); TVA, avian receptor for EnvA.
In contrast to a local injection, a global injection is defined as body-wide delivery, often via the blood-stream (for example, via tail vein infusion in rodents or retro-orbital injections in mice). Using global injections of recombinant AAVs (for example, the engineered PHP.eB strain)17, infection may spread into sparse or anatomically distributed cell types, which is often difficult to achieve without multiple injections, especially in larger animals17,18. Modification of viral capsid proteins has been used to promote such broad viral infections16,20. Further work is needed to establish definitively the ability of such peripherally administered AAV particles to infect large percentages of cells within the CNS. As an extension to the global approach, other ‘semi-global’ routes of administration exist, such as delivery of viral vectors into the cerebrospinal fluid via lumbar or intracerebroventricular injection.
Global injections may also enable localized infection with minimally invasive or non-invasive approaches by coupling the injection to a chemical/biophysical method, such as focused ultrasound, which helps to overcome anatomic barriers to productive infection21. Using this latter approach, one can target a globally injected AAV to an anatomically restricted locus within the CNS by selectively permeabilizing the blood–brain barrier. Global injections therefore need not be global in their resulting gene expression patterns.
A key caveat in the delivery of viral vector genomes is therefore the potentially far-reaching (or restricted) effect of particle delivery modality (local versus global) on genetic access — although the initial site of injection is important to know, where the viral genomes may ultimately end up (and thus are expressed) will determine the results and the conclusions drawn from the experimental observations. Most neuroscience studies use local viral injections, but the remaining principles below apply to both delivery methods.
Tropism.
Where does the virus go? Viral tropism is the capacity of a virus particle to infect a given cell type. It is a key feature of all viruses, determining which cells in the body are infected (cellular entry) and what the delivered viral genome can do in a given tissue (for example, drive transgene expression). To productively infect a cell, both viral particle attachment and entry, coupled with expression of the viral genome, are required. Most viruses used by neuroscientists are capable of infecting neurons — some with great selectivity compared with non-neuronal cells — and are therefore considered ‘neurotropic’. In parallel, increasing attention is being given to generating virus particles that infect other cell types as described in the next section.
To enter a given cell, all virus particles must bind to a specific receptor, which can be a protein, carbohydrate or lipid, depending on the virus. Engagement of the specific receptor triggers entry of the virus particle, often by endocytosis followed by delivery of the genome payload. Importantly, some virus particles use multiple receptors, and other virus particles from different families might use the same receptor3. A recently discovered example of this receptor requirement is the AAV receptor (AAVR). AAVs require AAVR, which is also referred to as KIAA0319L (a type 1 transmembrane protein), the normal function of which is unknown. Binding of an AAV particle to AAVR, through interactions with other glycoprotein and carbohydrate moieties, enables rapid endocytosis of the AAV particles into cells. AAV particles likely cannot enter cells that do not express AAVR. This is similarly the case with CAV, entry of which requires that cells express the coxsackievirus and adenovirus receptor (CAR)28. Expression of receptors on host tissues and cell types thus, in part, determines a virus particle’s capacity to infect a target population.
Of note, distinct AAV variants (called serotypes) use different carbohydrate and protein moieties to enter cells, resulting in preferential tropism29. Serotypes refer to subtypes of capsids (the proteinaceous shell of a viral particle) distinguished with antibodies. Distinct serotypes arise from differences in capsid proteins. To infect the brain, the capsid proteins most commonly used are derived from AAV1, AAV2, AAV5, AAV8 and AAV9 (ref.30). AAV2 is the most widely used; it is highly selective for neurons and shows limited spread upon intracerebral injection, making it ideal for many neuroscientific studies. Other AAV serotypes show greater spread and, thus, are more suitable for infecting larger brain regions, including those in non-human primates and even humans, whereas still others can infect non-neuronal cells in addition to neurons. The process of ‘pseudotyping’ an AAV, packaging an expression cassette flanked by AAV2 inverted terminal repeats into virions of other AAV serotypes, thus enables varying degrees of region-dependent viral spread around an injection site31.
Whereas cellular tropism depends on the presence or absence of a receptor (often called ‘susceptibility’), other parameters also contribute to tropism, including accessibility of the cell, extracellular defences and permissivity of the cell. Permissivity is the capacity of the cell to express the viral genome once the particle delivers the genome to the cell. Expression of virally encoded genes is essential for productive infection, and many strategies to express transgenes in given cell types of interest are available, such as cell type-specific20,32 or synthetic32 promoters, as well as constitutive promoters driving Cre- or Flp-dependent constructs7,9. In this Review, we define all of these parameters as ‘genetic access’.
A virus that is capable of entering a cell and expressing its gene products is thus said to have ‘genetic access’, a key property for recombinant vectors used to study the nervous system. But for genetic access to be useful, it needs to be selective; there are thus many cases in which restricted cellular entry is a feature, and not a bug.
Access.
What properties of neural cells can we leverage to gain selective genetic access? As genetic access is obligatorily dependent on viral particle attachment to cells coupled with cellular entry, genome delivery and genome expression, these processes represent at least two orthogonal entry points for engineering recombinant viruses targeted to defined cell types. An illustrative example of restricted genetic access is a comparison between the methods used for RbV and AAV.
Selective genetic access by RbV is attained at the level of receptor binding for cellular entry. RbV is a negative-sense, single-stranded RNA virus, and, unlike viruses with DNA genomes, cannot use intracellular molecular logic operations (for example, using Cre or Flp recombinases to activate/inhibit transgene expression). Rabies-based systems can achieve restricted genetic access via selective cellular entry. RbV encodes the receptor-binding G membrane protein, which is found in the virus particle envelope. Without the G membrane protein, an RbV particle cannot bind its receptor and attach to a cell. A G-protein-deleted RbV has been engineered (pseudotyped) to express the avian ASLV type A envelope protein (EnvA). EnvA binds the avian receptor TVA, which is not endogenously expressed in mammalian cells. Consequently, this pseudotyped virus particle cannot enter mammalian neurons unless they are engineered to heterologously express the avian receptor TVA6,33,34. Once the EnvA-pseudotyped RbV has entered a TVA-positive cell, the virus can express its transgene products (and/or react with any other gene products provided in trans, such as G protein).
Genetic access for an AAV, by contrast, depends both on cellular entry and on molecular logic operations. First, different AAV serotypes bind to different receptors, as discussed in the previous section. Second, once an AAV genome enters a cell’s nucleus, the single-stranded DNA genome must be converted into double-stranded DNA. Of note, this conversion to double-stranded DNA progresses slowly, and can be circumvented by use of a self-complementary AAV (scAAV). scAAV sidesteps the requirement for second-strand synthesis, and consequently has a much shorter time lag to expression35. A notable downside to this approach, however, is the size limitation of transgenes imposed by scAAV, which is approximately half that of standard AAVs.
Once an AAV is in double-stranded DNA form, it is a potential substrate for various Cre-On, Flp-On and related approaches7,8,36,37. For example, it is now straightforward to achieve exquisite neuronal cell type-specific transgene expression by injecting an AAV — whose transgene is expressed in a Cre-dependent manner — into a brain area of a mouse or rat harbouring Cre gene expression within a specific type of neuronal cell (termed a Cre-driver line). HSVs have also been used in very similar ways to achieve cell type-specific transgene expression38. Several approaches to generate Cre-dependent constructs are used. In the simplest form, a stop sequence, flanked by loxP sites, precedes the transgene of interest. Alternatively, double-floxed inverted open reading frame (DIO) or FLEX vectors are utilized (FIG. 3)7,39. Both strategies enable an irreversible, recombinase-dependent reaction to take place, which ultimately leads to constitutive expression of a given transgene. This has more recently been extended to other recombinases, such as Flp, Dre and VCre, and can be used intersectionally in various combinations to achieve refined access to specific cell types by virtue of numerous parameters8,37 (see below).
Fig. 3 |. Assessing a gene’s function within the CNS.
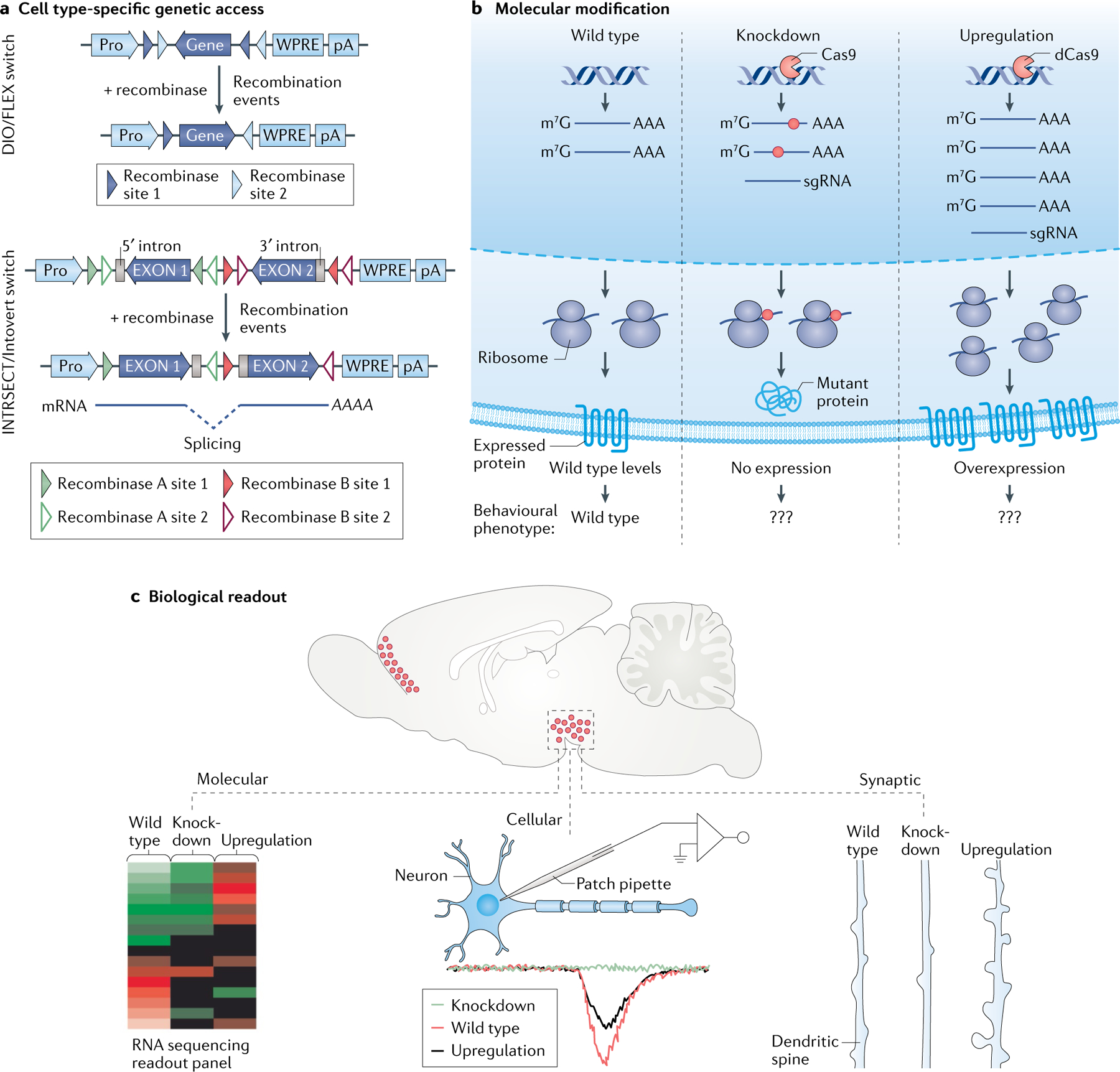
Theoretical framework for characterizing the role of a given gene in regulating molecular, cellular, synaptic, circuit and behavioural functions. a | To gain genetic access to a cell population in a cell type-specific fashion, single-recombinase (top) or dual-recombinase (bottom) ‘molecular logic’ can be used. The double-floxed inverted open reading frame (DIO)/flip-excision (FLEX; an irreversible, Cre-dependent molecular switch), often placed before a constitutive promoter (pro), allows for irreversible activation/expression of a cassette. When paired with cell type-specific expression of a recombinase (that is, Cre, Flp, Dre, VCre and so on) using a driver line, the DIO/FLEX switch is irreversibly activated, which leads to constitutive expression of the engineered transgene. This system thus benefits from the specificity of recombinase driver lines and the robust expression of constitutively active promoters (top). INTRSECT (a variant of the FLEX switch; INTRSECT constructs enable intersectional molecular logic through strategic use of introns) and Introvert (a variant of the FLEX switch, using a similar strategy to that of INTRSECT by strategically using introns)-based approaches can also be used to access a cell type using dual-recombinase logic (that is, based on the expression of two recombinases). These recombinases can be used to define any number of features of a cell population, allowing for increased specificity when genetically accessing a cell type (recombinases can be used to access a cell type based on marker gene expression, projection target or activity pattern). Introns are used in the INTRSECT/Introvert approaches to enable strategic inversions of split gene segments (so that the recombinase sites can be spliced out after recombination). This allows for the expansion of new, orthogonal molecular logic operations. Recombinase sites depicted are capable of recombining with one another only if they are of the same colour and shading (bottom). b | Molecular schemata for manipulations of gene expression. Among many options, a gene can be knocked down at the levels of DNA (using, for example, clustered regularly interspaced short palindromic repeats (CRISPR)) or RNA (microRNAs, newer RNA-targeting CRISPR systems). In the example shown, Cas9-mediated CRISPR knockdown is achieved via induction of an insertion-deletion in the genome. This genetic lesion is consequently manifested at the RNA level (represented in the middle sub-panel by a red dot), ultimately resulting in the formation of mutant (non-functional/inactive) protein. For upregulation of endogenous gene expression, a ‘dead’ Cas9 (dCas9) fusion construct enables increased transcription and, ultimately, more protein (represented in the right sub-panel). Triple question marks (???) represent the behavioural phenotype outcome being measured after genetic manipulations. c | Functional readouts after manipulating gene expression. Examples include gene expression adaptations revealed by RNA sequencing. This approach provides a readout of relative expression levels of wild-type, knockdown and upregulated genes (left). Alterations in synaptic strength can be determined using electrophysiology. The sample trace shows hypothetical responses from a neuron with a knockdown, wild-type or overexpressed gene product (middle). At the synaptic level, changes in dendritic spine number and morphology can be compared. These micro-scale adaptations may ultimately lead to observable changes in the animal’s circuit function and behaviour (right). AAA, 3′ poly(A) tail for mRNA; m7G, 7-methylguanosine (5′ cap for mRNA); sgRNA, single guide RNA (enables programmatic targeting of Cas proteins to a desired nucleic acid sequence); WPRE, woodchuck hepatitis virus post-transcriptional regulatory element.
It is likewise possible to target transgene expression to non-neuronal cell types in the nervous system. Astrocytes and oligodendrocytes can be infected by using specific AAV serotypes that infect these cell types (in addition to neurons), combined with a Cre-driver line selective for these glial subtypes. An alternative approach is to use such a glia-infecting AAV that drives transgene expression under the control of a promoter that is selective for these cell types40,41. Both strategies have proven successful for accessing astrocytes and oligodendrocytes, but there are still no viral species that are both highly infective of and selective for glia (for example, which do not also infect neurons). In contrast to astrocytes and oligodendrocytes, it has not yet been possible to gain efficient viral access to microglia because no available viral vector can infect this cell type in vivo to appreciable degrees. Endothelial cells in the nervous system have also recently become potential targets for viral vectors, in particular AAV-BR1 (refs42,43).
Restricted cellular entry (receptor usage) or transgene expression can be used to selectively access cell types by virtue of their expression of a principal marker gene7,8, connectivity25,44–46, activity34,47 or location in both space and time. There are a host of methods that combine viral and transgenic approaches to achieve these aims, many of which are beyond the scope of this Review and are described well elsewhere48,49. Regardless of how one gains genetic access to a given cell type, proper delivery of the virus particle to its target is essential.
Infectivity and toxicity.
How efficiently does a virus infect a cell and how harmful is it to the cell? In this section, we discuss trade-offs between a virus particle’s infectivity and cellular toxicity, largely avoiding a more detailed discussion of viral virulence (the property of causing disease), which is discussed elsewhere3.
Infectivity and toxicity are closely related but distinct entities. All cells have an intrinsic defence system that uses ‘pattern recognition receptors’ that bind to foreign proteins and nucleic acid. Binding to these receptors usually results in a cascade of cellular defensive responses such as cell death (apoptosis) and expression of cytokines such as interferon. Viral infection can also lead to more subtle, but significant, changes in a cell’s physiology. For example, viruses affect gene transcription — certain AAVs reduce the expression of neuronal nuclear protein (NeuN), which is often used as a neuronal marker, or induce ocular toxicity — and PRV can cause cellular fusion and subsequent electrophysiological changes50–52. These effects underscore the importance of choosing a viral vector wisely, fully understanding each of their potential effects and caveats, as well as always including experimental groups that control for viral infection per se.
The ideal viral vector would be highly infective, but minimally toxic. However, these parameters are often exceptionally difficult to decouple, thus determining the compromises one has to make in selecting the optimal virus. Many recombinant viruses, such as AAV, HSV, CAV and lentivirus, are used in neuroscience as ‘semi-inert’ vectors, having a reduced capacity to induce cell-intrinsic defences. As these are mutant viruses, care must be used in obtaining preparations that lack wild-type (replication-competent) virus and cellular debris53 (Box 1). By contrast, polysynaptic circuit-mapping tools need to spread beyond the initial site of infection (replicating even in postmitotic cells — neurons). Replication usually activates cellular defences, which often gives rise to pathogenic effects. Given the difficulty of decoupling efficient infectivity from virulence, there are no available polysynaptic tracers with minimal cytotoxicity. These replication-competent viruses, such as RbV, PRV (unrelated to RbV) and HSV-H129, rapidly replicate in neurons and move from cell to cell (via synaptic connections). As a result, infected animals will succumb to infection within days and, less frequently, weeks22,54,55.
Recent advances suggest that decoupling infectivity from cytotoxicity, within the setting of monosynaptic tracing, may soon become possible. RbV-based vectors have been identified or engineered that appear to have significantly reduced (or even minimal) cytotoxicity, notably CVS N2C and SAD B19 ΔG/ΔL double-mutants56,57. Similar advances have been made with PRV, where deletion of replication factor thymidine kinase or immediate early gene IE180 significantly reduces virulence and/or cytotoxicity22,44,58. Of note, CAV, HSV and AAV have also been engineered/applied for retrograde8,28,59 and/or monosynaptic anterograde60,61 tracing.
Expression dynamics.
When and for how long will your favourite transgene be expressed? Gene expression dynamics after a virus particle delivers its genome to cells is another important consideration for experimentalists. Different patterns of gene expression suit different experimental goals. The two key parameters for gene expression are the time to maximal expression (onset) and the length of time that expression is maintained (duration). Expression dynamics of different viral genomes vary broadly: some viral genomes are expressed quickly (and then decline, or become cytotoxic), whereas others are expressed more slowly and persist for a longer time. Knowledge of these patterns, and the dynamics for each viral vector (Table 1), is essential when selecting a virus. For example, if the goal is to rapidly and locally express a gene product (with less concern for long-term duration of expression), an excellent candidate is HSV62,63. This property of viral expression can be used, for example, to study the lasting influences of a transient change in gene expression early in development on functional outcomes in adulthood62. However, if one would like to gain local and long-term genetic access to a cell type (for longitudinal studies), with less concern for rapid expression, an AAV is well-suited64. A short time to maximal expression is a key consideration for studies in which layers of molecular and cellular adaptations may occur on rapid timescales — HSV is advantageous in these cases, given its rapid onset to maximal expression in vivo (~12 h) allowing one to more precisely relate the effect of the encoded transgene to rapid, downstream effects; by contrast, the time lag of 2–3 weeks required to achieve maximal expression by a standard AAV (not, for example, scAAV) allows more adaptations to intervene between transgene expression and downstream effects.
Of note, it is also possible to multiplex different viruses with other transgenic methods, often involving recombinases (constitutive or inducible). This enables creative ways to make brief windows of gene expression long (for example, by expressing Cre recombinase to activate a constitutively expressed locus) and to make long windows brief (for example, using small-molecule inducible proteins, such as Cre-ER (a tamoxifen-inducible Cre) or tTA (tetracycline transactivator)).
Functionality and uses of viral tools
Recombinant viruses have diverse utility in neuroscience, enabling studies at the molecular, cellular, circuit and whole-organismal levels. In the previous section, we outlined key viral properties to better understand the limitations and opportunities imposed by various viral reagents. Here, we apply these properties and principles by discussing some choice applications of recombinant viruses to dissecting different dimensions of neuronal or glial function in vivo.
Recording and modulating neurons.
Observation (imaging) and perturbation (modulation) are two of the key approaches to better understanding neural function. These studies, which record and manipulate neuronal activity, often require long-term access (for longitudinal studies), some degree of specificity (for cell type, projection and/or history of activity) and cellular health (to ensure that effects are not, in part, due to off-target or non-biological effects). Given these requirements, AAV has been the most popular viral vector for studies involving neural imaging and manipulation, due to its persistence of expression, ease of use and production, minimal cytotoxicity and ability to multiplex with other viral and transgenic approaches. AAVs have been used effectively to gain genetic access to neuronal populations by virtue of their expression of a marker gene, projection to a defined target region and history of activity. How is this done?
To gain access to a molecularly defined cell type, two principal modalities are currently available: promoter-specific elements and recombinase driver lines (that is, multiplexing with transgenic mice). Recombinase driver lines (most frequently, Cre-drivers), noted earlier, are by far the most popular, effective and reproducible approach, used in combination with a virus, to introduce a protein that modulates or tracks the activity of a given neuronal population. Such proteins include channelrhodopsin (ChR2), numerous other opsins and designer receptors exclusively activated by designer drugs (DREADDs) used for optogenetics or chemogenetics, respectively, as well as GCaMP and other indicator proteins used for fibre photometry or single-cell Ca2+ imaging in vivo. These approaches combine the specificity of recombinase expression with high levels of expression achieved by the use of constitutive promoters (for example, CAG, EF1a and so on). It remains essential, nonetheless, to validate the cellular specificity of transgene expression in each experimental system. With a few notable exceptions that often target broad subsets of neurons (for example, with the Syn1, Camk2a or Prs65 promoters), use of neural promoters to target specific subtypes of neurons have to date been disappointing. Promoters in these less successful instances have tended to have low expression or poor specificity for the chosen cell type. Nevertheless, engineered and synthetic promoters, as well as modified viral capsids, are being developed to gain more selective access to CNS cell types in the absence of genetically engineered animals, and offer future promise17,32,66–70.
A cell type can also be defined (and accessed) by virtue of its connectivity (see also the section below) and activity. AAVs are often multiplexed with gutted retrograde tracing viruses (for example, CAV46, PRV44, HSV8,71, RbV57 or even other AAV serotypes59) to enable long-term neural recording and manipulation. AAVs have also been used in tandem with activity-dependent promoters47 or Cre-driver lines72 to gain genetic access to neurons that were activated in a specific behavioural or physiologic setting73,74. Most importantly, these advances can be multiplexed to begin to define cell types in multiple dimensions75. The combination of orthogonal recombinases, as well as promoters that are cell type-selective and activity-dependent, with varied viral tools (that is, AAV for local delivery and other gutted vectors for retrograde tracing) will allow for imaging and manipulation of neural ensembles defined by multiple parameters. Indeed, this multidimensional definition may even prove essential for cell typing76.
Neural circuit mapping.
Structure within a neural network (connectivity) gives one context for understanding a neuron’s function. Towards this end, many circuit-mapping tools have been developed. These tools capitalize on advantageous properties of wild-type viruses — the ability to preferentially infect nerve terminals versus cell soma, move anterogradely or retrogradely within the cell (using the cell’s endogenous transport machinery) and spread at or near synapses in either direction, sometimes to more than one connected cell. Recombinant variants of these viruses were initially engineered to express fluorophores, but have now been repurposed to express an array of transgenes, helping to unify neuronal structure and function.
Depending on the circuit-mapping tool, different types of information can be gleaned (some examples are shown in FIG. 2). Recombinant viruses used for circuit mapping currently offer three broad classes of cellular access and labelling: circuit-specific5,8,24,25,44,59,77 (cell type to region, or vice versa), monosynaptic6,60,61,78,79 (cell type to cell type) or polysynaptic27,80–82 (cell type to cell types). Such approaches are also being used to map, with unprecedented detail, the topography of cells within a given brain region as relates to that cell’s afferent and efferent connections83. A key limitation of these approaches is the current inability to label an arbitrary number of upstream or downstream synapses in vivo for extended durations (although this can be done using carefully selected time points at which animals are euthanized). However, with rapid improvements in circuit-mapping technologies and small-molecule inducible gene expression systems, this could soon become a possibility. An enhanced ability to decouple infectivity from virulence in an engineered viral system would also greatly bolster these efforts.
Another approach for circuit mapping that converts structural information into nucleic acid form, termed ‘barcoding’, uses Sindbis virus to molecularly label nerve terminals. A virally encoded ‘barcode’ transgene is transmitted anterogradely to nerve terminals, and putative target brain regions are dissected and analysed for barcode transgene expression. Each injected viral particle contains a unique barcode sequence, such that the strength of connection can be inferred by the number of barcodes detected by sequencing across target brain regions84,85.
Although circuit mapping has enabled foundational studies elucidating the structure of neural networks, other key neuronal properties can be layered onto this information84–86. These include a neuron’s molecular composition22,36,54 (see below), recent activity pattern34 or projection to a defined output region79. Unifying these properties will begin to allow us to have a better understanding of how neural circuit architecture and composition influence functional output25,47,87,88.
Profiling gene function in the cell.
There are by now numerous studies — too many to cite here — that have utilized viral vectors to manipulate a gene of interest within a given brain region, or even a given cell type in a targeted brain region, to study the influence of that gene at the molecular, cellular, circuit and behavioural levels. A gene of interest might be selected based on a variant allele being associated with a neurological or psychiatric disorder, or based on altered expression within a gene network that is associated with that disorder. To study the function of a given gene of interest, the wild-type variant of that gene can be overexpressed, made constitutively active or engineered as a dominant negative or other mutant form of the gene (for example, a form associated with a human disease). The gene can be overexpressed on a wild-type or knockout background, knocked out by expression of Cre in a floxed mouse (or by expression of a microRNA in a wild-type mouse) and so on. Such multidimensional control of a gene can be achieved in a specific cell type by combining viral vectors whose transgenes are expressed in a Cre- and/or Flp-dependent manner in their respective lines, as stated earlier. These approaches are being used routinely today to establish the causal involvement of diverse types of genes in normal and pathophysiological phenomena (Fig. 3).
Two permutations of this overall experimental strategy warrant highlighting. First, viral-mediated overexpression of a gene can result in levels of the encoded RNA and protein that are an order of magnitude greater than those seen normally. Likewise, viral-mediated knockout of a gene can drive levels of that gene’s expression far below the lower range seen normally or pathologically. Driving gene expression to either extreme can produce artefactual results with misleading conclusions. The recent advent of CRISPR tools delivered to the brain with viral vectors makes it possible to overcome this confound of traditional viral vectors by activating or suppressing the activity of an endogenous gene in tunable ways, within the range of expression levels seen in vivo89.
Second, viral vectors are being used increasingly to characterize transcriptomic consequences of a gene’s manipulation at a genome-wide scale by performing high-throughput RNA sequencing (RNA-seq) on cells infected in vivo. In so doing, it becomes possible to provide empirical validation of gene co-expression networks via analyses that are being used more and more to identify driver or hub genes whose expression levels are correlated with large numbers of other genes in RNA-seq data sets. For example, overexpression or knockout of an identified hub gene within a given cell type and brain region has been shown to preferentially alter the expression levels of genes present within bioinformatically defined gene modules (or networks), compared with all other genes in the genome90–92. The availability of such RNA-seq data sets after viral manipulation of a gene also enables relating the altered molecular profile of the infected cells with altered cell morphology (for example, density of dendritic spines) and with altered functioning of those cells at the synaptic and circuit levels (FIG. 3). An important caveat of viral-based molecular profiling, however, is that viruses — even without expressing a transgene or deleting an endogenous gene — induce changes in a cell’s molecular composition, as noted earlier; it is therefore essential to perform proper controls to decouple the influence of a genetic lesion from that of the virus itself.
Future directions and conclusions
Viruses have been a spectacular boon for neuroscientists, with each of the several types of recombinant viruses used having significant advantages but also important limitations that make virus selection paramount. Neuroscientists have developed and applied an armamentarium of viruses out of a need for access — access that is easily achievable, rapid and selective for cell type, tunable and otherwise inert. Work continues to generate such ‘perfect’ viral vectors for neuroscience research.
The ideal recombinant virus would have several key properties, as alluded to throughout this Review. It would be easy to package at high titre with infinite payload (in vector core facilities using standardized viral preparation protocols), simple and minimally invasive to deliver, capable of infecting all cell types but only activated in desired cell types, completely non-toxic (and non-immunogenic), and tailorable and tunable in terms of its expression dynamics (that is, low versus high levels of transgene expression, rapid/slow onset and offset). This incomplete list hints at some of the advances that will be required in the coming years to optimize viral vectors for neuroscientific discovery in the laboratory, and, ultimately, utility within the clinic. To date, clinical trials with viral gene therapy for brain disorders have focused on the intracerebral delivery of an AAV or lentivirus to a brain region of interest but are still in the very early phases of development93–95.
Numerous new functionalities, with existing recombinant viruses, are possible in the laboratory and are currently in use. There is increasing modularity in the strategies used for viral vector engineering, including intersectional genetic strategies, which can in theory multiplex designer promoters with multiple orthogonal recombinase systems. These advances, in theory, make it possible to access a cell type by two or more parameters, such as a projection, a principal marker gene and a natural activity pattern in response to behaviourally relevant stimuli. However, there are many functionalities currently lacking from viral (or non-viral) approaches, which will need to be engineered moving forward.
Historically, viruses were first used in neuroscience for tracing and then, later, for transgene delivery (Box 1), taking advantage of key properties endowed by various wild-type viruses discovered in nature, such as polysynaptic spread. Only relatively recently did neuroscientists begin not only to engineer the payload of a virus (for example, to heterologously express a transgene) but also to impart new functionalities by tailoring the viral genome and the envelope/capsid. Exploring and exploit-ing this landscape has proven to be more of a challenge for some viruses (or functionalities) than others. In addition to rational design, which has guided viral engineering and use for the greater part of the past century or so, new approaches involving in vitro and in vivo evolution and machine learning-guided approaches96 promise to open up new avenues for engineering recombinant viral vectors. Advances made using these approaches will likely create a virtuous cycle between biological discovery and improved technology.
One of the most, if not the most, important considerations and caveats of using viral vectors in neuroscience is what we are ‘accessing’ in the first place. With the increased availability of techniques, allowing a high-dimensional investigation of neuronal populations (in terms of lineage, location, marker gene expression, connectivity and natural activity pattern), it becomes more or less clear which resolution is appropriate to define a given ‘cell type’. Indeed, the information needed to encode or define a particular cell population appears to be different, simply by virtue of where in the brain one looks76,88.
Regardless, continual advances in viral engineering will offer increased specificity in terms of how one defines and accesses a given cell type. It is expected that the field will increasingly be able to leverage the key properties (and limitations) of viruses known (and yet to be identified) to develop these functionalities moving forward.
Marker genes
Genes whose cell type-specific mRNA and/or protein expression can be used to identify a cell type.
Reporter genes
Genes whose expressed mRNA or protein is readily detectable and can be used to characterize another gene or gene product.
Baltimore classification
A classification scheme (conceived by virologist David Baltimore) that characterizes a virus based on its nucleic acid composition, in particular how the virus ultimately synthesizes mRNA.
Capsid
A proteinaceous shell that surrounds the viral genome.
Vectors
Vehicles used to transfer nucleic acid information into the cell.
Recombinase
An enzyme (for example, Cre or Flp) capable of recombining defined nucleic acid sequences, often used to activate or inactivate expression of a gene engineered to include sequences specific for that recombinase.
Pseudotyping
The process of heterologously expressing proteins on a virus’ capsid/envelope (often with the intention of gaining advantageous viral spread/entry properties).
Constructs
Designer DNA sequences.
Cre-driver line
A mouse or rat strain (for example, knock-in and BAC transgenic) engineered to drive expression of Cre recombinase in a cell type-specific pattern (often using a marker gene’s promoter or enhancer elements).
Acknowledgements
The authors thank L. Enquist (Princeton) for helpful discussions and comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Neuroscience thanks Eric Kremer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
References
- 1.Baltimore D Expression of animal virus genomes. Bacteriol. Rev 35, 235–241 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefkowitz EJ et al. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46, D708–D717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flint SJ, Racaniello VR, Rall GG, Skalka AM & Enquist LW Principles of Virology 4th edn Vol. 4 (ASM, 2015). [Google Scholar]
- 4.Seo JW et al. Positron emission tomography imaging of novel AAV capsids maps rapid brain accumulation. Nat. Commun 11, 2102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowland D et al. Distinct ventral pallidal neural populations mediate separate symptoms of depression. Cell 170, 284–297.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wall NR, Wickersham IR, Cetin A, De La Parra M & Callaway EM Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc. Natl Acad. Sci. USA 107, 21848–21853 (2010).This paper describes the first use of monosynaptic retrograde tracing from a genetically defined post-synaptic population, using a rabies-based system.
- 7.Atasoy D, Aponte Y, Su HH & Sternson SM A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci 28, 7025–7030 (2008).This paper applies the FLEX switch to AAVs, in tandem with Cre-driver lines, to achieve highly cell-selective yet robust gene expression.
- 8.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772 (2014).This paper, building on previous advances in transgenic reporter lines, describes a compact viral method for intersectional genetic cell type targeting.
- 9.Sohal VS, Zhang F, Yizhar O & Deisseroth K Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay MA, Glorioso JC & Naldini L Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med 7, 33–40 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Osakada F & Callaway EM Design and generation of recombinant rabies virus vectors. Nat. Protoc 8, 1583–1601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs A, Breakefield XO & Fraefel C HSV-1-based vectors for gene therapy of neurological diseases and brain tumors: part II. Vector systems and applications. Neoplasia 1, 402–416 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekstrand MI, Enquist LW & Pomeranz LE The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol. Med 14, 134–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zetsche B, Volz SE & Zhang F A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol 33, 139–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong DJ et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res 43, 6450–6458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen WE et al. Global representations of goal-directed behavior in distinct cell types of mouse neocortex. Neuron 94, 891–907.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol 34, 204–209 (2016).This paper uses AAV capsid variation to achieve broad gene expression patterns after a peripheral viral injection.
- 19.Challis C et al. Gut-seeded α-synuclein fibrils promote gut dysfunction and brain pathology specifically in aged mice. Nat. Neurosci 23, 327–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedbrook CN, Deverman BE & Gradinaru V Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci 41, 323–348 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Szablowski JO, Lee-Gosselin A, Lue B, Malounda D & Shapiro MG Acoustically targeted chemogenetics for the non-invasive control of neural circuits. Nat. Biomed. Eng 2, 475–484 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Pomeranz LE et al. Gene expression profiling with Cre-conditional pseudorabies virus reveals a subset of midbrain neurons that participate in reward circuitry. J. Neurosci 37, 4128–4144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomeranz LE, Reynolds AE & Hengartner CJ Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev 69, 462–500 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junyent F & Kremer EJ CAV-2—why a canine virus is a neurobiologist’s best friend. Curr. Opin. Pharmacol 24, 86–93 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Ekstrand MI et al. Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nectow AR, Ekstrand MI & Friedman JM Molecular characterization of neuronal cell types based on patterns of projection with Retro-TRAP. Nat. Protoc 10, 1319–1327 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Smith BN et al. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl Acad. Sci. USA 97, 9264–9269 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li SJ, Vaughan A, Sturgill JF & Kepecs A A viral receptor complementation strategy to overcome CAV-2 tropism for efficient retrograde targeting of neurons. Neuron 98, 905–917.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Murlidharan G, Samulski RJ & Asokan A Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci 7, 76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watakabe A et al. Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci. Res 93, 144–157 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Burger C et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther 10, 302–317 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Juttner J et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci 22, 1345–1356 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Callaway EM & Luo L Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci 35, 8979–8985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakurai K et al. Capturing and manipulating activated neuronal ensembles with CANE delineates a hypothalamic social-fear circuit. Neuron 92, 739–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarty DM, Monahan PE & Samulski RJ Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther 8, 1248–1254 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Poulin JF et al. Mapping projections of molecularly defined dopamine neuron subtypes using intersectional genetic approaches. Nat. Neurosci 21, 1260–1271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenno LE et al. Comprehensive dual- and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals. Neuron 107, 836–853.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias C et al. β-Catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai HC et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science 324, 1080–1084 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Jonquieres G et al. Recombinant human myelin-associated glycoprotein promoter drives selective AAV-mediated transgene expression in oligodendrocytes. Front. Mol. Neurosci 9, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai J et al. Hyperactivity with disrupted attention by activation of an astrocyte synaptogenic cue. Cell 177, 1280–1292.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korbelin J et al. A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol. Med 8, 609–625 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchio S, Sidman RL, Arap W & Pasqualini R Brain endothelial cell-targeted gene therapy of neurovascular disorders. EMBO Mol. Med 8, 592–594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaudhury D et al. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature 493, 532–536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nectow AR et al. Rapid molecular profiling of defined cell types using viral TRAP. Cell Rep 19, 655–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murugan M et al. Combined social and spatial coding in a descending projection from the prefrontal cortex. Cell 171, 1663–1677.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye L et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo L, Callaway EM & Svoboda K Genetic dissection of neural circuits. Neuron 57, 634–660 (2008).This paper, an update on the original landmark review, builds on the cutting-edge genetic strategies used to study neural circuits today.
- 49.Luo L, Callaway EM & Svoboda K Genetic dissection of neural circuits: a decade of progress. Neuron 98, 256–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He T et al. The influence of murine genetic background in adeno-associated virus transduction of the mouse brain. Hum. Gene Ther. Clin. Dev 30, 169–181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy KM, Tank DW & Enquist LW Pseudorabies virus infection alters neuronal activity and connectivity in vitro. PLoS Pathog 5, e1000640 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong W et al. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl Acad. Sci. USA 116, 5785–5794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beier KT et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc. Natl Acad. Sci. USA 108, 15414–15419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo L & Anderson DJ A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72, 938–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly RM & Strick PL Rabies as a transneuronal tracer of circuits in the central nervous system. J. Neurosci. Methods 103, 63–71 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Chatterjee S et al. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat. Neurosci 21, 638–646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reardon TR et al. Rabies virus CVS-N2c(DeltaG) strain enhances retrograde synaptic transfer and neuronal viability. Neuron 89, 711–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oyibo HK, Znamenskiy P, Oviedo HV, Enquist LW & Zador AM Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front. Neuroanat 8, 86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tervo DG et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016).This paper uses viral engineering to identify an effective retrograde-tracing AAV.
- 60.Zingg B et al. AAV-mediated anterograde transsynaptic tagging: mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron 93, 33–47 (2017).This paper identifies different pre-existing AAV serotypes that are capable of anterograde tracing.
- 61.Zingg B, Peng B, Huang J, Tao HW & Zhang LI Synaptic specificity and application of anterograde transsynaptic AAV for probing neural circuitry. J. Neurosci 40, 3250–3267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pena CJ et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356, 1185–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hultman R et al. Brain-wide electrical spatiotemporal dynamics encode depression vulnerability. Cell 173, 166–180.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaspar BK et al. Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc. Natl Acad. Sci. USA 99, 2320–2325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayat H et al. Locus coeruleus norepinephrine activity mediates sensory-evoked awakenings from sleep. Sci. Adv 6, eaaz4232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayford M et al. Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Nathanson JL et al. Short promoters in viral vectors drive selective expression in mammalian inhibitory neurons, but do not restrict activity to specific inhibitory cell-types. Front. Neural Circuits 3, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta P et al. Functional access to neuron subclasses in rodent and primate forebrain. Cell Rep 26, 2818–2832.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dittgen T et al. Lentivirus-based genetic manipulations of cortical neurons and their optical and electrophysiological monitoring in vivo. Proc. Natl Acad. Sci. USA 101, 18206–18211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dimidschstein J et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci 19, 1743–1749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatakis AM et al. A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron 80, 1039–1053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allen WE et al. Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee D, Hyun JH, Jung K, Hannan P & Kwon HB A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol 35, 858–863 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Wang W et al. A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol 35, 864–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim CK, Adhikari A & Deisseroth K Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci 18, 222–235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim EJ et al. Extraction of distinct neuronal cell types from within a genetically continuous population. Neuron 107, 274–282.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wickersham IR, Finke S, Conzelmann KK & Callaway EM Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49 (2007).This paper describes restricted retrograde tracing with a modified RbV variant.
- 78.Wickersham IR et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwarz LA et al. Viral-genetic tracing of the input–output organization of a central noradrenaline circuit. Nature 524, 88–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banfield BW, Kaufman JD, Randall JA & Pickard GE Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications. J. Virol 77, 10106–10112 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falkner AL et al. Hierarchical representations of aggression in a hypothalamic–midbrain circuit. Neuron 106, 637–648.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneeberger M et al. Regulation of energy expenditure by brainstem GABA neurons. Cell 178, 672–685.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beier KT et al. Topological organization of ventral tegmental area connectivity revealed by viral-genetic dissection of input–output relations. Cell Rep 26, 159–167.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen X et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 179, 772–786.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kebschull JM & Zador AM Cellular barcoding: lineage tracing, screening and beyond. Nat. Methods 15, 871–879 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Kebschull JM et al. High-throughput mapping of single-neuron projections by sequencing of barcoded RNA. Neuron 91, 975–987 (2016).This paper uses a modified Sindbis virus to profile projection neurons using sequencing, in place of classical imaging approaches.
- 87.Lerner TN, Ye L & Deisseroth K Communication in neural circuits: tools, opportunities, and challenges. Cell 164, 1136–1150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim DW et al. Multimodal analysis of cell types in a hypothalamic node controlling social behavior. Cell 179, 713–728.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yim YY, Teague CD & Nestler EJ In vivo locus-specific neuroepigenome editing. Nat. Rev. Neurosci 21, 471–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lorsch ZS et al. Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat. Neurosci 22, 1413–1423 (2019).This paper is a recent example of the use of viral vectors to modify a single gene locus, through the expression of CRISPR tools to effect locus-specific epigenome editing, in a targeted brain region and to study the downstream molecular, cellular and behavioural consequences.
- 91.Labonte B et al. Sex-specific transcriptional signatures in human depression. Nat. Med 23, 1102–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagot RC et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron 90, 969–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palfi S et al. Long-term follow-up of a phase I/II study of ProSavin, a lentiviral vector gene therapy for Parkinson’s disease. Hum. Gene Ther. Clin. Dev 29, 148–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hitti FL, Yang AI, Gonzalez-Alegre P & Baltuch GH Human gene therapy approaches for the treatment of Parkinson’s disease: an overview of current and completed clinical trials. Parkinsonism Relat. Disord 66, 16–24 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Kaplitt MG Gene-targeting approaches for movement disorders: recent advances. Curr. Opin. Neurol 32, 566–570 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Ogden PJ, Kelsic ED, Sinai S & Church GM Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 366, 1139–1143 (2019).This paper uses machine learning to guide AAV capsid design.
- 97.Bru T, Salinas S & Kremer EJ An update on canine adenovirus type 2 and its vectors. Viruses 2, 2134–2153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lundstrom K Viral vectors in gene therapy. Diseases 6, 42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dolan A, Jamieson FE, Cunningham C, Barnett BC & McGeoch DJ The genome sequence of herpes simplex virus type 2. J. Virol 72, 2010–2021 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McGeoch DJ et al. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol 69, 1531–1574 (1988). [DOI] [PubMed] [Google Scholar]
- 101.Morissette G & Flamand L Herpesviruses and chromosomal integration. J. Virol 84, 12100–12109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neve RL, Neve KA, Nestler EJ & Carlezon WA Jr. Use of herpes virus amplicon vectors to study brain disorders. Biotechniques 39, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Smith GA & Enquist LW A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl Acad. Sci. USA 97, 4873–4878 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klupp BG, Hengartner CJ, Mettenleiter TC & Enquist LW Complete, annotated sequence of the pseudorabies virus genome. J. Virol 78, 424–440 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Srivastava A, Lusby EW & Berns KI Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol 45, 555–564 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deyle DR & Russell DW Adeno-associated virus vector integration. Curr. Opin. Mol. Ther 11, 442–447 (2009). [PMC free article] [PubMed] [Google Scholar]
- 107.McCarty DM, Young SM Jr. & Samulski RJ Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet 38, 819–845 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Muesing MA et al. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313, 450–458 (1985). [DOI] [PubMed] [Google Scholar]
- 109.Parr-Brownlie LC et al. Lentiviral vectors as tools to understand central nervous system biology in mammalian model organisms. Front. Mol. Neurosci 8, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blomer U et al. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol 71, 6641–6649 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hioki H et al. Efficient gene transduction of neurons by lentivirus with enhanced neuron-specific promoters. Gene Ther 14, 872–882 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Conzelmann KK, Cox JH, Schneider LG & Thiel HJ Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology 175, 485–499 (1990). [DOI] [PubMed] [Google Scholar]
- 113.Walker PJ et al. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog 11, e1004664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dietzgen RG, Kondo H, Goodin MM, Kurath G & Vasilakis N The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res 227, 158–170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van den Pol AN et al. Viral strategies for studying the brain, including a replication-restricted self-amplifying δG vesicular stomatis virus that rapidly expresses transgenes in brain and can generate a multicolor golgi-like expression. J. Comp. Neurol 516, 456–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kebschull JM, Garcia da Silva P & Zador AM A new defective helper RNA to produce recombinant Sindbis virus that infects neurons but does not propagate. Front. Neuroanat 10, 56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hahn CS, Hahn YS, Braciale TJ & Rice CM Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc. Natl Acad. Sci. USA 89, 2679–2683 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ehrengruber MU Alphaviral gene transfer in neurobiology. Brain Res. Bull. 59, 13–22 (2002). [DOI] [PubMed] [Google Scholar]
- 119.Lustig S et al. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol 62, 2329–2336 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bredenbeek PJ, Frolov I, Rice CM & Schlesinger S Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol 67, 6439–6446 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Papez JW A proposed mechanism of emotion. 1937. J. Neuropsychiatry Clin. Neurosci 7, 103–112 (1995).This paper is the earliest to use a viral approach for neural circuit mapping.
- 122.Card JP et al. Pseudorabies virus infection of the rat central nervous system: ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci 13, 2515–2539 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Le Gal La Salle G et al. An adenovirus vector for gene transfer into neurons and glia in the brain. Science 259, 988–990 (1993). [DOI] [PubMed] [Google Scholar]
- 124.Akli S et al. Transfer of a foreign gene into the brain using adenovirus vectors. Nat. Genet 3, 224–228 (1993). [DOI] [PubMed] [Google Scholar]
- 125.Davidson BL, Allen ED, Kozarsky KF, Wilson JM & Roessler BJ A model system for in vivo gene transfer into the central nervous system using an adenoviral vector. Nat. Genet 3, 219–223 (1993). [DOI] [PubMed] [Google Scholar]
- 126.Kaplitt MG et al. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat. Genet 8, 148–154 (1994). [DOI] [PubMed] [Google Scholar]
- 127.Neve RL, Howe JR, Hong S & Kalb RG Introduction of the glutamate receptor subunit 1 into motor neurons in vitro and in vivo using a recombinant herpes simplex virus. Neuroscience 79, 435–447 (1997). [DOI] [PubMed] [Google Scholar]
- 128.Carlezon WA Jr et al. Sensitization to morphine induced by viral-mediated gene transfer. Science 277, 812–814 (1997). [DOI] [PubMed] [Google Scholar]
- 129.Song S et al. An HSV-1 vector containing the rat tyrosine hydroxylase promoter enhances both long-term and cell type-specific expression in the midbrain. J. Neurochem 68, 1792–1803 (1997). [DOI] [PubMed] [Google Scholar]
- 130.Carlezon WA Jr. et al. Regulation of cocaine reward by CREB. Science 282, 2272–2275 (1998). [DOI] [PubMed] [Google Scholar]
- 131.Kelz MB et al. Expression of the transcription factor δFosB in the brain controls sensitivity to cocaine. Nature 401, 272–276 (1999). [DOI] [PubMed] [Google Scholar]
- 132.Bolanos CA et al. Phospholipase Cγ in distinct regions of the ventral tegmental area differentially modulates mood-related behaviors. J. Neurosci 23, 7569–7576 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haberman RP, McCown TJ & Samulski RJ Inducible long-term gene expression in brain with adeno-associated virus gene transfer. Gene Ther 5, 1604–1611 (1998). [DOI] [PubMed] [Google Scholar]
- 134.Scammell TE et al. Focal deletion of the adenosine A1 receptor in adult mice using an adeno-associated viral vector. J. Neurosci 23, 5762–5770 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ahmed BY et al. Efficient delivery of Cre-recombinase to neurons in vivo and stable transduction of neurons using adeno-associated and lentiviral vectors. BMC Neurosci 5, 4 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Berton O et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 311, 864–868 (2006). [DOI] [PubMed] [Google Scholar]


